Summary
Huntington disease (HD)is a dominantly inherited neurodegenerative disorder caused by the expansion of a polyglutamine encoding CAG repeat in the huntingtin gene. Recently, it has been established that disease severity in HD is best predicted by the number of pure CAG repeats rather than total glutamines encoded. Along with uncovering DNA repair gene variants as trans-acting modifiers of HD severity, these data reveal somatic expansion of the CAG repeat as a key driver of HD onset. Using high-throughput DNA sequencing, we have determined the precise sequence and somatic expansion profiles of the HTT repeat tract of 68 HD-affected and 158 HD-unaffected African ancestry individuals. A high level of HTT repeat sequence diversity was observed, with three likely African-specific alleles identified. In the most common disease allele (30 out of 68), the typical proline-encoding CCGCCA sequence was absent. This CCGCCA-loss disease allele was associated with an earlier age of diagnosis of approximately 7.1 years and occurred exclusively on haplotype B2. Although somatic expansion was associated with an earlier age of diagnosis in the study overall, the CCGCCA-loss disease allele displayed reduced somatic expansion relative to the typical HTT expansions in blood DNA. We propose that the CCGCCA loss occurring on haplotype B2 is an African cis-acting modifier that appears to alter disease diagnosis of HD through a mechanism that is not driven by somatic expansion. The assessment of a group of individuals from an understudied population has highlighted population-specific differences that emphasize the importance of studying genetically diverse populations in the context of disease.
Keywords: Huntington disease, CAG repeat, CCGCCA loss, cis-acting modifier, African ancestry, genetically diverse
Using a high-throughput sequencing approach, we have contributed uniquely to the body of knowledge of Huntington disease (HD) and highlighted population-specific sequence data. We propose the CCGCCA loss is a cis-acting modifier of disease in African ancestry individuals that accelerates diagnosis through a mechanism that is not driven by somatic expansion.
Introduction
Huntington disease (HD; MIM: 143100) is a dominantly inherited neurodegenerative disorder, caused by expansion of the CAG repeat tract in exon 1 of the huntingtin (HTT; MIM: 613004) gene to 36 or more repeats.1 The inherited expanded CAG size in individuals affected with HD is 36 or more repeats, and inversely correlates with the age of onset (AoO).2,3 Thus, the longer the CAG repeat length inherited, the earlier the onset of HD symptoms.1 Expanded CAG repeat alleles are not only unstable in the germline, with a bias toward repeat length increases in successive generations,4 but are also unstable in somatic cells. Factors such as the length of the repeat, the age of the individual, and cell type affect the degree of somatic mosaicism observed.5, 6, 7, 8
The typical sequence of the HTT repeat tract is made up of a polyglutamine region (Q = glutamine) encoded by a number of pure CAG repeats in the first position (Q1 = CAG) and an intervening CAACAG sequence in the second position (Q2 = CAACAG). These are followed by the polyproline region (P = proline) encoded by the intervening CCGCCA sequence (P1 = CCGCCA), a stretch of pure CCG repeats (P2 = CCG), and lastly two downstream CCT repeats (P3 = CCT) (Figure 1). The HTT repeat tract has been reported to be a hotspot for variants8, 9, 10, 11 that alter the sequence relative to the reference. This may include duplication or loss of all or part of the intervening sequence between the pure CAG and CCG repeats (CAACAG-CCGCCA in typical alleles) and variation in the number of the downstream CCT repeats.8, 9, 10, 11, 12, 13
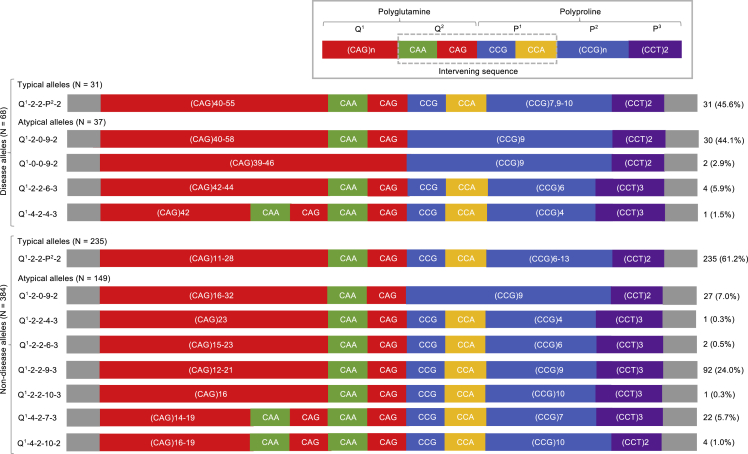
Figure 1.
The HTT disease and non-disease allele structures in African ancestry individuals
Schematic representation of the HTT disease and non-disease allele structures defined for this study. The typical allele structures were grouped together as Q1-2-2-P2-2, while the atypical allele structures are shown individually for deviations from the reference allele structure to be clearly demonstrated.
In other repeat expansion disorders such as spinocerebellar ataxia type 1 (SCA1), myotonic dystrophy type 1 (DM1), and fragile X syndrome, repeat stability and disease severity are affected by mutations within the repeat tract (i.e., atypical allele structures).14, 15, 16, 17, 18 In SCA1 and fragile X syndrome, the stabilizing interruptions have been identified on unexpanded alleles, while they are absent or very rare on expanded alleles.14,15 Mutations interrupting the HTT repeat tract may have a similar effect for HD.
A recent study of the HTT repeat tract in over 800 European individuals affected with HD revealed that atypical allele structures are more frequent than previously thought (∼8% of non-disease alleles and ∼3% of disease-associated alleles).8 Several studies have now shown that HD severity is best explained by the length of the pure CAG repeat tract (Q1) and not by the length of the polyglutamine tract encoded (Q1 + Q2).8,10,11 Given that somatic expansion of the CAG repeat is also best predicted by the number of pure CAG repeats, coupled with the observation that CAG length-independent variation in age at onset is associated with DNA repair gene variants, these data confirm that somatic expansion is a key driver of HD onset.8,10,11 Whether somatic expansion in African individuals affected with HD is modified by the same set of cis-acting sequence HTT repeat variants and trans-acting DNA repair gene variants or additional African-specific genetic variants is yet to be determined.
Although HD has been reported worldwide, there are distinct geographic differences in prevalence, with the lowest rates in African populations and those with African ancestry.19,20 These differences are particularly reflected in South Africa, where HD has been reported in three different groups (European ancestry, mixed ancestry, and African ancestry individuals) but at varying prevalence estimates (7.8, 2.2, and 0.5 per 100,000 individuals, respectively).21
In general, there is a greater amount of genetic diversity present in sub-Saharan African populations across the genome in comparison with all other populations.22 Specifically at the HTT locus, a large number of haplotypes have been previously defined, and, among South African non-disease and disease alleles, a unique C haplotype variant (C-SA) has been identified.23 It is therefore a reasonable expectation that there is more sequence diversity within the HTT repeat tract of African individuals. Sequence variation may have a similar modifier effect on the HD phenotype as seen in European ancestry individuals or provide unique insights into disease modification in African ancestry individuals. Using a high-throughput ultra-deep DNA sequencing assay specific for the HTT repeat tract,24 this study assessed HTT genetic diversity in a sample of South African HD-affected and -unaffected individuals. The results present the sequence variation identified in this complex region, background haplotypes, and the characterization of somatic expansion as potential genetic modifiers of the HD phenotype in individuals of African ancestry.
Materials and methods
Subjects
Blood DNA samples were sourced from the archives of the Division of Human Genetics, University of the Witwatersrand (Wits) and the National Health Laboratory Service (NHLS) in Johannesburg, South Africa, accumulated over the preceding 25 years. The study population comprised 68 unrelated individuals affected with HD and 158 unrelated individuals unaffected with HD, all of African ancestry (68 disease alleles, 384 non-disease alleles). All the individuals included in the present study were South African Bantu speakers from a small geographic area around Johannesburg (± 200 km). Population genetic structure is weak among South African Bantu speakers and is only relevant at a geographical scale, which is far larger than our study area.25 Our study population therefore constitutes a genetically homogenous Bantu-speaking population. Although only unrelated individuals were included in the sequence diversity assessment, HD-affected relatives of the probands were successfully sequenced to assess intergenerational instability. The affected individuals were originally referred for molecular diagnostic confirmation of disease status and, where available, AoO was patient reported after consultation with a medical geneticist or neurologist. The HD cohort comprised 42 females and 26 males, with an age of diagnosis ranging between 23 and 77 years; additional patient information is shown in the supplemental data (Table S1). Ethical approval for this study was obtained from the Human Research Ethics Committee (Medical), University of the Witwatersrand (certificate numbers M1704130, sub-study M110443).
HTT repeat tract sequence diversity
The HTT repeat tract sequencing followed an established ultra-deep high-throughput sequencing protocol developed to characterize the repeat tract precisely.24 Following sequencing on an Illumina MiSeq platform, genotyping was carried out using ScaleHD (v0.251) as previously described.8 The HTT repeat tract reference sequence is made up of a polyglutamine encoding region followed by the polyproline encoding region as shown in Figure 1. The allele structures were defined as either typical or atypical based on a comparison with the reference allele structure (LRG_763). The atypical alleles were defined based on deviations from the reference sequence as a result of variants within the repeat tract at the Q2, P1, and P3 regions.
HTT background haplotypes
The allele structures were investigated in the context of background haplotypes for the HTT locus spanning ∼196,063 kb on chromosome 4. Thirteen tag single nucleotide polymorphisms (tag-SNPs) were selected from previously studied haplotype SNPs,19 to define haplogroups A, B, and C, and the South African-specific haplogroup variant C-South Africa (C-SA) (Table S2).23 The tag-SNPs were genotyped using a MassARRAY System from Agena Bioscience and haplotypes constructed using manual and statistical phasing. Manual phasing was achieved using homozygous genotypes and repeat tract associations, while the statistical phasing was performed using PHASE (v2.1.1), which employs a Bayesian inference model.26,27 Two samples (one disease allele and three non-disease alleles) from the sequence diversity analysis were excluded due to unsuccessful tag-SNP genotyping. The LDhap tool from the LDlink suite was used to derive haplotype frequencies in the 1000 Genomes Project populations for the tag-SNPs used to define the most common disease haplotype.28
Quantification of HTT somatic expansion
The ratio of CAG repeat somatic expansions of disease-associated alleles was determined from the MiSeq read count distributions as described previously.8 The somatic expansion score was then calculated as the residuals of the log-transformed ratio of somatic expansion after adjusting for the effect of the inherited expanded CAG repeat length, age at sampling, and their interaction using multiple linear regression.8
Statistical analysis
Potential genetic modifiers of HD were investigated with multiple linear models in R (v3.4.3) using RStudio (v1.0.153). The lm function was used to determine associations between the HD phenotype and various explanatory variables: HTT repeat tract, background haplotypes and somatic expansion score. When studying the HD phenotype, the AoO of motor symptoms is the most well-defined and frequently used measure of disease severity.29 However, in our sample of HD-affected individuals, less than 50% had AoO information and, because the age of diagnosis (AoD) was available for all subjects and strongly correlated with AoO (r2 = 0.68, Figure S1), it was therefore used as a proxy for AoO. The assessment of the modifiers of the HD phenotype was conducted on the subset of HD-affected individuals with CAG repeat length between 39 and 52 repeats, as ≥53 CAG repeats violate linear model assumptions.29 As a result, four individuals were excluded from the analysis that had the following allele structures and haplotypes: two Q1-2-2-10-2 on haplotype C5, one Q1-2-2-7-2 on haplotype A4a, and one Q1-2-0-9-2 on haplotype B2. In the linear models, the reference for the allele structures and haplotypes was Q1-2-2-P2-2 and haplotype B2 respectively. To determine if the variation in the AoD was better explained by HTT allele structure or background haplotype, a goodness of fit test of the R-squared of the Q1-2-0-9-2 allele structure model and the haplotype B2 model in 5,000 bootstrapped samples was assessed. The estimated marginal mean AoD and expansion score were established using the emmeans function in R using RStudio.30
Results
HTT repeat tract sequence diversity
A total of 226 samples from individuals of African ancestry, 68 affected with HD and 158 unaffected (68 disease alleles and 384 non-disease alleles) were sequenced and genotyped. Seventeen different allele structures were identified and defined as either typical or atypical alleles (Table 1). The eight allele structures defined as typical had a variable number of CAG repeats and CCG repeats that ranged from six to 13 repeats. Nine allele structures were defined as atypical due to variants resulting in an apparent loss or duplication of the intervening sequences CAACAG (Q2 = 0 or 4) and CCGCCA (P1 = 0) and/or accompanied by an additional downstream CCT (P3 = 3) repeat. All the variants that resulted in the atypical alleles were synonymous and thus translated into huntingtin proteins with pure polyglutamine and pure polyproline regions. Of the 17 allele structures, three (one typical and two atypical) are unique to this study as they have not been previously described (asterisk in Table 1). Schematics for the disease and non-disease allele structures are shown in Figure 1.
Table 1.
Summary of African ancestry HTT disease and non-disease alleles
| Allele types | Allele structure nomenclature |
HTT repeat tract |
Allele occurrence |
Fisher exact test |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Q1 |
Q2 |
P1 |
P2 |
P3 |
Non-disease N = 384 |
Disease N = 68 |
||||||
| CAG | CAACAG | CCGCCA | CCG | CCT | n | % | n | % | p values | |||
| Typical alleles | Q1-2-2-6-2 | 14–17 | – | 2 | 2 | 6 | 2 | 11 | 2.9 | – | – | 0.384 |
| Q1-2-2-7-2 | 15–28 | 41–55 | 2 | 2 | 7 | 2 | 99 | 25.8 | 10 | 14.7 | 0.064 | |
| Q1-2-2-8-2 | 17 | – | 2 | 2 | 8 | 2 | 5 | 1.3 | – | – | 1 | |
| Q1-2-2-9-2 | 15–28 | 40 | 2 | 2 | 9 | 2 | 29 | 7.6 | 1 | 1.5 | 0.066 | |
| Q1-2-2-10-2 | 11–20 | 40–54 | 2 | 2 | 10 | 2 | 71 | 18.5 | 20 | 29.4 | 0.048 | |
| Q1-2-2-11-2 | 12–21 | – | 2 | 2 | 11 | 2 | 18 | 4.7 | – | – | 0.089 | |
| Q1-2-2-12-2 | 17 | – | 2 | 2 | 12 | 2 | 1 | 0.3 | – | – | 1 | |
| ∗Q1-2-2-13-2 | 17 | – | 2 | 2 | 13 | 2 | 1 | 0.3 | – | – | 1 | |
| Typical alleles subtotal | 235 | 61.2 | 31 | 45.6 | ||||||||
| Atypical alleles | Q1-2-2-4-3 | 23 | – | 2 | 2 | 4 | 3 | 1 | 0.3 | – | – | 1 |
| Q1-2-2-6-3 | 15–23 | 42–44 | 2 | 2 | 6 | 3 | 2 | 0.5 | 4 | 5.9 | 5.587 × 10−3 | |
| Q1-2-2-9-3 | 12–21 | – | 2 | 2 | 9 | 3 | 92 | 24.0 | – | – | 9.142 × 10−8 | |
| Q1-2-2-10-3 | 16 | – | 2 | 2 | 10 | 3 | 1 | 0.3 | – | – | 1 | |
| ∗Q1-4-2-4-3 | – | 42 | 4 | 2 | 4 | 3 | – | – | 1 | 1.5 | 0.154 | |
| Q1-4-2-7-3 | 14–19 | – | 4 | 2 | 7 | 3 | 22 | 5.7 | – | – | 0.059 | |
| ∗Q1-4-2-10-2 | 16–19 | – | 4 | 2 | 10 | 2 | 4 | 1.0 | – | – | 1 | |
| Q1-2-0-9-2 | 16–32 | 40–58 | 2 | 0 | 9 | 2 | 27 | 7.0 | 30 | 44.1 | 3.119 × 10−13 | |
| Q1-0-0-9-2 | – | 39–46 | 0 | 0 | 9 | 2 | – | – | 2 | 2.9 | 0.022 | |
| Atypical alleles subtotal | 149 | 38.8 | 37 | 54.4 | ||||||||
The novel allele structures unique to this study are indicated by an asterisk (∗). The most common non-disease and disease allele structures are indicated in underlined italics. The statistically significant frequency differences between the non-disease and disease alleles are indicated in italics (non-disease alleles: Q1-2-2-10-2 p = 0.048 and disease alleles: Q1-2-2-6-3 p = 5.587 × 10−3, Q1-2-2-9-3 p = 9.142 × 10−8 and Q1-2-0-9-2 p = 3.119 × 10−13).
The most common disease allele Q1-2-0-9-2 (30 out of 68 = 44.1%) had an atypical structure defined by a CCGCCA loss (P2 = 0). Although also present in unaffected individuals, it represented a much smaller proportion of the non-disease alleles (27 out of 384 = 7.0%). The most common non-disease allele Q1-2-2-7-2 (99 out of 384 = 25.8%) had a typical structure, with variability occurring only in the length of the CAG repeat.
When comparing disease and non-disease alleles, one typical allele structure, Q1-2-2-10-2, occurred more frequently in the non-disease alleles. Among the atypical alleles, the frequency of four structures differed significantly between the disease and non-disease alleles. Three of these, Q1-2-2-6-3, Q1-2-0-9-2, and Q1-0-0-9-2, were more frequent in disease alleles, while one atypical allele structure, Q1-2-2-9-3, was more frequent in non-disease alleles (Fisher exact p values in Table 1).
The comparison of these African alleles with the European alleles previously described (746 disease alleles)8 revealed differences in the structures defined and their frequencies. Among European HTT alleles, 92.2% of the non-disease and 97.2% of disease alleles had a typical allele structure,8 compared with the African HTT alleles where only 61.2% of non-disease alleles were typical and 45.6% of disease alleles were atypical (non-disease, 235 out of 384 > 688 out of 746, Fisher exact test p < 2 × 10−16; versus disease alleles, 31 out of 68 > 725 out of 746, Fisher exact test p < 2 × 10−16). The most common allele structure, Q1-2-2-7-2 (typical allele structure), was the same for both non-disease and disease alleles in European individuals and in the African non-disease alleles. However, the most common African disease allele structure, Q1-2-0-9-2 (i.e., CCGCCA loss, P2 = 0), was atypical and reportedly rare (non-disease alleles, 30 out of 746 = 4.0%; disease alleles, 0 out of 746 = 0%) among European disease alleles.8
A particularly interesting case of intergenerational instability was identified in association with the most common African disease allele structure, Q1-2-0-9-2, when relatives of the proband were assessed. In this case, we observed a paternal transmission of 43 CAG repeats, which resulted in an increase to 73 CAG repeats in the child affected with HD.
HTT haplogroup/haplotype diversity
Background haplotypes were constructed for 224 individuals with African ancestry (67 disease alleles and 381 non-disease alleles). Sixteen different haplotypes were identified across the four previously defined haplogroups (A, B, C, and C-SA) as well as an “other” haplogroup (Table 2). The “other” category was applied when the composition of tag-SNP alleles did not fall into any of the previously defined haplogroups/haplotypes.
Table 2.
Summary of the HTT haplogroups/haplotypes and associated allele structures in disease and non-disease alleles
| Haplogroups | Haplotypes | Allele structures |
Non-disease |
Disease |
||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| A | ∗A2a | Q1-2-2-7-2 | – | – | 1 | 1.5 |
| ∗A2b | Q1-2-2-7-2 | 13 | 3.4 | 1 | 1.5 | |
| A4a | Q1-2-2-7-2 Q1-2-2-12-2 Q1-2-2-13-2 |
5 1 1 |
1.3 0.3 0.3 |
3 – – |
4.5 – – |
|
| A4b | Q1-2-2-7-2 | 2 | 0.5 | 5 | 7.5 | |
| A6 | Q1-2-2-7-2 | 34 | 8.9 | – | – | |
| B | B1 | Q1-2-2-9-2 | 1 | 0.3 | 1 | 1.5 |
| B2 |
Q1-2-0-9-2 Q1-0-0-9-2 Q1-4-2-4-3 |
25 – – |
6.6 – – |
29 2 1 |
43.3 3.0 1.5 |
|
| C | C2 | Q1-4-2-7-3 Q1-2-2-8-2 |
21 5 |
5.5 1.3 |
– – |
– – |
| C4 | Q1-2-2-9-2 | 21 | 5.5 | 1 | 1.5 | |
| C4c | Q1-2-2-6-2 | 11 | 2.9 | – | – | |
| C5 | Q1-2-2-10-2 Q1-2-2-11-2 Q1-2-2-10-3 Q1-2-0-9-2 Q1-4-2-10-2 |
69 18 1 2 4 |
18.1 4.7 0.3 0.5 1.0 |
19 – – – – |
28.4 – – – – |
|
| C8 | Q1-2-2-9-2 | 7 | 1.8 | – | – | |
| C-SA | C3 | Q1-2-2-10-2 Q1-2-2-9-3 |
1 90 |
0.3 23.6 |
– – |
– – |
| C9 | Q1-2-2-6-3 Q1-2-2-7-2 |
2 7 |
0.5 1.8 |
4 – |
6.0 – |
|
| C10 | Q1-2-2-4-3 | 1 | 0.3 | – | – | |
| Other | O | Q1-2-2-7-2 Q1-2-2-9-3 Q1-2-2-10-2 |
36 2 1 |
9.4 0.5 0.3 |
– – – |
– – – |
| Total | #381 | 100.0 | 67 | 100.0 | ||
The two haplotypes that had not been previously identified in African ancestry individuals are indicated by an asterisk (∗). The most common non-disease and disease haplogroup/haplotype are indicated in underlined italics. The most common disease allele structure Q1-2-0-9-2 (29 out of 67 = 43.3%) is indicated in italics. Two samples (one disease allele and three non-disease alleles) from the sequence diversity analysis presented in Table 1 were excluded due to unsuccessful tag-SNP genotyping (#).
The largest proportion of non-disease alleles occurred on haplogroup C (159 out of 381 = 41.7%) and, within haplogroup C, haplotype C5 was the most common (94 out of 381 = 24.8%) (Table 2). For disease alleles, the largest proportion occurred on haplogroup B (33 out of 67 = 49.3%) and, within haplogroup B, haplotype B2 was the most common (32 out of 67 = 47.8%). The most common allele structure (Q1-2-0-9-2) in the disease alleles, characterized by the CCGCCA loss, was found exclusively on haplotype B2 and was therefore further assessed to determine whether it was African specific.
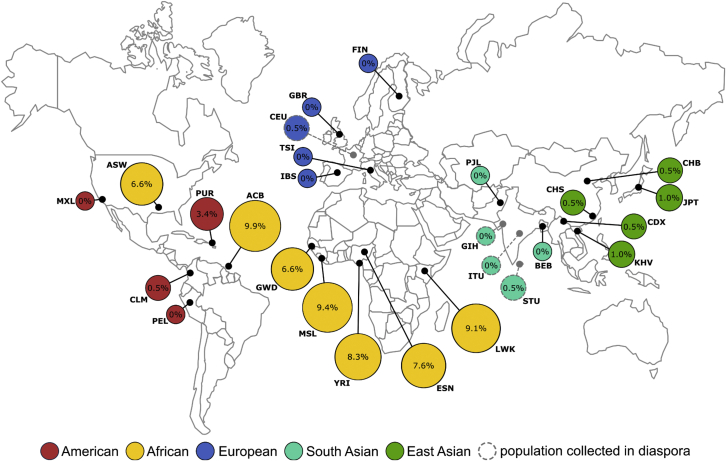
Haplotype B2 was found to be the most frequent in the seven African and African ancestry populations of the 1000 Genomes Project (Figure 2). The frequency ranged from 6.6% in Americans of African Ancestry in Southwest US (ASW), to 9.9% in the African Caribbean in Barbados (ACB). Among the non-disease alleles included in the present study, a comparable frequency of 6.6% was identified for haplotype B2. Apart from Puerto Rico (PUR), where its frequency was 3.4%, haplotype B2 was rare (frequency ≤ 1%) in all the other non-African populations. This indicates that, although this analysis was only conducted in non-disease alleles, haplotype B2 was revealed to be of African origin and largely African specific.
Figure 2.
Frequency of the HTT haplotype B2 in the populations of the 1000 Genomes Project
The African B2 haplotype was defined by SNPs rs2857936-rs762855-rs4690073 as described by Baine et al.23 The haplotype frequencies were obtained using the LDhap tool from the LDlink suite (ldlink.nci.nih.gov).28 Haplotype B2 was shown to have the highest frequencies among the African and African ancestry populations, ranging between 6.6% and 9.9%. Outside of the continental African populations, Puerto Rico (American) had the highest frequency of haplotype B2 (3.4%), followed by the five East Asian populations (range from 0.5% to 1.0%). The Columbian (American), Utah residents (European), and Sri Lankan (South Asian) populations had low frequencies (0.5%), and B2 was not detected in the rest of the populations analyzed. The results were comparable with the frequency of B2 in the African ancestry non-disease alleles included in this study. This indicates that, although this analysis was only conducted in non-disease alleles, haplotype B2 may be of an African origin and an African-specific haplotype.
CAG somatic expansion
The modifiers of the ratio of somatic CAG expansion of disease-associated alleles were assessed through the inclusion of the following explanatory variables; inherited expanded CAG repeat length, the age at sampling, and their interaction (Table S3 Model 1). The inherited expanded CAG repeat length and the age at sampling were shown to have a highly significant association with the ratio of somatic CAG expansions of the disease-associated allele observed in blood DNA (p < 2 × 10−16). A larger effect was observed for the inherited expanded CAG repeat length with every additional CAG repeat resulting in an increase of 0.131 (p = 8 × 10−16) in the ratio of somatic expansions; while every year delay in the age at sampling increased the ratio of somatic expansion by 0.008 (p = 1.8 × 10−3). In line with previous studies,7,8 the inherited CAG repeat length was shown to be the primary driver of the ratio of somatic expansion.
Moreover, a highly significant association (p < 2 × 10−16) was identified between allele structures and the ratio of somatic expansion (Figure S2; Table S3, Model 2). In addition to the CAG repeat, the disease allele structures, Q1-0-0-9-2 (p = 7.7 × 10−4), Q1-2-0-9-2 (p = 1 × 10−5), and Q1-2-2-6-3 (p = 0.014) were each shown to have a significant association with the ratio of somatic expansion. Individuals with these disease allele structures had a mean decrease in somatic expansion by 0.26, 0.17, and 0.15, respectively. Thus, individuals with the typical allele structure (Q1-2-2-P2-2) had a significantly higher ratio of somatic expansion overall, while individuals with disease alleles characterized by the loss of CCGCCA sequence had the lowest ratio of somatic expansion.
Potential modifiers of the HD phenotype
HTT repeat tract modification
A highly significant negative association (p = 3 × 10−14) was detected between the CAG repeat and the AoD, accounting for approximately 60% of the variation in the AoD (Figure S3). The degree of variation in AoD explained by CAG is directly comparable with the degree of variation in AoO explained by CAG in European ancestry populations (Figure S4), further highlighting the clinical utility of AoD. The other components of the HTT repeat tract were assessed individually for their association with the AoD (in years) (Table S4, Model 1). In addition to the CAG repeat length (2.9 years earlier, p = 6 × 10−12), the CCGCCA sequence was shown to have a significant association with the AoD (4.0 years earlier for loss of CCGCCA, p = 7 × 10- 4). Thus, each additional CAG repeat and loss of the CCGCCA sequence resulted in an earlier AoD in individuals affected with HD. Although not surprisingly, given the very small sample size (n = 24), not statistically significant, a similar trend of decreased AoO for individuals with the CCGCCA-loss allele was observed (Figure S5; Table S4, Models 2 and 3).
The association with each allele structure (all components of the repeat tract together) on the AoD was also assessed. A highly significant correlation was identified between the inherited CAG repeat length within each of the allele structures and the AoD (p = 1 × 10−10) (Figure 3A). The CCGCCA-loss allele structure (Q1-2-0-9-2) was the only allele structure that had a detectable significant association with the AoD in comparison with the grouped typical allele structures (7.1 years earlier, p = 8 × 10−4) (Table S4, Model 4).
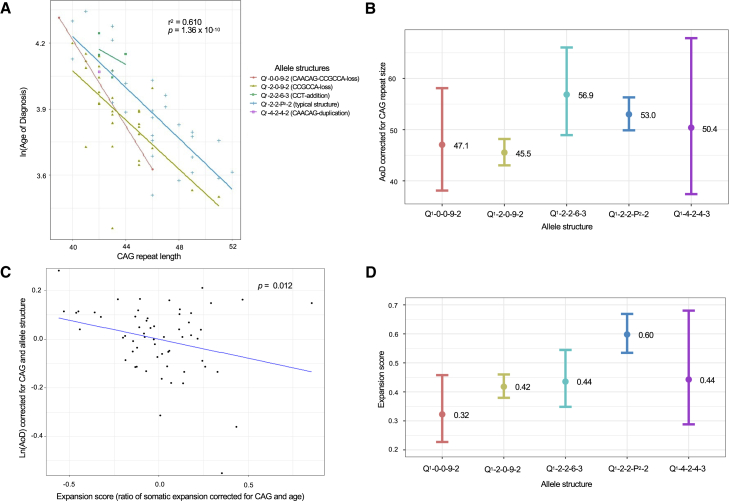
Figure 3.
The HTT allele structure associated with age at HD diagnosis and somatic expansion of the HD allele in blood DNA in African ancestry individuals
(A) Linear regression analysis testing the association between the log transformed AoD and the inherited CAG repeat length for each disease allele structure revealed a significant association (r2 = 0.61, p = 1.36 × 10−10). The Q1-0-0-9-2 and Q1-2-0-9-2 disease allele structures characterized by the loss of one or more of the intervening sequences had the earliest AoD.
(B) The estimated marginal mean AoD for the disease allele structures, corrected for repeat size. The Q1-2-0-9-2 allele structure had the earliest mean AoD (n = 30, 45.5 years: 95% CI = 43.0–48.2), followed by Q1-0-0-9-2 (n = 2, 47.1 years: 95% CI = 38.1–58.1), Q1-4-2-4-3 (n = 1, 50.4 years: 95% CI = 37.4–67.9), Q1-2-2-P2-2 (n = 31, 53.0 years: 95% CI = 49.9–56.3), and Q1-2-2-6-3 (n = 4, 56.9 years: 95% CI = 48.9–66.0).
(C) Linear regression analysis testing the association between the log transformed AoD (corrected for CAG repeat length and allele structure) and expansion score. Overall, a significant association (p = 0.012) was identified.
(D) The estimated marginal mean expansion score for the allele structures, corrected for CAG repeat length and age at sampling. The Q1-0-0-9-2 (n = 2, 0.32: 95% CI = 0.227–0.458) and Q1-2-0-9-2 (n = 30, 0.42: 95% CI = 0.380–0.460) allele structures had the lowest mean expansion score followed by Q1-2-2-6-3 (n = 4, 0.44: 95% CI = 0.348–0.545), Q1-4-2-4-3 (n = 1, 0.44: 95% CI = 0.288–0.680), and Q1-2-2-P2-2 (n = 31, 0.60: 95% CI = 0.535–0.669).
The estimated marginal mean AoD for each disease allele confirmed that individuals with the commonest African allele structure, Q1-2-0-9-2, have the earliest mean AoD of 45.5 years, while individuals with the Q1-2-2-6-3 allele structure had the most delayed mean AoD of 56.9 years (Figure 3B).
HTT haplogroup/haplotype modification
Haplogroup A, C, and haplogroup variant C-SA were shown to have a significant positive association with the HD phenotype (delayed the AoD) when compared with the most common haplogroup B (Table S4, Model 5). Individuals with an expanded HTT allele occurring on haplogroup B had a significantly earlier AoD compared with haplogroup C: 6.2 years (p = 0.022); haplogroup A, 8.6 years (p = 0.014); and haplogroup C variant C-SA, 11.8 years (p = 0.012).
Individuals with an expanded HTT allele on haplotype B2 had a significantly earlier AoD compared with the other haplotypes: A4a, 16.8 years (p = 0.018); C5, 7.8 years (p = 6.8 × 10−3); A4b, 9.1 years (p = 0.029); C9, 12.3 years (p = 7.9 × 10−3); and B1, 22.3 years (p = 0.019) (Table S4, Model 6). The estimated marginal mean AoD for each disease haplotype confirmed that haplotype B2 had the earliest AoD of 45.5 years (n = 29, 95% confidence interval [CI] = 43.0–48.1), while individuals with haplotype B1 had the most delayed mean AoD of 65.5 years (n = 1, 95% CI = 48.6–88.2) (Figure S6).The earliest mean AoD in individuals with haplotype B2 was the same for individuals with the most common allele structure, Q1-2-0-9-2, as these alleles occurred exclusively on the haplotype background B2.
To assess whether the allele structure itself or another variant on haplotype B2 was a more likely explanation for the disease-hastening effect detected, a goodness of fit test on 5,000 bootstrapped samples was conducted. The assessment of the CCGCCA-loss allele structure (Q1-2-0-9-2) compared with haplotype B2 as a better explanation of the earlier AoD revealed neither to have more of a significant association (Figure S7). There was no statistical indication that the haplotype B2 was more strongly associated with the AoD than the CCGCCA-loss allele structure (Q1-2-0-9-2).
CAG somatic expansion modification
The effect of the ratio of somatic expansion on the AoD was then considered through the assessment of the expansion score. The results revealed a highly significant correlation (p = 1.296 × 10−9) and an R-square value of 0.63. The inherited CAG repeat length, disease allele structures Q1-0-0-9-2 and Q1-2-0-9-2, and the expansion score were all shown to have a significant association with the AoD (Table 3, Model 1). Every CAG repeat increase resulted in an earlier AoD by 3.5 years (p = 2 × 10−11), while the allele structures Q1-2-0-9-2 and Q1-0-0-9-2 resulted in an earlier AoD by 10.2 years (p = 4 × 10−5) and 11.5 years (p = 0.034) respectively, compared with the grouped typical allele structure Q1-2-2-P2-2. Lastly, every unit increase in the expansion score resulted in an earlier AoD by 10.6 years (p = 0.012).
Table 3.
Multiple linear models testing the association between the HD phenotype and various explanatory variables
| Model | r2 | p value for model |
Parameter values |
||||
|---|---|---|---|---|---|---|---|
| Sample size | Explanatory variable | Effect in years | p value for explanatory variable | ||||
| 1 | Ln (AoD)∼ CAG + allele structures + expansion score | 0.625 | 1.296 × 10−9 | 60 | CAG | −3.504 | 1.56 × 10−11 |
| 2 | Q1-0-0-9-2 | −11.491 | 0.034 | ||||
| 30 | Q1-2-0-9-2 | −10.180 | 4.20 × 10−5 | ||||
| 4 | Q1-2-2-6-3 | −0.840 | 0.846 | ||||
| 1 | Q1-4-2-4-3 | −5.903 | 0.411 | ||||
| Expansion score | −10.600 | 0.012 | |||||
| 2 | Ln (AoD)∼ CAG + haplotypes + expansion score | 0.664 | 2.989 × 10−8 | 60 | CAG | −3.665 | 8.93 × 10−11 |
| 1 | A2a | 2.050 | 0.784 | ||||
| 1 | A2b | 11.664 | 0.163 | ||||
| 1 | A4a | 12.985 | 0.137 | ||||
| 4 | A4b | 15.765 | 3.28 × 10−3 | ||||
| 1 | B1 | 29.224 | 3.37 × 10−3 | ||||
| 1 | C4 | 2.773 | 0.719 | ||||
| 16 | C5 | 14.250 | 1.41 × 10−4 | ||||
| 4 | C9 | 11.517 | 9.37 × 10−3 | ||||
| Expansion score | −12.090 | 7.81 × 10−3 | |||||
The statistically significant explanatory variables are indicated in italics. Model 1. Linear model testing the association of the CAG repeat length, allele structure and expansion score on the AoD, relative to the grouped typical allele structure Q1-2-2-P2-2. The R-square and p values of the overall model show a significant association (r2 = 0.63, p = 1 × 10−9), the CAG repeat length, allele structures Q1-0-0-9-2 and Q1-2-0-9-2, and expansion score had a significant association. Model 2. Linear model testing the association of the CAG repeat length, background haplotype, and expansion score on the AoD, relative to the most common haplotype B2. The R-square and p values of the overall model show a significant association (r2 = 0.66, p = 3 × 10−8), and the CAG repeat length; haplotypes A4b, B1, C5, and C9; and expansion score had a significant association.
Similarly, when the background haplotype was considered in the assessment, a highly significant correlation (p = 3 × 10−8) and an R-square value of 0.66 was identified (Table 3, Model 2). Every CAG repeat increase resulted in an earlier AoD by 3.7 years, while the background haplotypes A4b, B1, C5, and C9 resulted in a delayed AoD by 15.8 years (p = 3 × 10−5), 29.2 years (p = 3 × 10−3), 14.3 years (p = 1 × 10−4), and 11.5 years (p = 9 × 10−3) respectively, compared with the background haplotype B2. Lastly, every unit increase in the expansion score resulted in an earlier AoD by 12.1 years (p = 8 × 10−3). The association of the expansion score with the AoD, corrected to CAG repeat size and allele structure, revealed an overall significant negative correlation (p = 0.012), illustrating the expansion score result observed in Table 3, Model 1 (Figure 3C).
The estimated marginal mean expansion scores for the disease allele structures confirmed that the largest mean expansion score was identified in the grouped typical allele structure Q1-2-2-P2-2 at 0.60, while the lowest expansion scores were associated with the atypical allele structures Q1-2-0-9-2 at 0.42 and Q1-0-0-9-2 at 0.32 (Figure 3D). Thus, although somatic expansion was shown to be significantly associated with the AoD, overall, nonetheless individuals with the commonest African Q1-2-0-9-2 allele structure that had the earliest AoD also had one of the lowest expansion scores in blood DNA. The earlier AoD seen in these individuals could thus not be attributed to somatic expansion in blood DNA.
Discussion
This study set out to characterize the HTT repeat tract sequence in African ancestry HD disease and non-disease alleles, and ultimately assess potential cis-acting genetic modifiers of the HD phenotype. A large amount of sequence diversity was observed with 17 different allele structures identified: eight were defined as typical (variation only in the number of CAG/CCG repeats), while nine were atypical (variation present throughout the HTT repeat tract). Less variation was identified in the non-disease alleles, with typical allele structures being more frequent, while atypical allele structures were more frequently observed in disease alleles.
Across the non-disease alleles, the typical allele structure Q1-2-2-7-2 was the most common. This allele structure has been previously shown to be the most common in both European ancestry non-disease (∼92%) and disease alleles (∼97%).8 In contrast, the atypical allele structure Q1-2-0-9-2, characterized by the CCGCCA loss, was the most common (∼44%) in African disease alleles. Although this allele structure has been previously identified in European ancestry individuals, it is very rare, especially among individuals affected with HD (0 out of 746).8
Three of the 17 allele structures identified in the disease and non-disease alleles were unique to this study (Table 1). This is possibly due to these allele structures being very rare in previously studied populations or, more likely, specific to African ancestry individuals. The differences between atypical allele frequencies in an African population and those recently reported European alleles (European atypical non-disease ∼ 8%, disease ∼ 3%; versus African atypical non-disease ∼ 39%, disease ∼ 54%)8 highlight the importance of research across different populations to improve understanding of the full range of diversity.
Analysis of the broader HTT locus in individuals of African ancestry revealed that the largest proportion of non-disease alleles occurred on haplogroup C and haplotype C5, while the largest proportion of disease alleles occurred on haplogroup B and haplotype B2 (Table 2). A comparison of the European ancestry haplotypes revealed the largest proportion of non-disease alleles occur on haplogroup C, while the largest proportion of disease alleles occurred on haplogroup A.19
The most common disease allele structure, characterized by the CCGCCA loss, occurred exclusively on haplotype B2. Although haplotype B2 has been identified in individuals of European ancestry, it is rare and differs by at least one tag-SNP (J.A. Collins, personal communication; M.R. Hayden, personal communication; G.E.B. Wright, personal communication).31 The assessment of haplotype B2 in other populations worldwide, showed that it is frequent (≥6.6%) in African populations and rare (≤1%) in non-African populations. The presence of haplotype B2 at a frequency of 3.4% among Puerto Ricans (Figure 2) is in line with the fact that ∼10% of the genome of these individuals is of African ancestry.32 The higher frequency in the African populations provides support for haplotype B2 being African specific and of African origin.
We have also identified the presence of haplotype variants A2a and A2b in two of our African individuals affected with HD, suggesting that, although rare, European high-risk haplotypes are present in African ancestry individuals. Prior to this study, A2a and A2b were described to be absent from East Asian and African ancestry populations.19 The presence of these haplotypes is potentially a result of admixture with European populations. Alternatively, these haplotypes may have been present in ancestral African populations and increased in frequency in European populations due to population bottlenecks arising during migration out of Africa.
Recent data have confirmed somatic expansion of the HTT CAG repeat as a potential driver of HD severity.8 In European ancestry individuals affected with HD, individual-specific rates of somatic expansion in blood DNA are inversely correlated with AoO, and positively correlated with disease progression. Here, we have demonstrated that, overall, there is a significant inverse association between individual-specific levels of somatic expansion in blood DNA and AoD as a proxy for age at onset in an African ancestry HD cohort. Whether individual-specific rates of somatic expansion in African individuals affected with HD are driven by the same set of DNA repair gene variants as observed in European populations8,10,11 is yet to be determined. However, given the higher genetic diversity observed in African populations, it seems likely that additional African-specific genetic variants may be in operation.
It has also recently been determined that HD severity is best explained by the length of the pure CAG repeat tract (Q1) and not by the length of the polyglutamine tract encoded (Q1 + Q2).8,10,11 Since the degree of somatic expansion is also best predicted by pure CAG length (Q1),8 these data suggest that somatic CAG expansion is potentially more important in relation to disease severity and progression than the number of glutamines encoded in the inherited allele. As all of the CAACAG duplications observed previously in the European ancestry population were present on a typical CCGCCA polyproline encoding background,8 the data presented here do not alter the interpretation of the primary effect of the CAACAG duplication. However, since the very rare CAACAG loss is observed on alleles both with and without the CCGCCA sequence in European ancestry populations,8,10,11,33 it is possible that some of the effects attributed to the CAACAG loss might be due to and/or exacerbated by the CCGCCA loss. Indeed, even after correcting for the number of pure CAG repeats, loss of the CAACAG sequence was still associated with worse HD outcomes.8 Unfortunately, the number of individuals with the double loss of the CAACAG and CCGCCA sequences (3 out of 746), versus those with only the CAACAG loss (4 out of 746) and those with only the CCGCCA loss (0 out of 746), precludes a reanalysis of our previously published data.8
Only two disease alleles lacking the CAACAG sequence (Q2 = 0) were detected in this study, precluding an assessment of the impact of this structure on HD severity. Rather, we determined that individuals carrying disease allele structures characterized by loss of the CCGCCA sequence (P1 = 0) had an earlier AoD by 4.0 years compared with individuals with the CCGCCA sequence (P1 = 2). Significant associations were also identified when comparing the disease allele structure Q1-2-0-9-2, characterized by loss of the CCGCCA sequence, with the reference allele structure, Q1-2-2-P2-2, with individuals having an earlier AoD by 7.1 years. One limitation of our study is that we were not able to obtain detailed clinical information on our HD cohort, and the widely used measure of AoO was only available for a small subset. Clearly, future studies would be facilitated by more in-depth phenotyping. Nonetheless, the robust and highly significant genetic associations we have revealed here confirm that AoD is a clinically meaningful measure capable of providing meaningful insights into HD biology.
The CCGCCA loss is thus proposed as a cis-acting modifier of the HD phenotype in African ancestry individuals. Very recently, an exome sequencing strategy applied to a cohort of HD individuals of European ancestry with either extreme early or extreme late AoO relative to their measured CAG length confirmed effects for the duplication and loss of the CAACAG sequence.34 Interestingly, these analyses also revealed 2 out of 213 individuals with extreme early onset with the Q1-2-0-9-2 structure. This structure was not observed in 206 individuals in the extreme late cohort, nor in 746 individuals in our unselected European ancestry HD cohort.8 These data suggest that the Q1-2-0-9-2 structure is over-represented in an extreme early cohort relative to an unselected cohort (2 out of 213 versus 0 out of 746, p = 0.049, Fisher’s exact test), and in an extreme early cohort relative to a combined unselected/extreme late cohort (2 out of 213 versus 0 out of 952, p = 0.033, Fisher’s exact test). These data thus suggest that the CCGCCA loss may also be a cis-acting modifier of HD motor onset in European individuals affected with HD.
Since inter-locus CAG repeat length instability is modified by the flanking sequence,35 it seems plausible that polymorphisms within the sequence could mediate changes in somatic instability. Previous inter-locus analyses of the relative expandability of multiple disease-associated CAG•CTG repeats (HD, DM1, SCA1, 2, 3, 7, etc.) have revealed associations between higher repeat instability and higher guanine and cytosine (GC) content in the immediate DNA flanking the CAG•CTG repeat.35 It thus seems that a reasonable extension of this observation might be that genetic variants that alter the GC content of the flanking DNA between alleles at one locus might similarly drive differences in somatic instability. Our data support this model, in that the CCGCCA loss was associated with altered somatic expansion scores. However, contrary to the prediction that higher GC content in the flanking sequence, as mediated by the CCGCCA loss, would increase expandability, we found loss of the CCGCCA sequence was actually associated with lower levels of somatic expansion. Thus, unless this effect is reversed in the critical brain regions, we speculate that the disease-accelerating association of the CCGCCA loss is mediated by a pathway other than somatic instability.
As the CCGCCA loss is a synonymous variant that does not alter the coding potential of the pure polyglutamine or pure polyproline tract, there is no obvious mechanism by which this variant could affect the amino sequence of the HTT protein. The total number of prolines encoded by the Q1-2-0-9-2 alleles is 11, exactly the same as that encoded by the most common typical expanded allele structure, Q1-2-2-7-2, in European ancestry populations. Combined with the observation that number of prolines encoded by the variable CCG repeat (P2) has not been revealed as a modifier of HD onset (model 1 Table S4 and Panegyres et al.36), it is unlikely that the phenotypic consequence of the CCGCCA loss is mediated simply by the number of proline in the HTT protein.
As has previously been speculated for the residual modifying effect of the CAACAG sequence (Q2) after correcting for pure CAG length,8 the effect of the CCGCCA loss could be driven by mechanisms that effect the efficiency of HTT transcription, mRNA folding or splicing, or canonical and/or repeat-associated non-AUG (RAN) translation.37, 38, 39, 40 In particular, the CAACAG-CCGCCA intervening sequence lies at a key position in the HTT mRNA that demarcates the boundary between the a long CAG hairpin that is observed in expanded disease-associated alleles, but not in non-disease-associated alleles.37, 38, 39, 40 The CCGCCA effects on mRNA folding in this region could affect RAN translation, which has recently been shown to be highly sensitive to repeat sequence variation at the ATXN8 locus.41 Instead, there may be effects on protein translation. Polyproline regions are known to stall translation,42 an effect that might be further modulated by the relative frequency of CCA and CCG proline tRNAs with potential downstream consequences on HTT protein folding. Alternatively, it is possible there is an effect mediated by a linked variant.43
In other repeat expansion disorders such as SCA1, SCA2 and DM1, interruptions in the repeat tract have been shown to be associated with the disease phenotype. In SCA1, interruptions in the repeat tract confer increased stability, delay AoO, and slow down the rate of aggregation.44 In SCA2, CAA interruptions were shown to be associated with a parkinsonism disease phenotype,45 while in individuals affected with DM1 carrying repeat interruptions there was a later AoO than expected for the repeat length and a reduced level of somatic expansion.46 The CCG and CGG interruptions have been shown to have a stabilizing effect in the blood and often lead to milder symptoms.47
Although the CCGCCA loss was not associated with an increased level of somatic expansion in blood DNA, we did identify a relatively rare large germline expansion where a paternal transmission of 43 CAG repeats to 73 CAG repeats resulted in juvenile HD (JHD) in an HD family carrying the CCGCCA-loss disease allele. Approximately 80% of JHD cases are the result of a paternal transmission, which can be attributed to substantial increases in repeat length occurring during male gametogenesis.6,48,49 A previous case report showed the CCGCCA loss on haplogroup B was associated with a very unusual paternal transmission of 26 CAG repeats to 44 CAG repeats in the child.50 These data suggest that the CCGCCA-loss allele may be associated with higher rates of germline expansion, as has also been proposed for CAACAG loss alleles.
The analysis of background haplotypes revealed that disease allele structures (Q1-0-0-9-2 and Q1-2-0-9-2) characterized by the CCGCCA loss were both present on haplotype B2, as well as being negatively associated with the HD phenotype, compared with haplotypes A4a, A4b, B1, C5, and C9. Haplotype B2 can thus be designated a high-risk haplotype (for early diagnosis) in African ancestry individuals due to its virtually complete association with the CCGCCA loss in disease alleles. The CCGCCA loss and haplotype B2 effects could not be separated out as there is no statistical indication that the earlier AoD exhibited in these individuals is better explained by the CCGCCA loss allele structure or haplotype B2. It is thus possible that the CCGCCA loss is in linkage disequilibrium with another variant on haplotype B2 that affects disease biology. For instance, a linked promoter or enhancer variant might affect HTT transcription rates.
Although HD has been extensively studied in European ancestry individuals, the allele sequence diversity within the HTT repeat tract in African ancestry individuals has not been previously described. Substantial diversity, shown by the presence of predominantly atypical allele structures, is reported. Intriguingly, the most common HD disease allele structure in an African ancestry HD population in South Africa is characterized by the loss of the CCGCCA sequence. This CCGCCA-loss allele structure is associated with an earlier AoD (by 7.1 years) among South African affected individuals of African ancestry, and possibly earlier age at motor onset among European individuals affected with HD.34 Among the HD alleles of African ancestry we have analyzed, this CCGCCA-loss allele structure occurs exclusively on haplotype B2, which we propose as a high-risk haplotype in African ancestry individuals. Despite our observation that overall somatic expansion had a significant inverse association with the HD phenotype in African individuals, in general, the CCGCCA-loss allele structure had the lowest ratio of somatic expansion in blood DNA, suggesting that the disease-accelerating association of the CCGCCA-loss allele is not mediated by an increase in somatic expansion. We propose the CCGCCA-loss allele occurring on haplotype B2 is a cis-acting modifier of HD in our African ancestry individuals that accelerates disease diagnosis through a mechanism that is not driven by somatic instability. Further larger studies in well phenotyped African and European ancestry populations will be required to determine whether the associations observed here are driven directly by the CCGCCA loss and/or by broader haplotype effects. Importantly, this study represents a single African population and thus further ascertainment of African individuals affected with HD and studies of non-disease alleles in Africa are warranted. Nonetheless, these findings already contribute uniquely to the body of knowledge of HD and provide population-specific sequence data for individuals previously understudied.
Data and code availability
The HTT repeat tract was genotyped from the MiSeq reads generated using ScaleHD (v0.251) (https://github.com/helloabunai/ScaleHD). The HTT repeat tract sequence alignments were visualized in Tablet (v1.17.08.17) (https://ics.hutton.ac.uk/tablet/). Statistical analyses were undertaken in R (v3.4.3) (https://www.r-project.org) using RStudio (v1.0.153) (https://www.rstudio.com). The dataset and code supporting the current study have not been deposited in a public repository as broad ethical consent has not been granted as the study participants were selected retrospectively from banked samples but is available from the corresponding author on request.
Acknowledgments
We are grateful to the individuals affected and unaffected with HD who were included in the study. This work is supported by grants to J.D. from the National Research Foundation (UID: 115568), the Faculty Research Committee, and the National Health Laboratory Service Research Trust (94639). This work was also supported by a self-initiated Research Trust Grant to A.K. from the South African Medical Research Council and a travel grant from the National Research Foundation (UID: 110654) and funding to D.G.M. from the CHDI Foundation.
Declaration of interests
Within the last 5 years, D.G.M. has been a scientific consultant and/or received an honoraria/stock options/grants from AMO Pharma, Charles River, LoQus23, Small Molecule RNA, Triplet Therapeutics, and Vertex Pharmaceuticals. D.G.M. also had research contracts with AMO Pharma and Vertex Pharmaceuticals. The other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2022.100130.
Web resources
LDhap tool, https://ldlink.nci.nih.gov/?tab=ldhap
OMIM, https://www.omim.org
Supplemental information
References
- 1.The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Walker F.O. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 3.Roos R.A.C. Huntington's disease: a clinical review. Orphanet J. Rare Dis. 2010;5:40–48. doi: 10.1186/1750-1172-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duyao M., Ambrose C., Myers R., Novelletto A., Persichetti F., Frontali M., Folstein S., Ross C., Franz M., Abbott M., et al. Trinucleotide repeat length instability and age of onset in huntington's disease. Nat. Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy L., Evans E., Chen C.M., Craven L., Detloff P.J., Ennis M., Shelbourne P.F. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum. Mol. Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 6.Telenius H., Kremer B., Goldberg Y.P., Theilmann J., Andrew S.E., Zeisler J., Adam S., Greenberg C., Ives E.J., Clarke L.A., et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat. Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 7.Veitch N.J., Ennis M., McAbney J.P., US-Venezuela Collaborative Research Project. Monckton D.G., Project T.U.-V.C.R. Inherited CAG·CTG allele length is a major modifier of somatic mutation length variability in Huntington disease. DNA Repair. 2007;6:789–796. doi: 10.1016/j.dnarep.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Ciosi M., Maxwell A., Cumming S.A., Hensman Moss D.J., Alshammari A.M., Flower M.D., Durr A., Leavitt B.R., Roos R.A.C., et al. TRACK-HD Team A genetic association study of glutamine-encoding DNA sequence structures, somatic CAG expansion, and DNA repair gene variants, with Huntington disease clinical outcomes. EBioMedicine. 2019;48:568–580. doi: 10.1016/j.ebiom.2019.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu S., Fimmel A., Fung D., Trent R.J. Polymorphisms in the CAG repeat-A source of error in Huntington disease DNA testing. Clin. Genet. 2000;58:469–472. doi: 10.1034/j.1399-0004.2000.580607.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee J.M., Correia K., Loupe J., Kim K.H., Barker D., Hong E.P., Chao M.J., Long J.D., Lucente D., Vonsattel J.P.G., et al. CAG repeat not polyglutamine length determines timing of huntington’s disease onset. Cell. 2019;178:887–900.e14. doi: 10.1016/j.cell.2019.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright G.E.B., Collins J.A., Kay C., McDonald C., Dolzhenko E., Xia Q., Bečanović K., Drögemöller B.I., Semaka A., Nguyen C.M., et al. Length of uninterrupted CAG repeats, independent of polyglutamine size, results in increased somatic instability and hastened age of onset in Huntington disease. Am. J. Hum. Genet. 2019;104:1116–1126. doi: 10.1016/j.ajhg.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldberg Y.P., McMurray C.T., Zeisler J., Almqvist E., Sillence D., Richards F., Gacy A.M., Buchanan J., Telenius H., Hayden M.R. Increased instability of intermediate alleles in families with sporadic Huntington disease compared to similar sized intermediate alleles in the general population. Hum. Mol. Genet. 1995;4:1911–1918. doi: 10.1093/hmg/4.10.1911. [DOI] [PubMed] [Google Scholar]
- 13.Gellera C., Meoni C., Castellotti B., Zappacosta B., Girotti F., Taroni F., DiDonato S. Errors in Huntington disease diagnostic test caused by trinucleotide deletion in the IT15 gene. Am. J. Hum. Genet. 1996;59:475–477. [PMC free article] [PubMed] [Google Scholar]
- 14.Chung M.-Y., Ranum L.P., Duvick L.A., Servadio A., Zoghbi H.Y., Orr H.T. Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat. Genet. 1993;5:254–258. doi: 10.1038/ng1193-254. [DOI] [PubMed] [Google Scholar]
- 15.Eichler E.E., Holden J.J., Popovich B.W., Reiss A.L., Snow K., Thibodeau S.N., Richards C.S., Ward P.A., Nelson D.L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- 16.Kraus-Perrotta C., Lagalwar S. Expansion, mosaicism and interruption: mechanisms of the cag repeat mutation in spinocerebellar ataxia type 1. Cerebellum Ataxias. 2016;3:1–11. doi: 10.1186/s40673-016-0058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., McFarland K.N., Landrian I., Wu S.S., Bower M., Hutter D., Bushara K., Teive H.A.G., Ashizawa T. Identifying novel interruption motifs in spinocerebellar ataxia type 10 expansions. Neurol. Clin. Neurosci. 2014;2:38–43. [Google Scholar]
- 18.Musova Z., Mazanec R., Krepelova A., Ehler E., Vales J., Jaklova R., Prochazka T., Koukal P., Marikova T., Kraus J., et al. Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am. J. Med. Genet. 2009;149A:1365–1374. doi: 10.1002/ajmg.a.32987. [DOI] [PubMed] [Google Scholar]
- 19.Warby S.C., Visscher H., Collins J.A., Doty C.N., Carter C., Butland S.L., Hayden A.R., Kanazawa I., Ross C.J., Hayden M.R. HTT haplotypes contribute to differences in Huntington disease prevalence between europe and East Asia. Eur. J. Hum. Genet. 2011;19:561–566. doi: 10.1038/ejhg.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pringsheim T., Wiltshire K., Day L., Dykeman J., Steeves T., Jette N. The incidence and prevalence of huntington's disease: a systematic review and meta-analysis. Mov. Disord. 2012;27:1083–1091. doi: 10.1002/mds.25075. [DOI] [PubMed] [Google Scholar]
- 21.Baine F.K., Krause A., Greenberg L.J. The frequency of Huntington disease and Huntington disease-like 2 in the South African population. Neuroepidemiology. 2016;46:198–202. doi: 10.1159/000444020. [DOI] [PubMed] [Google Scholar]
- 22.Choudhury A., Ramsay M., Hazelhurst S., Aron S., Bardien S., Botha G., Chimusa E.R., Christoffels A., Gamieldien J., Sefid-Dashti M.J., et al. Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nat. Commun. 2017;8:2062–2112. doi: 10.1038/s41467-017-00663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baine F.K., Kay C., Ketelaar M.E., Collins J.A., Semaka A., Doty C.N., Krause A., Greenberg L.J., Hayden M.R. Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes. Eur. J. Hum. Genet. 2013;21:1120–1127. doi: 10.1038/ejhg.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciosi M., Cumming S.A., Alshammari A.M., Symeonidi E., Herzyk P., McGuinness D., Galbraith J., Hamilton G., Monckton D.G. Library preparation and MiSeq sequencing for the genotyping-by-sequencing of the Huntington disease HTT exon one trinucleotide repeat and the quantification of somatic mosaicism. Protoc. Exch. 2018;2 [Google Scholar]
- 25.Sengupta D., Choudhury A., Fortes-Lima C., Aron S., Whitelaw G., Bostoen K., Gunnink H., Chousou-Polydouri N., Delius P., Tollman S., et al. Genetic substructure and complex demographic history of South African Bantu speakers. Nat. Commun. 2021;12:2080–2113. doi: 10.1038/s41467-021-22207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stephens M., Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J.M., Ramos E.M., Lee J.H., Gillis T., Mysore J.S., Hayden M.R., Warby S.C., Morrison P., Nance M., Ross C.A., et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78:690–695. doi: 10.1212/WNL.0b013e318249f683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenth R. 2018. Estimated Marginal Means, Aka Least-Squares Means. [Google Scholar]
- 31.Kay C., Collins J.A., Wright G.E.B., Baine F., Miedzybrodzka Z., Aminkeng F., Semaka A.J., McDonald C., Davidson M., Madore S.J., et al. The molecular epidemiology of Huntington disease is related to intermediate allele frequency and haplotype in the general population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018;177:346–357. doi: 10.1002/ajmg.b.32618. [DOI] [PubMed] [Google Scholar]
- 32.The 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Findlay Black H., Wright G.E.B., Collins J.A., Caron N., Kay C., Xia Q., et al. Frequency of the loss of CAA interruption in the HTT CAG tract and implications for Huntington disease in the reduced penetrance range. Genet. Med. 2020;22:2108–2113. doi: 10.1038/s41436-020-0917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAllister B., Donaldson J., Binda C.S., Powell S., Chughtai U., Edwards G., Stone J., Lobanov S., Elliston L., Schuhmacher L.-N., et al. Exome sequencing of individuals with Huntington’s disease implicates FAN1 nuclease activity in slowing CAG expansion and disease onset. Nat. Neurosci. 2022;25:446–457. doi: 10.1038/s41593-022-01033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brock G.J., Anderson N.H., Monckton D.G. Cis-acting modifiers of expanded CAG/CTG triplet repeat expandability: associations with flanking GC content and proximity to CpG islands. Hum. Mol. Genet. 1999;8:1061–1067. doi: 10.1093/hmg/8.6.1061. [DOI] [PubMed] [Google Scholar]
- 36.Panegyres P.K., Beilby J., Bulsara M., Toufexis K., Wong C. A study of potential interactive genetic factors in huntington’s disease. Eur. Neurol. 2006;55:189–192. doi: 10.1159/000093867. [DOI] [PubMed] [Google Scholar]
- 37.Busan S., Weeks K.M. The role of context in RNA structure: flanking sequences reconfigure CAG motif folding in huntingtin exon 1 transcripts. Biochemistry. 2013;52:8219–8225. doi: 10.1021/bi401129r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neueder A., Landles C., Ghosh R., Howland D., Myers R.H., Faull R.L.M., Tabrizi S.J., Bates G.P. The pathogenic exon 1 HTT protein is produced by incomplete splicing in huntington’s disease patients. Sci. Rep. 2017;7:1307–1310. doi: 10.1038/s41598-017-01510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krauss S., Griesche N., Jastrzebska E., Chen C., Rutschow D., Achmüller C., Dorn S., Boesch S.M., Lalowski M., Wanker E., et al. Translation of HTT mRNA with expanded CAG repeats is regulated by the MID1–PP2A protein complex. Nat. Commun. 2013;4:1511–1519. doi: 10.1038/ncomms2514. [DOI] [PubMed] [Google Scholar]
- 40.Bañez-Coronel M., Ayhan F., Tarabochia A.D., Zu T., Perez B.A., Tusi S.K., Pletnikova O., Borchelt D.R., Ross C.A., Margolis R.L., et al. RAN translation in Huntington disease. Neuron. 2015;88:667–677. doi: 10.1016/j.neuron.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez B.A., Shorrock H.K., Banez-Coronel M., Zu T., Romano L.E., Laboissonniere L.A., Reid T., Ikeda Y., Reddy K., Gomez C.M., et al. CCG• CGG interruptions in high-penetrance SCA8 families increase RAN translation and protein toxicity. EMBO Mol. Med. 2021;13:e14095. doi: 10.15252/emmm.202114095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pavlov M.Y., Watts R.E., Tan Z., Cornish V.W., Ehrenberg M., Forster A.C. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. USA. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becanović K., Nørremølle A., Neal S.J., Kay C., Collins J.A., Arenillas D., Lilja T., Gaudenzi G., Manoharan S., Doty C.N., et al. A SNP in the HTT promoter alters NF-kB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci. 2015;18:807–816. doi: 10.1038/nn.4014. [DOI] [PubMed] [Google Scholar]
- 44.Menon R.P., Nethisinghe S., Faggiano S., Vannocci T., Rezaei H., Pemble S., Sweeney M.G., Wood N.W., Davis M.B., Pastore A., Giunti P. The role of interruptions in polyQ in the pathology of SCA1. PLoS Genet. 2013;9:e1003648. doi: 10.1371/journal.pgen.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim J.-M., Hong S., Kim G.P., Choi Y.J., Kim Y.K., Park S.S., Kim S.E., Jeon B.S. Importance of low-range CAG expansion and CAA interruption in SCA2 parkinsonism. Arch. Neurol. 2007;64:1510–1518. doi: 10.1001/archneur.64.10.1510. [DOI] [PubMed] [Google Scholar]
- 46.Pešović J., Perić S., Brkušanin M., Brajušković G., Rakočević-Stojanović V., Savić-Pavićević D. Repeat interruptions modify age at onset in myotonic dystrophy type 1 by stabilizing DMPK expansions in somatic cells. Front. Genet. 2018;9:601. doi: 10.3389/fgene.2018.00601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cumming S.A., Hamilton M.J., Robb Y., Gregory H., McWilliam C., Cooper A., Adam B., McGhie J., Hamilton G., Herzyk P., et al. De novo repeat interruptions are associated with reduced somatic instability and mild or absent clinical features in myotonic dystrophy type 1. Eur. J. Hum. Genet. 2018;26:1635–1647. doi: 10.1038/s41431-018-0156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merritt A.D., Conneally P.M., Rahman N.F., Drew A.L. Excerpta Medica Foundation; 1969. Juvenile Huntington’s Chorea. [Google Scholar]
- 49.Nance M.A., Myers R.H. Juvenile onset huntington's disease-clinical and research perspectives. Ment. Retard. Dev. Disabil. Res. Rev. 2001;7:153–157. doi: 10.1002/mrdd.1022. [DOI] [PubMed] [Google Scholar]
- 50.Houge G., Bruland O., Bjørnevoll I., Hayden M.R., Semaka A. De novo Huntington disease caused by 26–44 CAG repeat expansion on a low-risk haplotype. Neurology. 2013;81:1099–1100. doi: 10.1212/WNL.0b013e3182a4a4af. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HTT repeat tract was genotyped from the MiSeq reads generated using ScaleHD (v0.251) (https://github.com/helloabunai/ScaleHD). The HTT repeat tract sequence alignments were visualized in Tablet (v1.17.08.17) (https://ics.hutton.ac.uk/tablet/). Statistical analyses were undertaken in R (v3.4.3) (https://www.r-project.org) using RStudio (v1.0.153) (https://www.rstudio.com). The dataset and code supporting the current study have not been deposited in a public repository as broad ethical consent has not been granted as the study participants were selected retrospectively from banked samples but is available from the corresponding author on request.