Abstract
The persistence of latent reservoir of the human immunodeficiency virus (HIV) is currently the major challenge in curing HIV infection. After HIV infects the human body, the latent HIV is unable to be recognized by the body’s immune system. Currently, the widely adopted antiretroviral therapy (ART) is also unble to eliminate it, thus hindering the progress of HIV treatment. This review discusses the existence of latent HIV vault for HIV treatment, its formation and factors affecting its formation, cell, and tissue localization, methods for detection and removing latent reservoir, to provide a comprehensive understanding of latent HIV vault, in order to assist in the future research and play a potential role in achieving HIV treatment.
Keywords: reservoir of latent HIV, formation, cell reservoir, tissue reservoir, detection, remove
Introduction
HIV is a global health problem. At present, nearly 37 million people around the world are living with HIV infection (Ortblad et al., 2013). HIV evades the host immunity in many ways after infection (Theys et al., 2018; Beitari et al., 2019; Yin et al., 2020), among which the most common is through the establishment of latent reservoir of the HIV (Sengupta and Siliciano, 2018; Pedro et al., 2019). After HIV infects activated CD4+T lymphocytes, it produces a large number of viral RNA and viral proteins, which are eventually recognized and killed by the host immune system (van Zyl et al., 2018). Subsequently, some HIV after integration into the host cells mainly exist in CD4+T cells of resting memory, and constitute the latent reservoir of HIV (Bruner et al., 2019). These cells carry the integrated latent protovirus and exist through homeostasis or antigen-driven proliferation (Shan et al., 2017; Rezaei et al., 2018). At present, ART control the HIV level in people living with HIV (PWLH) below detection line, which plays an important role in the process of antiviral therapy (Lu et al., 2018). However, due to the existence of a latent HIV reservoir, ART cannot eliminate the latent virus in the reservoir, and HIV will rebound once the treatment is stopped (Xiao et al., 2019; Cohn et al., 2020). HIV cannot be completely cured, although some breakthroughs have been made in the research field of latent HIV reservoir. However, its specific mechanism of action is not completely clear. This review provides insight into the formation, location, detection and elimination of latent HIV reservoir.
Formation of latent HIV reservoir and its influencing factors
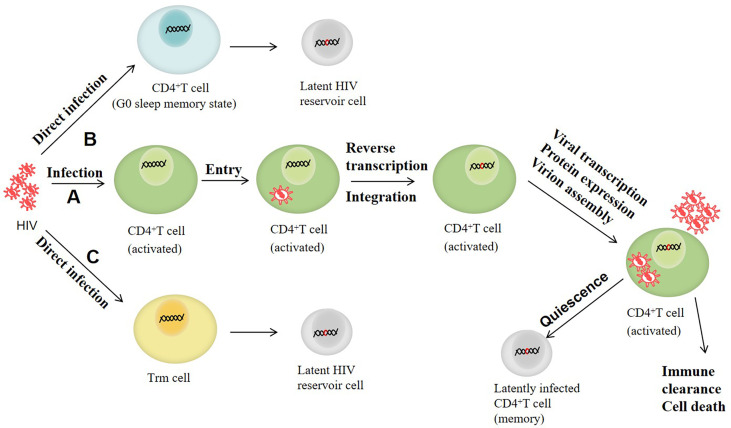
The reservoir of latent HIV can be established in the early stage of infection and is mainly composed of CD4+T cells with resting memory (Moranguinho and Valente, 2020). After HIV enters the human body, it mainly infects human CD4+T lymphocytes. The RNA is first reversely transcribed into HIV DNA, which is then integrated into the DNA of CD4+T cells, part of which transforms into a resting state in oder to inhibit the viral gene expression. The HIV in these resting CD4+T cells becomes the latent HIV, which is the main formation mode of the latent HIV reservoir at present (García et al., 2020). In addition, with the advent of research, other mechanisms for the establishment of latent HIV reservoirs have also been proposed. For instance, HIV can directly infect CD4+T cells that revert to a G0 dormant memory state, thus enabling the virus to enter latency (García et al., 2018). It has been suggested that latency may be established by direct infection of resting memory CD4+T cells (Trm cells) (García et al., 2020; O’Neil et al., 2021). Selective reverse transcriptional products tyrosine aminotransferase (Tat) and negative factor (Nef) exist in Trm cells (Schulze-Gahmen and Hurley, 2018), and can induce cell activation so that the virus genome can be integrated into the cell genome (Donahue et al., 2013; Pinto et al., 2020). Moreover, it has also been found that although Trm cells are resistant to HIV compared with activated CD4+T cells, mild stimulation of chemokine CC ligand 19 (CCL19) and chemokine CC ligand 21 (CCL21) and cytokines interleukin (IL)-4 and interleukin (IL)-7 can promote the direct infection of resting CD4+T cells with HIV without inducing significant T cell activation (Saleh et al., 2007; Wu, 2010; López-Huertas et al., 2019). The establishment and maintenance of latent infection may be influenced by a variety of factors, including the availability of host transcription factors, epigenetic modifications, HIV Tat protein defects, integration sites and directions, and post-transcriptional regulatory mechanisms. After the reservoir of latent HIV is established, the transcription level of HIV in the reservoir is very low, and almost no virus is produced as there is no viral protein, and the latent infected cells will not be affected by cytotoxicity, nor will they be recognized by the immune system, so that they can exist stably for a long time. Figure 1 shows the main formation process of latent HIV reservoir.
Figure 1.
The formation process of latent HIV reservoir. (A) HIV mainly infects human CD4+T lymphocytes. When it enters the cell, the RNA is first reversely transcribed into HIV DNA, which is then incorporated into the DNA of CD4+T cells. Some CD4+T cells with integrated HIV DNA are converted into a resting state, and the HIV in the resting CD4+T becomes latent HIV. (B) HIV directly infects CD4+T cells that revert to a G0 dormant memory state, thus enabling the virus to enter latency. (C) HIV establishes incubation by directly infecting resting memory CD4+T cells (Trm cells).
The factors affecting the formation of latent HIV reservoir can be generally divided into two aspects: inhibiting the formation of latent HIV reservoir and promoting the formation of latent HIV reservoir. B-cell lymphoma 2 (BCL2) is a key regulatory molecule of lymphoid tissue homeostasis, affecting HIV homeostasis in infected lymphocytes. BCL2 can directly bind Casp8p41 and prevent the latter from binding and activating Bcl-2 homolog antagonist/killer (BAK), thereby inducing apoptosis (Cummins et al., 2016). It has been found that the expression of Casp8p41 in resting memory CD4+T cells is negatively correlated with the absolute CD4+T count. BCL2 can inhibit the formation of latent HIV reservoirs depending on the enhanced cytotoxicity of Casp8p41 (Cummins et al., 2017). In addition, the purified Tat protein inhibits the establishment of latent HIV reservoir without changing the susceptibility of cells to HIV (Donahue et al., 2012).
On the contrary, some substances may also promote the formation of latent HIV reservoirs. Programmed cell death protein 1 (PD-1) and lymphocyte activation gene 3 (LAG-3) were initially identified as markers of HIV-infected cells (Fromentin et al., 2016). PD-1 plays an active role in silencing HIV transcription (Boussiotis and Patsoukis, 2022). A recent study reported that the TRIM28 protein helps in binding a chemical marker called SUMOylation to the cell’s gene regulators and inhibits the activity of HIV genes, thereby inhibiting its expression and promoting latency (Ma et al., 2019). The binding of interferon-γ-induced protein (IP) -10 to C-X-C chemokine receptor 3 (CXCR3) has also been reported to enhance latent infection of resting CD4+T cells against HIV (Wang et al., 2021). IP-10 stimulation promotes cofilin activity and actin dynamics, thereby promoting HIV entry and DNA integration, suggesting that IP-10 is also a key factor in the formation of latent HIV reservoir, and targeting IP-10 therapy may be a potential strategy in inhibiting latent HIV infection (Lei et al., 2019; Wang et al., 2021). In addition, cell-to-cell contact between infected cells and uninfected cells is also a key feature in the formation of an HIV latent virus reservoir (Pedro et al., 2019; Okutomi et al., 2020). Cell-to-cell contact enhances the susceptibility of resting CD4+T cells to HIV (Agosto et al., 2018). Previous studies have shown that monocytes or myeloid dendritic cells (mDCs) co-culturing with activated HIV-infected T cells may facilitate their transition to the activated latent state, highlighting the role of intercellular contact in the establishment of HIV latent reservoir. Furthermore, cytokines are also important factors in the formation of HIV latent virus reservoir (Vandergeeten et al., 2012). For instance, IL-10, IL-8 and transforming growth factor –β (TGF-β) can produce long-term latent infectious cells by reducing T cell activation, which has been proved in in vitro experiments (Wilson and Brooks, 2011; Travis and Sheppard, 2014; Morris et al., 2017).
The cell reservoir of HIV
Currently, CD4+T cells of resting memory constitute the major reservoir of latent HIV, which has widely been recognized by researchers (Campbell et al., 2018; Kwon et al., 2020; Moranguinho and Valente, 2020). However, a study reported that the rate of virus recurrence after the termination of ART therapy is much higher than the replication rate of CD4+T cells, indicating the presence of other reservoirs of latent HIV in addition to CD4+T cells (Chun et al., 2000). It has been proved that a variety of cells in circulating blood can also exist in the form of a latent reservoir of HIV after infection, which is associated with a variety of diseases and affect the development of HIV (Burdo, 2019; Kristoff et al., 2019; Veenhuis et al., 2019).
CD4+T lymphocytes of resting memory
CD4+T cells of resting memory are stable reservoirs of latent HIV infection (García et al., 2018). A previous study stated that there was no significant loss of integrated HIV DNA in resting memory T cells over time, with a half-life of about 25 years (Murray et al., 2014). Resting memory T cells can be divided into different subtypes, including primitive T cells (TN) and memory T cells (TM) (Terahara et al., 2019). TM cells are divided into central memory T cells (TCM), transitional memory T cells (TTM), effector memory T cells (TEM) and stem cell memory T cells (TSCM) (Corneau et al., 2017; Gálvez et al., 2021). Viral DNA has been detected in all of the above mentioned resting CD4+T cell subpopulations in HIV patients (Zerbato et al., 2016), suggesting that CD4+T lymphocytes of resting memory may be major hosts of latent viral infection.
Studies have confirmed that TM is an important part of the reservoir of latent HIV. In terms of function, TSCM cells exhibit higher response ability upon stimulation by homologous antigen (Lugli et al., 2013). Moreover, some TSCM cells express CC chemokine receptor 5 (CCR5) and C-X-C chemokine receptor 4 (CXCR4), the main co-receptors for HIV entry, and making them susceptible to HIV infection (Tabler et al., 2014). In addition, TSCM cells have a very long half-life and thus form the most stable part of the latent reservoir (Cartwright et al., 2016). TSCM cells can produce highly differentiated cells, such as TCM and TEM, while TTM are intermediate phenotypes between TCM and TEM cells, each of which maintains its latent reservoir (Kulpa et al., 2019). As mentioned above, in addition to the virus exposure, there are many other factors influencing the latent infection of the resting CD4 + T cells, as some of the negative control cells activate the immune factors. Previous study reported that some factors in TEM, TTM, and TCM are more active, which affect the reservoir of lurking in the formation these cells (Kwon et al., 2020).
Compared with TM, viral DNA could be detected in TN cells despite the low frequency of HIV infection (Gibellini et al., 2017). At the same time, data have shown that infected TN cells are treated with drugs that reverse latency. The number of extracellular virions produced by TN in each infected cell was the same as that of TM cells, suggesting that TN cells with latent infection may also be the important source of the virus after treatment interruption or failure, that is, they are important hosts of latent HIV infection, and should not be ignored because of their low infection frequency (Zerbato et al., 2019).
Mononuclear macrophages
In addition to resting memory CD4+T cells, myeloid cells, especially mononuclear macrophages, are currently considered to be important reservoirs of the latent HIV (Kumar et al., 2014). Mononuclear macrophages are considered to be early targets of HIV infection, because both CD4 receptors and CCR5 or CXCR4 co-receptors express on their surfaces (Lee et al., 1999). It has been shown that mononuclear macrophages play a key role in the innate immune response to pathogens, viral persistence, and viral library formation (Abreu et al., 2019; Kruize and Kootstra, 2019). Monocytes from bone marrow circulate in the blood and migrate to tissues to differentiate into various types of macrophages. Although monocytes rapidly differentiated into macrophages, studies have suggested that HIV has been detected in monocytes (Saini and Potash, 2014). In addition, macrophages have a long life span, found in almost all tissue in the body. Macrophages are relatively resistant to HIV-induced apoptosis, and can remain in antiviral treatment, so they are considered to play a key role in the establishment and persistence of latent HIV reservoir (Kruize and Kootstra, 2019). Brown et al. designed a long-term cultured in vitro model of macrophages infected with green fluorescent protein (GFP) labeled recombinant HIV. The results revealed that macrophages can establish incubation periods in vitro (Brown et al., 2006).
A quantitative virus outgrowth assay (QVOA) was used to measure the myeloid cells of latent infection in the simian immunodeficiency virus (SIV)-rhesus monkey model. The findings revealed that mononuclear macrophages with latent infection were detected in blood Broncho-alveolar lavage fluid, lung, spleen and brain (Abreu et al., 2019a; Abreu et al., 2019b), indicating that these cells persist during SIV infection and may act as latent viral reservoirs during antiretroviral therapy. Moreover, the isolated viruses produced by macrophages can infect activated CD4+T cells, suggesting that latent infected macrophages can re-infect after treatment interruption.
Dendritic cells
Dendritic cells (DCs) are a heterogeneous group of antigen-presenting cells, playing an important role in immune response (Worbs et al., 2017; Balan et al., 2019; Yin et al., 2021). DCs are divided into myeloid dendritic cells (mDCs) and plasmacytoid dendritic cells (pDCs) based on maturity and origin (Verna et al., 2021). mDCs and pDCs, in an in vitro experiment were found to have different susceptibility to HIV (Smed-Sörensen et al., 2005; Groot et al., 2006).
Low levels of preHIV can be detected in DCs, suggesting that DCs may play a role in the HIV reservoir (Pope et al., 1995). Through the formation of infectious or virologic synapses (McDonald et al., 2003), DCs will transfer the infection to antigen-specific CD4+T cells upon HIV encounter (Loré et al., 2005), thereby weakening the establishment of anti-HIV immune response. In addition, HIV can appear in DCs and fuse with T cell membranes (Yu et al., 2008). Subsequently, DCs may be a potential target for HIV infection and latency due to surface pattern recognition receptor interactions with pathogens (Cardinaud et al., 2017).
Follicular dendritic cells (FDCs) present in secondary lymphoid tissue are the major sites of HIV infection during antiretroviral therapy (Olivetta et al., 2020; Ollerton et al., 2020). The virus is captured as an immune complex on its surface, the resulting complex is highly infectious to CD4+T cells (Keele et al., 2008). A previous study reported that FDCs in mice can retain the captured virus particles in an infected state for at least 9 months in vivo (Smith et al., 2001). Data also suggested that HIV captured by FDCs is capable of replication and exhibit greater genetic diversity than viruses found in other tissues or cells, and hence is an important host for an infectious and diverse group of HIV (Keele et al., 2008).
Hemopoietic progenitor cells
Since hematopoietic progenitor cells (HPCs) express HIV receptors, long-term infection of HPCs may also be an important factor in the residual HIV after treatment (Carter et al., 2011). It has been confirmed that different subtypes of HIV can infect HPCs in vivo or in vitro (Li et al., 2017). To study latent infection in HPCs, Carter et al. (2010) conducted experiments using cells with different infection states. the findings revealed that viral gene expression was induced upon the treatment of latent HPCs with cytokines, stimulated the differentiation of bone marrow cell lines (granulocyte-macrophage colony-stimulating factor GM-CSF and tumor necrosis factor TNF-α), suggesting that HIV could infect HPCs and cause both active and latent infections.
Studies have shown that CD34+HPCs expressing CD4 CCR5 or CXCR4 and other receptors and co-receptors are associated with the susceptibility of these cells to HIV (Carter et al., 2010; McNamara et al., 2013). In addition, McNamara et al. (2013) assessed HIV infection of CD34+HPCs in 9 HIV patients who received ART and had no detectable viral load for at least 6 months. In four of the nine patients, preHIV genomes were detected in CD34 cells at a frequency of 3-40 genomes per 10,000 cells, suggesting that HPCs can serve as a reservoir of latent HIV.
Astrocyte
HIV can invade the central nervous system (CNS), causing neuroinflammatory immune activation and neurodegenerative alterations, resulting in HIV-related neurocognitive impairment (Knight et al., 2018; Knight et al., 2020). Astrocytes are the most abundant cell type in CNS and play a vital role in maintaining the CNS homeostasis and regulating blood flow in response to injury and diseases (Guttenplan and Liddelow, 2019).
Due to the absence of CD4 receptors, astrocytes lead to restricted HIV infection. Although the proportion of HIV binding to astrocytes is low, HIV DNA and RNA have been found in astrocytes using in-situ hybridization laser capture anatomy and nested polymerase chain reaction (PCR) (Churchill et al., 2006; Chong et al., 2018), suggesting that astrocytes can be infected by HIV. In addition, studies using human astrocytes and human peripheral blood mononuclear cell chimeric model further confirmed that astrocytes are the host of HIV (Lutgen et al., 2020).
HIV infection is found to be acquired through pH-dependent endocytosis, which consumes most of the virus particles, however, pH-dependent endocytosis still may be an important pathway for HIV to establish incubation in astrocytes (Chauhan and Khandkar, 2015). Previous studies reported that Tat protein expression can affect HIV infection and latency establishment in astrocytes. Tat protein promotes the formation of HIV latency by inducing tri-methylation of histone H3 on Lys27 (H3K27me3) expression in astrocytes. A decrease in Tat protein expression decreases the formation of latent HIV infected cells (Chauhan and Khandkar, 2015). Meanwhile, the in vitro use of latency reversing agents (LRAs) has further identified astrocytes as part of the reservoir of latent HIV (Schneider et al., 2015).
Other cells
In addition to the aforementioned cells, several other cells types are also reported to form latent reservoirs of HIV. Macrophages and microglia in the central nervous system are the main antigen-presenting cells that can be infected with HIV. Previously, researchers detected HIV DNA in these pair of cell isolated from the brain tissue of five dead individuals, proving them as the main cellular hosts for latent HIV (Thompson et al., 2011). At the same time, microglia can be sustained for a long time and can proliferate in situ, and hence proved to be a key drive for viral reservoirs (Wallet et al., 2019).
Epithelial cells are also susceptible to HIV. Renal tubular epithelial cell co-cultured with infected T cells becomes susceptible to HIV. HIV DNA and RNA were detected in renal tubular epithelial cells by in situ hybridization of biopsies collected from patients with HIV-associated nephropathy (Katuri et al., 2019). In addition, it has been reported that integrated HIV DNA can be detected after in vitro infection of liver cell lines and primary liver cells, and the release of infectious viruses was also found in the liver epithelium (Ganesan et al., 2018).
Tissue reservoir of HIV
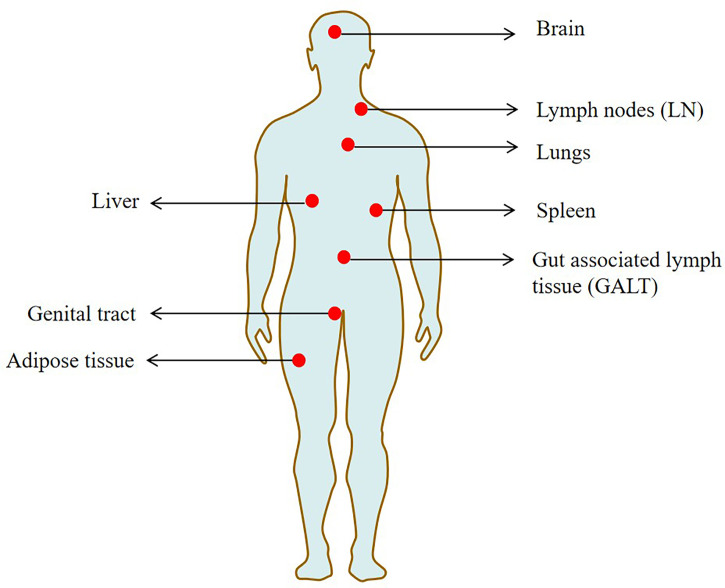
Studying the latent HIV reservoir in tissues is challenging due to the difficulty of tissue sampling. In recent years, the research on the latent HIV reservoir in tissue has mainly focused on the autopsy of non-humans primates and humans (Cohn et al., 2020). Lymph nodes (LN) and gut-associated lymph tissue (GALT) were found to be the major tissue reservoirs rich in viruses and with high frequency of infected cells (Churchill et al., 2016). In the course of HIV infection, B cell follicles in the lymphoid structure actively reject effector CD8+T cells to maintain normal B cell function, thereby providing favorable conditions for the formation of latent HIV reservoir (Connick et al., 2014; Fukazawa et al., 2015). Previous studies suggested that the gut may be the body’s largest reservoir of HIV. In addition, by examining the vaults of SIV-infected rhesus monkeys, the researchers found that the vast majority (>98%) of storage stocks are present in the gut (Estes et al., 2017).
Previously, a human autopsy study revealed that the HIV provirus was detected in 28 tissues, including the liver, spleen, genital tract and brain (Chaillon et al., 2020). Several major HIV reservoir cells, such as resting memory CD4+T cells, dendritic cells, macrophages and microglia, are widely located in these tissues (Honeycutt et al., 2017; Wallet et al., 2019). Some of the studied sites are termed sanctuary sites, which are protected from ART penetration (the brain and testis) and pose additional challenges for HIV treatments (Fletcher et al., 2014).
Tissue macrophages such as those found in seminal vesicles, urethra, adipose tissue and liver tissue are considered to be important hosts of HIV, including macrophages (Deleage et al., 2011; Ganor et al., 2013; Damouche et al., 2015). In addition, infected macrophages have been found at low but detectable frequencies in lung and duodenal tissue of patients on ART with undetectable plasma viruses (Cribbs et al., 2015). The reproductive tract is also rich in macrophages and may be an important reservoir of latent HIV, where antiretroviral drugs are difficult to enter. Study has found that the male reproductive tract may also exist in the latent HIV (Ganor et al., 2019). Additionally, tissue resident memory CD4+ T cells in the female genital tract (particularly the cervix) are highly enriched with HIV DNA (Cantero-Pérez et al., 2019). Figure 2 shows the main tissue reservoirs of HIV.
Figure 2.
The main tissue locations of latent HIV reservoir.
Detection of latent HIV reservoirs
At present, detection methods for finding accurate, sensitive, and scalable reservoirs of latent HIV are important in the treatment of reservoirs. Virus outgrowth assay (VOA) is considered the gold standard for the quantification of latent HIV reservoir. It measures the replicability of the original virus by diluting resting CD4+T cells and activating the intracellular viral gene expression, resulting in inducing the release of HIV from latent infected cells (Wang et al., 2018). However, this method is time-consuming, expensive, and requires a large amount of blood culture. Moreover, single stimulation is not sufficient to activate all latent viruses, which underestimates the size of latent HIV reservoir (Massanella et al., 2018; Wonderlich et al., 2019). Detection of HIV DNA based on PCR is relatively simple and rapid method for the detection of the latent reservoir, and provides a supplement for the VOA test (Ho et al., 2013). Quantitative real-time polymerase chain reaction (qPCR) for detection of HIV DNA to determine the amount of HIV DNA carried (Thomas et al., 2019), digital PCR (dPCR) for detection of HIV DNA for absolute quantitative HIV (Henrich et al., 2017), and Alu-polymerase chain reaction (Alu-PCR) for detection of HIV integration the amount of DNA is used to evaluate the size of the latent reservoir of HIV (Lada et al., 2018). PCR has made rapid development in the determination of latent reservoir of HIV, however, the size of the reservoir may be overestimated because PCR cannot distinguish intact from defective original virus (Ho et al., 2013).
To counter the above mentioned challenges, several innovative approaches have been developed for detecting latent HIV reservoirs including intact proviral DNA assay (IPDA), which differentiates intact and defective viruses, and screen different defective viruses by plasmid control (Gaebler et al., 2021). It was found that 90% of the defects in the viruses occurred in the encapsulation signal (ψ) and env regions. With the detection of ψ and env regions by dPCR, complete viruses and defective viruses could be identified (Bruner et al., 2019). Tat/Rev induced restricted dilution test (TILDA) is used to measure the frequency of CD4+T cells of latent HIV proto-virus. Researchers discovered that changing the preamplification settings did not affect on the Tat/Rev multiple splicing RNA assay, confirming the stability of the assay and supporting its adaptability to limited modifications to ensure better clinical use in latent HIV reservoirs (Bertoldi et al., 2020; Lungu et al., 2020). ISH and flow cytometry, have a higher sensitivity to replication of the original virus, and can phenotype host cells (Deleage et al., 2018; Pardons et al., 2019). There was a study that described how to use this technology to address some of the major questions remaining in the HIV feld in the era of ART.They discussed how CD4+T cell responses to HIV antigens, both following vaccination and HIV infection, can be characterized by measurement of cytokine mRNAs. They also described how their development of a dual HIV mRNA/protein assay (HIVRNA/Gag assay) enables high sensitivity detection of very rare HIV-infected cells and aids investigations into the translation competent latent reservoir in the context of HIV cure (Baxter et al., 2017). The emergence and development of these detection techniques provide strong evidence for the study of latent HIV reservoir. Table 1 summarizes the testing methods for latent HIV reservoirs.
Table 1.
The different detection methods of latent HIV reservoir.
| Assay | Advantages and disadvantages | References | |
|---|---|---|---|
| Viral outgrowth assay (VOA) | The replicable proto-virus was measured by diluting resting CD4+T cells to activate intracellular viral gene expression and induce the release of HIV from latent infected cells | The gold standard for measuring latent HIV reservoirs But it is expensive, time-consuming, requires a lot of blood culture and underestimates the size of the latent reservoir |
(Wonderlich et al., 2019) (Massanella et al., 2018) (Fun et al., 2017) (Badia et al., 2018) |
| PCR | Detection of HIV DNA using primers/probes | Simple, fast, and provides a supplement for VOA experiment But it’s possible to overestimate the size of the repository |
(Ho et al., 2013) |
| qPCR | Testing for HIV DNA to determine the amount of HIV DNA carried | (Thomas et al., 2019) (Vandergeeten et al., 2014) |
|
| ddPCR | Detection of HIV DNA for absolute HIVquantification | (Henrich et al., 2017) (Lada et al., 2018) |
|
| Alu-PCR | The amount of integrated HIV DNA was measured to evaluate the size of the latent reservoir | (Vandergeeten et al., 2014) (Lada et al., 2018) |
|
| Intact proviral DNA assay (IPDA) |
Complete proviruses were measured by multiplex digital PCR, and different defective proviruses were verified by plasmid control | Fast and able to distinguish between intact and defective proviruses But it didn’t screen the whole genome |
(Bruner et al., 2019) |
| TILDA | CD4+T cell frequency of latent HIV proto-virus was measured by Tat/Rev induced restricted dilution test | High sensitivity and stability But the transcripts measured may come from defective provirus genomes |
(Bertoldi et al., 2020) (Frank et al., 2019) |
| ISH and flow cytometry |
mRNA and viral proteins were measured after T cell activation | It has high sensitivity and can phenotype host cells But it can not prove that RNA or proteins are made by replicating protoviruses |
(Baxter et al., 2017) (Deleage et al., 2018) (Pardons et al., 2019) |
Strategies for removing latent HIV reservoirs
Shock and kill strategies
Shock and kill therapy use drugs to activate the gene transcription (shock) of HIV lurking in cells, and then kill the virus (kill) through the body’s immune system, ART or other intervention methods to eliminate the latent HIV. It is one of the most effective methods to remove the latent HIV (Sadowski and Hashemi, 2019; Nixon et al., 2020). LRAs are used for activation of viral transcription, the production of viral proteins, release of virus particles, and, effectively eliminating latent HIV in combination with ART (Spivak and Planelles, 2018; Delannoy et al., 2019). Therefore, LRAs play an important role in this process. It has been found that many substances can be used as LRAs to activate latent HIV reservoir, such as: 1) cytokines or receptor agonists, such as IL2, IL7, IL15, toll-like receptor 2,3(TLR2, 3) agonists, etc. The drugs were found to reactivate the expression of HIV, but they did not eliminate the latent infection cells, or affect the size of the HIV reservoir (Rochat et al., 2017; Madrid-Elena et al., 2018). 2) Epigenetic modification enzyme inhibitors, such as histone deacetylase inhibitors (HDACi) histone methyltransferase inhibitors (HMTi), and DNA methylation inhibitors (DNMTi), etc. HDACi and HMTi have been shown to reactivate HIV expression to some extent in vitro and in vivo (Archin et al., 2017). In addition, DNA methylation inhibitor 5-AzadC can also induce HIV expression in vitro, while its oxidation analog 5-AzaC is unable to do so (Bouchat et al., 2016). 3) Cellular signaling modulators, such as protein PKC receptor agonist prostratin and bryostatin, activate the protein kinase C (PKC) pathway to release NF-κB and positive transcription elongation factor b (pTEFb) from inactive complexes and increase pTEFb expression, ultimately leading to HIV reactivation (Wang et al., 2017; Schwartz et al., 2017). In addition, antioxidants, AKT regulators, protein phosphatase 1, and many other substances induce the expression of HIV proto-virus (Chirullo et al., 2013; Doyon et al., 2014; Smith et al., 2015; Tyagi et al., 2015), however, their specific mechanism of action remains unclear and needs further study. Table 2 summarizes commonly used latent reversal agents.
Table 2.
The commonly used latent reversal agents.
| Category | Examples | Mechanism of action | References |
|---|---|---|---|
| Cytokines or receptor agonists | IL2、IL7、IL15、TLR2,3 | Reactivation of HIV expression | (Rochat et al., 2017) (Rasmussen et al., 2016) |
| Epigenetic modification enzyme inhibitors | HDACi、HMTi、DNMTi | Reactivation of HIV expression in vitro and in vivo | (Archin et al., 2017) (Bernhard et al., 2011) |
| Cell signal regulator | Prostra、BryostatinC | Activation of PKC pathway results in the release of NF-κB and pTEFb from inactive complex and increases pTEFb expression |
(Wang et al., 2017) (Schwartz et al., 2017) |
| Others | |||
| Antioxidant | Auranofn (AF) | The mechanism of action is still unclear |
(Chirullo et al., 2013) |
| AKT regulator | Disulfram | (Doyon et al., 2014) | |
| Protein phosphatase 1 | SMAPP1 | (Smith et al., 2015) (Tyagi et al., 2015) |
|
Schwartz et al (Schwartz et al., 2017) investigated the reactivation potential of compounds releasing active pTEFb in combination with PKC agonists. The combination of HMBA/BETi and PKC agonists led to strong synergistic activation of HIV expression in several in vitro post-integrated latency cell line models (Darcis et al., 2015). Continuous treatment with demethylation agents (5-AZADC) and clinically tolerated HDACi was also shown to be more effective than corresponding concurrent treatment in inducing HIV gene expression in vitro and ex vivo ( Bouchat et al., 2016). These datas demonstrate the importance of treatment schedules in combination with LRAs for HIV activation.
The ability of LRAs to activate in vitro is correlated with the size of the HIV reservoir.However, some patients have very low or extremely high reactivation relative to the size of their reservoir (Darcis et al., 2017). Timely administration of LRAs in reactivation trials and a better understanding of the variability of reactivation in patients are important.A defective Cas9 (dCas9) protein fused to activators may be a new tool to reactivate potentially infected cells. CRISPR/dCas9 may be used to reactivate latent HIV in vitro experiments (Zhang et al., 2015). Similarly, CRISPR/dCas9 synergistically activates HIV when used in combination with HDAC inhibitors and PKC activators (Limsirichai et al., 2016).
Exosomes, as a way of material and information transmission between cells, are the key features in viral infection (Chen et al., 2021). Research studies on the activation of latent HIV reservoir have found exosomes to induce the activation of resting CD4+T cells infected with HIV through different mechanisms. For example, exosomes from HIV-infected cells have been shown to activate resting CD4+T lymphocytes via ADAM17 and tumor necrosis factor-α (TNF-α) dependent mechanisms (Arenaccio et al., 2014; Arenaccio et al., 2015). In addition, HIV-coded Tat protein is an effective viral transcription transactivator. Tat protein is encapsulated in exosomes for targeted delivery to latently infected CD4+T lymphocytes, which reactivate the virus. When combined with an HIV LRA, the expression of HIV mRNA increases by >30-fold (Tang et al., 2018).
Despite the reactivation of latent HIV, LRAs alone cannot significantly reduce the size of the reservoir, suggesting that kill is needed to destroy HIV-infected cells after shock therapy (Castro-Gonzalez et al., 2018; Sadowski and Hashemi, 2019; Maina et al., 2021). ART is an important method used to reduce the activated virus, and HIV-specific CD8+T cells play a key role in eradicating HIV hosts. However, the immune system of ART patients is unable to produce sufficient anti-HIV cytotoxic CD8+T cell response, therefore, it is unable to eliminate the activated cells in large numbers. Consequently, it is necessary to enhance cellular or humoral mediated immune response to promote cell apoptosis, and eliminate the latent HIV (Kim et al., 2018).
Block and lock
Contrary to the shock and kill strategy, the block and lock strategy permanently silence the original HIV to prevent the virus from rebounding. The block and lock strategy prevents the transcription and reactivation of HIV in the latent infection cells to inhibit the emergence of the latent virus (Moranguinho and Valente, 2020; Vansant et al., 2020). HIV transcription is a complex process involving many virus proteins and cytokines, including Tat proteins to induce viral transcription extension, host transcription factors, transcription suppressors, etc. Alterations in any of these factors may silence HIV transcription (Vansant et al., 2020). In addition, chromatin and epigenetic landscape and HIV integration sites also play important roles in viral transcription (Pearson et al., 2008; Tyagi et al., 2010).
To address these potential targets affecting latent HIV transcription and silencing, several block and lock strategies have been proposed: 1) Inhibition of Tat protein expression through didehydro-cortistatin A (dCA). dCA is an effective inhibitor of Tat protein, which can block the transcription and reactivation of HIV by LRAs in CD4+T cells. It has been found that dCA treatment can reduce or delay virus recurrence. Meanwhile, dCA has a strong specificity for Tat protein and plays an important role in inhibiting virus reactivation (Kessing et al., 2017). 2) Inhibition of HIV integration and relocalization of the residual original virus by LEDGINs. Lens epithelium-derived growth factor p75 (LEDGF/p75) is a determinant of HIV integration site selection and affects the transcription status of the original virus (Debyser et al., 2018). LEDGINs are small molecular inhibitors of the interaction between LEDGF/p75 and integrase (Christ et al., 2010), which can impede HIV integration and the catalytic activity of HIV integrase (IN), to increase the proportion of protovirus with transcriptional silencing phenotype (Symons et al., 2018). 3)Small interfering RNA (siRNA) is used to maintain the repressive heterochromatic landscape at the HIV5’ LTR promoter (Ahlenstiel et al., 2015). siRNA silenced HIV genes transcription by recruiting Argonaute1(AGO1), histone deacetylase 1, and histone methyltransferase (Méndez et al., 2018). 4) Promoting chromatin transcription complex (FACT) is also one of the important regulatory factors of HIV transcription. Curaxins CBL0100, an anticancer drug, was discovered to suppress HIV replication and reactivation by inhibiting RNAPII-mediated transcriptional elongation in a Tat-dependent manner (Jean et al., 2017). 5) Mammalian target of rapamycin (mTOR) inhibitors, hinders HIV reactivation by down-regulating cyclin-dependent kinase 9 (CDK9) phosphorylation and blocking NF-κB signal transduction (Besnard et al., 2016).
In addition, it has been found that heat shock protein 90(HSP90) inhibitors, Jak (Janus kinase)-STAT inhibitors, kinase inhibitors and bromodomain protein 4(BRD4) regulators also play corresponding roles in inhibiting the latent HIV transcription and reactivation (Vansant et al., 2020). Although many compounds are still in the early stages of HIV research, their emergence offers new possibilities for cure. Figure 3 shows the main process of shock and kill, and block and lock to eliminate the latent reservoir of HIV.
Figure 3.
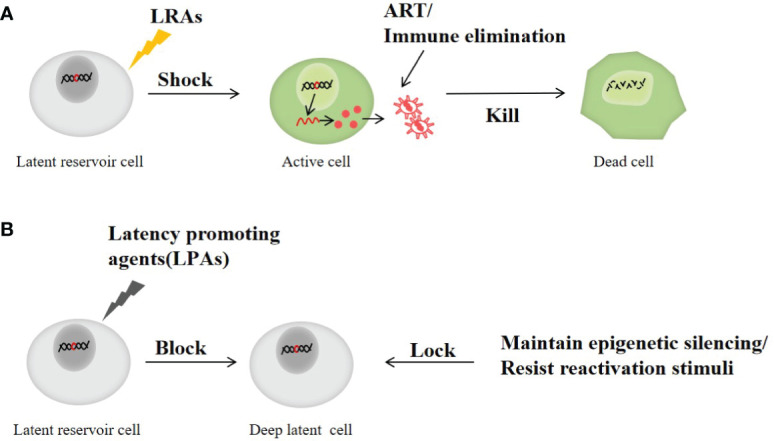
The main process of shock and kill and block and Lock therapy to clear latent reservoir of HIV. (A) Shock and kill strategy: the latent HIV is activated with a LRAs and then killed by ART or immunotherapy. (B) Block and lock strategy: by preventing the transcription and reactivation of HIV in latent infection cells to inhibit the emergence of latent virus and prevent virus rebound.
Immunization therapy
When the latent HIV is activated by LRAs, immunotherapy can kill or eliminate HIV-infected cells. Moreover, immunotherapy can stimulate the natural immunity of HIV-infected cells (Ferrari et al., 2017). The immune response can be enhanced by regulating cytokines and interleukins (Pankrac et al., 2017). IL-15 enhances the activity of natural killer cells (NK), thereby enhancing cell elimination after LRAs treatment (Garrido et al., 2018). Subsequently, a polo-like kinase 1 (PLK1) inhibitor can also enhance the anti-HIV effect of DCs (Gringhuis et al., 2017).
Broadly neutralizing antibodies (bNAbs) target specific proteins outside the HIV, thereby reducing infectivity. bNAbs can significantly reduce viral load (Parsons et al., 2018). In addition, bNAbs can also be used in combination with shock and kill therapy to eliminate infected cells. When LRAs activate potentially infected cells, bNAbs eliminate them (Martinez-Navio et al., 2019). Antibody therapy can also promote apoptosis of activated cells by targeting surface markers. In addition, the conjugation of antibodies with cytotoxic compounds or drugs can be used to target and kill specific cell types recognized by antibodies. Antibodies used in this class of antibody drug conjugate (ADC), including bNAbs, recognize HIV gag or HIV env gene products and, may provide effective killing of latent infected cells (Dan et al., 2018).
At the same time, the CD8+T cells isolated from HIV patients can modify their specificity in vitro to produce anti-HIV cytotoxic T lymphocytes (CTL) response and are amplified and reintroduced into patients, which can also promote their immune response to the reactivated cells, to eliminate the latent infection cells (Yang et al., 2016).
In addition to the above strategies, some other methods against latent HIV reservoir have been proposed, such as interferon (IFN) combined with ART therapy, stem cell transplantation (Kuritzkes, 2016), gene editing including CRISPR (Das et al., 2019; Herrera-Carrillo et al., 2020; Atkins et al., 2021), zinc-finger nucleases (Wayengera, 2011; Ahlenstiel et al., 2020) etc. Target cells can be induced to develop HIV resistance through gene editing, such as gene editing to remove the CCR5 gene, and reduce the binding of HIV to CD4 cells (Yang et al., 2016; Schwartz et al., 2017). Each method has its own characteristics and plays a certain role in potentially controlling the latent virus reservoir.
Discussion
The establishment of a latent HIV reservoir is a complex process, and can exist in a variety of cells and tissues throughout an infected person’s body, making it difficult to detect and eliminate. With the deepening of HIV research, the existence of latent HIV reservoir has gained extensive attention. Subsequently, great progress has been made in the research of latent HIV reservoir. Many methods for the detection and elimination of latent HIV reservoir have been proposed.
At present, activating latent HIV to kill, is still the key to eliminate the reservoir of latent HIV. Therefore, people are committed to finding safe and efficient activators to activate the latent HIV. Recently, exosomes have gradually become the focus of HIV clinical research. Previous studies have found that exosomes can activate latent HIV reservoir cells through various mechanisms, and various substances carried by exosomes play an important role in the occurrence and development of HIV, which has broad application prospects in the clearance of latent HIV reservoir and the treatment of HIV (Barclay et al., 2017; Chandra et al., 2021). In addition, clinical research proved that traditional Chinese medicine plays an irreplaceable role in the treatment of HIV (Li et al., 2020; Qian et al., 2021). Our study found that traditional Chinese medicine (TCM) and its related components can also activate the latent HIV, providing a new direction for TCM treatment of HIV. At the same time, new mechanisms for activating latent HIV reservoir have been proposed, which provide the possibility for the elimination of latent reservoir.
However, research on latent HIV reservoir is still in the initial stage, and further exploration is needed to better understand the intricacies of possible mechanism, location, detection, and clearance methods, to achieve a complete elimination of latent HIV reservoirs.
Author contributions
JC, TZ and YZ conceived of the presented idea. SL, HC and DC researched on the background of the study. CL and WL critically reviewed the manuscript. All authors contributed to and approved the final manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (81603552), the Natural Science Foundation of Beijing (7212172), the Pilot project of public welfare development and reform of Beijing Municipal Medical Research Institutes (2019-6) and the Special project for the construction of high-level health technical personnel in Beijing Health System (2022-2-024).
Acknowledgments
The authors would like to thank all the investigators and the staff of the Second People’s Hospital in Lianyungang City, China, for the provision of clinical isolates.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abreu C., Shirk E. N., Queen S. E., Beck S. E., Mangus L. M., Pate K. A.M., et al. (2019). Brain macrophages harbor latent, infectious simian immunodeficiency virus. Aids 33 Suppl 2 (Suppl 2), e01659–19 S181–s188. doi: 10.1097/qad.0000000000002269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu C. M., Veenhuis R. T., Avalos C. R., Graham S., Parrilla D. R., Ferreira E. A., et al. (2019. a). Myeloid and CD4 T cells comprise the latent reservoir in antiretroviral therapy-suppressed SIVmac251-infected macaques. mBio 10 (4):e00065-19. doi: 10.1128/mBio.01659-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu C. M., Veenhuis R. T., Avalos C. R., Graham S., Queen S. E., Shirk E. N., et al. (2019. b). Infectious virus persists in CD4(+) T cells and macrophages in antiretroviral therapy-suppressed simian immunodeficiency virus-infected macaques. J. Virol. 93 (15):e00065–19. doi: 10.1128/jvi.00065-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosto L. M., Herring M. B., Mothes W., Henderson A. J. (2018). HIV-1-Infected CD4+ T cells facilitate latent infection of resting CD4+ T cells through cell-cell contact. Cell Rep. 24 (8), 2088–2100. doi: 10.1016/j.celrep.2018.07.079 [DOI] [PubMed] [Google Scholar]
- Ahlenstiel C., Mendez C., Lim S. T., Marks K., Turville S., Cooper D. A., et al. (2015). Novel RNA duplex locks HIV-1 in a latent state via chromatin-mediated transcriptional silencing. Mol. Ther. Nucleic Acids 4 (10), e261. doi: 10.1038/mtna.2015.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlenstiel C. L., Symonds G., Kent S. J., Kelleher A. D. (2020). Block and lock HIV cure strategies to control the latent reservoir. Front. Cell Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.00424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archin N. M., Kirchherr J. L., Sung J. A., Clutton G., Sholtis K., Xu Y., et al. (2017). Interval dosing with the HDAC inhibitor vorinostat effectively reverses HIV latency. J. Clin. Invest. 127 (8), 3126–3135. doi: 10.1172/jci92684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenaccio C., Anticoli S., Manfredi F., Chiozzini C., Olivetta E., Federico M.. (2015). Latent HIV-1 is activated by exosomes from cells infected with either replication-competent or defective HIV-1. Retrovirology 12, 87. doi: 10.1186/s12977-015-0216-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenaccio C., Chiozzini C., Columba-Cabezas S., Manfredi F., Affabris E., Baur A., et al. (2014). Exosomes from human immunodeficiency virus type 1 (HIV-1)-infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a nef- and ADAM17-dependent mechanism. J. Virol. 88 (19), 11529–11539. doi: 10.1128/jvi.01712-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins A. J., Allen A. G., Dampier W., Haddad E. K., Nonnemacher M. R., Wigdahl B.. (2021). HIV-1 cure strategies: why CRISPR? Expert Opin. Biol. Ther. 21 (6), 781–793. doi: 10.1080/14712598.2021.1865302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badia R., Ballana E., Castellví M., García-Vidal E., Pujantell M., Clotet B., et al. (2018). CD32 expression is associated to T-cell activation and is not a marker of the HIV-1 reservoir. Nat. Commun. 9 (1), 2739. doi: 10.1038/s41467-018-05157-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balan S., Saxena M., Bhardwaj N. (2019). Dendritic cell subsets and locations. Int. Rev. Cell Mol. Biol. 348, 1–68. doi: 10.1016/bs.ircmb.2019.07.004 [DOI] [PubMed] [Google Scholar]
- Barclay R. A., Schwab A., DeMarino C., Akpamagbo Y., Lepene B., Kassaye S., et al. (2017). Exosomes from uninfected cells activate transcription of latent HIV-1. J. Biol. Chem. 292 (28), 11682–11701. doi: 10.1074/jbc.M117.793521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter A. E., Niessl J., Morou A., Kaufmann D. E. (2017). RNA Flow cytometric FISH for investigations into HIV immunology, vaccination and cure strategies. AIDS Res. Ther. 14 (1), 40. doi: 10.1186/s12981-017-0171-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitari S., Wang Y., Liu S. L., Liang C. (2019). HIV-1 envelope glycoprotein at the interface of host restriction and virus evasion. Viruses 11 (4):311. doi: 10.3390/v11040311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard W., Barreto K., Saunders A., Dahabieh M. S., Johnson P., Sadowski I.. (2011). The Suv39H1 methyltransferase inhibitor chaetocin causes induction of integrated HIV-1 without producing a T cell response. FEBS Lett. 585 (22), 3549–3554. doi: 10.1016/j.febslet.2011.10.018 [DOI] [PubMed] [Google Scholar]
- Bertoldi A., D'Urbano V., Bon I., Verbon A., Rokx C., Boucher C., et al. (2020). Development of c-TILDA: A modified TILDA method for reservoir quantification in long term treated patients infected with subtype c HIV-1. J. Virol. Methods 276, 113778. doi: 10.1016/j.jviromet.2019.113778 [DOI] [PubMed] [Google Scholar]
- Besnard E., Hakre S., Kampmann M., Lim H. W., Hosmane N. N., Martin A., et al. (2016). The mTOR complex controls HIV latency. Cell Host Microbe 20 (6), 785–797. doi: 10.1016/j.chom.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchat S., Delacourt N., Kula A., Darcis G., Van Driessche B., Corazza F, et al. (2016). Sequential treatment with 5-aza-2'-deoxycytidine and deacetylase inhibitors reactivates HIV-1. EMBO Mol. Med. 8 (2), 117–138. doi: 10.15252/emmm.201505557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussiotis V. A., Patsoukis N. (2022). Effects of PD-1 signaling on immunometabolic reprogramming. Immunometabolism 4(2):e220007. doi: 10.20900/immunometab20220007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A., Zhang H., Lopez P., Pardo C. A., Gartner S.. (2006). In vitro modeling of the HIV-macrophage reservoir. J. Leukoc. Biol. 80 (5), 1127–1135. doi: 10.1189/jlb.0206126 [DOI] [PubMed] [Google Scholar]
- Bruner K. M., Wang Z., Simonetti F. R., Bender A. M., Kwon K. J., Sengupta S., et al. (2019). A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566 (7742), 120–125. doi: 10.1038/s41586-019-0898-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo T. H. (2019). Editor’s commentary for special issue: “The role of macrophages in HIV persistence”. J. Neuroimmun. Pharmacol. 14 (1), 2–5. doi: 10.1007/s11481-019-09836-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G. R., Bruckman R. S., Chu Y. L., Trout R. N., Spector S.A.. (2018). SMAC mimetics induce autophagy-dependent apoptosis of HIV-1-Infected resting memory CD4+ T cells. Cell Host Microbe 24 (5), 689–702.e7. doi: 10.1016/j.chom.2018.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantero-Pérez J., Grau-Expósito J., Serra-Peinado C., Rosero D. A., Luque-Ballesteros L., Astorga-Gamaza A., et al. (2019). Resident memory T cells are a cellular reservoir for HIV in the cervical mucosa. Nat. Commun. 10 (1), 4739. doi: 10.1038/s41467-019-12732-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinaud S., Urrutia A., Rouers A., Coulon P. G., Kervevan J., Richetta C., et al. (2017). Triggering of TLR-3, -4, NOD2, and DC-SIGN reduces viral replication and increases T-cell activation capacity of HIV-infected human dendritic cells. Eur. J. Immunol. 47 (5), 818–829. doi: 10.1002/eji.201646603 [DOI] [PubMed] [Google Scholar]
- Carter C. C., McNamara L. A., Onafuwa-Nuga A., Shackleton M., Riddell J., Bixby D., et al. (2011). HIV-1 utilizes the CXCR4 chemokine receptor to infect multipotent hematopoietic stem and progenitor cells. Cell Host Microbe 9 (3), 223–234. doi: 10.1016/j.chom.2011.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C. C., Onafuwa-Nuga A., McNamara L. A., Riddell J., Bixby D., Savona M. R., et al. (2010). HIV-1 infects multipotent progenitor cells causing cell death and establishing latent cellular reservoirs. Nat. Med. 16 (4), 446–451. doi: 10.1038/nm.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright E. K., Palesch D., Mavigner M., Paiardini M., Chahroudi A., Silvestri G.. (2016). Initiation of antiretroviral therapy restores CD4+ T memory stem cell homeostasis in simian immunodeficiency virus-infected macaques. J. Virol. 90 (15), 6699–6708. doi: 10.1128/jvi.00492-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Gonzalez S., Colomer-Lluch M., Serra-Moreno R. (2018). Barriers for HIV cure: The latent reservoir. AIDS Res. Hum. Retroviruses 34 (9), 739–759. doi: 10.1089/aid.2018.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillon A., Gianella S., Dellicour S., Rawlings S. A., Schlub T. E., De Oliveira M. F., et al. (2020). HIV Persists throughout deep tissues with repopulation from multiple anatomical sources. J. Clin. Invest. 130 (4), 1699–1712. doi: 10.1172/jci134815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra P. K., Rutkai I., Kim H., Braun S. E., Abdel-Mageed A. B., Mondal D., et al. (2021). Latent HIV-exosomes induce mitochondrial hyperfusion due to loss of phosphorylated dynamin-related protein 1 in brain endothelium. Mol. Neurobiol. 58 (6), 2974–2989. doi: 10.1007/s12035-021-02319-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A., Khandkar M. (2015). Endocytosis of human immunodeficiency virus 1 (HIV-1) in astrocytes: a fiery path to its destination. Microb. Pathog. 78, 1–6. doi: 10.1016/j.micpath.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li C., Li R., Chen H., Chen D., Li W. (2021). Exosomes in HIV infection. Curr. Opin. HIV AIDS 16 (5), 262–270. doi: 10.1097/coh.0000000000000694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirullo B., Sgarbanti R., Limongi D., Shytaj I. L., Alvarez D., Das B., et al. (2013). A candidate anti-HIV reservoir compound, auranofin, exerts a selective 'anti-memory' effect by exploiting the baseline oxidative status of lymphocytes. Cell Death Dis. 4 (12), e944. doi: 10.1038/cddis.2013.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong P. F., Kira R., Mori H., Okumura A., Torisu H., Yasumoto S., et al. (2018). Clinical features of acute flaccid myelitis temporally associated with an enterovirus D68 outbreak: Results of a nationwide survey of acute flaccid paralysis in Japan, august-December 2015. Clin. Infect. Dis. 66 (5), 653–664. doi: 10.1093/cid/cix860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ F., Voet A., Marchand A., Nicolet S., Desimmie B. A., Marchand D., et al. (2010). Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 6 (6), 442–448. doi: 10.1038/nchembio.370 [DOI] [PubMed] [Google Scholar]
- Chun T. W., Davey R. T., Jr., Ostrowski M., Shawn Justement J., Engel D., Mullins J. I., et al. (2000). Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6 (7), 757–761. doi: 10.1038/77481 [DOI] [PubMed] [Google Scholar]
- Churchill M. J., Deeks S. G., Margolis D. M., Siliciano R. F., Swanstrom R.. (2016). HIV Reservoirs: what, where and how to target them. Nat. Rev. Microbiol. 14 (1), 55–60. doi: 10.1038/nrmicro.2015.5 [DOI] [PubMed] [Google Scholar]
- Churchill M. J., Gorry P. R., Cowley D., Lal L., Sonza S., Purcell D. F., et al. (2006). Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J. Neurovirol. 12 (2), 146–152. doi: 10.1080/13550280600748946 [DOI] [PubMed] [Google Scholar]
- Cohn L. B., Chomont N., Deeks S. G. (2020). The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe 27 (4), 519–530. doi: 10.1016/j.chom.2020.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connick E., Folkvord J. M., Lind K. T., Rakasz E. G., Miles B., Wilson N. A., et al. (2014). Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J. Immunol. 193 (11), 5613–5625. doi: 10.4049/jimmunol.1401161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneau A., Cosma A., Even S., Katlama C., Le Grand R., Frachet V., et al. (2017). Comprehensive mass cytometry analysis of cell cycle, activation, and coinhibitory receptors expression in CD4 T cells from healthy and HIV-infected individuals. Cytom. B Clin. Cytom. 92 (1), 21–32. doi: 10.1002/cyto.b.21502 [DOI] [PubMed] [Google Scholar]
- Cribbs S. K., Lennox J., Caliendo A. M., Brown L. A., Guidot D.M.. (2015). Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res. Hum. Retroviruses 31 (1), 64–70. doi: 10.1089/aid.2014.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins N. W., Sainski A. M., Dai H., Natesampillai S., Pang Y. P., Bren G. D., et al. (2016). Prime, shock, and kill: Priming CD4 T cells from HIV patients with a BCL-2 antagonist before HIV reactivation reduces HIV reservoir size. J. Virol. 90 (8), 4032–4048. doi: 10.1128/jvi.03179-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins N. W., Sainski-Nguyen A. M., Natesampillai S., Aboulnasr F., Kaufmann S., Badley A.D.. (2017). Maintenance of the HIV reservoir is antagonized by selective BCL2 inhibition. J. Virol. 91 (11), e00012–17. doi: 10.1128/jvi.00012-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damouche A., Lazure T., Avettand-Fènoël V., Huot N., Dejucq-Rainsford N., Satie A. P., et al. (2015). Adipose tissue is a neglected viral reservoir and an inflammatory site during chronic HIV and SIV infection. PloS Pathog. 11 (9), e1005153. doi: 10.1371/journal.ppat.1005153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan N., Setua S., Kashyap V. K., Khan S., Jaggi M., Yallapu M. M., et al. (2018). Antibody-drug conjugates for cancer therapy: Chemistry to clinical implications. Pharm. (Basel) 11 (2), 32. doi: 10.3390/ph11020032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcis G., Bouchat S., Kula A., Van Driessche B., Delacourt N., Vanhulle C., et al. (2017). Reactivation capacity by latency-reversing agents ex vivo correlates with the size of the HIV-1 reservoir. Aids 31 (2), 181–189. doi: 10.1097/qad.0000000000001290 [DOI] [PubMed] [Google Scholar]
- Darcis G., Kula A., Bouchat S., Fujinaga K., Corazza F., Ait-Ammar A., et al. (2015). An in-depth comparison of latency-reversing agent combinations in various in vitro and ex vivo HIV-1 latency models identified bryostatin-1+JQ1 and ingenol-B+JQ1 to potently reactivate viral gene expression. PLoS Pathog. 11 (7), e1005063. doi: 10.1371/journal.ppat.1005063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A. T., Binda C. S., Berkhout B. (2019). Elimination of infectious HIV DNA by CRISPR-Cas9. Curr. Opin. Virol. 38, 81–88. doi: 10.1016/j.coviro.2019.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debyser Z., Vansant G., Bruggemans A., Janssens J., Christ F.. (2018). Insight in HIV integration site selection provides a block-and-Lock strategy for a functional cure of HIV infection. Viruses 11 (1), 12. doi: 10.3390/v11010012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy A., Poirier M., Bell B. (2019). Cat and mouse: HIV transcription in latency, immune evasion and Cure/Remission strategies. Viruses 11 (3), 269. doi: 10.3390/v11030269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleage C., Chan C. N., Busman-Sahay K., Estes J. D. (2018). Next-generation in situ hybridization approaches to define and quantify HIV and SIV reservoirs in tissue microenvironments. Retrovirology 15 (1), 4. doi: 10.1186/s12977-017-0387-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleage C., Moreau M., Rioux-Leclercq N., Ruffault A., Jégou B., Dejucq-Rainsford N.. (2011). Human immunodeficiency virus infects human seminal vesicles in vitro and in vivo. Am. J. Pathol. 179 (5), 2397–2408. doi: 10.1016/j.ajpath.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue D. A., Bastarache S. M., Sloan R. D., Wainberg M. A. (2013). Latent HIV-1 can be reactivated by cellular superinfection in a tat-dependent manner, which can lead to the emergence of multidrug-resistant recombinant viruses. J. Virol. 87 (17), 9620–9632. doi: 10.1128/jvi.01165-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue D. A., Kuhl B. D., Sloan R. D., Wainberg M. A. (2012). The viral protein tat can inhibit the establishment of HIV-1 latency. J. Virol. 86 (6), 3253–3263. doi: 10.1128/jvi.06648-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon G., Sobolewski M. D., Huber K., McMahon D., Mellors J. W., Sluis-Cremer N.. (2014). Discovery of a small molecule agonist of phosphatidylinositol 3-kinase p110α that reactivates latent HIV-1. PloS One 9 (1), e84964. doi: 10.1371/journal.pone.0084964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes J. D., Kityo C., Ssali F., Swainson L., Makamdop K. N., Del Prete G. Q., et al. (2017). Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 23 (11), 1271–1276. doi: 10.1038/nm.4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G., Pollara J., Tomaras G. D., Haynes B. F. (2017). Humoral and innate antiviral immunity as tools to clear persistent HIV infection. J. Infect. Dis. 215 (suppl_3), S152–s159. doi: 10.1093/infdis/jiw555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C. V., Staskus K., Wietgrefe S. W., Rothenberger M., Reilly C., Chipman J. G., et al. (2014). Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. U.S.A. 111 (6), 2307–2312. doi: 10.1073/pnas.1318249111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank I., Acharya A., Routhu N. K., Aravantinou M., Harper J. L., Maldonado S., et al. (2019). A Tat/Rev induced limiting dilution assay to measure viral reservoirs in non-human primate models of HIV infection. Sci. Rep. 9 (1), 12078. doi: 10.1038/s41598-019-48354-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromentin R., Bakeman W., Lawani M. B., Khoury G., Hartogensis W., DaFonseca S., et al. (2016). CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PloS Pathog. 12 (7), e1005761. doi: 10.1371/journal.ppat.1005761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y., Lum R., Okoye A. A., Park H., Matsuda K., Bae J. Y., et al. (2015). B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat. Med. 21 (2), 132–139. doi: 10.1038/nm.3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fun A., Mok H. P., Wills M. R., Lever A. M. (2017). A highly reproducible quantitative viral outgrowth assay for the measurement of the replication-competent latent HIV-1 reservoir. Sci. Rep. 7, 43231. doi: 10.1038/srep43231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Falcinelli S. D., Stoffel E., Read J., Murtagh R., Oliveira T. Y., et al. (2021). Sequence evaluation and comparative analysis of novel assays for intact proviral HIV-1 DNA. J. Virol. 95 (6), e01986–20. doi: 10.1128/jvi.01986-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gálvez C., Grau-Expósito J., Urrea V., Clotet B., Falcó V., Buzón M. J., et al. (2021). Atlas of the HIV-1 reservoir in peripheral CD4 T cells of individuals on successful antiretroviral therapy. mBio 12 (6), e0307821. doi: 10.1128/mBio.03078-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan M., Poluektova L. Y., Kharbanda K. K., Osna N. A. (2018). Liver as a target of human immunodeficiency virus infection. World J. Gastroenterol. 24 (42), 4728–4737. doi: 10.3748/wjg.v24.i42.4728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganor Y., Real F., Sennepin A., Dutertre C. A., Prevedel L., Xu L., et al. (2019). HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 4 (4), 633–644. doi: 10.1038/s41564-018-0335-z [DOI] [PubMed] [Google Scholar]
- Ganor Y., Zhou Z., Bodo J., Tudor D., Leibowitch J., Mathez D., et al. (2013). The adult penile urethra is a novel entry site for HIV-1 that preferentially targets resident urethral macrophages. Mucosal Immunol. 6 (4), 776–786. doi: 10.1038/mi.2012.116 [DOI] [PubMed] [Google Scholar]
- García M., Buzón M. J., Benito J. M., Rallón N. (2018). Peering into the HIV reservoir. Rev. Med. Virol. 28 (4), e1981. doi: 10.1002/rmv.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M., López-Fernández L., Mínguez P., Morón-López S., Restrepo C., Navarrete-Muñoz M. A., et al. (2020). Transcriptional signature of resting-memory CD4 T cells differentiates spontaneous from treatment-induced HIV control. J. Mol. Med. (Berl) 98 (8), 1093–1105. doi: 10.1007/s00109-020-01930-x [DOI] [PubMed] [Google Scholar]
- Garrido C., Abad-Fernandez M., Tuyishime M., Pollara J. J., Ferrari G., Soriano-Sarabia N., et al. (2018). Interleukin-15-Stimulated natural killer cells clear HIV-1-Infected cells following latency reversal ex vivo. J. Virol. 92 (12), e00235–18. doi: 10.1128/jvi.00235-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini L., Pecorini S., De Biasi S., Bianchini E., Digaetano M., Pinti M., et al. (2017). HIV-DNA Content in different CD4+ T-cell subsets correlates with CD4+ cell : CD8+ cell ratio or length of efficient treatment. Aids 31 (10), 1387–1392. doi: 10.1097/qad.0000000000001510 [DOI] [PubMed] [Google Scholar]
- Gringhuis S. I., Hertoghs N., Kaptein T. M., Zijlstra-Willems E. M., Sarrami-Forooshani R., Sprokholt J. K., et al. (2017). HIV-1 blocks the signaling adaptor MAVS to evade antiviral host defense after sensing of abortive HIV-1 RNA by the host helicase DDX3. Nat. Immunol. 18 (2), 225–235. doi: 10.1038/ni.3647 [DOI] [PubMed] [Google Scholar]
- Groot F., van Capel T. M., Kapsenberg M. L., Berkhout B., de Jong E.C.. (2006). Opposing roles of blood myeloid and plasmacytoid dendritic cells in HIV-1 infection of T cells: transmission facilitation versus replication inhibition. Blood 108 (6), 1957–1964. doi: 10.1182/blood-2006-03-010918 [DOI] [PubMed] [Google Scholar]
- Guttenplan K. A., Liddelow S. A. (2019). Astrocytes and microglia: Models and tools. J. Exp. Med. 216 (1), 71–83. doi: 10.1084/jem.20180200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich T. J., Hatano H., Bacon O., Hogan L. E., Rutishauser R., Hill A., et al. (2017). HIV-1 persistence following extremely early initiation of antiretroviral therapy (ART) during acute HIV-1 infection: An observational study. PloS Med. 14 (11), e1002417. doi: 10.1371/journal.pmed.1002417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Carrillo E., Gao Z., Berkhout B. (2020). CRISPR therapy towards an HIV cure. Brief Funct. Genomics 19 (3), 201–208. doi: 10.1093/bfgp/elz021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeycutt J. B., Thayer W. O., Baker C. E., Ribeiro R. M., Lada S. M., Cao Y., et al. (2017). HIV Persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat. Med. 23 (5), 638–643. doi: 10.1038/nm.4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. C., Shan L., Hosmane N. N., Wang J., Laskey S. B., Rosenbloom D. I., et al. (2013). Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155 (3), 540–551. doi: 10.1016/j.cell.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean M. J., Hayashi T., Huang H., Brennan J., Simpson S., Purmal A., et al. (2017). Curaxin CBL0100 blocks HIV-1 replication and reactivation through inhibition of viral transcriptional elongation. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katuri A., Bryant J. L., Patel D., Patel V., Andhavarapu S., Asemu G., et al. (2019). HIVAN associated tubular pathology with reference to ER stress, mitochondrial changes, and autophagy. Exp. Mol. Pathol. 106, 139–148. doi: 10.1016/j.yexmp.2018.12.009 [DOI] [PubMed] [Google Scholar]
- Keele B. F., Tazi L., Gartner S., Liu Y., Burgon T. B., Estes J. D., et al. (2008). Characterization of the follicular dendritic cell reservoir of human immunodeficiency virus type 1. J. Virol. 82 (11), 5548–5561. doi: 10.1128/jvi.00124-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessing C. F., Nixon C. C., Li C., Tsai P., Takata H., Mousseau G., et al. (2017). In vivo suppression of HIV rebound by didehydro-cortistatin a, a "Block-and-Lock" strategy for HIV-1 treatment. Cell Rep. 21 (3), 600–611. doi: 10.1016/j.celrep.2017.09.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Anderson J. L., Lewin S. R. (2018). Getting the "Kill" into "Shock and kill": Strategies to eliminate latent HIV. Cell Host Microbe 23 (1), 14–26. doi: 10.1016/j.chom.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A. C., Brill S. A., Queen S. E., Tarwater P. M., Mankowski J.L.. (2018). Increased microglial CSF1R expression in the SIV/Macaque model of HIV CNS disease. J. Neuropathol. Exp. Neurol. 77 (3), 199–206. doi: 10.1093/jnen/nlx115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight A. C., Brill S. A., Solis C. V., Richardson M. R., McCarron M. E., Queen S. E., et al. (2020). Differential regulation of TREM2 and CSF1R in CNS macrophages in an SIV/macaque model of HIV CNS disease. J. Neurovirol. 26 (4), 511–519. doi: 10.1007/s13365-020-00844-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristoff J., Rinaldo C. R., Mailliard R. B. (2019). Role of dendritic cells in exposing latent HIV-1 for the kill. Viruses 12 (1), 32. doi: 10.3390/v12010037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruize Z., Kootstra N. A. (2019). The role of macrophages in HIV-1 persistence and pathogenesis. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulpa D. A., Talla A., Brehm J. H., Ribeiro S. P., Yuan S., Bebin-Blackwell A. G., et al. (2019). Differentiation into an effector memory phenotype potentiates HIV-1 latency reversal in CD4(+) T cells. J. Virol. 93 (24), e00969–19. doi: 10.1128/jvi.00969-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Abbas W., Herbein G. (2014). HIV-1 latency in monocytes/macrophages. Viruses 6 (4), 1837–1860. doi: 10.3390/v6041837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzkes D. R. (2016). Hematopoietic stem cell transplantation for HIV cure. J. Clin. Invest. 126 (2), 432–437. doi: 10.1172/jci80563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K. J., Timmons A. E., Sengupta S., Simonetti F. R., Zhang H., Hoh R, et al. (2020). Different human resting memory CD4(+) T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci. Transl. Med. 12 (528), eaax6795. doi: 10.1126/scitranslmed.aax6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lada S. M., Huang K., VanBelzen D. J., Montaner L. J., O'Doherty U., Richman D.D.. (2018). Quantitation of integrated HIV provirus by pulsed-field gel electrophoresis and droplet digital PCR. J. Clin. Microbiol. 56 (12), e01158–18. doi: 10.1128/jcm.01158-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B., Sharron M., Montaner L. J., Weissman D., Doms R.W.. (1999). Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc. Natl. Acad. Sci. U.S.A. 96 (9), 5215–5220. doi: 10.1073/pnas.96.9.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Yin X., Shang H., Jiang Y. (2019). IP-10 is highly involved in HIV infection. Cytokine 115, 97–103. doi: 10.1016/j.cyto.2018.11.018 [DOI] [PubMed] [Google Scholar]
- Li X., Li H., Li C., Xia W., Li A., Li W.. (2020). Traditional Chinese medicine can improve the immune reconstruction of HIV/AIDS patients. AIDS Res. Hum. Retroviruses 36 (4), 258–259. doi: 10.1089/aid.2019.0274 [DOI] [PubMed] [Google Scholar]
- Limsirichai P., Gaj T., Schaffer D. V. (2016). CRISPR-mediated activation of latent HIV-1 expression. Mol. Ther. 24 (3), 499–507. doi: 10.1038/mt.2015.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Zhao J., Cheng L., Jiang Q., Kan S., Qin E., et al. (2017). HIV-1 infection depletes human CD34+CD38- hematopoietic progenitor cells via pDC-dependent mechanisms. PLoS Pathog. 13 (7), e1006505. doi: 10.1371/journal.ppat.1006505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Huertas M. R., Morín M., Madrid-Elena N., Gutiérrez C., Jiménez-Tormo L., Santoyo J., et al. (2019). Selective miRNA modulation fails to activate HIV replication in in vitro latency models. Mol. Ther. Nucleic Acids 17, 323–336. doi: 10.1016/j.omtn.2019.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loré K., Smed-Sörensen A., Vasudevan J., Mascola J. R., Koup R.A.. (2005). Myeloid and plasmacytoid dendritic cells transfer HIV-1 preferentially to antigen-specific CD4+ T cells. J. Exp. Med. 201 (12), 2023–2033. doi: 10.1084/jem.20042413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli E., Gattinoni L., Roberto A., Mavilio D., Price D. A., Restifo N. P., et al. (2013). Identification, isolation and in vitro expansion of human and nonhuman primate T stem cell memory cells. Nat. Protoc. 8 (1), 33–42. doi: 10.1038/nprot.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lungu C., Procopio F. A., Overmars R. J., Beerkens R. J.J., Voermans J. J.C., Rao S., et al. (2020). Inter-laboratory reproducibility of inducible HIV-1 reservoir quantification by TILDA. Viruses 12 (9). doi: 10.3390/v12090973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgen V., Narasipura S. D., Barbian H. J., Richards M., Wallace J., Razmpour R., et al. (2020). HIV Infects astrocytes in vivo and egresses from the brain to the periphery. PloS Pathog. 16 (6), e1008381. doi: 10.1371/journal.ppat.1008381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D. Y., Wu H. Y., Yarla N. S., Xu B., Ding J., Lu T. R.. (2018). HAART in HIV/AIDS treatments: Future trends. Infect. Disord. Drug Targets 18 (1), 15–22. doi: 10.2174/1871526517666170505122800 [DOI] [PubMed] [Google Scholar]
- Madrid-Elena N., García-Bermejo M. L., Serrano-Villar S., Díaz-de Santiago A., Sastre B., Gutiérrez C., et al. (2018). Maraviroc is associated with latent HIV-1 reactivation through NF-κB activation in resting CD4(+) T cells from HIV-infected individuals on suppressive antiretroviral therapy. J. Virol. 92 (9), e01931–17. doi: 10.1128/jvi.01931-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maina E. K., Adan A. A., Mureithi H., Muriuki J., Lwembe R.M.. (2021). A review of current strategies towards the elimination of latent HIV-1 and subsequent HIV-1 cure. Curr. HIV Res. 19 (1), 14–26. doi: 10.2174/1570162x18999200819172009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Navio J. M., Fuchs S. P., Pantry S. N., Lauer W. A., Duggan N. N., Keele B. F., et al. (2019). Adeno-associated virus delivery of anti-HIV monoclonal antibodies can drive long-term virologic suppression. Immunity 50 (3), 567–575.e5. doi: 10.1016/j.immuni.2019.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massanella M., Yek C., Lada S. M., Nakazawa M., Shefa N., Huang K, et al. (2018). Improved assays to measure and characterize the inducible HIV reservoir. EBioMedicine 36, 113–121. doi: 10.1016/j.ebiom.2018.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Yang T., Luo Y., Wu L., Jiang Y., Song Z., et al. (2019). TRIM28 promotes HIV-1 latency by SUMOylating CDK9 and inhibiting p-TEFb. Elife 8. doi: 10.7554/eLife.42426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Wu L., Bohks S. M., KewalRamani V. N., Unutmaz D., Hope T. J., et al. (2003). Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 300 (5623), 1295–1297. doi: 10.1126/science.1084238 [DOI] [PubMed] [Google Scholar]
- McNamara L. A., Onafuwa-Nuga A., Sebastian N. T., Riddell J., Bixby D., Collins K.L. (2013). CD133+ hematopoietic progenitor cells harbor HIV genomes in a subset of optimally treated people with long-term viral suppression. J. Infect. Dis. 207 (12), 1807–1816. doi: 10.1093/infdis/jit118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez C., Ledger S., Petoumenos K., Ahlenstiel C., Kelleher A. D.. (2018). RNA-Induced epigenetic silencing inhibits HIV-1 reactivation from latency. Retrovirology 15 (1), 67. doi: 10.1186/s12977-018-0451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. H., 3rd, Valente S. T. (2020). Block-And-Lock: New horizons for a cure for HIV-1. Viruses 12 (12). doi: 10.3390/v12121443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. H., 3rd, Nguyen T., Nwadike A., Geels M. L., Kamp D. L., Kim B. R., et al. (2017). Soluble factors secreted by endothelial cells allow for productive and latent HIV-1 infection in resting CD4(+) T cells. AIDS Res. Hum. Retroviruses 33 (2), 110–120. doi: 10.1089/aid.2016.0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. M., Zaunders J. J., McBride K. L., Xu Y., Bailey M., Suzuki K., et al. (2014). HIV DNA Subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J. Virol. 88 (6), 3516–3526. doi: 10.1128/jvi.03331-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon C. C., Mavigner M., Sampey G. C., Brooks A. D., Spagnuolo R. A., Irlbeck D. M., et al. (2020). Systemic HIV and SIV latency reversal via non-canonical NF-κB signalling in vivo. Nature 578 (7793), 160–165. doi: 10.1038/s41586-020-1951-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okutomi T., Minakawa S., Hirota R., Katagiri K., Morikawa Y.. (2020). HIV Reactivation in latently infected cells with virological synapse-like cell contact. Viruses 12 (4):417. doi: 10.3390/v12040417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivetta E., Chiozzini C., Arenaccio C., Manfredi F., Ferrantelli F., Federico M.. (2020). Extracellular vesicle-mediated intercellular communication in HIV-1 infection and its role in the reservoir maintenance. Cytokine Growth Factor Rev. 51, 40–48. doi: 10.1016/j.cytogfr.2019.12.006 [DOI] [PubMed] [Google Scholar]
- Ollerton M. T., Berger E. A., Connick E., Burton G. F. (2020). HIV-1-Specific chimeric antigen receptor T cells fail to recognize and eliminate the follicular dendritic cell HIV reservoir in vitro. J. Virol. 94 (10):e00190–20. doi: 10.1128/jvi.00190-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil T. R., Hu K., Truong N. R., Arshad S., Shacklett B. L., Cunningham A. L., et al. (2021). The role of tissue resident memory CD4 T cells in herpes simplex viral and HIV infection. Viruses 13 (3):359. doi: 10.3390/v13030359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortblad K. F., Lozano R., Murray C. J. (2013). The burden of HIV: insights from the global burden of disease study 2010. Aids 27 (13), 2003–2017. doi: 10.1097/QAD.0b013e328362ba67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankrac J., Klein K., Mann J. F. S. (2017). Eradication of HIV-1 latent reservoirs through therapeutic vaccination. AIDS Res. Ther. 14 (1), 45. doi: 10.1186/s12981-017-0177-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardons M., Baxter A. E., Massanella M., Pagliuzza A., Fromentin R., Dufour C., et al. (2019). Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PloS Pathog. 15 (2), e1007619. doi: 10.1371/journal.ppat.1007619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons M. S., Cromer D., Davenport M. P., Kent S. J. (2018). HIV Reactivation after partial protection by neutralizing antibodies. Trends Immunol. 39 (5), 359–366. doi: 10.1016/j.it.2017.12.006 [DOI] [PubMed] [Google Scholar]
- Pearson R., Kim Y. K., Hokello J., Lassen K., Friedman J., Tyagi M., et al. (2008). Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82 (24), 12291–12303. doi: 10.1128/jvi.01383-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedro K. D., Henderson A. J., Agosto L. M. (2019). Mechanisms of HIV-1 cell-to-cell transmission and the establishment of the latent reservoir. Virus Res. 265, 115–121. doi: 10.1016/j.virusres.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D. O., DeMarino C., Vo T. T., Cowen M., Kim Y., Pleet M. L., et al. (2020). Low-level ionizing radiation induces selective killing of HIV-1-Infected cells with reversal of cytokine induction using mTOR inhibitors. Viruses 12 (8):885. doi: 10.3390/v12080885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope M., Gezelter S., Gallo N., Hoffman L., Steinman R. M.. (1995). Low levels of HIV-1 infection in cutaneous dendritic cells promote extensive viral replication upon binding to memory CD4+ T cells. J. Exp. Med. 182 (6), 2045–2056. doi: 10.1084/jem.182.6.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z., Zhang Y., Xie X., Wang J. (2021). Efficacy and safety of traditional Chinese herbal medicine combined with HAART in the treatment of HIV/AIDS: A protocol for systematic review and meta-analysis. Med. (Baltimore) 100 (52), e28287. doi: 10.1097/md.0000000000028287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T. A., Tolstrup M., Søgaard O. S. (2016). Reversal of latency as part of a cure for HIV-1. Trends Microbiol. 24 (2), 90–97. doi: 10.1016/j.tim.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Rezaei S. D., Lu H. K., Chang J. J., Rhodes A., Lewin S. R., Cameron P. U.. (2018). The pathway to establishing HIV latency is critical to how latency is maintained and reversed. J. Virol. 92 (13):e02225–17. doi: 10.1128/jvi.02225-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh S., Solomon A., Wightman F., Xhilaga M., Cameron P. U., Lewin S. R. (2007). Promising role of toll-like receptor 8 agonist in concert with prostratin for activation of silent HIV. J. Virol. 91 (4):e02084–16. doi: 10.1128/jvi.02084-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Hashemi F. B. (2019). Strategies to eradicate HIV from infected patients: elimination of latent provirus reservoirs. Cell Mol. Life Sci. 76 (18), 3583–3600. doi: 10.1007/s00018-019-03156-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini M., Potash M. J. (2014). Chronic, highly productive HIV infection in monocytes during supportive culture. Curr. HIV Res. 12 (5), 317–324. doi: 10.2174/1570162x1205141121100659 [DOI] [PubMed] [Google Scholar]
- Saleh S., Solomon A., Wightman F., et al. (2007). CCR7 ligands CCL19 and CCL21 increase permissiveness of resting memory CD4+ T cells to HIV-1 infection: a novel model of HIV-1 latency. Blood 110 (13), 4161–4164. doi: 10.1182/blood-2007-06-097907 [DOI] [PubMed] [Google Scholar]
- Schneider M., Tigges B., Meggendorfer M., Ziegenhain C., Brack-Werner R.. (2015). A new model for post-integration latency in macroglial cells to study HIV-1 reservoirs of the brain. Aids 29 (10), 1147–1159. doi: 10.1097/qad.0000000000000691 [DOI] [PubMed] [Google Scholar]
- Schulze-Gahmen U., Hurley J. H. (2018). Structural mechanism for HIV-1 TAR loop recognition by tat and the super elongation complex. Proc. Natl. Acad. Sci. U.S.A. 115 (51), 12973–12978. doi: 10.1073/pnas.1806438115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C., Bouchat S., Marban C., Gautier V., Van Lint C., Rohr O., et al. (2017). On the way to find a cure: Purging latent HIV-1 reservoirs. Biochem. Pharmacol. 146, 10–22. doi: 10.1016/j.bcp.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Sengupta S., Siliciano R. F. (2018). Targeting the latent reservoir for HIV-1. Immunity 48 (5), 872–895. doi: 10.1016/j.immuni.2018.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., Deng K., Gao H., Xing S., Capoferri A. A., Durand C. M., et al. (2017). Transcriptional reprogramming during effector-to-Memory transition renders CD4(+) T cells permissive for latent HIV-1 infection. Immunity 47 (4), 766–775.e3. doi: 10.1016/j.immuni.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smed-Sörensen A., Loré K., Vasudevan J., Louder M. K., Andersson J., Mascola J. R., et al. (2005). Differential susceptibility to human immunodeficiency virus type 1 infection of myeloid and plasmacytoid dendritic cells. J. Virol. 79 (14), 8861–8869. doi: 10.1128/jvi.79.14.8861-8869.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B. A., Gartner S., Liu Y., Perelson A. S., Stilianakis N. I., Keele B. F., et al. (2001). Persistence of infectious HIV on follicular dendritic cells. J. Immunol. 166 (1), 690–696. doi: 10.4049/jimmunol.166.1.690 [DOI] [PubMed] [Google Scholar]
- Smith K. A., Lin X., Bolshakov O., Griffin J., Niu X., Kovalskyy D., et al. (2015). Activation of HIV-1 with nanoparticle-packaged small-molecule protein phosphatase-1-Targeting compound. Sci. Pharm. 83 (3), 535–548. doi: 10.3797/scipharm.1502-01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak A. M., Planelles V. (2018). Novel latency reversal agents for HIV-1 cure. Annu. Rev. Med. 69, 421–436. doi: 10.1146/annurev-med-052716-031710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons J., Cameron P. U., Lewin S. R. (2018). HIV integration sites and implications for maintenance of the reservoir. Curr. Opin. HIV AIDS 13 (2), 152–159. doi: 10.1097/coh.0000000000000438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabler C. O., Lucera M. B., Haqqani A. A., McDonald D. J., Migueles S. A., Connors M., et al. (2014). CD4+ memory stem cells are infected by HIV-1 in a manner regulated in part by SAMHD1 expression. J. Virol. 88 (9), 4976–4986. doi: 10.1128/jvi.00324-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Lu H., Dooner M., Chapman S., Quesenberry P. J., Ramratnam B.. (2018). Exosomal tat protein activates latent HIV-1 in primary, resting CD4+ T lymphocytes. JCI Insight 3 (7):e95676. doi: 10.1172/jci.insight.95676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terahara K., Iwabuchi R., Hosokawa M., Nishikawa Y., Takeyama H., Takahashi Y., et al. (2019). A CCR5(+) memory subset within HIV-1-infected primary resting CD4(+) T cells is permissive for replication-competent, latently infected viruses in vitro. BMC Res. Notes 12 (1), 242. doi: 10.1186/s13104-019-4281-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theys K., Libin P., Pineda-Peña A. C., Nowé A., Vandamme A. M., Abecasis A. B.. (2018). The impact of HIV-1 within-host evolution on transmission dynamics. Curr. Opin. Virol. 28, 92–101. doi: 10.1016/j.coviro.2017.12.001 [DOI] [PubMed] [Google Scholar]
- Thomas J., Ruggiero A., Procopio F. A., Pantaleo G., Paxton W. A., Pollakis G.. (2019). Comparative analysis and generation of a robust HIV-1 DNA quantification assay. J. Virol. Methods 263, 24–31. doi: 10.1016/j.jviromet.2018.10.010 [DOI] [PubMed] [Google Scholar]
- Thompson K. A., Cherry C. L., Bell J. E., McLean C. A. (2011). Brain cell reservoirs of latent virus in presymptomatic HIV-infected individuals. Am. J. Pathol. 179 (4), 1623–1629. doi: 10.1016/j.ajpath.2011.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis M. A., Sheppard D. (2014). TGF-β activation and function in immunity. Annu. Rev. Immunol. 32, 51–82. doi: 10.1146/annurev-immunol-032713-120257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M., Iordanskiy S., Ammosova T., Kumari N., Smith K., Breuer D., et al. (2015). Reactivation of latent HIV-1 provirus via targeting protein phosphatase-1. Retrovirology 12, 63. doi: 10.1186/s12977-015-0190-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M., Pearson R. J., Karn J. (2010). Establishment of HIV latency in primary CD4+ cells is due to epigenetic transcriptional silencing and p-TEFb restriction. J. Virol. 84 (13), 6425–6437. doi: 10.1128/jvi.01519-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandergeeten C., Fromentin R., Chomont N. (2012). The role of cytokines in the establishment, persistence and eradication of the HIV reservoir. Cytokine Growth Factor Rev. 23 (4-5), 143–149. doi: 10.1016/j.cytogfr.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandergeeten C., Fromentin R., Merlini E., Lawani M. B., DaFonseca S., Bakeman W., et al. (2014). Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large-cohort studies. J. Virol. 88 (21), 12385–12396. doi: 10.1128/jvi.00609-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vansant G., Bruggemans A., Janssens J., Debyser Z. (2020). Block-And-Lock strategies to cure HIV infection. Viruses 12 (1):84. doi: 10.3390/v12010084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zyl G., Bale M. J., Kearney M. F. (2018). HIV Evolution and diversity in ART-treated patients. Retrovirology 15 (1), 14. doi: 10.1186/s12977-018-0395-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenhuis R. T., Clements J. E., Gama L. (2019). HIV Eradication strategies: Implications for the central nervous system. Curr. HIV/AIDS Rep. 16 (1), 96–104. doi: 10.1007/s11904-019-00428-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verna G., Liso M., Cavalcanti E., Bianco G., Di Sarno V., Santino A., et al. (2021). Quercetin administration suppresses the cytokine storm in myeloid and plasmacytoid dendritic cells. Int. J. Mol. Sci. 22 (15):8349. doi: 10.3390/ijms22158349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallet C., De Rovere M., Van Assche J., Daouad F., De Wit S., Gautier V., et al. (2019). Microglial cells: The main HIV-1 reservoir in the brain. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Lu P., Qu X., Shen Y., Zeng H., Zhu X., et al. (2017). Reactivation of HIV-1 from latency by an ingenol derivative from euphorbia kansui. Sci. Rep. 7 (1), 9451. doi: 10.1038/s41598-017-07157-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Simonetti F. R., Siliciano R. F., Laird G. M. (2018). Measuring replication competent HIV-1: advances and challenges in defining the latent reservoir. Retrovirology 15 (1), 21. doi: 10.1186/s12977-018-0404-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yin X., Ma M., Ge H., Lang B., Sun H., et al. (2021). IP-10 promotes latent HIV infection in resting memory CD4(+) T cells via LIMK-cofilin pathway. Front. Immunol. 12. doi: 10.3389/fimmu.2021.656663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayengera M. (2011). Proviral HIV-genome-wide and pol-gene specific zinc finger nucleases: usability for targeted HIV gene therapy. Theor. Biol. Med. Model. 8, 26. doi: 10.1186/1742-4682-8-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. B., Brooks D. G. (2011). The role of IL-10 in regulating immunity to persistent viral infections. Curr. Top. Microbiol. Immunol. 350, 39–65. doi: 10.1007/82_2010_96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonderlich E. R., Subramanian K., Cox B., Wiegand A., Lackman-Smith C., Bale M. J., et al. (2019). Effector memory differentiation increases detection of replication-competent HIV-l in resting CD4+ T cells from virally suppressed individuals. PLoS Pathog. 15 (10), e1008074. doi: 10.1371/journal.ppat.1008074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs T., Hammerschmidt S. I., Förster R. (2017). Dendritic cell migration in health and disease. Nat. Rev. Immunol. 17 (1), 30–48. doi: 10.1038/nri.2016.116 [DOI] [PubMed] [Google Scholar]
- Wu Y. (2010). Chemokine control of HIV-1 infection: beyond a binding competition. Retrovirology 7, 86. doi: 10.1186/1742-4690-7-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Q., Guo D., Chen S. (2019). Application of CRISPR/Cas9-based gene editing in HIV-1/AIDS therapy. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Buisson S., Bossi G., Wallace Z., Hancock G., So C., et al. (2016). Elimination of latently HIV-infected cells from antiretroviral therapy-suppressed subjects by engineered immune-mobilizing T-cell receptors. Mol. Ther. 24 (11), 1913–1925. doi: 10.1038/mt.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X., Chen S., Eisenbarth S. C. (2021). Dendritic cell regulation of T helper cells. Annu. Rev. Immunol. 39, 759–790. doi: 10.1146/annurev-immunol-101819-025146 [DOI] [PubMed] [Google Scholar]
- Yin X., Langer S., Zhang Z., Herbert K. M., Yoh S., König R., et al. (2020). Sensor sensibility-HIV-1 and the innate immune response. Cells 9 (1). doi: 10.3390/cells9010254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. J., Reuter M. A., McDonald D. (2008). HIV Traffics through a specialized, surface-accessible intracellular compartment during trans-infection of T cells by mature dendritic cells. PLoS Pathog. 4 (8), e1000134. doi: 10.1371/journal.ppat.1000134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbato J. M., McMahon D. K., Sobolewski M. D., Mellors J. W., Sluis-Cremer N.. (2019). Naive CD4+ T cells harbor a Large inducible reservoir of latent, replication-competent human immunodeficiency virus type 1. Clin. Infect. Dis. 69 (11), 1919–1925. doi: 10.1093/cid/ciz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbato J. M., Serrao E., Lenzi G., Kim B., Ambrose Z., Watkins S. C., et al. (2016). Establishment and reversal of HIV-1 latency in naive and central memory CD4+ T cells in vitro. J. Virol. 90 (18), 8059–8073. doi: 10.1128/jvi.00553-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Yin C., Zhang T., Li F., Yang W., Kaminski R., et al. (2015). CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 5, 16277. doi: 10.1038/srep16277 [DOI] [PMC free article] [PubMed] [Google Scholar]