Abstract
Atlantic Bonefish (Albula vulpes) are economically important due to their popularity with recreational anglers. In the State of Florida, USA, bonefish population numbers declined by approximately 60% between the 1990s and 2015. Habitat loss, water quality impairment, chemical inputs, and other anthropogenic factors have been implicated as causes, but the role of pathogens has been largely overlooked, especially with respect to viruses. We used a metagenomic approach to identify and quantify viruses in the blood of 103 A. vulpes sampled throughout their Western Atlantic range, including populations in Florida that have experienced population declines and populations in Belize, Mexico, Puerto Rico, and The Bahamas that have remained apparently stable. We identified four viruses, all of which are members of families known to infect marine fishes (Flaviviridae, Iflaviridae, Narnaviridae, and Nodaviridae), but all of which were previously undescribed. Bonefish from Florida and Mexico had higher viral richness (numbers of distinct viruses per individual fish) than fish sampled from other areas, and bonefish from the Upper Florida Keys had the highest prevalence of viral infection (proportion of positive fish) than fish sampled from any other location. Bonefish from Florida also had markedly higher viral loads than fish sampled from any other area, both for a novel narnavirus and for all viruses combined. Bonefish viruses may be indicators of environmentally driven physiological and immunological compromise, causes of ill health, or both.
Supplementary information
The online version contains supplementary material available at 10.1007/s10641-022-01306-9.
Keywords: Albulidae, Health, Disease, Epidemiology, Recreational fisheries, Conservation
Introduction
Bonefish (Albula spp.) are a complex of morphologically similar benthivorous marine fishes with a circumtropical distribution (Colborn et al. 1997; Pickett et al. 2020). Bonefish are physiologically adapted to high-speed burst swimming (Murchie et al. 2011). This trait and their preference to forage in expansive shallow coastal habitats (“flats”) have made them a prized recreational sportfish (Adams 2017). Catch-and-release angling for bonefish and associated tourism generate critical revenue for coastal communities around the Western Atlantic tropical region. For example, annual recreational fishing for bonefish and other flats species has yielded the following most recently estimated economic benefits: USD $45 million into Mexico in 2019 (Palomo and Perez 2021), USD $50 million into Belize in 2013 (Fedler 2014), USD $169 million into The Bahamas in 2018 (Fedler 2019), and USD $465 million into the Florida Keys in 2012 (Fedler 2013). Bonefish are also important for maintaining ecological connectivity among near-shore marine habitats (Murchie et al. 2019; Perez et al. 2019; Santos et al. 2019), and they represent a small but important human food source in some regions (Filous et al. 2020; Rennert et al. 2019). Ensuring the sustainability of bonefish populations through research, management, and conservation is therefore of high ecological and economic importance.
One of the largest bonefish fisheries has traditionally existed in and around Southern Florida and the Florida Keys, USA (Ault et al. 2008). However, Florida’s bonefish population declined significantly beginning in the 1980s (Rehage et al. 2019; Santos et al. 2019). For example, anglers and fishing guides reported a 56% decline in number and a 45% decline in size of bonefish caught in South Florida from the mid-1980s to 2015 (Rehage et al. 2019), and tournament fishing records indicate a 32% decrease in the number of fish caught per boat per day and a 17% decrease in size of the largest fish caught per tournament during this same time period (Boucek et al. 2022). This decline began in the Middle and Lower Keys, followed by Biscayne Bay, as confirmed by multiple data sources (Boucek et al. 2022; Kroloft et al. 2019; Rehage et al. 2019; Santos et al. 2019). Although comparable data from Belize, Mexico, Puerto Rico and The Bahamas have not yet been analyzed, expert knowledge based on interactions with long-time fishing guides and anglers indicates stable populations in these areas prior to and during this study (A. Adams, pers. obs.). Factors implicated in the Florida bonefish decline include habitat loss, changes in food availability, water quality impairments, weather and hydrological events, organic waste and chemicals from coastal communities and agriculture, and overexploitation (Brownscombe et al. 2019; Kroloft et al. 2019). Despite ongoing research and recent signs of possible recovery (Boucek et al. 2022), the causes remain poorly understood.
Emerging pathogens are responsible for some of the most widespread and impactful diseases of wildlife (Cunningham et al. 2017; Hassell et al. 2021; McCallum 2012; Scheele et al. 2019), including diseases of wild fishes (Chapman et al. 2021). For example, wild stocks of Mediterranean Dusky Grouper (Epinephelus marginatus) and Golden Grouper (E. costae) have experienced declines from repeated lethal epidemics of viral nervous necrosis, caused by the endemic red-spotted grouper nervous necrosis virus (Nodaviridae: Betanodavirus) (Boukedjouta et al. 2020), cyprinid herpesviruses (Alloherpesviridae: Cyprinivirus) likely caused the collapse of the Common Carp (Cyprinus carpio) fishery in the Upper Mississippi River, USA (Gibson-Reinemer et al. 2017), and viral hemorrhagic septicemia virus (Rhabdoviridae: Novirhabdovirus) has emerged globally, causing widespread mortality and population declines in both freshwater and marine systems, including economically important recreational fisheries (Escobar et al. 2018).
Recent research has begun to explore disease as a potential cause of, or contributor to, bonefish declines. Dunn et al. (Dunn et al. 2020) found that the composition of bacterial communities on bonefish gills differed between fish sampled in the Florida Keys and apparently stable populations in The Bahamas. The study also documented an increased abundance of potentially pathogenic bacteria on the gills of Florida bonefish. Bacteria are, however, difficult to categorize as pathogenic based on broad taxonomic assignments obtained through 16 S rRNA-based amplicon metabarcoding (“microbiome”) analyses (Schlaberg 2019). Viruses offer an intriguing complementary system because they are strictly dependent on the host intracellular molecular machinery for reproduction (Koonin et al. 2021) and because they are more likely to emerge in wildlife than bacteria or eukaryotic microbes (Cleaveland et al. 2001, 2007; Woolhouse et al. 2005).
In this study, we used metagenomic methods to identify viruses in Atlantic Bonefish across their Caribbean range and to examine spatial differences in infection rates and intensities. Metagenomic methods for virus discovery are “agnostic,” in that they employ massively parallel but random sequencing of nucleic acids from biological samples and compare those sequences to databases without a priori knowledge of what viruses might be present (Munang’andu et al. 2017; Taş et al. 2021; Zhang et al. 2019). These methods have shed important light on viruses of marine fishes (Geoghegan et al. 2018, 2021; Parry et al. 2020), showing that some wild marine fish are naturally infected with distant relatives of medically important viruses, including some of the most feared human viruses, such as the ebolaviruses (Filoviridae) (Shi et al. 2018), the coronaviruses (Coronaviridae) (Miller et al. 2021), and the influenza viruses (Orthomyxoviridae) (Parry et al. 2020). We applied these methods to blood samples collected non-lethally from bonefish in Florida where population declines have occurred and to apparently stable populations in Belize, Mexico, Puerto Rico, and The Bahamas. We sought to test the hypotheses that bonefish from impacted populations in Florida would host more viruses and at higher intensities of infection than bonefish from elsewhere.
Methods
Field sampling
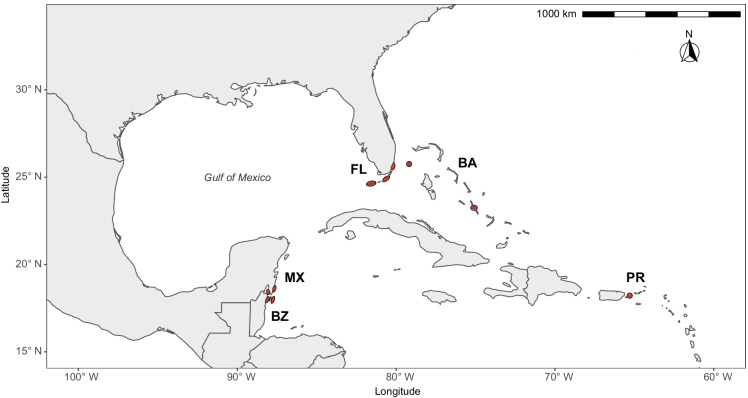
Bonefish were sampled from five areas in the Caribbean: Belize, Florida, Mexico, Puerto Rico, and The Bahamas (Fig. 1). Bonefish were sampled by angling (with spinning and fly-fishing tackle) and by netting (except in Florida), as previously described (Danylchuk et al. 2007; Perez et al. 2019), as required by local laws, regulations, and permits. Sampling occurred in Belize near the island of Ambergris Caye, Belize District. Sampling occurred in Florida from Biscayne Bay in the north to as far south as Key West. Sampling in Mexico occurred near the village of Xcalak in the State of Quintana Roo on the Yucatan Peninsula. Sampling in Puerto Rico occurred near the municipal island of Culebra, approximately 27 km east of the Puerto Rican mainland. Sampling in The Bahamas occurred on the islands of Bimini and Long Island. For purposes of analysis, we defined 10 locations based on physiography and political boundaries: Bay and Ocean in Belize (reflecting physiographic differences to the west and east of Ambergris Caye, respectively); Biscayne Bay, Upper Keys, and Lower Keys in Florida; Bay and Ocean in Mexico (reflecting physiographic differences to the west and east of Xcalak, respectively); Culebra in Puerto Rico; and Bimini and Long Island in The Bahamas.
Fig. 1.
Map of sampling areas: Belize (BZ), Florida, USA (FL), Mexico (MX), Puerto Rico (PR), and The Bahamas (TB). Ellipses indicate locations within which bonefish were sampled (see Online Resource 1)
Once a fish was captured, a blood sample of volume no more than 1% of fish mass (Lawrence et al. 2020) was collected from the caudal vein of the un-anesthetized fish using a 20-gauge, 1.5-in needle and a 5-ml syringe, then transferred immediately to an evacuated plasma preparation tube (PPT) containing K2EDTA anticoagulant designed for molecular analysis (Becton, Dickinson and Company, Franklin Lakes, NJ), and inverted repeatedly. All fish were measured (fork length) and released at their point of capture. All procedures, from capture to release, lasted between 1 and 30 min per fish, which minimized duration of handling and ensured that samples were collected before capture-induced stress could potentially affect viral replication (Gadan et al. 2013; Polinski et al. 2021). Blood samples were stored on ice in the field and then centrifuged at 3200 × g for 15 min within 6 h of collection. Plasma samples were then transferred in 0.5-ml aliquots to sterile 1.2-ml cryovials and mixed with an equal volume of 2× DNA/RNA Shield (Zymo Research, Irvine, CA) to preserve nucleic acids under field conditions and during transport. Tubes containing plasma were stored at −20° C and then shipped on ice to the University of Wisconsin-Madison and stored at −80° C until processing.
Sequencing, bioinformatics, and statistical analyses
Plasma samples were sequenced as previously described (Bennett et al. 2020; Sibley et al. 2016). Briefly, samples were centrifuged for 10 min at 10,000 × g to pellet cellular debris, and total nucleic acids were extracted from 200 µl of supernatant using the QIAamp MinElute Virus Spin Kit (Qiagen, Hilden, Germany), omitting carrier RNA. The Superscript IV system (Thermo Fisher, Waltham, MA, USA) with random hexamers was used to reverse transcribe RNA to cDNA, and cDNA libraries were prepared using the Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA). Libraries were sequenced on a MiSeq instrument using 150 × 150 cycle V2 paired-end sequencing chemistry (Illumina, San Diego, CA, USA), and sequencing adapters were removed from the resulting reads by on-board Illumina processing software.
Reads were quality screened using CLC Genomics Workbench v20.0.1 (CLC Bio, Aarhaus, Denmark). Reads were truncated at the first occurrence of a base with a Phred (quality) score of 30 or lower, and reads shorter than 50 bases in length were excluded from further analyses. Reads matching known reagent contaminants were removed by subtractive mapping. Reads attributable to teleost fish were removed by subtractive mapping to the assembled Common Carp (Cyprinus carpio) genome (accession LN590720.1) at a length fraction of 0.7 and a similarity fraction of 0.8. Remaining reads were subjected to de novo assembly using the SPAdes assembler v3.12.0 (Bankevich et al. 2012). Contiguous sequences (contigs) shorter than 500 nucleotides were discarded, and remaining contigs were screened for redundancy using cd-hit (Li and Godzik 2006).
A multi-stage homology search approach was then used to identify contigs representing putative viruses. First, we used the blastx algorithm (Altschul et al. 1990) implemented in CLC Genomics Workbench to assign sequence homology at the protein level to viruses in the GenBank database. Contigs matching known viral proteins with an E value less than 10−4 were subjected to further investigation (Viljakainen et al. 2018). Then, putative viral contigs were compared individually at the amino acid and nucleotide levels to viruses in GenBank (Sayers et al. 2019) using the tblastx and blastn algorithms, respectively. After a contig was confirmed to represent a virus, the bonefish read set containing the highest number of reads mapping to that contig with full coverage was selected and subjected to de novo assembly using SPAdes. Finally, NOVOPlasty (Dierckxsens et al. 2017) was applied to extend each viral contig to its maximum attainable length.
Protein coding regions of putative viral contigs were identified and checked manually using ORFfinder (Wheeler et al. 2003). Nearest known relatives of detected viruses were identified by comparison of the longest available portion of the viral replicase gene in GenBank using the blastx algorithm. Phylogenies of viruses were inferred using the same replicase gene sequences aligned to those of their nearest neighbors using a codon-based version of the MUSCLE algorithm (Edgar 2004), with the Gblocks algorithm applied to remove poorly aligned regions (Talavera and Castresana 2007), implemented in TranslatorX (Abascal et al. 2010). Alignments were checked and minor manual adjustments made as needed. Maximum likelihood phylogenetic trees were then inferred from each alignment, and 1000 bootstrap replicates were used to assess statistical support for clades using PhyML 3.0 (Guindon et al. 2010; Guindon and Gascuel 2003) with smart model selection (Lefort et al. 2017). Trees were displayed using FigTree v1.4.4 (Rambaut 2016).
Viral loads were quantified using normalized read depths, as previously described (Negrey et al. 2020). Briefly, reads from each bonefish were mapped to the replicase gene sequence of each virus at a stringency of 90%. Mapped reads were then normalized for sequencing depth and target sequence length, expressed as log10 viral reads per million total reads per kilobase of target sequence (Log10 (vRPM/kb + 1)), which correlates with quantitative real-time polymerase chain reaction data (Toohey-Kurth et al. 2017). For analyses of positivity or negativity, bonefish with a value of ≥ 1 to a given virus by this metric, over a read length of ≥ 50 bases, were classified as positive for that virus.
Inferential statistical analyses were performed using R v4.0.5 (R Core Team 2021). Viral richness (number of viruses detected in a fish), prevalence (proportion of fish in a population positive for each individual virus or for any virus), and loads (normalized read depths for each individual virus or for all viruses combined in a fish) were analyzed as outcome variable in generalized linear mixed effects models (GLMER) in the R package LME4 (Bates et al. 2015). Models were optimized for the empirical distributions of their associated data (see “Results” section) and were fitted with sampling area as a fixed effect and the following as random effects: location, sampling month, sampling year, and fish length. This approach allowed us to examine the effect of sampling area on viral outcomes while controlling for spatial and temporal autocorrelation. Overall significance of sampling area was computed by comparison to a null model using likelihood ratio tests. In instances where small sample sizes and/or low viral prevalence precluded this approach, we assessed variation in viral outcomes among sampling locations and in relation to fish length using Kruskal-Wallis tests and Spearman’s rank-order correlation, respectively.
Results
Fish and viruses
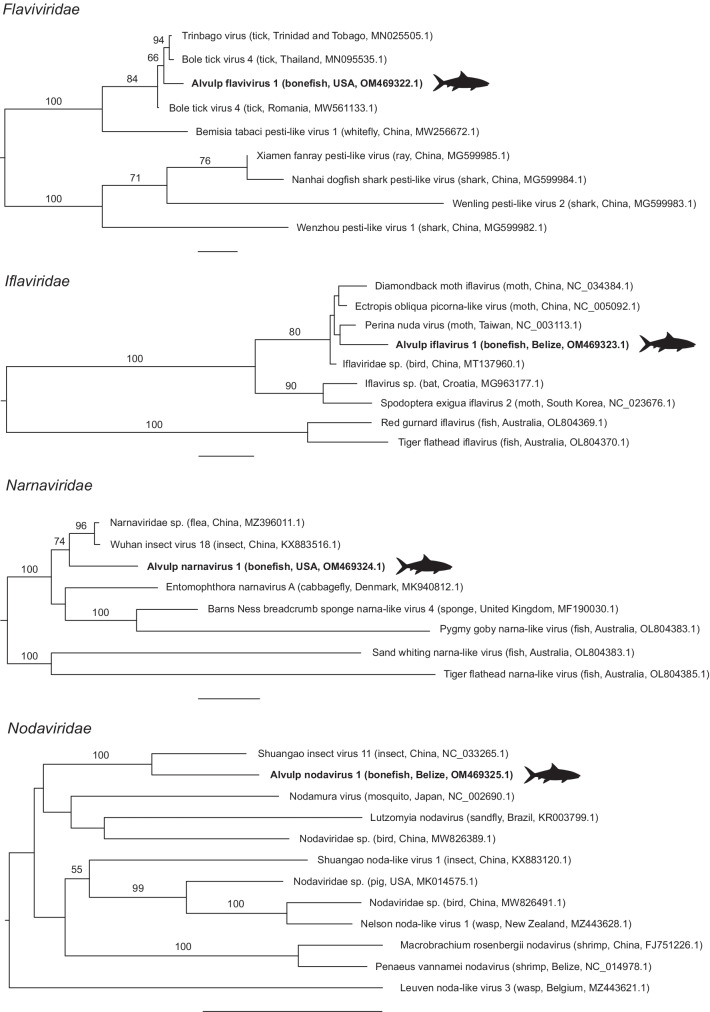
We sampled and analyzed 103 bonefish, including 18 from Belize, 54 from Florida, 10 from Mexico, 7 from Puerto Rico, and 14 from The Bahamas (Online Resource 1 and 2). No fish sampled had any outward signs of illness (e.g., visible lesions or behavioral deficits). Metagenomic sequencing of these 103 bonefish yielded a total of 859,875,768 sequence reads and 27,967,969,413 bases of total data. The average number of reads per bonefish passing quality filters was 1,666,426. Overall, we assembled a total of 413,666 contigs with an average length of 1060 nucleotides. Among these contigs, we identified four viruses in the families Flaviviridae, Iflaviridae, Narnaviridae, and Nodaviridae: alvulp flavivirus 1 (AvFLV-1); alvulp iflavirus 1 (AvIFV-1); alvulp narnavirus 1 (AvNAV-1); and alvulp nodavirus 1 (AvNOV-1), respectively (GenBank accession numbers OM469322–OM469325). Each virus is 78–81% similar at the amino acid level to known viruses in GenBank within the regions of the RNA-dependent RNA polymerase genes sequenced (Table 1). Phylogenetic trees show each virus to be a novel member of a taxon that has previously been identified in marine fishes (Fig. 2).
Table 1.
Viruses identified in Atlantic Bonefish (Albula vulpes)
| Virus1 | Accession | Genome | Family | Closest relative2 | % ID3 |
|---|---|---|---|---|---|
| AvFLV-1 | OM469322 | +ssRNA | Flaviviridae | Trinbago virus (tick, Trinidad and Tobago, MN025505.1) | 80.94 |
| AvIFV-1 | OM469323 | +ssRNA | Iflaviridae | Perina nuda virus (moth, Taiwan, NC_003113.1) | 81.29 |
| AvNAV-1 | OM469324 | +ssRNA | Narnaviridae | Wuhan insect virus 18 (insect, China, KX883516.1) | 78.64 |
| AvNOV-1 | OM469325 | dsRNA | Nodaviridae | Shuangao insect virus 11 (insect, China, NC_033265.1) | 78.42 |
1AvFLV-1, alvulp flavivirus 1; AvIFV-1, alvulp iflavirus 1; AvNAV-1, alvulp narnavirus 1; AvNOV-1, alvulp nodavirus 1
2Closest relative in the GenBank database (source, country, and accession in parentheses)
3Percent amino acid identity to the closest relative in the GenBank database
Fig. 2.
Maximum likelihood phylogenetic trees of bonefish viruses and their relatives in four virus families. Virus names are followed (in parentheses) by host, country, and GenBank accession number. Viruses identified in the present study are in bold with silhouettes. Numbers beside branches are bootstrap values based on 1000 replicates; only values ≥ 50% are shown. Scale bars = 0.5 substitutions per site
Viral richness
A generalized linear mixed effects regression model (GLMER) with the Poisson error family and the Laplace approximation method was the best fit for examining the influence of geography on viral richness. Viral richness was significantly higher in Florida and Mexico than in other areas (Fig. 3; Online Resource 11; Chi-squared = 16.30, df = 4, P = 0.002). Viral richness did not differ significantly among locations within areas (Fig. 3; Online Resource 1; Kruskal-Wallis Chi-squared = 9, df = 9, P = 0.44). However, the Upper Florida Keys had the highest richness of any location in the study, with 3 out of the 4 viruses detected (Online Resource 1). Viral richness for other locations ranged from zero to 2 (Fig. 3; Online Resource 1). Fish length was not significantly associated with viral richness (Kruskal-Wallis Chi-squared = 1.113, df = 91, P = 0.57).
Fig. 3.
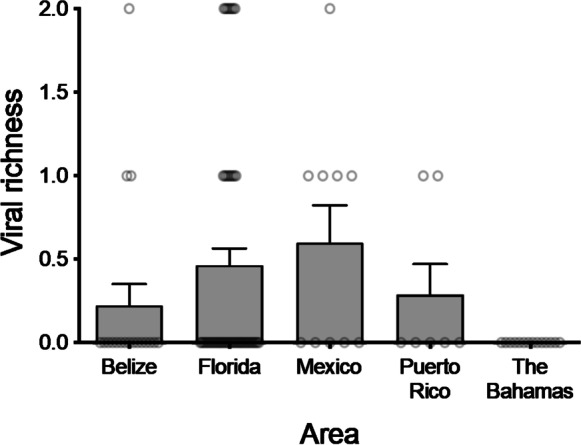
Viral richness by area. Richness is the number of viruses detected in an individual bonefish. Columns indicate mean richness and error bars indicate standard errors of the mean. Values for individual fish are represented by open circles. Sample sizes of fish are 18 (Belize), 54 (Florida), 10 (Mexico), 7 (Puerto Rico), and 14 (The Bahamas). Location-specific sample sizes and richness values are given in Online Resource 2
Viral prevalence
Generalized linear mixed effects models were the best fit for examining the influence of geography on infection, with the status of each fish (positive or negative for a given virus or for any virus) coded as a binomial response variable and area and location as predictors. The prevalence of AvNAV-1 varied among areas, with fish from Florida and Mexico having significantly higher prevalence than fish from all other locations (29.6% and 50.0%, respectively) (Fig. 4; Online Resource 1; Chi-squared = 21.07, df = 4, P < 0.001). AvFLV-1 displayed a similar but not statistically significant pattern (Fig. 4; Online Resource 1; Chi-squared = 7.96, df = 4, P = 0.09). Prevalence of AvIFV-1 and AvNOV-1 did not differ significantly among areas (Fig. 4; Online Resource 1; Chi-squared = 2.04, df = 4, P = 0.73). AvNOV-1 was detected in only 3 fish from Belize (one from the bay side and two from the ocean side). Prevalence of any virus, however, did differ among areas, with Mexico and Florida having the highest prevalence of any virus and The Bahamas having zero prevalence of all viruses (Fig. 4; Online Resource 1; Chi-squared = 15.71, df = 4, P = 0.003).
Fig. 4.
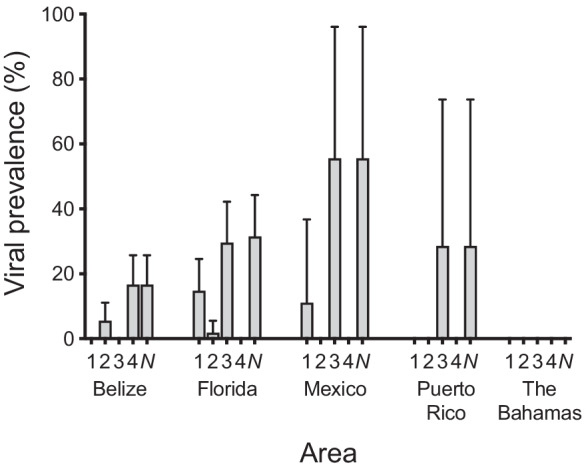
Viral prevalence by area. Prevalence is the proportion of individual bonefish in which a given virus is detected. Columns indicate the prevalence of each virus and error bars indicate standard errors of the mean. Viruses are (1) alvulp flavivirus 1 (AvFLV-1); (2) alvulp iflavirus 1 (AvIFV-1); (3) alvulp narnavirus 1 (AvNAV-1); (4) alvulp nodavirus 1 (AvNOV-1); and (N) any viruses (i.e., at least one of the four aforementioned viruses). Blank columns indicate lack of detection of a virus in that area. Prevalence values are given in Online Resource 1
Locations within areas also differed statistically with respect to viral prevalence (Online Resource 1). Specifically, fish from the Upper Florida Keys and both locations in Mexico had significantly higher prevalence of AvNAV-1 than fish from other locations (50.0%, Kruskal-Wallis Chi-squared = 20.31, df = 9, P = 0.02). Similarly, fish from the Upper Florida Keys had higher prevalence of infection with any virus (57.1%) than fish from any other location in the study; however, this relationship was marginally non-significant (Fig. 4; Online Resource 1, Kruskal-Wallis Chi-squared = 16.83, df = 9, P = 0.051). Fish length was not significantly associated with positivity for each individual virus (Kruskal-Wallis Chi-squared = 0.058–0.097, df = 91, P = 0.78–0.81) or with positivity for any virus (Kruskal-Wallis Chi-squared = 0.871, df = 91, P = 0.35). All AvFLV-1-positive fish (n = 9) were also AvNAV-1-positive, revealing a significant association between these two viruses (Fisher’s exact P < 0.001) in the positive direction (i.e., 14 AvNAV-1-positive fish were AvFLV-1-negative). No other associations between viruses were significant (Fisher’s exact P > 0.86 in all cases).
Viral load
A GLMER model with a Gaussian error distribution family and a log transformed response variable was the best fit for examining the influence of geography on viral load (both individual viruses and all viruses combined). AvNAV-1 loads were 12 times higher in fish from Florida than in fish from Puerto Rico, the area with the next-highest load of this virus (Fig. 5; Online Resource 1; Chi-squared = 6.40, df = 2, P = 0.04). Loads of AvFLV-1 were 9 times higher in fish from Florida than in fish from Mexico, the area with the next-highest load of this virus, but this was not statistically significant likely due to low prevalence of AvFLV-1 (Fig. 5; Online Resource 1; Chi-squared = 0.34, df = 1, P = 0.56). Loads of AvIFV-1 did not differ significantly among areas (Fig. 5; Online Resource 1; Kruskal-Wallis Chi-squared = 1.00, df = 1, P = 0.32). Loads of all viruses combined were higher in fish from Florida than in fish from elsewhere, likely due to the influence of AvNAV-1, but this trend was marginally non-significant (Fig. 5; Online Resource 1; Chi-squared = 7.22, df = 3, P = 0.06).
Fig. 5.
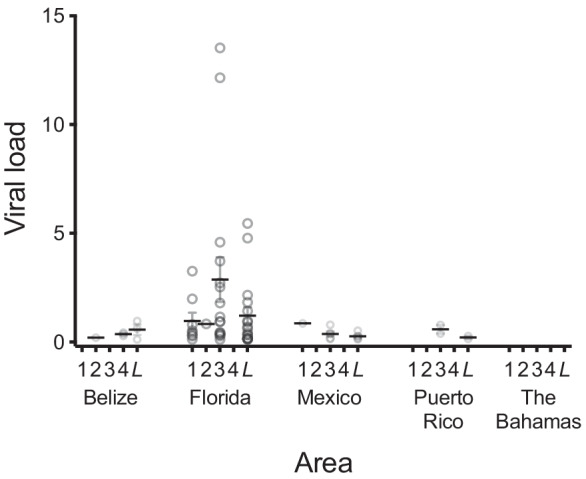
Viral load by area. Viral load is the intensity of infection of an individual fish with a given virus, expressed as the normalized number of sequence reads mapping to a given virus on a log10 scale (see text for further explanation), here multiplied by 1000 for clarity. Values for individual fish are represented by open circles, horizontal lines indicate means, and error bars indicate standard errors of the mean. Only virus-positive fish are included. Viruses are (1) alvulp flavivirus 1 (AvFLV-1); (2) alvulp iflavirus 1 (AvIFV-1); (3) alvulp narnavirus 1 (AvNAV-1); (4) alvulp nodavirus 1 (AvNOV-1); and (L) all viruses combined. Blank columns indicate lack of detection of a virus in that area. The two highest data points in Florida (column 3) are the outliers described in the main text. Viral load values are given in Online Resource 1
Loads of AvNAV-1 were statistically significantly higher in the Lower Florida Keys and Biscayne Bay than in fish from elsewhere (Kruskal-Wallis Chi-squared = 11.815, df = 5, P = 0.037). Fish from the Upper Florida Keys had noticeably higher loads of AvFLV-1 and AvIFV-1 than fish from any other location in the study (Fig. 5), but this trend was not significant (Kruskal-Wallis Chi-squared = 0.91, df = 1, P = 0.55), likely due to the low prevalence of this virus (Online Resource 1). Interestingly, two fish from the Lower Florida Keys had conspicuously higher viral loads than any other fish analyzed (Fig. 5). These outliers had loads of AvNAV-1 of approximately 1 × 10−1 and of all viruses combined of approximately 5 × 10−1 (likely due to the influence of AvNAV-1). Exclusion of these outliers from statistical analyses did not change the significance of any results reported above. Fish length was not significantly associated with loads of any individual virus (Spearman’s r = −0.50–0.30, df = 22, P = 0.18–0.99), but fish length was positively associated with the load of all viruses combined (Spearman’s r = 0.39, df = 24, P = 0.049).
Discussion
We identified four previously undescribed viruses in blood plasma from 103 bonefish sampled across their western Atlantic range. Bonefish from Florida and Mexico had higher viral richness than fish sampled from other areas, and bonefish from the Upper Florida Keys had higher prevalence of infection with any virus than fish from other sampled locations. Bonefish from Florida also had markedly higher loads of AvNAV-1 and of all viruses combined than fish sampled from any other region. Two fish from the Lower Florida Keys had loads of AvFLV-1 approximately ten times higher than the next most intensely infected fish in the study (Fig. 5). In aggregate, these results support the hypothesis that bonefish from populations in Florida where population declines have occurred harbor more viruses and at higher viral loads than bonefish in Belize, Mexico, Puerto Rico, and The Bahamas, where bonefish populations appear to have remained stable (Perez et al. 2021).
We also observed differences in metrics of viral infection among locations within areas. Bonefish from the Upper Florida Keys had higher overall viral prevalence and higher loads of AvNAV-1 than fish from the two other Floridian locations. Because viral load is measured only for infected individuals, it is more robust to bias than other measures that rely on classifying individuals as positive or negative. Viral load may also be more closely linked to the physiological and immunological state of a fish than other measures, because viral loads often increase rapidly when host responses that would normally suppress virus replication become impaired (Chen et al. 2014; Collet 2014; Munang’andu and Evensen 2019). We interpret these trends with caution, however, because of limited geographic resolution in data on bonefish population declines, limited sample sizes in our study, our cross-sectional study design, and imperfect knowledge about anti-viral defenses in A. vulpes. For example, declines appear to have occurred more markedly in the Florida Keys than in Biscayne Bay, which is not reflected in our findings.
The clinical effects of the viruses identified are unknown; they may result from compromised physiology and immune function, cause ill health, or both. None of the bonefish sampled had clinical signs (e.g., visible lesions or behavioral deficits), suggesting that the viruses identified are not severe pathogens. Indeed, some relatives of these viruses appear to be benign commensals (Geoghegan et al. 2021; Shi et al. 2018). Other relatives of these viruses are, however, known fish pathogens, although we caution against inferring pathogenic potential from phylogenetic relatedness. The family Nodaviridae, to which AvNOV-1 belongs, contains the causative agents of viral nervous necrosis and viral encephalopathy and retinopathy, which affect nearly 200 marine fish species and cause lethal epidemics in approximately 1/3 of those species (Bandin and Souto 2020). The family Flaviviridae, to which AvFLV-1 belongs, contains Cyclopterus lumpus virus, associated with an emerging liver disease of Lumpfish (Cyclopterus lumpus) that causes over 50% mortality in aquaculture settings (Skoge et al. 2018). Viruses within the family Narnaviridae, to which AvNAV-1 belongs, have been found at elevated prevalence in dead and dying farmed Atlantic Salmon, although their role as pathogens remains unclear (Bateman et al. 2021). Narnaviruses may actually infect symbionts of their apparent vertebrate hosts, such as internal parasites or disease-associated fungi, perhaps explaining observed associations with disease (Fay et al. 2022; Geoghegan et al. 2021). Viruses within the family Iflaviridae, to which AvIFV-1 belongs, have been found in pooled tissue samples of Tiger Flathead (Neoplatycephalus richardsoni) and Red Gurnard (Chelidonichthys cuculus) from coastal waters of New South Wales, Australia (Geoghegan et al. 2021). Like the narnaviruses, the iflaviruses may infect endoparasites and thus indirectly reflect parasite loads, which appears to be the case in mice (Fay et al. 2022).
Certain patterns we have documented remain unexplained. For example, we detected AvNOV-1 only in Belize. AvNOV-1 could be a virus with a restricted geographic range, or it could occur elsewhere at low prevalence or below the limit of detection of our assay (Toohey-Kurth et al. 2017). We similarly have no clear explanation for why some metrics of infection in fish from Mexico are comparable to levels in fish from Florida. Our sampling locations in Mexico bore little resemblance to those in Florida with respect to the degree or extent of anthropogenic disturbance. In Mexico at the time of sampling, we did observe a distinct overabundance of brown macroalga of the genus Sargassum, which has been responsible for harmful algal blooms across the Atlantic Basin in recent decades (Fidai et al. 2020; Langin 2018; Lapointe et al. 2021). We observed bonefish actively avoiding decaying algal mats, perhaps in response to hypoxia, high microbial concentrations, or chemical byproducts of algal decomposition. Such observations are anecdotal, of course, but potential links between bonefish health and large-scale ecological changes merit consideration (Alfonso et al. 2021; Shultz et al. 2016). Our finding of a weak but significantly positive relationship between the combined load of all viruses and fish length could indirectly reflect an association between viral infection and fish age (Rennert et al. 2019), as has been documented for other wildlife species (Negrey et al. 2020). Finally, the strong positive association between AvNAV-1 and AvFLV-1 might reflect similar transmission dynamics or biological interaction between these viruses in the host.
Consistent with the findings presented here, we previously documented that gill microbiomes of Florida bonefish have an overabundance of potentially pathogenic bacteria compared to fish from other Western Atlantic populations (Dunn et al. 2020). This finding could reflect elevated levels of microbial contamination in the waters of Florida, such as from sewage or agricultural runoff (see below). Viral diseases of ectothermic vertebrates are also associated with changes in the structure of host microbiomes (Campbell et al. 2019), suggesting possible synergies among classes of infectious agents. In this light, we note that we did not examine viral communities in the water at our sampling locations (due to logistic constraints of sampling seawater for viruses; Bofill-Mas and Rusiñol 2020). Therefore, we could not test the hypothesis that increased prevalence and loads of viruses in Florida bonefish resulted from increased viral concentrations in the marine environment. However, bonefish are known to move locally and sometimes over long distances (Boucek et al. 2019; Murchie et al. 2013, 2015; Santos et al. 2019), such that associating the virome of an individual fish with the environment at its precise site of capture would likely offer only limited information about the origins of its infections.
Our choice of blood for analysis restricted our findings to viruses present in this body compartment. Blood can be collected from fish non-lethally (Lawrence et al. 2020), which is requisite for an economically and socially important (and legally protected in some areas) species such as A. vulpes. Moreover, viremia (viruses in blood) reflects systemic infection (Munang’andu and Evensen 2019). Prior studies of marine fish viromes have analyzed organ tissues from commercially harvested fish (Geoghegan et al. 2018, 2021; Shi et al. 2018). This methodological difference and our focus on a single fish species may explain why we documented a lower diversity of viruses than in these previous studies. Similarly, these previous studies were conducted in Asia and Oceania, where fish viromes likely differ from those in North America. This biogeographic difference may explain why the viruses we identified in Atlantic Bonefish cluster separately from viruses in fish from China and Australia (Fig. 2). Finally, because our study was cross-sectional, we could not examine the stability of viromes over time. Follow-up studies involving repeated sampling of populations or, ideally, individual fish that have been marked, tagged, and released (Perez et al. 2019) would likely prove informative.
Chronic environmental stressors such as rising water temperature and declining water quality can compromise fish physiology and immune function (Gadan et al. 2013; Grant et al. 2003; Inendino et al. 2005; Snieszko 1974). In turn, compromised fish physiology and immune function can impair host immune responses and increase susceptibility to viral infection and replication (Gadan et al. 2013; Grant et al. 2003; Inendino et al. 2005). Florida’s marine environments have experienced substantial environmental degradation. Freshwater flows that reach southern Florida’s coastal waters are high in nutrients and coliform bacteria from outdated wastewater infrastructure and agricultural and urban runoff (Glibert et al. 2021; Herren et al. 2021; Lapointe and Clark 1992; Liang et al. 2013). Pharmaceutical residues, hormones, heavy metals, and other contaminants related to urbanization and industrialization also impact these waters (Carnahan et al. 2008; Durán-Álvarez et al. 2021; Ng et al. 2021; Singh et al. 2010). These stressors are likely affecting Florida’s bonefish populations, including their health (Brownscombe et al. 2019; Kroloft et al. 2019). Lack of reported bonefish illness or mortality events (“fish kills”) does not preclude health declines, because an ill or weakened bonefish would quickly succumb to predators (Danylchuk et al. 2007; Moxham et al. 2019).
We suggest that future research investigate not only the pathogenicity of the viruses we have identified (and others yet to be discovered), but also their interactions with anthropogenic stressors. Should such links be identified, the solutions are likely to require an “one health” mindset in which particular environmental drivers are identified and acted upon with the expectation of multiple benefits (Danovaro et al. 2021; Schweizer et al. 2022). Ecosystem-level improvements to water quality, for example, would likely have broad benefits beyond safeguarding bonefish health, ideally leading to improved human health and ecosystem integrity. Such considerations should form the basis for evidence-based ecological and social-ecological management, with the goal of improving the health and sustainability of bonefish populations and the ecosystems they inhabit (Holder et al. 2020; Hughes 2015; Hyder et al. 2020).
Supplementary Information
Below is the link to the electronic supplementary material.
(XLSX 13.2 KB)
(DOCX 21.0 KB)
Acknowledgements
We are grateful to the fisheries authorities in Florida, Belize, Mexico, Puerto Rico, and The Bahamas for permission to conduct field sampling of bonefish and for their support of scientific approaches to fisheries management. We also thank the mangers of the marine protected areas in which we worked and local fishing guides for providing invaluable information about bonefish distributions. Omar Arceo and Roberto Herrera provided indispensable assistance with bonefish collection in Belize and Mexico, respectively. We thank the owners and staff of El Pescador Lodge and Villas in Belize and Acocote Eco Inn in Mexico for kindly providing logistical and in-kind support.
Author contribution
Tony Goldberg and Aaron Adams contributed to study conception and design. Lewis Campbell, Nicholas Castillo, Addiel Perez, Juan Jacobo Schmitter-Soto, Sahar Chauffour Mejri, Ross Boucek, Rolando Santos Corugo, and Jennifer Rehage contributed to fish sampling and data collection in the field. Christopher Dunn processed bonefish samples for metagenomic sequencing. Goldberg and Campbell conducted bioinformatics and statistical analyses and wrote the manuscript. All authors contributed to interpretation, writing, reviewing, and editing, and all authors read and approved the final manuscript.
Funding
This research was funded by the Bonefish & Tarpon Trust. Additional support came from the National Science Foundation under grant no. HRD-1547798 awarded to Florida International University as part of the Centers of Research Excellence in Science and Technology (CREST) Program.
Data availability
Genetic sequences of viruses are available in GenBank, accession numbers OM469322–OM469325.
Declarations
Ethics approval
All fish sampling was performed under scientific collection permits from Biscayne National Park (BISC-2021-SCI-0010), Everglades National Park (EVER-2021-SCI-0025), Dry Tortugas National Park (DRTO-2021-SCI-0007), the Belize Fisheries Department (036 − 19), Instituto de Biodiversidad y Áreas Naturelles Protegidas del Estado de Quintana Roo (IBANQROO/DANP/DANPZS/0018/V-2019), the Mexican Comisión Nacional de Pesca (PPF/DGOPA-053/15), the Departmento de Recursos Naturales y Ambiantes de Puerto Rico (218-IC-083), and the Department of Marine Resources of The Bahamas (MA&MR/FIS/17). All fish sampling was approved by the Institutional Animal Care and Use Committees of Florida International University (protocol 21–058) and the University of Wisconsin-Madison (protocol V006191). This article does not contain any studies involving human participants.
Competing interests
The authors declare no competing interests. Aaron Adams is a Guest Editor of this special issue, but he had no involvement in the peer review of this article and had no access to information regarding its peer review.
Footnotes
The original online version of this article was revised: Figures 3 and 4 were duplicated in error. The correct Fig. 3 is now given.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
9/12/2022
A Correction to this paper has been published: 10.1007/s10641-022-01342-5
References
- Abascal F, Zardoya R, Telford M. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams AJ. Guidelines for evaluating the suitability of catch and release fisheries: Lessons learned from Caribbean flats fisheries. Fish Res. 2017;186:672–680. doi: 10.1016/j.fishres.2016.09.027. [DOI] [Google Scholar]
- Alfonso S, Gesto M, Sadoul B. Temperature increase and its effects on fish stress physiology in the context of global warming. J Fish Biol. 2021;98(6):1496–1508. doi: 10.1111/jfb.14599. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ault JS, et al. et al. Population dynamics and resource ecology of Atlantic tarpon and bonefish. In: Ault JS, et al.et al., editors. Biology and Management of the World Tarpon and Bonefish Fisheries. Boca Raton: CRC Press; 2008. pp. 131–146. [Google Scholar]
- Bandin I, Souto S (2020) Betanodavirus and VER disease: a 30-year research Review. Pathogens 9(2). 10.3390/pathogens9020106 [DOI] [PMC free article] [PubMed]
- Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman AW, et al. Descriptive multi-agent epidemiology via molecular screening on Atlantic salmon farms in the northeast Pacific Ocean. Sci Rep. 2021;11(1):3466. doi: 10.1038/s41598-020-78978-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1):1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- Bennett AJ, et al. Relatives of rubella virus in diverse mammals. Nature. 2020;586(7829):424–428. doi: 10.1038/s41586-020-2812-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bofill-Mas S, Rusiñol M. Recent trends on methods for the concentration of viruses from water samples. Curr Opin Environ Sci Health. 2020;16:7–13. doi: 10.1016/j.coesh.2020.01.006. [DOI] [Google Scholar]
- Boucek RE, Lewis JP, Stewart BD, Jud ZR, Carey E, Adams AJ. Measuring site fidelity and homesite-to-pre-spawning site connectivity of bonefish (Albula vulpes): using mark-recapture to inform habitat conservation. Environ Biol Fish. 2019;102(2):185–195. doi: 10.1007/s10641-018-0827-y. [DOI] [Google Scholar]
- Boucek R et al (2022) Using recreational tournament records to construct a 52-year time-series of the Florida Keys recreational Bonefish fishery. Environ Biol Fishes (this issue)
- Boukedjouta R, Pretto T, Abbadi M, Biasini L, Toffan A, Mezali K. Viral encephalopathy and retinopathy is endemic in wild groupers (genus Epinephelus spp.) of the Algerian coast. J Fish Dis. 2020;43(7):801–812. doi: 10.1111/jfd.13181. [DOI] [PubMed] [Google Scholar]
- Brownscombe JW, et al. Bonefish in South Florida: status, threats and research needs. Environ Biol Fish. 2019;102:pages329–348. doi: 10.1007/s10641-018-0820-5. [DOI] [Google Scholar]
- Campbell LJ, Garner TWJ, Hopkins K, Griffiths AGF, Harrison XA. Outbreaks of an emerging viral disease covary with differences in the composition of the skin microbiome of a wild United Kingdom amphibian. Front Microbiol. 2019;10:1245. doi: 10.3389/fmicb.2019.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnahan EA, Hoare AM, Hallock P, Lidz BH, Reich CD. Distribution of heavy metals and foraminiferal assemblages in sediments of Biscayne Bay, Florida, USA. J Coastal Res. 2008;24(1):159–169. doi: 10.2112/06-0666.1. [DOI] [Google Scholar]
- Chapman JM, Kelly LA, Teffer AK, Miller KM, Cooke SJ. Disease ecology of wild fish: opportunities and challenges for linking infection metrics with behaviour, condition, and survival. Can J Fish Aquat Sci. 2021;78(8):995–1007. doi: 10.1139/cjfas-2020-0315. [DOI] [Google Scholar]
- Chen YM, Wang TY, Chen TY. Immunity to betanodavirus infections of marine fish. Dev Comp Immunol. 2014;43(2):174–183. doi: 10.1016/j.dci.2013.07.019. [DOI] [PubMed] [Google Scholar]
- Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356(1411):991–999. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaveland S, Haydon DT, Taylor L. Overviews of pathogen emergence: which pathogens emerge, when and why? Curr Top Microbiol Immunol. 2007;315:85–111. doi: 10.1007/978-3-540-70962-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colborn J, Crabtree RE, Shaklee JB, Pfeiler E, Bowen BW. The evolutionary enigma of bonefishes (Albula spp.): cryptic species and ancient separations in a globally distributed shorefish. Evolution. 1997;55(4):807–820. doi: 10.1554/0014-3820(2001)055[0807:TEEOBA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Collet B. Innate immune responses of salmonid fish to viral infections. Dev Comp Immunol. 2014;43(2):160–173. doi: 10.1016/j.dci.2013.08.017. [DOI] [PubMed] [Google Scholar]
- Cunningham AA, Daszak P, Wood JLN (2017) One health, emerging infectious diseases and wildlife: two decades of progress? Philos Trans R Soc Lond B Biol Sci 372(1725). 10.1098/rstb.2016.0167 [DOI] [PMC free article] [PubMed]
- Danovaro R, Aronson J, Cimino R, Gambi C, Snelgrove PVR, Van Dover C. Marine ecosystem restoration in a changing ocean. Restor Ecol. 2021;29(S2):e13432. doi: 10.1111/rec.13432. [DOI] [Google Scholar]
- Danylchuk SE, Danylchuk AJ, Cooke SJ, Goldberg TL, Koppelman J, Philipp DP. Effects of recreational angling on the post-release behavior and predation of bonefish (Albula vulpes): the role of equilibrium status at the time of release. J Exp Mar Biol Ecol. 2007;346:127–133. doi: 10.1016/j.jembe.2007.03.008. [DOI] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45(4):e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CD, et al. Bacterial communities on the gills of bonefish (Albula vulpes) in the Florida Keys and The Bahamas show spatial structure and differential abundance of disease–associated bacteria. Mar Biol. 2020;167:85. doi: 10.1007/s00227-020-03698-7. [DOI] [Google Scholar]
- Durán-Álvarez JC, Mejia-Almaguer D, del Campo MN. Occurrence, spatiotemporal distribution and environmental fate of pharmaceutical residues in urban estuaries. In: Durán-Álvarez JC, Jiménez-Cisneros B, editors. Pharmaceuticals in Marine and Coastal Environments. Amsterdam: Elsevier; 2021. pp. 27–89. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar LE, Escobar-Dodero J, Phelps NBD. Infectious disease in fish: global risk of viral hemorrhagic septicemia virus. Rev Fish Biol Fish. 2018;28(3):637–655. doi: 10.1007/s11160-018-9524-3. [DOI] [Google Scholar]
- Fay EJ et al (2022) Natural rodent model of viral transmission reveals biological features of virus population dynamics. J Exp Med 219(2). 10.1084/jem.20211220 [DOI] [PMC free article] [PubMed]
- Fedler T (2013) Economic impact of the florida keys flats fishery. vol Report to Bonefish & Tarpon Trust, Miami
- Fedler A (2014) Economic impact of flats fishing in Belize. vol Report to Bonefish & Tarpon Trust, Miami, p Available at: https://www.bonefishtarpontrust.org/downloads/researchreports/stories/2013-belize-economic-study.pdf. Accessed 06/21/2022
- Fedler A (2019) The 2018 economic impact of flats fishing in The Bahamas. vol Report to Bonefish & Tarpon Trust, p Available at: https://www.bonefishtarpontrust.org/downloads/research-reports/stories/bahamas-flats-economic-impact-report.pdf. Accessed 06/21/2022
- Fidai YA, Dash J, Tomkins EL, Tonon T. A systematic review of floating and beach landing records of Sargassum beyond the Sargasso Sea. Environ Res Commun. 2020;2(12):122001. doi: 10.1088/2515-7620/abd109. [DOI] [Google Scholar]
- Filous A, et al. Population dynamics of roundjaw bonefish Albula glossodonta at a remote coralline atoll inform community-based management in an artisanal fishery. Fish Manag Ecol. 2020;27(2):200–214. doi: 10.1111/fme.12399. [DOI] [Google Scholar]
- Gadan K, Sandtro A, Marjara IS, Santi N, Munang’andu HM, Evensen O. Stress-induced reversion to virulence of infectious pancreatic necrosis virus in naive fry of Atlantic salmon (Salmo salar L.) PLoS ONE. 2013;8(2):e54656. doi: 10.1371/journal.pone.0054656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, Di Giallonardo F, Cousins K, Shi M, Williamson JE, Holmes EC. Hidden diversity and evolution of viruses in market fish. Virus Evol. 2018;4(2):vey031. doi: 10.1093/ve/vey031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, et al. Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 2021;7(1):veab005. doi: 10.1093/ve/veab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson-Reinemer DK, Chick JH, VanMiddlesworth TD, VanMiddlesworth M, Casper AF. Widespread and enduring demographic collapse of invasive common carp (Cyprinus carpio) in the Upper Mississippi River System. Biol Invasions. 2017;19(6):1905–1916. doi: 10.1007/s10530-017-1405-5. [DOI] [Google Scholar]
- Glibert PM, Heil CA, Madden CJ, Kelly SP (2021) Dissolved organic nutrients at the interface of fresh and marine waters: flow regime changes, biogeochemical cascades and picocyanobacterial blooms—the example of Florida Bay, USA. Biogeochemistry. 10.1007/s10533-021-00760-4
- Grant EC, Philipp DP, Inendino KR, Goldberg TL. Effects of temperature on the susceptibility of largemouth bass to largemouth bass virus. J Aquat Anim Health. 2003;15:215–220. doi: 10.1577/H03-009. [DOI] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hassell JM, et al. Towards an ecosystem model of infectious disease. Nat Ecol Evol. 2021;5(7):907–918. doi: 10.1038/s41559-021-01454-8. [DOI] [PubMed] [Google Scholar]
- Herren LW, Brewton RA, Wilking LE, Tarnowski ME, Vogel MA, Lapointe BE. Septic systems drive nutrient enrichment of groundwaters and eutrophication in the urbanized Indian River Lagoon, Florida. Mar Pollut Bull. 2021;172:112928. doi: 10.1016/j.marpolbul.2021.112928. [DOI] [PubMed] [Google Scholar]
- Holder PE, et al. Preparing for a changing future in recreational fisheries: 100 research questions for global consideration emerging from a horizon scan. Rev Fish Biol Fish. 2020;30(1):137–151. doi: 10.1007/s11160-020-09595-y. [DOI] [Google Scholar]
- Hughes RM. Recreational fisheries in the USA: economics, management strategies, and ecological threats. Fish Sci. 2015;81(1):1–9. doi: 10.1007/s12562-014-0815-x. [DOI] [Google Scholar]
- Hyder K, Maravelias CD, Kraan M, Radford Z, Prellezo R. Marine recreational fisheries — current state and future opportunities. ICES J Mar Sci. 2020;77(6):2171–2180. doi: 10.1093/icesjms/fsaa147. [DOI] [Google Scholar]
- Inendino KR, Grant EC, Philipp DP, Goldberg TL. Effects of factors related to water quality and population density on the sensitivity of juvenile largemouth bass to mortality induced by viral infection. J Aquat Anim Health. 2005;17:304–312. doi: 10.1577/H04-028.1. [DOI] [Google Scholar]
- Koonin EV, Dolja VV, Krupovic M, Kuhn JH. Viruses defined by the position of the virosphere within the replicator space. Microbiol Mol Biol Rev. 2021;85(4):e0019320. doi: 10.1128/MMBR.00193-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroloft EKN, Heinen JT, Braddock KN, Rehage JS, Santos RO. Understanding the decline of catch-and-release fishery with angler knowledge: a key informant approach applied to South Florida bonefish. Environ Biol Fish. 2019;102(2):319–328. doi: 10.1007/s10641-018-0812-5. [DOI] [Google Scholar]
- Langin K. Seaweed masses assault Caribbean islands. Science. 2018;360(6394):1157–1158. doi: 10.1126/science.360.6394.1157. [DOI] [PubMed] [Google Scholar]
- Lapointe BE, Clark MW. Nutrient inputs from the watershed and coastal eutrophication in the Florida Keys. Estuaries. 1992;15(4):465–476. doi: 10.2307/1352391. [DOI] [Google Scholar]
- Lapointe BE, et al. Nutrient content and stoichiometry of pelagic Sargassum reflects increasing nitrogen availability in the Atlantic Basin. Nat Commun. 2021;12(1):3060. doi: 10.1038/s41467-021-23135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MJ, et al. Best practices for non-lethal blood sampling of fish via the caudal vasculature. J Fish Biol. 2020;97(1):4–15. doi: 10.1111/jfb.14339. [DOI] [PubMed] [Google Scholar]
- Lefort V, Longueville JE, Gascuel O. SMS: smart model selection in PhyML. Mol Biol Evol. 2017;34(9):2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Liang Z, et al. Impact of mixed land-use practices on the microbial water quality in a subtropical coastal watershed. Sci Total Environ. 2013;449:426–433. doi: 10.1016/j.scitotenv.2013.01.087. [DOI] [PubMed] [Google Scholar]
- McCallum H. Disease and the dynamics of extinction. Philos Trans R Soc Lond B Biol Sci. 2012;367(1604):2828–2839. doi: 10.1098/rstb.2012.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AK, et al. Slippery when wet: cross-species transmission of divergent coronaviruses in bony and jawless fish and the evolutionary history of the Coronaviridae. Virus Evol. 2021;7(2):veab050. doi: 10.1093/ve/veab050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxham EJ, Cowley PD, Bennett RH, von Brandis RG. Movement and predation: a catch-and-release study on the acoustic tracking of bonefish in the Indian Ocean. Environ Biol Fish. 2019;102(2):365–381. doi: 10.1007/s10641-019-00850-1. [DOI] [Google Scholar]
- Munang’andu HM, Mugimba KK, Byarugaba DK, Mutoloki S, Evensen O. Current advances on virus discovery and diagnostic role of viral metagenomics in aquatic organisms. Front Microbiol. 2017;8:406. doi: 10.3389/fmicb.2017.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munang’andu HM, Evensen O. Correlates of protective immunity for fish vaccines. Fish Shellfish Immunol. 2019;85:132–140. doi: 10.1016/j.fsi.2018.03.060. [DOI] [PubMed] [Google Scholar]
- Murchie KJ, Cooke SJ, Danylchuk AJ, Suski CD. Estimates of field activity and metabolic rates of bonefish (Albula vulpes) in coastal marine habitats using acoustic tri-axial accelerometer transmitters and intermittent-flow respirometry. J Exp Mar Biol Ecol. 2011;396(2):147–155. doi: 10.1016/j.jembe.2010.10.019. [DOI] [Google Scholar]
- Murchie KJ, et al. Movement patterns of bonefish (Albula vulpes) in tidal creeks and coastal waters of Eleuthera, The Bahamas. Fish Res. 2013;147:404–412. doi: 10.1016/j.fishres.2013.03.019. [DOI] [Google Scholar]
- Murchie KJ, et al. Defining adult bonefish (Albula vulpes) movement corridors around Grand Bahama in the Bahamian Archipelago. Environ Biol Fish. 2015;98(11):2203–2212. doi: 10.1007/s10641-015-0422-4. [DOI] [Google Scholar]
- Murchie KJ, Haak CR, Power M, Shiply ON, Danylchuk AJ, Cooke SJ. Ontogenetic patterns in resource use dynamics of bonefish (Albula vulpes) in The Bahamas. Environ Biol Fish. 2019;102:117–127. doi: 10.1007/s10641-018-0789-0. [DOI] [Google Scholar]
- Negrey JD, et al. Demography, life-history trade-offs, and the gastrointestinal virome of wild chimpanzees. Philos Trans R Soc Lond B Biol Sci. 2020;375(1811):20190613. doi: 10.1098/rstb.2019.0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng B, et al. Understanding the occurrence and distribution of emerging pollutants and endocrine disruptors in sensitive coastal South Florida Ecosystems. Sci Total Environ. 2021;757:143720. doi: 10.1016/j.scitotenv.2020.143720. [DOI] [PubMed] [Google Scholar]
- Palomo L, Perez AU. 2019 economic impact of flats fishing in Quintana Roo, México. Miami: Bonefish & Tarpon Trust; 2021. [Google Scholar]
- Parry R, Wille M, Turnbull OMH, Geoghegan JL, Holmes EC (2020) Divergent influenza-like viruses of amphibians and fish support an ancient evolutionary association. Viruses 12(9). 10.3390/v12091042 [DOI] [PMC free article] [PubMed]
- Perez AU, Schmitter-Soto JJ, Adams AJ, Heyman WD. Connectivity mediated by seasonal bonefish (Albula vulpes) migration between the Caribbean Sea and a tropical estuary of Belize and Mexico. Environ Biol Fish. 2019;102(2):197–207. doi: 10.1007/s10641-018-0834-z. [DOI] [Google Scholar]
- Perez AU, Schmitter-Soto JJ, Adams AJ. The first estimate of interjurisdictional population dynamics for bonefish, Albula vulpes, a shared resource in the western Caribbean. Environ Biol Fish. 2021;104(3):341–356. doi: 10.1007/s10641-021-01081-z. [DOI] [Google Scholar]
- Pickett BD, Wallace EM, Ridge PG, Kauwe JSK. Lingering taxonomic challenges hinder conservation and management of global bonefishes. Fisheries. 2020;45(7):347–358. doi: 10.1002/fsh.10438. [DOI] [Google Scholar]
- Polinski MP, et al. Innate antiviral defense demonstrates high energetic efficiency in a bony fish. BMC Biol. 2021;19(1):138. doi: 10.1186/s12915-021-01069-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2021. [Google Scholar]
- Rambaut A (2016) FigTree, v1.4.3. Available from http://tree.bio.ed.ac.uk/software/figtree/. Accessed 04/02/2022
- Rehage JS, et al. How has the quality of bonefishing changed over the past 40 years? Using local ecological knowledge to quantitatively inform population declines in the South Florida flats fishery. Environ Biol Fish. 2019;102(2):285–298. doi: 10.1007/s10641-018-0831-2. [DOI] [Google Scholar]
- Rennert JJ, Shenker JM, Angulo-Valdés JA, Adams AJ. Age, growth, and age at maturity of bonefish (Albula species) among Cuban habitats. Environ Biol Fish. 2019;102(2):253–265. doi: 10.1007/s10641-018-0836-x. [DOI] [Google Scholar]
- Santos RO, et al. Linking bonefish (Albula vulpes) populations to nearshore estuarine habitats using an otolith microchemistry approach. Environ Biol Fish. 2019;102(2):267–283. doi: 10.1007/s10641-018-0839-7. [DOI] [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I. GenBank. Nucleic Acids Res. 2019;47(D1):D94–D99. doi: 10.1093/nar/gky989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheele BC, et al. Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science. 2019;363(6434):1459–1463. doi: 10.1126/science.aav0379. [DOI] [PubMed] [Google Scholar]
- Schlaberg R. Microbiome diagnostics. Clin Chem. 2019;66(1):68–76. doi: 10.1373/clinchem.2019.303248. [DOI] [PubMed] [Google Scholar]
- Schweizer M, et al. Fish health in the Nidda as an indicator for ecosystem integrity: a case study for Central European small streams in densely populated areas. Environ Sci Eur. 2022;34(1):10. doi: 10.1186/s12302-021-00584-x. [DOI] [Google Scholar]
- Shi M, et al. The evolutionary history of vertebrate RNA viruses. Nature. 2018;556(7700):197–202. doi: 10.1038/s41586-018-0012-7. [DOI] [PubMed] [Google Scholar]
- Shultz AD, Zuckerman ZC, Suski CD. Thermal tolerance of nearshore fishes across seasons: implications for coastal fish communities in a changing climate. Mar Biol. 2016;163:83. doi: 10.1007/s00227-016-2858-2. [DOI] [Google Scholar]
- Sibley SD, et al. Novel reovirus associated with epidemic mortality in wild largemouth bass (Micropterus salmoides) J Gen Virol. 2016;97(10):2482–2487. doi: 10.1099/jgv.0.000568. [DOI] [PubMed] [Google Scholar]
- Singh SP, Azua A, Chaudhary A, Khan S, Willett KL, Gardinali PR. Occurrence and distribution of steroids, hormones and selected pharmaceuticals in South Florida coastal environments. Ecotoxicology. 2010;19(2):338–350. doi: 10.1007/s10646-009-0416-0. [DOI] [PubMed] [Google Scholar]
- Skoge RH, Brattespe J, Økland AL, Plarre H, Nylund A. New virus of the family Flaviviridae detected in lumpfish (Cyclopterus lumpus) Arch Virol. 2018;163(3):679–685. doi: 10.1007/s00705-017-3643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snieszko SF. The effects of environmental stress on outbreaks of infectious diseases of fishes. J Fish Biol. 1974;6(2):197–208. doi: 10.1111/j.1095-8649.1974.tb04537.x. [DOI] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56(4):564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Taş N, de Jong AEE, Li Y, Trubl G, Xue Y, Dove NC. Metagenomic tools in microbial ecology research. Curr Opin Biotechnol. 2021;67:184–191. doi: 10.1016/j.copbio.2021.01.019. [DOI] [PubMed] [Google Scholar]
- Toohey-Kurth K, Sibley SD, Goldberg TL. Metagenomic assessment of adventitious viruses in commercial bovine sera. Biologicals: J Int Association Biol Stand. 2017;47:64–68. doi: 10.1016/j.biologicals.2016.10.009. [DOI] [PubMed] [Google Scholar]
- Viljakainen L, Holmberg I, Abril S, Jurvansuu J. Viruses of invasive Argentine ants from the European Main supercolony: characterization, interactions and evolution. J Gen Virol. 2018;99(8):1129–1140. doi: 10.1099/jgv.0.001104. [DOI] [PubMed] [Google Scholar]
- Wheeler DL, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31(1):28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YZ, Chen YM, Wang W, Qin XC, Holmes EC. Expanding the RNA virosphere by unbiased metagenomics. Annual Rev Virol. 2019;6(1):119–139. doi: 10.1146/annurev-virology-092818-015851. [DOI] [PubMed] [Google Scholar]
- Woolhouse MEJ, Haydon D, Antia R. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol Evol. 2005;20(5):238–244. doi: 10.1016/j.tree.2005.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 13.2 KB)
(DOCX 21.0 KB)
Data Availability Statement
Genetic sequences of viruses are available in GenBank, accession numbers OM469322–OM469325.