Abstract
Purpose:
Tyrosine kinase inhibitors (TKI) have poor efficacy in patients with glioblastoma (GBM). Here, we studied whether this is predominantly due to restricted blood–brain barrier penetration or more to biological characteristics of GBM.
Patients and Methods:
Tumor drug concentrations of the TKI sunitinib after 2 weeks of preoperative treatment was determined in 5 patients with GBM and compared with its in vitro inhibitory concentration (IC50) in GBM cell lines. In addition, phosphotyrosine (pTyr)-directed mass spectrometry (MS)-based proteomics was performed to evaluate sunitinib-treated versus control GBM tumors.
Results:
The median tumor sunitinib concentration of 1.9 μmol/L (range 1.0–3.4) was 10-fold higher than in concurrent plasma, but three times lower than sunitinib IC50s in GBM cell lines (median 5.4 μmol/L, 3.0–8.5; P = 0.01). pTyr-phosphoproteomic profiles of tumor samples from 4 sunitinib-treated versus 7 control patients revealed 108 significantly up- and 23 downregulated (P < 0.05) phosphopeptides for sunitinib treatment, resulting in an EGFR-centered signaling network. Outlier analysis of kinase activities as a potential strategy to identify drug targets in individual tumors identified nine kinases, including MAPK10 and INSR/IGF1R.
Conclusions:
Achieved tumor sunitinib concentrations in patients with GBM are higher than in plasma, but lower than reported for other tumor types and insufficient to significantly inhibit tumor cell growth in vitro. Therefore, alternative TKI dosing to increase intratumoral sunitinib concentrations might improve clinical benefit for patients with GBM. In parallel, a complex profile of kinase activity in GBM was found, supporting the potential of (phospho)proteomic analysis for the identification of targets for (combination) treatment.
Translational Relevance.
In GBM, restricted delivery of tyrosine kinase inhibitors (TKI) through the blood–brain barrier (BBB) to the target sites has been proposed as a pivotal reason for their limited activity in this disease thus far. We found that treatment of patients with GBM with the TKI sunitinib at the standard dose of 50 mg every day resulted in an insufficient drug tumor concentration to exert antitumor activity, indicating that the BBB indeed plays a clinically significant role in drug resistance. Alternative, high TKI dosing strategies may be needed to increase exposure and thereby improve efficacy in these patients. In addition, the feasibility of phosphoproteomic profiling of tumors from treated and control patients may lead to target discovery for (combination) treatment of patients with GBM in future studies.
Introduction
Despite currently available therapies, the prognosis and outcome of patients with glioblastoma (GBM) are poor. Clinical trials evaluating tyrosine kinase inhibitors (TKI) for recurrent GBM, including TKIs with a broad kinase inhibition profile such as sunitinib and vandetanib, resulted in minimal improvement in progression-free survival and no overall survival benefit, despite their significant clinical benefit in other malignancies (1–4). TKIs inhibit intracellular signaling, that is, tyrosine kinases, that exert a variety of biological activities in GBM, including cell proliferation and migration (5, 6). Whether their disappointing efficacy in GBM is due to restricted drug delivery of TKIs or a result of differential biological characteristics compared with other malignancies is unknown, but local TKI concentration reached upon treatment in the tumor may be hampered by the blood–brain barrier (BBB; ref. 7). While TKIs inhibit their target kinases at low concentrations, multiple other kinases that are involved in (GBM) tumor growth may be inhibited at higher concentrations (8, 9).
In addition, it is anticipated that the biological activity of specific kinases driving the growth of GBM in an individual patient is one of the factors that will determine response to treatment with TKIs (5). Analyzing tyrosine-phosphorylated proteins by mass spectrometry (MS)-based tyrosine-phosphoproteomics can provide insight in the effect of TKIs on cellular signaling (10–13). Initial promise has been generated on the identification of ALK-, ROS-, and PDGFRα-mediated non—small cell lung cancer (NSCLC) subtypes by this high-throughput method (14).
Based on these considerations, we hypothesized that in an individual patient both the achievable local GBM TKI concentrations and the specific activity of GBM-related cellular kinases, driving its malignant behavior, will influence potential treatment benefit.
To address aforementioned issues regarding (in)efficacy of TKI treatment in GBM, seeking to gain more insight in their activity at the tumor level in patients, we performed a pilot study of preoperative treatment with sunitinib in newly-diagnosed patients. Sunitinib tumor concentrations and its in vitro efficacy in GBM-cell lines were determined to study to what extent clinically achieved tumor concentrations are sufficient for inhibition of tumor cell proliferation. In addition, the effects of sunitinib treatment on tyrosine kinase signaling activities were analyzed.
Patients and Methods
Study design
This interventional pilot study was performed in Amsterdam UMC, location VUmc, Cancer Center Amsterdam (Amsterdam, the Netherlands) between January 2015 and October 2016, with preoperative, off-label use of the TKI sunitinib.The primary objective was to determine sunitinib tumor and cerebrospinal fluid (CSF) concentrations in patients with newly-diagnosed GBM, who were treated for 10 to 14 days prior to surgical resection (Supplementary Fig. S1). Based on previous studies reporting tumor concentrations, we aimed to include 5 patients to obtain reliable estimates of sunitinib tumor concentrations (15, 16). Tumor and CSF concentrations were compared with plasma concentrations in these patients and the in vitro sensitivity of GBM tumor cell lines. Secondary objectives were to determine the feasibility of tyrosine phosphoproteomic profiling in these tumors and to evaluate the effect of sunitinib treatment on tyrosine kinase signaling activity by MS-based tyrosine phosphoproteomics. Phosphoproteomic profiles of sunitinib-treated patients were compared with those obtained in resection specimens of a control group of patients with newly-diagnosed GBM without preoperative treatment.
The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines and approved by the Institutional Review Board (medical ethics committee) of Amsterdam UMC, location VUmc. All treated patients provided written study-specific informed consent. The patients who participated as a control group for the secondary objectives signed a general informed consent form from the Department of Neurosurgery of Amsterdam UMC, location VUmc, for use of their resected tissue for experimental purposes. This trial is registered within the Netherlands Trial Register (www.trialregister.nl, NL4609) and via www.ClinicalTrials.gov, identifier NCT02239952.
Patients
Eligible patients for preoperative treatment were adults (>18 years) with a newly-diagnosed brain tumor on initial MRI, suggesting resectable GBM according to interpretation of an expert neuro-radiologist. Main inclusion and exclusion criteria are described in Supplementary Methods.
Study treatment and pharmacokinetic sampling
Patients awaiting surgery received 2 weeks of treatment with the oral TKI sunitinib (50 mg once daily) prior to the initial resection of the tumor (Supplementary Fig. S1). The last dose of sunitinib was administered in the morning on the day of surgery. Venous blood sampling, for pharmacokinetic measurements of sunitinib (trough levels), was performed on day 8, the day before surgery, and approximately 5 days after surgery. Tumor tissue samples (approximately 1 cm3) and at least 30 μL of CSF were obtained during the tumor resection.
Tumor tissue preprocessing
Immediately after tumor resection, tumor tissue was placed in an aluminium tissue container, snap-frozen in liquid nitrogen, and stored at −80°C until further processing. Next, multiple 10-μm cryosections of frozen tissue were made for the lysis procedure as well as for hematoxylin and eosin (H&E) staining to determine tumor cell content. In general, approximately 50% of the tumor tissue was used for determination of tumor drug concentrations. From the remaining tissue, cryosections were made for MS-based phosphoproteomics as described before (17) and further detailed in Supplementary Methods. Minimal protein input was set at 5.0 mg per tumor.
Drug concentration measurements of sunitinib and its active metabolite
Concentrations of sunitinib in tumor tissue, CSF, and plasma were determined using LC-MS/MS as described previously (16), with further details in Supplementary Methods.
In vitro evaluation of sunitinib activity
Four GBM cell lines, U87, U251, T98G, and U138, were used to determine in vitro cell proliferation and sunitinib IC50 concentrations. The HT29 colorectal cell line was included as a positive control for comparison with prior experiments and response to sunitinib was assessed by MTT assay as described previously (details in Supplementary Methods; refs. 18, 19).
MS-based tyrosine phosphoproteomics
Peptides were separated by an Ultimate 3000 nanoLC-MS/MS system (Dionex LC-Packings) coupled online to a Q HF Exactive mass spectrometer (Thermo Fisher Scientific) as previously described (20, 21). MS/MS spectra were searched against the Uniprot human reference proteome FASTA file (release March 2017, 42161 entries, no fragments) using MaxQuant1.5.4.1 (22). Downstream data analysis and identification of phosphopeptides, phosphosites, and proteins is further described in Supplementary Methods.
Analysis of kinase activity and enrichment of posttranslational modification signatures
Ranking of the top 20 activated kinases in samples from control and sunitinib-treated patients and visualization of kinase-substrate networks of individual samples was performed using Integrative Inferred Kinase Activity (INKA) analysis as described before (12). Differentially activated kinases were identified and significance was determined with a Mann–Whitney U test. Identification of “outlier kinases” in individual samples based on INKA scores was done by application of Tukey rule, i.e., values differ from the median more than 1.5 times the interquartile range, and further filtering for an INKA score >10.
Posttranslational modifications (PTM) signature enrichment analysis (PTM-SEA) was performed as described elsewhere (ref. 23; for details, see Supplementary Methods).
Data availability statement
The MS proteomics data are available and were provided to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019038 (24).
Results
Patient accrual and characteristics
Six patients with newly diagnosed GBM signed informed consent and started preoperative sunitinib treatment, of whom 5 were evaluable for the primary analysis. One patient was replaced upon withdrawal of informed consent after 1 week of treatment. Median duration of preoperative sunitinib treatment was 13 days (range 11–18; Table 1). Study treatment was well tolerated and no grade 3 to 4 adverse events were observed. Tumor resection was performed without perioperative complications in all patients. GBM diagnosis was histopathologically confirmed for all patients. Nine patients with GBM undergoing surgical resection were included as a control group (Supplementary Table S1).
Table 1.
Sunitinib concentrations in tumor tissue, CSF, and plasma.
| Patient ID | Treatment duration (days) | Tumor weight (mg) | Tumor tissue (μmol/L) | CSF (μmol/L) | Plasma (μmol/L) |
|---|---|---|---|---|---|
| SUN-01 | 14 | 16.7 | 3.0 | 0.013 | 0.105 |
| SUN-02 | 11 | 7.9 | 1.5 | 0.014 | 0.111 |
| SUN-03 | 18 | 16.0 | 3.4 | 0.015 | 0.194 |
| SUN-04 A | 13 | 24.8 | 1.9 | 0.023 | 0.479 |
| SUN-04 B | 12.5 | 1.0 | |||
| SUN-04 C | 18.6 | 2.2 | |||
| SUN-05 | 12 | 17.0 | 1.4 | NA | 0.255 |
| Median | 13 | 16.7 | 1.9 | 0.015 | 0.194 |
Note: Sunitinib concentrations in micromoles per liter in tumor tissue, CSF, and plasma in 5 patients with GBM after a median of 13 days of preoperative study treatment. SUN-04 A, B, and C are different parts (biological replicates) of the same tumor.
Sunitinib concentrations in GBM resection specimens, CSF, and plasma
Sunitinib concentrations in tumor tissue and plasma were determined in all 5 patients. CSF could be collected during surgery in 4 of these patients. Median sunitinib tumor concentration was 1.9 μmol/L (range 1.0–3.4), 10-fold higher than the median sunitinib plasma concentration on the day of surgery (0.194 μmol/L, range 0.105–0.479). In CSF the median concentration was 0.015 μmol/L (range 0.013–0.023; Table 1). No relation between sunitinib tumor accumulation and use of dexamethasone and anticonvulsants was found (Supplementary Table S1).
Sunitinib sensitivity of GBM cell lines
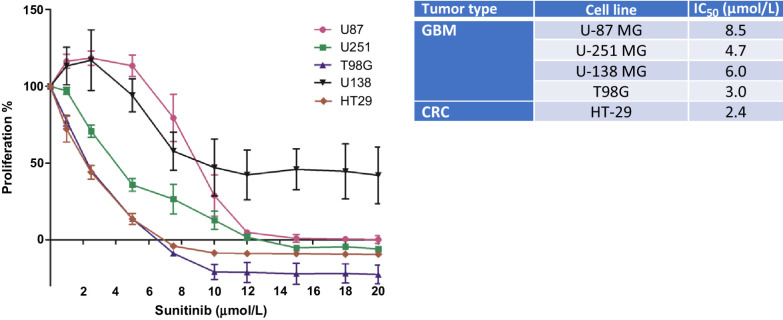
Sensitivity of GBM cell lines U87, U251, T98G, and U138, and the colorectal cell line HT29 as a positive control was tested for a concentration range from 0 to 20 μmol/L of sunitinib incubated for 72 hours. Exposure to sunitinib resulted in a heterogeneous response in the GBM cell lines, with a median IC50 value of 5.4 μmol/L (range 3.0–8.5 μmol/L), shown in Fig. 1. The IC50 concentrations for the GBM cell lines were significantly higher (2.8-fold) than the detected intratumoral sunitinib concentrations in patients as described above (P = 0.01). As a positive control, the colorectal cancer cell line was most sensitive with an IC50 similar to prior experiments and other cell lines previously tested (25).
Figure 1.
Sunitinib effect in vitro. Proliferation (MTT) assay of GBM and colorectal cancer cell lines incubated with increasing sunitinib concentrations, showing the percentage of proliferation compared with untreated controls (left) and calculated IC50 in micromoles per liter (right). Sunitinib inhibited proliferation of GBM tumor cells in vitro at concentrations 2.8 times higher than achieved intratumorally in patients (Table 1). GBM, glioblastoma; CRC, colorectal cancer.
Tyrosine phosphoproteomics of GBM resection specimens
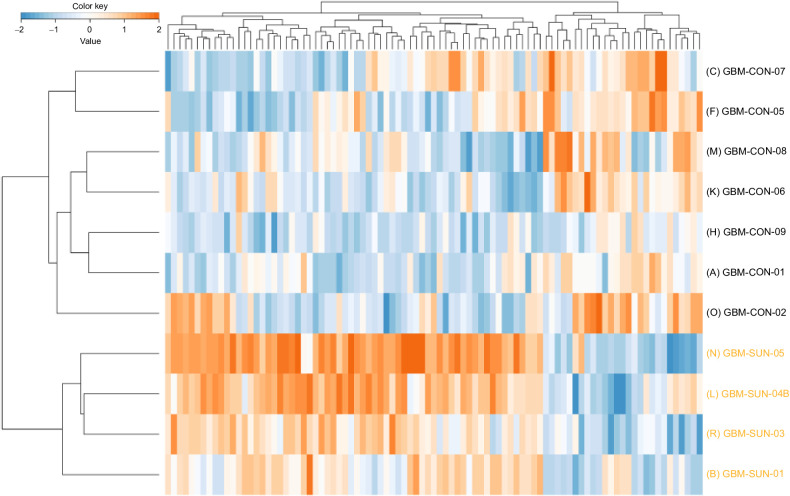
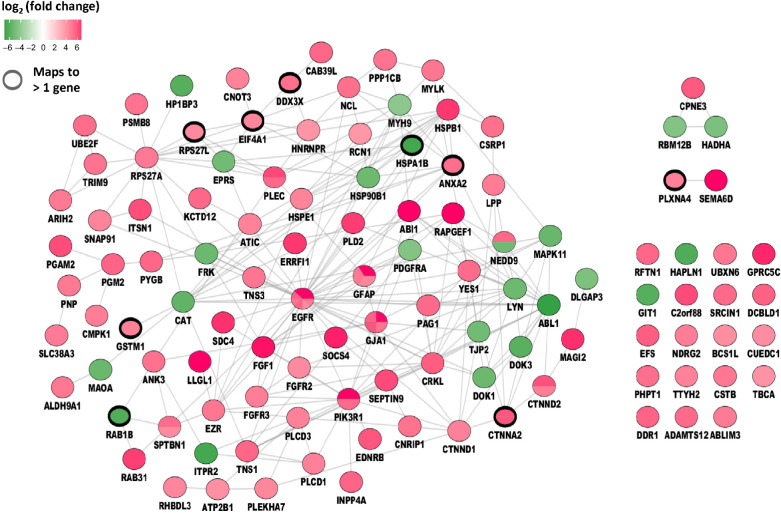
Phosphotyrosine (pTyr)-phosphoproteomics was successfully performed in 12 of 15 tumor samples, of which 11 were suitable for comparative analyses between sunitinib-pretreated and control samples. Three samples (2 control and 1 sunitinib-treated patient) were excluded for downstream (comparative) analysis because of much lower protein input and significantly less phosphopeptide identifications (P = 0.03; Supplementary Table S1A). One patient (SUN-wk1) who withdrew consent after 1 week of sunitinib treatment was analyzed for potential drug targets only. For the 12 tumor samples with 5-mg protein input 1,557 ± 329 (SD) phoshopeptides per sample were identified out of 6,767 ± 837 of the total number of peptides per sample (23% phosphopeptide enrichment). A median of 1,411 phosphosites were identified per sample with 91% phosphorylation on tyrosine residues, indicating succesful pTyr enrichment (Supplementary Table S1B). All identified and quantified phosphopeptides and sites can be found in Supplementary Tables S2 and S3. Unsupervised hierarchical clustering of identified phosphopeptides did not differentiate between tumor samples from treated versus untreated samples (Supplementary Fig. S2). Supervised clustering of the differentially detected tyrosine-phosphopeptides (cut-off P < 0.05 without additional filtering) revealed a clear separation between sunitinib-treated and control tissues (Fig. 2). Between the four sunitinib-treated and seven control tissues, 108 phosphopeptides were up- and 23 significantly downregulated (cut-off P < 0.05). Figure 3 shows a protein interaction network of these differentially regulated phosphopeptides (listed in Supplementary Table S4) mapped to proteins, showing an EGFR-centered signaling network with upregulation of multiple other kinases.
Figure 2.
Supervised clustering analysis of the tyrosine phosphoproteome. Supervised hierarchical clustering of differential phosphopeptides (P < 0.05) identified by tyrosine-phosphoproteomics shows separation of sunitinib-treated and control tumor tissues from 11 patients with glioblastoma. The heatmap shows the relative phosphopeptide intensities (z-score) in these samples based on log10-transformed values (orange, high abundance; blue, low abundance). SUN, sunitinib-treated; CON, control.
Figure 3.
Protein interaction network. Protein interaction network of differential phosphopeptides (P < 0.05) between sunitinib-treated and control samples. Regulated phosphopeptides are mapped to proteins and visualized as protein interaction network. Green, Down- and Red, Upregulated in sunitinib-treated patients. Colored subdivisions indicate identification of multiple upregulated phosphopeptides mapping to the same protein. Note EGFR was identified with three upregulated phosphopeptides in the sunitinib-treated group.
Analysis of active kinases and biological pathways and processes
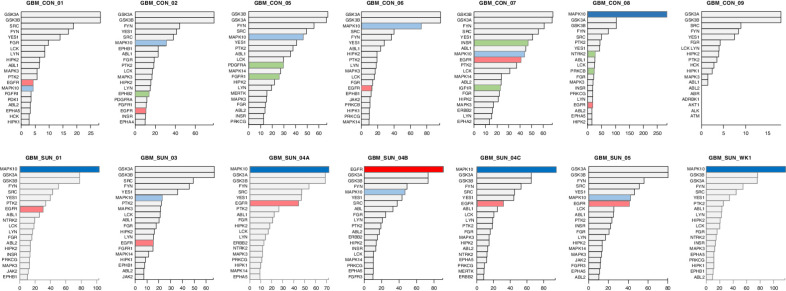
In general the top 20 active kinases that were identified by INKA analysis of individual GBM samples were mostly comparable between different GBM samples, including GSK3A and 3B, MAPK10, YES, FYN, and SRC (Fig. 4). In three treated and one untreated tumor sample MAPK10 and in one treated tumor sample EGFR stood out as the highest active kinase. For this tumor sample (SUN 04-B), an EGFR mutation was detected [EGFR (c.865G>A; p.A289T)] and potential EGFR amplification (Fig. 4). Comparative analysis of the kinase activities identified in both groups showed differential and upregulated activity of FGFR3 (P = 0.036) and EGFR (P = 0.047) in sunitinib-treated samples (Supplementary Fig. S3A). Activity of the known sunitinib targets KDR (VEGFR2), PDGFRA, and PDGFRB was low and not significantly different between the groups. Outlier analysis of kinase activity scores, as a potential strategy to identify drug targets in individual tumors, revealed nine outlier kinases in four tumors. These were MAPK10 (INKA score > 250 in CON-08 vs. median score < 50 for other 10 samples), PRKCB, NTRK2 and NTRK3, INSR and IGF1R, PDGFRA and FGFR1, and EPHB2 (Supplementary Fig. S3B).
Figure 4.
Ranking of top 20 active kinases in tumors from 11 patients with glioblastoma. Ranked kinase activities in 7 control and 4 sunitinib-treated tumors. For each tumor, bar graphs depict kinase ranking based on combined INKA scores of kinase- and substrate-centric analysis of tyrosine-phoshoproteomics (12). For patient SUN-04, three biological replicates have been analyzed. Top bars of potential hyperactive kinases are highlighted by dark coloring (MAPK10 blue, EGFR red). Note the very high INKA score for MAPK10 in patient CON-08. Light blue and red colored bars indicate lower ranked MAPK10 and EGFR kinase activities, respectively. Green colors highlight kinases with “outlier activity”, i.e., higher activity in one versus the remaining 10 tumors (see Supplementary Fig. S3). SUN, sunitinib-treated; CON, control.
Based on PTM-SEA analysis involving 558 PTM signatures, three strongly significantly enriched phosphosite signatures were identified in sunitinib-treated compared to control samples (23). Two of these were based on Phosphosite-Plus (PSP) EGFR and SRC kinase activity signatures and one was a PSP-derived EGF perturbation signature. Two moderately significant NetPath derived signatures corresponding to EGFR and Notch signaling were also found (Supplementary Fig. S4).
Discussion
Failure to achieve therapeutic TKI drug levels in brain tumors may be an underlying reason for their relatively poor efficacy. We found that treatment of patients with GBM with the TKI sunitinib at the standard dose of 50 mg every day resulted in a significantly lower median tumor concentration (1.9 μmol/L) than needed for antitumor activity in vitro as determined by proliferation assays with four GBM cell lines and also lower compared with previously reported drug concentrations in other tumors (Supplementary Table S5; refs. 16, 19). These findings are in accordance with the publicly available data on sunitinib sensitivity of the Sanger Institute (Hinxton, England). In their experimental design of studying cancer cell line sensitivity to sunitinib, it was found that the geometric mean of the IC50 for sunitinib was 11.6 μmol/L for 398 cancer cell lines, while for the 11 GBM cell lines a higher geometric mean of the IC50 of 18.7 μmol/L was detected (https://www.cancerrxgene.org/compound/Sunitinib/5/overview/ic50), supporting our finding that GBM cells are less sensitive to sunitinib than other tumor cells in general.
The 10-fold higher sunitinib concentrations in GBM tumor tissue compared with plasma in these patients are a result of cellular accumulation of sunitinib due to its physico-chemical characteristics (19, 26). The lower sunitinib concentration in CSF compared with tumor tissue and plasma is most likely due to a significantly lower protein concentration in CSF, which we did not measure and should therefore be interpreted with caution. Protein binding of drugs including sunitinib affects the free drug concentration and therefore the free drug concentration is assumed to be a more accurate measure of the potentially available active drug. However, one should realize that it is the intratumoral intracellular unbound drug that needs to interact with the target for its efficacy. Unfortunately, it is yet not possible to quantify intracellular free drug concentrations without interfering with intracellular drug distribution. Our data on drug distribution in patients with GBM provide additional insight into the currently scarcely available data, while being aware of the fact that the translation from preclinical in vitro and clinical plasma concentrations to intratumor concentrations should be taken with caution (27).
While it is known that the BBB is being disrupted by GBM tumor growth, it presumably still forms a barrier for adequate drug delivery. Brain tumor capillaries are composed of continuous and nonfenestrated capillaries and have high expression of drug efflux transporters, resulting in a hurdle for optimal drug delivery (28). Several TKIs have been shown to be both substrates as well as inhibitors (at higher concentrations) of these drug efflux transporters (29). A clear difference between drug availability of TKIs was shown between flank and orthotopic GBM models explaining the difference in response to treatment for several targeted agents (30–32). The GBM sunitinib concentrations we found here are nearly five times lower than the recently reported tumor concentrations in tumor biopsies of patients with other solid malignancies (9.0–9.5 μmol/L) indicative for hampered tissue penetration through the BBB (Supplementary Table S5; refs. 16, 19). Limited data are available on GBM TKI concentrations in patients, but Plotkin and colleagues reported comparable sunitinib concentrations (converted from ng/g to mmol/L) of 0.45 μmol/L (range 0.14–2.28) in tumor tissue and 0.15 μmol/L (range 0.0825–0.24) in the paired blood samples from these patients with GBM (33). The lack of clinical activity of sunitinib in GBM was shown by Neijns and colleagues in which sunitinib-treated patients with a recurrent GBM had no tumor response nor clinical benefit (2). In addition, compared with its concentration in other malignancies, the relatively low sunitinib concentrations in GBM (we found here) provide clinical evidence of prior preclinical findings that the BBB plays a significant role in the resistance of GBM to TKIs (28, 30–32). Taken together, these preclinical and clinical data indicate that GBM is difficult to treat with sunitinib, not only due to BBB-mediated reduced drug penetration but also due to a lower intrinsic sensitivity of GBM (28). Alternative treatment schedules or dosing to reach higher tumor concentrations have not been considered although several clinically approved TKIs have been evaluated for their potential benefit in GBM (34). Based on the above-mentioned findings one may hypothesize that higher dosing of TKIs will lead to increased exposure and thereby may improve antitumor activity, but at the same time such an approach is difficult to implement in the clinic due to increased toxicity (35). Still, some examples of alternative high dosing TKI strategies with manageable toxicity and promising antitumor activity have been reported (36–38). Based on these findings, a phase II/III clinical trial to evaluate this strategy in patients with recurrent GBM (NCT03025893) is ongoing with sunitinib dosing of 700 mg once every 2 weeks. Alternative approaches such as intrathecal TKI injection may also be considered to optimize local exposure, but this has been studied only in a preclinical model with nano-particulated, water-soluble erlotinib (39).
Besides achieved drug concentrations at the target site, the signaling context or network in tumors is essential for TKI efficacy as well (40, 41). In this study, a comparable number of phosphopeptides as were previously reported by Johnson and colleagues from 7 untreated patients with GBM identified including similar phospopeptide-related pathways as Src and Yes (42). Despite the small number of patients, supervised clustering of the GBM tissue tyrosine-phosphoproteome profiles revealed a clear separation between 4 sunitinib-treated versus 7 untreated patients. This was based on a relatively high number of upregulated phosphopeptides in the sunitinib-treated group (108 vs. 23 downregulated phosphopeptides; cut-off P < 0.05), resulting in an EGFR-centered signaling network (Fig. 3). These included phosphopeptides mapped to proteins which may potentially serve as targets for (combination) therapy, such as EGFR and FGFR, but we stress that the small sample size of the 2 groups calls for cautious interpretation of this comparative analysis.
In this small group of patients no relation between tumor concentration and phosphopeptide regulation was found. Several preclinical studies showed differential effects of TKI treatment on orthotopic versus flank implanted tumors and suggested a relation with achieved intratumor drug concentrations (32, 43). Single-cell phosphoproteomics revealed alteration of protein signaling without genetic changes may cause resistance as early as 2.5 days after start of treatment with an mTOR-inhibitor in a patient-derived in vivo GBM model (44).
In the per sample analysis, MAPK10 was identified as the highest ranking kinase in four tumors using INKA analysis (12). This included sample CON-08 with, compared with the other 10 samples, outlier kinase activity of MAPK10 with a very high INKA score suggestive of amplification (Fig. 4). However, no normal DNA was available for validation by whole-exome sequencing. MAPK10, also known as JNK3, regulates several physiologic neuronal functions including apoptosis (45, 46). Moreover, JNK kinases are known to be expressed and activated in the majority of GBMs and their activation is associated with promotion of GBM tumor growth (47, 48). Several selective small-molecule JNK3 inhibitors have been reported (49), but to the best of our knowledge, no studies currently evaluate their activity in GBM. Notable kinase activity of INSR/IGF1R was found in another individual sample. Dual inhibition of these kinases has shown preclinical activity in GBM (50). Taken together, these data show a complex profile of kinase activity in GBM, supporting the potential of (phospho)proteomic analysis for a better understanding of tumorbiology as well as for the identification of targets for (combination) treatment.
In conclusion, we found in patients with GBM clinical evidence for insufficient intratumoral concentrations of sunitinib to exert its antitumor activity, which may partly explain its intrinsic BBB-mediated resistance and may be overcome by high-dosed, alternatively scheduled administration. In parallel, a complex profile of kinase activity in GBM was found, supporting the potential of (phospho)proteomic analysis for the identification of targets for (combination) treatment.
Supplementary Material
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
M. Labots reports personal fees from Bristol-Myers Squibb and MSD outside the submitted work. G.J. Peters reports personal fees from Taiho Pharmaceuticals, Clear Creek Bio, Isofol Medical AB, and Ellipses Pharma and grants and personal fees from Rexahn Pharma outside the submitted work. H.M.W. Verheul reports nonfinancial support from Pfizer outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
M.E. van Linde: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Labots: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–review and editing. C.G. Brahm: Formal analysis, investigation, visualization, methodology, writing–review and editing. K.E. Hovinga: Investigation, writing–review and editing. P.C. De Witt Hamer: Conceptualization, supervision, investigation, visualization, methodology, writing–review and editing. R.J. Honeywell: Formal analysis, investigation, methodology, writing–review and editing. R. de Goeij-de Haas: Formal analysis, investigation, methodology, writing–review and editing. A.A. Henneman: Formal analysis, investigation, visualization, methodology, writing–review and editing. J.C. Knol: Formal analysis, investigation, methodology, writing–review and editing. G.J. Peters: Supervision, methodology, writing–review and editing. H. Dekker: Data curation, software, formal analysis, investigation, visualization, methodology, writing–review and editing. S.R. Piersma: Data curation, software, formal analysis, investigation, methodology, writing–review and editing. T.V. Pham: Data curation, software, validation, investigation, methodology, writing–review and editing. W.P. Vandertop: Conceptualization, resources, supervision, writing–review and editing. C.R. Jiménez: Conceptualization, resources, software, supervision, validation, investigation, visualization, methodology, writing–review and editing. H.M.W. Verheul: Conceptualization, resources, formal analysis, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. De Witt Hamer PC. Small molecule kinase inhibitors in glioblastoma: a systematic review of clinical studies. Neuro Oncol 2010;12:304–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Neyns B, Sadones J, Chaskis C, Dujardin M, Everaert H, Lv S, et al. Phase II study of sunitinib malate in patients with recurrent high-grade glioma. J Neurooncol 2011;103:491–501. [DOI] [PubMed] [Google Scholar]
- 3. Lee EQ, Kaley TJ, Duda DG, Schiff D, Lassman AB, Wong ET, et al. A multicenter, phase II, randomized, noncomparative clinical trial of radiation and temozolomide with or without vandetanib in newly diagnosed glioblastoma patients. Clin Cancer Res 2015;21:3610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol 2014;71:1437–44. [DOI] [PubMed] [Google Scholar]
- 5. Nakada M, Kita D, Watanabe T, Hayashi Y, Hamada J. Mechanism of chemoresistance against tyrosine kinase inhibitors in malignant glioma. Brain Tumor Pathol 2014;31:198–207. [DOI] [PubMed] [Google Scholar]
- 6. Pearson JRD, Regad T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct Target Ther 2017;2:17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fokas E, Steinbach JP, Rodel C. Biology of brain metastases and novel targeted therapies: time to translate the research. Biochim Biophys Acta 2013;1835:61–75. [DOI] [PubMed] [Google Scholar]
- 8. Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig PA, et al. The target landscape of clinical kinase drugs. Science 2017;358:eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rovithi M, Verheul HMW. Pulsatile high-dose treatment with antiangiogenic tyrosine kinase inhibitors improves clinical antitumor activity. Angiogenesis 2017;20:287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cutillas PR. Role of phosphoproteomics in the development of personalized cancer therapies. Proteomics Clin Appl 2015;9:383–95. [DOI] [PubMed] [Google Scholar]
- 11. Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 2010;3:ra3. [DOI] [PubMed] [Google Scholar]
- 12. Beekhof R, van Alphen C, Henneman AA, Knol JC, Pham TV, Rolfs F, et al. INKA, an integrative data analysis pipeline for phosphoproteomic inference of active kinases. Mol Syst Biol 2019;15:e8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jimenez CR, Verheul HM. Mass spectrometry-based proteomics: from cancer biology to protein biomarkers, drug targets, and clinical applications. Am Soc Clin Oncol Educ Book 2014:e504–10. [DOI] [PubMed] [Google Scholar]
- 14. Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190–203. [DOI] [PubMed] [Google Scholar]
- 15. Petty WJ, Dragnev KH, Memoli VA, Ma Y, Desai NB, Biddle A, et al. Epidermal growth factor receptor tyrosine kinase inhibition represses cyclin D1 in aerodigestive tract cancers. Clin Cancer Res 2004;10:7547–54. [DOI] [PubMed] [Google Scholar]
- 16. Labots M, Pham TV, Honeywell RJ, Knol JC, Beekhof R, de Goeij-de Haas R, et al. Kinase inhibitor treatment of patients with advanced cancer results in high tumor drug concentrations and in specific alterations of the tumor phosphoproteome. Cancers 2020;12:330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Labots M, van der Mijn JC, Beekhof R, Piersma SR, de Goeij-de Haas RR, Pham TV, et al. Phosphotyrosine-based-phosphoproteomics scaled-down to biopsy level for analysis of individual tumor biology and treatment selection. J Proteomics 2017;162:99–107. [DOI] [PubMed] [Google Scholar]
- 18. Gotink KJ, Rovithi M, de Haas RR, Honeywell RJ, Dekker H, Poel D, et al. Cross-resistance to clinically used tyrosine kinase inhibitors sunitinib, sorafenib and pazopanib. Cell Oncol 2015;38:119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gotink KJ, Broxterman HJ, Labots M, de Haas RR, Dekker H, Honeywell RJ, et al. Lysosomal sequestration of sunitinib: a novel mechanism of drug resistance. Clin Cancer Res 2011;17:7337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Piersma SR, Knol JC, de Reus I, Labots M, Sampadi BK, Pham TV, et al. Feasibility of label-free phosphoproteomics and application to base-line signaling of colorectal cancer cell lines. J Proteomics 2015;127:247–58. [DOI] [PubMed] [Google Scholar]
- 21. van der Mijn JC, Labots M, Piersma SR, Pham TV, Knol JC, Broxterman HJ, et al. Evaluation of different phospho-tyrosine antibodies for label-free phosphoproteomics. J Proteomics 2015;127:259–63. [DOI] [PubMed] [Google Scholar]
- 22. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- 23. Krug K, Mertins P, Zhang B, Hornbeck P, Raju R, Ahmad R, et al. A curated resource for phosphosite-specific signature analysis. Mol Cell Proteomics 2019;18:576–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vizcaino JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Rios D, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 2014;32:223–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gotink KJ, Broxterman HJ, Honeywell RJ, Dekker H, de Haas RR, Miles KM, et al. Acquired tumor cell resistance to sunitinib causes resistance in a HT-29 human colon cancer xenograft mouse model without affecting sunitinib biodistribution or the tumor microvasculature. Oncoscience 2014;1:844–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Honeywell RJ, Kathmann I, Giovannetti E, Tibaldi C, Smit EF, Rovithi MN, et al. Epithelial transfer of the tyrosine kinase inhibitors erlotinib, gefitinib, afatinib, crizotinib, sorafenib, sunitinib, and dasatinib: implications for clinical resistance. Cancers 2020;12:3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heffron TP. Challenges of developing small-molecule kinase inhibitors for brain tumors and the need for emphasis on free drug levels. Neuro Oncol 2018;20:307–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Tellingen O, Yetkin-Arik B, de Gooijer MC, Wesseling P, Wurdinger T, de Vries HE. Overcoming the blood-brain tumor barrier for effective glioblastoma treatment. Drug Resist Updat 2015;19:1–12. [DOI] [PubMed] [Google Scholar]
- 29. Deng J, Shao J, Markowitz JS, An G. ABC transporters in multi-drug resistance and ADME-Tox of small molecule tyrosine kinase inhibitors. Pharm Res 2014;31:2237–55. [DOI] [PubMed] [Google Scholar]
- 30. Parrish KE, Cen L, Murray J, Calligaris D, Kizilbash S, Mittapalli RK, et al. Efficacy of PARP inhibitor rucaparib in orthotopic glioblastoma xenografts is limited by ineffective drug penetration into the central nervous system. Mol Cancer Ther 2015;14:2735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pokorny JL, Calligaris D, Gupta SK, Iyekegbe DO Jr, Mueller D, Bakken KK, et al. The efficacy of the Wee1 inhibitor MK-1775 combined with temozolomide is limited by heterogeneous distribution across the blood-brain barrier in glioblastoma. Clin Cancer Res 2015;21:1916–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laramy JK, Kim M, Gupta SK, Parrish KE, Zhang S, Bakken KK, et al. Heterogeneous binding and central nervous system distribution of the multitargeted kinase inhibitor ponatinib restrict orthotopic efficacy in a patient-derived xenograft model of glioblastoma. J Pharmacol Exp Ther 2017;363:136–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Plotkin S, Supko JG, Curry WT. Concentrations of Sunitinib after oral administration in patients with high grade glioma [abstract]. Neuro-oncol 2011;13:iii41–68. [Google Scholar]
- 34. Abbruzzese C, Matteoni S, Signore M, Cardone L, Nath K, Glickson JD, et al. Drug repurposing for the treatment of glioblastoma multiforme. J Exp Clin Cancer Res 2017;36:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lankheet NAG, Desar IME, Mulder SF, Burger DM, Kweekel DM, van Herpen CML, et al. Optimizing the dose in cancer patients treated with imatinib, sunitinib and pazopanib. Br J Clin Pharmacol 2017;83:2195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rovithi M, Gerritse SL, Honeywell RJ, Ten Tije AJ, Ruijter R, Peters GJ, et al. Phase I dose-escalation study of once weekly or once every two weeks administration of high-dose sunitinib in patients with refractory solid tumors. J Clin Oncol 2019;37:411–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu Y, Du Y, Liu H, Ma T, Shen Y, Pan Y. Study of efficacy and safety of pulsatile administration of high-dose gefitinib or erlotinib for advanced non-small cell lung cancer patients with secondary drug resistance: a single center, single arm, phase II clinical trial. Thorac Cancer 2016;7:663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morikawa A, de Stanchina E, Pentsova E, Kemeny MM, Li BT, Tang K, et al. Phase I study of intermittent high-dose lapatinib alternating with capecitabine for HER2-positive breast cancer patients with central nervous system metastases. Clin Cancer Res 2019;25:3784–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim DH, Choi YJ, Sung KJ, Yoo SA, Sung YH, Kim JK, et al. Efficacy of nano-particulated, water-soluble erlotinib against intracranial metastases of EGFR-mutant lung cancer. Mol Oncol 2018;12:2182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sharma S, Petsalaki E. Large-scale datasets uncovering cell signalling networks in cancer: context matters. Curr Opin Genet Dev 2019;54:118–24. [DOI] [PubMed] [Google Scholar]
- 41. Sever R, Brugge JS. Signal transduction in cancer. Cold Spring Harb Perspect Med 2015;5:a006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johnson H, White FM. Quantitative analysis of signaling networks across differentially embedded tumors highlights interpatient heterogeneity in human glioblastoma. J Proteome Res 2014;13:4581–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Randall EC, Emdal KB, Laramy JK, Kim M, Roos A, Calligaris D, et al. Integrated mapping of pharmacokinetics and pharmacodynamics in a patient-derived xenograft model of glioblastoma. Nat Commun 2018;9:4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wei W, Shin YS, Xue M, Matsutani T, Masui K, Yang H, et al. Single-cell phosphoproteomics resolves adaptive signaling dynamics and informs targeted combination therapy in glioblastoma. Cancer Cell 2016;29:563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell 2000;103:239–52. [DOI] [PubMed] [Google Scholar]
- 46. Nakano R, Nakayama T, Sugiya H. Biological properties of JNK3 and its function in neurons, astrocytes, pancreatic beta-cells and cardiovascular cells. Cells 2020;9:1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Antonyak MA, Kenyon LC, Godwin AK, James DC, Emlet DR, Okamoto I, et al. Elevated JNK activation contributes to the pathogenesis of human brain tumors. Oncogene 2002;21:5038–46. [DOI] [PubMed] [Google Scholar]
- 48. Cui J, Han SY, Wang C, Su W, Harshyne L, Holgado-Madruga M, et al. c-Jun NH(2)-terminal kinase 2alpha2 promotes the tumorigenicity of human glioblastoma cells. Cancer Res 2006;66:10024–31. [DOI] [PubMed] [Google Scholar]
- 49. Koch P, Gehringer M, Laufer SA. Inhibitors of c-Jun N-terminal kinases: an update. J Med Chem 2015;58:72–95. [DOI] [PubMed] [Google Scholar]
- 50. Gong Y, Ma Y, Sinyuk M, Loganathan S, Thompson RC, Sarkaria JN, et al. Insulin-mediated signaling promotes proliferation and survival of glioblastoma through Akt activation. Neuro Oncol 2016;18:48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data are available and were provided to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD019038 (24).