Abstract
There is currently an increasing demand for the characterization of endophytic bacteria isolated from different parts of plants (rhizosphere, roots, fruit, leaf) in order to improve the organic agriculture practices. The current research was performed to identify both rhizospheric bacteria isolated from the rhizosphere of Ficus carica in three different sites in the north of Tunisia and endophytic bacteria isolated from dried figs. We then characterized them for a diversity of plant growth-promoting (PGP) activities. A collection of 120 isolates from rhizospheric soil and 9 isolates from dried figs was obtained and purified. 16SrDNA gene amplification of rhizospheric bacteria revealed significant diversity and allowed for the assigning of the isolates to 6 phyla: Gammaproteobacteria, Alphaproteobacteria, Betaproteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes. Representative strains of the collection (90 strains) were tested for numerous PGP activities and resistance to abiotic stresses. The most common PGP trait for all bacteria from the three regions was siderophore production (62%), followed by cellulase (38%), then protease activity (37%), then by lipases activity (17%) and lastly by solubilization of phosphates (9%). Twenty -three strains that showed most PGP traits were selected, 8 strains presented 12 or more, and 15 strains displayed between 7 and 11 of 17 PGP activities. The majority of the isolates manifested a possible adaptation to abiotic stress and unfavorable environments. PCR-DGGE analysis of soil rhizosphere of the three sites allowed also for the acquisition of a Cluster analysis of rhizospheric bacterial communities. Our current study identified and characterized for the first time in Tunisia rhizospheric and endophytic bacteria from dried fruit of Ficus carica.
Keywords: Dried figs, Endophytic, Ficus carica, Plant growth promoting rhizobacteria (PGPR), Rhizosphere
1. Introduction
More than 800 species of trees, epiphytes, and shrubs belong to the genus Ficus (Moraceae), which is one of the most numerous angiosperm genera on the planet (Singh et al., 2011, Badgujar et al., 2014). Among them, the fig tree (Ficus carica) is a common fruit species in Mediterranean countries, and its fruit, which possesses high nutritive and pharmaceutical values, can be eaten fresh or dried (Yang et al., 2009, Lazreg et al., 2011).
Mediterranean countries are among the world’s largest producers of fresh figs (Sadder and Ateyyeh, 2006, Barolo et al., 2014). Fresh figs are available only in one season, so during the rest of the year the fruit is marketed for consumption after drying. The two most common types of dried figs are sun-dried figs and tray-dried figs (Crisosto et al., 2010). Dried figs are a good source of carbohydrates, sugars, proteins, vitamin A, vitamin C, potassium, calcium and iron; in addition, they are fat free, sodium free and cholesterol free (Vinson, 1999, Slatnar et al., 2011). They also contain high amounts of polyphenols and crude fiber (Mawa et al., 2013). Dried figs are used as a food supplement by diabetics and, because of the high amount of sugars they contain, it is also consumed as a sweet (Badgujar et al., 2014). In Tunisia dried figs (called chriha in the Tunisian dialect) are often used as a tonic (Le Floc’h, E., , 1983, Leporatti and Ghedira, 2009). In biotechnology dried figs can be used as an alternative natural support for the immobilization of yeast cells and also as a biocatalyst in brewing (Bekatorou et al., 2002). During storage, dried figs form sugar efflorescence in which yeast frequently occurs; in fact, a new species of yeast (saccharomyces delphensis) was isolated from South African dried figs (Lansky and Paavilainen, 2010). These days dried figs are collected from shops in order to investigate the contamination of fungi (Saadullah and Abdullah, 2015).
Bacterial endophytes of dried fruit of Ficus carica are poorly documented and previous studies have focused only on root-associated endophytes with plant growth-promoting effects (Lai et al., 2006, Choubane et al., 2016). In Tunisia, people planted fig trees in ancient times and its distribution extends from the north to the south of the country; that is to say, it grows both in cold, humid regions and in hot, dry regions (Mars et al., 2009). In more recent times, with the aim to increase crop yield, people have excessively adopted the use of agrochemical products, such as herbicides, insecticides, fungicides, fertilizers, which is having a negative influence on the environment as well as on human health. Due to the problems arising from the use of agrochemicals products, scientists are exploring alternative biological methods. Among them, microorganisms known as “Plant Growth Promoting Rhizobacteria” (PGPR) are very promising, as they can be used as biofertilizer to ameliorate plant growth or improve crop production (Agbodjato et al., 2016). PGPR is able to facilitate the absorption and availability of nutrients via several mechanisms, such as phosphate solubilization, nitrogen fixation and phytohormones production (Arora et al., 2012). Several genera were described to be PGPR such as: Pseudomonas, Azospirillum, Azotobacter, Klebsiella, Enterobacter, Alcaligenes, Arthrobacter, Burkholderia, Bacillus, and Serratia (Prasad et al., 2019). In addition, biofertilization is considered as a green tool to increase harvest and improve the quality of figs, making it a good substitute to chemical fertilizer (Mishra et al., 2013). A study was conducted in Egypt to investigate the combined effects of organic manure and bio-N-fertilization on the growth and yield of fig trees. The results showed that poultry manure + Azotobacter and poultry manure + Azospirillum treatments resulted in the best growth, productivity and fruit quality (Osman and Abd El-Rhman, 2010).
The aims of the current research were to: (i) isolate and identify endophytic bacteria from dried fruit of Ficus carica, (ii) isolate and identify bacteria from the rhizosphere of Ficus carica in three different regions in the north of Tunisia, (iii) evaluate the bacterial community dynamics in the rhizosphere of three regions using PCR-DGGE analysis, (iv) identify plant growth-promoting bacteria (PGP) traits and abiotic stress resistance.
2. 2.Materials and methods
2.1. Soil sampling
Three farms in Tunisia were used to gather the samples; their geographic locations were along a gradient from 36° to 37° N latitude and from 9° to 11° E longitude. The rhizosphere soil of three fig trees of similar age was separately sampled at a depth of 30 cm. The soil surrounding the roots was collected after removing the roots. The sampling was conducted under sterile conditions. Then the samples were stored at −20 °C.
2.2. Collection of plant material
Thirty samples of dried figs were taken from two southern regions of Tunisia: Tataouine and Djerba. Fifteen figs from Djerba (tradionnally were sun -dried and soaked in olive oil) (Mars et al., 2008) and 15 figs from Tataouine (tradionnally were sun-dried) (Mars et al., 2008, Nikolidaki et al., 2017). Then the samples were stored in Ziplock bags at room temperature (20 °C).
2.3. Bacteria isolation from Ficus carica rhizosphere
Bacteria isolation was carried out by the suspension of one gram of rhizosphere soil in 9 ml of sterile physiological solution, then the tubes for each sample were shacked for 24 h at 400 rpm. Then, suspensions were diluted with physiological water in tenfold series and plated on PCA (Plate Count Agar), TSA (Tryptic Soy Agar), and KB (King'B agar) media. After cultivating for 5 days at 30 °C, different colonies were picked and plated in triplicate (Ferjani et al., 2015).
2.4. Bacteria isolation from dried figs
The surface of the collected samples was cleaned with 2 percent sodium hypochlorite for 1 min before being washed with sterile distilled water. In the next step the samples were crushed with sterile equipment. The samples were placed in a sterile stomacher bag (10 g for one sample), 190 ml physiological saline peptone solution was added, then the samples were deposited in a stomacher (Ntuli et al., 2017). The suspensions were then tenfold diluted with physiological water, plated on TSA, and cultured at 30 °C for seven days (Alvarez-Pérez et al., 2012).
2.5. DNA extraction and PCR amplification of 16 SrRNA and identification of isolates
Using a boiling lysis method previously described by Ferjani et al. (2015) genomic DNA was recovered from bacterial isolates. The 16S rRNA genes were amplified using PCR with 1 U of DreamTaq Green DNA Polymerase (Fermentas), primers forward 8F (AGAGTTTGATCMTGGCTCAG) and reverse1507R (TACCTTGTTACGACTT). Federici et al. (2011) defined the cycle parameters. The PCR products were cleaned using the Sigma-Aldrich GenElute PCR Clean-Up kit before being transferred for sequencing. The BLAST tool (https://www.ncbi.nlm.nih.gov/BLAST/) was used to determine which isolates were present. The neighbor-joining phylogenetic tree was produced by MEGA v7.0 (Kumar et al., 2016) with bootstrap values based on 1000 replications. Escherichia coli was used as outgroup.
2.6. Soil DNA Extraction, PCR-DGGE analysis of bacterial communities.
The PowerSoil DNA Isolation kit was used to extract DNA from soil samples (MoBio Laboratories Inc). PCR amplification was performed as previously described by Federici et al. (2011) in a final volume of 50 μL using primers forward 341F (ATTACCGCGGCTGCTGG) and reverse 534R (ATTACCGCGGCTGCTGG). Electrophoresis in 2 percent (w/v) agarose gels colored with ethidium bromide was used to examine PCR products. Federici et al. (2011) previously reported on bacterial community profiling using DGGE analysis. On a 6 percent polyacrylamide-bis-acrylamide (37.5:1) gel with a 40–60 percent urea-formamide denaturing gradient, the amplification products were resolved. At 60 °C, the gel was run at 100 V for 16 h. Gels were stained with SBYR Gold (Invitrogen) for 30 min after electrophoresis and photographed under UV light.The GelJ v.2.0 program was used to analyze digitalized DGGE fingerprints (Heras et al., 2015). The dendrograms were generated using a UPGMA and Dice's coefficient of similarity (Federici et al., 2011). To analyze the bacterial diversity present in the rhizosphere of Ficus carica in three regions in northern Tunisia, the Shannon– Wiener (H), evenness (E), richness (S), and Simpson (D) indices were computed.
2.7. Plant growth promoting (PGP) traits and abiotic stress tolerance
Eighty-two identified bacterial strains were tested for several PGP activities and resistance to numerous abiotic stresses. Resistance to osmotic stress was tested by adjoining 30 % of polyethylene glycol (PEG 8000) into tryptic soy broth (TSB) medium. Salt resistance was detected by adding 10 %, 13 %, 16 %, 20 % of NaCl to the culture media and cultivated at 30 °C for five days.
In order to study the tolerance to pH, culture medium was adjusted either by a solution of NaOH (3 M) to reach the target values of 10 and 12 or by a concentrated HCl solution (12 N) to attain the values of 3 and 4. The ability to grow at 45 °C, 50 °C and 55 °C was assessed in TSA by incubating at each temperature for five days (Ferjani et al., 2015). Mineral phosphate solubilization was checked by the appearance of a clear halo around the bacterial colonies grown on Pikovskaya medium (Nautiyal, 1999). The strains were spotted on CMC agar and skimmed milk media to evaluate Cellulase and protease activities separately, as described by Wang et al. (2015). Lipase activity was tested using the medium detailed by Omidvari (2008). The strains were deposited on this specific medium and incubated at 27 °C for 48 h. The observation of depositions around the bacterial colonies proved the presence of activity. The production of siderophore was identified on CAS (chrome azurols) agar medium as detailed by Wang et al. (2015). The color change around the colonies from blue into orange was noted as positive ones (Perez-Miranda et al., 2007).
3. Results
3.1. Cultivable bacteria isolated from Ficus carica rhizosphere
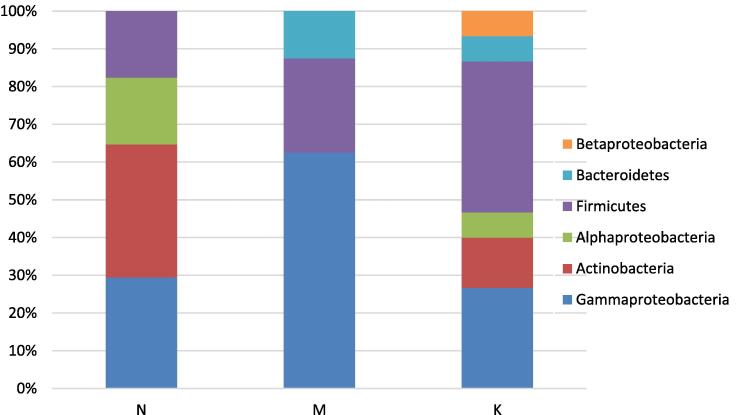
A total of 120 bacterial strains associated with Ficus carica rhizosphere were isolated from three sampling sites in different geographic locations in the north of Tunisia. Among them, 82 isolates were identified using partial 16S rRNA gene sequencing. A high diversity of bacterial communities associated with Ficus carica rhizosphere soil was detected, especially in two regions of Beni Khiar and Kerkouane. Phylogenetic analysis of the isolates demonstrated that species were divided into six phyla, 17 different bacterial genera (Fig. 1), indicating a large genetic diversity in Ficus carica rhizosphere. Bacillus and Pseudomonas were the most frequently observed genus in the collection at 25 % and 22.5 % respectively. The phylogenetic result also showed a slight predominance of Gram-positive bacteria (52.5 %) belonging to Firmicutes (27.5 %), Actinobacteria (20 %) and Bacteroidetes (5 %). For Gram-negative bacteria (47.5 %), Gammaproteobacteria (35 %), Alphaproteobacteria (10 %) and Betaproteobacteria (2.5 %) were present (Fig. 2).
Fig. 1.
Diversity of rhizospheric bacteria of Ficus carica. Phylogenetic association of the isolates from rhizosphere of F. carica at genus levels. N = Nabeul; M = Mateur; K = Kerkouane.
Fig. 2.
Diversity of rhizospheric bacteria of Ficus carica. Phylogenetic association of the isolates from rhizosphere of F. carica at class levels. N = Nabeul; M = Mateur; K = Kerkouane.
3.2. Cultivable bacteria isolated from dried fruit of Ficus carica
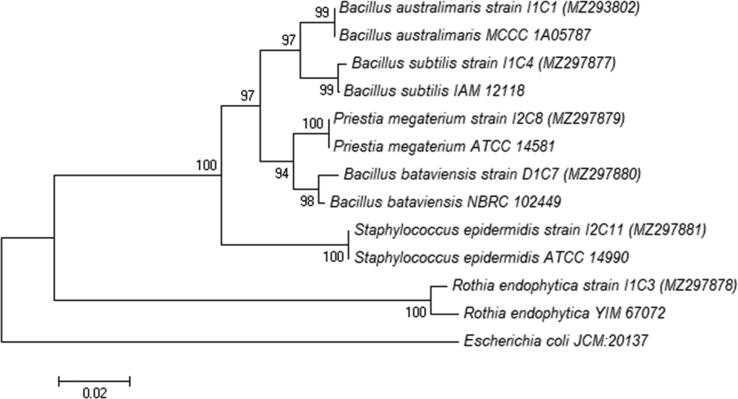
Nine bacterial strains were isolated from dried figs. A low diversity of bacterial communities associated with dried figs was observed, with Bacillus being the most frequent genus detected. BLAST analysis of the 16SrDNA sequences revealed that isolate IC1 has 100 % homology with Bacillus australimaris MCCC 1A05787(NR _148787.1), isolate IC8 with Priestia megaterium ATCC 14581(CP035094.1), IC11 with Staphylococcus epidermidis NBRC 100,911 (AP019721.1); 99 % homology of isolate IC4 with Bacillus subtilis IAM 12118(MK267098.1), isolate DC7 with Bacillus bataviensis NBRC 102,449 (NR_114093.1); and 98 % homology of isolate IC3 with Rothia endophytica DSM 26247(NR_109752.1). The 16S rDNA sequences were placed in the GenBank database, below the accession numbers MZ293802 for I1C1, MZ297877 for I1C4, MZ297879 for I2C8, MZ297880 for D1C7, MZ297881 for I2C11 and MZ297878 for I1C3 (Fig. 3).
Fig. 3.
The phylogenetic relationship between strains IC1, IC8, IC4, DC7, IC11, and IC3 using Neighbor-joining tree based on 16S rDNA sequences. At the nodes, bootstrap values greater than 70 percent (reported as percentages of 1000 replications) are provided.
3.3. Study of bacterial communities by DGGE
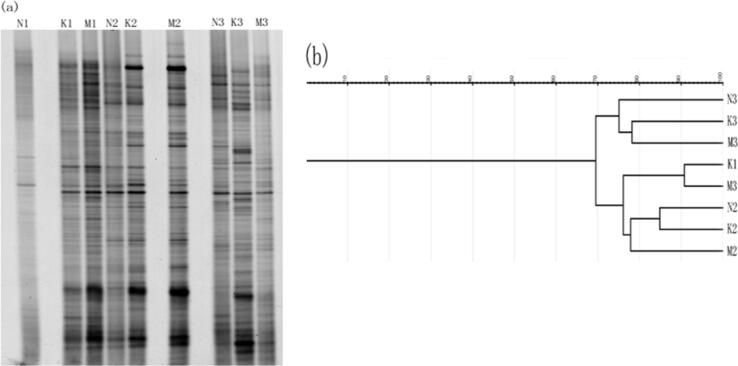
Denaturing gradient gel electrophoresis (DGGE) analysis was used to determine the bacterial diversity of soil rhizosphere samples, with each band on the gel representing a separate bacterial community (Muyzer et al., 1993). The DGGE profiles were reproducible between triplicate independent DNA extractions from the combined samples of the rhizosphere soil of F. carica from Nabeul (N), Kerkouane (K) and Mateur (M) (Fig. 4(a)). A Cluster analysis of bacterial populations obtained from DGGE profiles fixed on the median similarity matrix (UPGMA dendrogram, Fig. 4(b)) was also possible using PCR-DGGE analysis. For three regions, community diversity indexes were determinated: Nabeul (N), Kerkouane (K) and Mateur (M) (Fig. 5). The rhizosphere soil collected from the region of Kerkouane samples displayed the largest number of bands (richness score), the highest Shannon–Weaver index and also the highest Simpson index values. The value of evenness was quite constant and was defined by low E values (varying from 0.221 to 0.261).
Fig. 4.
(a) DGGE analysis of PCR products from DNA extracted from Ficus carica soil rhizosphere samples in three Tunisian regions. (b) Dendrogram illustrating the genetic similarity of soil rhizosphere after UPGMA cluster analysis with GelJ.
Fig. 5.
Diversity indices between soil rhizosphere samples of Ficus carica in three regions in Tunisia. S: Richness, H: Shannon–Weaver index, D: Simpson index of dominance, E: evenness.
3.4. PGP potential and abiotic stress of bacteria isolates
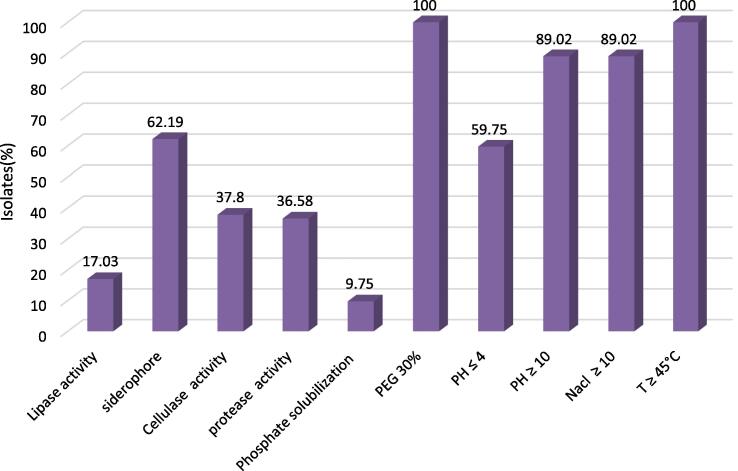
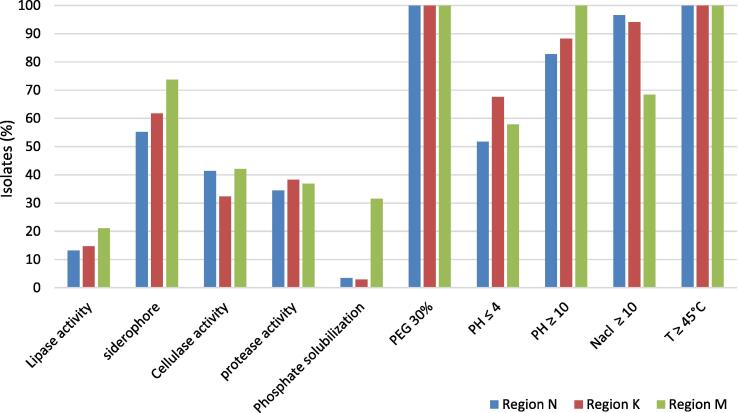
Eighty-two isolates were characterized for various PGP activities. Most isolates demonstrated several PGP traits, which can promote plant growth in different ways. The most common PGP trait for all bacteria from the three regions was siderophore production (62 %), followed by cellulase (38 %), then protease activity (37 %), then by lipases activity (17 %) and lastly by solubilization of phosphates (9 %). Besides, more studies were completed to assess the resistance of these strains to abiotic stresses (Fig. 6).
Fig. 6.
PGP activity and abiotic stress tolerance of the isolates from rhizosphere of Ficus carica.
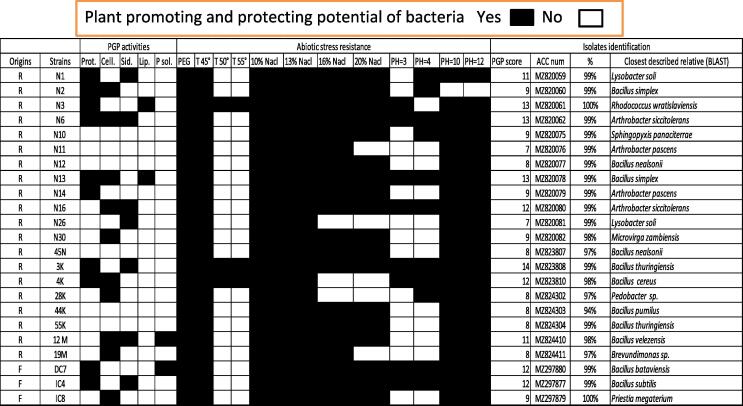
The results revealed that all strains showed stress resistance, and they all could grow in the presence of a significant concentration of PEG. All strains were capable to grow at 45 °C; among those, 27 % could also grow at 50 °C, and only 13 % could tolerate higher temperatures (55 °C). 89 % of isolates showed moderate tolerance to salt (10 % NaCl), 72 % could grow in 13 % NaCl, 52 % could exist in 16 % NaCl, and only 30 % could tolerate 20 % NaCl. The ability of the isolates to grow in a broad pH range was also tested. It was found that 85 % of the strains were facultative alkalophiles, which are capable to grow (until pH = 12), while 63 % of them could grow in acidic media (pH = 4). 29 % of the isolates were facultative acidophiles, which are capable to grow at pH = 3. The details of PGP traits and abiotic stress of the isolated strains were presented in Fig. 7. The most potentially beneficial rhizobacteria were represented in Fig. 8, which show their biocontrol traits. 23 selected strains showed most PGP traits, 8 strains presented 12 or more, and 15 strains displayed between 7 and 11 of 17 PGP activities. None of the isolates possess all PGP traits. The majority of the isolates manifested a possible adaptation to abiotic stress and unfavorable environments. All strains exhibited resistance to low moisture, and all strains could grow at 45 °C. All strains could exist in media with 10 % NaCl added, 18 strains could tolerate 20 % NaCl added, and the majority of the strains could grow in basic media (pH = 10 and pH = 12). Seven strains manifested siderophore activity, 10 strains displayed a positive protease activity, 10 strains presented cellulase activity and 2 strains had lipase activity. Also, only 2 strains among these selected bacteria showed phosphate solubilization activity.
Fig. 7.
Percentage of the strains associated with the rhizosphere of Ficus carica showing PGP traits and abiotic stress of isolated from three regions (Nabeul, Kerkouane and Mateur).
Fig. 8.
PGP potential of the isolates is demonstrated as a PGP score, determinated by the sum of the number of PGP activity by each strain.R = rhizosphere;F = fig; Prot. = protease;activity;Cell. = cellulose activity; Sid. = siderophore production; Lip. = Lipase.
4. Discussion
To our knowledge the identification and the characterization of rhizospheric and endophytic bacteria from dried fruit of Ficus carica is studied for the first time in Tunisia. Rhizospheric bacteria species were isolated in non-specific media in order to obtain the greatest number of genera as possible. In this research, we characterized the biodiversity via PCR-16S rRNA analysis and then sequencing partial 16 S rRNA genes. We found that the cultivable bacterial community was principally constituted of Pseudomonas, Bacillus, Arthrobacter, Lysobacter, Microbacterium and Stenotrophomonas. Among them, the genera Pseudomonas and Bacillus were the most frequent endophytic bacteria for their PGP activities as described in the literature (Zhang et al., 2017, Chaudhary and Sharma, 2019). It is well-known that soil microbial communities are one of the most significant reservoirs of biological diversity in the world receiving no less than one quarter of the living organisms on earth (Decaëns et al., 2006, Tibbett et al., 2020). 17 different genera have been identified by this study, and the high genetic diversity is probably due to multitude elements (Macdonald and Singh, 2014, Nwachukwu and Babalola, 2021). PGPR are agriculture bioresources that enhance both plant growth and productivity, these Plant microbiomes have a direct impact on noxious microbes in the rhizosphere, and many beneficial microorganisms isolated from soil could boost the defensive capacity of the plant (Zamioudis and Pieterse, 2012, Babalola et al., 2021). The studies of bacteria strains naturally associated with dried fruit are limited (Nyanga et al., 2007, Ribeiro et al., 2014) and the fruit flora commonly includes yeasts and molds (Qadri et al., 2015). For dried figs, natural sun-drying had a lot of disadvantages as a result of the uncontrolled time and temperature, as well as the lack of meshes. All these factors can generate infection by fungi (Trucksess and Scott, 2008). Aspergillus spp., Fusarium spp., and Penicillium species are the largest fungal contaminants in dried figs (Turkoz Bakirci, 2020, Javanmard, 2010). The study of bacterial communities in dried figs has not been previously explored. In our study, 9 bacteria were isolated from dried figs, and Bacillus was the most frequent genus detected. In fact, in order to investigate the safety of dried fruits this genus was found in dried figs: Bacillus Cereus (Akbas and Ozdemir, 2008) and Bacillus subtilis (Hamanaka et al., 2002). The genus Bacillus is also very common in dried vegetables (Kudjawu et al., 2011). We identified Rothia endophytica, which has been isolated from healthy Maize roots (Elbahnasawy et al., 2021) and from wheat (Yousaf et al., 2017). We also found Staphylococcus epidermidis, which has been detected in the nectar of Nicotiana glauca (Fridman et al., 2012).
After its initial adoption in 1993, DGGE has been universally employed in recent years to investigate microbial communities (Muyzer et al., 1993, Ferreira et al., 2008). Genetic diversity was evaluated as bands on DGGE profile. To evaluate differences in the bacterial population in soils, the Shannon–Wiener index of diversity (H), richness (S), and evenness (E) were used.
Few studies have applied the index of diversity to compare between same-plant rhizosphere soils in different regions. To our knowledge, rhizosphere soil of ficus carica has not been investigated by PCR-DGGE analysis to assess bacterial community dynamics. The DGGE profile of ficus carica rhizosphere from the region of Kerkouane was the most diverse followed by that of Mateur, with the least diverse being from Nabeul the values of richness (S) and Shannon–Wiener index (H) of each soil confirms the previous result. Some studies reported the study of rhizospheric bacteria using PCR-DGGE analysis in different plant bacteria in avocado (Yang et al., 2001), and peach palm (Almeida et al., 2005). The dendrogram representing the bacterial communities at different sampling dates were drawn (Fig. 5b) and N3, K3, M3 were grouped together with an average similarity of 75 %. K1, M3, N2, K2, M2 were in the same group with 76 % similarity. This result demonstrated that there is not a huge difference between the three regions concerning the bacterial community analysis.
The potential PGPR of rhizospheric and endophytic bacteria displayed numerous plant growth-promoting activities, such as siderophore production, phosphate solubilization, cellulase activity and protease activity (Kumar et al., 2014, Vinayarani and Prakash, 2018). In this research, we tested our isolates for various plant growth promoting properties, and we observed that Bacillus then Pseudomonas gave positive reactions for phosphate solubilization, production of siderophore, protease, cellulose and lipase. The role of siderophore is to chelate and to extract ferric iron from mineral or organic complexes present in the environment, and thus make it accessible to microorganisms and plants (Neilands, 1995, Ghosh et al., 2020). The production of siderophores is an ecological advantage in the rhizosphere and many rhizobacteria acquire this ability especially the following genera: Pseudomonas, Bacillus, Rhodococcus, and Enterobacter (Tian et al., 2009, Kour et al., 2019). Only few rhizobacteria displayed the ability to solubilize phosphate. Some studies reported that the activities of different soil enzymes were severely lowered in saline soils, which influenced the capacity of the soil (Batra and Manna, 1997, Dagar et al., 2004). Bacillus and Pedobacter Pseudomonas displayed the most efficiency in phosphate solubilization in our collection. Azospirillum, Burkholderia, Pseudomonas, Rhizobium and Enterobacter are reported in the literature as having highly efficient PSB (De Zutter et al., 2021). Phosphate solubilizing bacteria (PSB) are microorganisms that represent many functional characteristics associated to abilities of phosphate solubilization, they could show an encouraging substitute method to decrease the need of phosphate mineral fertilization to improve soil management and sustainable agriculture (Calvo et al., 2014, Alori et al., 2017, Amy et al., 2022). Other cell wall-degrading enzymes were also explored. We observed that 37 %, 36 %, and 17 % of the strains isolated from the rhizosphere have protease, cellulose and lipase activity, respectively. The rhizosphere bacterial species, such as Pseudomonas, Bacillus, were active against soil fungi by producing lytic enzymes, such as cellulase, protease, and lipase (Mabood et al., 2014, Chandran et al., 2021). In particular, Pseudomonas is known to produce lytic enzymes. These strains were active at suppressing fungal plant pathogens in tomatoes (Lugtenberg et al., 2001). Also, cell wall-degrading enzymes are produced by Bacillus spp such as proteases and chitinases. (El-Bendary et al., 2016, Schönbichler et al., 2020). The main factors limiting the biological activity of bacteria in the soil are: water deficit, extreme pH and temperatures, and nutrient deficiencies (Pandey et al., 2017). The frequent interactions between these different constraints influence the growth and survival capacity of microorganisms in arid soils (Lal et al., 2007). Our bacterial strains were tested for resistance to numerous abiotic stresses to assess their adaptability to all these naturals constraints. All the strains could grow at 45 °C, and even 13 % could tolerate a higher temperature of 55 °C; most Pseudomonas strains present this ability (Scheldeman et al., 2006). Also, it has been reported that Bacillus thermoamylovorans can survive at up to 60 °C (Choonut et al., 2020). All the strains isolated could grow on a dehydrated medium with a concentration of 30 % PEG 8000. It has been proven that inoculation with rhizobacteria allows an improvement in the water status of plants under limited irrigation conditions (Eid et al., 2021). The percentage decreases proportionally with the increase of the salt concentration; the tolerance to salinity is 89 %, 72 %, 52 % and 30 % for a concentration of NaCl of 10 %, 13 %, 16 % and 20 %, respectively. Salt stress tolerance is an attractive PGPR trait and is important for several soil types that suffer from salinity problems (Etesami and Maheshwari, 2018).
The study of abiotic stress tolerance for bacteria seems very interesting, for it can aid in the understanding of the adaptation mechanisms of microorganisms living in an environment and the selection of a large number of PGPR to improve the performance of plant tolerance under environmental stress conditions. Bacterial endophytes were known to be a source of new secondary metabolites for a potential therapeutic purpose (Tan and Zou, 2001, Shukla et al., 2014).
5. Conclusion
The results of this study might allow for the selection of several candidate strains that could be applied to promote plant growth when they are exposed to nutrient or environmental stress. Rhizospheric and endophytic bacteria from dried fruit of Ficus carica showed numerous PGP activities that potentially enhance plant growth. The selected strains will be further tested in future in vivo against phytopathogens. These plant growth-promoting bacteria may have a good impact in organic agriculture practices.
CRediT authorship contribution statement
Lamis Abid: Conceptualization, Methodology, Writing – original draft. Marwa Smiri: . Ermanno Federici: Conceptualization, Methodology, Resources. Bart Lievens: Methodology, Resources. Mohamed Manai: Supervision, Resources. Yunjun Yan: Resources. Najla Sadfi-Zouaoui: Supervision, Methodology, Resources.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by Tunisian Ministry of Higher Education and Scientific Research, (LR16ES05) and CSC scholarship. We thank Dr. Maroua Oueslati. (Laboratoire de Mycologie, Pathologies et Biomarqueurs (LR16ES05), Faculté des Sciences de Tunis) for helping in data analysis and for her comments on the manuscript.
Footnotes
Peer review under responsibility of King Saud University.
References
- Agbodjato N., Noumavo P., Adjanohoun A., Agbessi L., Baba-Moussa L. Synergistic Effects of Plant Growth Promoting Rhizobacteria and Chitosan on in vitro seeds germination, greenhouse growth, and nutrient uptake of Maize (Zea mays L.) Biotechnol. Res. Int. 2016;20(1):1–11. doi: 10.1155/2016/7830182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbas M.Y., Ozdemir M. Application of gaseous ozone to control populations of Escherichia coli, bacillus cereus and bacillus cereus spores in dried figs. Food Microbiol. 2008;25(2):386–391. doi: 10.1016/j.fm.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Almeida C.V., Yara R., Almeida M. Fungos endofıticos isolados de ápices caulinares de pupunheira cultivada in vivo e in vitro. Pesq agropec bras. 2005;40:467–470. doi: 10.1590/S0100-204X2005000500007. [DOI] [Google Scholar]
- Alori E.T., Glick B.R., Babalola O.O. Microbial Phosphorus Solubilization and Its Potential for Use in Sustainable Agriculture. Front. Microbiol. 2017;8:971. doi: 10.3389/fmicb.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Pérez S., Herrera C.M., de Vega C. Zooming-in on floral nectar: a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol. Ecol. 2012;80:591–602. doi: 10.1111/j.1574-6941.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- Amy C., Avice J.C., Laval K., Bressan M. Are native phosphate solubilizing bacteria a relevant alternative to mineral fertilizations for crops? Part I. when rhizobacteria meet plant P requirements. Rhizosphere. 2022;21 doi: 10.1016/j.rhisph.2022.100476. [DOI] [Google Scholar]
- Arora N.K., Tewari S., Singh S., Lal N. In: Bacteria in Agrobiology: Stress Management. Maheshwari D.K., editor. Springer; Berlin Heidelberg: 2012. PGPR for Protection of Plant Health Under Saline Conditions; pp. 239–258. [Google Scholar]
- Babalola O.O., Emmanuel O.C., Adeleke B.S., Odelade K.A., Nwachukwu B.C., Ayiti O.E., Adegboyega T.T., Igiehon N.O. Rhizosphere Microbiome Cooperations: Strategies for Sustainable Crop Production. Curr. Microbiol. 2021;78(4):1069–1085. doi: 10.1007/s00284-021-02375-2. [DOI] [PubMed] [Google Scholar]
- Badgujar H.B., Patel V.V., Bandivdekar A.H., Mahajan R.T. Traditional uses, phytochemistry and pharmacology of Ficus carica: A review. Pharm. Biol. 2014;52:1487–1503. doi: 10.3109/13880209.2014.892515. [DOI] [PubMed] [Google Scholar]
- Barolo M.I., Ruiz Mostacero N., López, S.N., Ficus carica L. (Moraceae): an ancient source of food and health. Food Chem. 2014;164:119–127. doi: 10.1016/j.foodchem.2014.04.112. [DOI] [PubMed] [Google Scholar]
- Batra L., Manna M.C. Dehydrogenase activity and microbial biomass carbon in salt- affected soils of semi- arid regions. Soil. Res. Rehabil. 1997;11(3):293–303. doi: 10.1080/15324989709381481. [DOI] [Google Scholar]
- Bekatorou A., Sarellas A., Ternan N.G., Mallouchaos A., Komaitis M., Koutinas A.A., Kanellaki M. Low-temperature brewing using yeast immobilized on dried figs.J. Agri. Food Chem. 2002;50:7249–7257. doi: 10.1021/jf020291q. [DOI] [PubMed] [Google Scholar]
- Calvo P., Nelson L., Kloepper J.W. Agricultural uses of plant biostimulants. Plant Soil. 2014;383:3–41. doi: 10.1007/s11104-014-2131-8. [DOI] [Google Scholar]
- Chandran H., Meena M., Swapnil P. Plant Growth-Promoting Rhizobacteria as a Green Alternative for Sustainable Agriculture. Sustainability. 2021;13:10986. doi: 10.3390/su131910986. [DOI] [Google Scholar]
- Chaudhary P., Sharma A. Response of nanogypsum on the performance of plant growth promotory bacteria recovered from nanocompound infested agriculture feld. Environ.Ecol. 2019;37(1B):363–372. [Google Scholar]
- Choonut A., Prasertsan P., Klomklao S., Sangkharak K. Bacillus thermoamylovorans-related strain isolated from high temperature sites as potential producers of medium-chain-length polyhydroxyalkanoate (mcl-PHA) Curr. Microbiol. 2020;77:3044–3056. doi: 10.1007/s00284-020-02118-9. [DOI] [PubMed] [Google Scholar]
- Choubane S., Cheba B.A., Benourrad A. Screening and Phenotypic Diversity of Amylase Producing Rhizospheric Bacteria from Some North African Plants. Proc. Technol. 2016;22:1197–1204. doi: 10.1016/J.PROTCY.2016.01.168. [DOI] [Google Scholar]
- Crisosto C.H., Bremer V., Ferguson L., Crisosto G.M. Evaluating quality attributes of four fresh fig (Ficus carica L.) cultivars harvested at two maturity stages. HortScience. 2010;45:707–710. doi: 10.21273/HORTSCI.45.4.707. [DOI] [Google Scholar]
- Dagar J.C., Tomar O.S., Kumar Y., Yadav R.K. Growing three aromatic grasses in different alkali soils in semi-arid regions of northern India. Land Degrad. Dev. 2004;15(2):143–151. [Google Scholar]
- De Zutter N., Ameye M., Debode J., De Tender C., Ommeslag S., Verwaeren J., VermeirDe Gelder L. Shifts in the rhizobiome during consecutive in planta enrichment for phosphate-solubilizing bacteria differentially affect maize P status. Microb. Biotechnol. 2021;14:1594–1612. doi: 10.1111/1751-7915.13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaëns T., Jiménez J.J., Gioia C., Measey G.J., Lavelle P. The values of soil animals for conservation biology. Eur. J. Soil Biol. 2006;42:S23–S38. doi: 10.1016/j.ejsobi.2006.07.001. [DOI] [Google Scholar]
- Eid A.M., Fouda A., Abdel-Rahman M.A., Salem S.S., Elsaied A., Oelmüller R., Hijri M., Bhowmik A., Elkelish A., Hassan S.E.D. Harnessing Bacterial Endophytes for Promotion of Plant Growth and Biotechnological Applications: An Overview. Plants. 2021;10:935. doi: 10.3390/plants10050935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbahnasawy M.A., Shehabeldine A.M., Khattab A.M., Amin B.H., Hashem A.H. Green biosynthesis of silver nanoparticles using novel endophytic Rothia endophytica: Characterization and anticandidal activity. J. Drug Deliv. Sci. Technol. 2021;62 doi: 10.1016/J.JDDST.2021.102401. [DOI] [Google Scholar]
- El-Bendary M.A., Hamed H.A., Moharam M.E. Potential of Bacillus isolates as bio-control agents against some fungal phytopathogens. Biocatal. Agric. Biotechnol. 2016;5:173–178. doi: 10.1016/j.bcab.2016.02.001. [DOI] [Google Scholar]
- Etesami H., Maheshwari D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018;156:225–246. doi: 10.1016/j.ecoenv.2018.03.013. [DOI] [PubMed] [Google Scholar]
- Federici E., Pepi M., Esposito A., Scargetta S., Fidati L., Gasperini S., Cenci G., Altieri R. Two-phase olive mill waste composting: community dynamics and functional role of resident microbiota. Bioresour. Technol. 2011;102:10965–10972. doi: 10.1016/j.biortech.2011.09.062. [DOI] [PubMed] [Google Scholar]
- Ferjani R., Marasco R., Rolli E., Cherif H., Cherif A., Gtari M., Boudabous A., Daffonchio D., Ouzari H.-I. The date palm tree rhizosphere is a niche for plant growth promoting bacteria in the oasis ecosystem. Biomed Res. Int. 2015;2015:1–10. doi: 10.1155/2015/153851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira E.P.d.B., Dusi A.N., Xavier G.R., Rumjanek N.G. Rhizosphere bacterial communities of potato cultivars evaluated through PCR-DGGE profiles. Pesqui Agropecuaria Bras. 2008;43(5):605–612. [Google Scholar]
- Fridman S., Izhaki I., Gerchman Y., Halpern M. Bacterial communities in floral nectar. Environ. Microbiol. Rep. 2012;4:97–104. doi: 10.1111/j.1758-2229.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- Ghosh S.K., Bera T., Chakrabarty A.M. Microbial siderophore - a boon to agricultural sciences. Biol. Control. 2020;144:104214. [Google Scholar]
- Hamanaka D., Uchino T., Hu W., Yasunaga E. Efects of infrared radiation on inactivation of Bacillus subtilis spore and Aspergillus niger spore. J. Jpn. Soc. Agric. Mach. 2002;64(6):69–75. doi: 10.11357/jsam1937.64.6_69. [DOI] [Google Scholar]
- Heras J., Dominguez C., Mata E., Pascual V., Lozano C., Torres C., Zarazaga M. GelJ a tool for 479 analyzing DNA fingerprint gel images. BMC. Bioinform. 2015;16(270) doi: 10.1186/s43141-020-00114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanmard M. Occurrence of Mould Counts and Aspergillus Species in Iranian Dried Figs at Different Stages of Production and Processing. J. Agric. Sci. Technol. 2010;12:331–338. [Google Scholar]
- Kour D., Rana K.L., Yadav N., Yadav A.N., Singh J., Rastegari A.A., Saxena A.K. In: Recent Advancement in White Biotechnology Through Fungi, Volume 2: Perspective for Value-added Products and Environments. Yadav A.N., Singh S., Mishra S., Gupta A., editors. Springer International Publishing; Cham: 2019. Agriculturally and industrially important fungi: current developments and potential biotechnological applications; pp. 1–64. [DOI] [Google Scholar]
- Kudjawu B.D., Sakyi-Dawson E., Amoa-Awua W.K. The microbiota of dried traditional vegetables produced in the Sudan Savannah and Guinea Savannah agro-ecological zones of Ghana. Int. Food Res. J. 2011;18(1):101–107. [Google Scholar]
- Kumar S.P., Hariprasad P., Singh S.B., Gowtham H.G., Niranjana S.R. Structural and functional diversity of rhizobacteria associated with Rauwolfia spp. across the Western Ghat regions of Karnataka, India. World J. Microbiol. Biotechnol. 2014;30(1):163–173. doi: 10.1007/s11274-013-1435-9. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.-A., Kämpfer P., Arun A.B., Shen F.-T., Huber B., Rekha P.D., Young C.-C. Deinococcus ficus sp. nov., isolated from the rhizosphere of Ficus religiosa L. Int. J. Syst. Evol. Microbiol. 2006;56(4):787–791. doi: 10.1099/ijs.0.64007-0. [DOI] [PubMed] [Google Scholar]
- Lal R., Sulaimanov M., Stewart B., Hansen D., Doraiswamy P. first ed. Taylor-Francis; New York: 2007. Climate change and terrestrial C sequestration in Central Asia. [Google Scholar]
- Lansky E.P., Paavilainen H.M. CRC Press; New York: 2010. Figs: the genus Ficus. [Google Scholar]
- Lazreg A.H., Gaaliche B., Fekih A., Mars M., Aouni M., Pierre C.J., Said K. In vitro cytotoxic and antiviral activities of Ficus carica latex extracts. Nat. Prod. Res. 2011;25(3):310–319. doi: 10.1080/14786419.2010.528758. [DOI] [PubMed] [Google Scholar]
- Le Floc’h, E., Imprimerie Officielle de la République Tunisienne; Tunis: 1983. Contribution a‘ une étude ethnobotanique de la flore Tunisienne. [Google Scholar]
- Leporatti M.L., Ghedira K. Comparative analysis of medicinal plants used in traditional medicine in Italy and Tunisia. J. Ethnobiol. Ethnomed. 2009;5:31–39. doi: 10.1186/1746-4269-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugtenberg B.J., Dekkers L., Bloemberg G.V. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 2001;39(1):461–490. doi: 10.1146/annurev.phyto.39.1.461. [DOI] [PubMed] [Google Scholar]
- Mabood F., Zhou X., Smith D.L. Microbial signaling and plant growth promotion. Can. J. Plant Sci. 2014;94:1051–1063. doi: 10.4141/cjps2013-148. [DOI] [Google Scholar]
- Macdonald C., Singh B. Harnessing plant-microbe interactions for enhancing farm productivity. BIOENG. 2014;5(1):5–9. doi: 10.4161/bioe.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars M., Chatti K., Saddoud O., Salhi Hannachi A., Trifi M., Marrakchi M. Fig cultivation and genetic ressources in Tunisia, an overview. Acta Hortic. 2008;798:27–32. doi: 10.17660/ActaHortic.2008.798.2. [DOI] [Google Scholar]
- Mars M., Gaaliche B., Ouerfelli I., Chouat S. Systèmes de production et ressources génétiques du figuier (Ficus carica L.) à Djebba et Kesra, deux villages de montagne au nord-ouest de la Tunisie. Revue des Régions Arides. 2009;22(1):33–45. [Google Scholar]
- Mawa S., Husain K., Jantan I. Ficus carica L. (Moraceae): Phytochemistry, Traditional Uses and Biological Activities. Evid. -based Complement. Altern. Med. 2013;2013:1–8. doi: 10.1155/2013/974256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra D.J., Singh R., Mishra U.K., Kumar S.S. Role of bio-fertilizer in organic agriculture: a review. Res. J Recent Sci. 2013;2:39–41. [Google Scholar]
- Muyzer G., De Waal E.C., Uitterlinden A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999;170(1):265–270. doi: 10.1016/S0378-1097(98)00555-2. [DOI] [PubMed] [Google Scholar]
- Neilands J.B. Siderophores: Structure and function of microbial iron transport compounds. J. Biol. Chem. 1995;270(45):26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- Nikolidaki E.K., Chiou A., Christea M., Gkegka A.P., Karvelas M., Karathanos V.T. Sun dried Corinthian currant (Vitis vinifera L., var. Apyrena) simple sugar profile and macronutrient characterization. Food Chem. 2017;221:365–372. doi: 10.1016/j.foodchem.2016.10.070. [DOI] [PubMed] [Google Scholar]
- Ntuli V., Chatanga P., Kwiri R., Gadaga H.T., Gere J., Matsepo T., Potloane R.P. Microbiological quality of selected dried fruits and vegetables in Maseru. Lesotho. Afr. J. Microbiol. Res. 2017;11(5):185–193. [Google Scholar]
- Nwachukwu B.C., Babalola O.O. Perspectives for sustainable agriculture from the microbiome in plant rhizosphere. Plant Biotechnol. Rep. 2021;15(3):259–278. [Google Scholar]
- Nyanga L.K., Nout M.J.R., Gadaga T.H., Theelen B., Boekhout T., Zwietering M.H. Yeasts and lactic acid bacteria microbiota from masau (Ziziphus mauritiana) fruits and their fermented fruit pulp in Zimbabwe. Int. J. Food Microbiol. 2007;120(1–2):159–166. doi: 10.1016/j.ijfoodmicro.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Omidvari M. Tehran University; Iran: 2008. Biological control of Fusarium solani, the causal agent of damping off, by fluorescent pseudomonads and studying some of their antifungal metabolite productions on it. M.S. thesis. [Google Scholar]
- Osman S.M., Abd El-Rhman I.E. Effect of organic bio-N-fertilization on growth, productivity of fig tree (Ficus carica, L.) Res. J. Agric. & . Biol. 2010;6(3):319–328. [Google Scholar]
- Perez-Miranda S., Cabirol N., George-Tellez R., Zamudio-Rivera L.S., Fernandez F.J. O-CAS, a fast and universal method for siderophore detection. J. Microbio. Meth. 2007;70(1):127–131. doi: 10.1111/jam.13025. [DOI] [PubMed] [Google Scholar]
- Pandey P., Irulappan V., Bagavathiannan M.V., Senthil-Kumar M. Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front. Plant Sci. 2017;8:537. doi: 10.3389/fpls.2017.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M., Srinivasan R., Chaudhary M., Choudhary M., Jat L.K. Plant Growth Promoting Rhizobacteria (PGPR) for Sustainable Agriculture: Perspectives and Challenges. Woodhead Publishing; 2019. pp. 129–157. [Google Scholar]
- Qadri O.S., Yousuf B., Srivastava A.K. Fresh-cut fruits and vegetables: critical factors influencing microbiology and novel approaches to prevent microbial risks—a review. Cogent Food Agric. 2015;1(1):1121606. [Google Scholar]
- Ribeiro C., Freixo R., Silva J., Gibbs P., Morais A.M.M.B., Teixeira P. Dried fruit matrices incorporated with the probiotic strains Lactobacillus plantarum. Int. J. Food Stud. 2014;3:69–73. doi: 10.7455/ijfs/3.1.2014.a6. [DOI] [Google Scholar]
- Saadullah A.A.M., Abdullah S.K. Contamination of Dried Figs with Fungi and Aflatoxigenic Potential of Some Isolates of Aspergillus Section Flavi. J. Biol. Agric. Healthc. 2015;5(2):76–80. [Google Scholar]
- Sadder M.T., Ateyyeh A.F. Molecular assessment of polymorphism among local Jordanian genotypes of the common fig (Ficus carica L.) Sci. Hortic. 2006;107(4):347–351. [Google Scholar]
- Scheldeman P., Herman L., Foster S., Heyndrickx M. Bacillus sporothermodurans and other highly heat-resistant spore formers in milk. J. Appl. Microbiol. 2006;101(3):542–555. doi: 10.3389/fmicb.2020.00521. [DOI] [PubMed] [Google Scholar]
- Schönbichler A., Díaz-Moreno S.M., Srivastava V., McKee L.S. Exploring the potential for fungal antagonism and cell wall attack by Bacillus subtilis natto. Front. Microbiol. 2020;11:521. doi: 10.3389/fmicb.2020.00521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S.T., Habbu P.V., Kulkarni V.H., Jagadish K.S., Pandey A.R., Sutariya V.N. Endophytic microbes: a novel source for biologically/pharmacologically active secondary metabolites. Asian J Pharmacol Toxicol. 2014;2:1–16. [Google Scholar]
- Singh D., Singh B., Goel R.K. Traditional uses, phytochemistry and pharmacology of Ficus religiosa: A review. J. Ethnopharmacol. 2011;134(3):565–583. doi: 10.1016/j.jep.2011.01.046. [DOI] [PubMed] [Google Scholar]
- Slatnar A., Klancar U., Stampar F., Veberic R. Effect of drying of figs (Ficus carica L.) on the contents of sugars, organic acids, and phenolic compounds. J. Agric. Food Chem. 2011;59:11696–11702. doi: 10.1021/jf202707y. [DOI] [PubMed] [Google Scholar]
- Tan R.X., Zou W.X. Endophytes: a rich source of functional metabolites. Nat. Prod. Rep. 2001;18(4):448–459. doi: 10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- Tian F., Ding Y., Zhu H., Yao L., Du B. Genetic diversity of siderophore-producing bacteria of tobacco rhizosphere. Braz. J. Microbiol. 2009;40(2):276–284. doi: 10.1590/s1517-838220090002000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbett M., Fraser T.D., Duddigan S. Identifying potential threats to soil biodiversity. PeerJ. 2020;8:e9271. doi: 10.7717/peerj.9271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucksess M.W., Scott P.M. Mycotoxins in botanicals and dried fruits: A review. Food Addit. Contam. Part A Chem. Anal. 2008;25:181–192. doi: 10.1080/02652030701567459. [DOI] [PubMed] [Google Scholar]
- Turkoz Bakirci G. Investigation of aflatoxins levels in commercial dried figs from westernTurkey. Int. Food. Res. 2020;27:245–251. [Google Scholar]
- Vinayarani G., Prakash H.S. Growth promoting rhizospheric and endophytic bacteria from Curcuma longa L. as biocontrol agents against rhizome rot and leaf blight diseases. Plant Pathol. J. 2018;34(3):218–235. doi: 10.5423/PPJ.OA.11.2017.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson J.A. The functional food properties of figs. Cereal Foods World. 1999;44:82–87. doi: 10.1007/978-94-007-2534-8_51. [DOI] [Google Scholar]
- Wang X., Mavrodi D.V., Ke L., Mavrodi O.V., Yang M., Thomashow L.S. Biocontrol and plant growth-promoting activity of rhizobacteria from Chinese fields with contaminated soils. Microb. Biotechnol. 2015;8(3):404–418. doi: 10.1111/1751-7915.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.H., Crowley D.E., Menge J.A. 16S rDNA fingerprinting of rhizosphere bacterial communities associated with healthy and Phytophtora infected avocado roots. FEMS Microbiol. Ecol. 2001;35:129–136. doi: 10.1111/j.1574-6941.2001.tb00796.x. [DOI] [PubMed] [Google Scholar]
- Yang X.M., Yu W., Ou Z.P., Ma H.L., Liu W.M., Ji X.L. Antioxidant and immunity activity of water extract and crude polysaccharide from Ficus carica L. fruit. Plant Foods Hum. Nutr. 2009;64(2):167–173. doi: 10.2147/CCID.S80906. [DOI] [PubMed] [Google Scholar]
- Yousaf M., Rehman Y., Hasnain S. High-yielding wheat varieties harbour superior plant growth promoting bacterial endophytes. Appl. Food Biotechnol. 2017;4(3):143–154. doi: 10.1094/MPMI-06-11-0179. [DOI] [Google Scholar]
- Zamioudis C., Pieterse C.M.J. Modulation of host immunity by beneficial microbes. Mol. Plant- Microbe Interact. 2012;25(2):139–150. doi: 10.1094/MPMI-06-11-0179. [DOI] [PubMed] [Google Scholar]
- Zhang X.X., Wang R.J., Fan C.B., Liu G., Pu S.Z. A highly selective fluorescent sensor for Cd2+ based on a new diarylethene with a 1,8-naphthyridine unit. Dyes Pigm. 2017;139:208–217. doi: 10.1016/j.dyepig.2016.12.023. [DOI] [Google Scholar]