Abstract
Fibroblasts are highly dynamic cells that play a central role in tissue repair and fibrosis. However, the mechanisms by which they contribute to both physiologic and pathologic states of extracellular matrix deposition and remodeling are just starting to be understood. In this review article, we discuss the current state of knowledge in fibroblast biology and heterogeneity, with a primary focus on the role of fibroblasts in skin wound repair. We also consider emerging techniques in the field, which enable an increasingly nuanced and contextualized understanding of these complex systems, and evaluate limitations of existing methodologies and knowledge. Collectively, this review spotlights a diverse body of research examining an often-overlooked cell type - the fibroblast - and its critical functions in wound repair and beyond.
Keywords: Wound healing, fibrosis, regeneration, fibroblasts, fibroblast heterogeneity
Talbott et al discuss the current state of knowledge in fibroblast biology and heterogeneity, primarily focusing on fibroblasts in skin wound repair. They also consider emerging techniques in the field and evaluate limitations of existing methodologies and knowledge.
Introduction
Broadly, fibroblasts are the cells of the body’s connective tissue responsible for producing and remodeling extracellular matrix (Lynch and Watt, 2018). Fibroblasts are not merely passive “bystander” cells, but rather are a highly active cell type with diverse and dynamic functions throughout the body and its development. Fibroblasts play critical roles in development/growth, homeostasis, and the injury response; they are vital in forming and maintaining the structure of virtually every organ, and are key contributors to the repair process and formation of new tissue following tissue damage (Plikus et al., 2021). In recent years, it has become increasingly well understood that fibroblasts are an extremely heterogeneous cell type, exhibiting significant phenotypic and functional variability between and even within tissues (Griffin et al., 2020; Lynch and Watt, 2018). These cells are also highly plastic (LeBleu and Neilson, 2020), making their study both challenging and fascinating, as they are capable of contributing to a wide array of cellular and tissue-level processes through highly varied mechanisms.
Fibroblasts have been most deeply studied in the skin, as they play an especially critical role in this organ’s physiologic and pathologic functioning (desJardins-Park et al., 2018; Gurtner et al., 2008). As the body’s largest organ and its first line of defense against the external environment, the skin must meet an unusually high demand for structural resiliency. The skin’s cells must be able to repair not only the day-to-day wear and tear caused by constant exposure to environmental stressors (e.g., sun damage) but also more significant wounds resulting from burns, lacerations, or other traumatic injuries. Wound repair is a complex, multi-stage, highly orchestrated process in which fibroblasts play multiple critical roles, via both signaling to other key wound cell types and direct closure and filling-in of the defect/injury site (Gurtner et al., 2008).
Adult wound healing results in formation of a scar, which consists of nonfunctional fibrotic tissue. Scarring is just one (acute) example of fibrosis, a process that can occur anywhere in the body and which results in replacement of native tissue with dense connective tissue, ultimately leading to loss of normal tissue function. Collectively, fibroses are estimated to be responsible for up to 45% of all deaths in the industrialized world (Henderson et al., 2020). As the primary downstream mediators of scarring and fibrosis, fibroblasts are an active target of research seeking to improve wound healing and organ fibrosis outcomes. While this review primarily focuses on fibroblasts’ roles in the skin and its wound healing, insights into fibroblast biology and heterogeneity gained from the skin may ultimately translate to other organs and fibrotic processes.
Overview of wound repair
As the body’s main external barrier, the skin is a frequent site of tissue damage. Following injury, the skin undergoes a complex wound repair reaction involving many cell types, cytokines, and molecular mediators (Eming et al., 2014). Skin wound repair involves interactions of multiple cell types, with fibroblasts playing key roles later in healing. Following hemostasis, during which the intrinsic and extrinsic coagulation pathways are initiated to prevent blood loss, immune cells are recruited into the injury site, where they help to sterilize the wound and remove debris (Eming et al., 2014). The release of chemokines by platelets and early inflammatory cells in the wound attracts macrophages (derived from circulating monocytes), one of the key immune players during skin repair. They, too, engulf necrotic cellular debris and pathogenic material from the wound site. A vital feature of macrophages is their dramatic ability to tailor their responses to specific environmental stimuli, displaying diversity and plasticity in the response to tissue damage (Gordon and Taylor, 2005). Wound macrophages were once classically divided into two main subsets – M1 (“classically activated”) macrophages, implicated in pro-inflammatory events; and M2 (“alternative activated”) macrophages, thought to be anti-inflammatory and pro-regenerative (Krzyszczyk et al., 2018) – and the deletion of specific macrophage populations was shown to dramatically alter the outcome of fibrotic reactions (Lucas et al., 2010). However, our modern understanding of macrophage and monocyte heterogeneity highlights that these cells, like fibroblasts, exhibit considerable plasticity and heterogeneity in function and origins, with some subtypes specific to particular disease states (Auffray et al., 2007; Hanna et al., 2011; Kohyama et al., 2009; Okabe and Medzhitov, 2014; Satoh et al., 2013; Satoh et al., 2017).
The early recruitment and activity of inflammatory cells and platelets activates and directs the migration of fibroblasts to the wound site around days 5–7 (Bussone, 2017). Fibroblasts are the primary cell type responsible for synthesizing and depositing new ECM to repair the skin’s structural framework (Bussone, 2017). Subsets of fibroblasts recruited to the wound differentiate into myofibroblasts under the influence of mechanical tension and cytokines such as TGF-β. Myofibroblasts are responsible for wound contracture following injury. While in rare instances (such as fetal wound healing (desJardins-Park et al., 2019)) native-like ECM is regenerated, in the context of typical postnatal wound repair, profound activation of fibroblasts by the wound cytokine/inflammatory milieu leads to deposition of excess, poorly-ordered matrix, resulting in fibrosis (scarring) (Seki et al., 2007; Szabo et al., 2007). Although communication between macrophages and fibroblasts has been implicated in driving both regenerative and fibrotic wound outcomes, the specific signaling pathways and spatiotemporal coordination involved in this cell crosstalk and its divergent influences remain underexplored.
Circulating fibroblasts (also known as fibrocytes), a group of bone marrow-derived mesenchymal progenitor cells (Abe et al., 2001), have been reported to migrate to the injury bed in response to cytokine stimuli and differentiate into contractile fibroblasts under the influence of IL-4, IL-13, and interferon (IFN)-ɣ (Shao et al., 2008), but these likely represent a small minority of all fibroblasts in skin wound healing (Grieb et al., 2011). A similar phenomenon has been described in several other tissues including liver, lung, heart, blood vessels, and cornea (Haudek et al., 2006; Lassance et al., 2018; Mori et al., 2005; Phillips et al., 2004; Reich et al., 2013; Yan et al., 2016). Although fibrocytes appear to arise from a hematopoietic lineage, their exact progenitor is unknown and the stimuli for fibrocyte recruitment and differentiation are not well defined (de Oliveira and Wilson, 2020). Given the limited knowledge into fibrocyte origins and characteristics, the extent of their functional contribution to acute fibrosis is unclear.
Finally, during the last stage of wound repair, the remodeling phase, fibroblasts continue to crosslink and turn over the initially deposited provisional ECM. They replace some collagen type III with collagen type I and structurally modify the initially deposited granulation tissue, causing strengthening and stiffening of the ECM over time to ultimately form the mature scar (Hinz, 2007). Remodeling is the longest and least well understood phase of wound repair and can last from weeks to months or even years.
Overview of modern wound healing research
Given the complexities of the wound repair process, it is unsurprising that a wide range of experimental models exist to study the biology of wound healing. These range from simple in vitro models involving isolated wound cell types, to more complex three-dimensional in vitro models incorporating multiple cell types, to in vivo animal models of wound repair with varying degrees of faithfulness to human wound biology (Table 1).
Table 1:
Comparison of models for studying wound repair.
| Model | Strengths | Limitations |
|---|---|---|
| In vitro monolayer models | • Ease of use • Amenable to genetic manipulation |
• Does not reconstitute the complex 3D environment and cell-cell and cell-matrix interactions of the skin. • Fibroblasts undergo pro-fibrotic shift in vitro |
| Three-dimensional culture systems (spheroid, organotypic, ex vivo) | • Ease of use • Amenable to genetic manipulation and tuning of native-like substrate properties (e.g., stiffness) • In ex vivo culture, contain all relevant cell types in native organization |
• Most models do not fully reconstitute all cell-cell and cell-matrix interactions, or functionality, of the skin. • Fibroblasts undergo pro-fibrotic shift in vitro. • May require use of exogenous growth factors or chemical inhibitors in culture media. |
| Mouse excisional/incisional in vivo wound models | • Ease of use, replicability • Genetically dissectible using transgenic animals • Allows for lineage tracing of fibroblast populations. • Can test potential anti-fibrotic agents. |
• Loose skinned animal model with a panniculus carnosus muscle, which is not analogous with human wound healing. |
| Wound-induced hair neogenesis (WIHN) | • In vivo model of skin regeneration, with recovery of sparse follicles in the scar center. | • No known correlates outside of rodents. • Limited degree of regeneration, with follicles emerging against a scar background. |
| Mouse xenograft in vivo models | • Ease of use • Provides initial understanding into human wound healing. • Can test potential anti-fibrotic agents. |
• Limited insight into immune-fibroblast interactions. • Surgical expertise required to overcome rejection of human skin grafts. • Donor tissue availability for xenografting. |
| Porcine in vivo models | • Anatomically and physiologically similar to human skin (especially red Duroc pigs) | • Expensive • Transgenic animals are limited • Experimental procedures require surgical and anesthetic expertise. |
In vitro models
In vitro platforms of varying complexity have been used over many years to study the behavior of defined wound cell populations. Fibroblast in vitro studies are limited by the fact that fibroblasts undergo significant, pro-fibrotic changes, with broad shifts in marker expression, and adopt a mechanically-activated, myofibroblast phenotype over time in culture independent of other stimuli (Baranyi et al., 2019; Bayreuther et al., 1988; Masur et al., 1996; Walmsley et al., 2015). Three-dimensional (3D) models are of growing interest and may more accurately recapitulate in vivo biology (Duval et al., 2017), often incorporating multiple cell types (though none replicate all relevant wound healing cell types). Layered spheroids (Ebner-Peking et al., 2021) (most commonly applied to the study of skin cancers (Klicks et al., 2019; Randall et al., 2018)) are low cost and relatively reproducible, but lack the air-liquid interface (ALI) inherent to the native skin environment (Klicks et al., 2017). More commonly used are layered models consisting of cells grown on a preformed scaffold. While these models can incorporate ALI culture and the scaffolds can be tuned to the mechanical and chemical properties of skin, they are limited by fibroblasts’ tendency to contract and/or degrade the scaffold over time (Randall et al., 2018), which may suggest that fibroblasts are still supra-physiologically “activated” in these models. The most truly biomimetic models are arguably organotypic culture with ex vivo culture of intact skin biopsies, which theoretically contain all relevant cell types in their native organization; however, these suffer from practical issues such as donor-to-donor variability and availability of samples, and lack an in vivo-like mechanical environment (Randall et al., 2018). Overall, in vitro fibroblast studies are severely hampered by their inability to simultaneously recapitulate multiple key factors that contribute to skin healing in vivo, including physical tension, inflammation, epithelial-mesenchymal signaling, and hemostasis (Liang et al., 2007).
In vivo models
Animal models are ultimately critical to capture the spatial and temporal complexity of wound repair and other fibrotic processes. Mice are the most often-used animal model for the study of wound healing and fibroses (with other rodents, including rats, less frequently used), with various models aiming to recapitulate excisional healing, incisional healing, ear and scalp wounds, chronic pressure ulcers, and hypertrophic scarring. The primary criticism/limitation of mice as a model for human wound repair is that – because of their loose skin and subcutaneous panniculus carnosus muscle, which largely lacks an analog in humans (Wong et al., 2011b) – unencumbered mouse wounds on the dorsal or ventral surfaces will contract rapidly and heal in a fraction of the time as that of tight-skinned humans (whose skin is under greater tension and adhered to underlying tissue) (Galiano et al., 2004). Contraction also results in a substantially smaller scar, with up to 90% of the wound area closing by contraction (Zomer and Trentin, 2018), rather than the entire wound being filled by formation of new (granulation) tissue (Galiano et al., 2004). One approach to circumvent this issue is to physically prevent wound contraction using a silicone splint affixed to the skin around the wound, resulting in healing through granulation and re-epithelialization of the wound bed in a process and on a timeline that better mimics human healing (Chen et al., 2013; Galiano et al., 2004; Wong et al., 2011b).
Alternatively, full-thickness wounds may be produced on the dorsal tail skin; these do not exhibit contraction and instead heal by secondary intention, with epithelium advancing over granulation tissue. Because of their relatively delayed healing sequence, dorsal tail wounds have been proposed as an experimental model for delayed wound closure (Falanga et al., 2004). Other rodent models, such as the rabbit ear punch model, rely on underlying cartilage as a “natural splint” to prevent wound contraction (Masson-Meyers et al., 2020). This wound model has also been used extensively to study healing mechanisms in MRL/MpJ “superhealer” mice, which display relatively rapid re-epithelialization of ear punch wounds and regenerative healing with little fibrosis compared to wild type mice (Heydemann, 2012). (In contrast to their ear punch regeneration, splinted dorsal wounds in MRL/MpJ heal via scarring, similar to wild type mice (Colwell et al., 2006).) It should be noted, however, that site-specific differences in skin architecture between these distinct wound models may limit translational relevance to the dorsal and ventral skin surfaces.
A popular unsplinted wound model is the wound-induced hair follicle neogenesis (WIHN) model, in which very large wounds (≥2.25 cm2 in adult mice) heal via substantial contraction with re-epithelialization in the wound center; these wounds regrow sparse hair follicles in the center of the wound (Ito et al., 2007; Wang et al., 2015), though on a background of fibrotic/scar-like ECM (Mascharak et al., 2022), indicating that WIHN healing likely does not represent complete wound regeneration.
While the above-mentioned models all employ excisional wounds, incisional wound models are also used (Ansell et al., 2014); simple incisions in mice tend to heal with extremely fine/minimal scars, but mechanically-loaded wound models have been developed which, by applying tension across mouse incisional wounds, approximate the mechanical stress experienced by healing human wounds and yield hypertrophic scarring resembling that commonly observed in humans (Aarabi et al., 2007). Parabiosis models represent another useful tool in the study of mouse wound healing, in which the blood supply of two mice is surgically joined to establish cross-circulation (Wong et al., 2011b); such models have been used to establish roles for circulating versus local factors and cells in wound repair (Hamou et al., 2009; Song et al., 2010; Wong et al., 2011b).
While mouse studies have played an important role in informing our current understanding of fibrotic disease mechanisms, including the role of key molecular pathways (e.g., PDGF, TGF-β, and Wnt signaling), cell types (e.g., alpha-smooth muscle actin [α-SMA]+ myofibroblasts), and cellular interactions (e.g., fibroblast-inflammatory cell and fibroblast-epithelial cell crosstalk), to date, few effective antifibrotic therapies have translated from mouse studies. Much of this disparity relates to fundamental structural and functional differences in mouse and human skin (Figure 1). In addition to the biomechanical differences between mice and humans (tight versus loose skin) outlined above, mouse skin is substantially thinner than human skin (<25 μm versus >100 μm), contains relatively less reticular (deep) dermal tissue, and lacks the rete ridges and (outside of the paws) sweat glands that are found in human skin (Zomer and Trentin, 2018). Further, mouse skin contains a much higher density of hair follicles than most of the human body (implying a higher density of stem cells and progenitors, which may have important repercussions for injury repair outcomes). Mouse wounds are also immunologically distinct from human wounds, containing relatively greater proportions of lymphocytes (including γδ T-cells, which are absent from human skin (Li et al., 2018)) and unique cytokines (including chemokine [C-C motif] ligand 6 [Ccl6], a critical mediator of fibroblast mechanotransduction signaling in scars and tumors (Coelho et al., 2007; Demircioglu et al., 2020)). Several of these structural and functional differences relate to cell and molecular processes implicated in WIHN, a process without an analog in humans (Gay et al., 2013).
Figure 1: Differences in the architecture of mouse versus human skin.
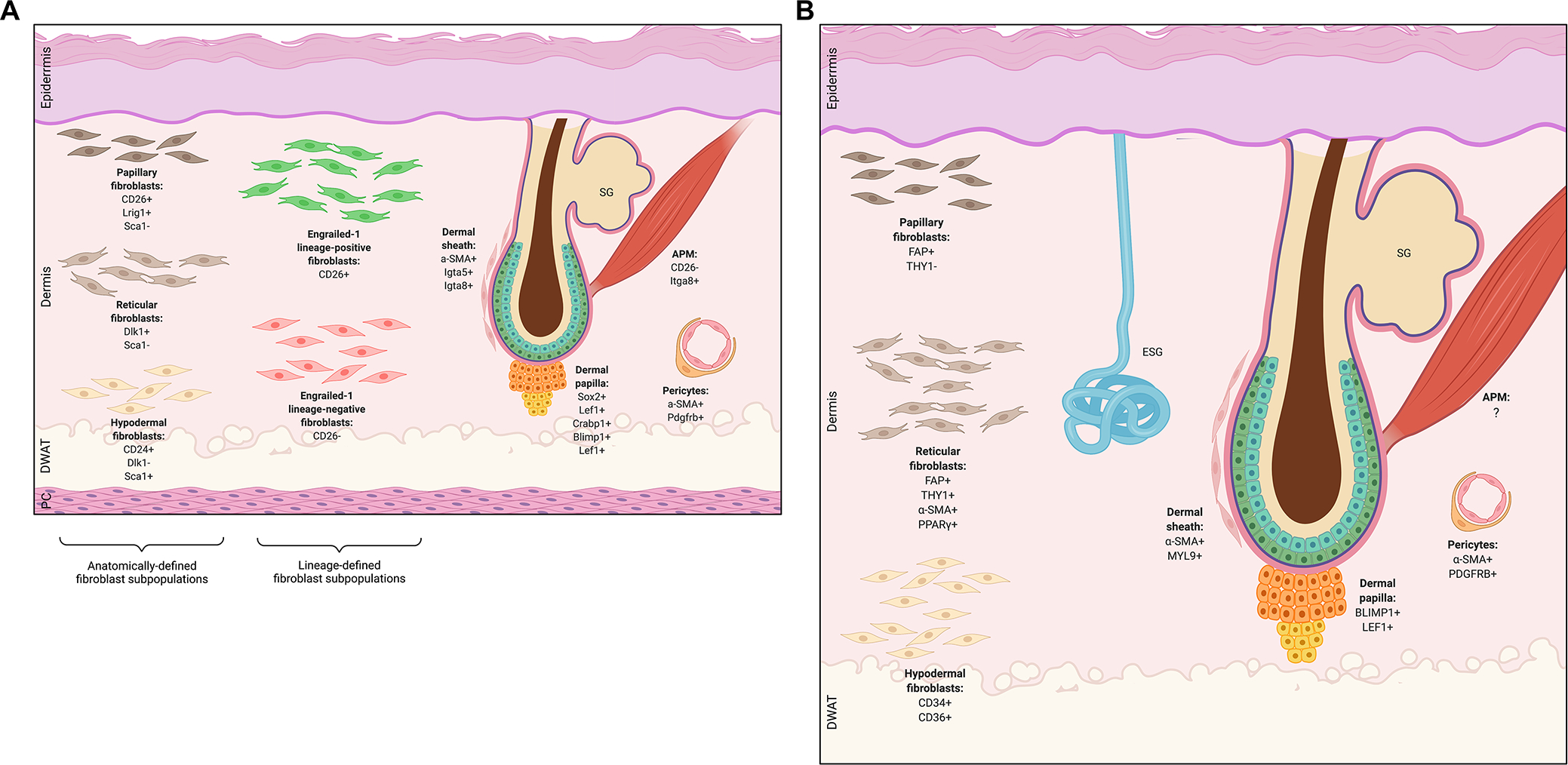
A) Depiction of mouse skin structure, which includes a panniculus carnosus (PC) muscle layer under the hypodermis. B) Depiction of human skin structure. Human skin differs from mouse skin in that it is thicker overall, has sparser hair follicles (HF), and contains eccrine sweat glands (ESG) which are not found in mice outside the skin of the paws. For both mouse and human skin schematics, various mesenchymal subpopulations and characteristic markers are shown, including microanatomically defined fibroblast lineages (papillary, reticular, hypodermal fibroblasts), functionally defined fibroblast lineages (Engrailed-1 lineage-positive and lineage-negative fibroblasts), HF-associated fibroblasts (dermal sheath, papilla, arrector pili), and pericytes. SG, sebaceous gland; DWAT, dermal white adipose tissue; APM, arrector pili muscle.
Due to these fundamental limitations of mouse wound healing research, large animal models also play an important role in discovery and translation. Pigs have the most anatomically and physiologically similar skin to humans, with similar epidermal and dermal thickness, relative hairlessness, and, importantly, tight/fixed skin with similar mechanical properties to humans (Masson-Meyers et al., 2020). Red Duroc pigs in particular are thought to be the ideal animal model for human wound healing, as they exhibit human-like wound healing and have a similar propensity for scarring (Wong et al., 2011b). Porcine tissues have also been shown to have similar responses to substances such as growth factors (Swindle et al., 2012), an important translational consideration. Key limitations of pigs, as with any large animal model, are that they are substantially more expensive to maintain, experimental procedures are inherently more complicated (typically requiring trained anesthesia/surgery personnel), reagents and methods are much less well developed than for mice (Zomer and Trentin, 2018), and although limited transgenic porcine models have been generated, these remain far from widespread application (Crociara et al., 2019; Yang and Wu, 2018). While these barriers are collectively prohibitive for the use of pigs in early-stage discovery, their biological similarities to humans make swine a critical model for filling the “translational gap” between mouse research and human clinical trials.
Xenotransplantation models can also be valuable tools for wound healing research. In these models, which have been used since at least the 1970s, human skin is transplanted onto mice, then wounded and its healing studied (Demarchez et al., 1986; Reed and Manning, 1973). These models are highly useful as they allow for the study of human cells’ response to wounding (Borrelli et al., 2021). However, it can be challenging to separate contributions of human cells within the graft from those of mouse cells in the surrounding tissue; some studies have suggested that mouse cells deposit the early granulation tissue that initially fills the xenograft wound (which is then re-invaded and replaced by human fibroblasts from the graft), though this may depend on the precise nature (e.g., thickness) of the grafted skin (Rossio-Pasquier et al., 1999). Another fundamental limitation is that xenograft recipients are necessarily immune-compromised (typically nude/athymic/T cell-deficient mice) to prevent rejection (Demarchez et al., 1986), which could alter the inflammatory response to wounding.
Finally, numerous models exist for the study of abnormal or impaired wound healing issues common in humans, such as chronic wounds due to diabetes, pressure, ischemia, or reperfusion, which do not naturally occur in animals (Grada et al., 2018). The study of impaired diabetic wound repair (which leads to chronic/non-healing wounds in humans) commonly employs different diabetic mouse models. While these exhibit delayed wound healing, they largely fail to recapitulate all aspects of diabetic wound repair, likely because human diabetes is typically multifactorial (compared to mouse diabetes models relying on single-gene manipulation) and, in fact, genes altered in diabetes models may independently affect wound repair (e.g., leptin may alter wound angiogenesis) (Fang et al., 2010; Grada et al., 2018).
Review of current literature
What is a fibroblast?
Overview of fibroblasts
Fibroblasts were initially reported in the 1800s, first by Virchow who described them as “spindle-shaped cells of the connective tissue,” and later by Ziegler who was the first to use the term “fibroblast” to describe the cells that deposit new connective tissue in wounds (Molenaar, 2003; Muhl et al., 2020b; Plikus et al., 2021). Fibroblasts are the most common cells in connective tissue throughout the body and are responsible for producing, depositing, and integrating the proteins of the ECM (Lynch and Watt, 2018), which consists of both fibrous/structural proteins and “ground substance,” a hydrated gel of molecules such as glycosaminoglycans (GAGs) and proteoglycans (Kendall and Feghali-Bostwick, 2014; Ravikanth et al., 2011). While largely nonproliferative in the absence of stimulation (e.g., injury) (LeBleu and Neilson, 2020), fibroblasts are far from functionally quiescent. In both homeostasis and tissue repair/fibrosis, fibroblasts not only secrete but also continuously turn over and remodel the ECM, via production of enzymes that cross-link (e.g., lysyl oxidase [Lox]) or promote or inhibit the degradation of (e.g., matrix metalloproteinases [MMPs] or tissue inhibitors of metalloproteinases [TIMPs], respectively) ECM proteins (Kendall and Feghali-Bostwick, 2014; Lu et al., 2011; Shaw and Rognoni, 2020). Fibroblasts are also a non-terminally differentiated and relatively plastic cell type and, beyond their direct functions in producing and maintaining ECM, are critical to a host of other processes such as inflammation, cancer progression, and angiogenesis (Kendall and Feghali-Bostwick, 2014; Shaw and Rognoni, 2020).
Experimental definitions of fibroblasts
Experimentally, fibroblasts can be defined in several ways, each with advantages and shortcomings. Classically, fibroblasts were considered to be cells that adhered in culture (Griffin et al., 2020; Lynch and Watt, 2018), and were isolated based on their relatively easy propagation on passaging (Plikus et al., 2021); however, other cell types can also adhere to tissue culture plastic, including macrophages and endothelial cells (Fleit et al., 1984; Kelley et al., 1987; Relou et al., 1998). Morphologically, fibroblasts are typically elongated, spindle-shaped cells, but upon activation (e.g., adherence to plastic substrate, wound stimuli) can spread out and become stellate (Ravikanth et al., 2011).
Unfortunately, no universal fibroblast markers have been identified; while some markers are considered “typical” of fibroblasts (for instance, ECM components such as vimentin or procollagen Iα2 chain), these are neither specific nor necessarily expressed by all fibroblasts, though some context-specific fibroblast markers have been defined (e.g., cardiac fibroblasts are thought to express discoidin domain receptor 2 [DDR2]) (Lynch and Watt, 2018). Many studies do use common “fibroblast markers” such as platelet-derived growth factor receptor alpha (PDGFRA) to prospectively define or isolate fibroblasts (Buechler et al., 2021; Driskell et al., 2013; Muhl et al., 2020b; Philippeos et al., 2018). However, this marker is not specific to fibroblasts (e.g., also expressed by cells of the central nervous system (Zhu et al., 2014)) and, critically, we and others have found that not all fibroblasts – whether in the dermis or other tissues – express PDGFRA (Biasin et al., 2020; Guerrero-Juarez et al., 2019; Mascharak et al., 2021a; Xie et al., 2018). A contrasting approach to defining fibroblasts is lineage exclusion, in which, rather than relying on positive selection based on defined markers, fibroblasts are isolated based on their lack of expression of hematopoietic and other non-fibroblast cell lineage markers (Walmsley et al., 2015); of course, a possible drawback of this approach is that it could fail to effectively exclude every non-fibroblast cell and thus be overly inclusive.
Transcriptomic analysis (e.g., scRNA-seq) allows fibroblasts to be classified based on broader patterns of gene expression, rather than positive or negative expression of a single marker. Fibroblasts can generally be distinguished by transcriptomic patterns corresponding to their function (i.e., a transcriptomic profile dominated by ECM genes), and may also express “typical” fibroblast markers such as CD34 and Pdgfra (Muhl et al., 2020b). However, large-scale transcriptomic studies have found that there is no single fibroblast transcriptomic identity or marker; considerable transcriptional diversity exists among fibroblasts (particularly between different organs), with significant variability in expression between different fibroblast clusters in terms of specific types and quantities of ECM genes/modifiers, as well as other genes, expressed (Kendall and Feghali-Bostwick, 2014; Muhl et al., 2020b). These studies suggest that panels of multiple markers, rather than a single or small number of genes, may be needed to transcriptomically define the fibroblast identity.
Defining fibroblasts across organs
Fibroblast transcriptional identities are particularly variable between different organs (Muhl et al., 2020b). A recent large-scale transcriptomic profiling of fibroblasts from 16 mouse tissues by Buechler et al. revealed various functional fibroblast identities (e.g., adventitial, parenchymal, alveolar), each with distinct expression patterns of known fibroblast-associated genes; thus, fibroblast specialization across organs was reflected in differential expression of a limited set of core, defining signaling pathways (e.g., nuclear factor kappa B [NF-kB], tumor necrosis factor [TNF], Wnt) (Buechler et al., 2021). However, while single-cell transcriptomic analysis allows for more nuanced classification and identification of fibroblasts, even this method may miss some heterogeneity among fibroblasts; for instance, a single transcriptionally-defined population could contain multiple epigenetically distinct subpopulations that might respond differently to a given stimulus (Lynch and Watt, 2018). Further, experimentally, relying on transcriptomic profiles to define fibroblast identity precludes prospective isolation of fibroblasts or fibroblast subtypes on this basis, as transcriptomic identity does not necessarily correlate directly to surface marker profiles or other phenotypic features that can be leveraged experimentally. Overall, the existing body of research demonstrates that many challenges remain in the classification of fibroblasts – including the lack of a unified definition of a fibroblast – and highlights the importance of incorporating multiple analytical lenses and balancing broad versus restrictive definitions of this diverse cell type.
Fibroblast heterogeneity in fibrosis
Over 160 years after Virchow first described apoptosis-resistant, matrix-producing cells of the connective tissue (Molenaar, 2003), the scientific community has come to appreciate the remarkable functional heterogeneity and organ-specific specialization of fibroblasts. Fibroblast heterogeneity can be broadly described according to developmental origin and spatial location.
Fibroblast heterogeneity according to developmental origin
The majority of fibroblasts in the body are embryonically derived from paraxial and lateral plate mesoderm precursors (Thulabandu et al., 2018), while fibroblasts in craniofacial tissues derive from neural crest mesenchyme (Wong et al., 2006) – thus, a first level of fibroblast heterogeneity is introduced during development. These differing embryonic origins may in turn influence fibrotic behavior in postnatal life: neural crest-derived, Wnt1 lineage-positive fibroblasts (WPFs) mediate scarless healing of the oral mucosa, while scarring healing in the skin of the trunk is accomplished by paraxial mesoderm-derived, Engrailed-1 (En1) lineage-positive fibroblasts (EPFs) in the dorsal skin (Rinkevich et al., 2015), and lateral plate mesoderm-derived, Paired related homeobox 1 (Prrx1) lineage-positive fibroblasts (PPFs) in the ventral skin (Leavitt et al., 2020) (Figure 1). Whereas EPFs and PPFs are responsible for the vast majority of fibrosis following injury, radiation, and tumor desmoplasia, their counterpart fibroblast populations – En1 and Prrx1 lineage-negative fibroblasts (ENFs and PNFs, respectively) – are non-fibrogenic and permit regenerative healing (Jiang et al., 2018; Mascharak et al., 2021a) (Figure 2). This behavior is cell-intrinsic: EPFs or PPFs transplanted into the normally non-scarring oral mucosa retain their fibrogenic behavior in response to wounding, while non-scarring WPFs, ENFs, or PNFs transplanted into dorsal or ventral skin retain their non-fibrogenic phenotype (Mascharak et al., 2021a; Rinkevich et al., 2015). It was recently shown that certain postnatal dermal fibroblasts in mice re-activate En1 expression in response to wound mechanical forces, and that blocking this reactivation via mechanotransduction inhibition yields ENF-mediated wound regeneration with recovery of hair follicles, glands, and normal matrix ultrastructure (Mascharak et al., 2021a). Thus, embryonically-defined fibroblast lineage hierarchies may be continuously reinforced through postnatal reactivation of developmental gene programs.
Figure 2: Fibroblast heterogeneity in mouse skin wound healing.
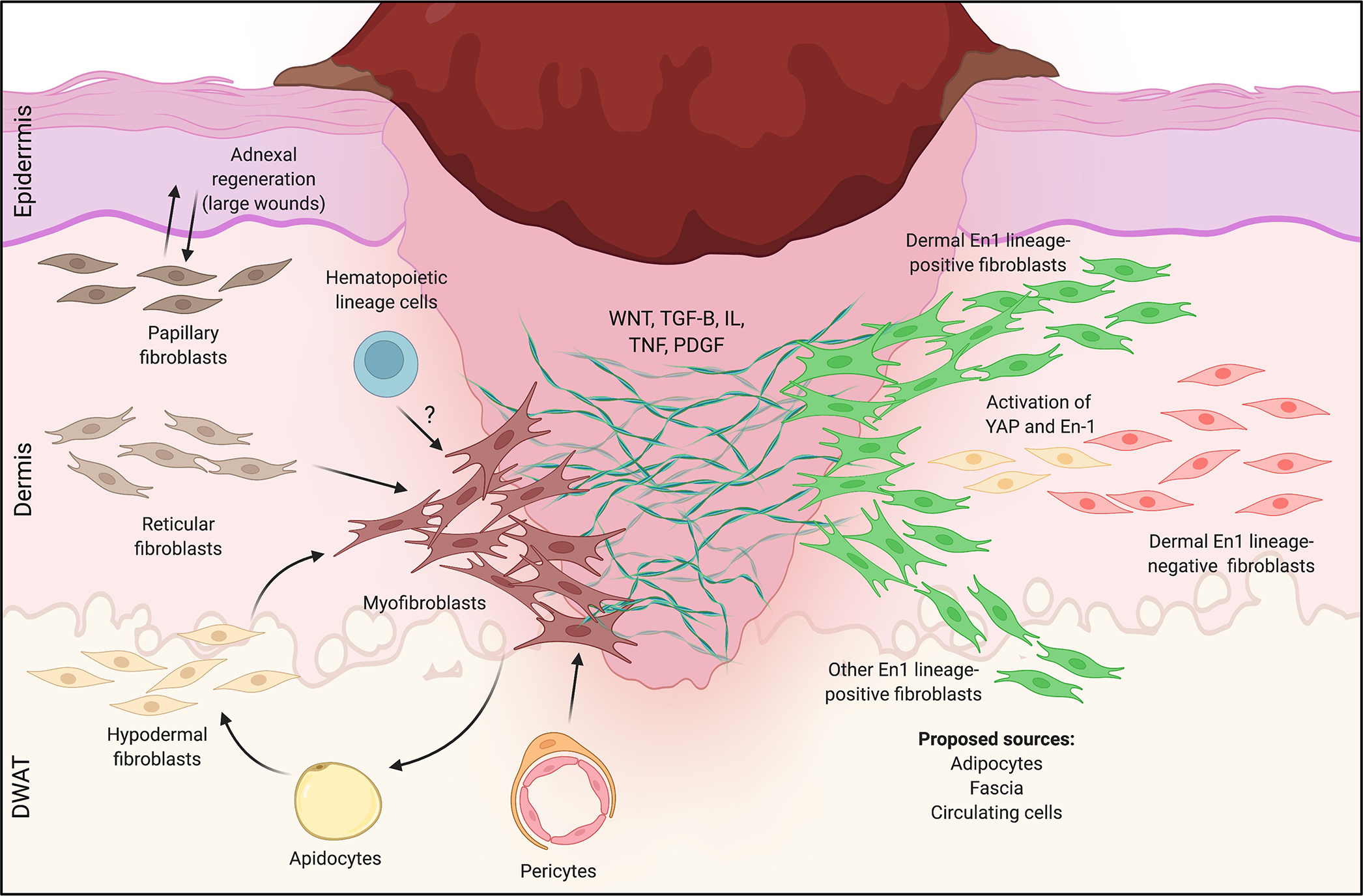
Left: Heterogeneous responses to wounding for the three microanatomically-defined lineages of fibroblasts (papillary, reticular, hypodermal). In small wounds, reticular (middle) and hypodermal (bottom) lineages dominate, giving rise to scarring myofibroblasts. In larger wounds (e.g., WIHN), papillary fibroblasts (top) preferentially interact with epidermal elements to drive sparse adnexal regeneration, and a subset of reticular layer-derived myofibroblasts regenerates adipocytes. Pericytes (bottom middle) and possibly circulating hematopoietic cells (“fibrocytes”, top middle) may also generate scarring myofibroblasts. Right: Heterogeneity in wound healing responses by Engrailed-1 (En-1) lineage-positive (green, EPF) and -negative (red, ENF) fibroblasts. EPFs are responsible for the vast majority of dorsal skin scarring in response to wounding, while ENFs do not contribute to fibrosis; a subset of ENFs reactivates En-1 to contribute postnatally-derived scarring EPFs. Proposed sources of scarring EPFs include the dermis (all layers), adipose tissue, circulating cells, and fascia. How these different lineages of fibroblasts (papillary, reticular, adipose, EPFs/ENFs, etc.) overlap and/or give rise to one another remains unclear.
In addition to the aforementioned fibrogenic and non-fibrogenic (EPF/ENF and PPF/PNF) lineages, skin fibroblasts have also been differentiated into papillary, reticular, and hypodermal subpopulations on the basis of lineage, surface marker expression, and location within the mouse dermis (Driskell et al., 2013; Driskell and Watt, 2015; Phan et al., 2020) (Figure 1). Several mesenchymal cell subpopulations have also been identified in close association with hair follicles, including dermal papillae fibroblasts (which are Sox2/Lef1/Crabp1+ and induce hair follicle formation and cycling), dermal sheath fibroblasts (which are α-SMA/Itga5/Itga8+ and harbor self-renewing hair follicle dermal stem cells), and arrector pili fibroblasts (which are CD26−Itga8+ and involved in piloerection); these hair follicle-associated fibroblasts develop from papillary dermal condensates via Wnt and Shh pathway activation (Lim et al., 2018; Noramly et al., 1999; Phan et al., 2020; Zhang et al., 2009). Consistent with their superficial location within the dermis, papillary fibroblasts (CD26+Lrig+Sca1−) descended from Blimp1 lineage-positive, Dlk1−Lrig+ progenitors are closely associated with the overlying epithelium (Driskell et al., 2013). Supporting fibroblasts’ anatomically-specific roles within the dermis, transplantation of mixed dermal cell suspensions lacking papillary fibroblasts fails to reconstitute either papillary dermal ECM or hair follicles, highlighting the importance of these specific fibroblasts for hair follicle formation (Driskell et al., 2013). In contrast, reticular (Dlk1+Sca1−) and hypodermal (CD24+Dlk1−Sca1+) fibroblasts, which are descended from Blimp1 lineage-negative, Dlk1+ progenitors, reside deeper within the dermis and contribute to the vast majority of early fibrosis and adipocyte regeneration during wound healing (Driskell et al., 2013; Plikus et al., 2017) (Figure 2). By dominating the wound repair process and effectively excluding papillary dermal fibroblasts (which typically do not participate until later in healing, where they play a more minor role), these deeper skin fibroblast populations thereby preclude hair follicle regeneration and instead promote formation of the “bare area” characteristic of a scar.
Single-cell sequencing supports that distinct – yet non-exclusive and highly dynamic – transcriptomic and epigenomic profiles exist for papillary versus reticular lineage fibroblasts (Foster et al., 2021; Thompson et al., 2021). For example, CD26 has been reported as a marker for pro-regenerative papillary fibroblasts in neonatal healing and WIHN, while Dlk1 is expressed by reticular fibroblasts; however, scATAC-seq of neonatal mouse wounds revealed that chromatin regions encoding both CD26 (Dpp4) and Dlk1 are accessible across multiple fibroblast clusters, rather than exclusive to one (Thompson et al., 2021). Further, CD26 is also expressed by 94% of EPFs (which are found across all dermal layers) and by En1-reactivating reticular fibroblasts in the dorsal skin in response to wound mechanical forces (Jiang et al., 2018; Mascharak et al., 2021a; Rinkevich et al., 2015). Such discrepancies highlight the difficulty of reconciling fibroblast heterogeneity simultaneously through micro-anatomical and lineage-based lenses, and these comparisons may be further confounded by differences in the experimental model systems used (e.g., unstented large wounds in WIHN versus smaller stented wounds). It will be important for future studies to establish how much of fibroblasts’ developmentally-encoded behavior (e.g., EPFs’ propensity for scarring, papillary fibroblasts’ association with follicular development and WIHN) depends on experimental context (size of the wound relative to total body surface area, age and breed of mouse, presence or lack of stenting mechanical force, etc.).
Fibroblast heterogeneity according to spatial location
Fibroblasts from different body sites have shown to display distinct gene expression profiles, highlighting fibroblast topographical heterogeneity. Such position-dependent differences in gene expression profiles (e.g., pattern formation, cell-cell signaling, matrix remodeling) along the anterior/posterior, proximal/distal, and dermal/non-dermal axes have found to reflect Hox gene expression (Rinn et al., 2008). The differentiation and specialization of mesenchymal progenitors over the course of development, and their interactions with developing endoderm- and ectoderm-derived structures, in turn yields fibroblast “organotypicity,” or organ-specific fibroblast heterogeneity.
A recent transcriptomic profiling of mouse heart, skeletal muscle, colon, and bladder, found that fewer than 20% of fibroblast-enriched genes were shared between these four organs (Muhl et al., 2020b). Differences in expression were particularly evident for genes comprising the matrisome, supporting the concept that fibroblast ECM production is tailored to the unique properties of each organ. Nonetheless, overarching commonalities may exist between fibroblasts of distinct origins: recently, a fibroblast transcriptional “atlas,” built from 230,000 fibroblasts across 17 mouse and human tissues in homeostatic and perturbed states (such as infection, injury, and fibrosis), revealed ten conserved gene transcriptional clusters reflecting differential enrichment of NF-κB, TNF, Ccl19, and Wnt signaling pathway genes (Buechler et al., 2021). Interestingly, two of these clusters – characterized by Col15a1 and Pi16 expression, respectively – were each represented in nearly all assayed tissues. These clusters were proposed by the authors to represent two “universal” fibroblast phenotypes: one basement membrane-secreting (expressing Col15a1, Col4a1, and Hspg2), and one with a “resource” stem-like phenotype (expressing Pi16, Dpp4, and Ly6c1). Thus, it is possible that transcriptional “tailoring” of organ-specific fibroblast subtypes occurs within the context of limited fundamental, conserved fibroblast gene expression programs that persist across diverse tissues and conditions.
Myofibroblast heterogeneity
In response to Wnt, TGF-β, interleukin, TNF, and PDGF signaling within the wound environment, fibroblasts from various niches (including the dermis, perivascular region, and adipose tissue) transition into myofibroblasts, dramatically increasing their expression of ECM and contractile proteins (such as α-SMA – encoded by the Acta2 gene – and myosins) (Klinkhammer et al., 2018; Murdaca et al., 2014; Piersma et al., 2015) (Figure 2). Like their non-activated fibroblast counterparts in homeostasis, myofibroblasts are heterogeneous and can be further segmented by scRNA-seq into transcriptionally distinct subpopulations. For example, scRNA-seq of mouse wound Acta2+ myofibroblasts revealed 12 populations, which could be further classified into Pdgfrahigh/Pdgfrblow (nine subclusters, including Crabp1+ subpopulations located under epithelium and Crabp1− subpopulations in the deep dermis) and Pdgfralow/Pdgfrbhigh (three subclusters, evenly distributed throughout the dermis) subtypes (Plikus et al., 2017). Another study that combined scRNA-seq with genetic lineage tracing differentiated mouse α-SMA/collagen type I (col-I)+ myofibroblasts into two subtypes – so-called “adipocyte precursors” (CD26highCD29+) and CD29high cells (CD26lowCD29high) – with unique transcriptomes and varying propensities for regenerating adipose tissue in concert with CD301b+ macrophages (Shook et al., 2018). Thus, dermal myofibroblasts likely also represent a heterogeneous cell population with diverse origins, phenotypes, and microenvironmental influences.
Human skin fibroblast heterogeneity
Efforts to fully characterize dermal fibroblast heterogeneity in humans are complicated by limited tissue availability, donor site-specific differences in mechanics and skin structure (as well as individual donor-to-donor variability), and generally poor correlation with known mouse fibroblast markers (Harn et al., 2021; Philippeos et al., 2018). Nonetheless, cell surface markers have been proposed to distinguish papillary (FAP+Thy1−) and reticular (FAP+Thy1+) fibroblasts in human skin (Korosec et al., 2019) (Figure 1). Like their counterparts in the mouse dermis, FAP+Thy1− human papillary fibroblasts were found to be incapable of regenerating adipocytes, while FAP+Thy1+ human reticular fibroblasts expressed high levels of ACTA2, PPARG, and CD36 and readily differentiated into adipocytes, consistent with the pro-fibrotic reticular fibroblast phenotype previously reported in mice. Single-cell transcriptomic profiling has also revealed five transcriptionally distinct fibroblast subpopulations present in human skin, including one localized to the upper dermis (COL6A5+), one in the deep dermis (CD36+ preadipocytes), and three in the reticular dermis (including two previously uncharacterized populations) (Philippeos et al., 2018). A separate study distinguished two major fibroblast clusters (one SFRP2/DPP4+ and one FMO2/LSP1+), with five minor clusters (including one expressing CRABP1) (Tabib et al., 2018). It should be noted, however, that these studies utilized cells derived from unwounded skin, indicating that these fibroblast subdivisions may or may not be relevant in the context of wounding; further, the fibroblast populations’ distinct gene expression signatures were largely lost following cell culture (Tabib et al., 2018; Walmsley et al., 2015), again highlighting a fundamental limitation of the in vitro study of fibroblasts. Transcriptomic profiling of scar, keloid, and scleroderma skin tissues has reportedly reproduced papillary, reticular, mesenchymal, and pro-inflammatory fibroblast subpopulations, with POSTN+ “mesenchymal” fibroblasts implicated in keloid formation (Deng et al., 2021); however, these subtype designations were based in gene expression signatures derived from unwounded skin (Solé-Boldo et al., 2020), further underscoring a need for additional studies specifically characterizing the heterogeneity of fibrotic skin fibroblasts.
Visceral organ fibroblast heterogeneity
Substantial intra-organ heterogeneity also exists in fibroblasts of the visceral organs (e.g., liver, lung, and colon). The liver is capable of a remarkable degree of regeneration, but chronic injury – for instance, prolonged chemical, autoimmune, or malignant insult – can result in cirrhosis, a fibrotic process mediated by myofibroblasts. The cellular origins of scarring liver myofibroblasts are controversial; these pro-fibrotic cells have alternately been proposed to be derived from resident hepatic stellate cells (expressing vitamin A metabolism-related proteins)(Bataller and Brenner, 2005; Geerts, 2001) or portal fibroblasts (Col15a1/elastin/Entpd2+) (Beaussier et al., 2007; Wells et al., 2004), bone marrow-derived mesenchymal cells (Forbes et al., 2004; Russo et al., 2006), or epithelial-to-mesenchymal transition of liver hepatocytes (though recent fate mapping results contest this hypothesis) (Chu et al., 2011; Zeisberg and Duffield, 2010).
The differing contributing populations identified by these studies may be the product of differences in experimental models: myofibroblasts responding to carbon tetrachloride hepatotoxic injury were, reportedly, primarily of stellate cell origin (Zhang et al., 2016), while myofibroblasts generated following cholestatic injury from bile duct ligation were found to originate from portal fibroblasts (Dobie et al., 2019). In an acute non-alcoholic steatohepatitis (NASH) mouse model, Dobie et al. revealed central vein-associated hepatic stellate cells to be the main culprit of liver fibrosis, with Lysophosphatidic Acid Receptor 1 (LPAR1) as a potential anti therapeutic target (Dobie et al., 2019). Stellate cells were further revealed to serve as the source of liver fibrosis via IL11/IL11Rα and TGF-β signaling in an additional scRNA-seq mouse atlas of NASH pathogenesis (Xiong et al., 2019). To obtain a better understanding of liver fibrosis pathogenesis, Ramachandran et al. characterized the transcriptome of 100,000 single human cells from healthy and cirrhotic human livers. Interactome analysis identified PDGFRα+, collagen-producing mesenchymal cells as a key contributor to fibrosis through several intra-scar pro-fibrogenic pathways including TNF Receptor Superfamily Member 12A (TNFRSF12A), PDGFR, and Notch signaling (Ramachandran et al., 2019). Single-cell RNA-seq has also revealed distinct transcriptomic profiles for stellate cells and portal fibroblasts (Dobie et al., 2019; Krenkel et al., 2019) as well as heterogeneity within the stellate cell population, with transcriptional profiles depending on distance from the central veins as well as injury state (Dobie et al., 2019). Such heterogeneity may have direct translational implications, as targeted inhibition of central vein-associated Lpar1+ stellate cells (the dominant injury-responsive cell in a study utilizing chemical liver injury) was shown to mitigate fibrosis (Dobie et al., 2019). Mechanical forces – specifically, Hippo signaling via Yes-associated protein (YAP) – have also been implicated in stellate cell-mediated fibrogenesis, and targeted pharmacological inhibition of YAP/TEAD was found to impede cirrhosis in a chemical liver injury model (Dechêne et al., 2010; Manmadhan and Ehmer, 2019; Mannaerts et al., 2015; Martin et al., 2016).
Like the liver, the lung parenchyma is capable of regeneration, due to its robust population of type II pneumocyte stem cells. However, loss of these stem cells or prolonged injury by agents such as bleomycin can instead induce pulmonary fibrosis, a pathologic process with progressive, devastating clinical consequences. Single-cell transcriptional profiling of normal and fibrotic (bleomycin-treated) mouse lungs has revealed multiple distinct cell subpopulations, including Acta2+ myofibroblasts, Col13a1 and Col14a1-expressing fibroblasts, lipofibroblasts (which are necessary for proper function of type II pneumocytes and clinically implicated in idiopathic pulmonary fibrosis [IPF]), mesothelial cells (which line the pleura), and mesenchymal progenitors (which give rise to the aforementioned cell types during lung morphogenesis), as well as a Pdgfrbhigh subpopulation that specifically emerges in fibrogenesis (El Agha et al., 2017a; Valenzi et al., 2019; Xie et al., 2018). Lineage-tracing experiments and in silico differentiation trajectory analysis also suggest that Tbx4 lineage-positive mesenchymal progenitors contribute to nearly all α-SMA+ myofibroblasts in adult lung injury (Arora et al., 2012; Xie et al., 2016), suggesting that, as in the skin, injury in the lung may induce reactivation of developmental pathways and contributions from multiple mesodermal lineages. As with dermal fibroblasts and stellate cells of the liver, Hippo mechanotransduction signaling has been shown to drive fibroblast activation, ECM deposition, and tissue stiffening in the lung in the context of bleomycin injury or IPF; accordingly, indirect inhibition of YAP/TAZ signaling (via fibroblast-specific dopamine receptor D1 antagonism) was found to ameliorate pulmonary fibrosis (Haak et al., 2019). Across multiple organs, activation of YAP/TAZ signaling promotes fibrosis, and NUAK family kinase 1 has been recently shown to act as a profibrotic kinase driving a positive feedback loop in concert with TGF-β and YAP/TAZ activation (Zhang et al., 2022).
In contrast to the liver and lungs, cardiac tissue does not have the ability to regenerate following acute or chronic injury after roughly the first week of life (Lam and Sadek, 2018) and instead responds with devastating fibrosis. Fibroblasts derived from epicardium (see section on epithelial-to-mesenchymal transition, below) appear to be the main effector cells of cardiac fibrosis and are activated following injury (Davis and Molkentin, 2014). In a similar approach to studies performed in the skin, lineage-tracing and single-cell -omics have been applied to healthy and fibrotic cardiac tissue to identify novel fibroblast subpopulations and lineages, though the functional importance of cardiac fibroblast heterogeneity is not yet clear (Ali et al., 2014; Muhl et al., 2020a; Wang et al., 2022). Such approaches have, however, successfully identified possible key molecular regulators of cardiac fibrosis. Single-cell profiling of infarcted cardiac tissue in mice revealed a fibrotic signature in Collagen Triple Helix Repeat Containing 1 (Cthrc1)-expressing fibroblasts within scars (Ruiz-Villalba et al., 2020). Furthermore, fibroblasts with a similar signature were identified in heart tissues from patients with myocardial infarcts and dilated cardiopathy (Ruiz-Villalba et al., 2020). The transcription factor Aebp1 has also been identified as a crucial cardiac fibrosis regulator in α-SMA+ myofibroblasts in human dilated cardiomyopathy and ischemic cardiomyopathy (Rao et al., 2021). Post-transcriptional control of cardiac wound healing by MBNL1 has recently been implicated in regulating fibroblast state plasticity during cardiac wound healing (Bugg et al., 2022). Suppression of the Hippo pathway has been found to play an antifibrotic role in the heart: Xiao et al. found that Lats1 and Lats2 mutant resting cardiac fibroblasts transitioned to a myofibroblast-like cell state (Xiao et al., 2019). In contrast, Lats1/2 deletion led to a relentless pro-fibrotic and pro-inflammatory cascade that ultimately resulted in organ failure. Reducing levels of YAP/TAZ effectors in Lats1/2 mutants attenuated the fibrotic phenotype following infarction. Thus, Hippo signaling appears to regulate cardiac fibroblast fate transition upon injury. Finally, recent evidence suggests that the oxygen sensing in the post-ischemic heart plays a critical role in regulating proliferation of cardiac fibroblasts, with Hif-1a opposing fibroblast proliferation through regulation of mitochondrial reactive oxygen species (ROS) (Janbandhu et al., 2022).
Fibroblast plasticity in fibrosis
While fibroblasts clearly exhibit heterogeneity by lineage, phenotypic markers, and transcriptomic profiles, the notion of distinct fibroblast identities is complicated by these cells’ plasticity and propensity for interconversion with other cell types. A canonical example of fibroblast plasticity potentially implicated in fibrosis is the phenomenon of epithelial-to-mesenchymal transition (EMT), which involves epithelial cells losing their typical cell-cell adhesions and apical-basal polarity to generate motile “fibroblastoid” cells (Kalluri and Neilson, 2003). Type I EMT refers to EMT that occurs during the process of development: for example, PDGF and TGF-β signaling in the developing heart induces expression of Sox9, Snail, and Slug, causing epicardial epithelial cells to convert into cardiac fibroblasts (Gittenberger-de Groot et al., 1998; von Gise and Pu, 2012). Type II EMT, in contrast, occurs during wound healing: in the heart, mirroring the developmental EMT process, EMT of epicardial cells following injury contributes fibroblasts and coronary smooth muscle cells to damaged cardiac tissue (Lepilina et al., 2006; Stone et al., 2016). Similarly, mechanical tension, NF-κB, TGF-β, and Wnt signaling may lead to generation of α-SMA+ myofibroblasts from injury-responsive epithelial progenitors in renal, liver, and pulmonary fibrosis (Wynn and Ramalingam, 2012). Targeting EMT may represent an anti-fibrotic strategy: EMT was reversed by using viral delivery of transcription factors to reprogram myofibroblasts into hepatocyte-like cells (expressing Foxa3, Gata4, Hnf1a, and Hnf4a) (Rezvani et al., 2016; Song et al., 2016) and cardiomyocytes (expressing Gata4, Mef2c, Tbx5), which could hold promise for ameliorating liver cirrhosis and cardiac fibrosis (Miyamoto et al., 2018). Although the transdifferentiation of epithelial cells to fibroblasts may represent the origin of a specific subset of myofibroblasts, many questions remain unanswered regarding EMT (Lovisa, 2021). The molecular pathways that contribute to the activation of EMT and this process’s orchestration at a transcriptional level remain to be elucidated, including during skin fibrosis.
Plasticity is also evident in the contributions of cells from various mesenchymal compartments to scarring. In the dermis, the lower (reticular) lineage of fibroblasts was initially implicated as the source of α-SMA+ myofibroblasts for fibrosis (Driskell et al., 2013); however, more recently, it was shown that all neonatal fibroblast populations defined by scATAC-seq have accessible chromatin for canonical myofibroblast markers (Acta2, Tagln, and Tgfbr2) (Thompson et al., 2021), suggesting the potential for greater plasticity than previously appreciated. Conversion of adipocytes, pericytes, fascial fibroblasts, and circulating hematopoietic cells into myofibroblasts has also been implicated in mouse skin and lung fibroses (Abe et al., 2001; Correa-Gallegos et al., 2019; Di Carlo and Peduto, 2018; Jiang et al., 2020; Shook et al., 2020). Furthermore, reversal of these cell-type transformations may be involved in regeneration of skin and lung parenchyma. In the context of WIHN, myofibroblasts were found to terminate their contractile behavior and convert into adipocytes in response to bone morphogenetic protein (BMP) ligands secreted by regenerated hair follicles (Plikus et al., 2017; Shook et al., 2018). Similarly, loss of myofibroblast-to-lipofibroblast conversion is a hallmark of IPF (El Agha et al., 2017b; Habiel and Hogaboam, 2017). It should be noted that reports of these cell-type interconversions have largely been derived from studies in highly-engineered mouse model systems; thus, while promising, further work is needed to establish their relevance to human tissue repair and fibrotic disease.
Another important functional consideration complicating fibroblast plasticity during skin repair is that the skin is naturally the first line of defense against environmental assaults. The skin must constantly respond to insults – whether significant injuries, akin to experimentally-induced or surgical wounds, or more minor tissue damage such as sun exposure over a human’s lifetime – to maintain its homeostatic functioning. Experimentally, this can be challenging as skin states may be significantly affected by model organisms’ behavioral tendencies, such as fighting or self-injury in mice. Furthermore, the biology of wound repair and skin can be impacted by tissue’s history of prior injury. Naik et al. illustrated that epidermal stem cells (EpSCs) have a prolonged memory to acute inflammation: these cells maintained increased chromatin accessibility of specific stress-response genes following injury, with some sites persisting up to 180 days after initial injury (Naik et al., 2017). Upon secondary insult, these “memory” genes were transcribed rapidly. Thus, the capacity of the EpSCs to “learn” from their prior experience of injury seems to prime skin to respond to subsequent injury more efficiently. Similarly, Gonzales et al. found that EpSCs developed epigenetic adaptations in wound repair-related genes following injury, allowing them to later transcribe stored wound memory genes involved in inflammation, cytoskeletal reorganization, and cell migration upon secondary injury for an accelerated wound healing response (Gonzales et al., 2021). It will be of interest for future studies to determine whether fibroblasts can similarly develop and maintain cellular “memory” to augment repeated responses to injury stimuli.
In attempting to reconcile fibroblast heterogeneity with plasticity, several themes emerge. First, developmental origin (e.g., lineage) and microenvironmental context (e.g., ECM stiffness, local inflammatory cytokines, cell-cell interactions) likely converge to determine fibroblast behavior. Second, unlike the unidirectional differentiation hierarchies of well-defined systems such as the hematopoietic cell lineage, fibroblasts and other mesenchymal populations seem to relate in a more complex, dynamic network that is highly context-dependent and variable by experimental model. The particular mode of tissue damage and specific model organism (e.g., neonatal versus aged mouse) may dictate which fibroblast subpopulations dominate the injury response and, accordingly, the final outcome of injury. One example of such context dependence in mice is the partially-regenerative healing by papillary fibroblasts observed in large contracting wounds (WIHN) compared to scarring healing by En1-activating reticular fibroblasts found in smaller stented wounds (Mascharak et al., 2021a; Rinkevich et al., 2015; Thompson et al., 2021). Third, importantly, fibroblast heterogeneity at rest does not correspond to fibroblast heterogeneity in injury; tissue damage may initiate multiple, distinct differentiation trajectories as various mesenchymal compartments and/or fibroblast subtypes are mobilized. Finally, molecular markers or cell subpopulations determined to have functional significance in lower organisms often do not have corollaries in human tissues, highlighting the critical role of model systems and organisms that more accurately approximate human fibrotic disease mechanisms.
Contributions from other cell types
The pivotal role of immune cells in fibrosis and wound repair has led to interest in interrogating and understanding the cross-talk between immune cells and fibroblasts, with a particular focus on macrophage-fibroblast interactions. Multiple studies have attempted to identify macrophage subpopulations capable of mitigating pro-fibrotic fibroblast activity. One macrophage-fibroblast co-culture study revealed that CD163 overexpression in human primary macrophages promoted a less-fibrotic fibroblast phenotype and supported a “pro-resolution” wound molecular milieu (Ferreira et al., 2020). Transplantation of a CD301b-expressing macrophage subpopulation into partially-healed (POD 3) mouse wounds was also found to increase fibroblast proliferation in and repopulation of wounds, consistent with these cells representing a reparative, anti-inflammatory macrophage phenotype (Shook et al., 2016). Collectively, the identification of signaling pathways governing fibroblast-immune cross-talk may ultimately allow for manipulation of the wound microenvironment toward skin regeneration. Canonical Wnt signaling has been implicated in fibrogenic myofibroblast activity in multiple tissues (Burgy and Königshoff, 2018; Piersma et al., 2015) and may also play a role in immune cell-fibroblast interactions in fibrosis. Gay et al. demonstrated that Wnt-related immune activity is associated with dermal scarring in mouse WIHN, with macrophages in late wounds found to phagocytize the dermal Wnt inhibitor SFRP4 to establish persistent Wnt activity and drive skin fibrosis (Gay et al., 2020). Hippo signaling may also be relevant to such bidirectional fibroblast-macrophage crosstalk, as recent studies suggest that YAP mechanotransduction signaling in fibroblasts elevates expression of colony stimulating factor 1 (Csf1), a lineage-specific growth factor for macrophages (Zhou et al., 2022; Zhou et al., 2018).
Another important factor to consider when studying fibroblast heterogeneity and wound healing is the role of hair follicle populations. The skin contains hair follicles, which are not static but rather actively cycle throughout the mammalian life cycle (Houschyar et al., 2020). This fact represents a potential confounding factor in experiments relating to the skin, as, for instance, wound-relevant biology can differ based on hair follicle stage (Matsuzaki and Yoshizato, 1998; Morgan, 2014). Further, hair follicles represent important stem and progenitor cell niches with a unique cellular composition compared to the rest of the skin; these cells are known to contribute to wound repair (Ito et al., 2005; Morgun and Vorotelyak, 2020). As humans are relatively hairless compared to mice, effects of hair follicle cells and molecular signaling on wound repair may be more pronounced in common (rodent) experimental models but relatively less relevant in human wound repair, posing a translational barrier. While studies have examined hair follicle cell populations at the single-cell level in both mice (Ge et al., 2020; Joost et al., 2016) and humans (Takahashi et al., 2020), these studies largely focused on development and/or homeostasis and few studies have looked explicitly or in depth at these cells, and their potential interactions with fibroblasts and other cell populations, in wound repair and fibrosis.
Although these studies are encouraging, such approaches are limited to the evaluation of the few already-known pathways and cell types that drive skin fibrosis. However, novel computational analytic methods may allow for broader interrogation of cell-cell interactions. A scRNA-seq tool called CellChat was recently developed to capture interactions between fibroblasts and other cell types in the skin during embryonic development and adult skin wound healing (Jin et al., 2021). This tool uses known ligand-receptor interactions to computationally predict interactions between cell populations based on single-cell transcriptomic datasets. In this study, 60 significant ligand-receptor pairs were identified in 25 cell groups in mouse wounded skin (POD 12). CellChat identified a complex, widespread, and highly influential network of TGF-β signaling involving wound fibroblasts, with multiple ligands identified as driving fibroblast-fibroblast, fibroblast-myeloid, and fibroblast-endothelial interactions. In contrast, outgoing non-canonical Wnt signaling was only identified in one population of fibroblasts and primarily drove fibroblast-to-fibroblast communication. In this study, CellChat analysis also highlighted that communication patterns between embryonic skin and adult wounds differed dramatically. While no signaling pathway was entirely unique to either embryonic skin or adult wound repair, the relative strength of each signaling network differed by context, with fibroblast growth factor (FGF), nerve growth factor (NGF), and Wnt signaling dominating embryonic skin development and TGF-β, galectin, and epidermal growth factor (EGF) predominating during adult wound repair. Overall, newly-developed scRNA-seq tools to assess global communication patterns among diverse wound cell types may be useful in identifying pathways for cell crosstalk that can be targeted to modulate fibrosis.
Future research directions
Molecular tools in the study of wound repair
Cutting-edge molecular tools continue to shed new light on the complex mechanisms of skin biology, wound repair, and fibrosis. Novel methods are being applied to study wound healing at multiple molecular “levels,” from epigenomic to transcriptomic to proteomic to tissue level (Figure 3). These include methods for characterizing the wound milieu at single-cell resolution and increasingly high-resolution methods for spatial interrogation of wound processes. However, these methods have key limitations to consider when interpreting and contextualizing data derived from them, which should be addressed in future studies to define fibroblast heterogeneity more universally.
Figure 3: Levels of molecular regulation of fibroblast behavior.
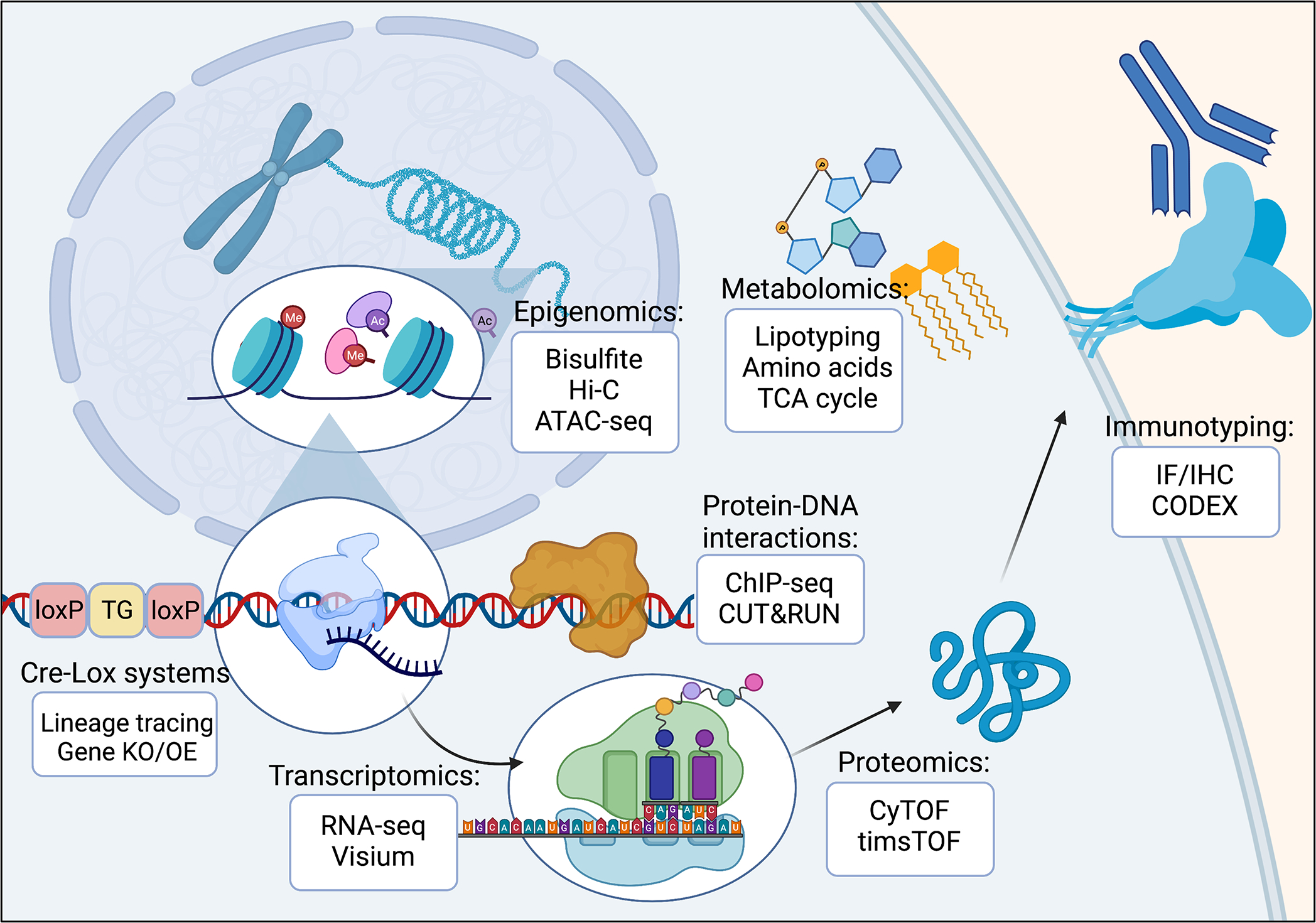
Fibroblast behavior depends on the complex interplay of many factors, including these cells’ epigenomic, transcriptomic, and proteomic profiles. Modern technologies are able to interrogate fibroblasts at each of these molecular levels to better understand fibroblast heterogeneity and functioning.
Species-specific differences
Regardless of the specific methods used for analysis, species-specific differences must always be considered and reconciled. Fibroblast subpopulations in homeostatic mouse skin have now been characterized by numerous studies using lineage-tracing and immunohistochemical techniques (Driskell and Watt, 2015). Several markers have been identified as discriminating papillary and reticular mouse skin fibroblasts, including Dpp4 (Korosec et al., 2019), netrin-1 (Ntn1), podoplanin (Pdpn), and matrix Gla protein (Mgp) (Janson et al., 2012; Nauroy et al., 2017; Reynolds et al., 2021; Sorrell and Caplan, 2009). However, attempts to apply these gene markers to human scRNA-seq datasets have displayed minimal cluster specificity (Vorstandlechner et al., 2020), highlighting the fact that fibroblast cluster-defining markers may be species-specific and cross-species conclusions must be tempered. Additionally, animal studies often rely on subjective analysis of histologic images, which can be prone to bias. Quantitative and increasingly automated image processing approaches may help to streamline and remove subjective bias from wound and fibrosis analyses; for instance, machine learning analysis of ECM and cellular interaction patterns, clinical datasets, and -omics data is increasingly being used to establish diagnostic and prognostic signatures of fibrotic disease (Mascharak et al., 2021a; Mäkelä et al., 2021; Showalter et al., 2021).
Differences in computational methods
A specific challenge of scRNA-seq, one of the most commonly used methods for interrogation of fibroblast heterogeneity, is low capture efficiency and high dropout rate, with relatively few mRNA molecules ultimately analyzed per captured cell (Liu and Trapnell, 2016). This leads to noisier and more variable data than prior methods such as bulk RNA-seq, which can provide substantial challenges for computational scRNA-seq analysis (Poirion et al., 2016). Initial data analysis requires clustering transcriptionally similar cells using computer-based methods (e.g., nearest-neighbor analysis); following the identification and annotation of cluster groups, distinct cluster gene signatures can be identified. However, resolution of individual clusters is highly subjective, depending on both the user and specific methods employed (Kharchenko, 2021). To date, there have been diverse efforts to cluster human and mouse fibroblast populations at the single-cell level, which have identified anywhere from three to twelve distinct fibroblast subpopulations (He et al., 2020; Solé-Boldo et al., 2020; Tabib et al., 2018); it is not yet clear how much of this variability results from true biological differences (e.g., wounded versus unwounded skin) and how much is a product of differing computational/analysis methods. Future methods to enable increased uniformity of cluster configuration between studies may aid in the increasingly robust identification of fibroblast subpopulations and integration of findings and conclusions across multiple studies.
Inference of cellular trajectories
A fundamental limitation of current sequencing methods is that protocols are designed to capture a static snapshot of cell states from a specific biological state or point in time (for instance, unwounded skin, or a given postoperative day in wound healing). This is clearly an issue for the study of a process like skin repair, which is highly dynamic, involves multiple biologically discrete but temporally overlapping stages, and may be misunderstood by simply analyzing a single timepoint. Multiple scRNA-seq analysis methods have been developed with the goal of deducing dynamic processes from these static/single-state datasets, including Monocle, a tool for establishing “trajectories” of changing gene expression among cells within a scRNA-seq dataset. However, these tools require caution in use and interpretation: for instance, they typically attempt to incorporate all analyzed cells within the same trajectories, possibly including cells that do not actually participate in the process of interest (Kharchenko, 2021). Further, even the most advanced computational methods cannot compensate for biological sampling limitations, such as a limited range of conditions or postoperative timepoints examined. Trajectory analysis methods may be most powerful when combined with experimental sampling across multiple distinct biological states, such as a recent study which employed multiple high-throughput methods (including scRNA-seq) combined with differentiation/trajectory analyses to establish dynamic trajectories of wound repair in both fibrosis and regeneration (Mascharak et al., 2022). Future longitudinal studies, incorporating samples collected at multiple timepoints and during distinct phases of dynamic processes such as wound repair, will enable more robust and powerful conclusions to be drawn from trajectory analyses and may provide a more nuanced understanding of molecular regulators that govern cell fate decisions in and outside the skin. Such approaches may also be bolstered by advancements in methods for real-time analysis of fibroblast and wound biology, such as improved in vitro models (e.g., functional organoid models with both epidermal and dermal components (Lee and Koehler, 2021; Lee et al., 2020)) and intravital microscopy (Jiang et al., 2020). Inference of cellular trajectories will also benefit from improved methods for lineage tracing, which conventionally rely on expression of reporter molecules under control of site-specific recombinases (e.g., Cre) but lack single-cell resolution. Genetic barcoding is a promising approach to bypass this limitation, whereby cell lineage may be linked to single-cell -omics data by expression of unique heritable genetic sequences (e.g., polylox and CRISPR barcodes) (Kalhor et al., 2018; Pei et al., 2017).
Loss of spatial information
The spatial organization of skin is critical to its function, with skin containing distinct anatomical layers (containing heterogeneous fibroblast populations) and defined structures such as hair follicles and glands (Driskell and Watt, 2015). Although existing single-cell analyses are useful in providing high-resolution molecular information from a cellular perspective, a significant drawback is that all spatial information is lost, as the tissue must be homogenized for individual cells to be processed through the sequencing pipeline. Thus, conclusions about the functional contributions of specific, isolated fibroblast subpopulations are inherently correlative. Transcriptomic and proteomic platforms such as Visium and CODEX, respectively, have introduced a spatial component to traditional RNA-sequencing or surface marker-based studies of cellular heterogeneity (Black et al., 2021; Foster et al., 2021; Goltsev et al., 2018). Such spatially-informed approaches may provide a deeper understanding of the mechanisms that govern fibroblast behavior during wound repair. To date, one study has examined the spatial transcriptomic profile of wounds, using Visium spatial transcriptomic sequencing (10x Genomics) to analyze mouse wounds over time (POD 2, 7, and 14) (Foster et al., 2021). The epidermal, dermal, and hypodermal layers were confirmed to cluster distinctly based on anatomically-specific transcriptional programs. Characteristic patterns of gene expression were confirmed for key known wound cell types within the spatial context of wounds; for example, Krt6b expression marked keratinocytes found superficially in the skin; Pdgfra was expressed by fibroblasts within the dermis; and Msr1 distinguished macrophages within the skin. At POD 14, there were clear spatial distinctions between transcriptomic profiles of the superficial versus deep regions of the dermis (e.g., Timp1 was expressed in the basal dermis and Thbs2 in the apical scar region). High-throughput spatial transcriptomic sequencing and other spatially-informed analyses (e.g., RNAscope multiplexed in situ hybridization) have yet to be widely applied in the fields of wound healing and fibroblast research, but they may hold promise for future studies examining spatially-restricted processes relevant to wound repair (e.g., epithelial-mesenchymal interactions at the epidermis-dermis junction, or hair follicle regeneration in wounds).
Multi-omic approaches
As is clear from a review of the literature, individual methods have advanced significantly and applications of novel techniques have meaningfully furthered the study of wound healing and fibroblast biology in recent years. However, the above challenges, and the lack of unified conclusions drawn from existing research (as evidenced by the relative dearth of translational advancements in the clinical wound healing space), underscore the hazards of overreliance on any single research tool or methodology in the complex study of fibroblast heterogeneity, wound repair, and fibrosis. Particularly promising moving forward are approaches that combine multiple analytic modalities, such as single-cell sequencing and spatial mapping or transcriptomic and epigenomic sequencing, as these can provide increased depth of insight compared to any single data type (Abbasi et al., 2020; Phan et al., 2021). Multi-omic approaches may be facilitated by tagging harvested cells with barcode oligonucleotide-labeled hashing antibodies, which allows for data to be linked, at the biological replicate level, across multiple assays (for example, paired scRNA-seq and scATAC-seq) (Foster et al., 2021; Mascharak et al., 2022; Thompson et al., 2021). This approach enables imputation of datasets onto one another and direct correlation of epigenomic, transcriptomic, proteomic, or other features, allowing for increasingly precise definition of fibroblast states (Foster et al., 2021; Granja et al., 2021; Mascharak et al., 2022).
Toward translation
This exploration of the current literature clearly shows that wound healing and fibroblast biology are an active and exciting area of research. New discoveries are regularly being made that shed light on the fundamental mechanisms of physiology and pathology in the skin. Overall, owing in large part to fundamental differences between model organism (e.g., mouse) and human biology, as well as limitations of individual methodologies and studies, translational progress has been modest. To date, no targeted molecular therapies to promote wound repair, prevent scarring, or drive wound regeneration exist (and therapeutic options for fibroses in other organs are similarly limited). However, taking a step back, patterns can be drawn from the robust body of wound healing and fibroblast literature that hold significant translational promise. While individual studies have critical limitations, a more robust approach may be to look for emergent patterns across multiple studies, which may represent more fundamental truths about the drivers of fibrosis.
A promising “case study” that speaks to this strategy is the example of tissue mechanics and mechanotransduction in scarring, which has emerged over numerous studies (Figure 4) (Barnes et al., 2018; Harn et al., 2019; Mascharak et al., 2021b; Ogawa, 2011). In vitro evidence has long supported the ability of stiff substrates, such as tissue culture plastic, to activate contractile and pro-fibrotic molecular fibroblast programming. In vivo, the importance of skin tension in scarring is supported by “experiments of nature” – for instance, early-gestation fetal skin has extremely low resting tension and heals without a scar (Hu et al., 2018; Larson et al., 2010), as do extremely loose-skinned Acomys (African spiny) mice (Seifert et al., 2012). It is also demonstrated by studies showing both that increasing tension across a wound increases scarring (Aarabi et al., 2007; Gurtner et al., 2011; He et al., 2021; Son and Hinz, 2021) and, conversely, that either decreasing tension or inhibiting cellular mechanical signaling (mechanotransduction) reduces scarring in wounds (Chen et al., 2021; Gurtner et al., 2011; Ma et al., 2018; Mascharak et al., 2021a; Wong et al., 2013; Wong et al., 2011a) as well as split-thickness skin grafts (Chen et al., 2022). Providing the ultimate translational validation, two randomized controlled clinical trials showed that tension offloading of human surgical wounds (following either abdominoplasty or scar revision operations) significantly reduced scarring (Lim et al., 2014; Longaker et al., 2014). The trajectory of knowledge in this “case study” – moving from in vitro evidence, to small and then large animal studies, ultimately to the clinic – highlights the possibilities for basic knowledge of fibroblast biology to ultimately pave the way for novel therapies. Further supporting a conserved role for mechanical signaling in fibrosis, mechanotransduction has been shown to be involved and/or dysregulated in other organs – including liver and lung – and in aging as well as injury states (Angelini et al., 2020; Dechêne et al., 2010; Haak et al., 2019; Manmadhan and Ehmer, 2019; Mannaerts et al., 2015; Martin et al., 2016), which further implies that mechanics-targeted approaches may be similarly effective in preventing fibrosis in diverse organs other than the skin.
Figure 4: Mechanisms of mechanotransduction signaling.
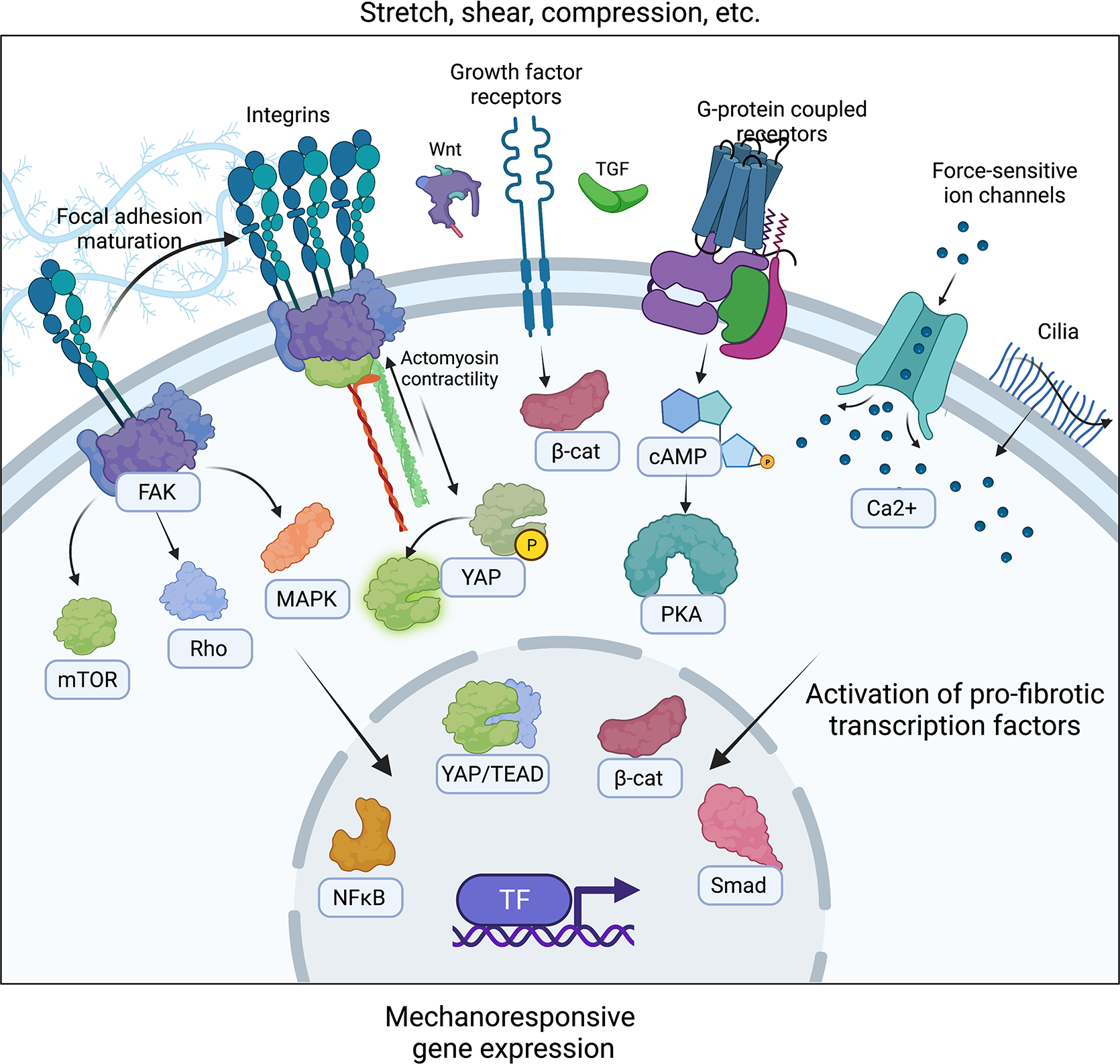
Schematic depicting the diverse mechanisms and signaling pathways/cascades by which fibroblasts and other mechanosensitive cell types may communicate changes in their mechanical environment to molecular phenotypic changes. Multiple levels of evidence exist to show that fibroblasts activate pro-fibrotic machinery in response to mechanical stress, and that fibroblast mechanical signaling can be targeted to mitigate fibrosis.
While no other pathway has been as thoroughly translated in the context of wound healing and scarring, similar patterns certainly emerge when the literature is taken as a whole – for instance, the importance of inflammation in fibrosis. This scenario may be more complex, as both “positive” (i.e., pro-regenerative/anti-fibrotic) and “negative” (pro-fibrotic) examples and forms of inflammation exist, precluding blanket inhibition of inflammation or ablation of immune cells as a viable target for blocking fibrosis. However, as studies continue to establish the precise mechanisms by which inflammatory cells communicate with other fibrosis-relevant cell types (including distinct subtypes of fibroblasts), and their dynamics throughout fibrosis development and resolution, similarly promising translational approaches may be identified which could be effective in targeting fibrosis in higher organisms and, ultimately, humans. It is critical for researchers to bear in mind not only how we can advance our fundamental understanding of molecular and cellular biology, but also how we can continually integrate and contextualize knowledge to draw increasingly powerful conclusions from the vast existing body of work.
Conclusions
Fibroblasts are a central cellular contributor to the skin and other organs, with dynamic roles in homeostasis, wound repair, and fibrosis. Long dismissed as relatively passive “bystander” or “filler” cells, fibroblasts are increasingly appreciated as critical mediators and governors of both physiologic and pathologic ECM deposition and remodeling. These cells have also been shown in recent years to possess impressive plasticity and heterogeneity, with phenotypically and functionally distinct subtypes of fibroblasts contributing to different anatomical sites and biological processes. Many key questions – including the full extent of fibroblast heterogeneity both in and outside of the skin, how fibroblast states change when perturbed (e.g., by injury), and how these cells are influenced by interactions with other skin and wound cell types – remain to be addressed, but emerging research techniques promise to shed further light on these fascinating cells in coming years.
Acknowledgments:
Funding: This work was supported by NIH R01-GM136659, NIH U24-DE029463, NIH R01-DE027346, the Wu Tsai Human Performance Alliance, the Hagey Laboratory for Pediatric Regenerative Medicine, the Gunn Olivier Fund, the Scleroderma Research Foundation, and the Pitch and Catherine Johnson Fund.
Footnotes
Declaration of interests: MTL is co-founder of, has an equity position in, and is on the Board of Directors of Neodyne Biosciences, Inc., which developed the embrace device. MTL, SM, and HET are inventors on patent 62/879,369 held by Stanford University that covers use of YAP inhibition for wound healing and patent application PCT/US2020/043717 that covers a machine-learning algorithm for analysis of connective tissue networks in scarring and chronic fibroses.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aarabi S, Bhatt KA, Shi Y, Paterno J, Chang EI, Loh SA, Holmes JW, Longaker MT, Yee H, and Gurtner GC (2007). Mechanical load initiates hypertrophic scar formation through decreased cellular apoptosis. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 21, 3250–3261. 10.1096/fj.07-8218com. [DOI] [PubMed] [Google Scholar]
- Abbasi S, Sinha S, Labit E, Rosin NL, Yoon G, Rahmani W, Jaffer A, Sharma N, Hagner A, Shah P, et al. (2020). Distinct Regulatory Programs Control the Latent Regenerative Potential of Dermal Fibroblasts during Wound Healing. Cell Stem Cell 27, 396–412 e396. 10.1016/j.stem.2020.07.008. [DOI] [PubMed] [Google Scholar]
- Abe R, Donnelly SC, Peng T, Bucala R, and Metz CN (2001). Peripheral blood fibrocytes: differentiation pathway and migration to wound sites. J Immunol 166, 7556–7562. 10.4049/jimmunol.166.12.7556. [DOI] [PubMed] [Google Scholar]
- Ali SR, Ranjbarvaziri S, Talkhabi M, Zhao P, Subat A, Hojjat A, Kamran P, Muller AM, Volz KS, Tang Z, et al. (2014). Developmental heterogeneity of cardiac fibroblasts does not predict pathological proliferation and activation. Circulation research 115, 625–635. 10.1161/CIRCRESAHA.115.303794. [DOI] [PubMed] [Google Scholar]
- Angelini A, Trial J, Ortiz-Urbina J, and Cieslik KA (2020). Mechanosensing dysregulation in the fibroblast: A hallmark of the aging heart. Ageing Res Rev 63, 101150. 10.1016/j.arr.2020.101150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansell DM, Campbell L, Thomason HA, Brass A, and Hardman MJ (2014). A statistical analysis of murine incisional and excisional acute wound models. Wound Repair Regen 22, 281–287. 10.1111/wrr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Metzger RJ, and Papaioannou VE (2012). Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet 8, e1002866. 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Fogg D, Garfa M, Elain G, Join-Lambert O, Kayal S, Sarnacki S, Cumano A, Lauvau G, and Geissmann F (2007). Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317, 666–670. 10.1126/science.1142883. [DOI] [PubMed] [Google Scholar]
- Baranyi U, Winter B, Gugerell A, Hegedus B, Brostjan C, Laufer G, and Messner B (2019). Primary Human Fibroblasts in Culture Switch to a Myofibroblast-Like Phenotype Independently of TGF Beta. Cells 8. 10.3390/cells8070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes LA, Marshall CD, Leavitt T, Hu MS, Moore AL, Gonzalez JG, Longaker MT, and Gurtner GC (2018). Mechanical Forces in Cutaneous Wound Healing: Emerging Therapies to Minimize Scar Formation. Adv Wound Care (New Rochelle) 7, 47–56. 10.1089/wound.2016.0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataller R, and Brenner DA (2005). Liver fibrosis. J Clin Invest 115, 209–218. 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayreuther K, Rodemann HP, Hommel R, Dittmann K, Albiez M, and Francz PI (1988). Human skin fibroblasts in vitro differentiate along a terminal cell lineage. Proc Natl Acad Sci U S A 85, 5112–5116. 10.1073/pnas.85.14.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussier M, Wendum D, Schiffer E, Dumont S, Rey C, Lienhart A, and Housset C (2007). Prominent contribution of portal mesenchymal cells to liver fibrosis in ischemic and obstructive cholestatic injuries. Lab Invest 87, 292–303. 10.1038/labinvest.3700513. [DOI] [PubMed] [Google Scholar]
- Biasin V, Crnkovic S, Sahu-Osen A, Birnhuber A, El Agha E, Sinn K, Klepetko W, Olschewski A, Bellusci S, Marsh LM, and Kwapiszewska G (2020). PDGFRα and αSMA mark two distinct mesenchymal cell populations involved in parenchymal and vascular remodeling in pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 318, L684–L697. 10.1152/ajplung.00128.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black S, Phillips D, Hickey JW, Kennedy-Darling J, Venkataraaman VG, Samusik N, Goltsev Y, Schürch CM, and Nolan GP (2021). CODEX multiplexed tissue imaging with DNA-conjugated antibodies. Nat Protoc 16, 3802–3835. 10.1038/s41596-021-00556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli MR, Shen AH, Griffin M, Mascharak S, Adem S, Deleon NMD, Ngaage LM, Longaker MT, Wan DC, and Lorenz HP (2021). A Novel Xenograft Model Demonstrates Human Fibroblast Behavior During Skin Wound Repair and Fibrosis. Adv Wound Care (New Rochelle). 10.1089/wound.2020.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechler MB, Pradhan RN, Krishnamurty AT, Cox C, Calviello AK, Wang AW, Yang YA, Tam L, Caothien R, Roose-Girma M, et al. (2021). Cross-tissue organization of the fibroblast lineage. Nature 593, 575–579. 10.1038/s41586-021-03549-5. [DOI] [PubMed] [Google Scholar]
- Bugg D, Bailey LRJ, Bretherton RC, Beach KE, Reichardt IM, Robeson KZ, Reese AC, Gunaje J, Flint G, DeForest CA, et al. (2022). MBNL1 drives dynamic transitions between fibroblasts and myofibroblasts in cardiac wound healing. Cell Stem Cell 29, 419–433.e410. 10.1016/j.stem.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgy O, and Königshoff M (2018). The WNT signaling pathways in wound healing and fibrosis. Matrix Biol 68–69, 67–80. 10.1016/j.matbio.2018.03.017. [DOI] [PubMed] [Google Scholar]
- Bussone G (2017). Subjectivity in primary headaches: insight the causes. Neurol Sci 38, 1–2. 10.1007/s10072-017-2949-y. [DOI] [PubMed] [Google Scholar]
- Chen JS, Longaker MT, and Gurtner GC (2013). Murine models of human wound healing. Methods Mol Biol 1037, 265–274. 10.1007/978-1-62703-505-7_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Henn D, Januszyk M, Barrera JA, Noishiki C, Bonham CA, Griffin M, Tevlin R, Carlomagno T, Shannon T, et al. (2022). Disrupting mechanotransduction decreases fibrosis and contracture in split-thickness skin grafting. Sci Transl Med 14, eabj9152. 10.1126/scitranslmed.abj9152. [DOI] [PubMed] [Google Scholar]
- Chen K, Kwon SH, Henn D, Kuehlmann BA, Tevlin R, Bonham CA, Griffin M, Trotsyuk AA, Borrelli MR, Noishiki C, et al. (2021). Disrupting biological sensors of force promotes tissue regeneration in large organisms. Nat Commun 12, 5256. 10.1038/s41467-021-25410-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu AS, Diaz R, Hui JJ, Yanger K, Zong Y, Alpini G, Stanger BZ, and Wells RG (2011). Lineage tracing demonstrates no evidence of cholangiocyte epithelial-to-mesenchymal transition in murine models of hepatic fibrosis. Hepatology 53, 1685–1695. 10.1002/hep.24206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho AL, Schaller MA, Benjamim CF, Orlofsky AZ, Hogaboam CM, and Kunkel SL (2007). The chemokine CCL6 promotes innate immunity via immune cell activation and recruitment. J Immunol 179, 5474–5482. 10.4049/jimmunol.179.8.5474. [DOI] [PubMed] [Google Scholar]
- Colwell AS, Krummel TM, Kong W, Longaker MT, and Lorenz HP (2006). Skin wounds in the MRL/MPJ mouse heal with scar. Wound Repair Regen 14, 81–90. 10.1111/j.1524-475X.2005.00092.x. [DOI] [PubMed] [Google Scholar]
- Correa-Gallegos D, Jiang D, Christ S, Ramesh P, Ye H, Wannemacher J, Kalgudde Gopal S, Yu Q, Aichler M, Walch A, et al. (2019). Patch repair of deep wounds by mobilized fascia. Nature 576, 287–292. 10.1038/s41586-019-1794-y. [DOI] [PubMed] [Google Scholar]
- Crociara P, Chieppa MN, Vallino Costassa E, Berrone E, Gallo M, Lo Faro M, Pintore MD, Iulini B, D’Angelo A, Perona G, et al. (2019). Motor neuron degeneration, severe myopathy and TDP-43 increase in a transgenic pig model of SOD1-linked familiar ALS. Neurobiol Dis 124, 263–275. 10.1016/j.nbd.2018.11.021. [DOI] [PubMed] [Google Scholar]
- Davis J, and Molkentin JD (2014). Myofibroblasts: trust your heart and let fate decide. J Mol Cell Cardiol 70, 9–18. 10.1016/j.yjmcc.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira RC, and Wilson SE (2020). Fibrocytes, Wound Healing, and Corneal Fibrosis. Invest Ophthalmol Vis Sci 61, 28. 10.1167/iovs.61.2.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechêne A, Sowa JP, Gieseler RK, Jochum C, Bechmann LP, El Fouly A, Schlattjan M, Saner F, Baba HA, Paul A, et al. (2010). Acute liver failure is associated with elevated liver stiffness and hepatic stellate cell activation. Hepatology 52, 1008–1016. 10.1002/hep.23754. [DOI] [PubMed] [Google Scholar]
- Demarchez M, Sengel P, and Prunieras M (1986). Wound healing of human skin transplanted onto the nude mouse. I. An immunohistological study of the reepithelialization process. Dev Biol 113, 90–96. 10.1016/0012-1606(86)90110-7. [DOI] [PubMed] [Google Scholar]
- Demircioglu F, Wang J, Candido J, Costa ASH, Casado P, de Luxan Delgado B, Reynolds LE, Gomez-Escudero J, Newport E, Rajeeve V, et al. (2020). Cancer associated fibroblast FAK regulates malignant cell metabolism. Nat Commun 11, 1290. 10.1038/s41467-020-15104-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng CC, Hu YF, Zhu DH, Cheng Q, Gu JJ, Feng QL, Zhang LX, Xu YP, Wang D, Rong Z, and Yang B (2021). Single-cell RNA-seq reveals fibroblast heterogeneity and increased mesenchymal fibroblasts in human fibrotic skin diseases. Nat Commun 12, 3709. 10.1038/s41467-021-24110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- desJardins-Park HE, Foster DS, and Longaker MT (2018). Fibroblasts and wound healing: an update. Regen Med 13, 491–495. 10.2217/rme-2018-0073. [DOI] [PubMed] [Google Scholar]
- desJardins-Park HE, Mascharak S, Chinta MS, Wan DC, and Longaker MT (2019). The Spectrum of Scarring in Craniofacial Wound Repair. Front Physiol 10, 322. 10.3389/fphys.2019.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Carlo SE, and Peduto L (2018). The perivascular origin of pathological fibroblasts. J Clin Invest 128, 54–63. 10.1172/JCI93558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobie R, Wilson-Kanamori JR, Henderson BEP, Smith JR, Matchett KP, Portman JR, Wallenborg K, Picelli S, Zagorska A, Pendem SV, et al. (2019). Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep 29, 1832–1847.e1838. 10.1016/j.celrep.2019.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, and Watt FM (2013). Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281. 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell RR, and Watt FM (2015). Understanding fibroblast heterogeneity in the skin. Trends Cell Biol 25, 92–99. 10.1016/j.tcb.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Duval K, Grover H, Han LH, Mou Y, Pegoraro AF, Fredberg J, and Chen Z (2017). Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology (Bethesda) 32, 266–277. 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Ebner-Peking P, Krisch L, Wolf M, Hochmann S, Hoog A, Vári B, Muigg K, Poupardin R, Scharler C, Schmidhuber S, et al. (2021). Self-assembly of differentiated progenitor cells facilitates spheroid human skin organoid formation and planar skin regeneration. Theranostics 11, 8430–8447. 10.7150/thno.59661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, Szibor M, Kosanovic D, Schwind F, Schermuly RT, et al. (2017a). Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis. Cell Stem Cell 20, 571. 10.1016/j.stem.2017.03.011. [DOI] [PubMed] [Google Scholar]
- El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, Szibor M, Kosanovic D, Schwind F, Schermuly RT, et al. (2017b). Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis. Cell Stem Cell 20, 261–273.e263. 10.1016/j.stem.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Martin P, and Tomic-Canic M (2014). Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med 6, 265sr266. 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falanga V, Schrayer D, Cha J, Butmarc J, Carson P, Roberts AB, and Kim SJ (2004). Full-thickness wounding of the mouse tail as a model for delayed wound healing: accelerated wound closure in Smad3 knock-out mice. Wound Repair Regen 12, 320–326. 10.1111/j.1067-1927.2004.012316.x. [DOI] [PubMed] [Google Scholar]
- Fang RC, Kryger ZB, Buck DW, De la Garza M, Galiano RD, and Mustoe TA (2010). Limitations of the db/db mouse in translational wound healing research: Is the NONcNZO10 polygenic mouse model superior? Wound Repair Regen 18, 605–613. 10.1111/j.1524-475X.2010.00634.x. [DOI] [PubMed] [Google Scholar]
- Ferreira DW, Ulecia-Morón C, Alvarado-Vázquez PA, Cunnane K, Moracho-Vilriales C, Grosick RL, Cunha TM, and Romero-Sandoval EA (2020). CD163 overexpression using a macrophage-directed gene therapy approach improves wound healing in ex vivo and in vivo human skin models. Immunobiology 225, 151862. 10.1016/j.imbio.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleit SA, Fleit HB, and Zolla-Pazner S (1984). Culture and recovery of macrophages and cell lines from tissue culture-treated and -untreated plastic dishes. J Immunol Methods 68, 119–129. 10.1016/0022-1759(84)90142-x. [DOI] [PubMed] [Google Scholar]
- Forbes SJ, Russo FP, Rey V, Burra P, Rugge M, Wright NA, and Alison MR (2004). A significant proportion of myofibroblasts are of bone marrow origin in human liver fibrosis. Gastroenterology 126, 955–963. 10.1053/j.gastro.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Foster DS, Januszyk M, Yost KE, Chinta MS, Gulati GS, Nguyen AT, Burcham AR, Salhotra A, Ransom RC, Henn D, et al. (2021). Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proc Natl Acad Sci U S A 118. 10.1073/pnas.2110025118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galiano RD, Michaels J, Dobryansky M, Levine JP, and Gurtner GC (2004). Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen 12, 485–492. 10.1111/j.1067-1927.2004.12404.x. [DOI] [PubMed] [Google Scholar]
- Gay D, Ghinatti G, Guerrero-Juarez CF, Ferrer RA, Ferri F, Lim CH, Murakami S, Gault N, Barroca V, Rombeau I, et al. (2020). Phagocytosis of Wnt inhibitor SFRP4 by late wound macrophages drives chronic Wnt activity for fibrotic skin healing. Sci Adv 6, eaay3704. 10.1126/sciadv.aay3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay D, Kwon O, Zhang Z, Spata M, Plikus MV, Holler PD, Ito M, Yang Z, Treffeisen E, Kim CD, et al. (2013). Fgf9 from dermal γδ T cells induces hair follicle neogenesis after wounding. Nat Med 19, 916–923. 10.1038/nm.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Tan SJ, Wang SH, Li L, Sun XF, Shen W, and Wang X (2020). Single-cell Transcriptome Profiling reveals Dermal and Epithelial cell fate decisions during Embryonic Hair Follicle Development. Theranostics 10, 7581–7598. 10.7150/thno.44306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerts A (2001). History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis 21, 311–335. 10.1055/s-2001-17550. [DOI] [PubMed] [Google Scholar]
- Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, and Poelmann RE (1998). Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res 82, 1043–1052. 10.1161/01.res.82.10.1043. [DOI] [PubMed] [Google Scholar]
- Goltsev Y, Samusik N, Kennedy-Darling J, Bhate S, Hale M, Vazquez G, Black S, and Nolan GP (2018). Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968–981.e915. 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales KAU, Polak L, Matos I, Tierney MT, Gola A, Wong E, Infarinato NR, Nikolova M, Luo S, Liu S, et al. (2021). Stem cells expand potency and alter tissue fitness by accumulating diverse epigenetic memories. Science 374, eabh2444. 10.1126/science.abh2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S, and Taylor PR (2005). Monocyte and macrophage heterogeneity. Nat Rev Immunol 5, 953–964. 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- Grada A, Mervis J, and Falanga V (2018). Research Techniques Made Simple: Animal Models of Wound Healing. J Invest Dermatol 138, 2095–2105.e2091. 10.1016/j.jid.2018.08.005. [DOI] [PubMed] [Google Scholar]
- Granja JM, Corces MR, Pierce SE, Bagdatli ST, Choudhry H, Chang HY, and Greenleaf WJ (2021). ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat Genet 53, 403–411. 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieb G, Steffens G, Pallua N, Bernhagen J, and Bucala R (2011). Circulating fibrocytes--biology and mechanisms in wound healing and scar formation. Int Rev Cell Mol Biol 291, 1–19. 10.1016/B978-0-12-386035-4.00001-X. [DOI] [PubMed] [Google Scholar]
- Griffin MF, desJardins-Park HE, Mascharak S, Borrelli MR, and Longaker MT (2020). Understanding the impact of fibroblast heterogeneity on skin fibrosis. Dis Model Mech 13. 10.1242/dmm.044164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nature communications 10, 650. 10.1038/s41467-018-08247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtner GC, Dauskardt RH, Wong VW, Bhatt KA, Wu K, Vial IN, Padois K, Korman JM, and Longaker MT (2011). Improving cutaneous scar formation by controlling the mechanical environment: large animal and phase I studies. Ann Surg 254, 217–225. 10.1097/SLA.0b013e318220b159. [DOI] [PubMed] [Google Scholar]
- Gurtner GC, Werner S, Barrandon Y, and Longaker MT (2008). Wound repair and regeneration. Nature 453, 314–321. 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- Haak AJ, Kostallari E, Sicard D, Ligresti G, Choi KM, Caporarello N, Jones DL, Tan Q, Meridew J, Diaz Espinosa AM, et al. (2019). Selective YAP/TAZ inhibition in fibroblasts via dopamine receptor D1 agonism reverses fibrosis. Sci Transl Med 11. 10.1126/scitranslmed.aau6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habiel DM, and Hogaboam CM (2017). Heterogeneity of Fibroblasts and Myofibroblasts in Pulmonary Fibrosis. Curr Pathobiol Rep 5, 101–110. 10.1007/s40139-017-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamou C, Callaghan MJ, Thangarajah H, Chang E, Chang EI, Grogan RH, Paterno J, Vial IN, Jazayeri L, and Gurtner GC (2009). Mesenchymal stem cells can participate in ischemic neovascularization. Plast Reconstr Surg 123, 45S–55S. 10.1097/PRS.0b013e318191be4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, Geissmann F, and Hedrick CC (2011). The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol 12, 778–785. 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harn HI, Chen CC, Wang SP, Lei M, and Chuong CM (2021). Tissue Mechanics in Haired Murine Skin: Potential Implications for Skin Aging. Front Cell Dev Biol 9, 635340. 10.3389/fcell.2021.635340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harn HI, Ogawa R, Hsu CK, Hughes MW, Tang MJ, and Chuong CM (2019). The tension biology of wound healing. Exp Dermatol 28, 464–471. 10.1111/exd.13460. [DOI] [PubMed] [Google Scholar]
- Haudek SB, Xia Y, Huebener P, Lee JM, Carlson S, Crawford JR, Pilling D, Gomer RH, Trial J, Frangogiannis NG, and Entman ML (2006). Bone marrow-derived fibroblast precursors mediate ischemic cardiomyopathy in mice. Proceedings of the National Academy of Sciences of the United States of America 103, 18284–18289. 10.1073/pnas.0608799103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, Kameyama N, Estrada Y, Der E, Krueger JG, et al. (2020). Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol 145, 1615–1628. 10.1016/j.jaci.2020.01.042. [DOI] [PubMed] [Google Scholar]
- He J, Fang B, Shan S, Xie Y, Wang C, Zhang Y, Zhang X, and Li Q (2021). Mechanical stretch promotes hypertrophic scar formation through mechanically activated cation channel Piezo1. Cell Death Dis 12, 226. 10.1038/s41419-021-03481-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson NC, Rieder F, and Wynn TA (2020). Fibrosis: from mechanisms to medicines. Nature 587, 555–566. 10.1038/s41586-020-2938-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydemann A (2012). The super super-healing MRL mouse strain. Front Biol (Beijing) 7, 522–538. 10.1007/s11515-012-1192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B (2007). Formation and function of the myofibroblast during tissue repair. J Invest Dermatol 127, 526–537. 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- Houschyar KS, Borrelli MR, Tapking C, Popp D, Puladi B, Ooms M, Chelliah MP, Rein S, Pförringer D, Thor D, et al. (2020). Molecular Mechanisms of Hair Growth and Regeneration: Current Understanding and Novel Paradigms. Dermatology 236, 271–280. 10.1159/000506155. [DOI] [PubMed] [Google Scholar]
- Hu MS, Borrelli MR, Hong WX, Malhotra S, Cheung ATM, Ransom RC, Rennert RC, Morrison SD, Lorenz HP, and Longaker MT (2018). Embryonic skin development and repair. Organogenesis 14, 46–63. 10.1080/15476278.2017.1421882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, and Cotsarelis G (2005). Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med 11, 1351–1354. 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Ito M, Yang Z, Andl T, Cui C, Kim N, Millar SE, and Cotsarelis G (2007). Wnt-dependent de novo hair follicle regeneration in adult mouse skin after wounding. Nature 447, 316–320. 10.1038/nature05766. [DOI] [PubMed] [Google Scholar]
- Janbandhu V, Tallapragada V, Patrick R, Li Y, Abeygunawardena D, Humphreys DT, Martin EMMA, Ward AO, Contreras O, Farbehi N, et al. (2022). Hif-1a suppresses ROS-induced proliferation of cardiac fibroblasts following myocardial infarction. Cell Stem Cell 29, 281–297.e212. 10.1016/j.stem.2021.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson DG, Saintigny G, van Adrichem A, Mahé C, and El Ghalbzouri A (2012). Different gene expression patterns in human papillary and reticular fibroblasts. J Invest Dermatol 132, 2565–2572. 10.1038/jid.2012.192. [DOI] [PubMed] [Google Scholar]
- Jiang D, Christ S, Correa-Gallegos D, Ramesh P, Kalgudde Gopal S, Wannemacher J, Mayr CH, Lupperger V, Yu Q, Ye H, et al. (2020). Injury triggers fascia fibroblast collective cell migration to drive scar formation through N-cadherin. Nature communications 11, 5653. 10.1038/s41467-020-19425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Correa-Gallegos D, Christ S, Stefanska A, Liu J, Ramesh P, Rajendran V, De Santis MM, Wagner DE, and Rinkevich Y (2018). Two succeeding fibroblastic lineages drive dermal development and the transition from regeneration to scarring. Nat Cell Biol 20, 422–431. 10.1038/s41556-018-0073-8. [DOI] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat Commun 12, 1088. 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, and Kasper M (2016). Single-Cell Transcriptomics Reveals that Differentiation and Spatial Signatures Shape Epidermal and Hair Follicle Heterogeneity. Cell Syst 3, 221–237.e229. 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Kalhor K, Mejia L, Leeper K, Graveline A, Mali P, and Church GM (2018). Developmental barcoding of whole mouse via homing CRISPR. Science 361. 10.1126/science.aat9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, and Neilson EG (2003). Epithelial-mesenchymal transition and its implications for fibrosis. J Clin Invest 112, 1776–1784. 10.1172/JCI20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JL, Rozek MM, Suenram CA, and Schwartz CJ (1987). Activation of human blood monocytes by adherence to tissue culture plastic surfaces. Exp Mol Pathol 46, 266–278. 10.1016/0014-4800(87)90049-9. [DOI] [PubMed] [Google Scholar]
- Kendall RT, and Feghali-Bostwick CA (2014). Fibroblasts in fibrosis: novel roles and mediators. Front Pharmacol 5, 123. 10.3389/fphar.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV (2021). The triumphs and limitations of computational methods for scRNA-seq. Nat Methods 18, 723–732. 10.1038/s41592-021-01171-x. [DOI] [PubMed] [Google Scholar]
- Klicks J, Maßlo C, Kluth A, Rudolf R, and Hafner M (2019). A novel spheroid-based co-culture model mimics loss of keratinocyte differentiation, melanoma cell invasion, and drug-induced selection of ABCB5-expressing cells. BMC Cancer 19, 402. 10.1186/s12885-019-5606-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klicks J, von Molitor E, Ertongur-Fauth T, Rudolf R, and Hafner M (2017). In vitro skin three-dimensional models and their applications. J Cell Biotech 3, 21–39. [Google Scholar]
- Klinkhammer BM, Floege J, and Boor P (2018). PDGF in organ fibrosis. Mol Aspects Med 62, 44–62. 10.1016/j.mam.2017.11.008. [DOI] [PubMed] [Google Scholar]
- Kohyama M, Ise W, Edelson BT, Wilker PR, Hildner K, Mejia C, Frazier WA, Murphy TL, and Murphy KM (2009). Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature 457, 318–321. 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosec A, Frech S, Gesslbauer B, Vierhapper M, Radtke C, Petzelbauer P, and Lichtenberger BM (2019). Lineage Identity and Location within the Dermis Determine the Function of Papillary and Reticular Fibroblasts in Human Skin. J Invest Dermatol 139, 342–351. 10.1016/j.jid.2018.07.033. [DOI] [PubMed] [Google Scholar]
- Krenkel O, Hundertmark J, Ritz TP, Weiskirchen R, and Tacke F (2019). Single Cell RNA Sequencing Identifies Subsets of Hepatic Stellate Cells and Myofibroblasts in Liver Fibrosis. Cells 8. 10.3390/cells8050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyszczyk P, Schloss R, Palmer A, and Berthiaume F (2018). The Role of Macrophages in Acute and Chronic Wound Healing and Interventions to Promote Pro-wound Healing Phenotypes. Front Physiol 9, 419. 10.3389/fphys.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam NT, and Sadek HA (2018). Neonatal Heart Regeneration: Comprehensive Literature Review. Circulation 138, 412–423. 10.1161/CIRCULATIONAHA.118.033648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson BJ, Longaker MT, and Lorenz HP (2010). Scarless fetal wound healing: a basic science review. Plast Reconstr Surg 126, 1172–1180. 10.1097/PRS.0b013e3181eae781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassance L, Marino GK, Medeiros CS, Thangavadivel S, and Wilson SE (2018). Fibrocyte migration, differentiation and apoptosis during the corneal wound healing response to injury. Exp Eye Res 170, 177–187. 10.1016/j.exer.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt T, Hu MS, Borrelli MR, Januszyk M, Garcia JT, Ransom RC, Mascharak S, desJardins-Park HE, Litzenburger UM, Walmsley GG, et al. (2020). Prrx1 Fibroblasts Represent a Pro-fibrotic Lineage in the Mouse Ventral Dermis. Cell Rep 33, 108356. 10.1016/j.celrep.2020.108356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBleu VS, and Neilson EG (2020). Origin and functional heterogeneity of fibroblasts. FASEB J 34, 3519–3536. 10.1096/fj.201903188R. [DOI] [PubMed] [Google Scholar]
- Lee J, and Koehler KR (2021). Skin organoids: A new human model for developmental and translational research. Exp Dermatol 30, 613–620. 10.1111/exd.14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Rabbani CC, Gao H, Steinhart MR, Woodruff BM, Pflum ZE, Kim A, Heller S, Liu Y, Shipchandler TZ, and Koehler KR (2020). Hair-bearing human skin generated entirely from pluripotent stem cells. Nature 582, 399–404. 10.1038/s41586-020-2352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, and Poss KD (2006). A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 127, 607–619. 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Li Y, Wu J, Luo G, and He W (2018). Functions of Vγ4 T Cells and Dendritic Epidermal T Cells on Skin Wound Healing. Front Immunol 9, 1099. 10.3389/fimmu.2018.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Park AY, and Guan JL (2007). In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc 2, 329–333. 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- Lim AF, Weintraub J, Kaplan EN, Januszyk M, Cowley C, McLaughlin P, Beasley B, Gurtner GC, and Longaker MT (2014). The embrace device significantly decreases scarring following scar revision surgery in a randomized controlled trial. Plast Reconstr Surg 133, 398–405. 10.1097/01.prs.0000436526.64046.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CH, Sun Q, Ratti K, Lee SH, Zheng Y, Takeo M, Lee W, Rabbani P, Plikus MV, Cain JE, et al. (2018). Hedgehog stimulates hair follicle neogenesis by creating inductive dermis during murine skin wound healing. Nature communications 9, 4903. 10.1038/s41467-018-07142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, and Trapnell C (2016). Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res 5. 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longaker MT, Rohrich RJ, Greenberg L, Furnas H, Wald R, Bansal V, Seify H, Tran A, Weston J, Korman JM, et al. (2014). A randomized controlled trial of the embrace advanced scar therapy device to reduce incisional scar formation. Plast Reconstr Surg 134, 536–546. 10.1097/PRS.0000000000000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovisa S (2021). Epithelial-to-Mesenchymal Transition in Fibrosis: Concepts and Targeting Strategies. Front Pharmacol 12, 737570. 10.3389/fphar.2021.737570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P, Takai K, Weaver VM, and Werb Z (2011). Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 3. 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas T, Waisman A, Ranjan R, Roes J, Krieg T, Müller W, Roers A, and Eming SA (2010). Differential roles of macrophages in diverse phases of skin repair. J Immunol 184, 3964–3977. 10.4049/jimmunol.0903356. [DOI] [PubMed] [Google Scholar]
- Lynch MD, and Watt FM (2018). Fibroblast heterogeneity: implications for human disease. The Journal of clinical investigation 128, 26–35. 10.1172/JCI93555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Kwon SH, Padmanabhan J, Duscher D, Trotsyuk AA, Dong Y, Inayathullah M, Rajadas J, and Gurtner GC (2018). Controlled Delivery of a Focal Adhesion Kinase Inhibitor Results in Accelerated Wound Closure with Decreased Scar Formation. J Invest Dermatol 138, 2452–2460. 10.1016/j.jid.2018.04.034. [DOI] [PubMed] [Google Scholar]
- Manmadhan S, and Ehmer U (2019). Hippo Signaling in the Liver - A Long and Ever-Expanding Story. Front Cell Dev Biol 7, 33. 10.3389/fcell.2019.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannaerts I, Leite SB, Verhulst S, Claerhout S, Eysackers N, Thoen LF, Hoorens A, Reynaert H, Halder G, and van Grunsven LA (2015). The Hippo pathway effector YAP controls mouse hepatic stellate cell activation. J Hepatol 63, 679–688. 10.1016/j.jhep.2015.04.011. [DOI] [PubMed] [Google Scholar]
- Martin K, Pritchett J, Llewellyn J, Mullan AF, Athwal VS, Dobie R, Harvey E, Zeef L, Farrow S, Streuli C, et al. (2016). PAK proteins and YAP-1 signalling downstream of integrin beta-1 in myofibroblasts promote liver fibrosis. Nat Commun 7, 12502. 10.1038/ncomms12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascharak S, desJardins-Park HE, Davitt MF, Griffin M, Borrelli MR, Moore AL, Chen K, Duoto B, Chinta M, Foster DS, et al. (2021a). Preventing Engrailed-1 activation in fibroblasts yields wound regeneration without scarring. Science 372. 10.1126/science.aba2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascharak S, desJardins-Park HE, Davitt MF, Guardino NJ, Gurtner GC, Wan DC, and Longaker MT (2021b). Modulating Cellular Responses to Mechanical Forces to Promote Wound Regeneration. Adv Wound Care (New Rochelle). 10.1089/wound.2021.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascharak S, Talbott HE, Januszyk M, Griffin M, Chen K, Davitt MF, Demeter J, Henn D, Bonham CA, Foster DS, et al. (2022). Multi-omic analysis reveals divergent molecular events in scarring and regenerative wound healing. Cell Stem Cell 29, 315–327.e316. 10.1016/j.stem.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson-Meyers DS, Andrade TAM, Caetano GF, Guimaraes FR, Leite MN, Leite SN, and Frade MAC (2020). Experimental models and methods for cutaneous wound healing assessment. Int J Exp Pathol 101, 21–37. 10.1111/iep.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masur SK, Dewal HS, Dinh TT, Erenburg I, and Petridou S (1996). Myofibroblasts differentiate from fibroblasts when plated at low density. Proc Natl Acad Sci U S A 93, 4219–4223. 10.1073/pnas.93.9.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki T, and Yoshizato K (1998). Role of hair papilla cells on induction and regeneration processes of hair follicles. Wound Repair Regen 6, 524–530. 10.1046/j.1524-475x.1998.60605.x. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Akiyama M, Tamura F, Isomi M, Yamakawa H, Sadahiro T, Muraoka N, Kojima H, Haginiwa S, Kurotsu S, et al. (2018). Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell 22, 91–103.e105. 10.1016/j.stem.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Molenaar JC (2003). [From the library of the Netherlands Journal of Medicine. Rudolf Virchow: Die Cellularpathologie in ihrer Begründung auf physiologische und pathologische Gewebelehre; 1858]. Ned Tijdschr Geneeskd 147, 2236–2244. [PubMed] [Google Scholar]
- Morgan BA (2014). The dermal papilla: an instructive niche for epithelial stem and progenitor cells in development and regeneration of the hair follicle. Cold Spring Harb Perspect Med 4, a015180. 10.1101/cshperspect.a015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgun EI, and Vorotelyak EA (2020). Epidermal Stem Cells in Hair Follicle Cycling and Skin Regeneration: A View From the Perspective of Inflammation. Front Cell Dev Biol 8, 581697. 10.3389/fcell.2020.581697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori L, Bellini A, Stacey MA, Schmidt M, and Mattoli S (2005). Fibrocytes contribute to the myofibroblast population in wounded skin and originate from the bone marrow. Exp Cell Res 304, 81–90. 10.1016/j.yexcr.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Muhl L, Genove G, Leptidis S, Liu J, He L, Mocci G, Sun Y, Gustafsson S, Buyandelger B, Chivukula IV, et al. (2020a). Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nature communications 11, 3953. 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhl L, Genové G, Leptidis S, Liu J, He L, Mocci G, Sun Y, Gustafsson S, Buyandelger B, Chivukula IV, et al. (2020b). Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun 11, 3953. 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdaca G, Spanò F, Contatore M, Guastalla A, and Puppo F (2014). Potential use of TNF-α inhibitors in systemic sclerosis. Immunotherapy 6, 283–289. 10.2217/imt.13.173. [DOI] [PubMed] [Google Scholar]
- Mäkelä K, Mäyränpää MI, Sihvo HK, Bergman P, Sutinen E, Ollila H, Kaarteenaho R, and Myllärniemi M (2021). Artificial intelligence identifies inflammation and confirms fibroblast foci as prognostic tissue biomarkers in idiopathic pulmonary fibrosis. Hum Pathol 107, 58–68. 10.1016/j.humpath.2020.10.008. [DOI] [PubMed] [Google Scholar]
- Naik S, Larsen SB, Gomez NC, Alaverdyan K, Sendoel A, Yuan S, Polak L, Kulukian A, Chai S, and Fuchs E (2017). Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480. 10.1038/nature24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauroy P, Barruche V, Marchand L, Nindorera-Badara S, Bordes S, Closs B, and Ruggiero F (2017). Human Dermal Fibroblast Subpopulations Display Distinct Gene Signatures Related to Cell Behaviors and Matrisome. J Invest Dermatol 137, 1787–1789. 10.1016/j.jid.2017.03.028. [DOI] [PubMed] [Google Scholar]
- Noramly S, Freeman A, and Morgan BA (1999). beta-catenin signaling can initiate feather bud development. Development 126, 3509–3521. 10.1242/dev.126.16.3509. [DOI] [PubMed] [Google Scholar]
- Ogawa R (2011). Mechanobiology of scarring. Wound Repair Regen 19 Suppl 1, s2–9. 10.1111/j.1524-475X.2011.00707.x. [DOI] [PubMed] [Google Scholar]
- Okabe Y, and Medzhitov R (2014). Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844. 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei W, Feyerabend TB, Rossler J, Wang X, Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C, et al. (2017). Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460. 10.1038/nature23653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QM, Fine GM, Salz L, Herrera GG, Wildman B, Driskell IM, and Driskell RR (2020). Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. eLife 9. 10.7554/eLife.60066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan QM, Sinha S, Biernaskie J, and Driskell RR (2021). Single-cell transcriptomic analysis of small and large wounds reveals the distinct spatial organization of regenerative fibroblasts. Exp Dermatol 30, 92–101. 10.1111/exd.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippeos C, Telerman SB, Oules B, Pisco AO, Shaw TJ, Elgueta R, Lombardi G, Driskell RR, Soldin M, Lynch MD, and Watt FM (2018). Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J Invest Dermatol 138, 811–825. 10.1016/j.jid.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RJ, Burdick MD, Hong K, Lutz MA, Murray LA, Xue YY, Belperio JA, Keane MP, and Strieter RM (2004). Circulating fibrocytes traffic to the lungs in response to CXCL12 and mediate fibrosis. The Journal of clinical investigation 114, 438–446. 10.1172/JCI20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piersma B, Bank RA, and Boersema M (2015). Signaling in Fibrosis: TGF-β, WNT, and YAP/TAZ Converge. Front Med (Lausanne) 2, 59. 10.3389/fmed.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Guerrero-Juarez CF, Ito M, Li YR, Dedhia PH, Zheng Y, Shao M, Gay DL, Ramos R, Hsi TC, et al. (2017). Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752. 10.1126/science.aai8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Wang X, Sinha S, Forte E, Thompson SM, Herzog EL, Driskell RR, Rosenthal N, Biernaskie J, and Horsley V (2021). Fibroblasts: Origins, definitions, and functions in health and disease. Cell 184, 3852–3872. 10.1016/j.cell.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirion OB, Zhu X, Ching T, and Garmire L (2016). Single-Cell Transcriptomics Bioinformatics and Computational Challenges. Front Genet 7, 163. 10.3389/fgene.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Dobie R, Wilson-Kanamori JR, Dora EF, Henderson BEP, Luu NT, Portman JR, Matchett KP, Brice M, Marwick JA, et al. (2019). Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 575, 512–518. 10.1038/s41586-019-1631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall MJ, Jüngel A, Rimann M, and Wuertz-Kozak K (2018). Advances in the Biofabrication of 3D Skin. Front Bioeng Biotechnol 6, 154. 10.3389/fbioe.2018.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M, Wang X, Guo G, Wang L, Chen S, Yin P, Chen K, Chen L, Zhang Z, Chen X, et al. (2021). Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level. Basic Res Cardiol 116, 55. 10.1007/s00395-021-00897-1. [DOI] [PubMed] [Google Scholar]
- Ravikanth M, Soujanya P, Manjunath K, Saraswathi TR, and Ramachandran CR (2011). Heterogenecity of fibroblasts. J Oral Maxillofac Pathol 15, 247–250. 10.4103/0973-029X.84516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed ND, and Manning DD (1973). Long-term maintenance of normal human skin on congenitally athymic (nude) mice. Proc Soc Exp Biol Med 143, 350–353. 10.3181/00379727-143-37318. [DOI] [PubMed] [Google Scholar]
- Reich B, Schmidbauer K, Rodriguez Gomez M, Johannes Hermann F, Gobel N, Bruhl H, Ketelsen I, Talke Y, and Mack M (2013). Fibrocytes develop outside the kidney but contribute to renal fibrosis in a mouse model. Kidney Int 84, 78–89. 10.1038/ki.2013.84. [DOI] [PubMed] [Google Scholar]
- Relou IA, Damen CA, van der Schaft DW, Groenewegen G, and Griffioen AW (1998). Effect of culture conditions on endothelial cell growth and responsiveness. Tissue Cell 30, 525–530. 10.1016/s0040-8166(98)80032-3. [DOI] [PubMed] [Google Scholar]
- Reynolds G, Vegh P, Fletcher J, Poyner EFM, Stephenson E, Goh I, Botting RA, Huang N, Olabi B, Dubois A, et al. (2021). Developmental cell programs are co-opted in inflammatory skin disease. Science 371. 10.1126/science.aba6500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezvani M, Español-Suñer R, Malato Y, Dumont L, Grimm AA, Kienle E, Bindman JG, Wiedtke E, Hsu BY, Naqvi SJ, et al. (2016). In Vivo Hepatic Reprogramming of Myofibroblasts with AAV Vectors as a Therapeutic Strategy for Liver Fibrosis. Cell Stem Cell 18, 809–816. 10.1016/j.stem.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, et al. (2015). Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science (New York, N.Y.) 348, aaa2151–aaa2151. 10.1126/science.aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, Ridky TW, Stadler HS, Nusse R, Helms JA, and Chang HY (2008). A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes & development 22, 303–307. 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossio-Pasquier P, Casanova D, Jomard A, and Démarchez M (1999). Wound healing of human skin transplanted onto the nude mouse after a superficial excisional injury: human dermal reconstruction is achieved in several steps by two different fibroblast subpopulations. Arch Dermatol Res 291, 591–599. 10.1007/s004030050460. [DOI] [PubMed] [Google Scholar]
- Ruiz-Villalba A, Romero JP, Hernandez SC, Vilas-Zornoza A, Fortelny N, Castro-Labrador L, San Martin-Uriz P, Lorenzo-Vivas E, Garcia-Olloqui P, Palacio M, et al. (2020). Single-Cell RNA Sequencing Analysis Reveals a Crucial Role for CTHRC1 (Collagen Triple Helix Repeat Containing 1) Cardiac Fibroblasts After Myocardial Infarction. Circulation 142, 1831–1847. 10.1161/CIRCULATIONAHA.119.044557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo FP, Alison MR, Bigger BW, Amofah E, Florou A, Amin F, Bou-Gharios G, Jeffery R, Iredale JP, and Forbes SJ (2006). The bone marrow functionally contributes to liver fibrosis. Gastroenterology 130, 1807–1821. 10.1053/j.gastro.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Satoh T, Kidoya H, Naito H, Yamamoto M, Takemura N, Nakagawa K, Yoshioka Y, Morii E, Takakura N, Takeuchi O, and Akira S (2013). Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature 495, 524–528. 10.1038/nature11930. [DOI] [PubMed] [Google Scholar]
- Satoh T, Nakagawa K, Sugihara F, Kuwahara R, Ashihara M, Yamane F, Minowa Y, Fukushima K, Ebina I, Yoshioka Y, et al. (2017). Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541, 96–101. 10.1038/nature20611. [DOI] [PubMed] [Google Scholar]
- Seifert AW, Kiama SG, Seifert MG, Goheen JR, Palmer TM, and Maden M (2012). Skin shedding and tissue regeneration in African spiny mice (Acomys). Nature 489, 561–565. 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, and Schwabe RF (2007). TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med 13, 1324–1332. 10.1038/nm1663. [DOI] [PubMed] [Google Scholar]
- Shao DD, Suresh R, Vakil V, Gomer RH, and Pilling D (2008). Pivotal Advance: Th-1 cytokines inhibit, and Th-2 cytokines promote fibrocyte differentiation. J Leukoc Biol 83, 1323–1333. 10.1189/jlb.1107782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw TJ, and Rognoni E (2020). Dissecting Fibroblast Heterogeneity in Health and Fibrotic Disease. Curr Rheumatol Rep 22, 33. 10.1007/s11926-020-00903-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook B, Xiao E, Kumamoto Y, Iwasaki A, and Horsley V (2016). CD301b+ Macrophages Are Essential for Effective Skin Wound Healing. J Invest Dermatol 136, 1885–1891. 10.1016/j.jid.2016.05.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook BA, Wasko RR, Mano O, Rutenberg-Schoenberg M, Rudolph MC, Zirak B, Rivera-Gonzalez GC, López-Giráldez F, Zarini S, Rezza A, et al. (2020). Dermal Adipocyte Lipolysis and Myofibroblast Conversion Are Required for Efficient Skin Repair. Cell Stem Cell 26, 880–895.e886. 10.1016/j.stem.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shook BA, Wasko RR, Rivera-Gonzalez GC, Salazar-Gatzimas E, Lopez-Giraldez F, Dash BC, Munoz-Rojas AR, Aultman KD, Zwick RK, Lei V, et al. (2018). Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362. 10.1126/science.aar2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter K, Spiera R, Magro C, Agius P, Martyanov V, Franks JM, Sharma R, Geiger H, Wood TA, Zhang Y, et al. (2021). Machine learning integration of scleroderma histology and gene expression identifies fibroblast polarisation as a hallmark of clinical severity and improvement. Ann Rheum Dis 80, 228–237. 10.1136/annrheumdis-2020-217840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solé-Boldo L, Raddatz G, Schütz S, Mallm JP, Rippe K, Lonsdorf AS, Rodríguez-Paredes M, and Lyko F (2020). Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun Biol 3, 188. 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son DO, and Hinz B (2021). A Rodent Model of Hypertrophic Scarring: Splinting of Rat Wounds. Methods Mol Biol 2299, 405–417. 10.1007/978-1-0716-1382-5_27. [DOI] [PubMed] [Google Scholar]
- Song G, Nguyen DT, Pietramaggiori G, Scherer S, Chen B, Zhan Q, Ogawa R, Yannas IV, Wagers AJ, Orgill DP, and Murphy GF (2010). Use of the parabiotic model in studies of cutaneous wound healing to define the participation of circulating cells. Wound Repair Regen 18, 426–432. 10.1111/j.1524-475X.2010.00595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Pacher M, Balakrishnan A, Yuan Q, Tsay HC, Yang D, Reetz J, Brandes S, Dai Z, Pützer BM, et al. (2016). Direct Reprogramming of Hepatic Myofibroblasts into Hepatocytes In Vivo Attenuates Liver Fibrosis. Cell Stem Cell 18, 797–808. 10.1016/j.stem.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Sorrell JM, and Caplan AI (2009). Fibroblasts-a diverse population at the center of it all. Int Rev Cell Mol Biol 276, 161–214. 10.1016/S1937-6448(09)76004-6. [DOI] [PubMed] [Google Scholar]
- Stone RC, Pastar I, Ojeh N, Chen V, Liu S, Garzon KI, and Tomic-Canic M (2016). Epithelial-mesenchymal transition in tissue repair and fibrosis. Cell Tissue Res 365, 495–506. 10.1007/s00441-016-2464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindle MM, Makin A, Herron AJ, Clubb FJ, and Frazier KS (2012). Swine as models in biomedical research and toxicology testing. Vet Pathol 49, 344–356. 10.1177/0300985811402846. [DOI] [PubMed] [Google Scholar]
- Szabo G, Mandrekar P, and Dolganiuc A (2007). Innate immune response and hepatic inflammation. Semin Liver Dis 27, 339–350. 10.1055/s-2007-991511. [DOI] [PubMed] [Google Scholar]
- Tabib T, Morse C, Wang T, Chen W, and Lafyatis R (2018). SFRP2/DPP4 and FMO1/LSP1 Define Major Fibroblast Populations in Human Skin. J Invest Dermatol 138, 802–810. 10.1016/j.jid.2017.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Grzenda A, Allison TF, Rawnsley J, Balin SJ, Sabri S, Plath K, and Lowry WE (2020). Defining Transcriptional Signatures of Human Hair Follicle Cell States. J Invest Dermatol 140, 764–773.e764. 10.1016/j.jid.2019.07.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Phan QM, Winuthayanon S, Driskell IM, and Driskell RR (2021). Parallel single cell multi-omics analysis of neonatal skin reveals transitional fibroblast states that restricts differentiation into distinct fates. J Invest Dermatol. 10.1016/j.jid.2021.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulabandu V, Chen D, and Atit RP (2018). Dermal fibroblast in cutaneous development and healing. Wiley Interdiscip Rev Dev Biol 7. 10.1002/wdev.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzi E, Bulik M, Tabib T, Morse C, Sembrat J, Trejo Bittar H, Rojas M, and Lafyatis R (2019). Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann Rheum Dis 78, 1379–1387. 10.1136/annrheumdis-2018-214865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, and Pu WT (2012). Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res 110, 1628–1645. 10.1161/CIRCRESAHA.111.259960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorstandlechner V, Laggner M, Kalinina P, Haslik W, Radtke C, Shaw L, Lichtenberger BM, Tschachler E, Ankersmit HJ, and Mildner M (2020). Deciphering the functional heterogeneity of skin fibroblasts using single-cell RNA sequencing. FASEB J 34, 3677–3692. 10.1096/fj.201902001RR. [DOI] [PubMed] [Google Scholar]
- Walmsley GG, Rinkevich Y, Hu MS, Montoro DT, Lo DD, McArdle A, Maan ZN, Morrison SD, Duscher D, Whittam AJ, et al. (2015). Live fibroblast harvest reveals surface marker shift in vitro. Tissue Eng Part C Methods 21, 314–321. 10.1089/ten.TEC.2014.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang Y, Ma H, Xie Y, Xu J, Near D, Wang H, Garbutt T, Li Y, Liu J, and Qian L (2022). Single-cell dual-omics reveals the transcriptomic and epigenomic diversity of cardiac non-myocytes. Cardiovasc Res 118, 1548–1563. 10.1093/cvr/cvab134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hsi TC, Guerrero-Juarez CF, Pham K, Cho K, McCusker CD, Monuki ES, Cho KW, Gay DL, and Plikus MV (2015). Principles and mechanisms of regeneration in the mouse model for wound-induced hair follicle neogenesis. Regeneration (Oxf) 2, 169–181. 10.1002/reg2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells RG, Kruglov E, and Dranoff JA (2004). Autocrine release of TGF-beta by portal fibroblasts regulates cell growth. FEBS Lett 559, 107–110. 10.1016/S0014-5793(04)00037-7. [DOI] [PubMed] [Google Scholar]
- Wong CE, Paratore C, Dours-Zimmermann MT, Rochat A, Pietri T, Suter U, Zimmermann DR, Dufour S, Thiery JP, Meijer D, et al. (2006). Neural crest-derived cells with stem cell features can be traced back to multiple lineages in the adult skin. J Cell Biol 175, 1005–1015. 10.1083/jcb.200606062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VW, Beasley B, Zepeda J, Dauskardt RH, Yock PG, Longaker MT, and Gurtner GC (2013). A Mechanomodulatory Device to Minimize Incisional Scar Formation. Adv Wound Care (New Rochelle) 2, 185–194. 10.1089/wound.2012.0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VW, Rustad KC, Akaishi S, Sorkin M, Glotzbach JP, Januszyk M, Nelson ER, Levi K, Paterno J, Vial IN, et al. (2011a). Focal adhesion kinase links mechanical force to skin fibrosis via inflammatory signaling. Nat Med 18, 148–152. 10.1038/nm.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VW, Sorkin M, Glotzbach JP, Longaker MT, and Gurtner GC (2011b). Surgical approaches to create murine models of human wound healing. J Biomed Biotechnol 2011, 969618. 10.1155/2011/969618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, and Ramalingam TR (2012). Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 18, 1028–1040. 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Hill MC, Li L, Deshmukh V, Martin TJ, Wang J, and Martin JF (2019). Hippo pathway deletion in adult resting cardiac fibroblasts initiates a cell state transition with spontaneous and self-sustaining fibrosis. Genes & development 33, 1491–1505. 10.1101/gad.329763.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Liang J, Liu N, Huan C, Zhang Y, Liu W, Kumar M, Xiao R, D’Armiento J, Metzger D, et al. (2016). Transcription factor TBX4 regulates myofibroblast accumulation and lung fibrosis. J Clin Invest 126, 3063–3079. 10.1172/JCI85328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Wang Y, Deng N, Huang G, Taghavifar F, Geng Y, Liu N, Kulur V, Yao C, Chen P, et al. (2018). Single-Cell Deconvolution of Fibroblast Heterogeneity in Mouse Pulmonary Fibrosis. Cell Rep 22, 3625–3640. 10.1016/j.celrep.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao XY, Ji Y, Li C, Guo L, et al. (2019). Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol Cell 75, 644–660 e645. 10.1016/j.molcel.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang Z, Jia L, and Wang Y (2016). Role of Bone Marrow-Derived Fibroblasts in Renal Fibrosis. Front Physiol 7, 61. 10.3389/fphys.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, and Wu Z (2018). Genome Editing of Pigs for Agriculture and Biomedicine. Front Genet 9, 360. 10.3389/fgene.2018.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M, and Duffield JS (2010). Resolved: EMT produces fibroblasts in the kidney. J Am Soc Nephrol 21, 1247–1253. 10.1681/ASN.2010060616. [DOI] [PubMed] [Google Scholar]
- Zhang CY, Yuan WG, He P, Lei JH, and Wang CX (2016). Liver fibrosis and hepatic stellate cells: Etiology, pathological hallmarks and therapeutic targets. World J Gastroenterol 22, 10512–10522. 10.3748/wjg.v22.i48.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, He X, Caldwell L, Goru SK, Ulloa Severino L, Tolosa MF, Misra PS, McEvoy CM, Christova T, Liu Y, et al. (2022). NUAK1 promotes organ fibrosis via YAP and TGF-β/SMAD signaling. Sci Transl Med 14, eaaz4028. 10.1126/scitranslmed.aaz4028. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, et al. (2009). Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev Cell 17, 49–61. 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Franklin RA, Adler M, Carter TS, Condiff E, Adams TS, Pope SD, Philip NH, Meizlish ML, Kaminski N, and Medzhitov R (2022). Microenvironmental Sensing by Fibroblasts Controls Macrophage Population Size. bioRxiv, 2022.2001.2018.476683. 10.1101/2022.01.18.476683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Franklin RA, Adler M, Jacox JB, Bailis W, Shyer JA, Flavell RA, Mayo A, Alon U, and Medzhitov R (2018). Circuit Design Features of a Stable Two-Cell System. Cell 172, 744–757 e717. 10.1016/j.cell.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Zhao X, Zheng K, Li H, Huang H, Zhang Z, Mastracci T, Wegner M, Chen Y, Sussel L, and Qiu M (2014). Genetic evidence that Nkx2.2 and Pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development 141, 548–555. 10.1242/dev.095323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomer HD, and Trentin AG (2018). Skin wound healing in humans and mice: Challenges in translational research. J Dermatol Sci 90, 3–12. 10.1016/j.jdermsci.2017.12.009. [DOI] [PubMed] [Google Scholar]


