ABSTRACT
Studies in a broad range of animal species have revealed phenotypes that are caused by ancestral life experiences, including stress and diet. Ancestral dietary macronutrient composition and quantity (over- and under-nutrition) have been shown to alter descendent growth, metabolism and behaviour. Molecules have been identified in gametes that are changed by ancestral diet and are required for transgenerational effects. However, there is less understanding of the developmental pathways altered by inherited molecules during the period between fertilization and adulthood. To investigate this non-genetic inheritance, we exposed great grand-parental and grand-parental generations to defined protein to carbohydrate (P:C) dietary ratios. Descendent developmental timing was consistently faster in the period between the embryonic and pupal stages when ancestors had a higher P:C ratio diet. Transcriptional analysis revealed extensive and long-lasting changes to the MAPK signalling pathway, which controls growth rate through the regulation of ribosomal RNA transcription. Pharmacological inhibition of both MAPK and rRNA pathways recapitulated the ancestral diet-induced developmental changes. This work provides insight into non-genetic inheritance between fertilization and adulthood.
Keywords: Diet, Nutrition, Epigenetics, Phenocopy
1. Introduction
Evidence accumulated across animal and plant research over the last 20 years has confirmed that the inherited determinants of an organism’s phenotype are more than just DNA [1]. Inheritance of RNA, protein and metabolites in the male or female gamete can influence various traits such as size, shape, behaviour and health [2,3]. Furthermore, ancestral environmental exposures can affect the levels and types of inherited molecules. The consequences of these molecules on descendent phenotype may range from short-term adaptation to a longer-term DNA mutation and selection [4,5], which can contribute to an individual’s disease risk [6].
Ancestral environmental changes that have been found to alter phenotypes of descendants include behavioural stress, toxin exposure and nutritional variation [1–3]. The latter is perhaps the most studied, and examples of ancestral diet altering descendent phenotype due to over- or under-nutrition have been documented in natural populations, including humans [7] and laboratory organisms such as Caenorhabditis elegans [8,9], Drosophila [10–14] and rodents [7]. In these studies, changes to descendent metabolism and growth often result in altered developmental timing, organ size, and body weight. For example, in Drosophila, paternal sugar exposure reprogrammed offspring lipid metabolism, leading to increased stored triglyceride levels [10]. Over 100 studies in rodents have also shown reprogramming of offspring feeding behaviour, birth and adult body weight, adiposity, insulin/glucose metabolism, hypertension etc., due to maternal or paternal obesity or starvation [7]. Another essential feature of developmental programming is that the effect on descendants is dependent on the time point of exposure to the environmental change [15]. Frequently, effects are noted in the offspring generation but are absent, weaker or even different in the grand-offspring generation [7]. This is due to offspring constantly experiencing the exposure directly, either across the placenta while in utero or as exposure to germ cells that will become offspring. The transmission mechanisms can also be indirect, for example, through faecal microbiota transfer, due to alterations in maternal care (including lactation). However, a major focus of the developmental programming field has been identifying molecules in the gametes that drive descendent phenotypes [2]. Epigenetic changes such as DNA methylation, histone modifications and non-coding RNAs have all been found to alter offspring gene expression resulting in developmental and adult phenotype differences. In the Drosophila system mentioned above [10], sugar exposure extensively altered repressive sperm histone methylation patterns in the exposed males, with fat metabolism genes being particularly affected and prone to subsequent transcriptional changes in offspring, but not grand offspring. In mammals, the physiological intimacy of gestation means that developmentally programmed offspring phenotypes are hard to prove due to gamete-derived molecules if the mother is the exposed parent. However, this is not the case when the father is the exposed parent, and there are strong examples of paternal diet- and stress-induced offspring phenotypes being due to alterations in sperm small non-coding RNAs [16–18].
In contrast to the abundance of studies that have searched for a causative epigenetic change in the gamete, there is less information on the post-fertilization mechanisms that bridge the reading of the inherited molecule and the ultimate cell, tissue or whole organism phenotypes. As mentioned, most developmental programming studies have reported metabolic phenotypes. Work by ourselves and others has suggested that Foxo1 mediates developmentally programmed changes to pancreas function in mammals- [19] and Myc-directed pathways [20]. However, less has been reported on changes to pathways that regulate offspring growth and developmental timing. Therefore, in this study, we aimed to understand the developmental pathways affected by ancestral diet at different offspring life stages.
In our previous study, we observed generational differences in reproductive fitness between isosequential Drosophila strains harbouring distinct mitochondrial DNAs (mitotypes) that were fed diets of varying protein to carbohydrate (P:C) ratios [21]. Large shifts in relative proportions of mitotypes within-population cages were seen in generations 1 to 4 but not in later generations. These differences in comparative mitotype fitness were reversible with a diet switch indicating that genetic selection was not responsible. We hypothesized that the within-strain generational differences could be due to transgenerational epigenetic inheritance effects. Notably, in our and other studies, effects accumulated across generations suggest that the fly’s diet, when it produced its gametes, was not the sole cause of the developmental differences [12,13]. We, therefore, designed our experiments to exclude parental effects by ensuring that all parents consumed the same diet, and variation in ancestral diet was limited to the great grand-parental and grand-parental generations (Figure 1). Our studies suggest that ancestral P:C diet-induced descendent developmental timing changes are due to alteration of MAPK signalling pathways in the period between the embryo and pupal stages. While theoretically possible, we suggest that it is exceedingly unlikely that independent mutations arose in the same pathways in three separate experimental blocks (i.e. completely separate multi-generational experiments).
Figure 1.
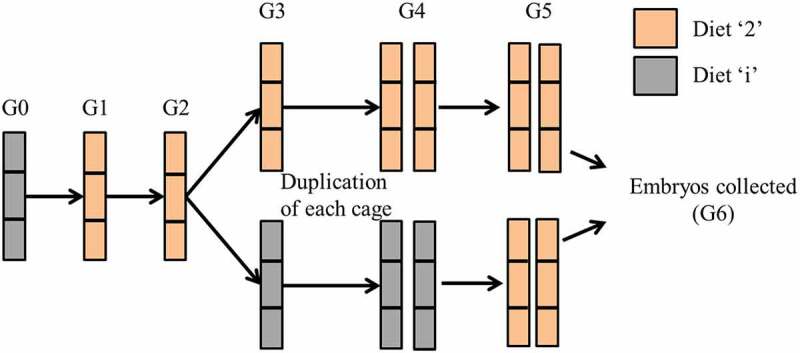
Overview of experimental design: The generational design allowed comparison of G6 embryos produced by great-grandparents and grandparents that differed in their dietary composition, with parents provided the same diet. Diets ‘4’ and ‘6’ were included as additional foods at G3, G4 and G5 only.
2. Materials and methods
(a) Diets and fly genotypes
We utilize four diets in this study that differ in their P:C ratios. Three of these diets, 1:2, 1:4 and 1:6 P:C (referred to as diet ‘2’, diet ‘4’, and diet ‘6’, respectively) are constructed diets, and one, Formula 4–24 instant Drosophila medium (Carolina biological supply company, referred to as diet ‘i’) is a pre-made diet. Diets 2, 4, and 6 were constructed as Aw et al. [21] described. We use the Alstonville; w1118 strain of flies used by Aw et al. [21], which were isosequential due to balancer chromosomes. The study of Aw et al. also confirmed that transgenerational effects of ancestral diet on Drosophila phenotype might vary with the genotype. When fed the 1:2 P:C diet in a population cage, we observed that the frequencies of these flies plateaued after generation 4 (Figure 1(a) [21]). As the four stains in the cage harboured distinct mitochondrial haplotypes but the same w1118 background, we were interested in exploring the possibility that this result was non-genetic in this follow-up study. The reproducibility of larval development including transgenerational effects has been established in a previous study. Aw et al. [21] reproduced the relative larval developmental time of the strains harbouring the Alstonville and Dahomey mitochondria first in an Oregon R background and second in a Canton S nuclear background.
To ensure the flies used in the current studies were isosequential and slightly deleterious mutations had not accumulated, three independently maintained populations of Alstonville, w1118 females, were each backcrossed to w1118/w1118 males for three generations before the generational experiment. The w1118/w1118 males were each sourced from three independent populations. White eye colour and mtDNA type were confirmed before and after each study to ensure no contamination with other fly strains in the laboratory [21]
(b) Generational experimental design
The experimental design differed from our previous study, which aimed to look at the frequency of flies in population cages [21]. We utilized a six-generation (G6) experimental design that controlled the number of generations in which flies were fed each diet (Figure 1). To determine the robustness of the results, we conducted three non-overlapping experimental blocks where the founding females for each block were derived from a different set of backcrossed lines. We then tested experimental replicates within blocks.
A single population cage was initially established with ~800 adult Alstonville flies fed diet ‘i’ (termed generation 0). Oviposition medium consisting of 10% treacle and 4% agar with a thin spread of baker’s yeast was placed into the cage in a small petri dish (3 cm in diameter). This medium was replaced daily for 3 d. On the third day, flies were provided with the oviposition medium for 6 h before removing the medium containing embryos. Embryos were collected from the agar plate onto a fine mesh screen using a water stream from a wash bottle. They were then washed with 4% bleach to remove microbes, thoroughly rinsed with distilled water, and transferred to 60 × 130 mm bottles containing food [22]. Embryos were placed into 12 bottles containing diet ‘2’ (generation 1) at ~200 embryos in each bottle, with four bottles per population cage. In each generation, four adult stock Alstonville and four adult stock Dahomey [21] males maintained on diet ‘i’ were ground in 1400 µl of ddH2O, and 140 µl of the solution was added to each bottle to standardize the microbiome. We did not evaluate microbial composition in this study. Aw et al. [21] found that dietary change from a 1:2 P:C diet to a 1:16 P:C diet does not significantly change the relative microbiota composition in Alstonville flies.
For all generations, day 0 was determined to be when the first flies eclosed. Flies from that day were discarded. Bottles were opened, and adult flies were allowed to populate the cages for 72 h. The bottles were then removed. Oviposition medium was placed in a small petri dish, embryos were collected as described previously, and established on the diet ‘2’ again (generation 2).
Embryos collected from generation 2 were used to seed bottles with diets ‘i’, ‘2’, ‘4’ or ‘6’ (generation 3). Embryos from generation 3 were moved onto the same diet as their parents (i.e. ‘i’ to ‘i’, ‘2’ to ‘2’ etc.; generation 4), and the number of cages was doubled at this generation from 3 to 6 cages. Finally, embryos from generation 4 were placed onto bottles containing diet ‘2’ (generation 5).
Embryos produced by generation 5 flies were collected within 45 minutes of laying to ensure single-celled zygotes were used for RNA-seq, RT-qPCR, and development assays. Collected embryos (generation 6) are referred to as their previous three ancestral generations. Those maintained consistently on diet ‘2’ are termed 222, while those fed different ancestral diets are termed ii2, 442, and 662 (Figure 2). We consider the ii2 diet to be the control diet.
Figure 2.
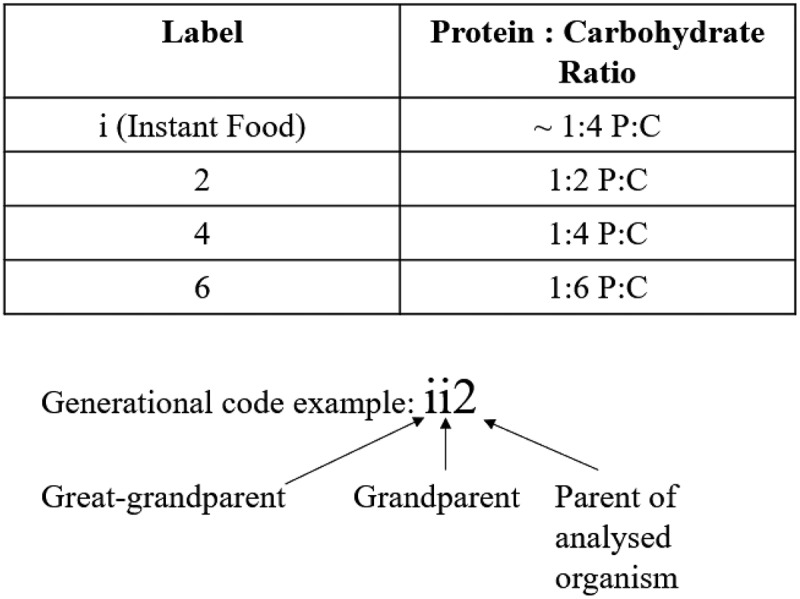
Diet designations: Each analysed organism is referred to by the diet treatment fed to the preceding three generations. The example refers to an organism whose: great grandparents ate the ‘i’ diet, grandparents ate the ‘i’ diet, and parents ate the ‘2’ diet, resulting in a label of ‘ii2’.
(c) Development time and adult weight
Development time of generation 6 embryos was conducted using diet ‘2’. It was measured as a count of adult flies eclosing (hatching) in 3d and the time to pupariation of G6 flies and G5 flies. These development time assays were conducted on three biological blocks and provided three independent measures. Pupariation time was also independently measured for larvae fed diets with the MAPK and ribosome inhibitors. Time to pupariation is calculated as the time in hours from the midpoint of the egg-laying period plus larval development time. Ten embryos per vial of the experimental diet were established with ten treatment blocks. Pupae were individually time-stamped on the side of the vial every 6 hours during daylight hours, and the mean development time of each vial was calculated. Pupariation was identified by observation of everted pupal spiracles.
For the eclosion studies, ten replicate bottles containing approximately 200 embryos were prepared for each diet as described for the generational experiment. Adult flies were cleared from bottles on the first day that eclosion was observed. Flies were then allowed to eclose for 72 h, after which they were collected and counted as the 3d eclosion period.
Adult weight of generation 6 flies was collected from development assays. Flies were initially weighed for wet weight, then incubated at 55°C for 72 h and weighed again for dry weight. Ten groups of five females were measured for each treatment in two independent blocks.
(d) Transcriptome analysis of embryos
Generation 6 embryos were snap-frozen in liquid nitrogen. RNA was isolated from ~20 embryos per cage using the PicoPure RNA isolation kit (Arcturus). DNAse treatment was performed using the RNase-free DNase set (Qiagen). RNAseq was performed at the Ramaciotti Centre for genomics (UNSW). RNA was extracted from five biological replicates of each 222 and ii2 diet group (10 samples in total) using the TruSeq Stranded mRNA-seq kit (Illumina, DA, USA). The samples were sequenced in one flowcell on the NextSeq 500, producing paired-end reads of 75bp.
Reads from the ten samples were mapped to the NCBI D. melanogaster genome GCF_000001215.4_Release_6_plus_ISO1_MT using the Subread package (v 1.6.3) [23]. Mapped reads were assigned to features using the featureCounts function of the Subread package, with an average 33.5 M reads assigned per sample. Lowly expressed genes (cpm < 0.5 in at least four samples) were filtered out. Differential expression analysis was performed using the R environment (R v3.6.3) and the Bioconductor packages edgeR v3.28.1 [24] and limma (voom) v3.42.2 [25]. A DGEList object was created using edgeR, and the data were normalized using the TMM method. This DGEList object was used as input in the voom analysis. The count data were log-transformed before applying the lmFit and eBayes functions to test for differential expression. The commands used to carry out the D.E. analysis are included in Supplementary DE_analysis.txt. Differential expression analysis with edgeR and limma (voom) produced 1550 and 1403 differentially expressed genes (adjusted p.value (Benjamini Hochberg) < 0.05). As 98% of the voom DEGs were also detected by edgeR we used this robust set of genes for downstream analysis. Functional analysis was performed using limma’s kegga and goana functions, the clustering functions within the STRINGdb package v1.26.0 [26] and the enricher function in the clusterProfiler package v3.8 [27].
(e) Pathway validation
Expression of genes of interest was performed using RT-qPCR from 5 replicates/ gene from each of 2 independent blocks (10 total) (Table S1). RNA was isolated using Trizol reagent, and cDNA was prepared using Superscript II RNase (ThermoFisher). Following Aw et al. [21], the expression of genes of interest was calculated relative to the average expression of housekeeping genes using the comparative cycle threshold (Ct) method. The efficiency of amplification was above 95%.
(f) Phenocopy of ancestral-diet induced effects on developmental timing with pharmacological inhibition of MAPK and ribosome biogenesis pathways.
Chemical inhibition of developmental pathways can lead to morphological differences resulting in a copy of a phenotype (phenocopy) distinct from their normal phenotype [21]. Chemical inhibitors can also provide partial inhibition, advantageous over knockouts when complete inhibition is lethal. In Drosophila, progression to the next larval stage is triggered by the larva attaining the necessary body weight. Reaching critical body weight activates a MAPK signalling cascade and upregulates ecdysone biosynthetic genes [28,29]. Furthermore, altered MAPK signalling may affect ribosomal RNA biogenesis and cellular growth [30].
To phenocopy the slower rate of development of flies fed the ii2 diet, we partially inhibited the MAPK and ribosome biogenesis pathways of the flies fed the 222 diet. Preliminary titrations showed that for the MAPK pathway, Egfr was inhibited with 100 μM Tyrphostin AG1478, and rolled was inhibited using 2.5 μM SCH772984. Ribosome biogenesis was inhibited at pol1 using 50 μM BMH-21. Each compound was administered in individual experiments. Development time and gene expression were then analysed.
(g) Statistical analyses
All data were analysed using JMP Pro 16.0.0 (SAS Institute 2021). The interquartile range (IQR) was calculated for all values from the same treatment group (IQR = Q3 – Q1, where Q3 and Q1 are the third and first quartiles of the fitted standard normal distribution). We considered data points smaller than Q1 – 1.5 IQR or greater than Q3 + 1.5 IQR as outliers, and they were removed. For development time, we include the mean development time of each vial. One-way ANOVAs investigated the effect of ancestral diet on time to pupation and 3d eclosion. Two-way ANOVAs of the log-transformed ΔCt data compared the expression of differentially expressed genes whose ancestors were raised on the ii2 and the 222 diets. The primary assumption of the pairwise approach is that the additive effect of concentration, gene, and replicate can be adjusted by subtracting the Ct number of the target gene from that of the reference gene [31]. Data from the distinct studies were combined as no significant block effects were detected. Differences in developmental time and expression were explored Post-hoc using two-tailed Student’s t-tests [21].
3. Results
(a) Development time and adult dry weight
Flies raised on the 222 dietary regimes developed significantly faster than all other treatments, with the ii2 and 442 regimes having the same rate of development and the 662 set having the slowest (Figure 3(a, b)). ANOVAs showed that ancestral diet significantly affected time to pupation (F3, 27.1 = 87.4, p < 0.0001) and 3 d eclosion (F3, 36 = 225.69, p < 0.0001). The effects on developmental timing were also seen when a single previous generation was exposed to a particular diet (Fig. S1). This indicated that a single generation could induce the ancestral diet effect rather than accumulative or exacerbated by an increased number of exposures.
Figure 3.
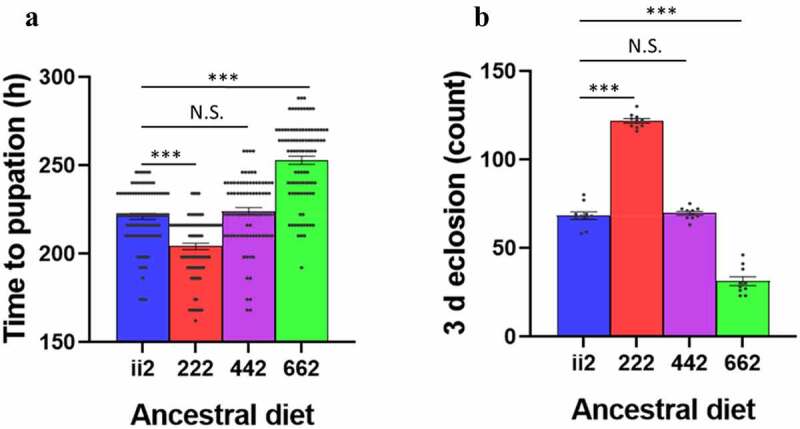
Development assays: Time to pupation (a) and 3 d eclosion (b) of the 222 flies was faster than that of the ii2 and 442 flies, while the 662 flies were the slowest to develop. Bars indicate mean values ± SEM. Post-hoc Student’s t-tests compared ii2 to all other diets in each assay. N.S. indicates not significant, *** indicates p < 0.001.
Adult dry-weight was measured to determine if the faster development of the 222 flies was perhaps a trade-off for lower body size, which is correlated with adult fecundity [32]. A Student’s t-test between ii2 and 222 flies showed no significant difference in adult female body weights (t18 = 0.17, p = 0.87).
(b) Transcriptome analysis of embryos
The ancestral diet-induced differences in developmental timing were evident in the period between the embryonic and pupal stages. Therefore, we performed RNA-seq on ii2 and 222 embryos to identify formative transcriptional signatures that could drive the differences in growth timing. A multidimensional scaling plot demonstrated that the five replicates of the 222 flies separated from the five replicates of the ii2 flies along with the main principal component consistent with a different transcriptional signature in these conditions (Figure 4(a)). Differential expression analysis with limma (voom) produced 1403 differentially expressed genes (DEGs) (adjusted p.value < 0.05). Eight hundred fifty-one of the voom DEGs were upregulated, and 552 were downregulated in the 222 samples compared to ii2, as illustrated by the volcano plot (Figure 4(b)). Pathway and network analysis of the DEGs revealed that growth-associated pathways such as ribosome biogenesis, MAPK signalling and Wnt signalling were higher in 222 compared to ii2. The top upregulated and downregulated KEGG pathways found using the kegga (limma package) are shown (Figure 4(c)). The KEGG pathway results are included in Supplementary Table S2. The interaction network between the genes in the Ribosome and MAPK signalling pathways was determined using STRINGdb (Figure 4(d)).
Figure 4.
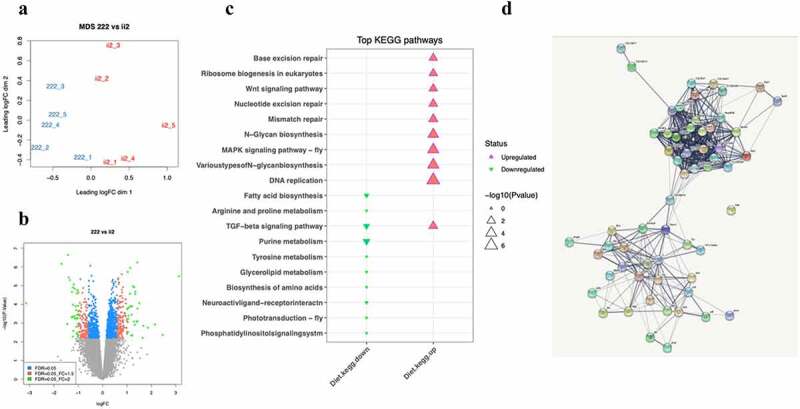
RNAseq analysis: (a) Multidimensional scaling (MDS) plot showing separation between samples (222 = 3 generations fed on 1:2 Protein: Carbohydrate diet, ii2 = change in ancestral diet). (b) Volcano plot showing gene expression changes associated with the 222 diet. (c) Top KEGG pathways as found using kegga (limma). (d) Genes involved in the Ribosome and MAPK signalling pathways had a fold change between the 222 diet and the ii2 diet with an FDR < 0.1. Edges indicate an interaction between genes.
(c) Pathway validation
We focused our subsequent studies on the MAPK and Ribosome Biogenesis development pathways. Diet plays an essential part in activating MAPK pathways [33]. Aldridge and Maggert [34] reported that ribosomal DNA (rDNA) copy number polymorphisms can be created by manipulating the diet of flies. To validate the RNAseq results, we compared the expression of four differentially expressed genes from the MAPK and Ribosome Biogenesis development pathways from embryos whose ancestors were raised on the ii2 and the 222 diets (Figure 5).
Figure 5.
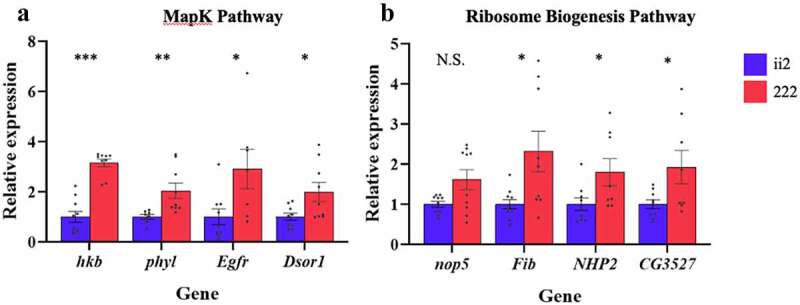
Pathway validation: Expression of MAPK signalling pathway genes (a) and Ribosome biogenesis genes (b) was higher for the 222 flies. Bars indicate relative expression ± SEM. N.S. indicates not significant * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
Two-way ANOVA comparing the expression of four MAPK genes hkb, phyl, Egfr, and Dsor1 genes in larvae fed two ancestral diets shows a significant effect of diet (F1, 65 = 22.52, p < 0.0001) but no significant effect of gene (F3, 65 = 0.24, p = 0.087) or a gene by diet interaction (F3, 65 = 0.37, p = 0.78). To further investigate these results, we conducted post hoc t-tests investigating the influence of ancestral diet on expression diet. The MAPK genes hkb, phyl, Egfr, and Dsor1 all showed significant effect of ancestral diet (t17 = 5.60, p < 0.0001; t15 = 3.72, p = 0.002, t14 = 2.68, p = 0.02; t16 = 2.63, p = 0.02, respectively).
Two-way ANOVA comparing the Ribosome Biogenesis genes nop5, Fib, NHP2, and CG3527 and two ancestral diets shows a significant effect of diet (F3, 65 = 22.52, p < 0.0001) no significant effect of gene (F3, 65 = 0.24, p = 0.87) or a diet by gene interaction (F3, 65 = 0.37, p = 0.78). Expression of three of the Ribosome Biogenesis genes: Fib, NHP2, and CG3527 were significantly affected by ancestral diet (t17 = 2.79, p = 0.01, t15 = 2.42, p = 0.03; t15 = 2.30, p = 0.04, respectively). The expression of nop5 was not significantly affected by diet (t18 = 1.90, p = 0.074).
(d) Phenocopy of ancestral diet induced effects on developmental timing with pharmacological inhibition of MAPK and ribosome biogenesis pathways
We were able to phenocopy the ancestral diet-induced effects on developmental timing through pharmacological inhibition of both MAPK proteins ERK1/2 and EGFR as well as ribosomal biogenesis (Fig. S2). These experiments confirmed the hierarchy of the two processes as inhibition of MAPK proteins affected rRNA transcript levels but not vice versa.
Partially blocking MAPK at EGFR with 100 μM Tyrphostin AG1478 in 222 flies showed reduced expression of seven of eight genes tested (Figure 6(a, b)). For the MAPK pathway ANOVA showed a significant effect of treatment (F1, 71 = 28.45, p < 0.0001), gene, (F3, 71 = 0.88, P = 0.46) and a treatment x gene interaction (F3, 71 = 0.85, P = 0.47). Post hoc t-tests showed significant effect of treatment for hkb, phyl, and Dsor1 (t17 = 2.83, p = 0.01; t18 = 3.20, p = 0.005; t18 = 3.67, p = 0.002, respectively), but treatment was not significant for Egfr (t18 = 1.5719, p = 0.13; Figure 6(a)). For the Ribosome biogenesis pathways ANOVA showed a significant effect of treatment (F1, 72 = 66.67, p < 0.0001) but no significant effect of gene or the interaction (F3, 72 = 0.22, p = 0.88, F3, 72 = 0.087, p = 0.97, respectively). Post hoc t-tests showed significant effects of treatment for nop5, Fib, NHP2, and CG3527 (t18 = 3.64, p = 0.002; t18 = 5.96, p < 0.001; t18 = 3.96, p = 0.0009; t18 = 3.64, p = 0.002, respectively; Figure 6(b)).
Figure 6.
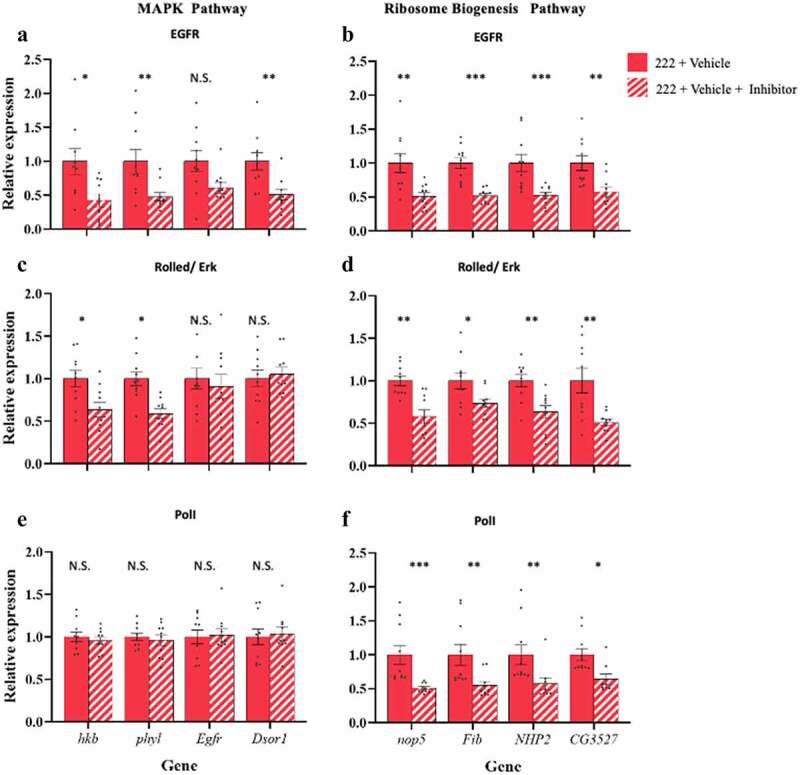
Phenocopy of ancestral diet induced transcriptional changes with pharmacological inhibition of target pathways: Blocking of MAPK with 100 μM Tyrphostin AG1478 (EGFR inhibitor, A, B) or 2.5 μM of SCH772984 (Rolled/Erk inhibitor, C, D), showed a reduction of MAPK gene expression at the point of inhibition and downstream (a, c) but also led to a reduction in expression of ribosome biogenesis pathway genes (b, d). Partially inhibiting ribosome biogenesis with 50 μM of BMH-21 (Pol 1 inhibitor, E, F) showed no change in MAPK expression (e). There was, however, a reduction in the expression of ribosome biogenesis pathway genes (f). Bars indicate relative expression ± SEM. Post-hoc Student’s t-tests N.S. indicates not significant, * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001.
Partial blocking of MAPK at ERK1/2 with 2.5 μM SCH772984 in 222 flies showed reduced expression of MAPK genes and the Ribosome biogenesis pathways downstream of the inhibitor (Figure 6(c,d)). ANOVA showed a significant effect of treatment (F1, 72 = 7.49, p = 0.01), but no significant effect of gene or the interaction (F3, 72 = 2.06, P = 0.11, F3, 72 = 2.19, P = 0.10, respectively). Post-hoc t tests showed significant effect of treatment for hkb and phyl, (t18 = 2.56, p = 0.02, t18 = 2.67, p = 0.02, respectively, Figure 6(c)), with no difference in expression upstream (t18 = 0.70, p = 0.49, t18 = 0.60, p = 0.56 for Egfr, and Dsor1, respectively, Figure 6(c)). As expected, Ribosome biogenesis pathways were down-regulated. ANOVA showed a significant effect of treatment (F1, 72 = 41.14, p < 0.0001), but no significant effect of gene or the interaction (F3, 72 = 1.42, p = 0.24, F3, 72 = 0.95, p = 0.42, respectively). Post-hoc t tests showed a significant effect of treatment for nop5, Fib, NHP2, and CG3527 (t18 = 3.98, p = 0.001, t18 = 2.48, p = 0.02, t18 = 3.06, p = 0.007, t18 = 3.26, p = 0.004, respectively; Figure 6(d)).
Partial inhibition of ribosome biogenesis with 50 μM of BMH-21 showed no inhibition of MAPK expression but reduced expression of ribosome biogenesis pathway genes (Figure 6(e,f)). There was no significant inhibition of MAPK pathway genes. ANOVA showed no significant effect of treatment, gene, or the interaction (F 1, 72 = 0.001, p = 0.97, F3, 72 = 0.05, p = 0.98, F3, 72 = 0.29, P = 0.83, respectively). Post-hoc t tests showed no significant effect of treatment for hkb, phyl, Egfr, and Dsor1 (t18 = 0.56, p = 0.59; t18 = 0.65, p = 0.52; t18 = 0.30, p = 0.77; t18 = 0.44, p = 0.66, respectively, Figure 6(e)). There was reduced expression of ribosome biogenesis pathway genes. ANOVA showed a significant effect of treatment (F1, 70 = 43.17, p < 0.0001), but no significant effect of gene or the interaction (F3, 70 = 0.97, p = 0.41, F3, 70 = 0.44, p = 0.73, respectively. Post-hoc t tests showed a significant effect of treatment for nop5, Fib, NHP2, and CG3527 (t18 = 4.46, p = 0.0003; t18 = 3.29, p = 0.004; t17 = 3.15, p = 0.006; t17 = 2.41, p = 0.03, respectively; Figure 6(f)).
4. Discussion
Here we have shown in a Drosophila model that the MAPK pathway is a mediator of grand-parental dietary effects on prepupal developmental timing. The overall impact of a higher ancestral P:C ratio (e.g. P:C 1:2) is faster development through the embryo to pupal stages than a lower ratio (e.g. P:C 1:6). This result was highly reproducible, seen in three independent experimental blocks, and induced when the grand-parental, or the grand-parental plus great grand-parental generation had the diets.
The study design differed from our previous investigations of multiple generational dietary exposures [21]. The nutritional formulations and use of the Alstonville strain were the same. Still, there were crucial differences, such as the number of generations fed a particular diet and the number of fly strains used. It was essential to change the study design to remove the influence of parental diet on offspring development which has been well described [12–14,35–38]. In comparison with our previous work, the close similarity of the effects of a two-generation and one-generation dietary exposure (Fig. S1) suggests that the new model is not reproducing the accumulative multiple generational effects [21]. This may be due to the cumulative impact occurring after more generations (it was previously seen after four generations on a diet). Otherwise, it could be linked to inter-strain competition, as previously seen in flies kept in cages containing 2–4 different Drosophila strains.
In invertebrates, dietary protein levels have been found to contribute to growth, fecundity and ageing [4,39,40]. Faster developmental timing is observed when a high P:C ratio is consumed by larvae [21], by only the parental generation [37] and now, in this study, only in the grand-parental generation. A recent study of the effects of an ancestral ‘rich’ relatively high protein vs ‘poor’ lower protein diet also observed a faster developmental timing in the grand-offspring of the ‘rich’ diet flies [41]. Interestingly, they observed a greater influence of grand-parental diet than parental diet on various offspring phenotypes, including larval to pupal developmental timing. However, the study did not investigate the underlying growth regulatory pathways affected by the different generational dietary exposures diet.
Diet is a strong determinant of developmental timing in animals [42,43]. In Drosophila, the effects of varying carbohydrates, proteins and lipids on the timing of the various developmental stages have been examined, and the importance of the insulin/TOR pathway for regulation has been highlighted [42]. In this pathway circulating levels of amino acids, carbohydrates and Drosophila insulin-like peptides are detected by cell-surface receptors, which activate intracellular networks of signalling molecules including PI3K/AKT, FOXO and TOR, which in turn regulate processes required for growth such as transcription, protein synthesis and autophagy [42]. However, it is essential to note that in our study, the developmental timing changes in offspring were due to their ancestors, not their diet. Thus, the flies on the ii2 or 222 diets are expected to have qualitatively equivalent circulating levels of amino acids and carbohydrates. Indeed, the RNA-seq analysis did not indicate that this crucial growth regulatory pathway is responsible for the observed ancestral diet effects.
While providing new insight into how transgenerational effects are mediated in the post-fertilization period, our study does not provide evidence on the molecular change in the gametes that drives the subsequent developmental changes. A previous transgenerational study in Drosophila found rDNA copy number reduction in offspring of flies that consumed a high protein diet [34]. We found no evidence for this genetic change with qPCR for rDNA copy number (t22 = 0.45, p = 0.67, ii2 vs 222 larvae) in our study, which points to an epigenetic source of the growth phenotype. Mechanistic studies of the genes identified here could link the observed phenotypes to an epigenetic change in the gametes. The prominence of the MAPK pathway in our study’s network analysis of embryonic transcriptomes makes it tempting to predict that the transgenerational phenotypes are caused by the inheritance of histone modifications or small non-coding RNA that modulates this pathway. Future sophisticated cell- and stage-specific gene deletions of MAPK genes are needed to dissect the mechanisms involved.
Corroboration of the results we observe in a second and third genetic background is required to test the generality of our results. Strain genetics influence various diet responses, including metabolism, offspring development time and transgenerational phenotypes [44–46]. The white mutation in Drosophila has received particular attention as it is now known to influence a wide variety of metabolic pathways [44,47]. However, we and others have abundant evidence that transgenerational effects of diet can be strong determinants of phenotype regardless of the presence [10,11,21,48–50] or absence of the white gene mutation [14,21]. Indeed, there are parallels between the Drosophila white mutation and the mouse agouti gene variants in that mutations initially selected as tools for genetics research have uncovered epigenetic inheritance mechanisms and have profound effects on organismal metabolism [51,52]. Nonetheless, it is unclear whether the non-genetic effects we have observed here result from diet alone or the interaction of diet with the white genetic background. Future work on this model will determine the genetic influences on this novel transgenerational effect.
Supplementary Material
Acknowledgement
This study was funded by an Australian Research Centre Discovery Project Grant (DP190102555). S.C. performed the RNA-seq bioinformatics analysis in the Systems Biology Initiative UNSW headed by Professor Marc Wilkins. S.C.’s work within the SBI was funded by the Australian Federal Government CRIS scheme and a UNSW RIS grant.
Funding Statement
This work was supported by the Australian Research Council [DP190102555].
Author contributions
SGT: Conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft, writing – review and editing.
NAY: Conceptualization, funding acquisition, methodology, project administration, supervision, writing – original draft, writing – review and editing.
SMC: Data curation, formal analysis, methodology, resources, software, visualization, writing – review and editing.
JCS: Conceptualization, methodology, writing – review and editing.
RGM: Review, writing and editing
NT: Funding acquisition, methodology, writing – review and editing.
MJM: Funding acquisition, methodology, writing – review and editing.
JWOB: Conceptualization, funding acquisition, methodology, project administrations, resources, software, supervision, writing – review and editing.
Data availability
The data supporting this study’s findings are available upon reasonable request from the corresponding author, JWOB.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Summary statement
Diet-induced changes in the timing of embryo to pupa development of descendant lineages are caused by alterations to MAPK signalling in Drosophila.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19336934.2022.2088032
References
- [1].Cavalli G, Heard E.. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571(7766):489–499. [DOI] [PubMed] [Google Scholar]
- [2].Perez MF, Lehner B. Intergenerational and transgenerational epigenetic inheritance in animals. Nat Cell Biol. 2019;21(2):143–151. [DOI] [PubMed] [Google Scholar]
- [3].Legoff L, D’Cruz SC, Tevosian S, et al. Transgenerational inheritance of environmentally induced epigenetic alterations during mammalian development. Cells. 2019;8(12):1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Auge GA, Leverett LD, Edwards BR, et al. Adjusting phenotypes via within- and across-generational plasticity. New Phytol. 2017;216(2):343–349. [DOI] [PubMed] [Google Scholar]
- [5].Prokopuk L, Western PS, Stringer JM. Transgenerational epigenetic inheritance: adaptation through the germline epigenome? Epigenomics. 2015;7(5):829–846. [DOI] [PubMed] [Google Scholar]
- [6].Nilsson EE, Sadler-Riggleman I, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet. 2018;4(2):dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Aiken CE, Ozanne SE. Transgenerational developmental programming. Hum Reprod Update. 2014;20(1):63–75. [DOI] [PubMed] [Google Scholar]
- [8].Kishimoto S, Uno M, Okabe E, et al. Environmental stresses induce transgenerationally inheritable survival advantages via germline-to-soma communication in Caenorhabditis elegans. Nat Commun. 2017;8:14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tauffenberger A, Parker JA. Heritable transmission of stress resistance by high dietary Glucose in Caenorhabditis elegans. PLoS Genet. 2014;10(5):e1004346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ost A, Lempradl A, Casas E, et al. Paternal diet defines offspring chromatin state and intergenerational obesity. Cell. 2014;159(6):1352–1364. [DOI] [PubMed] [Google Scholar]
- [11].Xia B, Gerstin E, Schones DE, et al. Transgenerational programming of longevity through E(z)-mediated histone H3K27 trimethylation in Drosophila. Aging (Albany NY). 2016;8(11):2988–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vijendravarma RK, Narasimha S, Kawecki TJ. Effects of parental larval diet on egg size and offspring traits in Drosophila. Biol Lett. 2010;6(2):238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Strilbytska O, Velianyk V, Burdyliuk N, et al. Parental dietary protein-to-carbohydrate ratio affects offspring lifespan and metabolism in Drosophila. Comp Biochem Physiol A: Mol Integr Physiol. 2020;241:110622. [DOI] [PubMed] [Google Scholar]
- [14].Emborski C, Mikheyev AS. Ancestral diet transgenerationally influences offspring in a parent-of-origin and sex-specific manner. Philos Trans R Soc Lond B Biol Sci. 2019;374(1768):20180181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lecomte V, Youngson NA, Maloney CA, et al. Parental programming: how can we improve study design to discern the molecular mechanisms? Bioessays. 2013;35(9):787–793. [DOI] [PubMed] [Google Scholar]
- [16].Klastrup LK, Bak ST, Nielsen AL. The influence of paternal diet on sncRNA-mediated epigenetic inheritance. Mol Genet Genomics. 2019;294(1):1–11. [DOI] [PubMed] [Google Scholar]
- [17].Gapp K, Jawaid A, Sarkies P, et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci. 2014;17(5):667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen Q, Yan MH, Cao ZH, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351(6271):397–400. [DOI] [PubMed] [Google Scholar]
- [19].Talchai SC, Accili D. Legacy effect of Foxo1 in pancreatic endocrine progenitors on adult beta-cell mass and function. Diabetes. 2015;64(8):2868–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ng SF, Lin RCY, Maloney CA, et al. Paternal high-fat diet consumption induces common changes in the transcriptomes of retroperitoneal adipose and pancreatic islet tissues in female rat offspring. FASEB J. 2014;28(4):1830–1841. [DOI] [PubMed] [Google Scholar]
- [21].Aw WC, Towarnicki SG, Melvin RG, et al. Genotype to phenotype: diet-by-mitochondrial DNA haplotype interactions drive metabolic flexibility and organismal fitness. PLoS Genet. 2018;14(11):e1007735. Internet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Clancy DJ, Kennington. A simple method to achieve consistent larval density in bottle cultures. Drosoph Inf Serv. 2001;84:168–169. [Google Scholar]
- [23].Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41(10):e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012;41(D1):D808–D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tennessen JM, Thummel CS. Coordinating growth and maturation - insights from Drosophila. Curr Biol. 2011;21(18):R750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rewitz KF, Yamanaka N, Gilbert LI, et al. The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science. 2009;326(5958):1403–1405. [DOI] [PubMed] [Google Scholar]
- [30].Sriskanthadevan-Pirahas S, Lee J, Grewal SS. The EGF/Ras pathway controls growth in Drosophila via ribosomal RNA synthesis. Dev Biol. 2018;439(1):19–29. [DOI] [PubMed] [Google Scholar]
- [31].Yuan JS, Reed A, Chen F, et al. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Yadav P, Sharma VK. Correlated changes in life history traits in response to selection for faster pre-adult development in the fruit fly Drosophila melanogaster. J Exp Biol. 2014;217(4):580–589. [DOI] [PubMed] [Google Scholar]
- [33].Slattery ML, Lundgreen A, Wolff RK. Dietary influence on MAPK-signaling pathways and risk of colon and rectal cancer. Nutr Cancer. 2013;65(5):729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Aldrich JC, Maggert KA. Transgenerational inheritance of diet-induced genome rearrangements in Drosophila. PLoS Genet. 2015;11(4):e1005148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guida MC, Birse RT, Dall’Agnese A, et al. Intergenerational inheritance of high fat diet-induced cardiac lipotoxicity in Drosophila. Nat Commun. 2019;10:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zajitschek F, Zajitschek S, Manier M. Paternal diet affects differential gene expression, but not sperm competition, in sons. Biol Lett. 2017;13(2):914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Matzkin LM, Johnson S, Paight C, et al. Preadult parental diet affects offspring development and metabolism in Drosophila melanogaster. Plos One. 2013;8(3):0059530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Valtonen TM, Kangassalo K, Polkki M, et al. Transgenerational effects of parental larval diet on offspring development time, adult body size and pathogen resistance in Drosophila melanogaster. Plos One. 2012;7(2):e31611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bruce KD, Hoxha S, Carvalho GB, et al. High carbohydrate-low protein consumption maximizes Drosophila lifespan. Exp Gerontol. 2013;48(10):1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lee KP, Simpson SJ, Clissold FJ, et al. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc Natl Acad Sci U S A. 2008;105(7):2498–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Deas JB, Blondel L, Extavour CG. Ancestral and offspring nutrition interact to affect life-history traits in Drosophila melanogaster. Proceedings of the Royal Society B-Biological Sciences. 2019; 286(1897): 20182778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Danielsen ET, Moeller ME, Rewitz KF. Nutrient signaling and developmental timing of maturation. Curr Top Dev Biol. 2013;105:37–67. [DOI] [PubMed] [Google Scholar]
- [43].Nagarajan S, Grewal SS. An investigation of nutrient-dependent mRNA translation in Drosophila larvae. Biol Open. 2014;3(11):1020–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sasaki A, Nishimura T, Takano T, et al. white regulates proliferative homeostasis of intestinal stem cells during ageing in Drosophila. Nat Metab. 2021;3(4):546–557. [DOI] [PubMed] [Google Scholar]
- [45].Wolff GL, Kodell RL, Moore SR, et al. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12(11):949–957. [PubMed] [Google Scholar]
- [46].Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132(8 Suppl):2393S–400S. [DOI] [PubMed] [Google Scholar]
- [47].Myers JL, Porter M, Narwold N, et al. Mutants of the white ABCG transporter in Drosophila melanogaster have deficient olfactory learning and cholesterol homeostasis. Int J Mol Sci. 2021;22(23). DOI: 10.3390/ijms222312967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vaziri A, Khabiri M, Genaw BT, et al. Persistent epigenetic reprogramming of sweet taste by diet. Sci Adv. 2020;6(46). DOI: 10.1126/sciadv.abc8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Xia B, de Belle JS. Transgenerational programming of longevity and reproduction by post-eclosion dietary manipulation in Drosophila. Aging (Albany NY). 2016;8(5):1115–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Buescher JL, Musselman LP, Wilson CA, et al. Evidence for transgenerational metabolic programming in Drosophila. Dis Model Mech. 2013;6(5):1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Waterland RA, Travisano M, Tahiliani KG, et al. Methyl donor supplementation prevents transgenerational amplification of obesity. Int J Obes (Lond). 2008;32(9):1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Blewitt M, Whitelaw E. The use of mouse models to study epigenetics. Cold Spring Harb Perspect Biol. 2013;5(11):a017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this study’s findings are available upon reasonable request from the corresponding author, JWOB.


