Abstract
Background
Adult survivors of childhood cancer are at increased risk of cardiac late effects.
Methods
Using whole-genome sequencing data from 1870 survivors of European ancestry in the St. Jude Lifetime Cohort (SJLIFE) study, genetic variants were examined for association with ejection fraction (EF) and clinically assessed cancer therapy–induced cardiac dysfunction (CCD). Statistically significant findings were validated in 301 SJLIFE survivors of African ancestry and 4020 survivors of European ancestry from the Childhood Cancer Survivor Study. All statistical tests were 2-sided.
Results
A variant near KCNK17 showed genome-wide significant association with EF (rs2815063-A: EF reduction = 1.6%; P = 2.1 × 10-8) in SJLIFE survivors of European ancestry, which replicated in SJLIFE survivors of African ancestry (EF reduction = 1.5%; P = .004). The rs2815063-A also showed a 1.80-fold (P = .008) risk of severe or disabling or life-threatening CCD and replicated in 4020 Childhood Cancer Survivor Study survivors of European ancestry (odds ratio = 1.40; P = .04). Notably, rs2815063-A was specifically associated among survivors exposed to doxorubicin only, with a stronger effect on EF (3.3% EF reduction) and CCD (2.97-fold). Whole blood DNA methylation data in 1651 SJLIFE survivors of European ancestry showed statistically significant correlation of rs2815063-A with dysregulation of KCNK17 enhancers (false discovery rate <5%), which replicated in 263 survivors of African ancestry. Consistently, the rs2815063-A was associated with KCNK17 downregulation based on RNA sequencing of 75 survivors.
Conclusions
Leveraging the 2 largest cohorts of childhood cancer survivors in North America and survivor-specific polygenomic functional data, we identified a novel risk locus for CCD, which showed specificity with doxorubicin-induced cardiac dysfunction and highlighted dysregulation of KCNK17 as the likely molecular mechanism underlying this genetic association.
Cancer treatment–induced cardiac dysfunction (CCD) is a leading cause of morbidity and mortality in adult survivors of childhood cancer (1). Survivors are 6 to 15 times more likely to develop congestive heart failure (CHF) compared with their siblings (2-5). Exposures to anthracycline chemotherapy and chest radiation are well-established dose-dependent risk factors for CCD and heart failure. Among survivors clinically assessed in the St. Jude Lifetime Cohort (SJLIFE) (4), those exposed to a cumulative anthracycline dose [expressed in doxorubicin equivalents (6)] of at least 250 mg/m2 (odds ratio [OR] = 2.7, 95% confidence interval [CI] = 1.1 to 6.9) or cardiac radiation of at least 15 Gy (OR = 1.9, 95% CI = 1.1 to 3.7) were more likely to have evidence of cardiomyopathy (ejection fraction [EF] < 50%) than unexposed survivors. Other chemotherapeutic agents that have been reported to be associated with cardiotoxicity include cyclophosphamide, ifosfamide, cytarabine, and cisplatin (7-9). Newer agents, such as tyrosine kinase inhibitors, have also been found to be cardiotoxic leading to development of left ventricular dysfunction and heart failure (10,11). Although there is a clear relationship between treatment exposures and CCD (4,5), interindividual variability in CCD risk suggests a potential role of genetic susceptibility.
Candidate gene and genome-wide association (GWA) studies for CCD among survivors have identified several single nucleotide polymorphisms (SNPs) although only a small number have been replicated in independent cohorts (12-18). Thus, the role of genetics in the pathophysiology of CCD in childhood cancer survivors requires additional investigation. To this end, we analyzed whole-genome sequencing (WGS) data with respect to EF as an intermediate phenotype of clinically assessed CCD in 1870 childhood cancer survivors of European ancestry from the SJLIFE. Statistically significant genetic associations were replicated in 2 independent cohorts of childhood cancer survivors, including 301 survivors of African ancestry from SJLIFE using EF and clinically assessed CCD and 4020 survivors of European ancestry from the Childhood Cancer Survivor Study (CCSS) for self-report of a diagnosis of CHF.
Methods
Study Population
The SJLIFE (19) is a retrospective cohort study with prospective clinical follow-up and ongoing enrollment of 5-year survivors of childhood cancer treated at St. Jude Children’s Research Hospital. The CCSS is a retrospectively constructed cohort study with prospective longitudinal follow-up surveys of 5-year survivors of childhood cancer treated at 31 institutions in the United States and Canada. The study methodologies and characteristics of SJLIFE (20,21) and CCSS (22-24) have been previously described and provided in Supplementary Methods (available online). The institutional review boards at St. Jude Children’s Research Hospital and each of the centers participating in the CCSS approved the study, and participants provided informed consent.
Ejection Fraction and CCD or CHF
Two-dimensional Doppler echocardiography was performed in SJLIFE using a VIVID-7 machine (General Electric Medical Systems, Milwaukee, WI, USA) with 3-dimensional imaging of left ventricular volumes to measure EF as a quantitative trait, according to the American Society of Echocardiography guidelines (25). Echocardiogram was performed on survivors during their SJLIFE campus visits. When multiple EF measurements were available, we used the lowest EF of each survivor to reflect the most severe form of CCD known for the survivor. In SJLIFE, the presence of CCD was clinically assessed based on EF and pharmacologic treatment. Using modifications of the National Cancer Institute’s Common Terminology Criteria for Adverse Events (CTCAE) version 4.03 (3,20), CCD was classified as moderate (grade 2: resting EF < 50%-40% or 10%-19% absolute drop from baseline), severe or disabling (grade 3: resting EF < 39%-20%, or >20% absolute drop from baseline, or medication initiated), life-threatening (grade 4: resting EF < 20%; refractory or poorly controlled heart failure; intervention such as ventricular assist device, intravenous vasopressor support; or heart transplant indicated), or fatal (grade 5: death). In CCSS, participants completed a multi-item questionnaire at baseline and follow-up surveys that included age at onset of CHF based on self-report of medical diagnosis (Have you ever been told by a doctor or other health-care provider that you have or have had heart failure?) and pharmacologic treatment. Using the CTCAE (3,20), CHF was graded as moderate (grade 2: self-reported CHF not requiring medication), severe or disabling (grade 3: cardiomyopathy of CHF requiring medication), life-threatening (grade 4: cardiac transplantation), or fatal (grade 5: death as a result of heart failure). Survivors in SJLIFE and CCSS are assessed and graded repeatedly for history of CCD and CHF based on clinical ascertainment at SJLIFE visits and longitudinal surveys, respectively, and for this study, we used the highest CTCAE grade of each survivor.
WGS and Phenotype Data in SJLIFE
Details of DNA sample extraction, WGS, quality control, mapping, variant identification, and annotation are described previously (26,27) and in Supplementary Methods (available online). Of the 2986 SJLIFE survivors with WGS data that passed quality control, 654 survivors were excluded because echocardiography was not performed (n = 490) and of missing cumulative anthracycline dose (n = 8) and average heart radiation dose (n = 156), leaving 2332 survivors evaluable for further analysis. Anthracycline doses were converted to doxorubicin equivalents as described earlier (6). Mean radiation dose to the heart in centigray was estimated using previously established methods by the radiation physicists at MD Anderson Cancer Center (Houston, TX, USA) (28). Radiation treatment details included energy source, tumor dose, and field locations. Scatter dose to the heart was estimated for each case, regardless of radiation site and target volume. Ancestry was classified based on genotype data as European and African in 1870 and 301 survivors, respectively (Supplementary Methods, available online).
Statistical Analyses
Considering the relatively small number of survivors with CCD, our discovery GWA analysis used EF as a continuous variable, a quantitative trait, and intermediate phenotype used in characterizing and monitoring CCD. Association analyses were conducted assuming an additive genetic model as described previously (26,27,29) and included sex, age at cancer diagnosis, age at last follow-up, cumulative anthracycline dose, average heart radiation dose, and the top 20 genotype-based principal components as covariates. Associations with a P value less than 5 x 10-8 were considered statistically significant at the genome-wide level. Additional details are provided in Supplementary Methods (available online). Next, we evaluated genome-wide significant association(s) identified in the EF analysis with clinically assessed CCD in SJLIFE survivors of European ancestry. We used the partial proportional-odds extension of cumulative logit models (30) estimating the odds of developing a CTCAE grade 2 or higher and grade 3 or higher CCD simultaneously (Supplementary Methods, available online). To further assess the associations of variants that met the statistical significance thresholds in the discovery analysis, we used 2 replication cohorts including 301 survivors of African ancestry from the SJLIFE (for EF and CCD) and an additional 4020 of European ancestry from the CCSS (for self-reported CHF). All statistical tests were 2-sided, and results with a P value less than .05 were considered statistically significant unless otherwise stated.
Clinical and Functional Characterization
We characterized the replicated risk locus by 1) performing conditional analysis; 2) assessing its potential pleotropic effects on other cardiovascular conditions; 3) examining its effect with respect to clinical and treatment factors; and 4) evaluating potential target gene(s) underlying the risk locus by utilizing the GTEx portal (31) (accessed on October 22, 2020), the eQTLGen Consortium (32), and survivor-specific DNA methylation and RNA-sequencing data (Supplementary Methods, available online).
Previously Reported SNPs Associated With CCD in Childhood Cancer Survivors
We searched the literature for SNPs previously reported to be associated with CCD in pediatric cancer survivors that were replicated in at least 1 independent sample. We found 8 SNPs for anthracycline-related cardiotoxicity (12-18,33,34). Details of studies reporting these SNPs and their analyses are provided in Supplementary Table 1 and Supplementary Methods (available online).
Results
Demographic and treatment characteristics of survivors in the discovery and replication cohorts are provided in Table 1 and Supplementary Table 2 (available online), respectively. In the discovery cohort, 227 (12.1%) developed a CTCAE grade 2 or higher CCD, and the median EF among survivors with and without CCD was 46% (range = 18%-63%) and 57% (range = 50%-77%), respectively. In SJLIFE and CCSS replication cohorts, 43 (14.3%) and 230 (5.7%) developed a CTCAE grade 2 or higher CCD and CHF, respectively. The lower prevalence of CHF in CCSS than the prevalence of CCD in SJLIFE is likely because of misclassification resulting from self-report, especially the grade 2 CHF, which is defined as the self-reported congestive heart failure not requiring medication.
Table 1.
Demographic and treatment characteristics of survivors of European ancestry in the discovery cohort of St. Jude Lifetime Cohort (SJLIFE)
| Variables | With CCD (n = 227) | Without CCD (n = 1643) | P a |
|---|---|---|---|
| Age at cancer diagnosis | <.001 | ||
| Median (range), y | 10.5 (0.0-20.5) | 6.9 (0.0-23.6) | |
| Age at last follow-up | <.001 | ||
| Median (range), y | 41.0 (20.2-64.4) | 36.2 (9.2-70.2) | |
| Ejection fraction | <.001 | ||
| Median (range), % | 46 (18-63) | 57 (50-77) | |
| Time from cancer diagnosis to echocardiogram showing the lowest EF | <.001 | ||
| Median (range), y | 28.9 (11.4-53.8) | 24.7 (7.7-49.8) | |
| Age at CCD | |||
| Median (range), y | 37.5 (20.2-61.6) | NA | NA |
| Sex, No. (%) | 0.04 | ||
| Male | 135 (59.5) | 851 (51.8) | |
| Female | 92 (40.5) | 792 (48.2) | |
| Anthracycline dose, No. (%) | <.001 | ||
| 0 mg/m2 | 64 (28.2) | 536 (32.6) | |
| 1-250 mg/m2 | 90 (39.6) | 875 (53.3) | |
| >250 mg/m2 | 73 (32.2) | 232 (14.1) | |
| Average heart radiation dose, No. (%) | <.001 | ||
| 0 Gy | 84 (37) | 758 (46.1) | |
| 1-25 Gy | 33 (14.5) | 320 (19.5) | |
| >25 Gy | 110 (48.5) | 565 (34.4) |
P values were obtained from 2-sided Mann-Whitney (continuous variables) and χ2 tests (categorical variables). CCD = cancer treatment–induced cardiac dysfunction; EF = ejection fraction; NA = not applicable.
Common variant GWA analysis in the discovery cohort of survivors of European ancestry identified a locus on 6p21.2 with genome-wide significance (rs2815063-A; EF reduction = 1.6%; P = 2.1 × 10-8) (Supplementary Figure 1 and Table 2, available online). The genomic inflation factor of 0.975 indicates no influence of population stratification on the association results. Variants showing suggestive associations (P < 1.0 × 10-5) for EF are provided in Supplementary Table 3 (available online). No statistically significant rare or low-frequency variants were identified; top 10 results are provided in Supplementary Table 4 (available online). The association between rs2815063-A and EF persisted, albeit with slight attenuation, after further adjustment for use of cardiac medications (Supplementary Methods, available online) by those with a CTCAE grade 3 or higher CCD (EF reduction = 1.1%; P = 7.5 × 10-6).
Table 2.
Genome-wide significant associations for ejection fraction among survivors from the St. Jude Lifetime Cohort (SJLIFE) study in the discovery and independent replication cohortsa
| SNPsb | Positionc | EA | NEA | SJLIFE discovery cohort (European descent) |
SJLIFE replication cohort (African descent) |
Genes (+/- 10 kb) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | BETA | SE | P d | EAF | BETA | SE | P d | |||||
| rs2815063 | 39294759 | A | C | 0.13 | −0.016 | 0.003 | 2.1 × 10-8 | 0.49 | −0.015 | 0.005 | .004 | KCNK17 |
| rs115537302 | 39274943 | A | C | 0.07 | −0.019 | 0.004 | 8.7 × 10-8 | 0.01 | −0.009 | 0.026 | .73 | NA |
| rs115057884 | 39289210 | C | T | 0.07 | −0.019 | 0.004 | 9.0 × 10-8 | 0.02 | −0.011 | 0.020 | .60 | KCNK17 |
| rs116181858 | 39261645 | G | C | 0.07 | −0.019 | 0.004 | 9.6 × 10-8 | 0.01 | −0.009 | 0.028 | .76 | NA |
| rs114566901 | 39261967 | C | A | 0.07 | −0.019 | 0.004 | 1.7 × 10-7 | 0.01 | −0.009 | 0.028 | .76 | NA |
Covariates included sex, age at cancer diagnosis, age at last follow-up, cumulative anthracycline dose, average heart radiation dose, and the top 20 genotype-based principal components. BETA = per allele change in ejection fraction; EA = effect allele; EAF = effect allele frequency; NA = not applicable; NEA = non-effect allele.
All SNPs shown are from the 6p21.2 locus.
Genomic positions are shown relative to GRCh38 (hg38).
P values were 2-sided based on linear regression.
The association between rs2815063-A and EF was replicated in SJLIFE survivors of African ancestry, with 1.5% reduction (P = .004) in EF (Table 2).
Association of the 6p21.2 Locus With CCD or CHF
In the discovery cohort, rs2815063-A showed statistically significant associations for increased cumulative odds of both a CTCAE grade 2 or higher (OR = 1.57; P = .002) and grade 3 or higher (OR = 1.80; P = .008) CCD (Table 3). Results from sensitivity analyses based on 2 separate logistic regression models assessing the association of rs2815063-A with a CTCAE grade 2 or higher and grade 3 or higher CCD/CHF in SJLIFE discovery and CCSS replication cohorts were also similar to those obtained from the cumulative logistic models (Supplementary Table 5, available online). The associations were not statistically significant among SJLIFE survivors of African ancestry (P > .60). Notably, among the 4020 CCSS survivors of European ancestry, the rs2815063-A showed statistically significant association with the cumulative odds of a self-reported CTCAE grade 3 or higher CHF (OR = 1.40; P = .04; Table 3). The results for a self-reported CTCAE grade 2 or higher CHF were also consistent, albeit not statistically significant (OR = 1.21; P = .18).
Table 3.
Association between rs2815063-A and CCD/CHF among survivors from the St. Jude Lifetime Cohort (SJLIFE) study and the Childhood Cancer Survivor Study (CCSS)a
| Cohorts | Analyses | Survivor cases |
Survivor controls |
Cumulative OR (95% CI) | P b | ||
|---|---|---|---|---|---|---|---|
| No. | EAF | No. | EAF | ||||
| SJLIFE discovery cohort European ancestry | CTCAE grade ≥ 2 | 227 | 0.18 | 1643 | 0.12 | 1.57 (1.19 to 2.09) | .002 |
| CTCAE grade ≥ 3 | 67 | 0.20 | 1643 | 0.12 | 1.80 (1.17 to 2.77) | .008 | |
| SJLIFE replication cohort African ancestry | CTCAE grade ≥ 2 | 43 | 0.51 | 258 | 0.49 | 1.16 (0.67 to 2.03) | .60 |
| CTCAE grade ≥ 3 | 20 | 0.50 | 258 | 0.49 | 1.10 (0.53 to 2.26) | .80 | |
| CCSS replication cohort | CTCAE grade ≥ 2 | 230 | 0.15 | 3790 | 0.13 | 1.21 (0.91 to 1.60) | .18 |
| European ancestry | CTCAE grade ≥ 3 | 163 | 0.15 | 3790 | 0.13 | 1.40 (1.02 to 1.92) | .04 |
Covariates included sex, age at cancer diagnosis, age at last follow-up, cumulative anthracycline dose, average heart radiation dose, and the top 20 genotype-based principal components. CI = confidence interval; CTCAE = National Cancer Institute’s Common Terminology Criteria for Adverse Events; EAF = effect allele (A) frequency; OR = odds ratio.
P values were 2-sided based on cumulative logistic regression.
Characterization of the 6p21.2 Locus
Conditional analysis did not show evidence of any secondary association signal or role of rare or low- frequency variants at the 6p21.2 locus, suggesting rs2815063-A as the variant best representing the observed association signal (Supplementary Results, available online). We did not observe pleiotropic effect of rs2815063-A with any of the 8 other cardiovascular conditions in survivors (Supplementary Table 6, available online). Slightly stronger associations of rs2815063-A with EF reduction were observed in survivors classified into high (EF reduction = 1.72%; P = 9.6 × 10-4) or moderate (EF reduction = 1.87%; P = 1.4 × 10-4) risk groups compared with those classified into low risk (EF reduction = 1.36%; P = .002) groups based on the International Guideline Harmonization Group guidelines (Supplementary Table 7, available online). However, no statistically significant association between rs2815063-A and EF (P = .21) was observed in survivors not exposed to either anthracyclines or chest radiation. Similarly, statistically significant and stronger associations between rs2815063-A and a CTCAE grade 2 or higher CCD were observed in survivors classified as high- and moderate-risk groups (OR = 1.57; P = 0.04; OR = 1.73; P = .02, respectively) but not in the low-risk group (OR = 1.30; P = .37). A borderline association was observed in the unexposed survivors (OR = 2.32; P = .06), although these estimates are likely unstable because of the small number of survivors with CCD (n = 16) in the analysis.
We observed similar, but slightly stronger, associations for rs2815063-A among male (EF reduction = 1.72%; P = 1.2 × 10-5; OR for CCD = 1.79; P = .008) than female (EF reduction = 1.47%; P = 2.8 × 10-4; OR for CCD = 1.46; P = .12) survivors. Among survivors exposed to anthracyclines but no heart radiotherapy, rs2815063-A showed supportive association (EF reduction = 1.35%; P = .005; OR for CCD = 1.59; P = .12) but not among those exposed to heart radiotherapy with no anthracyclines (EF reduction = 1.21%; P = .05; OR for CCD = 0.99; P = .98) (Table 4). Among survivors exposed to doxorubicin only, rs2815063-A showed greater associations (EF reduction = 3.33%; P = 5.1 × 10-5; OR for CCD = 2.97; P = .006). In contrast, among survivors exposed to daunorubicin only, the associations were not observed (EF reduction = 0.23%; P = .68; OR for CCD = 1.08; P = .93). Notably, among 338 CCSS survivors (37 with CTCAE grade ≥2 CHF) exposed to doxorubicin only, the rs2815063-A also showed stronger association (OR = 2.00; P = .09) compared with the result based on all survivors (OR = 1.21; P = .18) (Table 3).
Table 4.
Treatment-specific effects of the rs2815063-A with CCD in the discovery cohort including survivors of European descent from the St. Jude Lifetime Cohort
| Cardiotoxic exposures | Survivor cases, No. | Survivor controls, No. | Associations with EF |
Associations with CCD |
||
|---|---|---|---|---|---|---|
| BETA (SE) | P a | OR (95% CI) | P b | |||
| Anthracyclines only | 68 | 549 | −0.014 (0.005) | .005 | 1.59 (0.89 to 2.85) | .12 |
| Heart radiation only | 48 | 327 | −0.012 (0.006) | .05 | 0.99 (0.49 to 2.01) | .98 |
| Doxorubicin only | 53 | 250 | −0.033 (0.008) | <.001 | 2.97 (1.37 to 6.43) | .006 |
| Daunorubicin only | 13 | 249 | −0.002 (0.006) | .69 | 1.08 (0.23 to 5.10) | .93 |
P values were 2-sided based on linear regression. EF = ejection fraction; CCD = cancer treatment–induced cardiac dysfunction; BETA = effect size; SE = standard error; OR = odds ratio; CI = confidence interval.
P values were 2-sided based on logistic regression.
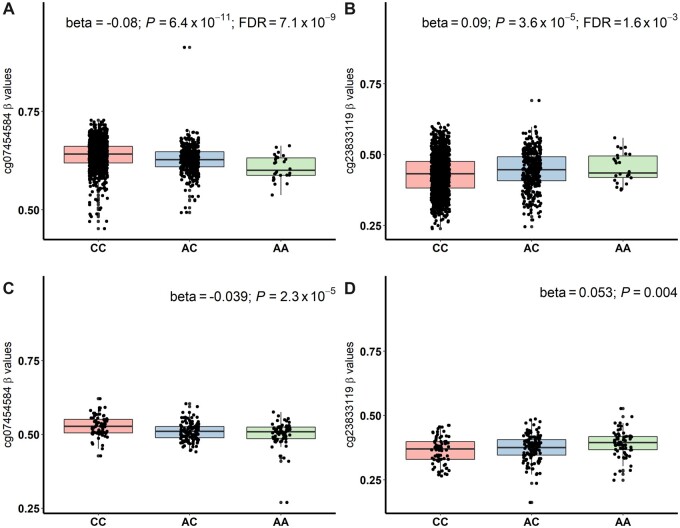
The rs2815063-A was found to regulate expression of KCNK17 and KCNK5 in multiple tissues based on the GTEx data (Supplementary Figure 2, available online). However, after accounting for multiple comparisons, differential expression of the KCNK5 with rs2815063-A was statistically significant only in the tibial nerve. In whole blood samples of 31 569 individuals in the eQTLGen Consortium (32), rs2815063-A also decreased expression of KCNK17 (z score = -16.0; P = 1.1 × 10-57). In the 1651 SJLIFE survivors of European ancestry, we found rs2815063-A was statistically significantly correlated (false discovery rate < 0.05) with differential methylation of 8 DNA methylation probes. Of these, 6 probes (cg26253826, cg20869405, cg13075951, cg23833119, cg24537809, and cg00925516) showed hypermethylation, and 2 (cg07454584 and cg04321126) were hypomethylated (Figure 1; Supplementary Table 8 and Supplementary Figure 3, available online). In 263 SJLIFE survivors of African ancestry, cg23833119 (P = .002) and cg07454584 (P = 3.1 × 10-5) also showed statistically significant correlation with rs2815063-A even after accounting for multiple testing of 8 DNA methylation probes. Both cg23833119 and cg07454584 overlap enhancers that cross talk with KCNK17 promoter and are located on intron 1 of KCNK17 and approximately 34 kb downstream of KCNK17, respectively. Consistently, we also observed statistically significant association between the rs2815063-A and downregulation of KCNK17 in survivors of European ancestry (normalized effect size = -0.50; P = .04) (Supplementary Figure 4, available online).
Figure 1.
Whole blood DNA methylation levels of cg07454584 (approximately 34 kb downstream of KCNK17) and cg23833119 (located on intron 1 of KCNK17) with respect to 3 genotypes of rs2815063 at the 6p21.2 locus. Boxplots in (A) and (B) show beta values (ratio of methylated to unmethylated probe intensities) of cg07454584 and cg23833119 among 1651 childhood cancer survivors of European ancestry, and those in (C) and (D) show results in 263 survivors of African ancestry. Results from linear regression models examining associations of cg07454584 and cg23833119 with rs2815063 (additively coded), adjusted for age at sample collection and sex are shown on top of each boxplot. P values were 2-sided based on linear regression. FDR = false discovery rate.
Associations of Previously Reported CCD SNPs
Among survivors of European ancestry in the discovery cohort, associations of 3 of the 10 SNPs were nominally statistically significant (P < .05) (Supplementary Table 9, available online). Among survivors exposed to anthracyclines and/or chest radiation, rs4149178-G on SLC22A7 was statistically significantly associated with increased EF (EF increase = 0.008; P = .008). Among survivors exposed to anthracyclines only, we observed a statistically significant interaction effect on EF between AA genotype of rs2232228 on HAS3 and exposure to more than 250 mg/m2 anthracyclines (EF reduction = 0.032; P = .04). Moreover, we also observed a statistically significant association between rs2229774-A on RARG and CCD (OR = 2.08; P = .04) along with a suggestive association for rs7853758-A on SLC28A3 (OR = 0.56; P = .09).
Discussion
By utilizing the 2 well-established cohorts of childhood cancer survivors in North America, we describe the largest gene discovery effort for CCD risk, leveraging the WGS and survivor-specific polygenomic functional data. We identified and replicated a novel locus at 6p21.2 in CCD among survivors of European ancestry and highlighted dysregulation of KCNK17 as the potential molecular mechanism underlying the 6p21.2 locus.
The prevalence of CCD was 59.5% among male survivors in SJLIFE discovery cohort, but the prevalence of CHF was only 41.7% in the CCSS, likely because of misclassification due to self-reported CHF in the CCSS affecting the CHF prevalence overall and by sex. The misclassification of CHF and/or its different timing of survey or assessment also likely resulted in attenuation of effect of rs2815063-A on CHF in the CCSS than that was based on clinically assessed CCD in SJLIFE. Compared with a CTCAE grade 2 or higher CCD, rs2815063-A showed greater effect on a CTCAE grade 3 or higher CCD, suggesting rs2815063-A may also be related to severity of CCD. The rs2815063-A was associated with EF among survivors exposed to cardiotoxic therapies but not in unexposed survivors; the associations with CCD/CHF were also consistent. Moreover, the rs2815063-A was associated with CCD among survivors exposed to anthracyclines alone, specifically among those exposed to doxorubicin alone with a greater effect (nearly threefold risk) than that observed in all survivors (1.6-fold risk). No associations were observed among survivors exposed to heart radiotherapy alone or daunorubicin alone. These data may suggest the specificity of rs2815063-A with doxorubicin-induced cardiac dysfunction, which was also supported by results in the CCSS. Considering significant association of high average heart radiation dose with prevalence of CCD in survivors of both European and African ancestries in SJLIFE and of CHF in CCSS, additional studies are required to further investigate the lack of association of rs2815063-A among survivors exposed to heart radiotherapy alone.
A 3-staged case-control genome-wide association study including 4100 dilated cardiomyopathy (DCM) cases (EF < 45% and absence of heart diseases; average age ranging from 47.3 to 56.6 years) and 7600 controls (free from cardiomyopathies, heart failure or heart diseases; average age ranging from 47.7 to 57.4 years) of European ancestry in the general population reported a genome-wide significant association for rs9262636 in HCG22 at chromosome 6p21.33 (35). The rs9262636 is located approximately 8.34 Mb away from the rs2815063 reported in this study and based on the 1000 Genomes European data; these 2 SNPs are not in linkage disequilibrium (r2 = 0.0001). Furthermore, rs9262636 was not associated with either EF (beta = 0.0006; P = .80) or CCD (OR = 0.89; P = .37) among survivors of European ancestry in this study. Another case-control study including 59 DCM patients and 39 DCM-free individuals with left ventricular biopsies and 150 DCM patients and 168 DCM-free individuals with peripheral blood reported a CpG site (cg18601596) near KCNK17 showing statistically significant association with dilated cardiomyopathy (36). The DCM patients had an EF between more than 45% and less than 55%, and DCM-free individuals were symptom-free patients after heart transplantation with normal systolic and diastolic function. Both DCM patients and DCM-free individuals were of European ancestry. However, cg18601596 was not statistically significantly associated with the rs2815063 genotypes (beta = 0.02; P = .19) among survivors of European ancestry in this study. These data, therefore, suggest the specific role of rs2815063-A in CCD among childhood cancer survivors.
DNA methylation results in survivors showed a correlation of rs2815063-A with dysregulation of KCNK17 enhancers. This finding was consistent with downregulation of KCNK17 expression with respect to the A allele among survivors and in the general population (32), suggesting KCNK17 as the potential target gene underlying the association of rs2815063-A with CCD in survivors. KCNK17 encodes a pH-sensitive cardiac 2-pore domain potassium channel (K2P) TASK-4. The protein TASK-4 has also been identified as a contributor to a progressive and severe cardiac conduction disorder combined with idiopathic ventricular fibrillation by whole exome sequencing (37). KCNK17 was strongly expressed in human Purkinje cells (about 12-fold) and the atrioventricular node (about sixfold) compared with the ventricles. Gain-of-function variants on KCNK17 have been associated with greater cardiac conduction compared with the other allele alone (37,38). Potassium channels represent the most important ion channels contributing to repolarization in cardiomyocytes, and abnormalities in repolarization have been reported to play roles in heart failure. Prolongation of action potential duration has been consistently shown to be associated with heart failure in humans and animals (39-42). Results from experimental models of heart failure suggested a substantial contribution of down regulation of potassium currents to the enhanced lability of the repolarization process in heart failure, predisposing to early after-depolarizations (42). Changes in action potential duration and QT interval can result in electromechanical dysfunction leading to channelopathies and associated heart failure (43). Moreover, it is increasingly recognized that mutations in ion channel genes can result in dilated cardiomyopathy with or without arrhythmias (44). However, additional research in survivor-specific human-induced pluripotent stem cell–derived cardiomyocytes and/or animal models are required to confirm KCNK17 as the target gene and understand the underlying molecular mechanisms in a tissue-specific manner.
Our study also provided support for 4 of the 8 previously reported CCD-associated SNPs in childhood cancer survivors, including those on RARG, HAS3, SLC22A7, and SLC28A3. Several factors may contribute to non-statistically significant results of previously reported CCD SNPs in our study. Most of the CCD SNPs were reported in studies that used candidate-gene approaches. Genetic association studies focusing on SNPs on candidate genes have not been very successful because of limited sample sizes and failure to account for population stratification and multiple hypotheses testing. Additional studies with larger sample sizes are needed to ensure that the previously reported SNPs that did not show associations with CCD in our study are not false positives.
Potential bias by only including survivors who are alive for DNA sample collection is a limitation of our study. However, the loss of survivors who died of cardiac causes before study participation would have biased our genetic associations toward the null. Compared with clinically assessed CCD in the SJLIFE discovery cohort, CHF in the CCSS replication cohort was defined based on self-report. This likely resulted in misclassification and attenuation of genetic effect in the CCSS. Accurate ascertainment of CHF in CCSS might have affected the strength of genetic association of rs2815063-A with CHF.
In conclusion, we used WGS data and conducted a GWA study of cardiac dysfunction in survivors of childhood cancer and identified a novel locus on 6p21.2. Notably, we identified specificity of this locus with doxorubicin-induced cardiac dysfunction with a nearly threefold increased risk of CCD. Survivor-specific polygenomic functional data suggested dysregulation of KCNK17 as potential molecular mechanism underlying the 6p21.2 locus. Future investigations are warranted to confirm our findings mechanistically in a tissue-specific manner.
Funding
The St. Jude Lifetime Cohort (SJLIFE; U01 CA195547: MMH and LLR, Principal Investigators) and the Childhood Cancer Survivor Study (CCSS; U24 CA55727; GTA Principal Investigator) are supported by the National Cancer Institute at the National Institutes of Health and the Cancer Center Support CORE grant (CA21765: C. Roberts, Principal Investigator). The CCSS original cohort genotyping was supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. This work is also supported by R01 CA261898 (YS and PWB, Principal Investigators), R01 CA216354 (YY and JZ, Principal Investigators) from the National Cancer Institute at the National Institutes of Health and the American Lebanese Syrian Associated Charities, Memphis, TN, USA.
Notes
Role of the funders: The funders of the study had no role in the design and conduct of the study; were not involved in collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Disclosures: The authors declare that there is no conflict of interest. KCO and LMM, who are JNCI Associate Editors and co-authors of this article, were not involved in the editorial review or decision to publish the manuscript.
Author contributions: Conceptualization, Supervision and Investigation (YS, GTA, YY); Resources and Project Administration (LLR, MMH, GTA, LMM, SB); Data Curation and Software (YS, MJE, KS); Formal Analysis and Visualization (YS); Funding Acquisition (YS, JZ, MMH, LLR, GTA, YY); Validation (NQ, QL, WQ, YShao, EP, HLM, JE, JRM); Methodology (YS, YY); Writing—original draft (YS); Writing—review & editing (all authors).
Data Availability
Aligned binary files for all the SJLIFE survivors and the joint genotype calls are accessible through the St. Jude Cloud (https://stjude.cloud). All data generated or analyzed during the study are included in the published article (and its supplementary files). Summary statistics from the GWAS discovery analysis are also available from the St. Jude Cloud (https://viz.stjude.cloud/community/cancer-survivorship-community~4/data).
Supplementary Material
Contributor Information
Yadav Sapkota, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Matthew J Ehrhardt, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA; Department of Oncology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Na Qin, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Zhaoming Wang, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA; Department of Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Qi Liu, School of Public Health, University of Alberta, Edmonton, AB, Canada.
Weiyu Qiu, School of Public Health, University of Alberta, Edmonton, AB, Canada.
Kyla Shelton, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Ying Shao, Department of Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Emily Plyler, Department of Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Heather L Mulder, Department of Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
John Easton, Department of Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
J Robert Michael, Department of Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Paul W Burridge, Department of Pharmacology, Northwestern University, Chicago, Il, USA.
Xuexia Wang, Department of Mathematics, University of North Texas, Denton, TX, USA.
Carmen L Wilson, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA.
John L Jefferies, Division of Cardiovascular Disease, The University of Tennessee Health Science Center, Memphis, TN, USA.
Eric J Chow, Clinical Research Division, Fred Hutchinson Cancer Research Center, WA, USA.
Kevin C Oeffinger, Department of Community and Family Medicine, Duke University, Durham, NC, USA.
Lindsay M Morton, Raditional Oncology Branch, National Cancer Institute, Bethesda, MD, USA.
Chunliang Li, Department of Tumor Cell Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Jun J Yang, Department of Pharmacy and Pharmaceutical Sciences, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Jinghui Zhang, Department of Computational Biology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Smita Bhatia, Institute of Cancer Outcomes and Survivorship, University of Alabama at Birmingham, Birmingham, AL, USA.
Daniel A Mulrooney, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA; Department of Oncology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Melissa M Hudson, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA; Department of Oncology, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Leslie L Robison, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Gregory T Armstrong, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA.
Yutaka Yasui, Department of Epidemiology and Cancer Control, St. Jude Children’s Research Hospital, Memphis, TN, USA.
References
- 1. Armstrong GT, Chen Y, Yasui Y, et al. Reduction in late mortality among 5-year survivors of childhood cancer. N Engl J Med. 2016;374(9):833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mulrooney DA, Yeazel MW, Kawashima T, et al. Cardiac outcomes in a cohort of adult survivors of childhood and adolescent cancer: retrospective analysis of the Childhood Cancer Survivor Study cohort. BMJ. 2009;339(1):b4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oeffinger KC, Mertens AC, Sklar CA, et al. ; for the Childhood Cancer Survivor Study. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006;355(15):1572-1582. [DOI] [PubMed] [Google Scholar]
- 4. Mulrooney DA, Armstrong GT, Huang S, et al. Cardiac outcomes in adult survivors of childhood cancer exposed to cardiotoxic therapy: a cross-sectional study. Ann Intern Med. 2016;164(2):93-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mulrooney DA, Hyun G, Ness KK, et al. Major cardiac events for adult survivors of childhood cancer diagnosed between 1970 and 1999: report from the Childhood Cancer Survivor Study cohort. BMJ. 2020;368:l6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feijen EA, Leisenring WM, Stratton KL, et al. Equivalence ratio for daunorubicin to doxorubicin in relation to late heart failure in survivors of childhood cancer. J Clin Oncol. 2015;33(32):3774-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pai VB, Nahata MC.. Cardiotoxicity of chemotherapeutic agents: incidence, treatment and prevention. Drug Saf. 2000;22(4):263-302. [DOI] [PubMed] [Google Scholar]
- 8. Goldberg MA, Antin JH, Guinan EC, et al. Cyclophosphamide cardiotoxicity - an analysis of dosing as a risk factor. Blood. 1986;68(5):1114-1118. [PubMed] [Google Scholar]
- 9. Quezado ZMN, Wilson WH, Cunnion RE, et al. High-dose ifosfamide is associated with severe, reversible cardiac dysfunction. Ann Intern Med. 1993;118(1):31-36. [DOI] [PubMed] [Google Scholar]
- 10. Cheng H, Force T.. Why do kinase inhibitors cause cardiotoxicity and what can be done about it? Prog Cardiovasc Dis. 2010;53(2):114-120. [DOI] [PubMed] [Google Scholar]
- 11. Kerkela R, Grazette L, Yacobi R, et al. Cardiotoxicity of the cancer therapeutic agent imatinib mesylate. Nat Med. 2006;12(8):908-916. [DOI] [PubMed] [Google Scholar]
- 12. Aminkeng F, Bhavsar AP, Visscher H, et al. ; for the Canadian Pharmacogenomics Network for Drug Safety Consortium. A coding variant in RARG confers susceptibility to anthracycline-induced cardiotoxicity in childhood cancer. Nat Genet. 2015;47(9):1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krajinovic M, Elbared J, Drouin S, et al. Polymorphisms of ABCC5 and NOS3 genes influence doxorubicin cardiotoxicity in survivors of childhood acute lymphoblastic leukemia (vol 16, pg 530, 2016). Pharmacogenomics J. 2017;17(1):107-107. [DOI] [PubMed] [Google Scholar]
- 14. Visscher H, Rassekh SR, Sandor GS, et al. ; for the CPNDS consortium. Genetic variants in SLC22A17 and SLC22A7 are associated with anthracycline-induced cardiotoxicity in children. Pharmacogenomics. 2015;16(10):1065-1076. [DOI] [PubMed] [Google Scholar]
- 15. Visscher H, Ross CJ, Rassekh SR, et al. ; for the CPNDS Consortium. Validation of variants in SLC28A3 and UGT1A6 as genetic markers predictive of anthracycline-induced cardiotoxicity in children. Pediatr Blood Cancer. 2013;60(8):1375-1381. [DOI] [PubMed] [Google Scholar]
- 16. Visscher H, Ross CJD, Rassekh SR, et al. ; for the Canadian Pharmacogenomics Network for Drug Safety Consortium. Pharmacogenomic prediction of anthracycline-induced cardiotoxicity in children. J Clin Oncol. 2012;30(13):1422-1428. [DOI] [PubMed] [Google Scholar]
- 17. Wang X, Liu W, Sun CL, et al. Hyaluronan synthase 3 variant and anthracycline-related cardiomyopathy: a report from the children’s oncology group. J Clin Oncol. 2014;32(7):647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang X, Sun CL, Quinones-Lombrana A, et al. CELF4 variant and anthracycline-related cardiomyopathy: a children’s oncology group genome-wide association study. J Clin Oncol. 2016;34(8):863-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hudson MM, Ness KK, Nolan VG, et al. Prospective medical assessment of adults surviving childhood cancer: study design, cohort characteristics, and feasibility of the St. Jude Lifetime Cohort study. Pediatr Blood Cancer. 2011;56(5):825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hudson MM, Ehrhardt MJ, Bhakta N, et al. Approach for classification and severity grading of long-term and late-onset health events among childhood cancer survivors in the St. Jude lifetime cohort. Cancer Epidemiol Biomarkers Prev. 2017;26(5):666-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howell CR, Bjornard KL, Ness KK, et al. Cohort profile: the St. Jude Lifetime Cohort Study (SJLIFE) for paediatric cancer survivors. Int J Epidemiol. 2021;50(1):39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robison LL, Armstrong GT, Boice JD, et al. The Childhood Cancer Survivor Study: a national cancer institute-supported resource for outcome and intervention research. J Clin Oncol. 2009;27(14):2308-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leisenring WM, Mertens AC, Armstrong GT, et al. Pediatric cancer survivorship research: experience of the Childhood Cancer Survivor Study. J Clin Oncol. 2009;27(14):2319-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robison LL, Mertens AC, Boice JD, et al. Study design and cohort characteristics of the childhood cancer survivor study: a multi-institutional collaborative project. Med Pediatr Oncol. 2002;38(4):229-239. [DOI] [PubMed] [Google Scholar]
- 25. Lang RM, Bierig M, Devereux RB, et al. Recommendations for chamber quantification: a report from the American Society of Echocardiography’s Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J Am Soc Echocardiogr. 2005;18(12):1440-1463. [DOI] [PubMed] [Google Scholar]
- 26. Sapkota Y, Cheung YT, Moon W, et al. Whole-genome sequencing of childhood cancer survivors treated with cranial radiation therapy identifies 5p15.33 locus for stroke: a report from the St. Jude Lifetime Cohort Study. Clin Cancer Res. 2019;25(22):6700-6708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sapkota Y, Wilson CL, Zaidi AK, et al. A novel locus predicts spermatogenic recovery among childhood cancer survivors exposed to alkylating agents. Cancer Res. 2020;80(17):3755-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stovall M, Weathers R, Kasper C, et al. Dose reconstruction for therapeutic and diagnostic radiation exposures: use in epidemiological studies. Radiat Res. 2006;166(1 pt 2):141-157. [DOI] [PubMed] [Google Scholar]
- 29. Sapkota Y, Qin N, Ehrhardt MJ, et al. Genetic variants associated with therapy-related cardiomyopathy among childhood cancer survivors of African ancestry. Cancer Res. 2021;81(9):2556-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peterson B, Harrell FE.. Partial proportional odds models for ordinal response variables. J Roy Stat Soc Ser C-Appl Stat. 1990;39(2):205-217. [Google Scholar]
- 31. Battle A, Brown CD, Engelhardt BE, et al. ; for the eQTL manuscript working group. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vosa U, Claringbould A, Westra HJ, et al. ; for the i2QTL Consortium. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet. 2021;53(9):1300-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schneider BP, Shen F, Gardner L, et al. Genome-wide association study for anthracycline-induced congestive heart failure. Clin Cancer Res. 2017;23(1):43-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wells QS, Veatch OJ, Fessel JP, et al. Genome-wide association and pathway analysis of left ventricular function after anthracycline exposure in adults. Pharmacogenet Genomics. 2017;27(7):247-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meder B, Ruhle F, Weis T, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J. 2014;35(16):1069-1077. [DOI] [PubMed] [Google Scholar]
- 36. Meder B, Haas J, Sedaghat-Hamedani F, et al. Epigenome-wide association study identifies cardiac gene patterning and a novel class of biomarkers for heart failure. Circulation. 2017;136(16):1528-1544. [DOI] [PubMed] [Google Scholar]
- 37. Friedrich C, Rinne S, Zumhagen S, et al. Gain-of-function mutation in TASK-4 channels and severe cardiac conduction disorder. Embo Mol Med. 2014;6(7):937-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chai S, Wan XP, Ramirez-Navarro A, et al. Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. J Clin Invest. 2018;128(3):1043-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Akar FG, Rosenbaum DS.. Transmural electrophysiological heterogeneities underlying arrhythmogenesis in heart failure. Circ Res. 2003;93(7):638-645. [DOI] [PubMed] [Google Scholar]
- 40. Aronson RS. Afterpotentials and triggered activity in hypertrophied myocardium from rats with renal-hypertension. Circ Res. 1981;48(5):720-727. [DOI] [PubMed] [Google Scholar]
- 41. Nuss HB, Kaab S, Kass DA, et al. Cellular basis of ventricular arrhythmias and abnormal automaticity in heart failure. Am J Physiol-Heart Circ Physiol. 1999;277(1):H80-H91. [DOI] [PubMed] [Google Scholar]
- 42. Tomaselli GF, Beuckelmann DJ, Calkins HG, et al. Sudden cardiac death in heart-failure - the role of abnormal repolarization. Circulation. 1994;90(5):2534-2539. [DOI] [PubMed] [Google Scholar]
- 43. Rahm AK, Lugenbiel P, Schweizer PA, et al. Role of ion channels in heart failure and channelopathies. Biophys Rev. 2018;10(4):1097-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Olson TM, Michels VV, Ballew JD, et al. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293(4):447-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Aligned binary files for all the SJLIFE survivors and the joint genotype calls are accessible through the St. Jude Cloud (https://stjude.cloud). All data generated or analyzed during the study are included in the published article (and its supplementary files). Summary statistics from the GWAS discovery analysis are also available from the St. Jude Cloud (https://viz.stjude.cloud/community/cancer-survivorship-community~4/data).