Abstract
Background
Carotenoid cleavage oxygenases (CCOs) include the carotenoid cleavage dioxygenase (CCD) and 9-cis-epoxycarotenoid (NCED), which can catalize carotenoid to form various apocarotenoids and their derivatives, has been found that play important role in the plant world. But little information of CCO gene family has been reported in litchi (Litchi chinensis Sonn.) till date.
Results
In this study, a total of 15 LcCCO genes in litchi were identified based on genome wide lever. Phylogeny analysis showed that LcCCO genes could be classified into six subfamilies (CCD1, CCD4, CCD7, CCD8, CCD-like, and NCED), which gene structure, domain and motifs exhibited similar distribution patterns in the same subfamilies. MiRNA target site prediction found that there were 32 miRNA target sites in 13 (86.7%) LcCCO genes. Cis-elements analysis showed that the largest groups of elements were light response related, following was plant hormones, stress and plant development related. Expression pattern analysis revealed that LcCCD4, LcNCED1, and LcNCED2 might be involving with peel coloration, LcCCDlike-b might be an important factor deciding fruit flavor, LcNCED2 and LcNCED3 might be related to flower control, LcNCED1 and LcNCED2 might function in fruitlet abscission, LcCCD4a1, LcCCD4a2, LcCCD1, LcCCD4, LcNCED1, and LcNCED2 might participate in postharvest storage of litchi.
Conclusion
Herein, Genome-wide analysis of the LcCCO genes was conducted in litchi to investigate their structure features and potential functions. These valuable and expectable information of LcCCO genes supplying in this study will offer further more possibility to promote quality improvement and breeding of litchi and further function investigation of this gene family in plant.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-03772-w.
Keywords: Litchi, CCO genes, Expression analysis, Flower control, Fruit development and maturation, Postharvest storage
Background
Carotenoids are isoprenoid-based compounds, also named as a kind of important natural pigments. Carotenoids can be found from archaea and eubacteria to eukaryotes (like animals, higher plants, fungi and algae), play vital roles in photosynthesis, signaling, antioxidant properties, electron transport, and light absorption [1–3]. Carotenoid cleavage oxygenase (CCO) is a type of important enzyme in the carotenoid metabolic pathway, which can catalyze carotenoid to various apocarotenoids and their derivatives to perform important biological functions in plants. The CCOs can be divided into two forms, one named as CCD (Carotenoid Cleavage Dioxygenase) and the other is NCED (9-cis-epoxycarotenoid dioxygenase) depending on whether the substrates are epoxidated [4].
The CCO gene family has been reported commonly involved in the formation of flavor and scent, coloration, even growth and development, ecological adaptation in plants through regulating the carotenoids pathway. In Arabidopsis thaliana, AtCCO genes family includes 4 CCD and 5 NECD genes [5]. At the beginning of the previous work, the CCO genes were divided into five categories, which included CCD1, CCD4, CCD7, CCD8, and NCED [6, 7]. Recently, another category of CCO genes called CCD-like (CCDL) was found in grape (Vitis vinifera), tomato (Solanum lycopersicum), apple (Malus × domestica) and Sugar cane (Saccharum) [8–11]. In general, different categories of CCO genes exhibit different roles. CCD1 can catalyze carotenoids into several metabolites like α-ionone, β-ionone, and geranylacetone, which play an important role in the formation of the flavor and scent of horticultural plants [12, 13]. CmCCD4a gene contributes to white color formation in chrysanthemum petalsonly (Chrysanthemum morifolium Ramat.) by catalyzing the carotenoids to colorless compound [14]. GmCCD4 in soybeans was also found to be a negative regulator of carotenoid content [15]. Natural Variation in CCD4 gene promoter is a major genetic determinant of natural variation in C30 apocarotenoids which is responsible for red coloration of citrus peel [16]. The CCD7 and CCD8 enzymes are involved in the biosynthesis of the strigolactone (a relatively novel apocarotenoid hormone), which could control shoot branching and reproductive development and regulate plant responses to drought and salt stress [7, 17–19]. NCED is the key enzyme for the biosynthesis of abscisic acid (ABA), which is closely involved in the fruit development, ripening and senescence. Such as FaNCED1 RNAi in strawberry (Fragaria × ananassa) fruits could decline the ABA content significantly and resulted in uncolored fruits [20]. The application of exogenous ABA could accelerate the accumulation of anthocyanin by increasing the expression of NCED genes to promote the coloration of strawberry [21], grape [22], sweet cherry [23] and litchi fruits [24]. Some reports showed that ABA might be correlate with the fruit abscission of the citrus, apple and litchi [44–46]. The ABA content increased by uniconazole spraying was helpful to the flower control and fruit retention of litchi [48, 49]. ABA was considered to play key role during the fruit senescence, which was the most important factor that deciding the shelf life of fruit [53, 54]. Additionally, ABA is also reported to be related to the bud dormancy, leaf abscission, and responses to diverse environmental stresses [25]. Together, these studies showed that CCO genes play an important role in plant world.
Litchi (Litchi chinensis Sonn.) is a member of the sapindaceae family and an important subtropical and tropical economic fruit which is famous by its attractive skin colour and exotic flavour. But there are still some challenges in the litchi planting industry, such as the peel coloration (‘stay green’ or pigmenting uneven problem in some varities like ‘Feizixiao’), fruit abscission, flowering control and postharvest preservation. The CCO gene family have been reported to be involved in important biological functions in the plants [1–3]. However, this gene family has not been identified in litchi. In this study, genome-wide identification of CCO gene family had been conducted, and their gene structure, domain, motif, phylogenetic relationship, miRNA target sites, cis-elements, 3D protein structure, and expression patterns were comprehensively analyzed. The study may provide a solid foundation for future functional studies of CCO genes in litchi and other fruit trees.
Results
Identification of LcCCO genes and their physicochemical properties
After homology search, a total of 15 LcCCO genes were identified in litchi. Physicochemical properties analysis found that the largest protein was LcCCD1, which contained 1434 amino acids, the smallest protein was LcCCD4c1, which contained 303 amino acids. MW of LcCCO proteins ranged from 16.25 kDa to 34.23 kDa, pI ranged from 5.54 to 8.15. Instability index analysis revealed that LcCCD1, LcCCDlike-a, LcCCDlike-b, LcCCD4c1, LcCCD4c2, and LcCCD8b were stable proteins(Instability index < 40), and the rest were unstable proteins. Aliphatic index of LcCCO proteins ranged from 75.61 to 84.62. Grand Average of Hydropathicity analysis showed that all LcCCO proteins were hydrophilic protein. Subcellular localization prediction exhibited that most of LcCCO proteins (11, 73.3%) were located in chloroplast, three LcCCO proteins (LcCCDlike-a, LcCCDlike-b, and LcCCD4c1) were located in cytoplasm, one LcCCO proteins (LcCCD8b) was located in mitochondrion (Table 1).
Table 1.
Basic information of LcCCO gene family
| LcCOO genes |
Gene ID in genome |
Genomic position |
Number of amino Acids (aa) |
MW (KDa) | pI | Instability index | Aliphatic index |
Grand average of hydropathicity |
Subcellular localization prediction |
|---|---|---|---|---|---|---|---|---|---|
| LcCCD1 | LITCHI004397.m1 | Chr14:923,933–940,844 | 1434 | 162.43 | 6.12 | 35.41 | 84.62 | -0.231 | Chloroplast |
| LcCCDlike-a | LITCHI017175.m1 | Chr1:43,820,531–43,837,433 | 1172 | 131.99 | 5.54 | 36.52 | 75.61 | -0.314 | Cytoplasm |
| LcCCDlike-b | LITCHI001770.m1 | Chr5:30,309,771–30,311,628 | 359 | 40.68 | 5.90 | 29.50 | 80.56 | -0.179 | Cytoplasm |
| LcCCD4 | LITCHI017848.m1 | Chr15:133,065–136,343 | 589 | 64.70 | 6.65 | 42.13 | 80.44 | -0.202 | Chloroplast |
| LcCCD4a1 | LITCHI000409.m1 | Chr5:11,437,371–11,439,125 | 584 | 65.72 | 7.23 | 49.31 | 80.77 | -0.142 | Chloroplast |
| LcCCD4a2 | LITCHI000415.m1 | Chr5;11,580,107–11,582,498 | 584 | 65.70 | 7.23 | 48.84 | 82.11 | -0.099 | Chloroplast |
| LcCCD4b | LITCHI000422.m1 | Chr5;11,719,278–11,721,297 | 577 | 64.97 | 8.15 | 46.41 | 81.56 | -0.182 | Chloroplast |
| LcCCD4c1 | LITCHI015832.m1 | Chr1;19,365,348–19,366,692 | 303 | 34.23 | 6.37 | 39.62 | 80.03 | -0.230 | Cytoplasm |
| LcCCD4c2 | LITCHI015831.m1 | Chr1;19,334,658–19,337,656 | 566 | 62.63 | 6.48 | 31.10 | 81.64 | -0.277 | Chloroplast |
| LcCCD7 | LITCHI006417.m1 | Chr11;1,151,592–1,157,255 | 628 | 70.57 | 7.27 | 49.75 | 80.51 | -0.269 | Chloroplast |
| LcCCD8a | LITCHI006516.m1 | Chr11;1,903,352–1,906,489 | 548 | 61.54 | 6.28 | 42.19 | 76.17 | -0.426 | Chloroplast |
| LcCCD8b | LITCHI017183.m1 | Chr1;43,942,769–43,947,076 | 566 | 62.63 | 6.48 | 31.10 | 81.64 | -0.277 | Mitochondrion |
| LcNCED1 | LITCHI012579.m1 | Chr2;12,609,603–12,614,092 | 596 | 66.73 | 6.82 | 47.43 | 77.03 | -0.389 | Chloroplast |
| LcNCED2 | LITCHI028785.m1 | Chr9;17,432,409–17,435,810 | 598 | 66.85 | 6.51 | 43.21 | 80.87 | -0.300 | Chloroplast |
| LcNCED3 | LITCHI015114.m1 | Chr1;8,869,747–8,872,765 | 601 | 67.21 | 6.95 | 40.23 | 82.63 | -0.331 | Chloroplast |
Phylogenetic analysis
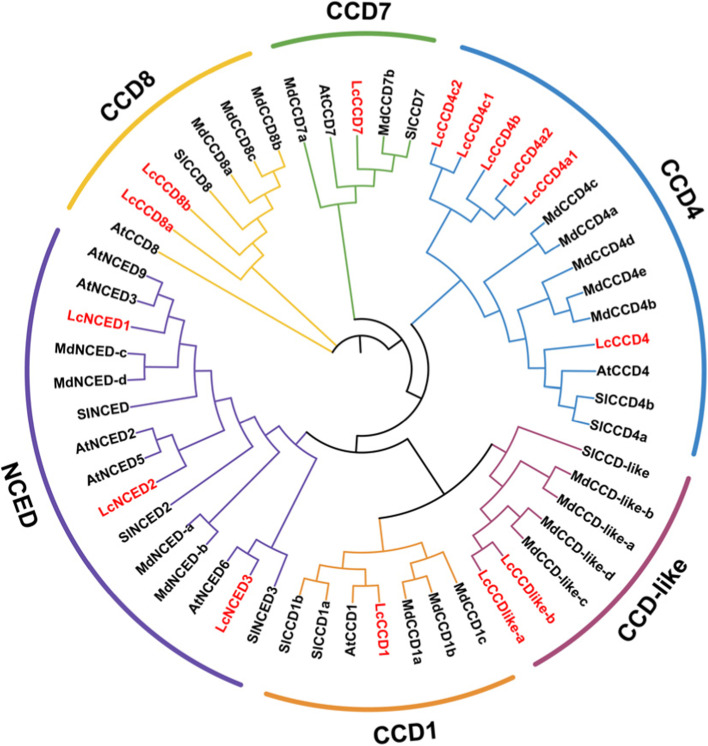
To better understand the evolutionary relationships among CCD proteins, maximum-likelihood (ML) methods were adopted to construct an unrooted phylogenetic tree which containing 55 CCO proteins from the following four species: Arabidopsis thaliana (9), Solanum lycopersicum (10), Malus × domestica (21) and Litchi chinensis Sonn (15) (Fig. 1). The result showed that the 55 CCD proteins could be divided into six subfamilies (CCD1, CCD4, CCD7, CCD8, CCD-Like, and NCED) (Fig. 1). CCD4 are the largest subfamily, including six members (LcCCD4, LcCCD4a1, LcCCD4a2, LcCCD4b, LcCCD4c1, and LcCCD4c2), while CCD7 is the smallest subfamily, just one member (LcCCD7). Compared to Arabidopsis thaliana, the number of LcCCO genes is about twice as much as the two formers.
Fig. 1.
Phylogenetic tree was constructed by ML method. Red character represented CCO proteins from Litchi chinensis Sonn., black character represented CCO proteins from Arabidopsis thaliana, Solanum lycopersicum, Malus × domestica
Gene structure, domain, motif and chromosomal arrangement analysis
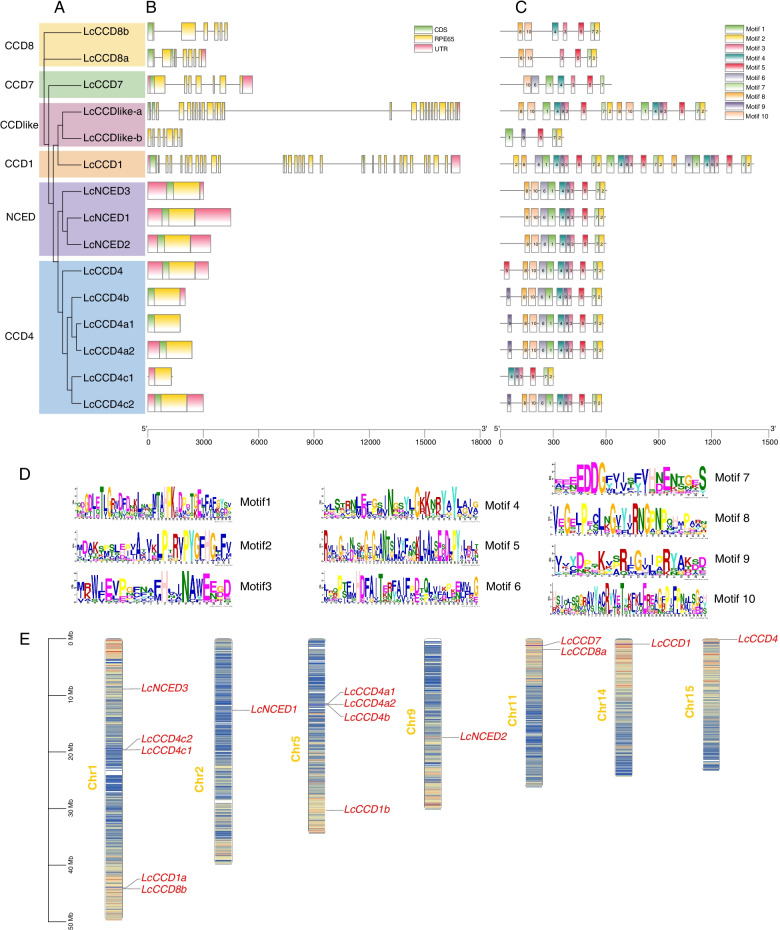
Gene structure analysis showed that the numbers of exon of LcCCO genes ranged from 1 to 32. CCD1 subfamily contained the largest number of exons, following were the CCD7 and CCD8 subfamily, CCD4 and NCED subfamily just had one exon. LcCCO genes in the same subfamilies displayed similar structure distribution patterns (Fig. 2B). Conserved domain analysis exhibited that all of LcCCO genes contained a RPE65 domain (Fig. 2B). Motif analysis showed that all of LcCCO genes had the motif 5 and motif 7, and like conserved domain, displayed similar patterns in the same subfamilies. Such as in NCED family, the distribution pattern of motifs of each member began with motif9, 10, 6, 8, 10, 6, 1, 4, 9, 3, 5, 7, and 2 from N-terminal to C-terminal (Fig. 2C-D). Chromosomal distribution analysis showed that LcCCO genes located in seven chromosomes (Fig. 2E). Chromosomes1 and 5 (Chr1 and Chr5) contained nine (60%) LcCCO genes (five and four respectively). Chr11 had two (13.3%) LcCCO genes. Chr2, 9, 14 and 5 carried only one (6.7%) LcCCO gene. LcCCDlike-b, LcNCED2, and LcCCD4 were located in the regions with high gene density.
Fig. 2.
Gene structure, conserved domain and motif of LcCCO genes. A Phylogenetic tree of LcCCO genes. B The distribution of gene structure and conserved domain. C The distribution of conserved motif, D motif elements. E Chromosomal distribution of LcCCO genes
Prediction of miRNA target site of LcCCO genes
MiRNA target site prediction showed that a total of 31 miRNA target sites could be found in 13 (86.7%) LcCCO genes with the exception of LcCCDlike-b and LcNCED3 (Table 2). Among all LcCCO genes, LcCCD1 and LcCCDlike-a existed the most miRNA target sites, which could be targeted by 10 and 7 miRNAs separately. LcCCD4c1 and LcCCD7 just had one miRNA target site. In the same subfamily, we found that some members could be targeted by a same miRNA. Such as LcCCD4a1, LcCCD4a2, LcCCD4b, and LcCCD4c2, which belonged to the CCD4 subfamily, could by targeted by Lc-miRN23 simultaneously. LcCCD1 and LcCCDlike-a, which belonged to CCD1 subfamily, could be targeted by Lc-miRN58 concurrently, but the LcCCDlike-a existed two different Lc-miRN58 target sites. More generally, LcCCO genes in the same or different subfamilies were targeted by different miRNAs. Such as LcNCED1, LcNCED2, and LcNCED3, which belonged to NCED subfamily, there were no common miRNA targets.
Table 2.
The potential miRNA target sites of LcCCO genes
| miRNA | Target | Expectation | miRNA Length |
Target_start | Target end | Inhibition | Multiplicity |
|---|---|---|---|---|---|---|---|
| Lc-miR408b/d/f | LcCCD1 | 3.5 | 20 | 2577 | 2597 | Cleavage | 1 |
| Lc-miR172h | LcCCD1 | 4 | 20 | 2445 | 2465 | Cleavage | 1 |
| Lc-miR408a/c/e | LcCCD1 | 4.5 | 20 | 2578 | 2598 | Cleavage | 1 |
| Lc-miR160c/d | LcCCD1 | 5 | 20 | 2057 | 2077 | Cleavage | 1 |
| Lc-miR172a/b/c/d/e/i/j | LcCCD1 | 5 | 20 | 2445 | 2465 | Cleavage | 1 |
| Lc-miRN19 | LcCCD1 | 5 | 20 | 3794 | 3814 | Cleavage | 1 |
| Lc-miRN58 | LcCCD1 | 5 | 21 | 1942 | 1963 | Cleavage | 1 |
| Lc-miRN49 | LcCCDlike-a | 4 | 21 | 2941 | 2962 | Cleavage | 2 |
| Lc-miRN49 | LcCCDlike-a | 4.5 | 21 | 1267 | 1288 | Cleavage | 2 |
| Lc-miR156g/l | LcCCDlike-a | 5 | 20 | 80 | 101 | Cleavage | 1 |
| Lc-miR397a/b | LcCCDlike-a | 5 | 20 | 1752 | 1772 | Cleavage | 2 |
| Lc-miR397a/b | LcCCDlike-a | 5 | 20 | 3351 | 3371 | Cleavage | 2 |
| Lc-miR397c/d | LcCCDlike-a | 5 | 19 | 1753 | 1772 | Cleavage | 2 |
| Lc-miR397c/d | LcCCDlike-a | 5 | 19 | 3352 | 3371 | Cleavage | 2 |
| Lc-miRN24 | LcCCDlike-a | 5 | 20 | 323 | 343 | Cleavage | 1 |
| Lc-miRN58 | LcCCDlike-a | 5 | 21 | 1331 | 1352 | Translation | 2 |
| Lc-miRN58 | LcCCDlike-a | 5 | 21 | 3005 | 3026 | Translation | 2 |
| Lc-miRN53 | LcCCD4 | 4.5 | 20 | 1148 | 1168 | Cleavage | 1 |
| Lc-miR166a | LcCCD4 | 5 | 20 | 1180 | 1200 | Translation | 1 |
| Lc-miRN19 | LcCCD4a1 | 4.5 | 20 | 821 | 841 | Cleavage | 1 |
| Lc-miRN23 | LcCCD4a1 | 4.5 | 21 | 704 | 725 | Cleavage | 1 |
| Lc-miRN56a/b | LcCCD4a1 | 4.5 | 20 | 1120 | 1140 | Cleavage | 1 |
| Lc-miRN19 | LcCCD4a2 | 4.5 | 20 | 821 | 841 | Cleavage | 1 |
| Lc-miRN23 | LcCCD4a2 | 4.5 | 21 | 704 | 725 | Cleavage | 1 |
| Lc-miRN56a/b | LcCCD4a2 | 4.5 | 20 | 1120 | 1140 | Cleavage | 1 |
| Lc-miRN56a/b | LcCCD4b | 4.5 | 20 | 1099 | 1119 | Cleavage | 1 |
| Lc-miRN23 | LcCCD4b | 5 | 21 | 683 | 704 | Cleavage | 1 |
| Lc-miRN26a/b | LcCCD4c1 | 4 | 20 | 882 | 902 | Cleavage | 1 |
| Lc-miRN26a/b | LcCCD4c2 | 3.5 | 20 | 1707 | 1727 | Cleavage | 1 |
| Lc-miR6833 | LcCCD4c2 | 4.5 | 20 | 450 | 470 | Cleavage | 1 |
| Lc-miR171b/c/d/g/h/j/o/sq | LcCCD4c2 | 5 | 20 | 472 | 492 | Cleavage | 1 |
| Lc-miRN23 | LcCCD4c2 | 5 | 21 | 692 | 713 | Cleavage | 1 |
| Lc-miRN45 | LcCCD4c2 | 5 | 20 | 1453 | 1473 | Translation | 1 |
| Lc-miR156c/r | LcCCD7 | 5 | 20 | 1518 | 1538 | Cleavage | 1 |
| Lc-miRN34 | LcCCD8a | 5 | 20 | 1518 | 1538 | Cleavage | 1 |
| Lc-miRN56a/b | LcCCD8a | 5 | 20 | 1621 | 1642 | Cleavage | 1 |
| Lc-miR166b/e/f/g/h/i/j/l/m/n/o | LcCCD8b | 4.5 | 20 | 824 | 844 | Cleavage | 1 |
| Lc-miR166c/k | LcCCD8b | 4.5 | 20 | 824 | 844 | Cleavage | 1 |
| Lc-miRN13 | LcCCD8b | 5 | 20 | 873 | 893 | Cleavage | 1 |
| Lc-miR156e | LcNCED1 | 4 | 19 | 667 | 686 | Cleavage | 1 |
| Lc-miR156a/b/o/p/q | LcNCED1 | 5 | 19 | 667 | 686 | Cleavage | 1 |
| Lc-miR156f | LcNCED1 | 5 | 20 | 667 | 687 | Cleavage | 1 |
| Lc-miR156k/s | LcNCED1 | 5 | 19 | 667 | 686 | Cleavage | 1 |
| Lc-miRN16a/b | LcNCED1 | 5 | 21 | 412 | 433 | Translation | 1 |
| Lc-miRN54a/b | LcNCED1 | 5 | 20 | 1016 | 1036 | Translation | 1 |
| Lc-miR395a/b/c | LcNCED2 | 4.5 | 20 | 948 | 968 | Cleavage | 1 |
| Lc-miRN24 | LcNCED2 | 5 | 20 | 687 | 707 | Cleavage | 1 |
| Lc-miRN45 | LcNCED2 | 5 | 20 | 1241 | 1261 | Cleavage | 1 |
| Lc-miRN53 | LcNCED2 | 5 | 20 | 1654 | 1674 | Translation | 1 |
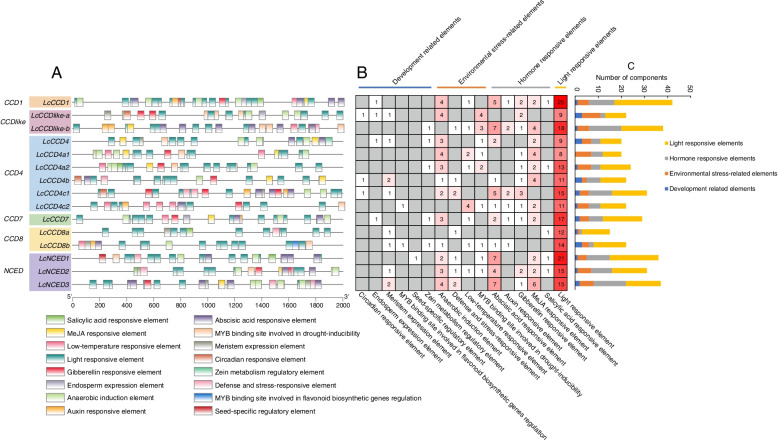
Cis-regulatory elements analysis of LcCCO genes
Cis-regulatory elements analysis found that a total of 411 cis-elements could be identified in the promoter region of LcCCO genes with the exception of common elements like TATA-box and CAAT-box and some unknown functional elements (Fig. 3 and Table S2). Among these elements of LcCCO genes, the largest group was light responsive related, included 213 (51.82%) elements, such as Box 4, GA-motif, MRE and G-box elements. The second largest group was about plant hormones related, comprised 103 (25.06%) elements, such as methyl jasmonate (MeJA) response elements (CGTCA-motif and TGACG-motif), salicylic acid (SA) response elements (TCA-element), gibberellin (GA) response elements (GARE-motif, TATC-box and P-box), abscisic acid (ABA) response elements (ABRE) and Auxin responsive element (TGA-element and AuxRR-core). ABA response elements were the largest group of plant hormones related cis-elements in the CCD1, CCD4, CCD7, CCD-like, and NCED subfamily. The third largest group was about stress related, embodied 70 (217.03%) elements, such as anaerobic induction element (ARE and GC-motif), defence and stress responsive elements (TC rich repeats) and low temperature responsive elements (LTR). The fourth largest group was about growth and development related, possessed 25 (6.08%) elements, such as endosperm expression (GCN4_motif), meristem expression (CAT-box), MYB binding site involved in flavonoid biosynthesis (MBSI) and seed specific regulatory element (RY-element).
Fig. 3.
Cis-regulatory element analysis of LcCCO genes in litchi. A The distribution of cis regulatory elements on the LcCCO gene promoter. B and C The statistics of cis regulatory elements of each LcCCO genes.
Structural features of LcCCO proteins
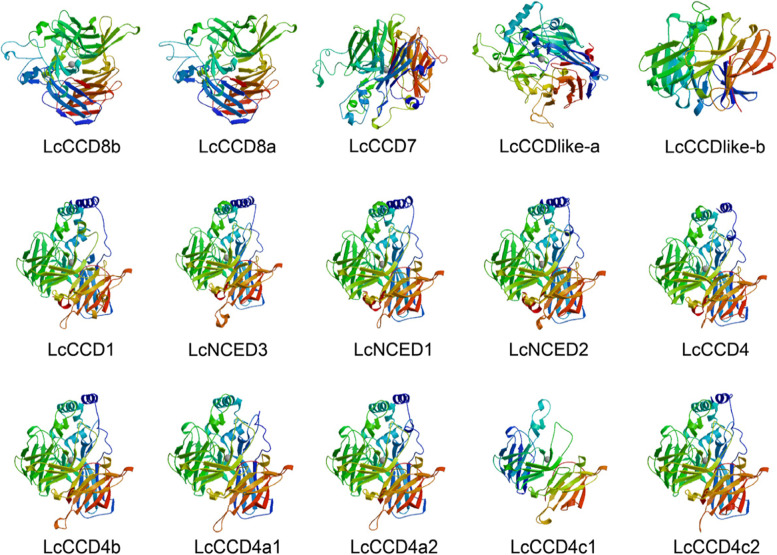
Secondary structures analysis showed that LcCCO proteins consisted an α-helix, extended chain and random coil. Random coiled amino acids occupied the largest proportion (> 50%), followed by α-helix (10.31% ~ 28.73%) and extended chain (16.28% ~ 28.08%) (Table S3). 3D structures prediction revealed that the structures of CCD8 subfamily were similar, the structures of CCD4 subfamily (excepted for LcCCD4c1 protein), NCED subfamily and LcCCD1 protein were similar (Fig. 4), suggested that they shared functionality.
Fig. 4.
Prediction of three-dimensional domain of LcCCO proteins (purple, blue, green, yellow, orange, red, N-terminal to C-terminal)
GO enrichment analysis of LcCCO genes
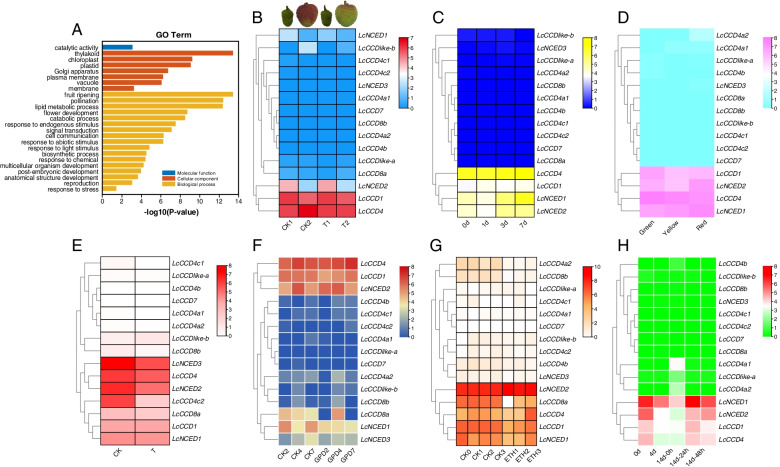
In order to predict the exact functions the litchi genes, GO enrichment analysis of LcCCO genes had been conducted in study. The result showed that the function of LcCCO genes functioned in moleculler function, cellular component and biological process (Fig. 5A and Table S4). When comes to the biological process, it was clearly that LcCCO genes were involving in the process of fruit ripening, pollination, flower development, catabolic process, response to endogenous stimulus, signal transduction, response to abiotic stimulus, response to light stimulus, reproduction so on.
Fig. 5.
The GO enrichment analysis and expression pattern analysis of LcCCO genes by RNA-seq data. A The GO enrichment analysis of LcCCO genes in litchi. B The expression of LcCCO genes during peel coloring of ‘Feizixiao’ litchi treated by exogenous CPPU, CK: control group, T: CPPU treatment group. CK1 and T1: Green stage (the peel just completely wraps the pulp, 35d after anthesis), CK2 and T2: The best edible stage of fruit (57d after anthesis). C The expression of LcCCO genes of ‘Feizixiao’ litchi on the 0, 1, 3 and 7 days after bags removed. 0d: completely green; 1d, only the stipe was colored, 3d: The peel was half colored, 7d: fully colored. D The expression of LcCCO genes of ‘Nuomici’ Litchi during three different development stages of fruit. Green: the peel is completely green; Yellow: peel yellow; Red: peel red. E The expression of LcCCO genes of the entire inflorescences samples of ‘Feizixiao’ litchi on 28 days after the uniconazole treatment. CK: control group; T: treatment group. F The expression of LcCCO genes of fruit samples of ‘Wuye’ litchi after 2, 4 and 7 days treated by girdling plus defoliation. CK: control group, GPD: girdling plus defoliation. G The expression of LcCCO genes of abscission zone samples of ‘Feizixiao’ litchi after 0, 1, 2, 3 days treated by exogenous ethephon. CK: control group, ETH: exogenous ethephon treatment. H The expression of LcCCO genes of the peel samples on 0d and 4d after stored at room temperature and 0 h, 24 h and 48 h stored at room temperature after precooling for 14 days
Expression patterns analysis of LcCCO genes by RNA-seq data
In order to investigate the potential function of LcCCO genes, the expression pattern of LcCCO genes related to peel coloration, fruit abscission, flowering control, and postharvest preservation of litchi were analysed based on the RNA-seq data supplied by our research group (not published) and other groups published online (Fig. 5B-G).
During the peel coloration inhibition experiment of ‘Feizixiao’ litchi induced by exogenous CPPU treatment (Fig. 5B and Table S5), compared to the complete green stage of fruit, LcCCD1, LcNCED1, and LcNCED2 exhibited down-regulated expression in the best edible stage (this stage of ‘Feizixiao’ litchi which existed ‘stay green’ phenomenon) between control and treatment groups, but much more obviously in the treatment groups (decreased by 1.49, 5.44, and 6.24 times in control groups and 1.33, 2.38, and 5.58 times in the treatment groups separately). LcCCDlike-b displayed up-regulated expression, increased by 74.88 times in control groups and 50.41 times in the treatment groups separately. LcCCD4 just showed up-regulated expression in the control groups, but no obvious change in the treatment groups. During the experiment of light-regulated anthocyanin biosynthesis in the peel of ‘Feizixiao’ litchi (Zhang et al., 2016a), LcCCD1, LcCCD4, LcNCED1, and LcNCED2 showed up-regulated expression and reached the peak on the third day or seventh day after bags removed. No apparent change found in others genes (Fig. 5C and Table S6). These results suggested that LcCCDlike-b, LcCCD1, LcCCD4, LcNCED1, and LcNCED2 might play an important role in the fruit maturation of ‘Feizixiao’ litchi.
Compared to ‘Feizixiao’ litchi, ‘Nuomici’ litchi fruit could complete coloring [28]. LcCCD1 and LcCCD4 exhibited down-regulated expression in the yellow and red stage of fruit, LcNCED1 showed up-regulated expression after green stage, and reached the peak in the red stage. LcNCED2 displayed down-regulated expression at yellow stage and up-regulated expression in the red stage (Fig. 5D and Table S7). These finding suggest that LcCCD1, LcCCD4, LcNCED1, and LcNCED2 might function during the fruit maturation of ‘Nuomici’ litchi.
Uniconazole treatment of litchi inflorescences can control flowering and improve fruit-setting in litchi [29]. LcCCD4, LcCCD4c2, LcCCD8a, LcNCED2, and LcNCED3 showed down-regulated expression obviously in the entire inflorescences after uniconazole spraying, decreased by 1.72, 1.53, 15.97, 2.80, and 3.18 times separately. No apparent change found in other genes (Fig. 5E and Table S8), indicated that the above genes might be involved in the flowering control and fruit-setting improvement of litchi.
In the fruitlet samples during fruit abscission of ‘Wuye’ litchi induced by girlding plus defoliation treatment [30], LcCCD1, LcCCD4a2, LcCCD8a, and LcNCED1 showed down-regulated expression on the second day after treatment, and LcCCD4a2 decreased most significantly (11.07 times). LcCCD4 and LcNCED2 displayed down-regulated expression on the fourth day after treatment, decreased by 1.15 and 1.13 times separately. LcNCED3 exhibited down-regulated expression on the seventh day after treatment, LcCCD4 showed up-regulated expression on the seventh day after treatment (Fig. 5F and Table S9). In the abscission zone samples during fruitlet abscission of ‘Feizixiao’ litchi caused by exogenous ethephon treatment [31], LcCCD4a2, LcCCD4b, LcCCD8a, and LcCCD8b showed down-regulated expression evidently on the first, second and third day. LcCCD1, LcCCD4, and LcNCED1 displayed down-regulated expression on the first and second day and up-regulated expression on the third day after treatment. LcNCED2 exhibited up-regulated expression evidently during the whole times (Fig. 5G and Table S10). These results indicated the above genes might be related in the fruitlet abscission of litchi.
In the peel samples of ‘Huaizhi’ litchi on 0d and 4d after stored at room temperature and 0 h, 24 h, and 48 h stored at room temperature after precooling for 14 days [32], LcCCD1, LcCCD4, LcNCED1, and LcNCED2 showed relative higher expression on 0d in the sample which stored at room temperature without precooling treatment, but expression inhibition could be found obviously in the samples which do the precooling treatment. All of the above four genes showed down-regulated expression on 4d after stored at room temperature without precooling treatment, up-regulated expression on 14 h and 48 h after stored at room temperature treated by precooling. It was interesting that LcCCD4a1 and LcCCD4a2 showed significantly up-regulated expression in 0 h stored at room temperature after precooling (Fig. 5H and Table S11). These data suggested that the above genes might be involved in the rapid fruit senescence induced by pre-cold storage.
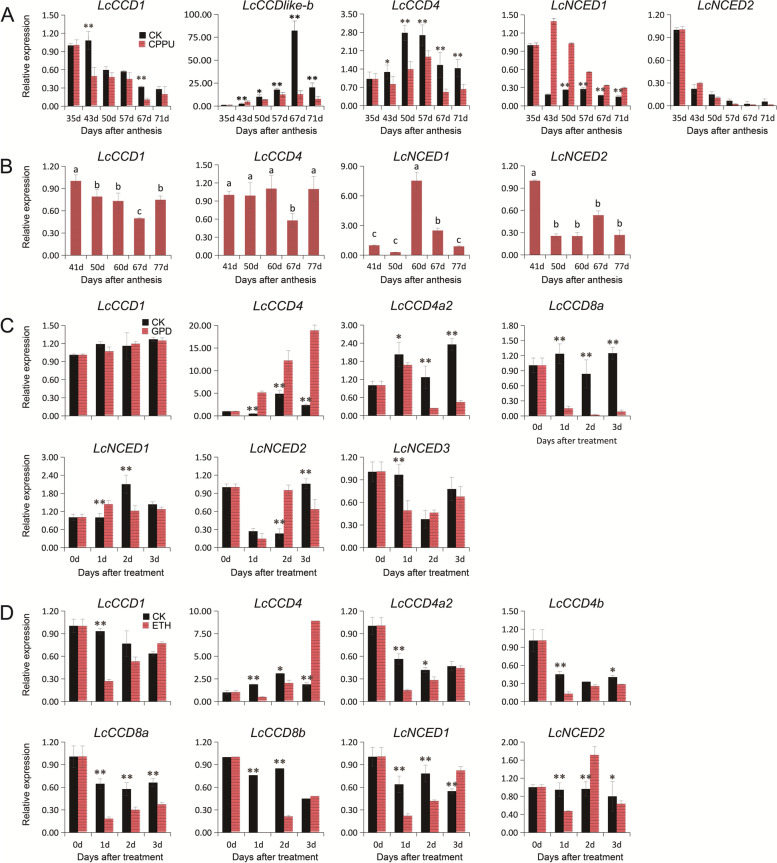
Identification of expression patterns of LcCCO genes by quantitative qRT-PCR
In order to further explore the potential function of LcCCO genes, the samples which involving in the inhibition of peel coloration of ‘Feizixiao’ litchi induced by exogenous CPPU, the natural peel coloration of ‘Nuomici’ litchi, fruitlet abscission of litchi produced by girdling plus defoliation treatment and exogenous ethephon treatment were collected. The expression patterns of these pivotal LcCCO genes obtained by the RNA-seq data were assessed by quantitative qRT-PCR (Fig. 6). The results showed that the expression pattern of most of LcCCO genes were consistent with their expression patterns in the RNA-seq data described above (Figs. 5B, D, F-G, and 6) excepted for the expression of LcNCED1 during the coloration of ‘Feizixiao’ litchi and the expression of LcCCD4 during the coloration of ‘Nuomici’ litchi (Figs. 5B, D, 6A-B). Interestingly, there were also some differences of the expression of LcCCO genes between the experiment of fruitlet abscission of ‘Feizixiao’ litchi and ‘Wuye’ litchi produced by girdling plus defoliation treatment (Fig. 6C). This might be caused by the variety differences between ‘Wuye’ litchi and ‘Feizixiao’ litchi. But there were still some LcCCO genes shared the similar expression patterns, Such as LcCCD4, LcCCD4a2, LcCCD8a, and LcNCED1.
Fig. 6.
The expression of LcCCO genes identified by by qPCR. A The expression of LcCCO genes of the peel tissues during fruit maturation of ‘Feizixiao’ litchi treated by exogenous CPPU after anthesis. 35d: Green stage (the peel just completely wraps the pulp), corresponding to the CK1 and T1, 57d: The best edible stage of fruit, corresponding to the CK2 and T2 in Fig. 5A. B The expression of LcCCO genes of the peel tissues during fruit natural maturation of ‘Nuomici’ litchi. C The expression of LcCCO genes of fruitlet during the fruitlet abscission of ‘Feizixiao’ litchi treated by girdling plus defoliation treatment. D The expression of LcCCO genes of abscission zone tissues during the fruitlet abscission of ‘Feizixiao’ litchi treated by exogenous ethephon
Discussion
Identification of LcCCO genes
Compared to the MYB, bZIP and bHLH gene family, CCO is a relatively small gene family in plant. In our study, a total of 15 LcCCO genes were identified in litchi and could be divided into six (CCD1, CCD4, CCD7, CCD8, CCD-like, and NCED) subfamilies based on the phylogenetic relationships analysis with Arabidopsis thaliana, Solanum lycopersicum, Malus × domestica and Litchi chinensis Sonn (Fig. 1), which was consistent with the previous work [10, 35]. Physicochemical properties analysis showed that the length of most of LcCCO proteins ranged from 500 to 600aa (Table 1), which displayed similarity with other plants [6, 36, 37]. RPE65 domain is a specific conserved domain in CCO protein, which is the key to the enzymatic oxidation activity cleavage of carotenoids [38]. Conserved domain analysis showed that all of LcCCO proteins contained a RPE65 domain and which exhibited similar distribution pattern in the same subfamily. Like distribution of RPE65 domain, gene structure and motif showed high similarity of distribution pattern in the same subfamily too (Fig. 2A-C). These results indicated that the genes in the same subfamily which held the similar function probably. Motif 5 and motif 7 were located in all LcCCO protein, implied that they were important characteristics and may be responsible for common functions between them. Subcellular localization analysis can help to understand the site where the protein will function. In the study, 11 (73.3%) LcCCO proteins were predicted to be located in the chloroplast, suggested that these genes might participate in chlorophyll photosynthesis. 3 (20.0%) LcCCO proteins located in cytoplasm and 1 (6.7%) LcCCO protein located in mitochondria (Table 1), indicated that these genes functioned in cytoplasm and mitochondria, and might not be involved in chlorophyll photosynthesis. These results are consistent with the previous study [35].
MiRNA is considered as a kind of post transcriptional regulator, and play a critical role during the development of plant [39, 39, 40]. In our result, 13 (86.7%) LcCCO genes obtained 31 miRNA target sites predicted combined with the litchi miRNAs described previously [41], suggested that post transcriptional regulation of LcCCO genes by miRNA might be functioning during the development of litchi. Cis-regulatory elements located in the gene promoter region which could regulated the gene expression on transcriptional level [42]. Cis-regulatory elements analysis found that a large number of cis-elements which involving in light responsive, plant hormones and stress (biological and abiotic stress related) and pant growth and development could be detected (Fig. 3A-C and Table S2). It was interesting that ABRE element which related to ABA response were the largest group in the plant hormones related cis-elements. These indicated that the transcription of LcCCO genes could be in response to light, plant hormones, biological and abiotic stress and pant growth and development. In order to investigate their function during the development period of litchi, RNA-seq data and quantitative qRT-PCR analyses related to peel coloration, flowering control, fruit abscission, and postharvest preservation were used to do further analysis.
LcCCO genes might be involved in the coloration and flavor of litchi
The colour of horticultural produce, is a key factor that can decide and enhance their economic value. The carotenoids metabolism pathway of CCD4 had been proved to be related to the color formation in plant species like chrysanthemum petalsonly and citrus by affecting the catalytic degradation pathway of carotenoids [14, 16]. ABA was considered as the critical hormone related to the coloration in plant, including strawberry [21], grape [22], sweet cherry [23] and litchi fruit [24]. NCED is a key regulator of ABA biosynthesis. FaNCED1 RNAi resulted in uncolored strawberry fruits by declining the ABA and anthocyanin content significantly (Fragaria × ananassa) [20]. ‘Feizixiao’litchi existed in ‘stayed green’category at the best edible stage. RNA-seq data and qRT-PCR analysis showed that the expression of LcCCD4, LcNCED1, and LcNCED2 could be inhibited obviously by exogenous CPPU treatment during fruit maturation of ‘Feizixiao’ litchi. LcCCD4 reached a peak at 50d and 57d after anthesis (the best edible stage) and then declined, LcNCED1 and LcNCED2 reached the peak at 43d and 35d after anthesis separately, but kept relative low expression (Figs. 5B, 6A, Table S5). RNA-seq data showed that LcCCD4, LcNCED1, and LcNCED2 exhibited up-regulated expression apparently during light-regulated anthocyanin biosynthesis to promote the coloration of ‘Feizixiao’ litchi after removing bag, and the TPM value of them was much higher than the experiment samples treated by CPPU (Fig. 5B-C and Table S5-6). Compared to ‘Feizixiao’ variety, ‘Nuomici’ litchi fruit could fulfil coloration. RNA-seq data and qRT-PCR analysis showed that LcCCD4 exhibited relative high expression before 67d after anthesis and reached the peak at 60d (yellow stage). LcNCED1 exhibited relative higher expression than ‘Feizixiao’ litchi samples described above and reached a peak at 60d after anthesis and then declined, but still kept relative higher expression at 67d (Red stage). LcNCED2 exhibited two peaks at 41d (green stage) and 67d (red stage) after anthesis (Figs. 5D, 6B and Table S7). Among the above results, the higher expression of LcCCD4, LcNCED1, and LcNCED2 during the early stage of fruit maturation of ‘Nuomici’ litchi might contribute to the carotenoid degradation and anthocyanin accumulation to complete coloration. The relative low expression of LcCCD4, LcNCED1, and LcNCED2 in ‘Feizixiao’ litchi might be the key factor to induce the ‘stay green’ phenomenon. In addition to participating in coloring, CCO genes liked CCD1 was reported to be involving in the formation of the flavor and scent in plants by catalyzing degradation of carotenoids [12, 43]. We could clearly find that LcCCD1 showed down expression during the fruit maturation in the both of ‘Feizixiao’ and ‘Nuomici’ varieties simultaneously, and LcCCDlike-b displayed down expression significantly in ‘Feizixiao’ variety but could not be detected in the ‘Nuomici’ variety (Fig. 5B, D and Table S5, S7). Although the dynamic of carotenoid content all showed a downtrend, but displayed some differences during fruit maturation of ‘Feizixiao’and ‘Nuomici’ varieties (Supplement Fig. 1). These implied that LcCCDlike-b might be an important factor which involving in the different flavor between these two varieties. But these conjectures need further works to confirmed them.
LcCCO genes might be involved in fruitlet abscission of litchi
ABA was also reported to be related to plant organ abscission [44, 45]. Litchi is an important subtropical and tropical economic fruit. It is reported that there are 3–5 fruit drop waves (I, II, III, IV, V) between different cultivars, and high ABA lever is regarded one of most important physiological reasons for the fruit drop wave II, III, and V [46]. Based on the public RNA-seq data and qRT-RCR analysis, we found that the expression of LcNCED1, LcNCED2, and other LcCCO genes were enhanced in fruitlet and abscission zone samples during fruitlet abscission induced by GPD and exogenous ETH treatment in ‘Feizixiao’ variety separately (Figs. 5F-G, 6C-D and Table S9-10). These implied that LcNCED1 and LcNCED2 might function in accelerating the ABA content to promote the fruitlet abscission of litchi, the function of other LcCCO genes needed further work to investigate. Contrary to the result of ‘Feizixiao’ variety, the expression of LcNCED1, LcNCED2 other CCO genes in ‘Wuye’ variety were inhibited in the fruitlet samples during fruitlet abscission induced by GPD treatment (Fig. 5F and Table S9). The differences of gene expression between ‘Wuye’ variety and ‘Feizixiao’ variety treated by GPD, which might be caused by the difference between these two varieties, but needed further works to demonstrate this conjecture.
LcCCO genes might be involved in flower control of litchi
Easily flowering and without controlling of the inflorescence development and flowering can lead to low fruit-setting percentage or even zero yield by consuming excessive amounts of accumulated nutrients during the production of litchi [47]. Uniconazole spraying which can regulate the changes of endogenous hormone levels (reducing GA content and increased ABA content) to function in the flower control and fruit retention of litchi, is considered as an effective chemical method [48, 49]. Another research proved that uniconazole was a strong competitive inhibitor of ABA 8'-hydroxylase to effectively inhibit ABA catabolism in Arabidopsis plants [50]. In this study, the RNA-seq data published by Wei et al. [29] showed that LcNCED2 and LcNCED3 displayed low expression in litchi inflorescences treated by uniconazole (Fig. 5E and Table S8). These suggested that the ABA accumulation is the result of the joint action of biosynthesis regulated by LcNCED2 and LcNCED3 and catabolism inhibition by uniconazole. The later might be much more important during the flowering controlling to improve the litchi fruit-setting. but also need more experiment to confirm it.
LcCCO genes might be involved in the postharvest preservation of litchi
Litchi is a non-climacteric tropical and subtropical fruit which is highly perishable, and typical symptoms is pericarp browning and loss of flavor after harvest. The shelf life of litchi fruit could be prolonged up to a month storage in cold environment, but the fruit senescence of fruits which were at ambient temperatures after pre-cold storage treatment could be accelerated, only 1–2 days, much less than the fruit under ambient conditions, which is approximately 4–6 days [32, 51, 52]. ABA was considered as one of key factors which contributed the fruit senescence [53, 54]. In this study, The RNA-seq data published by Yun et al. [32] showed that LcCCD4a1 and LcCCD4a2 displayed significantly up-regulated expression at 0 h, LcCCD1, LcCCD4, LcNCED1, and LcNCED2 displayed up-regulation at 14 h and 48 h stored at room temperature after treated by precooling (Fig. 5H and Table S11). These data suggested that the above genes might be involved in the rapid fruit senescence induced by pre-cold storage by enhanced the ABA content and accelerate the carotenoid degradation, but need further experiments to confirmed it.
Conclusions
In conclusion, CCO gene family were identified comprehensively in litchi. A total of 15 LcCCO genes were identified and could be classified into five subfamilies based on the phylogenetic relationships with other several species. And then the physicochemical properties, the distribution of gene structure, conserved domain and motif, cis-elements, miRNA target sites, 3D protein structure were further analysed. In addition, RNA-seq data and qRT-PCR analysis revealed that LcCCO genes might be related to the peel coloration, fruit flavor, flower control, fruit abscission and postharvest storage of litchi. Our result not only will help lay the foundation for the function identification of CCO gene family in litchi, but also will help us understand the role of this gene family in other plant species.
Methods
Identification of LcCCO genes
The genome and gene annotation files of litchi was supplied by College of Horticulture, South China Agricultural University, Guangzhou, China (data unpublished yet). The CCO protein sequences of Arabidopsis thaliana were downloaded from TAIR database (https://www.arabidopsis.org/). Homology search was conducted by the TBtools software [26], and then confirmed the RPE65 (retinal pigment epithelial membrane protein) domain used the Pfam database (http://pfam.xfam.org/) and Hmmer2.3 database (http://hmmer.janelia.org/). The sequences did not contain the RPE65 domain would be deleted, and the rest proteins were considered as the litchi CCO (LcCCO) proteins. Physicochemical properties of LcCCO proteins like molecular weight (MW), Pi, instability index, aliphatic index was analyzed by ExPASY website (http://web.expasy.org/protparam/), Subcellular localization prediction was conducted by the BUSCA website (http://busca.biocomp.unibo.it/), signal peptide and transmembrane structure were predicted by the MBC website (http://cello.life.nctu.edu.tw).
Phylogenetic analysis
The CCO protein sequences of Arabidopsis thaliana, Solanum lycopersicum, Malus × domestica and Litchi chinensis Sonn. were used to do phylogenetic analysis. Phylogenetic tree was conducted by Clustal X2 and MEGA 6 software using maximum-likelihood (ML) methods with the following parameter settings: poisson model, partial deletion and 1000 bootstrap replicates. At last, the LcCCO proteins were renamed depended on which subfamily they belonged to.
Gene structure, conserved domain, conserved motif and chromosomal arrangement analysis
The structure information of LcCCO genes can be acquired by the gff file of litchi genome. The conserved domain identification was conducted by NCBI cd-search website (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and Pfam website (http: //www.sanger.ac.uk/software/Pfam/). The motif analysis was executed by MEME tools (http://meme-suite.org/tools/meme). Phylogenetic tree, gene structure, conserved domain and motif of LcCCO genes were displayed by TBtools software [26].
miRNA target site prediction
Litchi miRNAs sequences could be obtained from the previous works (Ma et al., 2018) and the LcCCO genes sequences were adopted to do the miRNA target site prediction by the psRNATarget website (https://www.zhaolab.org/psRNATarget/analysis?function=3) with default parameter settings.
Cis-regulatory elements analysis
The 2000 bp upstream sequences from the translation initiation codon ‘ATG’ of LcCCO genes were extracted by the TBtools software [26] and then used to do the cis-regulatory elements analysis by Plant Care database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
3D protein structure analysis
Firstly, protein secondary structure of LcCCO proteins were predicted by GOR IV (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html), and then 3D protein structure was predicted by the SWISS-MODEL online tools (https://swissmodel.expasy.org/).
Gene Ontology (GO) enrichment analysis
Firstly, all litchi genes was blasted to the uniprot_sprot.fasta file downlorded from Swissprot database (https://www.uniprot.org/). Then GO annotation and enrichment analysis was conducted by the TBtools software [26].
Expression analysis of LcCCO genes by RNA-seq data
RNA-seq data in this study used for the expression analysis including the following seven sets of data (The first set data was home data, RNA-seq was conduct by the paied-end sequencing based on Illumina platform. Other six sets data were publically available): (1) The peel samples of complete green stage (the peel just completely wraps the pulp) and the best edible stage of ‘Feizixiao’ variety with three biological repetitions, which obtained from the inhibition of the fruit coloration experiment by exogenous N-(2-Chloro-4-pyridyl)-N’-phenylurea (CPPU) (not published yet). (2) The peel samples of coloration of ‘Feizixiao’ litchi on 0d, 1d, 3d and 7d after removed bag, accession number is SRA312830 (https://www.ncbi.nlm.nih.gov/sra/?term=SRX1445119) [27]. (3) Three stages of peel samples of ‘Nuomici’ litchi: green, yellow and red peel stage, accession number is SRP047115 (https://www.ncbi.nlm.nih.gov/sra/?term=SRP047115) [28]. (4) The entire inflorescences samples on 28 days after the uniconazole treatment, accession number is SRP092890 (https://www.ncbi.nlm.nih.gov/sra/?term=SRP092890) [29]. (5) The fruit samples on 2d, 4d and 7d after fruit abscission of ‘Wuye’ litchi caused by girdling plus defoliation treatment (GPD), accession number is SRA234477 (https://www.ncbi.nlm.nih.gov/sra/?term=SRA234477) [30]. (6) The abscission zone samples on 0d, 1d, 2d and 3d after fruit abscission of ‘Feizixiao’ litchi caused by exogenous ethephon (ETH), accession number is SRP173341 (https://www.ncbi.nlm.nih.gov/sra/?term=SRP173341) [31]. (7) The peel samples on 0d and 4d after stored at room temperature and 0 h, 24 h and 48 h stored at room temperature after precooling for 14 days, accession number is SRA247016 (https://www.ncbi.nlm.nih.gov/sra/?term=SRA247016) [32]. More detailed plant material information were listed in Table 3. The transcripts per kilobase million (TPM) value of LcCCO genes were calculated by the Salmon software [33]. The log2 value of TPM were used to draw heatmaps by TBtools software [26], and if the TPM values of were less than 1 in any group samples (such as in control group or treatment group), they will be discarded without further analysis. The P value was conducted by the edgeR tools of Cloud platform OmicShare of Genedenovo Biotechnology Co., Ltd (Guangzhou, China) (https://www.omicshare.com/tools/).
Table 3.
Plant materials used for RNA-Seq and qRT-PCR analysis
| Set of RNA-Seq data | Variety | Sample ID and | Samples | Treatment | Platform | LibraryLayout | Accesion number | Data sources |
|---|---|---|---|---|---|---|---|---|
| Plant materials for RNA-Seq analysis | ||||||||
| 1 |
‘Feizixiao’ (27-year-old) |
CK1_1, CK1_2,CK1_3, CK2_1, CK2_2,CK2_3, T1_1, T1_2, T1_3, T2_1, T2_1, and T2_1. | CK1 and T1: 35d Green stage (the peel just completely wraps the pulp, 35d after anthesis); CK2 and T2: 57d (The best edible stage of fruit) after anthesis. | 35 days after anthesis treated with 4 mg/L CPPU. | ILLUMINA | PAIRED | Home data | |
| 2 |
‘Feizixiao’ (8-year-old) |
0d (SRR2952606),1d (SRR2954687), 3d (SRR2954690) and 7d (SRR2954691). | 0d (completely green), 1d (only the stipe was colored), 3d (The peel was half colored), and 7d (fully colored).(mix sample of 3 biological replicates). | Fruit clusters were bagged at 42 days after anthesis and removed after 2 weeks. | ILLUMINA | PAIRED | SRA312830 | Zhang et al., 2016 |
| 3 |
‘Nuomici’ (Adult tree) |
Green (SRX700596),Yellow (SRX700598), and Red (SRX700599). | Green,Yellow and Red represented 52, 62 and 72 days after anthesis separately. (mix sample of 3 biological replicates) | Normal condition. | ILLUMINA | SINGLE | SRP047115 | Lai et al., 2015 |
| 4 |
‘Feizixiao’ (10-year-old) |
CK (SRX2336010) and T (SRX2934817). | CK and T reprensted 0 and 28 days after the uniconazole treatment. (mix sample of 2 biological replicates). | Inflorescence length of about 15 cm treated with 50 ppm Uniconazole. | ILLUMINA | SINGLE | SRP092890 | Wei et al., 2015 |
| 5 |
‘Wuye’ ( 9-year-old) |
CK2 (SRX847812), CK4 (SRX847822), CK7 (SRX847823), GPD2 (SRX847824), GPD4 (SRX847825), and GPD7 (SRX847826). | GPD2, GPD4, and GPD7 represented 2, 4 and 7 days after treatment. (Mix sample). | Treated with girdling followed by defoliation (GPD treatment) at 35 days after anthesis | ILLUMINA | SINGLE | SRA234477 | Li et al., 2015 |
| 6 |
‘Feizixiao’ (9-year-old) |
CK0 (SRX5126892),CK1 (SRX5126893), CK2 (SRX5126894), CK3 (SRX5126895), ETH1 (SRX5126896), ETH2 (SRX5126897), and ETH3(SRX5126898). | CK0,CK1,CK2, and CK3 represented 0, 1,2 and 3 days after treatment | Treatment 250 mg/L ethephon solution at 25 days after anthesis. | ILLUMINA | SINGLE | SRP173341 | Li et al., 2015 |
| 7 |
‘Feizixiao’ (Adult tree) |
0d (SRX968371), 4d (SRX968373), 14d-0 h (SRX968375), 14d-24 h (SRX968377), and 14d-48 h (SRX968379). | Samples after harvest on 0d and 4d after stored at room temperature and 0 h, 24 h, and 48 h stored at room temperature after precooling for 14 days. (1 biological replicate). | Samples after harvest. | ILLUMINA | SINGLE | SRA247016 | Yun et al., 2015 |
| Plant materials for qRT-PCR analysis | ||||||||
| 1 |
‘Feizixiao’ (27-year-old) |
35d, 43d, 50d, 57d, 67d, and 71d. | Peel (3 biological replicates). | 35 days after anthesis treated with 4 mg/L CPPU. | Home data | |||
| 2 |
‘Nuomici’ (27-year-old) |
41d, 50d, 60d, 67d, and 77d. | Peel (3 biological replicates). | Normal condition, ( sample follected at 41 days after anthesis). | Home data | |||
| 3 |
‘Feizixiao’ (27-year-old) |
0d, 1d, 2d, and 3d. | Fruitlet (3 biological replicates). | Girdling plus defoliation treatment (GPD) at 35 days after anthesis. | Home data | |||
| 4 |
‘Feizixiao’ (27-year-old) |
0d, 1d, 2d, and 3d. | Abscission zone tissues (3 biological replicates). | 250 mg/L Exogenous ethephon (ETH) at 35 days after anthesis. | Home data | |||
Expression analysis of LcCCO genes identified by quantitative qRT-PCR
To further investigate the function of LcCCO genes, four sets of litchi samples with 3 biological replicates in each group were collected for the quantitative qRT-PCR analysis as followed: (1) The peel samples of different development stages (on 35d (complete green stage, corresponed to the T1 and CK1 treatment used in the RNA-seq data), 43d, 50d, 57d (best edible stage, corresponed to the T2 and CK2 treatment used in the RNA-seq data), 67d and 71d after anthesis) of ‘Feizixiao’ litchi fruit after exogenous CPPU treatment. (2) The peel samples of different development stages of ‘Nuomici’litchi (on 41d (green stage), 50d (early yellow stage), 60d (late yellow stage), 67d (best edible stage, red stage) and 77d after anthesis, corresponed to the RNA-seq data above, including green, yellow and red stage). (3) The fruitlet samples of different stage on 0d, 1d, 2, d and 3d of ‘Feizixiao’ litchi after GPD treatment. (4) The Absicssion zone samples of different stage on 0d, 1d, 2, d and 3d of ‘Feizixiao’ litchi after exogenous ETH treatment. More detailed plant material information were listed in Table 3. Total RNA extracted by the RNA Kit RNAiso Plus (#9108) and Fruit-mate (#9192) from Takara Biomedical Technology (Beijing) Co., Ltd, China. The quantity and quality of RNA were conducted by the NanoPhotometer spectrophotometer (#Nano-600) from Jinpeng Analytical Instrument Co., Ltd (Shanghai), China). Reverse transcription and qRT-PCR conducted by the following kits: HiScript III 1st Srand cDNA Synthesis Kit (R312) and ChamQ Universal SYBR qRT-PCR Master Mix (Q711) from Vazyme Biotech Co.,Ltd, Nanjing, China separately. EF-1α and GAPDH genes were used as reference genes [34]. All the primers were listed in Table S1. The 2−ΔΔCt method was adopted to do result calculation. All the samples were set three technical repetitions. T-test was adopted to do difference analysis.
Supplementary Information
Additional file 1. Supplement Figure 1. The dynamic of carotenoid content during fruit maturation of litchi. A: ‘Feizixiao’ variety; B: ‘Nuomici’ variety. Different letters indicated statistically differences between days of after anthesis using one-way ANOVA with the SAS test (P <0.05). Supplement Table S1. Primers of selected LcCCO genes in litchi and reference genes. Supplement Table S2. Cis-acting element information in the promoter region of LcCCO genes in Litchi. Supplement Table S3. Two-dimensional structures of LcCCO proteins. Supplement Table S4. GO enrichment analysis of LcCCO genes. Supplement Table S5. The TPM value and differential expression analysis of LcCCO genes during pericarp coloring of ‘Feizixiao’ litchi treated by exogenous CPPU. Supplement Table S6. The TPM value and differential expression analysis of LcCCO genes of ‘Feizixiao’ litchi on the 0, 1, 3 and 7 days after bags removed (Zhang et al., 2016a). Supplement Table S7. The TPM value and differential expression analysis of LcCCO genes in ‘Nuomici’ Litchi during three different development stages of fruit (Lai et al., 2015). Supplement Table S8. The TPM value and differential expression analysis of LcCCO genes of the entire inflorescences samples of 'Feizixiao’ litchi on 28 days after the uniconazole treatment (Wei et al., 2017b). Supplement Table S9. The TPM value and differential expression analysis of LcCCO genes of fruit samples of ‘Wuye’ litchi after 2, 4 and 7 days treated by girdling plus defoliation(Li et al., 2015a). Supplement Table S10. The TPM value and differential expression analysis of LcCCO genes of abscission zone samples of ‘Feizixiao’litchi after 0, 1, 2, 3 days treated by exogenous ethephon (Li et al., 2015b). Supplement Table S11. The TPM value and differential expression analysis of LcCCO genes of the peel samples on 0d and 4d after stored at room temperature and 0h, 24h and 48h stored at room temperature after precooling for 14 days [32].
Acknowledgements
Not applicable.
Abbreviations
- CCO
Carotenoid cleavage oxygenase
- CCD
Carotenoid cleavage dioxygenase
- NCED
9-Cis-epoxycarotenoid dioxygenase
- CPPU
N-(2-Chloro-4-pyridyl)-N’-phenylurea
- ABA
Abscisic acid
- GPD
Girdling plus defoliation treatment
- ETH
Ethephon
- NCBI
National Center for Biotechnology Information
- MW
Molecular weight
- pI
Isoelectric point
- GRAVY
Grand average of hydropathicity
- qRT-PCR
Real-time polymerase chain reaction
- EF-1α
Elongation factor 1-alpha
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
Authors’ contributions
Xiao-Qi Yue: Methodology, Writing-review & editing. Yue Zhang: Writing—original draft, Investigation. Cheng-Kun Yang: Resources, Data curation, Validation. Feng Ding, Fu-Chu Hu and Xiang-He Huang: Resources, Review. Rui Xia: Review, Editing, Jian-Guo Li: Review, Editing, Kai-Bing Zhou: Review, Funding acquisition. Wu-Qiang Ma: Conceptualization, Writing—Review & Editing, Supervision, Funding acquisition. All authors read and approved the final manuscript.
Funding
This work supported by the Natural Science Foundation of Hainan Province (2019RC082 and 320QN191), Initial funding Hainan University introduces high-level personnel (KYQD(2R)1970), the National Natural Science Foundation of China (32060655 and 31960570).
Availability of data and materials
The RNA-seq data are obtained from NCBI (https://www.ncbi.nlm.nih.gov/Traces/study/), and the accession numbers are SRA312830, SRP047115, SRP092890, SRA234477, SRP173341 and SRA247016. The COO protein sequences The CCO protein sequences of Arabidopsis thaliana were downloaded from TAIR database (https://www.arabidopsis.org/). The litchi genome data used in this study had been deposited into CNGB Sequence Archive (CNSA, https://db.cngb.org/cnsa/) of China National GeneBank DataBase (CNGBdb) with accession number CNP0001024, which will be released after the publication of the litchi genome paper. Public access to all databases is open. Other data supporting the results of this article are included within the article and its additional files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wu-Qiang Ma, Email: wuqiangma@hainanu.edu.cn.
Kai-Bing Zhou, Email: kaibingzhou0528@163.com.
References
- 1.Kloer DP, Schulz GE. Structural and biological aspects of carotenoid cleavage. Cell Mol Life Sci. 2006;63:2291–2303. doi: 10.1007/s00018-006-6176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walter MH, Strack D. Carotenoids and their cleavage products: biosynthesis and functions. Nat Prod Rep. 2011;28(4):663–692. doi: 10.1039/c0np00036a. [DOI] [PubMed] [Google Scholar]
- 3.Sui X, Kiser PD, von Lintig J, Palczewski K. Structural basis of carotenoid cleavage: from bacteria to mammals. Arch Biochem Biophys. 2013;539(2):203–213. doi: 10.1016/j.abb.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, et al. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006;45(6):982–993. doi: 10.1111/j.1365-313X.2006.02666.x. [DOI] [PubMed] [Google Scholar]
- 5.Priya R, Sneha P, Dass JFP, Doss CGP, Manickavasagam M, Siva R. Exploring the codon patterns between CCD and NCED genes among different plant species. Comput Biol Med. 2019;114:103449. doi: 10.1016/j.compbiomed.2019.103449. [DOI] [PubMed] [Google Scholar]
- 6.Vallabhaneni R, Bradbury L, Wurtzel ET. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch Biochem Biophys. 2010;504(1):104–111. doi: 10.1016/j.abb.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang RK, Wang CE, Fei YY, Gai JY, Zhao TJ. Genome-wide identification and transcription analysis of soybean carotenoid oxygenase genes during abiotic stress treatments. Mol Biol Rep. 2013;40(8):4737–4745. doi: 10.1007/s11033-013-2570-y. [DOI] [PubMed] [Google Scholar]
- 8.Ahrazem O, Gómez-Gómez L, Rodrigo MJ, Avalos J, Limón MC. Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions. Int J Mol Sci. 2016;17(11):1781. doi: 10.3390/ijms17111781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Ding G, Gu T, Ding J, Li Y. Bioinformatic and expression analyses on carotenoid dioxygenase genes in fruit development and abiotic stress responses in Fragaria vesca. Mol Genet Genomics. 2017;292(4):895–907. doi: 10.1007/s00438-017-1321-5. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Zuo X, Shao H, Fan S, Ma J, Zhang D, et al. Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica) Plant Physiol Biochem. 2018;123:81–93. doi: 10.1016/j.plaphy.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Su W, Zhang C, Feng J, Feng A, You C, Ren Y, et al. Genome-wide identification, characterization and expression analysis of the carotenoid cleavage oxygenase (CCO) gene family in Saccharum. Plant Physiol Biochem. 2021;162:196–210. doi: 10.1016/j.plaphy.2021.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Sun Z, Hans J, Walter MH, Matusova R, Beekwilder J, Verstappen FWA, et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta. 2008;228(5):789–801. doi: 10.1007/s00425-008-0781-6. [DOI] [PubMed] [Google Scholar]
- 13.Ilg A, Beyer P, Al-Babili S. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 2009;276(3):736–747. doi: 10.1111/j.1742-4658.2008.06820.x. [DOI] [PubMed] [Google Scholar]
- 14.Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006;142(3):1193–1201. doi: 10.1104/pp.106.087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J, Yang S, Tang K, Li G, Gao X, Liu B, et al. GmCCD4 controls carotenoid content in soybeans. Plant Biotechnol J. 2021;19(4):801–813. doi: 10.1111/pbi.13506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, Zhu K, Sun Q, Zhang W, Wang X, Cao H, et al. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol Plant. 2019;12(9):1294–1307. doi: 10.1016/j.molp.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 17.Pasare SA, Ducreux LJM, Morris WL, Campbell R, Sharma SK, Roumeliotis E, et al. The role of the potato (Solanum tuberosum) CCD8 gene in stolon and tuber development. New Phytol. 2013;198(4):1108–1120. doi: 10.1111/nph.12217. [DOI] [PubMed] [Google Scholar]
- 18.Ha CV, Leyva-González MA, Osakabe Y, Tran UT, Nishiyama R, Watanabe Y, et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc Natl Acad Sci U S A. 2014;111(2):851–856. doi: 10.1073/pnas.1322135111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan X, Zheng H, Zhao J, Xu Y, Li X. ZmCCD7/ZpCCD7 encodes a carotenoid cleavage dioxygenase mediating shoot branching. Planta. 2016;243(6):1407–1418. doi: 10.1007/s00425-016-2479-5. [DOI] [PubMed] [Google Scholar]
- 20.Jia HF, Chai YM, Li CL, Lu D, Shen YY. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011;157(1):188–199. doi: 10.1104/pp.111.177311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Joyce DC. ABA effects on ethylene production, PAL activity, anthocyanin and phenolic contents of strawberry fruit. Plant Growth Regul. 2003;39(2):171–174. doi: 10.1023/A:1022539901044. [DOI] [Google Scholar]
- 22.Wheeler S, Loveys B, Ford C, Davies C. The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust J Grape Wine Res. 2010;15(3):195–204. doi: 10.1111/j.1755-0238.2008.00045.x. [DOI] [Google Scholar]
- 23.Shen X, Zhou K, Liu L, Zhang K, Yuan H, Liao X, et al. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.) Plant Cell Physiol. 2014;55(5):862–880. doi: 10.1093/pcp/pcu013. [DOI] [PubMed] [Google Scholar]
- 24.Singh SP, Saini MK, Singh J, Pongener A, Sidhu GS. Preharvest application of abscisic acid promotes anthocyanins accumulation in pericarp of litchi fruit without adversely affecting postharvest quality. Postharvest Biol Technol. 2014;96:14–22. doi: 10.1016/j.postharvbio.2014.05.005. [DOI] [Google Scholar]
- 25.Li S, Assmann SM, Albert R. Predicting essential components of signal transduction networks: a dynamic model of guard cell abscisic acid signaling. PLoS Biol. 2006;4(10):e31. doi: 10.1371/journal.pbio.0040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 27.Zhang HN, Li WC, Wang HC, Shi SY, Bo S, Liu LQ, et al. Transcriptome profiling of light-regulated anthocyanin biosynthesis in the pericarp of litchi. Front Plant Sci. 2016;7:963. doi: 10.3389/fpls.2016.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai B, Hu B, Qin YH, Zhao JT, Wang HC, Hu GB. Transcriptomic analysis of Litchi chinensis pericarp during maturation with a focus on chlorophyll degradation and flavonoid biosynthesis. BMC Genomics. 2015;16(1):225. doi: 10.1186/s12864-015-1433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei Y, Dong C, Zhang H, Zheng X, Shu B, Shi S, et al. Transcriptional changes in litchi (Litchi chinensis Sonn.) inflorescences treated with uniconazole. PLoS ONE. 2017;12(4):e0176053. doi: 10.1371/journal.pone.0176053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li C, Wang Y, Huang X, Li J, Wang H, Li J. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front Plant Sci. 2015;6:439. doi: 10.3389/fpls.2015.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Wang Y, Ying P, Ma W, Li J. Genome-wide digital transcript analysis of putative fruitlet abscission related genes regulated by ethephon in litchi. Front Plant Sci. 2015;6:502. doi: 10.3389/fpls.2015.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yun Z, Qu H, Hui W, Feng Z, Zhang Z, Duan X, et al. Comparative transcriptome and metabolome provides new insights into the regulatory mechanisms of accelerated senescence in litchi fruit after cold storage. Sci Rep. 2016;6:19356. doi: 10.1038/srep19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong HY, Chen JW, Li CQ, Lei C, Wu JY, Chen JY, et al. Selection of reliable reference genes for expression studies by reverse transcription quantitative real-time PCR in litchi under different experimental conditions. Plant Cell Rep. 2011;30(4):641–653. doi: 10.1007/s00299-010-0992-8. [DOI] [PubMed] [Google Scholar]
- 35.Wei Y, Wan H, Wu Z, Wang R, Ruan M, Ye Q, et al. A comprehensive analysis of carotenoid cleavage dioxygenases genes in Solanum Lycopersicum. Plant Mol Biol Rep. 2016;34(2):512–523. doi: 10.1007/s11105-015-0943-1. [DOI] [Google Scholar]
- 36.Zhang XH, Liu HQ, Guo QW, Zheng CF, Li CS, Xiang XM, et al. Genome-wide identification, phylogenetic relationships, and expression analysis of the carotenoid cleavage oxygenase gene family in pepper. Genet Mol Res. 2016;15(4):gmr8695. 10.4238/gmr.15048695. [DOI] [PubMed]
- 37.Zhang S, Guo Y, Zhang Y, Guo J, Li K, Fu W, et al. Genome-wide identification, characterization and expression profiles of the CCD gene family in Gossypium species. 3 Biotech. 2021;11(5):249. doi: 10.1007/s13205-021-02805-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohmiya A. Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Tissue Cult Lett. 2009;26(4):351–358. [Google Scholar]
- 39.Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289(1):3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 40.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 41.Ma W, Chen C, Liu Y, Zeng M, Meyers BC, Li J, et al. Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol. 2018;217(4):1535–1550. doi: 10.1111/nph.14934. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi-Shinozaki K, Shinozaki K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005;10(2):88–94. doi: 10.1016/j.tplants.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Ilg A, Beyer P, Al-Babili S. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 2010;276(3):736–747. doi: 10.1111/j.1742-4658.2008.06820.x. [DOI] [PubMed] [Google Scholar]
- 44.Cheng C, Zhang L, Yang X, Zhong G. Profiling gene expression in citrus fruit calyx abscission zone (AZ-C) treated with ethylene. Mol Genet Genomics. 2015;290(5):1991–2006. doi: 10.1007/s00438-015-1054-2. [DOI] [PubMed] [Google Scholar]
- 45.Lee Y, Do VG, Kim S, Kweon H, McGhie TK. Cold stress triggers premature fruit abscission through ABA-dependent signal transduction in early developing apple. PLoS One. 2021;16(4):e0249975. [DOI] [PMC free article] [PubMed]
- 46.Zhao M, Li J. Molecular events involved in fruitlet abscission in litchi. Plants (Basel) 2020;9(2):151. doi: 10.3390/plants9020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang X, Subhadrabandhu S, Mitra SK, Ben-Arie R, Stern RA. Origin, history, production and processing. In: Menzel CM, Waite GK, editors. Litchi and Longan: botany, production and uses. Oxfordshire: CABI Publishing; 2005. pp. 1–24. [Google Scholar]
- 48.Wang LX, Zeng LP, Xin-Guo LI. Effects of different concentrations of uniconazole and ethrel on blossom of litchi ‘Feizixiao’. Chin Hortic Abs. 2012;28(08):5–6. [Google Scholar]
- 49.Weicai LI, Zhang H, Shi S, Liu L, Shu B, Liang Q, et al. Effects of S-3307 and GA_3 on fluorescence characteristics of litchi leaves during floral induction. Chin J Trop Crops. 2014;35(12):2414–2419. [Google Scholar]
- 50.Saito S, Okamoto M, Shinoda S, Kushiro T, Koshiba T, Kamiya Y, et al. A plant growth retardant, uniconazole, is a potent inhibitor of ABA catabolism in Arabidopsis. Biosci Biotechnol Biochem. 2006;70(7):1731–1739. doi: 10.1271/bbb.60077. [DOI] [PubMed] [Google Scholar]
- 51.Hai L, Song L, You Y, Li Y, Duan X, Jiang Y, et al. Cold storage duration affects litchi fruit quality, membrane permeability, enzyme activities and energy charge during shelf time at ambient temperature. Postharvest Biol Technol. 2011;60(1):24–30. doi: 10.1016/j.postharvbio.2010.11.008. [DOI] [Google Scholar]
- 52.Wang H, Qian Z, Ma S, Zhou Y, Patrick JW, Duan X, et al. Energy status of ripening and postharvest senescent fruit of litchi (Litchi chinensis Sonn.) BMC Plant Biol. 2013;13:55. doi: 10.1186/1471-2229-13-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kou X, He Y, Li Y, Chen X, Feng Y, Xue Z. Effect of abscisic acid (ABA) and chitosan/nano-silica/sodium alginate composite film on the color development and quality of postharvest Chinese winter jujube (Zizyphus jujuba Mill. cv. Dongzao) Food Chem. 2019;270:385–394. doi: 10.1016/j.foodchem.2018.06.151. [DOI] [PubMed] [Google Scholar]
- 54.Xiong T, Tan Q, Li S, Mazars C, Galaud J-P, Zhu X. Interactions between calcium and ABA signaling pathways in the regulation of fruit ripening. J Plant Physiol. 2021;256:153309. doi: 10.1016/j.jplph.2020.153309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplement Figure 1. The dynamic of carotenoid content during fruit maturation of litchi. A: ‘Feizixiao’ variety; B: ‘Nuomici’ variety. Different letters indicated statistically differences between days of after anthesis using one-way ANOVA with the SAS test (P <0.05). Supplement Table S1. Primers of selected LcCCO genes in litchi and reference genes. Supplement Table S2. Cis-acting element information in the promoter region of LcCCO genes in Litchi. Supplement Table S3. Two-dimensional structures of LcCCO proteins. Supplement Table S4. GO enrichment analysis of LcCCO genes. Supplement Table S5. The TPM value and differential expression analysis of LcCCO genes during pericarp coloring of ‘Feizixiao’ litchi treated by exogenous CPPU. Supplement Table S6. The TPM value and differential expression analysis of LcCCO genes of ‘Feizixiao’ litchi on the 0, 1, 3 and 7 days after bags removed (Zhang et al., 2016a). Supplement Table S7. The TPM value and differential expression analysis of LcCCO genes in ‘Nuomici’ Litchi during three different development stages of fruit (Lai et al., 2015). Supplement Table S8. The TPM value and differential expression analysis of LcCCO genes of the entire inflorescences samples of 'Feizixiao’ litchi on 28 days after the uniconazole treatment (Wei et al., 2017b). Supplement Table S9. The TPM value and differential expression analysis of LcCCO genes of fruit samples of ‘Wuye’ litchi after 2, 4 and 7 days treated by girdling plus defoliation(Li et al., 2015a). Supplement Table S10. The TPM value and differential expression analysis of LcCCO genes of abscission zone samples of ‘Feizixiao’litchi after 0, 1, 2, 3 days treated by exogenous ethephon (Li et al., 2015b). Supplement Table S11. The TPM value and differential expression analysis of LcCCO genes of the peel samples on 0d and 4d after stored at room temperature and 0h, 24h and 48h stored at room temperature after precooling for 14 days [32].
Data Availability Statement
The RNA-seq data are obtained from NCBI (https://www.ncbi.nlm.nih.gov/Traces/study/), and the accession numbers are SRA312830, SRP047115, SRP092890, SRA234477, SRP173341 and SRA247016. The COO protein sequences The CCO protein sequences of Arabidopsis thaliana were downloaded from TAIR database (https://www.arabidopsis.org/). The litchi genome data used in this study had been deposited into CNGB Sequence Archive (CNSA, https://db.cngb.org/cnsa/) of China National GeneBank DataBase (CNGBdb) with accession number CNP0001024, which will be released after the publication of the litchi genome paper. Public access to all databases is open. Other data supporting the results of this article are included within the article and its additional files.