Abstract
The myelomonocytic receptor CD33 (Siglec-3) inhibits innate immune reactivity by extracellular V-set domain recognition of sialic acid (Sia)-containing “self-associated molecular patterns” (SAMPs). We earlier showed that V-set domain-deficient CD33-variant allele, protective against late-onset Alzheimer’s Disease (LOAD), is derived and specific to the hominin lineage. We now report multiple hominin-specific CD33 V-set domain mutations. Due to hominin-specific, fixed loss-of-function mutation in the CMAH gene, humans lack N-glycolylneuraminic acid (Neu5Gc), the preferred Sia-ligand of ancestral CD33. Mutational analysis and molecular dynamics (MD)-simulations indicate that fixed change in amino acid 21 of hominin V-set domain and conformational changes related to His45 corrected for Neu5Gc-loss by switching to N-acetylneuraminic acid (Neu5Ac)-recognition. We show that human-specific pathogens Neisseria gonorrhoeae and Group B Streptococcus selectively bind human CD33 (huCD33) as part of immune-evasive molecular mimicry of host SAMPs and that this binding is significantly impacted by amino acid 21 modification. In addition to LOAD-protective CD33 alleles, humans harbor derived, population-universal, cognition-protective variants at several other loci. Interestingly, 11 of 13 SNPs in these human genes (including CD33) are not shared by genomes of archaic hominins: Neanderthals and Denisovans. We present a plausible evolutionary scenario to compile, correlate, and comprehend existing knowledge about huCD33-evolution and suggest that grandmothering emerged in humans.
Keywords: CD33, pathogen, sialic acids, archaic genome, molecular dynamics simulation, phylogenetic analysis
Introduction
In keeping with the fundamental importance of reproduction for the process of biological evolution via natural selection, loss of fecundity generally coincides with the end of lifespan in almost all species studied to date. Humans and certain toothed whales like orcas are so far the only mammals known to manifest prolonged postreproductive lifespans under naturalistic conditions (Hawkes et al. 1998; Hawkes 2004, 2010; Johnstone and Cant 2010; Cant and Croft 2019; Khan, Kim, et al. 2020). One current explanation for such prolonged postreproductive survival is late life kin selection of grandmothers and other elderly caregivers of helpless young—apparently contrary to the concept of “antagonistic pleiotropy,” which posits that natural selection does not operate in late life (Williams 1957; Byars and Voskarides 2020). In this regard, we previously observed several commonly occurring, derived gene variants that directly or indirectly impact postreproductive, late life cognition, and are present only in human genomes and not in those of any other “great apes” (Schwarz et al. 2016). This was considered as genomic evidence for the evolution of human postmenopausal longevity (Hawkes 2016). Here, we further explore the human-specific, derived alleles of genes that protect against late life cognitive decline, and ask when and how these emerged in hominins?
One such interesting example is a human-specific, derived allele of CD33 associated with direct or indirect protection against late-onset Alzheimer’s Disease (LOAD) (Schwarz et al. 2016; Zhao 2019). In vertebrates, glycan-binding proteins of the immunoglobulin (Ig) superfamily called sialic acid (Sia)-binding Ig-like lectins (Siglecs) form a major component of the immune system (Läubli and Varki 2020). As the name indicates, Siglecs recognize Sias on cell surface or secreted glycoproteins and glycolipids. Siglec-3, commonly known as CD33, is the eponymous member of the rapidly evolving subgroup of Siglecs called CD33-related Siglecs (or CD33rSiglecs) (Freeman et al. 1995; Siddiqui et al. 2017). In contrast, other Siglecs (Siglecs 1, 2, 4, and 15) show evolutionary conservation (Bornhöfft et al. 2018). CD33 is a type-I transmembrane protein with an amino terminal Ig-like V-set domain followed by one Ig-like C2-set domain proximal to the transmembrane region (Freeman et al. 1995). Its cytoplasmic tail contains immunoregulatory signaling motifs called immunoreceptor tyrosine-based inhibitory motif (ITIM)s, which upon ligand binding to the extracellular V-set domain, undergo phosphorylation and recruit effector molecules like tyrosine phosphatases, SHP-1/2, which inhibit the cellular immune responses. Human CD33 (huCD33) binds α2-3- and α2-6-linked N-acetylneuraminic acid (Neu5Ac), the predominant Sia in humans, associated either with N- and O-glycosylated molecules or sialylated glycolipids (gangliosides). More recently, a CD33 ligand composed of sialylated keratan sulfate chains carried on a minor isoform/glycoform of RPTPζ (phosphacan) was found in the extracellular milieu of the human brain parenchyma (Gonzalez-Gil et al. 2022).
HuCD33 undergoes alternative splicing, resulting in two isoforms—full-length CD33M containing the ligand-binding V-set domain and truncated D2-CD33 (or CD33m) lacking this domain (Hernandez-Caselles et al. 2006). The elimination of the terminal V-set domain is mediated through differential splicing affected by two coinherited single nucleotide polymorphisms (SNPs) at positions rs3865444 in huCD33 promoter and rs12459419 located within exon 2 (Malik et al. 2013). The two isoforms, CD33M and D2-CD33, differ not only in their molecular weights, but also in their cellular localization and functionality which are associated with Sia-interacting V-set domain (Schwarz et al. 2016; Siddiqui et al. 2017; Saha et al. 2019).
HuCD33 is extensively studied for its role in different immune responses, under both normal and pathophysiological conditions including cancers (Lamba et al. 2009; Hollingworth et al. 2011; Naj et al. 2011; Bradshaw et al. 2013; Malik et al. 2013). Furthermore, the microglial expression of CD33 is linked with neurological pathologies like LOAD. Incidence of LOAD has been strongly associated with varied expression of CD33 isoforms in the brain of affected individuals (Hollingworth et al. 2011; Naj et al. 2011), where the LOAD-protective CD33 allele increases the ratio of D2–CD33 isoform relative to CD33M. CD33 is reported in almost all vertebrates, including nonhuman primates (Bornhöfft et al. 2018; Khan, de Manuel, et al. 2020). Although there is often high similarity in the sequence and overall genomic location, CD33 has undergone various species-specific changes. For example, murine CD33 which shows about 54% identity with huCD33 V-set and 72% identity with C2 domain, has markedly different Sia-binding and cellular expression patterns from huCD33 protein (Brinkman-Van der Linden et al. 2003). CD33 expression has greatly diverged in humans even in comparison to our closest living evolutionary relatives, the great apes. Examination of CD33 in peripheral blood showed significantly increased production of CD33M in human monocytes relative to those of chimpanzees (Schwarz et al. 2016). Furthermore, the abundance of CD33 was also markedly higher in the human brain. Interestingly, although LOAD-associated neurological pathologies, for example, buildup of Aβ proteins, hyperphosphorylated tau proteins as neurofibrillary tangles, have been observed in aged nonhuman primate brains, AD has largely been regarded as a uniquely human disease (Edler et al. 2017, 2020). Interspecies variations in CD33 have also been studied in other apes like gorilla and bonobo, in comparison to huCD33 (Padler-Karavani et al. 2014).
The presence of two physiologically significant isoforms, their distinct cellular localization, and association with uniquely human pathologies like LOAD have made huCD33 a target of much evolutionary interest. The Sia-binding V-set domain of CD33rSiglecs including CD33 itself shows high sequence variability among different species, often making it difficult to identify their orthologs. The selective pressure for this accelerated evolution of the V-set domains has been attributed to the evasion of infectious pathogens that exploit these human innate receptors. The surfaces of each vertebrate cell are layered with tens to hundreds of million Sia-terminating glycans, forming as “self-associated molecular patterns” (SAMPs), which prevent erroneous activation of innate immune responses against the body’s own cells (Varki 2011). However, several human pathogens, for example, Neisseria gonorrhoeae, N. meningitidis, Haemophilus influenzae, Escherichia coli K1, Group B Streptococcus (GBS), and Trypanosoma cruzi cloak themselves with sialoglycans, effectively mimicking host SAMPs, and thereby avoiding the immune response (Varki and Gagneux 2012). Conversely, other infectious agents like influenza virus recognize SAMPs and utilize them as receptors to initiate binding and subsequent infections (Tortorici et al. 2019). CD33 has also been shown to interact with Hepatitis B viral surface sialoglycans, thereby impacting its pathogenesis (Tsai et al. 2021). SAMPs and their interacting partners, Siglecs (primarily the V-set domains) are therefore continually evolving to maintain their distinctive “self-recognition” properties, whereas also avoiding exploitation by Sia-cloaked pathogens and parasites—an example of the “Red Queen Effect” (Varki and Angata 2006).
In this work, with a focus on CD33, a postreproductive cognitive health-associated human protein, we attempt to explore the evolutionary pressures that selected for unique changes in huCD33. Using human-specific pathogens like N. gonorrhoeae, GBS, and E. coli K1, we demonstrate the differential impact of these mutations on the bacterial interactions with huCD33. We also determine the effect of these mutations on huCD33–sialoglycan binding and identify that the amino acid at position 21 within the V-set domain plays a critical role in Sia-specificity of human and chimpanzee CD33. Last but not least, we extend our study to archaic hominin genomes and show that the human-specific CD33 mutations (except the presence of truncated isoform) are shared evolutionary changes of human, Neanderthal, and Denisovan common ancestor. We also expanded our analysis to include other human-specific derived genomic changes associated with the cognitive health of postreproductive human grandmothers and other elderly caregivers. Finally, we draw an evolutionary scenario to connect the current knowledge of CD33 sialoglycan recognition and pathogen engagement to propose a role for the infectious pathogens as key selective agents in human-specific CD33 evolution, generating new alleles protective against infections, that could secondarily have come under selection for their protective effects against cognitive pathologies like LOAD.
Results
Sequences of huCD33 Extracellular Domains Show Many Changes Distinct from Closely Related Great Apes
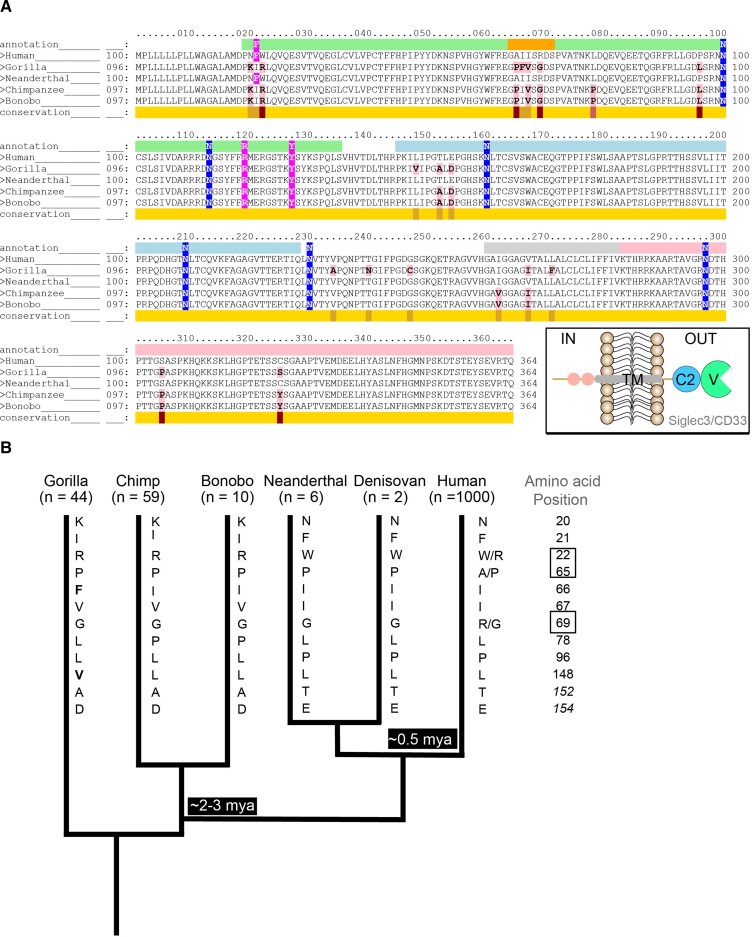
Previous investigations have identified unique properties of huCD33 that influence the functionality in humans. The presence of a huCD33 V-set truncated isoform as well as its overall expression difference in microglia has been associated with the protection against the occurrence of neurological pathologies like LOAD in humans. Like other CD33rSiglecs, CD33 immunomodulatory roles depend both on its ligand-interacting extracellular domains and signaling motif-containing cytoplasmic tail. To gain a comprehensive understanding of different CD33 domain variations, we compared the amino acid residues of full-length CD33 from human and related nonhuman primates including chimpanzee, gorilla, and bonobo (fig. 1A). Although the regions encoding the C2-set domain and cytoplasmic tail are highly conserved, the amino acid residues within huCD33 V-set domain differ from their nonhuman counterparts in as many as ten positions. Since different amino acid residues in Sia-binding V-set domain could potentially impact huCD33–sialoglycan interactions and subsequent downstream signaling pathways, we further examined the overall frequency of these changes (fig. 1B). We analyzed human sequences from the 1000 Genome database (Consortium et al. 2015) and compared them with 44 gorilla, 59 chimpanzee, and 10 bonobo sequences (Prado-Martinez et al. 2013; Xue et al. 2015; de Manuel et al. 2016). Most of these amino acid residues (except at positions 66 and 148) are conserved in all the great apes and appeared to have changed only in the human lineage. Interestingly, the amino acid residues at positions 66 and 148 in huCD33 are isoleucine and leucine respectively, similar to that of chimpanzee and bonobo. The corresponding amino acids in its more distant evolutionary relative, gorilla, are phenylalanine (Phe) (at position 66) and valine (at position 148). The presence of the same amino acid in human, chimpanzee, and bonobo at these positions suggests a more ancient occurrence of these two changes, possibly before the divergence of chimpanzee about 6–8 Ma. In contrast, it has previously been shown that the two linked SNPs, resulting in the splicing of the V-set truncated isoform represent a derived evolutionary modification of the CD33 proteins in humans and are absent in chimpanzees (Schwarz et al. 2016).
Fig. 1.
Human-specific changes in CD33 are primarily present in the Sia-binding V-set domain. (A) Comparison of amino acid sequences of CD33 from humans and “great apes” was performed using Conformational Analysis Tools software. The Great ape genomes included in the analysis are gorilla, chimpanzee, and bonobo and were compared against the human protein as the template. The conservation of the sequence is indicated with yellow being the most and red being the least conserved regions. Amino acids that are different from huCD33 are highlighted in pink. Amino acids encoding the different CD33 domains are indicated above the sequence with different colors corresponding to schematic in inset, namely, V-set domain in green, C2 domain in light blue, transmembrane domain in gray, and cytoplasmic end in light pink. The flexible C–C′ loop is indicated in orange. Amino acids that are in contact with Neu5Ac in huCD33 are highlighted in magenta and N-glycosylation sites in blue. (B) Phylogenetic analysis of the evolution of the extracellular domains of CD33 proteins from human, Great apes, and archaic genomes. The number of genomes (n) for each group included in the analysis is indicated. Human and great ape CD33 sequences were compared with six Neanderthal and two Denisovan genomes. Amino acid changes present in huCD33 were also present in the ancient genomes. The positions of the amino acids that are different between human and the apes are mentioned, and the identity of the amino acid present in the corresponding positions for each group is indicated by the single letter abbreviations along the branch. Amino acids at positions 152 and 154 are within the C2 domain of CD33 protein and italicized. Polymorphisms within the human population at positions 22, 65, and 69 of CD33 protein are indicated. Amino acids in gorilla CD33 at positions 66 and 148 are different from other apes and are bold. Possible timeline for the diversion of the hominin lineage is indicated in the tree. Length of the branches in the tree is not to scale.
To further understand the selection pressure, we calculated the nonsynonymous to synonymous substitution rate ratio (omega, ω = d[N]/d[S]) for the CD33 V-set domains of human and other great apes. The omega value of CD33 V-set domain is greater than >1 (ω = 1.49), which reflects V-set domain evolution under positive selection. Subsequently, we also analyzed the Ka/Ks ratios of exon 2 sequences in every species. Except for gorilla, the other two great apes (chimpanzee and bonobo) showed Ka/Ks ratios greater than one indicating that high Ka/Ks ratio of exon 2 is not an accidental event but an evolutionary phenomenon. Taken together, these results demonstrate that CD33 in humans has been rapidly evolving under positive selection, distinct from its orthologs in the great apes.
Archaic Neanderthal and Denisovan Genomes Share Most huCD33 Protein Changes, Except for the SNPs for the LOAD-Protective Allele
Divergence of humans from other ancient hominin lineage such as Neanderthals and Denisovans has been estimated to date back ∼0.5 Ma (Green et al. 2010). Although full-length CD33 itself is an ancient molecule, we noted that the AD-protective CD33 truncated isoform is recently derived in humans, postdating our divergence from Neanderthals and Denisovans (Schwarz et al. 2016). Since huCD33 extracellular domains showed high accumulation of changes compared with the great apes, we wanted to determine if these changes were present in the common ancestor of the hominin lineage. We, therefore, compared CD33 protein sequences from six Neanderthal and two Denisovan archaic genomes obtained from the Max Planck Institute for Evolutionary Anthropology (Prufer et al. 2014) (http://cdna.eva.mpg.de) with the corresponding human sequences of the 1000 Genome database (fig. 1B). Interestingly, all the amino acid residues in huCD33 that are different from the great apes are present in the ancient genomes, suggesting their occurrence in a common ancestor. These observations thus suggest that the complete loss of Sia-binding V-set domain is the latest evolutionary modification of huCD33, likely succeeding the individual amino acid changes within its extracellular domain.
A Single Amino Acid Change Facilitated CD33 Engagement to the Uniquely Human Pathogen Neisseria gonorrhoeae
In addition to microglial expression in the brain, CD33 is also present on tissue macrophages and peripheral blood monocytes (Schwarz et al. 2016). These cells are important components of innate immune responses throughout the body, including the reproductive tract. The human female reproductive tract is also a unique niche for the microbiome, which can be invaded by important pathogens like N. gonorrhoeae (Ng). Ng is a uniquely human infectious agent, responsible for the second most prevalent, sexually transmitted infection causative for the disease gonorrhea in human populations. Gonorrhea affects both males and females and if untreated, can have detrimental effects on reproductive health (Edwards and Apicella 2004). We have previously shown that Ng interacts with huCD33 but not the chimpanzee ortholog (Landig et al. 2019). The bacterium is incapable of endogenous Neu5Ac synthesis, but instead scavenges the molecule from its host (Parsons et al. 1988; Apicella et al. 1990). Once inside the female reproductive tract, Ng utilizes the host sugar nucleotide CMP-Neu5Ac from its microenvironment to transfer Neu5Ac onto its own bacterial lipooligosaccharide. Sialylated Ng then successfully interacts with several human Siglecs including 3 (CD33), 5, 9, 11, 14, and 16 (Landig et al. 2019). However, unlike other Siglec interactions, Ng binding to CD33 appears to be entirely Sia dependent. Interestingly, of all the Neisseria species currently known, only Ng and N. meningitidis are pathogenic to humans and both are thought to be evolutionarily young compared with others (Caugant and Brynildsrud 2020). Since reproductive health/success of an organism is the key determinant of Darwinian fitness, we hypothesized that highly infectious disease like gonorrhea could potentially impact the evolution of humans, mediated through binding immune-modulating proteins like CD33.
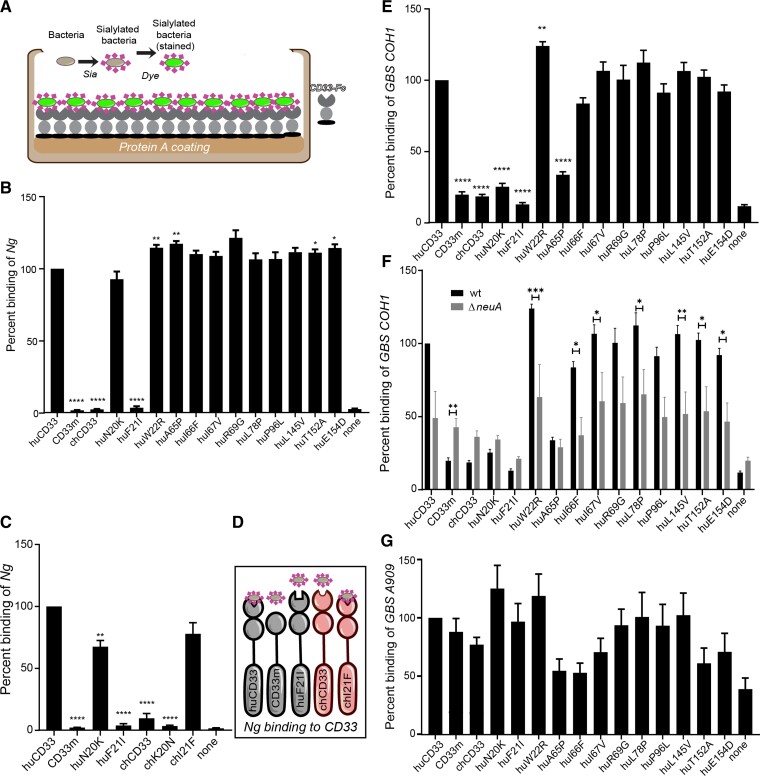
To explore our hypothesis, we examined the binding of sialylated Ng to different recombinant CD33 protein mutants, each containing the two extracellular domains with an amino acid residue changed from human to chimpanzee at the corresponding positions identified in figure 1B. Fluorescently labeled Ng was allowed to interact with human recombinant Fc-chimeric constructs of the CD33 proteins that were immobilized onto protein A-coated plates (fig. 2A and B). Sia-dependence of the interaction was confirmed by comparing binding with bacteria grown in the presence and absence of CMP-Neu5Ac (supplementary fig. S1A, Supplementary Material online). We observed a significant reduction in bacterial binding to chimpanzee CD33 (chCD33) compared with human protein containing both V- and C2-domains. However, in the absence of the V-set domain in the truncated form of huCD33 (CD33m), bacterial binding was lost. Except for the residue at position 21, all the other amino acid alterations from human to chimpanzee CD33 maintained high bacterial binding. In fact, changing the amino acid residues at positions 22, 65 (of the V-set domain), 152, and 154 (of C2 domain) increased the binding significantly compared with wildtype huCD33. In contrast, mutating the amino acid at position 21 from human to chimpanzee residue completely abolished huCD33 binding of sialylated Ng. Interestingly, mutating the chimpanzee CD33 amino acid at position 21 to its corresponding human residue enabled Ng to now engage chimpanzee CD33 (fig. 2C and D). Considering that Ng and its closest relative meningococcus are both uniquely human pathogens thought to have evolved from commensal Neisseria (Seifert 2019), our data suggest important implications of CD33 amino acid change at position 21 on Ng–huCD33 interaction and their mutual evolution.
Fig. 2.
Human-specific amino acid changes in CD33 affects bacterial binding. (A) Schematic of the ELISA-based assay using recombinant CD33-Fc chimeric proteins immobilized on protein A coated plates used to determine binding of the sialylated bacteria is shown. (B) Binding of fluorescently labeled Neisseria gonorrhoeae (Ng) was determined. The position of the amino acid different from the wildtype huCD33 protein is indicated below each bar in the x-axis. The bacterial binding to each individual CD33 mutant was normalized to the binding of wildtype huCD33 for that assay. “None” indicates no protein control for the background bacterial binding to the plate. (C) Binding of Ng to immobilized recombinant CD33 proteins containing the corresponding amino acid mutation (position 20 or 21) either in human or chimp CD33 protein backbone. (D) Pictorial summary of Ng binding to CD33 wild type and mutant proteins. (E) Binding of GBS COH1 strain to different CD33 mutant proteins in an ELISA-based assay with immobilized recombinant CD33 proteins. (F) Sialic acid dependence of the binding was determined using wildtype and ΔneuA mutant strains of COH1. (G) Interaction of CD33 proteins among different GBS strains was compared using A909 and COH1 strains. ‘hu’ indicates the corresponding amino acid change in huCD33 backbone and ‘ch’ using chimp CD33. The graphs show the cumulative result from three independent experiments, each done in triplicate. Statistical analysis was performed in Prism software using one-way ANOVA with Durrett post-comparison test. *<0.01, **<0.001, ***<0.0001.
Many Amino Acid Changes in CD33 Extracellular Domains Impact GBS Engagement
Although the association of Neisseria with CD33 is a case of Sia-mediated interaction, there are other examples of human pathogens that engage Siglecs in Sia-independent manner. One such example is GBS, which has been widely studied for its various Siglec interactions (Carlin et al. 2007). GBS is an encapsulated pathogen commonly associated with pneumonia, sepsis, and meningitis in infants and neonates. It comprises nine serologically distinct groups (Ia, Ib, and II–VIII), differing in their capsular sialoglycan structures, but all containing α2-3-linked terminal Neu5Ac. Certain GBS strains have been shown to bind human Siglecs 5 and 7 in a Sia-independent manner through cell wall anchored β-protein (Fong et al. 2018), whereas some Sia-dependent binding was observed for CD33 and Siglec-9. Human Siglec-9 binding is also thought to be partially Sia independent. Interestingly, some GBS strains are also known to interact with nonhuman primate Siglecs, for example, Siglec-9 from chimpanzee (Padler-Karavani et al. 2014). Since infections by GBS mostly impact newborns and infants, we hypothesized that it could also play a role in overall Siglec evolution in humans. Similar to the Ng-CD33 binding assay (as in fig. 2A), we examined the interactions between the recombinant CD33 proteins and GBS group III strain, COHI (fig. 2E). Although the bacteria bound strongly with full-length extracellular domains of huCD33, the binding was significantly reduced in the truncated human isoform (CD33m) and the chimpanzee protein. Like Ng, GBS COHI interaction was also markedly disrupted by amino acid changes at position 21. Additionally, changing the residues at positions 20 and 65 from human to chimpanzee significantly reduced the bacterial interaction with CD33. However, GBS COHI engagement with the CD33 mutants was not entirely Sia dependent (fig. 2F). Using GBS COHIΔneuA, a mutant strain lacking its sialyltransferase enzyme (NeuA) and hence incapable of surface sialylation, we observed that about 50% of the bacterial binding to CD33 could be attributed to Sia-independent interactions. Interestingly, the CD33 binding profile of COHI was not uniform for the other serogroups of GBS, for example, GBS group Ia strain, A909 (fig. 2G). None of the amino acid changes showed significant effects on CD33 interaction with GBS A909, relative to the wildtype human protein. Even the truncated huCD33 isoform (CD33m) displayed similar binding suggesting that the CD33 binding for A909 is primarily Sia independent. Unlike Ng and GBS, we did not observe any differential sialoglycan binding with E. coli K1, another uniquely human pathogen of newborn infants, which contains Sia polymers on its surface (supplementary fig. S1B, Supplementary Material online). Altogether, the data demonstrate the diverse nature of CD33-interactions in just three major pathogens and suggest an impact of uniquely human pathogens in the evolution of CD33 ligand-binding domain.
Ancestral Sialoglycan Preference of CD33 is Disrupted by Amino Acid Change at Position 21
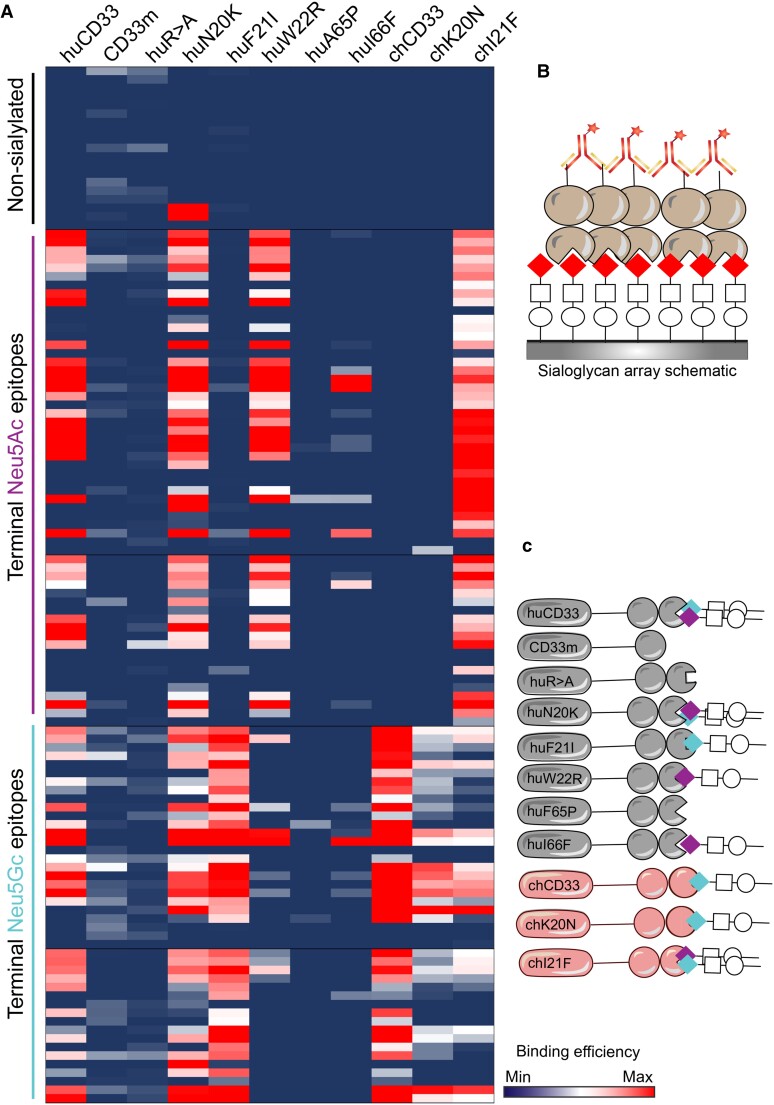
A key change in the evolution of humans was the loss of CMP-Neu5Ac hydroxylase (CMAH), the enzyme that converts CMP-Neu5Ac to CMP-Neu5Gc resulting in a primarily Neu5Ac-rich sialome in humans, unlike any other Old-World primates, which express both Neu5Ac and Neu5Gc. This change is dated to ∼2–3 Ma when human ancestors were evolving from ancestral hominins. Since we observed numerous changes mainly in huCD33 V-set domain, which is critical in sialoglycan interaction and therefore important for its downstream signaling pathways, we wanted to specifically understand the effect on CD33 sialoglycan interactions. We used a microarray of chemoenzymatically synthesized glycans with defined structures, terminally capped with either Neu5Ac or Neu5Gc in different glycosidic linkages (fig. 3A and B) and examined their relative interactions with recombinant, soluble CD33 proteins (fig. 3). HuCD33 with V- and C2- domains bound to both Neu5Ac and Neu5Gc-terminating sialoglycans and showed maximum binding when the Sia was α2-6-linked to an underlying lactose or lactosamine glycan (supplementary fig. S2, Supplementary Material online). Most of this binding was lost in the truncated huCD33 lacking the Sia-binding V-set domain, indicating that the interactions are Sia dependent. Conversely, the chimpanzee protein (which is identical to the bonobo orthologs and differs by only two amino acids from the gorilla) demonstrated a strong preference toward Neu5Gc-terminating sialoglycans and showed almost no binding for Neu5Ac-epitopes (fig. 3A and 3C). Considering the varied sialoglycan profiles of the two organisms, these distinct binding preferences of human and chimpanzee CD33 are interesting and suggest functional implications of the evolutionary changes in their extracellular domains.
Fig. 3.
Single amino acid changes affect CD33 sialoglycan binding. (A) Sialoglycan-binding profile of purified, soluble, recombinant CD33 proteins was determined using a sialoglycan microarray containing defined, chemically synthesized glycans (experiment schematic shown in B). Nonsialylated, Neu5Ac- (indicated in purple on the left) and Neu5Gc- (indicated in blue) terminating glycans were grouped together in the heatmap as shown in the left. Each column indicates the binding profile of the protein indicated on the top and each row represent a distinct glycan. Blue indicates no binding and red indicates very strong binding preferences characterized by an average relative fluorescence unit (RFU) of more than 90th percentile. The result of the heatmap is summarized in the cartoon (C) with the colored diamonds corresponding to the termination Sia and supplementary figure S2, Supplementary Material online. The names of the individual glycans are presented in supplementary file S1, Supplementary Material online.
Because the impact of amino acid residue at position 21 was most pronounced in both of our bacterial-CD33 binding assays (fig. 2B and D), we next examined the influence of this change on CD33–sialoglycan binding (fig. 3 and supplementary fig. S2, Supplementary Material online). Indeed, changing the amino acid at position 21 completely altered Sia-epitope preference of CD33 for both human and chimpanzee. The presence of human amino acid residue at position 21 enabled strong binding of Neu5Ac-epitopes by chCD33, unlike its entirely Neu5Gc-preferring wildtype counterpart (fig. 3A and C). On the other hand, the chimpanzee amino acid at the same position in huCD33 abolished its Neu5Ac binding. To determine if the Sia-binding changes are specific for position 21 and not an arbitrary effect of any amino acid change in V-set domain, we also looked at the Sia-epitopes of position 20 amino acid substitutions. Unlike position 21, amino acid modifications at position 20 did not have any major impact on the Sia-binding of CD33, which maintained the overall wildtype profile. Interestingly, modifications at position 22 demonstrated Neu5Ac-prefered binding for huCD33, whereas chimpanzee amino acid residues at 65 and 66 of huCD33 almost abolished any sialoglycan binding. Altogether, the data emphasized the importance of different amino acid changes in huCD33 V-set domain in its sialoglycan binding and identified the amino acid at position 21 to be critical in the functionality of CD33 protein.
Molecular Dynamics Simulations Provide Structural Insights for the Differences in Sia-Binding Preference Between Human and Chimpanzee CD33
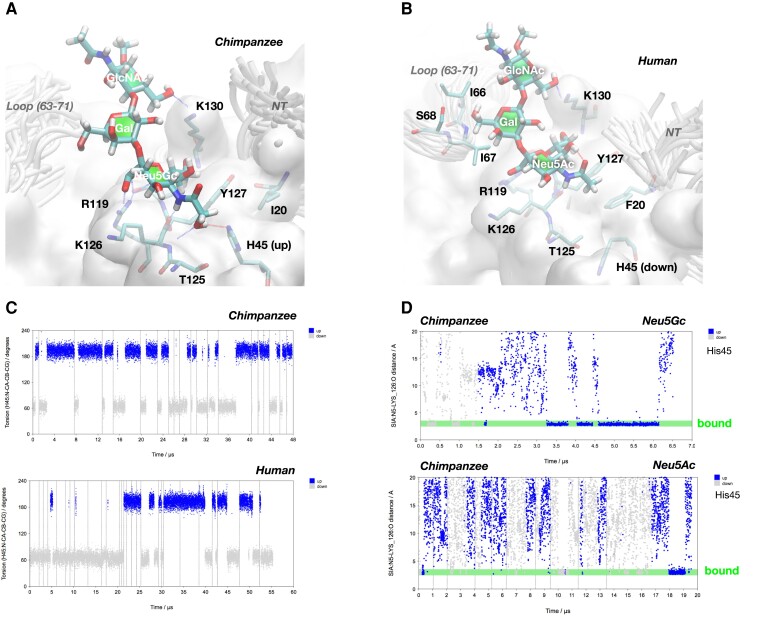
We performed an extensive theoretical investigation based on molecular dynamics (MD) simulations. A detailed analysis of several available crystal structures of huCD33 revealed that the V-set domain is dynamic. For example, the C–C′ loop as well as the side chains of phenylalanine at position 21 (Phe21) and histidine at position 45 (His45) are resolved in two different conformations in PDB entry 5ihb (supplementary fig. S3, Supplementary Material online). Of all the amino acids that differ between human and chimpanzee, only the side chain of Phe21 is in direct contact with a bound Neu5Ac residue in the crystal structures of huCD33 (through the methyl group at position 5). Based on the assumption that Neu5Gc binds to the same binding site as Neu5Ac, the change in binding preference from Neu5Gc (in chimpanzee) to Neu5Ac (in human) cannot be explained by a simple I21F mutation. Both amino acids have hydrophobic side chains that cannot establish favorable interactions with the polar glycolyl group. Consequently, there is probably a more complex reason for the shift of binding preference. Based on data derived from 47 MD simulations covering an accumulated timescale of more than 100 µs, we conclude that in chCD33 His45 adopts mainly the “up” conformation (compare fig. 4A), which allows favorable hydrogen bonding with the glycolyl group. MD simulations (as well as X-ray crystallography) show that in huCD33 His45 can also exist in the “up” conformation (compare figs. 4C and supplementary fig. S4, Supplementary Material online), which would be compatible with favorable Neu5Gc binding. However, when His45 is in the “down” conformation Phe21 can stack partly with tyrosine (Tyr) at position 127 (fig. 4B) forming a small hydrophobic pocket, which allows the methyl group of Neu5Ac to bind favorably. To demonstrate if the binding affinity difference between Neu5Ac and Neu5Gc may be indeed correlated to the up/down conformational equilibrium of His45, we performed a series of MD simulations of chCD33 on the microsecond timescale where Neu5AcOMe or Neu5GcOMe molecules are present in the solution. The lifetimes of the complexes spontaneously formed during the MD with Neu5Gc are on average much longer when His45 is “up” (fig. 4D top, supplementary fig. S4, Supplementary Material online). In contrast, the lifetimes of the complexes spontaneously formed with Neu5Ac are much shorter independent of the conformational state of His45 (fig. 4D bottom), which would explain the lack of measurable binding affinity of Neu5Ac to chCD33. In summary, our extensive MD simulations—including unbiased simulation of carbohydrate binding and unbinding events—could provide a reasonable explanation for a change in binding specificity that is likely to be caused by an alteration of the protein–ligand interaction pattern remote from the mutated amino acid.
Fig. 4.
Structural modeling to understand the differential binding preference of human and chimpanzee CD33 proteins. (A) 3D model of the complex between Neu5Gcα2–3Galβ1–4GlcNAcβOMe and chCD33. The increased affinity of Neu5Gc may be explained by intermolecular hydrogen bonds involving the OH-group of Gc. It should be noted that the number of favorable interactions is maximal when His45 is in “up” conformation. (B) 3D model of the complex between Neu5Acα2–3Galβ1–4GlcNAcβOMe and huCD33. The methyl group of Ac is located in a small hydrophobic pocket formed by the side chains of Tyr127 and Phe20. It should be noted that His45 is in “down” conformation because otherwise—in the conformation shown—the bulky side chain of Phe20 would overlap partly with His45 in “up” conformation. (C) MD of His45 side chain orientation. Accumulated MD trajectories of torsion angle N–Cα–Cβ–Cγ are shown. The “up” conformation is present when torsion values are fluctuating around 200° and the “down” conformation is characterized by values around 70°. For chimpanzee, it can be observed reproducibly that simulations started with His45 in “down” conformation undergo a transition to the “up” conformation on the microsecond timescale. In contrast the “down” conformation appears to be more stable in huCD33, which would make Neu5Ac binding more likely. (D) MD simulation of unbiased binding and unbinding events of Neu5Gc (top) and Neu5Ac (bottom) to chCD33. For Neu5Gc the lifetime of the complex is significantly longer when His45 is in “up” conformation, as can be seen from the 6.5 µs MD simulation shown on the top. Also, for Neu5Ac multiple binding and unbinding events occurred on a timescale of about 20 µs, however, in general (with one exception), the lifetimes of the complexes formed are significantly shorter than for Neu5Gc.
Human-Specific Polymorphisms in Cognitive Health-Related Genomic Variants are Present in All Human Populations
In an earlier study, we observed several genes, directly associated with neurodegenerative diseases or correlated with aggravation of the cognitive decline in aged-population, are derived alleles in humans (Schwarz et al. 2016). Increasing evidence of correlation between cognitive health and nonneurological, metabolic conditions, for example, diabetes (Peila et al. 2002, Munshi 2017) suggest that such derived alleles could be important in the maintenance of cognitive health in human grandparents. Here, we expanded this list of cognition-protective gene variants through the literature and database (https://alzoforum.org) searches (Finch and Sapolsky 1999; Altshuler et al. 2000; Nakajima et al. 2004; Thompson et al. 2004; Vander Molen et al. 2005; Helgason et al. 2007; Wen et al. 2007; Carrasquillo et al. 2011; Naj et al. 2011; Raj et al. 2014; Xu et al. 2015; Liu et al. 2018; Rathore et al. 2018) to include additional gene variants, namely BINI, ARID5B, PICALM, PILRA. Supplementary table S1, Supplementary Material online describes the characteristics of 13 human genes that are implicated in diseases including dementia, cardiovascular diseases, hypertension, and AD. Although some of these physiological abnormalities like salt retention, hypertension, diabetes, appear nonneurological, they have been associated with the aggravation of the pathologies resulting in late life cognitive decline (Iadecola 2013). Notably, the derived alleles are common and found in globally diverse human populations, indicating that they predate the common ancestor of modern humans (supplementary table S2, Supplementary Material online).
SNPs Associated with Human-Specific Cognitive-Protective Alleles are Unusual in their Absence in the Archaic Hominin Genomes
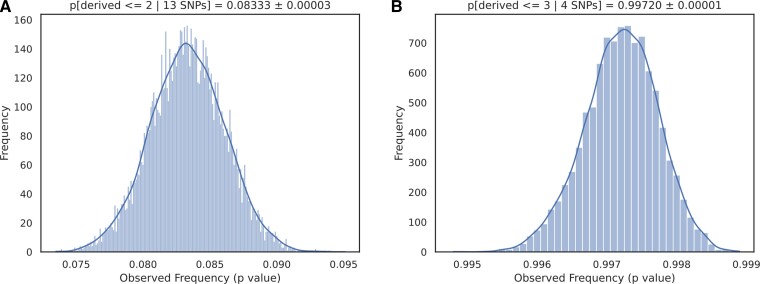
With the availability of genomes from extinct archaic hominins (Reich et al. 2010; Meyer et al. 2012; Prufer et al. 2014), a set of SNPs can be assessed as to whether their protective phenotypes arose recently in the evolutionary history of anatomically modern humans. We previously showed that many other human–chimpanzee differences were shared with archaic hominins (Denisovan/Neanderthal) genomes; for example, genomic changes in CD33rSiglecs (Khan, de Manuel, et al. 2020; Khan, Kim, et al. 2020). To gain similar insights about the evolutionary origin of these cognitive-protective loci, we analyzed the Neanderthal and Denisovan reference genomes and compared them with modern human sequences. Analysis of the 1000 Genomes dataset shows the presence of protective alleles in human populations with variable frequency (table 1). Analysis of the available genomic data from Neanderthal and Denisovan genomes showed that only two derived variants (rs2975760 and rs2588969; table 1) are present in these archaic genomes, suggesting the remaining 11 derived, protective variants arose after the divergence of modern and archaic hominins ∼0.5 Ma (Green et al. 2010; Mendez et al. 2016). This is in striking contrast to most human–chimpanzee genomic differences in which the archaic hominins are similar to humans. In fact, the majority of the Sia-related genes lack positive selection signatures and rather show neutral evolution in the modern human lineage (Moon et al. 2018). To more formally assess whether the high frequency, global distribution, and recent origin observed for 11 of the 13 SNPs is unusual, we performed a resampling analysis of variants in the genome. Variants in the 1000 genomes dataset that met the following criteria were considered: (1) present in both Altai Neanderthal and Denisovan minimal filters, (2) derived in at least one modern individual from nonadmixed African populations, (3) called in both archaic samples, and (4) have an ancestral allele matching the reference or alternative allele. To eliminate any bias in the analysis and match the allele frequency (AF) of these SNPs compared with that of any random SNPs, we first matched our universe of SNPs to the 13 SNPs of interest by AF, ± 2 derived haplotypes (fig. 5). Resampling was then performed by drawing a SNP from each of the 13 matched sets and assessing how many derived alleles were observed, resulting in a P-value = 0.08333 ± 0.00003. As a less conservative estimate, directly sampling from the universe of SNPs and estimating the probability of observing at most two derived SNPs and a mean AF as large as the empirical variants of interest produced a highly significant P-value = 0.00487 (supplementary fig. S5, Supplementary Material online). Repeating either analysis on the set of other SIGLEC-related SNPs indicates that they are consistent with a random draw from the genome (Khan, de Manuel, et al. 2020). Regardless of the individual limitations, taken together our phylogenetic analyses demonstrate the unique patterns of allele frequencies in worldwide populations distribution of these 13 late life cognitive decline-linked SNPs (fig. 5). Interestingly, coinherited CD33 SNPs associated with the cognitive health in LOAD are present only in modern human genomes (Schwarz et al. 2016). A noteworthy example in our list is the human gene encoding the protein, apolipoprotein E (APOE), involved in fat metabolism in mammals. APOE gene exists in three allelic variants (E2, E3, and E4) where APOE4 is associated with high risk of LOAD and other allele like APOE2 is protective against the cognitive decline in elderly caregivers (Reiman et al. 2020). Interestingly the presence of APOE4 is also correlated with the protection from severe diarrhea in children (Oriá et al. 2005). Although conclusive determination of the positive selection of these alleles in modern human requires further analysis, our data suggest that the evolutionary origin of most of these cognitive health-protective changes followed after the divergence of modern humans from archaic genomes. This is also supported by the presence of grandparents, uniquely in humans. Regardless, the process of evolutionary emergence of each of these alleles is likely to be distinct and deserves further investigation.
Table 1.
Gene Variants Directly or Indirectly Affecting Cognitive Function.
| Gene | Associated Diseasea | SNP ID | hg19 Position | Allele | Derived Global AFb | Archaic Genotype | |
|---|---|---|---|---|---|---|---|
| Altai | Denisovan | ||||||
| AGT | Sodium retention | rs699 | 1:230845794 | G | 0.295 | 0/0 | 0/0 |
| A | |||||||
| BIN1 | AD | rs7561528 | 2:127889637 | A | 0.8 | 0/0 | 0/0 |
| G | |||||||
| SGC2 | Hypertension | rs1017448 | 2:224466344 | T | 0.879 | 0/0 | 0/0 |
| C | |||||||
| CAPN10 | Type II Diabetes | rs2975760 | 2:241531163 | C | 0.882 | 1/1 | 1/1 |
| T | |||||||
| PPARG | Type II Diabetes | rs1801282 | 3:12393125 | C | 0.070 | 0/0 | 0/0 |
| G | |||||||
| CYP3A5 | Sodium retention | rs776746 | 7:99270539 | T | 0.621 | 0/0 | 0/0 |
| C | |||||||
| ARID5B | AD | rs2588969 | 10:63611354 | A | 0.532 | 0/0 | 1/1 |
| C | |||||||
| SPON1 | Dementia | rs2618516 | 11:14021639 | C | 0.341 | 0/0 | 0/0 |
| T | |||||||
| PICALM | AD | rs10792832 | 11:85867875 | G | 0.313 | 0/0 | 0/0 |
| A | |||||||
| rs3851179 | 11:85868640 | C | 0.315 | 0/0 | 0/0 | ||
| T | |||||||
| APOE | LOAD | rs429358 | 19:45411941 | C | 0.849 | 0/0 | 0/0 |
| T | |||||||
| rs7412 | 19:45412079 | C | 0.075 | 0/0 | 0/0 | ||
| T | |||||||
| CD33 | LOAD | rs3865444 | 19:51727962 | C | 0.211 | 0/0 | 0/0 |
| A | |||||||
| PILRA | AD | rs1859788 | 7:99971834 | G | 0.341 | − | − |
| A | |||||||
| TCFLC2 | Type II Diabetes | rs7903146 | 10:114758349 | T | 0.772 | − | − |
| C | |||||||
| CD33 | LOAD | rs12459419 | 19:51728477 | C | 0.211 | − | − |
| T | |||||||
Note.—Allele has the derived allele as the lower, bolded entry. Archaic genotypes are reported for SNPs passing all quality filters.
See supplementary table 1, Supplementary Material online for details and the primary literature citations.
See supplementary table 2, Supplementary Material online for the global population distribution.
Fig. 5.
Resampling analysis with matched AF SNPs from 1000 genomes variants. Frequency distribution of SNPs with similar properties to LOAD-protective set (A) and other SIGLEC SNPs (B).
Discussion
Fossil evidence and genomic comparisons leave little doubt about the fact that our species evolved from an African hominin. However, the detailed understanding of modern human origins is plagued by numerous uncertainties, with regard to the identity of the ancestral lineage and precise geographic locations. The evolution of modern humans was accompanied by many anatomical and behavioral changes, but increasing evidence suggests it also included uniquely human-derived modifications in the genome compared with the archaic genomes (Neanderthal/Denisovan) or the genome of the great apes (Schwarz et al. 2016; Khan, de Manuel, et al. 2020). Taken together with our previous study (Schwarz et al. 2016), we have identified many such human-specific genes associated with cognitive health of grandmothers and other human elders who are often involved in the caregiving of the young. These findings, which appear paradoxical to the concept of senescence due to antagonistic pleiotropy, have lent much additional support to the “Grandmother hypothesis” (Hawkes et al. 1998) bolstering the case for the selection of human female postreproductive survival and the existence of grandmothers. Unlike in any other mammals (except orcas and some other toothed whales), the occurrence of this prolonged postreproductive life span in humans has stirred scientific interest. Although deciphering the precise evolutionary course of any gene/protein is challenging and the proposed schemes/players are not entirely verifiable, here we attempt to compile the current evolutionary and experimental findings of one such protein associated with late life cognitive decline: CD33.
A ratio of high wildtype huCD33 and a low truncated isoform of CD33 have been implicated in the progression of LOAD associated with the cognitive health of elderly population. In contrast, LOAD is unknown in chimpanzees, although evidence of LOAD pathologies has been observed in some chimpanzee brains. We found that huCD33, which is highly expressed in microglia of the human but not chimpanzee brain, recognizes Neu5Ac—the predominant Sia synthesized in humans—as SAMPs. In contrast, our closest evolutionary relative, the apes and other old-world primates contain both Neu5Ac and Neu5Gc. We found that the ancestral form of CD33 in chimpanzees and other great apes selectively recognizes only Neu5Gc-glycans as SAMPs (fig. 3). Notably, Neu5Gc—the ligand recognized by chCD33 is rare in chimpanzee brain, and there is also significantly less chCD33 protein compared with CD33 in humans (Schwarz et al. 2016). On the other hand, SNPs resulting in the truncated CD33 have only been observed in the human genome and not any of the archaic or great ape genomes. We also find that the truncated huCD33 does not interact with Sia (fig. 3). Taken together, these observations suggest that full-length CD33–Sia interactions are stronger in human brain compared with chimpanzee and the human-specific SNPs in CD33 resulting in the truncated protein abolish this interaction.
The CD33–Sia (i.e., protein–ligand) interaction in the human brain is important, given the immunoinhibitory function of CD33 in the microglia and the overall roles of microglia in the maintenance of brain homeostasis, including the immune surveillance to clear damaged cells and amyloid plaques comprising of β-amyloid (Aβ) peptides (Colonna and Butovsky 2017). Extracellular accumulation of Aβ peptides and intracellular phosphorylation of the tau proteins are pathophysiology associated with AD brains. Microglial expression of high full-length CD33 protein and/or low truncated CD33m isoform indicates increased interactions with the Sia-ligands and hence, inhibition of the immune activation of microglia mediated by CD33 cytosolic ITIM (or ITIM-like) motifs and therefore, low phagocytosis of the damaged proteins, including amyloid plaques rich in Sia residues (Szumanska et al. 1987; Salminen and Kaarniranta 2009). This is further evident in studies with CD33 null mice where reduced expression of CD33 in the microglia and lower number of CD33 positive microglial cells are correlated with lower brain accumulation of insoluble Aβ (Griciuc et al. 2013). Similar observations were made with CD33 knockout in human microglia and macrophages (Wißfeld et al. 2021), where CD33 absence resulted in increased amyloid plaque clearance. In fact, CD33 is among one of the most common genes linked with AD in genome-wide studies (Naj et al. 2011) and is reviewed as potential candidate for therapeutic interventions against LOAD (Zhao 2019; Griciuc et al. 2020). Notably, cognitive health has also been associated with changes in the inflammatory activity in the brain (Simen et al. 2011; Tangestani Fard and Stough 2019), with increased neuroinflammation observed in aged brain and cognitive pathologies like AD (Eikelenboom and van Gool 2004; Eikelenboom et al. 2006). Altogether, these findings also excite the evolutionary interest in the genomic changes of immunomodulatory proteins like CD33 and their role in the cognitive health in human, especially elders.
In addition to the brain microglia, CD33 is an important immune modulator of hematopoietic cells where this innate receptor prevents the immune activation of the peripheral macrophages against SAMPs on the surface of self-cells. The absence of the ligand-binding V-set domain in truncated CD33m, therefore, prevents the suppression of macrophage phagocytic activity against the self-cells and contributes to more active immune cells. In this regard, it is important to note that unlike full-length CD33, CD33m isoform is not expressed on the cell surface, but instead stored intracellularly (Siddiqui et al. 2017; Saha et al. 2019). Therefore, the question remains what could have led to the selection of the truncated isoform of huCD33 that does not interact with Sia? In this regard, CD33 on macrophages plays crucial roles in different immune responses as well as during infections. HuCD33 has also recently been shown to be involved in immunomodulation during infection with hepatitis B virus (Tsai et al. 2021). Our previous and current data show that uniquely human pathogens like Neisseria and GBS display Neu5Ac, that is, recognized as “self” by human but not chimp CD33 (Landig et al. 2019). In the current work, we further found that the Sia-binding-domain-depleted, truncated huCD33 isoform does not bind and thus escaped exploitation by sialylated pathogens (fig. 2). This suggests that this truncated CD33 may have been an adaptation to counter the CD33-exploiting, immune-evasive behavior of pathogens like Neisseria and GBS.
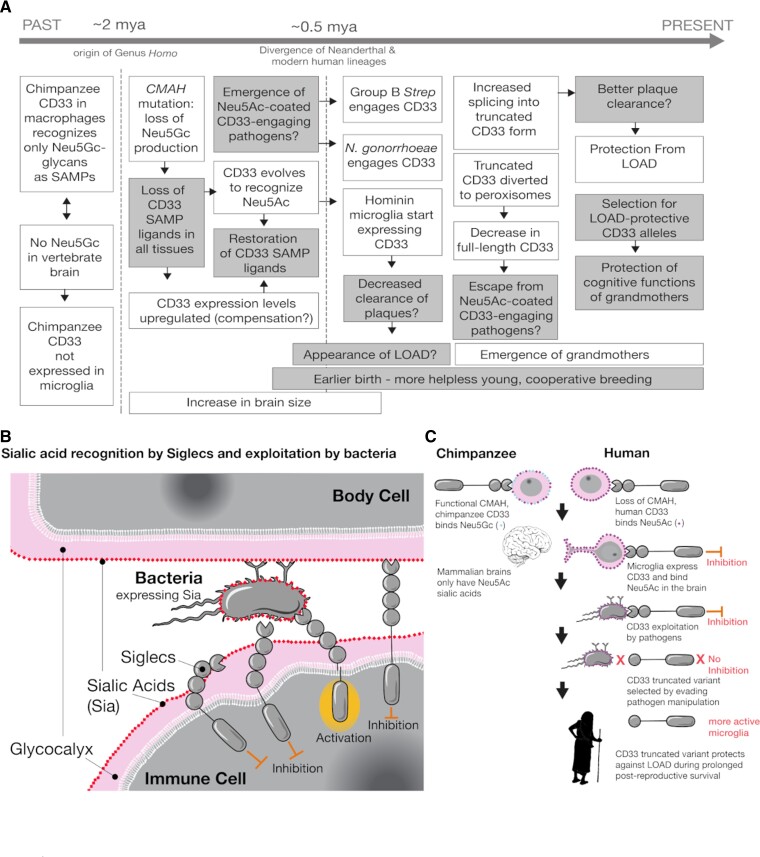
Taking together all currently available experimental data (including this study), we attempt to draw a plausible evolutionary scenario for CD33 protein evolution in humans and present in the context of relevant evolutionary events (fig. 6A). We hypothesize that the scarcity of the strongly preferred Neu5Gc ligand of ancestral CD33 in the brains of chimpanzee (and other great apes) was associated with low microglial expression. Subsequent hominin loss of CMAH (i.e., complete loss of Neu5Gc ligand) could then have been selected for the upregulation of CD33 levels perhaps to compensate for the loss of ligands, a change to Neu5Acbinding preference, and functional recruitment of CD33 to human microglia. Alongside the microglial CD33, the corresponding changes in the tissue macrophage proteins might have facilitated the emergence of Neu5Ac-coated pathogens (for example, N. gonorrhoeae and GBS) that evolved “molecular mimicry” of Neu5Ac-SAMP ligands to manipulate the immune response (fig. 6B). Appearance of the truncated isoform lacking the ligand-binding domain (CD33m), then probably allowed CD33 to escape the immune evasion by these sialylated pathogens (fig. 2). This selection pressure to stop manipulation by sialylated pathogens could have also altered splicing toward a higher level of truncated CD33, which also gets diverted to peroxisomes (Siddiqui et al. 2017). Although the significance of this diversion is unclear, decrease of full-length CD33 would facilitate escape from Neu5Ac-coated, CD33-engaging pathogens.
Fig. 6.
Scenario for evolution of huCD33 in relationship to cell surface sialic acids, infectious disease, brain microglia, and cognitive maintenance of grandmothers and other elderly caregivers. (A) This schematic presentation combines the known/likely facts (white boxes, including data from this manuscript) as well as suggested possibilities (gray boxes) into the most likely evolutionary scenario for human-specific evolution of CD33. Starting from the left, the likely chronological order of occurrence is indicated (by arrowheads) with the approximate timeline on the top, along the dotted lines. “?” indicates our reasonable assumption leading to the event. (B) Pictorial summary of the Siglec interactions with sialylated bacteria and the effect in the immune modulation. (C) Occurrence of key evolutionary events presented in the chronological order based on the currently available data. See text for further discussion.
Finally, sometime during the last 2 million years, increased brain size (both relative and absolute) must have resulted in an increasingly prolonged period of development and maturation (Charnov et al. 2001). The selective forces underlying this increase remain to be identified and could range from ecological, social, and technocultural, or more likely complex combinations thereof, including important contributions from language, a diagnostic feature of our species without clearly determined age of appearance. This also resulted in more helpless young and would have greatly benefited from cooperative breeding and caregiving (allomothering). It has been proposed that the origin of grandmothers may date back to the beginning of the genus Homo and may have directly contributed to altered anatomy and life history in our genus, long before the advent of Homo sapiens. One important effect of grandmother subsidies for the young could have been its contribution to the reduction in inter-birth interval, characteristic of our species, as compared with living great apes. Another great impact of allomothering by grandmothers and postreproductive elders would have been the transmission of knowledge and norms, which crucially rely on the capacity for language in our species and remains without demonstrated counterpart in any other species (fig. 6C). Importantly, both types of contribution would also have helped reduce extrinsic mortality, which would have in turn led to further increases in longevity. However, the value of postmenopausal grandmothers and other elderly caregivers would then have been seriously blunted by the appearance of LOAD or any other cognitive decline. The synthesis of the truncated isoform of CD33 protects from Neisseria during reproductive age and a higher ratio of truncated to full-length isoforms correlates to a decrease of LOAD in grandmothers. However, a small amount of the full-length isoform remains, likely to downregulate hyper-inflammation that might arise during prolonged absence of SAMP recognition. Notably, when an elderly caregiver gets LOAD, not only are the evolutionary benefits of the individual lost, but this also presents an increased burden on younger group members to care for that elder individual. Under this proposed scenario, the current state in the evolution of huCD33 protein represents a trade-off between the evolutionary response to exploitation by pathogens in early life and cognitive maintenance in postreproductive late life. A similar evolutionary scenario appears to underly the case of the human APOE gene where variants include both risk alleles (APOE4) and protective alleles (APOE2 and APOE3) for cardiovascular disease and LOAD (Reiman et al. 2020). In this instance, the ancestral APOE4 allele is associated with increased risks of loss of cognitive functions and the derived alleles may serve to protect the cognition of the elderly caregivers. Interestingly, the APOE4 allele is also correlated with the protection from severe diarrhea in early years of life (Oriá et al. 2005). Given these examples, it remains to be seen how widespread this pattern is wherein variants conveying survival advantages in early life coexist with other variants that protect cognition late in life, a case of coexisting variants with opposite patterns of antagonistic pleiotropy.
Materials and Methods
Bacterial Culture and Cell Lines
The bacterial strains were N. gonorrhoeae F62ΔlgtD (generous gift from Sanjay Ram, University of Massachusetts Worcester), GBS strains (generous gifts from Victor Nizet, University of California San Diego). Neisseria were grown overnight on chocolate II agar and GBS on Todd Hewitt agar at 37 °C and 5% CO2 from respective frozen glycerol stocks. Before assay, GBS was grown in Todd Hewitt broth. The E. coli K1 strain was grown in LB. HEK293A or U937 cells in DMEM or RPMI1640 media with 10% FCS, respectively, were used.
Sialylation of Neisseria
Following overnight growth, Neisseria were grown in GC broth with IsoVitaleX, with or without 30 µM CMP-Neu5Ac (Nacalai USA. Inc.) until OD600 of 0.4–0.5.
Bacterial Staining
The bacteria were washed with prewarmed Hanks′ Balanced Salt solution (HBSS) and stained with 2 µM SYTO13 (Thermo Scientific) for 30 min at 37 °C and shaking at 200 rpm in dark. The stained bacteria were washed with HBSS and resuspended to a final concentration of OD600 = 1/ml in HBSS for the binding assay.
Generation of CD33 Mutant Proteins
A genomic fragment (1228 bp) of human or chimpanzee CD33(M), including the first four exons (two Ig domains) was fused with pcDNA3.1(−) containing C-terminal FLAG(EK) sequence followed by hIgG1-Fc genomic fragment (hinge+2 Ig-like domains) and described elsewhere (Angata and Varki 2000; Angata et al. 2002). The mutant variants were made from either construct using Q5 site-directed mutagenesis kit (NEB) according to the manufacturer’s instructions (supplementary table S3, Supplementary Material online). Mutagenesis primers listed were designed using NEBaseChanger software.
Truncated CD33_EK_Fc Construction
Total mRNA was isolated from U937 cells using Oligotex Direct mRNA Mini Kit (QIAGEN). CD33m was PCR amplified by SuperScript III One-Step RT-PCR (Invitrogen) and primers 5'-TTATATGCTAGCGCCACCATGCCGCTGCTGCTACTGCTGC-3', NheI site underlined and 5'-GCGCGCGATATCATGAACCACTCCTGCTCTGGTCTCTTG-3', EcoRV site underlined. PCR products were run on 2% agarose gel and 396 bp band corresponding to CD33m excised and cut with NheI/EcoRV. Digested bands were subcloned into pcDNA3.1(−) containing a C-terminal FLAG (EK) sequence followed by a hIgG1-Fc genomic fragment (hinge + 2 Ig-like domains).
Purification of CD33 Mutants
Transfection supernatants were spun down at 500 g for 5 min to remove cellular debris. The pH of the supernatant was adjusted to 8.0 for optimal binding of protein A-sepharose beads to hIgG-Fc fusion proteins. Protein A-Sepharose 4 Fast Flow suspension (GE Healthcare) was washed with Tris-Buffered Saline (TBS) pH 8.0, and 1:500 ratio of beads:media added to the supernatant and incubated for 24 h at 4 °C. Thereafter, supernatants with beads were transferred to disposable columns until all liquid has run thru. Beads were washed thrice with TBS and proteins eluted directly in 0.3 ml of 1 M Tris–HCl pH 8.0 using 0.1 M Glycine Buffer pH 2.8. Each eluate was centrifuged in Amicon Ultra-15 filter unit—Molecular Weight Cut-Off (MWCO) 30 K for full-length variants (MWCO 10 K for CD33m) and washed with TBS. After the final wash, each retentate was recovered from the column and stored at −80 °C.
Binding Assay
Bacterial binding was done with recombinant Fc-chimeric proteins of CD33. Briefly, protein A coated black 96-well plate (Pierce, Thermo Scientific) was washed with TBS containing 0.05% Tween 20 and coated with 200 ng/well of the respective CD33 protein. SYTO13 (Thermo Scientific) stained bacteria were incubated in the wells for 30 min at 37 °C and 5% CO2 without shaking. Fluorescence measurements to indicate bound bacteria were done at 488 nm (excitation) and 530 nm (emission). Data were analyzed using excel and Prism software.
Evolutionary Analysis and Detection of Positive Selection
CD33 protein-coding sequences were aligned in CLUSTALW program implemented in MEGA7 and back translated to obtain a codon alignment. The phylogenetic tree of CD33 protein-coding sequences was reconstructed with neighbor-joining method, which was implemented in MEGA7 (fig. 1), 1000 bootstrap replicates (Kumar et al. 2016). The unrooted neighbor-joining tree was used for the subsequent analysis.
To assess the significance of the relative absence of LOAD-protective SNPS in archaic genomes, a resampling analysis was performed from SNPS in the 1000 genomes variant call set. Variants were filtered based on archaic quality filters and grouped by global allele frequencies before randomly selecting SNPS based on the query AF. The frequency of observing these random SNPS in archaic genomes was used to estimate an empirical probability.
Nonsynonymous/synonymous substitution ratios (ω = dN/dS, or Ka/Ks) have become a useful means for quantifying the impact of natural selection on molecular evolution. In general, the ratio ω = dN/dS is less than one if the gene is undergoing purifying selection, equal to one if the gene is evolving neutrally, and greater than one if positive selection has accelerated the fixation of nonsynonymous substitutions that resulted in amino acid changes. The pair-wise computation of Ka/Ks between V-set exon of each species was performed using the program DnaSp v.0 6.0. The initial unrooted tree fed to the program in the format of Newick was: ([Chimpanzee5:0.00000000, Bonobo:0.00000000]:0.00222522, Gorilla:0.00979959, Human:0.01351877).
Molecular Simulation
Starting structures of the V-type domain (residues 18–142) of huCD33 were built based on PDB entries 5j0b (A chain) and 6d49. The initial 3D models of chCD33 were built by swapping residues: N20K, F21I, W22R, A65P, I67V, R69G (in 6d49), L78P, P96L. The side chain of His45 was modeled in two conformations (compare fig. 4): ‘down’ (as in PDB entry 6d49) and ‘up’ (as present in PDB entries 5ihb or 5j06 chains A). Additionally, systems were built that contain five molecules of Neu5GcαOMe or Neu5AcαOMe distributed in the simulation box which allowed to simulate binding events. In total, 27 MD trajectories were sampled for huCD33 and 20 for chCD33, most of them covering a microsecond timescale (compare supplementary fig. S4, Supplementary Material online).
Sialoglycan Microarray
The sialoglycan microarray experimental method was adopted from the literature reported earlier (Meng et al. 2018; Lu et al. 2019). Sialoglycans were printed in quadruplets on NHS-functionalized glass slides (PolyAn 3D-NHS; catalog# PO-10400401) and then blocked, washed, and dried. The slides were rehydrated using Ovalbumin-PBS (1%, w/v) and then incubated with CD33 proteins followed by probing with secondary Cy3-conjugated goat antihuman IgG. The slides were scanned at wavelength 532 nm and heatmap was plotted.
Supplementary Material
Acknowledgments
We are very thankful to Dr Sanjay Ram (University of Massachusetts, Worcester) and Dr Victor Nizet (University of California, San Diego) for the bacterial strains that have greatly benefited the project. This work was supported by National Institute of Health grant R01GM32373 and Cure Alzheimer's Fund grant (to A.V.).
Contributor Information
Sudeshna Saha, Departments of Medicine, Pathology, Anthropology and Cellular and Molecular Medicine, Center for Academic Research and Training in Anthropogeny and Glycobiology Research and Training Center, University of California San Diego, San Diego, CA 92093, USA.
Naazneen Khan, Departments of Medicine, Pathology, Anthropology and Cellular and Molecular Medicine, Center for Academic Research and Training in Anthropogeny and Glycobiology Research and Training Center, University of California San Diego, San Diego, CA 92093, USA.
Troy Comi, Department of Genetics, Princeton University, Princeton, NJ 08544, USA.
Andrea Verhagen, Departments of Medicine, Pathology, Anthropology and Cellular and Molecular Medicine, Center for Academic Research and Training in Anthropogeny and Glycobiology Research and Training Center, University of California San Diego, San Diego, CA 92093, USA.
Aniruddha Sasmal, Departments of Medicine, Pathology, Anthropology and Cellular and Molecular Medicine, Center for Academic Research and Training in Anthropogeny and Glycobiology Research and Training Center, University of California San Diego, San Diego, CA 92093, USA.
Sandra Diaz, Departments of Medicine, Pathology, Anthropology and Cellular and Molecular Medicine, Center for Academic Research and Training in Anthropogeny and Glycobiology Research and Training Center, University of California San Diego, San Diego, CA 92093, USA.
Hai Yu, Department of Chemistry, University of California Davis, Davis, CA 95616, USA.
Xi Chen, Department of Chemistry, University of California Davis, Davis, CA 95616, USA.
Joshua M Akey, Department of Genetics, Princeton University, Princeton, NJ 08544, USA.
Martin Frank, Biognos AB, Gothenburg SE-402 74, Sweden.
Pascal Gagneux, Departments of Medicine, Pathology, Anthropology and Cellular and Molecular Medicine, Center for Academic Research and Training in Anthropogeny and Glycobiology Research and Training Center, University of California San Diego, San Diego, CA 92093, USA.
Ajit Varki, Departments of Medicine, Pathology, Anthropology and Cellular and Molecular Medicine, Center for Academic Research and Training in Anthropogeny and Glycobiology Research and Training Center, University of California San Diego, San Diego, CA 92093, USA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability
The data for the resampling analysis are available on Code Ocean (DOI:10.24433/CO.9371212.v1; https://codeocean.com/capsule/2131051/tree).
References
- Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl M-C, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, et al. 2000. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 26:76–80. [DOI] [PubMed] [Google Scholar]
- Angata T, Kerr SC, Greaves DR, Varki NM, Crocker PR, Varki A. 2002. Cloning and characterization of human Siglec-11. A recently evolved signaling molecule that can interact with SHP-1 and SHP-2 and is expressed by tissue macrophages, including brain microglia. J Biol Chem. 277:24466–24474. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. 2000. Cloning, characterization, and phylogenetic analysis of siglec-9, a new member of the CD33-related group of siglecs. Evidence for co-evolution with sialic acid synthesis pathways. J Biol Chem. 275:22127–22135. [DOI] [PubMed] [Google Scholar]
- Apicella MA, Mandrell RE, Shero M, Wilson ME, Griffiss JM, Brooks GF, Lammel C, Breen JF, Rice PA. 1990. Modification by sialic acid of Neisseria gonorrhoeae lipooligosaccharide epitope expression in human urethral exudates: an immunoelectron microscopic analysis. J Infect Dis. 162:506–512. [DOI] [PubMed] [Google Scholar]
- Bornhöfft KF, Goldammer T, Rebl A, Galuska SP. 2018. Siglecs: a journey through the evolution of sialic acid-binding immunoglobulin-type lectins. Dev Comp Immunol. 86:219–231. [DOI] [PubMed] [Google Scholar]
- Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, et al. 2013. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 16:848–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman-Van der Linden EC, Angata T, Reynolds SA, Powell LD, Hedrick SM, Varki A. 2003. CD33/Siglec-3 binding specificity, expression pattern, and consequences of gene deletion in mice. Mol Cell Biol. 23:4199–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byars SG, Voskarides K. 2020. Antagonistic pleiotropy in human disease. J Mol Evol. 88:12–25. [DOI] [PubMed] [Google Scholar]
- Cant MA, Croft DP. 2019. Life-history evolution: grandmothering in space and time. Curr Biol. 29:R215–R218. [DOI] [PubMed] [Google Scholar]
- Carlin AF, Lewis AL, Varki A, Nizet V. 2007. Group B streptococcal capsular sialic acids interact with siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol. 189:1231–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Sando SB, Aasly JO, et al. 2011. Replication of EPHA1 and CD33 associations with late-onset Alzheimer’s disease: a multi-centre case-control study. Mol Neurodegener. 6:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant DA, Brynildsrud OB. 2020. Neisseria meningitidis: using genomics to understand diversity, evolution and pathogenesis. Nat Rev Microbiol. 18:84–96. [DOI] [PubMed] [Google Scholar]
- Charnov EL, Turner TF, Winemiller KO. 2001. Reproductive constraints and the evolution of life histories with indeterminate growth. Proc Natl Acad Sci U S A. 98:9460–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Butovsky O. 2017. Microglia function in the central nervous system during health and neurodegeneration. Annu Rev Immunol. 35:441–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GP, Auton A, Brooks LD, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Manuel M, Kuhlwilm M, Frandsen P, Sousa VC, Desai T, Prado-Martinez J, Hernandez-Rodriguez J, Dupanloup I, Lao O, Hallast P, et al. 2016. Chimpanzee genomic diversity reveals ancient admixture with bonobos. Science 354:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edler MK, Munger EL, Meindl RS, Hopkins WD, Ely JJ, Erwin JM, Mufson EJ, Hof PR, Sherwood CC, Raghanti MA. 2020. Neuron loss associated with age but not Alzheimer’s disease pathology in the chimpanzee brain. Philos Trans R Soc Lond B Biol Sci. 375:20190619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edler MK, Sherwood CC, Meindl RS, Hopkins WD, Ely JJ, Erwin JM, Mufson EJ, Hof PR, Raghanti MA. 2017. Aged chimpanzees exhibit pathologic hallmarks of Alzheimer’s disease. Neurobiol Aging. 59:107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JL, Apicella MA. 2004. The molecular mechanisms used by Neisseria gonorrhoeae to initiate infection differ between men and women. Clin Microbiol Rev. 17:965–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eikelenboom P, van Gool WA. 2004. Neuroinflammatory perspectives on the two faces of Alzheimer’s disease. J Neural Transm (Vienna). 111:281–294. [DOI] [PubMed] [Google Scholar]
- Eikelenboom P, Veerhuis R, Scheper W, Rozemuller AJ, van Gool WA, Hoozemans JJ. 2006. The significance of neuroinflammation in understanding Alzheimer’s disease. J Neural Transm (Vienna). 113:1685–1695. [DOI] [PubMed] [Google Scholar]
- Finch CE, Sapolsky RM. 1999. The evolution of Alzheimer disease, the reproductive schedule, and apoE isoforms. Neurobiol Aging. 20:407–428. [DOI] [PubMed] [Google Scholar]
- Fong JJ, Tsai CM, Saha S, Nizet V, Varki A, Bui JD. 2018. Siglec-7 engagement by GBS β-protein suppresses pyroptotic cell death of natural killer cells. Proc Natl Acad Sci U S A. 115:10410–10415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SD, Kelm S, Barber EK, Crocker PR. 1995. Characterization of CD33 as a new member of the sialoadhesin family of cellular interaction molecules. Blood. 85:2005–2012. [PubMed] [Google Scholar]
- Gonzalez-Gil A, Porell RN, Fernandes SM, Maenpaa E, Li TA, Li T, Wong PC, Aoki K, Tiemeyer M, Yu ZJ, et al. 2022. Human brain sialoglycan ligand for CD33, a microglial inhibitory Siglec implicated in Alzheimer’s disease. J. Biol Chem. 298(6):101960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A, Federico AN, Natasan J, Forte AM, McGinty D, Nguyen H, Volak A, LeRoy S, Gandhi S, Lerner EP, et al. 2020. Gene therapy for Alzheimer’s disease targeting CD33 reduces amyloid beta accumulation and neuroinflammation. Hum Mol Genet. 29:2920–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE. 2013. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron 78:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes K. 2004. Human longevity: the grandmother effect. Nature 428:128–129. [DOI] [PubMed] [Google Scholar]
- Hawkes K. 2010. Colloquium paper: how grandmother effects plus individual variation in frailty shape fertility and mortality: guidance from human–chimpanzee comparisons. Proc Natl Acad Sci U S A. 107(Suppl 2):8977–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes K. 2016. Genomic evidence for the evolution of human postmenopausal longevity. Proc Natl Acad Sci U S A. 113:17–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes K, O’Connell JF, Jones NG, Alvarez H, Charnov EL. 1998. Grandmothering, menopause, and the evolution of human life histories. Proc Natl Acad Sci U S A. 95:1336–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Pálsson S, Thorleifsson G, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, et al. 2007. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 39:218–225. [DOI] [PubMed] [Google Scholar]
- Hernandez-Caselles T, Martinez-Esparza M, Perez-Oliva AB, Quintanilla-Cecconi AM, Garcia-Alonso A, Alvarez-Lopez DM, Garcia-Penarrubia P. 2006. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J Leukoc Biol. 79:46–58. [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. 2011. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 43:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadecola C. 2013. The pathobiology of vascular dementia. Neuron 80:844–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RA, Cant MA. 2010. The evolution of menopause in cetaceans and humans: the role of demography. Proc Biol Sci. 277:3765–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, de Manuel M, Peyregne S, Do R, Prufer K, Marques-Bonet T, Varki N, Gagneux P, Varki A. 2020. Multiple genomic events altering hominin SIGLEC biology and innate immunity predated the common ancestor of humans and archaic hominins. Genome Biol Evol. 12:1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Kim SK, Gagneux P, Dugan LL, Varki A. 2020. Maximum reproductive lifespan correlates with CD33rSIGLEC gene number: implications for NADPH oxidase-derived reactive oxygen species in aging. FASEB J. 34:1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba JK, Pounds S, Cao X, Downing JR, Campana D, Ribeiro RC, Pui CH, Rubnitz JE. 2009. Coding polymorphisms in CD33 and response to gemtuzumab ozogamicin in pediatric patients with AML: a pilot study. Leukemia 23:402–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landig CS, Hazel A, Kellman BP, Fong JJ, Schwarz F, Agarwal S, Varki N, Massari P, Lewis NE, Ram S, et al. 2019. Evolution of the exclusively human pathogen Neisseria gonorrhoeae: human-specific engagement of immunoregulatory Siglecs. Evol Appl. 12:337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Läubli H, Varki A. 2020. Sialic acid-binding immunoglobulin-like lectins (Siglecs) detect self-associated molecular patterns to regulate immune responses. Cell Mol Life Sci. 77:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dai X, Tao W, Liu H, Li H, Yang C, Zhang J, Li X, Chen Y, Ma C, et al. 2018. APOE influences working memory in non-demented elderly through an interaction with SPON1 rs2618516. Hum Brain Mapp. 39:2859–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu N, Ye J, Cheng J, Liu C-C, Yao W, Yan J, Khan N, Yi W, Varki A, Cao H. 2019. Redox-controlled site-specific α2-6-sialylation. J Am Chem Soc. 141:4547–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, Estus S. 2013. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 33:13320–13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez FL, Poznik GD, Castellano S, Bustamante CD. 2016. The divergence of neandertal and modern human Y chromosomes. Am J Hum Genet. 98:728–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C, Sasmal A, Zhang Y, Gao T, Liu CC, Khan N, Varki A, Wang F, Cao H. 2018. Chemoenzymatic assembly of mammalian O-mannose glycans. Angew Chem Int Ed Engl. 57:9003–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JM, Aronoff DM, Capra JA, Abbot P, Rokas A. 2018. Examination of signatures of recent positive selection on genes involved in human sialic acid biology. G3 (Bethesda) 8:1315–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi MN. 2017. Cognitive dysfunction in older adults with diabetes: what a clinician needs to know. Diabetes Care 40:461–467. [DOI] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, et al. 2011. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 43:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T, Wooding S, Sakagami T, Emi M, Tokunaga K, Tamiya G, Ishigami T, Umemura S, Munkhbat B, Jin F, et al. 2004. Natural selection and population history in the human angiotensinogen gene (AGT): 736 complete AGT sequences in chromosomes from around the world. Am J Hum Genet. 74:898–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oriá RB, Patrick PD, Zhang H, Lorntz B, de Castro Costa CM, Brito GA, Barrett LJ, Lima AA, Guerrant RL. 2005. APOE4 protects the cognitive development in children with heavy diarrhea burdens in Northeast Brazil. Pediatr Res. 57:310–316. [DOI] [PubMed] [Google Scholar]
- Padler-Karavani V, Hurtado-Ziola N, Chang YC, Sonnenburg JL, Ronagh A, Yu H, Verhagen A, Nizet V, Chen X, Varki N, et al. 2014. Rapid evolution of binding specificities and expression patterns of inhibitory CD33-related Siglecs in primates. FASEB J. 28:1280–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons NJ, Patel PV, Tan EL, Andrade JR, Nairn CA, Goldner M, Cole JA, Smith H. 1988. Cytidine 5′-monophospho-N-acetyl neuraminic acid and a low molecular weight factor from human blood cells induce lipopolysaccharide alteration in gonococci when conferring resistance to killing by human serum. Microb Pathog. 5:303–309. [DOI] [PubMed] [Google Scholar]
- Peila R, Rodriguez BL, Launer LJ, Honolulu-Asia AS. 2002. Type 2 diabetes, APOE gene, and the risk for dementia and related pathologies: the Honolulu-Asia Aging Study. Diabetes 51:1256–1262. [DOI] [PubMed] [Google Scholar]
- Prado-Martinez J, Sudmant PH, Kidd JM, Li H, Kelley JL, Lorente-Galdos B, Veeramah KR, Woerner AE, O’Connor TD, Santpere G, et al. 2013. Great ape genetic diversity and population history. Nature 499:471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, Rothamel K, Stranger BE, Bennett DA, Evans DA, et al. 2014. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet. 23:2729–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathore N, Ramani SR, Pantua H, Payandeh J, Bhangale T, Wuster A, Kapoor M, Sun Y, Kapadia SB, Gonzalez L, et al. 2018. Paired immunoglobulin-like type 2 receptor alpha G78R variant alters ligand binding and confers protection to Alzheimer’s disease. PLoS Genet. 14:e1007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Arboleda-Velasquez JF, Quiroz YT, Huentelman MJ, Beach TG, Caselli RJ, Chen Y, Su Y, Myers AJ, Hardy J, et al. 2020. Exceptionally low likelihood of Alzheimer’s dementia in APOE2 homozygotes from a 5,000-person neuropathological study. Nat Commun. 11:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Siddiqui SS, Khan N, Verhagen A, Jiang W, Springer S, Ghosh P, Varki A. 2019. Controversies about the subcellular localization and mechanisms of action of the Alzheimer’s disease-protective CD33 splice variant. Acta Neuropathol. 138:671–672. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. 2009. Siglec receptors and hiding plaques in Alzheimer’s disease. J Mol Med. 87:697–701. [DOI] [PubMed] [Google Scholar]
- Schwarz F, Springer SA, Altheide TK, Varki NM, Gagneux P, Varki A. 2016. Human-specific derived alleles of CD33 and other genes protect against postreproductive cognitive decline. Proc Natl Acad Sci U S A. 113:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert HS. 2019. Location, location, location-commensalism, damage and evolution of the pathogenic neisseria. J Mol Biol. 431:3010–3014. [DOI] [PubMed] [Google Scholar]
- Siddiqui SS, Springer SA, Verhagen A, Sundaramurthy V, Alisson-Silva F, Jiang W, Ghosh P, Varki A. 2017. The Alzheimer’s disease-protective CD33 splice variant mediates adaptive loss of function via diversion to an intracellular pool. J Biol Chem. 292:15312–15320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simen AA, Bordner KA, Martin MP, Moy LA, Barry LC. 2011. Cognitive dysfunction with aging and the role of inflammation. Ther Adv Chronic Dis. 2:175–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumanska G, Vorbrodt AW, Mandybur TI, Wisniewski HM. 1987. Lectin histochemistry of plaques and tangles in Alzheimer’s disease. Acta Neuropathol. 73:1–11. [DOI] [PubMed] [Google Scholar]
- Tangestani Fard M, Stough C. 2019. A review and hypothesized model of the mechanisms that underpin the relationship between inflammation and cognition in the elderly. Front Aging Neurosci. 11:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. 2004. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 75:1059–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici MA, Walls AC, Lang Y, Wang C, Li Z, Koerhuis D, Boons G-J, Bosch B-J, Rey FA, de Groot RJ, et al. 2019. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 26:481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai TY, Huang MT, Sung PS, Peng C-Y, Tao M-H, Yang H-I, Chang W-C, Yang A-S, Yu C-M, Lin Y-P, et al. 2021. SIGLEC-3 (CD33) serves as an immune checkpoint receptor for HBV infection. J Clin Invest. 131:141965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Molen J, Frisse LM, Fullerton SM, Qian Y, Del Bosque-Plata L, Hudson RR, Di Rienzo A. 2005. Population genetics of CAPN10 and GPR35: implications for the evolution of type 2 diabetes variants. Am J Hum Genet. 76:548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. 2011. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 21:1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Angata T. 2006. Siglecs—the major subfamily of I-type lecins. Glycobiology 16:1R–27R. [DOI] [PubMed] [Google Scholar]
- Varki A, Gagneux P. 2012. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci. 1253:16–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen G, Wessel J, Zhou W, Rao F, Stridsberg M, Mahata SK, Gent PM, Das M, Cooper RS, Rao F, et al. 2007. An ancestral variant of Secretogranin II confers regulation by PHOX2 transcription factors and association with hypertension. Hum Mol Genet. 16:1752–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GC. 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11:398–411. [Google Scholar]
- Wißfeld J, Nozaki I, Mathews M, Raschka T, Ebeling C, Hornung V, Brüstle O, Neumann H. 2021. Deletion of Alzheimer’s disease-associated CD33 results in an inflammatory human microglia phenotype. Glia 69:1393–1412. [DOI] [PubMed] [Google Scholar]
- Xu W, Tan L, Yu JT. 2015. The role of PICALM in Alzheimer’s disease. Mol Neurobiol. 52:399–413. [DOI] [PubMed] [Google Scholar]
- Xue Y, Prado-Martinez J, Sudmant PH, Narasimhan V, Ayub Q, Szpak M, Frandsen P, Chen Y, Yngvadottir B, Cooper DN, et al. 2015. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science 348:242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L. 2019. CD33 in Alzheimer’s disease – biology, pathogenesis, and therapeutics: a mini-review. Gerontology 65:323–331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data for the resampling analysis are available on Code Ocean (DOI:10.24433/CO.9371212.v1; https://codeocean.com/capsule/2131051/tree).