Abstract
In the presence of tyrosine, the TyrR protein of Escherichia coli represses the expression of the tyrP gene by binding to the double TyrR boxes which overlap the promoter. Previously, we have carried out methylation, uracil, and ethylation interference experiments and have identified both guanine and thymine bases and phosphates within the TyrR box sequences that are contacted by the TyrR protein (J. S. Hwang, J. Yang, and A. J. Pittard, J. Bacteriol. 179:1051–1058, 1997). In this study, we have used missing contact probing to test the involvement of all of the bases within the tyrP operator in the binding of TyrR. Our results indicate that nearly all the bases within the palindromic arms of the strong and weak boxes are important for the binding of the TyrR protein. Two alanine-substituted mutant TyrR proteins, HA494 and TA495, were purified, and their binding affinities for the tyrP operator were measured by a gel shift assay. HA494 was shown to be completely defective in binding to the tyrP operator in vitro, while, in comparison with wild-Type TyrR, TA495 had only a small reduction in DNA binding. Missing contact probing was performed by using the purified TA495 protein, and the results suggest that T495 makes specific contacts with adenine and thymine bases at the ±5 positions in the TyrR boxes.
Regulation of gene expression in bacteria requires the binding of the regulatory protein to a specific DNA target which is termed an operator. In Escherichia coli K-12, the transcriptional expression of the tyrP gene, which codes for the tyrosine-specific transporter, is repressed by the TyrR regulatory protein in the presence of the cofactor tyrosine (21, 24). The tyrP operator partially overlaps the −35 region of the tyrP promoter and contains two adjacent TyrR binding sites, which are termed TyrR boxes (13). Each of the TyrR boxes is related to the palindrome TGTAAAN6TTTACA (consensus sequence). The upstream box of tyrP, which has a high degree of homology with the TyrR box consensus sequence and which binds TyrR in the absence of cofactors in vitro, has been referred to as the strong box (4, 13). The downstream TyrR box, which is less homologous to the TyrR box consensus sequence and binds TyrR only in the presence of the cofactors tyrosine and ATP and an adjacent strong TyrR box, has been referred to as a weak box (4, 13). The TyrR-mediated repression of tyrP expression requires the binding of TyrR protein to both the strong and weak TyrR boxes (4).
Although extensive studies have been carried out to elucidate the mechanism of TyrR-mediated repression and activation at tyrP as well as other promoters of the TyrR regulon, relatively little is known about the specific interactions between the TyrR protein and its DNA targets. The TyrR system differs from those of other classic repressors like TrpR in a number of ways: (i) the binding of the cofactor tyrosine causes the protein to self-associate to form a hexamer rather than directly modulating the tertiary structure of the DNA-binding domain; (ii) the hexamer but not the dimer is able to bind to a combination of strong and weak boxes; and (iii) the differences between a strong and a weak TyrR box reside in the central six bases (AT rich in strong boxes) and in the overall agreement with the consensus sequence. Therefore it is of considerable interest to map the important contact sites in both boxes. In a previous study, we have carried out in vitro methylation, uracil, and ethylation interference experiments and in vivo mutational studies to probe the bases and phosphates of the tyrP operator which are important for TyrR binding (11). We have shown that the guanine residues at the ±8 positions, the thymine residues at the ±7 positions, and the thymine residues at the +5 positions (in the top strand) in both TyrR boxes play a key role in making contacts with the TyrR protein (11). A number of contacts with the phosphate backbone have also been found at the end of and within the central regions of the TyrR boxes (11).
The DNA-binding domain of TyrR is located near the carboxyl-terminal end of the protein and is composed of a classic helix-turn-helix (HTH) motif (10, 27). An alanine scan of the HTH motif of the TyrR protein has identified amino acid residues in both helices, whose side chains are believed to be involved either in maintaining the correct conformation of the DNA-binding motif or in binding to the TyrR boxes (27). The residue arginine-484 in the first helix and the residues histidine-494, threonine-495, asparagine-499, and arginine-502 in the second helix are important for TyrR-mediated repression of tyrP expression (11, 27), and by using the coordinates of the lambda Cro and catabolite gene activator protein (CAP), they have been predicted to face DNA (2, 7, 16, 19). The side chains of these amino acid residues have thus been assumed to make contacts with the tyrP operator DNA.
The previous modification interference experiments did not allow the examination of any involvement of cytosine or adenine residues exposed in the major groove of the tyrP operator in the interactions with the TyrR protein. In this paper, we describe the use of missing contact probing (9) to test the involvement of all of the bases in the tyrP operator in TyrR binding. We also report an analysis of specific amino acid-base contacts between the TyrR protein and the tyrP operator, using missing contact probing with the mutant TyrR protein TyrR-TA495.
MATERIALS AND METHODS
Chemicals and Enzymes.
The enzymes used in this study were all purchased commercially. Dimethyl sulfate, formic acid (88%), hydrazine, and piperidine were purchased from Dupont-NEN. [α-32P]dATP and [α-32P]dGTP (3000 Ci/mmol; 10 mCi/ml) were purchased from Amersham.
Preparation of the mutant TyrR proteins.
Plasmids which allow expression of the mutant TyrR proteins TyrR-TA495 and TyrR-HA494 from the T7 promoter were constructed as described below. The M13tg131 derivatives which carry the mutant tyrR genes coding for TyrR-TA495 and TyrR-HA494 (11) were each mutagenized by using the oligonucleotide 5′CGCATGGGATCCTTCACC3′ to create a BamHI site immediately upstream of the start codon of tyrR. The resultant phagemids were each digested with BamHI, and the 2.3-kb DNA fragments containing the mutant tyrR genes were purified and ligated into the expression vector pET15b (Novagen, Madison, Wis.). The resultant pET15b derivatives were each transformed into E. coli BL21(DE3) (Novagen). The mutant TyrR proteins were overexpressed and purified as N-terminal fusions with an oligohistidine tag. The conditions and procedures for overexpression and purification of the mutant TyrR proteins were set up as described in the technical manual supplied by Novagen.
Preparation of labelled DNA fragments.
The 151-bp BamHI-HindIII fragment containing the tyrP regulatory region was generated by PCR using mpMU330 as a template. Following sequence verification, this PCR fragment was cloned into the BamHI and HindIII sites of pUC19. The tyrP fragment was labelled at the top (or the bottom) strand by digesting the pUC19 derivative with BamHI (or HindIII) and by filling in the restriction end with the Klenow fragment of DNA polymerase in the presence of [α-32P]dGTP (or [α-32P]dATP). The labelled DNA was precipitated with ethanol and digested with HindIII (or BamHI). The labelled tyrP fragment was then purified on a 5% polyacrylamide gel.
Missing contact probing.
Depurination (A+G) and depyrimidation (C+T) reactions were carried out as described by Brunelle and Schleif (9). Each reaction mixture contained approximately 105 cpm of the end-labelled tyrP fragment. The modified DNA fragments were each incubated with the wild-type or the mutant TyrR protein (TA494) in a footprinting buffer containing 5 mM Tris-HCl (pH 7.6), 80 mM KCl, 8 mM MgCl2, 1 mM dithiothreitol, 0.2 mM ATP, 1 mM tyrosine, and 4% (vol/vol) glycerol. The concentration of the TyrR protein was determined empirically such that the amounts of bound and unbound fractions of the modified DNA were approximately equal. Following incubation for 20 min at 37°C, the TyrR-DNA complexes were separated from the free DNA by electrophoresis on a 5% polyacrylamide gel in 50 mM Tris-borate (pH 7.0)–1 mM MgCl2–0.2 mM ATP–1 mM tyrosine at 4°C. The gel was autoradiographed for 2 h at 4°C, and DNA bands corresponding to the bound and unbound fractions were eluted from the gel by a gel eluter (The Australian Chromatography Company–Hoefer). About 3 × 104 cpm of the DNA sample recovered from the bound or unbound fraction was resuspended in 30 μl of 0.5 M piperidine and heated for 20 min at 95°C. After freeze-drying, the piperidine-treated DNA samples were electrophoresed on a 6% polyacrylamide sequencing gel.
Gel shift assay.
The end-labelled tyrP fragment described above was used in a gel shift assay to determine the binding affinities of the wild-type and mutant TyrR proteins for the tyrP operator. The binding reactions were carried out at 37°C for 20 min in the footprinting buffer (as described above) in the presence of 0.5 pmol of the tyrP fragment and various nanomolar amounts of the wild-type and mutant TyrR proteins. The samples were then analyzed on a 5% polyacrylamide gel.
RESULTS
Effects of depurination and depyrimidation of the tyrP operator on binding by the wild-type TyrR protein.
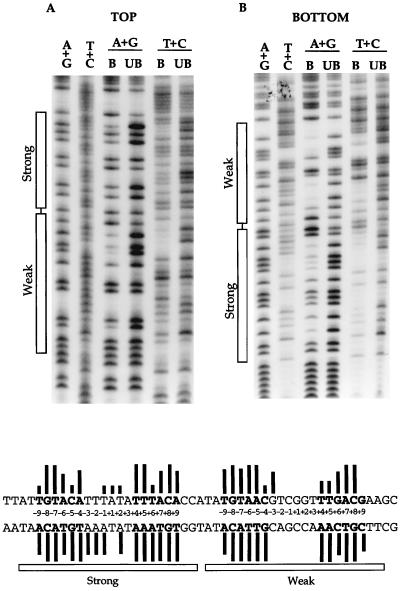
The missing contact experiments were performed by using a 151-bp BamHI-HindIII fragment carrying the tyrP operator region. The 3′ end of either the top or the bottom strand of this DNA fragment was labelled with 32P by filling in the restriction end with Klenow enzyme in the presence of [α-32P]dGTP or [α-32P]dATP. Each of the labelled fragments was then subjected to A+G depurination or T+C depyrimidation under conditions where, on average, each molecule contained less than one missing base (see Materials and Methods). The modified DNA fragments were each mixed with wild-type TyrR protein in the presence of ATP and tyrosine to allow the formation of protein-DNA complexes, and the reaction mixtures were then subjected to a mobility gel shift assay. The bound and free DNA fractions were recovered from the gel, cleaved at the phosphate backbone where a base had been eliminated, and examined on a sequencing gel.
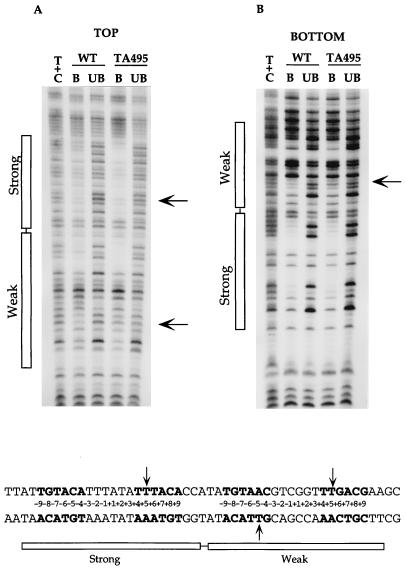
The important base contacts for TyrR on the top and bottom strands are shown in Fig. 1, and the data are also quantitatively summarized below the autoradiographs. Data from the top strand (Fig. 1A) indicate that depurination or depyrimidation at any one of the positions of the palindromic sequences, −8G, −7T, −6A, −4A, +4T, +5T, +7A, +8C, and +9A of the strong box and −9T, −8G, −7T, −6A, −5A, −3G, +6G, +7A, and +8C of the weak box, significantly interferes with the binding of the TyrR protein. In addition, weak but detectable signals were also identified from the bases at the palindromic positions −9T, −5C, and +6T of the strong box and at positions −4C, +4T, and +5T of the weak box. By examining the pattern of the bottom strand (Fig. 1B), it can be seen that removal of any base in the palindromic sequence of either the strong or the weak box can interfere with the binding of the TyrR protein. The depurination in the central region of the strong box also had a significant effect on the binding of the TyrR protein. At the center of the strong box, effects of missing contact were identified at positions −1T, +1A, and +2T in the top strand and positions −3A, −2A, −1A, and +2A in the bottom strand. In contrast, at the center of the weak box, only one effect was found at position −3G in the top strand. Several sites showing stronger bands in the bound rather than the free DNA fractions were observed at positions which flank the palindromic sequences of each of the two TyrR boxes, and therefore it appears that the removal of a base at any of these positions favors the formation of a protein-DNA complex.
FIG. 1.
Effects of depurination or depyrimidation on binding of the wild-type TyrR protein to the tyrP operator. Boxes indicate the regions corresponding to the strong and weak TyrR boxes. The first two lanes in each autoradiogram display the Maxam-Gilbert sequencing ladders. A+G and T+C, depurination and depyrimidation reactions, respectively; B, bound lane derived from the TyrR-DNA complex; UB, unbound lane derived from the free DNA. Results shown are from experiments using the DNA fragment labelled in the top (A) and bottom (B) strands. The sequence shown at the bottom corresponds to the sequence shown in the autoradiograms. The relative strength of interference due to the removal of any base is illustrated by the length of the respective bar. The palindromic sequences of the strong and the weak boxes are shown in boldface, and the positions in the boxes are indicated by the numbers between the two strands.
Determination of binding affinities of the mutant TyrR proteins HA494 and TA495 for the tyrP operator by mobility gel shift assay.
The amino acid residues histidine-494 and threonine-495 are situated at positions 1 and 2 in the second helix of the HTH DNA-binding motif of TyrR (27). These residues have been studied by mutational analysis and are believed to interact with specific bases within the TyrR boxes (11, 27). The alanine-substituted mutants HA494 and TA495 have previously been constructed, and it has been shown that HA494 was completely unable to repress the tyrP promoter, while TA495 was only partially defective in repression (11). In order to study the effects of these substitutions in vitro, TyrR proteins with these changes were purified as N-terminal fusions with oligohistidine domains by using pET15b as the expression vector (for details, see Materials and Methods). As the N-terminal domain of TyrR is primarily involved in interactions with RNA polymerase in transcription activation (25, 26), the histidine tag at the N terminus of TyrR is not expected to affect the DNA-binding function of the protein. The mutant proteins with oligohistidine domains were therefore used directly in mobility gel shift assays without protease cleavage.
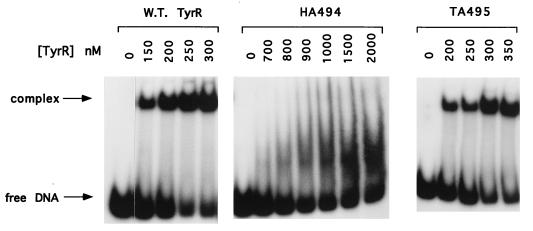
In this experiment, the 151-bp tyrP operator fragment described in the previous section was used. Approximately 0.5 pmol of the end-labelled fragment was incubated with various amounts of each of the mutant TyrR proteins as well as the wild-type TyrR protein in the presence of tyrosine and ATP. Under this assay condition, the TyrR proteins are expected to form hexamers in solution (14, 22). With the wild-type TyrR protein, the amount of the retarded band representing the TyrR-DNA complex increases as the TyrR concentration is increased (Fig. 2). Judged by the concentration of the wild-type TyrR, at which half of the total DNA is present in the retarded complex, the dissociation constant is estimated to be approximately 200 nM. For the mutant protein HA494, no retarded band was observed at a concentration of 700 nM (Fig. 2). Increasing the protein concentration to 2 μM did not improve the specific binding, indicating that the affinity of TyrR protein for the tyrP operator was lost when histidine-494 was replaced by alanine. For the mutant protein TA495, the specific binding affinity for the tyrP operator was only slightly decreased (Fig. 2). The alanine substitution at position 495 caused an increase in the dissociation constant of TyrR from 200 nM to slightly less than 300 nM. These in vitro results are in agreement with the previous in vivo observation that histidine-494 plays a critical role in DNA binding whereas the involvement of threonine-495 in DNA binding is less important (11).
FIG. 2.
Gel shift assay. The gel shift assay was carried out by using the 151-bp tyrP fragment in the presence of the wild-type TyrR protein, mutant protein HA494, or mutant protein TA495. Various concentrations of the proteins are shown above the autoradiograms.
Rationale for missing contact probing with the mutant TyrR protein TA495.
Missing contact probing has been successfully used in determining specific amino acid-base contacts (8, 9). In this method, an amino acid residue which is known to contact DNA is replaced with a smaller amino acid such as alanine. Unlike the wild-type protein, the alanine-substituted protein is unable to discriminate between the wild-type DNA and the DNA with a base missing at the site contacted by the original amino acid residue. Therefore a comparison of the footprinting patterns generated in the presence of the wild-type protein and the mutant protein may allow the identification of an individual interaction between a specific base and an amino acid residue.
A critical aspect of this method is that the alanine mutant must still retain its ability to bind DNA even though the formation of the protein-DNA complex requires an increased concentration of this protein. As shown in the previous section, unlike the mutant protein HA494, which was completely inactive in DNA binding, the mutant protein TA495, in spite of a weakened binding affinity for the tyrP operator, was still able to form a protein-DNA complex. Therefore TA495 was chosen for use in the missing contact probing experiments.
Depurination probing.
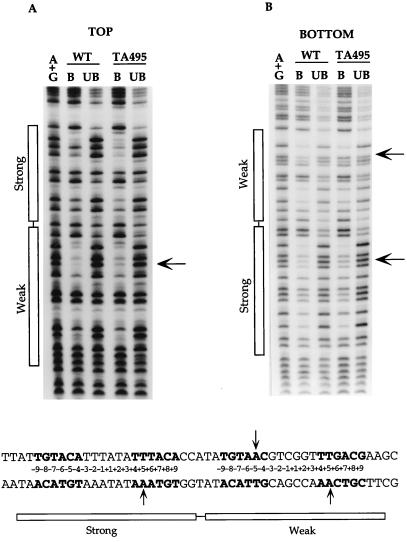
The missing contact experiments were carried out as described above. Results from the depurination (A+G) analysis are shown in Fig. 3. A visual comparison of the footprinting patterns generated in the presence of the wild-type and mutant proteins identified three possible contact sites. These are the adenine residues at the +5 positions of both the strong and weak TyrR boxes in the bottom strand as well as the adenine residue at the −5 position of the weak box in the top strand.
FIG. 3.
Depurination (A+G) analysis using the wild-type TyrR protein and the mutant TyrR protein TA495. Designations are as described in the legend to Fig. 1. Arrows indicate positions where interference is significant with the wild-type TyrR protein but is weaker or has disappeared with the mutant protein TA495. A summary of the data is shown below the autoradiograms.
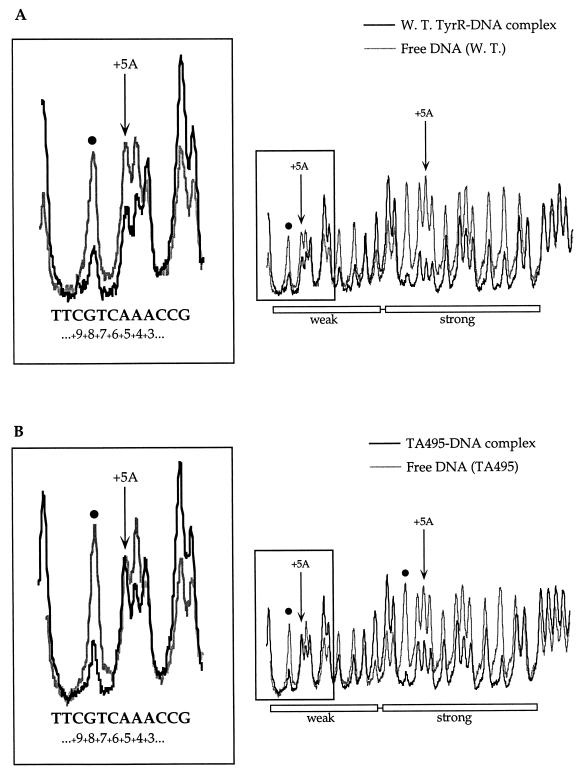
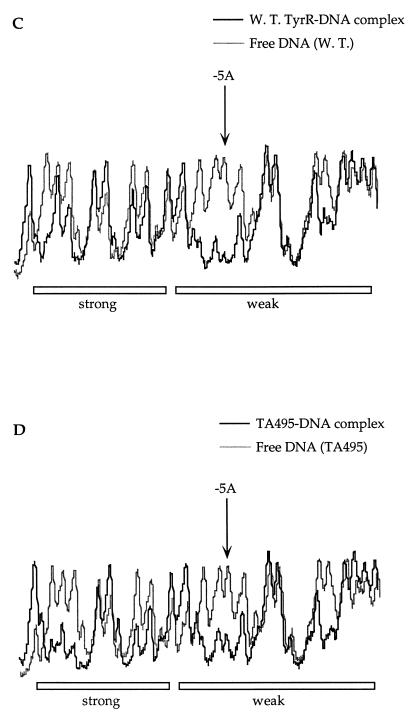
To obtain a better resolution of the results, densitometric scans of the autoradiographs were performed. The scan data shown in the boxes in Fig. 4A and B are focused on a small region including positions +9 and +3 of the bottom strand in the weak box. In this region, only the purine residues G--AAA--G can be seen. The fragment derived from the deletion of guanine at position 8 shows a similar displacement to the unbound fraction in both the wild-type (Fig. 4A) and mutant (Fig. 4B) proteins. Although the displacement is less, the same consistent effect is seen with the fragment arising from depurination at position +4. The adenine at position +3 has been shown not to be important for TyrR binding (11), and this is confirmed by inspection of the results involving both the wild-type and mutant proteins, in which there is an almost equal distribution of DNA in both the bound and free fractions. If one now considers the adenine at position +5, a quite different picture emerges. Whereas the binding of the wild-type protein to the DNA is severely affected by depurination at this position, in the case of the mutant, the same fragment is equally distributed between the bound and free fractions. These results suggest a possible specific interaction between threonine-495 of the TyrR protein and the adenine at position +5 of in the weak box.
FIG. 4.
Densitometric scans of the data shown in Fig. 3. Dark and light lines, scans of the protein-DNA complex from the B lanes and of the free DNA from the UB lanes, respectively. Arrows indicate positions where interference is seen for wild-type TyrR but is weaker or has disappeared for TA495. The locations of the strong and weak TyrR boxes are indicated. (A) Samples from the depurination reactions carried out in the presence of wild-type TyrR protein and the tyrP fragment labelled in the bottom strand. (B) Samples from the depurination reactions carried out in the presence of the TA495 protein and the tyrP fragment labelled in the bottom strand. (C) Samples from the depurination reactions carried out in the presence of wild-type TyrR protein and the tyrP fragment labelled in the top strand. (D) Samples from the depurination reactions carried out in the presence of the TA495 protein and the tyrP fragment labelled in the top strand.
The data from this scan also show significant but less-marked changes at position +5 in the bottom strand of the strong box (Fig. 4A and B) and at position −5 in the top strand of the weak box (Fig. 4C and D). The lesser effects might be considered a result of the involvement of these particular bases in either making more than one contact with the protein or directing a local DNA distortion. The position −5 in the bottom strand of the strong box is occupied by a guanine residue and shows no effect (Fig. 4A and B).
Depyrimidation probing.
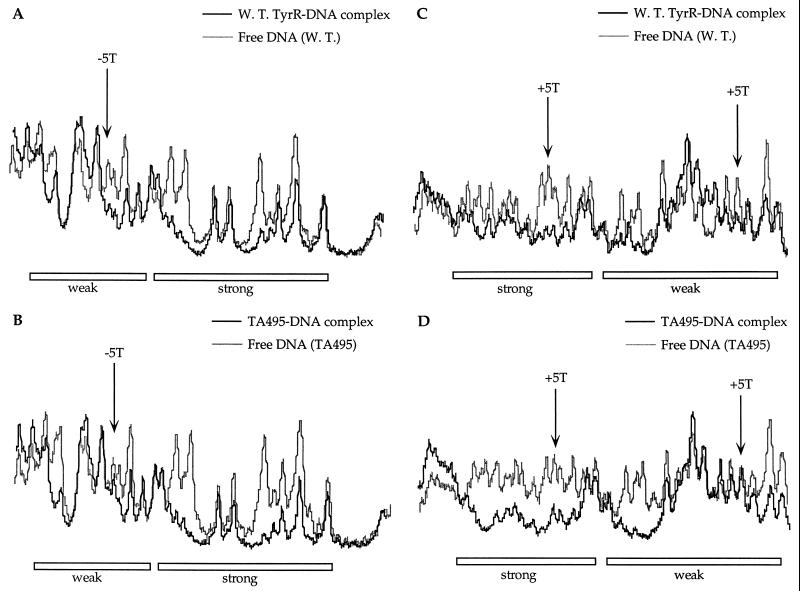
The results of depyrimidation probing and the data from densitometric scans of the autoradiographs are shown in Fig. 5 and 6, respectively. In the bottom strand, depyrimidation of the thymine residue at position −5 of the weak box has a greater effect on the binding of the wild-type protein than on the binding of the mutant protein TA495 (Fig. 6A and B). This indicates that unlike the wild-type protein, the mutant TA495 protein cannot discriminate between the presence and absence of a thymine at this position. In the top strand, deletion of thymine at the +5 positions of both the strong and weak boxes also resulted in a similar effect, but the effect of missing contact is shown to be more remarkable in the weak box than in the strong box (Fig. 6C and D).
FIG. 5.
Depyrimidation (C+T) analysis using the wild-type TyrR protein and the mutant TyrR protein TA495. Designations are as described in the legend to Fig. 1.
FIG. 6.
Densitometric scans of protein-DNA complexes (B lanes) and free DNA (UB lanes) in Fig. 5. (A) Samples from the depyrimidation reactions carried out in the presence of the wild-type TyrR protein and the tyrP fragment labelled in the bottom strand. (B) Samples from the depyrimidation reactions carried out in the presence of the TA495 protein and the tyrP fragment labelled in the bottom strand. (C) Samples from the depyrimidation reactions carried out in the presence of the wild-type TyrR protein and the tyrP fragment labelled in the top strand. (D) Samples from the depyrimidation reactions carried out in the presence of the TA495 protein and the tyrP fragment labelled in the top strand.
DISCUSSION
Implications of the results from missing contact footprinting using wild-type TyrR protein.
Previous studies of the tyrP operator have involved site-directed mutagenesis of the strong TyrR box and methylation and ethylation interference and uracil substitution experiments to investigate the binding of the TyrR protein to both boxes. The mutagenesis studies have implicated the invariant (G · C)(C · G)8 symmetrical base pairs as playing a critical role (11) (the nomenclature for the symmetrical base pairs is as follows: the letters in the first set of parentheses denote the base pair in the left arm of the palindrome, and the letters in the second set of parentheses represent the base pair in the right arm of the palindrome; the number indicates the symmetrical positions). Results from these studies have also shown that the (T · A)(A · T)7 symmetrical base pairs are of major importance in TyrR binding (11). Methylation interference studies have confirmed the importance of the guanine residues at the ±8 positions in both boxes and have indicated that the guanine residues at the −5 position in the strong box and the −4 position in the weak box are also necessary for TyrR binding (11). Uracil substitution experiments have revealed important van der Waals interactions with thymines at positions −7 and +5 of both boxes and weaker interactions at the +7 positions of both boxes and at the −5 position of the weak box (11). Ethylation interference experiments have indicated that contacts between the TyrR protein and the phosphate backbone occur both at the ends and in the central regions of the two boxes (11). The limitation of these methods was their inability to detect interactions with adenine residues via the major groove and to identify any interactions with cytosine residues. The missing contact studies reported in this paper overcome these difficulties. Although we cannot rule out the possibility that some structural changes caused by the removal of certain bases are responsible for differences in TyrR binding, it seems likely, taking into account our previous genetic results, that the majority of the bases in the palindromic arms of each of the TyrR boxes are involved in contacts with the TyrR protein. In addition to confirming previous observations, this work also shows that elimination of cytosine residues at the ±8 positions from either box has a major effect on binding between the TyrR protein and the DNA (Fig. 1). Similarly, removal of adenine residues from their positions within the palindromic arms of each of the boxes causes major shifts of the tyrP fragments towards the unbound fraction (Fig. 1). Since these effects were not seen with the previously reported methylation interference studies, we must assume that these interactions with adenine occur via the major groove at the N7 or the NH6 functional group. Removal of the nonconsensus base C at the −5 position of the strong box and at the −4 position of the weak box has only slight effects on binding, as does the removal of the consensus base T at the +6 position of the strong box (Fig. 1). At first, the failure to observe a more significant effect at the −5 position in the top strand of the strong box was unexpected, since mutational studies had suggested that the (C · G)(G · C)5 base pairs were an efficient replacement for the (A · T)(T · A)5 base pairs at these symmetrical positions (11). However, a strong effect is seen at −5 in the bottom strand. This may imply that the importance of (C · G)(G · C)5 base pairs shown by mutational studies could be due to a critical role that is played by G at this position. Methylation interference showed that the G at −5 in the strong box is a contact site (11). The results from missing contact probing with the mutant protein TA495 described in this paper suggest that the −5G in the strong box is contacted by an amino acid residue(s) other than T495.
The strong TyrR boxes of various genes of the TyrR regulon contain AT-rich sequences between their palindromic arms (18), and it is of interest that, in the case of the strong box of tyrP, removal of any of the four bottom-strand adenine residues in this location has a significant effect on binding (Fig. 1). No effect or interaction is seen with equivalent positions in the weak box. A strong TyrR box can be bound either by a TyrR dimer or by a TyrR hexamer, but a weak TyrR box can be bound by the hexamer only when a strong box is nearby and on the same face of the helix. Ethylation interference studies have already shown that the central region of the strong box appears to be more intimately associated with the protein than the corresponding region of the weak box (11). The depurination studies confirm this important difference, with depurination of the central region of the strong box, but not the weak box, significantly affecting binding. Subtle differences can also be seen in the relative effects at positions +4 and +5 of the strong and weak boxes. In addition, there is a strong effect at −3 in the top strand of the weak box which is not present in the strong box. Taken together, these may indicate that although, in general, similar bases in both boxes are involved in critical interactions with the TyrR protein, subtle but important differences exist in the relationship of the protein to each box.
Removal of bases from the +9 position of the strong box and the −9 position of the weak box had a major impact on TyrR binding (Fig. 1). These two positions are located in the interior region of the operator, and their removal may have an effect on the structure of the operator. Previous studies involving ethylation interference (11) and fluorescent probes (5) have indicated that base pairs at positions ±9 are in close proximity to the TyrR protein, but mutational studies have suggested that these base pairs play a less significant role in TyrR binding (11).
Deletion of bases immediately outside the palindromic arms of the two boxes appears to facilitate binding of the TyrR protein to the operator (Fig. 1A and B). Similar effects were also seen when the internal bases +3T of the top strand of the weak box and −1G of the bottom strand of the weak box were removed (Fig. 1A and B). In all these cases we assume that the deletion alters DNA structure in a way which enhances the binding affinity of TyrR, perhaps by increasing flexibility.
Specific interactions between threonine-494 of TyrR and base pairs at positions ±5 of the TyrR boxes.
From the results of missing contact probing with the mutant TyrR protein TA494, it appears that the threonine at position 495 in TyrR makes specific contacts with adenine and thymine residues at the ±5 positions in the TyrR boxes. The weak box contains symmetrical base pairs (A · T)(T · A) at positions ±5, whereas the strong box contains asymmetrical base pairs (A · T)(G · C) at the equivalent positions.
Previous genetic studies have shown that changing the threonine residue at position 494 into a serine causes a slight increase in the DNA-binding activity of the TyrR protein (27), whereas an alanine replacement at this position results in a significant reduction in the binding activity (11). This indicates that it is the hydroxyl group but not the methyl group of T495 that plays an important role in DNA binding. Direct contacts can occur through hydrogen bonding between the hydroxyl group of T494 and the N6 of the adenine or the O4 of thymine.
Youderian and coworkers have carried out extensive genetic studies to investigate the importance of the threonine residue at position 81 of the Trp repressor in DNA binding (6, 17). Their results have suggested a direct contact between T81 of the Trp repressor and the symmetrical base pairs (G · C)(C · G)3 and (A · T)(T · A)4 of the trp operator. In this case, it has been proposed that the hydroxyl group of T81 forms a hydrogen bond with the O4 group of the thymine residues at the ±4 positions and that the methyl group makes a hydrophobic contact with the C-5 atom of the cytosine residues at the ±3 positions (6).
A direct contact between a threonine residue in the DNA-binding domain of a regulatory protein and A · T base pairs in an operator has also been found in the PurR-DNA crystal structure (20). In this complex, the Oγ of T15, which is the first residue in the recognition helix, hydrogen bonds to the O4 of thymine at position 7′ via a water interface, and the Oγ of T16 hydrogen bonds simultaneously to the N6 of adenine and the O4 of thymine at position 6.
A model of specific interactions has been proposed for the invariant (G · C)(G · C)8 base pairs, where the side chains of H494 and R484 of TyrR donate hydrogen bonds to the O6 and N7 of the guanine, respectively (11). This proposal, together with the assumption of the specific contact of T495 as described above, allows the prediction of the orientation of the recognition helix of the TyrR protein on the 22-bp palindromic sequence of the TyrR boxes. This prediction positions the recognition helices of a TyrR dimer on the operator in same orientation as those for the λ repressor (12), the λ Cro repressor (3, 7), CAP (19), and the 434 repressor (1) and in the opposite orientation to those for the Lac repressor (15), the PurR repressor (20), and the Tet repressor (23).
ACKNOWLEDGMENTS
This work was supported by a grant from the Australian Research Council. J. S. Hwang is the recipient of an Overseas Postgraduate Research Scholarship from the Australian Government and a Malaysian Alumni Melbourne University Postgraduate Scholarship.
We thank Jing Hong An, Yan Jiang, and Thu Betteridge for technical assistance and Barrie Davidson and coworkers for wild-type TyrR protein.
REFERENCES
- 1.Aggarwal A K, Rodgers D W, Drottar M, Ptashne M, Harrison S C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 2.Albright R A, Matthews B W. Crystal structure of lambda-Cro bound to a consensus operator at 3.0 Å resolution. J Mol Biol. 1998;280:137–151. doi: 10.1006/jmbi.1998.1848. [DOI] [PubMed] [Google Scholar]
- 3.Albright R A, Matthews B W. How Cro and lambda-repressor distinguish between operators: the structural basis underlying a genetic switch. Proc Natl Acad Sci USA. 1998;95:3431–3436. doi: 10.1073/pnas.95.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews A E, Lawley B, Pittard A J. Mutational analysis of repression and activation of the tyrP gene in Escherichia coli. J Bacteriol. 1991;173:5068–5078. doi: 10.1128/jb.173.16.5068-5078.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey M, Hagmar P, Millar D P, Davidson B E, Tong G, Haralambidis J, Sawyer W H. Interaction between the Escherichia coli regulatory protein TyrR and DNA: a fluorescence footprinting study. Biochemistry. 1995;34:15802–15812. doi: 10.1021/bi00048a026. [DOI] [PubMed] [Google Scholar]
- 6.Bass S, Sorrells V, Youderian P. Mutant Trp-repressors with new DNA-binding specificities. Science. 1988;242:240–245. doi: 10.1126/science.3140377. [DOI] [PubMed] [Google Scholar]
- 7.Brennan R G, Roderick S L, Takeda Y, Matthews B W. Protein-DNA conformational changes in the crystal structure of a λ Cro-operator complex. Proc Natl Acad Sci USA. 1990;87:8165–8169. doi: 10.1073/pnas.87.20.8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunelle A, Schleif R. Determining residue-base interactions between AraC protein and araI DNA. J Mol Biol. 1989;209:607–622. doi: 10.1016/0022-2836(89)90598-6. [DOI] [PubMed] [Google Scholar]
- 9.Brunelle A, Schleif R F. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci USA. 1987;84:6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Somerville R L. The TyrR protein of Escherichia coli, analysis by limited proteolysis of domain structure and ligand-mediated conformational changes. J Biol Chem. 1993;268:5040–5047. [PubMed] [Google Scholar]
- 11.Hwang J S, Yang J, Pittard A J. Critical base pairs and amino acid residues for protein-DNA interaction between the TyrR protein and tyrP operator of Escherichia coli. J Bacteriol. 1997;179:1051–1058. doi: 10.1128/jb.179.4.1051-1058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jordan S R, Pabo C O. Structure of the lambda complex at 2.5 Å resolution: details of the repressor-operator interaction. Science. 1988;242:893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- 13.Kasian P A, Davidson B E, Pittard J. Molecular analysis of the promoter operator region of the Escherichia coli K-12 tyrP gene. J Bacteriol. 1986;167:556–561. doi: 10.1128/jb.167.2.556-561.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwok T, Yang J, Pittard A J, Wilson T J, Davidson B E. Analysis of an Escherichia coli mutant TyrR protein with impaired capacity for tyrosine-mediated repression, but still able to activate at ς70 promoters. Mol Microbiol. 1995;17:471–481. doi: 10.1111/j.1365-2958.1995.mmi_17030471.x. [DOI] [PubMed] [Google Scholar]
- 15.Lehming N, Sartorius J, Oehler S, von Wilcken-Bergmann B, Müller-Hill B. Recognition helices of lac and λ repressors are oriented in opposite directions and recognize similar DNA sequences. Proc Natl Acad Sci USA. 1988;85:7947–7951. doi: 10.1073/pnas.85.21.7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson G, Wilson C, Gunasekera A, Ebright Y W, Ebright R E, Berman H M. Structure of the CAP-DNA complex at 2.5 Å resolution: a complete picture of the protein-DNA interface. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 17.Pfau J, Arvidson D N, Youderian P. Mutants of Escherichia coli Trp repressor with changes of conserved, helix-turn-helix residue threonine 81 have altered DNA-binding specificities. Mol Microbiol. 1994;13:1001–1012. doi: 10.1111/j.1365-2958.1994.tb00491.x. [DOI] [PubMed] [Google Scholar]
- 18.Pittard A J, Davidson B E. TyrR protein of Escherichia coli and its role as repressor and activator. Mol Microbiol. 1991;5:1585–1592. doi: 10.1111/j.1365-2958.1991.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 19.Schultz S C, Smelds G C, Steitz T A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90°. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 20.Schumacher M A, Choi K Y, Zalkin H, Brennan R G. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by α helices. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 21.Whipp M J, Halsall D M, Pittard A J. Isolation and characterization of an Escherichia coli mutant defective in tyrosine- and phenylalanine-specific transport systems. J Bacteriol. 1980;143:1–7. doi: 10.1128/jb.143.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson T J, Maroudas P, Howlett G J, Davidson B E. Ligand-induced self-association of the Escherichia coli regulatory protein TyrR. J Mol Biol. 1994;238:309–318. doi: 10.1006/jmbi.1994.1294. [DOI] [PubMed] [Google Scholar]
- 23.Wissmann A, Baumeister R, Müller G, Hecht B, Helbl V, Pfleiderer K, Hillen W. Amino acids determining operator binding specificity in the helix-turn-helix motif of Tn10 Tet repressor. EMBO J. 1991;10:4145–4152. doi: 10.1002/j.1460-2075.1991.tb04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wookey P J, Pittard J, Forrest S M, Davidson B E. Cloning of the tyrP gene and further characterization of the tyrosine-specific transport system in Escherichia coli K-12. J Bacteriol. 1984;160:169–174. doi: 10.1128/jb.160.1.169-174.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Camakaris H, Pittard A J. Mutations in the tyrR gene of Escherichia coli which affect TyrR-mediated activation but not TyrR-mediated repression. J Bacteriol. 1993;175:6372–6375. doi: 10.1128/jb.175.19.6372-6375.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, Camakaris H, Pittard A J. Further genetic analysis of the activation function of the TyrR regulatory protein of Escherichia coli. J Bacteriol. 1996;178:1120–1125. doi: 10.1128/jb.178.4.1120-1125.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Ganesan S, Sarsero J, Pittard A J. A genetic analysis of various functions of the TyrR protein of Escherichia coli. J Bacteriol. 1993;175:1767–1776. doi: 10.1128/jb.175.6.1767-1776.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]