Abstract
Cancer cells modulate their metabolic activities to adapt to their growth and proliferation. Despite advances in breast cancer biology having led to the widespread use of molecular targeted therapy and hormonal drugs, the molecular mechanisms in metabolism related to the regulation of breast cancer cell proliferation are still poorly understood. Here, we investigate the possible role of SHMT2, a key enzyme in serine metabolism, in breast cancer. Firstly, SHMT2 is found highly expressed in both breast cancer cells and tissues, and patients with high expression of SHMT2 have a worse prognosis. Moreover, the intervention of SHMT2 by either knockdown or over-expression in vitro induces the effect on breast cancer proliferation. Mechanistically, RNA-seq shows that over-expression of SHMT2 affect multiple signaling pathways and biological process in breast cancer cells. Furthermore, we confirm that SHMT2 promotes breast cancer cell growth through MAPK and VEGF signaling pathways. Finally, we verify the role of SHMT2 in promoting breast cancer growth in the xenograft tumor model. Our results indicate that SHMT2 plays a critical role in regulating breast cancer growth through MAPK, and VEGF signaling pathways, and maybe serve as a therapeutic target for breast cancer therapy.
Keywords: SHMT2, breast cancer, serine/glycine metabolism, VEGF, MAPK
Introduction
Breast cancer is by far the most common malignancy in women and poses a major threat to women’s health worldwide [1]. Despite advances in breast cancer biology has led to the widespread use of molecular targeted therapy and hormonal drugs [2], the molecular mechanisms associated with proliferative regulation of breast cancer cells are still poorly understood as the complexity and heterogeneity of breast cancer. Thus, the identification of novel targeted therapies for breast cancer is an urgent need.
Reprogrammed metabolism is considered a hallmark of cancer [3]. In addition to aerobic glycolysis [4] and glutamine metabolism [5], carbon metabolism is also one of the main metabolic pathways of tumor cells, which is mainly provided by serine/glycine metabolism [6]. Serine hydroxymethyltransferase (SHMT) is a key enzyme in serine/glycine metabolism. Two SHMT genes, SHMT1 and SHMT2, have been identified in the human genome, in cytosolic and mitochondrial forms of SHMT [7]. It has been suggested that cytosolic SHMT may catalyze the conversion of glycine to serine, whereas glycine synthesis from serine occurs in the mitochondria [8].
SHMT2, not SHMT1 was discovered to be over-expression in various tumors and correlate with cancer progression and poor prognosis [9]. In addition to the oncogenic role of the isozyme widely studied, some specific mechanisms of SHMT2 in human carcinogenesis have been identified. For instance, in human glioblastoma multiforme SHMT2 activity limits that of pyruvate kinase and reduces oxygen consumption, triggering a metabolic state that confers a profound survival advantage to cells in poorly vascularized tumor regions [10]. STAT3/SHMT2/PKM2 loop in prostatic carcinoma cells can regulate a metabolic shift in response to inflammation at the early stages of cancer progression [11]. Moreover, transcription factor NRF2 [12] and histone lysine demethylase KDM4C [13] activating ATF4 modulate the expression of the key serine/glycine biosynthesis enzyme genes including SHMT2 to support glutathione and nucleotide production. Studies on noncoding RNAs in tumors have been increasing in recent years, and it has also been reported that SHMT2 could be directly and negatively regulated by miR-615-5p in HCC cells [14]. Above all some mechanisms have been studied, most of the research is working on addressing how SHMT2 is regulated, however, the downstream of SHMT2 is little studied.
Regarding breast cancer, SHMT2 was also found to be over-expression in breast cancer cells, and the expression level of SHMT2 was positively correlated with breast cancer grade [15]. In breast cancer, HIF1α and MYC cooperate to drive SHMT2 up-regulation, which leads to an increased concentration of nicotinamide adenine dinucleotide phosphate (NADPH) and enhanced redox balance; this in turn facilitates cancer cell growth under hypoxic conditions [16]. However, it is unknown that the downstream pathway regulated by SHMT2 pathway in tumor growth is involved in breast cancer. Our results showed that SHMT2 was highly expressed in both breast cancer cells and tissues, and patients with high expression of SHMT2 had a worse prognosis. We then explored that SHMT2 regulated breast cancer proliferation in vitro. We further confirmed that SHMT2 promoted growth through MAPK, apoptosis, and VEGF signaling pathways. Finally, we verified the role of SHMT2 in promoting breast cancer growth again in the xenograft tumor model. Our findings provide new insights into the biological role of SHMT2 in breast cancer and suggest that SHMT2 may be a new therapeutic target for breast cancer.
Materials and methods
Cell lines and specimen
Human breast cancer cell lines (HCC1806, MDA-MB-231, MCF-7, ZR-75-1, BT549) and normal breast cell lines (MCF-10A) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). BT549, HCC1806, and MDA-MB-231 are triple-negative breast cell lines. And MCF-7 and ZR-75-1 are hormone receptor-positive. The cells were cultured as monolayers in RPMI-1640 culture medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 μg/ml penicillin, and 100 μg/ml streptomycin and maintained in an incubator with a humidified atmosphere of 95% air and 5% CO2 at 37°C. All cell lines had been authenticated by tandem repeat fingerprinting (STR) by Beijing Yuewei Gene Technology Co., Ltd. within 6 months from we get them and tested for mycoplasma contamination every 3 months using MycoAlert (C0297M, biyotime, Shengzhen).
140 paraffin-embedded breast cancer patient tumors corresponding to adjacent normal tissues were collected from Sun Yat-sen University Cancer Center (Guangzhou, China). The study was approved by the ethics committee of Sun Yat-sen University Cancer Center. Informed consent and agreement were obtained from all the patients. Detailed clinical and pathologic information was available for all cases. The pathological TNM status was assessed according to the seventh edition of the American Joint Committee on Cancer.
Reagents and antibodies
Antibodies against SHMT2 (A1215), GAPDH (A19056), β-actin (AC026), VEGF (A12303), PEDF (A3475), were obtained from ABclonal Technology Co., Ltd. (ABclonal, Wuhan, China). Antibodies against JNK (AF6318), P38 (AF6456), ERK (BF8004), P-JNK (Thr183+Tyr185)(AF3318), P-P38 (Thr180/Tyr182) (AF4001), P-ERK (Thr202/Tyr204) (AF1015), cytochrome-C (AF0146), PARP (DF7198), Cleaved-PARP (Asp214) (AF7023), Cleaved-Caspase 3 (Asp175) (AF7022), Cleaved-Caspase 9 (Asp353) (AF5240), BAX (AF0120), Bcl-2 (AF6139), Goat Anti-Rabbit IgG (H+L) HRP (S0001) and Goat Anti-Mouse IgG (H+L) HRP (S0002) were purchased from Affinity Biosciences (Affbiotech, Jiangsu, China). ERK MAPK inhibitor FR180204 (HY-12275), p38 MAPK inhibitor SB202190 (HY-10295) were purchased from the Med Chem Express (MCE).
Preparation of shRNA or expression plasmid
The SHMT2 shRNA (HSH109479) and the overexpression plasmid of SHNT2 (p-SHMT2) (T3093) were purchased from GeneCopoeia, Inc. (Guangzhou, China). The sequence of the human SHMT2-specific shRNA was 5’-TCTGAACAACAAGTACTCGG-3’ and 5’-TCTCAGGATCACTGTCCGAC-3’, and the scramble shRNA was 5’-GGCTCCGAACGGGTCACGATT-3’. For the in vitro delivery of the shRNA and plasmid into tumor cells, the sequences were first encapsulated into Lipofectamine 3000 (L3000150, Thermo Scientific), that had been validated by many analyses by dissolving in Opti-MEM. The knockdown efficiency was validated by western blot.
Transient transfection
A total of 2 × 105 breast cancer cells were seeded into each well of a six-well tissue culture plate (Nunc). The next day (when the cells were 70-80% confluent), the culture medium was aspirated, and the cell monolayer was washed with prewarmed sterile phosphate-buffered saline (PBS). The cells were transfected with the shRNA or plasmid at the indicated dose using Lipofectamine 3000 (L3000150) (Thermo Scientific). The cells were harvested after 48 h of transfection, and western blot analyses or other experiments were performed.
Western blot analysis
Total proteins were extracted using RIPA lysis buffer. The western blot analysis was conducted as described [11]. The following primary antibodies were used: SHMT2, GAPDH, β-actin, VEGF, PEDF, JNK, P38, ERK, P-JNK, P-P38, P-ERK, PARP, Cleaved-PARP, Cleaved-Caspase 3, Cleaved-Caspase 9, BAX, Bcl-2.
Immunohistochemistry
Tissue samples including breast cancer tissue samples and mice tissues were preserved in paraffin and cut into 4-μm slices. Breast cancer specimens were stained with SHMT2. Mice tissues were stained with SHMT2, P-P38, P-ERK, Ki-67, and MVD. The Immunohistochemical process was according to a previously described method [11].
MTT assay
Cells plated in 96-well plates (1000 cells/well) were transfected with control or specific shRNA. 48 h later, 10 ul MTT reagent (5 mg/ml) was added to a single well and treated for 2.0 h. Then MTT was discarded and 150 ul Dimethyl Sulphoxide (DMSO) was added to a single well. Finally, the OD value at 490 nm was measured. Regarding cell viability, cells treated with only transfection reagent were used as the blank control, The percent of cell viability is the ratio of OD value of other groups to that of the blank group.
Colony formation assay
Cells were transfected with SHMT2 shRNA or plasmid for 24 h, trypsinized, and re-suspended as single cells. The cells (1 × 103/ml) were then mixed in 21 ml of 1640 culture medium containing 5% FBS. The cultures were maintained in a 37°C/5% CO2 incubator for 12-14 days. The cell clones were then washed three times with phosphate-buffered saline (PBS), fixed in methanol for 10 minutes, and stained with Crystal Violet for 10 minutes at room temperature. The dye was washed off, and the colonies that contained more than 50 cells were counted.
RNA sequencing
RNA was extracted from the vector group and the SHMT2 OVER ZR-75-1 cells using the RNeasy Kit (Qiagen) according to the manufacturer’s protocol. RNA quality control was performed using an Agilent Bioanalyzer. RNA-seq libraries were generated using Illumina TruSeq mRNA-stranded kits following Illumina protocols. Libraries were quantitated using an Agilent bioanalyzer, and the pooled libraries were sequenced with an Illumina HiSeq 4000 system using Illumina reagents and protocols. Abundance quantifications were imported into R software, and a gene expression matrix was constructed using the R Bioconductor package (41). Count values summarized were analyzed using the DESeq2 algorithm. Differential expression was defined at a threshold of FDR = 0.05 and absolute log fold change > 1.
Apoptosis assay
Cells were stained with Annexin V-FITC and PI (propidium iodide) using an Annexin V-FITC/PI-staining kit (Genechem, Shanghai, China). Analysis was performed using Multicycle for Windows (Beckman Coulter).
Immunofluorescence
The treated cells were seeded in 6 well Culture plate cover with a glass slide (24 mm × 24 mm) at a density of 1 × 105 cells per well. After 48 h, the cells were washed with PBS, fixed with 4% paraformaldehyde solution, and permeabilized with 0.1% Triton X-100. The cells were incubated with a rabbit anti-cytochrome-c antibody and then incubated with a Goat anti-Rabbit IgG (H+L), Alexa Fluor 568 (A-11036, Thermo Scientific). The nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI), and the cells were examined under a fluorescence microscope.
The concentrations of VEGF and PEDF were determined by ELISA
HCC1806 and ZR-75-1 cells were seeded in 6-well plates and treated with SHMT2 shRNA or overexpression plasmids for 48 hours. The VEGF and PEDF levels in the culture supernatant were quantified using a VEGF Immunoassay Kit (ml064281) (Enzyme-linked Biotechnology, Shanghai, China) and a Chemokine PEDF ELISA Kit (ab213815) (Abcam, Shanghai, China) according to the manufacturer’s protocols.
Angiogenesis experiment
Assessment of endothelial tube formation under treatment with 20 ng/ml of human recombinant IL6, G-CSF, PGF, or 100 ng/ml of ANG-2 (#200 06, #300 23, #100 06, #130 07, Peprotech, Rocky Hill, NJ, USA) was performed in a 6-well µ angiogenesis plate (#89646, ibid GmbH, Planegg, Germany). 2 × 105 HUVECs were seeded onto pre-plated 10 µl of growth factor reduced Matrigel® matrix (#356231, Corning Inc., Corning, NY, USA) in 70 µl of DMEM supplemented with 5% FCS and 10% of EGM™2 MV and the respective growth factor. After 11 h of incubation, the cells were stained with 4 µg/ml calcein-AM (#17783, Merck) in Hanks’ balanced salt solution (HBSS, #14175-053, Gibco) for 30 min at 37°C. Images were analyzed with the free AngioTool software.
Primary human umbilical vein endothelial cells (HUVECs, pooled, ATCC® #PCS-100-013™) were grown in an endothelial growth medium (EGM™2 MV; #CC-3202, Lonza, Basel, Switzerland) and only early passages (< p6) were used.
Xenograft tumor model
All animal maintenance and operational procedures were carried out with the principles of the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. Meanwhile, this study was approved by the Animal Care and Ethics Committee of Sun Yat-Sen University Cancer Center. We purchased female nude mice (Balb/c) aged 5 weeks from Beijing Vital River Laboratory Animal Technology Co., Ltd. Randomization was conducted and mice were treated by an unblinded manner. For xenograft tumor formation, 8 × 106 HCC1806 cells with shSHMT or p-SHMT or shRNA were respectively suspended in 100 ul PBS and injected subcutaneously into the right flank of each mouse (5 or 6 in each group). After two weeks of injection, the tumor volume was calculated as V = width2 × length and recorded every three days for four weeks. At the end point, the mice were sacrificed by CO2 inhalation, and tumors were taken from mice for weighing and photographing.
Statistical analysis
All results were presented as the mean ± SD. We repeated all the experiments at least three times. GraphPad Prism 6 (San Diego, CA, USA) and SPSS software package (standard version 18.0; SPSS, Chicago, IL) were used for statistical analysis. Two-tailed Student’s t test was used to determine differences between each group. Kaplan Meier methods were used to compute the survival analyses. The experimental data are presented as the mean ± SD. P-values are denoted as follows: *P < 0.05, ***P < 0.001.
Results
SHMT2 is increased in breast cancer cells and tissue, and is associated with a poor prognosis
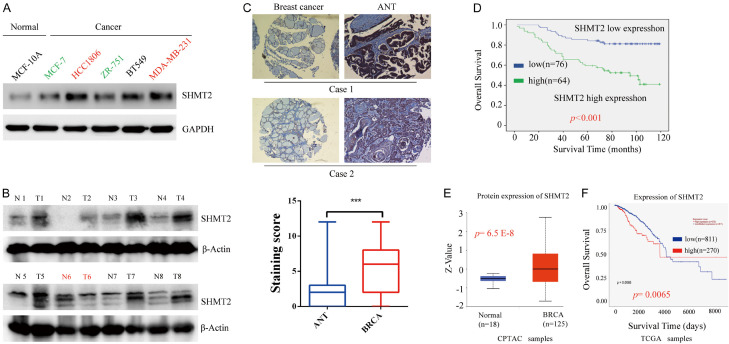
We first detected the expression of SHMT2 protein in a human normal mammary cell line (MCF-10A) and breast cancer cell lines (MCF-7, BT549, HCC1806, ZR-75-1) by western blotting. High SHMT2 protein is expressed in breast cancer cell lines while faint expression in a normal cell line (Figure 1A). Moreover, the expression of SHMT2 in triple-negative breast cell lines including HCC1806, MDA-MB-231, and BT549 cell lines is higher than that in non-triple negative breast cancer MCF7 and ZR-751 (Figure 1A). Moreover, SHMT2 expression was found in breast cancer tissue (Figure 1B), as well as in the breast cancer cell lines. In immunohistochemical staining, as shown in Figure 1C, positive staining of SHMT2 was observed in breast cancer tissue but not in adjacent normal breast tissue. Analysis of breast cancer expression profile data from the TCPAC database also confirmed our findings. Compared with normal breast tissue, SHMT2 expression was significantly up-regulated in cancer tissue (Figure 1E). Overall, it suggests that SHMT2 might be a potential biomarker of breast cancer.
Figure 1.
SHMT2 is highly expressed in breast cancer and associated with a poor prognosis. A. The expression of SHMT2 protein in various Breast cancer cell lines (MCF-1, MDA-MB-231, HCC1806, ZR-75-1, BT549) and a normal breast cell line (MCF-10A) was analyzed by Western blotting. B. The expression of SHMT2 protein was tested by Western blotting in 8 breast cancer tissue and matched para-cancer tissue. C. Representative pictures of SHMT2 by immunohistochemistry were shown in breast cancer tissues and matched para-carcinoma tissues. Original magnification, 200 ×. D. Kaplan-Meier method was used to compute survival analysis comparing patients with high (n = 64) and low (n = 76) expression. E. With CPTAC data, the expression of SHMT2 in breast cancer tissues compared with normal breast tissues. F. Survival analysis was further conducted. ***P < 0.001, Student’s t-test. The data are presented as the mean ± SD of three tests. T, breast cancer tissue; N, matched para-carcinoma tissue.
Furthermore, research was on larger sample size. We explored the expression of SHMT2 in 140 breast cancer tissues and its relationship with clinic pathologic features. The overall survival curves revealed an unfavorable prognosis for the high-SHMT2 group compared to the low-SHMT2 group (Figure 1D). A similar result was obtained from the TCPAC and TCGA data (Figure 1F).
Up-regulation of SHMT2 promotes cell growth in breast cancer
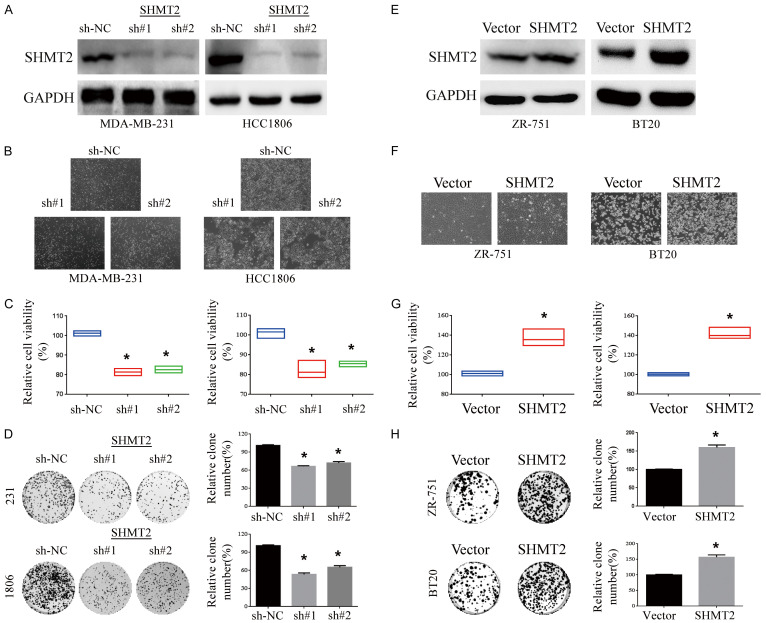
To further explore the role of SHMT2 in breast cancer, we observe whether SHMT2 affects the proliferation of breast cancer. The effect of SHMT2 intervention was better demonstrated by knocking down SHMT2 in an over-expressed cell line and over-expressing SHMT2 in a slightly lower-expressed cell line. Thus, SHMT2 was knockout in HCC1806 cells with high expression, while SHMT2 was over-expressed in ZR-751 cells with low expression (Figure 2A, 2E). Microscopically, the cell density and the total number of SHMT2-inhibited cells were decreased (Figure 2B), while that increased significantly in breast cells with SHMT2 over-expression (Figure 2F). SHMT2 knockdown inhibited cell viability (Figure 2C), and SHMT2-overexpressing cell line activated cell viability (Figure 2G). In colony formation assay, SHMT2 knockdown inhibited cell clonogenicity (Figure 2D) and vice versa (Figure 2H).
Figure 2.
Up-regulation of SHMT2 promotes cell growth in breast cancer. SHMT2 protein was detected by western blot in HCC1806 with SHMT2 knockdown (A) and ZR-75-1 (E) with SHMT2 over-expression. And the cell lines HCC1806 (B) and ZR-75-1 (F) were photographed. The cell viability of breast cancer cell lines HCC1806 (C) and ZR-75-1 (G) were determined by MTT assay 48 h after transfection. The effect of down-regulation (D) or up-regulation (H) of SHMT2 on tumor cell colony formation was analyzed by plate clone formation assay. The relative colony formation rates in HCC1806 (D) and ZR-75-1 (H) cells were calculated. *P < 0.05, Student’s t-test. The data are presented as the mean ± SD of three tests.
SHMT2 knockdown activates caspase dependent apoptotic pathway
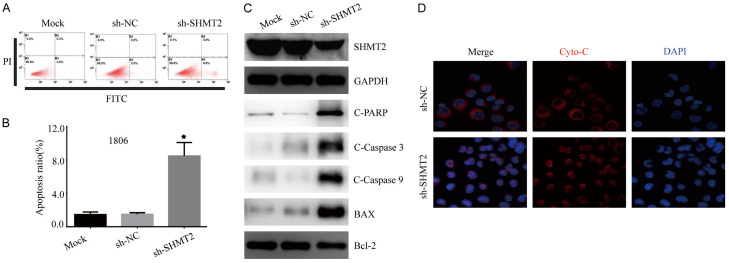
We also assessed the proportion of apoptosis in HCC1806 cells transfected with sh-SHMT2 or sh-NC by Annexin V/PI staining-based FACS analysis. SHMT2 knockdown enhanced apoptosis in HCC1806 cells (Figure 3A, 3B). Detected by western blotting, SHMT2 knockdown significantly activated apoptosis-associated transcription factors including cleaved-PARP, cleaved-caspase-9, cleaved-caspase-3, BAX, and Bcl-2 protein in HCC1806 cells (Figure 3C).
Figure 3.
SHMT2 knockdown activates caspase dependent apoptotic pathway. A. Apoptosis rate was detected by annexin-V-FITC and PI double-stained cells combined with flow cytometry fluorescence separation in an HCC1806 cell line with SHMT2 knockdown. B. The apoptosis rate of each group was quantitatively analyzed. *P < 0.05, Student’s t-test. The data are presented as the mean ± SD of three tests. C. Western blot analysis of caspase-dependent proteins in HCC1806 with SHMT2 knockdown. D. Representative images of immunofluorescence to monitor cytochrome-c in HCC1806 with SHMT2 knockdown. Original magnification, 200 ×.
SHMT2 affects mitochondrial function and the oxidative phosphorylation process [17]. Mitochondria-mediated apoptosis pathway plays an important role in the cell death process [18]. As cytochrome-c release from mitochondria to the cytoplasm is an important event in the mitochondria dependent apoptosis pathway [19], we performed an immunofluorescence imaging (IF) analysis to monitor changes in the subcellular localization of cytochrome-c in HCC1806 cells. SHMT2 knockdown triggered the release of cytochrome-C from the mitochondria and occurred frequently together with apoptotic bodies and the condensation of nuclear chromatin after DAPI staining (Figure 3D). These results suggest that blocking SHMT2 may activate mitochondrial apoptotic signaling pathways.
SHMT2 over-expression affects multiple signaling pathways and biological processes in breast cancer cell
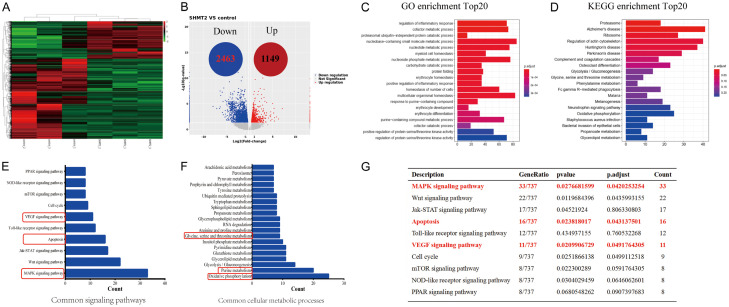
To clarify the regulatory mechanism of SHMT2 in promoting breast cancer growth, we used RNA-seq (Figure 4A) to explore the regulatory factors in ZR-75-1 with over-expressed SHMT2 (Figure 2F). A total of 3,612 genes with differential expression were profoundly altered by SHMT2 over-expression, including 1,149 up-regulated genes and 2,463 down-regulated genes (Figure 4B). Further GO analysis revealed that over-expression of SHMT2 significantly affected multiple biological processes, including regulation of inflammatory response, immune response regulation, ubiquitin-proteasome independent protein lysis process, nucleotide metabolism, etc (Figure 4C). In addition according to KEGG enrichment analysis, over-expression of SHMT2 significantly impacted proteasome signaling pathways, ribosomal genesis, glycolysis/gluconeogenesis, serine/glycine metabolism, oxidative phosphorylation, and other important signaling pathways and biochemical metabolic process (Figure 4D).
Figure 4.
SHMT2 over-expression affects multiple signaling pathways and biological processes in the breast cancer cells. A. Clustering results of differentially expressed genes comparing ZR-75-1 cells with SHMT2 over-expression with the control group cells. B. Volcanic map of differentially expressed mRNAs. C. GO enrichment histogram of differentially expressed mRNA. D. The methodology of KEGG is conducted according to the literature reported by Kanehisa [20]. KEGG pathway enrichment bars of differentially expressed mRNA. E. Common signaling pathways are closely associated with cell proliferation and survival. F. SHMT2 over-expression affects a variety of biochemical metabolic processes in breast cancer cells. G. SHMT2 over-expression affects the MAPK pathway, apoptosis pathway, and VEGF pathway.
Next, we focused on the analysis of common signaling pathways and common intracellular biochemical metabolic processes affecting cell survival and proliferation.
We found that SHMT2 over-expression significantly affected the MAPK pathway, WNT pathway, Jak-STAT pathway, apoptosis pathway, VEGF, and mTOR pathway (Figure 4E). In addition, SHMT2 over-expression also significantly affects intracellular oxidative phosphorylation, purine and pyrimidine metabolism, glycolysis, triglyceride metabolism, serine/glycine metabolism, and RNA metabolism and other important biochemical metabolic processes (Figure 4F). SHMT2 over-expression affects the MAPK pathway, apoptosis pathway, and VEGF pathway (Figure 4G). These results suggest that SHMT2 can affect cell growth and proliferation in multiple pathways, the high expression of SHMT2 in breast cancer cells is very important for cell proliferation and growth.
SHMT2 promotes breast cancer cell proliferation by activating MAPK pathway
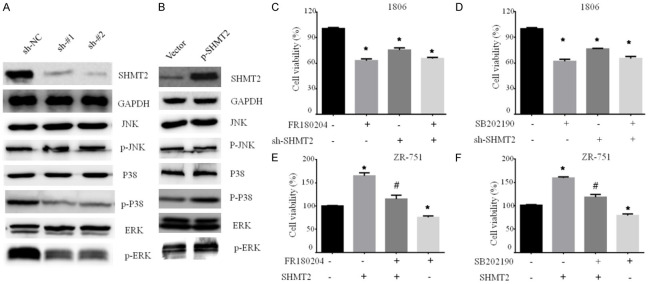
As result of RNA-Seq, SHMT2 over-expression affected MAPK signaling pathway. To further identify it, we analyzed the activities of several pro-survival proteins by western blot. SHMT2 knockdown dramatically suppressed the phosphorylation of ERK1/2 and p38, whereas total JNK, total ERK1/2, and total P38 protein did not change (Figure 5A). Conversely, SHMT2 over-expression increased the phosphorylation of ERK1/2 and p38, suggesting an activation of the MAPK signaling pathways (Figure 5B).
Figure 5.
SHMT2 promotes breast cancer cell proliferation by activating MAPK pathway. Western blotting analysis of proteins with phosphorylation or not related in MAPK signaling pathway in breast cancer cells with SHMT2 knockdown (A) and over-expression (B). Cell viability was analyzed by MTT assay in HCC1806 cells with SHMT2 knockdown treated with an ERK inhibitor FR180204 (C) or a p38 inhibitor SB202190 (D). ZR-75-1 cells with SHMT2 overexpression were treated with an ERK inhibitor FR180204 (E) or a p38 inhibitor SB202190 (F). # represents the comparison between inhibitor plus SHMT2 over-expression group and SHMT2 over-expression group. * represents the comparison between treatment group and control group. #, *P < 0.05. Student’s t-test. The data are presented as the mean ± SD. The error bars show the SD.
As the above result shows, SHMT2 activated MAPK signaling pathway. Furthermore, we confirmed whether the ERK/p38 MAPK signaling pathway mediated by SHMT2 is involved in the regulation of cell growth. We analyzed the effect of ERK and p38 specific inhibitors (FR180204 and SB202190) on cell viability in breast cancer cells with SHMT2 knockdown or over-expression. Cell viability was inhibited by FR180204, SB202190, and SHMT2 knockdown. The inhibition degree by SHMT2 knockdown was smaller than that of FR180204 and SB202190. However, the addition of SHMT2 shRNA did not enhance the effect of FR180204 or SB202190 on cell viability (Figure 5C, 5D). SHMT2 over-expression significantly increased cell viability (Figure 5E). Treatment with ERK or P38 inhibitors partly attenuated the effect on cell viability caused by SHMT2 over-expression (Figure 5E, 5F). Our results indicated that SHMT2 partially modulates MAPK signaling to regulate breast tumor cell growth.
SHMT2 regulates the VEGF signaling pathway in breast cells
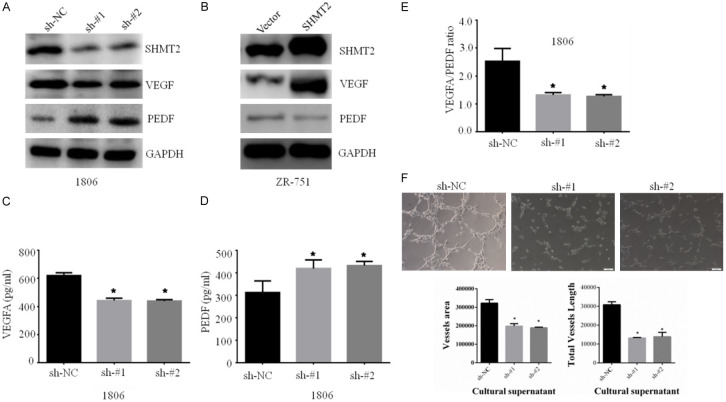
The proteins of VEGF and PEDF mediated angiogenesis play an important role in tumor growth and cancer cell proliferation [21,22]. And SHMT2 over-expression affected VEGF signaling pathway from the RNA-seq result, which promoted us to investigate further. The western blotting results confirmed that SHMT2 knockdown decreased VEGF protein expression and up-regulated PEDF protein (Figure 6A). In contrast, SHMT2 over-expression had the opposite result (Figure 6B). And similar results were obtained by ELISA (Figure 6C, 6D). Consequently, the ratio of VEGF with PEDF dramatically decreased after SHMT2 knockdown (Figure 6E). Finally, in the angiogenesis experiment, SHMT2 knockdown suppressed angiogenesis, which was embodied in decreased vessel area and length (Figure 6F).
Figure 6.
SHMT2 regulates the VEGF signaling pathway in breast cells. Western blotting analysis of VEGF and PEDF proteins in MAPK signaling pathway in breast cancer cells with SHMT2 knockdown (A) and overexpression (B). VEGF and PEDF proteins in supernatant detected by ELISA in breast cancer cells with SHMT2 knockdown (C) and over-expression (D). Subsequently, the ratio of VEGF and PEDF was calculated from ELISA results in breast cells with sh-SHMT2 (E). Finally, an angiogenesis experiment was conducted in breast cells with sh-SHMT2, and vessel area and length were calculated (F). *P < 0.05, Student’s t-test. The data are presented as the mean ± SD of three tests.
SHMT2 promotes tumor growth in vivo
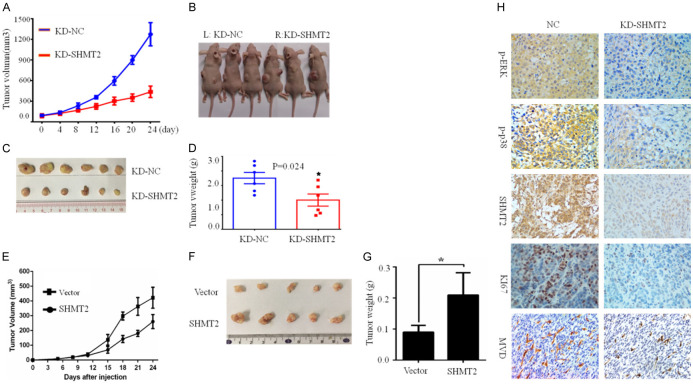
Our results revealed that SHMT2 promoted breast cancer cell growth in vitro, furthermore, we verified the role of SHMT2 in breast cancer in vivo. Xenograft experiment was conducted by subcutaneously injecting HCC1806 cells with SHMT2 knockdown (KD-SHMT2) and control cells (KD-NC), and BT549 cells with SHMT2 over-expression (SHMT2) and control cells (Vector). Tumor volumes were calculated and growth curves were plotted (Figure 7A, 7E). SHMT2 knockdown significantly suppressed tumor volume (Figure 7A, 7C) and weight (Figure 7D) compared with that in the control group, while SHMT2 over-expression significantly increased tumor volume (Figure 7E, 7F) and weight (Figure 7G).
Figure 7.
Knockdown of SHMT2 inhibits breast tumor growth in vivo. HCC1806 cells with SHMT2 knockdown (KD-SHMT2) and control cells (KD-NC), while BT549 cells (SHMT2) with SHMT2 over-expression and control cells (Vector) were injected into the left and right back subcutaneously in 100 ul PBS. The average tumor volume reached 100 mm3 and tumor volume was measured. Xenograft tumors were measured with vernier calipers, tumor volumes were calculated and growth curves were plotted (A, E). After 7 measurements, the mice were sacrificed and photographed (B, F). The xenograft was removed and photographed (C) and weighed (D, G). Immunohistochemical staining (H) detected the protein expression levels of P-ERK, P-P38, KI67, CD31, and SHMT2 in tumor tissues, *P < 0.05, Student’s t-test. The data are presented as mean ± SD from two independent experiments. N = 5 or 6/group, magnification 200 ×.
The results above in vitro indicated that SHMT2 regulated MAPK, VEGF signaling pathway, we used immune-histochemical staining to detect proteins related to the signaling pathway. When SHMT2 expression was decreased, p-ERK, p-p38, ki67, and MCD (micro-vessel density) was inhibited (Figure 7E).
Discussion
In this study, we evaluated the biological role, molecular mechanisms, and clinical significance of SHMT2 in breast cancer carcinogenesis. As a key enzyme in serine metabolism, SHMT2 catalyzes the reversible reaction of serine to glycine by transferring the β-carbon from serine to tetrahydrofolate, providing one-carbon unit for the de novo synthesis of purine. SHMT2 has been implicated in cancer growth, progression, metastasis, and drug resistance [15,23-25]. However, the biological roles and clinical significance of SHMT2 and its precise molecular mechanisms in breast cancer have not been reported.
We demonstrated the high expression of SHMT2 in breast cancer cells, tumor tissues, and breast cancer samples compared to normal cells and normal human breast tissues. To evaluate our hypothesis that SHMT2 plays a potential oncogenic role in breast cancer, we performed in vitro studies to investigate regulatory mechanisms. Our results revealed that SHMT2 knockdown inhibited cell viability, clonogenicity, and angiogenesis and induced apoptosis in vitro. Moreover, SHMT2 over-expression had the opposite effect. Finally, we demonstrated the SHMT2 mediated tumor growth in the xenograft mouse model.
Our results showed that patients with high expression of SHMT2 predicted poor prognosis, suggesting a potential oncogenic role of SHMT2 in breast cancer. The results are consistent with previous studies that SHMT2 is expressed at a high level in various tumors and indicates a poor prognosis [15,23,24,26].
Apoptosis resistance is a protective mechanism of tumor survival and plays an important role in the response of breast cancer to chemotherapy and radiotherapy [27]. In this study, we showed that SHMT2 knockdown confers apoptosis along with activation of caspase proteins and release of cytochrome C from the mitochondria to cytosol, a mitochondria-mediated apoptosis event. However, it is little known how SHMT2 regulate apoptosis mediated by mitochondria, which is an interesting question for us to study in the future.
VEGF is known as one of the principal initiators in the development and progression of vascularization, then impacts tumor angiogenesis, tumor growth promotion, and survival [28-30]. Besides VEGF, the pigment epithelium-derived factor (PEDF) is also involved in angiogenesis, an anti-angiogenic factor with anti-tumor properties counterbalancing the effect of VEGF [31]. Moreover, some studies in vivo and in vitro show an association between an increase in the VEGF/PEDF ratio and a bad prognosis in nasopharyngeal carcinoma [32] and lung adenocarcinoma [33]. Our result also showed that SHMT2 knockdown in breast cancer cell lines led to a significant reduction in the VEGF/PEDF ratio at the protein levels, suggesting that SHMT2 at least partially targets VEGF signaling. At present, no studies have reported that SHMT2, which mediated serine metabolism and one-carbon unit metabolism can affect VEGF signaling pathway and angiogenesis. Our findings suggest that SHMT2 may be involved in the regulation of tumor angiogenesis. This is a novel biological function of SHMT2. However, the molecular mechanism of SHMT2 regulating VEGF and angiogenesis needs to be further researched.
In our study, SHMT2 regulated the phosphorylation level of p38 and ERK. p38 and ERK inhibitors significantly antagonized the up-regulation or knockdown of SHMT2 affecting breast cell viability. It suggests that SHMT2 can promote breast cancer cell proliferation by activating the MAPK pathway. It has been reported that SHMT2 regulates AKT/MTOR signaling pathways to facilitate hepatocyte proliferation and regeneration [34]. These results indicate that SHMT2 can regulate the cell survival signaling pathway. Moreover, we demonstrated that the knockdown of SHMT2 markedly inhibited tumor growth in the xenograft model, confirming the role of SHMT2 in tumor growth and survival. Immunohistochemistry on the xenograft tumors showed that SHMT2 knockdown inhibited p-ERK, p-p38 proteins involved in the MAPK signaling pathway, proliferation index Ki67, and microvessel density marker CD31. It’s been verified that SHMT2 regulated the MAPK, and VEGF signaling pathways.
SHMT2 has been reported to affect the ROS/JNK/P53 [35] and Akt/mTOR [34] pathways through ROS or glycine production. Our study proved that SHMT2 affects intracellular ROS levels (Supplementary Figure 1), and ROS has been reported to affect vascular proliferation [36] and the MAPK signaling pathways [37,38]. Changes in intracellular ROS levels may be the reason why SHMT2 affects the VEGF and MAPK signaling pathways.
In conclusion, we demonstrated that SHMT2 plays a critical role in human breast cancer growth by simultaneously regulating multiple signaling pathways, such as the ERK/p38 MAPK, VEGF/PEDF, and mitochondrial apoptosis pathway. Our study also demonstrated that high SHMT2 expression independently predicted worse overall survival in patients with breast cancer. Our finding suggests that SHMT2 could therefore serve as a potential prognostic biomarker and therapeutic target for breast cancer therapy.
Acknowledgements
We apologize to the investigators whose studies we could not include due to reference number constraints. We thank Dr. Xinhua Xie from the Department of Breast Surgery, Sun-Yat sen University Cancer Center, Guangzhou, China for kindly Providing Breast cancer clinical samples and cell lines. This work was supported by the funds from the National Natural Science Foundation of China (81773103 (LHX), 82102838 (XW), 82103437 (WY)), Natural Science Foundation of Guangdong Province (2017A030313617 (LHX)), The Youth Innovative Talents Project of Guangdong Province (2018KQNCX049 (XSHY)), and the Youth scientific research project of Innovation Strong College of Guangzhou University of Chinese Medicine (2019QN09 (XSHY)), Chinese Postdoctoral Science Foundation (2021M693649 (WY)).
IHC analysis of specimen tissues in this study was conducted in compliance with the Declaration of Helsinki and approved by the research medical ethics committee of Sun Yat-sen University. Informed consent was obtained from all subjects. All animal maintenance and operational procedures were carried in accordance with the animal licence protocol approval by Animal Care and Ethics Committee of Sun-Yat Sen University Cancer Center.
Disclosure of conflict of interest
None.
Abbreviations
- SHMT2
Serine hydroxymethyltransferase 2
- VEGF
Vascular Endothelial Growth Factor
- PEDF
Pigment Epithelium Derived Factor
- MAPK
Mitogen-activated protein kinase
- mTOR
Mammalian target of rapamycin
- PLP
pyridoxal 5’-phosphate
- THF
tetrahydrofolate
- ELISA
Enzyme Linked Immunosorbent Assay
- MTT
3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide
- OS
Overall Survival
Supporting Information
References
- 1.Vagia E, Mahalingam D, Cristofanilli M. The landscape of targeted therapies in TNBC. Cancers (Basel) 2020;12:916. doi: 10.3390/cancers12040916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu G, Dustin D, Fuqua SA. Targeted therapy for breast cancer and molecular mechanisms of resistance to treatment. Curr Opin Pharmacol. 2016;31:97–103. doi: 10.1016/j.coph.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrow TA, Brenner AA, Whitehead VM, Chen XN, Duncan RG, Korenberg JR, Shane B. Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J Biol Chem. 1993;268:11910–11916. [PubMed] [Google Scholar]
- 8.Tramonti A, Paiardini A, Paone A, Bouzidi A, Giardina G, Guiducci G, Magnifico MC, Rinaldo S, McDermott L, Menendez JA, Contestabile R, Cutruzzola F. Differential inhibitory effect of a pyrazolopyran compound on human serine hydroxymethyltransferase-amino acid complexes. Arch Biochem Biophys. 2018;653:71–79. doi: 10.1016/j.abb.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Zeng Y, Zhang J, Xu M, Chen F, Zi R, Yue J, Zhang Y, Chen N, Chin YE. Roles of mitochondrial serine hydroxymethyltransferase 2 (SHMT2) in human carcinogenesis. J Cancer. 2021;12:5888–5894. doi: 10.7150/jca.60170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR, Shelton LM, Gui DY, Kwon M, Ramkissoon SH, Ligon KL, Kang SW, Snuderl M, Vander Heiden MG, Sabatini DM. SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature. 2015;520:363–367. doi: 10.1038/nature14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marrocco I, Altieri F, Rubini E, Paglia G, Chichiarelli S, Giamogante F, Macone A, Perugia G, Magliocca FM, Gurtner A, Maras B, Ragno R, Patsilinakos A, Manganaro R, Eufemi M. Shmt2: a stat3 signaling new player in prostate cancer energy metabolism. Cells. 2019;8:1048. doi: 10.3390/cells8091048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, Cantley LC. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet. 2015;47:1475–1481. doi: 10.1038/ng.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao E, Ding J, Xia Y, Liu M, Ye B, Choi JH, Yan C, Dong Z, Huang S, Zha Y, Yang L, Cui H, Ding HF. KDM4C and ATF4 cooperate in transcriptional control of amino acid metabolism. Cell Rep. 2016;14:506–519. doi: 10.1016/j.celrep.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song J, Kim D, Chun CH, Jin EJ. MiR-370 and miR-373 regulate the pathogenesis of osteoarthritis by modulating one-carbon metabolism via SHMT-2 and MECP-2, respectively. Aging Cell. 2015;14:826–837. doi: 10.1111/acel.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, Chen Z, Xue D, Zhang Q, Liu X, Luh F, Hong L, Zhang H, Pan F, Liu Y, Chu P, Zheng S, Lou G, Yen Y. Prognostic and therapeutic value of mitochondrial serine hydroxyl-methyltransferase 2 as a breast cancer biomarker. Oncol Rep. 2016;36:2489–2500. doi: 10.3892/or.2016.5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, Qing G, Liu Z, Simon MC, Rabinowitz JD, Thompson CB. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014;4:1406–1417. doi: 10.1158/2159-8290.CD-14-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tramonti A, Paiardini A, Paone A, Bouzidi A, Giardina G, Guiducci G, Magnifico MC, Rinaldo S, McDermott L, Menendez JA, Contestabile R, Cutruzzola F. Differential inhibitory effect of a pyrazolopyran compound on human serine hydroxymethyltransferase-amino acid complexes. Arch Biochem Biophys. 2018;653:71–79. doi: 10.1016/j.abb.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Xiong S, Mu T, Wang G, Jiang X. Mitochondria-mediated apoptosis in mammals. Protein Cell. 2014;5:737–749. doi: 10.1007/s13238-014-0089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santucci R, Sinibaldi F, Cozza P, Polticelli F, Fiorucci L. Cytochrome c: an extreme multifunctional protein with a key role in cell fate. Int J Biol Macromol. 2019;136:1237–1246. doi: 10.1016/j.ijbiomac.2019.06.180. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Z, Dong Y, Lu J, Yang F, Zheng Y, Yang H. Role of hypoxia in inhibiting dendritic cells by VEGF signaling in tumor microenvironments: mechanism and application. Am J Cancer Res. 2021;11:3777–3793. [PMC free article] [PubMed] [Google Scholar]
- 22.Becerra SP, Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–271. doi: 10.1038/nrc3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TH, Vemu PL, Hoy GE, Boudjadi S, Chatterjee B, Shern JF, Khan J, Sun W, Barr FG. Serine hydroxymethyltransferase 2 expression promotes tumorigenesis in rhabdomyosarcoma with 12q13-q14 amplification. J Clin Invest. 2021;131:e138022. doi: 10.1172/JCI138022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo L, Zheng Y, Lin Z, Li X, Li X, Li M, Cui L, Luo H. Identification of SHMT2 as a potential prognostic biomarker and correlating with immune infiltrates in lung adenocarcinoma. J Immunol Res. 2021;2021:6647122. doi: 10.1155/2021/6647122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie M, Pei DS. Serine hydroxymethyltransferase 2: a novel target for human cancer therapy. Invest New Drugs. 2021;39:1671–1681. doi: 10.1007/s10637-021-01144-z. [DOI] [PubMed] [Google Scholar]
- 26.Ji L, Tang Y, Pang X, Zhang Y. Increased expression of Serine Hydroxymethyltransferase 2 (SHMT2) is a negative prognostic marker in patients with hepatocellular carcinoma and is associated with proliferation of HepG2 cells. Med Sci Monit. 2019;25:5823–5832. doi: 10.12659/MSM.915754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saatci O, Kaymak A, Raza U, Ersan PG, Akbulut O, Banister CE, Sikirzhytski V, Tokat UM, Aykut G, Ansari SA, Dogan HT, Dogan M, Jandaghi P, Isik A, Gundogdu F, Kosemehmetoglu K, Dizdar O, Aksoy S, Akyol A, Uner A, Buckhaults PJ, Riazalhosseini Y, Sahin O. Targeting lysyl oxidase (LOX) overcomes chemotherapy resistance in triple negative breast cancer. Nat Commun. 2020;11:2416. doi: 10.1038/s41467-020-16199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kong D, Zhou H, Neelakantan D, Hughes CJ, Hsu JY, Srinivasan RR, Lewis MT, Ford HL. VEGF-C mediates tumor growth and metastasis through promoting EMT-epithelial breast cancer cell crosstalk. Oncogene. 2021;40:964–979. doi: 10.1038/s41388-020-01539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song Y, Zeng S, Zheng G, Chen D, Li P, Yang M, Luo K, Yin J, Gu Y, Zhang Z, Jia X, Qiu N, He Z, Li H, Liu H. FOXO3a-driven miRNA signatures suppresses VEGF-A/NRP1 signaling and breast cancer metastasis. Oncogene. 2021;40:777–790. doi: 10.1038/s41388-020-01562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang YQ, Chen WL, Zhang F, Wei XL, Zeng D, Liang YK, Wu JD, Zhang LY, Guo CP, Zeng HC, Hao SS, Li RH, Huang WH, Zhang GJ. Over-expression of both VEGF-C and Twist predicts poor prognosis in human breast cancer. Clin Transl Oncol. 2019;21:1250–1259. doi: 10.1007/s12094-019-02051-9. [DOI] [PubMed] [Google Scholar]
- 31.Becerra SP, Notario V. The effects of PEDF on cancer biology: mechanisms of action and therapeutic potential. Nat Rev Cancer. 2013;13:258–271. doi: 10.1038/nrc3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi D, Guo W, Chen W, Fu L, Wang J, Tian Y, Xiao X, Kang T, Huang W, Deng W. Nicotine promotes proliferation of human nasopharyngeal carcinoma cells by regulating alpha7AChR, ERK, HIF-1alpha and VEGF/PEDF signaling. PLoS One. 2012;7:e43898. doi: 10.1371/journal.pone.0043898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivera-Perez J, Monter-Vera M, Barrientos-Alvarado C, Toscano-Garibay JD, Cuesta-Mejias T, Flores-Estrada J. Evaluation of VEGF and PEDF in prostate cancer: a preliminary study in serum and biopsies. Oncol Lett. 2018;15:1072–1078. doi: 10.3892/ol.2017.7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang M, Yuan F, Bai H, Zhang J, Wu H, Zheng K, Zhang W, Miao M, Gong J. SHMT2 promotes liver regeneration through glycine-activated Akt/mTOR pathway. Transplantation. 2019;103:e188–e197. doi: 10.1097/TP.0000000000002747. [DOI] [PubMed] [Google Scholar]
- 35.Wu H, Bai H, Duan S, Yuan F. Downregulating serine hydroxymethyltransferase 2 deteriorates hepatic ischemia-reperfusion injury through ROS/JNK/P53 signaling in mice. Biomed Res Int. 2019;2019:2712185. doi: 10.1155/2019/2712185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukai T, Ushio-Fukai M. Cross-talk between NADPH oxidase and mitochondria: role in ROS signaling and angiogenesis. Cells. 2020;9:1849. doi: 10.3390/cells9081849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong L, Lei Y, Liu Y, Tan F, Li S, Wang X, Xu M, Cai W, Du B, Xu F, Zhou Y, Han H, Sun H, Qiu L. Vaccarin prevents ox-LDL-induced HUVEC EndMT, inflammation and apoptosis by suppressing ROS/p38 MAPK signaling. Am J Transl Res. 2019;11:2140–2154. [PMC free article] [PubMed] [Google Scholar]
- 38.Yang G, Chang CC, Yang Y, Yuan L, Xu L, Ho CT, Li S. Resveratrol alleviates rheumatoid arthritis via reducing ROS and inflammation, inhibiting MAPK signaling pathways, and suppressing angiogenesis. J Agric Food Chem. 2018;66:12953–12960. doi: 10.1021/acs.jafc.8b05047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.