Significance
Uterine receptivity is indispensable for normal embryo implantation. Uterine receptivity is mainly dominated by ovarian estrogen and progesterone through their respective nuclear receptors ER and PR, expressed in the endometrium. However, how their homeostasis is maintained at protein level has been less studied. Here, we showed that the deficiency of uterine Mitogen-activated protein kinase p38 alpha (P38α) derailed normal uterine receptivity establishment, mainly due to the significant down-regulation of the PR protein rather than its transcript in the uterine stromal cells. Further exploration uncovered that the ubiquitin ligase Ube3c was afforded responsibility for PR polyubiquitination and protein degradation and that P38α could phosphorylate the Ube3c to restrain PR protein polyubiquitination. Our finding revealed a regulation manner for uterine progesterone responsiveness.
Keywords: uterine receptivity, P38α, progesterone receptor, Ube3c
Abstract
Estrogen and progesterone specify the establishment of uterine receptivity mainly through their respective nuclear receptors, ER and PR. PR is transcriptionally induced by estrogen–ER signaling in the endometrium, but how the protein homeostasis of PR in the endometrium is regulated remains elusive. Here, we demonstrated that the uterine-selective depletion of P38α derails normal uterine receptivity ascribed to the dramatic down-regulation of PR protein and disordered progesterone responsiveness in the uterine stromal compartment, leading to defective implantation and female infertility. Specifically, Ube3c, an HECT family E3 ubiquitin ligase, targets PR for polyubiquitination and thus proteasome degradation in the absence of P38α. Moreover, we discovered that P38α restrains the polyubiquitination activity of Ube3c toward PR by phosphorylating the Ube3c at serine741 . In summary, we provided genetic evidence for the regulation of PR protein stability in the endometrium by P38α and identified Ube3c, whose activity was modulated by P38α-mediated phosphorylation, as an E3 ubiquitin ligase for PR in the uterus.
In mammals, the punctual occurrence of embryo implantation during early pregnancy, whereby the embryo and the maternal uterus are in close contact and establish a direct physiological connection, determines the normal ongoing embryo development and the final pregnancy outcome (1). The synchronization of blastocyst activation and the establishment of uterine receptivity is indispensable for successful implantation (2). Uterine receptivity establishment is mainly coordinated by ovarian estrogen and progesterone, involving the specific spatiotemporal expression of a series of signal molecules (3, 4). In mice, estrogen and progesterone spatiotemporally regulate the proliferation and differentiation of uterine epithelial and stromal cells, which control the adaptation of the uterus to ensure a propitious uterine receptive state for blastocyst implantation (5, 6). Estrogen and progesterone function mainly through their nuclear receptors: the estrogen receptor (ERα/ERβ) and the progesterone receptor (PRA/PRB) (7, 8). Evidence from knockout mice has shown that PR is critical for uterine function as PR-null female mice are unable to support embryo implantation (9). To date, a large number of PR regulated downstream genes have been identified, revealing the essential role of progesterone-dependent pathways in regulating implantation and subsequent decidualization (10). Meanwhile, some molecules, such as Steroid receptor coactivators(SRCs) and Polycomb complex protein Bmi1, have been demonstrated to act as cofactors for PR during transcription (11, 12). Besides these, more regulation manners for the precise modulation of PR activity during early pregnancy remain to be explored for comprehensively understanding the mechanism of PR functions.
In order to make corresponding differentiation adapted to pregnancy, the states and functions of uterine cells show dramatic changes under the stimulation of various extracellular signals, like hormones and embryonic signals. As one of the dominant pathways involved in external signal transduction, the function of the P38 MAPK serine/threonine protein kinase family during embryo implantation is far from clear. The P38 MAPK family contains four members: P38α (Mapk14), P38β (Mapk11), P38γ (Mapk12), and P38δ (Mapk13) (13). Although they share approximately 60% of the amino acid sequence, they are encoded by different genes and exhibit distinct expression patterns. Most cell types show a high level of P38α expression, while expressions of other P38 MAPKs are tissue-specific (14). Phosphorylation of target proteins by P38 MAPK affects various aspects of their functions, including protein stability, DNA binding, subcellular localization, and protein–protein interactions (15, 16). P38α was initially identified as a protein kinase related to stress and inflammation, and inflammatory-like reactions were also considered to exist in the uterus during embryo implantation (2, 17).
Previous studies using the P38 MAPK inhibitor SB203580 have suggested that the induction of the COX2-PPARδ inflammatory signal requires P38 MAPK activation during decidualization (18). Therefore, to further explore the potential role of uterine P38α in peri-implantation events, we employed a genetic mouse model with uterine-specific deletion of P38α and provided evidence that mice with P38α deficiency in the uterus showed disrupted uterine receptivity and hindered embryo implantation due to the decreased PR protein level and defective PR transcriptional activity. Furthermore, the E3 ubiquitin ligase Ube3c was identified to promote the polyubiquitination of PR at the position 416, 441, and 445 lysine residue, resulting in proteasome-mediated degradation, and this activity was inhibited by the P38α-regulated phosphorylation of Ube3c at the position 741 serine residue.
Results
Mice with Uterine P38α Deletion Show Compromised Fertility Due to Impaired Implantation.
To gain insight into the expression pattern of P38α and its activated form phospho-P38α in the peri-implantation uteri, we performed in situ hybridization (ISH) and immunohistochemistry (IHC), and the results showed that the messenger RNA (mRNA) and protein of P38α were spatiotemporally expressed in the peri-implantation uterus (Fig. 1A and SI Appendix, Fig. S1A). On D1 (the day when the vaginal plug is seen), the localization of P38α was observed in luminal and glandular epithelial cells, accompanied with a weak expression in the subepithelial uterine stromal cells. On D4, when the uteri were stimulated by the rising progesterone produced by the newly formed corpora luteum from D3, P38α was expressed extensively in the subepithelial uterine stromal region. With the onset of implantation on D5, the expression of P38α expanded to the stromal cells surrounding the implanting embryo, accompanied with higher protein levels. In the wake of the initiation of decidualization on D6, P38α was observed in the decidualizing cells. Its expression remained highly immense along with the progression of endometrium-decidual transformation until the decidua was fully developed on D8 (Fig. 1A and SI Appendix, Fig. S1A). However, other members of the P38 MAPK family exhibited a relatively low expression level in stromal cells throughout the peri-implantation period (SI Appendix, Fig. S2 A–C). We noticed that the expression pattern of phospho-P38 in the peri-implantation uterus overlapped with P38α in a spatiotemporal manner (SI Appendix, Fig. S1B), suggesting that P38α was activated during the stage of uterine receptivity establishment and decidual development.
Fig. 1.
P38α is indispensable for normal implantation. (A) ISH of Mapk14 in D1, D4, D5, D6, and D8 uteri. The pink site indicates the location of Mapk14. Positive signals were not detected in the uterus labeled with the Sense probe. Le, luminal epithelium; S, stroma; Bl, blastocyst; Em, embryo (Scale bars, 100 μm). (B) qRT-PCR analysis to reveal the knockout efficiency of Mapk14 in P38αf/f and P38αd/d uteri on D4. Data shown represent the means ± SEM; ***P < 0.001. (C and D) IHC and Western blot analysis revealed the efficient ablation of P38α and p-P38 at protein level (Scale bars, 100 μm). (E) Average litter sizes in P38αf/f and P38αd/d mice. Number within the bar indicates the number of mice tested. (F) Gross morphological implantation sites in P38αf/f mice compared with P38αd/d mice as determined by blue dye injection on D5. The unimplanted embryos were recovered from P38αd/d uteri (Scale bars, 100 μm). (G) Average number of implantation sites in P38αf/f and P38αd/d mice on D5 morning of pregnancy. Mice that failed to recover any embryos were excluded in statistical analysis. Number within the bar indicates the number of mice tested. (H) Morphological implantation sites in P38αf/f mice compared with P38αd/d mice as determined by blue dye injection on D6. (I) Average number of implantation sites in P38αf/f and P38αd/d mice on D6 morning of pregnancy. Number within the bar indicates the number of mice tested. **P < 0.01. (J) Representative hematoxylin and eosin staining of cross-sections of P38αf/f and P38αd/d implantation sites on D5 (Scale bars, 100 μm). (K) IHC staining of COX2 in P38αf/f and P38αd/d uteri on D5 of pregnancy (Scale bars, 100 μm).
Given that the systemic P38α-null mice are embryonic lethal (19), we generated a mouse model harboring a uterine-specific deletion of Mapk14 (hereafter referred to as P38αd/d) by crossing Mapk14-loxp mice (hereafter referred to as P38αf/f) with progesterone receptor (Pgr)–driven Cre (Pgr-Cre) mice to examine the potential physiological functions of P38α in mouse uteri. The knockout efficiency was confirmed by qRT-PCR, IHC, and western blot (WB) (Fig. 1 B–D). To assess the physiological relevance of P38α in female reproduction, we examined pregnancy outcomes in P38αf/f and P38αd/d female mice crossed with wild-type fertile male mice of the same genetic background. We found that female mice of both genotypes mated normally and formed vaginal plugs. However, P38αd/d female mice were completely infertile (Fig. 1E). Then we compared the implantation status in P38αf/f and P38αd/d female mice. P38αf/f mice showed normal embryo implantation on the D5 morning, which was visualized by a tail vein injection of blue dye. All P38αd/d female mice exhibited implantation failure with morphologically normal blastocysts recovered from the uterus (Fig. 1 F and G). In addition, we found an impaired implantation event with fewer implantation sites displaying a faint blue dye reaction in knockout mice even when analyzed on D6 (Fig. 1 H and I). Furthermore, the blastocyst was observed suspended in the P38αd/d uterine lumen, while implanted embryos were detected in P38αf/f mice on D5 and D6 (Fig. 1J and SI Appendix, Fig. S3A). The weak expression of uterine COX2 around the blastocyst on D5 and D6 further confirmed the abnormal blastocyst attachment reaction and subsequent decidual initiation in P38αd/d uteri (Fig. 1K and SI Appendix, Fig. S3B). These findings indicated that P38α is essential for normal on-time embryo implantation in mice.
We further noted that P38α deficiency greatly hampers the process of decidualization. The number of implantation sites still displayed a significant decline in P38αd/d mice on D8 (SI Appendix, Fig. S3 C and D). In addition, the morphology of the P38αd/d implantation sites appeared to be smaller than that in P38αf/f mice, consisting with a dramatic decrease in the average weight of implantation sites on D8 in P38αd/d mice (SI Appendix, Fig. S3 E and F). These results implied severe defects in the formation of decidual mass in the uteri of pregnant P38αd/d mice, leading to embryo growth retardation and mortality.
These data demonstrated that the pregnancy failure in P38αd/d mice was due to defects that existed around the time of peri-implantation, suggesting the importance of P38α signaling in normal pregnancy, particularly during the process of implantation.
Uterine P38α Deficiency Weakens Progesterone Responsiveness Resulting in Derailed Uterine Receptivity.
Since the embryos in P38αd/d female mice were wild type without genetic defects, we determined that the derailed uterine receptivity should be the main cause of the implantation failure in knockout mice. To reveal the molecular basis of uterine defects in the absence of P38α, we performed RNA sequencing (RNA-seq) to compare the differentially expressed genes in P38αf/f and P38αd/d mouse uteri on D4 when the uteri were in the receptive state (Fig. 2A). We noticed that progesterone-responsive genes in uterine stromal cells, such as nuclear receptor subfamily 2 group F member 2 (Nr2f2) and homeobox A10 (Hoxa10), were significantly reduced in the P38αd/d uteri, while estrogen-regulated genes such as lactoferrin (Ltf) and mucin 1 (Muc1) were aberrantly induced (Fig. 2B), suggesting a disturbed balance of progesterone and estrogen signal. We also confirmed these changes using the qRT-PCR analysis (Fig. 2 C and D). Consistently, the abnormally high-expressed Muc1 was observed in the epithelia (Fig. 2E), and Hoxa10 and Hand2 were expressed at a lower level in the stromal compartment (Fig. 2 F and G). When the mouse uteri were in receptivity on D4, stromal cells displayed intense cell proliferation with cell cycle quiescence in epithelia regulated by the progesterone signal. In control mice, the Ki67-positive proliferative cells were exclusively in the stromal cells with no proliferation in the epithelia, but in P38αd/d uteri, the epithelia displayed an anomalously robust proliferation accompanied with a dramatically decreased stromal growth (Fig. 2H). These cellular defects indicated an abnormal uterine receptivity upon P38α deficiency, coupled with largely demolished normal gene expressions. The observations above suggest that uterine P38α is essential for progesterone’s effect during the establishment of uterine receptivity.
Fig. 2.
P38α depletion derails uterine receptivity accompanying decreased expression of P4-responsive genes. (A) Volcano plots showing the differentially expressed genes in P38αf/f and P38αd/d uteri on D4 of pregnancy. (B) Heat map showing the expression level of differentially expressed genes in P38αf/f and P38αd/d uteri on D4 of pregnancy. (C and D) qRT-PCR analysis of implantation-related marker gene expression in P38αf/f and P38αd/d uteri on D4 of pregnancy. Data shown represent the means ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001. (E–G) IHC (E and G) and ISH (F) staining of implantation-related marker gene expression in P38αf/f and P38αd/d uteri on D4 of pregnancy (Scale bars, 100 μm). (H) The proliferation of uterine cells was compared in P38αf/f and P38αd/d mice by IHC staining for Ki67 (Scale bars, 100 μm).
Deletion of P38α Exerts No Apparent Adverse Effects on Ovarian Progesterone and Estrogen Biosynthesis.
Since the Pgr-Cre mouse model could also drive conditional gene loss in the developing corpus luteum (20) and the uterine function during the peri-implantation period is under the profound influence of the ovarian steroid hormones, progesterone and estrogen, we next checked progesterone synthesis in the ovary. It was uncovered that the localization and expression of the key steroid biosynthetic enzymes, cytochrome P450 cholesterol side-chain cleavage enzyme (P450SCC) and 3β-hydroxysteroid dehydrogenase (3β-HSD), were unaffected in the ovary after P38α deletion (SI Appendix, Fig. S4A). This finding was consistent with the observation of comparable serum progesterone (P4) and 17β-estradiol (E2) on D4 (SI Appendix, Fig. S4 B and C). These results suggested that the efficient deletion of P38α exerts no apparent adverse effects on ovarian progesterone and estrogen biosynthesis.
P38α Is Required for the Stability of PR Protein in Uterine Stromal Cells.
Since the contribution of the progesterone ligand level to lower progesterone responsiveness in P38αd/d uteri has been excluded, we next analyzed the mRNA and protein level of hormone receptor ERα and PR on D4. We found that the mRNA levels of estrogen receptor α (Esr1) and the progesterone receptor (Pgr) were comparable in uteri even in the absence of P38α (Fig. 3 A and B). In addition, the mRNA localization of Pgr in both the epithelium and stroma of P38αd/d uteri was similar to that in P38αf/f female mice (Fig. 3C). However, the protein level of PR was significantly reduced in P38α-deleted uteri, while the estrogen receptor α (ERα) expression displayed no difference (Fig. 3D). The IHC staining analysis further revealed that while ERα was comparably expressed in P38αf/f and P38αd/d uteri, a significantly decreased expression of PR was observed in P38αd/d uterine stroma, with a slightly diminished epithelial PR protein (Fig. 3E).
Fig. 3.
Loss of P38α compromises PR protein level rather than mRNA level in uterine stromal cells. (A and B) qRT-PCR analysis of Esr1 and Pgr expression in P38αf/f and P38αd/d uteri on D4. NS, not significant. (C) ISH of Pgr in P38αf/f and P38αd/d uteri on D4 of pregnancy (Scale bars, 100 μm). (D and E) Western blot and IHC staining show the protein level of ERα and PR in P38αf/f and P38αd/d uteri on D4 of pregnancy (Scale bars, 100 μm). (F) qRT-PCR analysis of Pgr expression in P38αf/f and P38αd/d ovariectomized mouse uteri treated with E2 for the indicated times. NS, not significant. (G and H) Western blot and IHC staining were performed to compare the protein level of PR in P38αf/f and P38αd/d ovariectomized mouse uteri when treated with E2 for the indicated times (Scale bars, 100 μm).
The lack of progesterone responsiveness in FKBP52-deficient mouse models can be amended by exogenous high-level progesterone (21). Next, we also tried to amend the decreased progesterone responsiveness and implantation defects with an exogenous progesterone supplement on D3 and D4 in P38αd/d mice. Nevertheless, none of the tested P38αd/d female mice showed an embryo implantation site as visualized by the blue dye method. In the meantime, morphologically normal blastocysts were recovered by flushing the P38αd/d uteri on D5 (SI Appendix, Fig. S4 D and E).
PR is a well-characterized transcriptional target gene of estrogen–ER signaling in uterine stroma (22). To further confirm that the hampered PR expression in D4 uterine stroma was due to the reduced PR protein level upon P38α deletion, we employed an ovariectomized mouse model treated with a single dose of E2. This treatment induced a parallel level of Pgr mRNA in both P38αf/f and P38αd/d uteri (Fig. 3F). However, the protein level of PR was significantly lower in P38αd/d uteri than in P38αf/f uteri, observed from 6 h after E2 treatment continuing to 12 h (Fig. 3G). IHC staining analysis also revealed reduced stromal PR proteins in P38αd/d uteri after E2 stimulation (Fig. 3H). These findings indicated that defective progesterone responsiveness resulting from P38α deficiency is mainly due to the reduced PR proteins in stromal cells.
To explore whether the active form of P38, phospho-P38, takes part in regulating the PR protein level, we employed the P38 MAPK inhibitor SB203580 or the P38α MAPK-specific inhibitor PH797804 to block the kinase activity of total P38 or only P38α in the cultured stromal cells. These two inhibitors had no effects on the expression of P38 and P38α. When P38 MAPK activity was blocked by SB203580 or PH797804 in uterine stromal cells in vitro, the mRNA level of PR remained unchanged (SI Appendix, Fig. S5A), while the PR protein level exhibited a sharp decrease after 12 h of inhibitor treatment (SI Appendix, Fig. S5B), which was consistent with the in vivo observation and suggested that the PR protein level was maintained by P38α at the posttranscriptional level.
To determine whether the decreased level of PR protein was due to decreased translation or increased degradation, the protein synthesis inhibitor cycloheximide (CHX), which blocks the translation of new proteins, was used with the P38 inhibitors. We observed that in the presence of CHX, the inhibition of P38α MAPK signaling still induced a sharp decrease in PR protein levels (Fig. 4A). In addition, the half-life of PR protein in uterine stromal cells treated with SB203580 or PH797804 was shortened from 6 h to 2 h (Fig. 4 B and C). Furthermore, we found that the addition of the proteasome inhibitor MG-132 can effectively block the degradation of PR resulting from the inhibition of P38α MAPK pathway (Fig. 4D). These results suggested that the P38α MAPK signaling pathway can protect the PR protein from proteasome-mediated degradation.
Fig. 4.
Inhibition of P38α MAPK reduces the half-life of PR protein. (A) Western blot analysis of PR in CHX (50 μg/mL)-treated P38αf/f and P38αd/d uterine stromal cells with or without SB203580/PH797804 treatment. (B and C) The half-life of PR in isolated uterine stromal cells when treated with the vehicle/SB203580/PH797804 for 12 h, followed by treatment with cycloheximide (50 μg/mL) for the indicated times. Veh, vehicle. (D) Western blot analysis of PR in the isolated uterine stromal cells in the presence of proteasome inhibitor MG-132 in the indicated treatment group.
Ube3c Promotes the Degradation of PR Protein through Polyubiquitination.
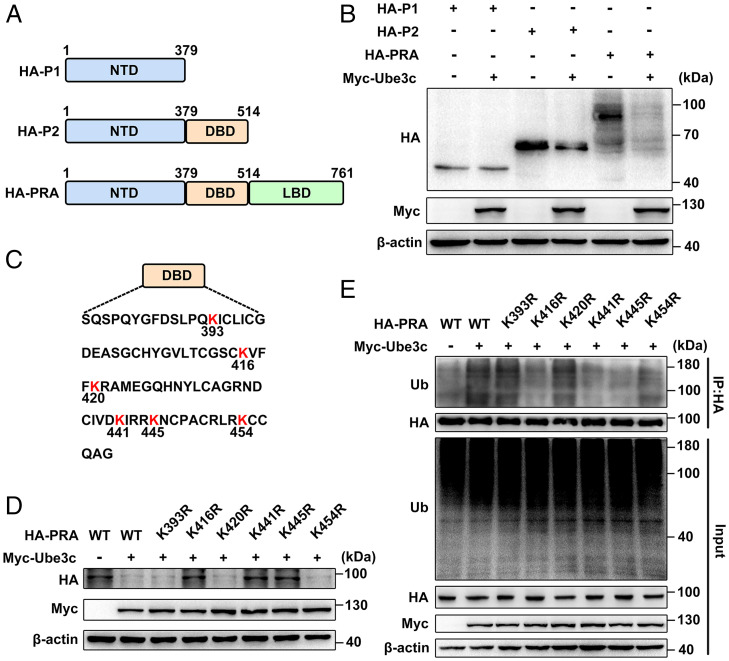
To further test the regulatory mechanism of reduced PR protein stability in P38αd/d uteri, we employed mass spectrometry (MS) to screen the PR-interacting E3 ubiquitin ligases that can promote PR protein degradation in D4 uteri. We compared the differentially interacting proteins of PR in both P38f/f and P38αd/d uteri and excluded the nonspecific background by comparing with the IgG control (Fig. 5A). Ube3c was identified as one PR-interacting E3 ligase specifically detected in P38αd/d uteri. We first checked the expression status of Ube3c and found that both the mRNA and protein were expressed at a comparable level in P38f/f and P38αd/d uteri (Fig. 5 B and C). Interestingly, when Ube3c and PR were simultaneously expressed, Ube3c could significantly improve the degradation efficiency of PR protein (Fig. 5D). Further analysis revealed that Ube3c could increase the PR polyubiquitination level (Fig. 5E). As the predominant functional isoform of PR in the uteri, PRA mainly consists of three domains: the N-terminal domain (NTD), the DNA-binding domain (DBD), and the ligand-binding domain (LBD) (Fig. 6A). To explore the ubiquitination site catalyzed by Ube3c in PRA protein, hemagglutinin (HA)-labeled PRA and two truncated forms (P1 and P2) were exogenously expressed in 293T cells along with Myc- tagged Ube3c. Obviously, Ube3c can interact with full-length PRA or P2 fragments and induce their degradation (Fig. 6B and SI Appendix, Fig. S6 A–C). However, there is no interaction between P1 and Ube3c, consistent with the resistant down-regulation of P1 by Ube3c. These results indicated that the interaction with Ube3c occurs in the DBD domain of PRA, and there are six lysine residues (K393, K416, K420, K441, K445, and K454) in this region (Fig. 6C). Point mutation was performed in all six lysine residues one by one to screen out the sites recognized by Ube3c. Results showed that only the K416, K441, and K445 site mutations abolished the degrading effects by Ube3c (Fig. 6D). In agreement with this finding, the polyubiquitination of the K416R, K441R, and K445R forms of PR by Ube3c was significantly weakened (Fig. 6E). Altogether, we found that Ube3c modifies the K416, K441, and K445 sites on PRA with polyubiquitination to promote proteasome-mediated protein degradation.
Fig. 5.
Ube3c promotes PR protein degradation via polyubiquitination. (A) Venn diagrams showing the number of proteins that interact with PR in P38αf/f and P38αd/d uteri on D4 of pregnancy. (B and C) qRT-PCR and Western blot analysis showing the expression pattern of Ube3c in P38αf/f and P38αd/d uteri on D4 of pregnancy. NS, not significant. (D and E) Western blot analysis revealed the stability (D) and ubiquitination (E) of HA-PRA with or without overexpression of Myc-Ube3c in 293T cells. IP, immunoprecipitation; Ub, ubiquitin.
Fig. 6.
Ube3c mediates PR ubiquitination at K416, K441 and K445. (A) Domain architecture of PR protein. (B) Western blot analysis showing the protein level of HA-P1, HA-P2, and HA-PRA with or without Myc-Ube3c in 293T cells. (C) Lysine (red K) sites in DBD domain of PR. (D and E) Western blot and immunoprecipitation revealed the stability (D) and polyubiquitination (E) of different point mutation forms of HA-PRA.
P38α Regulates the Catalytic Activity of Ube3c through Phosphorylation at S741.
To explore the molecular link between P38α and Ube3c, we first exogenously overexpressed P38α under the conditions in which Ube3c promotes PRA degradation in 293T cells and found that P38α can effectively inhibit the degradation of PRA promoted by Ube3c, while P38α kinase SB203580 hindered this function (Fig. 7A). Since previous studies have suggested that Ube3c may be a phosphorylated substrate of P38α (23), we first used an antibody against the pan-phosphorylated serine substrate of P38 MAPK (p-Ser) to detect whether Ube3c can be modified in the presence of activated P38α. We found that Ube3c can be phosphorylated by P38α on the serine residues (Fig. 7B). We also used NetPhos-3.1 (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1) to predict that there are four potential serine sites (S355, S380, S486, and S741) in Ube3c that can be phosphorylated by P38 MAPK (Fig. 7C). These serine sites were then mutated to alanine to abolish their phosphorylation modification, and the results showed that the Ube3c point mutant form S741A could no longer be phosphorylated by activated P38α (Fig. 7D). In addition, P38α lost the ability to inhibit S741A-induced degradation of PRA protein (Fig. 7E). Overall, our results revealed that in the presence of P38α, Ube3c was phosphorylated at serine 741 and lost its activity to catalytic PRA polyubiquitination and degradation. Conversely, catalytically unrestrained Ube3c promoted PRA degradation through the ubiquitin–proteasome pathway when P38α was ablated.
Fig. 7.
P38α regulates the catalytic activity of Ube3c. (A) Western blot analysis showing the weakened catalytic activity of Myc-Ube3c result from the overexpression of P38α in 293T cells. (B) Immunoprecipitation and Western blot assay to analyze the p-Ser in Ube3c. (C) Potential serine sites in Ube3c that can be phosphorylated by P38α. (D and E) Western blot analysis revealed the p-Ser level (D) and catalytic activity (E) of different point mutation forms of Myc-Ube3c.
Discussion
In mammals, the steroid hormone progesterone profoundly affects the function of the endometrium through the intracellular receptor PR and plays a central role in the establishment and maintenance of pregnancy (10). Plentiful research has shown that disordered progesterone-responsive signaling in the uterus is harmful to female fertility. Defective progesterone responsiveness or progesterone resistance, which is a common clinical pathological manifestation of gynecological diseases, has played a huge role in promoting the occurrence and development of endometriosis and endometrial cancer (24, 25). Therefore, it is imperative to decipher the potential molecular pathways regulating PR function in the uterus.
Previous studies have shown that a variety of posttranslational modifications can regulate the function of PR protein (26). The phosphorylation of PR has been most well studied among various posttranslational modifications (27). Moreover, Protein inhibitor of activated STAT protein 1/3 (PIAS1/3) serves as the small ubiquitin-like modifier (SUMO) E3 ligase and regulates PR protein transcriptional activity through SUMO modification at K388 (28, 29). Acetylation at K638, K640, and K641 is also influential in regulating PR function (30, 31). A previous study also provided genetic evidence that the ubiquitination of PR regulated by Bmi1 did not lead to PR degradation but was essential for progesterone activity and embryo implantation (12).
In the present study, we found that mice lacking P38α in the uterus showed a reduced protein level of stromal PR, which resulted in a disordered uterine receptivity and defective embryo implantation. Interestingly, the PR protein level in the epithelia did not display an obvious down-regulation in the P38αd/d uteri, and some progesterone target genes in the epithelia, such as IHH, were also expressed at a comparable level. These observations confirmed the specific regulation of stromal PR but not the epithelial PR by P38α, and the underlying mechanism for this cell–context regulation of PR stability by P38α needs further exploration. Several negative regulators for PR protein stability and activity have been reported, such as SPOP, BRCA1, and CUEDC2, which can increase the polyubiquitination of PR and induce PR degradation (32–34). All this evidence was from PR-positive cancer cells, but not under physiological circumstances. Thus, the mouse with uterine P38α deficiency was a valuable model to explore the regulation of PR protein stability in the uterine cells, especially the endometrial stromal cells.
Utilizing the uteri of the P38αd/d mice, we uncovered that the E3 ligase Ube3c was responsible for PR polyubiquitination and degradation and that its activity was regulated by P38 phosphorylation. Consistent with other members of the HECT E3 ligase, the HECT domain is the center of the ubiquitination modification function of Ube3c (35). A previous study with Ube3c reported that the ∼50 residues (aa 693–743) preceding the HECT domain are essential for the catalytic activity and stability of the HECT domain of Ube3c (36). Consistently, we found that P38α can phosphorylate Ube3c at S741, which is also localized in this reported regulatory region, to inhibit the activity of HECT domain, thereby reducing the ubiquitination and increasing the stability of PR protein. One manner of polyubiquitination-regulated protein stability is through substrate modification, such as the phosphorylation residue as a label for recognition and degradation by the Cullin–Ring family E3 ligase (37). Another manner relies on the regulated activity of the E3 ligase or its related molecule; for example, the phosphorylation of the E3 ligase can both increase or decrease enzyme activity, dependent on the phosphorylation residue and other cellular context character. The ubiquitin E3 ligase Parkin, RNF43, and GARU have been reported to be regulated in this manner (38–40). Our study also provided more evidence for this regulation manner; the molecular mechanism of this inhibitory phosphorylation for enzyme activity needs further exploration. In the further detailed analysis, we found that the ubiquitination modification sites of the PR protein recognized by Ube3c are K416, K441, and K445, which can be equally ubiquitinated by the E3 ubiquitin ligase HERC4 in human stromal cells (41), indicating the conserved regulatory roles of K416, K441, and K445 for the stability of the PR protein.
In summary, our study provided the genetic evidence for the indispensable function of P38α MAPK in guaranteeing normal implantation by maintaining a proper PR protein level. We found that P38α-mediated phosphorylation of S741 site ahead of the HECT domain of Ube3c, acted as a brake for the catalytic activity of Ube3c, ensuring the stability and transcription activity of the PR protein to maintain the normal progesterone responsiveness in the uterus. P38α ablation induced the down-regulation of the PR protein and decreased progesterone responsiveness in the uterus, leading to defective embryo implantation (Fig. 8).
Fig. 8.
Illustrative model of uterine P38α function during uterine receptivity establishment. In the presence of P38α, the E3 ubiquitin ligase Ube3c was phosphorylated at Ser741, which down-regulated its polyubiquitination activity toward PR and thus maintained the proper PR level in the stroma cell. The deficiency of P38α led to reduced Ube3c phosphorylation, which displayed aberrant high activity to induce the PR polyubiquitination and proteasome-mediated protein degradation.
Materials and Methods
Animals and Treatments.
P38αf/f mice were generated as previously described (42). Uterine-specific mutant mice were generated by crossing P38αf/f mice with Pgr-Cre mice (20). All mice were housed in the animal care facility, according to the institutional guidelines for the care and use of laboratory animals. Female mice were mated with fertile wild-type male mice to induce pregnancy (D1 = the day when the vaginal plug was seen). To examine implantation, pregnant mice were killed in the morning of D5 to D8. Implantation sites were visualized by an intravenous injection of Chicago blue dye solution, and the number of implantation sites, demarcated by distinct blue bands, was recorded. Mice that failed to recover any embryos were excluded in statistical analysis. Mouse blood samples were collected on the D4 morning, and serum progesterone and 17β-estradiol levels were measured by radioimmunoassay. For exogenous progesterone treatment, each mouse was subcutaneously injected with 2 mg in 0.1 mL sesame oil at 10:00 AM from D3 to D4, and then the pregnant mice were killed on D5. To test the ovarian hormonal influence on uteri, mice were ovariectomized and rested for 10 d, then injected with oil, 17β-estradiol (100 ng/0.1 mL per mouse), or progesterone (2 mg/0.1 mL per mouse). Mice were killed at indicated times and the uteri processed for immunostaining and mRNA analysis using qRT-PCR.
Primary Uterine Stromal Cell Culture.
Murine primary uterine stromal cells were isolated and cultured as previously described (43). The uteri of pseudopregnant mice on D4 were cut into small pieces (3–4 mm). Those tissue pieces were first digested in HBSS containing 6 mg/mL Dispase (Gibco) and 25 mg/mL trypsin (Sigma) and then incubated in HBSS containing 0.5 mg/mL collagenase (Gibco). The digested cells were passed through a 70 μm filter to obtain the stromal cells. Cells were plated at 5 × 105 cells per 60 mm dish or the corresponding numbers according to dish area and cultured with phenol red–free DMEM and Ham F-12 nutrient mixture (1:1; DMEM/F12; Gibco) containing 10% charcoal-stripped FBS (Biological Industries) and antibiotics. Two hours later, the medium was replaced with fresh medium and cultured overnight at 37 °C in an atmosphere of 5% CO2/95% air. SB203580 (LC Laboratories) and PH797804 (Selleck Chemicals) were first dissolved in DMSO, then diluted in a culture medium with working concentrations at 10 μM and 1 μM, respectively.
Construction of the Overexpressed Plasmids.
PCR was used to generate the full-length coding region of mouse PRA or Ube3c from uterine complementary DNA. The amplified DNA fragments were cloned into the corresponding overexpression vectors to form pCMV-HA-PRA, pCMV-Myc-Ube3c plasmids. The point mutation plasmids were constructed using the Fast Site-Directed Mutagenesis Kit (Tiangen). All the constructs were sequenced to confirm identity. Empty vector was performed as a control.
Immunoprecipitant MS.
Protein lysis from P38αf/f and P38αd/d D4 uteri were incubated with PR (Cell Signaling Technology) overnight at 4 °C. Then, the immunoprecipitants were separated in SDS-PAGE and visualized using Coomassie brilliant blue stain. Proteins were isolated, digested, and labeled and finally subjected to liquid chromatography-MS in the School of Life Sciences, Xiamen University.
RNA-Seq and Bioinformatic Analysis.
Total RNA was extracted from D4 uteri of P38αf/f and P38αd/d using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. Purified RNA was prepared and subjected to RNA sequencing using BGISEQ-500 platform (China, BGI). RNA-seq raw data were initially filtered to obtain clean data after quality control by Trimgalore. High-quality clean data were aligned to the mouse reference genome (mm10) using STAR. Differential expression genes were normalized to fragments per kilobase of exon model per million mapped reads (RPKM) using the EdgeR 3.9 package in R with the criteria of fold change significantly greater than 2 or less than 0.5 and P < 0.05. The visualization of RNA-Seq data were done by ggplot2 package in R.
ISH.
ISH was performed as previously described (44). Mouse-specific isotopes or digoxygenin-labeled antisense RNA probes for Mapk14, Mapk11, Mapk12, Mapk13, and Hoxa10 were used for hybridization. Cryosections hybridized with sense probes served as negative controls.
Immunostaining.
Uterine and ovarian tissues were fixed in 10% neutral buffered formalin. Then, 5 μm paraffin sections were used for IHC. Rehydrated sections were subjected to microwave heating in sodium citrate buffer for antigen retrieval, followed by incubation with 5% BSA for 1 h at room temperature. The P38α MAPK, phospho-P38 MAPK, COX2, P450SCC, 3β-HSD, Muc1, Hand2, ERα, PR, and Ki67 antibodies were used, respectively, and detailed information for antibodies is provided in SI Appendix, Table 1.
Western Blot Analysis.
Protein extraction and Western blot analysis were performed as described previously (43). P38α MAPK, phospho-P38 MAPK, ERα, PR, phospho-MAPK/CDK substrates, HA, Myc, Ube3c, ubiquitin, and β-actin antibodies were used, respectively. β-actin served as a loading control.
Immunoprecipitation.
Corresponding plasmids were transfected into 293T cells and the proteins of 293T cells were lysed in RIPA lysis buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS. Subsequently, protein A/G (commercial reagent and is recombinant fusion protein that combines IgG binding domains of both Protein A and Protein G) magnetic beads coupled with HA or Myc antibodies were incubated with protein lysates overnight at 4 °C. Immunoprecipitated proteins were separated by SDS-PAGE and detected by immunoblotting using antibodies specific to HA, Myc, p-Ser, and ubiquitin. MG-132 was used when detecting ubiquitin.
qRT-PCR.
Total RNA was isolated from uterine tissues or cells using TRIzol reagent. A total of 1 μg RNA was used to synthesize cDNA. The expression levels of different genes were validated by RT-PCR analysis with SYBR Green (Takara) on an ABIQ5 system. All the RT-PCR experiments were repeated at least 3 times, and the expression values were normalized against GAPDH. All PCR primers are listed in SI Appendix, Table 2.
Statistical Analysis.
Statistical analysis was performed using the GraphPad Prism program. Comparison of means was performed using the independent-samples Student t test. The data are shown as means ± SEM.
Supplementary Material
Acknowledgments
We are grateful to Professor Francesco DeMayo (National Institute of Environmental Health Sciences) for providing us with the Pgr-Cre/+ mice and Professor Jiahuai Han (Xiamen University) for providing the p38α and Mkk6 plasmids. This work was supported by the National Key R&D program of China (2021YFC2700302 to H.W.), the National Natural Science Foundation of China (81830045 and 82030040 to H.W., 81971388 to S.K.), and the Natural Science Foundation of Fujian Province (2020J01016 to S.K., 2020J01176 to Z.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206000119/-/DCSupplemental.
Data Availability
The sequencing data generated in this study have been deposited in the Gene Expression Omnibus database under accession code GSE199500 (45). Source data are provided with this paper.
References
- 1.Wang H., Dey S. K., Roadmap to embryo implantation: Clues from mouse models. Nat. Rev. Genet. 7, 185–199 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Zhang S., et al. , Physiological and molecular determinants of embryo implantation. Mol. Aspects Med. 34, 939–980 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marquardt R. M., Kim T. H., Shin J. H., Jeong J. W., Progesterone and estrogen signaling in the endometrium: What goes wrong in endometriosis? Int. J. Mol. Sci. 20, 3822 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasquez Y. M., DeMayo F. J., Role of nuclear receptors in blastocyst implantation. Semin. Cell Dev. Biol. 24, 724–735 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hantak A. M., Bagchi I. C., Bagchi M. K., Role of uterine stromal-epithelial crosstalk in embryo implantation. Int. J. Dev. Biol. 58, 139–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huet-Hudson Y. M., Andrews G. K., Dey S. K., Cell type-specific localization of c-myc protein in the mouse uterus: Modulation by steroid hormones and analysis of the periimplantation period. Endocrinology 125, 1683–1690 (1989). [DOI] [PubMed] [Google Scholar]
- 7.Hamilton K. J., Hewitt S. C., Arao Y., Korach K. S., Estrogen hormone biology. Curr. Top. Dev. Biol. 125, 109–146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lydon J. P., DeMayo F. J., Conneely O. M., O’Malley B. W., Reproductive phenotpes of the progesterone receptor null mutant mouse. J. Steroid Biochem. Mol. Biol. 56, 67–77 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Lydon J. P., et al. , Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 9, 2266–2278 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Wu S. P., Li R., DeMayo F. J., Progesterone receptor regulation of uterine adaptation for pregnancy. Trends Endocrinol. Metab. 29, 481–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cha J., Sun X., Dey S. K., Mechanisms of implantation: Strategies for successful pregnancy. Nat. Med. 18, 1754–1767 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin Q., et al. , Polycomb subunit BMI1 determines uterine progesterone responsiveness essential for normal embryo implantation. J. Clin. Invest. 128, 175–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulthard L. R., White D. E., Jones D. L., McDermott M. F., Burchill S. A., p38(MAPK): Stress responses from molecular mechanisms to therapeutics. Trends Mol. Med. 15, 369–379 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuadrado A., Nebreda A. R., Mechanisms and functions of p38 MAPK signalling. Biochem. J. 429, 403–417 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Roux P. P., Blenis J., ERK and p38 MAPK-activated protein kinases: A family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68, 320–344 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarubin T., Han J., Activation and signaling of the p38 MAP kinase pathway. Cell Res. 15, 11–18 (2005). [DOI] [PubMed] [Google Scholar]
- 17.He B., et al. , Blastocyst activation engenders transcriptome reprogram affecting X-chromosome reactivation and inflammatory trigger of implantation. Proc. Natl. Acad. Sci. U.S.A. 116, 16621–16630 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scherle P. A., Ma W., Lim H., Dey S. K., Trzaskos J. M., Regulation of cyclooxygenase-2 induction in the mouse uterus during decidualization. An event of early pregnancy. J. Biol. Chem. 275, 37086–37092 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Adams R. H., et al. , Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol. Cell 6, 109–116 (2000). [PubMed] [Google Scholar]
- 20.Soyal S. M., et al. , Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis 41, 58–66 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Tranguch S., et al. , FKBP52 deficiency-conferred uterine progesterone resistance is genetic background and pregnancy stage specific. J. Clin. Invest. 117, 1824–1834 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurita T., et al. , Regulation of progesterone receptors and decidualization in uterine stroma of the estrogen receptor-alpha knockout mouse. Biol. Reprod. 64, 272–283 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Dumont A. A., Dumont L., Berthiaume J., Auger-Messier M., p38α MAPK proximity assay reveals a regulatory mechanism of alternative splicing in cardiomyocytes. Biochim. Biophys. Acta Mol. Cell Res. 1866, 118557 (2019). [DOI] [PubMed] [Google Scholar]
- 24.McKinnon B., Mueller M., Montgomery G., Progesterone resistance in endometriosis: An acquired property? Trends Endocrinol. Metab. 29, 535–548 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Bulun S. E., et al. , Endometriosis. Endocr. Rev. 40, 1048–1079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdel-Hafiz H. A., Horwitz K. B., Post-translational modifications of the progesterone receptors. J. Steroid Biochem. Mol. Biol. 140, 80–89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Treviño L. S., Weigel N. L., Phosphorylation: A fundamental regulator of steroid receptor action. Trends Endocrinol. Metab. 24, 515–524 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones M. C., et al. , Regulation of the SUMO pathway sensitizes differentiating human endometrial stromal cells to progesterone. Proc. Natl. Acad. Sci. U.S.A. 103, 16272–16277 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Man J. H., et al. , PIAS3 induction of PRB sumoylation represses PRB transactivation by destabilizing its retention in the nucleus. Nucleic Acids Res. 34, 5552–5566 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung H. H., Sze S. K., Tay A. S., Lin V. C., Acetylation at lysine 183 of progesterone receptor by p300 accelerates DNA binding kinetics and transactivation of direct target genes. J. Biol. Chem. 289, 2180–2194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daniel A. R., et al. , The progesterone receptor hinge region regulates the kinetics of transcriptional responses through acetylation, phosphorylation, and nuclear retention. Mol. Endocrinol. 24, 2126–2138 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao K., et al. , Tumor suppressor SPOP mediates the proteasomal degradation of progesterone receptors (PRs) in breast cancer cells. Am. J. Cancer Res. 5, 3210–3220 (2015). [PMC free article] [PubMed] [Google Scholar]
- 33.Calvo V., Beato M., BRCA1 counteracts progesterone action by ubiquitination leading to progesterone receptor degradation and epigenetic silencing of target promoters. Cancer Res. 71, 3422–3431 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Zhang P. J., et al. , CUE domain containing 2 regulates degradation of progesterone receptor by ubiquitin-proteasome. EMBO J. 26, 1831–1842 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh S., Ng J., Sivaraman J., Exploring the “Other” subfamily of HECT E3-ligases for therapeutic intervention. Pharmacol. Ther. 224, 107809 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Singh S., Sivaraman J., Crystal structure of HECT domain of UBE3C E3 ligase and its ubiquitination activity. Biochem. J. 477, 905–923 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Petroski M. D., Deshaies R. J., Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Avraham E., Rott R., Liani E., Szargel R., Engelender S., Phosphorylation of Parkin by the cyclin-dependent kinase 5 at the linker region modulates its ubiquitin-ligase activity and aggregation. J. Biol. Chem. 282, 12842–12850 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Nemoto K., et al. , Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation. Nat. Commun. 8, 1004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsukiyama T., et al. , A phospho-switch controls RNF43-mediated degradation of Wnt receptors to suppress tumorigenesis. Nat. Commun. 11, 4586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang P., et al. , SOX4 facilitates PGR protein stability and FOXO1 expression conducive for human endometrial decidualization. eLife 11, e72073 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishida K., et al. , p38alpha mitogen-activated protein kinase plays a critical role in cardiomyocyte survival but not in cardiac hypertrophic growth in response to pressure overload. Mol. Cell. Biol. 24, 10611–10620 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He H., et al. , Rbbp7 is required for uterine stromal decidualization in mice. Biol. Reprod. 93, 13 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Lu J., et al. , A positive feedback loop involving Gcm1 and Fzd5 directs chorionic branching morphogenesis in the placenta. PLoS Biol. 11, e1001536 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. W. Gene expression in D4 WT and P38α KO uteri. GEO Data Base. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE199500. Deposited 27 March 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data generated in this study have been deposited in the Gene Expression Omnibus database under accession code GSE199500 (45). Source data are provided with this paper.