Abstract
Two genes encoding functional RNase H (EC 3.1.26.4) were isolated from a gram-positive bacterium, Bacillus subtilis 168. Two DNA clones exhibiting RNase H activities both in vivo and in vitro were obtained from a B. subtilis DNA library. One (28.2 kDa) revealed high similarity to Escherichia coli RNase HII, encoded by the rnhB gene. The other (33.9 kDa) was designated rnhC and encodes B. subtilis RNase HIII. The B. subtilis genome has an rnhA homologue, the product of which has not yet shown RNase H activity. Analyses of all three B. subtilis genes revealed that rnhB and rnhC cannot be simultaneously inactivated. This observation indicated that in B. subtilis both the rnhB and rnhC products are involved in certain essential cellular processes that are different from those suggested by E. coli rnh mutation studies. Sequence conservation between the rnhB and rnhC genes implies that both originated from a single ancestral RNase H gene. The roles of bacterial RNase H may be indicated by the single rnhC homologue in the small genome of Mycoplasma species.
RNase H (EC 3.1.26.4) endonucleolytically cleaves RNA in RNA-DNA hybrid molecules (26). This activity is present in almost all organisms, from viruses to humans (3). An RNase H gene that encodes bacterial RNase HI (rnhA) (2) was first cloned from Escherichia coli K-12 by measurement of biochemical activity (21, 31). Subsequently, use of conditional lethal E. coli rnhA mutants, very sensitive to the residual levels of RNase H activity in vivo (18), permitted isolation of a second RNase H gene (rnhB) from E. coli that encodes bacterial RNase HII (13).
Isolation of rnhA genes from Salmonella typhimurium LT2 (18), Thermus thermophilus HB8 (17), Mycobacterium smegmatis (34), and the yeast Saccharomyces cerevisiae (18) has been reported. The three-dimensional structure has been determined by X-ray crystallographic analysis for E. coli RNase HI (25, 41), the heat-stable RNase HI from T. thermophilus (12), and the retroviral homologue RNase H domain of human immunodeficiency virus (HIV) reverse transcriptase (28). Extensive mutagenesis of E. coli RNase HI (6, 9, 22–24) has been carried out, and a detailed mechanism for the enzyme’s catalytic reaction has been proposed (24). Based upon studies of various E. coli rnhA mutants, physiological roles of RNase HI (rnhA) in DNA replication (1, 4, 16, 27, 35), repair (15), and transcription (36) have been proposed. In contrast to the highly active RNase HI (rnhA), constituting more than 90% of the total RNase H activity of E. coli (3), RNase HII (rnhB), which exhibits only 0.4% of the activity of wild-type RNase HI with poly(rA) · poly(dT) used as a substrate (13), has not been studied in any detail.
By computer-assisted searches of the complete genomes of bacteria, a clearly recognizable homologue of RNase HI (rnhA) cannot be found in the Archaebacteria or Mycoplasma genomes. In contrast, ubiquitous RNase HII (rnhB) homologues have been recognized in Archaebacteria (33, 42). The lack of obvious bacterial RNase HI (rnhA) or RNaseHII (rnhB) homologues in the genomes of Mycoplasma species (8, 11), along with the viability at low temperature of an E. coli mutant strain that lacks both RNase HI and RNase HII (12a), suggests that RNase H is dispensable for cell viability or can be replaced by another enzyme possessing exonuclease activity (29).
We attempted to isolate a functional RNase H gene(s) from the gram-positive bacterium Bacillus subtilis by screening a DNA library. Two RNase H genes are present in the B. subtilis genome, including the newly characterized rnhC gene. Mutational analysis indicated that loss of rnhB and rnhC renders B. subtilis unable to grow, suggesting essential roles for these RNase H genes in cell viability. Discovery of the B. subtilis rnhC gene allowed computational identification of the rnhC homologue in the Mycoplasma genomes, where no RNase H homologue had yet been reported (8, 11).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Preparation and transformation of competent E. coli cells were by the method of Mandel and Higa (32). Preparation and transformation of competent B. subtilis cells were as previously described (40). Luria-Bertani (LB) broth was used for growth of E. coli. Antibiotic medium 3 (Difco Laboratories, Detroit, Mich.) was used for growth of B. subtilis. Bacteria were grown at 37°C unless otherwise mentioned. Antibiotic resistance gene cassettes were prepared from the E. coli plasmids listed in Table 1. All plasmids were purified by CsCl-ethidium bromide ultracentrifugation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmida | Genotype or insert | Reference or source |
|---|---|---|
| B. subtilis strains | ||
| 1A1 (= 168 trpC2) | trpC2 | BGSCd |
| CU741 | trpC2 leuC7 | 14 |
| OA101 | Prototroph isolated from CU741 | 14 |
| BEST23 | CU741 plus rnhC151::cat | pMIB15-N1 × CU741: Cm |
| BEST138 | OA101 plus rnhB21::neo | pMIB21LNEO × OA101: Nm |
| BEST206 | 1A1 plus ypdQ44::spc | pRNHA-4 × 1A1: Sp |
| BEST218 | 1A1 plus rnhB21::neo | BEST138 × 1A1: Nm |
| BEST220 | 1A1 plus rnhC151::cat | BEST21 × 1A1: Cm |
| BEST207 | ypdQ44::spc rnhB21::neo | BEST23 × BEST206: Cm |
| BEST208 | ypdQ44::spc rnhC151::cat | BEST138 × BEST206: Nm |
| E. coli strains | ||
| JA221 | F−hsdR hsdM+ trp leu lacY recA1 | 13 |
| MIC3037b | F−rnhA339::cat recC271 hsdR hsdM+ trp leu | 15 |
| MIC2067b | F−rnhA339::cat rnhB716::kam | This study |
| MIC1021b | F−rnhA-91 recB270 | 16 |
| Plasmids | ||
| pRNHA-1 | ypdQ (0.474-kb insert) in pGEM4 | This study |
| pUC2.2 | ypdQ (2.2-kb insert) in pUC18 | N. Ohtani |
| pRNHA-4 | ypdQ44::spc in pUC18 | This study |
| pMIB21 | rnhB in pBR322 | This study |
| pMIB21LNEO | rnhB21::neo in pBR322 | This study |
| pMIB15 | rnhC in pBR322 | This study |
| pMIB15-N1 | rnhC151::cat in pBR322 | This study |
| pGEM4 | 13 | |
| pBEST6 | Chimeric plasmid pBR322 and pGEM4 | This studyc |
| pBEST517A | spc cassette | 12a |
| pBEST512 | neo cassette | 14 |
| pBEST4F | cat cassette | 20 |
Selection conditions for both E. coli and B. subtilis were 5-μg/ml chloramphenicol (Cm) and 50-μg/ml spectinomycin (Sp). Selection for neomycin resistance transformants was on 25-μg/ml kanamycin (Km) for E. coli and 5-μg/ml neomycin (Nm) for B. subtilis.
Temperature-sensitive growth phenotype. Details for construction of MIC2067 will be published elsewhere.
AvaI-BglI fragment of pBR322 and PvuII-BglI fragment of pGEM4 were ligated to give multiple cloning sites to the pBR322 replicon.
Bacillus Genetic Stock Center (Ohio State University, Columbus).
In vitro DNA manipulations.
Type II restriction enzymes and T4 DNA ligase were obtained from Toyobo (Tokyo, Japan), except for NotI (Takara Shuzo, Kyoto, Japan). A Takara exonuclease III (ExoIII) deletion kit was purchased from Takara. DNA manipulations in vitro were done according to procedures described in reference 38 or the manufacturers’ instructions, unless specified otherwise. Southern hybridization was carried out with nylon membranes (Nytran 13N; Schleicher & Schuell, Dassel, Germany), as previously described (14).
Complementation of conditional-lethal E. coli RNase H mutants.
Three E. coli mutants—MIC3037, MIC1021, and MIC2067—form colonies at 30°C but not at 42°C. The temperature-dependent growth defect of MIC3037 and MIC1021 was explained as the adverse effect of recBC and rnhA double mutations at restrictive temperature (16). This temperature-sensitive phenotype was relieved by introduction of the recBC or rnhA gene (16, 17). The temperature-sensitive growth of MIC2067 resulted from the rnhA and rnhB double mutation and was relieved only by delivering plasmids that carry an RNase H gene (12a), such as the following: pSK760, rnhA from E. coli (21); pMIC27, rnhB from E. coli (13); pMY2051, RNH1 from S. cerevisiae (18); pRET4, rnhA from T. thermophilus (17). The versatility of the cloning system using MIC3037 and MIC2067 is demonstrated in this study. MIC1021 was used only when chloramphenicol was needed as a plasmid selection marker.
RNase H activity in vitro.
A 0.5-ml overnight culture of E. coli was harvested in a 1.5-ml microcentrifuge tube. The pellet was suspended in 50 μl of 10 mM Tris-HCl (pH 7.5)–1 mM EDTA, sonicated, and centrifuged. The supernatant was transferred to a fresh tube and adjusted to 40 mM Tris-HCl (pH 6.8)–1% sodium dodecyl sulfate (SDS)–50 mM dithiothreitol–5% (vol/vol) glycerol in 100 μl. Ten microliters was boiled for 3 min immediately before loading on an SDS-polyacrylamide gel containing poly([32P]rA) · poly(dT), and the renaturation gel assay was carried out as described previously (13).
Construction of a B. subtilis DNA library.
Genomic DNA for library construction or analysis by conventional gel electrophoresis was prepared by a liquid isolation method (37). Agarose (1.0%, wt/vol) in TAE solution (50 mM Tris-acetate [pH 8.0], 1.0 mM EDTA) was used for conventional gel electrophoresis at room temperature.
DNA fragments (>4 kb) of a partial Sau3A digest of strain OA101 genomic DNA were isolated from a low-melting-point agarose gel and ligated in the BamHI site of plasmid pBR322 (19). Competent MIC3037 cells were incubated under transformation conditions (18) and spread on LB plates containing ampicillin (100 μg/ml) at 30°C. Transformants that were tetracycline sensitive were grown in LB medium containing ampicillin at 30°C. Aliquots of 96 cultures were collected and each was designated a “Bsu club.” Thirty-two Bsu clubs containing 2,955 independent colonies were obtained. Based on an insert size of 5 kb (data not shown), the library should contain, on average, three copies of each DNA sequence (30).
Determination of the nucleotide sequence.
To obtain deletion clones of plasmid pMIB15-3 or pMIB21F at average intervals of 250 bp from the T7 promoter, the ExoIII digestion method (10) was applied. The sequences of the two clones were determined by dideoxy chain termination sequencing with 35S-labeled nucleotides with the T7 promoter-primer as the sequence primer. Alignment of sequences was done with GENETYX, version 7.0.
Overexpression of RNase H enzymes in E. coli.
For B. subtilis RNase HII expression in E. coli, the 984-bp EcoRI-BamHI segment from a deletion derivative of pMIB21F (see Fig. 2) was inserted in the XbaI site 25 bp downstream of the T7 promoter of pBEST6, yielding pMIB2106. The plasmid was introduced into E. coli BL21(DE3) (39) and selected by ampicillin (100 μg/ml). The resultant transformant was grown in LB medium containing ampicillin at 37°C. Isopropyl-β-d-thiogalactopyranoside was added at a final concentration 0.4 mM to the culture when it was at an approximate A590 of 0.5, with incubation continuing for 4 h. Aliquots were removed at hourly intervals and analyzed by SDS-polyacrylamide gel electrophoresis.
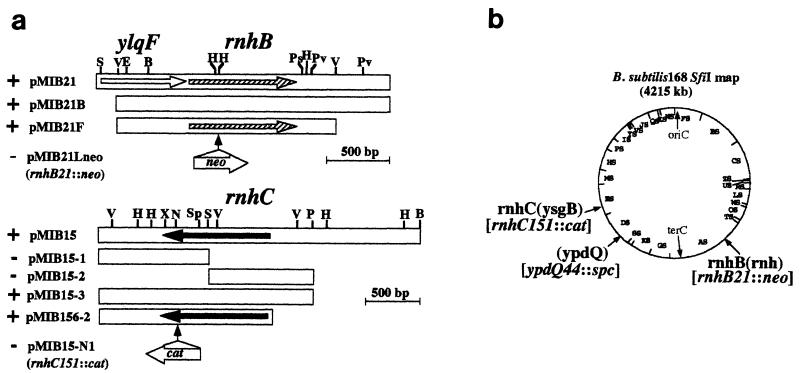
FIG. 2.
Cloned fragment carrying RNase H genes from B. subtilis 168. (a) Plus and minus signs indicate temperature-sensitive complementation of the E. coli rnh-deficient mutants MIC3037 and MIC2067 and RNase H activity in the gel assay. Restriction enzymes are as follows: B, BamHI; E, EcoRI; H, HindIII; N, NotI; Ps, PstI; Pv, PvuII; S, SmaI; Sp, SphI; V, EcoRV; X, XhoI. Antibiotic resistance genes were inserted in the site indicated by the vertical arrow. Scales are 500 bp for each clone. All fragments are oriented from left (distal to oriC) to right (distal to terC). (b) Locations of the three genes cloned in this study in the B. subtilis physical map (19). Gene names cited in reference 30 are in parentheses. Mutations are shown in brackets.
RESULTS
Cloning of two functional RNase H genes from B. subtilis 168.
The B. subtilis DNA library was constructed in E. coli MIC3037 (rnhA339::cat rnhB+ recC271) and subdivided into 32 groups designated Bsu clubs (see Materials and Methods). The library was appropriate for direct screening of the RNase H gene(s) by temperature-sensitive complementation assay and activity in the gel assay, because the host strain, MIC3037, did not grow at the restricted temperature (42°C) and lacks the major E. coli RNase HI (rnhA) activity, as shown in lane 2 of Fig. 1. Promoters of B. subtilis genes normally function in E. coli. Therefore, no special sequences were employed for gene expression. By the two independent screenings described below, two clones were positive for both assays.
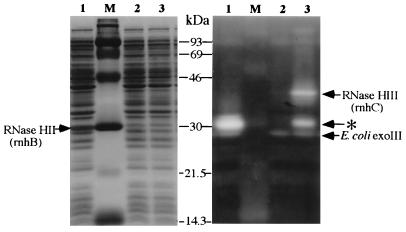
FIG. 1.
Renaturation gel assay for B. subtilis RNase HII (rnhB) and RNaseHIII (rnhC) expressed in E. coli. Lysates from MIC3037 strains carrying pMIB21 (lane 1), pBR322 (lane 2), and pMIB15 (lane 3) were run. Preparation of lysates and assays in the gel are described in Materials and Methods. Cleared areas in the righthand panel are positions where RNase H proteins degraded radiolabeled substrate. MIC3037 (rnhA mutant) lacks major RNase HI activities, and RNase HII (rnhB) activities cannot be detected by this assay (13). Only activities by ExoIII (E. coli exoIII) are visible and indicated. The band in lane 3 indicated by an asterisk may be a degradative product of RNase HIII (rnhC). Size markers (lane M) are as follows: 93, phosphorylase b; 69, bovine serum albumin; 46, ovalbumin; 30, carbonic anhydrase; 21.5, trypsin inhibitor; 14.3, lysozyme.
(i) Each Bsu club was spread after appropriate dilution on an LB plate containing ampicillin (100 μg/ml), and colonies were selected at 42°C. Plasmid DNAs isolated from candidate colonies were analyzed by digestion with HindIII, EcoRI, and PstI. A total of 100 independent clones were classified into 14 groups. The 14 representative clones were examined for RNase H activity in the renaturation gel assay and for the ability to suppress temperature-sensitive growth of MIC2067 (rnhA339::cat rnhB716::kam). One clone, pMIB15, exhibited an RNase H activity of approximately 40 kDa (Fig. 1) and suppressed the temperature-sensitive phenotype of strain MIC2067 (Fig. 2).
(ii) Lysed Bsu clubs were examined directly in the gel assay for screening of RNase H activity. When activity was detected, all 96 colonies in the club were examined to isolate the positive clone. After screening of the 32 Bsu clubs, RNase H activity was detected in 2 clubs, from which three independent clones were obtained. Only one clone, pMIB21, giving an approximately 30-kDa product in the gel assay (Fig. 1), also suppressed the temperature-sensitive phenotypes of both MIC3037 and MIC2067 at 42°C (Fig. 2). The other two clones did not complement the temperature-sensitive growth of any E. coli mutants.
Deletion analysis of the insert in pMIB21 or pMIB15 located the RNase H gene as shown for pMIB21F and pMIB156-2 (Fig. 2a).
Nucleotide and amino acid sequences of the rnhB gene.
The nucleotide sequence of the insert (1,685 bp) of pMIB21F was determined as described in Materials and Methods (data not shown). One open reading frame (ORF) was found that encodes a protein of 255 amino acid residues (calculated molecular mass, 28,204 Da; pI, 5.52), consistent with the estimated size of the RNase H activity (approximately 30 kDa) from the gel assay (Fig. 1). Plasmid pMIB2106, described in Materials and Methods, overexpressed the gene product in E. coli BL21(DE3) (data not shown). The expressed protein was subjected to automated sequence analysis in a gas-phase protein sequencer (model 477A; Perkin-Elmer Applied Biosystems), and the N-terminal 15 amino acids, MNTLTVKDIKDRLQE, were identical to those predicted from the nucleotide sequence (Fig. 3). Searches of the B. subtilis genome database (33a) revealed an ORF designated rnh (accession no. BG12666) encoded by nucleotides 1,676,850 to 1,677,614 (30). The rnh gene had a similarity of 63% (amino acid) to that of E. coli RNase HII (rnhB). Consequently, the rnh gene is designated rnhB, encoding B. subtilis RNase HII.
FIG. 3.
Alignment of the two RNase H sequences of B. subtilis 168. The sequence comparison between B. subtilis rnhB and rnhC is shown. Identical amino acids are boxed, with overall similarity of 20.0%. The four conserved motifs suggested for the rnhB (33, 42) are indicated as I through IV.
Nucleotide and amino acid sequences of the rnhC gene.
The nucleotide sequence of the insert (1,248 bp) of pMIB156-2 (Fig. 2a) was also determined. One ORF was found that encodes a protein of 313 amino acid residues (calculated molecular mass, 33,915 Da; pI, 10.07), consistent with the estimated size of the RNase H activity (approximately 40 kDa) from the gel assay (Fig. 1). The ORF corresponds to the ysgB gene (accession no. BG12324) encoded by nucleotides 2,925,133 to 2,926,071 of the B. subtilis genome; the function of ysgB is unknown (30). The amino acid sequence is shown in Fig. 3. The ysgB gene product had little similarity to E. coli RNase HI (rnhA). Although the overall similarity between B. subtilis rnhB and the ysgB gene is 20.0%, there are a few well-conserved regions between the two, as indicated in Fig. 3. The ysgB gene, therefore, is designated rnhC, encoding B. subtilis RNase HIII.
Overexpression of the RnhC product in E. coli BL21(DE3) with a pBEST6 vector was unsuccessful. However, a similar expression plasmid constructed with a PCR-amplified DNA fragment overproduced a product whose amino acid sequence was identical to that of the ysgB gene (35a).
RNase HI (rnhA) homologue in the B. subtilis genome.
On searching the entire B. subtilis genome for proteins related to E. coli RNaseHI (rnhA), a single sequence with 29% homology, ypdQ (accession no. BG11608), encoded by nucleotides 2,309,611 to 2,310,056 of the B. subtilis genome (30) was found. The ypdQ gene was cloned as a 474-bp DNA fragment amplified by using the forward primer 5′-ACCTCGCCATTAGGATGAAC and the reverse primer 5′-TGCAGCCAAAAAAATGATACC from genomic DNA of B. subtilis 1A1. PCR was performed in 20 cycles with a GeneThermoUnit GTU1605 (Taitech, Tokyo, Japan). The PCR fragment was inserted in pGEM4, resulting in pRNHA-1, but this plasmid was unable to suppress the temperature-sensitive growth of any of the E. coli rnh mutants in Table 1. Possibly this protein is not expressed well in E. coli mutants and/or has lower levels of RNase H activity than that needed to give suppression in vivo. This is consistent with the observation that no rnhA homologue was obtained in screening for the Bsu clubs. The ypdQ gene encodes a protein of 132 amino acid residues (calculated molecular mass, 14,529 Da; pI, 5.60). The product expressed in E. coli is being characterized (35a).
RNase H genes are conserved in related B. subtilis species.
Southern hybridization to SfiI and NotI fragments by three clones, pRNHA-1, pMIB21F, and pMIB15-3 (data not shown), placed these clones in the B. subtilis physical map shown in Fig. 2b. The same DNA probes gave clear signals to genomic DNAs from closely related B. subtilis species, B. subtilis W23 and Bacillus natto IFO1212 (data not shown). The results indicated that all three genes are conserved in these two strains.
RNase H-deficient mutant of B. subtilis.
Mutations of each of the three B. subtilis genes were constructed in E. coli plasmids. The ypdQ gene (rnhA homologue) was disrupted by insertion of a spectinomycin resistance gene cassette (spc) in the unique BsmI site of pUC2.2, resulting in pRNHA-4. The spc cassette was prepared by SmaI digestion of pBEST517A (Table 1). Similarly, the rnhB gene in pMIB21F was disrupted by insertion of a neomycin resistance gene cassette (neo) in the internal HindIII site, resulting in pMIB21Lneo (Fig. 2a). The neo cassette gene was prepared by HindIII digestion from pBEST512 (14). The rnhC gene in pMIB156-2 was disrupted by insertion of a chloramphenicol resistance gene cassette (cat) in the unique NotI site after being blunt ended, resulting in pMIB15-N1 (Fig. 2a). The cat gene was prepared by SmaI digestion of pBEST4F (20).
Mutations carried in these plasmids were introduced into the B. subtilis genome by gene replacement (40). Single mutants were selected by appropriate antibiotic resistance: spectinomycin for the ypdQ mutant, neomycin for the rnhB mutant, and chloramphenicol for the rnhC mutant at 30°C (Table 1). The genomic structures of these mutants were verified by Southern analysis by using the parental plasmids as probes (data not shown). The rnhC mutant BEST220 (rnhC151::cat) gave colonies slightly smaller than the other two single mutants.
Construction of double mutants by genetic crosses of the single mutants was successful except for rnhB and rnhC. Attempts to introduce the rnhB21::neo mutation of BEST218 into BEST220 (rnhC151::cat) resulted in no neomycin-resistant transformants. Other markers unrelated to RNase H, proB or leuB, could be introduced into BEST220, giving approximately 103 transformants per microgram of donor DNA. In contrast, introduction of the rnhC151::cat mutation of BEST220 into BEST218 (rnhB21::neo) resulted in strains that formed very tiny colonies on selection plates containing chloramphenicol at frequencies similar to those of proB or leuB transfer. However, these tiny colonies did not produce viable colonies when restreaked on fresh plates, even in the absence of antibiotics. Introduction of the ypdQ mutation gave no phenotype, regardless of what other mutations were present in the recipient strain. All of the viable mutants were able to grow in the temperature range of 25 to 50°C, as does the wild-type strain. The results are interpreted to indicate that loss of both RNase HII (rnhB) and RNase HIII (rnhC) renders B. subtilis inviable. From the formation of colonies of inviable cells in the BEST220 × BEST218 cross, it can be interpreted that low levels of RnhC allow for consecutive cell divisions. The number of cell divisions may be enough to form visible (tiny) colonies. In the other cross, BEST218 × BEST220, RnhB was rapidly degraded or diluted out before producing sufficient numbers of cells.
rnhC homologues in the database.
On searching the current database for sequences related to B. subtilis rnhC, homologues were detected in various bacterial and eukaryotic genomes. However, most had been already designated rnh or rnhB in the database due to the slight similarity between rnhB and rnhC, as indicated in Fig. 3, with the exception of three genes. Two of these were from Mycoplasma species, the putative gene from Mycoplasma genitalium (accession no. MG199) (8, 33a) and the putative gene from Mycoplasma pneumoniae (accession no. C09_orf143b) (11, 33a), and the third was from the hyperthemophilic bacterium Aquifex aeolicus (accession no. aq_1768) (5, 33a). Their alignment with B. subtilis rnhC is shown in Fig. 4. As the rnhB gene in A. aeolicus was already reported (accession no. aq_1955) (5), it seems likely that this hyperthermophilic bacterium has an rnhC homologue and an rnhB homologue, although no obvious rnhA homologue was detected. It should be mentioned that an rnhC homologue was not found in the E. coli genome (33a).
FIG. 4.
Newly identified sequences that are similar to B. subtilis RNase HIII (rnhC). The three ORFs homologous to the B. subtilis rnhC sequence are aligned. The four conserved regions shown in Fig. 3 (I through IV) are indicated. Accession numbers are in the text. The numbers of N-terminal amino acid residues omitted in the alignment are shown in parentheses.
DISCUSSION
Two distinctive RNase H genes of B. subtilis 168.
Two clones encoding functional RNase H genes were isolated from a strain of the gram-positive bacterium B. subtilis. The two genes, rnhB and rnhC, were obtained based on the abilities to complement E. coli RNase H mutants in vivo (Fig. 2a) and to exhibit RNase H activity in vitro (Fig. 1). The temperature-sensitive E. coli strain MIC2067 newly adopted in this study was more specific for in vivo RNase H activity than MIC2067 newly adopted in this study was more specific for in vivo RNase H activity than MIC3037 (12a), and the MIC strains in Table 1 can discriminate the rnh genes in vivo if the genes are correctly expressed. In addition to these functional B. subtilis RNase H genes, a gene, ypdQ, that has some similarity to rnhA in the B. subtilis genome was also identified. However, the ypdQ clone was negative for all RNase H criteria. It remains to be demonstrated whether the failure of the ypdQ gene to suppress the temperature-sensitive phenotypes of MIC strains is due to either incorrect expression or low activity in E. coli. Alternatively, as YpdQ lacks the basic protruding region characteristic of E. coli RNase HI (25, 41), it may require an additional polypeptide for RNase H activity, similar to that observed for RNase H of HIV reverse transcriptase (28). It may be worth testing the ypdQ homologues observed in related B. subtilis species for enzyme activity.
Are RNase H proteins essential for B. subtilis?
Because the E. coli K-12 genome seems not to have an rnhC homologue, rnhA and rnhB double mutants such as MIC2067 completely abolish RNase H. Although MIC2067 shows a temperature-sensitive growth phenotype, double mutants of a certain E. coli genetic background grow normally (12a). In contrast, the rnhB and rnhC double mutant of B. subtilis 168 is unable to form viable colonies. These observations suggest that RNase H-requiring biological processes in B. subtilis are different from those suggested for E. coli (1, 4, 15, 16, 27, 35, 36). Distinctive pI values of the two B. subtilis RNase H genes (5.5 for rnhB and 10.1 for rnhC), compared with those of E. coli (9.7 for rnhA and 6.9 for rnhB), may imply different roles or substrate recognition properties in vivo. Alternative explanations include the acquisition of a suppressor mutation in laboratory strains of E. coli resulting in complete loss of RNase H proteins. Isolation of a B. subtilis rnhB and rnhC double mutant that has a suppressor mutation(s) is underway.
The requirement for RNase H in B. subtilis raises the question of whether RNase H is dispensable for bacteria. Complete DNA sequences are known for two Mycoplasma genomes, in which no homologue of rnhA and rnhB was reported, indicating that RNase H is dispensable (29). However, the presence of homologues to the newly characterized rnhC in these Mycoplasma species (Fig. 4) may imply that the single bacterial RNase H of the smallest genomes performs a function similar to the two RNase H proteins of B. subtilis.
The genome of A. aeolicus, a hyperthermophilic hydrogen-oxidizing bacterium, has both rnhB and rnhC but lacks an rnhA homologue. The bacterium grows at 95°C, similar to primordial forms of life (5). This suggests that the two rnh genes of A. aeolicus represent an ancestral structure of the two present B. subtilis RNase H genes.
ACKNOWLEDGMENTS
We thank A. Ogura, K. Matsui, K. Fujita, and M. Murayama for technical help. We especially thank N. Ohtani for kindly communicating data prior to publication.
REFERENCES
- 1.Asai T, Kogoma T. D-loops and R-loops: alternative mechanisms for the initiation of chromosome replication in Escherichia coli. J Bacteriol. 1994;176:1807–1812. doi: 10.1128/jb.176.7.1807-1812.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carl P L, Bloom A D, Crouch R J. Isolation and mapping of a mutation in Escherichia coli with altered levels of ribonuclease H. J Bacteriol. 1972;144:28–35. doi: 10.1128/jb.144.1.28-35.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crouch R J. Ribonuclease H: from discovery to 3D structure. New Biol. 1990;2:771–777. [PubMed] [Google Scholar]
- 4.Dasgupta S, Masukata H, Tomizawa J. Multiple mechanisms for initiation of ColE1 DNA replication: DNA synthesis in the presence and absence of ribonuclease H. Cell. 1987;24:1113–1122. doi: 10.1016/0092-8674(87)90597-6. [DOI] [PubMed] [Google Scholar]
- 5.Deckert G, Warren P V, Gaasterland T, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 6.Doi N, Yomo T, Itaya M, Yanagawa H. Insertion of foreign random sequences of 120 amino acid residues into an active enzyme. FEBS Lett. 1998;427:51–54. doi: 10.1016/s0014-5793(96)01522-0. [DOI] [PubMed] [Google Scholar]
- 7.Frank P, Braunshofer C, Wintrersburger U. Yeast RNase H(35) is the counterpart of the mammalian RNase HI, and is evolutionary related to prokaryotic RNase HII. FEBS Lett. 1998;421:23–26. doi: 10.1016/s0014-5793(97)01528-7. [DOI] [PubMed] [Google Scholar]
- 8.Fraser C M, Gocayne J D, White O, et al. The minimal genome complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 9.Haruki M, Noguchi E, Akasako A, Oobatake M, Itaya M, Kanaya S. A novel strategy for stabilization of Escherichia coli ribonuclease HI involving a screen for an intragenic suppressor of carboxyl-terminal deletions. J Biol Chem. 1994;269:26904–26911. [PubMed] [Google Scholar]
- 10.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 11.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa K, Okumura M, Katayanagi K, Kimura S, Kanaya S, Nakamura H, Morikawa K. Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution. J Mol Biol. 1993;230:529–542. doi: 10.1006/jmbi.1993.1169. [DOI] [PubMed] [Google Scholar]
- 12a.Itaya, M. Unpublished data.
- 13.Itaya M. Isolation and characterization of a second ribonuclease H (RNase HII) of Escherichia coli K12 encoded by the rnhB gene. Proc Natl Acad Sci USA. 1990;87:8587–8591. doi: 10.1073/pnas.87.21.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itaya M. Stability and asymmetric replication of the Bacillus subtilis 168 chromosome. J Bacteriol. 1993;175:741–749. doi: 10.1128/jb.175.3.741-749.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itaya M, Crouch R J. A combination of RNase H (rnh) and recBCD or sbcB mutations in E. coli K-12 adversely affects growth. Mol Gen Genet. 1991;227:424–432. doi: 10.1007/BF00273933. [DOI] [PubMed] [Google Scholar]
- 16.Itaya M, Crouch R J. Correlation of activity with phenotypes of Escherichia coli partial function mutants of rnh, the gene encoding RNase H. Mol Gen Genet. 1991;227:433–437. doi: 10.1007/BF00273934. [DOI] [PubMed] [Google Scholar]
- 17.Itaya M, Kondo K. Molecular cloning of a ribonuclease H (RNase HI) gene from an extreme thermophile, Thermus thermophilus HB8: a thermostable RNase H can functionally replace the Escherichia coli enzyme in vivo. Nucleic Acids Res. 1991;19:4443–4449. doi: 10.1093/nar/19.16.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itaya M, McKelvin D, Chatterjie S K, Crouch R J. Selective cloning of genes encoding RNase H from Salmonella typhimurium, Saccharomyces cerevisiae and Escherichia coli rnh mutant. Mol Gen Genet. 1991;227:438–445. doi: 10.1007/BF00273935. [DOI] [PubMed] [Google Scholar]
- 19.Itaya M, Tanaka T. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene-directed mutagenesis method. J Mol Biol. 1991;220:631–648. doi: 10.1016/0022-2836(91)90106-g. [DOI] [PubMed] [Google Scholar]
- 20.Itaya M, Yamaguchi I, Kobayashi K, Endo T, Tanaka T. The blasticidin S resistance gene (bsr) selectable in a single copy state in the Bacillus subtilis chromosome. J Biochem (Tokyo) 1990;107:799–801. doi: 10.1093/oxfordjournals.jbchem.a123128. [DOI] [PubMed] [Google Scholar]
- 21.Kanaya S, Crouch R J. DNA sequence of the gene coding for Escherichia coli ribonuclease H. J Biol Chem. 1983;258:1276–1281. [PubMed] [Google Scholar]
- 22.Kanaya S, Katsuda C, Kimura S, Nakai T, Kitakuni E, Nakamura H, Katayanagi K, Morikawa K, Ikehara M. Stabilization of Escherichia coli ribonuclease H by introduction of an artificial disulfide bond. J Biol Chem. 1991;266:6038–6044. [PubMed] [Google Scholar]
- 23.Kanaya S, Nakai C, Konishi A, Inoue H, Ohtsuka E, Ikehara M. A hybrid ribonuclease H. A novel RNA cleaving enzyme with sequence-specific recognition. J Biol Chem. 1992;267:8492–8498. [PubMed] [Google Scholar]
- 24.Kashiwagi T, Jeanteur D, Haruki M, Katayanagi M, Kanaya S, Morikawa K. Proposal for new catalytic roles for two invariant residues in Escherichia coli ribonuclease HI. Protein Eng. 1996;9:857–867. doi: 10.1093/protein/9.10.857. [DOI] [PubMed] [Google Scholar]
- 25.Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, Matsuzaki T, Morikawa K. Three dimensional structure of ribonuclease H from E. coli. Nature. 1990;347:306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- 26.Keller W, Crouch R J. Degradation of RNA.DNA hybrids by ribonuclease H and DNA polymerases of cellular and viral origin. Proc Natl Acad Sci USA. 1972;69:3360–3364. doi: 10.1073/pnas.69.11.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kogoma T, Subia N L, Meyenberg K. Function of ribonuclease H in initiation of DNA replication in Escherichia coli K-12. Mol Gen Genet. 1985;200:103–109. doi: 10.1007/BF00383320. [DOI] [PubMed] [Google Scholar]
- 28.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 29.Kooin E V, Mushegian A R, Bork P. Non-orthologous gene displacement. Trends Genet. 1996;12:334–336. [PubMed] [Google Scholar]
- 30.Kunst F, Ogasawara N, Albertini A M, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 31.Maki H, Horiuchi T, Sekiguchi M. Structure and expression of the dnaQ and the RNase H genes of Escherichia coli: overlap of the promoter regions. Proc Natl Acad Sci USA. 1983;80:7137–7141. doi: 10.1073/pnas.80.23.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandel M, Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970;53:159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- 33.Mian S I. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic Acids Res. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Microbial Genome WWW Home Page. [Online.] National Institute of Genetics. http://ddbjs4d.genes.nig.ac.jp:8880/ [29 August 1998, last date accessed.]
- 34.Mizrahi V, Huberts P, Dawes S S, Dudding L R. A PCR method for the sequence analysis of the gyrA, polA, and rnhA gene segments from mycobacteria. Gene. 1993;136:287–290. doi: 10.1016/0378-1119(93)90481-h. [DOI] [PubMed] [Google Scholar]
- 35.Ogawa T, Pickett G G, Kogoma T, Kornberg A. RNase H confers specificity in the dnaA-dependent initiation of replication at the unique origin of the Escherichia coli chromosome in vivo and in vitro. Proc Natl Acad Sci USA. 1984;81:1040–1044. doi: 10.1073/pnas.81.4.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Ohtani, N., and S. Kanaya. Unpublished data.
- 36.Quinones A, Kucherer C, Piechocki R, Messer W. Reduced transcription of the rnh gene in Escherichia coli mutants expressing the SOS regulon constitutively. Mol Gen Genet. 1987;206:95–100. doi: 10.1007/BF00326542. [DOI] [PubMed] [Google Scholar]
- 37.Saito H, Miura K. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Studier F W, Moffat B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T, Kawano N. Cloning vehicles for the homologous Bacillus subtilis host-vector system. Gene. 1980;10:131–136. doi: 10.1016/0378-1119(80)90130-4. [DOI] [PubMed] [Google Scholar]
- 41.Yang W, Hendrickson W A, Crouch R J, Satow Y. Structure of ribonuclease H phased at 2 Ä resolution by MAD analysis. Science. 1990;249:1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y-B, Ayalew S, Lacks S A. The rnhB gene encoding RNase HII of Streptococcus pneumoniae and evidence of conserved motifs in eukaryotic genes. J Bacteriol. 1997;179:3828–3836. doi: 10.1128/jb.179.12.3828-3836.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]