Abstract
RNA-RBP interaction is important in immune regulation and implicated in various immune disorders. The differentiation of proinflammatory T cell subset TH17 and its balance with regulatory T cell (Treg) generation is closely related to autoimmune pathogenesis. The roles of RNA-RBP interaction in regulation of TH17/Treg differentiation and autoinflammation remain in need of further investigation. Here we report that lncRNA-GM polarizes TH17 differentiation but inhibits iTreg differentiation by reducing activity of Foxo1, a transcriptional factor that is important in inhibiting TH17 differentiation but promoting Treg generation. lncRNA-GM–deficient mice were protected from experimental autoimmune encephalomyelitis. Mechanistically, lncRNA-GM directly binds to cytoplasmic Foxo1, thus inhibiting its activity through blocking dephosphorylation of Foxo1 by phosphatase PP2A to promote Il23r transcription. The human homolog of lncRNA-GM (AK026392.1) also polarizes human TH17 differentiation. Our study provides mechanistic insight into the interaction of lncRNA and transcriptional factor in determining T cell subset differentiation during T cell–mediated autoimmune diseases.
LncRNA-GM promotes TH17 differentiation but inhibits Treg generation to exacerbate autoimmune inflammation by targeting Foxo1.
INTRODUCTION
Autoimmune diseases are serious pathological conditions that are characterized by the attack of healthy host cells by autoimmune cells. So far, the effective therapeutic strategies to treat autoimmune diseases are lacking due to limited understanding of its cellular and molecular mechanisms (1). T cell dysfunction is closely related to a variety of autoimmune pathologies such as multiple sclerosis (MS) (2, 3). However, the molecular basis for dysregulation of T cell differentiation in autoimmune diseases remains unclear. Identification of previously unidentified mechanisms underlying T cell–mediated autoimmune inflammation will add insight into the mechanisms that underlie the development of autoimmunity and may suggest possible drug targets for treatment of autoimmune disease.
T helper 17 (TH17) cells are potent triggers of tissue inflammation and are associated with many autoimmune diseases including psoriasis, MS, and rheumatoid arthritis. TH17 cells are differentiated from naïve CD4+ T cells stimulated by interleukin-6 (IL-6) and transforming growth factor–β (TGF-β), these TH17 cells are regarded as nonpathogenic. When these nonpathogenic TH17 cells are further stimulated with additional cytokines, IL-23 and/or IL-1β, they exhibit pathogenic potential that is crucial for mediating autoimmune inflammation (4–6). In addition to TH17 cells, inflammatory TH1 cells are also important in tissue inflammation (7). In contrast, regulatory T (Treg) cells, which are commonly divided into thymus-derived Treg [natural Treg (nTreg)] cells and peripherally induced Treg (iTreg) with highly expression of CD25 and transcription factor, Foxp3 (forkhead box P3), are the major subset for inhibiting excessive inflammation and maintaining immune tolerance (8).
The dynamic balance between the proinflammatory T cell subset TH17 and the Treg cells is essential for the regulation of autoimmunity and inflammation. The disordered TH17/Treg cell balance in vivo is closely related to the pathogenesis and progress of a variety of autoimmune diseases (9). Transcription factors are required for the control of TH17 and Treg cell differentiation. Retinoic acid-related orphan receptor gammat (RORγt) and Foxp3 determine differentiation of CD4+ T cell into TH17 and Treg cells, respectively. Signal transducers and activators of transcription (STAT3) and STAT5 are involved in the regulation of TH17/Treg cell balance (10). In addition, forkhead box O1 (Foxo1) has been recently identified as a key transcription factor to balance TH17/Treg cell differentiation, through promoting Foxp3 expression in Treg cells while suppressing Il17a and Il23r expression in TH17 cells (11, 12). The activity of Foxo1 is tightly regulated by Akt-mediated phosphorylation, which induces its nuclear export and represses the transcription of Foxo1-regulated genes (13). Although Foxo1 has been found to be important in control of TH17/Treg cell balance, how naïve CD4+ T cells integrate diverse signals under complex immune microenvironment to regulate the activity of Foxo1 and TH17/Treg cell balance needs further investigations.
RNA binding proteins (RBPs) are involved in various physiological and pathological conditions via mediating posttranscriptional regulation of gene expression (14). The dysregulation of RBPs is correlated with many human diseases, such as autoimmune and inflammatory diseases (15). The location and activity of RBPs are regulated by both coding and noncoding RNA, which is important for regulation of immune cell development and function (15). mRNA-interacting RBPs are essential for pre-mRNA processing, mRNA stability, and mRNA decay and potently regulate T cell activation and inflammation. For example, Act1 functions as an RBP by interacting with the inflammatory mRNAs and mediating mRNA stabilization, to inhibit IL-17 inflammatory signaling and ameliorate skin inflammation (16). Notably, noncoding RNAs interaction with RBP play important roles in various immunological processes via regulation of gene transcription or translation (17, 18). However, the expression and function of long noncoding RNA (lncRNA) that are highly specific to TH17 cells and the function of the related lncRNA-RBP complexes remain to be identified.
LncRNA-GM, a new lncRNA that we identified in virus-infected macrophages, is important in the regulation of type I interferon (IFN) production in the antiviral innate immune response by interacting with glutathione S-transferase M1 (GSTM1) (19). Notably, lncRNA-GM is highly expressed in the spleen and lymph nodes (19), suggesting that it may have a potential role in regulation of lymphocyte function in addition to its antiviral activity. Here, we show that lncRNA-GM, relatively highly expressed in TH17 cells, promotes TH17 cell differentiation and TH17-mediated inflammatory pathology during autoimmune diseases. Furthermore, lncRNA-GM directly targets Foxo1 and promotes phosphorylation of Foxo1 and thus relieving Il23r from Foxo1-mediated transcriptional inhibition and facilitating the development of TH17 cells. The homolog of lncRNA-GM in human T cells exerts a similar role in promoting TH17 cell differentiation. Thus, our findings provide a new insight to lncRNA-RBP interaction for the regulation of TH17-driven tissue inflammation and autoimmune diseases.
RESULTS
lncRNA-GM expression is increased in TH17 cells
As the expression and function of lncRNA are highly cell type specific (18), we detected the expression patterns of lncRNA-GM in different immune cells. In addition to monocytes/macrophages, lncRNA-GM was also highly expressed in CD4+ T cells (fig. S1A). We further examined its expression in differentiated CD4+ T subsets and found it was especially increased in TH17 cells as compared to TH1 and iTreg cells (Fig. 1A). Especially, lncRNA-GM exhibited higher expression under optimal pathogenic TH17 polarizing conditions (IL-6 + TGF-β + IL-1β + IL-23) (20), compared to nonpathogenic (IL-6 + TGF-β) and pathogenic (IL-6 + IL-1β + IL-23) conditions (Fig. 1A). In addition, the expression of lncRNA-GM was similar in nTreg and iTreg cells (Fig. 1B). Consistently, the human homolog of lncRNA-GM (AK026392.1, h-lncRNA-GM) was highly expressed in human TH17 cells derived from peripheral blood naïve CD4+ T cells (Fig. 1C). Absolute copy number analysis revealed that lncRNA-GM owned about 60 transcript copies per cell in TH17 cells (Fig. 1D and fig. S1B), to a similar level as other lncRNAs in immune cells (21, 22). Thus, lncRNA-GM is expressed in TH17 cells at a relatively high level.
Fig. 1. lncRNA-GM is selectively up-regulated in TH17 cells.
(A) Quantitative polymerase chain reaction (qPCR) analysis of lncRNA-GM mRNA expression in TH1, TH2, nonpathogenic TH17 (IL-6 + TGF-β), pathogenic TH17 (IL-6 + IL-1β + IL-23), optimal pathogenic TH17 (IL-6 + TGF-β + IL-1β + IL-23) cells, and iTreg cells from mice spleen (n = 3 to 4). (B) qPCR of nTreg cells (CD4+ CD25+) in thymocytes and iTreg cells in splenocytes for lncRNA-GM mRNA expression (n = 3 to 4). (C) qPCR analysis of human lncRNA-GM mRNA expression in TH1, TH2, TH17, and iTreg cells from human peripheral blood mononuclear cells (n = 3). (D) Copy number analysis of lncRNA-GM determined by qPCR in TH1 and TH17 (optimal pathogenic induction) cells (n = 3). (E) Chromatin immunoprecipitation (ChIP)–qPCR analysis of the enrichment of H3K9ac, H3K27ac, and H4K4me3 in lncRNA-GM promoter region in naïve T, TH1, TH17, and iTreg cells (n = 3 to 6). (F) qPCR analysis of lncRNA-GM mRNA expression in TH17 cells treated with 20 μM CPI-637 (CPI; n = 4). Results are presented as means ± SD (A to F). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. NS, not significant; DMSO, dimethyl sulfoxide.
To explore why lncRNA-GM is highly expressed in TH17 cells, we investigated whether its expression was regulated by epigenetic modifications. Given that histone modifications including H3K9ac, H3K27ac, and H3K4me3 are transcriptional activation markers for gene expression, which are important for CD4+ T subsets differentiation (23), we first determined whether lncRNA-GM expression was regulated by histone modifications in TH17 cells. By chromatin immunoprecipitation (ChIP)–quantitative polymerase chain reaction (qPCR) assay in naïve T, TH1, TH17, and iTreg cells, we found that the H3K9ac modification, but not H3K4me3 and H3K27ac modifications, was obviously enriched at the promoter region of lncRNA-GM in TH17 cells (Fig. 1E). Furthermore, pharmacological inhibition of H3K9 acetyltransferase with the treatment of specific inhibitor CPI-637 substantially suppressed lncRNA-GM expression in TH17 cells (Fig. 1F). The above results suggested that lncRNA-GM is epigenetically increased by H3K9ac in TH17 cells.
lncRNA-GM promotes TH1/TH17 cell differentiation but inhibits iTreg cell differentiation
We next wondered whether lncRNA-GM could play a role in T cell development or function and generated lncRNA-GM–deficient (lncRNA-GM−/−) mice (19). We found that the proportion of peripheral T cells and B cells was similar between lncRNA-GM−/− mice and wild-type (WT) littermates (fig. S2, A and B). The development of CD4+ T cells and CD8+ T cells was intact in lncRNA-GM−/− mice (fig. S2, C to E). The percentage of nTreg (CD4+CD25+Foxp3+) cells in the thymus of lncRNA-GM−/− mice was comparable to that in WT mice (fig. S2F). Besides, the proliferation of T cells from WT and lncRNA-GM−/− mice was comparable after ligation of T cell receptor (TCR; fig. S3, A and B). Thus, lncRNA-GM did not affect the development or proliferation of peripheral CD4+ T cells.
The high expression of lncRNA-GM in TH17 cells promoted us to investigate the role of lncRNA-GM in CD4+ T cell differentiation and function. We found that the ratio of effector/memory T cells (CD44hi CD62Llo) in lncRNA-GM−/− mice was lower than that in WT littermates, but the ratio of naïve CD4+ T (CD44lo CD62Lhi) cells was similar (Fig. 2A). Next, we measured the proportions of IFN-γ+, IL-4+, IL-17A+, or Foxp3+ T cells within splenic CD4+ T cells and found that the proportions of IFN-γ+ TH1 and IL-17A+ TH17 cells were reduced, while Foxp3+ Treg cells were increased in lncRNA-GM−/− CD4+ T cells in vivo (Fig. 2B). Furthermore, the differentiations of IFN-γ+ TH1 cells and pathogenic TH17 cells (IL-6 + IL-1β + IL-23 or optimal IL-6 + TGF-β + IL-1β + IL-23 induction) were reduced, while Foxp3+ Treg cells were increased in lncRNA-GM−/− CD4+ T cells in vitro. Moreover, there was no significant difference in the frequency of nonpathogenic TH17 cells (IL-6 + TGF-β induction) and IL-4+ TH2 cells between WT and lncRNA-GM−/− mice (Fig. 2, C and D). In addition, overexpression of lncRNA-GM by lentivirus significantly increased TH17 cell differentiation in both WT and lncRNA-GM−/− naïve T cells in vitro (Fig. 2E). These data indicate that lncRNA-GM might promote the differentiation of TH17 cells and inhibit the differentiation of iTreg cells.
Fig. 2. lncRNA-GM promotes TH17 cell differentiation but inhibits Treg cell differentiation in vivo and in vitro.
(A) Flow cytometric analysis of CD44+ and CD62L+ in splenic CD4+ T cells from WT and lncRNA-GM−/− mice (n = 4). (B and C) Flow cytometric analysis IFN-γ+, IL-4+, IL-17A+, and Foxp3+ cells in splenic CD4+ T cells (B) and differentiated TH1, TH2, nonpathogenic TH17 (IL-6 + TGF-β), pathogenic TH17 (IL-6 + IL-1β + IL-23), optimal pathogenic TH17 (IL-6 + TGF-β + IL-1β + IL-23), and iTreg cells for 3 days (C) (n = 4 to 5). (D) Quantification of IFN-γ+, IL-4+, IL-17A+, and Foxp3+ T cells in differentiated TH1, TH2, nonpathogenic TH17, pathogenic TH17, optimal pathogenic TH17, and iTreg cells (n = 3 to 5). (E) Flow cytometric analysis and quantification of IL-17A+ cells transfected with control lentivirus and lncRNA-GM overexpression lentivirus in TH17 cells (n = 4). Results are presented as means ± SD (A, B, D, and E). One of three representative experiments is shown. *P < 0.05; **P < 0.01; ***P < 0.001.
Consistently, silencing of human lncRNA-GM with small interfering RNA (siRNA) in human peripheral blood naïve CD4+ T cells significantly suppressed expression of TH17 signature genes, such as IL17, IL17F, IL21, IL23R, and BATF and promoted expression of Treg-specific genes, such as FOXP3, IL10, IL10RA, and IL10RB (fig. S4, A and B). Therefore, lncRNA-GM can control TH17/Treg cell balance by polarizing TH17 cell differentiation and suppressing iTreg cell induction.
lncRNA-GM promotes TH1/TH17 and inhibits iTreg differentiation by activating mTORC1
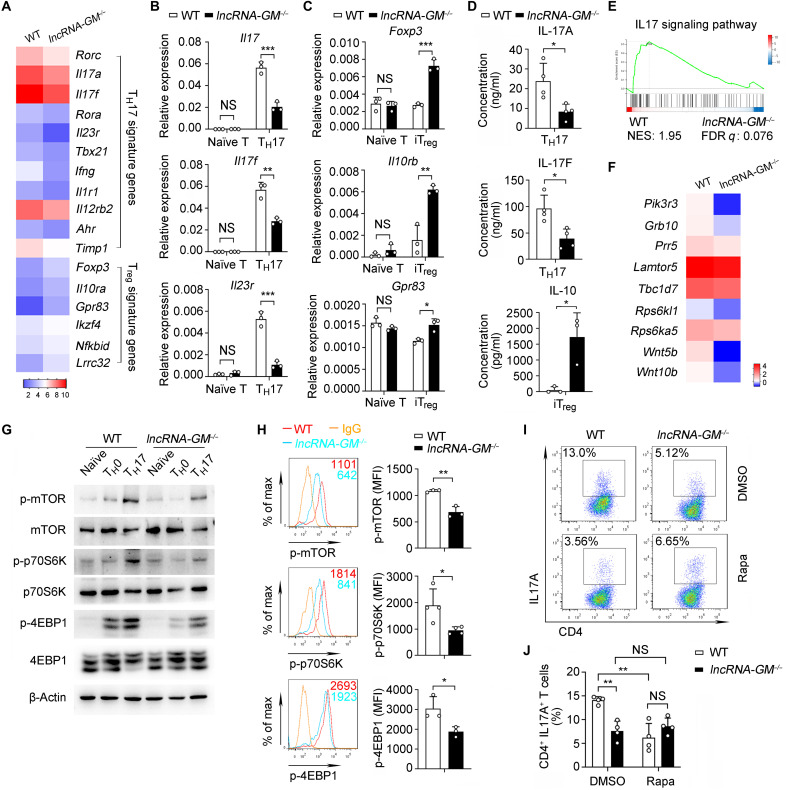
To investigate the molecular mechanism by which lncRNA-GM promotes TH17 cell differentiation, we induced TH17 cells under optimal pathogenic polarizing conditions in vitro. We performed RNA sequencing (RNA-seq) analysis of the differentiated lncRNA-GM−/− and WT TH17 cells. RNA-seq analysis revealed lower expression of TH17 signature genes in lncRNA-GM−/− TH17 cells, such as Rorc, Il17a, Il17f, Rora, Il23r, Tbx21, etc., as compared to that in WT TH17 cells. Meanwhile, Treg-specific genes such as Foxp3, Il10ra, Gpr83, and Lrrc32 in lncRNA-GM−/− TH17 cells were up-regulated (Fig. 3A). qPCR analysis confirmed that lncRNA-GM deficiency notably down-regulated Il17, Il17f, Il23r, Rorc, and Tbx21 expression in TH17 cells (Fig. 3B and fig. S5A) and up-regulated Foxp3, Il10rb, and Gpr83 expression in iTreg cells (Fig. 3C). Furthermore, lncRNA-GM−/− TH17 cells showed lower IL-17A and IL-17F production than WT cells, while lncRNA-GM−/− iTreg cells produced more IL-10 (Fig. 3D). In accordance, gene set enrichment analysis showed that the down-regulated genes in lncRNA-GM−/− TH17 cells were related to IL17 signaling pathway (Fig. 3E). These data further indicate that lncRNA-GM can polarize TH17 cell differentiation but inhibit iTreg cell generation.
Fig. 3. lncRNA-GM promotes TH17 cell differentiation by activating mTORC1 signaling.
(A) Heatmap showing the representative up-regulated or down-regulated genes from WT and lncRNA-GM−/− TH17 cells (fold change > 1.5, P < 0.05). (B) qPCR analysis of Il17, Il17f, and Il23r mRNA expression in naïve T and TH17 cells (n = 3). (C) qPCR analysis of Foxp3, Il10rb, and Gpr83 mRNA expression in naïve T and iTreg cells (n = 3). (D) Cytometric bead array analysis of IL-17A and IL-17F in TH17 cell supernatants and IL-10 in iTreg cell supernatants from WT and lncRNA-GM−/− mice (n = 3 to 4). (E) Gene set enrichment analysis of TH17 cells from WT and lncRNA-GM−/− mice. (F) Heatmap showing down-regulated mTOR pathway–related genes in lncRNA-GM−/− TH17 cells (P < 0.05). (G) Immunoblot analysis of mTORC1 and downstream signaling pathway in WT and lncRNA-GM−/− naïve T, TH0 (α-CD3/CD28), and TH17 (α-CD3/CD28 plus IL-6 + TGF-β + IL-1β + IL-23) cells for 3 days. (H) Phosphorylation of mTOR, p70S6K, and 4EBP1 was measured by flow cytometry from WT and lncRNA-GM−/− TH17 cells (n = 3 to 4). (I and J) Flow cytometric analysis (I) and quantification (J) of CD4+ IL-17A+ cells in WT and lncRNA-GM−/− TH17 cells after treatment with DMSO and 50 nM rapamycin (Rapa; n = 4). Naïve CD4+ T cells were cultured in vitro under optimal pathogenic TH17 cell polarizing conditions for 3 days. Results are presented as means ± SD (B, C, D, H, and J). One of three representative experiments is shown. *P < 0.05; **P < 0.01; ***P < 0.001. FDR, false discovery rate. NES, normalized enrichment scores; MFI, mean fluorescence intensity.
Mammalian target of rapamycin (mTOR) signaling has been shown to play widespread roles in various biological processes, such as metabolic regulation, protein biosynthesis, autophagy, cell survival, etc. (24, 25). mTOR can form two distinct protein complexes: the rapamycin-sensitive mTORC1 (mTOR complex 1) and the rapamycin-insensitive mTORC2 (mTOR complex 2). Notably, mTOR is essential for CD4+ T cell differentiation both in vitro and in vivo. mTORC1 is essential for TH1 and TH17 cell differentiation, whereas mTORC2 promotes TH2 cell differentiation. In addition, mTORC1/2 signaling acts as a negative regulator for iTreg generation but a positive regulator of nTreg function in homeostasis (26–28). Intriguingly, by analysis of RNA-seq data, we found that the expression of mTOR pathway–related genes, such as Lamtor5, Tbc1d7, Grb10, and Prr5, was significantly decreased in lncRNA-GM−/− TH17 cells (Fig. 3F). To assess whether lncRNA-GM promotes TH17 cell differentiation by activating mTORC1 signaling, we detected the activation of mTORC1 and downstream signaling in TH17 cells (IL-6 + TGF-β + IL-1β + IL-23 induction). Both immunoblot analysis and flow cytometric assay showed that TH17 cell polarization–induced phosphorylation of mTOR and activation of mTORC1 pathway (phosphorylation of p70S6K and 4EBP1) were remarkably inhibited by lncRNA-GM deletion (Fig. 3, G and H). Furthermore, inhibition of mTORC1 signaling with rapamycin blocked TH17 cell differentiation in WT CD4+ T cells (Fig. 3, I and J, and fig. S5B), while activation of mTORC1 signaling with MHY1485 reversed the decreased TH17 cell differentiation in lncRNA-GM−/− CD4+ T cells (fig. S5, C and D). Meanwhile, we also observed that mTORC1 and downstream signaling p70S6K and 4EBP1 were down-regulated in lncRNA-GM−/− TH1 cells comparing to that in WT TH1 cells (fig. S5, E and F) and treatment with rapamycin blocked TH1 cell differentiation in WT CD4+ T cells (fig. S5G). Moreover, lncRNA-GM deficiency also inhibited phosphorylation of mTORC1 and its downstream signal p70S6K and 4EBP1 in lncRNA-GM−/− iTreg cells (fig. S5, H and I), and rapamycin promoted iTreg cell differentiation in WT and lncRNA-GM−/− CD4+ cells (fig. S5J). However, lncRNA-GM deficiency did not affect the activation of mTOR signaling in nTreg cells (fig. S5K).
The downstream signals triggered by IL-6, IL-2, and TGF-β are important for the differentiation of TH17 cells (29). However, deficiency of lncRNA-GM did not influence the activation of Stat3, Smad2, and Stat5 induced by IL-6, IL-2, and TGF-β, respectively (fig. S5L), indicating that the TH17-polarizing role of lncRNA-GM is not dependent on IL-6/IL-2/TGF-β signaling pathways. Collectively, these data suggest that lncRNA-GM promotes TH1/TH17 and inhibits iTreg cell differentiation by activating mTORC1 signaling.
lncRNA-GM promotes TH17 differentiation independent of in cis and GSTM1
LncRNAs can affect the expression of their adjacent genes through in cis mode (30). However, qPCR analysis revealed that the expression of lncRNA-GM adjacent genes Haus3 and Mxd4 in TH17 cells and iTreg cells was similar between WT and lncRNA-GM−/− mice (fig. S6, A and B), indicating that lncRNA-GM regulates TH17/Treg cell balance independent of its in cis function.
Our previous study showed that lncRNA-GM in macrophages promoted TANK-binding kinase 1 (TBK1) activation to exert antiviral function by binding to GSTM1, a glutathione S-transferase (19). So, we explored whether lncRNA-GM promoted TH17 cell differentiation by binding to GSTM1. However, the public database in BioGPS showed that the expression of GSTM1 in T cells is markedly lower than its expression in macrophages (fig. S6C). Consistently, the mRNA level of GSTM1 in CD4+ T cells was just about 1 of 150 of those in macrophages, with its protein level also extremely low in naïve T and TH17 cells (fig. S6, D and E). In addition, our RNA immunoprecipitation (RIP)–qPCR analysis demonstrated that lncRNA-GM did not bind to GSTM1 in TH17 cells (fig. S6F). Thus, lncRNA-GM promotes TH17 cell differentiation independent of GSTM1.
lncRNA-GM binds to Foxo1 and inhibits Foxo1 dephosphorylation to activate mTORC1
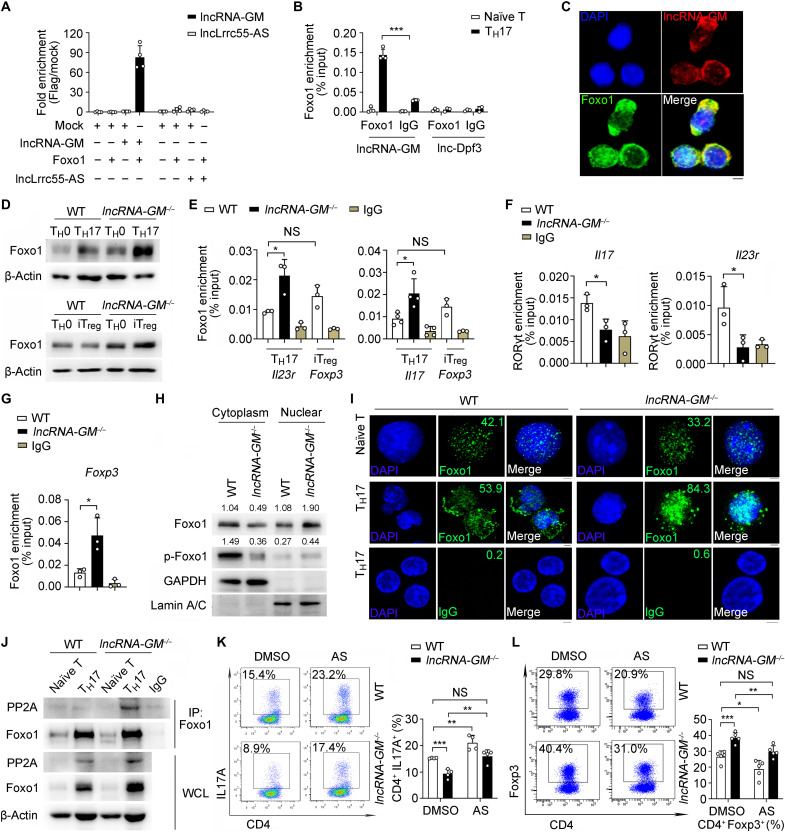
RNA-RBP interaction plays essential role in regulation of T cell differentiation and autoinflammation (31, 32). To figure out the mechanism for lncRNA-GM in regulation of mTORC1 activation, we screened for lncRNA-GM binding partners via RIP-qPCR assay (fig. S7A). Of 11 candidate TH17 cell–related proteins (transcription factor, receptor, mTOR-associated protein, etc.), there was potent binding between lncRNA-GM and Foxo1, a member of forkhead box O (Foxo) transcription factor family (Fig. 4A). We further confirmed lncRNA-GM directly bound to Foxo1 in the cytoplasm of differentiated TH17 cells by RIP-qPCR (Fig. 4B) and RNA–fluorescence in situ hybridization (FISH; Fig. 4C). There was nearly no interaction between Foxo1 with negative control [Mock plasmids or immunoglobulin G (IgG)] or two other lncRNAs, lncLrrc55-AS (21) and lnc-Dpf3 (33), which had been previously reported to bind transcriptional factors and were also expressed in T cells, indicating that Foxo1 interacted with lncRNA-GM in a specific manner (Fig. 4, A and B). Therefore, we identified Foxo1 as a lncRNA-GM–interacting RBP. Intriguingly, lncRNA-GM deletion resulted in a remarkable increase of the Foxo1 protein level (Fig. 4D) and Foxo1 mRNA expression (fig. S7B) in both TH17 and iTreg cells. However, the levels of Foxo1 mRNA or protein in TH1 and TH2 cells were similar between WT and lncRNA-GM−/− littermates (fig. S7, C and D). These results indicate that lncRNA-GM directly binds to Foxo1 and reduces Foxo1 expression in TH17 and iTreg cells.
Fig. 4. lncRNA-GM directly interacts with Foxo1 and attenuates Foxo1 dephosphorylation by PP2A.
(A and B) RIP-qPCR analysis of lncRNA-GM and lncLrrc55-AS immunoprecipitated by Flag antibody in HEK293T (A) and lncRNA-GM and lnc-Dpf3 immunoprecipitated by Foxo1 or IgG in T cells (B) (n = 3 to 4). (C) RNA FISH analysis of lncRNA-GM (red) and Foxo1 (green) in TH17 cells. DNA (blue) was stained with 4′,6-diamidino-2-phenylindole (DAPI). Scale bar, 2.5 μm. (D) Immunoblot analysis of Foxo1 levels in TH17 and iTreg cells. (E and F) ChIP-qPCR analysis of the recruitment of Foxo1 to Il23r and Il17 promoter regions (E) and RORγt to Il23r and Il17 promoter regions (F) in TH17 cells and the recruitment of Foxo1 to Foxp3 promoter regions in WT iTreg cells (n = 3 to 4). (G) ChIP-qPCR analysis of the recruitment of Foxo1 to Foxp3 promoter regions in iTreg cells (n = 3). (H) Immunoblot analysis of Foxo1 and p-Foxo1 (Ser256) levels in cytoplasm and nuclear extracts from TH17 cells. (I) Immunofluorescence analysis of Foxo1 (green) or IgG (green) in naïve T or TH17 cells. DNA (blue) was stained with DAPI. Scale bar, 2.5 μm. (J) Immunoblot analysis of PP2A in T cells after IP with anti-Foxo1 antibody. (K and L) Flow cytometric analysis of IL-17A+ cells in TH17 (K) after treatment with DMSO or 50 nM AS1842856 (AS) and Foxp3+ cells in iTreg (L) after treatment with 200 nM AS1842856 (n = 4 to 5). Results are presented as means ± SD (A, B, E, F, G, K, and L). One of three representative experiments is shown. *P < 0.05; **P < 0.01; ***P < 0.001.
Because Foxo1 is involved in TH17 and Treg cell development and function (11), Foxo1 could directly bind to the Foxp3 promoter region and promote Foxp3 expression (34). Meanwhile, Foxo1 binds to the Il23r promoter to inhibit TH17 differentiation (12). Therefore, we sought to determine whether the transcriptional activity of Foxo1 depends on its interaction with lncRNA-GM. ChIP-qPCR showed that the enrichment of Foxo1 at Il23r and Il17 gene promoter was increased in lncRNA-GM−/− TH17 cells, which was accompanied by reduced binding of RORγt at Il23r and Il17 gene promoter (Fig. 4, E and F). These results suggested that lncRNA-GM promotes TH17 cell differentiation by targeting Foxo1 and relieving the suppressive effect of Foxo1 on Il23r and Il17. Consistently, enrichment of Foxo1 at Foxp3 promoter was also increased in lncRNA-GM−/− iTreg cells (Fig. 4G). Furthermore, we did not detect binding of Foxo1 on the Tbx21 promoter region in TH1 cells (fig. S7E), which was consistent with the previous observations (35, 36). Dephosphorylation has been shown to mediate nuclear retention of Foxo1 (13, 37). We examined Foxo1 and phosphorylated (p-) Foxo1 protein level in nuclear and cytoplasm and found that Foxo1 phosphorylation in cytoplasm was significantly reduced and its nuclear accumulation was increased in lncRNA-GM−/− TH17 cells (Fig. 4H). We also observed that Foxo1 nuclear accumulation was increased in lncRNA-GM−/− TH17 cells but not in naïve T cells by confocal microscopy (Fig. 4I). Therefore, lncRNA-GM deficiency promotes dephosphorylation and nuclear retention of Foxo1 and consequently enhances Foxo1-dependent inhibition of Il23r and Il17 gene transcription.
Protein phosphatase 2 (PP2A), an important Foxo1 phosphatase, dephosphorylates Foxo1 in cytoplasm and plays a critical role in TH17 cell development (38, 39). We found that phosphatase PP2A directly bound to Foxo1 in TH17 cells, and this interaction was significantly increased in lncRNA-GM−/− TH17 cells (Fig. 4J). Furthermore, treatment of Foxo1 inhibitor AS1842856 could efficiently increase the differentiation of TH17 cells and decrease the differentiation of iTreg cells (Fig. 4, K and L). Foxo1 inhibition also reversed TH17 cell differentiation that was suppressed by lncRNA-GM deletion (Fig. 4K). Foxo1 could indirectly activate adenosine monophosphate-activated protein kinase (AMPK) and inhibit mTORC1 activity, which were also up-regulated by TCR signaling of T cell activation (11, 40). In the presence of anti-CD3 and anti-CD28 stimulation, phosphorylation of Foxo1 (Ser256) and mTORC1 signaling were pronouncedly reduced in lncRNA-GM−/− CD4+ T cells compared with that in WT CD4+ T cells (fig. S7F). Consistently, Foxo1 inhibition efficiently increased the activation of mTOR signaling in both WT and lncRNA-GM−/− TH17 cells (fig. S7G). Together, these data suggested that during TH17 cell differentiation, lncRNA-GM binds to transcriptional factor Foxo1 and reduces its dephosphorylation by PP2A for preventing Foxo1 from nuclear translocation and transcription suppression, thus consequently resulting in mTORC1 activation and promotion of TH17 differentiation.
lncRNA-GM–deficient mice attenuates tissue inflammation and autoimmune diseases in vivo
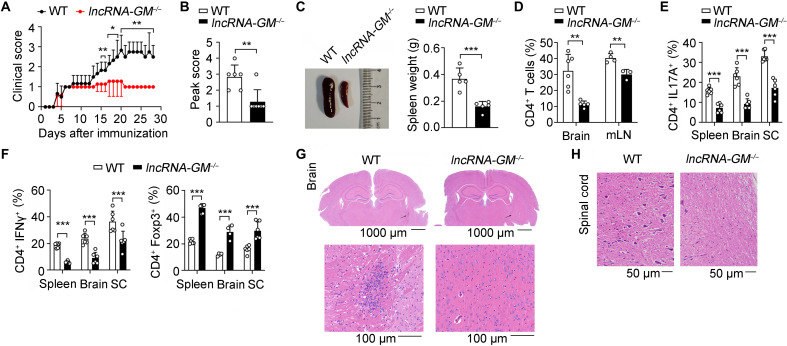
MS is a chronic, organ-specific T cell–mediated autoimmune disease, characterized by demyelination and axonal loss (41). Given that lncRNA-GM promoted TH17 cell differentiation, we tested the in vivo role of lncRNA-GM in regulating a TH17-dependent autoimmune disease model, experimental autoimmune encephalomyelitis (EAE), an established mouse model of human MS. lncRNA-GM–deficient mice showed milder disease severity with lower clinical score, peak score compared to WT mice (Fig. 5, A and B). In addition, lncRNA-GM−/− mice had smaller spleen and less spleen weight (Fig. 5C), accompanied by substantially reduced infiltration of CD4+ T cells and monocytes in brain and mesenteric lymph node (Fig. 5D and fig. S8, A and B). The numbers of neutrophils (CD11b+Ly6Ghi) and B cells (B220+) were comparable between lncRNA-GM−/− and control mice (fig. S8, C and D). Especially, the infiltration of TH17 and TH1 cells into the brain, spinal cord, and spleen was decreased, while the infiltration of Treg cells was increased in lncRNA-GM−/− mice with EAE (Fig. 5, E and F, and fig. S8E). Histological analysis showed less mononuclear cell infiltration in the brain and spinal cord of lncRNA-GM−/− mice (Fig. 5, G and H).
Fig. 5. Deficiency of lncRNA-GM attenuates pathogenesis of active EAE.
(A and B) Mean clinical score (A) and peak clinical score (B) of WT and lncRNA-GM−/− mice after induction of EAE with MOG35–55 and pertussis toxin (n = 6). (C to H) At day 28 after EAE induction, WT and lncRNA-GM−/− mice were analyzed for spleen size and weight (n = 5) (C), quantification of CD4+ T cells in the brain and mesenteric lymph node (mLN; n = 3 to 6) (D), quantification of IL-17A+, IFN-γ+, and Foxp3+ T cells in the brain, spleen, and spinal cord (SC; n = 4 to 6) (E and F), and histology of hematoxylin and eosin–stained brain (G) and spinal cord (H). Results are presented as means ± SD (A, B, C, D, E, and F). One of three representative experiments is shown. *P < 0.05; **P < 0.01; ***P < 0.001.
As we previously demonstrated that lncRNA-GM enhanced type I IFN production in macrophages (19) and type I IFNs (IFN-α and IFN-β) was shown to ameliorate pathological inflammation of central nervous system (CNS) diseases (42–44), we tested whether lncRNA-GM prevents autoimmune inflammation via regulating type I IFN production. We found no difference of sera IFN-α or Ifna4 and Ifnb1 mRNA levels between WT and lncRNA-GM−/− mice (fig. S8, F to H). Thus, lncRNA-GM promotes progress of EAE independent of type I IFN production.
To further confirm the T cell–intrinsic regulatory role of lncRNA-GM in the pathogenesis of EAE, we adoptively transferred Rag2−/− mice with WT or lncRNA-GM−/− CD4+ T cells before EAE induction. Compared to the recipient Rag2−/− mice that were transferred with WT CD4+ T cells, the mice receiving lncRNA-GM−/− CD4+ T cells displayed remarkably decreased severity and reduced clinical scores (Fig. 6, A and B), accompanied by reduced CD4+ and CD8+ T cell proportions in the spleen (Fig. 6C and fig. S9A) and less TH17/TH1 cells but more Treg cells in CNS (Fig. 6D and fig. S9, B to D). Therefore, lncRNA-GM mediates autoimmune inflammation by polarizing TH17 cell differentiation but inhibiting Treg induction.
Fig. 6. Deficiency of lncRNA-GM reduces severity of EAE by T cell adoptive transfer.
(A and B) Mean clinical score (A) and peak clinical score (B) of Rag2−/− recipients of WT and lncRNA-GM−/− CD4+ T cells after induction of EAE (n = 7). (C and D) At day 28 after EAE induction, Rag2−/− recipient mice were analyzed for quantification of CD4+ and CD8+ T cells in the spleen (n = 4) (C) and quantification of CD4+ IL-17A+, IFN-γ+, and Foxp3+ T cells in the spleen, brain, and spinal cord (n = 3 to 6) (D). Results are presented as means ± SD (A to D). One of three representative experiments is shown. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
DISCUSSION
Hyperactivity of proinflammatory CD4+ T cells causes autoimmunity and inflammation, whereas Treg cells inhibit autoinflammation and maintain immune homeostasis (45). The molecular mechanisms governing the balance between TH17/Treg cells during T cell–mediated autoinflammation remains unclear. In this study, we demonstrated that lncRNA-GM, with a specific higher expression in TH17 cells, directly binds to Foxo1 and forms a new RNA-RBP complex to facilitate TH17 cell differentiation and promote the development of autoimmune diseases. We also found human lncRNA-GM promotes TH17 cell differentiation and inhibits Treg cell induction, indicating that lncRNA-GM may serve as a potential therapeutic target for the treatment of TH17-mediated autoimmune diseases.
RNA-RBP interactions are critical for gene expression and function of the immune system. RBPs interact with RNAs with specific sequences or structures to control RNA stability, translation, and translocation (14). Especially, RBPs are found to widely interact with noncoding RNAs such as lncRNAs, small nuclear RNAs, and transfer RNAs. Among them, lncRNA interaction with RBPs has been shown to play critical roles in T cell differentiation. For example, Linc-CCR2-5′ binds to GATA binding protein-3 (GATA-3) and is required for the TH2-specific gene expression and TH2 cell migration (46). LincRNA-MAF-4 is exclusively expressed in TH1 cells and promotes the development of TH1 cells by inhibiting the function of the transcription factor c-MAF4 (47). Long noncoding RNA nuclear-enriched abundant transcript 1 (lncRNA NEAT1), specifically expressed in TH17 cells, promotes TH17 cell differentiation by binding STAT3 and inhibiting its degradation (48). LncRNA Flicr (Foxp3 long intergenic noncoding RNA) in Treg cells negatively regulates Foxp3 transcription and inhibits Treg cell differentiation (49). Different from these findings, we report that lncRNA-GM specifically binds to transcription factor Foxo1, forming a new RNA-RBP complex to regulate the TH17/Treg cell differentiation balance and promote autoimmune pathology. We previously showed lncRNA-GM play a pivotal role in antiviral activity of macrophage via binding to GSTM1 and reducing GSTM1-mediated S-glutathionylation of TBK1 (19). In our current study, we reveal that lncRNA-GM interacts with Foxo1 to repress Foxo1 transcriptional activity, leading to TH17/Treg cell imbalance. We demonstrate that lncRNA-GM promotes TH17 cell differentiation by targeting Foxo1 and relieving the suppressive effect of Foxo1 on Il23r and Il17 and inhibits iTreg cell differentiation by regulating transcription of Foxp3. Therefore, our work broadened the cell type–specific immunoregulatory function of lncRNA-GM in the pathogenesis of inflammatory autoimmune diseases, and it will be valuable to further investigate the regulation and function of lncRNA-GM in other pathological conditions in the future.
Compared with mRNAs, most of the lncRNAs are expressed at relatively low levels in cells (50). LncRNAs that display low cellular abundance but play significant biological and immunological functions have been widely reported. For example, lnc13, which has only 10 molecules per cell, potently represses inflammatory gene expression via binding to heterogeneous nuclear ribonucleoprotein D (hnRNPD) and regulates the susceptibility to celiac disease (51), and lincRNA-EPS (long intergenic noncoding RNA-erythroid prosurvival), which has 11 copy numbers per cell, plays an important role in inhibition of inflammatory responses via controlling nucleosome positioning and repressing gene transcription (22). Furthermore, other lncRNAs with low copies including Gas5 (with glucocorticoid receptor) (52), gadd7 (with TDP-43) (53), and PANDA (with NF-YA) (54) have been also reported to own key biological roles. Therefore, the regulatory role of lncRNA in inflammatory and immune responses largely depends on their protein, DNA, or RNA binding activity and does not necessarily require a large cellular abundance. Other than the copy number of RNA, the specific RNA secondary structure and multiple protein-binding motif sequence might play more important roles in the interaction of lncRNA with the target proteins. In this study, we show that lncRNA-GM, which contains ~60 copy number per TH17 cell, promotes TH17 cell polarization via binding to Foxo1 and mediating its dephosphorylation and nuclear export, thus adding a new way of moderately expressed lncRNA for regulating immune responses.
Furthermore, the expression of lncRNA is relatively tissue and cell specific. Abnormal expression of lncRNA in CNS can lead to the occurrence of various neurological disorders. It was shown that some lncRNAs (such as lncRNA-Malat1 and lnc-DC) expressions were dysregulated in the serum of patients with MS (55). In addition, high expression of lncRNAs was also found in some patients with other diseases such as asthma, myasthenia gravis, and thymoma (56, 57). We clarified the role of lncRNA-GM in TH17 cell–driven inflammatory autoimmune diseases. The expression of lncRNA-GM was tissue specific with high expression in the spleen and lymph nodes and cell type specific with exclusively high expression in TH17 cells. Consistently, the homolog of lncRNA-GM in human was also highly expressed in TH17 cells from human peripheral blood mononuclear cells, indicating that lncRNA-GM homolog expression may also be helpful for predication of T cell–associated autoimmune diseases.
TH17 cell differentiation and function are determined by defined cytokine milieus and distinct signals induced by cytokines. IL-6–STAT3 and TGF-β–Smad2/3 signaling pathways promote TH17 cell differentiation, whereas IL-2–STAT5 signaling inhibits TH17 cell differentiation (5, 6, 29). In addition, T cells depend on mTOR activity for their development, activation, and differentiation. In our study, we identified that lncRNA-GM promotes TH1/TH17 cell differentiation and inhibits iTreg cell differentiation by enhancing the mTORC1 signaling pathway. Foxo1 is known as a rheostat that maintains mTORC1 activity. On one hand, under the conditions of oxidative stress or growth factor limitation, Foxo1 promotes Sestrin3 (Sesn3) expression that further inhibits mTORC1 activity in tuberous sclerosis complex 2 (TSC2)–dependent manner (58). On the other hand, Foxo1 indirectly activates AMPK by induction of Sesn3, the activated AMPK could also inhibit mTORC1 signaling (40). In addition, AMPK can be also activated by liver kinase B1 (LKB1), which is independent of Foxo1 activity (40). Therefore, in our study, the observation that Foxo1 inhibitor AS1842856 could partially rescue TH17 differentiation in lncRNA-GM−/− mice might due to compensative effect of AMPK activation induced by other factors, such as LKB1 and TSC2. Overall, our data indicated that lncRNA-GM directly binds to Foxo1 and promotes TH17 cell differentiation, which is mainly depending on activation of mTORC1 pathway.
As previously described, transcription factor Foxo1 is critical for regulation of TH17/Treg cell balance (59). Our data presented a previously unknown mechanism for the regulation of Foxo1 activity by lncRNA-GM–mediated phosphorylation. LncRNA-GM directly interacts with Foxo1 and blocks its dephosphorylation mediated by PP2A, enhancing its nuclear export. Intriguingly, PP2A is reported to decrease the activation of Smad2 but increase the activation of Smad3 for differentiation of TH17 (39). Our data further provide evidence for the positive role of PP2A in TH17 cell differentiation through regulating Foxo1 phosphorylation. However, the mechanism of PP2A in regulating TH17/Treg cell differentiation and the related inflammatory autoimmune diseases remain to be further clarified in detail. In conclusion, we illustrate a previously unknown mechanism of lncRNA-GM in the pathogenesis of TH17/Treg cell imbalance–associated autoimmune diseases, providing a potential therapeutic strategy for control of autoimmune inflammation.
MATERIALS AND METHODS
Mice
C57BL/6J mice were from Joint Ventures Sipper BK Experimental Animal Company (Shanghai, China), and Rag2−/− [B6(Cg)-Rag2tm1.1Cgn/J, 008449] mice were obtained from The Jackson Laboratory. lncRNA-GM−/− mice were prepared as described previously (19). All animal experiments were conformed to the National Institute of Health Guide for the Care and Use of Laboratory Animals, with the approval of the Scientific Investigation Board of Second Military Medical University, Shanghai.
In vitro cell proliferation assay
Purified CD4+ T cells or CD8+ T cells were labeled with CellTrace Violet (Thermo Fisher Scientific) and stimulated with anti-mouse CD3 (5 μg/ml) and anti-mouse CD28 (2 μg/ml) for 72 hours. Cell proliferation of CD4+ or CD8+ T cells was determined by flow cytometry.
Flow cytometry
For surface staining, cells were stained with the appropriate antibodies in phosphate-buffered saline for 20 min at room temperature. For intracellular staining, cells were fixed with 4% paraformaldehyde and then stained with specific antibodies in 1× fixation and permeabilization solution (BD Biosciences). Foxp3 was stained with a Foxp3 transcription factor staining buffer set (eBioscience), following the manufacturer’s instructions. Stained cells were detected with an LSRII or a Fortessa flow cytometer (BD Biosciences) and analyzed using FACS (fluorescence-activated cell sorting) Diva software (BD Biosciences) or FlowJo software (Tree Star).
Cytometric bead array analysis
For detection of IL-17A, IL-17F, and IL-10 concentrations in TH17 or iTreg cell supernatants, the mouse IL-17A Flex Set, IL-17F Flex Set, or IL-10 Flex Set (BD Biosciences) was used following the manufacturers’ instructions.
Immunoblot and immunoprecipitation analysis
A total of 1 × 106 cells were lysed in 1× Cell Lysis Buffer (Cell Signaling Technology) to obtain whole-cell lysates. Equalized whole-cell lysates were used for immunoblot analysis as previously reported (60). Antibodies were used for immunoblot analysis as follows: anti–p-mTOR (Ser2448), anti-mTOR, anti-Foxo1, anti–p-Foxo1 (Ser256), anti–4E-BP1, anti–p-4E-BP1 (Thr37/46), anti–p-p70S6K (Thr421/Ser424), anti-p70S6K, anti–p-Stat3 (Tyr705), anti-Stat3, anti–p-Smad2 (Ser465/467), anti-Smad2, anti–p-Stat5 (Tyr694), anti-Stat5, anti-Lamin A/C (4C11), and anti–PP2A-A were obtained from Cell Signaling Technology; anti–glyceraldehyde phosphate dehydrogenase and anti-GSTM1 were from ProteinTech; anti–β-actin was from Santa Cruz Biotechnology. The nucleus and cytoplasm proteins were extracted according to the manufacturer’s instructions by NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific).
For immunoprecipitation, 5 × 106 T cells were harvested and then lysed in phosphatase extraction buffer [20 mM imidazole-HCl, 2 mM EDTA, and 2 mM EGTA (pH 7.0)]. Foxo1 was immunoprecipitated using anti-Foxo1 antibody (Cell Signaling Technology) and protein A agarose. Beads were washed four times in 1× tris-buffered saline, and the proteins were determined by immunoblot analysis.
RNA interference
For interference of human lncRNA-GM homolog, human CD4+ T cells were purified from peripheral blood. CD4+ T cells were transfected with 200 nM siRNA-AK026392.1 using the P3 Primary Cell 4D-Nucleofector X Kit reagent (Lonza) following the manufacturer’s instructions. TH17 cell differentiation medium was applied 24 hours after transfection. mRNA expression was analyzed by qPCR 3 days later. Two different set of siRNAs were used for AK026392.1 interference: (i) GCC CUC AAU UCU UGA AGC ATT and (ii) GGU ACA UCU GUA GCC AAU UTT. Control siRNAs were (i) UUC UCC GAA CGU GUC ACG UTT and (ii) ACG UGA CAC GUU CGG AGA ATT.
RIP assay
RIP assay was used as previously described (19). A total of 5 × 106 T cells were harvested and lysed with Cell Lysis Buffer (Cell Signaling Technology) and supplemented with protease inhibitor cocktail (Sigma-Aldrich) and ribonuclease inhibitor (Thermo Fisher Scientific), incubated on ice for 30 min, and then centrifuged. Cell lysates were collected and then incubated with GSTM1 (Proteintech), Foxo1 (Cell Signaling Technology) antibody or control antibody IgG (Cell Signaling Technology) at 4°C for overnight. Protein G magnetic beads (Cell Signaling Technology) were added to each sample and incubated for 3 hours at 4°C. Beads were washed four times in wash buffer [20 mM tris-HCl (pH 7.0), 1 mM EDTA, 0.5% NP-40, and 100 mM NaCl] and then subjected to RNA extraction.
RNA FISH and immunofluorescence microscopy
Cy3-conjugated lncRNA-GM probes were designed by the Stellaris FISH Probe Designer (Biosearch Technologies). RNA FISH was performed as described previously (19). TH17 cells were fixed with fixation/permeabilization solution (eBioscience) for 15 min at room temperature, followed by hybridization with probes according to the Stellaris RNA FISH protocol.
For immunofluorescence analysis, naïve T and TH17 cells were fixed with fixation/permeabilization solution for 15 min at room temperature and then followed by incubation with anti-FOXO1A antibody (Abcam) or control antibody IgG (Cell Signaling Technology) and Alexa Fluor 647 anti-rabbit secondary antibody (Thermo Fisher Scientific). Images were obtained with a laser scanning confocal microscope (Leica TCS SP8) and analyzed by the LAS X software version 2.0.2.15022.
ChIP-qPCR assay
T cells were harvested and cross-linked according to the ChIP Assay Kit (Millipore) protocol. ChIP assay was performed with anti-Foxo1 antibody, anti-H3K9Ac antibody, anti-H3K27Ac antibody, anti-H3K4me3 antibody or control normal rabbit IgG antibody (Cell Signaling Technology), anti-RORγt (AFKJS-9) antibody (Thermo Fisher Scientific), and normal rat IgG antibody (Santa Cruz Biotechnology). The primers for qPCR were listed as follows: lncRNA-GM, GAA TTT TGT GGC AGC TCA GC (forward) and ACA CAC AAT AGC CTT GGC TG (reverse); Il23r, CAC CAT TCG CCC TCA AGA AC (forward) and CGT CTC TGG AGG TCA TGG TT (reverse); Il17, AGC TCC CAA GAA GTC ATG CT (forward) TAC GTC AAG AGT GGG TTG GG (reverse); Foxp3, GTT CCT GCT GTC AGG GTA GC (forward) and GTT CTT GTC AGA GGC AGG CT (reverse).
EAE induction and T cell adoptive transfer
The EAE mouse model was induced as described previously (61). Briefly, female mice at the age of 12 weeks were immunized with 300 mg of MOG35–55 (Sangon) in Freund’s Complete Adjuvant (Thermo Fisher Scientific) and treated with 200 ng of pertussis toxin (Gibco) intraperitoneally on days 0 and 2.
For T cell adoptive transfer, CD4+ T cells were collected from WT and lncRNA-GM−/− mice and then intravenously transferred into Rag2−/− mice at 5 × 106 per mouse. The recipient Rag2−/− mice were immunized with MOG35–55 and treated with pertussis toxin at days 0 and 2. Clinical scores were evaluated according to the following standards: 0, normal; 1, limp tail; 2, limp tail, impaired righting reflex, and paresis of one limb; 3, hindlimb paralysis; 4, hindlimb and forelimb paralysis; and 5, moribund.
For flow cytometric analysis of brain and spinal cord mononuclear cells, single-cell suspensions from brain and spinal cord were poured into 35% Percoll (GE Healthcare) reagent. The cells at the bottom layer were then collected and used immediately for antibody staining.
Lentiviral infection
The cDNA for lncRNA-GM (19) was amplified by PCR, inserted into pRRLSIN-cPPT-SFFV-EGFP-MCS-SV40-puromycin (GV655) lentiviral vector (Genechem, China), and then subjected to lentiviral packaging and purification. Naïve CD4+ T cells were stimulated with anti-mouse CD3 (5 μg/ml) and anti-mouse CD28 (2 μg/ml) antibodies for 24 hours. The activated CD4+ T cells (5 × 105) were then pun infected for 90 min at 1500 rpm and polybrene (10 μg/ml) and incubated at 37°C for 24 hours. Later, the virus supernatants were removed, and medium containing IL-6 (30 ng/ml), h-TGF-β1 (3 ng/ml), IL-1β (10 ng/ml), and IL-23 (20 ng/ml) was added for TH17 cell induction.
Statistical analysis
All the experiments were independently repeated at least three times. Statistical analysis was performed using the software GraphPad Prism 8.0. The data were assessed by unpaired two-tailed Student’s t test. P ≤ 0.05 was considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001).
Acknowledgments
We thank M. Yang and T. Chen (Second Military Medical University) for the helpful suggestions and technical assistance.
Funding: This work was supported by grants from the National Key R&D Program (2018YFA0507401), National Natural Science Foundation of China (81788101, 81801577, and 31870909), CAMS Innovation Fund for Medical Sciences (2021-1-I2M-017), Young Elite Scientists Sponsorship Program by CAST (2020QNRC001), and China National Postdoctoral Program for Innovative Talents (BX2021046).
Author contributions: X.C. designed and supervised the research. Y.C. and K.C. performed the experiments. J.L., X.Z., H.Z., Y.W., Z.L., Y.L., S.L., S.X.L., and N.L. contributed reagents and analytical tools. Y.C., K.C., and X.C. analyzed the data and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-seq data are deposited in NCBI GEO under accession code GSE182253.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S9
Original data
REFERENCES AND NOTES
- 1.Fugger L., Jensen L. T., Rossjohn J., Challenges, progress, and prospects of developing therapies to treat autoimmune diseases. Cell 181, 63–80 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Chataway J., Tackling progression in multiple sclerosis. Lancet Neurol. 17, 489–491 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Theofilopoulos A. N., Kono D. H., Baccala R., The multiple pathways to autoimmunity. Nat. Immunol. 18, 716–724 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaublomme J. T., Yosef N., Lee Y., Gertner R. S., Yang L. V., Wu C., Pandolfi P. P., Mak T., Satija R., Shalek A. K., Kuchroo V. K., Park H., Regev A., Single-cell genomics unveils critical regulators of Th17 cell pathogenicity. Cell 163, 1400–1412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghoreschi K., Laurence A., Yang X.-P., Tato C. M., McGeachy M. J., Konkel J. E., Ramos H. L., Wei L., Davidson T. S., Bouladoux N., Grainger J. R., Chen Q., Kanno Y., Watford W. T., Sun H.-W., Eberl G., Shevach E. M., Belkaid Y., Cua D. J., Chen W., O’Shea J. J., Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature 467, 967–971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee Y., Awasthi A., Yosef N., Quintana F. J., Xiao S., Peters A., Wu C., Kleinewietfeld M., Kunder S., Hafler D. A., Sobel R. A., Regev A., Kuchroo V. K., Induction and molecular signature of pathogenic TH17 cells. Nat. Immunol. 13, 991–999 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stienne C., Michieletto M. F., Benamar M., Carrie N., Bernard I., Nguyen X. H., Lippi Y., Duguet F., Liblau R. S., Hedrick S. M., Saoudi A., Dejean A. S., Foxo3 transcription factor drives pathogenic T helper 1 differentiation by inducing the expression of eomes. Immunity 45, 774–787 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakaguchi S., Yamaguchi T., Nomura T., Ono M., Regulatory T cells and immune tolerance. Cell 133, 775–787 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Littman D. R., Rudensky A. Y., Th17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Zhu J., T helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harb. Perspect. Biol. 10, a030338 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedrick S. M., Michelini R. H., Doedens A. L., Goldrath A. W., Stone E. L., FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12, 649–661 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu C., Yosef N., Thalhamer T., Zhu C., Xiao S., Kishi Y., Regev A., Kuchroo V. K., Induction of pathogenic TH17 cells by inducible salt-sensing kinase SGK1. Nature 496, 513–517 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo C. T., Li M. O., Foxo transcription factors in T cell biology and tumor immunity. Semin. Cancer Biol. 50, 13–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner M., Diaz-Munoz M. D., RNA-binding proteins control gene expression and cell fate in the immune system. Nat. Immunol. 19, 120–129 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Gebauer F., Schwarzl T., Valcárcel J., Hentze M. W., RNA-binding proteins in human genetic disease. Nat. Rev. Genet. 22, 185–198 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Herjan T., Hong L., Bubenik J., Bulek K., Qian W., Liu C., Li X., Chen X., Yang H., Ouyang S., Zhou H., Zhao J., Vasu K., Cockman E., Aronica M., Asosingh K., Licatalosi D. D., Qin J., Fox P. L., Hamilton T. A., Driscoll D., Li X., IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat. Immunol. 19, 354–365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun X., Haider Ali M. S. S., Moran M., The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem. J. 474, 2925–2935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y. G., Satpathy A. T., Chang H. Y., Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 18, 962–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Wang P., Zhang Y., Xu J., Li Z., Li Z., Zhou Z., Liu L., Cao X., Decreased expression of the host long-noncoding RNA-GM facilitates viral escape by inhibiting the kinase activity TBK1 via S-glutathionylation. Immunity 53, 1168–1181.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Ni L., Wan S., Zhao X., Ding X., Dejean A., Dong C., Febrile Temperature critically controls the differentiation and pathogenicity of T helper 17 cells. Immunity 52, 328–341.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., Li M., Xue Y., Li Z., Wen W., Liu X., Ma Y., Zhang L., Shen Z., Cao X., Interferon-inducible cytoplasmic lncLrrc55-AS promotes antiviral innate responses by strengthening IRF3 phosphorylation. Cell Res. 29, 641–654 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atianand M. K., Hu W., Satpathy A. T., Shen Y., Ricci E. P., A.-Dominguez J. R., Bhatta A., Schattgen S. A., McGowan J. D., Blin J., Braun J. E., Gandhi P., Moore M. J., Chang H. Y., Lodish H. F., Caffrey D. R., Fitzgerald K. A., A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chisolm D. A., Weinmann A. S., Connections between metabolism and epigenetics in programming cellular differentiation. Annu. Rev. Immunol. 36, 221–246 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Kim J., Guan K. L., mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21, 63–71 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Mossmann D., Park S., Hall M. N., mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer 18, 744–757 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Delgoffe G. M., Pollizzi K. N., Waickman A. T., Heikamp E., Meyers D. J., Horton M. R., Xiao B., Worley P. F., Powell J. D., The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 12, 295–303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgoffe G. M., Kole T. P., Zheng Y., Zarek P. E., Matthews K. L., Xiao B., Worley P. F., Kozma S. C., Powell J. D., The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity 30, 832–844 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H., Long L., Zhou P., Chapman N. M., Chi H., mTOR signaling at the crossroads of environmental signals and T-cell fate decisions. Immunol. Rev. 295, 15–38 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu R., Hatton R. D., Weaver C. T., The Th17 family flexibility follows function. Immunol. Rev. 252, 89–103 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batista P. J., Chang H. Y., Long noncoding RNAs: Cellular address codes in development and disease. Cell 152, 1298–1307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heissmeyer V., Vogel K. U., Molecular control of Tfh-cell differentiation by Roquin family proteins. Immunol. Rev. 253, 273–289 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Ivanov P., Paul A., Post-transcriptional regulatory networks in immunity. Immunol. Rev. 253, 253–272 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J., Zhang X., Chen K., Cheng Y., Liu S., Xia M., Chen Y., Zhu H., Li Z., Cao X., CCR7 chemokine receptor-inducible lnc-Dpf3 restrains dendritic cell migration by inhibiting HIF-1α-mediated glycolysis. Immunity 50, 600–615.e15 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Ouyang W., Beckett O., Ma Q., Paik J. H., DePinho R. A., Li M. O., Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11, 618–627 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Rao R. R., Li Q., Bupp M. R. G., Shrikant P. A., Transcription factor Foxo1 represses T-bet-mediated effector functions and promotes memory CD8+ T cell differentiation. Immunity 36, 374–387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng Y., Kerdiles Y., Chu J., Yuan S., Wang Y., Chen X., Mao H., Zhang L., Zhang J., Hughes T., Deng Y., Zhang Q., Wang F., Zou X., Liu C.-G., Freud A. G., Li X., Caligiuri M. A., Vivier E., Yu J., Transcription factor Foxo1 is a negative regulator of natural killer cell maturation and function. Immunity 42, 457–470 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen J., Lu Y., Tian M., Huang Q., Molecular mechanisms of FOXO1 in adipocyte differentiation. J. Mol. Endocrinol. 62, R239–R253 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Yan L., Lavin V. A., Moser L. R., Cui Q., Kanies C., Yang E., PP2A regulates the pro-apoptotic activity of FOXO1. J. Biol. Chem. 283, 7411–7420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Q., Jin X., Zheng M., Rohila D., Fu G., Wen Z., Lou J., Wu S., Sloan R., Wang L., Hu H., Gao X., Lu L., Phosphatase PP2A is essential for TH17 differentiation. Proc. Natl. Acad. Sci. U.S.A. 116, 982–987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hay N., Interplay between FOXO, TOR, and Akt. Biochim. Biophys. Acta 1813, 1965–1970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh J., Vidal-Jordana A., Montalban X., Multiple sclerosis: Clinical aspects. Curr. Opin. Neurol. 31, 752–759 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Prinz M., Schmidt H., Mildner A., Knobeloch K.-P., Hanisch U.-K., Raasch J., Merkler D., Detje C., Gutcher I., Mages J., Lang R., Martin R., Gold R., Becher B., Brück W., Kalinke U., Distinct and nonredundant in vivo functions of IFNAR on myeloid cells limit autoimmunity in the central nervous system. Immunity 28, 675–686 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Rasouli J., Casella G., Ishikawa L. L. W., Thome R., Boehm A., Ertel A., Melo-Silva C. R., Mari E. R., Porazzi P., Zhang W., Xiao D., Sigal L. J., Fortina P., Zhang G.-X., Rostami A., Ciric B., IFN-β acts on monocytes to ameliorate CNS autoimmunity by inhibiting proinflammatory cross-talk between monocytes and Th cells. Front. Immunol. 12, 679498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khorooshi R., Morch M. T., Holm T. H., Berg C. T., Dieu R. T., Draeby D., Issazadeh-Navikas S., Weiss S., Lienenklaus S., Owens T., Induction of endogenous Type I interferon within the central nervous system plays a protective role in experimental autoimmune encephalomyelitis. Acta Neuropathol. 130, 107–118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee G. R., The balance of Th17 versus Treg cells in autoimmunity. Int. J. Mol. Sci. 19, 730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu G., Tang Q., Sharma S., Yu F., Escobar T. M., Muljo S. A., Zhu J., Zhao K., Expression and regulation of intergenic long noncoding RNAs during T cell development and differentiation. Nat. Immunol. 14, 1190–1198 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranzani V., Rossetti G., Panzeri I., Arrigoni A., Bonnal R. J., Curti S., Gruarin P., Provasi E., Sugliano E., Marconi M., De Francesco R., Geginat J., Bodega B., Abrignani S., Pagani M., The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 16, 318–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shui X., Chen S., Lin J., Kong J., Zhou C., Wu J., Knockdown of lncRNA NEAT1 inhibits Th17/CD4+ T cell differentiation through reducing the STAT3 protein level. J. Cell. Physiol. 234, 22477–22484 (2019). [DOI] [PubMed] [Google Scholar]
- 49.Zemmour D., Pratama A., Loughhead S. M., Mathis D., Benoist C., Flicr, a long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc. Natl. Acad. Sci. U.S.A. 114, E3472–E3480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Statello L., Guo C. J., Chen L. L., Huarte M., Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C.-Rubio A., F.-Jimenez N., Kratchmarov R., Luo X., Bhagat G., Green P. H., Schneider R., Kiledjian M., Bilbao J. R., Ghosh S., A long noncoding RNA associated with susceptibility to celiac disease. Science 352, 91–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kino T., Hurt D. E., Ichijo T., Nader N., Chrousos G. P., Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3, ra8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X., Li D., Zhang W., Guo M., Zhan Q., Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. EMBO J. 31, 4415–4427 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hung T., Wang Y., Lin M. F., Koegel A. K., Kotake Y., Grant G. D., Horlings H. M., Shah N., Umbricht C., Wang P., Wang Y., Kong B., Langerød A., B.-Dale A.-L., Kim S. K., van de Vijver M., Sukumar S., Whitfield M. L., Kellis M., Xiong Y., Wong D. J., Chang H. Y., Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet. 43, 621–629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shaker O. G., Mahmoud R. H., Abdelaleem O. O., Ibrahem E. G., Mohamed A. A., Zaki O. M., Abdelghaffar N. K., Ahmed T. I., Hemeda N. F., Ahmed N. A., Mansour D. F., LncRNAs, MALAT1 and lnc-DC as potential biomarkers for multiple sclerosis diagnosis. Biosci. Rep. 39, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu L., Jiang J., Yin Y., Hu B., LncRNA XLOC_003810 modulates thymic Th17/Treg balance in myasthenia gravis with thymoma. Clin. Exp. Pharmacol. Physiol. 47, 989–996 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Qiu Y. Y., Wu Y., Lin M. J., Bian T., Xiao Y. L., Qin C., LncRNA-MEG3 functions as a competing endogenous RNA to regulate Treg/Th17 balance in patients with asthma by targeting microRNA-17/ RORγt. Biomed. Pharmacother. 111, 386–394 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Chen C. C., Jeon S. M., Bhaskar P. T., Nogueira V., Sundararajan D., Tonic I., Park Y., Hay N., FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and rictor. Dev. Cell 18, 592–604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu C., Chen Z., Xiao S., Thalhamer T., Madi A., Han T., Kuchroo V., SGK1 governs the reciprocal development of Th17 and regulatory T cells. Cell Rep. 22, 653–665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K., Liu J., Liu S., Xia M., Zhang X., Han D., Jiang Y., Wang C., Cao X., Methyltransferase SETD2-mediated methylation of STAT1 is critical for interferon antiviral activity. Cell 170, 492–506.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Yang M., Chen T., Li X., Yu Z., Tang S., Wang C., Gu Y., Liu Y., Xu S., Li W., Zhang X., Wang J., Cao X., K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8+ T cell activation. Nat. Immunol. 16, 1253–1262 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S9
Original data