Abstract
Purpose:
We sought to determine whether the detection of circulating tumor DNA (ctDNA) in samples of patients undergoing chemotherapy for advanced leiomyosarcoma (LMS) is associated with objective response or survival.
Experimental Design:
Using ultra–low-passage whole-genome sequencing (ULP-WGS) of plasma cell-free DNA from patients treated on a prospective clinical trial, we tested whether detection of ctDNA evaluated prior to the start of therapy and after two cycles of chemotherapy was associated with treatment response and outcome. Associations between detection of ctDNA and pathologic measures of disease burden were evaluated.
Results:
We found that ctDNA was detectable by ULP-WGS in 49% patients prior to treatment and in 24.6% patients after two cycles of chemotherapy. Detection of pretreatment ctDNA was significantly associated with a lower overall survival [HR, 1.55; 95% confidence interval (CI), 1.03–2.31; P = 0.03] and a significantly lower likelihood of objective response [odds ratio (OR), 0.21; 95% CI, 0.06–0.59; P = 0.005]. After two cycles of chemotherapy, patients who continued to have detectable levels of ctDNA experienced a significantly worse overall survival (HR, 1.77; 95% CI, 1–3.14; P = 0.05) and were unlikely to experience an objective response (OR, 0.05; 95% CI, 0–0.39; P = 0.001).
Conclusions:
Our results demonstrate that detection of ctDNA is associated with outcome and objective response to chemotherapy in patients with advanced LMS. These results suggest that liquid biopsy assays could be used to inform treatment decisions by recognizing patients who are likely and unlikely to benefit from chemotherapy.
See related commentary by Kasper and Wilky, p. 2480
Translational Relevance.
In this study, we found that patients with advanced leiomyosarcoma (LMS) who had high levels of circulating tumor DNA (ctDNA) prior to the start of treatment and after receiving two cycles of chemotherapy had a significantly worse overall survival than patients with low ctDNA. Furthermore, patients with low ctDNA were significantly more likely to experience an objective response with additional cycles of chemotherapy. These studies demonstrate that ctDNA can be used for risk stratification in future clinical trials for patients with LMS and by providers and their patients to decide on the utility of treatment, with chemotherapy known to bear a significant risk of toxicity.
Introduction
Leiomyosarcoma (LMS) is a malignant mesenchymal tumor arising from the smooth muscle lineage (1). It is the most commonly occurring soft tissue sarcoma in adults. LMS is frequently a clinically aggressive disease, and patients are at high risk for relapse after initial complete resection of the tumor (1). Metastatic LMS is incurable with current systemic antitumor therapies. Doxorubicin- or gemcitabine-based regimens have demonstrated benefit by extending progression-free survival (PFS) for some patients and are standard therapies for patients with metastatic disease (2–4). Although patients frequently experience toxicity during treatment with these agents, it remains unclear how to identify the patients that are most likely to benefit from therapy. The lack of prognostic biomarkers is an obstacle to more rational clinical decision making for patients with progressive LMS.
Recent studies have shown that circulating tumor DNA (ctDNA) is often detected in the plasma of patients with advanced solid malignancies (5, 6). Several studies show that ctDNA levels correlate with prognosis and that changes in ctDNA levels can be associated with response to therapy (7–9). In previous work, we demonstrated that ctDNA can be detected and quantified in patients with LMS and found that ctDNA levels correlate with tumor size and tracked longitudinally with changes in disease burden (10). However, this cohort was a small, clinically heterogenous population and associations with ctDNA levels and tumor burden were descriptive. To determine whether ctDNA levels correlate with treatment response and outcome, we utilized well-annotated banked plasma samples collected from patients with advanced LMS who were treated on a prospective clinical trial (SARC021) designed to test the efficacy of a novel chemotherapy combination in patients with relapsed soft-tissue sarcomas (11).
Materials and Methods
Patient cohort
Our cohort consisted of patients who had previously been enrolled in the prospective SARC021 study with a diagnosis of LMS confirmed by central pathology review (11). Eligible patients were 15 years of age or older with a diagnosis of an advanced unresectable or metastatic soft-tissue sarcoma, of intermediate or high grade, for which no standard curative therapy was available. The SARC021 trial was an open-label, randomized, phase III, multicenter trial that compared treating patients with soft-tissue sarcomas with either doxorubicin alone or with the combination of doxorubicin and evofosfamide (11). The study showed that outcomes did not differ between the arms of the trial, failing to demonstrate a benefit from the addition of evofosfamide (11). LMS was the most common diagnosis enrolled on the trial (36% of the entire cohort).
Patients enrolled on SARC021 could submit optional peripheral blood samples drawn prior to therapy, after two cycles of chemotherapy and at the time of subsequent disease evaluations for future studies. As dictated by the trial protocol, blood was collected in EDTA tubes and plasma was isolated by centrifugation and stored at −80°C at the Nationwide Children's Hospital/RINCH biorepository. All patients that were evaluated for ctDNA had provided signed written informed consent for sample banking at the time of enrollment to SARC021 (11). The study was conducted in accordance with Declaration of Helsinki and that the studies were approved or deemed nonhuman subject research by an institutional review board.
Patient selection for ctDNA studies
Patients diagnosed with LMS were included in the analytical cohort if they had plasma samples collected prior to initiating therapy. A subset of patients had a plasma sample collected after completion of two cycles of chemotherapy. Clinical data, including the extent of disease and clinical outcomes, were obtained from the SARC021 study database (11). These clinical data included: primary tumor location, extent of disease, tumor grade, tumor stage, tumor size at various evaluation time-points, number and location of metastatic sites, vital status, time to first occurrence of progression or death, and tumor response (defined as stable disease, partial response, complete response, or disease progression).
Sample preparation
Frozen plasma samples were shipped to Dana-Farber Cancer Institution (DFCI) for ctDNA analysis on dry ice. After thawing, cell-free DNA was extracted using the QIAamp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer's instructions. Plasma sample volumes received by DFCI ranged from 0.4 to 2.0 mL.
ULP-WGS and ctDNA quantification
Extracted DNA was quantified using a Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific). High-molecular weight DNA contamination of cell-free DNA was determined by Bioanalyzer (Agilent Technologies) and size selection was performed if necessary (AMPure XP beads; Beckman Coulter). Up to 40 ng of cell-free DNA was used for KAPA Hyper library preparation (Kapa Biosystems). Libraries were assessed for quality by a Bioanalyzer followed by quantification using the MiSeq Nano Flow cell (Illumina). Barcoded libraries were pooled and sequenced on a HiSeq 2500 in the rapid run mode (Illumina) to a targeted coverage of 0.2× (actual range, 0.06× to 0.23×). Sequencing results were demultiplexed, aligned and processed using the Picard tools, BWA alignment tool and the GATK tool (12–14). Analysis of the ULP-WGS data was performed using the Broad Institutes ichorCNA algorithm with manual curation of results to confirm tumor percentages (15). Previous studies have shown that ULP-WGS can be used to identify ctDNA in patients with copy-number altered tumors (15, 16). Serial dilution experiments validated that this approach can detect and accurately quantify ctDNA when constituting as little as 3% of a cell-free DNA sample (15).
Copy-number analysis
Copy-number segments and estimates of tumor fraction and ploidy were generated by ichorCNA (15). The log2 ratio values of segments were adjusted for tumor fraction and ploidy such that the data were consistent across samples. GISTIC2.0 was used to determine gene-level copy-number analyses (17). For copy neutral segments predicted by ichorCNA, the log2 ratio was set to zero. The amplification/deletion log2 ratio threshold used for GISTIC was 0.3. Significant gains and losses were determined with a false discovery rate of 0.25. This analysis was performed on plasma samples that were positive for detectable levels of ctDNA. We explored associations between common CNVs seen in the cohort and outcome. We also compared CNVs seen in our cohort to those described previously (18–20).
Statistical analysis
The primary explanatory variable of interest is “ctDNA positivity,” which was coded as positive or negative based on presence or absence of detectable levels of ctDNA in the sample. As the threshold for ctDNA detection in ichorCNA is 3%, any sample estimated by ichorCNA to have less than 3% ctDNA was considered below the level of detection and coded as negative or undetectable. The ctDNA level in each sample with detectable levels was also recorded as a percent of the total cell-free DNA sample as reported by ichorCNA. PFS was measured from randomization to documented disease progression or death, whichever occurred first. Patients who were alive without a documented progression were censored at the time of their last disease evaluation. Overall survival (OS) was measured from date of randomization to death.
Differences in ctDNA levels prior to therapy were compared with a Wilcoxon rank-sum test. The associations between baseline ctDNA status (detectable vs. undetectable) and patient/disease characteristics were evaluated using a two-sample t test for continuous variables and a chi-square test (or Fisher exact test if chi-square assumptions were violated) for categorical variables. Spearman correlation coefficient was used to evaluate the association between tumor size and ctDNA detection and between treatment response and ctDNA levels. Differences in proportions between groups were estimated with an odds ratio (OR). The Kaplan–Meier estimator was used to estimate OS and PFS. Comparison of OS and PFS between groups of interest (e.g., patients with or without detectable ctDNA) was performed with a log–rank test. Cox models were used to generate estimates of the HR and corresponding 95% confidence intervals (CI) for time-to-event data.
A landmark approach was used to analyze outcomes for patients with detectable versus undetectable ctDNA levels after two cycles of chemotherapy. The analysis of tumor response only included patients who did not have a response after the completion of two cycles of chemotherapy and who had ctDNA measured after two cycles of chemotherapy. Specifically, patients who had a response before the completion of two cycles of chemotherapy were not part of this analysis. This analysis compared the post-cycle two response rates between patients with detectable or undetectable ctDNA levels in blood samples collected after the completion of two cycles of chemotherapy. The comparison OS data between patients with and without detectable ctDNA after the completion of two cycles of chemotherapy only included patients who were alive (OS) at the completion of two cycles of chemotherapy. Similarly, the comparison of the PFS between the groups only included patients who were alive and did not have progressive disease at the time of completion of two cycles of chemotherapy. The time-to-event clock for the landmark analyses started at the completion of two cycles of chemotherapy. All P values are one-sided and a P value of <0.05 was considered statistically significant. All statistical analyses were performed with SAS version 9.4 and R version 3.6.1.
Data availability
Data were generated by the authors and available on request.
Results
Detection of ctDNA in patients with progressive LMS
Out of the 230 LMS patients enrolled in the SARC021 study (11), pretreatment plasma was available from 98 patients. Post-treatment serial plasma samples obtained after completion of cycle 2 of chemotherapy were available for 69 of the 98 patients with pretreatment samples. Twenty patients had no posttreatment evaluation and 9 patients had posttreatment ctDNA evaluation at later time-points including cycle 5 week 3, cycle 6 week 3, and end of therapy. We analyzed ctDNA in all 98 pretreatment samples and in 85 posttreatment samples (Supplementary Table S1). All 69 samples taken after two cycles of chemotherapy were included in the analyses of the posttreatment time-point. Heterogeneity of the other posttreatment timepoint data limited the ability for a formal statistical analysis. The descriptive characteristics of our analytic cohort and the patients with LMS excluded from our study (due to unavailability of plasma for ctDNA studies) are provided in Table 1. Treatment response rates for our analytic cohort were similar to the LMS cohort excluded from this study (Table 1) and to the overall SARC021 cohort regardless of histologic diagnosis (CR rate of 1% and PR rate of 22%; ref. 11). The cohort of patients with plasma samples available for this analysis do not significantly differ from patients without plasma samples with regard to the majority of characteristics. However, the analytical cohort had a significant overrepresentation of patients with >5 metastatic sites (54.1% vs. 35.0%, respectively; P = 0.005).
Table 1.
Baseline characteristics of patients with LMS with and without ctDNA evaluation.
| Patients | |||
|---|---|---|---|
| Without ctDNA evaluation | With ctDNA evaluation | ||
| Characteristic | N = 120 | N = 98 | P |
| Age at diagnosis, years | |||
| Median (range) | 57.2 (33–81) | 59.6 (28–83) | 0.08 |
| Sex | |||
| Male | 37 (31%) | 35 (35.7%) | 0.45 |
| Female | 83 (69%) | 63 (64.3%) | |
| Primary tumor location | |||
| Extremity | 13 (10.8%) | 18 (18.4%) | 0.11 |
| Other | 107 (89.2%) | 80 (81.6%) | |
| Extent of disease | |||
| Locally advanced | 6 (5%) | 3 (3.1%) | 0.52 |
| Distant metastatic | 114 (95%) | 95 (96.9%) | |
| Grade at entry | |||
| Low/intermediate grade | 49 (41%) | 29 (30%) | 0.09 |
| High grade | 71 (59%) | 69 (70%) | |
| Previous radiotherapy | |||
| No | 88 (73.3%) | 71 (72.4%) | 0.88 |
| Yes | 32 (26.7%) | 27 (25.6%) | |
| Stage | |||
| I | 7 (5.8%) | 4 (4.1%) | 0.72 |
| II | 30 (25%) | 20 (20.4%) | |
| III | 41 (34.2%) | 33 (33.7%) | |
| IV | 40 (33.3%) | 39 (39.8%) | |
| Unknown | 2 (1.7%) | 2 (2%) | |
| Previous adjuvant/neoadjuvant | |||
| Yes | 14 (11.7%) | 9 (9.2%) | 0.36 |
| No | 106 (88.3%) | 89 (90.8%) | |
| No. of sites at entry | |||
| ≥5 | 42 (35%) | 53 (54.1%) | 0.005 |
| <5 | 78 (65%) | 45 (45.9%) | |
| Best response | |||
| CR | 1 (0.8%) | 0 | 0.53 |
| PR | 21 (17.5%) | 23 (23.5%) | |
| SD | 73 (61%) | 55 (56.1%) | |
| PD | 16 (13.3%) | 15 (15.3%) | |
| NE | 9 (8%) | 5 (5.1%) | |
Abbreviations: CR, complete response; NE, non-evaluable; PD, progressive disease; PR, partial response; SD, stable disease.
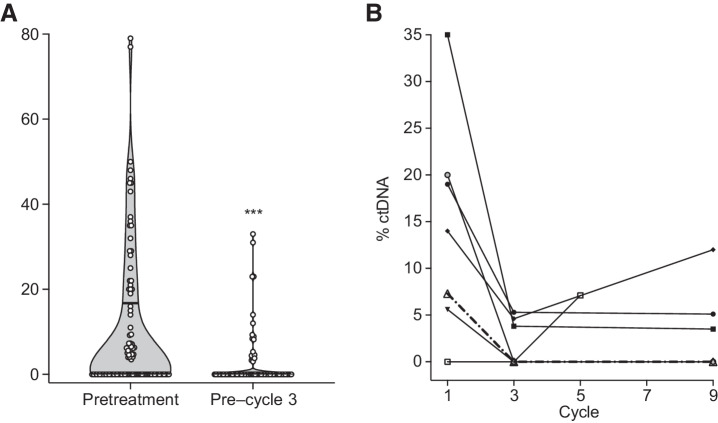
We detected ctDNA with ULP-WGS in pretreatment plasma samples of 48 of 98 (49.0%) patients and in 17 of 69 (24.6%) patients with a sample collected after two cycles of chemotherapy. The range of ctDNA levels, expressed as the percentage of circulating tumor DNA from total cell free DNA, was significantly higher in pretreatment samples (range 0–79%, median 0%) compared with samples taken after two cycles of chemotherapy (range 0% to 33%, median 0%, P ≤ 0.001; Fig. 1A). In those with samples collected at additional timepoints, ctDNA levels changed dynamically throughout the course of the patient's enrollment on study (Fig. 1B).
Figure 1.
Detection of circulating tumor DNA at pretreatment and after two cycles of chemotherapy. A, Violin plot showing ctDNA content as a percentage of total cell-free DNA extracted from plasma of patients with advanced LMS. For both pretreatment and post–cycle 2 levels, the median ctDNA level is zero (below the limit of detection), with the upper quartiles indicated by a lateral solid line within each of the violin plots. The ctDNA content of each sample is indicated by a white circle. ctDNA levels are significantly higher in pretreatment samples compared with post–cycle 2 levels (Mann–Whitney test; ***, P < 0.001). B, Spaghetti plot demonstrating changes in ctDNA levels over time in a subset of patients with three or more serial samples (at least one of which is above the level of detection).
ctDNA association with clinical features of LMS
We found that detectable levels of ctDNA prior to the start of treatment was significantly associated with patients who had larger primary tumors (75% of patients with tumors >10 cm vs. 28% with tumors < 10 cm, respectively; P < 0.001; Table 2; Supplementary Table S2), and had more than five sites of metastatic disease (73.9% vs. 26.1%, respectively; P < 0.001; Table 2). We also observed that patients who had previously received radiation therapy were less likely to have detectable levels of ctDNA prior to starting chemotherapy in the trial (19% vs. 36%, respectively; P = 0.06; Table 2). There was no significant difference in the ctDNA detection rate at the time of enrollment by treatment arm (Table 2) with 39.6% for those assigned to doxorubicin alone having detectable ctDNA versus 44.0% for those assigned to doxorubicin with evofosfamide (P value = 0.66).
Table 2.
Association between ctDNA detection and clinical features in patients with LMS.
| ctDNA positive | ctDNA negative | ||
|---|---|---|---|
| Characteristic | N = 48 | N = 50 | P value |
| Age (mean) | 60 | 59.1 | |
| ≥65 years | 19 (40%) | 16 (32%) | 0.72 |
| <65 years | 29 (60%) | 34 (68%) | |
| Sex | |||
| Male | 17 (35%) | 18 (36%) | 0.95 |
| Female | 31 (65%) | 32 (64%) | |
| Number of sites at entry | |||
| >5 sites | 35 (73%) | 18 (36%) | <0.001 |
| <5 sites | 13 (27%) | 32 (64%) | |
| Tumor size | |||
| ≥10 cm | 36 (75%) | 14 (28%) | <0.001 |
| <10 cm | 12 (25%) | 36 (72%) | |
| Stage | |||
| I–II | 9 (19%) | 15 (30%) | 0.24 |
| III–IV | 39 (81%) | 33 (66%) | |
| Grade | |||
| Low/intermediate grade | 18 (38%) | 11 (22%) | 0.09 |
| High grade | 30 (62%) | 39 (78%) | |
| Primary tumor location | |||
| Extremity | 5 (10%) | 13 (26%) | 0.07 |
| Other | 43 (90%) | 37 (74%) | |
| Treatment | |||
| Doxorubicin | 19 (40%) | 22 (44%) | 0.66 |
| Doxorubicin + evofosfamide | 29 (60%) | 28 (56%) | |
| Previous RT | |||
| Yes | 9 (19%) | 18 (36%) | 0.06 |
| No | 39 (81%) | 32 (64%) | |
| Extent of disease | |||
| Locally advanced | 1 (2%) | 2 (4%) | 1 |
| Distant metastatic | 47 (98%) | 48 (96%) | |
Abbreviation: RT, radiotherapy.
Pretreatment ctDNA levels are associated with outcomes in patients treated with chemotherapy
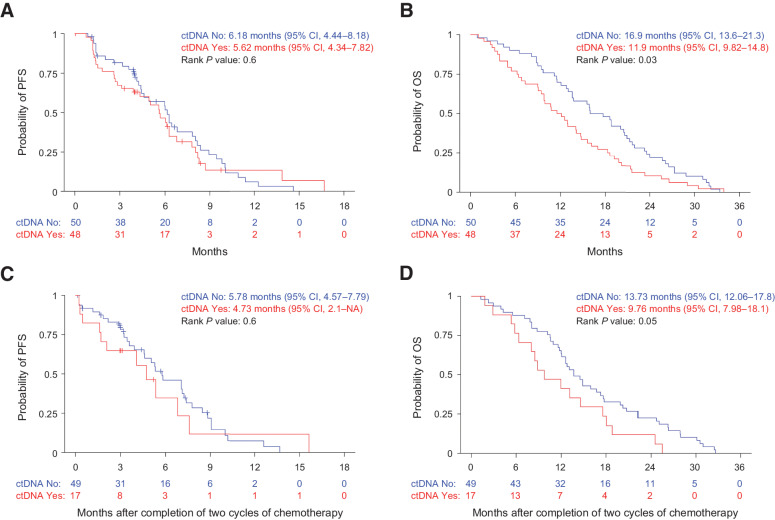
We found that patients with undetectable levels of ctDNA at baseline had a higher objective response rate than patients with detectable levels of ctDNA (37.5% response rate vs. 11%, respectively; P = 0.004; Table 3; Supplementary Table S3) and detection of ctDNA was associated with a significantly lower likelihood of objective response (OR, 0.21; 95% CI, 0.06–0.59; P = 0.005). There was no significant difference in PFS between the patients with baseline detectable ctDNA compared with those without detectable ctDNA (HR, 1.13; 95% CI, 0.71–1.8; P = 0.6; Fig. 2A). However, there was a statistically significant difference in OS between the groups. The 1-year OS rate was 70% (95% CI, 58.4%–83.9%) for patients with nondetectable ctDNA and 50% (95% CI, 37.7%–66.3%) for patients with detectable ctDNA (HR, 1.55; 95% CI, 1.03–2.31; P = 0.034; Fig. 2B).
Table 3.
Association between ctDNA detection and objective clinical response in patients with LMS.
| Clinical response | ctDNA positive | ctDNA negative | P value |
|---|---|---|---|
| Pretreatment | N = 45a | N = 48a | 0.004 |
| Partial response | 5 | 18 | |
| No response | 40 | 30 | |
| Posttreatment | N = 17 | N = 51 | 0.002 |
| Partial response | 0 | 19 | |
| No response | 17 | 32 |
aData on response available for only 93/98 patients.
Figure 2.
Shorter OS in patients with detectable circulating tumor DNA. A, PFS and (B) OS by pretreatment ctDNA detection in patients with advanced, unresectable, or metastatic LMS. C, PFS and (D) OS by ctDNA detection after completion of two cycles of chemotherapy in patients with advanced, unresectable, or metastatic LMS.
On-therapy ctDNA levels are associated with objective responses and survival
Studies have shown that ctDNA levels can change significantly after the initiation of effective therapy (6–8, 16, 21–23). To determine whether ctDNA levels obtained after the initiation of chemotherapy were associated with response to treatment and survival, we examined data from the 69 patients with plasma samples collected after two cycles of chemotherapy (Fig. 1A). There was no difference in the percent of patients with detectable levels of ctDNA after two cycles of chemotherapy whether patients were treated with doxorubicin alone or doxorubicin with evofosfamide (23% vs. 26% respectively; P = 0.83). Among 17 patients with detectable ctDNA after cycle 2, all of them (100%) had detectable ctDNA in their pretreatment sample. Among 52 patients with negative post-cycle 2 samples, 37 of them (71%) were negative at baseline and 15 (29%) were positive at baseline.
Next, we evaluated whether ctDNA detection after two cycles of chemotherapy was associated with treatment response. This analysis was restricted to those patients who did not have an objective response or progression prior to cycle 2 (n = 68). It was found that none of 17 patients (0%) with detectable ctDNA (post-cycle 2) had a subsequent objective response whereas 19 of 51 patients (37%) with undetectable ctDNA experienced a response (P = 0.002; Table 3; Supplementary Table S3). This equated to a significantly lower likelihood of response for patients with detectable ctDNA after two cycles of treatment (OR = 0.05; 95% CI, 0%–0.39%; P = 0.001). Interestingly, of the 19 patients with ctDNA levels below 3% (undetectable ctDNA) who experienced a partial response, 14 responses were observed radiologically 2 to 4 months after the post-cycle 2 plasma was collected. One-year PFS in patients with detectable ctDNA was also lower than patients with undetectable ctDNA but the difference in did not reach significance (HR, 1.2; 95% CI, 0.6–2.37; P = 0.61; Fig. 2C). However, patients with detectable ctDNA had a significantly lower 1-year survival compared with patients with undetectable ctDNA (HR, 1.77; 95% CI, 1–3.14; P = 0.049; Fig. 2D).
Recurrent somatic variants are detectable in the ctDNA of patients with LMS and correlate with survival
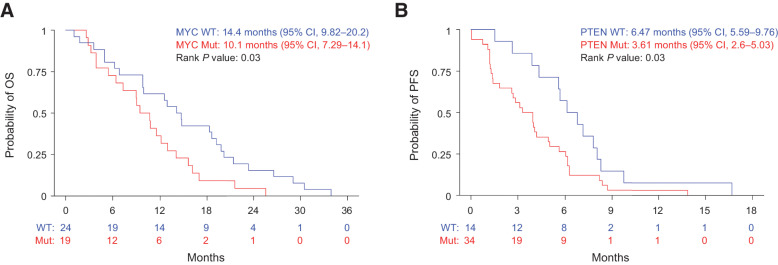
Analysis of the copy-number alterations from patients with detectable ctDNA at the time of study entry demonstrated recurrent events in genes reported to be frequently altered in LMS (Supplementary Fig. S1; refs. 18–20). We estimated PFS and OS based on the presence or absence of the commonly copy-number altered genes including gains of MYOCD and MYC and deletions of RB1, PTEN, and TP53. Of these alterations, we found that copy gains of MYC were significantly associated with a worse 1-year OS (HR, 1.94; 95% CI, 1.06–3.56; P = 0.03; Fig. 3A) and deletions of PTEN were associated with a worse 1-year PFS (HR, 2.05; 95% CI, 1.07–3.92; P = 0.03; Fig. 3B; Supplementary Tables S4 and S5).
Figure 3.
Somatic gain of MYC and loss of PTEN are associated with worse OS and PFS, respectively, in patients with detectable circulating tumor DNA. A, OS in patients with copy-gain of MYC and (B) PFS in patients with copy-loss of PTEN among patients with detectable ctDNA prior to therapy.
Discussion
Over 50% of patients diagnosed with LMS will develop progressive metastatic disease and the prognosis for these patients is poor (1, 24, 25). Front-line treatment for these patients utilizes one of a small number of accepted chemotherapy regimens designed to improve outcomes (2). However, the expected objective response rate of treatment with either a doxorubicin- or gemcitabine-based regimen is only 15% to 20% and prolonged disease control is achieved in only 5% to 10% of patients (2, 26). Furthermore, these agents may have significant toxicities, some associated with cumulative dose and others that are less predictable. Because of an absence of reliable biomarkers that predict which patients are most likely to respond to specific therapies, treatment, typically with either an anthracycline or gemcitabine-based chemotherapy is often offered to patients who recur after primary resection or who present with metastatic disease. Once initiated, therapy often continues until patients experience further progression or after side effects become intolerable. Evaluation of response to therapy is currently measured by radiologic imaging but objective responses typically manifest after four to six cycles of treatment (11). The absence of prognostic biomarkers and early indicators of treatment response make it challenging for oncologists and patients to decide whether to undergo cytotoxic chemotherapy and to decide how long they should continue therapy before moving on to other regimens or experimental agents. We believe that biomarkers that identify patients who are more likely to respond to chemotherapy or robust measures of treatment response early in therapy would greatly enhance clinical decision making in patients with advanced LMS.
In previous work, we have demonstrated that the detection of ctDNA in patients with LMS is feasible using a low-cost sequencing approach that identified the presence of segmental copy-number variants, characteristic of the somatic landscape of LMS, in cell-free DNA isolated from peripheral blood samples (10). Although this assay has limited sensitivity, being unable to detect ctDNA when constituting less than 3% of a cell-free DNA sample, this limit of detection has proven to be a convenient threshold for differentiating patients with high levels of ctDNA from those with low levels (10, 15, 16, 21). In the case of osteosarcoma and Ewing sarcoma, the distinction between high and low levels of ctDNA is prognostic (21). In this study, we took advantage of a unique collection of plasma samples banked from patients enrolled in a prospective study of patients with LMS undergoing treatment with doxorubicin (11). These samples were not collected for the express purpose of ctDNA studies and therefore were unlikely to be optimally handled for the goals of our project. Nevertheless, they provide the first insight, that we are aware of, into the potential clinical value of liquid biopsy assays in the management of patients with progressive and metastatic LMS. In this study, patients received either doxorubicin alone or doxorubicin plus evofosfamide. The addition of evofosfamide did not significantly impact the PFS or OS for patients treated on SARC021, including the patients with a diagnosis of LMS (11). We found that ctDNA levels were not significantly different between patients with LMS who were treated on either arm of the study. This provided us the advantage of being able to study the entire cohort of patients with LMS enrolled on this trial who had plasma samples available in the SARC biorepository.
Applying ULP-WGS to plasma samples collected from patients with LMS prior to the start of SARC021 trial therapy, we found that approximately half had ctDNA levels above the limit of detection of this assay at baseline. This is consistent with previous observations in other sarcomas, including Ewing sarcoma and osteosarcoma (21). We also found that ctDNA levels were associated with other clinical features of aggressive disease including primary tumor size and the presence of numerous metastases. Not only was pretreatment ctDNA detection significantly associated with overall survival, but we also found that patients with undetectable ctDNA levels by ULP-WGS were more likely to experience an objective response to chemotherapy than patients with detectable levels of ctDNA. Although it remains unclear why patients with treatment refractory tumors have higher levels of ctDNA in their blood, we hypothesize that ctDNA shed is associated with more aggressive biology and a higher burden of microscopic and gross metastatic disease. We also found that the detection of somatic copy-number gains of MYC and copy-number loss of PTEN, in patients with detectable levels of ctDNA, was associated with a worse OS and PFS, respectively. These findings suggest that pretreatment ctDNA levels may be informative for patients and providers contemplating the initiation of a chemotherapy regimen.
Another unique feature of this cohort was the availability of serial plasma samples from a large proportion of our patients after the initiation of therapy, something that was not available in our previous sarcoma-focused analysis of ctDNA in Ewing sarcoma and osteosarcoma (21). In this study, we observed that ctDNA levels declined in patients with LMS after initiation of therapy, a trend frequently observed across other solid tumor malignancies (6–8, 16, 21–23, 27). In fact, only 26% of patients with available blood samples had detectable levels of ctDNA by ULP-WGS after two cycles of chemotherapy. However, patients who continued to have detectable levels of ctDNA were extremely unlikely to experience an objective response to chemotherapy and had a significantly shorter survival compared with patients without detectable ctDNA. Furthermore, the majority of patients with undetectable ctDNA levels who experienced an objective response did not demonstrate radiographic evidence of that response for another two to four cycles of therapy. These data demonstrate that ctDNA levels after the initiation of therapy could be a useful response-based biomarker of tumor sensitivity to chemotherapy and could help clinicians and patients decide whether to continue to chemotherapy, especially in the context of patients experiencing adverse events.
We believe that the findings in this study have important implications for the care of patients with progressive and metastatic LMS. However, this analysis also had limitations including the fact that plasma samples were not optimally collected or processed for liquid biopsy studies, which may have diminished our ability to detect ctDNA in some patients. Future studies designed to take advantage of well-established collection and processing techniques would optimize the utility of peripheral blood samples collected from patients with LMS (28–30). Furthermore, serial sampling was largely restricted to the collection of a peripheral blood sample after two cycles of therapy. Numerous studies demonstrate that ctDNA levels change quickly after the first dose of therapy in some cancers (6–8, 16, 21–23, 27). Collection of more frequent blood samples during therapy would be expected to help identify the optimal timepoint for differentiating patients with chemosensitive disease from those with refractory tumors. Another limitation was that the SARC021 study did not include the uterus as an option for coding the site of origin for patients with any soft-tissue sarcoma enrolled to the study, including patients with LMS. Therefore, we were unable to distinguish patients who had primary uterine LMS from patients with nonuterine LMS coded as having “other” as the primary site of disease. The uterus is a common site of LMS and may respond differently to chemotherapy (1). Future studies will be needed to determine whether ctDNA levels differ in patients with uterine LMS. Finally, the size of our cohort likely limited our ability to fully evaluate the association of pre- and on-treatment ctDNA levels with PFS in patients with advanced LMS. Larger, prospective studies could also help determine whether ctDNA assessment can supplement existing radiologic measurements of response, such as RECIST, or be combined with emerging radiomic techniques to better predict responses (31).
To our knowledge, this is currently the largest cohort of patients with LMS evaluated for the presence of ctDNA while undergoing treatment with chemotherapy. This study identifies, for the first time, a significant association between the detection of ctDNA with outcome and the likelihood of experiencing an objective response to treatment, especially when measured after initiation of therapy. This study justifies a more comprehensive and prospective study of liquid biopsies in this disease which would be expected to lead to the incorporation of ctDNA studies in the routine care of patients with LMS.
Supplementary Material
Acknowledgments
This study was supported by a Sarcoma Alliance for Research through Collaboration (SARC) Demonstration Project Grant and the LMSARC research fund (to B.D. Crompton), by the Väre Foundation (to L.M. Madanat-Harjuoja), the Päivikki and Sakari Sohlberg Foundation (to L.M. Madanat-Harjuoja), and the Pediatric Research Foundation (to L. Diller). The Jill Effect, The Catherine England Leiomyosarcoma Fund (to S. George). We would like to thank the physicians and patients who participated in the SARC021 trial and generously contributed samples and data to this effort.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
W.D. Tap reports other support from Sarcoma Alliance for Research through Collaboration (SARC) during the conduct of the study as well as personal fees from Eli Lilly, EMD Serono, Mundipharma, C4 Therapeutics, Daiichi Sankyo, Blueprint, Agios Pharmaceuticals, NanoCarrier, Deciphera, Adcendo, Ayala, Kowa, Servier, Bayer, Epizyme, Cogent, Medpactor, Foghorn, and Amgen outside the submitted work; in addition, W.D. Tap has a patent for Companion Diagnostic for CDK4 inhibitors - 14/854,329 pending to MSK/SKI and a patent for Enigma and CDH18 as companion Diagnostics for CDK4 inhibition - SKI2016-021-03 pending to MSK/SKI, is on the scientific advisory boards of Certis Oncology Solutions and Innova Therapeutics, owns shares in Certis Oncology Solutions and Atropos Therapeutics, and is a co-founder of Atropos Therapeutics. K.V. Ballman reports other support from SARC (nonprofit) during the conduct of the study as well as personal fees from Takeda, Agenus, Janssen, Lilly, Sanofi, and Ariad outside the submitted work; in addition, K.V. Ballman has a patent for Prostate Cancer Gene Signature issued and with royalties paid. S. George reports grants from Blueprint Medicines, Deciphera, ARIAD, Daiichi Sankyo, Pfizer, Bayer, Merck, Eisai, Springworks, Blueprint Medicines, Immunicum, and Kayothera and other support from WCG outside the submitted work. B.D. Crompton reports grants from SARC during the conduct of the study; in addition, B.D. Crompton's spouse reports employment with Acceleron. No disclosures were reported by the other authors.
Authors' Contributions
L.M. Madanat-Harjuoja: Data curation, formal analysis, visualization, writing–original draft, writing–review and editing. K. Klega: Data curation, formal analysis, visualization, writing–original draft, project administration, writing–review and editing. Y. Lu: Formal analysis, writing–original draft, writing–review and editing. D.S. Shulman: Formal analysis, supervision, writing–original draft, writing–review and editing. A.R. Thorner: Investigation, writing–original draft, writing–review and editing. A. Nag: Investigation, writing–original draft, writing–review and editing. W.D. Tap: Resources, formal analysis, writing–review and editing. D.K. Reinke: Resources, data curation, writing–original draft, writing–review and editing. L. Diller: Funding acquisition, writing–review and editing. K.V. Ballman: Formal analysis, supervision, visualization, writing–original draft, writing–review and editing. S. George: Conceptualization, supervision, funding acquisition, writing–original draft. B.D. Crompton: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, writing–original draft, writing–review and editing.
References
- 1. George S, Serrano C, Hensley ML, Ray-Coquard I. Soft tissue and uterine leiomyosarcoma. J Clin Oncol 2018;36:144–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Seddon B, Strauss SJ, Whelan J, Leahy M, Woll PJ, Cowie F, et al. Gemcitabine and docetaxel versus doxorubicin as first-line treatment in previously untreated advanced unresectable or metastatic soft-tissue sarcomas (GeDDiS): a randomised controlled phase 3 trial. Lancet Oncol 2017;18:1397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Maki RG, Wathen JK, Patel SR, Priebat DA, Okuno SH, Samuels B, et al. Randomized phase II study of gemcitabine and docetaxel compared with gemcitabine alone in patients with metastatic soft tissue sarcomas: results of sarcoma alliance for research through collaboration study 002 [corrected]. J Clin Oncol 2007;25:2755–63. [DOI] [PubMed] [Google Scholar]
- 4. Judson I, Verweij J, Gelderblom H, Hartmann JT, Schöffski P, Blay J-Y, et al. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: a randomised controlled phase 3 trial. Lancet Oncol 2014;15:415–23. [DOI] [PubMed] [Google Scholar]
- 5. Abbou SD, Shulman DS, DuBois SG, Crompton BD. Assessment of circulating tumor DNA in pediatric solid tumors: the promise of liquid biopsies. Pediatr Blood Cancer 2019;66:e27595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol 2016;34:3375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kurtz DM, Scherer F, Jin MC, Soo J, Craig AFM, Esfahani MS, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol 2018;36:2845–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hemming ML, Klega KS, Rhoades J, Ha G, Acker KE, Andersen JL, et al. Detection of circulating tumor DNA in patients with leiomyosarcoma with progressive disease. JCO Precis Oncol 2019;2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tap WD, Papai Z, Van Tine BA, Attia S, Ganjoo KN, Jones RL, et al. Doxorubicin plus evofosfamide versus doxorubicin alone in locally advanced, unresectable or metastatic soft-tissue sarcoma (TH CR-406/SARC021): an international, multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2017;18:1089–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H, Durbin R.: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 2017;8:1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Klega K, Imamovic-Tuco A, Ha G, Clapp AN, Meyer S, Ward A, et al. Detection of somatic structural variants enables quantification and characterization of circulating tumor DNA in children with solid tumors. JCO Precis Oncol 2018;2018:PO.17.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Network CGAR. Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell 2017;171:950–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chudasama P, Mughal SS, Sanders MA, Hübschmann D, Chung I, Deeg KI, et al. Integrative genomic and transcriptomic analysis of leiomyosarcoma. Nat Commun 2018;9:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Larramendy ML, Kaur S, Svarvar C, Böhling T, Knuutila S. Gene copy number profiling of soft-tissue leiomyosarcomas by array-comparative genomic hybridization. Cancer Genet Cytogenet 2006;169:94–101. [DOI] [PubMed] [Google Scholar]
- 21. Shulman DS, Klega K, Imamovic-Tuco A, Clapp A, Nag A, Thorner AR, et al. Detection of circulating tumour DNA is associated with inferior outcomes in Ewing sarcoma and osteosarcoma: a report from the Children's Oncology Group. Br J Cancer 2018;119:615–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dawson S-J, Tsui DWY, Murtaza M, Biggs H, Rueda OM, Chin S-F, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199–209. [DOI] [PubMed] [Google Scholar]
- 23. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med 2015;7:302ra133. [DOI] [PubMed] [Google Scholar]
- 24. Lusby K, Savannah KB, Demicco EG, Zhang Y, Ghadimi MP, Young ED, et al. Uterine leiomyosarcoma management, outcome, and associated molecular biomarkers: a single institution's experience. Ann Surg Oncol 2013;20:2364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xue M, Chen G, Dai J, Hu J. Development and validation of a prognostic nomogram for extremity soft tissue leiomyosarcoma. Front Oncol 2019;9:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akin S, Dizdar O, Karakas Y, Turker A, Kars A. Ifosfamide and doxorubicin in the treatment of advanced leiomyosarcoma. Curr Probl Cancer 2018;42:344–9. [DOI] [PubMed] [Google Scholar]
- 27. Chicard M, Colmet-Daage L, Clement N, Danzon A, Bohec M, Bernard V, et al. Whole-exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clin Cancer Res 2018;24:939–49. [DOI] [PubMed] [Google Scholar]
- 28. Kang Q, Henry NL, Paoletti C, Jiang H, Vats P, Chinnaiyan AM, et al. Comparative analysis of circulating tumor DNA stability In K3EDTA, Streck, and CellSave blood collection tubes. Clin Biochem 2016;49:1354–60. [DOI] [PubMed] [Google Scholar]
- 29. van Dessel LF, Beije N, Helmijr JCA, Vitale SR, Kraan J, Look MP, et al. Application of circulating tumor DNA in prospective clinical oncology trials - standardization of preanalytical conditions. Mol Oncol 2017;11:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. Arch Pathol Lab Med 2018;142:1242–53. [DOI] [PubMed] [Google Scholar]
- 31. Spinnato P, Kind M, Le Loarer F, Bianchi G, Colangeli M, Sambri A, et al. Soft tissue sarcomas: the role of quantitative MRI in treatment response evaluation. Acad Radiol 2021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were generated by the authors and available on request.