A small-molecule approach to degrade tumor cell–surface PD-L1 overcomes adaptive immune resistance during MAPK inhibitor therapy, showcasing control of PD-L1 protein stability as an emerging combinatorial strategy.
Abstract
MAPK inhibitor (MAPKi) therapy in melanoma leads to the accumulation of tumor-surface PD-L1/L2, which may evade antitumor immunity and accelerate acquired resistance. Here, we discover that the E3 ligase ITCH binds, ubiquitinates, and downregulates tumor-surface PD-L1/L2 in MAPKi-treated human melanoma cells, thereby promoting T-cell activation. During MAPKi therapy in vivo, melanoma cell–intrinsic ITCH knockdown induced tumor-surface PD-L1, reduced intratumoral cytolytic CD8+ T cells, and accelerated acquired resistance only in immune-competent mice. Conversely, tumor cell–intrinsic ITCH overexpression reduced MAPKi-elicited PD-L1 accumulation, augmented intratumoral cytolytic CD8+ T cells, and suppressed acquired resistance in BrafV600MUT, NrasMUT, or Nf1MUT melanoma and KrasMUT-driven cancers. CD8+ T-cell depletion and tumor cell–intrinsic PD-L1 overexpression nullified the phenotype of ITCH overexpression, thereby supporting an in vivo ITCH–PD-L1–T-cell regulatory axis. Moreover, we identify a small-molecular ITCH activator that suppresses acquired MAPKi resistance in vivo. Thus, MAPKi-induced PD-L1 accelerates resistance, and a PD-L1–degrading ITCH activator prolongs antitumor response.
Significance:
MAPKi induces tumor cell–surface PD-L1 accumulation, which promotes immune evasion and therapy resistance. ITCH degrades PD-L1, optimizing antitumor T-cell immunity. We propose degrading tumor cell–surface PD-L1 and/or activating tumor-intrinsic ITCH as strategies to overcome MAPKi resistance.
This article is highlighted in the In This Issue feature, p. 1825
INTRODUCTION
Disrupting PD-L1 interaction with PD-1 rejuvenates antitumor immunity and elicits clinical antitumor responses across a wide range of cancer types (1). Cancer cells can express robust surface levels of PD-L1 to tolerize tumor-specific T cells, but the regulation of PD-L1 protein levels on the tumor cell surface is poorly understood. Dedifferentiated or quasi-mesenchymal tumor cells upregulate PD-L1/L2 (or vice versa) and induce an immune-suppressive microenvironment, including expansion of M2-like macrophages and regulatory T cells as well as depletion of CD8+ T cells (2). Targeted therapy, including MAPK inhibitor (MAPKi) therapy in melanoma, leads to dedifferentiation, PD-L1/2 upregulation, and resistance (3), and both MAPKi treatment and mesenchymal signatures are associated with innate anti–PD-1 resistance (4, 5).
Tumor cell expression of PD-L1 is induced by transcriptional mechanisms—for example, in response to inflammatory cytokines such as IFNγ or TNFα—or by natural selection via immune editing, for example, gene amplification of PD-L1 (6) and structural variation of the 3′ untranslated region (7). The tumor cell–intrinsic control of PD-L1 protein stability also regulates antitumor immunity (8–11). Our previous work showed that clinical melanoma induces PD-L1/L2 protein levels early on MAPKi therapy (3), suggesting a mechanism of immune evasion that reduces the efficacy of targeted therapy and that may be ameliorated by combining anti–PD-1/L1 therapy. Indeed, in murine models of BrafMUT and NrasMUT melanoma, the introduction of high mutational burden (and presumably neoantigen burden) extends the durability of MAPKi responses in a fashion dependent on CD8+ T cells (12, 13), and swiftly sequencing anti–PD-1/L1 ahead of combination with MAPKi therapy strongly suppressed acquired MAPKi resistance (13).
To understand how dynamic PD-L1/L2 protein levels are regulated via protein–protein interactions in MAPKi-resistant melanoma cells, we took two complementary affinity purification–mass spectrometry approaches. From these data, we prioritized a common hit, the E3 ligase ITCH, for validation as a PD-L1–interacting partner, determined its regulation of PD-L1 stability at the tumor cell surface, and characterized ITCH-mediated PD-L1 polyubiquitination and lysosomal targeting followed by degradation. We also investigated the impact of PD-L1–ITCH interaction on T-cell activation in cocultures and on in vivo MAPKi resistance using multiple melanoma models in syngeneic and immune-compromised hosts. In additional mechanistic studies, we dissected how perturbing tumor-intrinsic ITCH expression during MAPKi treatment impacts the tumor immune microenvironment and investigated the critical contribution of CD8+ T cells to the tumor-extrinsic function of ITCH expressed by melanoma and other MAPK-addicted cancer cells. Lastly, we provided proof-of-concept data for the feasibility of identifying an ITCH activator that can suppress acquired MAPKi resistance in vivo.
RESULTS
PD-L1/L2—Highly Expressed by MAPKi-Adapted Melanoma—Interacts with ITCH
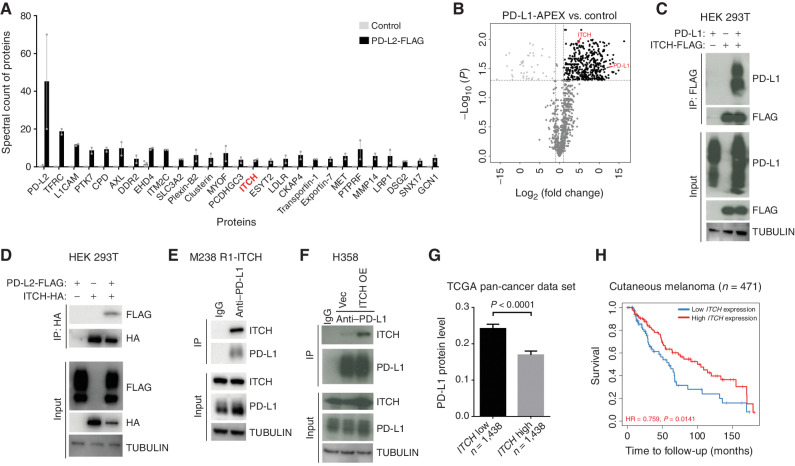
Using a human melanoma cell line (M229 R5; ref. 14) with acquired resistance to a BRAFV600MUT inhibitor (vemurafenib), we engineered via lentiviral stable transduction PD-L2-FLAG expression and performed anti-FLAG immunoprecipitation followed by mass spectrometry. Among the top hits is an E3 ligase, ITCH (Fig. 1A), which is not known to regulate antitumor immunity or therapy resistance. Because artifactual protein interactions can occur or physiologic interactions can be lost after cell membrane solubilization, we surveyed the in situ neighborhood protein interactome of PD-L1 in live melanoma cells. We engineered M229 R5 via lentiviral stable transduction to express the bacterial biotinylation enzyme (APEX2) fused to the C-terminus of PD-L1. Mass spectrometry quantification of biotin-labeled proteins in cells treated (vs. not treated) with the APEX2 activator H2O2 identified ITCH as a PD-L1 proximity interaction partner (Fig. 1B).
Figure 1.
The E3 ligase ITCH interacts with PD-L1 and PD-L2. A, Spectral count of indicated proteins from shotgun mass spectrometry–based analysis of anti-FLAG immunoprecipitates from the M229 R5 cell line engineered to express PD-L2-FLAG (n = 2) or empty vector (control, n = 4). Average total spectral count in samples: 5,156 (control), 4,229 (anti-FLAG), mean ± SEMs. Proteins are shown with confidence score for interaction [Significance Analysis of INTeractome (SAINT)] higher than 0.95. B, Proteins identified in proximity to PD-L1 in the M229 R5 cell line stably expressing PD-L1–APEX2 (H2O2 treated vs. not treated, n = 3). Black dots, proteins significantly enriched in H2O2-treated cell. C, Immunoprecipitation (IP) of HEK 293T cells (transfected with PD-L1 and ITCH-FLAG, alone or together) using anti-FLAG M2 beads, followed by Western blots (WB) of indicated proteins. TUBULIN, loading control. D, IP of HEK 293T cells (transfected with PD-L2-FLAG and/or ITCH-HA) using anti-HA beads, followed by WBs of indicated proteins. TUBULIN, loading control. E, IP of the human melanoma cell line M238 R1 stably expressing ITCH using anti–PD-L1 antibody (rabbit-IgG as control), followed by WBs of indicated proteins. TUBULIN, loading control. F, IP of the human non–small cell lung carcinoma cell line H358, with or without ITCH overexpression (OE), using anti–PD-L1 (rabbit-IgG as control) followed by WBs of indicated proteins. TUBULIN, loading control. Vec, empty vector. G, PD-L1 protein levels compared by splitting 7,194 patient-derived tumors with matched RNA-seq data and PD-L1 protein levels (measured by reverse-phase protein array from 32 TCGA cancer types) into top versus bottom 20% of ITCH RNA expression levels. P value, Student t test. H, Kaplan–Meier survival curve of 471 cutaneous melanoma patients from TCGA data set with high (top 20%) versus low (bottom 20%) ITCH RNA expression levels. Time of diagnosis is set as the starting time for follow-up. P value, log-rank test.
To corroborate the hypothesis that PD-L1 or PD-L2 physically interacts with ITCH, we used HEK 293T cells to overexpress PD-L1, ITCH-FLAG, or PD-L1 + ITCH-FLAG (Fig. 1C) or PD-L2-FLAG, ITCH-HA, or PD-L2-FLAG + ITCH-HA (Fig. 1D). After immunoprecipitation against the FLAG epitope, we detected PD-L1 only when PD-L1 and ITCH-FLAG were cotransfected (Fig. 1C). After immunoprecipitation against the HA epitope, we detected PD-L2-FLAG only when PD-L2-FLAG and ITCH-HA were cotransfected (Fig. 1D). To corroborate the interaction of PD-L1 with ITCH in additional cancer cell lines, we used stable lentiviral transduction to express ITCH in another human melanoma cell line with acquired resistance to a BRAFV600MUT inhibitor (M238 R1; ref. 14) and a human non–small cell lung carcinoma cell line, H358. We chose H358 based on its relatively high basal level of PD-L1 protein expression despite its lack of detectable PD-L2 protein expression by flow cytometry. Indeed, when we immunoprecipitated against endogenous PD-L1, we detected exogenous (Fig. 1E and F) or endogenous (Fig. 1F) ITCH. Given the evidence of physical interaction between ITCH and PD-L1, we evaluated the pan-cancer relationship between bulk tumor ITCH RNA expression levels and PD-L1 protein levels from The Cancer Proteome Atlas (TCPA) data set (15). Consistent with the E3 ligase ITCH being a potential negative regulator of PD-L1, we observed that higher expression of ITCH RNA is associated with lower PD-L1 protein levels (Fig. 1G; Supplementary Fig. S1A), and their levels are anticorrelated (Supplementary Fig. S1B). Consistent with the T cell–suppressive role of PD-L1, we detected positive correlations between ITCH expression and CD8+ T-cell infiltration levels by analyzing The Cancer Genome Atlas (TCGA) skin melanoma RNA sequencing (RNA-seq) data (Supplementary Fig. S1C). Accordingly, in TCGA skin melanoma (Fig. 1H; Supplementary Fig. S1D) and renal clear-cell carcinoma (Supplementary Fig. S1E), tumors with higher ITCH RNA levels are associated with improved survival, suggesting that higher ITCH protein levels via interaction with and degradation of PD-L1/L2 may enhance CD8+ T-cell activity/persistence and tumor immune surveillance.
ITCH Ubiquitinates and Downregulates Tumor-Surface PD-L1
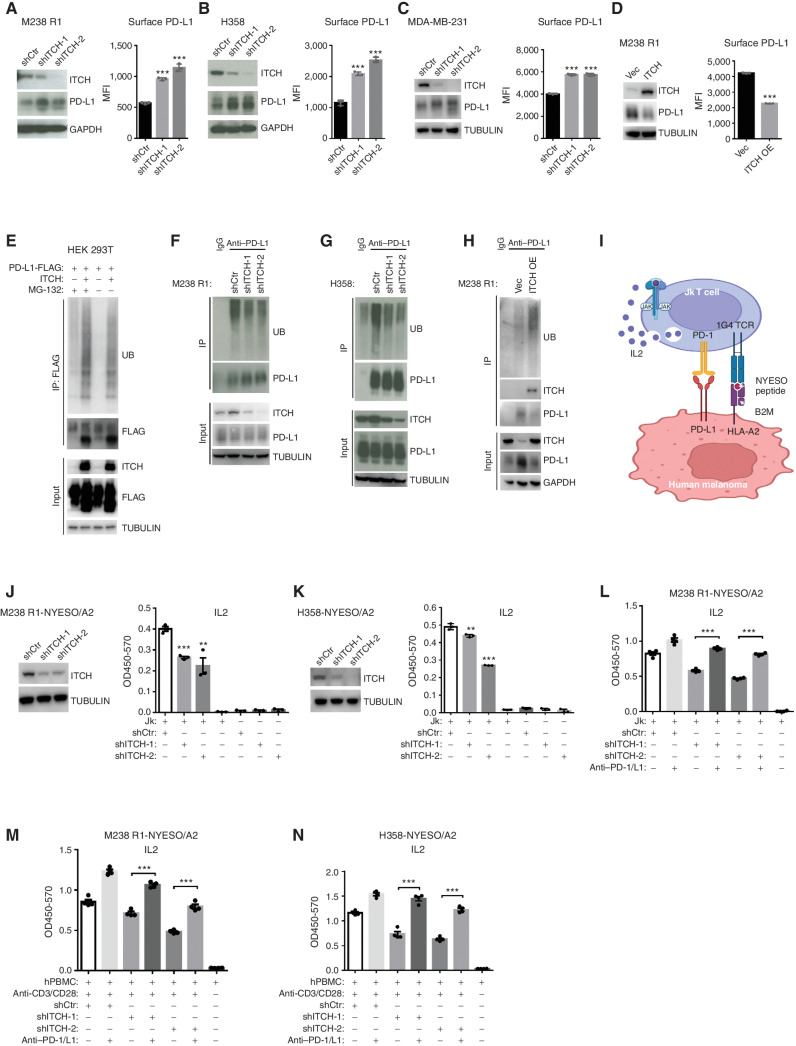
We then experimentally tested the hypothesis that ITCH, as an E3 ligase, downregulates the total and cell-surface levels of PD-L1/L2 in cancer cells via ubiquitination. ITCH knockdown in M238 R1, H358, and MDA-MB-231 (a mesenchymal breast cancer cell line with abundant PD-L1 expression) by two independent short hairpin RNAs (shRNA) increased the total and cell-surface levels of PD-L1 (Fig. 2A–C). ITCH knockdown increased the cell-surface level of PD-L2 to a lesser degree (vs. PD-L1) in M238 R1 and elicited no effect in MDA-MB-231 (PD-L2 is undetectable in H358; Supplementary Fig. S2A). Conversely, ITCH overexpression in M238 R1 reduced PD-L1 and PD-L2 total or cell-surface levels (Fig. 2D; Supplementary Fig. S2B). Measurements by Western blots and flow cytometry were confirmed by the immunofluorescent visualization of PD-L1 with either ITCH knockdown or overexpression (Supplementary Fig. S2C). Moreover, we showed that ITCH overexpression accelerated PD-L1 internalization from the cell surface (Supplementary Fig. S2D) and enhanced PD-L1 colocalization with a lysosomal marker, LAMP-1 (Supplementary Fig. S2E). Furthermore, treatment with a lysosomal inhibitor (chloroquine), but not a proteasome inhibitor (MG-132), rescued downregulation of cell-surface PD-L1 levels (Supplementary Fig. S2F) and total PD-L1 levels (Supplementary Fig. S2G) elicited by ITCH overexpression, suggesting that ITCH-dependent PD-L1 polyubiquitination marks PD-L1 for internalization followed by lysosomal degradation. Using real-time PCR, we showed that ITCH knockdown did not alter the PD-L1 mRNA levels (Supplementary Fig. S2H). Using HEK 293T cells, we observed that cotransfection of ITCH with either PD-L1-FLAG (Fig. 2E) or PD-L2-FLAG (Supplementary Fig. S2I) led to the polyubiquitination of PD-L1 or PD-L2, respectively. Mass spectrometry analysis identified lysine 46 (K46) and K162 of PD-L1 as major ubiquitination sites caused by ITCH overexpression in HEK 293T cells (Supplementary Fig. S2J and S2K). By analyzing ubiquitin branching or linkage patterns, we found that ITCH cotransfection with PD-L1 in HEK 293T cells increased the level of K63-linked ubiquitin (Supplementary Fig. S2L and S2M), which is thought to control protein endocytosis, trafficking, and lysosomal degradation (16). Because PD-L1 and PD-L2 polyubiquitination was readily detectable in this overexpression system, we sought to detect the polyubiquitination of endogenous PD-L1 in M238 R1 and H358. We found that endogenous PD-L1 was polyubiquitinated in both cancer cell lines tested (Fig. 2F and G). Furthermore, knockdown of endogenous ITCH reduced PD-L1 polyubiquitination (Fig. 2F and G), whereas overexpression of ITCH increased PD-L1 polyubiquitination (Fig. 2H). Thus, in cancer cells that display upregulated PD-L1 (and PD-L2) protein levels as the result of a spontaneous mesenchymal phenotype or BRAFV600MUT inhibitor–induced mesenchymal transition/resistance, ITCH mediates polyubiquitination and reduces total and cell-surface protein levels of PD-L1 (and PD-L2).
Figure 2.
ITCH polyubiquitinates PD-L1 and promotes T-cell activation by downregulating tumor cell–surface levels of PD-L1. A–C, Left, Western blots (WB) of M238 R1 (A), H358 (B), or MDA-MB-231 (C) cells stably expressing control shRNA (shCtr) or ITCH-targeting shRNAs (shITCH-1 and shITCH-2). GAPDH or TUBULIN, loading control. Right, cell-surface levels of PD-L1 were measured by cell-surface staining and flow cytometry analysis (right). MFI, mean fluorescence intensity. Mean ± SEMs (n = 3). D, Left, WBs of M238 R1 cells stably expressing empty vector (Vec) or overexpressing (OE) ITCH. TUBULIN, loading control. Right, cell-surface levels of PD-L1 were measured by cell-surface staining and FACS analysis. Mean ± SEMs (n = 3). E, HEK 293T cells transiently expressing PD-L1-FLAG, with or without ITCH cotransfection, were pretreated with or without MG-132 (20 μmol/L) for 4 hours, followed by anti-FLAG immunoprecipitation (IP) and detection of ubiquitin (UB) by WBs. TUBULIN, loading control. F and G, M238 R1 (F) or H358 (G) cells expressing shCtr or ITCH shRNAs were pretreated with MG-132 (20 μmol/L) for 4 hours, followed by anti–PD-L1 IP and UB detection by WBs. TUBULIN, loading control. H, M238 R1 cells expressing Vec or OE ITCH were pretreated with MG-132 (20 μmol/L) for 4 hours followed by anti–PD-L1 IP and then WB detection of UB. GAPDH, loading control. I, Schematic of the coculture assay used in J–L. J and K, Left, WBs of indicated proteins in NYESO-HLA-A2–expressing M238 R1 (J) or H358 (K) cells with shCtr or ITCH shRNA stable expression. TUBULIN, loading control. Right, IL2 production was measured by ELISA after coculture experiments with indicated cell lines. Mean ± SEMs (n = 3). L, IL2 production was measured by ELISA after coculture experiment with indicated cell lines and treatment. Anti–PD-1 and anti–PD-L1 antibodies were added to the culture media at the final concentration of 10 μg/mL (each antibody) and preincubated with Jk T cells for 30 minutes before coculture with target cells. Mean ± SEMs (n = 4). M and N, IL2 production measured by ELISA after coculture of human PBMCs (hPBMC) with M238 R1 (M) or H358 (N) cell lines. Anti-CD3 and anti-CD28 antibodies were added at the final concentration of 1 μg/mL (each antibody) to activate hPBMCs. Anti–PD-1 and anti–PD-L1 antibodies were added at the final concentration of 10 μg/mL (each antibody). Mean ± SEMs (n = 4). P values, Student t test. **, P < 0.01; ***, P < 0.001.
Tumor-Intrinsic ITCH Loss of Function Antagonizes T Cells via PD-L1 Stabilization
To address whether the ITCH–PD-L1 physical and functional interaction constitutes a signaling axis that regulates cancer antigen–specific T cells, we established a coculture assay. Using lentiviral stable transduction, we engineered M238 R1 and H358 to express the cancer testis antigen NY-ESO-1, fused with HLA-A2 and B2M. We then used a Jurkat T-cell line (abbreviated as Jk) engineered with a T-cell receptor (TCR) clonotype (1G4) specific to NYESO-HLA-A2 (Fig. 2I; ref. 17). The fusion of the NYESO peptide with a flexible linker, β2 microglobulin, another flexible linker, and then the heavy chain of HLA-A2 makes up a single-chain trimer that enhances peptide antigen occupancy and presentation to CD8+ T cells (18). We first confirmed that Jk T cells upregulated PD-1 expression after treatment with anti-CD3 antibody or after antigen-specific stimulation by coculture with MAPKi-resistant melanoma cells (Supplementary Fig. S2N). Then, we observed that coculture of M238 R1-NYESO/A2 or H358-NYESO/A2 with Jk T cells, but not cancer cells or Jk T cells alone, resulted in IL2 secretion (Fig. 2J–L). Importantly, ITCH knockdown, which upregulated cancer cell–surface levels of PD-L1 and PD-L2, reduced IL2 secretion (Fig. 2J–L). Moreover, cotreatment with PD-1– plus PD-L1–blocking antibodies completely reversed this drop in IL2 secretion (Fig. 2L), indicating that ITCH regulates CD8+ T-cell activation via ITCH-mediated regulation of PD-L1 (and PD-L2) levels. We confirmed this result by separate coculture assays using, instead of Jk T cells, primary human peripheral blood mononuclear cells (PBMC) activated by anti-CD3 and anti-CD28 antibodies (Fig. 2M and N).
ITCH Loss of Function Accelerates MAPKi Resistance by Immune Evasion
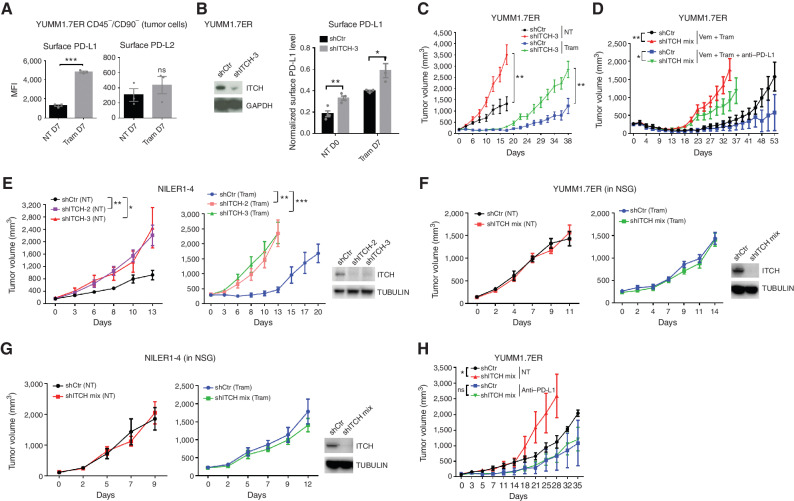
We then evaluated the functional impact of tumor cell–intrinsic ITCH in the context of an immune microenvironment in vivo and of MAPKi-induced PD-L1 upregulation in mouse tumor cells. We used a syngeneic BrafV600E murine melanoma model (YUMM1.7ER) with high mutational burden caused by UV mutagenesis (19). We first showed that 7 days of MEK inhibitor (MEKi; trametinib at 1 mg/kg/day) treatment in vivo upregulated tumor cell–surface PD-L1 protein (Fig. 3A; Supplementary Fig. S3). In this early window of therapy in vivo, we also tested the impact of Itch knockdown on tumor cell–surface levels of PD-L1 (Fig. 3B). We observed that, without treatment, ITCH knockdown upregulated the tumor cell–surface levels of PD-L1 to a level similar to that induced by 7 days of trametinib treatment. Moreover, ITCH knockdown in trametinib-treated tumors further upregulated PD-L1 on the tumor cell surface. Thus, in mouse melanoma cells in vivo, both basal (i.e., no treatment) and therapy-induced levels of tumor cell–surface PD-L1 are subject to ITCH control. Importantly, ITCH knockdown accelerated basal tumor growth, which is consistent with lower ITCH levels being associated with worse survival in patients with cutaneous melanoma (Fig. 1H), as well as the onset of acquired resistance to MEK inhibition (Fig. 3C). Because the standard-of-care MAPKi treatment for patients with BRAFV600MUT melanoma is a combination of BRAFV600MUT inhibitor and MEKi and because the triplet of BRAFV600MUT inhibitor and MEKi plus anti–PD-L1 has been approved clinically (20), we evaluated the effect of tumor cell–intrinsic ITCH knockdown on the onset of acquired resistance (Fig. 3D). ITCH knockdown also accelerated the development of acquired resistance to doublet and triplet therapies (Fig. 3D). To validate these tumor growth and therapy resistance phenotypes, we used another syngeneic murine melanoma model driven by NrasQ61R (NILER1-4) with a high mutational burden caused by UV mutagenesis (12). In accordance with findings in BrafV600E melanoma, we found that ITCH knockdown accelerated both tumor growth without treatment and the development of trametinib (3 mg/kg/day) resistance (Fig. 3E).
Figure 3.
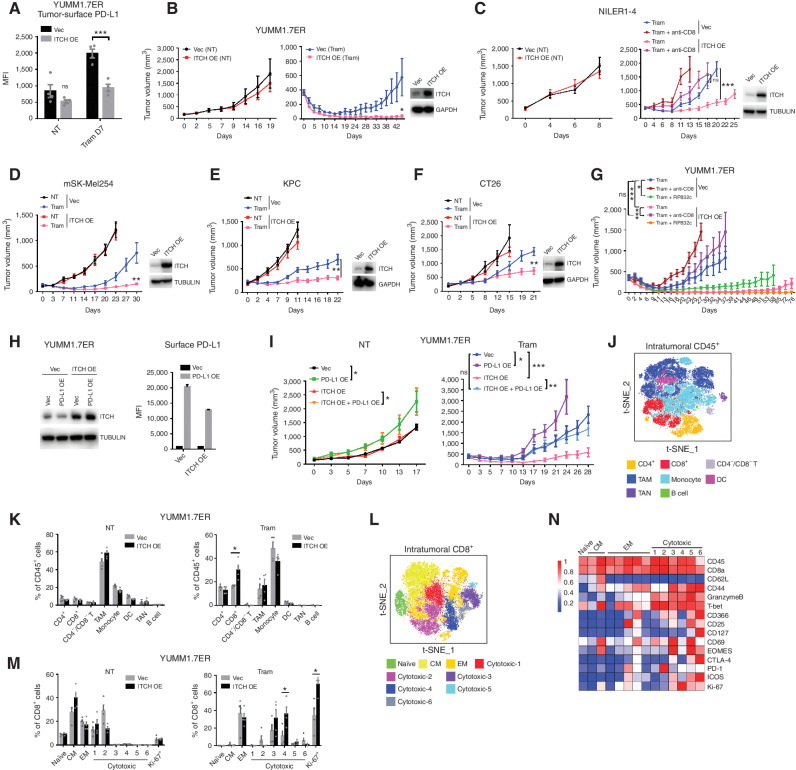
ITCH limits MAPKi-elicited accumulation of tumor cell–surface PD-L1 and suppresses resistance only in immune-competent hosts. A, FACS analysis of YUMM1.7ER tumor-surface PD-L1/L2 levels with or without 7 days of trametinib (1 mg/kg/d) treatment [Tram D7 or no treatment (NT) D7]. Subcutaneously growing tumors were dissociated into single cells. Tumor cells were stained and gated as the CD45/CD90 double-negative population. See Supplementary Fig. S3 for an example of a gating strategy. Mean ± SEMs (n = 3). MFI, mean fluorescence intensity. B, Left, WBs of YUMM1.7ER cells stably expressing control or ITCH shRNA. GAPDH, loading control. Right, FACS analysis of tumor-surface PD-L1 from shCONTROL (shCtr) and ITCH-knockdown (shITCH-3) YUMM1.7ER tumors under NT on D0 or Tram on D7 (1 mg/kg/day). Average PD-L1 expression of the CD45+ population was used as an internal control to normalize measurements of tumor-surface PD-L1 levels on different days. Mean ± SEMs (n = 4 for NT D0 groups and n = 3 for Tram D7 groups). C, Growth curves of shCtr and shITCH-3 YUMM1.7ER tumors under NT or Tram (1 mg/kg/day) treatment in C57BL/6 mice. Mean ± SEMs (n = 6 for NT groups and n = 8 for Tram groups). D, Growth curves of shCtr and ITCH-knockdown (shITCH mix) YUMM1.7ER tumors on the BRAFV600MUT inhibitor vemurafenib (Vem) + Tram treatment or Vem + Tram + anti–PD-L1 (Vem, 50 mg/kg/day; Tram, 0.3 mg/kg/day; anti–PD-L1, 200 μg/mouse twice weekly) in C57BL/6 mice. Mean ± SEMs (n = 7 for Vem + Tram groups and n = 9 for Vem + Tram + anti–PD-L1 groups). shITCH mix, admixture of lentiviruses of shITCH-2 and shITCH-3 to achieve higher knockdown efficiency. E, Left or middle, growth curves of shCtr and shITCH-mix NILER1-4 tumors on NT or Tram (3 mg/kg/d) treatment in C57BL/6 mice. Mean ± SEMs (n = 6 for NT groups and n = 10 for Tram groups). Right, WBs of the stable cell lines used for tumor engraftment. TUBULIN, loading control. F and G, Left or middle, growth curves of shCtr and shITCH-mix YUMM1.7ER (F) or NILER1-4 (G) tumors on NT or Tram (1 or 3 mg/kg/day) treatment in immune-deficient NSG mice. Mean ± SEMs (n = 7 for NT groups and n = 8 for Tram groups). Right, WBs of the stable cell lines used for tumor engraftment. TUBULIN, loading control. H, Growth curves of shCtr and shITCH-mix YUMM1.7ER tumors on NT or anti–PD-L1 treatment in C57BL/6 mice. Treatment started at tumor size ∼50 mm3. Mean ± SEMs (n = 6 for NT groups and n = 8 for anti–PD-L1 groups). P values, Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
The tumor growth and therapy resistance phenotypes resulting from ITCH knockdown are dependent on T, B, or natural killer (NK) cells, as we observed no differences in the growth curves of either YUMM1.7ER or NILER1-4 tumors, with or without trametinib treatment (1 or 3 mg/kg/day, respectively), in NOD scid gamma (NSG) mice (Fig. 3F and G). In addition, the tumor growth and therapy resistance phenotypes resulting from ITCH knockdown are not tumor cell–intrinsic, as we observed no differences in the short-term and long-term growth rates of the YUMM1.7ER or NILER1-4 cell line in vitro without or with trametinib treatment (Supplementary Fig. S4A and S4B). Because tumor cell–intrinsic ITCH knockdown accelerated tumor growth through tumor cell–extrinsic or immune mechanisms, we tested whether this effect is due to the induction of tumor cell–surface PD-L1 levels in vivo. To do so, we treated syngeneic tumor-bearing mice with an anti–PD-L1 antibody (without cotreatment with a MEKi; Fig. 3H) to neutralize PD-L1 tumor cell–surface upregulation mediated by tumor cell–intrinsic ITCH knockdown (Fig. 3B). As expected, ITCH knockdown and anti–PD-L1 treatment, respectively, accelerated and decelerated YUMM1.7ER tumor growth (Fig. 3H). Importantly, in the presence of both tumor cell–intrinsic ITCH knockdown and anti–PD-L1 treatment, we abolished the ability of ITCH knockdown to accelerate tumor growth in an immune-competent host (Fig. 3H).
Tumor-Intrinsic ITCH Loss of Function Creates a Protumorigenic Immune Microenvironment
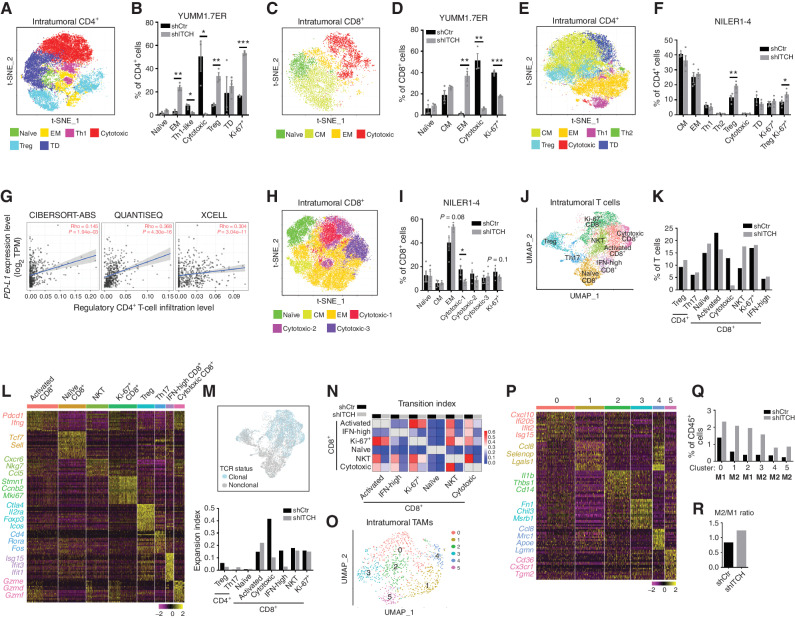
To investigate ITCH knockdown–induced changes in the tumor immune microenvironment, we profiled YUMM1.7ER shCONTROL and ITCH-knockdown tumors after 14 days of trametinib treatment (1 mg/kg/day) by performing cytometry by time of flight (CyTOF; Supplementary Fig. S5A). Analysis of CyTOF data from the dissociated tumors (n = 3 per group) showed that ITCH-knockdown (vs. shCONTROL) tumors harbored less CD4+ T-cell infiltration (Supplementary Fig. S5B). Subclustering analysis of intratumoral CD4+ T cells revealed that ITCH-knockdown tumors contained higher fractions of regulatory CD4+ T cells (Treg; CD4+FOXP3+) and effector/effector memory CD4+ T cells (EM; CD4+CD62L−CD44+) but lower fractions of cytotoxic CD4+ T cells (CD4+Granzyme B+) and T helper 1 (Th1)–like CD4+ T cells (CD4+T-bet+; Fig. 4A and B). Tregs also showed the highest Ki-67 expression across different CD4+ T-cell subclusters, indicating more proliferative Tregs in ITCH-knockdown tumors early on treatment (Supplementary Fig. S5C). Analysis of intratumoral CD8+ T cells showed that ITCH-knockdown tumors harbored much fewer cytotoxic CD8+ T cells (CD8+Granzyme B+) but more effector/EM CD8+ T cells (CD8+CD62L−CD44+). Also, the fraction of Ki-67+, proliferative CD8+ T cells was much lower in ITCH-knockdown (vs. shCONTROL) tumors (Fig. 4C and D; Supplementary Fig. S5D). We also used CyTOF to profile NILER1-4 shCONTROL versus ITCH-knockdown tumors 5 days after trametinib (3 mg/kg/day) treatment (Supplementary Fig. S5E). CyTOF analysis of dissociated NILER1-4 tumors (n = 4 per group) also showed that ITCH-knockdown (vs. shCONTROL) tumors contained higher fractions of regulatory CD4+ T cells and proliferating regulatory CD4+ T cells (Fig. 4E and F; Supplementary Fig. S5F). Tumor or antigen-presenting cells’ expression of PD-L1 has been shown to convert naïve CD4+ or Th1 cells to regulatory or suppressor CD4+ T cells (21, 22). Consistently, from an analysis of 471 clinical cutaneous melanoma samples using TIMER2.0, intratumoral PD-L1 expression was positively correlated with regulatory CD4+ T-cell infiltration (Fig. 4G). Analysis of intratumoral CD8+ T cells shows that ITCH-knockdown NILER1-4 tumors harbored fewer cytotoxic CD8+ T cells in general and cluster 1 (cytotoxic-1) in particular (Fig. 4H and I; Supplementary Fig. S5G). Also, the fraction of Ki-67+ CD8+ T cells trended lower in ITCH-knockdown (vs. shCONTROL) NILER1-4 tumors. Thus, CyTOF data derived from two murine melanoma models support the concept that tumor cell–expressed ITCH during MAPKi therapy suppresses the expansion of intratumoral regulatory CD4+ T cells and promotes the expansion of cytotoxic CD8+ T cells. These immune cellular effects are consistent with the impact of ITCH knockdown in upregulating PD-L1 tumor cell–surface levels in MAPKi-treated melanoma (Figs. 1–3) and in accelerating the development of MAPKi resistance only in immune-competent hosts (Fig. 3).
Figure 4.
Immune impacts of tumor-intrinsic ITCH entail CD8+ T cell–mediated suppression of MAPKi resistance. A, T-distributed stochastic neighbor embedding (t-SNE) map of intratumoral CD4+ T cells from shCONTROL and ITCH-knockdown tumors on trametinib treatment. YUMM1.7ER tumors were analyzed by CyTOF. Inferred cell types are indicated by clusters with distinct colors. TD, terminally differentiated. B, Fractions of indicated cell types in total CD4+ T cells from shCONTROL and ITCH-knockdown tumors (YUMM1.7ER), both on trametinib treatment. Mean ± SEMs (n = 3). C, As in A, except intratumoral CD8+ T cells. CM, central memory. D, As in B, except total CD8+ T cells. E, As in A, except n = 4 (NILER1-4). F, As in B, except n = 4 (NILER1-4). G, Spearman correlation score (Rho) between intratumoral PD-L1 expression and Treg infiltration was calculated by three different algorithms (CIBERSORT-ABS, QUANTISEQ, and XCELL). Rho > 0, positive correlation. P values, as indicated. TPM, transcripts per million. H, As in A, except intratumoral CD8+ T cells from NILER1-4 tumors (n = 4). I, As in B, except total CD8+ T cells and n = 4 (NILER1-4). J, Uniform manifold approximation and projection (UMAP) of intratumoral T cells (n = 8,405) analyzed by scRNA-seq (shCONTROL and ITCH-knockdown NILER1-4 tumors on trametinib treatment). Different cell clusters are denoted by distinct colors. NKT, natural killer T cell. K, Fractions of indicated cell types in total T cells from shCONTROL and ITCH-knockdown NILER1-4 tumors. L, Heat map showing expression levels of differentially expressed genes (rows) among different T-cell subpopulations (columns) in NILER1-4 tumors. Representative genes of each cluster are highlighted. M, UMAP in J colored by clonality based on scTCR-seq analysis (top). Clonal expansion indices of T-cell subpopulations (bottom) in NILER1-4 tumors. N, Transition indices between indicated CD8+ T-cell subpopulations in NILER1-4 tumors. O, UMAP of intratumoral TAMs (n = 991) in NILER1-4 tumors analyzed by scRNA-seq. Different cell clusters are denoted by distinct colors. P, As in L, except TAM subpopulations in NILER1-4 tumors. Q, Fractions of each TAM subpopulation in total CD45+ cells from shCONTROL and ITCH-knockdown NILER1-4 tumors, both on trametinib treatment. R, The ratios of M2-like TAMs to M1-like TAMs in shCONTROL and ITCH-knockdown NILER1-4 tumors, both on trametinib treatment. P values, Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To dissect the impact of ITCH knockdown on the tumor immune microenvironment comprehensively, we performed an analysis of single-cell RNA sequencing (scRNA-seq) data coupled to single-cell TCR sequencing (scTCR-seq) data to evaluate deeper immune phenotypic alterations in shCONTROL vs. ITCH-knockdown NILER1-4 tumors early on MEKi treatment (Supplementary Fig. S5E). Four tumors per group were dissociated and combined into one sample to sort for the CD45+ population. A total of 15,532 CD45+ cells were identified in scRNA-seq data analysis, and immune cell clusters were annotated by key lineage markers (Supplementary Fig. S6A and S6B). ITCH-knockdown tumors showed higher fractions of tumor-associated macrophages (TAM) and neutrophils (TAN) but a lower fraction of T cells among CD45+ cells (Supplementary Fig. S6C). Subclustering of the T-cell population identified eight subpopulations based on differentially expressed genes (Fig. 4J–L). Consistent with CyTOF-based findings, ITCH-knockdown tumors harbored a higher fraction of regulatory CD4+ T cells (cluster 4) but lower fractions of activated and cytotoxic CD8+ T cells (Fig. 4K and L). Moreover, regulatory CD4+ T cells in ITCH-knockdown tumors were more proliferative and less exhausted with higher Mki67 and lower Lag3 expression (Supplementary Fig. S6D). In addition, in ITCH-knockdown tumors, CD8+ T cells were less active and cytotoxic based on the lower expression of Ifng, Pdcd1, Lag3, Prf1, Gzmb, Klrd1, and Klrc1 (Supplementary Fig. S6D). From single-cell TCR sequencing (scTCR-seq) data analysis, we identified a total of 1,724 TCR clonotypes with unique α and β chain pairs. Four hundred twenty-four of these clonotypes were represented by two or more cells, which defined 3,248 clonal T cells (Fig. 4M). We then analyzed the clonal expansion of different T-cell subpopulations and observed that shITCH (vs. shCONTROL) tumors harbored less expansion of cytotoxic and IFNhi CD8+ T cells (Fig. 4M). Additional analysis revealed a consistent pattern in shITCH (vs. shCONTROL) tumors of lower transition indices between pairs of CD8+ T-cell subpopulations, indicating that reduced ITCH expression in tumor cells blunted phenotypic conversions among functional T-cell subsets and blocked naïve T-cell activation (Fig. 4N).
Because the TAM population nearly tripled in size with ITCH knockdown, we defined six subclusters based on differential gene expression (Fig. 4O and P). We identified clusters 0 and 2 as M1-like TAMs because of high expression of proinflammatory cytokines and/or M1 markers such as Cxcl10, Ifi205, Il1b, and Thbs1. Clusters 1, 3, 4, and 5 were identified as M2-like TAMs, as they expressed highly anti-inflammatory or protumorigenic cytokines and/or M2 markers such as Ccl8, Selenop, Fn1, Chil3, Mrc1, Apoe, Lgmn, Tgm2 (Fig. 4P). Importantly, we found that ITCH-knockdown tumors contained not only higher fractions of every TAM subpopulation (Fig. 4Q) but also a higher M2-to-M1 ratio (Fig. 4R). Furthermore, analysis of gene expression levels showed that TAM subpopulations in ITCH-knockdown tumors tend to express lower levels of proinflammatory cytokines such as Il1b and Tnf and cytotoxic genes such as Gzmb and Prf1 but higher levels of M2 macrophage markers such as Mrc1, Cd209a, and Chil3 as well as protumorigenic cytokines such as Ccl6, Ccl8, Ccl9, Tgfb1, and Tgfbi (Supplementary Fig. S6E). To validate the modulation in TAMs by ITCH knockdown observed early during MAPKi treatment in NILER1-4 tumors, we analyzed scRNA-seq data derived from whole YUMM1.7ER tumors (i.e., non-CD45+–sorted; Supplementary Fig. S5A). We were able to detect 886 TAMs in total. Reclustering of the TAM population identified six subclusters (Supplementary Fig. S6F and S6G). We defined clusters 2 and 5 as M1-like TAMs based on their expression of proinflammatory cytokines and M1-polarization–related genes such as Cxcl9, Cxcl10, Malat1, and Neat1. Clusters 0, 1, 3, and 4 were identified as M2-like TAMs based on the expression of anti-inflammatory cytokines or protumorigenic cytokines and M2 markers such as Mgl2, Ccl9, Cxcl2, Lgmn, Selenop, Ccl5, Ccl8, Apoe, Chil3 (Supplementary Fig. S6G). Consistent with findings from the NILER1-4–derived scRNA-seq data, ITCH knockdown resulted in much higher levels of M2-like TAMs (clusters 0, 1, 3) within early-on MAPKi-treated YUMM1.7ER tumors (Supplementary Fig. S6H). The ratio of M2 to M1 TAMs in ITCH-knockdown tumors was ∼2-fold higher than in shCONTROL tumors (Supplementary Fig. S6I). Finally, TAM clusters in ITCH-knockdown tumors tended to express lower levels of proinflammatory cytokines (Il1b and Tnf) and cytotoxic genes (Prf1 and Gzmb) but higher levels of the M2 macrophage marker Mrc1 and protumorigenic cytokines (Ccl6, Ccl8, and Ccl9; Supplementary Fig. S6J).
ITCH Suppresses MAPKi Resistance by PD-L1 Degradation and CD8+ T-cell Expansion
Based on the above findings, we hypothesized that ITCH overexpression in tumor cells should suppress acquired MAPKi resistance. We engineered YUMM1.7ER cells to express empty vector or harbor ITCH overexpression; these two sublines grew at indistinguishable rates in vitro with or without MEKi (trametinib) treatment (Supplementary Fig. S7A). After implanting these two cell lines into syngeneic mice and allowing tumors to grow exponentially (reaching ∼400 mm3), flow cytometry analysis of tumors with no treatment versus trametinib treatment (0.45 mg/kg/day × 7 days) showed that, as expected (Fig. 3A), in vivo MEK inhibition induced the tumor cell–intrinsic surface level of PD-L1 and, importantly, ITCH overexpression reduced the tumor cell–intrinsic surface level of PD-L1 (after MEKi treatment) to the level observed in treatment-naïve tumors (Fig. 5A). Furthermore, ITCH overexpression in YUMM1.7ER tumor cells did not inhibit tumor growth in the absence of MAPKi treatment (Fig. 5B), likely because basal PD-L1 expression is already low (Fig. 5A). However, as hypothesized, after MAPKi treatment of established (∼400 mm3) tumors, ITCH overexpression strongly suppressed resistance development (Fig. 5B). At the last follow-up (day 60) of MAPKi-treated tumors with ITCH overexpression, we did not observe any case of acquired resistance, with 6 of 10 tumors displaying complete responses. We tested this hypothesis further in four additional syngeneic tumor models with variable levels of trametinib responsiveness (13); these included NILER1-4 (NrasMUT melanoma), mSK-Mel254 (Nf1−/− melanoma), KPC [KrasMUT pancreatic ductal adenocarcinoma (PDAC)], and CT26 (KrasMUT colorectal carcinoma; Fig. 5C–F). In every additional model tested, tumor cell–intrinsic ITCH overexpression suppressed acquired MAPKi resistance.
Figure 5.
ITCH suppresses MAPKi resistance by PD-L1 degradation and CD8+ T-cell expansion. A, FACS analysis of YUMM1.7ER [vector-only (VEC); ITCH overexpression (OE)] tumor-surface PD-L1 levels with or without 7 days of trametinib (0.45 mg/kg/day) treatment [Tram D7 or no treatment (NT)]. Subcutaneously growing tumors were dissociated into single cells. Tumor cells were stained and gated as the CD45/CD90 double-negative population. Mean ± SEMs (n = 4). MFI, mean fluorescence intensity. B, Left and middle, growth curves of Vec and ITCH OE YUMM1.7ER tumors on NT or Tram (0.45 mg/kg/day) treatment in C57BL/6 mice. Mean ± SEMs (n = 8 for NT and n = 10 for Tram groups). Right, Western blots (WB) of the stable cell lines used for tumor engraftment. GAPDH, loading control. C, Left and middle, growth curves of Vec and ITCH OE NILER1-4 tumors on NT or Tram (3 mg/kg/day), alone or in combination with anti-CD8 (200 μg per mouse twice weekly) treatment, in C57BL/6 mice. Mean ± SEMs (n = 7–10). Right, WBs of the stable cell lines used for tumor engraftment. TUBULIN, loading control. D, Left, growth curves of Vec and ITCH OE mSK-Mel254 tumors on NT or Tram (3 mg/kg/day) treatment in C57BL/6 mice. Mean ± SEMs (n = 8–10). Right, WBs of the stable cell lines used for tumor engraftment. TUBULIN, loading control. E, Left, growth curves of Vec and ITCH OE KPC tumors on NT or Tram (3 mg/kg/day) treatment in C57BL/6 mice. Mean ± SEMs (n = 10). Right, WBs of the stable cell lines used for tumor engraftment. GAPDH, loading control. F, Left, growth curves of Vec and ITCH OE CT26 tumors on NT or Tram (5 mg/kg/day) treatment in BALB/c mice. Mean ± SEMs (n = 9–11). Right, WBs of the stable cell lines used for tumor engraftment. GAPDH, loading control. G, Growth curves of Vec and ITCH OE YUMM1.7ER tumors on Tram (0.45 mg/kg/day), alone or in combination with anti-CD8 (200 μg per mouse twice weekly) or RP832c (10 mg/kg/day, D0–D7) treatment, in C57BL/6 mice. Mean ± SEMs (n = 8 for Tram and n = 10 for Tram + anti-CD8 and Tram + RP832c groups). H, YUMM1.7ER Vec and ITCH OE stable lines were engineered to express Vec or PD-L1 (PD-L1 OE). WBs (left) and FACS analysis (right) of the total and cell-surface PD-L1 of the indicated double-transduced stable lines. TUBULIN, loading control. I, Growth curves of engrafted tumors (derived from stable lines in H) on NT or Tram (0.45 mg/kg/day) treatment in C57BL/6 mice. Mean ± SEMs (n = 7–10). J, T-distributed stochastic neighbor embedding (t-SNE) map of intratumoral CD45+ cells from Vec and ITCH OE YUMM1.7ER tumors on NT and trametinib treatment and analyzed by CyTOF. Inferred cell types are denoted by distinct colors. DC, dendritic cell. K, Fractions of indicated cell types in CD45+ cells from Vec and ITCH OE YUMM1.7ER tumors on NT and trametinib treatment. Mean ± SEMs (n = 4). L, As in J, except for CD8+ T cells. CM, central memory. M, As in K, except for CD8+ T cells. N, Heat map showing scaled mean expression levels of indicated protein markers in different cell clusters of CD8+ T cells in YUMM1.7ER tumors. P values, Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
Based on prior in vitro and in vivo analyses of an ITCH–PD-L1–T-cell regulatory axis (Figs. 2–4), we also hypothesized that suppression of acquired MAPKi resistance by ITCH overexpression requires CD8+ T cells. Importantly, in both syngeneic models tested, systemic CD8 neutralization and depletion of CD8+ T cells completely rescued or reversed the suppression of MEKi resistance caused by tumor-intrinsic ITCH overexpression (Fig. 5C and G). Moreover, as tumor cell–intrinsic ITCH knockdown led to M2 TAM polarization (Fig. 4; Supplementary Fig. S6), we tested whether targeting M2-like TAMs by a peptide agonist of CD206 (RP832c) could suppress trametinib resistance (13, 23). Although it was difficult to discern an added resistance-suppressive effect of RP832c on ITCH overexpressing tumors, RP832c treatment (dosed daily over the first 7 days) phenocopied the resistance-suppressive effect of ITCH overexpression (Fig. 5G). Prior analyses supported the notion that ITCH modulates T-cell functions in vitro (Fig. 2J–N) or tumor growth in immune-competent hosts (Fig. 3H) via regulating PD-L1. To support this concept further, we hypothesized that tumor cell–intrinsic overexpression of PD-L1 on top of ITCH overexpression (Fig. 5H) in YUMM1.7ER cells would nullify the resistance-suppressive phenotype of ITCH overexpression. Indeed, while ITCH overexpression suppressed acquired trametinib resistance, combined overexpression of ITCH and PD-L1 reversed the ITCH-mediated resistance-suppressive phenotype and restored the resistance kinetics to that observed without ITCH overexpression (Fig. 5I).
Next, we performed CyTOF to analyze the tumor immune microenvironments before and after MAPKi treatment of mice-bearing tumors without or with tumor-intrinsic ITCH overexpression (n = 4 tumors per group; Supplementary Fig. S7B). CD45+ immune cells were gated out for subclustering analysis according to their expression of immune cell lineage markers. Eight different cell populations were identified, and we found that the fraction of CD8+ T cells in the CD45+ cell population under trametinib treatment was higher in tumors with ITCH overexpression. (Fig. 5J and K; Supplementary Fig. S7C). Subclustering analysis of the CD8+ T-cell population identified nine distinct functional subpopulations. On trametinib treatment (but not before treatment), ITCH overexpression enhanced CD8+ T-cell proliferation (fraction of Ki-67+ cells) and increased the fraction of the most proliferative, cytotoxic CD8+ T cells (cluster cytotoxic-4, Ki-67 highest; Fig. 5L–N). Subclustering analysis of the CD4+ T-cell population showed 6 different subpopulations, and ITCH overexpression also increased CD4+ T-cell proliferation (fraction of Ki-67+ cells; Supplementary Fig. S7D–S7F). These CyTOF findings mirror our previous findings with tumor cell–ITCH knockdown (Fig. 4) and the positive clinical correlation between ITCH expression and CD8+ T-cell infiltration (Supplementary Fig. S1C). These CyTOF findings also support the in vivo phenotype of tumor-intrinsic ITCH overexpression, namely, MAPKi-resistance suppression that is dependent on PD-L1 downregulation (Fig. 5I) and CD8+ T-cell expansion (Fig. 5C and G).
Identification of a Small-Molecular ITCH Activator that Suppresses Acquired MAPKi Resistance
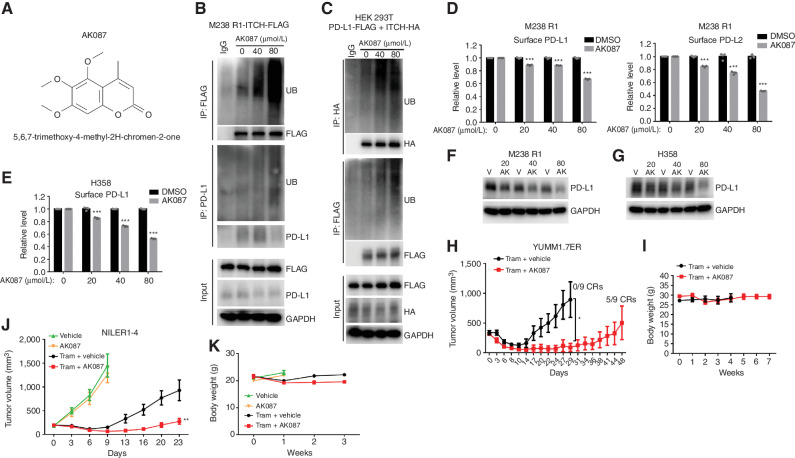
We then sought to provide in vivo proof-of-principle data supportive of the translational potential of our findings by identifying a small-molecular ITCH activator. First, we queried a small-molecule screen intended to identify ITCH inhibitors (24) for potential ITCH activators. One compound, AK087 (Fig. 6A), displayed an ability to enhance (140%) the autoubiquitination of ITCH in an assay of reconstituted E1, E2, E3 (ITCH), and ubiquitin (tagged with horseradish peroxidase) recombinant proteins. AK087 dose dependently enhanced the poly/autoubiquitination of ITCH-FLAG expressed in a human melanoma cell line adapted to BRAF inhibition (M238 R1) while enhancing the polyubiquitination of endogenous PD-L1 (Fig. 6B). In human HEK 293T cells coexpressing PD-L1-FLAG and ITCH-HA, we observed similar findings (Fig. 6C). Moreover, in human melanoma cells with upregulated surface PD-L1/L2 (M238 R1) and human lung cancer cell line with upregulated surface PD-L1 (H358), AK087 dose dependently reduced the tumor cell–surface (Fig. 6D and E) and total (Fig. 6F and G) levels of PD-L1/L2. Lastly and importantly, AK087 cotreatment (dosed subcutaneously at 10 mg/kg/day for 17 days) with trametinib suppressed acquired MAPKi resistance in mice bearing YUMM1.7ER melanoma tumors, which was associated with five of nine complete responses (vs. zero of nine complete responses without AK087 cotreatment) and no appreciable body weight loss or superficial evidence of toxicities (Fig. 6H and I). We observed consistent findings using AK087 in the NILER1-4 melanoma model (Fig. 6J and K).
Figure 6.
AK087 is an ITCH activator that downregulates tumor cell–surface PD-L1/L2 and suppresses MAPKi resistance in vivo. A, Structure and chemical name of AK087. B, M238 R1 cells with ITCH-FLAG overexpression were treated with 0, 40, or 80 μmol/L of AK087 for 6 days and then treated with MG-132 (20 μmol/L) for 4 hours, followed by anti-FLAG or anti–PD-L1 immunoprecipitation (IP) and Western blot (WB) detection of indicated proteins [ubiquitin (UB)]. GAPDH, loading control. C, HEK 293T cells cotransfected with PD-L1-FLAG and ITCH-HA were treated with 0, 40, or 80 μmol/L of AK087 for 6 days and then treated with MG-132 (20 μmol/L) for 4 hours, followed by anti-HA or anti-FLAG IP and WB detection of indicated proteins. GAPDH, loading control. D, M238 R1 cell-surface levels of PD-L1 (left) and PD-L2 (right) as measured by cell-surface staining and FACS analysis after 0, 20, 40, or 80 μmol/L AK087 treatment for 6 days. Mean ± SEMs (n = 3). E, As in D, except for H358 (PD-L2 is undetectable in H358). F and G, WBs of total PD-L1 protein levels in M238 R1 (F) and H358 (G) cells after 6 days of treatment with vehicle (V or DMSO) or 20, 40, or 80 μmol/L of AK087 (AK). GAPDH, loading control. H, Growth curves of YUMM1.7ER tumors on trametinib (Tram; 0.45 mg/kg/day) in combination with daily vehicle (4% Tween80 + 8% DMSO in double-distilled water) or AK087 (10 mg/kg/day, from D0–D17) treatment in C57BL/6 mice. Mean ± SEMs (n = 9). CR, complete response. I, Weekly body weights of mice in H. J, Growth curves of NILER1-4 tumors on daily vehicle (4% Tween80 + 8% DMSO in double-distilled water), AK087 (10 mg/kg/day, from D0–D9), Tram (2 mg/kg/day) in combination with daily vehicle or AK087 (10 mg/kg/day, from D0–D18) treatment in C57BL/6 mice. Mean ± SEMs (n = 9–10). K, Weekly body weights of mice in J. P values, Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
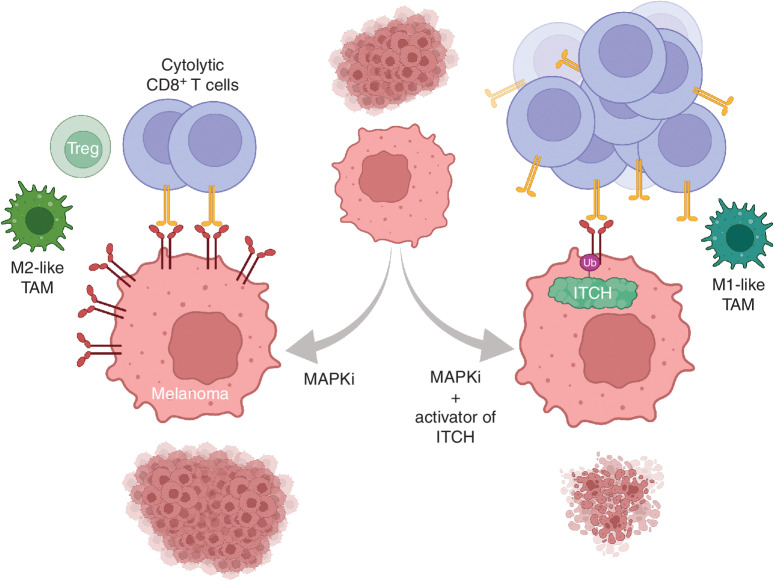
Harnessing antitumor immunity based on the release of a key immune-checkpoint interaction between PD-L1 and PD-1 has revolutionized oncology therapy. The PD-L1 expression level in tumor cells is highly dynamic and regulated as concerted responses to alterations in tumor-intrinsic cell states and immunologic cues. Regulation at posttranslational levels can occur via ubiquitination, compartmentalization, glycosylation, palmitoylation, and phosphorylation. The current study supports a model (Fig. 7) in which the E3 ligase ITCH mediates the polyubiquitination of PD-L1 and downregulates tumor cell–surface PD-L1 levels (via ubiquitin-directed lysosomal degradation) in MAPKi-adapted melanoma cells and potentially in other biological contexts such as quasi-mesenchymal tumor cell states that contribute to therapy resistance and metastatic potential. Our prior study (3) demonstrated PD-L1/L2 upregulation as a recurrent and early MAPKi response in clinical melanoma as well as in cell line and in vivo syngeneic models of melanoma. By downregulating surface PD-L1 in MAPKi-treated melanoma, tumor cell–intrinsic ITCH promotes tumor immune surveillance by CD8+ T cells and may also ameliorate immune-suppressive microenvironmental features such as protumorigenic macrophages and Treg differentiation (Fig. 7). Hence, we nominate tumor cell–surface PD-L1 stability and tumor-intrinsic ITCH activity as MAPKi cotargets. We propose to develop pharmacologic strategies to destabilize cell-surface PD-L1 and/or to activate ITCH function within melanoma cells in order to develop combinatorial strategies to prevent adaptive immune resistance and thereby acquired MAPKi resistance.
Figure 7.
Proposed combinatorial strategy to reduce immune-mediated MAPKi resistance. MAPKi therapy of melanoma elicits tumor cell–surface PD-L1/L2 accumulation, which evades tumor antigen–specific cytolytic CD8+ T cells and potentially alters the phenotype or differentiation of intratumoral immune cell types such as Tregs and TAMs. This immune evasion or immune-suppressive tumor microenvironment reduces the durability of MAPKi responses, especially in tumors with high mutational or neoantigen burdens. ITCH, as an E3 ligase that ubiquitinates tumor cell–surface PD-L1/L2 and targets them for internalization and lysosomal degradation, can be activated pharmacologically during the early phase of MAPKi therapy to enhance tumor rejection by cytolytic CD8+ T cells. Subsequent immunologic memory may suppress acquired MAPKi resistance driven by nonimmune or genetic mechanisms. Strategies alternative to ITCH activation may involve proteolysis-targeting chimeras against PD-L1/L2 or depletion of Tregs or M2-like TAMs.
What immunologic factors dictate the durability of clinical responses to MAPKi remain ill-defined. The ways in which syngeneic murine melanoma, in response to MAPKi therapy, undergoes “immunogenic” cell death (25) and evades antitumor immunity (this study) directly regulate the durability of MAPKi responses, as do tumor mutational burden (this study and our prior study of NrasMUT melanoma; ref. 12). MAPKi therapy in BrafV600MUT melanoma (without mutational burden) elicits pyroptotic cell death and tumor necrosis. The release of the chromatin protein HMGB1 by necrotic cells can trigger protumorigenic inflammation associated with TAMs (26, 27). Thus, our additional findings that ITCH loss of function leads to M2-like TAM polarization and that pharmacologic targeting of M2-like TAMs phenocopies ITCH gain of function (in suppressing resistance) suggest an alternative immune-based strategy to enhance the durability of MAPKi responses. ITCH loss of function also induces the levels of Tregs. As pharmacologic strategies targeting Tregs advance to the clinic (28, 29), the repertoire of immune-based strategies to enhance the durability of MAPKi responses may also expand.
The simultaneous combination of anti–PD-L1 with a BRAFV600MUT inhibitor and MEKi (so-called “triplet” therapy), in early clinical data, appears beneficial and has been approved for patients with BRAFV600MUT melanoma (20). Retrospective clinical data analysis and in vivo therapeutic modeling showed that a lead-in regimen of anti–PD-1/L1 (± anti–CTLA-4) before MAPKi combination augments the efficacy of triplet therapy by enhancing MAPKi durability (and overcoming innate resistance to immune-checkpoint blockade; ref. 13). In syngeneic melanoma models, this sequential combinatorial regimen appears to maximize antitumor immunity by remodeling a similar network of innate and adaptive immune cells that is downstream of tumor cell–intrinsic ITCH overexpression during the early window of MAPKi therapy.
A still maturing but promising area of cancer therapeutics development lies in proteolysis-targeting chimeras (PROTAC) that induce targeted protein degradation by the ubiquitin–proteasome pathway (30). PROTACs serve as bridges that bring together a protein being targeted for degradation and a non-native or nonphysiologic E3 ligase. Here, we provided a proof-of-concept alternative approach of using a small-molecular E3 ligase activator to target a natural E3 substrate for degradation. MAPKis remain noncurative for a large portion of patients with BRAFV600MUT cutaneous melanoma and experimental for the majority of patients with MAPK-addicted cancers (e.g., KRASMUT PDAC). Thus, direct or indirect PD-L1 degraders should be codeveloped with MAPKi and, potentially, immune-checkpoint inhibitor therapies.
METHODS
Cell Lines
All human and mouse cancer cell lines were routinely tested for Mycoplasma and profiled and identified by RNA-seq and the GenePrint 10 System (Promega, B9510) at periodic intervals during the course of this study. Early passages of cells stored in liquid nitrogen were thawed and used for experiments within a month of in vitro culture. The human cell lines HEK 293T (ATCC, CRL-3216, RRID:CVCL_0063), M229 R5 (RRID:CVCL_IM73), M238 R1 (RRID:CVCL_IM74), H358 (ATCC, CRL-5807, RRID:CVCL_1559), and MDA-MB-231 (ATCC, CRM-HTB-26, RRID:CVCL_0062) and mouse cell lines mSK-Mel254, KPC, and CT26 (ATCC, CRL-2639, RRID:CVCL_7254) were maintained in high-glucose DMEM (Omega Scientific, DM-22) with 10% heat-inactivated FBS (Omega Scientific, FB-02) and 2 mmol/L glutamine. PLX4032 (1 μmol/L; LC Laboratories, LC-V-2800) was added in the culture medium of M229 R5 and M238 R1. The YUMM1.7ER cell line was maintained in DMEM/F12 with 10% heat-inactivated FBS and 2 mmol/L glutamine. The NILER1-4 cell line was cultured in high glucose DMEM with 20% heat-inactivated FBS and 2 mmol/L glutamine. Jk T cells were cultured in the RPMI 1640 medium (Gibco, 21875034) with 10% heat-inactivated FBS, 10 mmol/L HEPES, 1 mmol/L sodium pyruvate, 2 mmol/L glutamine, and 50 μmol/L beta-mercaptoethanol. Human PBMCs (ATCC, PCS-800-011) were maintained in the RPMI 1640 medium with 10% heat-inactivated FBS, 10 mmol/L HEPES, 1 mmol/L sodium pyruvate, and 2 mmol/L glutamine. All cell lines were maintained in a humidified, 5% CO2 incubator.
Constructs and Engineered Cell Lines
For overexpression constructs, cDNAs of human and mouse PD-L1, PD-L2, and/or ITCH were subcloned into the lentivirus vector cPPT-puro (Addgene, 12252, RRID:Addgene_12252; GFP replaced with puromycin resistance gene), with or without C-terminal tag (FLAG or HA) as indicated. The APEX2 coding sequence was fused to the C-terminus of human PD-L1 with the linker peptide GGGGSGGGGS and subcloned into the lentivirus vector pLV-puro (Addgene, 85132, RRID:Addgene_85132). shRNAs were constructed into the lentivirus vector pLKO.1-puro (Addgene, 8453, RRID:Addgene_8453). Stable lines were selected by adding 10 μg/mL puromycin in the culture medium 48 hours after lentiviral infection. shRNA-targeting sequences are as follows:
Human ITCH sh1: CGAAGACGTTTGTGGGTGATT
Human ITCH sh2: GCCTATGTTCGGGACTTCAAA
Mouse ITCH sh2: GCAGCAGTTTAACCAGAGATT
Mouse ITCH sh3: AATCCAGACCACCTGAAATAC
Control shRNA: CCTAAGGTTAAGTCGCCCTCG
Immunoprecipitation–Mass Spectrometry Sample Tryptic Digestion
PD-L2 with C-terminal FLAG tag, vector FLAG immunoprecipitation samples (for PD-L2 interactome analysis), or PD-L1 C-terminal FLAG-tag cotransfected with/without ITCH (for PD-L1 ubiquitination analysis) was subjected to immunoprecipitation. After elution in buffer (0.1 mol/L glycine-HCl, pH 3.0), eluates were reduced and alkylated by sequentially incubating with 5 mmol/L TCEP and 10 mmol/L iodoacetamide (chloroacetamide, for ubiquitination analysis) for 30 minutes at room temperature in the dark. The samples were then incubated overnight at 37°C with Lys-C and trypsin protease at a ratio of 1:100. Peptide digests were desalted using Pierce C18 tips (100 μL bed volume, 87784), dried by vacuum centrifugation, and reconstituted in 5% formic acid.
APEX2-Based Proximity Labeling
PD-L1–APEX2-expressing cells were cultured as previously described. Biotin-phenol (500 μmol/L) was added to the media and incubated at 37°C for 30 minutes. The peroxidase reaction was activated by adding H2O2 (no H2O2 was added to the negative control) to 1 mmol/L and incubating at room temperature for 1 minute. The reaction was quenched by washing cells three times with a quencher-containing PBS (10 mmol/L sodium azide, 5 mmol/L Trolox, 10 mmol/L sodium ascorbate). Cells were harvested by trypsinization and then flash-frozen in liquid nitrogen.
Streptavidin Pulldown
Cells were lysed in RIPA buffer (50 mmol/L Tris-HCl pH 7.5, 150 mmol/L NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% TritonX-100) supplemented with the protease inhibitor cocktail (Roche, 11836153001) and Benzonase (1 μL of 250U/μL; SIGMA, E8263-5KU) and incubated at 37°C for 20 minutes. Lysates were clarified by centrifugation and quantitated using the Pierce 660 nm protein assay, and then 1 mg of protein was incubated with 300 μL of high-capacity streptavidin beads (Thermo Fisher, 88816) for each sample at room temperature for 1 hour. Streptavidin beads were then washed three times with RIPA buffer, once with 1 mol/L KCl, once with 2 mol/L Urea in 25 mmol/L Tris-HCl pH = 8.0, and three more times with RIPA buffer. Bound proteins were then reduced, alkylated, and digested on beads with Lys-C and trypsin. The supernatant from the on-bead digestion was then transferred to another tube, bound to SP3/CMMB beads by the addition of acetonitrile to a concentration of 95%, and eluted in 0.1% formic acid.
LC-MS Data Acquisition
Samples were loaded onto a 75 μm × 25 cm homemade C18 column connected to a nanoflow Dionex Ultimate 3000 UHPLC system and fractionated online using a 140-minute gradient of increasing acetonitrile delivered at a 200 nL/min flow rate. An Orbitrap Fusion Lumos Tribrid mass spectrometer was used for data acquisition using the data-dependent acquisition (DDA) mode. Full mass spectrometer scans were acquired at 120K resolution with the automatic gain control (AGC) target set to 2e5 and a maximum injection time set to 100 ms. MS/MS scans were collected at 15K resolution after isolating precursors with an isolation window of 1.6 m/z and high-energy collisional dissociation (HCD)–based fragmentation using 35% collision energy. For DDA, a 3-second cycle time was used to acquire MS/MS spectra corresponding to peptide targets from the preceding full mass spectrometer scan. Dynamic exclusion was set to 25 seconds.
Database Search
MS/MS database searching was performed using MaxQuant (1.6.17.0, RRID:SCR_014485) against the human reference proteome from EMBL (RRID:SCR_014042; UP000005640_9606 HUMAN Homo sapiens, 20,600 entries, released in April 2020). The search included carbamidomethylation on cysteine as a fixed modification and methionine oxidation and N-terminal acetylation as variable modifications; identification of ubiquitination sites was searched with di-Gly modification as a variable modification in addition to the aforementioned ones. The digestion mode was set to trypsin and allowed a maximum of two missed cleavages. The precursor mass tolerances were to 20 and 4.5 ppm for the first and second searches, respectively, whereas a 20-ppm mass tolerance was used for fragment ions. Data sets were filtered at 1% FDR at both the PSM and protein levels. Peptide quantitation was performed using MaxQuant's LFQ mode. Visualization of the ubiquitinated peptide-spectrum match was conducted by parsing MaxQuant search results through PDV (31).
Statistical Analysis of Proteomics Data
The PD-L2-FLAG immunoprecipitation data set was searched with MaxQuant, and the resulting MS/MS spectral count information was used with SAINTexpress (v3.6.3, RRID:SCR_018562) to generate a protein interaction confidence score. MSStats (3.10, RRID:SCR_014353) was used to analyze the MaxQuant LFQ data in the PD-L1-APEX2 proximity labeling experiment to statistically assess protein enrichment. Equalized medians were used for normalization, and the Tukey median polish method was used for protein summarization. P values for t tests were corrected for multiple hypothesis testing using the Benjamini–Hochberg adjustment.
Western Blot and Immunoprecipitation
Cells were lysed in IP lysis buffer (Thermo Fisher Scientific, 87787) with protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific, 78440) for immunoprecipitation and Western blotting. Dynabeads (Thermo Fisher Scientific, 88847) preincubated with antibody or anti-FLAG M2 beads (Thermo Fisher Scientific, A36797) were used to immunoprecipitate proteins of interest based on the manufacturer's protocol. Antibodies used in immunoprecipitation and Western blot are as follows: TUBULIN (Sigma-Aldrich, T9026, RRID:AB_477593), ITCH (BD, 611198, RRID:AB_398732), HA (CST, 3724S, RRID:AB_1549585), FLAG (CST, 14793S, RRID:AB_2572291), PD-L1 (CST, 13684S, RRID:AB_2687655), GAPDH (CST, 5174S, RRID:AB_10622025), and UBIQUITIN (CST, 3933S, RRID:AB_2180538; CST, 3936S, RRID:AB_331292).
Cell-Surface PD-L1 and PD-L2 Detection
For cultured cell lines, cells were detached from the culture dish using the trypsin-free detachment solution Accutase (BioLegend, 423201) to preserve cell-surface proteins. Cells (5 × 105) were stained for either APC-anti–PD-L1 (1 μL/sample, BioLegend, 329708, RRID:AB_940360), PE-anti–PD-L2 (1 μL/sample, BioLegend, 329606, RRID:AB_1089019), or both followed by flow cytometry analysis to quantify the mean fluorescence intensity (MFI). For subcutaneous tumor models, tumors were dissociated to single-cell suspensions using a tumor dissociation kit and gentleMACS Octo Dissociator (Miltenyi Biotec). Cells (1 × 106) were incubated with 20% of FBS in PBS with 25 μg/mL of anti-mouse CD16/CD32 (clone 2.4G2) antibody (Invitrogen, 14-0161-86, RRID:AB_467135) at 4°C for 10 minutes to minimize nonspecific binding prior to surface staining with BV510-anti-CD45 (1 μg/mL, BioLegend, 103138, RRID:AB_2563061), BV421-anti-CD90 (2 μg/mL, BioLegend, 328122, RRID:AB_2561420), APC-anti–PD-L1 (2 μg/mL, BioLegend, 124312, RRID:AB_10612741), PE-anti–PD-L2 (2 μg/mL, BioLegend, 107206, RRID:AB_2162011), and PerCP-anti-TER119 (2 μg/mL, BioLegend, 116226, RRID:AB_893635) at room temperature for 20 minutes, followed by 7AAD (10 μL in 500 μL PBS per sample, Beckman Coulter, A07704) staining for 5 minutes on ice. Live tumor cells were gated as the BV510−/BV421−/PerCP− population (CD45−/CD90−), and MFIs of APC (PD-L1) and PE (PD-L2) were measured by flow cytometry analysis. Average PD-L1 expression of the CD45+ population was used as an internal control to normalize measurements of tumor-surface PD-L1 levels on different days.
Ubiquitination Assay
The PD-L1 ubiquitination assay was performed following the protocol of the Signal-Seeker Ubiquitination Detection Kit (Cytoskeleton, BK161). In brief, cells were treated with 20 μmol/L MG-132 (Selleck, S2619) for 4 hours, followed by cell lysis with protease and deubiquitination inhibitors. Cell lysates were purified by passing through a filter, and immunoprecipitation was performed using Dynabeads (preincubated with anti–PD-L1) or anti-FLAG M2 beads. Ubiquitination on the target protein was detected by Western blot using anti-UBIQUITIN antibody (CST, 3933S, RRID:AB_2180538; CST, 3936S, RRID:AB_331292).
PD-L1 Internalization Assay
The assay was performed as described previously (8). In brief, cell-surface PD-L1 was labeled with unconjugated anti–PD-L1 (BioLegend, 329703, RRID:AB_940362) for 1 hour on ice and washed twice to remove unbound antibody. Cells were resuspended in the culture medium on ice, and a baseline sample was removed and kept on ice. Cells were incubated at 37°C in a water bath and removed at the indicated times followed by immediate dilution in ice-cold PBS to stop further endocytosis. Cells were washed twice, and the remaining cell-surface–bound anti–PD-L1 antibodies were stained with Alexa Fluor-488–conjugated anti-mouse secondary antibody (Invitrogen, A11001, RRID:AB_2534069) for 30 minutes on ice. Finally, samples were washed twice and analyzed by flow cytometry.
Microscopy and Fluorescent Imaging
For imaging by confocal microscopy, cells were plated in an 8-chamber cell culture slide at 50% to 80% confluence. Anti–PD-L1 (BioLegend, 329703, RRID:AB_940362) was added at a 1:100 dilution to label cell-surface PD-L1 at room temperature for 20 minutes. Cells were washed 3 times with PBS, and the sample wells for 0-minute internalization were fixed by methanol for 20 minutes at −20°C. The sample wells for PD-L1 internalization were transferred into a 37°C cell culture incubator with the full medium for 30 minutes, followed by PBS wash 3 times and fixation by methanol for 20 minutes at −20°C. Then, the standard immunofluorescence staining protocol was applied with Alexa Fluor-647–conjugated anti-LAMP1 (1:100, 4°C overnight; CST, 73589S) and Alexa Fluor-488-conjugated anti-mouse secondary antibody (1:200, room temperature, 1 hour; Invitrogen, A11001, RRID:AB_2534069) for the samples before and after PD-L1 internalization. The slides were mounted in an antifade solution with DAPI (Invitrogen, P36935). Confocal images were taken by ZEISS LSM 880 63X (oil) objective at the University of California, Los Angeles (UCLA) CHS confocal microscopy core.
For the general immunofluorescence imaging of PD-L1 expression in cell lines, cells were plated in an 8-chamber cell culture slide at 50% to 80% confluence. Standard immunofluorescence staining protocol was performed with methanol fixation (−20°C, 20 minutes) and Alexa Fluor-555–conjugated anti–PD-L1 (1:50, 4°C overnight; CST, 40216, RRID:AB_2799172). The slides were mounted in antifade solution with DAPI (Invitrogen, P36935), and images were taken by a Zeiss microscope (AXIO Imager A1, 40× objective) mounted with a charge-coupled device camera (Retiga EXi QImaging).
Real-Time PCR
One million cells were subjected to total RNA extraction, reverse transcription, and cDNA quantification using the SYBR Green method by the MyiQ Real-Time PCR Detection System (Bio-Rad). The relative expression of PD-L1 was calculated using the delta-Ct method and normalized to TUBULIN levels. The sequence of PCR primers used was as follows:
PD-L1-F: TGCCGACTACAAGCGAATTACTG
PD-L1-R: CTGCTTGTCCAGATGACTTCGG
TUBULIN-F: GCACGATGGATTCGGTTAGGTC
TUBULIN-R: TCGGCTCCCTCTGTGTAGTGG
Coculture Assay
Target cell lines expressing NYESO-HLA-A2 were plated on 24-well plates at a concentration of 1 × 105 per well. Twelve to 16 hours later, media were changed, and Jk T cells expressing TCR (1G4) or human PBMCs (ATCC, PCS-800-011) were added to the culture wells at a concentration of 1 × 106/mL (400 μL) for 24 hours. Anti-CD3 (Invitrogen, 16-0037-81, RRID:AB_468854) and anti-CD28 (Invitrogen, 16-0289-81, RRID:AB_468926) were added into the culture media at a final concentration of 1 μmol/L (when using human PBMCs). Anti–PD-1 (BioLegend, 329925, RRID:AB_11147369) and anti–PD-L1 (BioLegend, 329715, RRID:AB_11149486), when applicable, were added in the culture media at a final concentration of 10 μmol/L. Media were harvested after coculture and diluted from 1/10 to 1/50 and subjected to ELISA to detect IL2 production (BioLegend, 431804).
Subcutaneous Syngeneic Tumor Models
Animal research in this study was approved by the local Animal Research Committee. C57BL/6 (RRID:IMSR_JAX:000664Info), BALB/c (RRID:IMSR_JAX:000651), and NSG (RRID:IMSR_JAX:005557) mice were obtained from the Radiation Oncology breeding colony at UCLA. Female mice were used at 6 to 8 weeks of age. All animal experiments were conducted according to the guidelines approved by the UCLA Animal Research Committee. For subcutaneous tumor models, C57BL/6, BALB/c, or NSG (YUMM1.7ER, NILER1-4, mSK-Mel254, KPC, and CT26) mice were injected on both flanks with 1 million cells per injection. Tumors were measured with a caliper every 2 or 3 days, and tumor volumes were calculated using the formula (length × width2)/2. Once tumors reached a size of 100 to 150 mm3, mice were assigned randomly into experimental groups. Special mouse diets (for C57BL/6, BALB/c, or NSG) were generated by incorporating trametinib (LC Laboratories, T-8123) to achieve daily dosing of mice at 0.45, 1, or 3 mg/kg/day or PLX4032 (LC Laboratories, LC‑V‑2800) 50 mg/kg/day plus trametinib 0.3 mg/kg/day (for the BRAF inhibitor + MEKi combination) to facilitate daily drug dosing and to reduce animal stress (TestDiet). Anti–PD-L1 (200 μg/mouse; Bio X Cell, BE0101, RRID: AB_10949073) was intraperitoneally administered twice per week. Anti-CD8 (200 μg/mouse; Bio X Cell, BE0117, RRID:AB_10950145) was intraperitoneally administered twice per week starting from 1 day before trametinib treatment. RP832c was subcutaneously administrated daily (10 mg/kg; Riptide Bioscience) from days 0 to 7 simultaneously with starting trametinib treatment. AK087 (Specs, 42718300) was dissolved in the vehicle (4% Tween80, 8% DMSO in double-distilled water) and subcutaneously administered near the tumor daily (10 mg/kg) starting with trametinib treatment but only from days 1 to 17. Tumors were excised from mice, minced, and digested to single-cell suspensions using a tumor dissociation kit and gentleMACS Octo Dissociator (Miltenyi Biotec), sorted (by 7-AAD; BD Pharmingen, 51-68981E), and prepared for scRNA-seq and/or CyTOF analysis.
Mass Cytometry of Murine Tissues
Cells (2 × 106 or fewer) were incubated with 20% FBS in PBS with 25 μg/mL of anti-mouse CD16/CD32 (eBioscience, 14-0161-86, RRID: AB_467135) antibody at 4°C for 10 minutes to minimize nonspecific binding prior to surface staining with an antibody cocktail at 4°C for 30 minutes in a 50 μL volume. Cells were incubated with 2.5 μmol/L 194Pt monoisotopic cisplatin (Fluidigm, 201194) at 4°C for 1 minute. Cells were then washed twice with FACS buffer and barcoded using palladium metal barcoding reagents according to the manufacturer's protocol (Fluidigm). Subsequently, fixation and permeabilization were performed using the FOXP3 fix and permeabilization kit according to the manufacturer's protocol (eBioscience, 00-5523-00). Cells were then subjected to an intracellular staining antibody cocktail [FOXP3 (Invitrogen, 14-5773-82, RRID: AB_467576), Ki-67 (Invitrogen, 14-5698-82, RRID: AB_10854564), granzyme B (Fluidigm, 3171002B, RRID: AB_2687652), T-bet (BioLegend, 644802, RRID: AB_1595503), iNOS (Fluidigam, 3161011B), and EOMES (Invitrogen 14-4875-82, RRID:AB_11042577)] for 30 minutes at room temperature. Subsequently, cells were washed twice with FOXP3 permeabilization buffer, washed twice with FACS buffer, and incubated overnight in 1.6% PFA in PBS with 100 nmol/L iridium nucleic acid intercalator (Fluidigm, 201192A). Finally, cells were washed twice with PBS with 0.5% BSA, filtered, and washed twice with water with 0.1% BSA prior to analysis. Samples were analyzed using a Helios mass cytometer based on the Helios 6.5.358 acquisition software (Fluidigm).
CyTOF Data Analysis
All the samples were preprocessed by CATALYST (RRID:SCR_017127), including normalization, debarcoding, and compensation. The normalized flow cytometry standard (fcs) files were then uploaded into Cytobank (RRID:SCR_014043; ref. 32), and data were gated to exclude beads and to include only live, single cells. CD8+ and CD4+ T cells were gated from the CD45+CD3+ populations, and data were downloaded separately into individual files for each sample, respectively. We applied Cytofkit (33) to perform the t-distributed stochastic neighbor embedding (t-SNE) analysis separately on the manually gated CD4+ and CD8+ populations from tumor samples. We selected 5,000 events/sample (all events if fewer than 5,000) to ensure equal representation of cells across samples. For CD4+ T cells, 12 markers, including CD44, CD62L, CD25, CD69, CD366, FOXP3, PD-1, CTLA-4, ICOS, EOMES, T-bet, and Ki-67, were used to cluster the cell populations. For CD8+ T cells, CD44, CD62L, CD25, CD69, CD366, granzyme B, PD-1, CTLA-4, ICOS, EOMES, T-bet, and Ki-67 were used. We chose 1,000 iterations, perplexity of 30, and theta of 0.5 as the standard t-SNE parameters. Mean intensity values of markers in each cluster were calculated and visualized via heat maps. Cells were assigned to different populations on the basis of the local gradient expression of known markers. Numbers of cells and percentages of different immune cell subsets were calculated for each sample.
Single-Cell 5′ Gene Expression and V(D)J Sequencing
Three or four different tumors were dissociated to single-cell suspensions using a tumor dissociation kit (Miltenyi Biotec, 130-095-929, RRID:SCR_020285) and gentleMACS Octo Dissociator (Miltenyi Biotec, 130-095-937). Equal numbers of cells per tumor were pooled together (2 × 106 total per sample). Cells were incubated with 20% FBS in PBS with 25 μg/mL of anti-mouse CD16/CD32 antibody (eBioscience, 14-0161-86, RRID: AB_467135) at 4°C for 10 minutes to minimize background antibody binding. Then cells were stained with BV510-anti-CD45 (1 μg/mL, BioLegend, 103138, RRID:AB_2563061) and PerCP-anti-TER119 (2 μg/mL, BioLegend, 116226, RRID:AB_893635) at room temperature for 20 minutes, followed by 7AAD (10 μL in 500 μL PBS per sample, Beckman Coulter, A07704) staining for 5 minutes on ice. Cells after staining were sorted by the BD FACSAria II sorting system to harvest the BV510 (CD45)-positive and PerCP (TER119, 7AAD)-negative populations. Cells recovered were subjected to the 10X Genomics standard protocol for coupled scRNA-seq and scTCR-seq library preparation using Chromium Next GEM Single-Cell 5′ Library and Gel Bead Kit v1.1 (10x Genomics, 1000167) and V(D)J Enrichment Kit for Mouse T Cells (10X Genomics, 1000071). Libraries were sequenced by the NovaSeq 6000 S2 flow cell with 2 × 50 reads targeting a minimum of 20,000 read pairs per cell for the scRNA-seq library and 5,000 read pairs per cell for the scTCR-seq library.
Analysis of scRNA-seq Data
Alignment to GRCm38 reference genome, barcode, and unique molecular identifier (UMI) counting was performed using Cell Ranger (10X Genomics, v2.1.0, RRID:SCR_017344). The Seurat package (RRID:SCR_007322; ref. 34) was used for downstream analysis. Cells with fewer than 200 genes detected or greater than 20% mitochondrial RNA content were excluded from further analysis. Raw UMI counts were normalized to UMI count per million total counts and log-transformed. Variable genes were detected based on average expression and dispersion for each data set independently. We then used the CellCycleScoring function to calculate scores of the S and G2–M cell-cycle phases for each cell. Single cells from different conditions were integrated into a single assay based on variable genes identified from each sample. We then used the ScaleData function to calculate scaled z-scores of each variable gene in the integrated assay and regress out the effect of the number of genes per cell, mitochondrial RNA content, and cell-cycle scores (S phase score and G2–M phase score). This scaled data set was then used for principal component analysis (PCA) of cells. Clusters and uniform manifold approximation and projection (UMAP) projections were generated based on the top 30 PCA dimensions. Clusters were annotated based on the expression of known marker genes, including Cd14 (myeloid), Igtam, Csf1r (monocyte/macrophage), Flt3 (dendritic cell), S100a8, S100a9 (neutrophil), Ncr1 (NK cell), Cd19, Cd79a (B cell), Cd3d, Cd3e, and Cd3g (T cell). Cell clusters coexpressing markers of multiple cell types were defined as doublets and excluded from further analysis. We next isolated the monocyte/macrophage and T-cell populations identified from the broad clustering analysis and performed reclustering analysis on them separately. Cells were reclustered as described above, and functional subpopulations were inferred and annotated by identifying differentially expressed marker genes with log fold changes higher than 0.4 using MAST (RRID:SCR_016340) in the FindAllMarkers function. The M2/M1-like TAM ratios in the macrophage population were calculated as the ratio of proportions between the inferred anti-inflammatory and proinflammatory subpopulations.
Analysis of scTCR-seq Data
Alignment to the GRCm38 reference genome and TCR contig annotation was performed by the Cell Ranger vdj pipeline (10X Genomics, v2.1.0, RRID:SCR_017344). For the TCR clonotype analysis, only cells assigned with both productive TRA and TRB sequences were kept for further analysis. If one cell had two or more TRA–TRB pairs identified, the pair with higher UMIs was considered the dominant TRA–TRB pair in the corresponding cell and used in the analysis. We defined each unique TRA–TRB pair as a clonotype. The clonal status of TCR clones was characterized as nonclonal (n = 1) and clonal (n ≥ 2) based on their cell numbers. The TCR clonotype of each cell was further linked to inferred functional subsets based on the barcode information. We used the STARTRAC package (35) to estimate the expansion and transition index of distinct T-cell subsets.
Analysis of Patient-Derived, Public Data Sets
Using Web server TIMER2.0 (http://timer.comp-genomics.org/; ref. 36), we split a data set of 471 cutaneous melanoma patients or 533 renal clear-cell carcinoma patients from the TCGA database into top n% patients with high intratumoral ITCH expression and bottom n% patients with low intratumoral ITCH expression to generate patient survival curve (n% indicated in figure legends). The difference between groups was calculated by both the Cox proportional hazards model and the log-rank test. A significant difference was defined as P < 0.05. Analysis of correlations between intratumoral PD-L1 expression and Treg infiltration or between ITCH RNA levels and CD8+ T-cell infiltration levels in tumor-derived transcriptome data was also performed by TIMER2.0 with algorithms CIBERSORT-ABS (37), QUANTISEQ (38), XCELL (39), TIMER (40), or EPIC (41). A data set of 471 cutaneous melanoma patient samples was analyzed with tumor purity adjustments. A significant difference for Spearman correlation analysis was defined as P < 0.05.
For pan-cancer correlation analysis between ITCH mRNA levels and PD-L1 protein levels, level 3 normalized mRNA expression data were downloaded from the TCGA data portal (RRID:SCR_003193) (https://portal.gdc.cancer.gov/) and level 4 normalized PD-L1 protein expression data were downloaded from the TCPA database (https://tcpaportal.org/tcpa/download.html). Two data sets were merged by matching the sample names, and the Spearman correlation score was calculated (n = 7,194). Tumor samples were ranked by ITCH mRNA expression levels and split into the top (ITCH high) and bottom (ITCH low) 20% (or 50%, as indicated in the figure legends) groups to compare PD-L1 protein levels.
Data Availability
Raw sequencing files of scRNA-seq and scTCR-seq are available at the Gene Expression Omnibus (RRID:SCR_005012; GSE199733). Raw files of the mass spectrometry data and mass cytometry data are available at FlowRepository (RRID:SCR_013779; http://flowrepository.org/) using the experiment ID: FR-FCM-Z566. Raw files of mass spectrometry data are available at the MassIVE database (RRID:SCR_013665; https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with data set ID: MSV000089180. Any additional information regarding data reported in this article is available from the corresponding author upon request. There are no restrictions on data availability. No custom code was used to analyze data in this study.
Supplementary Material
Acknowledgments
We thank Dr. Keith Ansell (LifeArc) for data sharing, Drs. Guideng Li and David Baltimore (Caltech) for sharing the NYESO-HLA-A2 construct and engineered Jk T-cell line (1G4), and Dr. ChunYing Song for technical assistance. This research was supported by grants (to R.S. Lo) from the NIH (1R01CA176111A1, 1R21CA215910-01, R21CA255837-01, and 1P01CA168585), the Melanoma Research Alliance (MRA; Team Science Awards), and the V Foundation for Cancer Research (Translational Award). Additional funding was provided by the MRA Dermatology Fellows Award (to Z. Yang and S. Liu), the Jonsson Comprehensive Cancer Center (JCCC) Postdoctoral Seed Grant (to Z. Yang), and JCCC Postdoctoral Fellowships (to Z. Yang and S. Liu). We also acknowledge the support (to R.S. Lo) of Mary Tanner and Maurizio Grimaldi as well as the Ressler Family Foundation. G. Moriceau was supported by funding from the NIH (1P01CA168585 Core C) and the MRA (Young Investigator Award). J. Wohlschlegel was supported by the NIH (R01 GM089778). CyTOF was performed in the UCLA JCCC Flow Cytometry Core Facility that is supported by NIH award P30 CA016042, the JCCC, the David Geffen School of Medicine at UCLA, the UCLA Chancellor's Office, and the UCLA Vice Chancellor's Office of Research. We thank Dr. Xinmin Li (Director) and the Technology Center for Genomics and Bioinformatics at UCLA for excellent technical support.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
R.S. Lo reports grants from Merck, Pfizer, and OncoSec and nonfinancial support for clinical trial from Bristol Myers Squibb. R.S. Lo and Z. Yang report being coinventors on a patent for U.S. Provisional Application Serial No. 63/364,010. No disclosures were reported by the other authors.
Authors’ Contributions
Z. Yang: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y. Wang: Resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. S. Liu: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. W. Deng: Resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. S.H. Lomeli: Resources, investigation, methodology, writing–review and editing. G. Moriceau: Resources, investigation, methodology, writing–review and editing. J. Wohlschlegel: Resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. M. Piva: Resources, investigation, visualization, writing–review and editing. R.S. Lo: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019;20:69–84. [DOI] [PubMed] [Google Scholar]
- 3. Song C, Piva M, Sun L, Hong A, Moriceau G, Kong X, et al. Recurrent tumor cell-intrinsic and -extrinsic alterations during MAPKi-induced melanoma regression and early adaptation. Cancer Discov 2017;7:1248–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hugo W, Shi H, Sun L, Piva M, Song C, Kong X, et al. Non-genomic and immune evolution of melanoma acquiring MAPKi resistance. Cell 2015;162:1271–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hugo W, Zaretsky JM, Sun L, Song C, Homet-Moreno B, Hu-Lieskovan S, et al. Genomic and transcriptomic features of resistance and sensitivity to anti–PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goodman AM, Piccioni D, Kato S, Boichard A, Wang HY, Frampton G, et al. Prevalence of PDL1 amplification and preliminary response to immune checkpoint blockade in solid tumors. JAMA Oncol 2018;4:1237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3′-UTR disruption in multiple cancers. Nature 2016;534:402–6. [DOI] [PubMed] [Google Scholar]
- 8. Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 2017;549:101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang X, Zhang Q, Lou Y, Wang J, Zhao X, Wang L, et al. USP22 deubiquitinates CD274 to suppress anticancer immunity. Cancer Immunol Res 2019;7:1580–90. [DOI] [PubMed] [Google Scholar]
- 10. Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, et al. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 2017;549:106–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2018;553:91–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong A, Piva M, Liu S, Hugo W, Lomeli SH, Zoete V, et al. Durable suppression of acquired MEK inhibitor resistance in cancer by sequestering MEK from ERK and promoting antitumor T-cell immunity. Cancer Discov 2021;11:714–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang Y, Liu S, Yang Z, Algazi AP, Lomeli SH, Wang Y, et al. Anti–PD-1/L1 lead-in before MAPK inhibitor combination maximizes antitumor immunity and efficacy. Cancer Cell 2021;39:1375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010;468:973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods 2013;10:1046–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Erpapazoglou Z, Walker O, Haguenauer-Tsapis R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells 2014;3:1027–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bethune MT, Gee MH, Bunse M, Lee MS, Gschweng EH, Pagadala MS, et al. Domain-swapped T cell receptors improve the safety of TCR gene therapy. Elife 2016;5:e19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hansen T, Yu YY, Fremont DH. Preparation of stable single-chain trimers engineered with peptide, beta2 microglobulin, and MHC heavy chain. Curr Protoc Immunol 2009; Chapter 17:Unit17.5. [DOI] [PubMed] [Google Scholar]
- 19. Wang J, Perry CJ, Meeth K, Thakral D, Damsky W, Micevic G, et al. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res 2017;30:428–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gutzmer R, Stroyakovskiy D, Gogas H, Robert C, Lewis K, Protsenko S, et al. Atezolizumab, vemurafenib, and cobimetinib as first-line treatment for unresectable advanced BRAF(V600) mutation-positive melanoma (IMspire150): primary analysis of the randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2020;395:1835–44. [DOI] [PubMed] [Google Scholar]
- 21. Amarnath S, Mangus CW, Wang JC, Wei F, He A, Kapoor V, et al. The PDL1-PD1 axis converts human TH1 cells into regulatory T cells. Sci Transl Med 2011;3:111ra20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, et al. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med 2009;206:3015–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jaynes JM, Sable R, Ronzetti M, Bautista W, Knotts Z, Abisoye-Ogunniyan A, et al. Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci Transl Med 2020;12:eaax6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G, et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis 2014;5:e1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Erkes DA, Cai W, Sanchez IM, Purwin TJ, Rogers C, Field CO, et al. Mutant BRAF and MEK inhibitors regulate the tumor immune microenvironment via pyroptosis. Cancer Discov 2020;10:254–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002;418:191–5. [DOI] [PubMed] [Google Scholar]
- 27. Vakkila J, Lotze MT. Inflammation and necrosis promote tumour growth. Nat Rev Immunol 2004;4:641–8. [DOI] [PubMed] [Google Scholar]
- 28. Solomon I, Amann M, Goubier A, Vargas FA, Zervas D, Qing C, et al. CD25-Treg-depleting antibodies preserving IL-2 signaling on effector T cells enhance effector activation and antitumor immunity. Nat Cancer 2020;1:1153–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol 2020;21:298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schapira M, Calabrese MF, Bullock AN, Crews CM. Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov 2019;18:949–63. [DOI] [PubMed] [Google Scholar]
- 31. Li K, Vaudel M, Zhang B, Ren Y, Wen B. PDV: an integrative proteomics data viewer. Bioinformatics 2019;35:1249–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom 2010; Chapter 10:Unit10.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen H, Lau MC, Wong MT, Newell EW, Poidinger M, Chen J. Cytofkit: a bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput Biol 2016;12:e1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, et al. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 2018;564:268–72. [DOI] [PubMed] [Google Scholar]
- 36. Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Finotello F, Mayer C, Plattner C, Laschober G, Rieder D, Hackl H, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med 2019;11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016;17:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Racle J, de Jonge K, Baumgaertner P, Speiser DE, Gfeller D. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife 2017;6:e26476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing files of scRNA-seq and scTCR-seq are available at the Gene Expression Omnibus (RRID:SCR_005012; GSE199733). Raw files of the mass spectrometry data and mass cytometry data are available at FlowRepository (RRID:SCR_013779; http://flowrepository.org/) using the experiment ID: FR-FCM-Z566. Raw files of mass spectrometry data are available at the MassIVE database (RRID:SCR_013665; https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with data set ID: MSV000089180. Any additional information regarding data reported in this article is available from the corresponding author upon request. There are no restrictions on data availability. No custom code was used to analyze data in this study.