Abstract
A cluster of genes for the anaerobic degradation of benzoate has been described for the phototrophic bacterium Rhodopseudomonas palustris. Here we provide an initial analysis of the regulation of anaerobic benzoate degradation by examining the contributions of two regulators: a new regulator, BadR, encoded by the benzoate degradation gene cluster, and a previously described regulator, AadR, whose gene lies outside the cluster. Strains with single mutations in either badR or aadR grew slowly on benzoate but were relatively unimpaired in growth on succinate and several intermediates of benzoate degradation. A badR aadR double mutant was completely defective in anaerobic growth on benzoate. Effects of the regulators on transcriptional activation were monitored with an R. palustris strain carrying a chromosomal fusion of ′lacZ to the badE gene of the badDEFG operon. This operon encodes benzoyl-coenzyme A (benzoyl-CoA) reductase, an unusual oxygen-sensitive enzyme that catalyzes the benzene ring reduction reaction that is the rate-limiting step in anaerobic benzoate degradation. Expression of badE::′lacZ was induced 100-fold when cells grown aerobically on succinate were shifted to anaerobic growth on succinate plus benzoate. The aadR gene was required for a 20-fold increase in expression that occurred in response to anaerobiosis, and badR was responsible for a further 5-fold increase in expression that occurred in response to benzoate. Further studies with the badE::′lacZ fusion strain grown with various kinds of aromatic acids indicated that BadR probably responds to benzoyl-CoA acting as an effector molecule. Sequence information indicates that BadR is a member of the MarR family of transcriptional regulators. These studies expand the range of functions regulated by MarR family members to include anaerobic aromatic acid degradation and provide an example of a MarR-type protein that acts as a positive regulator rather than as a negative regulator, as do most MarR family members. AadR resembles the Escherichia coli Fnr regulator in sequence and contains cysteine residues that are spaced appropriately to serve in the capacity of a redox-sensing protein.
The anaerobic degradation of aromatic compounds by bacteria has been studied in a sustained way only in the last decade. A major theme that has emerged is that diverse aromatic compounds, including aromatic hydrocarbons, chlorinated aromatics, phenols, and plant-derived lignin monomers, are metabolized to form benzoate or, more often, benzoyl- coenzyme A (benzoyl-CoA) as a common intermediate (12, 13). Benzoate and benzoyl-CoA are then degraded to central biosynthetic intermediates by a series of reactions that involve aromatic ring reduction, ring modification, and ring cleavage as critical steps. The two organisms that have been most studied with respect to anaerobic benzoate degradation are Rhodopseudomonas palustris, a photoheterotrophic bacterium, and Thauera aromatica, a denitrifier. These two species have slightly different benzoate degradation pathways that diverge in the steps following ring reduction (12, 19). Studies of benzoate degradation enzymes have been aided by the recent cloning and sequencing of clusters of benzoate degradation genes from each organism (2, 8).
In spite of good progress on the enzymology of anaerobic benzoate degradation, very little is known about how this process is regulated in either R. palustris or T. aromatica. Both organisms are facultative anaerobes that can degrade some aromatic compounds under aerobic as well as anaerobic conditions. In the presence of oxygen, aromatic rings are hydroxylated by oxygenases and, hence, aerobic degradation proceeds by a biochemical strategy completely different from that used anaerobically. Thus, R. palustris and T. aromatica must regulate the expression of aromatic compound degradation genes in response not only to aromatic carbon source availability but also to oxygen levels.
To date, just one gene, aadR from R. palustris, has been identified as being involved in the regulation of anaerobic aromatic acid degradation. Strains with mutations in aadR grow extremely slowly on benzoate and not at all on 4-hydroxybenzoate. The aadR mutation has little, if any, effect on growth on nonaromatic carbon sources and no obvious effect on other aspects of metabolism (6). AadR is similar in its deduced amino acid sequence to Fnr, a well-studied regulator of anaerobic respiration in Escherichia coli (36), and this similarity has led to the suggestion that the role of AadR is to regulate the expression of anaerobic aromatic compound degradation genes in response to oxygen. However, this notion has not been shown directly, and it is equally possible, based on available evidence, that AadR is primarily a sensor of aromatic compounds.
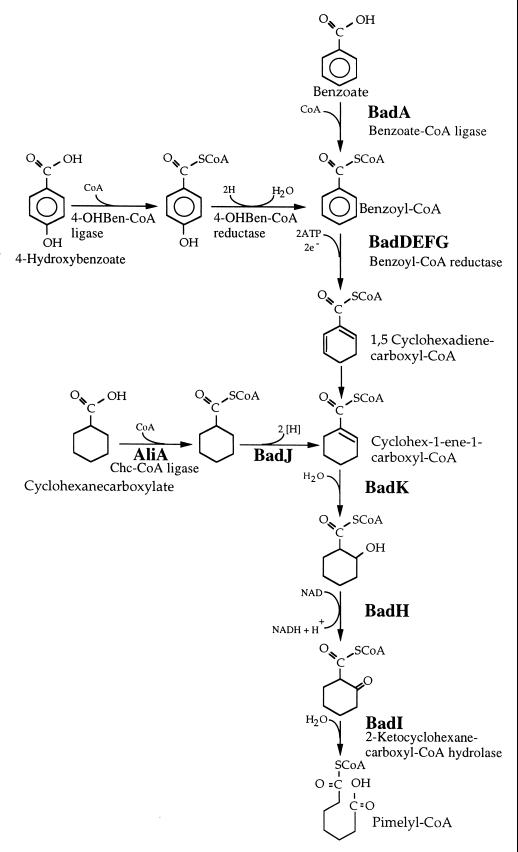
In this study, we evaluated the effect of aadR on anaerobic benzoate degradation at the transcriptional level by monitoring the expression of the badDEFG operon by using a badE::′lacZ fusion as a reporter. The badDEFG operon encodes benzoyl-CoA reductase, an unusual iron-sulfur flavoprotein that catalyzes the ring reduction step that is critical for the anaerobic degradation of benzoate (8) (Fig. 1). The reaction catalyzed is a strongly endergonic two-electron reduction that is energized by the hydrolysis of two molecules of ATP (1). Benzoyl-CoA reductase is extremely sensitive to oxygen. Because of this fact and because the reductase catalyzes an energy-requiring reaction, one would expect the badDEFG operon to be tightly regulated.
FIG. 1.
Anaerobic benzoate degradation pathway and reactions involved in the conversion of 4-hydroxybenzoate (4-OHBen) and cyclohexanecarboxylate (Chc) to intermediates of benzoate degradation by R. palustris (8). The product of benzoyl-CoA reduction is uncertain but is probably one or more of three cyclohexadienecarboxyl-coenzyme A isomers (10, 17), just one of which is shown. SCoA, coenzyme A.
We also examined the contribution of badR, a recently described gene (8), to the expression of benzoyl-CoA reductase and characterized the effects of various carbon sources on badDEFG operon expression.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used are described in Table 1. R. palustris cultures were grown anaerobically in defined mineral medium (16) prepared as described previously (11). Carbon sources were added at the time of inoculation to a final concentration of 3 mM, except for succinate, which was added to 10 mM. Cultures were incubated at 30°C and illuminated with a 40-W incandescent light bulb. E. coli cultures were grown in Luria broth at 37°C. The growth of the cultures was monitored by measuring the A660 spectrophotometrically. Antibiotics were used at the following concentrations (in micrograms per milliliter): for R. palustris, gentamicin at 100 and kanamycin at 100; and for E. coli, ampicillin at 100, gentamicin at 10, and kanamycin at 100.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− λ−recA1 Δ(lacZYA-argF)U169 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 φ80dlacZΔM15 | GIBCO-BRL |
| S17-1 λpir | S17-1 lysogenized with λpir phage | 33 |
| R. palustris | ||
| CGA009 | Wild-type strain; spontaneous Cmr derivative | 16 |
| CGA606 | badE::′lacZ; Kmr | 8 |
| CGA607 | badE::′lacZ aadR; Kmr | This study |
| CGA609 | badR; Gmr | This study |
| CGA610 | badE::′lacZ badR; Kmr Gmr | This study |
| CGA611 | aadR badE::′lacZ badR; Kmr Gmr | This study |
| CGA615 | aadR badR; Gmr | This study |
| CGA101 | aadR | 6 |
| Plasmids | ||
| pUC19 | High-copy-number cloning vector; Apr | 38 |
| pCF116 | Mobilizable suicide vector; sacB; Kmr | 9 |
| pGMΩ1 | Contains the Gmr cassette | 32 |
| pPE408 | pJQ200KS; sacB suicide vector containing badE::′lacZ | 8 |
| pPE600 | pUC19 with a 1,873-bp fragment containing badR; Apr | This study |
| pPE601 | pPE600 with a Gmr cassette at the MscI site in badR | This study |
| pPE602 | pCF116 containing badR::Gmr inserted at SacI-XbaI sites | This study |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance; Cmr, chloramphenicol resistance.
Cloning and DNA manipulation.
Standard protocols were used for cloning and transformations (31). Plasmid DNA was purified with a QIAprep spin miniprep kit (QIAGEN Inc., Chatsworth, Calif.). DNA fragments were purified from agarose gels with a GeneClean spin kit (Bio 101, Inc., La Jolla, Calif.). Chromosomal DNA was purified by a variation of the method of Saito and Miura (30) as described previously (7).
Mutant construction.
A badR mutant was generated by gene replacement with a cloned copy of badR that had been interrupted with a gentamicin resistance (Gmr) cassette. To generate this construct, a 1,873-bp fragment containing badR was PCR amplified from cosmid pPE304 (8) with primers BADR#1 (5′-CGGGATCCGATCAGGTCTTCGAACTGC-3′) and BADR#2 (5′-CGGGATCCCCAACCAGGCTCATCTTGG-3′), which incorporated BamHI sites (underlined) at the ends of the PCR product. The PCR product was then cloned into the BamHI site of pUC19 to generate pPE600. The cloned badR gene was then interrupted at a unique MscI site with the Gmr cassette from pGMΩ1 (32). The badR::Gmr construct was subcloned into pCF116 (9), which contains the sacB gene, for counterselection. The resulting plasmid, pPE602, was mated into R. palustris, and exconjugants were screened as described previously (7) to identify recombinants harboring the disrupted badR gene. A badC mutant was constructed similarly. Briefly, badC was PCR amplified and cloned, and the Gmr cassette from pGMΩ1 (32) was cloned into an XmaI site within badC. The badC::Gmr construct was subcloned into a sacB-containing vector and mated into R. palustris. R. palustris strains with the badE::′lacZ aadR genotype were constructed similarly by mating pPE408 (8) into aadR mutant CGA101 (6). Southern hybridization with a Genius kit (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) and PCR analysis were carried out to verify mutants.
Primer extension.
Primer extension analysis was used to determine the start site for the transcription of badR. An avian myeloblastosis virus reverse transcriptase primer extension system was used according to the protocol supplied by the manufacturer (Promega Corp.). The primer used to map the badR start site (5′-GCAACATATTTGCGCATTGGTAG-3′) was complementary to nucleotides 81 to 103 of badR. Primer extension products were analyzed on a 6% polyacrylamide gel next to a sequence ladder generated with the same primer. Sequencing reactions were performed with the fmol DNA sequencing system from Promega. RNA was prepared with TRI-REAGENT (Molecular Research Center Inc., Cincinnati, Ohio) according to the protocol supplied by the manufacturer. To improve cell lysis, ground glass was added to cells resuspended in TRI-REAGENT, and the cell suspensions were sonicated.
Sequence analysis.
DNA sequences were analyzed with GENE Inspector, version 1.0.1 (Textco Inc., West Lebanon, N.H.). Similar sequences were identified from the SWISS-PROT and GENPEPT databases by use of the BLAST network service at the National Center for Biotechnology Information (Bethesda, Md.) and the BLOCK SEARCH program (14). The GAP and PILEUP programs from the University of Wisconsin Genetics Computer Group software package, version 8.0, were used to make sequence comparisons and alignments.
Measurement of β-galactosidase activity.
β-Galactosidase activity was measured by a variation of the method of Miller (23). Cells in the logarithmic phase of growth were harvested, washed in Z buffer, and sonicated. Cell extract and Z buffer were combined to a volume of 1 ml, and 0.2 ml of a 4-mg/ml solution of o-nitrophenylgalactopyranoside was added to start the reactions. The rate of increase in the A420 due to o-nitrophenol formation was measured spectrophotometrically. Activity was calculated with a millimolar extinction coefficient of 4.5 for o-nitrophenol at 420 nm.
Protein concentrations of cells extracts were determined with a protein assay kit from Bio-Rad, Richmond, Calif.
Nucleotide sequence accession numbers.
The DNA sequences of aadR and badR have been assigned GenBank accession no. M92426 and U75363, respectively.
RESULTS
Molecular characteristics of badR.
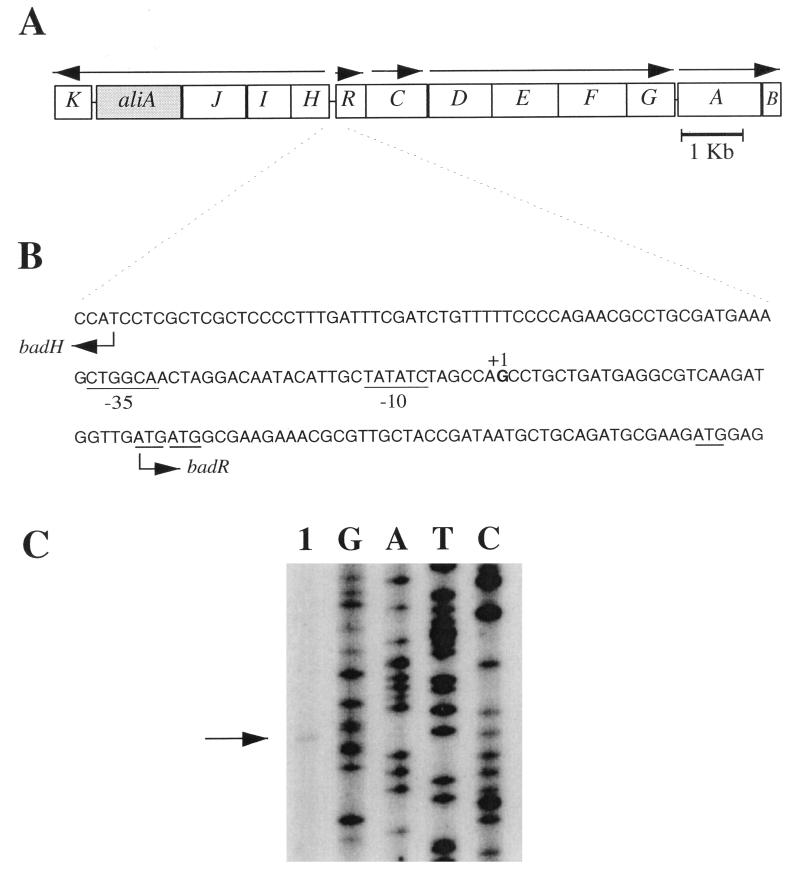
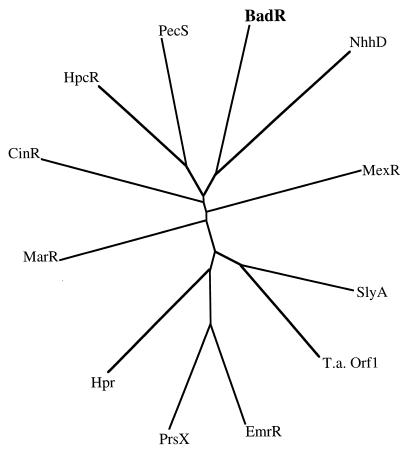
The benzoic acid degradation (bad) gene cluster (Fig. 2A) from R. palustris includes a 528-bp open reading frame, designated badR (8), that is predicted to encode a protein with similarity to members of the MarR family of transcriptional regulators (35). When the BLOCK SEARCH program (14) was applied to the BadR protein, the helix-turn-helix motif associated with the MarR family (5, 35) was evident. GAP alignments of BadR with other MarR family members revealed that BadR is most similar to HpcR, a negative regulator of homoprotocatechuate degradation in E. coli (29), with 26% sequence identity and 46% similarity. A phylogenetic tree showing the relatedness of BadR to characterized members of the MarR family is shown in Fig. 3.
FIG. 2.
(A) Map of the bad (benzoic acid degradation) gene cluster from R. palustris. Arrows indicate probable transcriptional units. The genes encode enzymes of benzoate degradation, as indicated in Fig. 1. The location on the R. palustris chromosome of aadR with respect to the bad gene cluster has not been determined but is at least 30 kb away. (B) Nucleotide sequence of the badR promoter region showing the start site of badR transcription (+1) and the putative −10 and −35 regions. Three possible ATG start codons for badR are underlined. (C) Mapping of the badR transcriptional start site by primer extension. RNA was isolated from benzoate-grown cells (lane 1). A sequence ladder generated with the same primer is shown. The position of the primer extension product is indicated by the arrow.
FIG. 3.
Phylogenetic tree of members of the MarR family of regulatory proteins constructed with the AllAll program from the Computational Biochemistry Research Group (4a). The proteins included, their accession numbers, their sources, and the systems regulated are as follows: BadR (U75363), R. palustris, this study; NhhD (D67027), Rhodococcus rhodochrous, nitrile hydratase expression (18); MexR (U23763), Pseudomonas aeruginosa, multidrug resistance (27); SlyA (P40676), Salmonella typhimurium, hemolysin production (24); T.a. Orf1 (AJ001830), T. aromatica, open reading frame adjacent to genes encoding 4-hydroxybenzoyl-coenzyme A reductase (3); EmrR (P24201), E. coli, multidrug resistance (20); PrsX (P42192), E. coli, complementation of MarR mutation (35); Hpr (P11065), Bacillus subtilis, protease production and sporulation (25); MarR (P27245), E. coli, multiple antibiotic resistance (4); CinR (U64802), Butyrivibrio fibrisolvens, release of cinnamate from plant material (5); HpcR (S56952), E. coli, homoprotocatechuate degradation (29); and PecS (P42195), Erwinia chrysanthemi, pectinase and cellulase production (28). All proteins on the tree, with the exception of T.a. Orf1, have been directly demonstrated to have a regulatory function.
The transcriptional start site of badR was located by primer extension to 27 bp upstream of the first of the three possible ATG initiation codons (Fig. 2). The badR promoter region contained elements typical of a ς70 promoter. These include a sequence (TATATC) 6 bp upstream of the +1 transcriptional start site that matched the E. coli ς70 −10 consensus sequence (TATAAT) at four of six positions and a sequence (CTGGCA) starting 31 bp upstream of the +1 site that matched the E. coli ς70 −35 consensus sequence (TTGACA) at four of six positions.
Characterization of a badR mutant.
To determine whether badR is involved in anaerobic benzoate degradation, a badR mutant of R. palustris was constructed by gene replacement as described in Materials and Methods. The mutant grew slowly on benzoate relative to the wild-type parent but was unimpaired in growth on carbon sources, including cyclohex-1-ene-1-carboxylate, which are metabolized via the lower part of the benzoate pathway (Fig. 1 and Table 2). These results suggested that BadR might play a role in regulating an early step of benzoate degradation.
TABLE 2.
Anaerobic rates of growth of badR and aadR mutants
| Carbon source | Anaerobic rates of growth of the following straina:
|
|||
|---|---|---|---|---|
| Wild type | badR | aadR | badR aadR | |
| Benzoate | 12 (1) | 29 (6) | 58 (18) | NG |
| Cyclohex-1-ene-1-carboxylate | 8 (2) | 7 (1) | 28 (1) | 18 (2) |
| Succinate | 6 (1) | 6 (1) | 10 (3) | 11 (3) |
Reported as the doubling times in hours. Numbers are the averages of two or more growth experiments; standard deviations are given in parentheses. NG, no growth.
The possibility that the phenotype of the badR mutant was due to a polar effect on the expression of badC, the immediate downstream gene, can be discounted because we have mapped a separate transcriptional start site for badC (data not shown). Also, unlike the badR mutant, which grew slowly on benzoate, a badC mutant that we constructed was completely unable to grow on benzoate. The badC mutant also grew much more slowly than the badR mutant on succinate. The deduced BadC protein resembles an NADPH:quinone oxidoreductase (8). Its role in anaerobic aromatic compound degradation has not yet been determined.
Effect of BadR on gene expression.
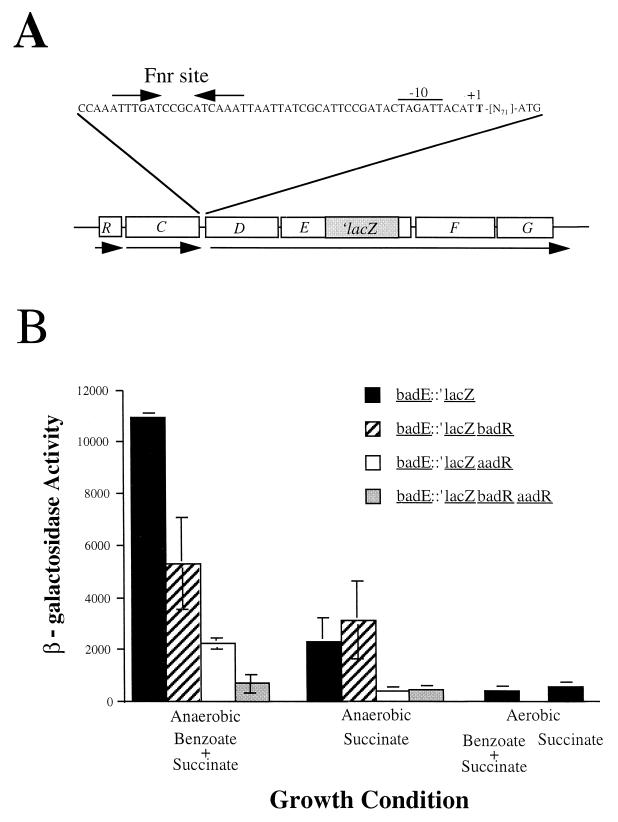
We tested the possible role of BadR in regulating one or both of the first two steps of anaerobic benzoate degradation by examining the effect of the badR mutation on benzoate-coenzyme A (benzoate-CoA) ligase synthesis and expression of the benzoyl-CoA reductase operon. Immunoblot analysis showed that the badR mutant synthesized wild-type levels of benzoate-CoA ligase, indicating that BadR is not required for the expression of badA, the gene that encodes benzoate-CoA ligase (data not shown). Expression of the benzoyl-CoA reductase operon was tested by measuring the levels of β-galactosidase activity from R. palustris strains carrying a chromosomal transcriptional fusion of badE to ′lacZ (badE::′lacZ) (Fig. 4A). In a wild-type background (strain CGA606), the activity of the badE::′lacZ fusion was induced about fivefold by anaerobic growth in the presence of benzoate (Fig. 4B). In a badR background (strain CGA610), however, benzoate did not induce increased lacZ expression from the badE::′lacZ fusion. The levels of β-galactosidase activity in the badR badE::′lacZ strain were approximately the same in cells grown on benzoate plus succinate as in cells grown on succinate alone (Fig. 4B). This result suggests that BadR regulates gene expression in response to benzoate as a carbon source.
FIG. 4.
(A) Map of the badDEFG chromosomal region of strain CGA606 containing the badE::′lacZ fusion. The badDEFG start site of transcription (+1), putative −10 region, and Fnr consensus binding sequence are indicated. (B) Expression of β-galactosidase from a chromosomally encoded badE::′lacZ fusion in wild-type (strain CGA606), badR mutant (strain CGA610) aadR mutant (strain CGA607), and badR aadR double-mutant (strain CGA615) backgrounds. Cells were grown on 10 mM succinate in the presence or absence of 3 mM benzoate. Activities are expressed as nanomoles of product formed per minute per milligram of protein. Values are the averages of data obtained from three or more separately prepared cell extracts. Standard deviations are represented by error bars.
Although benzoate was the best inducer of the badE::′lacZ fusion, growth in the presence of 4-hydroxybenzoate, which is also processed through benzoyl-CoA (Fig. 1), also elicited induced levels of β-galactosidase activity (6,838 nmol of product/min/mg of protein). The nonmetabolizable compound 3-chlorobenzoate did not serve as an inducer (3,016 nmol of product/min/mg of protein) when it was supplied in combination with succinate.
AadR, a second regulator required for full expression of the benzoyl-CoA reductase operon.
To test the possible involvement of AadR in the expression of benzoyl-CoA reductase, we introduced the badE::′lacZ fusion into an aadR mutant strain of R. palustris to generate strain CGA607. The aadR mutation caused a reduction in badE::′lacZ expression regardless of the carbon source used for growth (Fig. 4B). Moreover, the level of badE expression in the aadR mutant grown anaerobically on succinate was reduced to the level seen for wild-type cells grown aerobically on succinate (Fig. 4B). Although the introduction of the aadR mutation caused an overall reduction in the level of badE expression in cells grown anaerobically, the cells still retained their ability to induce badE expression when grown anaerobically in the presence of benzoate, in comparison with cells grown on succinate only. These results are consistent with the idea that AadR mediates gene expression in response to anaerobiosis.
BadR and AadR have additive effects on benzoyl-CoA reductase expression.
A badR aadR double mutant (strain CGA611) grown anaerobically on succinate had the same low levels of badE::′lacZ expression as an aadR single mutant grown under the same conditions. The double mutant differed from the aadR single mutant, however, in being unable to activate badE::′lacZ expression in response to benzoate, since β-galactosidase activities in cells grown on benzoate plus succinate were just as low as those in cells grown on succinate alone (Fig. 4B). This result indicates that BadR and AadR have independent effects on badE expression and that these effects are additive. The importance of these two regulators is underscored by the finding that a badR aadR mutant of R. palustris, in contrast to a badR or an aadR single mutant, was completely defective in anaerobic growth on benzoate (Table 2).
DISCUSSION
Our results indicate that the regulatory protein BadR is required for the induction of benzoyl-CoA reductase expression in response to aromatic compounds and that AadR modulates the levels of expression in response to anaerobiosis. Together, these two regulators account for the approximately 100-fold induction of badE::′lacZ expression that occurs when R. palustris cells grown aerobically on succinate are shifted to anaerobic growth with benzoate.
BadR is most closely related to members of the MarR family of bacterial regulatory proteins (Fig. 3). MarR, the best-characterized member of this family, negatively regulates the expression of the antibiotic resistance genes marAB (4). An inducer of these genes, 2-hydroxybenzoate (salicylate), has been shown to bind to MarR and, by inhibiting its ability to bind to the marAB operator region, to allow transcription of the marAB operon (21). In addition to MarR, several other proteins in this family are likely to respond to aromatic compounds (35). These include other regulators of antibiotic resistance, such as MexR (27) and EmrR (20), which has also been shown to respond to salicylate; HpcR, which regulates genes involved in the metabolism of homoprotocatechuate (29); and CinR, which responds to cinnamoyl esters (5). PecS regulates the expression of virulence factors, perhaps in response to plant-derived phenolic compounds (28).
The description of BadR expands the range of functions regulated by MarR family members to include anaerobic aromatic compound degradation. It also adds to the MarR family another regulator that is likely to respond to an aromatic effector molecule. BadR may respond to either benzoate or benzoyl-CoA to induce the expression of the benzoyl-CoA reductase operon. Of the two compounds, benzoyl-CoA is the more likely effector because (i) benzoyl-CoA accumulated to high levels in the badE::′lacZ reporter strain that was used to monitor gene expression (8); (ii) several compounds, such as 4-hydroxybenzoate, that have been found to induce badE expression are metabolized by R. palustris to form benzoyl-CoA, but not free benzoate, as an intermediate; and (iii) aromatic compounds, such as 3-chlorobenzoate, that are structurally similar to benzoate but that are not metabolized to benzoyl-CoA do not induce badE::′lacZ expression.
BadR differs from many members of the MarR family in that it activates, rather than represses, gene expression. However, SlyA (24) and NhhD (18) are two other MarR family members that have also been shown to regulate gene expression in a positive manner. MexR has both positive and negative regulatory effects on expression (27). No work has yet been reported on the mechanism responsible for transcriptional activation by any of these proteins. It is likely that BadR binds directly to the badD promoter region to activate gene expression, but this notion has yet to be demonstrated.
The activation of badDEFG operon expression mediated by AadR in response to anaerobiosis makes sense because the AadR protein has the signature characteristics of a redox-sensing protein. It has the four cysteine residues, three at its N terminus and one near the center of the protein, that are conserved among most Fnr family members and that are required for the E. coli Fnr protein to be active (6, 22). Recent data indicate that the cysteines coordinate a 4Fe-4S center in Fnr that disassembles on exposure to oxygen, rendering the protein inactive, and then reassembles when the protein is rereduced (15). The reduced form of the protein forms dimers that bind DNA. The promoter region of the badDEFG operon includes an Fnr consensus binding sequence (TTGAT-N4-ATCAA) (34) centered at −39.5 relative to the transcriptional start site (Fig. 4A) (8). It is likely that the AadR protein responds to anaerobiosis by assuming an active conformation that is proficient in binding to the R. palustris badDEFG promoter to activate transcription. We cannot rule out, however, the possibility that the effect of AadR on transcription is indirect and occurs through the activation of yet another regulatory protein.
AadR is the first example of an Fnr family member reported to be involved in regulating aromatic compound degradation in response to oxygen. From what is known so far, AadR is quite specialized in the target genes that it regulates relative to other Fnr family members (37). It does not appear, for example, to control genes required for photosynthesis, as does the FnrL protein from the related phototroph, Rhodobacter sphaeroides (40), a species that does not grow on aromatic compounds. Many Fnr proteins, including those from Rhodobacter sphaeroides and another phototroph of the same genus, Rhodobacter capsulatus, regulate genes for anaerobic respiration (39). The wild-type strain of R. palustris used in our work is unable to grow by anaerobic respiration in the dark with a variety of electron acceptors, including nitrate and dimethyl sulfoxide.
This study has provided the first information about how anaerobic benzoate degradation is regulated in one organism. It has also provided an initial analysis of a new regulatory protein, BadR, and has improved understanding of the role of AadR in the physiology of R. palustris. However, it has also shown that understanding of the regulation of anaerobic benzoate degradation is far from complete. For example, several portions of the anaerobic benzoate degradation pathway, including the initial benzoate activation step and the lower portion of the pathway, do not appear to be regulated by BadR. We know, however, that the production of enzymes in addition to benzoyl-CoA reductase is regulated in response to aromatic compounds. Benzoate-CoA ligase is synthesized in 10-fold higher amounts by cells grown anaerobically in the presence of aromatic acids than by cells grown on succinate (7, 16). Levels of the three enzymes that catalyze the conversion of cyclohex-1-ene-1-carboxyl coenzyme A to pimelyl-coenzyme A are also higher in benzoate-grown cells (26). How the genes encoding these other benzoate degradation enzymes are regulated is an open question since, based on sequence analysis, the bad gene cluster does not include other genes that seem likely to have a regulatory function.
ACKNOWLEDGMENTS
This work was supported by the Division of Energy Biosciences, Department of Energy (grant DE-FG02-95ER20184), and by the U.S. Army Research Office (grant DAAG55-98-1-0188).
We thank Jane Gibson, Rebecca Parales, and Abel Ferrández for helpful discussions.
REFERENCES
- 1.Boll M, Fuchs G. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur J Biochem. 1995;234:921–933. doi: 10.1111/j.1432-1033.1995.921_a.x. [DOI] [PubMed] [Google Scholar]
- 2.Breese K, Boll M, Alt-Mörbe J, Schägger H, Fuchs G. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium Thauera aromatica. Eur J Biochem. 1998;256:148–154. doi: 10.1046/j.1432-1327.1998.2560148.x. [DOI] [PubMed] [Google Scholar]
- 3.Breese K, Fuchs G. 4-Hydroxybenzoyl-CoA reductase (dehydroxylating) from the denitrifying bacterium Thauera aromatica. Prosthetic groups, electron donor, and genes of a member of the molybdenum-flavin-iron-sulfur proteins. Eur J Biochem. 1998;251:916–923. doi: 10.1046/j.1432-1327.1998.2510916.x. [DOI] [PubMed] [Google Scholar]
- 4.Cohen S P, Hächler H, Levy S B. Genetic and functional analysis of the multiple antibiotic resistance (mar) locus in Escherichia coli. J Bacteriol. 1993;175:1484–1492. doi: 10.1128/jb.175.5.1484-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Computational Biochemistry Research Group. [On line.] AllAll program. cbrg.inf.ethz.ch./section3_1.html. Swiss Federal Institute of Technology, Zurich, Switzerland. [13 July 1998, last date accessed.]
- 5.Dalrymple B P, Swadling Y. Expression of a Butyrivibrio fibrisolvens E14 gene (cinB) encoding an enzyme with cinnamoyl ester hydrolase activity is negatively regulated by the product of an adjacent gene (cinR) Microbiology. 1997;143:1203–1210. doi: 10.1099/00221287-143-4-1203. [DOI] [PubMed] [Google Scholar]
- 6.Dispensa M, Thomas C T, Kim M-K, Perrotta J A, Gibson J, Harwood C S. Anaerobic growth of Rhodopseudomonas palustris on 4-hydroxybenzoate is dependent on AadR, a member of the cyclic AMP receptor protein family of transcriptional regulators. J Bacteriol. 1992;174:5803–5813. doi: 10.1128/jb.174.18.5803-5813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egland P G, Gibson J, Harwood C S. Benzoate-coenzyme A ligase, encoded by badA, is one of three ligases able to catalyze benzoyl-coenzyme A formation during anaerobic growth of Rhodopseudomonas palustris on benzoate. J Bacteriol. 1995;177:6545–6551. doi: 10.1128/jb.177.22.6545-6551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egland P G, Pelletier D A, Dispensa M, Gibson J, Harwood C S. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc Natl Acad Sci USA. 1997;94:6484–6489. doi: 10.1073/pnas.94.12.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuqua C W, Winans S C. A luxR-luxI-type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson K J, Gibson J. Potential early intermediates in anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl Environ Microbiol. 1992;58:696–698. doi: 10.1128/aem.58.2.696-698.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harwood C S, Gibson J. Anaerobic and aerobic metabolism of diverse aromatic compounds by the photosynthetic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol. 1988;54:712–717. doi: 10.1128/aem.54.3.712-717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harwood C S, Gibson J. Shedding light on anaerobic benzene ring degradation: a process unique to prokaryotes? J Bacteriol. 1997;179:301–309. doi: 10.1128/jb.179.2.301-309.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heider J, Fuchs G. Anaerobic metabolism of aromatic compounds. Eur J Biochem. 1997;243:577–596. doi: 10.1111/j.1432-1033.1997.00577.x. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff S, Henikoff J G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991;19:6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khoroshilova N, Popescu C, Münck E, Beinert H, Kiley P J. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim M-K, Harwood C S. Regulation of benzoate-CoA ligase in Rhodopseudomonas palustris. FEMS Microbiol Lett. 1991;83:199–204. [Google Scholar]
- 17.Koch J, Eisenreich W, Bacher A, Fuchs G. Products of enzymatic reduction of benzoyl-CoA, a key reaction in anaerobic aromatic metabolism. Eur J Biochem. 1993;221:649–661. doi: 10.1111/j.1432-1033.1993.tb17593.x. [DOI] [PubMed] [Google Scholar]
- 18.Komeda H, Kobayashi M, Shimizu S. Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction products in Rhodococcus rhodocrous. Proc Natl Acad Sci USA. 1996;93:4267–4272. doi: 10.1073/pnas.93.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laempe D, Eisenreich W, Bacher A, Fuchs G. Cyclohexa-1,5-diene-1-carboxyl-CoA hydratase, an enzyme involved in anaerobic metabolism of benzoyl-CoA in the denitrifying bacterium Thauera aromatica. Eur J Biochem. 1998;255:618–627. doi: 10.1046/j.1432-1327.1998.2550618.x. [DOI] [PubMed] [Google Scholar]
- 20.Lomovskaya O, Lewis K, Matin A. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol. 1995;177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin R G, Rosner J L. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melville S B, Gunsalus R P. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem. 1990;265:18733–18736. [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 24.Oscarsson J, Mizunoe Y, Uhlin B E, Haydon D J. Induction of haemolytic activity in Escherichia coli by the slyA gene product. Mol Microbiol. 1996;20:191–199. doi: 10.1111/j.1365-2958.1996.tb02500.x. [DOI] [PubMed] [Google Scholar]
- 25.Perego M, Hoch J A. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production in Bacillus subtilis. J Bacteriol. 1988;170:2560–2567. doi: 10.1128/jb.170.6.2560-2567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrotta J A, Harwood C S. Anaerobic metabolism of cyclohex-1-ene-1-carboxylate, a proposed intermediate of benzoate degradation, by Rhodopseudomonas palustris. Appl Environ Microbiol. 1994;60:1775–1782. doi: 10.1128/aem.60.6.1775-1782.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Praillet T, Nasser W, Robert-Baudouy J, Reverchon S. Purification and functional characterization of PecS, a regulator of virulence-factor synthesis in Erwinia chrysanthemi. Mol Microbiol. 1996;20:391–402. doi: 10.1111/j.1365-2958.1996.tb02626.x. [DOI] [PubMed] [Google Scholar]
- 29.Roper D I, Fawcett T, Cooper R A. The Escherichia coli C homoprotocatechuate degradative operon: hpc gene order, direction of transcription and control of expression. Mol Gen Genet. 1993;237:241–250. doi: 10.1007/BF00282806. [DOI] [PubMed] [Google Scholar]
- 30.Saito H, Miura K I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 32.Schweizer H P. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 33.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 34.Spiro S, Gaston K L, Bell A I, Roberts R E, Busby S J W, Guest J R. Interconversion of the DNA-binding specificities of two related transcriptional regulators, CRP and FNR. Mol Microbiol. 1990;4:1831–1838. doi: 10.1111/j.1365-2958.1990.tb02031.x. [DOI] [PubMed] [Google Scholar]
- 35.Sulavik M C, Gambino L F, Miller P F. The MarR repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli: prototypic member of a family of bacterial regulatory proteins involved in sensing phenolic compounds. Mol Med. 1995;1:436–446. [PMC free article] [PubMed] [Google Scholar]
- 36.Unden G, Schirawski J. The oxygen-responsive transcriptional regulator FNR of Escherichia coli: the search for signal and reactions. Mol Microbiol. 1997;25:205–210. doi: 10.1046/j.1365-2958.1997.4731841.x. [DOI] [PubMed] [Google Scholar]
- 37.VanSpanning R J M, DeBoer A P N, Reijnders W N M, Westerhoff H V, Stouthamer A H, VanDerOost J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol Microbiol. 1997;25:893–907. doi: 10.1046/j.1365-2958.1997.2801638.x. [DOI] [PubMed] [Google Scholar]
- 38.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains; nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 39.Zeilstra-Ryalls J H, Gabbert K, Mouncey N J, Kaplan S, Kranz R G. Analysis of the fnrL gene and its function in Rhodobacter capsulatus. J Bacteriol. 1997;179:7264–7273. doi: 10.1128/jb.179.23.7264-7273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeilstra-Ryalls J H, Kaplan S. Aerobic and anaerobic regulation in Rhodobacter sphaeroides 2.4.1: the role of the fnrL gene. J Bacteriol. 1995;177:6422–6431. doi: 10.1128/jb.177.22.6422-6431.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]