Abstract
High-throughput lysis and proteolytic digestion of biopsy-level tissue specimens is a major bottleneck for clinical proteomics. Here we describe a detailed protocol of pressure cycling technology (PCT)-assisted sample preparation for proteomic analysis of biopsy tissues. A piece of fresh frozen or formalin-fixed paraffin-embedded tissue weighing ~0.1–2 mg is placed in a 150 μL pressure-resistant tube called a PCT-MicroTube with proper lysis buffer. After closing with a PCT-MicroPestle, a batch of 16 PCT-MicroTubes are placed in a Barocycler, which imposes oscillating pressure to the samples from one atmosphere to up to ~3,000 times atmospheric pressure. The pressure cycling schemes are optimized for tissue lysis and protein digestion, and can be programmed in the Barocycler to allow reproducible, robust and efficient protein extraction and proteolysis digestion for mass spectrometry-based proteomics. This method allows effective preparation of not only fresh frozen and formalin-fixed paraffin-embedded tissue, but also cells, feces and tear strips. It takes ~3 h to process 16 samples in one batch. The resulting peptides can be analyzed by various mass spectrometry-based proteomics methods. We demonstrate the applications of this protocol with mouse kidney tissue and eight types of human tumors.
Subject terms: Proteomic analysis, Mass spectrometry
High-throughput lysis and proteolytic digestion of biopsy-level tissue specimens is a major bottleneck for clinical proteomics. This protocol describes pressure cycling technology (PCT)-assisted sample preparation of biopsy tissues.
Introduction
Tissues are common specimens in biomedical research and applications with histomorphological examination of clinical tissue specimens as the major approach to diagnose most cancers. Emerging omics technologies allow comprehensive investigation of molecular regulation in tissue specimens, offering systematic insights to disease pathogenesis in addition to uncovering potential therapeutic targets. Mass spectrometry (MS)-based quantitative proteomics allows for the identification and quantification of thousands of proteins expressed in various tissue specimens, offering invaluable information for disease diagnosis and treatment1–3. For instance, quantitative proteomics of tumor and cognate nontumor tissues can identify candidate tumor-specific proteins as biomarkers4–7. Moreover, in formalin-fixed paraffin-embedded (FFPE) specimens where viruses and bacteria are disinfected for safe proteomic analysis, quantitative proteomics has been applied in coronavirus disease 2019 (COVID-19) autopsies to understand host responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) (ref. 8).
For analysis of large cohorts of clinical samples, reproducible and rapid sample preparation are inseparable technical requirements. Since each clinical sample is unique, valuable and not renewable, it is essential that the extraction of peptides from minute amounts of tissues should be thorough, high throughput and highly reproducible, which is difficult to achieve with other available methods such as filter-aided sample preparation (FASP)9, direct tissue proteomics10, single-pot, solid-phase-enhanced sample-preparation11,12 and high hydrostatic pressure13, in particular for challenging tissues such as punched FFPE tissues. The pressure cycling technology (PCT)-assisted proteomic sample preparation method generates alternating high pressure and applies it to samples in pressure-resistant tubes, called PCT-MicroTubes (Fig. 1). The cycling pressure promotes tissue lysis and enzymatic reactions14. PCT is implemented in a Barocycler (Fig. 1), which introduces these periodic pressure oscillations, accelerating proteolysis through alternating ultrahigh (up to 45,000 psi) and mild pressure, which can obtain reproducibly high yields of peptides from tissue biopsies. As a machine-controlled process with a consistent reaction volume, PCT allows for repeatable preparation of hundreds to thousands of samples, minimizing technical variation and batch effects. Optimized PCT can homogenize minute amounts of tissues (0.2–2 mg) and cells (~50,000–500,000 cells) and extracts sufficient numbers of peptides for multiple proteome analyses15. However, with this method, some hard and relatively large tissues remain incompletely lysed. Shao et al. introduced a miniaturized and disposable mechanical tissue homogenizer, the PCT-MicroPestle (Fig. 1)16. Compared with the conventional PCT method15, PCT-MicroPestle workflow increased the yield of extracted peptides by 20–40% when applied to different types of tissue samples. After lysis and digestion, extracted peptides can be directly injected into a high-performance liquid chromatograph (HPLC) coupled to MS for proteomic analysis, or tagged with labeling reagents before LC–MS analysis, such as tandem mass tag17 and isobaric tags for relative and absolute quantitation18.
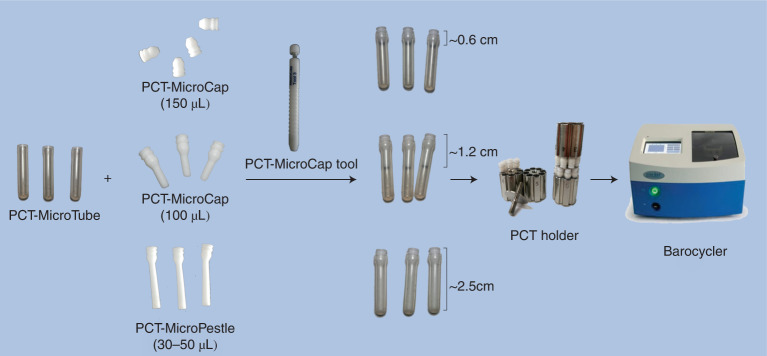
Fig. 1. PCT-assisted sample preparation equipment setup.
Photos of the PCT-MicroTubes, the PCT-MicroCaps, the PCT-MicroPestle, the PCT-MicroCap tool for handling the caps, the PCT holder and the barocycler.
MS is the principal technology of quantitative proteomics. There are two main MS-based proteomic strategies: targeted and untargeted proteomics. Selective or multiple reaction monitoring19 and parallel reaction monitoring20,21 are the two most common targeted MS acquisition modes, which offer superior reproducibility and sensitivity by reducing the interference of ions other than the targeted peptide precursor ions. For untargeted proteomics, data-dependent acquisition (DDA) and data-independent acquisition (DIA)22–24 are widely used to discover potential biomarkers in clinical specimens.
Here we present an optimized workflow for high-throughput proteome analysis of tissues using PCT-assisted tissue sample preparation (Fig. 2). As little as 0.1 mg of tissue or 5,000 cells could be effectively processed using this protocol. This method also enables analysis of other clinical biospecimens beyond tissues, including cells, feces and tear strips (Fig. 2a). This protocol directly processes the specific strips that are a widely used tool to collect tear samples in the clinic to obtain information about eye and systemic health. Furthermore, the workflow offers a high peptide yield and a high degree of reproducibility25. It is especially suitable for large clinical cohort studies where limiting amounts of patient tissue samples are available for analysis.
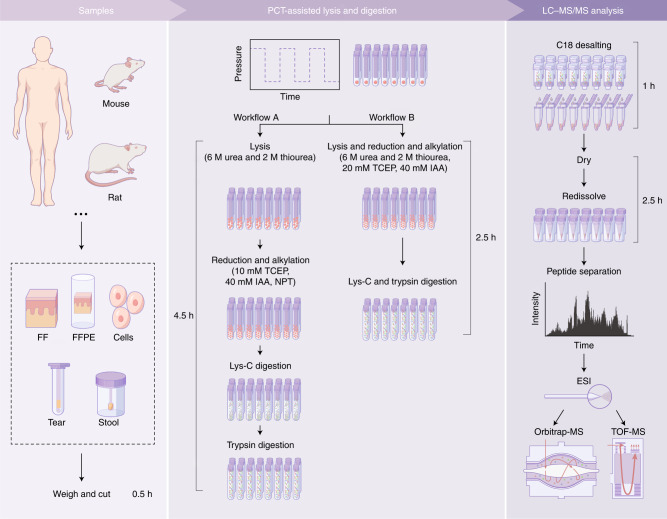
Fig. 2. Workflow and experimental design for PCT-assisted sample preparation.
Various types of samples can be processed by this protocol using two workflows. Workflow A comprises four major steps, including tissue lysis, protein reduction and alkylation, Lys-C digestion and trypsin digestion. A simplified workflow B can achieve comparable performance. The resultant peptides are cleaned using C18 cartridges before LC–MS/MS analysis. NPT, normal pressure and temperature; ESI, electrospray ionization; TOF-MS, time-of-flight mass spectrometry.
Development of the protocol
The original PCT protocol for processing fresh frozen (FF) tissue biopsies allows for the preparation of up to six samples within 6–8 h (ref. 4), and improved the peptide yield by ~60% compared with the conventional in-solution method. We then evaluated the minimal sample amount requirement for robust PCT-assisted sample preparation, and found that ~50,000 human cells and 0.2–0.5 mg of wet mouse or human tissue are sufficient to produce enough peptides for multiple MS analyses15. We further developed a MicroPestle device that increased the yield of extracted peptides by 20–40% when applied to different types of tissue samples16. Improvements to the Barocycler (Fig. 1) instrument and protocol optimization allowed for the simultaneous processing of 16 samples26. The protocol was further optimized to permit analysis of FFPE tissues27. Recently, we improved the method to accelerate and simplify some procedures and to generate enhanced peptide yield, as described as the second-generation PCT method25.
Here in this detailed protocol, two sample preparation schemes are provided. The first scheme is a modified conventional PCT-based method comprising four main steps (Fig. 2, workflow A). We use 6 M urea and 2 M thiourea as the lysis buffer to extract the proteins assisted by PCT. After tissue lysis and protein extraction, we then reduce and alkylate proteins with 10 mM Tris (2-carboxyethyl) phosphine (TCEP) and 40 mM iodoacetamide (IAA) on a thermomixer at atmosphere. Next, Lys-C (enzyme-to-substrate ratio, 1:40) and trypsin (enzyme-to-substrate ratio, 1:50) are sequentially added to the foregoing mixture in two steps for PCT-assisted enzymatic digestion. The other scheme is a two-step PCT-based method (Fig. 2, workflow B) that combines PCT-assisted lysis, reduction and alkylation in the first step, and PCT-assisted Lys-C/trypsin enzymatic digestion in the second step. The optimal enzyme-to-substrate ratios of Lys-C and trypsin are 1:80 and 1:20, respectively. This procedure exhibits higher throughput and peptide yield with digestion efficiency comparable to the conventional four-step method. Digestion efficiency is evaluated by peptide and protein identifications, and by the percentage of missed cleavages. After both workflows, the prepared peptides are desalted using C18 solid phase extraction (SPE) columns before MS analysis.
Applications of the method
This protocol has been developed to enable the analysis of minute amounts of clinical specimens, including tissues, cells, feces and tear strips, among others (Fig. 2). These samples can come from different species, including humans, mice and rats. Using this PCT-based protocol, we can generate ~50 µg of peptides from 0.5 mg FF or FFPE tissue. From this amount of tissue, the total protein amount is estimated to be ~100 µg, so with a total volume of 50 µL during the lysis procedure, the protein concentration is ~2 µg/µL which is empirically optimal for in-solution digestion. If the tissue is >2 mg, the concentration of proteins, even if fully solubilized, will go beyond 8 µg/µL, which is not empirically optimal for effective proteolytic digestion. Therefore, the PCT-based digestion efficiency decreases when applied to larger amounts (>0.5 mg tissue) of samples15 or in the case of prolonged lysis. Moreover, PCT-based digestion is not recommended for fluid samples such as plasma, serum and urine unless they are solidified in papers, after which this protocol can also process these samples.
With the advantage of smaller sample size requirement, high degree of reproducibility and high throughput with minimal hands-on time, this protocol is suitable for use in a variety of biomedical applications with a focus on tissue proteomics4,5,8. This protocol is essentially designed for using minute amounts of tissue samples, and this ability could also be exploited to understand tissue heterogeneity. For example, if the research objective is to compare the proteomes among different layers of a skin specimen, this PCT-assisted sample preparation method allows effective proteomic analysis of each layer of the skin after dissection of small (0.1 mg) tissue samples. In some cases, we have also successfully performed proteomics analysis for tissue samples smaller than 0.1 mg. On the other hand, if the research goal is to obtain a comprehensive and stable proteomic profiling of a highly heterogeneous tissue such as a skin of various distinct layers, one may process multiple pieces of these tissues with the protocol and mix the resultant peptides for LC–MS/MS analysis to obtain a more homogeneous representation of the proteome.
Considerations for experimental design
One of the major advantages of the PCT-based workflow is the small sample consumption, so a typical tissue biopsy weighing 0.5–1 mg is sufficient for peptide preparation. To achieve consistent sample quality, FF tissue samples should be stored at −80 °C or in liquid nitrogen, whereas FFPE tissues can be stored at room temperature (RT, 20–25 °C). Before storage in −80 °C or liquid nitrogen, the FF tissue specimens should be washed with 4 °C PBS three times to minimize any blood contamination. Cell samples are collected and stored as dry pellets at −80 °C or in liquid nitrogen. Tear strips are stored at −20 °C or −80 °C within 1 h after collection. Feces samples are scooped from the middle part of the feces, where urine and water are not mixed, placed in a centrifuge tube and stored at −80 °C within 1 h after collection.
For a large clinical cohort, quality control (QC) during sample preparation and data acquisition is necessary. Currently, the maximum capacity of the MicroTube Cartridges that can be used in the PCT instrument is 16. Therefore, it is recommended to include a QC sample that is identical or similar to the biopsy sample in every batch. This QC sample is for evaluating technical variation during sample preparation. We also recommend a pooled peptide mixture for QC during MS data acquisition.
Advantages and limitations
Compared with other protocols for proteomic sample preparation, the major benefits of this protocol include the ability of processing minute amounts of compact tissues with maximized peptide yield, reproducibility of the workflow, semi-automation and robustness. These are essential for high-throughput quantification of large-scale cohort studies. Once the tissue is placed inside the PCT-MicroTube with lysis buffer, all reactions occur in the same tube, and there is no evidence that a specific group of proteins are lost using this protocol. Our data have shown that the percentage of membrane proteins is higher with this protocol compared with conventional in-solution digestion4. Recent proteomic investigations of >20 types of human tissues prepared by this protocol identified >10,000 proteins of all Gene Ontology annotations, and showed no bias in certain protein groups28,29. Proteomic analysis of samples prepared by this method reduces artificial protein modifications including deamidation30 and scrambled disulfides31.
This protocol is applicable to all types of FF and FFPE tissue samples, but not necessarily the best choice for all of them. It is suitable for minute amount of tissues of a weight between 0.1 and 2 mg and particularly beneficial for compact tissues that can hardly be solubilized for in-solution proteolytic digestion. FFPE slides can also be effectively processed by other protocols such as FASP9; however, the loss of charged hydrophilic peptides is inevitable during sodium dodecyl sulfate removal in the FASP protocol32,33. Tissues prepared by laser-assisted microdissection can also be effectively analyzed using this protocol34–36. Tissues heavier than 2 mg cannot be effectively processed by this protocol unless divided into aliquots of the optimal weight15,16. The protocol has marginal benefits to liquid samples such as plasma, serum and urine unless they are solidified in papers.
The PCT-assisted method is currently semi-automated and needs to be further developed into full automation. The pressure and cycling scheme have been optimized, and work pretty well for most tissue samples we have processed, as we demonstrate in this protocol paper. One could also try to further optimize these parameters, including the reaction temperature, although this is not required. The lysis buffer could be also altered according to the tissue type. Compared with other tissue lysis and proteolytic digestion methods, a major hurdle for wide applications of this method is the requirement of a special instrument called Barocycler (Fig. 1), the availability of which is increasing owing to its unique benefits in the proteomic analysis of minute amounts of tissues as described above. Once the instrument is set up, cost is no longer a limitation.
Materials
Biological materials
-
Tissue samples (mouse kidney samples were obtained from a 6-month-old C57BL6 male mouse; the collection and testing of 32 FFPE human tissue samples were obtained from 16 patients; and tissue was stored at −80 °C)
Caution
All experiments should be performed according to institutional and national regulations. The procedures in this protocol were reviewed and approved by the Ethics Committee of Harbin Medical University Cancer Hospital and Medical Ethical Committee of Westlake University. Written informed consent was obtained from each patient.
Reagents
-
Heptane (Sigma-Aldrich, cat. no. 246654-2L)
Caution
Heptane is a health hazard level 5 compound. Use the solution in a fume hood, and handle with gloves.
Ethanol (EtOH; Sinopharm Chemical Reagent limited corporation, cat. no. 10009218)
-
Formic acid (FA; Fisher Scientific, cat. no. A117-50)
Caution
FA is a health hazard level 3 compound with a flammability level of 3. Avoid contact with skin and eyes, and avoid inhalation. Use the solution in a fume hood, and handle with gloves.
Trizma Base (Sigma-Aldrich, cat. no. T6791-500G)
-
HCl, 1 mmol, pH 3 (Hualishi, cat. no. HLS HCL001C; SCR, cat. no. 10011008)
Caution
HCl is a health hazard level 3 compound. Avoid contact with skin and eyes, and avoid inhalation. Use the solution in a fume hood, and handle with gloves.
Phosphate-buffered saline (PBS; Hyclone, cat. no. SH30256.01)
-
Urea (Sigma-Aldrich, cat. no. U1250-1KG)
Caution
Urea is a health hazard level 2 compound. Avoid contact with skin and eyes, and avoid inhalation.
-
Thiourea (Sigma-Aldrich, cat. no. T8656-500G)
Caution
Thiourea is a health hazard level 2 compound. Avoid contact with skin and eyes, and avoid inhalation.
Ammonium bicarbonate (ABB; General-Reagent, cat. no. G12990A)
-
TCEP (Damas-Beta, cat. no. 61820E)
Caution
TCEP is a health hazard level 5 compound. Avoid contact with skin and eyes, and avoid inhalation.
-
IAA (Sigma-Aldrich, cat. no. 16125-25G)
Caution
IAA is a health hazard level 3 compound. Avoid contact with skin and eyes, and avoid inhalation.
Lys-C (Hualishi, cat. no. HLS LYS001C)
Trypsin (Hualishi, cat. no. HLS TRY001C)
-
Trifluoroacetic acid (TFA; Fisher Chemical, T/3258/PB05)
Caution
TFA is a health hazard level 3 compound with a flammability level of 3. Avoid contact with skin and eyes, and avoid inhalation. Use the solution in a fume hood, and handle with gloves.
-
Methanol (MeOH; Sinopharm Chemical Reagent limited corporation, cat. no. 10014118)
Caution
MeOH is a health hazard level 1 compound. Avoid contact with skin and eyes, and avoid inhalation.
-
Acetonitrile (ACN; General-Reagent, cat. no. G80988B; Fisher Chemical, cat. no. A955-4)
Caution
ACN is a health hazard level 4 compound. Avoid contact with skin and eyes, and avoid inhalation.
Water (Fisher Chemical, cat. no. W6-4)
Internal retention time (iRT) normalization kit (iRT kit, Biognosys AG, cat. no. Ki-3002-1)
Equipment
Tubes (microtubes, 1.5 mL, 2 mL; Axygen, cat. no. MCT-150-C, MCT-200-C)
Milli-Q (MERCK, Milli-Q Direct)
Analytical balance (Mettler Toledo, cat. no. XS205)
Pipettes (Eppendorf)
Pipette tips (Axygen, cat. no. T-1000-B, T-200-Y, T-300)
Mini vacuum pump (Kylin-Bell, cat. no. GL-810)
Thermo shaker (Allsheng, cat. no. MS-100)
Barocycler 2320-EXT (Pressure BioSciences, Inc.)
PCT-MicroCap Tool, PCT-MicroTube (150 µL), PCT-MicroPestle, PCT-MicroCaps (100 µL, 150 µL) (Pressure BioSciences)
Rex laboratory pH meter (INESA, cat. no. PHS-3C)
Vortex (Allsheng, cat. no. MTV-1)
Centrifuge (Eppendorf, cat. no. 5430R)
Low-speed benchtop centrifuge (Hangzhou Allsheng Instruments, cat. no. Mini-6KS)
SOLAµ SPE HRP plates (Thermo Scientific, cat. no. C42520A1A)
Speedvac vacuum concentrator (Thermo Scientific, cat. no. SPD2020)
ScanDrop2 (AnalytikJena)
Vial caps (Thermo Fisher Scientific Bermen, cat. no. C5000-54B)
Vials (Thermo Fisher Scientific Bermen, cat. no. C4000-11)
Nano-flow analytical columns (home-made, 1.9 µm, 120 Å, 150 mm × 75 µm)
Trap columns for Nano LC system (Thermo Fisher Scientific, cat. no. 164946, 3 µm, 100 Å, 20 mm × 75 µm; cat. no. 160454, 3 µm, 100 Å, 5 mm × 300 µm)
Nano-flow LC system (Thermo Fisher Scientific, DIONEX UltiMate 3000 RSLCnano System; Bruker Daltonics, nanoElute)
Q-Oribtrap MS system (Thermo Fisher Scientific, Q Exactive Hybrid Quadrupole-Orbitrap)
Tims-Q/TOF MS system (Bruker Daltonics, timsTOF Pro)
Software
Proteome Discoverer v2.5.0.400 (Thermo Fisher Scientific)
pFind v3.1.5 (Institute of Computing Technology, Chinese Academy of Sciences)
DIA-NN v1.8 (https://github.com/vdemichev/diann)
Reagent setup
Pretreatment solvents for processing FFPE samples
The dewaxing solvent is 100% (vol/vol) heptane. The gradient rehydration solvents are 100% (vol/vol) EtOH, 90% (vol/vol) EtOH in water and 75% (vol/vol) EtOH in water. The solvent for complete rehydration and acidic hydrolysis is 0.1% (vol/vol) FA in water. All solvents are of analytical grade. The dewaxing solvent and rehydration solvent are stable at RT for up to 1 month with the cap closed properly. The solvent for basic hydrolysis is 0.1 M Tris–HCl (pH 10.0).
Caution
Tris–HCl is a health hazard level 2 compound. Avoid contact with skin and eyes, and avoid inhalation.
Critical
100% heptane should be stored strictly away from moisture and light.
Critical
0.1 M Tris–HCl (pH 10.0) should be freshly made each time right before usage because of its instability.
Pretreatment solvents for processing FF samples
The washing solvents are 70% (vol/vol) EtOH in water, 85% (vol/vol) EtOH in water and 100% (vol/vol) EtOH. All solvents are of analytical grade. The pretreatment solvents for FF samples are stable at RT for up to 1 month when sealed properly.
Stock solution for lysis, reduction and alkylation buffers
Stock solution is 0.1 M ABB (pH 8). It can be stored at 4 °C for up to a week. Check its pH before usage.
Lysis buffer
For FF tissues, FFPE tissues, cells and feces, the lysis buffer contains 6 M urea and 2 M thiourea in 0.1 M ABB. For tear strips, the lysis buffer contains 8 M urea in 0.1 M ABB.
Critical
Lysis buffer can be aliquoted and stored at −80 °C for up to 1 month after it has been freshly prepared. Avoid multiple freeze–thaw cycles when using lysis buffer.
Reduction and alkylation reagent stocks
The reduction stocks for PCT-assisted lysis are 200 mM TCEP and 100 mM TCEP in 0.1 M ABB, respectively. The alkylation stock for PCT-assisted lysis is 800 mM IAA.
Critical
Reduction and alkylation stocks can be divided into aliquots and stored at −80 °C for up to 1 month. Avoid multiple freeze–thaw cycles when using them. IAA stocks should be kept strictly away from light.
Lys-C and trypsin stocks
The Lys-C stock used in this study is 0.25 mg/mL by dissolving 50 µg Lys-C in 200 µL 1 mM HCl. The trypsin stock used in this study is 0.25 mg/mL by dissolving 100 µg trypsin in 400 µL 1 mM HCl.
Critical
Enzyme stocks can be divided into aliquots and stored at −80 °C for up to 1 month. Avoid multiple freeze–thaw cycles when using them.
Desalting solvents
The activation solvent is 100% (vol/vol) MeOH. The equilibration solvents are 80% (vol/vol) ACN/0.1% (vol/vol) TFA in water, and 2% (vol/vol) ACN/0.1% (vol/vol) TFA in water. The washing solvent is 2% (vol/vol) ACN/0.1% (vol/vol) TFA in water. The elution solvent is 60% (vol/vol) ACN/0.1% (vol/vol) TFA in water. All solvents are of analytical grade.
Critical
Desalting solvents can be prepared in a big stock and are stable at RT for up to 2 months when properly sealed.
Buffers for UltiMate 3000 HPLC–MS/MS
Buffer A and sample loading buffer consist of 2% (vol/vol) ACN/0.1% (vol/vol) FA in water. Buffer B consists of 98% (vol/vol) ACN/0.1% (vol/vol) FA in water. HPLC buffer stocks (buffer A and buffer B) are stable for up to 2 weeks at RT if kept in darkness. All solvents for HPLC–MS/MS are of HPLC or MS grade.
Buffers for nanoElute instrument HPLC–MS/MS
Buffer A and sample loading buffer consist of 0.1% (vol/vol) FA in water. Buffer B consists of 0.1% (vol/vol) FA in ACN. HPLC buffer stocks (buffer A and buffer B) are stable for up to 2 weeks at RT if kept in darkness. All solvents for HPLC–MS/MS are of HPCL or MS grade.
Equipment setup
PCT setup for four-step PCT-assisted lysis and digestion
| PCT scheme | Parameters | Setting |
|---|---|---|
| Lysis | Pressure | 45 kpsi |
| Cycles | 90 (60 for cell samples) | |
| On time | 25 s | |
| Off time | 10 s | |
| Temperature | 30 °C | |
| Lys-C digestion | Pressure | 20 kpsi |
| Cycles | 45 | |
| On time | 50 s | |
| Off time | 10 s | |
| Temperature | 30 °C | |
| Trypsin digestion | Pressure | 20 kpsi |
| Cycles | 90 | |
| On time | 50 s | |
| Off time | 10 s | |
| Temperature | 30 °C |
PCT setup for two-step PCT-assisted lysis and digestion
| PCT scheme | Parameters | Setting |
|---|---|---|
| Lysis | Pressure | 45 kpsi |
| Cycles | 90 (60 for cell samples) | |
| On time | 25 s | |
| Off time | 10 s | |
| Temperature | 30 °C | |
| Lys-C and trypsin digestion | Pressure | 20 kpsi |
| Cycles | 120 | |
| On time | 50 s | |
| Off time | 10 s | |
| Temperature | 30 °C |
Nano-flow HPLC setup for UltiMate 3000
The sample is loaded on the trap column at a flow rate of 6 µL/min, where the injection volume is 2 µL and the sample temperature is 7 °C
- The reference separation gradient at a flow rate of 300 nL/min is given below. Buffer A is 2% (vol/vol) ACN/0.1% (vol/vol) FA in water and buffer B is 98% (vol/vol) ACN/0.1% (vol/vol) FA in water. All reagents are of HPLC or MS grade
Time (min) Buffer B (%) 0 7 4 7 94 30 95 80 99 80 100 7 105 7
Nano-flow HPLC setup for nanoElute
The sample is loaded on the trap column at 217.5 bar, where the injection volume is 0.8 µL and the sample temperature is 7 °C
- The reference separation gradient at a flow rate of 300 nL/min is given below. Buffer A is 0.1% (vol/vol) FA in water, and buffer B is 0.1% (vol/vol) FA in ACN. All reagents are of HPLC or MS grade. The trap column is equilibrated by ten column volumes of buffer A at 217.5 bar, and separation column is equilibrated by four column volumes of buffer A at 500 bar
Time (min) Buffer B (%) 0 5 50 27 60 40 62 80 65 80
DDA-MS method
DDA-MS parameters used for nano-flow LC–MS/MS analysis are given below.
| Parameter | Value |
|---|---|
| Polarity | Positive |
| Spectrum data type | Centroid |
| Full MS | |
| Microscans | 1 |
| Resolution | 60,000 |
| AGC target | 3 × 106 |
| Maximum IT | 80 ms |
| Number of scan ranges | 390–1,010 m/z |
| dd-MS2 | |
| Microscans | 1 |
| Resolution | 30,000 |
| AGC target | 1 × 105 |
| Maximum IT | 80 ms |
| TopN | 20 |
| (N)CE | 27 |
| Dynamic exclusion | 20 s |
PulseDIA-PASEF method
PulseDIA-PASEF parameters used for nano-flow LC-MS/MS analysis are given below.
| Parameter | Value |
|---|---|
| Polarity | Positive |
| Ion mobility mode | Enable TIMS |
| Mode | |
| Scan range mass | 100–1,700 |
| Scan range 1/k0 | 0.7–1.3 |
| Resolution | 60,000 |
| Save spectra | Save frame spectra |
| Absolute threshold | 10 |
| Mobilogram peak detection | |
| Intensity threshold | 5,000.00 |
| MS/MS | |
| Number of PASEF MS/MS scans | 10 |
| Charge range | 0–5 |
| Scheduling | |
| Target intensity | 20,000 |
| Intensity threshold | 2,500 |
| Active exclusion | True |
| Release after | 0.40 min |
PulseDIA-PASEF MS isolation window settings are provided in Supplementary Data 3 and 4.
Procedure
Critical
To briefly spin down during the procedure, we always use a low-speed benchtop centrifuge at a low spin rate at RT.
Critical
In the following steps, if the temperature is not specified, the temperature is RT (20–25 °C) by default.
Critical
The procedures for handling the PCT-MicroTubes, PCT-MicroCaps and PCT-MicroPestles with the PCT-MicroCap tool are illustrated in Supplementary Video 1.
Sample pretreatment
Timing 2 h for FFPE, 0.5 h for FF, cells, feces and tear strips
- Dependent on the sample type (FFPE, FF, cells, feces or tear strips) to be processed, follow the procedure A–E as appropriate before performing PCT-assisted lysis and digestion. It is also important to prepare a QC sample that is relevant for the experiment. Refer to Box 1 for a method to prepare a tissue sample for QC.
-
(A)FFPE samples
-
(i)Dewaxing. Punch a core of FFPE tissue (~0.5 mg, 1 mm3) or cut a slice (25 mm2 × 20 µm) and place it in a 1.5 mL tube. Add 1 mL of heptane, vortex on a Thermo Shaker at 800 rpm for 10 min at 25 °C. Discard the heptane using a gel loading tip and a vacuum pump. Repeat once.Critical stepFor slice samples, centrifuging at 12,000g for 5 min is recommended before each solvent discard to minimize sample loss.
-
(ii)Rehydration. Rehydrate the dewaxed sample in a sequential EtOH series (100% EtOH, 90% EtOH and 75% EtOH) as follows. Add 1 mL of 100% EtOH into the tube, and vortex on a Thermo Shaker at 800 rpm for 5 min at 25 °C. Discard the supernatant. Repeat this rehydration procedure with 1 mL of 90% EtOH, followed by rehydration with 1 mL of 75% EtOH.Critical stepFor slice samples, centrifugation at 12,000g for 5 min is recommended before each solvent discard to minimize sample loss.
-
(iii)Complete rehydration and acidic hydrolysis. Add 200 µL of 0.1% FA to the 1.5 mL tube containing the tissue sample, cap the tube and vortex on a Thermo Shaker at 600 rpm for 30 min at 30 °C. Discard the supernatant.Critical stepFor slice samples, centrifugation at 12,000g for 5 min is recommended before each solvent discard to minimize sample loss.
-
(iv)Basic hydrolysis. Wash the tissue sample once with 0.1 M Tris–HCl (pH 10.0) by adding 200 µL of 0.1 M Tris–HCl to the tube. Carefully transfer the tissue into a new PCT-MicroTube using a pair of pointed forceps, ensuring that all tissue pieces are transferred. Add 12.5 µL 0.1 M Tris–HCl into the PCT-MicroTube, and close the PCT-MicroCap (100 µL cap size). Place the PCT-MicroTube in a fresh 1.5 mL tube, and cap the tube, then gently vortex on a Thermo Shaker at 600 rpm for 30 min at 95 °C. Immediately cool the sample on ice. Briefly spin down the 1.5 mL tube to bring the evaporated liquids down to the bottom of the PCT-MicroTube.Critical stepFor slice samples, centrifugation at 12,000g for 5 min is recommended before each solvent discard to minimize sample loss.Critical step0.1 M Tris–HCl (pH 10.0) should be freshly prepared, and the pH value should be accurately measured before basic hydrolysis. Heating should be no longer than 45 min. Substantial protein degradation was observed at heating for 45 min. Immediate cooling on ice is essential.Critical stepAlways briefly spin down the tube before opening the PCT-MicroCap of the PCT-MicroTube.
-
(i)
-
(B)FF samplesCriticalThe following steps are for FF tissue samples embedded in optimal cutting temperature (OCT) compound. For FF samples without OCT, the following steps could be skipped.
-
(C)Cell samples
-
(i)Wash cell pellets with 4 °C PBS. Centrifuge at 300g for 5 min at 4 °C. Discard the supernatant. Repeat three times.
-
(ii)Add 30 µL of lysis buffer (6 M urea and 2 M thiourea), vortex for 30 s by pipetting up and down gently. Transfer the cell suspension to a PCT-MicroTube using pipettes.
-
(i)
-
(D)Feces
-
(i)Wash ~200–300 mg feces with 500 µL of 4 °C PBS, centrifuge at 500g for 5 min at 4 °C and collect the supernatant. Repeat three times. Combine all the supernatant together.
-
(ii)Add 100 µL of the supernatant to a PCT-MicroTube. Place the PCT-MicroTube into a 1.5 mL tube, and centrifuge at 20,000g for 20 min at 4 °C. Discard the supernatant, and retain the precipitate.Critical stepThere is great heterogeneity in fecal protein content between different people. After this pretreatment, we generally use 50 µg protein for digestion.
-
(i)
-
(E)Tear strips
-
(i)Cut 3 mm2 tear strips using sterilized tweezers and scissors.Critical stepCut the tear strips to cover the wet length.
-
(i)
-
(A)
- Perform PCT-assisted lysis and digestion using either the four-step (option A) or the two-step (option B) procedure. Option B is preferred since it is the faster option to complete.
-
(A)Four-step PCT-assisted lysis and digestionTiming ~4.5 h per set of 16 samples
-
(i)Add 30 µL of lysis buffer (6 M urea and 2 M thiourea) to the sample in the PCT-MicroTube.
-
(ii)Close the PCT-MicroPestle in each PCT-MicroTube with a PCT-MicroCap Tool, then place the PCT-MicroTubes into a PCT holder and screw the PCT holder together to securely fasten the PCT-MicroTubes in place. Apply cycling pressure to the PCT holder containing the samples with the lysis scheme for 90 cycles (see ‘Equipment setup’).Critical stepEnsure the tissue samples are located directly below the PCT-MicroPestles.Critical stepEnsure the PCT-MicroTubes are located symmetrically in balance samples in the upper and lower parts of the PCT holder to avoid PCT-holder deformation due to uneven force under high pressure.
-
(iii)Remove the PCT-MicroTubes from the PCT holder after the pressure cycling reaction has finished and briefly spin down the PCT-MicroTubes by placing them in 1.5 mL tubes. Then open the PCT-MicroPestles using the PCT-MicroCap Tool to remove each PCT-MicroPestle and add 5 µL of 100 mM TCEP stock (final concentration 10 mM) and 2.5 µL of 800 mM IAA stock (final concentration 40 mM) to bring the reaction volume to 50 µL at this step. Close each PCT-MicroTube with a PCT-MicroCap (100 µL cap size) using the PCT-MicroCap Tool. Place each PCT-MicroTube to a fresh 1.5 mL tube, cap the tube and incubate in the dark for 30 min with gentle vortexing (800 rpm) on a Thermo Shaker at 25 °C.Critical stepAlways remember to briefly spin down the PCT-MicroTube in a 1.5 mL tube before opening the PCT-MicroPestle to minimize sample loss.
-
(iv)Briefly spin down the PCT-MicroTubes in the 1.5 mL tubes. Open the 1.5 mL tube caps, and take out the PCT-MicroTubes. First, remove each 100 µL PCT-MicroCap and add 45 µL of 0.1 M ABB to the PCT-MicroTube. Second, add 5 µL of 0.25 mg/mL Lys-C at an enzyme-to-substrate ratio of 1:40 (wt/wt)—for instance, 2.5 µg of Lys-C for 100 µg of proteins to be digested—into the lysate.Critical stepAlways remember to briefly spin down the PCT-MicroTube in a 1.5 mL tube before opening the PCT-MicroCap to minimize sample loss.Critical stepIn cases where the volume of lysis or the protein amount is different, adjust the volume of 0.1 M ABB accordingly to top up the reaction to a final volume of 100 µL.Critical stepAdding proteolytic enzyme requires a priori knowledge of the protein amount. However, we do not recommend direct measurement of protein concentration using the tissue sample being analyzed in this experiment because measuring protein concentration leads to significant sample loss. Protein yield could be estimated from the literature since the protocol is standardized and reproducible. Otherwise, a pilot experiment could be performed to obtain the protein yield from a particular type of sample. The final peptide yield is ~50% of the protein yield.
-
(v)Close the PCT-MicroCaps (100 µL cap size), place the PCT-MicroTubes into the PCT holder in balance and screw the PCT holder to avoid PCT-holder deformation due to uneven force under high pressure. Apply pressure cycling to the sample holder with Lys-C digestion scheme for 45 cycles (see ‘Equipment setup’).
-
(vi)Briefly spin down the PCT-MicroTubes, and open the PCT-MicroCaps as above. First, add 46 µL of 0.1 M ABB. Second, add 4 µL of 0.25 mg/mL trypsin at an enzyme-to-substrate ratio of 1:50 (wt/wt)—for instance, 2 µg of trypsin for 100 µg of proteins to be digested—into the lysate solution.Critical stepAlways remember to briefly spin down the PCT-MicroTube in a 1.5 mL tube before opening the PCT-MicroCap to minimize sample loss.Critical stepFor trypsin digestion, the urea concentration should be <1.6 M.Critical stepIn cases where the protein amount is different, adjust the volume of 0.1 M ABB accordingly to top up the reaction to a final volume of 150 µL.
-
(vii)Close the PCT-MicroCaps (150 µL cap size). Place the tubes into the PCT holder in balance, and screw the holder. Apply pressure cycling to the sample holder with trypsin digestion scheme for 90 cycles (see ‘Equipment setup’).
-
(viii)Remove the PCT-MicroTubes from the PCT holder after reaction has finished. Briefly spin down the PCT-MicroTubes. Open the PCT-MicroCap and transfer the samples to a 1.5 mL tube using a pipette. Add 18 µL of 10% TFA into the lysate solution (yielding a final TFA concentration of 1%) to stop the Lys-C and trypsin digestion.
-
(ix)Ensure the final pH is 2–3; otherwise, adjust the pH to 2–3 with 10% TFA. Centrifuge the solution at 12,000g for 5 min, and collect the supernatant (~160 µL).Pause pointPeptide solution can be stored at −20 °C for weeks and at −80 °C for months.
-
(i)
-
(B)Two-step PCT-assisted lysis and digestionTiming ~2.5 h per set of 16 samples
-
(i)Add 30 µL of lysis buffer (6 M urea and 2 M thiourea) to PCT-Microtubes containing tissue samples. We recommend the lysis buffer contain 8 M urea if tear strips are used for the sample.
-
(ii)Add 5 µL of 200 mM TCEP stock (final concentration 20 mM in the final volume of 50 µL) and 2.5 µL of 800 mM IAA stock (final concentration 40 mM in the final volume of 50 µL) to each PCT-MicroTube.
-
(iii)Close each PCT-MicroTube with a PCT-MicroPestle. Place the tubes into the PCT holder, and screw the PCT holder together.Critical stepEnsure the tissue samples are located directly below the PCT-MicroPestle.
-
(iv)Apply pressure cycling to the sample holder with the lysis scheme for 90 cycles (see ‘Equipment setup’).Critical stepEnsure the PCT-Microtubes are located symmetrically in balance in the upper and lower parts of the PCT holder to avoid PCT-holder deformation due to uneven force under high pressure.
-
(v)Remove the PCT-MicroTubes from the holder after the PCT-assisted reaction has finished. Briefly spin down the PCT-MicroTubes, then remove the PCT-MicroPestles. First, add 75 µL of 0.1 M ABB to each PCT-MicroTube. Second, add 5 µL of 0.25 mg/mL Lys-C at an enzyme-to-substrate ratio of 1:80 (wt/wt) (for instance, 1 µg of Lys-C for 80 µg of proteins to be digested). Third, add 20 µL of 0.25 mg/mL trypsin at an enzyme-to-substrate ratio of 1:20 (wt/wt) (for instance, 5 µg of trypsin for 100 µg of proteins to be digested).Critical stepAlways remember to spin down the PCT-MicroTubes before opening the PCT-MicroCap to minimize sample loss.Critical stepMake sure the working concentration for urea during enzymatic digestion is <1.6 M.Critical stepIn cases where the volume of lysis or the protein amount is different, adjust the volume of 0.1 M ABB accordingly to top up the reaction to a final volume of 150 µL.
-
(vi)Close each PCT-MicroTube with a PCT-MicroCap (150 µL cap size). Place the PCT-MicroTubes into a PCT holder, and screw the holder together. Apply pressure cycling to the sample holder with the Lys-C and trypsin digestion scheme for 120 cycles (see ‘Equipment setup’).
-
(vii)Remove the PCT-MicroTubes from the holder after the PCT-assisted reaction has finished. Briefly spin down the PCT-MicroTubes, and then open the PCT-MicroCaps. Transfer the sample to a 1.5 mL tube, and add 18 µL of 10% TFA into the lysate solution (for a final concentration of 1%) to stop the Lys-C and trypsin digestion.
-
(viii)Ensure the final pH is 2–3; otherwise, adjust the pH to 2–3 with TFA. Centrifuge the solution at 12,000g for 5 min, and collect the supernatant (~160 µL).Pause pointPeptide solution can be stored at −20 °C for weeks and at −80 °C for months.
-
(i)
-
(A)
Box 1 QC samples.
Take a piece of tissue (for example, our laboratory frequently uses FF mouse liver) weighing ~0.5 mg from the PCT-QC sample for each batch. We recommend the inclusion of an independent PCT-QC sample in each batch of a PCT-assisted experiment followed by MS analysis, to allow evaluation of the protein lysis and digestion efficacy and reproducibility.
-
2
Prepare MS-QC samples by taking peptides from the same or similar tissue samples and mixing them into a pooled peptide sample. MS-QC samples should be injected at least twice per week.
Desalting peptides
Timing 1 h per set of 16 samples
-
3
Prepare and label one new SPE HRP column for each sample.
Critical step
The size of the SPE HRP column is selected on the basis of the amount of proteins. For 0.5 mg mouse kidney tissues and 5 × 105 cells, we recommend columns with the specification of 2 mg/1 mL 96-well plate.
-
4
Place each C18 Micro Spin column in a fresh 2 mL tube, then condition the column with 200 µL of 100% (vol/vol) MeOH by centrifugation at 120g for 30 s. Repeat this step once. Discard the filtrate.
-
5
Change a new set of 2 mL tubes. Condition each C18 Micro Spin column with 200 µL of 80% (vol/vol) ACN/0.1% TFA (vol/vol) in water by centrifugation at 120 g for 30 s. Repeat this step once. Discard the filtrate.
-
6
Change a new set of 2 mL tubes. Equilibrate each C18 Micro Spin column with 200 µL of 2% (vol/vol) ACN/0.1% (vol/vol) TFA in water by centrifugation at 120g for 30 s. Repeat this step twice. Discard the filtrate.
-
7
Change a new set of 2 mL tubes. Load the peptide supernatant from Step 2 A or B (~160 µL) to each C18 Micro Spin column by centrifugation at 60g for 2 min. Reload the distillate once, and repeat this step.
-
8
Wash each C18 Micro Spin column with 200 µL of 2% (vol/vol) ACN/0.1% (vol/vol) TFA in water by centrifugation at 120g for 30 s at RT. Repeat this step eight to ten times. Change the set of tubes for collecting filtrate when needed (the maximal capacity of the 2 mL tube is 600 µL).
-
9
Change to a new set of 1.5 mL tubes, and label them properly. Place each C18 Micro Spin column in its corresponding tube. Elute the desalted peptides with 200 µL of 60% (vol/vol) ACN/0.1% (vol/vol) TFA in water by centrifugation at 60g for 2 min at RT. All solvents used for desalting are of analytical grade.
Further steps before MS analysis
Timing 2.5 h per set of 16 samples
-
10
Dry the desalted peptides in a SpeedVac at 45 °C according to the manufacturer’s protocol.
Pause point
Dried peptides can be stored at −80 °C for at least 1 year.
-
11
Dissolve the dried peptides in 30 µL of MS buffer A by vortexing for 3 min.
-
12
Measure the peptide concentration using a NanoDrop 1000 spectrophotometer with the option of ‘protein A280’ function (1 AU = 1 mg/mL).
Critical step
The peptides can be labeled with stable isotope chemical tags such as tandem mass tag and isobaric tags for relative and absolute quantitation. Please refer to the respective manuals for this optional procedure. For label-free experiments, peptides can be injected into instruments directly after being dissolved in MS buffer A.
-
13
Adjust the peptide concentration to 0.25 µg/µL using MS buffer A. Vortex for 1 min.
-
14
Centrifuge the solution at 12,000g for 15 min, and transfer 10 µL of the supernatant containing MS-ready peptide solution to the MS sample vials.
LC-MS data acquisition
Timing 24 h per set of 16samples for 60 min LC gradient
-
15
Place the samples in an LC autosampler cooled to 4–8 °C.
-
16
Build the acquisition batch including the sample name, acquisition method, vial position, injection volume and other parameters.
Critical step
Each acquisition batch should contain experimental samples in addition to one PCT-QC sample and one MS-QC sample.
-
17
Analyze the peptide samples using the preferred LC–MS/MS methods according to the experimental design.
Critical step
It is recommended to analyze the missed cleavage rate of peptide samples from a few randomly selected samples using DDA-MS as a QC, before performing quantitative proteomic analysis.
Data analysis
Timing <1 d
-
18We typically analyze DDA-MS data using pFind37 v3.1.5 for missed cleavage rate evaluation. Typical parameters are presented in the table below, and other parameters are kept as default.
Parameter Value MS data MS date format RAW MS instrument HCD-FTMS Identification Enzyme Trypsin_P KR P C Open search False Fixed modification Carbamidomethyl[C] Variable modification Oxidation[M] Peptides FDR 1% Proteins FDR 1% -
19We typically analyze DDA-MS data using Proteome Discoverer v2.5.0.400 for proteome profiling. Typical parameters selected in the home page are presented in the table below, and other parameters are kept as default.
Parameter Value Processing Workflow ProcessingWF_QExactive\PWF_QE_Precursor_Quan_and_LFQ_MPS_SequestHT_Percolator.pdProcessingWF Consensus Workflow ConsensusWF\CWF_Comprehensive_Enhanced Annotation_LFQ_and_Precursor_Quan.pdConsensusWE -
20We typically analyze DIA data using DIA-NN38 v1.8. Typical parameters are presented in the table below, and other parameters are kept as default.
Parameter Value Output Precursor FDR % 1.0 Algorithm Mass accuracy 20 MS1 accuracy 10 Protein inference Off Quantification Robust LC (high precision) Critical step
Several other suitable software tools can also be used for the data analysis24. For PulseDIA analysis, the quantitative values for each injection should be combined as the final quantitative values39.
Troubleshooting
Troubleshooting advice can be found in Table 1.
Table 1.
Troubleshooting table
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 2A(ii), 2A(v), 2A(vii), 2B(iv), 2B(vi) | PCT-MicroPestle or PCT-MicroCap cannot be closed properly | The volume of the solution exceeds the limit of the PCT-MicroTube. This most likely occurs in cell samples when PBS is not completely removed | Remove some buffer. For cell samples, minimize the residual PBS solution |
| PCT holder cannot be screwed properly | Frayed edges for PCT-MicroPestle or PCT-MicroCap due to excessive use | Replace with a new PCT-MicroPestle or PCT-MicroCap | |
| PCT-MicroPestles or PCT-MicroCaps are not closed tightly | Replace with a new PCT-MicroPestle or PCT-MicroCap | ||
| The thread for the PCT holder is damaged | Replace it with a new PCT holder | ||
| The PCT-MicroTubes are not placed in balance or there is an asymmetric number of samples in the upper and lower parts of the PCT holder | Adjust the number of samples in upper and lower parts of the PCT holder | ||
| 10 | Some peptide samples are dried slower than usual | Errors occurred in SpeedVac | Check the vacuum of the SpeedVac |
| Tube cap interference from the other samples | Adjust the cap position of the relevant samples, and dry again | ||
| During desalting, the washing buffer does not completely pass through the column before elution, so the eluent’s volume is larger than usual | Continue to dry samples under vacuum | ||
| 12 | Low peptide recovery (<20 µg for tissue, cell lines and tear strips) | Sample loss during transfer | Remember to always spin down before opening the PCT-MicroPestle or the PCT-MicroCap |
| Sample loss during desalting | Check the C18 protocol. Pay special attention to ensure the solvents are properly prepared and used |
Timing
All times listed below are approximate for a set of 16 samples.
Step 1, sample pretreatment: 2 h for FFPE, 0.5 h for FF, cells, feces and tear strips
Step 2, two-step PCT-assisted lysis and digestion: 2.5 h; four-step PCT-assisted lysis and digestion: 4.5 h
Steps 3–9, desalting peptides: 1 h
Steps 10–14, further steps before MS analysis: 2.5 h
Steps 15–17, LC-MS data acquisition for 60 min LC gradient: 24 h
Steps 18–20, data analysis: <1 d
Anticipated results
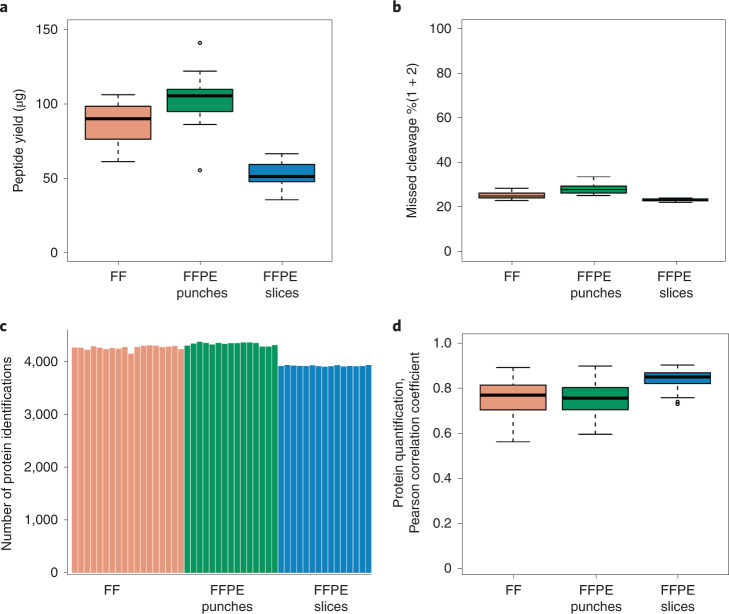
The protocol was tested and evaluated using 18 FF tissue blocks, 15 FFPE tissue punches and 15 FFPE tissue slices from a mouse kidney with full process replicates. The weight of each FF tissue block and FFPE tissue punch was ~1 mg (0.8–1.2 mg). The thickness of each FFPE tissue slice (25 mm2) was 20 µm. Together, these samples were digested using the two-step PCT-assisted preparation method in <1 d. Similar peptide yields were obtained from the same tissue types (Fig. 3a), with the coefficient of variation of 16%, 18% and 17% representing the biological variations for FF, FFPE punches, and FFPE slices respectively. The 48 peptide samples from mouse kidney were analyzed by DDA-MS with a 90 min effective LC gradient on a nanoflow DIONEX UltiMate 3000 RSLCnano System coupled to a Q Exactive HF hybrid Quadrupole-Orbitrap. The detailed parameters of the methodology are shown in ‘Equipment setup’, and the representative extracted ion chromatograms are shown in Supplementary Fig. 1. On the basis of the annotated SwissProt Mouse (16,985 protein sequences, downloaded on 9 February 2018) fasta database, the raw data were analyzed using pFind37 v3.1.5 for missed cleavage rate and Proteome Discoverer v2.5.0.400 for proteome profiling. The median values of peptide missed cleavage percentages of FF, FFPE punches and FFPE slices were 25.25%, 28.04% and 23.44%, respectively (Fig. 3b). About 4,000 proteins were identified with high protein false discovery rate (FDR) confidence in each sample (Fig. 3c and Supplementary Data 1), with median Pearson correlation coefficient (r) values of 0.77, 0.75 and 0.85 for FF, FFPE punches and FFPE slices, respectively (Fig. 3d).
Fig. 3. Demo analysis of mouse kidney tissue samples.
a, Peptide yield of 18 FF, 15 FFPE punch (0.8–1.2 mg for each sample) and 15 FFPE slice (20 µm) mouse kidney samples. b, Percentages of peptides with one and two missed cleavages. c, Number of protein identifications for each tissue sample with 500 ng peptide injection. d, Pearson correlation coefficient (r) of proteins quantified by Proteome Discoverer v2.5.0.400 with high protein FDR confidence.
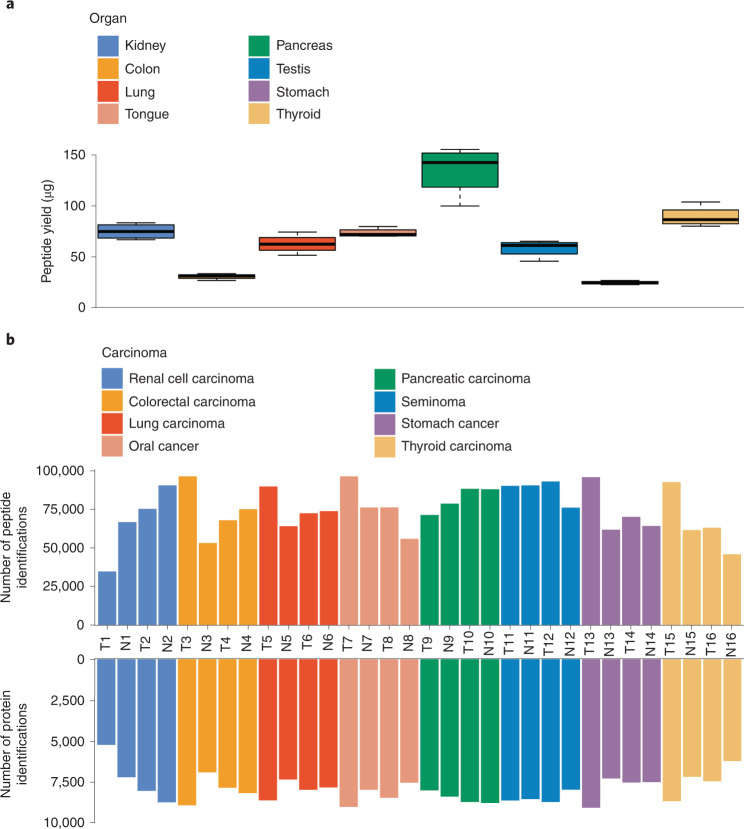
We also applied this protocol to eight types of nontumorous tissue, including kidney, colon, lung, tongue, pancreas, testis, stomach and thyroid. We obtained a peptide yield of 25 µg (stomach) to 142 µg (pancreas) per milligram of tissue (Fig. 4a). Next, we applied this protocol to 32 FFPE tissue samples (tumor and nontumorous pairs) from these eight types of tissue, followed by PulseDIA-PASEF analysis containing two MS runs of 60 min LC effective LC gradient on a nanoElute System coupled to a timsTOF Pro mass spectrometer. The detailed parameters of the methodology are shown in ‘Equipment setup’, and the representative extracted ion chromatograms are shown in Supplementary Fig. 2. The PulseDIA-PASEF data were analyzed using DIA-NN (v1.8) and a home-built library containing 418,459 peptides and 13,553 proteins. For the 32 FFPE tissue samples, the median value of peptide identifications was 75,069 (interquartile range (IQR): 24,534; Q1: 64,034; Q3: 88,568) and that of protein identifications was 7,991 (IQR: 1126; Q1: 7493; Q3: 8619) (Fig. 4b, Supplementary Data 2).
Fig. 4. Application of the PCT-assisted sample preparation protocol for human tumors.
a, Peptide yield of eight types of tissue (0.8–1.2 mg) as shown in boxplots by summarizing data from four replicates. b, The numbers of peptide and protein identifications from 32 FFPE samples (tumor and nontumor pairs) collected from 16 patients with cancer. These samples are from eight types of tissues. T, tumor tissue; N, non-tumorous tissue.
Taken together, both sets of results reveal that the protocol described here enables robust, high-throughput, high-yield and reproducible proteomic sample preparation from small amounts of samples for immediate use in proteomic analysis.
Supplementary information
Supplementary Figs. 1 and 2.
Experimental data from Fig. 2. Proteomic data of mouse kidney samples in the form of FF, FFPE punch and FFPE slice.
Experimental data from Fig. 3. Proteomic data of 32 independent FFPE tissue samples (benign and tumor pairs) from eight types of cancer tissues.
PulseDIA-PASEF MS isolation window: part 1 setting.
PulseDIA-PASEF MS isolation window: part 2 setting.
The procedures for handling the PCT-MicroTubes, MicroCaps, MicroPestles with the PCT tool.
Acknowledgements
This work is supported by grants from the National Key R&D Program of China (2021YFA1301601 and 2020YFE0202200), Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (LR19C050001), National Natural Science Foundation of China (81972492) and National Science Fund for Young Scholars (21904107).
Author contributions
T.G. and X.C. designed the studies. X.C., X.Y., W.L., C.C., H.G., J.Y. and L.X. participated in the development of this protocol. X.C., C.L., R.S, L.Q. and L.Y. performed the PCT-based sample preparation. X.C performed the LC–MS/MS analysis. X.C., Z.X. and W.G. analyzed the data. X.C., T.G. and Y.Z. wrote the manuscript with input from all co-authors. T.G. and Y.Z. supervised the project.
Peer review
Peer review information
Nature Protocols thanks Louise Bundgaard and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
The raw MS data of this study have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository40 with the dataset identifier PXD030645. Project accession of the mouse kidney data: IPX0003852001. Project accession of the cancer tissue data: IPX0003852002.
Competing interests
Y.Z. and T.G. are shareholders of Westlake Omics Inc. C.W., W.G., X.Y., W.L., C.C., H.G., J.Y. and L.X. are employees of Westlake Omics Inc. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Guo, T. et al. Nat. Med. 21, 407–413 (2015): 10.1038/nm.3807
Zhu, Y. et al. Mol. Oncol. 13, 2305–2328 (2019): 10.1002/1878-0261.12570
Shao, W. et al. Nat. Commun. 10, 2524 (2019): 10.1038/s41467-019-10513-5
Charmpi, K. et al. Genome Biol. 21, 302 (2020): 10.1186/s13059-020-02188-9
Nie, X. et al. Cell 184, 775–791.e714 (2021): 10.1016/j.cell.2021.01.004
Contributor Information
Yi Zhu, Email: zhuyi@westlake.edu.cn.
Tiannan Guo, Email: guotiannan@westlake.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41596-022-00727-1.
References
- 1.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Y, Aebersold R, Mann M, Guo T. SnapShot: clinical proteomics. Cell. 2021;184:4840–4840.e1. doi: 10.1016/j.cell.2021.08.015. [DOI] [PubMed] [Google Scholar]
- 3.Xiao Q, et al. High-throughput proteomics and AI for cancer biomarker discovery. Adv. Drug Deliv. Rev. 2021;176:113844. doi: 10.1016/j.addr.2021.113844. [DOI] [PubMed] [Google Scholar]
- 4.Guo T, et al. Rapid mass spectrometric conversion of tissue biopsy samples into permanent quantitative digital proteome maps. Nat. Med. 2015;21:407–413. doi: 10.1038/nm.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, et al. Identification of protein abundance changes in hepatocellular carcinoma tissues using PCT-SWATH. Proteomics Clin. Appl. 2019;13:1700179. doi: 10.1002/prca.201700179. [DOI] [PubMed] [Google Scholar]
- 6.Eckert MA, et al. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569:723–728. doi: 10.1038/s41586-019-1173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hood BL, et al. Proteomic analysis of formalin-fixed prostate cancer tissue. Mol. Cell Proteomics. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Nie X, et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184:775–791.e14. doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 10.Hwang SI, et al. Direct cancer tissue proteomics: a method to identify candidate cancer biomarkers from formalin-fixed paraffin-embedded archival tissues. Oncogene. 2007;26:65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- 11.Hughes CS, et al. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014;10:757. doi: 10.15252/msb.20145625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughes CS, et al. Single-pot, solid-phase-enhanced sample preparation for proteomics experiments. Nat. Protoc. 2019;14:68–85. doi: 10.1038/s41596-018-0082-x. [DOI] [PubMed] [Google Scholar]
- 13.Fowler CB, et al. Elevated hydrostatic pressure promotes protein recovery from formalin-fixed, paraffin-embedded tissue surrogates. Lab Invest. 2008;88:185–195. doi: 10.1038/labinvest.3700708. [DOI] [PubMed] [Google Scholar]
- 14.Powell BS, Lazarev AV, Carlson G, Ivanov AR, Rozak DA. Pressure cycling technology in systems biology. Methods Mol. Biol. 2012;881:27–62. doi: 10.1007/978-1-61779-827-6_2. [DOI] [PubMed] [Google Scholar]
- 15.Shao S, et al. Minimal sample requirement for highly multiplexed protein quantification in cell lines and tissues by PCT-SWATH mass spectrometry. Proteomics. 2015;15:3711–3721. doi: 10.1002/pmic.201500161. [DOI] [PubMed] [Google Scholar]
- 16.Shao S, et al. Reproducible tissue homogenization and protein extraction for quantitative proteomics using MicroPestle-assisted pressure-cycling technology. J. Proteome Res. 2016;15:1821–1829. doi: 10.1021/acs.jproteome.5b01136. [DOI] [PubMed] [Google Scholar]
- 17.Thompson A, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 18.Ross PL, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallien S, et al. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell Proteomics. 2012;11:1709–1723. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 23.Powell K. Technology to watch in 2018. Nature. 2018;553:531–534. doi: 10.1038/d41586-018-01021-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhang F, Ge W, Ruan G, Cai X, Guo T. Data-independent acquisition mass spectrometry-based proteomics and software tools: a glimpse in 2020. Proteomics. 2021;20:1900276. doi: 10.1002/pmic.201900276. [DOI] [PubMed] [Google Scholar]
- 25.Gao H, et al. Accelerated lysis and proteolytic digestion of biopsy-level fresh-frozen and FFPE tissue samples using pressure cycling technology. J. Proteome Res. 2020;19:1982–1990. doi: 10.1021/acs.jproteome.9b00790. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Guo T. High-throughput proteomic analysis of fresh-frozen biopsy tissue samples using pressure cycling technology coupled with SWATH Mass spectrometry. Methods Mol. Biol. 2018;1788:279–287. doi: 10.1007/7651_2017_87. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y, et al. High-throughput proteomic analysis of FFPE tissue samples facilitates tumor stratification. Mol. Oncol. 2019;13:2305–2328. doi: 10.1002/1878-0261.12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu T, et al. DPHL: a DIA pan-human protein mass spectrometry library for robust biomarker discovery. Genomics Proteomics Bioinformatics. 2020;18:104–119. doi: 10.1016/j.gpb.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge W, et al. Computational optimization of spectral library size improves DIA-MS proteome coverage and applications to 15 tumors. J. Proteome Res. 2021;20:5392–5401. doi: 10.1021/acs.jproteome.1c00640. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Chen Y, Yu C. Fast and efficient non-reduced Lys-C digest using pressure cycling technology for antibody disulfide mapping by LC–MS. J. Pharm. Biomed. Anal. 2016;129:203–209. doi: 10.1016/j.jpba.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Nie S, Greer T, Huang X, Zheng X, Li N. Development of a simple non-reduced peptide mapping method that prevents disulfide scrambling of mAbs without affecting tryptic enzyme activity. J. Pharm. Biomed. Anal. 2022;209:114541. doi: 10.1016/j.jpba.2021.114541. [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Burchmore R, Jonsson NN, Johnson PCD, Eckersall PD. Technical report: In-gel sample preparation prior to proteomic analysis of bovine faeces increases protein identifications by removal of high molecular weight glycoproteins. J. Proteomics. 2022;261:104573. doi: 10.1016/j.jprot.2022.104573. [DOI] [PubMed] [Google Scholar]
- 33.Wu C, et al. Coupling suspension trapping-based sample preparation and data-independent acquisition mass spectrometry for sensitive exosomal proteomic analysis. Anal. Bioanal. Chem. 2022;414:2585–2595. doi: 10.1007/s00216-022-03920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xuan Y, et al. Standardization and harmonization of distributed multi-center proteotype analysis supporting precision medicine studies. Nat. Commun. 2020;11:5248. doi: 10.1038/s41467-020-18904-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunt AL, et al. Extensive three-dimensional intratumor proteomic heterogeneity revealed by multiregion sampling in high-grade serous ovarian tumor specimens. iScience. 2021;24:102757. doi: 10.1016/j.isci.2021.102757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee S, et al. Molecular analysis of clinically defined subsets of high-grade serous ovarian cancer. Cell Rep. 2020;31:107502. doi: 10.1016/j.celrep.2020.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li D, et al. pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics. 2005;21:3049–3050. doi: 10.1093/bioinformatics/bti439. [DOI] [PubMed] [Google Scholar]
- 38.Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai X, et al. PulseDIA: data-independent acquisition mass spectrometry using multi-injection pulsed gas-phase fractionation. J. Proteome Res. 2021;20:279–288. doi: 10.1021/acs.jproteome.0c00381. [DOI] [PubMed] [Google Scholar]
- 40.Ma J, et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1 and 2.
Experimental data from Fig. 2. Proteomic data of mouse kidney samples in the form of FF, FFPE punch and FFPE slice.
Experimental data from Fig. 3. Proteomic data of 32 independent FFPE tissue samples (benign and tumor pairs) from eight types of cancer tissues.
PulseDIA-PASEF MS isolation window: part 1 setting.
PulseDIA-PASEF MS isolation window: part 2 setting.
The procedures for handling the PCT-MicroTubes, MicroCaps, MicroPestles with the PCT tool.
Data Availability Statement
The raw MS data of this study have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository40 with the dataset identifier PXD030645. Project accession of the mouse kidney data: IPX0003852001. Project accession of the cancer tissue data: IPX0003852002.