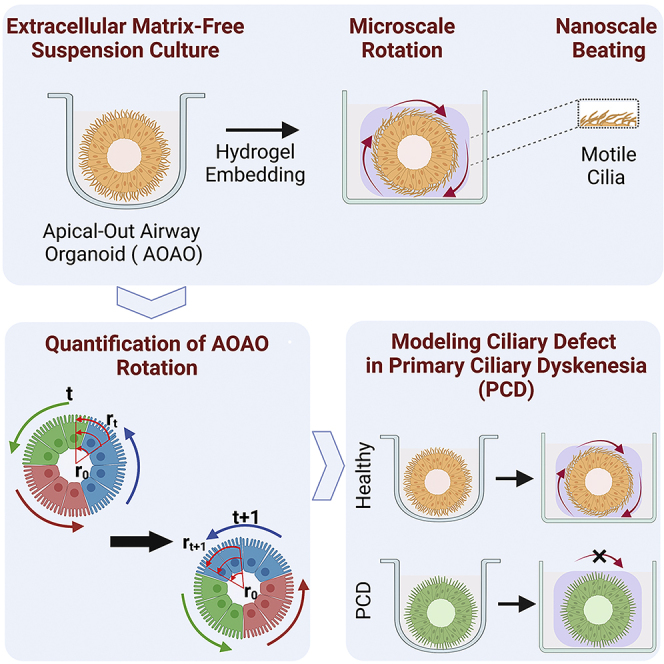
Summary
Motile cilia project from the airway apical surface and directly interface with inhaled external environment. Owing to cilia’s nanoscale dimension and high beating frequency, quantitative assessment of their motility remains a sophisticated task. Here we described a robust approach for reproducible engineering of apical-out airway organoid (AOAO) from a defined number of cells. Propelled by exterior-facing cilia beating, the mature AOAO exhibited stable rotational motion when surrounded by Matrigel. We developed a computational framework leveraging computer vision algorithms to quantify AOAO rotation and correlated it with the direct measurement of cilia motility. We further established the feasibility of using AOAO rotation to recapitulate and measure defective cilia motility caused by chemotherapy-induced toxicity and by CCDC39 mutations in cells from patients with primary ciliary dyskinesia. We expect our rotating AOAO model and the associated computational pipeline to offer a generalizable framework to expedite the modeling of and therapeutic development for genetic and environmental ciliopathies.
Subject areas: Molecular physiology, Bioengineering, Cell biology
Graphical abstract
Highlights
-
•
Apical-out airway organoid (AOAO) exhibits cilia-powered rotation in 3D
-
•
Cilia motility is directly correlated with AOAO rotational speed
-
•
The AOAO rotation reveals defective cilia motility under pathological conditions
Molecular physiology; Bioengineering; Cell biology.
Introduction
Motile cilia are specialized, highly conserved organelles that project from the luminal epithelial surface lining the respiratory tract, middle ear cavity, fallopian tube, and brain ventricles (Breunig et al., 2010; Bustamante-Marin and Ostrowski, 2017; Faubel et al., 2016; Gurr et al., 2009; Koyama et al., 2019; Luo et al., 2017; Lyons et al., 2006; Tilley et al., 2015). Motile cilia have the typical 9 + 2 microtubule architecture with a central pair of microtubule singlets surrounded by nine outer microtubule doublets (Gueron and Levit-Gurevich, 2001; Ishikawa, 2017). Motile cilia function as mechanical nanomachines that generate high-speed beating motion from cycles of unidirectional sliding of outer doublet microtubules, which are powered by dynein molecular motors (Gueron and Levit-Gurevich, 1999, 2001; Satir et al., 2014). Coordinated cilia beating serves critical functions in facilitating the directional transport of luminal substances, such as mucus in the respiratory tract and fertilized egg in the fallopian tube (Bustamante-Marin and Ostrowski, 2017; Koyama et al., 2019; Lyons et al., 2006; Tilley et al., 2015).
Abnormal cilia motility (ciliopathies) can result from genetic disorders affecting the structure or function of motile cilia, such as primary ciliary dyskinesia (PCD), which lead to devastating consequences, including chronic infection in the lung and ear, laterality defects, infertility, and rarely abnormal accumulation of cerebrospinal fluid in the brain (Barbato et al., 2009; Bush et al., 2007; Leigh et al., 2009). There are no therapeutic cures that can reverse the defects in cilia motility or halt the progression of diseases caused by genetic cilia abnormalities in PCD (Barbato et al., 2009; Bush et al., 2007; Leigh et al., 2009). Acquired motile ciliopathies may result from inhaled or ingested ciliotoxic chemicals. These chemicals have been extensively implicated to compromise cilia motility in the middle ear (Agius et al., 1995; Pezzoli et al., 2017), fallopian tube (Knoll et al., 1995; Talbot and Riveles, 2005), and brain ventricles (Liu et al., 2014; Omran et al., 2017). In the respiratory system alone, cilia dysfunction is a pathological finding observed in several chronic diseases, especially cigarette smoke related, which together affect over 35 vmillion Americans (Tilley et al., 2015; Yaghi and Dolovich, 2016). Thus, understanding the fundamental mechanisms regulating cilia motility under various pathophysiological conditions is of pivotal importance.
Airway organoids engineered from patient stem cells are a promising model for investigating respiratory pathophysiology, including ciliopathies. The airway epithelium is polarized with cilia beating and mucus secretion taking place on its apical surface that directly interacts with the inhaled air. However, the nanoscale dimension (200-300 nm diameter) of cilia combined with their high beating frequency (10-40 Hz) makes the measurement of cilia motility and function a challenging task. Over the past decades, high-speed video microscopy has emerged as a powerful tool for quantitative imaging of individual cilium motion in live cells and tissues. However, it requires a highly specialized microscopic setup and cumbersome analytical process (Dimova et al., 2005; Schipor et al., 2006), and is difficult to scale up to a high-throughput format. Thus, biomedical advancements are needed to enable comprehensive assessment and investigation of cilia pathophysiology and to promote effective therapeutic development.
Here we describe a suspension, hydrogel-free culture strategy for reproducible engineering of apical-out airway organoids from a defined number of cells. Importantly, powered by cilia beating on its exterior surface, the apical-out organoid rotated in soft supporting material, which inspired the use of organoid rotation as a functional readout of respiratory cilia motility. We developed a computational framework that leveraged computer vision algorithms to reliably calculate the angular speed of apical-out organoid rotation and correlated it with the direct measurement of cilia motility. To assess such correlation, we analyzed organoids treated with known chemical modulators of cilia motility as well as those engineered from bronchial epithelial cells derived from healthy and PCD patients.
Results
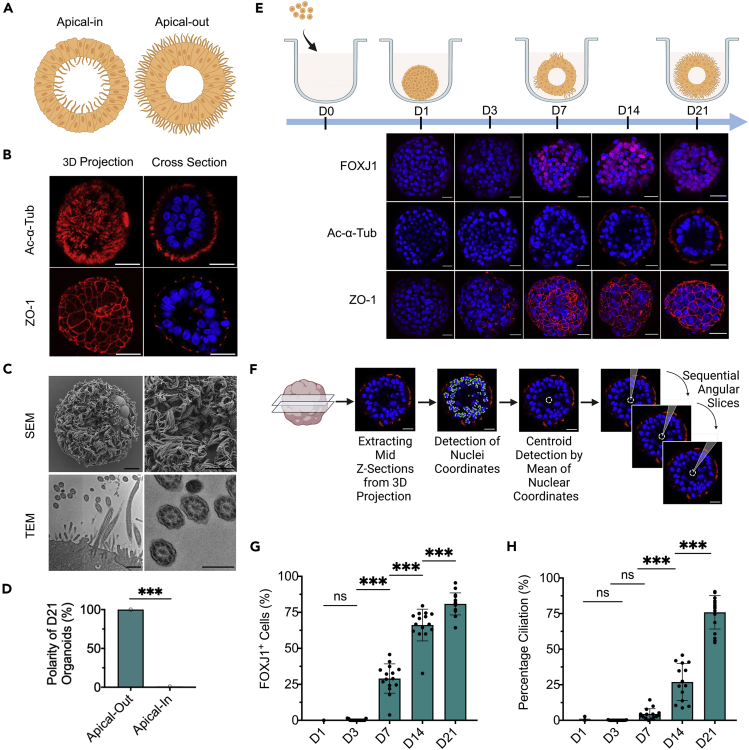
Engineering apical-out airway organoid
The interaction between epithelial cells and their surrounding extracellular matrix (ECM) plays instrumental roles in determining tissue polarity. Apical-in organoids are typically produced from airway epithelial cells in ECM-embedded culture, leading to the recognition of the organoid’s exterior surface that faces the ECM to be basal-lateral and its interior surface to be apical (Barkauskas et al., 2017; Dye et al., 2015; McCauley et al., 2017; Rock et al., 2009; Sachs et al., 2019). Here we assessed whether the removal of ECM support during airway organoids biogenesis from a defined number of human airway basal stem cells (hABSCs) can reverse the apical-basal recognition and epithelial polarity (Figure 1A). Bronchus-derived hABSCs were expanded in two-dimensional (2D) culture using the expansion medium formulated based on Bronchial Epithelial Cell Growth Medium (Levardon et al., 2018). To enable airway organoid formation, defined number (500) of hABSCs, dissociated from 3D expansion, were allowed to aggregate together on top of a cell-repellent surface in 96-well plate with no ECM support. Following overnight suspension culture in differentiation medium (PneumaCult-ALI), we observed effective spheroid formation followed by differentiation into ciliated airway organoid with apical-out polarity (cilia beating on the outer surface) by the end of week 3 (Video S1). This was referred to as Apical-Out Airway Organoid (AOAO). Compared to using the expansion medium for hABSC cell aggregation followed by transitioning to differentiation medium, the use of differentiation medium for both initial cell aggregation and subsequent differentiation was essential for maintaining spheroid tissue integrity (Figure S1).
Figure 1.
Characterization of the engineered AOAOs
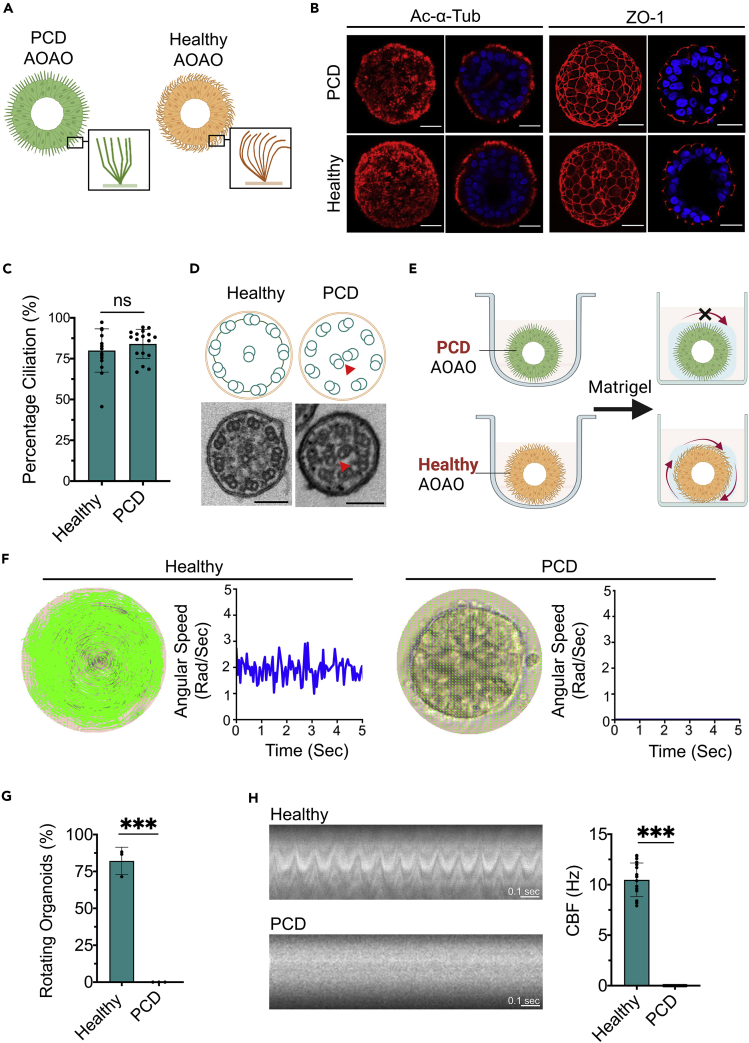
(A) Diagram of apical-in versus apical-out airway organoids.
(B) Immunofluorescence images of day 21 AOAOs stained with markers of cilia (Ac-α-Tub) and tight junction (ZO-1). Scale bar, 25 μm.
(C) SEM (scale bar, 10 μm) and TEM (scale bar, 800 nm (left), 400 nm (right)) images of AOAOs.
(D) Quantification of the percentage of day 21 (D21) organoids with apical-out versus apical-in epithelial polarity indicated by apical Ac-α-Tub localization.
(E) Time-series characterization of AOAO maturation by immunostaining of FOXJ1 (nuclear marker of ciliated cells), Ac-α-Tub, and ZO-1. Scale bar, 25 μm.
(F) Diagram showing the approach for assessing percentage ciliation by quantifying surface coverage of Ac-α-Tub expression.
(G and H) Time-series quantification of FOXJ1+ cell abundance (G) and percentage ciliation (H) in AOAOs. All data represent means ± SD from ≥15 organoids. ∗∗∗p < 0.001, Unpaired t-test/one-way ANOVA with Tukey’s multiple comparisons test.
To characterize epithelial polarity in the resulting day 21 AOAOs, we performed immunofluorescence staining of key polarity markers of airway epithelium and observed highly selective localization of ciliary Acetylated-alpha-Tubulin (Ac-α-Tub) on the organoid outer surface. Consistent with this orientation, epithelial tight junction protein, Zona Occludens-1 (ZO-1), formed highly organized intercellular junctions underneath the apical surface (Figure 1B). Using scanning (SEM) and transmission electron microscopy (TEM), we observed dense motile cilia covering the organoid outer surface with typical 9 + 2 microtubule organization, further verifying the apical-out epithelial polarity (Figure 1C). Next, we assessed the consistency of epithelial polarity in day 21 organoids resulting from continuous 3D suspension culture by examining Ac-α-Tub localization on the organoid’s exterior versus interior surface and observed 100% apical-out polarity (Figure 1D).
To track temporal dynamics of ciliogenesis and epithelial polarization, AOAOs were harvested on day 1, 3, 7, 14, and 21 of suspension differentiation, and evaluated for ciliated cell nuclear marker Forkhead Box J1 (FOXJ1), Ac-α-Tub, and ZO-1 (Figures 1E, S2, S3, and S4). FOXJ1+ ciliated cells emerged as early as day 7 and their abundance gradually increased to 81 ± 8% on day 21 (Figure 1G). We then calculated the percentage ciliation by quantifying cilia coverage on the organoid’s exterior surface. To do this, mid-Z-sections were selected from confocal z stack images of each organoid whole-mount stained for Ac-α-Tub and Hoechst-33342 (nuclei). The centroid of each Z section was identified using k-means clustering on nuclei coordinates. Angular slices of 1 degree from the centroid were assessed regarding their overlapping with ciliary Ac-α-Tub expression on the organoid edge. Finally, the percentage ciliation was calculated by normalizing the number of angular slices containing Ac-α-Tub fluorescence signal by 360 (Figure 1F). Applying this analytical pipeline to time-series images of AOAOs, we observed a steady increase in percentage ciliation over time, reaching 76 ± 12% on day 21 (Figure 1H), which echoed the gradual increase in FOXJ1+ ciliated cell abundance (Figure 1G). The observed percentage ciliation remained stable between day 21 and day 28 (Figure S5), the time interval for most subsequent experiments.
Besides ciliated cells, other cell types of airway epithelium, such as basal cells, club cells, and goblet cells, were not identified in the AOAOs differentiated using our standard condition (Figures S6–S8). The native human airway is known to undergo goblet cell hyperplasia and mucus hypersecretion following stimulation with cytokines, such as Interleukin 13 (IL-13) (Feldman et al., 2019). To test this possibility in AOAO differentiation, IL-13 (5 ng/mL) was supplemented to the differentiation medium over the entire differentiation process, which led to massive induction of goblet cells in the resulting AOAOs when analyzed on day 21 (Figure S8), suggesting responsiveness of the presented AOAO model to inflammatory stimuli.
Assessing reversibility of apical-out airway organoid epithelial polarity
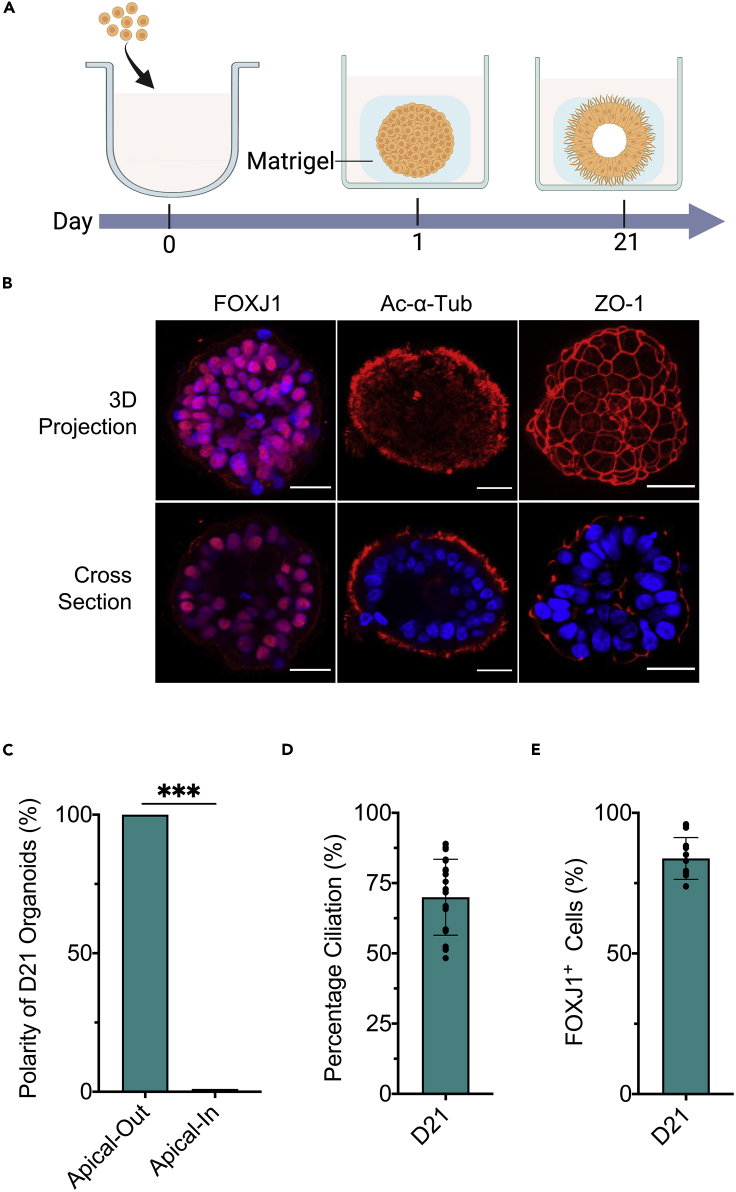
Upon demonstrating the ECM-free, suspension culture as a driver for establishing consistent apical-out airway polarity, we next investigated the stability of such epithelial polarity when the surrounding extracellular environment changed. To do this, we transitioned hABSC aggregates, following only 1-day suspension culture, into Matrigel-embedded culture and continued the differentiation until day 21. To our surprise, as indicated by FOXJ1, Ac-α-Tub, and ZO-1 expression (Figure 2A), all organoids subjected to this two-phase culture procedure (1 day in suspension followed by 20 days in Matrigel embedding) remained exhibiting homogeneous apical-out polarity (Figures 2B, 2C, and S9). Furthermore, these organoids from two-phase culture underwent robust ciliogenesis leading to day 21 ciliated cell abundance (FOXJ1+, 83 ± 7%, Figure 2D) and percentage ciliation (70 ± 14%, Figure 2E) that was not statistically different from that of standard AOAOs that have only experienced suspension culture (p = 0.2964 for ciliated cell abundance; p = 0.1448 for percentage ciliation). These results imply that airway epithelial polarity was effectively established within the first day of 3D suspension culture and remained stable even after being transitioned to ECM-supported culture. During the transition from suspension to Matrigel-embedded culture, we also observed sporadic merging of individual hABSC aggregates into larger organoid bodies, where Ac-α-Tub expression can be found on both the interior and exterior surfaces (Figure S10).
Figure 2.
Assessing epithelial polarity reversibility in organoids transitioned from suspension to Matrigel-embedded culture
(A) Human ABSC aggregates were formed in ECM-free suspension culture for 1 day and then transferred into ECM-rich, Matrigel-embedded culture and maintained for an additional 20 days.
(B) Immunofluorescence images of day 21 organoids stained for FOXJ1, Ac-α-Tub, and ZO-1. Scale bar, 25 μm.
(C) Quantification of the percentage of day 21 (D21) organoids with apical-out versus apical-in epithelial polarity indicated by apical Ac-α-Tub localization.
(D and E) Quantification of FOXJ1+ cell abundance (D) and percentage ciliation (E) in day 21 (D21) organoids. Data represent means ± SD from ≥15 organoids. ∗∗∗p < 0.001, Unpaired t-test.
Developing computer vision algorithms to assess apical-out airway organoid rotation
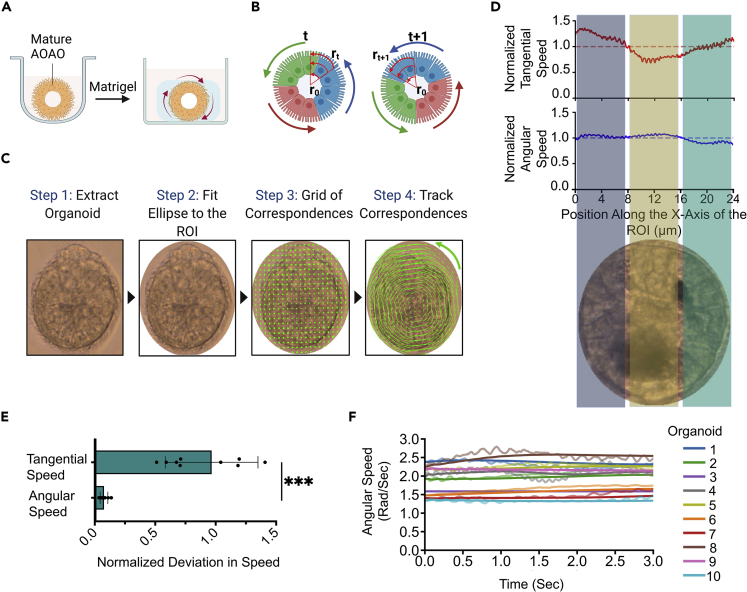
Intriguingly, the beating motion of exterior-facing cilia endows motility to the AOAO, which can be readily observed in suspension culture (Video S1). Here we investigated the possibility of stabilizing such cilia-powered AOAO motility by providing a 3D surrounding material support for cilia to propel against. To do this, mature AOAOs (between day 21 and day 28 of suspension differentiation) were embedded within the Matrigel matrix (Figure 3A), which effectively enabled the AOAOs to adopt stable rotational motion (Video S2), offering a unique opportunity to transform nanoscale, high-frequency cilia motility into microscale, low-frequency organoid rotation.
Figure 3.
Enabling and quantifying AOAO rotation
(A) Diagram for enabling consistent AOAO rotational motion via matrix embedding.
(B) Diagram depicting the computational method for calculating the organoid’s angular rotational motion: r0, the center of the organoid; rt, the position of the correspondence at time t; and rt+1, the position of the correspondence at time t+1.
(C) Stepwise description of the computational framework used to calculate correspondence movement.
(D) The tangential speed (μm/s) and angular speed (rad/s) normalized by their mean values for a representative organoid. The X axis is the position of correspondences along the X axis of the ROI. The speed of correspondences on the organoid, perpendicular to every position on the X axis was projected onto the X axis and averaged to obtain the tangential or angular speed for that position. The velocities were then normalized by their mean values. The color shaded regions represent the region of organoids which was used to calculate the tangential and angular speed.
(E) Deviation in the angular and tangential speed with respect to their mean values of 10 representative organoids from three independent replicates. The deviation was calculated by taking the mean squared difference between the speed profile and its mean value. The deviation was then normalized by the mean value.
(F) The instantaneous angular speed profile of 10 representative organoids from three independent replicates. The running mean (window = 5) of instantaneous angular speed was shown as the solid line. ∗∗∗p < 0.001, Unpaired t-test.
To reliably quantify the rotational motion of AOAOs, we developed computer vision-based motion tracking algorithms (Bouguet, 2000; Bradski, 2000). From video recordings of AOAO rotation (Video S2), we detected the center of each organoid (r0) and the position of the correspondence being tracked (rt) and used these vectors to determine the distance of the correspondence from the center. The change in the position of correspondence (rt+1) was used in the next step to calculate the distance covered by the correspondence (Figure 3B). To quantify the rotational motion, we identified the region of interest (ROI) by fitting an ellipse to the organoid to suppress the surrounding background. We generated a grid of correspondences in the ROI which were then tracked by the tracking algorithm. The distance covered by correspondences was then divided by the time taken to determine tangential speed (Figure 3C).
The tangential speed calculated above was dependent on the distance of the correspondence being tracked from the AOAO center. This led to a large variation in measurements obtained at different regions of the same organoid. For example, the organoid’s tangential speed profile had a parabolic shape with minimum at the central region and maximum at the periphery (Figures 3D and S11A). This was owing to the correspondences close to the organoid center not covering a large distance in comparison to those at the periphery, which as a result yielded a lower tangential speed. To overcome this constraint, we further calculated the angular speed of each correspondence, which became independent of its exact position within the organoid, by dividing the tangential speed by the distance of each correspondence from the organoid center (Figure S11B). By assuming the rotation axis being parallel to the viewing axis, the angular speed of the entire AOAO was determined by taking the mean angular speed of all correspondences being tracked (Video S3). To compare the tangential and angular speed profiles across the entire length of the AOAO, we calculated the mean squared deviation of the speed from its mean value and then normalized it by the mean value (Figure 3E). The deviation in tangential speed was 2-fold greater than that in angular speed. Thus, to ensure consistency in measuring AOAO rotational motion, we utilized the angular speed as the main readout. Furthermore, to detect the time-dependent variability in tracking AOAO rotation, we visualized the instantaneous angular speed of 10 representative AOAOs and demonstrated consistent rotational motion throughout the entire recorded time period (Figure 3F). Finally, the long timescale stability of the computer vision algorithms is demonstrated by the analysis of 5-min video recordings of AOAO rotation (Figure S12 and Video S4).
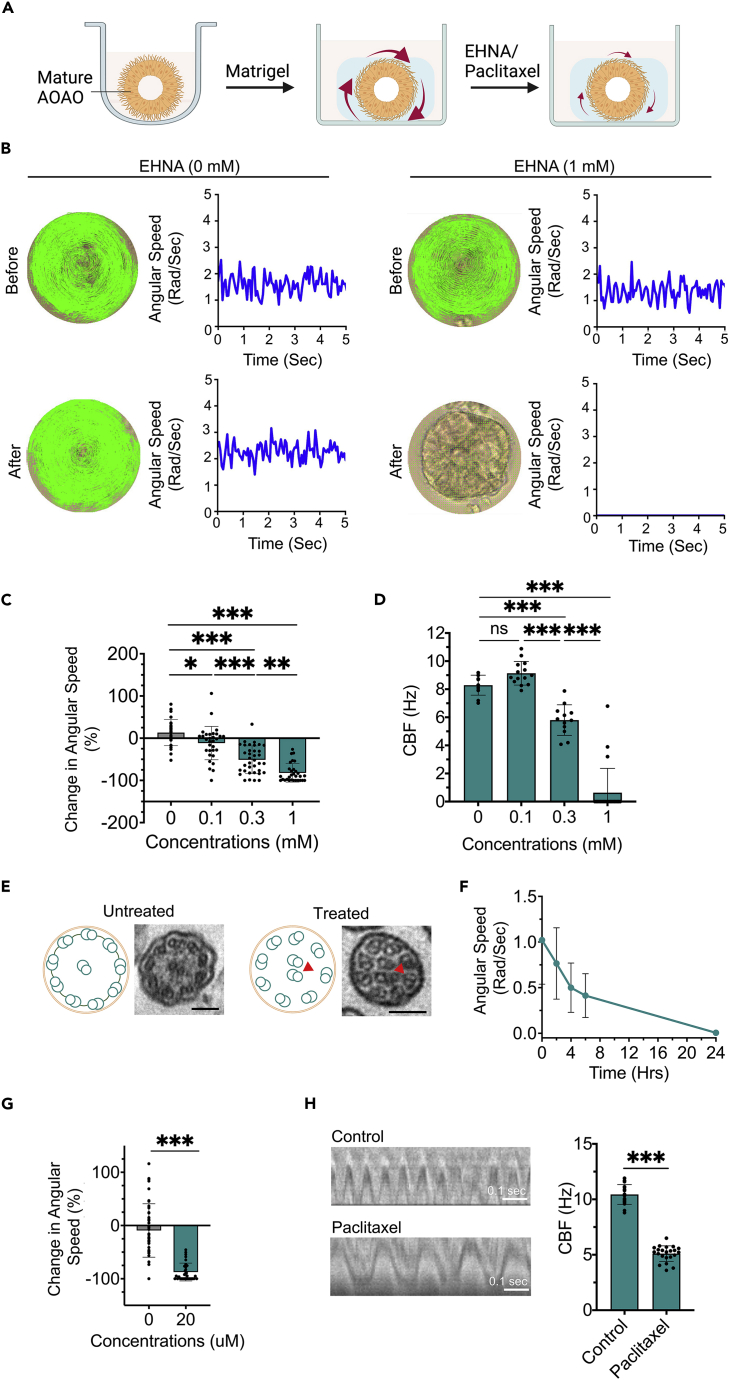
Assessing drug-induced inhibition of cilia motility and apical-out airway organoid rotation
To assess the correlation between cilia motility and cilia-powered AOAO rotational motion, we applied known chemical inhibitors of cilia motility to Matrigel-embedded, mature AOAOs (Figure 4A). EHNA (erythro-9-(2-hydroxy-3-nonyl)adenine) is an inhibitor of dynein, the molecular motor that powers axonemal doublet microtubule sliding and thus cilia beating (Bouchard et al., 1981; Schliwa et al., 1984). We used the computer vision-based motion tracking algorithms developed above to compute the angular speed of the same organoid before and after EHNA treatment (Figure 4B and Video S5). We introduced EHNA at a range of concentrations (0, 0.1, 0.3, and 1 mM) to mature AOAOs for 2 h, and observed an EHNA-dose-dependent reduction in organoid angular speed (Figure 4C) without changes in organoid size distribution (Figure S13). In parallel, we confirmed the inhibitory effect of EHNA on cilia beating frequency (CBF) using kymography analysis (Figure 4D).
Figure 4.
Characterization of AOAO rotation and cilia motility in the presence of cilia beating inhibitors
(A) Diagram showing AOAO rotational motion in response to EHNA or paclitaxel treatment.
(B) Computational methods used for calculating organoid angular speed following 1 mM EHNA treatment for 2 h.
(C) Quantification of organoid angular speed following 2-h treatment of EHNA at various concentrations.
(D) CBF analyzed via conventional kymographs following 1, 0.3, and 0.1 mM EHNA treatment for 2 h.
(E) TEM imaging of ciliary ultrastructure with and without 24-h paclitaxel (20 μM) treatment. Scale bar, 100 nm. Arrowheads indicated mislocated microtubules.
(F) Time-series analysis of paclitaxel’s effect on AOAO angular speed.
(G) Change in organoid angular speed in the presence or absence of 20 μM paclitaxel treatment for 24 h.
(H) CBF quantification via kymograph analysis in the presence or absence of 20 μM Paclitaxel for 24 h. Data represent means ± SD from three independent replicates with ≥10 organoids. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Unpaired t-test/one-way ANOVA with Tukey’s multiple comparisons test.
Paclitaxel is a chemotherapeutic agent that stabilizes microtubule structures and thus interferes with microtubule-dependent mitosis, cell migration, and cilia beating (Orr et al., 2003; Schiff et al., 1979; Zhu and Chen, 2019). Treatment of mature AOAOs with paclitaxel (20 μM) for 24 h led to abnormalities in ciliary ultrastructure (Figure 4E), which is in line with prior studies (Boisvieux-Ulrich et al., 1989; Shinohara et al., 2015). We then incubated the Matrigel-embedded AOAOs with 20 μM paclitaxel, monitored them periodically for 24 h, and observed paclitaxel-induced, progressive reduction of organoid angular speed (Figures 4F and 4G), without changes in organoid size distribution (Figure S14). Consistent with this, 24-h paclitaxel treatment dramatically reduced CBF shown by kymography analysis (Figure 4H). In our study, although EHNA and paclitaxel-induced inhibition of both cilia motility and AOAO rotation, they had no effect on the percentage of FOXJ1+ ciliated cells and cell viability within AOAOs (Figures S15 and S16). Altogether, our findings validated the correlation between the angular speed of the AOAO and cilia motility on its exterior surface.
Modeling and characterization of genetic ciliopathy using apical-out airway organoids
Primary ciliary dyskinesia (PCD) is a collection of genetic disorders involving abnormal motile cilia ultrastructure and function (Antony et al., 2013; Blanchon et al., 2012; Brennan et al., 2021; Dutcher and Brody, 2020; Horani et al., 2016). Mutations in the CCDC39 gene cause inner dynein arm defects and axonemal disorganization in cilia and have been associated with PCD (Blanchon et al., 2012; Ma et al., 2019; Oda et al., 2014). Using hABSCs carrying mutations in the CCDC39 gene, we intended to assess whether AOAOs can be effectively generated from PCD-bearing cells and whether the PCD-associated ciliary defects can be recapitulated by the AOAO rotational motion. We expanded hABSCs isolated from healthy and PCD (with CCDC39 mutations) patients and transitioned them for AOAO formation via 3D suspension culture (Figure 5A). Following 21 days of differentiation in suspension, as indicated by Ac-α-Tub and ZO-1 expression, airway organoids engineered from both healthy and PCD cells underwent effective epithelial differentiation with consistent apical-out polarity (Figure 5B). Furthermore, we observed comparable percentage ciliation on the apical surface of healthy and PCD organoids (p = 0.3212), indicating that the CCDC39 mutations did not affect basic ciliogenesis (Figures 5C and S17). However, as expected, PCD organoids exhibited defects in ciliary ultrastructure as indicated by TEM, showing a surrounding microtubule pair being mislocated to the center, compared to the normal 9 + 2 ciliary ultrastructure observed in healthy organoids (Figure 5D). Building on these morphological findings, we went on to assess and compare the rotational motion of PCD and healthy AOAOs by transferring them, following maturation, from 3D suspension culture to Matrigel embedding (Figure 5E). Consistent with defective ciliary structures, although of comparable size distribution (Figure S18), none of the embedded PCD AOAOs were able to rotate, as compared to over 75% of the embedded healthy AOAOs showing stable rotational motion (Figures 5F and 5G, and Video S6). Lastly, we performed CBF analysis and did not observe obvious cilia motility in PCD organoids as compared to robust cilia beating in healthy organoids (Figure 5H). These findings further validated our AOAO model and its associated computational analysis pipeline as effective tools for modeling and assessing human motile ciliopathy.
Figure 5.
Modeling PCD-associated ciliary defects using AOAO rotation
(A) Diagram showing organoids engineered from hABSCs derived from PCD and healthy patients.
(B) Imaging of day 21 AOAOs (PCD and healthy) stained for Ac-α-Tub and ZO-1. Scale bar, 25 μm.
(C) Quantification of percentage ciliation in PCD and healthy AOAOs.
(D) TEM images of healthy and PCD AOAOs showing cilia ultrastructural defects in the PCD organoids (arrowheads). Scale bar, 100 nm.
(E) Diagram showing PCD and healthy AOAO’s rotational motion.
(F) Computational methods used for calculating angular velocities of healthy and PCD AOAOs.
(G) Rotation analysis of Matrigel-embedded, healthy, and PCD AOAOs.
(H) Kymograph-based CBF quantification of healthy and PCD AOAOs. ∗∗∗p < 0.001, Unpaired t-test.
Discussion
In this study, we developed a robust approach to engineer apical-out airway organoids (AOAOs) with cilia beating taking place at the organoid’s exterior surface, which effectively propelled the organoid to rotate when placed in 3D Matrigel support. We have established robust tangential speed calculation algorithms and thereby a direct correlation between cilia motility and AOAO rotational motion. Our experimental results and computational tools will facilitate respiratory injury assessment with improved efficacy and will support the development of high-throughput assays for personalized disease management and therapeutic screening, benefiting patients suffering from respiratory diseases resulting from defective cilia function. Given the high-level similarity between motile cilia in different organ systems, we also expect the wide applicability of our apical-out organoid model and its associated computational tools beyond the respiratory system.
In the native human lung, the apical airway surface is exposed directly to the external environment and therefore is the main interface interacting with respiratory pathogens, such as bacteria and viruses. The apical-out conformation enabled by the AOAO engineering described here will allow the introduction of respiratory pathogens and pollutants directly to the apical airway surface in a non-invasive, repetitive manner with unprecedented convenience. The use of suspension culture to produce apical-out polarity in airway organoid is consistent with prior observations that transition from Matrigel-embedded to suspension culture promoted epithelial polarity to switch from apical-in to apical-out conformation in intestinal and distal lung organoids (Co et al., 2019; Li et al., 2020; Salahudeen et al., 2020). Despite the polarity reversal reported in these prior studies, here we show that the apical-out polarity established within the first day of suspension culture of hABSC aggregates cannot be reversed by subsequent switching to Matrigel-embedded culture that usually favors apical-in polarity.
The cellular composition of AOAO is predominantly ciliated cells, while the epithelial cell types are more diverse (including basal, goblet, and club cells) in Matrigel-embedded apical-in airway organoid culture or air-liquid interface culture (Barkauskas et al., 2017; Dye et al., 2015; McCauley et al., 2017; Rock et al., 2009; Sachs et al., 2019). This suggests the impact of substrate presence in modulating airway lineage specification and stem cell maintenance, which warrants further investigation. Nonetheless, we show that goblet cells can be induced in AOAO culture if pro-inflammatory cytokine is introduced during organoid differentiation, recapitulating the inflammatory response observed in the native human airway.
The human lung is equipped with approximately three billion motile cilia, which are tiny hair-like protrusions (100-nm diameter) that constantly sweep mucus in the cephalic direction (Bustamante-Marin and Ostrowski, 2017). Such nano-scale dimension combined with high beating frequency (10-20 Hz) makes the measurement of cilia motility a challenging task requiring specialized high-speed video cameras at high magnification (Dimova et al., 2005; Schipor et al., 2006). As our work reveals, powered by cilia beating on its exterior surface, the AOAO exhibited persistent rotational motion when surrounded by the Matrigel matrix. This novel phenomenon tackles the experimental and computational challenges associated with characterizing cilia motility and its pathophysiology, by enabling the conversion of nanoscale, high-frequency cilia beating into microscale, low-frequency AOAO rotation. The stable, directional motion of the cilia-powered AOAO rotation suggests coordinated cilia motility on the AOAO surface and offers an effective means to reveal and investigate the mechanisms underlying the coordinated, functional output of a given motile cilia population, either within an individual ciliated cell or within a multicellular tissue such as the AOAO.
This study delivered a complete computational pipeline for quantifying organoid rotational motion from video data acquired using a basic microscopic setup by leveraging the most recent developments in computer vision, specifically the tracking algorithms to conduct real-time analysis of video data. We have developed and deployed a computational framework that was able to extract significant features (such as rotational motion, percentage ciliation, and CBF) for the characterization of airway organoids and their pathophysiology. The framework is generalizable and allows high-throughput feature extraction from video data, which enables the processing of large quantities of data and robust statistical comparison. To overcome the challenge of analyzing the 3D organoid rotation from a 2D microscope view, we assumed the organoid rotation axis to be parallel to the microscope viewing axis. In this work, as a proof of concept for cilia motility analysis via AOAO rotation, we recorded and analyzed the angular speed of the same AOAOs before and after the treatment with cilia beating inhibitors (EHNA and paclitaxel). In particular, we observed EHNA dose-dependent reduction of CBF that was paralleled by gradual deceleration of AOAO rotation. This suggests the feasibility of using AOAO rotation analysis to reveal mild to severe defects of motile cilia beating.
Finally, our AOAO model recapitulates defective cilia motility under pathophysiological conditions and allows cilia injury assessment using patient-derived cells. We successfully engineered AOAOs from hABSCs derived from the PCD patient carrying CCDC39 mutations and recapitulated the PCD-specific disease phenotype via analysis of AOAO rotational motion. This paves the way for using the AOAO rotation as a robust functional readout for developing effective assays for personalized disease management and therapeutic screening.
Limitations of the study
There are some limitations to this study. First, while the 3D organoid rotation assay offers effective means for assessing human motile ciliopathy, it still requires 3D hydrogel embedding. In addition, when transforming the organoid’s tangential speed into angular speed, we assumed the rotation axis to be parallel to the viewing axis of the microscope, in order to overcome the challenge of analyzing 3D organoid rotation from a 2D microscopic view. Finally, while we have established the mature AOAOs to be composed mostly of ciliated cells, further investigation is needed to characterize the identity of the remaining, non-ciliated cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-ZO-1 | Invitrogen | Cat# 33-9100; RRID: AB_2533147 |
| Mouse Anti-Acetylated-α-Tubulin | Sigma-Aldrich | Cat# T6793; RRID:AB_477585 |
| Mouse Anti-MUC5AC | Thermo Fisher Scientific | Cat# MA5-12178; RRID: AB_10978001 |
| Mouse Anti-TRP63 | Biocare Medical | Cat# cm163a; RRID: AB_10582730 |
| Mouse Anti-FOXJ1 | Thermo Fisher Scientific | Cat# 14-9965-82; RRID: AB_1548835 |
| Donkey Anti-Mouse Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202; RRID: AB_141607 |
| Donkey Anti-Mouse Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A-31571; RRID: AB_162542 |
| Chemicals, peptides, and recombinant proteins | ||
| A8301 (inhibitor of TGF-β type 1 receptor kinase) | Sigma-Aldrich | Cat# SML0788-5MG |
| BEBM™ Bronchial Epithelial Cell Growth Basal Medium | Lonza | Cat# CC-3171 |
| BEGM™ Bronchial Epithelial Cell Growth Medium SingleQuots™ Supplements and Growth Factors | Lonza | Cat# CC-4175 |
| Bovine Collagen, Type I | Adbanced BioMatrix | Cat# 5225 |
| CHIR99021 (activator of WNT pathway) | Reprocell | Cat# 04000402 |
| Dulbecco’s Phosphate-Buffered Saline (DPBS), 1X without calcium and magnesium | Corning | Cat# 21-031-CV |
| DMH-1 (inhibitor of BMP4/SMAD signaling) | Tocris Bioscience | Cat# 4126 |
| EHNA, hydrochloride (inhibitor of Dynein) | Millipore Sigma | Cat# 32-463-010MG |
| Growth factor reduced Matrigel | Corning | Cat# CB 40230 |
| Heparin solution | Stemcell Technologies | Cat# 07980 |
| HyClone™ FetalClone™ I Serum (a fetal bovine serum alternative) | GE Healthcare | Cat# SH30080.03 |
| Hydrocortisone stock solution | Stemcell Technologies | Cat# 07925 |
| Paclitaxel (stabilizing microtubules) | Cayman Chemicals | Cat# 10461 |
| PneumaCult™-ALI Medium | Stemcell Technologies | Cat# 05001 |
| Penicillin-Streptomycin | Lonza Walkersville Inc. | Cat# 17-602E |
| RPMI 1640 with L-glutamine | Corning | 10-040-CV |
| Y27632 (inhibitor of Rho-associated protein kinase) | Cayman Chemical | 129830-38-2 |
| 0.25% Trypsin-EDTA | Gibco™ | 25200056 |
| Critical commercial assays | ||
| ReadyProbes™ Cell Viability Imaging Kit, Blue/Green | Invitrogen | Cat# R37609 |
| Deposited data | ||
| Codes for angular speed calculations and the corresponding requirements | This paper | https://github.com/BaratiLab/organoid_tracking.git |
| Experimental models: Cell lines | ||
| Bronchus-derived human airway basal stem cells | Lonza | Cat# CC-2541 |
| Airway basal stem cells derived from de-identified lung tissues of healthy donor or donor with primary ciliary dyskinesia (carrying mutations in CCDC39 (c.830_831delCA (p.Thr277Argfs∗3) and c.1871_1872delTA (p.Ile624Lysfs∗3)) | Washington University | N/A |
| Software and algorithms | ||
| GraphPad Prism | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Imaris (v9.0.1) | Oxford Instruments Group | https://imaris.oxinst.com/ |
| Other | ||
| 96-well cell-repellent microplate with lid | GreinerBio-One | Cat# 655970 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xi Ren (xiren@cmu.edu).
Materials availability
This work did not generate new unique reagents and all materials in this study are commercially available.
Experimental model and subject details
Cells
Bronchus-derived human airway basal stem cells (hABSCs) were purchased from Lonza. Additional hABSCs were obtained from surgical excess of de-identified tissues of healthy lung donors or donors carrying mutations in CCDC39 gene (c.830_831delCA (p.Thr277Argfs∗3) and c.1871_1872delTA (p.Ile624Lysfs∗3)) with permission of the institutional review board at Washington University in Saint Louis. The hABSCs were cultured in 804G-conditioned medium coated culture vessels in bronchial epithelial cell growth medium (BEGM) supplemented with 1 μM A8301, 5 μM Y27632, 0.2 μM of DMH-1, and 0.5 μM of CHIR99021 at 37°C with 5% CO2 (Levardon et al., 2018).
Method details
Culture of airway basal cells
Bronchus-derived human airway basal stem cells (hABSCs) were purchased from Lonza. Additional hABSCs were obtained from surgical excess of de-identified tissues of healthy lung donors or donors carrying mutations in CCDC39 gene (c.830_831delCA (p.Thr277Argfs∗3) and c.1871_1872delTA (p.Ile624Lysfs∗3)) with permission of the institutional review board at Washington University in Saint Louis. The hABSCs were cultured in 804G-conditioned medium coated culture vessels in bronchial epithelial cell growth medium (BEGM) supplemented with 1 μM A8301, 5 μM Y27632, 0.2 μM of DMH-1, and 0.5 μM of CHIR99021 at 37°C with 5% CO2 (Levardon et al., 2018).
Differentiation of airway basal cells into apical-out airway organoids
Human ABSCs (P2) were trypsinized and resuspended (5000 cells/ml) in differentiation medium (PneumaCult-ALI Medium) supplemented with 10 μM Y27632. 100 μL of resuspended hABSCs were placed per well in a 96-well cell-repellent microplate (GreinerBio-One, 655970). The cultures were maintained at 37°C with 5% CO2 for 21-28 days. To assess how IL-13 modulated organoid maturation, IL-13 (5 ng/mL) was supplemented to the differentiation medium during the entire differentiation period.
To assess organoid polarity in response to two-phase (ECM-deprived and then ECM-supported) culture, day-1 organoids formed in 96-well cell-repellent microplate were collected, resuspended in 40% (vol/vol) growth factor reduced (GFR) Matrigel and added to a new well plate that has been pre-coated with 40% GFR-Matrigel. The culture was then proceeded for an additional 20 days in differentiation medium.
Immunofluorescence staining
Airway organoids were collected from the 96-well cell-repellent microplate and fixed with 4% paraformaldehyde (PFA) for 1 hour at 4°C. The fixed organoids were washed with PBS with 0.1% Tween-20 and permeabilized with 1% Triton-X for 1 hour before incubating with primary antibodies diluted in 1% bovine serum albumin (BSA) overnight. Next, the organoids were washed with PBS with 0.1% Tween20 and incubated with secondary antibodies. The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) before capturing z-stacks of stained organoids on a Zeiss LSM 700 laser scanning confocal microscope.
Scanning electron microscopy
The apical-out airway organoids (AOAOs) were fixed with 2.5% glutaraldehyde in 0.01 M PBS (pH 7.4) for 1 hour at room temperature. The organoids were washed 3 times in 0.01 M PBS and then post-fixed with aqueous 1% osmium tetroxide for 1 hour at 4°C. Next, the organoids were rinsed 3 times in 0.01 M PBS before dehydrating in a graded series of 30%, 50%, 70%, and 90% ethanol, followed by 3 changes in 100% ethanol. The organoids were further dehydrated in hexamethyldisilazane for 15 minutes and allowed to air dry. The fixed and dehydrated organoids were mounted on studs and sputter-coated with 5 nm gold-palladium alloy prior to imaging with JEOL JSM 7800.
Transmission electron microscopy
The AOAOs were rinsed in 0.01 M PBS and fixed with 2.5% glutaraldehyde in 0.01M PBS (pH 7.4) for 1 hour at room temperature. The organoids were washed 3 times in 0.01M PBS and then post-fixed with aqueous 1% osmium tetroxide containing 1% potassium ferricyanide for 1 hour at 4°C. Next, the organoids were rinsed 3 times in 0.01 M PBS before dehydrating in a graded series of 30%, 50%, 70%, and 90% ethanol, followed by 3 changes in 100% ethanol. The organoids were washed in Polybed 812 epoxy resin for 3 times for 1 hour each before polymerizing at 37°C overnight and then for additional 48 hours at 60°C. Finally, the prepared organoid samples were sectioned at 60 nm, placed on copper grids, and imaged with JEM 1400 Flash TEM.
Calculating percentage ciliation in AOAOs
Z-stack images of AOAOs, stained with Acetylated-α-Tubulin (Ac-α-Tub) and DAPI, were acquired using a Zeiss LSM 700 laser scanning confocal microscope. For percentage ciliation calculations, 3-4 cross-sections per organoids were selected from mid-z-stacks. The pixel coordinates of the edges of DAPI stained nuclei were used to determine the centroid of the apical-out organoid. By using k-means (k=1) clustering on the edge coordinates of DAPI-stained nuclei, we determined the centroid of the organoid in a robust and unsupervised manner (Virtanen et al., 2020). From the calculated centroid, each organoid cross-section was divided into 1-degree angular segments (Figure 1F). Finally, the presence of Ac-α-Tub immunofluorescence signal was detected in each angular segment. The angular segments containing the Ac-α-Tub signal were recorded and used to calculate the percentage ciliation of the organoid (Pedregosa et al.).
Calculating abundance of ciliated cells in AOAOs
Z-stack images of AOAOs, stained for the presence FOXJ1 using Anti-FOXJ1 antibody and for nuclei using DAPI, were acquired using a Zeiss LSM 700 laser scanning confocal microscope. For calculating the percentage abundance of FOXJ1+ ciliated cells, the mid-cross section of each organoid was selected from the z-stacks. The number of FOXJ1+ and DAPI+ cells were calculated using the Spots tool in the IMARIS software. The percentage of ciliated cells was calculated by normalizing the number of FOXJ1+ cells by the DAPI+ total cell number.
Matrigel embedding of AOAOs
Mature AOAOs at day-21 to day-28 of differentiation were collected together and embedded in Matrigel matrices. For Matrigel embedding, collected AOAOs were resuspended in 40% (vol/vol) GFR-Matrigel in differentiation medium, which was kept on a heat plate set to 37°C for 10 minutes to enable effective gelation. Upon matrix gelation, differentiation medium was added to the top of the AOAO-containing gel matrices. All matrix-embedded AOAOs were maintained at 37°C with 5% CO2. The next day, 30-seconds video recordings of AOAOs were captured using EVOS FL Auto 2 Imaging System.
Angular speed calculation from video data
The video recordings of AOAOs were preprocessed by cropping to the region of interest containing the organoid, using Gaussian blur to reduce the noise, and smoothing variations in contrast to improve the performance of the tracking algorithm (Lu et al., 2005; Ravichandran et al.). Since the organoids generally have a spheroid shape, an ellipse was fit to the region of interest to mask out the surrounding region of the organoid. From the first frame of the video, a grid of equispaced correspondences was selected (Figure 3C). The correspondences were 5 pixels apart in each direction. These correspondences were tracked for the duration of the video by using the Lukas-Kanade (LK) tracking algorithm implementation from OpenCV python package (Bouguet, 2000; Bradski, 2000; Fu and Kovesi, 2010). The distance covered by each correspondence was measured and converted to the tangential speed (Sun et al., 2010). The correspondence which did not move, hence were marking the background, was filtered out. The tangential speed was converted to angular speed for each organoid by dividing by the distance of correspondence from the center (Figure S11). To eliminate the error accumulation by the LK tracking algorithm over time, we recomputed the correspondences every 25 frames. The angular speed for each organoid was the average angular speed of all correspondences for the entire duration of the video. The difference in tangential and angular speed was quantified by calculating the mean normalized deviation of speed (tangential and angular) from the mean (Figure 3E) using the following equation:
Finally, in this work, we assumed that the organoid rotation axis was parallel to the viewing axis of the microscope. This assumption enabled us to transform the tangential speed (μm/sec) of the organoid into angular speed (rad/sec) by dividing with its distance from the center of the organoid.
Treating AOAOs with paclitaxel or EHNA for angular speed analysis
Mature AOAOs (day-21 to day-28 of differentiation) were embedded in Matrigel for two days before treatment with desired chemical inhibitors of cilia motility. For paclitaxel, AOAOs were treated with paclitaxel (20 μM, diluted in differentiation medium) for 24 hours, with the control group being treated with an equal concentration of dimethyl sulfoxide (DMSO). For EHNA, AOAOs were treated with EHNA (0.1, 0.3, or 1 mM) for 2 hours, with the control group being treated with an equal concentration of phosphate buffer saline (PBS). Following chemical treatment of desire time periods, 30-seconds video recordings of AOAOs, pre- and post-treatment, were captured using EVOS FL Auto 2 Imaging System.
Imaging the AOAO cilia beating for kymographs generation and cilia beating frequency (CBF) calculation
Mature AOAOs (day-21 to day-28 of differentiation) were transferred to a 1.5-mL Eppendorf tube and kept on ice. Cold collagen type 1 was neutralized with the neutralization solution, added to the organoids at a final concentration of 2 mg/mL, and the entire AOAO-collagen mixture was transferred to glass bottom region of a Mattek dish. The Mattek dish containing organoids in collagen was kept on ice for additional 10 minutes until the organoids settle down to the bottom of the dish. The Mattek dish was then kept on a heat plate set to 37°C for 10 minutes. 1 mL of differentiation medium was placed in the dish before capturing video recording of cilia beating using Zeiss AxioObserver Z1 microscope with a 100X, 1.45 NA objective and pco.edge 5.5 camera. The high-speed video recordings of cilia beating were preprocessed using the previously described method to smoothen noise and variations in contrast. Additionally, the Contrast Limited Adaptive Histogram Equalization (CLAHE) function in Python v. 3.7 software was used to improve the contrast of the cilia with respect to the background (Lu et al., 2005; Reza, 2004). From preprocessed video, the region of interest on organoid surface containing cilia was cropped. The normal vector with respect to the organoid surface for each region of interest was calculated. The pixel intensity along the normal vector was then mean pooled for each frame, thereby generating the kymograph for ciliary motion. The peaks in the kymograph were counted and divided by the duration of the video to obtain the CBF value for the organoid. At least 10 kymographs were generated per organoid and the average value represented the CBF of the organoid.
Quantification of cell viability in AOAOs after EHNA and paclitaxel treatment
Mature AOAOs (day-21 to day-28 of differentiation) were treated with 1 mM EHNA for 2 hours or 20 μM paclitaxel for 24 hours. Mature AOAOs fixed with 4% PFA for 1 hour at 4°C were used as a dead cell control. NucBlue Live (stains all cells, live and dead) and NucGreen Dead (stains only dead cells) reagents were then added to AOAOs at two drops per mL for 1 hour. Stained AOAOs were transferred to a 1.5-mL Eppendorf tube and kept on ice. Cold collagen type 1 was neutralized with the neutralization solution, added to the organoids at a final concentration of 2 mg/mL, and the entire AOAO-collagen mixture was transferred to glass-bottom region of a Mattek dish. The Mattek dish containing organoids in collagen was kept on ice for an additional 10 minutes until the organoids settle down to the bottom of the dish. The Mattek dish was then kept on a heat plate set to 37°C for 10 minutes and 1 mL of differentiation medium was placed in the dish. Z-stack images of AOAOs were acquired using a Zeiss LSM 700 laser scanning confocal microscope. For quantification of cell viability, three mid-cross sections of each organoid were selected from the z-stacks. The number of NucBlue Live+ total cells and NucGreen Dead+ dead cells were calculated using ImageJ. The number of NucGreen Dead+ cells normalized by the that of the NucBlue Live+ total cells was used for calculating the percentage of dead cells.
Calculation of organoid size
To calculate the area of the AOAOs analyzed for rotational motion, the first frame of each video recording was used. From this image, the correspondences on the outer surface of the AOAO (n > 30) were manually selected. These correspondences were selected such that they comprise of and represent the periphery of the AOAO. The selected correspondences were then treated as vertices of an irregular polygon. The representation of AOAO as a polygon accounts for any variability on the shape of the organoid. Finally, the area from the selected correspondences was calculated using the shoelace algorithm (Gauss’s area formula). This method calculates the area as a scalar with units in pixel squared. In the analyzed images, the image dimensions were 1024x1024 pixels.
Quantification and statistical analysis
Quantitative data were displayed as means ± s.d. Statistical significances were determined using one-way analysis of variance (ANOVA) with post-hoc Tukey’s Test and unpaired t-test. Statistical analyses were performed using GraphPad. The level of significance was set at ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Graphics
Schematics were created using Microsoft PowerPoint and Biorender. Plots were prepared using GraphPad.
Acknowledgments
This work was supported by the Department of Defense Peer-Reviewed Medical Research Program W81XWH2110183 (X.R. and A.B.F.), and the DSF Charitable Foundation (X.R.). We are grateful to Misti West for laboratory management, to Ming Sun and Mara L. Sullivan from the Center for Biologic Imaging at the University of Pittsburgh for assistance with TEM sample processing, and to Kian Patel for proofreading the article.
Author contributions
† These authors contributed equally. P.W., P.Y., A.B.F., and X.R. contributed to the overall design of the project, data interpretation, and preparation of the article. P.W. and P.Y. performed the experiments and analyzed the data. L.A.P. and S.C.W. performed image acquisition regarding cilia beating. D.B.S and J.M.F. performed electron microscopy studies. E.R.J. assisted with figure preparation. S.L.B., A.H., and J.X. provided patient-derived human airway basal stem cells and advised on experiments related to these cells.
Declaration of interests
A patent has been filed by Carnegie Mellon University on the technology described in this study.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104730.
Contributor Information
Amir Barati Farimani, Email: barati@cmu.edu.
Xi Ren, Email: xiren@cmu.edu.
Supplemental information
Data and code availability
-
•
The data reported in this paper will be shared by the lead contact, Xi Ren (xiren@cmu.edu).
-
•
The codes used for angular speed calculations and the corresponding requirements to run are publicly available on GitHub through the link, https://github.com/BaratiLab/organoid_tracking.git.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Agius A.M., Wake M., Pahor A.L., Smallman L.A. Smoking and middle ear ciliary beat frequency in otitis media with effusion. Acta Otolaryngol. 1995;115:44–49. doi: 10.3109/00016489509133345. [DOI] [PubMed] [Google Scholar]
- Antony D., Becker-Heck A., Zariwala M.A., Schmidts M., Onoufriadis A., Forouhan M., Wilson R., Taylor-Cox T., Dewar A., Jackson C., et al. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum. Mutat. 2013;34:462–472. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbato A., Frischer T., Kuehni C.E., Snijders D., Azevedo I., Baktai G., Bartoloni L., Eber E., Escribano A., Haarman E., et al. Primary ciliary dyskinesia: a consensus statement on diagnostic and treatment approaches in children. Eur. Respir. J. 2009;34:1264–1276. doi: 10.1183/09031936.00176608. [DOI] [PubMed] [Google Scholar]
- Barkauskas C.E., Chung M.I., Fioret B., Gao X., Katsura H., Hogan B.L.M. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchon S., Legendre M., Copin B., Duquesnoy P., Montantin G., Kott E., Dastot F., Jeanson L., Cachanado M., Rousseau A., et al. Delineation of CCDC39/CCDC40 mutation spectrum and associated phenotypes in primary ciliary dyskinesia. J. Med. Genet. 2012;49:410–416. doi: 10.1136/jmedgenet-2012-100867. [DOI] [PubMed] [Google Scholar]
- Boisvieux-Ulrich E., Laine M.C., Sandoz D. In vitro effects of taxol on ciliogenesis in quail oviduct. J. Cell. Sci. 1989;92:9–20. doi: 10.1242/jcs.92.1.9. [DOI] [PubMed] [Google Scholar]
- Bouchard P., Penningroth S.M., Cheung A., Gagnon C., Bardin C.W. erythro-9-[3-(2-Hydroxynonyl)]adenine is an inhibitor of sperm motility that blocks dynein ATPase and protein carboxylmethylase activities. Proc. Natl. Acad. Sci. USA. 1981;78:1033–1036. doi: 10.1073/pnas.78.2.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouguet J.-Y. Intel Corporation, Microprocessor Research Labs; 2000. Pyramidal Implementation of the Lucas Kanade Feature Tracker. [Google Scholar]
- Bradski G. The OpenCV library. Dr. Dobb's J. Softw. Tools. 2000;25:120–125. [Google Scholar]
- Brennan S.K., Molter D., Menezes M., Dunsky K., Leonard D., Lieu J., Hirose K., Hazan G., Horani A., Ferkol T., Brody S.L. Implementation of a screening tool for primary ciliary dyskinesia (PCD) in a pediatric otolaryngology clinic. Int. J. Pediatr. Otorhinolaryngol. 2021;142:110586. doi: 10.1016/j.ijporl.2020.110586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig J.J., Arellano J.I., Rakic P. Cilia in the brain: going with the flow. Nat. Neurosci. 2010;13:654–655. doi: 10.1038/nn0610-654. [DOI] [PubMed] [Google Scholar]
- Bush A., Chodhari R., Collins N., Copeland F., Hall P., Harcourt J., Hariri M., Hogg C., Lucas J., Mitchison H.M., et al. Primary ciliary dyskinesia: current state of the art. Arch. Dis. Child. 2007;92:1136–1140. doi: 10.1136/adc.2006.096958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante-Marin X.M., Ostrowski L.E. Cilia and mucociliary clearance. Cold Spring Harb. Perspect. Biol. 2017;9:a028241. doi: 10.1101/cshperspect.a028241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co J.Y., Margalef-Català M., Li X., Mah A.T., Kuo C.J., Monack D.M., Amieva M.R. Controlling epithelial polarity: a human enteroid model for host-Pathogen interactions. Cell Rep. 2019;26:2509–2520.e4. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova S., Maes F., Brewster M.E., Jorissen M., Noppe M., Augustijns P. High-speed digital imaging method for ciliary beat frequency measurement. J. Pharm. Pharmacol. 2005;57:521–526. doi: 10.1211/0022357055777. [DOI] [PubMed] [Google Scholar]
- Dutcher S.K., Brody S.L. HY-DIN' in the cilia: discovery of central pair-related mutations in primary ciliary dyskinesia. Am. J. Respir. Cell. Mol. Biol. 2020;62:281–282. doi: 10.1165/rcmb.2019-0316ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D., et al. In vitro generation of human pluripotent stem cell derived lung organoids. Elife. 2015;4 doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubel R., Westendorf C., Bodenschatz E., Eichele G. Cilia-based flow network in the brain ventricles. Science. 2016;353:176–178. doi: 10.1126/science.aae0450. [DOI] [PubMed] [Google Scholar]
- Feldman M.B., Wood M., Lapey A., Mou H. SMAD signaling restricts mucous cell differentiation in human airway epithelium. Am. J. Respir. Cell. Mol. Biol. 2019;61:322–331. doi: 10.1165/rcmb.2018-0326OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S.C., Kovesi P. Proceedings of the 2010 International Conference on Digital Image Computing: Techniques and Applications. IEEE Computer Society; 2010. Robust extraction of optic flow differentials for surface reconstruction; pp. 468–473. [Google Scholar]
- Gueron S., Levit-Gurevich K. Energetic considerations of ciliary beating and the advantage of metachronal coordination. Proc. Natl. Acad. Sci. USA. 1999;96:12240–12245. doi: 10.1073/pnas.96.22.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueron S., Levit-Gurevich K. Springer; 2001. Energetic Considerations of Ciliary Beating. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurr A., Stark T., Pearson M., Borkowski G., Dazert S. Vol. 266. European Archives of Oto-Rhino-Laryngology; 2009. p. 1865. (The Ciliary Beat Frequency of Middle Ear Mucosa in Children with Chronic Secretory Otitis Media). [DOI] [PubMed] [Google Scholar]
- Horani A., Ferkol T.W., Dutcher S.K., Brody S.L. Genetics and biology of primary ciliary dyskinesia. Paediatr. Respir. Rev. 2016;18:18–24. doi: 10.1016/j.prrv.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T. Axoneme structure from motile cilia. Cold Spring Harb. Perspect. Biol. 2017;9:a028076. doi: 10.1101/cshperspect.a028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll M., Shaoulian R., Magers T., Talbot P. Ciliary beat frequency of hamster oviducts is decreased in vitro by exposure to solutions of mainstream and sidestream cigarette Smoke1. Biol. Reprod. 1995;53:29–37. doi: 10.1095/biolreprod53.1.29. [DOI] [PubMed] [Google Scholar]
- Koyama H., Shi D., Fujimori T. Biophysics in oviduct: Planar cell polarity, cilia, epithelial fold and tube morphogenesis, egg dynamics. Biophys. Physicobiol. 2019;16:89–107. doi: 10.2142/biophysico.16.0_89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh M.W., Pittman J.E., Carson J.L., Ferkol T.W., Dell S.D., Davis S.D., Knowles M.R., Zariwala M.A. Clinical and genetic aspects of primary ciliary dyskinesia/Kartagener syndrome. Genet. Med. 2009;11:473–487. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levardon H., Yonker L.M., Hurley B.P., Mou H. Expansion of airway basal cells and generation of polarized epithelium. Bio. Protoc. 2018;8:e2877. doi: 10.21769/BioProtoc.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Yang N., Chen J., Huang X., Zhang N., Yang S., Liu G., Liu G. Next-Generation Porcine intestinal organoids: an apical-out organoid model for swine enteric virus infection and immune response investigations. J. Virol. 2020;94:e01006-20. doi: 10.1128/JVI.01006-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Jin X., Prasad R.M., Sari Y., Nauli S.M. Three types of ependymal cells with intracellular calcium oscillation are characterized by distinct cilia beating properties. J. Neurosci. Res. 2014;92:1199–1204. doi: 10.1002/jnr.23405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Xie W., Pei J., Huang J. 2005. Dynamic Texture Recognition by Spatio-Temporal Multiresolution Histograms. [Google Scholar]
- Luo W., Yi H., Taylor J., Li J.-d., Chi F., Todd N.W., Lin X., Ren D., Chen P. Cilia distribution and polarity in the epithelial lining of the mouse middle ear cavity. Sci. Rep. 2017;7:45870. doi: 10.1038/srep45870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons R.A., Saridogan E., Djahanbakhch O. The reproductive significance of human Fallopian tube cilia. Hum. Reprod. Update. 2006;12:363–372. doi: 10.1093/humupd/dml012. [DOI] [PubMed] [Google Scholar]
- Ma M., Stoyanova M., Rademacher G., Dutcher S.K., Brown A., Zhang R. Structure of the decorated ciliary doublet microtubule. Cell. 2019;179:909–922.e12. doi: 10.1016/j.cell.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley K.B., Hawkins F., Serra M., Thomas D.C., Jacob A., Kotton D.N. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell. 2017;20:844–857.e6. doi: 10.1016/j.stem.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Yanagisawa H., Kamiya R., Kikkawa M. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science. 2014;346:857–860. doi: 10.1126/science.1260214. [DOI] [PubMed] [Google Scholar]
- Omran A.J.A., Saternos H.C., Althobaiti Y.S., Wisner A., Sari Y., Nauli S.M., AbouAlaiwi W.A. Alcohol consumption impairs the ependymal cilia motility in the brain ventricles. Sci. Rep. 2017;7:13652. doi: 10.1038/s41598-017-13947-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr G.A., Verdier-Pinard P., McDaid H., Horwitz S.B. Mechanisms of Taxol resistance related to microtubules. Oncogene. 2003;22:7280–7295. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., Blondel, M., Prettenhofer, P., Weiss, R., Dubourg, V., et al. Scikit-learn: machine learning in Python. Mach. Learn. Python, 6.
- Pezzoli M., Lofaro D., Oliva A., Orione M., Cupi D., Albera A., Bongioannini G., Albera R. Effects of smoking on eustachian tube and hearing. Int. Tinnitus. J. 2017;21:98–103. doi: 10.5935/0946-5448.20170019. [DOI] [PubMed] [Google Scholar]
- Ravichandran, A., Chaudhry, R., and Vidal, R. View-Invariant Dynamic Texture Recognition Using a Bag of Dynamical Systems. 7. [DOI] [PubMed]
- Reza A.M. Realization of the contrast limited adaptive Histogram equalization. J. VLSI Signal Process. Syst. Signal Image Video Technol. 2004;38:35–44. [Google Scholar]
- Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., Hogan B.L.M. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. USA. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs N., Papaspyropoulos A., Zomer-van Ommen D.D., Heo I., Böttinger L., Klay D., Weeber F., Huelsz-Prince G., Iakobachvili N., Amatngalim G.D., et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019;38:e100300. doi: 10.15252/embj.2018100300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen A.A., Choi S.S., Rustagi A., Zhu J., van Unen V., de la O S.M., Flynn R.A., Margalef-Català M., Santos A.J.M., Ju J., et al. Progenitor identification and SARS-CoV-2 infection in human distal lung organoids. Nature. 2020;588:670–675. doi: 10.1038/s41586-020-3014-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir P., Heuser T., Sale W.S. A structural basis for how motile cilia beat. Bioscience. 2014;64:1073–1083. doi: 10.1093/biosci/biu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff P.B., Fant J., Horwitz S.B. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- Schipor I., Palmer J.N., Cohen A.S., Cohen N.A. Quantification of ciliary beat frequency in sinonasal epithelial cells using differential interference contrast microscopy and high-speed digital video imaging. Am. J. Rhinol. 2006;20:124–127. [PubMed] [Google Scholar]
- Schliwa M., Ezzell R.M., Euteneuer U. erythro-9-[3-(2-Hydroxynonyl)]adenine is an effective inhibitor of cell motility and actin assembly. Proc. Natl. Acad. Sci. USA. 1984;81:6044–6048. doi: 10.1073/pnas.81.19.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara K., Chen D., Nishida T., Misaki K., Yonemura S., Hamada H. Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev. Cell. 2015;35:236–246. doi: 10.1016/j.devcel.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Sun D., Roth S., Black M.J. 2010. Secrets of Optical Flow Estimation and Their Principles. Paper presented at: 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition. [Google Scholar]
- Talbot P., Riveles K. Smoking and reproduction: the oviduct as a target of cigarette smoke. Reprod. Biol. Endocrinol. 2005;3:52. doi: 10.1186/1477-7827-3-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilley A.E., Walters M.S., Shaykhiev R., Crystal R.G. Cilia dysfunction in lung disease. Annu. Rev. Physiol. 2015;77:379–406. doi: 10.1146/annurev-physiol-021014-071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaghi A., Dolovich M.B. Airway epithelial cell cilia and obstructive lung disease. Cells. 2016;5:40. doi: 10.3390/cells5040040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Chen L. Progress in research on paclitaxel and tumor immunotherapy. Cell. Mol. Biol. Lett. 2019;24:40. doi: 10.1186/s11658-019-0164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data reported in this paper will be shared by the lead contact, Xi Ren (xiren@cmu.edu).
-
•
The codes used for angular speed calculations and the corresponding requirements to run are publicly available on GitHub through the link, https://github.com/BaratiLab/organoid_tracking.git.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.