ABSTRACT
At a time when antibiotic resistance is seemingly ubiquitous worldwide, understanding the mechanisms responsible for successful emergence of new resistance genes may provide insights into the persistence and pathways of dissemination for antibiotic-resistant organisms in general. For example, Escherichia coli strains harboring a class A β-lactamase-encoding gene (blaCTX-M-15) appear to be displacing strains that harbor a class C β-lactamase gene (blaCMY-2) in Washington State dairy cattle. We cloned these genes with native promoters into low-copy-number plasmids that were then transformed into isogenic strains of E. coli, and growth curves were generated for two commonly administered antibiotics (ampicillin and ceftiofur). Both strains met the definition of resistance for ampicillin (≥32 μg/mL) and ceftiofur (≥16 μg/mL). Growth of the CMY-2-producing strain was compromised at 1,000 μg/mL ampicillin, whereas the CTX-M-15-producing strain was not inhibited in the presence of 3,000 μg/mL ampicillin or with most concentrations of ceftiofur, although there were mixed outcomes with ceftiofur metabolites. Consequently, in the absence of competing genes, E. coli harboring either gene would experience a selective advantage if exposed to these antibiotics. Successful emergence of CTX-M-15-producing strains where CMY-2-producing strains are already established, however, requires high concentrations of antibiotics that can only be found in the urine of treated animals (e.g., >2,000 μg/mL for ampicillin, based on literature). This ex vivo selection pressure may be important for the emergence of new and more efficient antibiotic resistance genes and likely for persistence of antibiotic-resistant bacteria in food animal populations.
IMPORTANCE We studied the relative fitness benefits of a cephalosporin resistance enzyme (CTX-M-15) that is displacing a similar enzyme (CMY-2), which is extant in E. coli from dairy cattle in Washington State. In vitro experiments demonstrated that CTX-M-15 provides a significant fitness advantage, but only in the presence of very high concentrations of antibiotic that are only found when the antibiotic ampicillin, and to a lesser extent ceftiofur, is excreted in urine from treated animals. As such, the increasing prevalence of bacteria with blaCTX-M-15 is likely occurring ex vivo. Interventions should focus on controlling waste from treated animals and, when possible, selecting antibiotics that are less likely to impact the proximal environment of treated animals.
KEYWORDS: antibiotic, antimicrobial, resistance, β-lactamase, bla CMY-2 , bla CTX-M-15 , bla KPC-3 , competition, hydrolysis, antibiotic resistance, fitness
INTRODUCTION
The seemingly ubiquitous distribution of antibiotic-resistant bacteria across different host populations (humans, domestic animals, wildlife) and diverse environments presents a significant challenge to developing focused and effective mitigation measures (1–4). While antibiotic selection pressure is critical to the success of antibiotic-resistant bacteria, the extensive distribution of these organisms where selection pressure is unlikely to occur highlights the extent and apparent ease with which these organisms are disseminated. The widespread distribution of antibiotic resistance also presents a challenge when trying to identify the most important dissemination pathways, because it is difficult to infer directionality of movement after the fact.
One way to pierce this fog of ubiquity is to focus on the mechanisms by which novel antibiotic resistance genes emerge in new populations. Emergence occurs in two different contexts. In the first, a resistance gene enters a population where there are no other genes conferring similar resistance. When bacteria harboring the new gene are exposed to a selective antibiotic with sufficient concentration and frequency, the novel gene is likely to rapidly spread among proximal hosts and adjacent populations. The recent emergence of mcr-1, presumably aided by wide-scale selection from the use of colistin in food animal populations (5, 6), may be a recent example of such as scenario.
Another scenario involves emergence of novel resistance genes in populations where similar resistance genes are already present. For example, the rapid worldwide expansion of the bacteria harboring the cephalosporin resistance gene blaCTX-M (encoding a β-lactamase) occurred in the face of competition against bacteria that produce similar but distinct β-lactamase enzymes (e.g., CMY-2) (7). We surmise that successful emergence events in the face of competition offer unique opportunities to identify key factors that are necessary for the success of novel genes but that are also likely to be important to dissemination and persistence of antibiotic resistance in general. That is, studying such events is likely to provide insight into how to mitigate antibiotic resistance beyond what can be learned by simply documenting the presence and abundance of antibiotic resistance genes using cross-sectional surveillance studies.
An example of emergence in the presence of extant antibiotic resistance genes is under way for Escherichia coli found in dairy cattle located in Washington State (USA), where the primary antibiotic selection pressure is limited to an aminopenicillin (ampicillin) and a third-generation cephalosporin (ceftiofur) (8). Before 2009, a class C β-lactamase (CMY-2, encoded by blaCMY-2) was present in 96.9% of ceftiofur-resistant E. coli isolates that were collected from dairy herds in Washington State. CTX-M is a class A β-lactamase that was not documented until 2011, when the prevalence was 14.9% for E. coli. When the investigators sequenced 99 blaCTX-M-positive isolates from this study, the most prevalent genes were blaCTX-M-15 (50.5%), blaCTX-M-27 (26.3%), blaCTX-M-14 (16.2%), and blaCTX-M-65 (5.1%) (9). Another study of seven commercial dairies in Washington State in 2015 found that 38.1% of cattle fecal samples were PCR positive for blaCMY-2 and 26.2% were positive for blaCTX-M (10). Because both enzymes (CMY-2 and CTX-M) convey resistance to third-generation cephalosporins, the change in prevalence is consistent with CTX-M becoming more prevalent because it provides a greater benefit or reduced fitness cost relative to CMY-2.
The goal of this project was to identify the factors that are likely to contribute to the emergence of E. coli strains that harbor blaCTX-M when other strains with blaCMY-2 are present in the population. For this work, we selected blaCTX-M-15 as the representative gene, and we included carbapenem resistance (KPC-3, encoded by blaKPC-3) to provide an additional basis for comparison, but also to ask if such a gene could be competitive in a host population where carbapenem antibiotics are not used (11). To address these questions, we compared fitness costs and benefits using cloned genes under the control of native promoters that were introduced into isogenic strains of E. coli for growth curve comparisons. We also assessed the kinetics of protein synthesis and the efficiency of enzymatic hydrolysis of ampicillin, ceftiofur, and ceftiofur metabolites (desfuroylceftiofur [DFC], DFC-cysteine, and DFC-dimer) (12).
RESULTS
The outcome of our experiments could vary if there are different variants of promoter regions for the genes that we cloned. To minimize the chances of interference with the promoter region, we cloned the entire leader sequence for each gene, and we included the insertion sequences upstream from the leader sequences that encompass the promoter region (accession numbers ON412782, ON412783, and ON412784). To evaluate global variation in these leader sequences, we compared the leader sequences and 50 bases of enzyme coding region against the National Center for Biotechnology Information (NCBI) GenBank database (13). We restricted these queries to E. coli only, with 100% coverage. For blaCTX-M-15, blaCMY-2, and blaKPC-3, this resulted in sequences of 305, 373, and 232 bp, respectively. Using default parameters, identical matches were evident for 84.7% (848/1,001) of blaCTX-M-15 sequences, for 91% (374/411) of blaCMY-2 sequences, and for 95.5% (21/22) of blaKPC-3 sequences (accessed 5 May 2022). For all comparisons, fewer than 0.6% of sequences retrieved by the BLASTN algorithm matched by less than 98%. Consequently, there is some variation in the leader sequences of these genes, but the variants that we cloned are representative of the majority of what has been documented in GenBank.
Relative growth advantage in the absence of antibiotic.
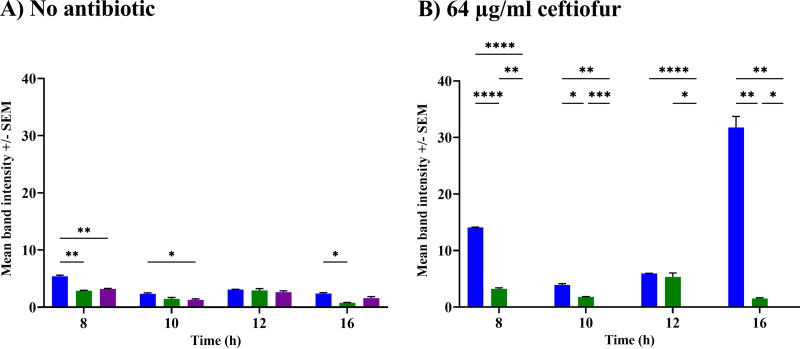
For culture experiments in the presence or absence of antibiotics, we used cloned genes under the control of native promoters that encoded tag-free enzymes (i.e., no His or FLAG epitopes). We pooled results from 15 growth curve control experiments (no antibiotic) (see Fig. S1 in the supplemental material) and found no significant differences between strains harboring blaCTX-M-15 (with an area under the curve [AUC] of 4.74) and blaCMY-2 (AUC, 4.73; P = 0.9) and between the strain harboring blaKPC-3 (AUC, 5.17) and the strain with the pMMB207ΔblaTEM-1 control plasmid (AUC, 5.2; P = 0.15). Strains with blaKPC-3 or a control plasmid both grew significantly better than the strains with blaCTX-M-15 or blaCMY-2 (P < 0.0001), consistent with a lower fitness cost associated with carriage of blaKPC-3 compared to that for the other two genes in the absence of antibiotic selection. To determine if differences in protein synthesis could explain the relative growth advantage of the blaKPC-3-expressing strain, we constructed additional blaCTX-M-15, blaCMY-2, and blaKPC-3 strains that included a common epitope (FLAG) for detection by Western blotting. In the absence of antibiotic, all three enzymes were synthesized constitutively, with small differences depending on the clone and time point (Fig. 1A).
FIG 1.
Relative protein production of CMY-2 (blue), CTX-M-15 (green) and KPC-3 (purple) by isogenic strains of E. coli with or without exposure to ceftiofur. Average normalized densitometry values (3 independent replicates; ± SEM) for Flag-tagged recombinant CMY-2, CTX-M, and KPC-3 after 8, 10, 12, 14, or 16 h of culture with no antibiotic (A) or with 64 μg/mL of ceftiofur (B). Significant differences were found, based on two-way ANOVAs and Tukey multiple-comparison test: *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.0001.
Relative growth advantage in the presence of antibiotics.
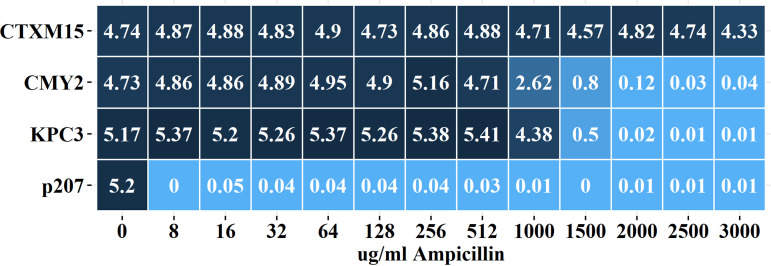
When cultured with ampicillin, all three enzymes were protective against ampicillin at a concentration of 512 μg/mL (AUC, >4.7) (Fig. 2) with growth from both CMY-2- and KPC-3-producing strains decreasing in the presence of 1,000 μg/mL ampicillin (AUC, 2.62 and 4.38, respectively) and mostly unable to grow in the presence of 1,500 μg/mL (AUC, ≤0.8). In contrast, the CTX-M-15-producing strain exhibited robust growth (AUC, >4.3) in the presence of the highest concentration tested (3,000 μg/mL) (Fig. 2; see also Fig. S2).
FIG 2.
Average area under the curve (AUC) values for E. coli cultures exposed to ampicillin. Heatmap depicts average AUC for growth curves from CTX-M-15-, CMY-2-, and KPC-3-producing strains of E. coli and for the plasmid-only negative-control strain (p207) in the presence of no antibiotic (3 independent replicates) or 8 to 3,000 μg/mL (3 independent replicates). See Fig. S2 in the supplemental material for original growth curves and P values for multiple comparison tests.
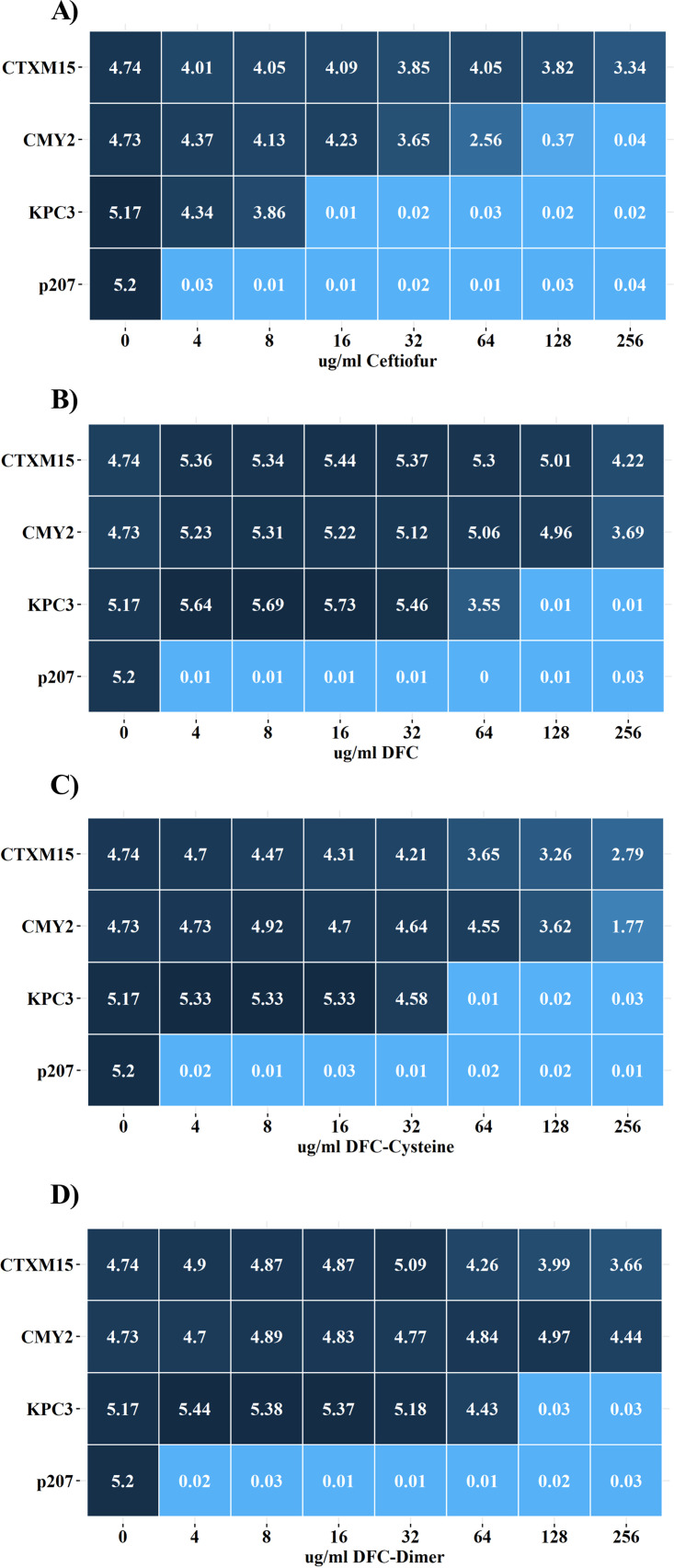
When cultured with ceftiofur, strains producing CMY-2 or KPC-3 were severely compromised (AUC, 2.56 and 0.01) in the presence of 64 or 16 μg/mL of ceftiofur, respectively (Fig. 3A; see also Fig. S3). In contrast, the CTX-M-15-producing strain was only partially compromised (AUC, >3.3) with 256 μg/mL ceftiofur (Fig. 3A). Once administered, ceftiofur is rapidly metabolized into desfuroylceftiofur (DFC), DFC-cysteine, and DFC-dimer, and these compounds are the ones the bacteria are more likely to encounter (12, 14, 15). When cultured with DFC, CTX-M-15- and CMY-2-producing strains exhibited similar AUC values for concentrations up to 128 μg/mL, but the CTX-M-15 strains grew better at 256 μg/mL (AUC, 3.69 and 4.22, respectively) (Fig. 3B; see also Fig. S4). The blaKPC-3 strain had some difficulty growing with 64 μg/mL DFC (AUC, 3.55) and it was not able to grow at higher concentrations. Consequently, the CTX-M-15 enzyme conferred greater fitness at the highest concentration of DFC. In the presence of 64 μg/mL, the CMY-2 protein was significantly more abundant than CTX-M-15 at 8 and 16 h (Fig. 1B), consistent with the CTX-M-15 enzyme conferring greater fitness at the highest concentration of DFC.
FIG 3.
Average area under the curve (AUC) values for E. coli cultures exposed to ceftiofur and its metabolites. Heatmap depicts average AUC for growth curves from CTX-M-15-, CMY-2-, and KPC-3-producing strains of E. coli and for the plasmid-only negative-control strain (p207) in the presence of 0 to 256 μg/mL ceftiofur (A), DFC (B), DFC-cysteine (C), or DFC-dimer (D). Values represent averages (3 independent replicates). See Fig. S3 to S6 in the supplemental material for original growth curves and P values for multiple-comparison tests.
When cultured with DFC-cysteine, growth of the KPC-3-producing strain was equivalent to or better than the that of other strains with as much as 32 μg/mL but did not grow with 64 μg/mL (Fig. 3C; see also Fig. S5). The CMY-2-producing strain grew better than the CTX-M-15-producing strain in the presence of 8 to 128 μg/mL DFC-cysteine (Fig. 3C), but when cultured in 256 μg/mL DFC-cysteine, the CMY-2-producing strain exhibited a highly repeatable bimodal peak (see Fig. S5), with overall growth compromised compared with the CTX-M-15-producing strain (Fig. 3C). Consequently, the CTX-M-15 enzyme conferred greater fitness at the highest concentration of DFC-cysteine.
When cultured in concentrations up to 32 μg/mL, all three strains grew similarly in the presence of DFC-dimer (Fig. 3D; see also Fig. S6). The KPC-3-producing strain was able to grow in up to 64 μg/mL but with a noticeable decay in growth after 10 h (see Fig. S6). Between 64 and 256 μg/mL, the CMY-2-producing strain grew better than the CTX-M-15-producing strain (Fig. 3D). Thus, the CMY-2 enzyme has greater fitness at the highest concentration of DFC-dimer. The control strain (DH10B/pMMB207ΔblaTEM-1, labeled p207) did not grow with the lowest tested concentrations of ampicillin (8 μg/mL), ceftiofur (4 μg/mL), DFC (4 μg/mL), DFC-cysteine (4 μg/mL), or DFC-dimer (4 μg/mL) (Fig. 2; see also Fig. S2 to S6).
Antibiotic hydrolysis and degradation kinetics.
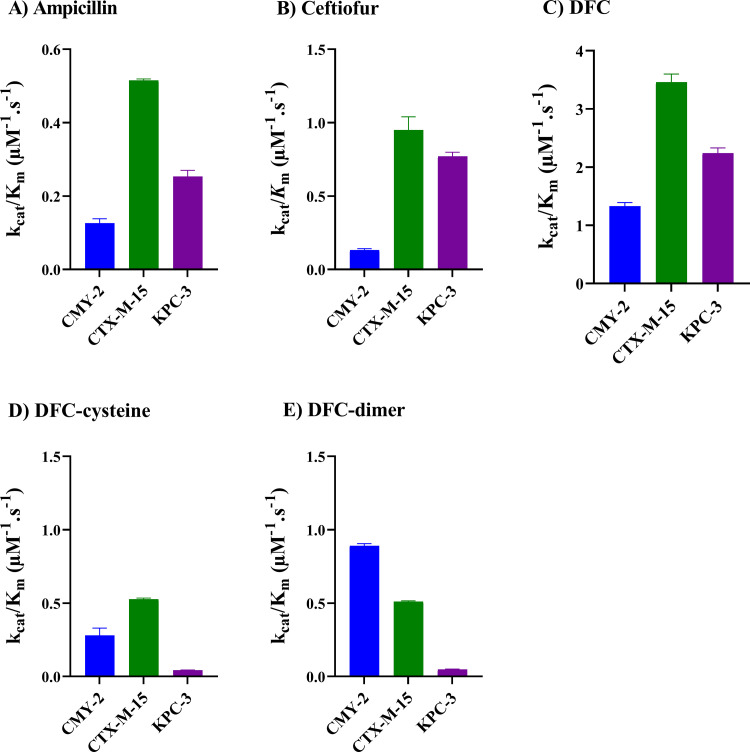
Using high-performance liquid chromatography (HPLC)-purified, tag-free recombinant proteins (CTX-M-15, KPC-3, and CMY-2) (see Fig. S7), we estimated the efficiency of antibiotic hydrolysis with a Michaelis-Menten model (16–18). All three enzymes were capable of hydrolyzing ceftiofur, DFC, DFC-cysteine, DFC-dimer, and ampicillin (see Fig. S8). The rank order for kcat/Km (catalytic efficiency) for ampicillin was CMY-2 > CTX-M-15 > KPC-3; for ceftiofur and DFC the order was CTX-M-15 > KPC-3 > CMY-2; for DFC-cysteine the order was CTX-M-15 > CMY-2 > KPC-3; and for DFC-dimer the order was CMY-2 > CTX-M-15 > KPC-3 (Fig. 4 and Table 1).
FIG 4.
Average kcat/Km values for hydrolysis of antibiotics. Values were estimated for hydrolysis of ampicillin (A), ceftiofur (B), DFC (C), DFC-cysteine (D), and DFC-dimer (E) using recombinant CMY-2, CTX-M-15, and KPC-3 enzymes. See Materials and Methods for additional details and Table 1 and Fig. S7 and S8 for supporting information.
TABLE 1.
Steady-state kinetics of hydrolysis for ceftiofur, DFC, DFC-cysteine, DFC-dimer, and ampicillin by recombinant CMY-2, CTX-M-15, and KPC-3a
| Substrate | β-Lactamase | Vmax (μmol/s) | Km (μM) | Kcat (s−1) | Kcat/Km (μM−1 s−1) |
|---|---|---|---|---|---|
| Ceftiofur | CMY-2 | 1.31 ± 0.03 | 994.1 ± 34.8 | 131.3 ± 9.4 | 0.132 ± 0.01 |
| CTX-M-15b | 3.14 ± 0.07 | 330.0 ± 14.1 | 313.7 ± 13.0 | 0.95 ± 0.09 | |
| KPC-3 | 1.92 ± 0.07 | 249.3 ± 20.7 | 192.6 ± 6.1 | 0.77 ± 0.029 | |
| DFC | CMY-2 | 1.80 ± 0.1 | 134.9 ± 7.8 | 180.0 ± 5.4 | 1.33 ± 0.06 |
| CTX-M-15b | 4.89 ± 0.2 | 141.2 ± 11.2 | 489.8 ± 16.2 | 3.46 ± 0.14 | |
| KPC-3 | 2.40 ± 0.03 | 107.1 ± 9.0 | 240.7 ± 8.2 | 2.24 ± 0.09 | |
| DFC-cysteine | CMY-2 | 0.124 ± 0.0 | 44.49 ± 2.0 | 12.4 ± 0.1 | 0.28 ± 0.05 |
| CTX-M-15 | 0.58 ± 0.03 | 111.2 ± 5.9 | 58.66 ± 4.5 | 0.527 ± 0.007 | |
| KPC-3 | 0.114 ± 0.01 | 260.5 ± 7.4 | 11.43 ± 0.7 | 0.043 ± 0.001 | |
| DFC-dimer | CMY-2 | 1.04 ± 0.0 | 116.8 ± 6.4 | 104.1 ± 9.0 | 0.891 ± 0.014 |
| CTX-M-15 | 1.17 ± 0.0 | 227.8 ± 11.6 | 117.2 ± 7.6 | 0.51 ± 0.006 | |
| KPC-3 | 0.454 ± 0.0 | 933.6 ± 55.2 | 45.42 ± 0.2 | 0.048 ± 0.003 | |
| Ampicillin | CMY-2 | 0.48 ± 0.01 | 387.4 ± 1.9 | 48.84 ± 2.3 | 0.126 ± 0.012 |
| CTX-M-15b | 1.01 ± 0.06 | 197.8 ± 13.0 | 101.9 ± 5.6 | 0.515 ± 0.004 | |
| KPC-3 | 0.72 ± 0.0 | 286.1 ± 13.8 | 72.36 ± 24.3 | 0.253 ± 0.017 |
All values are the averages of three independent replicates ± SEM.
Parameter estimates for CTX-M-15 were published by Ahmadvand et al. (48).
Based on the rank order for kcat/Km, the KPC-3-producing strain should grow better than the CMY-2-producing strain at higher concentrations of ceftiofur, but the observed order was reversed (Fig. 3A; see also Fig. S3). To determine if the inconsistency between the rank order for kcat/Km and growth curves was due to protein instability, we cultured isogenic strains that produced the beta-lactamase enzymes with a FLAG tag for detection by Western blotting. Cultures were incubated for 12 h before addition of kanamycin to stop protein synthesis. Cell pellets were prepared at different time points, and the abundance of protein, estimated by Western blot analysis, showed no difference in protein decay for CTX-M-15 (slope = −0.17, standard error [SE] = 0.02, n = 7) and CMY-2 (slope = −0.22, SE = 0.03, n = 8) (P = 0.2), while an approximate 5-fold difference was detected for degradation of KPC-3 (slope = −1.01, SE = 0.14, n = 8) (P < 0.0001).
Coculture competition assay.
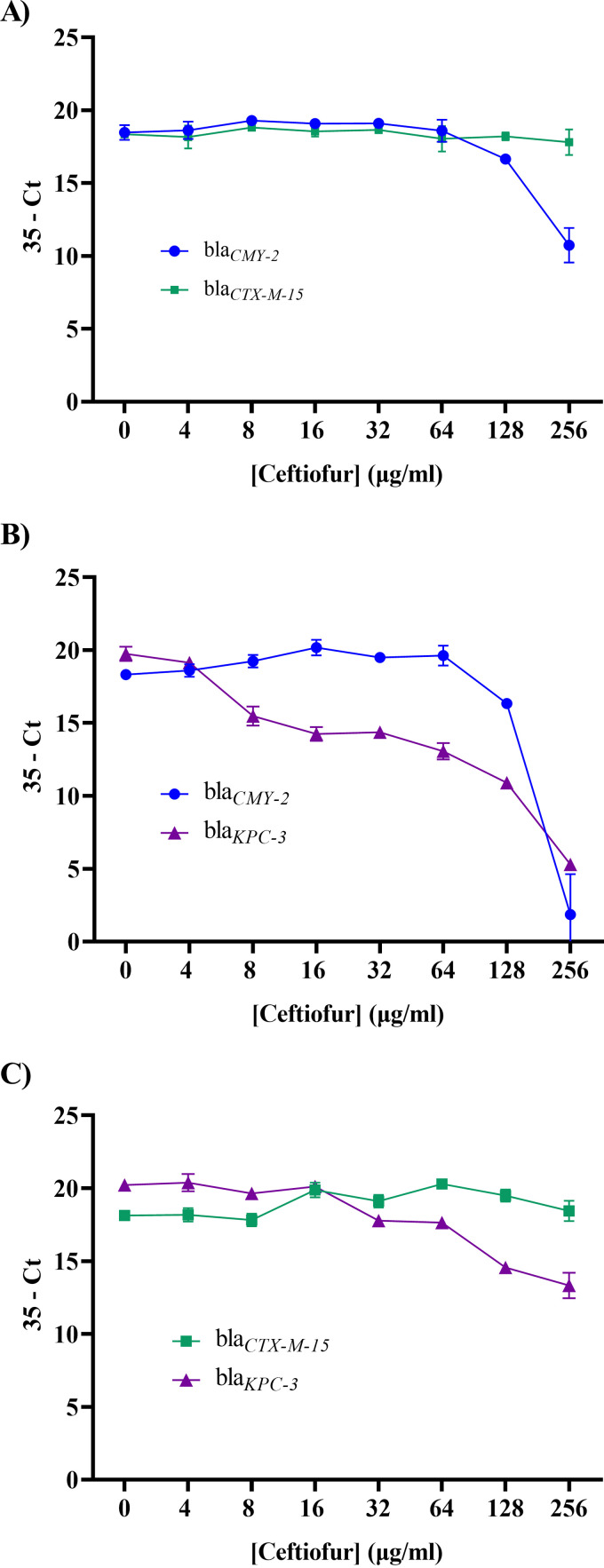
Coculture competition experiments are potentially problematic due to the presence of more than one extracellular β-lactamase enzyme confounding the findings. Nevertheless, it is possible that monocultures are not representative of competition outcomes either. To validate fitness conclusions from monoculture growth curves, we quantified gene copy number after pairwise coculture of the three isogenic strains with ceftiofur (the cycle threshold [CT] values are presented as [35 − estimated CT] for ease of interpretation). When the CTX-M-15-producing and CMY-2-producing strains were cultured together, the copy number for blaCMY-2 decreased at 128 and 256 μg/mL (Fig. 5A), consistent with reduced monoculture growth at higher concentrations of ceftiofur (Fig. 3; see also Fig. S3).
FIG 5.
Real-time PCR results for coculture competition assays. (A) Coculture of CMY-2- and CTX-M-15-producing strains. (B) Coculture of CMY-2- and KPC-3-producing strains. (C) Coculture of CTX-M-15- and KPC-3-producing strains. Each time point is the average ± SEM for three independent replicates.
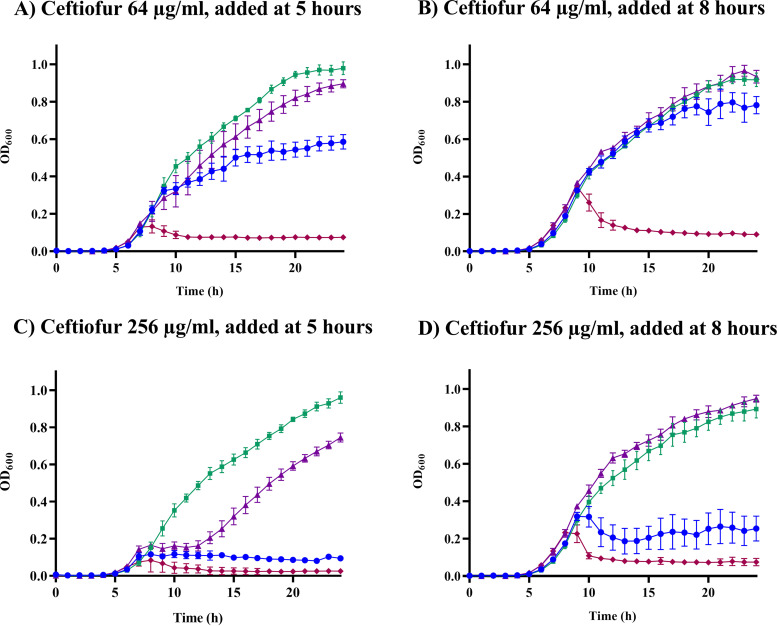
When the KPC-3-producing strain was cultured with the CMY-2-producing strain, the blaKPC-3 copy number decreased (Fig. 5B), but not as rapidly as might be expected given that the KPC-3-producing strain cannot grow as a monoculture in ≥16 μg/mL ceftiofur (Fig. 3; see also Fig. S3). The persistence of blaKPC-3 was more evident when this strain was cocultured with the CTX-M-15-producing strain (Fig. 5C). One potential explanation for the inconsistencies between monoculture growth curves and coculture quantitative PCR (qPCR) data is a spillover benefit from the accumulation of enzyme in the periplasmic space and the presence of extracellular CMY-2 or CTX-M-15 in the culture medium. To test this idea, monocultures of each strain were incubated for 5 or 8 h before the addition of ceftiofur. Later addition of 64 μg/mL ceftiofur allowed all three strains to grow robustly (Fig. 6A; see also Fig. S9), consistent with accumulation of increasing concentrations of bioavailable enzyme in the periplasmic space and culture medium prior to addition of antibiotic. Notably, unlike the CMY-2-producing strain, delaying the addition of 256 μg/mL ceftiofur by 8 h completely rescued the KPC-3-producing strain (Fig. 6B; see also Fig. S9).
FIG 6.
E. coli cultures with ceftiofur (64 or 256 μg/mL) added at 5 or 8 h postinoculation. Cultures were initiated with no antibiotic for the first 5 h (A and C) or for the first 8 h (B and D), before adding 64 μg/mL ceftiofur (A and B) or 256 μg/mL ceftiofur (C and D). Each time point is the average ± SEM for three independent replicates. See Fig. S9 in the supplemental material for additional results.
DISCUSSION
The recent shift in prevalence of blaCTX-M-15-positive strains of E. coli, compared to blaCMY-2-positive strains, found in dairy cattle in Washington State (8) offers an opportunity to explore the mechanisms of emerging antibiotic resistance. For a new antibiotic resistance gene to emerge in a population, it must offer a significant fitness advantage for the host bacteria compared to competing bacteria. Relative to conspecific bacteria, this advantage could be evident through (i) reduced fitness costs to the host bacteria in the absence of antibiotic exposure and/or (ii) increased fitness benefits for the host bacteria in the presence of antibiotics (19, 20). Importantly, if the only competing bacteria in a population are susceptible to an antibiotic that is being used in that population, then exposure to relatively low concentrations of the antibiotic may favor emergence of resistant strains. For this study, however, we focused on the conditions that are necessary for the emergence of a new antibiotic resistance trait in the face of competition with bacteria that harbor similar resistance traits (e.g., blaCTX-M-15 and blaCMY-2).
Fitness in the absence of antibiotics.
We found that all three enzymes are constitutively expressed in isogenic strains of E. coli when under the control of native promoters. Our host strain, E. coli DH10B, is closely related to wild-type strain E. coli MG1655 (21), which is known to harbor a rich array of transcription factors (22), one or more of which is presumably responsible for this constitutive expression. Without antibiotics, the average areas under the curve (AUC) for the KPC-3-producing strain were indistinguishable from those of the control strain that harbored a cloning plasmid that lacked a gene insert (AUC, 5.17 and 5.2, respectively) (see Fig. S1 in the supplemental material). The AUC was also indistinguishable between the CTX-M-15- and CMY-2-producing strains (AUC, 4.74 and 4.73, respectively), but was lower than the control strain AUC. Consequently, there is some fitness cost associated with production of CTX-M-15 and CMY-2, but this cost does not distinguish these two clones.
While these results suggest that the fitness cost in the absence of antibiotic selection pressure is not important for the increasing prevalence of E. coli strains that harbor blaCTX-M-15, there are caveats to this conclusion. All three genes considered in this study are found primarily on plasmids (23). As such, these genes are genetically linked to other traits that may add additional fitness costs or benefits, depending on the environmental context. Similar tradeoffs may occur when more than one plasmid is present. For example, Subbiah et al. (24) found a significant fitness cost for a blaCMY-2-bearing plasmid (peH4) by comparing growth of strain DH10B with and without this plasmid. This fitness cost, however, was largely ablated when a second plasmid (pTmpR) was simultaneously present in the cell. Others have shown the compensatory mutations between plasmids and hosts can ameliorate fitness costs (25). Nevertheless, while it is important that the host, plasmid, and cytosolic plasmid population can alter the fitness outcome for strains that harbor the genes studied herein, we expect that over time the predominate strains will converge on combinations of traits that, at a minimum, reduce fitness costs to a low level.
Fitness in the presence of antibiotics.
The aminopenicillin ampicillin (available since 1961) and the third-generation cephalosporin ceftiofur (available since 1988) are two of the most used β-lactam antibiotics on Washington State dairies (8). Penicillin is also widely used, but it can have 10-fold lower activity against E. coli compared with ampicillin (26) and therefore was not included in this study. Ampicillin is an injectable antibiotic that is used to treat metritis and pneumonia in cattle, and it is mostly excreted unchanged. The National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS) lists the MIC for resistant E. coli as ≥32 μg/mL (27), and one study showed that following intravenous (i.v.) administration (10 mg/kg of body weight), the maximum serum concentrations in calves was 59 μg/mL and the average concentration in urine was 2,132 μg/mL after 15 min (28). The catalytic efficiencies (kcat/Km) of the three β-lactamase enzymes studied here were modest (Fig. 4A; see also Table S1 in the supplemental material), but all three strains of E. coli were able to grow equally well with little inhibition in medium containing up to 512 μg/mL ampicillin. Importantly, between 1,000 and 3,000 μg/mL ampicillin, the CTX-M-15-producing strain was mostly uninhibited, whereas the other two strains had difficulty growing with 1,000 μg/mL, with little to no growth with higher tested concentrations (Fig. 2; see also Fig. S2). If ampicillin is important for the increasing prevalence of CTX-M-15-producing strains, this will only happen if the bacteria encounter sufficiently high concentrations of antibiotic, and this only appears feasible when the antibiotic is concentrated by the kidney and excreted in urine (28).
Ceftiofur is used to treat pneumonia, metritis, and foot rot in dairy cattle (12, 29–33). After administration, ceftiofur undergoes rapid conversion into desfuroylceftiofur (DFC), which then undergoes reversible conversion primarily into DFC-cysteine and also DFC-dimer (12, 14, 15). The NARMS MIC for ceftiofur-resistant strains of Enterobacteriaceae is ≥8 μg/mL (27), and the maximum serum and urine concentrations of ceftiofur compounds range from approximately 10 to 20 μg/mL and up to 330 μg/mL, respectively (34). The catalytic efficiency for hydrolysis varies depending on the compound, with monoculture growth curves reflecting the observed higher kcat/Km for CTX-M-15 when the substrate is ceftiofur, DFC, or DFC-cysteine (Fig. 4). CMY-2 exhibited a higher kcat/Km for DFC-dimer. For both CMY-2 and CTX-M-15 enzymes, the differences in growth curves occur with concentrations that are more than what would be encountered in serum but that are well within the expected concentrations of these compounds in urine. The exact effect may depend on the ratio of the different ceftiofur compounds and the mode of administration (i.v., intramuscular, or subcutaneous). Furthermore, ancillary data provided by Hornish and Kotarski (12) suggest that the ratio of DFC-dimer to DFC-cysteine can vary from 1:1.2 to 1:2, depending on the formulation of ceftiofur that is administered, but more data are needed to estimate the variance for these ratios.
Our kcat/Km findings for KPC-3 reflected the relative advantage of this enzyme compared to CMY-2 for ampicillin, ceftiofur, and DFC. While this was reflected by growth curves in the presence of 1,000 μg/mL ampicillin (Fig. 2; see also Fig. S2), the KPC-3-producing strain fared poorly at higher concentrations of ceftiofur and DFC. One potential explanation for this discrepancy is that the protein degrades nearly 5 times faster than the other two enzymes. Interestingly, qPCR data from coculture experiments with ceftiofur suggest that the KPC-3-producing strain is retained in the population longer than expected from the monoculture experiments (Fig. 5). This is likely attributable to the presence of extracellular CTX-M-15 or CMY-2 enzyme, which essentially rescues the KPC-3 by hydrolyzing antibiotic until the remaining concentration no longer overwhelms the KPC-3-producing strain. This effect of β-lactamase enzymes in culture supernatant is a well-known in vitro phenomenon that may function in vivo as well (35), although this combinatorial effect was curiously absent when the CTX-M-15-producing and CMY-2-producing strains were cocultured.
Relevance of in vitro findings to farm environments.
β-Lactams are mostly concentrated and eliminated by glomerular filtration, tubular secretion, or both (36, 37), and according to the estimates from the data presented herein, the published concentration of ampicillin in urine collected from only a few hours after antibiotic administration is well within the range that would favor CTX-M-15-producing strains over both CMY-2- and KPC-3 producing strains (>2,000 μg/mL) (28). While there are potential caveats with respect to the relative concentration of DFC-dimer (see above), a similar CTX-M-15 advantage may arise from ceftiofur and its metabolites in urine. Of course, the highest concentrations will be restricted to the first urinary events following antibiotic treatment, and depending on the rate of excretion, this may taper off quickly with time. Nevertheless, there remains a possibility that at least some portion of the environment proximal to the treated animals will be exposed to high concentrations of antibiotic residues that could selectively favor CTX-M-15-producing strains over CMY-2-producing strains, but only if certain conditions are met.
First, excreted antibiotics need to be bioavailable and to persist long enough to exert a change in the abundance of relevant antibiotic-resistant bacteria. To determine if antibiotics remain bioavailable in different soil types, Subbiah et al. (38) used in vitro experiments to assess the fate of a panel of antibiotics with different soil slurries. β-Lactam antibiotics (ampicillin, cephalothin, cefoxitin, and ceftiofur) were retained in the supernatant and remained bioavailable while, depending on the soil type, other antibiotics (e.g., oxytetracycline, ciprofloxacin, and neomycin) were removed from the aqueous phase. Others have shown that β-lactam antibiotics remain bioavailable in soil (38–40). Furthermore, Subbiah et al. (39) showed that a modest concentration of ceftiofur metabolites (~13 ppm) in urine remains bioavailable in a mixture of soil and calf feces for an average 2.7 days at 23°C and 23.3 days at 4°C.
To extend these findings to in vivo settings, Liu et al. (41) treated calves with ceftiofur and quantified changes in the abundance of resistant E. coli in both feces and pen soil. Treatment produced a 3-log10/g increase in the abundance of resistant bacteria within 10 days. This tapered off quickly in fresh calf feces, while population numbers remained high (~4.5 log10/g) in the soil for the remaining 20-day monitoring period. The investigators also showed that the urine effect on soil populations was approximately 10 times greater than the influence from feces of treated animals. Those authors further showed that the abundance of bacteria needed to colonize untreated calves with a 50% probability (i.e., ID50) was only 2.83 log10/g, an abundance well below the expected population density of resistant strains after the proximal environment is exposed to urine from treated calves. Finally, quantification of ceftiofur-resistant E. coli on 14 dairy farms in Washington State found a higher abundance of these organisms in calf pens and hospital pens, where antibiotic exposures are more likely compared to healthy adult animal housing pens (10).
Consequently, the conditions necessary for success of CTX-M-15-producing strains can be met, including high concentrations of antibiotic in urine, a significant effect of this urine on the abundance of resistant bacteria in the environment, and a relatively low threshold of abundance needed to colonize new hosts. We do not have complementary data from excretion of ampicillin, but given that it remains bioavailable in soil (38), it is conceivable that the outcome is analogous. Our inferences would benefit from more information about the concentration and composition of antibiotic residues in urine following administration of ampicillin and ceftiofur. While we did not include penicillin in our analysis because E. coli is innately resistant to therapeutic concentrations of penicillin, it is possible that penicillin excreted in urine occurs at a concentration sufficiently high to selectively favor strains of E. coli that produce β-lactamase enzymes. Finally, while we focused on CTX-M-15 for this analysis, it is possible that other CTX-M β-lactamases from our study population (9) convey different levels of fitness costs and benefits.
Despite these limitations and given the totality of our findings, the most parsimonious inference from this work is that there is a high probability that using ampicillin or ceftiofur selectively favors resistant bacteria in cattle and that high concentrations of antibiotic residues found in urine could be responsible for the growing prevalence of CTX-M-15-producing strains in dairy cattle populations. β-Lactams are used in other food animals (e.g., beef cattle, swine, poultry), and thus these findings may be relevant to other production environments.
Collectively, these findings also point to the importance of considering how to control the excreted antibiotics in production environments. For example, where economically feasible, treated animals should be isolated for a “washout” period (e.g., 10 days), and contaminated bedding and soil could be removed and treated separately from other farm waste. Composting waste, when done correctly, will destroy all mesophilic bacteria like E. coli and will likely destroy or at least render most antibiotics unavailable (42). It might also be possible to put additives in the housing of treated animals, such as biochar (43), to help sequester antibiotic residues. Antibiotic use practices could be adopted, when possible, to avoid using β-lactam antibiotics when suitable alternatives will work (44). As an example, injectable oxytetracycline is less likely to have an impact on the environment compared to ceftiofur (41). Mitigation of poultry may be more challenging than cattle in some regards. For cattle, the highest concentrations of β-lactam antibiotics are found in urea-based urine that impacts bacteria in the proximal environment. Depending on the temperature and time of year, these impacts may be ameliorated by unfavorable conditions for bacteria to proliferate (39). For poultry, however, antibiotics concentrated in the kidney are excreted with uric acid-based urine directly into the gastrointestinal tract, where bacteria reside at a near-ideal body temperature (39°C), and thus selection and proliferation of resistant bacteria may be more pronounced in poultry on a year-round basis.
Combining the in vitro results with previous published work in vivo supports the premise that high concentrations of antibiotics in urine are an important factor in the emergence of CTX-M-15-producing strains in cattle populations. All things being equal, however, this is probably the last wave of emerging β-lactamase genes that will colonize dairy populations. Our rationale for this conclusion is that emerging genes need to provide a significant fitness advantage to gain prevalence in the population. If CTX-M-15-producing strains are already capable of protecting bacteria from the highest expected concentrations of ampicillin and ceftiofur in these environments, then this is equivalent to an asymptotic function and there is little opportunity for bacteria with even more efficient enzymes to outcompete strains that produce CTX-M-15. Of course, widespread changes in practices on farms and coselection of different combinations of traits may render our prediction invalid.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains (see Table S1 in the supplemental material) were grown in Luria Bertani-Lennox (LB broth) medium (Difco Laboratories, Inc.) at 37°C with continuous shaking at 200 rpm and on LB agar plates at 37°C. Antibiotics were added to media as needed (carbenicillin [Carb], 100 μg/mL; ampicillin [Amp], 100 μg/mL; kanamycin [Kan], 100 μg/mL; chloramphenicol [Cm], 30 μg/mL; streptomycin [Str], 100 μg/mL). Unless otherwise noted, reagents and antibiotics were purchased from Sigma-Aldrich, Inc., and VWR International, respectively.
Deletion of blaTEM-1 from pMMB207 to make pMMB207ΔblaTEM-1.
We generated a pMMB207 variant by disrupting the native blaTEM-1 gene using inverse PCR, leaving the chloramphenicol gene (cmr) available for selection (designated pMMB207ΔblaTEM-1). Briefly, tail-to-tail primers (see Table S2) were used with inverse PCR to amplify the entire plasmid except the blaTEM-1 sequence and its promoter region. Q5 High-Fidelity DNA polymerase (New England Biolabs) was used for vector amplification, and PCR conditions included 98°C for 2 min and then 98°C (30 s), 60°C (25 s), and 72°C (360 s) for 35 cycles, with final extension at 72°C for 5 min. Product was cleaned using QIAquick PCR purification kit (Qiagen). Cleaned PCR product was then transformed into chemically competent, streptomycin-resistant E. coli DH10B per the manufacturer’s protocol via a 30-s heat shock using a 42°C water bath. Transformants were plated onto LB agar containing chloramphenicol (30 μg/mL) and streptomycin (100 μg/mL) for selection. Isolated clones were streaked onto LB agar containing only carbenicillin (100 μg/mL) as a negative control. Gene deletion was confirmed by PCR and sequencing (Eurofins Genomics) using primers p207_Tem-1_F and p207_Tem-1_R (see Table S2).
Cloning of blaCTX-M-15, blaCMY-2, and blaKPC-3.
Three strains of bacteria, blaCTX-M-15-positive AR-0044 (E. coli) (45), blaCMY-2-positive AR-0081 (Klebsiella pneumoniae) (45), and blaKPC-3-positive AR-0114 (E. coli) (46) were used for PCR templates (see Table S1). Genomic DNA was extracted from overnight cultures using the DNeasy UltraClean microbial kit (Qiagen) following the manufacturer’s instructions. All plasmid extractions were performed using the PureLink Quick plasmid miniprep kit (Invitrogen) following the manufacturer’s instructions. Platinum PCR Super mix (Invitrogen) was used to generate PCR amplicons containing each gene with its native promoter, using primers listed in Table S2. PCR conditions included 95°C for 2 min and then 95°C (30 s), 35 cycles with 60°C (for blaCMY-2 and blaKPC-3) or 62°C (for blaCTX-M-15) for 25 s, followed by 72°C (40 s), with a final extension at 72°C for 5 min. Resulting products were cloned by restriction digest (SacI and BamHI for blaCMY-2 and SacI and HindIII for both blaCTX-M-15 and blaKPC-3) (High-Fidelity polymerase; New England Biolabs), and ligation (T4 ligase; New England Biolabs Inc.) to pMMB207ΔblaTEM-1 following standard cloning techniques (47). These products were transformed into E. coli DH10B by a heat shock method. An empty pMMB207Δ blaTEM-1 was transformed into E. coli DH10B and used as a no-insert control. Transformants were selected on LB agar containing chloramphenicol (30 μg/mL) or carbenicillin (100 μg/mL). All conventional PCR for verification of constructs and correct gene insertion into pMMB207ΔblaTEM-1 used DreamTag Green PCR master mix (ThermoFisher Scientific). PCR products were confirmed by sequencing both strands using primers listed in Table S2.
Growth curve experiments.
To detect potential fitness costs and benefits, bacterial strains (DH10B/pMMB207ΔblaTEM-1::blaCMY-2, DH10B/pMMB207ΔblaTEM-1::blaCTX-M-15, DH10B/pMMB207ΔblaTEM-1::blaKPC-3, and DH10B/pMMB207ΔblaTEM-1) were incubated overnight at 37°C with continuous shaking (200 rpm) and typically in a volume of 5 mL LB broth without any antibiotic. Cultures were diluted to an optical density at 600 nm (OD600) of 0.8 and further diluted to 1:10,000 (104 cells) in 5 mL of fresh LB broth. In a 96-well plate, 100 μL of the final dilution was added with the appropriate antibiotic in 2-fold dilutions (from 4 to 256 μg/mL for ceftiofur sodium [Zoetis], DFC [Santa Cruz Biotechnology], DFC-cysteine [Toronto Research Chemicals], and DFC-dimer [Toronto Research Chemicals] and from 8 to 3,000 μg/mL for ampicillin [ThermoFisher Scientific]) or no antibiotic. The OD600 was measured every hour for 20 h by using an automated turbidimeter (Bioscreen C; Labsytems). Growth curves were summarized by calculating the AUC for each average growth curve using GraphPad Prism v. 9.3.1 (GraphPad Software). While we present results as average AUC values as heatmaps for the sake of brevity, the original growth curves are provided in the supplemental data.
Coculture competition assays.
We used pairwise coculture experiments to confirm relative fitness inferences from monoculture growth curves. Briefly, precultures of the strains were incubated at 37°C overnight with continuous shaking (200 rpm) and typically in a volume of 5 mL LB broth without any antibiotic. Precultures were diluted to an OD600 of 0.8 and further diluted to 1:10,000 (104 cells) in 5 mL of fresh LB broth with the appropriate antibiotic in 2-fold dilutions (from 4 to 256 μg/mL for ceftiofur, or no antibiotic). Cultures of the strains were then mixed at a ratio of 1:1, while monocultures of each strain were grown under the same conditions as growth controls. Initial proportions were confirmed by plating 25 μL of each culture onto LB agar plates containing 100 μg/mL of carbenicillin (triplicate). Cultures were incubated at 37°C with continuous shaking (200 rpm) for 12 h, and then 1.6 mL of each culture was collected in a 2-mL microcentrifuge tube for DNA extraction using the DNeasy UltraClean microbial kit (Qiagen, Germany) following manufacturer instructions. qPCR assays were performed in which a gene (blaCMY-2, blaCTX-M-15, and blaKPC-3) was amplified (three biological and two technical replicates) using SsoAdvanced universal SYBR green supermix (Bio-Rad Laboratories), 200 nM oligonucleotide primers (see Table S2), and a CFX96 real-time system (Bio-Rad Laboratories). The amplification efficiency and specificity of each primer pair were estimated in triplicate using a serially diluted pool of experimental genomic DNA samples. Because we used the same strain (E. coli DH10B) and the same plasmid (pMMB207Δ blaTEM-1), we assumed that the average plasmid copy number was similar for all strains. To invert the data for easier interpretation, the threshold cycle (CT) of 35 was used as our cutoff cycle, and the CT value of each target gene (blaCMY-2, blaCTX-M-15, and blaKPC-3) was subtracted from the CT cutoff value.
Time of ceftiofur addition.
To assess how bacterial cultures respond to the time when antibiotic exposure occurs, bacterial strains (DH10B/pMMB207ΔblaTEM-1::blaCMY-2, DH10B/pMMB207ΔblaTEM-1::blaCTX-M-15, DH10B/pMMB207ΔblaTEM-1::blaKPC-3, and DH10B/pMMB207ΔblaTEM-1) were incubated overnight at 37°C with continuous shaking (200 rpm) in a volume of 5 mL LB broth without any antibiotic. Cultures were diluted to an OD600 of 0.8 and further diluted to 1:10,000 (104 cells) in 5 mL broth. In a 96-well plate, 100 μL of the final dilution was added without any antibiotic. Cultures were incubated for 5 or 8 h before adding ceftiofur to each well in 2-fold dilutions (from 4 to 256 μg/mL) or with no antibiotic. Growth curve data were collected every hour (OD600) for 20 h by using an automated turbidimeter Bioscreen C (cell growth). These experiments were performed with three biological replicates and two technical replicates, and growth curves were summarized by calculating the area under the curve for each culture using GraphPad Prism.
Enzyme synthesis.
To identify differences in the kinetics of enzyme synthesis, we used similar methods as described above to generate recombinant CTX-M-15, CMY-2 and KPC-3 expression plasmids with native promoters, but with the addition of N-terminal FLAG (DYKDDDDK) epitope tags for each enzyme (see Table S2). Each assembled plasmid was then transformed into E. coli DH10B. A fresh colony of each construct (DH10B/pMMB207ΔblaTEM-1::blaCTXM-15-FLAG, DH10B/pMMB207ΔblaTEM-1::blaCMY-2-FLAG, and DH10B/pMMB207ΔblaTEM-1::blaKPC-3-FLAG) was inoculated separately into overnight cultures of 250 mL LB broth, with each flask containing a different concentration of ceftiofur. Aliquots (1 mL; two technical replicates) were collected from each flask at four different time points (8, 10, 12, and 16 h after corresponding antibiotic treatments and controls). Collected samples (three biological replicates per strain and antibiotic concentration) were pelleted by centrifugation at 4,000 × g (4°C for 2 min) in preparation for Western blotting.
Protein preparations were resuspended in hot 2× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer and transferred to a heat block (95°C for 10 min). Any kD Tris-glycine precast gels (Bio-Rad Laboratories) were used for SDS-PAGE protein separation (1× SDS-PAGE running buffer, 100 V, 0.31 A, 95-min run time). A Trans-Blot turbo transfer starter system (Bio-Rad Laboratories) was used to transfer protein onto 0.2-μm nitrocellulose membranes (Bio-Rad Laboratories). Ponceau S (ThermoFisher Scientific) staining was used to verify protein transfer prior to the addition of antibodies for protein detection. Anti-FLAG tag primary antibody (1:1,000; ThermoFisher Scientific) was used with secondary goat anti-mouse antibody (1:5,000; DyLight 488, conjugate) to detect CTX-M-15, CMY-2, and KPC-3 FLAG-tagged recombinant proteins. Anti-DnaK primary antibody (1:1,000; ThermoFisher Scientific) was used with secondary goat anti-rat antibody (1:5,000; DyLight 488, conjugate) to detect DnaK as a control. A ChemiDoc MP imaging system (Bio-Rad Laboratories) was used to detect fluorescent signals. Image Lab v.4.1 (Bio-Rad Laboratories) was used to collect densitometry data.
Recombinant constructs for kinetic studies.
To express and purify recombinant CTX-M-15, CMY-2, and KPC-3 for enzyme kinetics experiments, blaCTX-M-15, blaCMY-2, and blaKPC-3 (minus native promoter) were inserted into a high-copy-number pET200 TOPO vector without epitope tags. Blunt-end PCR products were generated using primers (see Table S2) with the following PCR conditions: 95°C for 2 min followed by 35 cycles of 95°C (30 s), 60°C (for blaCMY-2 and blaKPC-3) or 62°C (for blaCTX-M-15) (25 s), 72°C (40 s), and a final extension at 72°C for 5 min. Resulting products were cloned into a pET200 TOPO vector following the manufacturer’s recommendations and reagents (ThermoFisher Scientific). Plasmids were then transformed into chemically competent E. coli Top10 by the heat shock method. A negative control, no-gene insert pET200 was transformed into a Top10 E. coli strain. Transformants were selected on LB agar containing kanamycin (100 μg/mL). PCR was used to amplify inserted genes (using DreamTag green PCR master mix; ThermoFisher Scientific), and PCR products were confirmed by sequencing of both strands using primers listed in Table S2. Once confirmed, the vectors were extracted using PureLink Quick plasmid miniprep kit (Invitrogen) following the manufacturer instructions, and these were then transformed into E. coli BL21(DE3) for overexpression of the enzymes.
Protein expression.
E. coli-BL21(DE3) cells containing pET200 with the gene of interest were grown overnight at 37°C in LB medium containing kanamycin (100 μg/mL). Cultures were diluted 1:100 with fresh LB medium containing kanamycin (100 μg/mL), grown at 37°C to an OD600 of 0.5, and induced with a final concentration of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; ThermoFisher Scientific) at 30°C for 18 h. After induction, cells were harvested by centrifugation at 9,600 × g for 15 min at 4°C.
Protein purification.
Cells were resuspended in lysis buffer containing 20 mM Tris (pH 8.0) and sonicated 10 times with 30-s pulses (Branson Ultrasonics model 450 sonifier; Danbury CT). The lysate was cleared by centrifugation (31,400 × g, 4°C, 60 min). The extract was loaded at 7 mL/min onto an anion-exchange column (Hiprep diethylaminoethyl [DEAE] sepharose 16/10; GE Healthcare) that had been previously equilibrated with 20 mM Tris-HCl (pH 8.0). In the first step of purification, the enzymes (CTX-M-15 [48], CMY-2, and KPC-3) were collected in the flowthrough fraction. The enzymes were concentrated using an Amicron Ultra 15-10 K apparatus down to 10 mg/mL and applied to a preequilibrated hydroxyapatite column (Bio-Rad, Hercules, CA) while exchanging the buffer to 5 mM potassium phosphate, pH 6.8. Concentrated protein was applied to a hydroxyapatite column (HA; Bio-Rad) that was preequilibrated with buffer A (5 mM potassium phosphate, pH 6.8) and then eluted from the column with a linear gradient of buffer B (500 mM potassium phosphate, pH 6.8). The enzymes of interest were eluted in 50 mM phosphate and concentrated with a centrifugal concentrator (as described above) to 10 mg/mL before storage at −80°C. The final concentration of protein was determined in a Bradford protein assay (Bio-Rad Laboratories). SDS-PAGE was used to confirm the purity of the enzymes. Purity of enzymes was estimated to be >95% based on the single band identified at 31 kDa for CTX-M-15, 37 kDa for CMY-2, and 32 kDa for KPC-3. All chromatography steps were performed using an ÄKTA FPLC instrument (Cytiva).
Rates of enzyme degradation.
To determine the rates at which CTX-M-15, CMY-2 and KPC-3 degrade, single colonies of DH10B/pMMB207ΔblaTEM-1::blaCTXM-15-FLAG, DH10B/pMMB207ΔblaTEM-1::blaCMY-2-FLAG, and DH10B/pMMB207ΔblaTEM-1::blaKPC-3-FLAG were inoculated overnight into replicate 250-mL LB broth cultures with either ceftiofur (8 μg/mL) or no antibiotic. Cultures were allowed to grow for 12 h before kanamycin (100 μg/mL) was added to stop de novo protein synthesis. After 1-mL samples were collected from each flask, cultures were then incubated at 37°C with continuous shaking (200 rpm), and 1-mL samples were collected every 30 min for the first 2 h and then every 2 h for up to 12 h. Samples were pelleted by centrifugation (4,000 × g, 4°C, 2 min). Samples were collected from three biological replicates, and cell pellets were subjected to Western blot analysis as described above. Enzyme densitometry data were normalized by dividing by the densitometry values for matched DnaK data before being natural log transformed and averaged across replicates. These averages were regressed against natural log-transformed hours (1 to 10 or 12 h) after addition of kanamycin. Slopes from these models were then compared (Student’s t test) to test for significant differences in rates of degradation.
Enzyme kinetics.
To estimate catalytic efficiency, purified recombinant proteins were retrieved from the freezer and their purity and condition were reconfirmed by SDS-PAGE. All kinetic measurements were performed at 37°C. Antibiotic concentrations (0 to 3 mM) were prepared in 50 mM phosphate buffer, pH 7.0. Hydrolysis of ceftiofur, DFC, DFC-cysteine, DFC-dimer, and ampicillin was determined after combining 75 μL of purified recombinant enzyme (CTX-M-15, CMY-2, or KPC-3) (final concentration of 10 nM) with antibiotics in 96-well plates and monitoring changes in absorbance due to cleavage of the β-lactam ring. Monitored wavelengths included 289 nm for ceftiofur, 238 nm for hydrolyzed ceftiofur, 260 nm for DFC, DFC-cysteine, and DFC-dimer, 225 nm for hydrolyzed DFC, DFC-cysteine, and DFC-dimer, 220 nm for ampicillin, and 212 nm for hydrolyzed ampicillin. The reactions were performed in triplicate and monitored every 10 s for 30 min. Absorption data were recorded using a Tecan Spark instrument. The kinetic parameters (kcat and Km) for the hydrolysis of substrates were determined using the Michaelis-Menten equations (16–18) in GraphPad Prism.
Statistical analysis.
For most experiments, we used two technical and at least three biological replicates (technical replicates were averaged before statistical analyses). To test for fitness costs in the absence of antibiotics, we used 15 biological replicates (each with two technical replicates), and AUC values were compared using a one-way analysis of variance (ANOVA). Normality of the residues was confirmed with Shapiro-Wilk’s test (passed, P > 0.05), and equality of variances was confirmed with Bartlett’s test (passed, P > 0.05). All other analyses were performed using one- or two-way ANOVAs (alpha = 0.05) using GraphPad Prism v. 9.3.1. Tukey’s honestly significant difference or an uncorrected Fisher’s least significant difference multiple-comparison test were used for pairwise comparisons if the overall model was statistically significant.
Data availability.
The sequences determined in this project have been deposited with GenBank under accession numbers ON412782, ON412783, and ON412784.
ACKNOWLEDGMENTS
We thank L. Jones, C. Deobald, D. White, V. Vadyvaloo, and L. Knodler for technical assistance and C. McDowell for reviewing the manuscript. This project was supported in part by the National Institutes of Health National Institute of General Medical Sciences through institutional training grant award T32-GM008336, the M.J. Murdock Charitable Trust, the Poncin Scholarship Fund, and the Paul G. Allen School for Global Health. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture or any other entities affiliated with this work.
J.J.A., D.R.C. and M.A.D. conceived the experiments. J.J.A., P.A., J.H. and S.-Y.L. performed the experiments. J.J.A., J.L., E.L., C.K. and D.R.C. analyzed the results. J.J.A., P.A. and D.R.C. wrote the manuscript. All authors reviewed the manuscript.
The other authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Douglas R. Call, Email: drcall@wsu.edu.
Charles M. Dozois, INRS
REFERENCES
- 1.Aslam B, Khurshid M, Arshad MI, Muzammil S, Rasool M, Yasmeen N, Shah T, Chaudhry TH, Rasool MH, Shahid A, Xueshan X, Baloch Z. 2021. Antibiotic resistance: one health one world outlook. Front Cell Infect Microbiol 11:771510. 10.3389/fcimb.2021.771510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kraemer SA, Ramachandran A, Perron GG. 2019. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms 7:180. 10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fletcher S. 2015. Understanding the contribution of environmental factors in the spread of antimicrobial resistance. Environ Health Prev Med 20:243–252. 10.1007/s12199-015-0468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan SA, Imtiaz MA, Sayeed MA, Shaikat AH, Hassan MM. 2020. Antimicrobial resistance pattern in domestic animal-wildlife-environmental niche via the food chain to humans with a Bangladesh perspective: a systematic review. BMC Vet Res 16:302. 10.1186/s12917-020-02519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang R, Van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, Jin L, Zhang Q, Liu Y, Rieux A, Dorai-Schneiders T, Weinert LA, Iqbal Z, Didelot X, Wang H, Balloux F. 2018. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun 9:1179. 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkey PM, Jones AM. 2009. The changing epidemiology of resistance. J Antimicrob Chemother 64:i3–i10. 10.1093/jac/dkp256. [DOI] [PubMed] [Google Scholar]
- 8.Davis MA, Sischo WM, Jones LP, Moore DA, Ahmed S, Short DM, Besser TE. 2015. Recent emergence of Escherichia coli with cephalosporin resistance conferred by blaCTX-M on Washington State dairy farms. Appl Environ Microbiol 81:4403–4410. 10.1128/AEM.00463-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Afema JA, Ahmed S, Besser TE, Jones LP, Sischo WM, Davis MA. 2018. Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in Washington State. Appl Environ Microbiol 84:e02430-17. 10.1128/AEM.02430-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Zhao Z, Avillan JJ, Call DR, Davis M, Sischo WM, Zhang A. 2019. Dairy farm soil presents distinct microbiota and varied prevalence of antibiotic resistance across housing areas. Environ Pollut 254:113058. 10.1016/j.envpol.2019.113058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bush K, Jacoby GA. 2010. Updated functional classification of β-lactamases. Antimicrob Agents Chemother 54:969–976. 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornish RE, Kotarski SF. 2002. Cephalosporins in veterinary medicine: ceftiofur use in food animals. Curr Top Med Chem 2:717–731. 10.2174/1568026023393679. [DOI] [PubMed] [Google Scholar]
- 13.NCBI Resource Coordinators. 2016. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44:D7–D19. 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Peng H, Kong J, Zhao T, Zhang S, Cao X. 2018. Pharmacokinetic profile of ceftiofur hydrochloride injection in lactating Holstein dairy cows. J Vet Pharmacol Ther 41:301–306. 10.1111/jvp.12469. [DOI] [PubMed] [Google Scholar]
- 15.Beyer A, Baumann S, Scherz G, Stahl J, Von Bergen M, Friese A, Roesler U, Kietzmann M, Honscha W. 2015. Effects of ceftiofur treatment on the susceptibility of commensal porcine E. coli: comparison between treated and untreated animals housed in the same stable. BMC Vet Res 11:265. 10.1186/s12917-015-0578-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soeung V, Lu S, Hu L, Judge A, Sankaran B, Prasad BVV, Palzkill T. 2020. A drug-resistant beta-lactamase variant changes the conformation of its active-site proton shuttle to alter substrate specificity and inhibitor potency. J Biol Chem 295:18239–18255. 10.1074/jbc.RA120.016103. [DOI] [PubMed] [Google Scholar]
- 17.Brown NG, Shanker S, Prasad BV, Palzkill T. 2009. Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis. J Biol Chem 284:33703–33712. 10.1074/jbc.M109.053819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walkiewicz K, Benitez Cardenas AS, Sun C, Bacorn C, Saxer G, Shamoo Y. 2012. Small changes in enzyme function can lead to surprisingly large fitness effects during adaptive evolution of antibiotic resistance. Proc Natl Acad Sci USA 109:21408–21413. 10.1073/pnas.1209335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stalder T, Rogers LM, Renfrow C, Yano H, Smith Z, Top EM. 2017. Emerging patterns of plasmid-host coevolution that stabilize antibiotic resistance. Sci Rep 7:4853. 10.1038/s41598-017-04662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melnyk AH, Wong A, Kassen R. 2015. The fitness costs of antibiotic resistance mutations. Evol Appl 8:273–283. 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durfee T, Nelson R, Baldwin S, Plunkett G, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csörgo B, Pósfai G, Weinstock GM, Blattner FR. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol 190:2597–2606. 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao Y, Yurkovich JT, Seo SW, Kabimoldayev I, Dräger A, Chen K, Sastry AV, Fang X, Mih N, Yang L, Eichner J, Cho B-K, Kim D, Palsson BO. 2018. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res 46:10682–10696. 10.1093/nar/gky752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53:2227–2238. 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subbiah M, Top EM, Shah DH, Call DR. 2011. Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids. Appl Environ Microbiol 77:4486–4493. 10.1128/AEM.02788-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loftie-Eaton W, Bashford K, Quinn H, Dong K, Millstein J, Hunter S, Thomason MK, Merrikh H, Ponciano JM, Top EM. 2017. Compensatory mutations improve general permissiveness to antibiotic resistance plasmids. Nat Ecol Evol 1:1354–1363. 10.1038/s41559-017-0243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogers H, Mandelstam J. 1962. Inhibition of cell-wall-mucopeptide formation in Escherichia coli by benzylpenicillin and 6-[D(−)-α-aminophenylacetamido]penicillanic acid (ampicillin). Biochem J 84:299–303. 10.1042/bj0840299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Antimicrobial Resistance Monitoring System for Enteric Bacteria. 2019. Antibiotics tested by NARMS. https://www.cdc.gov/narms/antibiotics-tested.html. Accessed May 4, 2022.
- 28.Long PE, Guarnieri JA, Herbst DV, Teske RH. 1983. Disposition of ampicillin administered intravenously and intratracheally to young calves. J Vet Pharmacol Ther 6:273–279. 10.1111/j.1365-2885.1983.tb00002.x. [DOI] [PubMed] [Google Scholar]
- 29.Schuler AM, Rice C, Gorden PJ. 2017. Survey of treatment practices on Midwest dairy farms. Bovine Practitioner 51:190–199. [Google Scholar]
- 30.Mclaughlin CL, Stanisiewski EP, Risco CA, Santos JEP, Dahl GE, Chebel RC, Lagrow C, Daugherty C, Bryson L, Weigel D, Hallberg J, Lucas MJ. 2013. Evaluation of ceftiofur crystalline free acid sterile suspension for control of metritis in high-risk lactating dairy cows. Theriogenology 79:725–734. 10.1016/j.theriogenology.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 31.Sawant AA, Hegde NV, Straley BA, Donaldson SC, Love BC, Knabel SJ, Jayarao BM. 2007. Antimicrobial-resistant enteric bacteria from dairy cattle. Appl Environ Microbiol 73:156–163. 10.1128/AEM.01551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawant AA, Sordillo LM, Jayarao BM. 2005. A survey on antibiotic usage in dairy herds in Pennsylvania. J Dairy Sci 88:2991–2999. 10.3168/jds.S0022-0302(05)72979-9. [DOI] [PubMed] [Google Scholar]
- 33.Constable P, Pyörälä S, Smith G, Guardabassi L, Jensen L, Kruse H. 2016. Guidelines for antimicrobial use in cattle, p 143–160. In Guardabassi L, Jensen LB, Kruse H, Guide to Antimicrobial Use in Animals. Blackwell Publishing, Oxford, United Kingdom. [Google Scholar]
- 34.El-Gendy AAM, Tohamy MA, Ismail M. 2007. Comparative pharmacokinetic and renal clearance study of ceftiofur in cross breed Friesian and Buffalo calves. J Vet Med Res 17:69–77. 10.21608/jvmr.2007.77896. [DOI] [Google Scholar]
- 35.Brook I. 2009. The role of beta-lactamase-producing-bacteria in mixed infections. BMC Infect Dis 9:202. 10.1186/1471-2334-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levison ME, Levison JH. 2009. Pharmacokinetics and pharmacodynamics of antibacterial agents. Infect Dis Clin North Am 23:791–815. 10.1016/j.idc.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalhoff A. 1998. Pharmacokinetics of selected antibacterial agents. Antibiot Chemother (1971) 49:1–148. 10.1159/000059303. [DOI] [PubMed] [Google Scholar]
- 38.Subbiah M, Mitchell SM, Ullman JL, Call DR. 2011. β-Lactams and florfenicol antibiotics remain bioactive in soils while ciprofloxacin, neomycin, and tetracycline are neutralized. Appl Environ Microbiol 77:7255–7260. 10.1128/AEM.05352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subbiah M, Shah DH, Besser TE, Ullman JL, Call DR. 2012. Urine from treated cattle drives selection for cephalosporin resistant Escherichia coli in soil. PLoS One 7:e48919. 10.1371/journal.pone.0048919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cycoń M, Mrozik A, Piotrowska-Seget Z. 2019. Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front Microbiol 10:338. 10.3389/fmicb.2019.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J, Zhao Z, Orfe L, Subbiah M, Call DR. 2016. Soil-borne reservoirs of antibiotic-resistant bacteria are established following therapeutic treatment of dairy calves. Environ Microbiol 18:557–564. 10.1111/1462-2920.13097. [DOI] [PubMed] [Google Scholar]
- 42.Van Epps A, Blaney L. 2016. Antibiotic residues in animal waste: occurrence and degradation in conventional agricultural waste management practices. Curr Pollution Rep 2:135–155. 10.1007/s40726-016-0037-1. [DOI] [Google Scholar]
- 43.Mitchell SM, Subbiah M, Ullman JL, Frear C, Call DR. 2015. Evaluation of 27 different biochars for potential sequestration of antibiotic residues in food animal production environments. J Environ Chem Eng 3:162–169. 10.1016/j.jece.2014.11.012. [DOI] [Google Scholar]
- 44.Subbiah M, Mitchell SM, Call DR. 2016. Not all antibiotic use practices in food-animal agriculture afford the same risk. J Environ Qual 45:618–629. 10.2134/jeq2015.06.0297. [DOI] [PubMed] [Google Scholar]
- 45.CDC N, DHQP. Gram-negative carbapenemase detection, on CDC & FDA Antibiotic Resistance Isolate Bank. https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=9. Accessed May 4, 2022.
- 46.CDC N, DHQP. Enterobacterales Carbapenemase Diversity, on CDC & FDA Antibiotic Resistance Isolate Bank. https://wwwn.cdc.gov/ARIsolateBank/Panel/PanelDetail?ID=8. Accessed May 4, 2022.
- 47.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 48.Ahmadvand PA, Johannetsy J, Lewis JA, Call DR, Kang C. 2022. Characterization of interactions between CTX-M-15 and clavulanic acid, desfuroylceftiofur, ceftiofur, ampicillin, and nitrocefin. Int J Mol Sci 23:5229. 10.3390/ijms23095229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2 and Fig. S1 to S9. Download aem.00791-22-s0001.pdf, PDF file, 3.3 MB (3.3MB, pdf)
Data Availability Statement
The sequences determined in this project have been deposited with GenBank under accession numbers ON412782, ON412783, and ON412784.