Abstract
Round spermatid injection (ROSI) technique holds great promise for clinical treatment of a proportion of infertile men. However, the compromised developmental potential of ROSI embryos largely limits the clinical application, and the mechanisms are not fully understood. Here, we describe the transcriptome, chromatin accessibility, and DNA methylation landscapes of mouse ROSI embryos derived from early-stage round spermatids using a single-cell multiomics sequencing approach. By interrogating these data, we identify the reprogramming defects in ROSI embryos at the pronuclear stages, which are mainly associated with the misexpression of a cohort of minor zygotic genome activation genes. We screen a small compound, A366, that can significantly increase the developmental potential of ROSI embryos, in which A366 can partially overcome the reprogramming defects by amending the epigenetic and transcriptomic states. Collectively, our study uncovers the reprogramming defects in ROSI embryos for understanding the mechanisms underlying compromised developmental potential and offers an avenue for ROSI technique optimization.
The reprogramming defects of ROSI embryos uncovered by single-cell multiomics sequencing can be largely amended by A366.
INTRODUCTION
Azoospermia is a major factor leading to male infertility, which occurs in around 1% of the total population of men (1). In some infertile men, round spermatids are the most mature haploid cells visible during testicular biopsy, and the development of round spermatid injection (ROSI) can help them have their own offspring (2). Although there are studies about the clinical application of the ROSI technique, a major bottleneck limiting this technique is the compromised developmental potential of reconstructed embryos (3); therefore, there is great demand to elucidate the mechanisms underlying this phenotype in ROSI embryos.
In the process of spermiogenesis, haploid male germ cells undergo marked maturation processes, including histone-protamine exchange and chromatin condensation. Upon fertilization, extensive epigenetic reprogramming occurs to ensure the proper transmission of parental genetic or epigenetic information to zygote (4–6). Thus, the proper reprogramming of epigenome in male gametes is critical for correct epigenetic resetting upon fertilization (7) and the subsequent early embryonic development. Different from the conventional in vitro fertilization or intracytoplasmic sperm injection (ICSI) approach, ROSI uses round spermatids, and a proportion of the reconstructed embryos can develop to full term (8). Previous studies revealed that there were several stages of round spermatids during spermiogenesis (9, 10), and the early-stage round spermatids were proven to be suboptimal for successful ROSI (9). Hence, infertile men with serious spermiogenesis defects, such as early-stage round spermatid arrest, are difficult to be treated via ROSI technique for compromised developmental potential of the reconstructed embryos.
Breakthroughs have been achieved in delineating the epigenetic reprogramming landscapes during early embryonic development, such as the reprogramming of DNA methylation, histone modifications, and chromatin architecture (11–13). The interconnection among different epigenetic modifications and their cooperation with transcription factors instruct cell fate determination and the development of early embryos (14–17). Previous studies detected abnormal epigenetic modifications in the male pronucleus of ROSI embryos and aberrant chromosome segregation (18–20), indicating an association between epigenetic reprogramming defects and compromised developmental potential. However, it is still unclear how the reprogramming defects alter the cellular behaviors of ROSI embryos. It is presumed that incomplete traversal of epigenetic modifications, such as histone modifications, DNA methylation, or chromatin condensation, might affect the subsequent embryonic reprogramming process in ROSI embryos and thereby lead to the poor developmental potential (18, 19). In this regard, dissecting the epigenetic reprogramming roadmap in preimplantation ROSI embryos is prerequisite for the optimization of present ROSI approach using small compounds as those in other diseases (21).
With the advent of single-cell sequencing approaches, especially single-cell multiomics sequencing–based approaches (22, 23), it is now possible to simultaneously decipher the complex regulatory networks that control different molecular layers at single-cell resolution. In this study, we applied a single-cell multiomics sequencing approach to explore the molecular features of ROSI embryos. Comparative analysis of transcriptome, methylome, and chromatin accessibility uncovered the reprogramming defects in ROSI embryos reconstructed with early-stage round spermatids, which were associated with the misexpression of a cohort of minor ZGA (zygotic genome activation) genes. The treatment of ROSI embryos by A366, a selective euchromatic histone lysine methyltransferase 2 (Ehmt2, also known as G9A) inhibitor, could amend the expression of a cohort of minor ZGA genes to a certain extent and improve the developmental potential of ROSI embryos.
RESULTS
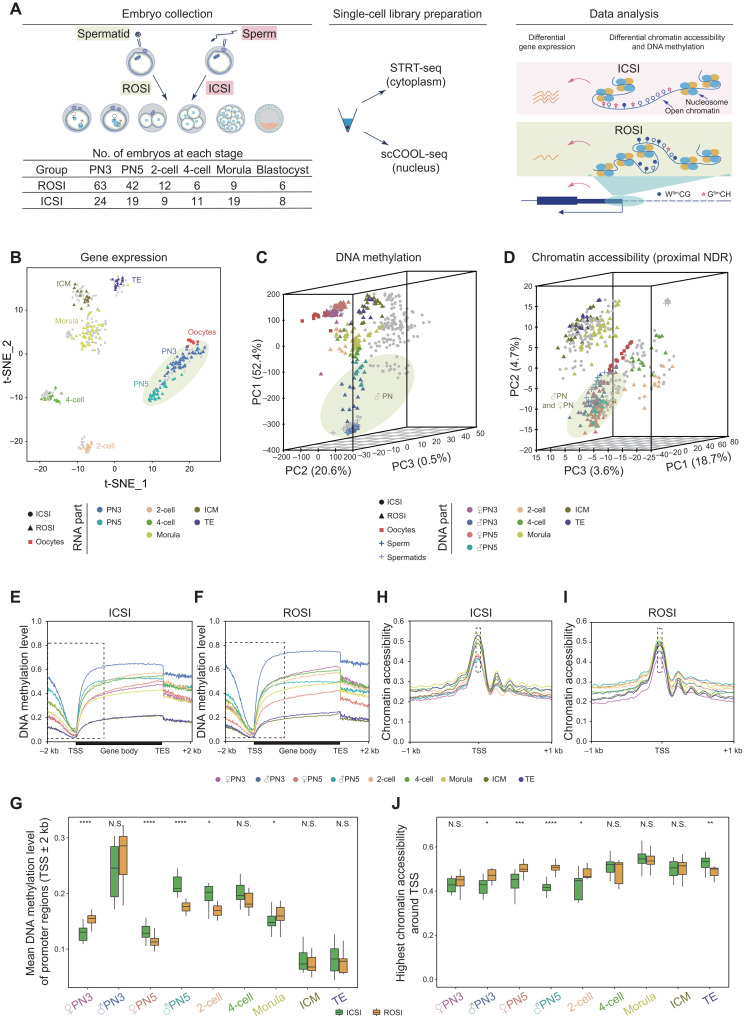
The overview of transcriptome, chromatin accessibility, and DNA methylation in the preimplantation mouse ROSI embryos
The mouse ROSI platform used in this study was established as previously described (24). Here, we used the nuclei of early-stage round spermatids (fig. S1, A and B) for intracytoplasmic injection to obtain ROSI embryos. Single-cell RNA sequencing (scRNA-seq) analysis revealed that the round spermatids used in this study were RS2 stage spermatids and were still at the early stage of spermiogenesis (fig. S1, C to E) (9). We then confirmed that the developmental progress of both ROSI and ICSI embryos was essentially synchronous upon fertilization (fig. S2, A to C). The blastocyst rate (14.10%) and live-birth rate (10.87%) of ROSI embryos generated on C57BL/6N × DBA/2N background were comparable to a previous study (fig. S2, D to G) (9) but lower than two previous studies (25, 26). The most likely explanation for this discrepancy was that we used much earlier round spermatids (fig. S1, C to E). In contrast to ROSI, for ICSI embryos, the blastocyst rate was 51.61%, and live-birth rate were 35.25%. Both ROSI and ICSI underwent similar micromanipulation process, and only the developmental stage of haploid male germ cells used was different. Similar results were observed when the embryos were constructed on B6D2F1 background (table S1). The immature epigenetic state of early-stage round spermatids might make them more resistant to epigenetic reprogramming upon fertilization (9, 19, 27). The weight of all alive fetuses and their placentas was comparable between ROSI and ICSI embryos (fig. S2, H and I). In addition, the weight and reproduction capability of adult mice obtained via ROSI or ICSI were also indistinguishable (fig. S2, J and K), indicating that the developmental defects underlying ROSI embryos were within a relatively narrow developmental time window, and if the embryos overcame that problem, then their subsequent development would be relatively normal.
Multiomics sequencing can simultaneously profile gene expression, DNA methylation, and chromatin accessibility at single-cell resolution. To explore the epigenetic reprogramming information of ROSI embryos, single-cell multiomics sequencing was performed on each individual cell of ROSI embryos. A total of 138 ROSI embryos were included in this study, with 90 ICSI embryos as the control to eliminate the effects coming from embryo micromanipulation technique itself, covering six critical stages of the preimplantation development, as well as the gametes, including sperm, round spermatids, and metaphase-II (MII) oocytes (Fig. 1A). In total, we sequenced 452 RNA samples and 493 DNA samples. After stringent quality control, 428 cells were retained with RNA data, and 439 cells were retained with DNA data for the subsequent analysis. An average of 9108 genes (fig. S3A), 16,549,572 GCH sites, and 1,871,398 WCG sites (see Materials and Methods for details, fig. S3, B to F, and table S2) were detected per cell. Data normalization was performed to eliminate the variations from the sequencing method itself. In addition, when dividing cells at each stage into three groups according to genomic coverage or sequencing depth, we found that the variance of genomic coverage or sequencing depth of single cells showed little influence on DNA methylation or chromatin accessibility (fig. S4, A to D). Unsupervised clustering analysis was then performed for each layer of data independently. t-distributed stochastic neighbor embedding (t-SNE) indicated that cells at the same developmental stage clustered more closely by gene expression (Fig. 1B and fig. S5, A and B). Principal components analysis (PCA) showed differential DNA methylation and chromatin accessibility profiles in ROSI embryos (Fig. 1, C and D, and fig. S5, C and D). Consistent results were obtained from both datasets regarding the inferred copy number variations (CNVs) or not, and the gender of embryos had no effect on these conclusions (figs. S6, A to K, and S7, A to D). We also focused on the epigenetic regulatory elements that were always associated with gene expression regulation. The global DNA methylation differences around transcription start sites (TSSs) were obvious at the pronuclear stages (PN3 and PN5) and became comparable when developed to blastocyst stage (Fig. 1, E and F). In detail, PN3 pronucleus of ROSI embryos showed higher DNA methylation level relative to the ICSI control, which was essentially consistent with the previous studies (18, 19), while PN5 pronucleus of ROSI embryos was hypomethylated (Fig. 1G). Meanwhile, as for the chromatin accessibility around TSSs, ROSI embryos showed higher chromatin accessibility from the pronuclear stages and became comparable when developed to 4-cell stage (Fig. 1, H to J). Together, these results indicated that the reprogramming of DNA methylation and chromatin accessibility was globally achieved during the preimplantation development of ROSI embryos but still exhibited certain variations as compared to ICSI embryos, especially at the pronuclear stages.
Fig. 1. The overview of transcriptome, chromatin accessibility, and DNA methylation in the preimplantation mouse ROSI embryos.
(A) Schematic illustration of the number of embryos and single-cell multiomics sequencing analyses used in this study. STRT-seq, single cell–tagged reverse transcription sequencing; scCOOL-seq, single-cell chromatin overall omics-scale landscape sequencing. (B) Dimensionality reduction analysis of gene expression data from single-cell multiomics sequencing using t-SNE. Each dot represents a single cell. ROSI samples are triangles and colored by developmental stages, while ICSI samples are in gray. Yellow-green circle indicates pronucleus. (C and D) Dimensionality reduction analysis of DNA methylation (C) and chromatin accessibility data (D) from single-cell multiomics sequencing using PCA. Each dot represents a single cell. ROSI samples are triangles and colored by developmental stages, while ICSI samples are in gray. Yellow-green circle indicates male pronucleus (C) and male and female pronuclei (D), respectively. (E and F) DNA methylation level along gene bodies, 2-kb upstream of TSSs and 2-kb downstream of transcription end sites (TESs) of all RefSeq genes in ICSI (E) and ROSI (F) embryos. (G) Boxplot showing DNA methylation level of promoter regions (TSSs ± 2 kb) at each stage. (H and I) Chromatin accessibility around TSSs (±1 kb) in ICSI (H) and ROSI (I) embryos. (J) Boxplot showing chromatin accessibility of TSSs at each stage. Unpaired two-tailed Student’s t test (G and J) is used for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. N.S., not significant.
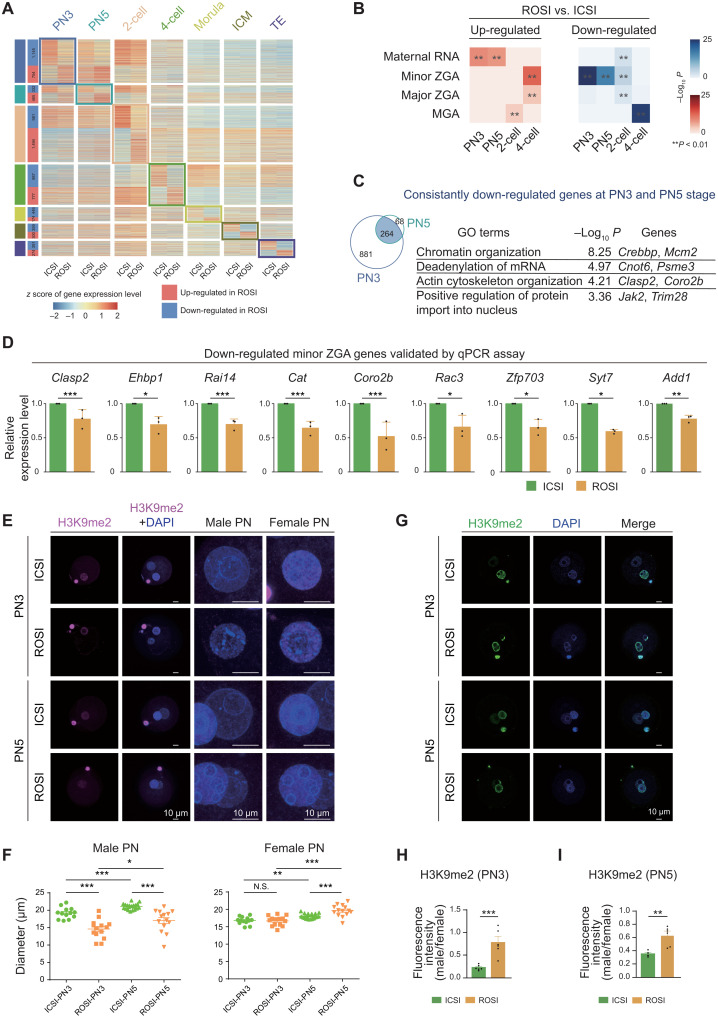
The misexpression of maternal to zygotic transition–associated genes in ROSI embryos
As aforementioned, the epigenetic reprogramming occurred in the pronuclei of ROSI embryos upon fertilization seemed to be distinct from that of ICSI control; in particular, the male pronucleus of ROSI embryos was with unique epigenetic characteristics. Consistent with this finding, 1145, 332, and 981 down-regulated differentially expressed genes (DEGs) between ROSI and ICSI embryos were detected early at the PN3, PN5, and 2-cell stages, respectively (Fig. 2A and table S3). Notably, we observed that, compared to the ICSI control, the up-regulated genes in ROSI embryos at the pronuclear stages (PN3 and PN5) were mainly enriched in maternal RNA, while the down-regulated genes were associated with the process of minor ZGA (Fig. 2B and fig. S8A) (28). Minor ZGA, which occurs at the mid-to-late stage in mouse one-cell embryos, is essential for the initiation of major ZGA and early embryonic development (29). Weighted correlation network analysis (WGCNA) results further confirmed the expression patterns of minor ZGA genes (fig. S8, B and C). Gene Ontology (GO) analysis of the down-regulated genes (264) at both PN3 and PN5 stages of ROSI embryos showed that these genes were enriched in processes such as “chromatin organization” and “actin cytoskeleton organization” (Fig. 2C and fig. S8, D and E). Quantitative polymerase chain reaction (qPCR) was performed to confirm the misexpression of minor ZGA genes. Among the 16 candidates, 13 minor ZGA genes (e.g., Clasp2, Ehbp1, Rai14, and Cat) (30) were validated to be lowly expressed in ROSI embryos at the PN3 stage, and the validation rate was 81.3% (Fig. 2D and fig. S8F). We found that certain genes of maternal RNA were also misexpressed at the pronuclear stages in ROSI embryos (fig. S8, G and H). Notably, the minor ZGA genes down-regulated in ROSI embryos were enriched in the chromatin and cytoskeleton organization processes, suggesting a likely relationship between misexpression of minor ZGA genes and chromatin/cytoskeletal organization defects (31). These results at the PN3 stage could be well reproduced using false discovery rate (FDR) analysis (table S3).
Fig. 2. The misexpression of maternal to zygotic transition–associated genes in ROSI embryos.
(A) Heatmap showing the DEGs at each stage. (B) Hypergeometric test analysis of DEGs between ROSI and ICSI embryos in “maternal RNA,” “minor ZGA,” “major ZGA,” and “MGA” (28) gene clusters. (C) Venn plot showing that 264 genes are consistently down-regulated at the PN3 and PN5 stages in ROSI embryos. Enriched GO terms and representative genes are listed. (D) qPCR results showing the mRNA levels of nine representative down-regulated minor ZGA genes (PN3) in three biological repeats. (E) Immunofluorescence of H3K9me2 in ROSI and ICSI embryos at the PN3 (top) and PN5 (bottom) stages. The magnified images are shown on the right. Scale bars, 10 μm. (F) Dot plots showing the diameter of ROSI and ICSI pronuclei in three biological repeats. Each dot corresponds to the diameter of each pronucleus. (G) Immunofluorescence of H3K9me2 in ROSI and ICSI embryos at the PN3 (top) and PN5 (bottom) stages. Scale bars, 10 μm. (H and I) Bar graph showing the fluorescence intensity of H3K9me2 in male pronucleus relative to female pronucleus at the PN3 (H) and the PN5 stages (I) in three biological repeats. Each dot corresponds to level in each individual embryo. Unpaired two-tailed Student’s t test (D, F, H, and I) is used for statistical analysis. *P < 0.05, **P < 0.01, and ***P < 0.001.
To test whether the chromatin/cytoskeletal organization was really aberrant at the pronuclear stages in ROSI embryos, the morphology of pronucleus of ICSI and ROSI embryos was analyzed. We found that the average diameter of male pronucleus in ROSI embryos was significantly less than that of ICSI control, and this phenotype was maintained to the PN5 stage (Fig. 2, E and F). Although the female pronucleus of ROSI embryos exhibited no obvious difference at the PN3 stage, it was found to be bigger than that of ICSI embryos at the PN5 stage (Fig. 2F). In addition, aberrant histone H3 lysine 9 dimethylation (H3K9me2) deposition, which was closely related to several chromatin features such as lamina-associated domains (32) and topologically associated domain boundaries (33), was observed in the 4′,6-diamidino-2-phenylindole (DAPI)–dense heterochromatin of male pronucleus in ROSI embryos (Fig. 2, G to I), consistent with a previous study (19). Together, our data illustrated that ROSI embryos had misexpression of maternal to zygotic transition–associated genes and exhibited the chromatin/cytoskeletal organization defects.
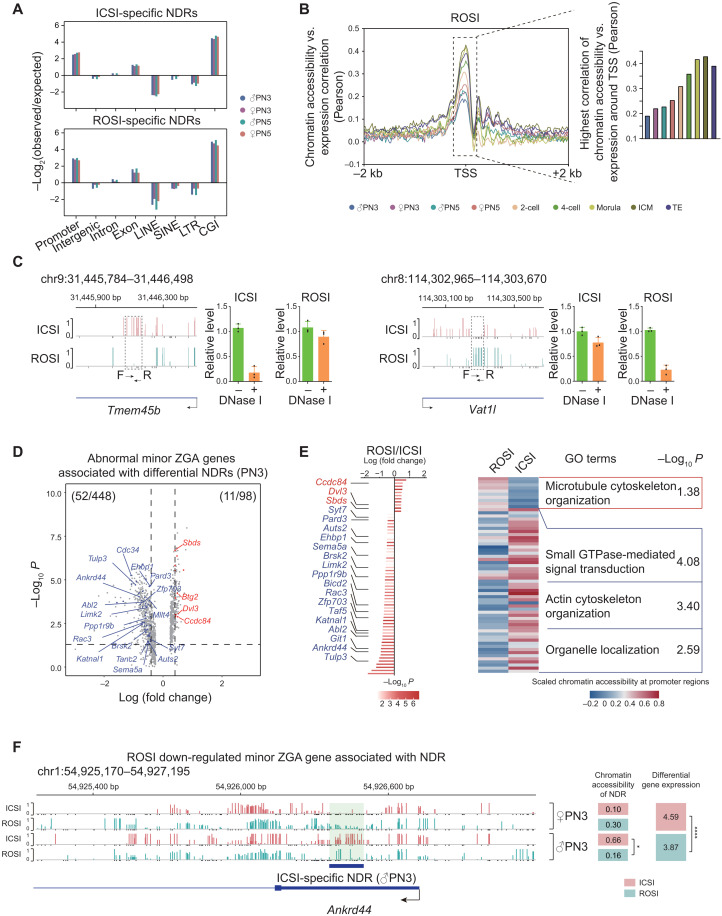
Chromatin accessibility dynamics in ROSI embryos and the association with misexpression of minor ZGA genes
Early mammalian embryonic development is accompanied with spatiotemporal chromatin reconfiguration, suggesting its potential regulatory function (11). On the basis of the stage-specific chromatin accessibility landscape, we found that negative regulation of microtubule depolymerization and protein localization to microtubule cytoskeleton–related terms were prominent in the male PN5 pronucleus of ROSI embryos, while nuclear membrane organization related terms were enriched at the PN3 stage (fig. S9A). We then analyzed the differences of chromatin accessibility between ROSI and ICSI embryos at the pronuclear stages. We found that both ROSI- and ICSI-specific nucleosome-depleted regions (NDRs) were enriched in the promoter regions (Fig. 3A), which were always correlated with gene expression in early embryos (14) (Fig. 3B and fig. S9, B to D). Low-input DNase I qPCR (liDNaseI-qPCR) was then performed to validate the sequencing findings. We found that three loci with low GCH methylation level were closed in both ROSI and ICSI embryos, while two loci with high GCH methylation level were open in both ROSI and ICSI embryos. In contrast, five NDRs were validated to be specifically closed in ROSI embryos, while six NDRs were specifically open in ROSI embryos. These results were mainly consistent with the sequencing findings (Fig. 3C and fig. S10, A to N).
Fig. 3. Chromatin accessibility dynamics in ROSI embryos and the association with misexpression of minor ZGA genes.
(A) Relative enrichment analysis of ICSI-specific (top) and ROSI-specific NDRs (bottom) in different genomic regions at the pronuclear stages (PN3 and PN5). (B) Pearson correlation of chromatin accessibility and gene expression level around promoter regions (TSSs ± 2 kb) of all RefSeq genes in ROSI embryos at each stage. Bar plot on the right showing the highest correlation around TSSs. (C) Two representative loci detected as ICSI-specific NDR (Tmem45b) (left) and as ROSI-specific NDR (Vat1l) (right) at 4-cell stage validated by liDNaseI-qPCR assay in three biological repeats. (D) Volcano plot showing the relationship between differential gene expression of minor ZGA genes and chromatin accessibility of proximal NDRs (PN3). Blue dots represent genes that are down-regulated in ROSI embryos and occupied with the ICSI specific-proximal NDRs, while red dots are vice versa. (E) Bar plot showing the differentially expressed minor ZGA genes [from (D)], while heatmap showing the chromatin accessibility of associated NDRs at the promoter regions in ROSI and ICSI embryos. Representative GO terms are shown on the right. GTPase, guanosine triphosphatase. (F) Genome browser view of the chromatin accessibility of ICSI-specific NDR around Ankrd44. The GCH sites with chromatin accessibility lower than 0.1 (but are detected) are shown as black bars. The chromatin accessibility of NDRs and the gene expression level in ROSI and ICSI embryos are listed on the right. *P < 0.05 and ****P < 0.0001.
We then focused on the minor ZGA genes that were misexpressed in ROSI embryos. Among the 448 down-regulated and 98 up-regulated minor ZGA genes at the PN3 stage, 52 and 11 genes had altered chromatin accessibility, respectively (Fig. 3D and table S4). Notably, the 52 down-regulated genes (e.g., Ankrd44) with less accessible chromatin were enriched in genes for biological processes such as “actin cytoskeleton organization” and “organelle localization” (Fig. 3, D to F). Consistently, the down-regulated minor ZGA genes in ROSI PN5 embryos were associated with the establishment of spindle localization (fig. S11A). These data suggested that abnormal chromatin accessibility in the pronuclei of ROSI embryos might be associated with the misexpression of minor ZGA genes critical for cytoskeletal organization. Further FDR analysis of the NDR data could also reproduce the main findings (table S4).
Transcription factors can exert their functions by binding to the enhancer regions and might be associated with chromatin accessibility changes in ROSI embryos (14). We used Hypergeometric Optimization of Motif EnRichment (HOMER) to identify the enriched binding motifs for transcription factors in the distal NDRs of ICSI embryos (fig. S11B). Consistent with previous reports (11, 15), the motif of Specificity Protein 1 (SP1) was enriched in the pronuclear stages. SP1 showed relative enrichment in ROSI-specific distal NDRs rather than ICSI-specific distal NDRs at the pronuclear stages (fig. S11C).
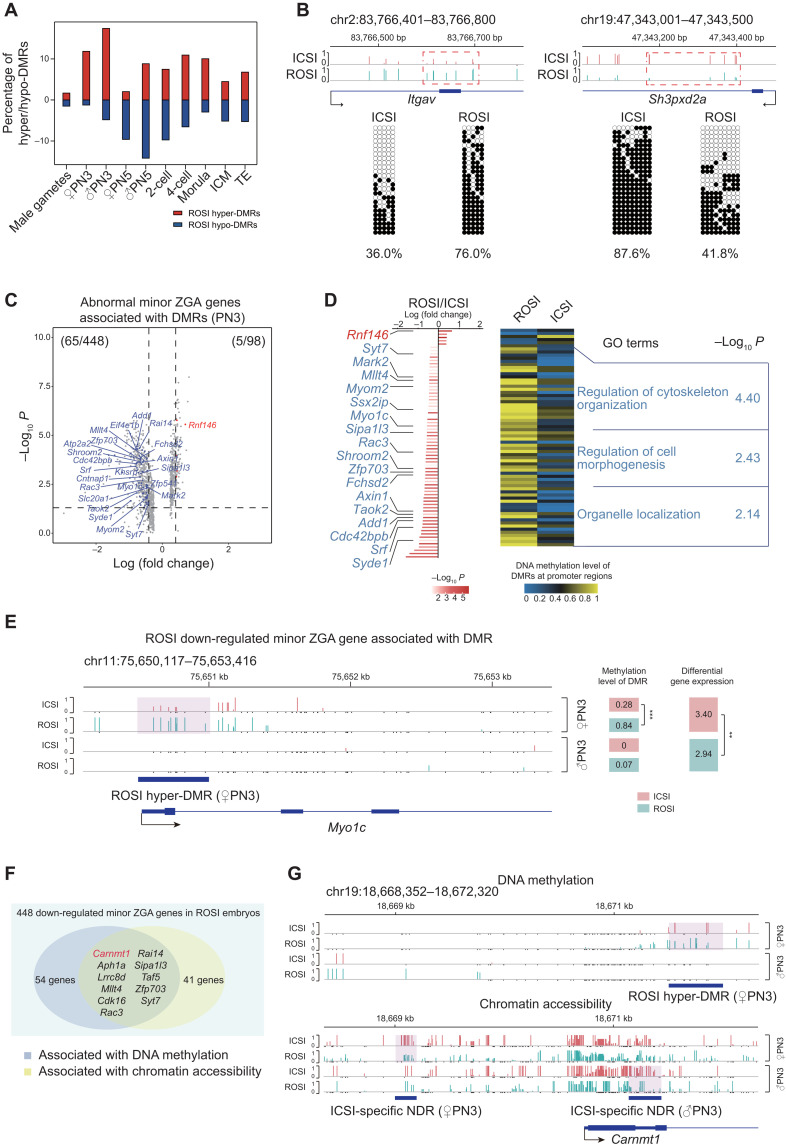
DNA methylation reprogramming alterations in ROSI embryos
The genome of mammalian embryos undergoes a stepwise DNA demethylation/remethylation process (34, 35), and ROSI embryos are always with aberrant DNA methylation state (18, 19). To dissect the DNA methylation–level reprogramming defects in ROSI embryos, we analyzed the layer of DNA methylation data. We found that both sperm and round spermatids used in this study were hypermethylated, while the MII oocyte was moderately methylated (fig. S12A), which was consistent with previous studies (36, 37). Upon fertilization, the male pronucleus of both ROSI and ICSI embryos underwent a similar round of extensive DNA demethylation. By the blastocyst stage, the global DNA methylation level became comparable (fig. S12A). Despite that the global demethylation was essentially achieved, the percentage of differentially methylated regions (DMRs) in the whole genome between ICSI and ROSI embryos increased markedly upon fertilization, and the most prominent alterations appeared at the pronuclear stages (PN3 and PN5) (Fig. 4A), which was also obvious in Fig. 1C. The differences persisted to subsequent stages (Fig. 4A), indicating the existence of differential DNA methylation reprogramming patterns between ICSI and ROSI embryos. Hyper-DMRs and hypo-DMRs at the pronuclear stages in ICSI or ROSI embryos were all enriched in the promoter, intron, and exon regions (fig. S12, B and C), indicating the likely correlation between DNA methylation changes and gene expression (fig. S12D) (17). Similarly, 10 DMRs were validated to be differentially methylated between ROSI and ICSI embryos using bisulfite genomic PCR-based Sanger sequencing (Fig. 4B and fig. S12E).
Fig. 4. DNA methylation reprogramming alterations in ROSI embryos.
(A) Bar plot showing the fractions of hyper-DMRs (red) and hypo-DMRs (blue) in total genomic titles with three WCG sites covered in ROSI embryos compared to ICSI embryos. (B) Two representative loci detected as ROSI hyper-DMR (left) and as ROSI hypo-DMR (right) at morula stage validated by bisulfite genomic PCR-based Sanger sequencing. (C) Volcano plot showing the relationship between differential expression of minor ZGA genes and DNA methylation level of DMRs at the promoter regions (PN3). Blue dots represent genes that are down-regulated in ROSI embryos and the associated DMRs are also hypermethylated, while red dots are vice versa. (D) Bar plot showing the differentially expressed minor ZGA genes associated with DMRs [from (C)], while heatmap showing the DNA methylation level of associated DMRs at the promoter regions in ROSI and ICSI embryos (PN3). Representative GO terms are shown on the right. (E) Genome browser view of DNA methylation level around the promoter region of Myo1c. WCG sites with DNA methylation level lower than 0.1 (but are detected) are shown as black bars. The DNA methylation level and gene expression level are shown on the right. **P < 0.01 and ***P < 0.001. (F) Venn plot showing 11 down-regulated minor ZGA genes associated with both chromatin accessibility and DNA methylation in ROSI embryos (PN3). (G) Genome browser views of DNA methylation level (top) and chromatin accessibility (bottom) of ROSI hyper-DMR or ICSI-specific NDR around the promoter region of Carnmt1.
To confirm whether DNA methylation alterations were also associated with the maternal to zygotic transition, we analyzed the DNA methylation levels of 448 minor ZGA genes that were down-regulated in ROSI embryos. We found that 65 genes were hypermethylated at the promoter regions. GO analysis of these hypermethylated minor ZGA genes could also enrich in the cytoskeleton-related terms (Fig. 4, C to E, and table S4). Using FDR, similar results could also be obtained (table S4). Notably, we found that the expression of 11 DEGs [including Carnmt1 (also known as 2410127L17Rik), Aph1a, Cdk16, Lrrc8d, Mllt4, Rac3, Rai14, Sipa1l3, Taf5, Zfp703, and Syt7] was associated with both DNA methylation and chromatin accessibility changes (Fig. 4, F and G). Among these genes, Rac3, encoding a nuclear receptor coactivator, is involved in cytoskeletal and nucleus organization (38).
A366 treatment improves the developmental capacity of ROSI embryos
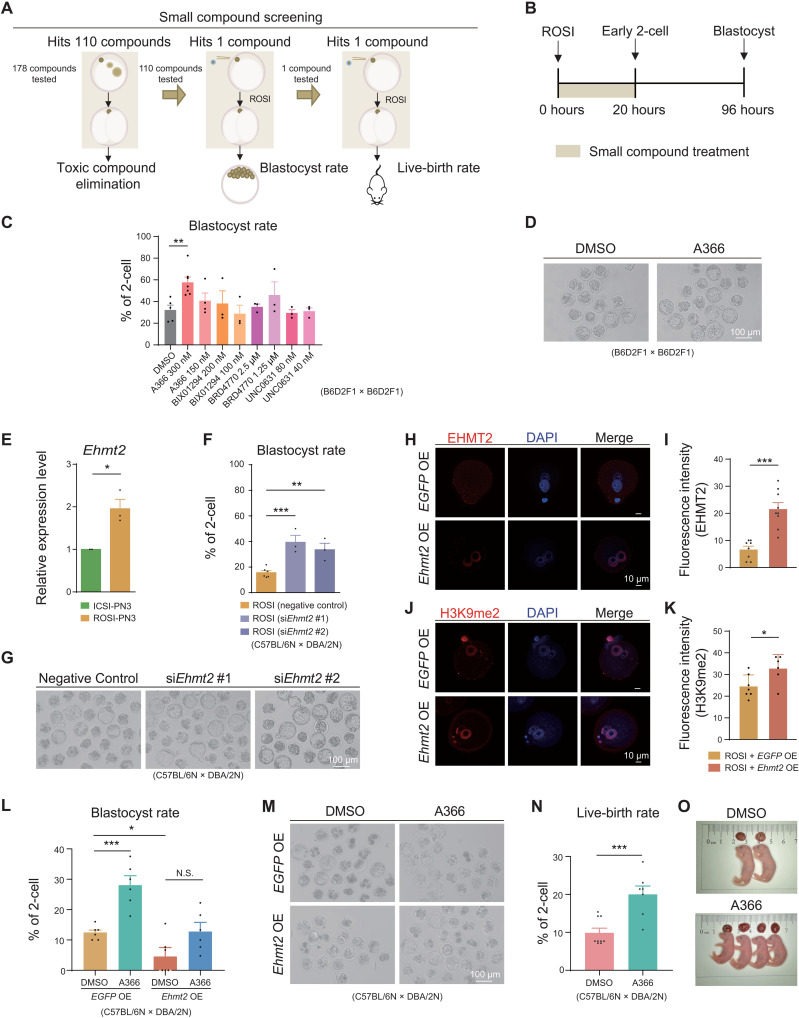
On the basis of the single-cell multiomics sequencing analyses and experimental data, the reprogramming defects of ROSI embryos were mainly detected at the pronuclear stages and exhibited as the changes of both transcriptome and epigenetic modifications. In support of this, certain epigenetic modifications (e.g., H3K9me2) of round spermatid could also pass to the male pronucleus after fertilization (Fig. 2, G to I). Hence, we asked whether there was previously unidentified small compound that could improve the developmental potential of ROSI embryos by targeting specific epigenetic modification. To this end, a library composed of 178 small compounds, targeting epigenetic modifications, including DNA methylation and histone modifications, were screened. We evaluated the final effect of these small compounds based on the developmental potential of ROSI embryos from three aspects: (i) nontoxic to the early embryos, (ii) improved rate of blastocyst development, and (iii) increased live-birth rate (Fig. 5A).
Fig. 5. A366 treatment improves the developmental capacity of ROSI embryos.
(A) Flowchart for small compound screening using ROSI embryos. (B) Schematic diagram showing the time frame for small compound treatment. (C and D) Blastocyst rate (C) of ROSI embryos treated by G9A inhibitors and representative ROSI embryos (D) treated by A366 at 94 hours after injection. Scale bar, 100 μm. DMSO, dimethyl sulfoxide. (E) Relative expression of Ehmt2 in ICSI and ROSI embryos (PN3) in three biological repeats. (F and G) Blastocyst rate (F) and representative ROSI embryos (G) after siEhmt2 injection at 94 hours after injection in three biological repeats. Scale bar, 100 μm. (H and I) Immunofluorescence (H) and statistical analysis (I) of EHMT2 in ROSI embryos that overexpressed Ehmt2 (Ehmt2 OE) and control group (EGFP OE) (PN5) in three biological repeats. Each dot corresponds to level in each individual embryo. Scale bars, 10 μm. (J and K) Immunofluorescence (J) and statistical analysis (K) of H3K9me2 in Ehmt2 OE-ROSI embryos and control group (PN5) in three biological repeats. Each dot corresponds to level in each individual embryo. Scale bars, 10 μm. (L and M) Blastocyst rate (L) and representative EGFP OE and Ehmt2 OE ROSI embryos (M) treated by A366 at 94 hours after injection in six biological repeats. Scale bar, 100 μm. (N and O) Live-birth rate (N) and representative alive fetuses of ROSI embryos (O) treated by A366 and control group. Unpaired two-tailed Student’s t test (C, E, F, I, K, L, and N) is used for statistical analysis. *P < 0.05, **P < 0.01, and ***P < 0.001.
Our analyses identified that the reprogramming defects mainly occurred before the 2-cell stage; therefore, small compound treatment was performed on ROSI embryos for 20 hours after injection of round spermatids when the embryos would reach the early 2-cell stage (Fig. 5B). We found that a series of H3K9me2 methyltransferase G9A (encoded by Ehmt2) inhibitors all showed a trend to increase the blastocyst rate of ROSI embryos such as A366 (39), BIX01294, and BRD4770 (Fig. 5C). Notably, A366 performed best among these candidates (P < 0.01) (Fig. 5, C and D), with an optimal treatment time frame (0 to 20 hours after injection) and dosage (300 nM) (fig. S13, A to C). Consistent with this finding, H3K9me2 deposition could be observed in the round spermatids used in ROSI assay and the male pronucleus of ROSI embryos (Fig. 2, G to I, and fig. S13D). In addition, Ehmt2, albeit not categorizing into the DEGs (fig. S13E), also exhibited a higher expression level in ROSI embryos at the PN3 stage (P < 0.05) (Fig. 5E).
To confirm the outcomes of aberrant H3K9me2 deposition and excessive Ehmt2 expression, we analyzed the developmental capacity of ROSI embryos after knockdown or overexpression of Ehmt2. Knockdown of Ehmt2 using small interfering RNA (siRNA) (fig. S13F) could markedly increase the blastocyst rate of ROSI embryos (36.30 and 35.30% for two Ehmt2 siRNA compared to 15.80% for control, P < 0.001 and P < 0.01, respectively; Fig. 5, F and G, and table S1). In contrast, when Ehmt2 were overexpressed, there was an increase in H3K9me2 deposition (Fig. 5, H to K, and fig. S13, G and H), and the blastocyst rate of ROSI embryos decreased in a dose-dependent manner (fig. S13I). However, simultaneous treatment by A366 in Ehmt2 overexpression group was unable to eliminate the Ehmt2 overexpression block on embryonic development (Fig. 5, L and M, and table S1). The treatment of ROSI embryos by A366 could lastly improve the live-birth rate by about twofold (18.70% versus 9.30%; P < 0.001) (Fig. 5, N and O, and table S1). Notably, the live offspring from ROSI embryos treated by A366 was healthy and fertile (fig. S13, J to L). In addition, the live-birth rate of in vivo fertilized zygotes treated with A366 was comparable to the control group, indicating the safety of A366 treatment (fig. S13M). Together, A366 treatment could significantly increase the blastocyst rate and full-term development of mouse ROSI embryos.
A366 treatment can partially overcome the reprogramming defects of ROSI embryos derived from early-stage round spermatids
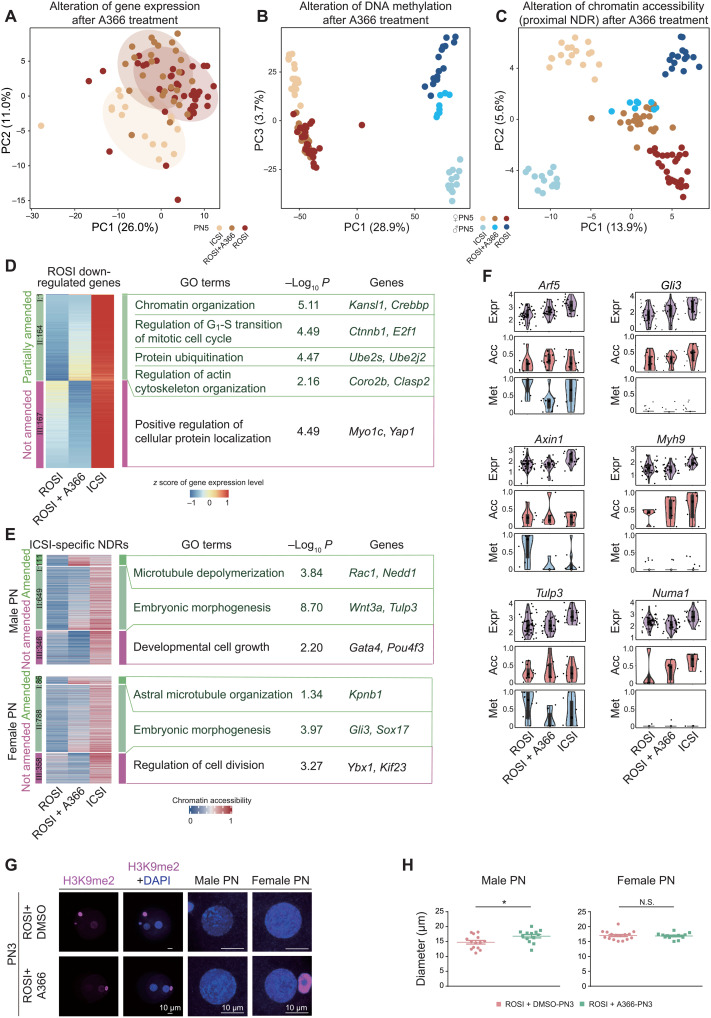
To illustrate how A366 improved the developmental potential of ROSI embryos, single-cell multiomics sequencing was performed on the PN5-stage ROSI embryos treated by A366. ROSI embryos without treatment and ICSI embryos were used as the controls. Dimensionality reduction analysis was performed to cluster cells based on the gene expression profiles. We found that ROSI and ICSI embryos could be separated into two independent cell clusters (Fig. 6A), indicating that the transcriptome of these two types of embryos already exhibited certain differences at the PN5 stage. Notably, A366 treatment could change the global gene expression patterns of ROSI embryos (Fig. 6A). Unbiased cell clustering was then performed using the DNA methylation and chromatin accessibility data (proximal NDR). Consistent with the transcriptome data, A366-treated ROSI embryos were reverted into an independent cellular state as seen from both male and female pronuclei (Fig. 6, B and C). These data were consistent with the finding that A366 could affect the developmental potential of ROSI embryos.
Fig. 6. A366 treatment can partially overcome the reprogramming defects of ROSI embryos derived from early-stage round spermatids.
(A to C) Dimensionality reduction analysis of gene expression (A), DNA methylation (B), and chromatin accessibility (C) of ICSI, ROSI, and A366-treated ROSI embryos based on DEGs, DMRs, and differential NDRs between ROSI and ICSI embryos (PN5) using PCA, respectively. Cells are colored by experimental group. (D and E) Heatmap showing the gene expression alterations of ROSI down-regulated genes (D) and the chromatin accessibility alterations of ICSI-specific NDRs (E) in A366-treated ROSI embryos (PN5). Representative GO terms and genes are shown on the right. (F) Violin plots showing the gene expression [log2(TPM/10 + 1), where TPM is transcripts per million, purple], chromatin accessibility (GCH methylation level, red) of associated proximal NDRs, and DNA methylation level (WCG methylation level, blue) of associated DMRs of representative minor ZGA genes (PN5). Regions with more than one WCG or GCH site covered in a single cell are used to calculate the methylation level. (G) Immunofluorescence of H3K9me2 in A366-treated ROSI embryos and control group (PN3). The magnified images are shown on the right. Scale bars, 10 μm. (H) Dot plots showing the diameter of male (left) and female (right) pronuclei in A366-treated ROSI embryos and control group (PN3) in three biological repeats. *P < 0.05, unpaired two-tailed Student’s t test. Each dot corresponds to the diameter of each pronucleus.
We then dissected the detailed molecular differences among ROSI, ROSI-treated (A366-treated), and ICSI embryos. To this end, the differences at the levels of gene expression, chromatin accessibility, and DNA methylation were first identified, respectively. According to the patterns after A366 treatment, they were divided into three categories (table S5): (i) fully amended, (ii) partially amended, and (iii) not amended. Among the 332 down-regulated and 465 up-regulated genes in ROSI embryos, the expression of 165 and 159 genes in each gene group could be amended by A366, respectively (Fig. 6D, fig. S14A, and table S5). Note that the down-regulated genes amended by A366 were mainly associated with actin cytoskeleton and chromatin organization (Fig. 6D), consistent with the defects observed at the pronuclear stage of ROSI embryos (Fig. 2, E and F). A366 treatment could also amend most of the chromatin accessibility of ICSI-specific proximal NDRs in both male pronucleus (760 of 1106) and female pronucleus (874 of 1232) of ROSI embryos (Fig. 6E, fig. S14B, and table S5). Notably, the neighboring genes of these amended ICSI-specific NDRs were enriched in the processes of microtubule depolymerization and embryonic morphogenesis in the male pronucleus and astral microtubule organization in the female pronucleus (Fig. 6E). In terms of DNA methylation, a similar amending effect could be observed in A366-treated embryos at hypermethylated DMRs and hypomethylated DMRs in ROSI embryos. Notably, the DNA methylation state of genes related to cytoskeletal organization could be amended in female pronucleus (fig. S14, C and D). Generally consistent results were obtained on the basis of FDR analysis (table S5). To determine whether the amending effect by A366 treatment was correlated with H3K9me2 modification, we analyzed the distribution of these DMRs on the H3K9me2 peaks. Given that there was sparse H3K9me2 signal in round spermatids, the analysis was performed using a previously reported H3K9me2 chromatin immunoprecipitation sequencing data in oocytes (40). Notably, in the female pronucleus, the hypermethylated DMRs amended by A366 were more significantly overlapped with H3K9me2 peaks than the ones that could not be amended (fig. S14E).
As aforementioned, both proximal NDRs and DNA methylation were associated with gene expression regulation. Therefore, we tried to determine which genes amended by A366 were associated with chromatin accessibility or DNA methylation changes at the promoter regions. We found that the expression of a subset of genes (46 of 165 up-regulated genes and 36 of 159 down-regulated genes) such as Myh9, Numa1, and Gli3 was associated with chromatin accessibility rather than DNA methylation at the promoter regions. In contrast, the gene expression of another subset of genes (82 of 165 up-regulated genes and 31 of 159 down-regulated genes) such as Arf5 and Axin1 was associated with DNA methylation. Tulp3 was associated with both chromatin accessibility and DNA methylation (Fig. 6F). In addition, A366 treatment could restore the morphology of male pronucleus in ROSI embryos (Fig. 6, G and H, and fig. S14, F and G). Together, the compromised developmental potential of ROSI embryos could be improved by A366 treatment, wherein A366 partially overcame the reprogramming defects detected at the pronuclear stages.
DISCUSSION
Correct epigenetic reprogramming in mammalian early embryos is critical for cell fate establishment and subsequent developmental potential. In this study, we constructed the single-cell multiomics landscapes of ROSI embryos in mice. In this context, we dissected the defects occurred in ROSI embryos at the levels of transcriptome, methylome, and chromatin accessibility. Notably, by interrogating these multiple layers of information, we uncovered the reprogramming defects in ROSI embryos, which were associated with the misexpression (down-regulation) of a cohort of minor ZGA genes. We then found that these defects could be overcome to a certain extent by A366 treatment, wherein A366 treatment could increase the overall developmental rate by about twofold. A previous study revealed that assisted reproductive technologies–associated DNA methylation variations could be largely resolved around birth (41). Similarly, the de novo chromosomal instability in early embryos could also be depleted from embryonic germ layers to a great extent (42, 43). Given that poor developmental outcomes were observed in the nonintervention ROSI group, it was most likely that the reprogramming defects uncovered in this study could not be eliminated spontaneously.
Reprogramming barriers exist in several kinds of reconstructed embryos or artificial pluripotent stem cells. For instance, somatic cell nuclear transfer (SCNT) embryos have DNA methylation and histone modification reprogramming barriers (44–46). Accordingly, the use of inhibitors or expression of key enzymes is effective in improving the development of SCNT embryos (44, 45). Notably, H3K9 methylation exerts important roles in pluripotency establishment (44, 47), especially that histone H3 lysine 9 trimethylation acts as a critical epigenetic barrier in mammalian SCNT reprograming process. Similarly, the previous studies also observed the existence of abnormal DNA methylation and H3K9 methylation at the pronuclear stages in ROSI embryos (18, 19); however, systematic delineation of the epigenetic abnormalities in ROSI embryos, especially upon fertilization, was still lacking. In this study, we uncovered the reprogramming defects in ROSI embryos reconstructed with early-stage round spermatids. A remarkable difference between ROSI and SCNT was that the developmental defects of SCNT embryos appeared at the onset of ZGA; thus, we speculated that the defects identified in ROSI embryos might not be severe enough to impede the full initiation of major ZGA (29, 48). The reprogramming defects in ROSI embryos were most likely correlated with the misexpression of a cohort of minor ZGA genes, but the causal relationship between these two events remained undetermined. Presumably, the aberrant epigenome of early-stage spermatids might determine the developmental fate of ROSI embryos, as exemplified by the deposition of H3K9me2 in male pronucleus. Our treatment of ROSI embryos with the EHMT2 inhibitor A366 could indeed improve the developmental potential, consistent with a previous finding that decreasing H3K9 methylation could increase the developmental potential of ROSI embryos (49). We do not rule out the possibility that there are other defects inherent to ROSI embryos. Even with A366 treatment, the developmental potential of ROSI embryos did not recover to the same level of ICSI controls, and the epigenome reprogramming was still not fully completed. With the development of techniques to measure histone modifications in extremely low input (50), it will be promising to find whether the reprogramming defects of ROSI embryos uncovered in this study might be determined by histone modifications or whether other histone or DNA modifications inherited from round spermatids act as other potential reprogramming defects. Here, note that because of the limit of cell numbers used in single-cell multiomics analyses in the present study, genomic coverage variations could be observed among individual cells.
Minor ZGA is important for the early embryonic development in mammals (29). A previous study revealed that cohesin impeded the minor ZGA in SCNT embryos, in which the three-dimensional (3D) chromatin architecture of the genome was aberrantly reprogrammed (31). The observed enrichment in chromatin-related processes and DEGs at the pronuclear stages suggests that ROSI embryos derived from the early-stage round spermatids might also have 3D chromatin architecture defects as that in SCNT embryos. Parental genomes undergo asymmetric reprogramming in various epigenetic features such as DNA methylation, chromatin accessibility, and H3K27me3 levels (51–53). Moreover, the reprogramming factors within male and female pronuclei are distinct (54). Although most of these modifications are resolved upon fertilization, parental-biased escaping in the process of reprogramming is critical for the establishment of allele-specific expression patterns. Further studies are required to understand more about how shuttling factors existed in the ROSI male pronucleus (derived from early-stage round spermatids) exchange information between the male and female pronuclei. It will also be interesting to validate whether similar reprogramming defects also exist in human ROSI embryos and whether it can be eliminated by A366 treatment.
MATERIALS AND METHODS
Animals
All the animal studies were performed in accordance with the ethical guidelines of South Medical University ethics committee (L2016149). All the animal research protocols were in accordance with the U.S. Public Health Service Policy on Use of Laboratory Animals and were approved by the Ethics Committee on Use and Care of Animals of Southern Medical University.
DBA/2N male mice were used for the collection of sperm and spermatids, while C57BL/6N female mice were used for the collection of oocytes. The mice were purchased from Beijing Vital River Laboratory Animal Technologies Co. Ltd., (Beijing, China). B6D2F1 female and male mice were used for the preliminary screening of small compound. ICR mice purchased from Guangdong Experimental Animal Center were used as embryo transfer recipients.
Collection of oocytes, sperm, and round spermatids
To obtain MII oocytes for ICSI/ROSI, B6D2F1 or C57BL/6N female mice were superovulated by injecting 8 U of pregnant mare serum gonadotropin each and followed by 8 U of human chorionic gonadotropin (hCG) 46 to 48 hours later. Mice were euthanized by cervical dislocation 13.5 hours after hCG injection. Cumulus-oocyte complexes were collected from the oviduct ampullae, and MII oocytes were then obtained by removing the cumulus cells in M2 medium (Sigma-Aldrich) containing hyaluronidase (1 mg/ml).
Sperm and early-stage round spermatids were collected from cauda epididymidis and testis, respectively. Fluorescence-activated cell sorting (FACS) was then used to isolate the haploid round spermatids, consistent with a previous study (9). Briefly, the suspension of testicular cells was obtained by mechanical isolation and enzymatic digestion. The cell suspension was filtered through a 40-mm nylon mesh and stained with Hoechst 33342 (2.5 mg/ml) for 15 min. The isolation of haploid round spermatids was performed using MoFlo XDP (Beckman).
ROSI and ICSI
ICSI (55) and ROSI (24) were performed as previously described with a slight modification. Briefly, for ROSI, the MII-stage oocytes were first placed in the balanced Ca2+-free Chatot-Ziomek-Bavister (CZB) medium containing 10 mM SrCl2 for 10 min for preactivation. The nucleus of round spermatid, which was separated from the cytoplasm by pipetting, was then microinjected into the MII oocyte in M2 medium (Sigma-Aldrich, M7167) containing cytochalasin B (5 μg/ml). The injected oocytes were further activated for 5 hours in the balanced Ca2+-free CZB medium containing 10 mM SrCl2 in an incubator (37°C, 5% CO2). After activation, the injected oocytes were transferred to the balanced potassium simplex optimized medium (KSOM) (Millipore, MR-020P-D) for further culture. For ICSI, the head of sperm was microinjected into an oocyte. The micromanipulation process was performed using PiezoXpert device (Eppendorf).
Embryo transfer
The 2-cell stage embryos derived by ROSI or ICSI were transferred into the oviducts of the pseudo-pregnant female mice. Caesarean section was performed at embryonic day 19.5, and the live pups were fostered by lactating ICR females.
Collection of mouse preimplantation embryos
Both ROSI and ICSI embryos with normal morphology at certain stage were collected for single-cell multiomics sequencing: 7.5 hours (PN3 zygote), 12 hours (PN5 zygote), 28 hours (2-cell), 46 hours (4-cell), 75 hours (morula), and 94 hours (blastocyst) after injection according to previous studies (15, 27, 56). The standard of pronuclear-stage embryos was mainly consistent with previous studies as described later (6, 57).
The physical isolation of male and female pronuclei at the PN3 and PN5 stages was performed as previously described (15). Briefly, zygotes were transferred into M2 medium containing cytochalasin B (5 μg/ml) at 7 and 11.5 hours after injection and cultured for 30 min. Zona pellucida was removed by a piezo-driven micromanipulator, and the pronuclei were isolated from the zygotes. Next, the pronuclei were placed into lysis buffer immediately for the preparation of single-cell DNA library. The remaining cytoplasm wrapped in the cytosol was collected into lysis buffer immediately for the preparation of scRNA library. The isolation of individual blastomere of other stage preimplantation embryos was performed as previously described (58). The embryos were then transferred to M2 medium to terminate the digestion. Single blastomere was isolated by gentle and repeated pipetting. When all of the blastomeres were separated, they were removed from the manipulation drops, washed three to five times in prewarmed phosphate-buffered saline (PBS) with 1% human serum album (HSA), and placed into lysis buffer immediately for the preparation of scRNA and DNA libraries.
Reverse transcription and qPCR
To analyze the gene expression in ICSI and ROSI embryos, the cDNA of 15 ICSI or ROSI zygotes was synthesized by a reverse transcription system and amplified using the Single Cell Squence Specific Amplification Kit (Vazyme, P621-01) according to the manufacturer’s instructions. To confirm the knockdown efficiency of Ehmt2 siRNA, the cDNA was prepared from 15 2-cell embryos. qPCR was then performed using 2× PCR master mix (GenStar, A301-10) on LightCycler96 system (Roche). The data were analyzed using the ΔΔCt method. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal control to normalize the expression of target genes. The primers were listed in table S7.
Immunofluorescence
Mouse embryos were usually fixed with 4% paraformaldehyde for 1 hour and then permeabilized with 0.5% Triton X-100 for 1 hour at room temperature. However, for the calculation of interpronuclear distance, geometric center distance, and diameters, the embryos were fixed and permeabilized using 4% paraformaldehyde containing 0.2% Triton X-100 for 20 min at room temperature (Figs. 2E and 6G and figs. S2A and S14F). The embryos were then blocked with 1% bovine serum albumin (Sigma-Aldrich) at room temperature for 1 hour and incubated with the primary antibodies against EHMT2 (1:400; Invitrogen, 435200) and H3K9me2 (1:400; Abcam, ab1220) at 4°C overnight. After washing three times with PBS containing 0.05% Tween 20, the embryos were incubated with the appropriate secondary antibodies at room temperature for 1 hour. The nuclei were stained with DAPI (Thermo Fisher Scientific, R37606). Images were captured under identical imaging conditions on the same confocal laser scanning microscope (Carl Zeiss LSM 880). Z projections were done by maximum intensity projections of the indicated Z-stacks. Quantitative fluorescence image analysis was performed using ImageJ (Fiji) software (https://imagej.net/software/fiji/downloads). The intensity of fluorescence was calculated from images of stacked or sectioned images after the signal areas were detected automatically in regions of interest.
The calculation of interpronuclear distance and geometric center distance between two pronuclei
The 3D immunofluorescence images of embryos at the pronuclear stages were reconstructed using Imaris software as previously described (59, 60). The interpronuclear distance was first calculated by Imaris 9.0.2 at the same stage (fig. S2A). Next, we compared the developmental progress between ROSI and ICSI embryos at the pronuclear stages according to the diameter of pronucleus. The largest cross section of pronucleus was selected in the Z-stack images, and the diameter of pronucleus was counted by the ZEN 2.1 (black). To eliminate the effect of reduced diameter of pronuclei in ROSI embryos on measuring interpronuclear distance, the geometric center distance between two pronuclei was reconstructed and calculated using Imaris 9.0.2 software as previously described (fig. S2C) (59, 60). The nuclear envelope of two pronuclei at the PN5 stage was close but not yet fused. Under this condition, the software used here could not identify them; thus, the geometric center distance between two pronuclei at the PN5 stage was calculated by adding the radius of two pronuclei together.
Low-input DNase I qPCR
ICSI or ROSI 4-cell embryos were used for liDNaseI-qPCR assay as previously described (15). In detail, the zona pellucida was removed using Acidic Tyrode’s Solution (pH 2.5) (Sigma-Aldrich, T1788). The embryos were then resuspended in 36 μl of lysis buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 3 mM MgCl2, and 0.1% Triton X-100] and incubated on ice for 5 min. DNase I [2 U/μl; New England Biolabs (NEB), M0303L] was added to the final concentration of 10 U/ml and incubated at 37°C for 5 min. The reaction was stopped by adding 80 μl of stop buffer [10 mM tris-HCl (pH 7.5), 10 mM NaCl, 0.15% SDS, and 10 mM EDTA] containing 2 μl of Proteinase K (20 mg/ml; Life Technologies). After that, 20 ng of circular carrier DNA was added, and the mixture was incubated at 50°C for 1 hour. DNA was purified by extraction with phenol-chloroform and precipitated by ethanol in the presence of linear acrylamide (Yeasen, 10408ES403). For qPCR validation of NDRs, the ethanol-precipitated DNA was dissolved in 15 μl of double distilled H2O. DNase I free control samples with the same number of embryos were treated by the same way without the addition of DNase I. The treated DNA was then used as the template for multiplexed preamplification with the primers for 15 cycles in a 50-μl volume using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711). The preamplified product (0.2 μl) was used for quantification with two technical replicates. The relative amount of intact DNA at NDRs amplifiable after DNase I treatment was quantified against DNase I free control samples. Three biological replicates were performed for each locus, and the primers were listed in table S7.
Bisulfite genomic PCR-based Sanger sequencing
Genomic DNA from ICSI or ROSI embryos at morula stage was purified using QIAamp DNA Micro kit (QIAGEN, 56304). Sodium bisulfite treatment of DNA was then performed using the EZ DNA methylation–direct kit (Zymo Research, D5021). PCR amplification was performed using hot start DNA polymerase (TAKARA) with specific primers for DMR regions of interest (table S7). The PCR product was gel-extracted, subcloned into the pClone007 Versatile Simple Vector (TSINGKE, 007VS), and sequenced. The results were analyzed using Quantification Tool for Methylation Analysis (QUMA, http://quma.cdb.riken.jp/).
Small compound screening and treatment
The epigenetic modification–related small compound library was from TargetMol Company (L1200) (table S6) and in-house chemical collections. The small compound was added to Ca2+-free CZB medium containing 10 mM SrCl2 for activation before and after ROSI injection as aforementioned. The injected oocytes were then cultured in the balanced KSOM medium containing small compound until 20 hours after injection and subsequently transferred to the fresh balanced KSOM medium for the subsequent culture.
In vitro transcription of mRNA and intracytoplasmic injection
The cDNAs of both Ehmt2 and EGFP were cloned into T7-driven vectors, respectively. The mRNAs were then transcribed using the mMESSAGE mMACHINE T7 Ultra Kit (Ambion, AM1344) according to the manufacturer’s instructions. The storage concentration of each mRNA was optimized to 1 μg/μl. Ehmt2 or EGFP mRNA was diluted to the injection concentration with nuclease-free water. The integrity of mRNA was confirmed by electrophoresis with formaldehyde gels. The injection of mRNA into MII oocytes was performed using a piezo-driven micromanipulator, and about 10 pl of mRNA was injected into each MII oocyte. The primer sequences were listed in table S7.
Knockdown of Ehmt2 in ROSI embryos
siRNA against mouse Ehmt2 or negative control was synthesized by Geneyuan Company. The siRNA was diluted to the injection concentration with nuclease-free water before injection. Each MII oocyte was injected with approximately 10 pl of siRNA using a piezo-driven micromanipulator. The siRNA was listed in table S7.
scRNA/DNA library preparation and sequencing
The cell lysis buffer mixture containing 4 U of ribonuclease (RNase) inhibitor (Takara, 2313B), 0.25% IGEPAL CA-630 (Sigma-Aldrich, I3021), and in vitro methylation mix (NEB, M0227L) were prepared. The DNA and RNA of each cell were then isolated by the nuclear separation mixture containing 0.2 μl of Dynabeads MyOne Carboxylic Acid (Invitrogen, 65011), 0.2% Tween 20 (Sigma-Aldrich, P1379), 1% Triton X-100 (Sigma-Aldrich, T8787), 4 U of RNase inhibitor, 50 mM dithiothreitol, and 2 μl of 5× SuperScript II first-strand buffer (Invitrogen, 18064071). Subsequently, for RNA part, scRNA-seq library was constructed using single cell–tagged reverse transcription sequencing method. The high-quality libraries were sequenced with 150–base pair (bp) pair-end reads on Illumina Hiseq Xten (Novogene). For DNA part, the magnetic beads containing cell nuclei (DNA) were treated with EZ-96 DNA Methylation-Direct MagPrep kit (Zymo Research, D5044) to complete the bisulfite conversion. Four rounds of amplification were then conducted by Klenow exo- (ENzymics, NG202) and scBS-seq-P5-N6-oligo1 (CTACACGACGCTCTTCCGATCTNNNNNN). The purified products were used to perform the second strand synthetization using scBS-seq-P7-N6-oligo2 (AGACGTGTGCTCTTCCGATCTNNNNN). Last, the DNA library was constructed with incorporated universal primers and index primers (NEB). After purification twice with 0.8× AMPure XP beads, the DNA library was checked for quality, and each cell was sequenced for about 3 Gb on Illumina Hiseq Xten platform (Novogene).
scRNA-seq data processing and normalization
scRNA-seq raw reads were processed as previously described (61). First, we used cell barcode sequence (8 bp) and read ID to extract the R1 and R2 reads. Template switch oligo sequence, polyadenylate tails, and low-quality bases were then removed to obtain clean reads. The clean R1 reads were aligned to mouse genome (mm10, UCSC version) using TopHat2 (v2.0.14) in single-end module with default parameters (62), and uniquely mapped reads according to unique molecular identifier (UMI) numbers were counted using HTSeq (63). Last, the category of UMIs for each gene was calculated as the transcript copy number, and gene expression level was shown as log2(TPM/10 + 1), where TPM (transcripts per million) was calculated as the number of UMIs of each gene divided by all UMIs in a given cell. Once the TPM was higher than 0, we considered that this gene was detected in each individual cell.
The numbers of genes and transcripts were calculated in each individual cell. In detail, for the male gamete dataset, cells with more than 1000 genes detected were retained for the subsequent analysis. For other stages, cells with more than 1000 genes and 10,000 UMI detected were retained for the subsequent analysis.
Dimensionality reduction analysis of scRNA-seq data and cell type identification
The R package Seurat (v3.1.2) (64) was used to perform dimensionality reduction and clustering analysis based on log2(TPM/10 + 1) expression matrix. Only the genes expressed in more than three cells were retained for downstream analysis. FindIntegrationAnchors (dims = 1:20) and IntegrateData function (dims = 1:20) were used to integrate two batches of scRNA-seq data (batch information was provided in table S2). Subsequently, PC1 to PC50 were selected to perform the RunPCA function, and PC1 to PC20 were selected to run the RunTSNE function to generate t-SNE plot. To identify the clusters of inner cell mass (ICM) and trophectoderm (TE), we extracted the cells at blastocyst stage to perform FindClusters function (resolution = 0.8) to obtain clusters 0 to 2. According to the expression levels of Cdx2, Sox2, Nanog, Sox17, Dppa1, Id2, and Gata3, we defined cluster 0 as ICM and clusters 1 and 2 as TE.
Integration analysis of round spermatids using scRNA-seq data
To double-check the developmental stage of the round spermatids used in this study, we used the mouse round spermatid dataset with similar scRNA library preparation and the same bioinformatics analysis pipeline for the integration analysis (9). Merge function in Seurat package (v3.1.2) was used to integrate different datasets, and PC1 to PC20 were selected to perform integration and Uniform Manifold Approximation and Projection analysis. All the expressed genes were used to perform K-means clustering analysis.
Identification of DEGs
To identify DEGs, we used the FindMarker function in Seurat (test.use = “LR”) based on log2(TPM/10 + 1) expression values. Likelihood ratio test was used to calculate P values. FDR was calculated as Benjamini-Hochberg (BH)–adjusted P values. Only genes with |fold change| > 1.5 and P < 0.05 were defined as DEGs. GO analysis was performed with Metascape (www.metascape.org) using default parameters.
Weighted correlation network analysis
To evaluate the expression patterns of minor ZGA genes, we selected the transcriptome data of seven stages of PN3, PN5, 2-cell, 4-cell, Morula, TE, and ICM to perform WGCNA (65). The power = 10 was used to execute the blockwiseModules function, and two kinds of modules were found from it.
Read mapping and quality control of single-cell DNA part data
For single-cell DNA part data (DNA methylation and chromatin accessibility), Trim:galore (v0.3.3), Cutadapt (v1.2.1), and FastQC (v0.11.7) were used to trim adapter sequences, six bases of random primer, and low-quality bases away from raw data. Cleaned reads were mapped to mouse reference genome mm10 (downloaded from the UCSC genome browser) with Bismark (v0.7.6) (66) in paired-end and nondirectional manner. The unmapped reads were then remapped to the same reference genome in single-end and nondirectional manner as previously described (14). After alignment, duplicated reads generated through PCR amplification were removed by Samtools (v0.1.18). According to our previous study (14), the methylation level of WCG (W = A/T) and GCH (H = A/C/T) sites were calculated as the ratio of methylated reads (reported as “C”) divided by sequencing depth (“C + T”) at 1× depth. The level of DNA methylation or chromatin accessibility of different genomic regions or global level was estimated by the mean methylation level of all WCG or GCH sites covered in the region or whole genome. Only cells with bisulfite conversion rate higher than 97%, genome coverage of more than 3%, more than 500,000 WCGs, and 5,000,000 GCH sites covered were retained for further analysis.
CNV analysis
After alignment, we used HMMcopy (67) to deduce the CNVs in each individual cell considering mappability and GC content of the genome in 1-Mb windows.
Genotype deducing
To deduce the genotype of embryo, we counted the reads number of autosome (here, we used “chr1”) and sex chromosome “X” and “Y.” As control, the genotype of MII oocyte was XX, while the genotype of sperm included X and Y. Notably, at the pronuclear stages (PN3 and PN5), if only the female pronucleus was retained after quality control (but the male pronucleus in the same embryo did not pass the quality control), then this embryo would be removed from the downstream sex analysis. The detailed gender information was included in the table S2.
Defining open chromatin regions
As mentioned in the experimental method, GCH sites in open chromatin regions would be methylated during in vitro methylation process. So, we used methylation level of GCH sites (1× depth) to define open chromatin regions (NDRs). Because of the sparse nature of single-cell genomic data, we aggregated all cells at the same stage from ICSI/ROSI-derived embryos to define NDRs as previously described (13, 23, 68). As reported in our previous study (14), we used a sliding window with 100-bp length at 20-bp step size to calculate the significance of difference (chi-square test, P ≤ 10−15) of GCH methylation level compared to the whole genome level. Only DMRs longer than 140 bp with more than five GCH sites covered were defined as NDRs. According to the distance to TSSs, NDRs were classified into two groups, named proximal (overlapped with promoters) NDRs, and distal (located at least 2-kb away from TSSs and within 100 kb) NDRs.
Estimation of methylation level
The global DNA methylation level or chromatin accessibility of each individual cell was estimated as the average methylation level of all the WCG or GCH sites covered at 1× depth. We also calculated the DNA methylation level and chromatin accessibility in different genomic regions, 500-bp nonoverlapping sliding windows and NDRs, which were determined on the basis of the mean methylation level of all WCG or GCH sites covered in that region. Only regions with more than three WCG or five GCH sites covered were retained for methylation level analysis, unless stated otherwise. All genomics annotations, including introns, exons, TSSs, and transcription end sites (TESs), were downloaded from the UCSC genome browser (mm10). Repeat regions were downloaded from RepeatMasker (mm10). Promoters were defined as regions 2-kb upstream and 2-kb downstream of TSSs. Gene bodies (intragenic region) were regions from TSSs to TESs, while intergenic regions were regions complementary to gene bodies in the genome.
Dimensionality reduction analysis of single-cell DNA part data
We calculated the chromatin accessibility of the proximal NDRs in each individual cell. Regions covered in at least 10% of cells were extracted. Before using “prcomp” to reduce the dimensionality, we first used the “ppca” method in pcaMethods package (69) to fill in the missing values. For dimensionality reduction analysis of DNA methylation data based on 500-bp tiles, we used tiles covered in at least 10% of cells and performed PCA after imputing the missing value. Only regions covered in at least 5% of cells were retained for dimensionality reduction analysis performed in A366-treated embryos, and the input to PCA was the matrix with inputting missing values of chromatin accessibility or DNA methylation level on differential NDRs or DMRs at the PN5 stage between ICSI and ROSI embryos.
Stage-specific proximal NDRs and ICSI- or ROSI-specific NDRs
Differential NDRs were calculated on the basis of merged NDRs between the comparison groups, which were called on the basis of aggregated data at the same stage from ICSI/ROSI-derived embryos. Meanwhile, regions were restricted on those which were simultaneously covered by at least 5% of cells or two cells in all comparison groups considering the coverage of single-cell data (22, 70). In detail, we extracted the global GCH methylation level of each individual cell from the chromatin accessibility of each NDR in that cell to scale the methylation level of NDRs to the genome-wide level. We then systematically searched for the stage-specific proximal NDRs at the pronuclear stages and from 2-cell to blastocyst stages. Only NDRs in one cell type with scaled difference higher than 0.1 to the maximum level of the other stages and with P < 0.05 were retained. Pearson correlation between the retained NDRs and corresponding gene expression levels was calculated, and only proximal NDRs with a correlation of >0.6 were lastly defined as the stage-specific proximal NDRs. ICSI- or ROSI-specific NDRs were those with the difference of scaled chromatin accessibility between ROSI and ICSI greater than 0.1 with P < 0.05, and FDR was calculated as BH-adjusted P values. In addition, the RNA expression patterns between ROSI and ICSI embryos were positively correlated with the chromatin accessibility changes between them.
Enrichment analysis of transcription factor motifs
Motif enrichment analysis was performed on all distal NDRs and differential distal NDRs between ROSI and ICSI embryos using HOMER with the parameters “len 8,10,12 -size given.” Only transcription factors with a TPM of ≥10 and motifs with P ≤ 10−10 in at least one stage in ROSI or ICSI embryos were retained for analysis.
Differentially methylated regions
For differential methylation analysis between ROSI and ICSI embryos, we aggregated all single-cell data at each stage (13, 23, 68). That meant that we defined the DNA methylation level of each WCG site by the mean level of that in all single cells derived by ICSI or ROSI at the same stage, which could circumvent the variance of genomic coverage among cells to a certain extent. We then calculated the DNA methylation level of 500-bp nonoverlapping tiles covered in two groups. Only tiles with methylation differences between two groups larger than 0.2 and with P < 0.05 were defined as DMRs. FDR was calculated as BH-adjusted P values. We defined DMR-associated genes when DMRs overlapped with promoter regions; meanwhile, methylation levels of DMRs and promoter regions varied inversely with expression levels.
Volcano plot
In fig. S8 (D and E), minor ZGA genes with higher expression levels in ROSI embryos (fold change > 1.5, P < 0.05, up-regulated) or in ICSI embryos (fold change > 1.5, P < 0.05, down-regulated) were colored in red or blue, respectively. Genes associated with ICSI or ROSI-specific proximal NDRs were colored in Fig. 3D and fig. S11A, with the blue dots representing genes whose expression levels were significantly lower in ROSI embryos and occupied with the ICSI-specific proximal NDRs (absolute GCH methylation difference > 0.1, P < 0.05), and the red dots were vice versa. Genes associated with DMRs were colored in Fig. 4C, with the red dots representing genes whose expression levels were significantly up-regulated in ROSI embryos, and the associated DMRs were also hypomethylated. The blue dots were vice versa (absolute DNA methylation difference > 0.2, P < 0.05).
Enrichment analysis of DMRs and NDRs
Enrichment analysis of DMRs and NDRs were calculated on the basis of the proportion of observed ratio to expected ratio as mentioned in a previous study (45). Briefly, the observed ratio was estimated by the number of DMRs overlapped with a genome context divided by the total number of this class of DMRs; the expected ratio was the number of 500-bp tiles overlapped with the genome context in the whole genome divided by the total number of 500-bp nonoverlapping genome tiles. The significance analysis of relative enrichment was performed by the chi-square test. For NDRs, “observed” represented NDRs located at different genomic regions divided by the total number of NDRs at that stage, while “expected” meant the whole genome 500-bp tiles overlapped with different genomic regions divided by the total number of 500-bp tiles.
Classification of DEGs, NDRs, and DMRs amended by A366 treatment
To define the DEGs, NDRs, and DMRs amended by A366 at the PN5 stage, we classified the DEGs, ICSI- or ROSI-specific NDRs, and DMRs into three categories according to the average level of each group as the following standards:
For ROSI up-regulated DEGs, ROSI hyper-NDRs, and ROSI hyper-DMRs, that is:
(i) Fully amended by A366: R > I ≥ R_A366.
(ii) Partially amended by A366: R > R_A366 > I.
(iii) Not amended by A366: R_A366 ≥ R > I.
For ROSI down-regulated DEGs, ROSI hypo-NDRs, and ROSI hypo-DMRs, that is:
(i) Fully amended by A366: R_A366 ≥ I > R;
(ii) Partially amended by A366: I > R_A366 > R;
(iii) Not amended by A366: I > R ≥ R_A366.
Statistical analysis
The statistical analysis of experimental data was performed using unpaired two-tailed Student’s t test to compare the differences between different groups with GraphPad Prism version 6.0.0. All the experimental data were from at least three independent biological repeats, and the data were presented as means ± SEM, unless stated otherwise. P < 0.05 was considered statistically significant.
For the statistical analysis of stage-specific proximal NDRs, ICSI- or ROSI-specific proximal NDRs were obtained by comparing the differences between different stages and samples with unpaired two-tailed Student’s t test. P < 0.05 was considered statistically significant. For the statistical analysis of DMRs, unpaired two-tailed Student’s t test was used to compare the differences between different stages and samples. P < 0.05 was considered statistically significant. Correlation and corresponding statistical significance were calculated on the basis of Pearson correlation coefficient in R language.
For the statistical analysis of DEGs, Likelihood ratio test was used to compare the differences between different stages and samples with R package Seurat. P < 0.05 was considered statistically significant.
Acknowledgments
We thank Beckman for supporting of the FACS.
Funding: This work was supported by the National Key R&D Program of China (2017YFA0105001 to X.-Y.Z., 2021YFA1102700 to L.L., 2020YFA0113200 to S.G., 2019YFA0801802 to M.W., and 2020YFA0113300 to M.W.), the Natural Science Foundation of China (82071711 to X.-Y.Z., 31671544 to X.-Y.Z., 31970787 to G.C., 32170869 to G.C., 32070833 to L.L., 82101745 to L.L., 31970814 to S.G., and 81901542 to M.W.), Key Research and Development Program of Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) (2018GZR110104002 to X.-Y.Z.), Clinical Innovation Research Program of Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) (2018GZR0201003 to X.-Y.Z.), Outstanding Scholar Program of Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory) (2018GZR110102004 to X.-Y.Z.), The Science and Technology Projects of Guangzhou City (201904020031 to X.-Y.Z.), the Guangdong Natural Science Foundation (2019A1515010446 to G.C.), Natural Science Funds for Distinguished Young Scholar of Guangdong province (2022B1515020110 to L.L.), the Natural Science Foundation of Shenzhen (JCYJ20210324120212033 to G.C. and JCYJ20180305163311448 to G.C.), SZU Top Ranking Project (86000000210), and the 2115 Talent Development Program of China Agricultural University.
Author contributions: X.-Y.Z., G.C., and L.L. conceived and supervised the project. J.W., C.Z., S.G., S.R., J.Z., L.M., M.C., K.S., M.W., C.L., Y.Z., and F.L. performed the experiments. With the help of A.P.H., X.B., and K.M., L.L., X.S., X.Y., and J.F. performed bioinformatics analysis. X.-Y.Z., G.C., L.L., J.W., S.G., and X.S. wrote the manuscript with the help from all the authors.
Competing interests: X.-Y.Z., J.W., S.R., C.Z., L.M., C.L., F.L., and G.C. are inventors on a patent application related to this work filed by the Southern Medical University (no. CN 202110542860.8, filed: 19 May 2021). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw sequencing datasets generated in the present study are deposited in the GSA (Genome Sequence Archive in BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences), under accession number (CRA004360, https://ngdc.cncb.ac.cn/gsa). The processed datasets generated in the present study are deposited in the OMIX (China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences), under accession number (OMIX355, https://ngdc.cncb.ac.cn/omix).
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S7
REFERENCES AND NOTES
- 1.Willott G. M., Frequency of azoospermia. Forensic Sci. Int. 20, 9–10 (1982). [DOI] [PubMed] [Google Scholar]
- 2.Gianaroli L., Selman H. A., Magli M. C., Colpi G., Fortini D., Ferraretti A. P., Birth of a healthy infant after conception with round spermatids isolated from cryopreserved testicular tissue. Fertil. Steril. 72, 539–541 (1999). [DOI] [PubMed] [Google Scholar]
- 3.Tanaka A., Nagayoshi M., Takemoto Y., Tanaka I., Kusunoki H., Watanabe S., Kuroda K., Takeda S., Ito M., Yanagimachi R., Fourteen babies born after round spermatid injection into human oocytes. Proc. Natl. Acad. Sci. U.S.A. 112, 14629–14634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer W., Niveleau A., Walter J., Fundele R., Haaf T., Demethylation of the zygotic paternal genome. Nature 403, 501–502 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J., Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 10, 475–478 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Santos F., Hendrich B., Reik W., Dean W., Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 241, 172–182 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Okada Y., Yamaguchi K., Epigenetic modifications and reprogramming in paternal pronucleus: Sperm, preimplantation embryo, and beyond. Cell. Mol. Life Sci. 74, 1957–1967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogura A., Matsuda J., Yanagimachi R., Birth of normal young after electrofusion of mouse oocytes with round spermatids. Proc. Natl. Acad. Sci. U.S.A. 91, 7460–7462 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y., Zheng Y., Gao Y., Lin Z., Yang S., Wang T., Wang Q., Xie N., Hua R., Liu M., Sha J., Griswold M. D., Li J., Tang F., Tong M. H., Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 28, 879–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M., Liu X., Chang G., Chen Y., An G., Yan L., Gao S., Xu Y., Cui Y., Dong J., Chen Y., Fan X., Hu Y., Song K., Zhu X., Gao Y., Yao Z., Bian S., Hou Y., Lu J., Wang R., Fan Y., Lian Y., Tang W., Wang Y., Liu J., Zhao L., Wang L., Liu Z., Yuan R., Shi Y., Hu B., Ren X., Tang F., Zhao X. Y., Qiao J., Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell 23, 599–614.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W., Li W., Li Y., Ma J., Peng X., Zheng H., Ming J., Zhang W., Zhang J., Tian G., Xu F., Chang Z., Na J., Yang X., Xie W., The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534, 652–657 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Liu X., Wang C., Liu W., Li J., Li C., Kou X., Chen J., Zhao Y., Gao H., Wang H., Zhang Y., Gao Y., Gao S., Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537, 558–562 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Zhu P., Guo H., Ren Y., Hou Y., Dong J., Li R., Lian Y., Fan X., Hu B., Gao Y., Wang X., Wei Y., Liu P., Yan J., Ren X., Yuan P., Yuan Y., Yan Z., Wen L., Yan L., Qiao J., Tang F., Single-cell DNA methylome sequencing of human preimplantation embryos. Nat. Genet. 50, 12–19 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Guo F., Li L., Li J., Wu X., Hu B., Zhu P., Wen L., Tang F., Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res. 27, 967–988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu F., Liu Y., Inoue A., Suzuki T., Zhao K., Zhang Y., Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 165, 1375–1388 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L., Guo F., Gao Y., Ren Y., Yuan P., Yan L., Li R., Lian Y., Li J., Hu B., Gao J., Wen L., Tang F., Qiao J., Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 20, 847–858 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Yuan P., Yan Z., Yang M., Huo Y., Nie Y., Zhu X., Qiao J., Yan L., Single-cell multiomics sequencing reveals the functional regulatory landscape of early embryos. Nat. Commun. 12, 1247 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurotaki Y. K., Hatanaka Y., Kamimura S., Oikawa M., Inoue H., Ogonuki N., Inoue K., Ogura A., Impaired active DNA demethylation in zygotes generated by round spermatid injection. Hum. Reprod. 30, 1178–1187 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Kishigami S., Van Thuan N., Hikichi T., Ohta H., Wakayama S., Mizutani E., Wakayama T., Epigenetic abnormalities of the mouse paternal zygotic genome associated with microinsemination of round spermatids. Dev. Biol. 289, 195–205 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Yamagata K., Suetsugu R., Wakayama T., Assessment of chromosomal integrity using a novel live-cell imaging technique in mouse embryos produced by intracytoplasmic sperm injection. Hum. Reprod. 24, 2490–2499 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Morel D., Jeffery D., Aspeslagh S., Almouzni G., Postel-Vinay S., Combining epigenetic drugs with other therapies for solid tumours - past lessons and future promise. Nat. Rev. Clin. Oncol. 17, 91–107 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Clark S. J., Argelaguet R., Kapourani C. A., Stubbs T. M., Lee H. J., Alda-Catalinas C., Krueger F., Sanguinetti G., Kelsey G., Marioni J. C., Stegle O., Reik W., scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argelaguet R., Clark S. J., Mohammed H., Stapel L. C., Krueger C., Kapourani C. A., Imaz-Rosshandler I., Lohoff T., Xiang Y., Hanna C. W., Smallwood S., Ibarra-Soria X., Buettner F., Sanguinetti G., Xie W., Krueger F., Gottgens B., Rugg-Gunn P. J., Kelsey G., Dean W., Nichols J., Stegle O., Marioni J. C., Reik W., Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576, 487–491 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kimura Y., Yanagimachi R., Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development 121, 2397–2405 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Ohta H., Sakaide Y., Wakayama T., Functional analysis of male mouse haploid germ cells of various differentiation stages: Early and late round spermatids are functionally equivalent in producing progeny. Biol. Reprod. 80, 511–517 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Zhu H., Sun H., Yu D., Li T., Hai T., Liu C., Zhang Y., Chen Y., Dai X., Li Z., Li W., Liu R., Feng G., Zhou Q., Transcriptome and DNA methylation profiles of mouse fetus and placenta generated by round spermatid injection. Front. Cell Dev. Biol. 9, 632183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C., Chen C., Liu X., Li C., Wu Q., Chen X., Yang L., Kou X., Zhao Y., Wang H., Gao Y., Zhang Y., Gao S., Dynamic nucleosome organization after fertilization reveals regulatory factors for mouse zygotic genome activation. Cell Res. , (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park S. J., Shirahige K., Ohsugi M., Nakai K., DBTMEE: A database of transcriptome in mouse early embryos. Nucleic Acids Res. 43, D771–D776 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B., Xu Q., Wang Q., Feng S., Lai F., Wang P., Zheng F., Xiang Y., Wu J., Nie J., Qiu C., Xia W., Li L., Yu G., Lin Z., Xu K., Xiong Z., Kong F., Liu L., Huang C., Yu Y., Na J., Xie W., The landscape of RNA Pol II binding reveals a stepwise transition during ZGA. Nature 587, 139–144 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Abe K. I., Funaya S., Tsukioka D., Kawamura M., Suzuki Y., Suzuki M. G., Schultz R. M., Aoki F., Minor zygotic gene activation is essential for mouse preimplantation development. Proc. Natl. Acad. Sci. U.S.A. 115, E6780–E6788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K., Wu D. Y., Zheng H., Wang Y., Sun Q. R., Liu X., Wang L. Y., Xiong W. J., Wang Q., Rhodes J. D. P., Xu K., Li L., Lin Z., Yu G., Xia W., Huang B., Du Z., Yao Y., Nasmyth K. A., Klose R. J., Miao Y. L., Xie W., Analysis of genome architecture during SCNT reveals a role of cohesin in impeding minor ZGA. Mol. Cell 79, 234–250.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Poleshko A., Smith C. L., Nguyen S. C., Sivaramakrishnan P., Wong K. G., Murray J. I., Lakadamyali M., Joyce E. F., Jain R., Epstein J. A., H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis. eLife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang Q., Ang J. Y. J., Lee A. Y., Cao Q., Li K. Y., Yip K. Y., Leung D. C. Y., G9a plays distinct roles in maintaining DNA methylation, retrotransposon silencing, and chromatin looping. Cell Rep. 33, 108315 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Gu T. P., Guo F., Yang H., Wu H. P., Xu G. F., Liu W., Xie Z. G., Shi L., He X., Jin S. G., Iqbal K., Shi Y. G., Deng Z., Szabo P. E., Pfeifer G. P., Li J., Xu G. L., The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Okano M., Bell D. W., Haber D. A., Li E., DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Zhang Z., Chen J., Liu W., Lai W., Liu B., Li X., Liu L., Xu S., Dong Q., Wang M., Duan X., Tan J., Zheng Y., Zhang P., Fan G., Wong J., Xu G. L., Wang Z., Wang H., Gao S., Zhu B., Stella safeguards the oocyte methylome by preventing de novo methylation mediated by DNMT1. Nature 564, 136–140 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Guo H., Zhu P., Wu X., Li X., Wen L., Tang F., Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 23, 2126–2135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Konstantinidis D. G., Pushkaran S., Johnson J. F., Cancelas J. A., Manganaris S., Harris C. E., Williams D. A., Zheng Y., Kalfa T. A., Signaling and cytoskeletal requirements in erythroblast enucleation. Blood 119, 6118–6127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pappano W. N., Guo J., He Y., Ferguson D., Jagadeeswaran S., Osterling D. J., Gao W., Spence J. K., Pliushchev M., Sweis R. F., Buchanan F. G., Michaelides M. R., Shoemaker A. R., Tse C., Chiang G. G., The histone methyltransferase inhibitor A-366 uncovers a role for G9a/GLP in the epigenetics of leukemia. PLOS ONE 10, e0131716 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeung W. K. A., Brind’Amour J., Hatano Y., Yamagata K., Feil R., Lorincz M. C., Tachibana M., Shinkai Y., Sasaki H., Histone H3K9 methyltransferase G9a in oocytes Is essential for preimplantation development but dispensable for CG methylation protection. Cell Rep. 27, 282–293.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Novakovic B., Lewis S., Halliday J., Kennedy J., Burgner D. P., Czajko A., Kim B., Sexton-Oates A., Juonala M., Hammarberg K., Amor D. J., Doyle L. W., Ranganathan S., Welsh L., Cheung M., McBain J., McLachlan R., Saffery R., Assisted reproductive technologies are associated with limited epigenetic variation at birth that largely resolves by adulthood. Nat. Commun. 10, 3922 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang M., Rito T., Metzger J., Naftaly J., Soman R., Hu J., Albertini D. F., Barad D. H., Brivanlou A. H., Gleicher N., Depletion of aneuploid cells in human embryos and gastruloids. Nat. Cell Biol. 23, 314–321 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Zamani Esteki M., Viltrop T., Tsuiko O., Tiirats A., Koel M., Noukas M., Zilina O., Teearu K., Marjonen H., Kahila H., Meekels J., Soderstrom-Anttila V., Suikkari A. M., Tiitinen A., Magi R., Koks S., Kaminen-Ahola N., Kurg A., Voet T., Vermeesch J. R., Salumets A., In vitro fertilization does not increase the incidence of de novo copy number alterations in fetal and placental lineages. Nat. Med. 25, 1699–1705 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Matoba S., Liu Y., Lu F., Iwabuchi K. A., Shen L., Inoue A., Zhang Y., Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 159, 884–895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao R., Wang C., Gao Y., Xiu W., Chen J., Kou X., Zhao Y., Liao Y., Bai D., Qiao Z., Yang L., Wang M., Zang R., Liu X., Jia Y., Li Y., Zhang Y., Yin J., Wang H., Wan X., Liu W., Zhang Y., Gao S., Inhibition of Aberrant DNA re-methylation improves post-implantation development of somatic cell nuclear transfer embryos. Cell Stem Cell 23, 426–435.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Matoba S., Wang H., Jiang L., Lu F., Iwabuchi K. A., Wu X., Inoue K., Yang L., Press W., Lee J. T., Ogura A., Shen L., Zhang Y., Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23, 343–354.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J., Liu H., Liu J., Qi J., Wei B., Yang J., Liang H., Chen Y., Chen J., Wu Y., Guo L., Zhu J., Zhao X., Peng T., Zhang Y., Chen S., Li X., Li D., Wang T., Pei D., H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 45, 34–42 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Aoki F., Worrad D. M., Schultz R. M., Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 181, 296–307 (1997). [DOI] [PubMed] [Google Scholar]
- 49.N. T. Minh, N. B. Tu, N. T. T. Tram, H.-T. Bui, N. Van Thuan, paper presented at the 6th International Conference on the Development of Biomedical Engineering, Vietnam, 24 September 2017. [Google Scholar]
- 50.Xiong H., Luo Y., Wang Q., Yu X., He A., Single-cell joint detection of chromatin occupancy and transcriptome enables higher-dimensional epigenomic reconstructions. Nat. Methods 18, 652–660 (2021). [DOI] [PubMed] [Google Scholar]
- 51.Eckersley-Maslin M. A., Alda-Catalinas C., Reik W., Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell Biol. 19, 436–450 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Chen Z., Djekidel M. N., Zhang Y., Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat. Genet. 53, 551–563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mei H., Kozuka C., Hayashi R., Kumon M., Koseki H., Inoue A., H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat. Genet. 53, 539–550 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Liu W., Yin J., Kou X., Jiang Y., Gao H., Zhao Y., Huang B., He W., Wang H., Han Z., Gao S., Asymmetric reprogramming capacity of parental pronuclei in mouse zygotes. Cell Rep. 6, 1008–1016 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Yoshida N., Perry A. C., Piezo-actuated mouse intracytoplasmic sperm injection (ICSI). Nat. Protoc. 2, 296–304 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Conine C. C., Sun F., Song L., Rivera-Perez J. A., Rando O. J., Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell 46, 470–480.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adenot P. G., Mercier Y., Renard J. P., Thompson E. M., Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 124, 4615–4625 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Yan L., Yang M., Guo H., Yang L., Wu J., Li R., Liu P., Lian Y., Zheng X., Yan J., Huang J., Li M., Wu X., Wen L., Lao K., Li R., Qiao J., Tang F., Single-cell RNA-seq profiling of human preimplantation embryos and embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Scheffler K., Uraji J., Jentoft I., Cavazza T., Monnich E., Mogessie B., Schuh M., Two mechanisms drive pronuclear migration in mouse zygotes. Nat. Commun. 12, 841 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugawara Y., Kamioka H., Honjo T., Tezuka K., Takano-Yamamoto T., Three-dimensional reconstruction of chick calvarial osteocytes and their cell processes using confocal microscopy. Bone 36, 877–883 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X., Zhu X., Li R., Yan J., Wei Y., Zhao Y., Wang W., Ren Y., Yuan P., Yan Z., Hu B., Guo F., Wen L., Tang F., Qiao J., Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell 20, 858–873.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S., Pyl P. T., Huber W., HTSeq--A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W. M. III, Hao Y., Stoeckius M., Smibert P., Satija R., Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langfelder P., Horvath S., WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ha G., Roth A., Lai D., Bashashati A., Ding J., Goya R., Giuliany R., Rosner J., Oloumi A., Shumansky K., Chin S. F., Turashvili G., Hirst M., Caldas C., Marra M. A., Aparicio S., Shah S. P., Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 22, 1995–2007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen C., Yu W., Alikarami F., Qiu Q., Chen C. H., Flournoy J., Gao P., Uzun Y., Fang L., Davenport J. W., Hu Y., Zhu Q., Wang K., Libbrecht C., Felmeister A., Rozich I., Ding Y. Y., Hunger S. P., Felix C. A., Wu H., Brown P. A., Guest E. M., Barrett D. M., Bernt K. M., Tan K., Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood 139, 2198–2211 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stacklies W., Redestig H., Scholz M., Walther D., Selbig J., pcaMethods—A bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–1167 (2007). [DOI] [PubMed] [Google Scholar]
- 70.Gkountela S., Castro-Giner F., Szczerba B. M., Vetter M., Landin J., Scherrer R., Krol I., Scheidmann M. C., Beisel C., Stirnimann C. U., Kurzeder C., Heinzelmann-Schwarz V., Rochlitz C., Weber W. P., Aceto N., Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176, 98–112.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Tables S1 to S7