Summary
Background
Understanding how SARS-CoV-2 affects respiratory centres in the brainstem may help to preclude assisted ventilation for patients in intensive care setting. Viral invasion appears unlikely, although autoimmunity has been implicated, the responsible antigens remain unknown. We previously predicted the involvement of three epitopes within distinct brainstem proteins: disabled homolog 1 (DAB1), apoptosis-inducing-factor-1 (AIFM1), and surfeit-locus-protein-1 (SURF1).
Methods
Here, we used microarrays to screen serum from COVID-19 patients admitted to intensive care and compared those with controls who experienced mild course of the disease.
Findings
The results confirm the occurrence of IgG and IgM antibodies against the hypothesised epitopes in COVID-19 patients. Importantly, while IgM levels were similar in both groups, IgG levels were significantly elevated in severely ill patients compared to controls, suggesting a pathogenic role of IgG.
Interpretation
The newly discovered anti-neuronal antibodies might be promising markers of severe disease and the targeted peptide epitopes might be used for targeted immunomodulation. Further work is needed to determine whether these antibodies may play a role in long-COVID.
Funding
AF, CF and PR received support from the German Research Foundation (grants FL 379/22-1, 327654276-SFB 1315, FR 4479/1-1, PR 1274/8-1). SH, DR, and DB received support from the Ministry of Economy, State of Mecklenburg Western Pomerania, Germany (grant COVIDPROTECT: “Optimisation of diagnostic and therapeutic pathways for COVID-19 patients in MV”). SH received support from the Research Group Molecular Medicine University of Greifswald (FVMM, seed funding FOVB-2021-01). AV received support from the Else Kröner Fresenius Foundation and the Alzheimer Research Initiative.
Keywords: Autoimmunity, Encephalitis, Brainstem, SARS-CoV-2, Epitopes, Long-COVID
Research in context.
Evidence before this study
The intense research effort into COVID-19 made clear that SARS-CoV-2 affects the nervous system and that immune-mediated mechanisms may be involved. Indeed, anti-neuronal autoantibodies have been found in cerebrospinal fluid from severely ill COVID-19 patients.
In addition, autoimmune mechanisms are thought to be involved in the long-lasting disabling cognitive impairment that might be associated with COVID-19, as an aspect of the so called Long-COVID.
Importantly, the neuronal proteins involved as autoantigens in COVID-19 and their epitopes are still unknown.
Added value of this study
Here, we show autoantibodies against three neuronal antigens that are known to play a role in central respiratory drive and reflex responses when lung function is impaired. We previously formulated and published exact predictions regarding these antigens and the epitopes involved, and the present data now confirm these hypotheses. Crucially, the IgG binding to the epitopes differentiate severely ill patients from mildly affected ones, indicating potential clinical relevance of the autoimmune response. In sum, autoimmune responses to these neuronal antigens appears to be a contributing factor in severe COVID-19. In addition, the three neuronal antigens that we identified are required for synaptic plasticity and higher cognition, and their involvement might also provide insights into long-hauling neurological consequences of the infection.
Implications of all the available evidence
In the present study, we identified both the neuronal proteins that are targeted by IgG autoantibodies in COVID-19 and their exact epitopic sequences. The present data not only suggest a potential mechanism for the damage to the brain structures mediating vital and cognitive functions, but they also provide a potential therapeutic approach. Further studies are needed to determine whether the epitopic peptides may be used as immunomodulators to restore tolerance for the autoantigens targeted by COVID-19 autoimmunity.
Alt-text: Unlabelled box
Introduction
The severe-acute-respiratory-syndrome-coronavirus-2 (SARS-CoV-2) disease (COVID-19) causes respiratory failure with reduced response to hypoxemia and dissociation between lung damage and respiratory function.1, 2, 3 These observations and the loss-of-taste and smell suggested early on during the pandemic that brainstem-allocated respiratory centres might be affected.4 Brainstem injury in COVID-19 is now documented with a variety of clinical manifestations including disorders of eye-movements, coordination, consciousness, as well as involuntary movements.5,6 Concomitant imaging findings range from normal, through isolated lesions, to extensive tissue loss and have been described as brainstem encephalitis, rhombencephalitis, or acute necrotising encephalopathy.5, 6, 7, 8
Causally, a direct cytopathic effect of SARS-CoV-2 appears unlikely because viral invasion is infrequent and does not elicit local reactive inflammation.9,10 Rather, evidence points to autoimmune mechanisms. First, immunomodulation is usually at least partially effective.5, 6, 7, 8,11,12 Second, cerebrospinal fluid (CSF) from patients with neurological manifestations contains high pro-inflammatory cytokine levels.13 Third, CSF-immunohystochemistry shows brainstem-binding antibodies (Abs).11,12,14 Nevertheless, diagnostic anti-neuronal Ab-panels are consistently negative.6,7,12,13 In summary, brainstem damage in COVID-19 is likely associated with auto-Abs but the antigens are unknown.
We previously identified three brainstem-related proteins (disabled-homolog-1, DAB1; mitochondrial apoptosis inducing factor 1, AIFM1; surfeit-locus-protein-1, SURF1) sharing three potentially immunogenic sequences of 6 amino-acids (aa), namely GSQASS, LNEVAK, and SAAEAS (Table 1; Figure 1, Panel a) with SARS-CoV-2.15 Therefore, we hypothesised the following: (1) COVID-19 patients with respiratory failure and/or acute neurologic manifestations have auto-Abs against these three proteins in serum and CSF. (2) The three shared hexapeptides are the immune dominant epitopes.15,16 Here, we show results from 23 critically ill COVID-19-patients (14 serum samples and 23 CSF-samples) all requiring intensive care and manifesting neurologic deficits (Table 2), and from 16 serum samples from mildly affected patients.
Table 1.
Hexapeptides shared between brainstem proteins and SARS-CoV-2. Alongside the 6-mers, the human and viral proteins in which they are contained are indicated. The hexapeptides have been previously identified as potential epitopes targeted by cross-reactive Abs following the immune response against the virus. (15). (UniProt: Universal Protein database. www.uniprot.org, accessed on 01/24/2022). *Including variants of concern B.1.617.2 (Protein Data Bank ID: 7W92. https://www.rcsb.org/structure/7W92 accessed on 01/24/2022) and B.1.1.529 (Protein Data Bank ID: 7QO7. https://www.rcsb.org/structure/7QO7 accessed on 01/24/2022).
| Predicted epitope | Human brainstem protein (epitope position) [UniProt link; Gene] | SARS-CoV2 protein (epitope position) [UniProt link; Gene] |
|---|---|---|
| GSQASS | Disabled homolog 1 (560-565) [https://www.uniprot.org/uniprotkb/O75553; DAB1] |
Nucleoprotein (179-184) [https://www.uniprot.org/uniprotkb/P0DTC9; N] |
| LNEVAK | Apoptosis-inducing factor 1, mitochondrial (601-606) [https://www.uniprot.org/uniprotkb/O95831; AIFM1] |
Spike glycoprotein* (1186-1191, S2-domain) [https://www.uniprot.org/uniprotkb/P0DTC2/entry; S] |
| SAAEAS | Surfeit locus protein 1 (47-52) [https://www.uniprot.org/uniprotkb/Q15526; SURF1] |
Nucleoprotein (250-255) [https://www.uniprot.org/uniprotkb/P0DTC9; N] |
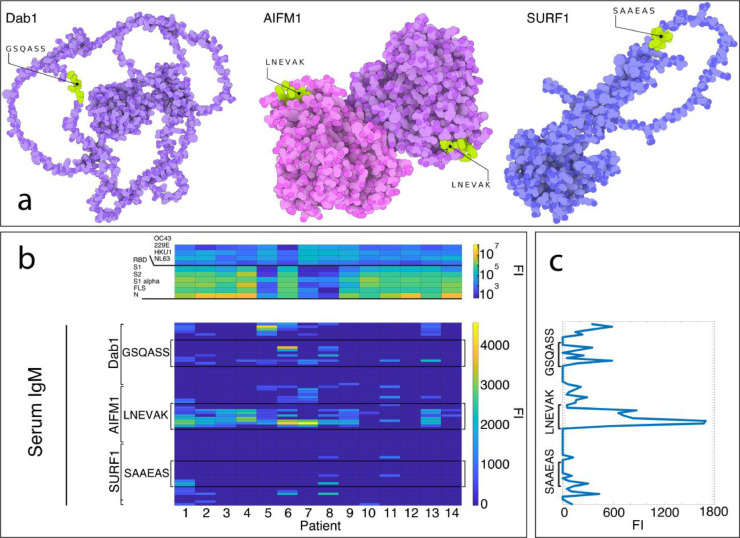
Figure 1.
Auto-Abs against DAB1, AIFM1, and SURF1 in sera of 14 severely affected COVID-19 patients. a) crystallography and modelled structures showing the predicted and confirmed epitopic sites exposed in known conformational states of the three target proteins. The sites are therefore freely accessible to auto-Abs for recognition and binding. AIFM1 is presented in its dimeric form. The 3D-illustrations were produced with The Protein Imager with crystallography data and structure predictions.58, 59, 60 b) Heatmaps showing in the serum IgG response (top heatmap) of 14 severely ill COVID-19 patients against proteins of SARS-CoV-2 and, as control for cross-reactivity, common cold coronaviruses (OC43, 229E, HKU1, NL63), and the IgM binding to the human epitopes from the autoantigens DAB1, AIFM1, and SURF1.15 c) immunofluorescence intensity averaged across the 14 patients and plotted against the predicted epitopes from DAB1, AIFM1, and SURF1. (FI=fluorescence intensity).
Table 2.
Demographical and clinical characteristics of patients and control population.
| Critically ill patients | Mildly affected patients | |
|---|---|---|
| General characteristics | N Mean±DS Range | N Mean±DS Range |
| Age | 23 69.8±10.9 48-88 | 16 51.8±10.1 29-64 |
| Gender | ||
| Man Women |
18(78.2%) 5(21.7%) |
9(56.25%) 7(43.75%) |
| Chronicdiseases | ||
|
7 (30.4%) 18(78.3%) 8 (34.8%) 3 (13.6%) 2(8.7%) 2 (8.7%) 4 (17.4%) |
0 3(18%) 0 0 0 0 0 |
| Neurological symptoms | ||
|
16(100%) 7(30%) 6(26%) 3(13%) 1(4%) 1(4%) 1(4%) 1(4%) |
_ |
| Days onset COVID-19/onset neurological symptoms | 23 18.6±12.9 0–43 | _ |
| Other COVID19 symptoms | ||
|
23(100%) 23(100%) 23(100%) |
7(44%) 5(31%) 0 |
| Days onset COVID-19/discharge/negativisation | 23 45.0±30.8 13–160 | 16 17.0±10.1 6–43 |
| Days onset COVID-19/admission to the hospital | 23 6.3±3.7 0–16 |
_ |
| Days onset COVID-19/admission to ICU | 23 7.2±3.5 2–16 |
_ |
| Days on ICU | 23 34.1±21.4 5–77 |
_ |
| PCO2 ICU admission | 23 37.9±13.1 28–74.6 | |
| PO2 ICU admission | 23 87.9±36.4 49.6–212 |
_ |
Methods
Ethics
Approval was obtained by the Ethics Committee of Charité (Number: EA2/066/20) Charité - University Medicine Berlin and by the Ethics Committee, Università di Brescia, Italy (Number: NP 4463). Informed consent was obtained from all patients.
Patients and Samples
Serum and CSF samples were obtained from 24 severely-ill hospitalised patients (14 serum samples, 22 CSF samples) who were admitted between April and December 2020 to the intensive care unit of the Charité University Hospital Berlin (Campus Benjamin Franklin), manifesting neurological deficits. Samples from one patient were lost in transit and not analysed. Mild disease serum samples were obtained from 16 COVID-19 patients who did not require hospitalisation, did not show respiratory distress or neurologic signs or symptoms, and did not present any respiratory or neurologic co-morbidities. Mildly affected patients were prospectively included between October 2020 and February 2021 at the University Hospital of Bari. Clinical characteristics of the participating patients can be found in Table 2. Samples were stored at -80°C until analyses and were heat-inactivated at 56°C for 30 min immediately before microarray assay.17
Peptide microarray screening
The amino-acid sequence of DAB1, AIFM1, and SURF1 containing the three immunological determinants (GSQAASS, LNEVAK, and SAAEAS, respectively) predicted in Lucchese & Flöel, 2020, and the 9 aa flanking the three 6-mer in both directions were elongated with GSGSGSG linkers and translated into overlapping peptides of 15 aa length with an overlap of 14 aa (Supplementary Table 2). Additionally the 3 predicted 6-mer were elongated with neutral GSGSGSG linkers at the C- and N-terminus and translated into overlapping peptides of 15 aa length with an overlap of 14 aa (Supplementary Table 3). The total number of peptides was 88. All of them were analysed in duplicates. 8 spots of HA control (YPYDVPDYAG) and 7 spots of Polio control (KEVPALTAVETGAT) were included on the array. CSF was applied undiluted (1:10 and 1:3 for two patients respectively due to lack of material)
The custom peptide microarray was pre-stained with the secondary and control Abs to investigate background interactions with the 88 different peptides that could interfere with the main assays. Secondary Abs: Goat anti-human IgG (Fc) DyLight680 (0.1 µg/ml) (Catalog Nr. SA5-10138; lot SC2357783) and goat anti-human IgM (µ chain) DyLight800 (0.2 µg/ml) (Item No. 609-145-007; lot 37641); 45 min staining in incubation buffer at RT. Control -Ab: Mouse monoclonal anti-HA (12CA5) DyLight800 (0.5 µg/ml) (PEPperPRINT Nr. PPC.038.003; Batch 02.10.2020); 45 min staining in incubation buffer at room temperature (RT).
Subsequent incubation of further custom peptide microarray copies with human cerebrospinal fluid (CSF) samples at dilutions of 1:10 (Patient 21 of the CSF Analysis, Figure 2) and 1:3 (Patient 22 of the CSF Analysis, Figure 2) in incubation buffer or neat was followed by staining with the secondary and control Abs as well as microarray read-out. The additional HA control peptides framing the peptide microarrays were simultaneously stained with the control -Ab as internal quality control to confirm the assay performance and the peptide microarray integrity.
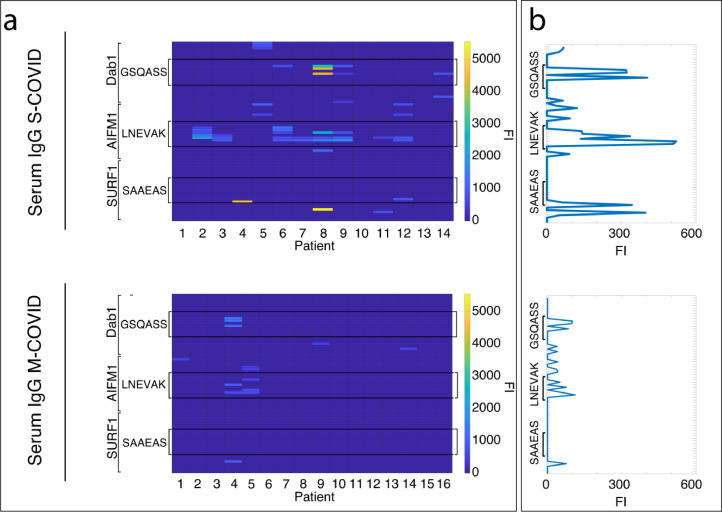
Figure 2.
Severely vs. mildly affected COVID-19 patients significantly differ in serum IgG binding against DAB1, AIFM1, and SURF1. a) Heatmaps showing on the top the serum IgG binding against the epitopes of the three neuronal autoantigens characterising severely affected COVID-19 patients (S-COVID), on the bottom the absence of IgG binding to same auto antigens in the majority of the mildly affected patients (M-COVID). b) The immunofluorescence intensity values presented in the heatmaps in panel a were averaged across patients within the two groups and plotted against the predicted epitopes from DAB1, AIFM1, and SURF1 in order to allow for direct comparison between severely and mildly affected patients. (FI=fluorescence intensity).
After 15 min pre-swelling in washing buffer (PBS, pH 7.4 with 0.05% Tween 20; 3 × 10 s after each assay) and 30 min incubation in blocking buffer (Rockland blocking buffer MB-070, item Nr. MB-070; 30 min before the first assay), the custom peptide microarray copy was initially incubated with the secondary and control Abs for 45 min at room temperature to analyse background interactions with the 88 different custom peptides that could interfere with the main assays (incubation buffer: washing buffer with 10% blocking buffer).
At scanning (Innopsys InnoScan 710-IR Microarray Scanner; scanning resolution of 20 µm; scanning gain of 50 at low laser power (680 nm, red) and 40 at high laser power (800 nm, green))gains of 50/40 (red/green), we did not observe any background interaction of the secondary or control Abs with the 88 different peptides of the microarray (see adjusted scan). Simultaneous staining with the control -Ab gave rise to the expected well-defined HA control spot pattern framing the peptide microarray, validating the overall peptide microarray integrity and assay performance.
Quantification of spot intensities and peptide annotation were based on 16-bit grey scale tiff files that exhibit a higher dynamic range than the 24-bit colorised tiff files included here. Microarray image analysis was done with PepSlide® Analyser. A software algorithm breaks down fluorescence intensities of each spot into raw, foreground and background signal, and calculates averaged median foreground intensities and spot-to-spot deviations of spot duplicates. Based on averaged median foreground intensities, intensity maps were generated and interactions in the peptide maps highlighted by an intensity colour code with red (IgG) or green (IgM) for high and white for low spot intensities. We tolerated a maximum spot-to-spot deviation of 40%, otherwise the corresponding intensity value was zeroed.
We further plotted averaged spot intensities of the assays to visualise overall spot intensities and signal-to-noise ratios. The intensity plots were correlated with peptide and intensity maps as well as with visual inspection of the microarray scans to identify IgG and IgM responses of the human CSF samples.
Anti-SARS-CoV-2 Abs screening: Corona Array – a bead-based multiplexed immunoassay
The Corona Array is based on the xMAP® technology (Luminex®),18 and is an in-house bead-based suspension array for simultaneous analysis of antibodies in a sample against different recombinant corona virus antigens.
Recombinant proteins
Ten recombinant His-tagged proteins were commercially purchased and used in this study, including 6 SARS-CoV-2 proteins, as well as 4 proteins from endemic human coronaviruses (Table S3).
Coupling of recombinant proteins to magnetic microspheres
Each recombinant protein was covalently coupled to MagPlex® magnetic microspheres (Luminex Corp., Austin, TX; beads) of unique color-codes (100 µg/1.25 × 107 beads). The coupling was carried out at RT following a standard carbodiimide coupling procedure described before.19,20 In brief, uncoupled beads from the bead-stock solution were resuspended (30 sec vortexing, 3 min ultrasonic bath, 30 sec vortexing) and transferred to a microcentrifuge tube (Protein LoBind; Eppendorf AG, Hamburg, Germany). After discarding the storage solution, the beads were washed three times with activation buffer (100 mM NaH2PO4, pH 6.2) to adjust the pH value for the activation reaction, and then incubated in the dark (900 rpm, 20 min, RT) with 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC; ThermoFisher Scientific, Karlsruhe, Germany; final concentration 50 mg/mL) and N-hydroxysulfosuccinimide (sulfo-NHS; ThermoFisher Scientific; final concentration: 50 mg/mL). The carboxyl groups of the beads were activated by EDC and form a sulfo-NHS-ester intermediate in the presence of sulfo-NHS. This is replaced in the next step by the primary amines of the recombinant proteins and a covalent amide bond is formed with the beads.19 To prepare for protein coupling, the activated beads were washed three times with coupling buffer (50 mM MES; 2-(N-morpholino) ethanesulfonic acid; pH 5.0; Sigma-Aldrich Chemie GmbH, Taufkirchen). Finally, the lyophilised recombinant proteins were dissolved in coupling buffer to a final concentration of 0.1 mg/mL, and then coupled to the beads by incubating them together in the dark (900 rpm, 2 h, RT). Protein-coupled beads were then washed six times with wash buffer (0.05% Tween20 in DPBS), resuspended in block store buffer (1% w/v bovine serum albumin (BSA)/0.05% v/v ProClin™ 300 in DPBS; Sigma), transferred into 15 mL reaction tubes (Protein LoBind), and stored in the dark overnight at 4 °C, where any remaining free carboxyl groups on the beads were blocked with BSA in the buffer.
The individual antigen-coupled bead populations were combined into the bead master mix later used in the Corona Array (10-plex assay). For that purpose, the antigen-coupled beads were resuspended again (30 s vortex, 3 min ultrasonic bath, 30 s vortex) and the bead solutions were combined and adjusted to 1250 beads/50 µL with bead buffer (50% LowCross-Buffer® (Candor Bioscience GmbH, Wangen)/50% block store buffer (1% BSA in DPBS). The aliquoted master mix was stored at 4 °C.
Efficiency assessment of coupling reaction (coupling control)
Before proceeding to the Corona Array, we assessed the coupling efficiency of the proteins to the beads. Since the recombinant proteins are all His-tagged, the coupling control was done by incubating the protein-coupled bead master mix with anti-His5 antibody (mouse anti-(H)5 antibody; Qiagen, Hilden, Germany; Cat. No. 34660; final concentration: 10 µg/mL) (900 rpm, 45 min, RT), while the bead buffer served as blank. After washing the beads with wash buffer to remove unbound antibodies, the beads were incubated (900 rpm, 30 min, RT) with a secondary anti-mouse RPE-conjugated detection antibody (Goat F(ab’)2 anti-Mouse IgG (H+L)-RPE; Dianova, Hamburg, Germany; Cat. No. 115-116-146; final concentration: 5 µg/mL). Finally, the beads were washed three times with wash buffer, and resuspended in sheath fluid (xMap Sheath Fluid, Luminex Corporation). The fluorescence signals were measured on a BioPlex® 200 system (Bio-Rad Laboratories) using the following measurement settings: Bead type: MagPlex® beads; beads: 100 beads/region; sample timeout: 60 s; sample volume: 80 μL; gate settings: 7,500–15,000 (BioPlex® Manager™ 5.0, Bio-Rad Laboratories).
Multiplex screening for IgG antibodies to SARS-CoV2 and endemic corona viruses in serum and liquor (Corona Array)
To exactly determine IgG titres against the different antigens tested, different seven-step dilution series were prepared for serum and liquor samples in bead buffer, based on the expected range of signals. The dilutions used were 1:20–1:312,500 (serum) and 1:1–1:12,500 (liquor). A plasma pool (prepared from plasma samples of corona vaccines at day 14 after the second vaccination; AICOVI; NCT04826770) was included on each plate for subsequent normalisation of data. After thoroughly re-suspending the bead master mix, it was added to the wells of half-well 96-well plates (Greiner Bio-One GmbH, Frickenhausen, Germany) at a final count of 1250 beads/50 µL/well. The bead storage solution was removed, and the diluted samples/plasma pool were transferred to the beads (50 µL/well). The plates were sealed with aluminium foil and incubated in the dark on a shaker (4 °C, 18 h, 900 rpm) to allow the binding of plasma/serum/liquor antibodies to protein-coupled beads.
Unbound antibodies were removed through three washing steps with 100 µL wash buffer/well. Bound IgG antibodies were detected by incubation with goat anti-human IgG RPE-conjugated detection antibody (R-PE AffiniPure F(ab')₂ Fragment Goat Anti-Human IgG, Fcγ fragment specific; Jackson ImmunoResearch; Cat. No.: 109-116-098; 50 µL/cavity, diluted in bead buffer to a final concentration of 5 µg/mL) in the dark (900 rpm, 90 min, RT).
After three additional washing steps with wash buffer to remove unbound detection antibodies, beads were resuspended in sheath fluid in the dark on a plate shaker (900 rpm, at least 1 min, RT) until measurement. The antigen-antibody reactions were read on the BioPlex® 200 system as described above. Signal detection on the BioPlex® 200 System includes determination of the autofluorescence of the beads and the fluorescence intensity of the bound detection antibodies. This allows the differentiation of the individual recombinant proteins, as well as quantification of antibody levels. The level of antibody response is given as median fluorescence intensity (MFI) of at least 100 beads per bead region measured.
Corona array data analysis
The xMAPr analysis tool (xMAPr 1.0; https://michalik.shinyapps.io/xMAPr_app/) was used to derive a relative antibody concentration from the multiple data points of the serum and CSF titrations. The relative antibody concentration was calculated using the signal intensity (MFI) and the dilution factor in a non-linear regression model. Only data sets with R² > 0.98 were used for further analysis. Response values obtained for samples on different plates were normalised using the plasma pool. An imputation was performed for all samples that failed the curve fitting algorithm based on a LOESS fit over all measurement of a single dilution. A limit of detection (LOD) for each antigen was determined by using the blank measurements and the LOD was set as mean MFI+3 × SD.
Statistics
All the microarray fluorescence intensity values below 500 for serum samples and below 100 for CSF samples were set to zero.21 The thresholded serum immunofluorescence data on the peptides in the supplementary table one entered as binary variable two identical (one for IgG data and one for IgM data) linear mixed models with the 4-level factor DETERMINANT (GSQASS, LNEVAK, SAAEAS, NONE) and the demographic covariate GENDER and AGE as fixed effect predictors, SUBJECT as random effect, and a logit link function. Fixed coefficients were used as post-hoc measure of single contrasts. The correlations were computed as one-tailed Kendall's tau. The immunofluorescence intensity indicating autoantibody binding to the predicted antigens in severely ill and control patients was compared with a Wilcoxon signed-rank test. The hypothesis of bivariate normality for the correlation analysis was rejected by way of the Kolmogorov-Smirnov test. Similarly, normality was rejected for the immunofluorescence intensity indicating autoantibody binding to the predicted antigens in severely ill and control patients, again with a KS-test. Given the very specific and novel research question of the present study an exact estimate of the expected effect size was not possible. Previous research found up autoantibodies in serum of up to 80% of hospitalised COVID-19 patients as compared to 5–10% in various type of controls.22 Since we were hypothesising a specific pathogenic role for the anti neuronal antibodies in severe COVID we adopted the high 80% estimate. A power analysis conducted with Gpower.23 showed that a significant difference for such proportions and alpha=0.05 could be found comparing two samples of minimum size n=13, which we adopted as minimum sample size for each group.
Role of funders
Funders did not have any role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Figure 1 shows the binding of serum IgMs from 14/23 critically ill patients along three 24-aa-long regions of DAB1, AIFM1, SURF1, respectively, each dissected into 15-aa-peptides with an overlap of 14 aa. Each region is centred on GSQASS, LNEVAK, and SAAEAS, respectively, with the addition of the 9 aa flanking the three 6-mers in both directions on the sequence of the human proteins and of GSGSGSG linkers (see Methods and Supplementary Table 1).
Of the 14 sera from critically ill patients, thirteen showed IgM binding to at least one of the 15-mers containing one of the three predicted epitopes (i.e. GSQASS, LNEVAK, and SAAEAS) and only in 2/14 patients auto-Abs exclusively against one single epitope were detectable.
The serum Ab-binding was higher for the peptides containing the whole 6-mers, independent of the position of the hexapeptide within the 15-mer for GSQASS and LNEVAK and, in the case of SAAEAS, mainly when it was at the N-terminus of the 15-mer (Figure 1, panel C). Indeed, containing one of the three determinants as compared with none was a significant predictor of IgM (generalised linear mixed model; F(3, 976)=25; p<0.001) binding. The effect was driven by LNEVAK, which was associated with higher IgM (fixed coefficient; t=−7; p<0.001) than the peptides containing none of the three 6-mers. The same analysis on IgG binding from serum of the severely affected patients (Figure 2, panel a, top heatmap) confirmed the immunogenicity of the predicted determinants as compared to the other peptides tested (generalised linear mixed model; F(3, 976)=11.2; p<0.001). It also confirmed that LNEVAK was driving the effect also for the IgG binding (fixed coefficient; t=−5.2 p<0.001). Moreover, the critically ill patient not showing IgM against any of the predicted epitopes showed significant IgG binding against two of them. Gender and age did not significantly affect the IgM or the IgG response.
When compared with the 16 sera from mildly affected COVID patients, the IgM binding was similar (data not shown), but the levels of IgG binding to the three predicted epitopes significantly differed (Figure 2) between the two groups (Wilcoxon signed-rank test; z=2.633; p=0.008) and 14 of the 16 mildly affected controls did not show any detectable IgG against the neuronal autoantigens.
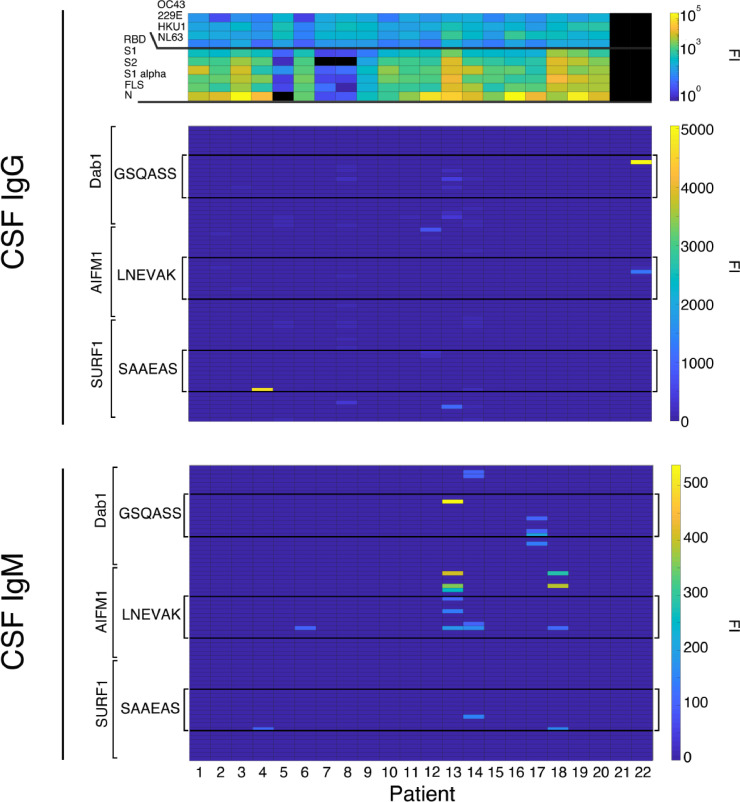
Overall, the Ab binding to the three antigens was higher for serum than CSF samples. Figure 3 shows the binding of CSF IgM and IgG from 22 patients, including patients 1–13 from the serum analysis. CSF from 11/22 patients contained either IgG (n=4), IgM (n=3), or both (n=3) binding at least one peptide containing one of the three predicted immune determinants (i.e. GSQAASS, LNEVAK, and SAAEAS) in its entirety. Only in 4 out of the 11 patients, auto-Abs bound to one single epitope; IgM and IgG in one case, only IgG in two, and only IgM in one. Among the remaining patients, one showed Abs against all three hexapeptides, and 6 patients against two. Of the 11 patients showing the predicted CSF-Abs, 4 were not included in the serologic analysis and therefore the total number of patients showing either systemic or intrathecal detectable Abs against the predicted epitopes was 18/23 (78%).
Figure 3.
Auto-Abs against DAB1, AIFM1, and SURF1 in CSF of 22 severely affected COVID-19 patients. Heatmaps showing in the upper panel the CSF IgG response against virus proteins of the different patients, in the middle panel the microarray CSF immunofluorescence CSF IgG binding to human peptides containing the sequences predicted by Lucchese and Flöel15 and in the lower panel the CSF IgM binding to the same sequences. (FI=fluorescence intensity).
Additionally, the microarrays containing the three predicted 6-mers elongated with neutral aa linkers (GSGSGSG, Table S1) showed serum IgG- and IgM-binding to all the three determinants in one patient (data not shown) and CSF IgG-binding to all the three, to two, and to one determinant in 1, 2, and 1 patient respectively (Figure S1). IgM was not detectable in the CSF in this analysis.
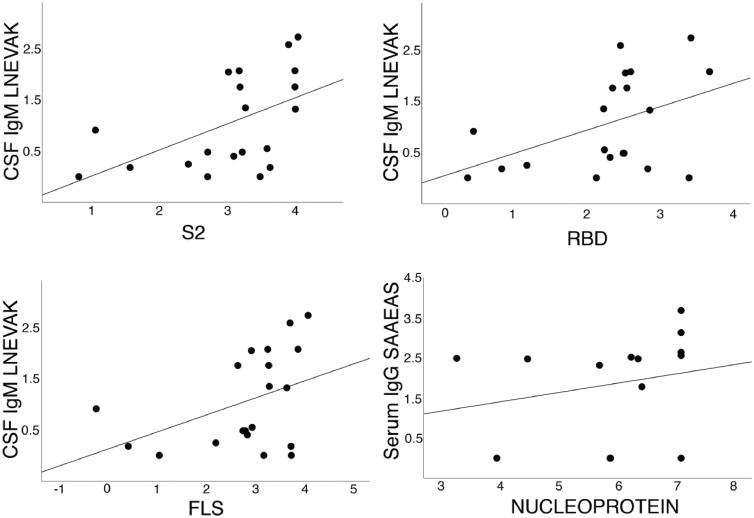
The levels of anti-neuronal Abs correlated with the immunity against SARS-CoV-2 confirming the association between autoimmunity and the infection in critically-ill patients. In particular, intrathecal IgM-binding to DAB1-peptides containing LNEVAK showed direct correlation with intrathecal humoral response against the spike S2 (Kendall's tau =0.398; p=0.007), RBD domain (Kendall's tau=0.345; p=0.017) and the full-length Spike (Kendall's tau=0.292; p=0.037) of the SARS-COV-2. Of note, the predicted immune determinant LNEVAK is part of the sequence of both DAB1 and the spike S2 domain (Table 1). Moreover, the serum IgG binding to SAAEAS-containing SURF1 peptides correlated positively with the serum response to the viral nucleoprotein (Kendall's tau=0.388, p=0.032). The nucleoprotein sequence contains SAAEAS (Table 1). The correlations are illustrated in Figure 4.
Figure 4.
Significant correlations between the immune response to SARS-CoV-2 proteins (S2 and RBD domains of the spike protein, full length spike protein, FLS, and nucleoprotein) and the epitopes of the human brainstem proteins DAB1 (epitope LNEVAK) and SURF1 (epitope SAAEAS), as detailed in the Results.
Discussion
Our study revealed Abs against novel brainstem neuronal antigens that may underlie the severe respiratory failure in COVID 19 patients with neurological complications. The results show that the newly discovered antibodies target the predicted shared epitopes of three proteins DAB1, AIFM1, and SURF1 that are shared with SARS-CoV2.15 These new auto-Abs show a correlation with anti-viral immune response and clinical severity.
DAB1 is required in the Reelin pathway and the two proteins need to be co-expressed.24 Reelin, and therefore DAB1 as well, is a specific marker of brainstem neurons regulating the response to hypoxemia.25 Such response is impaired in Reelin deficiency remarkably mirroring respiratory failure in COVID-19.1,25 Moreover, the neurons expressing Reelin and DAB1 also express the progesterone-receptor-membrane-component-1 (PRMC1), which promotes neurogenesis and neuroprotection.25,26 PRMC1-mediated neuroprotection of respiratory neurons damaged by autoimmunity might contribute to the sex disparity in COVID-19 mortality.27
AIFM1 and SURF1 mutations cause Leigh syndrome, which is characterised by a subacute necrotising encephalopathy mainly affecting specifically the brainstem and closely resembling the COVID-19 acute necrotising encephalopathy.8,28,29 Of relevance, death in Leigh syndrome often results from respiratory failure and mouse models show selective impairment of the respiratory centres in the brainstem.30 Peripheral neuropathy is also associated both with COVID-19 and with altered SURF1 and AIFM1.31,32 Additionally, AIFM1-dysfunction is associated with olfactory impairment.33 In summary, physiological functions and pathologic relevance of the proteins targeted by the newly discovered auto-Abs are consistent with the features of COVID-19. Autoimmune damage to these three proteins is also consistent with reports of COVID-19-severity correlating with anti-SARS-CoV-2-Ab and auto-Ab titres.34,35
The three epitopes (GSQASS from DAB1, LNEVAK from AIFM1, and SAAEAS from SURF1) are located in exposed regions of the respective protein and therefore available for antibody binding (Figure 1, Panel A). The Auto-Abs against the three proteins have high specificity, as indicated by the statistically significant stronger binding to the peptides containing the three 6-mers (see Figure 1, Panel B and Results) and by the IgG binding to 15-aa long peptides containing the 6-mer epitopes flanked by irrelevant GS repeats (Figure S1). Additionally, LNEVAK is present in the brain specific neuronal isoform of AIFM1 and absent from the isoform not expressed in normal brain tissue.36,37 This supports the idea that the auto-Abs selectively target the central nervous system.
The primary objective of the study was to demonstrate the occurrence of the hypothesised Abs in severe COVID-19. In addition, we showed significantly higher levels of IgG against the predicted neuronal antigens in severely affected COVID-19 patients in comparison with the mildly affected ones, who showed in the vast majority almost no detectable IgG binding (Figure 2). Remarkably, the two groups of patients showed a similar IgM profile. Indeed, IgM typically shows low-affinity for the antigen and auto-Abs of this class are usually considered not pathogenic.38 Conversely, high-affinity IgG auto-Abs typically result in pathologic processes functionally affecting the target antigens.39 Moreover, the three neuronal antigens DAB1, AIFM1, and SURF1 are intracellular proteins and the main mechanism of AB internalisation mediating intracellular immunity shows higher affinity for IgG.40 These data suggest that in critically ill COVID patients immature B-cells recognise the three predicted auto antigens with subsequent class-switch and affinity maturation resulting in potentially pathogenic high affinity IgG. The latter may then affect the three neuronal antigens DAB1, AIFM1, and SURF1 and associated brainstem and cognitive functions.
In the present work, we also identified the epitopes bound by the auto-Abs. These oligopeptides might be used for targeted immunomodulation instead of general immunosuppression in patients positive for the respective Auto-Abs, thus preserving anti-viral immunocompetence.41 To this purpose, replication of our findings and animal studies needs to confirm the pathogenic role of the novel Auto-Abs. Of note, previous research showing that pre-existing autoantibodies predispose patients to severe COVID-19 infection is coherent with our hypothesis and results, implicating a loss of self-tolerance as one of the mechanisms associated with severe disease.42,43
From a clinical point of view, the majority of our severely ill patients presented alteration of consciousness, mainly in form of delirium, and myoclonus (Table 2). Both clinical manifestations are indeed associated with brainstem damage, especially myoclonus in COVID-19, thus providing convincing clinical correlates for possible damage mediated by the autoantibodies targeting the brainstem that we present here.44, 45, 46
Interestingly, our results may also bear relevance for neurological sequelae of COVID, including memory and attention deficits, dysautonomia, delusion and psychosis. Specifically, both brainstem damage and autoimmunity have been implicated in long-COVID.9,47 Accordingly, DAB1 and the Reelin pathway are necessary for synaptic plasticity and they are altered in schizophrenia.48,49 Alteration of AIMF1 and SURF1 impairs neurogenesis and cognition.50,51 This suggests involvement of auto-Abs against DAB1, AIFM1, and SURF1 and potential benefits of immunomodulation in long-COVID. In such a scenario of autoimmune neurological long-COVID, severely affected patients would present a more intense autoantibody production whereas a more moderate, chronic autoimmune response might underlie long-COVID and minimal to no response might characterise patients without respiratory symptoms, like the mild cases reported here. Such model would be in accordance with previous research showing correlation of cognitive deficits in long-COVID with severity of respiratory symptoms in mildly affected patients.52 Further studies specifically focussing on long-COVID are now warranted. Moreover, the intense neuroinflammatory response associated to both severe COVID and long-COVID might promote autoimmunity given that inflammatory cascades can promote an increase of specificity of the autoimmune adaptive response.53, 54, 55, 56
This study tested a precise hypothesis and showed the occurrence of autoantibodies binding exactly to the predicted epitopes that are shared between neuronal antigens and SARS-CoV-2. However, there are some limitations. The small sample size did not allow for subgroup analysis and future studies should examine specific demographic and clinical risk factors for the occurrence of the autoantibodies we showed here. Severely and moderately affected Patients were recruited over an overlapping period of time across 2020–21 although with a certain offset. Although such an offset might be thought as a potential temporal confounder, the two main factors influencing the immune response against SARS-CoV in the general population, namely variant circulation and vaccination, could not possibly affect the comparison presented here. First, the epitopes we present here are highly conserved among variants (Table 1) and, second, the vaccination campaign began at the very end of the recruitment period of the present study.57 Finally, the cross-sectional nature of the present study, that focussed primarily on severe disease, cannot reveal the dynamics of the autoimmune response. Longitudinal studies are needed in this direction, especially focussing on long-COVID.
In conclusion, this study showed that critically ill patients present higher levels of IgG against the three neuronal proteins DAB1, AIFM1, and SURF1 as compared to mild disease. These auto-Abs appear to be promising biomarkers of severe COVID and thus might allow for early treatment of high-risk patients before intensive care is needed. Finally, future research on newly discovered auto-Abs in long-COVID may open the door to effective and safe treatment of long-hauling neuropsychiatric consequences of the SARS-CoV-2 pandemic, given the role that the newly identified auto antigens play in cognitive processes.
Contributors
Conceptualization: GL, AV, AF. Methodology: GL, AV, FB, CF, AF. Investigation: GL, AV, FB, DR, SH, BB, HP, CF, AF. Visualization: GL, AS. Funding acquisition: RF, CF, AF. Project administration: GL, AV, FB, CF, AF. Supervision: CF, AF. Writing – original draft: GL, AV, DR, SH. Writing – review & editing: GL, AV, FB, DR, SH, BB, AS, RF, HP, CF, AF. The underlying data were verified by GL, AV, FB, CF and AF. All authors read and approved the final version of the manuscript.
Data sharing statement
All data are available upon request. No restrictions apply.
Declaration of interests
The Universitätsmedizin Greifswald has filed a patent application on immunomodulatory peptides for the treatment of SARS-CoV-2 infection, on which GL and AF are named as inventors.
FB, AV, DR, SH, BB, AS, RF, HP declare that they have no competing interests. CF declares honoraria for lectures from Boehringer Ingelheim, Pfizer, and Bristol-Myers-Squibb.
Acknowledgements
German Research Foundation grant FL 379/22-1 (AF). German Research Foundation grant 327654276-SFB 1315 (AF). German Research Foundation grant FR 4479/1-1(CF). German Research Foundation grant PR 1274/8-1 (HP). COVIDPROTECT: “Optimization of diagnostic and therapeutic pathways for COVID-19 patients in MV” funded by Ministry of Economy, State of Mecklenburg Western Pomerania, Germany (SH, DR, BB). Research Group Molecular Medicine University of Greifswald seed funding FOVB-2021-01 (SV). Else Kröner Fresenius Foundation and Alzheimer Research Initiative (AV).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104211.
Contributor Information
Guglielmo Lucchese, Email: guglielmo.lucchese@uni-greifswald.de.
Agnes Flöel, Email: agnes.floeel@med.uni-greifswald.de.
Appendix. Supplementary materials
References
- 1.Herrmann J, Mori V, Bates JHT, Suki B. Modeling lung perfusion abnormalities to explain early COVID-19 hypoxemia. Nat Commun. 2020;11:4883. doi: 10.1038/s41467-020-18672-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gattinoni L, Coppola S, Cressoni M, Busana M, Rossi S, Chiumello D. COVID-19 does not lead to a “typical” acute respiratory distress syndrome. Am J Respir Crit Care Med. 2020;201:1299–1300. doi: 10.1164/rccm.202003-0817LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hossain R, Lazarus MS, Roudenko A, et al. CT scans obtained for nonpulmonary indications: associated respiratory findings of COVID-19. Radiology. 2020;296:E173–E179. doi: 10.1148/radiol.2020201743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nath A. Neurologic complications of coronavirus infections. Neurology. 2020;94:809–810. doi: 10.1212/WNL.0000000000009455. [DOI] [PubMed] [Google Scholar]
- 5.Chang P, Tasch ES, Rapoport LN, Bakhadirov K. A post-infectious steroid-responsive brainstem lesion associated with COVID-19. Neurohospitalist. 2021;11:152–155. doi: 10.1177/1941874420959544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khoo A, McLoughlin B, Cheema S, et al. Postinfectious brainstem encephalitis associated with SARS-CoV-2. J. Neurol. Neurosurg Psychiatry. 2020;91:1013. doi: 10.1136/jnnp-2020-323816. [DOI] [PubMed] [Google Scholar]
- 7.Jeanneret V, Winkel D, Risman A, Shi H, Gombolay G. Post-infectious rhombencephalitis after coronavirus-19 infection: a case report and literature review. J Neuroimmunol. 2021;357 doi: 10.1016/j.jneuroim.2021.577623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology. 2020;296:E119. doi: 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spudich S, Nath A. Nervous system consequences of COVID-19. Science. 2022;375:267. doi: 10.1126/science.abm2052. [DOI] [PubMed] [Google Scholar]
- 10.Lucchese G. Cerebrospinal fluid findings in COVID-19 indicate autoimmunity. Lancet Microbe. 2020;1:e242. doi: 10.1016/S2666-5247(20)30147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimaldi S, Lagarde S, Harlé JR, Boucraut J, Guedj E. Autoimmune encephalitis concomitant with SARS-CoV-2 infection: insight from 18F-FDG PET imaging and neuronal autoantibodies. J Nucl Med. 2020;61:1726. doi: 10.2967/jnumed.120.249292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delamarre L, Gollion C, Grouteau G, et al. COVID-19–associated acute necrotising encephalopathy successfully treated with steroids and polyvalent immunoglobulin with unusual IgG targeting the cerebral fibre network. J Neurol Neurosurg Psychiatry. 2020;91:1004. doi: 10.1136/jnnp-2020-323678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pilotto A, Masciocchi S, Volonghi I, et al. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) encephalitis is a cytokine release syndrome: evidences from cerebrospinal fluid analyses. Clin Infect Dis. 2021;73:e3019. doi: 10.1093/cid/ciaa1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franke C, Ferse C, Kreye J, et al. High frequency of cerebrospinal fluid autoantibodies in COVID-19 patients with neurological symptoms. Brain Behav Immun. 2021;93:415. doi: 10.1016/j.bbi.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucchese G, Flöel A. Molecular mimicry between SARS-CoV-2 and respiratory pacemaker neurons. Autoimmun Rev. 2020;19 doi: 10.1016/j.autrev.2020.102556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willers J, Lucchese A, Mittelman A, Dummer R, Kanduc D. Definition of anti-tyrosinase MAb T311 linear determinant by proteome-based similarity analysis. Exp Dermatol. 2005;14:543–550. doi: 10.1111/j.0906-6705.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 17.Pastorino B, Touret F, Gilles M, Luciani L, de Lamballerie X. Charrel RN evaluation of chemical protocols for inactivating SARS-CoV-2 infectious samples. Viruses. 2020;12:624. doi: 10.3390/v12060624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Angeloni S., Das S., Dunbar S., Stone V., Swift S. Vol. 4. Luminex Corporation; 2018. (xMAP® Cookbook. A Collection of Methods and Protocols for Developing Multiplex Assays with xMAP® Technology). [Google Scholar]
- 19.Meyer TC, Schmidt F, Steinke J, Bröker BM, Völker U, Michalik S. Technical report: xMAPr - High-dynamic-range (HDR) quantification of antigen-specific antibody binding. J Prot. 2020;212 doi: 10.1016/j.jprot.2019.103577. [DOI] [PubMed] [Google Scholar]
- 20.Meyer TC, Michalik S, Holtfreter S, et al. A comprehensive view on the human antibody repertoire against staphylococcus aureus antigens in the general population. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.651619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz T, Heiss K, Mahendran Y, et al. M. SARS-CoV-2 Proteome-Wide Analysis Revealed Significant Epitope Signatures in COVID-19 Patients. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.629185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang SE, Feng A, Meng W, et al. New-onset IgG autoantibodies in hospitalized patients with COVID-19. Nat Commun. 2021;12:5417. doi: 10.1038/s41467-021-25509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- 24.Deguchi K, Inoue K, Avila WE, et al. Reelin and disabled-1 expression in developing and mature human cortical neurons. J Neuropathol Exp Neurol. 2003;62:676–684. doi: 10.1093/jnen/62.6.676. [DOI] [PubMed] [Google Scholar]
- 25.Tan W, Sherman D, Turesson J, Shao XM, Janczewski WA, Feldman JL. Reelin demarcates a subset of pre-Bötzinger complex neurons in adult rat. J Comp Neurol. 2012;520:606–619. doi: 10.1002/cne.22753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X, Hu Y, Zhou H, et al. Inhibition of progesterone receptor membrane component-1 exacerbates neonatal hypoxic-ischemic cerebral damage in male mice. Exp Neurol. 2022;347 doi: 10.1016/j.expneurol.2021.113893. [DOI] [PubMed] [Google Scholar]
- 27.Thomas N, Gurvich C, Kulkarni J. Sex differences and COVID-19. Adv Exp Med Biol. 2021;1327:79–91. doi: 10.1007/978-3-030-71697-4_6. [DOI] [PubMed] [Google Scholar]
- 28.Lake NJ, Compton AG, Rahman S, Thorburn DR. Leigh syndrome: one disorder, more than 75 monogenic causes. Ann Neurol. 2016;79:190–203. doi: 10.1002/ana.24551. [DOI] [PubMed] [Google Scholar]
- 29.Wu X, Wu W, Pan W, Wu L, Liu K, Zhang HL. Acute necrotizing encephalopathy: an underrecognized clinicoradiologic disorder. Mediators Inflamm. 2015;2015 doi: 10.1155/2015/792578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana A, Zanella S, Koch H, et al. Fatal breathing dysfunction in a mouse model of Leigh syndrome. J Clin Invest. 2012;122:2359–2368. doi: 10.1172/JCI62923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michaelson NM, Malhotra A, Wang Z, et al. Peripheral neurological complications during COVID-19: a single center experience. J Neurol Sci. 2021;434 doi: 10.1016/j.jns.2021.120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pareyson D, Piscosquito G, Moroni I, Salsano E, Zeviani M. Peripheral neuropathy in mitochondrial disorders. Lancet Neurol. 2013;12:1011–1024. doi: 10.1016/S1474-4422(13)70158-3. [DOI] [PubMed] [Google Scholar]
- 33.Vaishnav RA, Getchell ML, Huang L, Hersh MA, Stromberg AJ, Getchell TV. Cellular and molecular characterization of oxidative stress in olfactory epithelium of Harlequin mutant mouse. J Neurosci Res. 2008;86:165–182. doi: 10.1002/jnr.21464. [DOI] [PubMed] [Google Scholar]
- 34.Legros V, Denolly S, Vogrig M, et al. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol. 2021;18:318–327. doi: 10.1038/s41423-020-00588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang EY, Mao T, Klein J, et al. Diverse functional autoantibodies in patients with COVID-19. Nature. 2021;595:283–288. doi: 10.1038/s41586-021-03631-y. [DOI] [PubMed] [Google Scholar]
- 36.Hangen E, De Zio D, Bordi M, et al. A brain-specific isoform of mitochondrial apoptosis-inducing factor: AIF2. Cell Death Differ. 2010;17:1155. doi: 10.1038/cdd.2009.211. [DOI] [PubMed] [Google Scholar]
- 37.Delettre C, Yuste VJ, Moubarak RS, Bras M, Robert N, Susin SA. Identification and characterization of AIFsh2, a mitochondrial apoptosis-inducing factor (AIF) isoform with NADH oxidase activity. J Biol Chem. 2006;281:18507. doi: 10.1074/jbc.M601751200. [DOI] [PubMed] [Google Scholar]
- 38.Lobo PI. Role of natural autoantibodies and natural IgM anti-leucocyte autoantibodies in health and disease. Front Immunol. 2016;6:7–198. doi: 10.3389/fimmu.2016.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elkon K, Casali P. Nature and functions of autoantibodies. Nat Clin Pract Rheumatol. 2008;4:491–498. doi: 10.1038/ncprheum0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng J, James LC. Intracellular antibody immunity and its applications. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montaño J, Garnica J, Santamaria P. Immunomodulatory and immunoregulatory nanomedicines for autoimmunity. Semin Immunol. 56:101535. [DOI] [PubMed]
- 42.Manry J, Bastard P, Gervais A, et al. The risk of COVID-19 death is much greater and age dependent with type I IFN autoantibodies. Proc Natl Acad Sci USA. 2022;119 doi: 10.1073/pnas.2200413119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benghanem S, Mazeraud A, Azabou E, et al. Brainstem dysfunction in critically ill patients. Crit Care. 2020;24:5. doi: 10.1186/s13054-019-2718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rábano-Suárez P, Bermejo-Guerrero L, Méndez-Guerrero A, et al. Generalized myoclonus in COVID-19. Neurology. 2020;95:e767–e772. doi: 10.1212/WNL.0000000000009829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shetty K, Jadhav AM, Jayanthakumar R, et al. Myoclonus-ataxia syndrome associated with COVID-19. J Mov Disord. 2021;14:153–156. doi: 10.14802/jmd.20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yong SJ. Persistent brainstem dysfunction in Long-COVID: a hypothesis. ACS Chem Neurosci. 2021;12:573. doi: 10.1021/acschemneuro.0c00793. [DOI] [PubMed] [Google Scholar]
- 48.Trotter J, Lee GH, Kazdoba TM, et al. DAB1 is required for synaptic plasticity and associative learning. J Neurosci. 2013;33:15652–15668. doi: 10.1523/JNEUROSCI.2010-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fatemi SH, Earle JA, McMenomy T. Reduction in Reelin immunoreactivity in hippocampus of subjects with schizophrenia, bipolar disorder and major depression. Mol Psychiatry. 5:654–571. [DOI] [PubMed]
- 50.Inak G, Rybak-Wolf A, Lisowski P, et al. Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. Nat Commun. 2021;12:1929. doi: 10.1038/s41467-021-22117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khacho M, Clark A, Svoboda DS, et al. Mitochondrial dysfunction underlies cognitive defects as a result of neural stem cell depletion and impaired neurogenesis. Hum Mol Genet. 2017;26:3327. doi: 10.1093/hmg/ddx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hampshire A, Trender W, Chamberlain SR, et al. Cognitive deficits in people who have recovered from COVID-19. EClinicalMedicine. 2021;39 doi: 10.1016/j.eclinm.2021.101044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kassianidis G, Siampanos A, Poulakou G, et al. Calprotectin and Imbalances between acute-phase mediators are associated with critical illness in COVID-19. Int J Mol Sci. 2022;23:4894. doi: 10.3390/ijms23094894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theoharides TC. Could SARS-CoV-2 spike protein be responsible for long-COVID syndrome? Mol Neurobiol. 2022;59:1850–1861. doi: 10.1007/s12035-021-02696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burbelo PD, Iadarola MJ, Keller JM, Warner BM. Autoantibodies targeting intracellular and extracellular proteins in autoimmunity. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.548469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fletcher AJ, James LC. Coordinated neutralization and immune activation by the cytosolic antibody receptor TRIM21. J Virol. 2016;90:4856–4859. doi: 10.1128/JVI.00050-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siddiqui S, Alhamdi HWS, Alghamdi HA. Recent chronology of COVID-19 pandemic. Front Public Health. 2022;10 doi: 10.3389/fpubh.2022.778037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomasello G, Armenia I, Molla G. The protein imager: a full-featured online molecular viewer interface with server-side HQ-rendering capabilities. Bioinformatics. 2020;36:2909–2911. doi: 10.1093/bioinformatics/btaa009. [DOI] [PubMed] [Google Scholar]
- 59.Bienert S, Waterhouse A, de Beer TA, et al. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45:D313–D319. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.