Abstract
Sustaining DNA damage response (DDR) signalling via retention of DDR factors at damaged sites is important for transmitting damage-sensing and repair signals. Herein, we found that DNA damage provoked the association of ribosomes with IRES region in lncRNA CTBP1-DT, which overcame the negative effect of upstream open reading frames (uORFs), and elicited the novel microprotein DNA damage-upregulated protein (DDUP) translation via a cap-independent translation mechanism. Activated ATR kinase-mediated phosphorylation of DDUP induced a drastic ‘dense-to-loose’ conformational change, which sustained the RAD18/RAD51C and RAD18/PCNA complex at damaged sites and initiated RAD51C-mediated homologous recombination and PCNA-mediated post-replication repair mechanisms. Importantly, treatment with ATR inhibitor abolished the effect of DDUP on chromatin retention of RAD51C and PCNA, thereby leading to hypersensitivity of cancer cells to DNA-damaging chemotherapeutics. Taken together, our results uncover a plausible mechanism underlying the DDR sustaining and might represent an attractive therapeutic strategy in improvement of DNA damage-based anticancer therapies.
INTRODUCTION
Genomic DNA in living organisms is highly vulnerable to exogenous and endogenous damage. To prevent damaged genomic DNA-induced genetic instability (1–3), diverse DNA damage response (DDR) mechanisms, such as canonical homologous recombination repair (HRR) and non-homologous end-joining (NHEJ) pathways and the post-replication repair (PRR) pathway, have evolved to counteract different types of DNA damage (4–6). The DDR pathway consists of sensors, transducers and effectors that sense DNA damage, propagate the signal, and initiate the appropriate responses, such as cell cycle arrest, DNA repair or apoptosis (7–9). For instance, following DNA damage, the rapidly phosphorylated histone H2AX, referred to as γ-H2AX, which is a prerequisite for the sustained retention of signalling and repair proteins, recruits ring-type ubiquitin ligase RNF8 and, together with E2 ubiquitin conjugase 13 (UBC13), promotes protein ubiquitination at DNA damage sites, which facilitates the assembly of the RAD18 complex that transmits DNA damage signals (10–13). Therefore, stabilisation or retention of sensors, transducers and effectors at DNA lesions contributes to DNA damage repair.
Recent studies have demonstrated that RAD18 exerts its DNA repair function through multiple mechanisms (13–16). It has been reported that RAD18 formed a complex with RAD6, and subsequently promoted monoubiquitylation of proliferating cell nuclear antigen (PCNA) at stalled replication forks to recruit DNA polymerase, consequently leading to translesion DNA synthesis (TLS)-mediated post-replication repair (PRR) of damaged DNA (17,18). Furthermore, several reports have shown that RAD18 also contributes to DNA double-strand break repair either through the 53BP1-directed NHEJ pathway (19) or through the recombinase RAD51 paralog RAD51C-mediated HRR pathway (13). Importantly, these studies showed that RAD18 plays critical roles in sustaining retention of DNA polymerase at stalled replication forks and retention of 53BP1 and RAD51C at DNA lesions (13,19–21).
In addition to the vital roles in accurate cell replication and maintenance of genomic stability and integrity, accumulating evidence indicates that the DDR is also involved in resistance to DNA damage-based chemo- and radiotherapy. For instance, RAD18 contributes to 5-fluorouracil (5-FU) resistance in colorectal cancer (22) and radiotherapy resistance in glioma (23). Therefore, further understanding of the molecular mechanism governing the DDR pathway activated by different types of DNA lesions would facilitate the identification of better targets to overcome resistance to DNA damage-based anti-tumour chemo- and radiotherapies.
In the present study, we identified a novel microprotein, DNA damage-upregulated protein (DDUP), encoded by long non-coding RNA (lncRNA) CTBP1 divergent transcript (CTBP1-DT; NR_033339.1), which was rapidly elevated in response to DNA damage via a post-transcriptional mechanism. We further demonstrated that DNA damage-induced DDUP upregulation and DDUP foci formation played a vital role in DNA damage repair via PRR- and HRR-repair mechanisms, thereby promoting resistance to platinum-based chemotherapy. Importantly, treatment with ATR inhibitor Berzosertib, an intravenously administered small molecule, inhibited the formation of DNA damage-induced DDUP foci, consequently leading to hypersensitivity of ovarian cancer cells to DNA-damaging chemotherapeutics. Taken together, our results uncover a plausible mechanism by which the DDR is sustained, and could assist the development of attractive therapeutic strategies for the treatment of genotoxic-resistant cancers.
MATERIALS AND METHODS
Cell lines and cell culture
The human HeLa cell line and human embryonic kidney 293T cell line were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and grown in DMEM medium (Gibco, Grand Island, NY, USA) supplemented with 5% FBS (Gibco, Grand Island, NY, USA). All the cell lines in this study underwent mycoplasma testing regularly, and were authenticated using short tandem repeat (STR) profiling at the medicine lab of Forensic Medicine Department of Sun Yat-Sen University (China).
Vectors, retroviral infection and transfection
The full length of CTBP1-DT was generated by subcloning the PCR-amplified human CTBP1-DT sequence and cloned into pSin-EF2-vector. The DDUP ORF with FLAG-tag expression plasmid was generated by cloning the full length of CTBP1-DT. The mutation constructs of 5′-ATG1mDDUP-3′, 5′-ATG2mDDUP-3′, 5′-ATG1/2mDDUP-3′ in which the putative ORF start codon was mutated to ATT in the CTBP1-DT-psin EF2 vector. DDUP/T174A and DDUP/T174D mutant were also generated using a QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, USA). The IRES, uORF and 5′UTR (5′-untranslated region) sequences of CTBP1-DT were obtained through chemical gene synthesis (Genewiz, Suzhou, China). mCherry-IRES-GFP, mCherry-uORF-GFP, mCherry-5′UTR-GFP and frames were amplified by overlap PCR and then cloned into the psin-EF2 vector at the EcoRI and BamHI sites. pDRGFP (Addgene #26475), pCBASceI (Addgene #26477), pcDNA3-ATR WT (Addgene #31611), pcDNA3-ATR-N (Addgene #53767), pcDNA3-ATR-M (Addgene #53768), pcDNA3-ATR-C (Addgene #53769) plasmids were purchased from Addgene (Cambridge, MA). Vectors were transfected using Lipofectamine 3000 (Thermo Fisher Scientific, CA) according to the manufacturer's instructions.
Immunoblotting analysis (IB)
IB was performed according to a standard protocol with the following antibodies: anti-phospho-Histone H2A.X Ser139 (#9718), anti-RAD18 (#9040), anti-PCNA (#13110), anti-ubiquityl-PCNA Lys164 (#13439), anti-DYKDDDDK Tag (#14793) were purchased from CST (Danvers, MA, USA); anti-ATR (ab2905), anti-phospho SQ/TQ (ab130947), anti-ATM (ab32420), anti-RAD51C (ab72063), anti-Ki67 (ab15580) were purchased from Abcam (Cambridge, MA). Anti-ATR pS428 (# 720107) was purchased from Thermo Fisher Scientific.
RNA extraction, reverse transcription and Polymerase Chain Reaction (PCR)
Total RNA was extracted from the indicated cell using Trizol (Life Technologies) reagent and reverse transcription of total mRNA was performed using a GoScript™ Reverse Transcription Mix kit (Promega, Beijing, China) according to the manufacturer's protocol. PCR was subsequently performed with the reverse-transcribed cDNA. Real-time q-PCR was performed and quantified in Bio-Rad CFX qRT-PCR detection system (Applied Biosystems Inc, CA, USA), using FastStart Universal SYBR Green Master (ROX; Roche, Toronto, CA). Expression data were normalized to the geometric mean of housekeeping gene GAPDH to control the variability in expression levels and calculated as 2−[(Ctof gene) − (Ctof GAPDH)], where Ct represents the threshold cycle for each transcript. The primers used in this study were listed in Supplementary Table S2.
Chromatin fraction
The indicated cells (1 × 106) were seeded in 10 cm dish and cultured overnight. Then cells were harvested after treatment with CPT or CDDP and resuspended in 200 μl of buffer A (10 mM KCl, 15 mM MgCl2, 10 mM HEPES, [pH 7.9], 0.34 mM sucrose, 1 mM dithiothreitol, 10% glycerol, 0.1% Triton X-100 and protease inhibitor mixture), and incubated for 10 min on ice. The cell pellet was collected at 1300 g for 4 min at 4°C. After being washed with buffer A, the cell pellet was lysed in 200 μl of lysis buffer (0.2 mM EGTA, 1 mM dithiothreitol, and protease inhibitor mixture). Insoluble chromatin was harvested by centrifugation (1700 g, 4 min, 4°C).
The chromatin fraction was then treated with DNAase (stem cell Technologies, CA) for 1 h and then incubated with anti-DDUP antibody (Sino Biological, China) overnight at 4°C. After then the supernatant was incubated with 20 μl of protein G-agarose beads overnight at 4°C. The agarose beads were washed six times with wash buffer (25 mM HEPES [pH 7.4], 0.5% NP-40, 1 mM EDTA, 150 mM NaCl, 2% glycerol, 1 mM PMSF). The precipitated components were subjected to LC–MS/MS or western blot analysis. The protein level in LC–MS/MS analysis were quantified by following thresholds, which were considered to be significantly differentially abundant: pFDR value <0.05, fold change ≥2.
Immunoprecipitation (IP)
The indicated cells were lysed in lysis buffer (25 mM HEPES [pH 7.4], 1% NP-40, 1 mM EDTA, 150 mM NaCl, 2% glycerol, 1 mM PMSF) and maintained on ice for 30 minutes. After centrifugation at 12 000 rpm for 10 min at 4°C, the supernatant was subjected to DDUP antibody-conjugated G-agarose beads and rotated overnight at 4°C. The immunoprecipitates were washed six times with wash buffer (25 mM HEPES [pH 7.4], 0.5% NP-40, 1 mM EDTA, 150 mM NaCl, 2% glycerol, 1 mM PMSF) and samples were ready for immunoblotting or re-immunoprecipitation.
In Supplementary Figure S5D, to remove Flag-tagged DDUP fragment-associated γ-H2AX or RAD18, the immuniprecipated complex, eluted from DDUP antibody-conjugated beads by glycine (0.2 M, pH 2.5–3.0) buffer and immediately neutralized with Tris buffer (1.0 M, pH 8.0–8.5) and diluted with lysis buffer, was then immunoprecipitated again using the anti-Flag antibody-conjugated beads overnight at 4°C. After centrifugation, the supernatant was subjected to immunoblotting analysis.
Recombinant DDUP protein, peptide synthesis and Anti-DDUP antibody preparation
The Recombinant DDUP and DDUP/T174D protein were obtained from Abclonal (Wuhan, China). Peptide synthesis and anti-DDUP preparation were performed by Sino Biological (Beijing, China). Western blotting was performed according to standard protocols. DDUP microprotein is a small protein of about 19.74 kDa, which can be separated by 16% Tricine SDS-PAGE (24). The DDUP antibody for western blot, IF staining, and immunohistochemistry used in this study was 1:1000, 1:200, and 1:100 dilution, respectively.
Far-western analysis
Immunoblotting was performed by using the proteins immunoprecipitated by anti-ATR (ab2905, Abcam), anti-RAD18 (ab188235, Abcam), anti-human gamma H2A.X (phosphor S139) (ab81299, Abcam) and anti-RAD51C (A302-645A, Bethyl Laboratories) antibodies. Briefly, the indicated cell lysates were separated by SDS-PAGE, and were transferred onto a PVDF membrane. The membrane was then preincubated in 10% skimmed milk at 4°C for 1 h. Then, the human recombinant DDUP protein (Abclonal, China) was added and further incubated for 18 h at 4°C. The membrane was then washed six times with TBST buffer and subjected to immunoblotting analysis by anti-DDUP antibody (Sino Biological, China).
Identification of new protein by MS analysis
Proteins were extracted and digested with trypsin gold mixture (#V5280, Promega, China). Liquid chromatography (LC) was used to fractionate each tryptic peptide mixture into 16 fractions. The components were analysed using a Q Exactive mass spectrometer (Thermo Fisher Scientific, CA). Three-frame translation of all non-coding RNAs from NCBI RefSeq-RNA sequences. The predicted protein sequences smaller than 50 amino acids were filtered. Combination of three-frame translated non-coding RNAs, Homo sapiens reviewed Swiss-Prot protein sequences and common contaminants sequences, a specific novel protein database (including reviewed proteins and predicted ORFs) was created. MaxQuant (version 1.5.7.4) (25,26) was used for MS data searches. The common search parameters included: enzyme, trypsin; fixed modification, carbamidomethyl (C); variable modifications, oxidation (M), Gln → pyro-Glu (N-terminus), and acetyl (protein N-terminus); Two missed cleavage sites were allowed. Criteria for confident identification with false discovery rate (FDR) filtering (1% at the peptide level and 1% at the protein level). Smith-Waterman analysis was conducted to rule out the new peptides which are similar to the RefSeq protein sequences (≤1 mismatch). To apply stringent quality control, only one unique peptide with at least nine residues were used for new protein identifications.
Polysome profiling by sucrose gradient ultracentrifugation
Cells were pre-treated with 100 μg/ml cycloheximide (CHX) for 10 min, followed by two ice-cold phosphate buffered saline washes and the addition of 1ml cell lysis buffer (10 mM Tris–HCl [pH 7.4], 5 mM MgCl2, 100 mM KCl, 1% Triton X-100). After a 30-min rotation to coat evenly at 4°C, cell lysates were scraped and transferred to pre-chilled 1.5 ml tubes. Cell debris were removed by centrifuging at 12 000 g at 4°C for 10 min, and then transferred supernatant to the surface of 10–50% gradient sucrose buffer (20 mM HEPES–KOH, 5 mM MgCl2, 100 mM KCl, either 10% or 50% sucrose (w/v)). Ultracentrifugation of the sucrose gradients was conducted using a SW-41/40 Ti rotor, 40 000 rpm for 2 h at 4°C. Gradients are fractionated using the Biocomp Piston Gradient Fractionator (Speed: 0.3 mm/s) and a 270 nM UV monitor. Sucrose gradient fractions were immediately extracted using a Trizol (Life Technologies) reagent according to the manufacturer's instructions.
Polysome profiling sequencing
After obtaining RNA from ribosome footprints, the ribosomal RNA (rRNA) was removed by Epicentre Ribo-zero rRNA Removal Kit (Epicentre, WI, USA) according to the manufacturer's instructions. Subsequently, the rRNA-depleted RNA was used to construct sequencing libraries by NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs, MA, USA) following the manufacturer's recommendations. The high-throughput sequencing was performed by Novogen (Novogen, Beijing, China) using Illumina HiSeq X Ten. Data were mapped to the reference genome using Bowtie2 (v2.2.3), the reads mapped to splice variants of one gene were summed. Genes with a fold change of > 2 and P < 0.01 in edgeR were identified as differential genes.
Single-cell gel electrophoresis assay
Comet assay was performed according to manufacturer's protocol (Trevigen, MD, USA). Briefly, cells treated with indicated agents were harvested, resuspended in pre-chill PBS at 1 × 105 cells per millilitre, mixed with melt agarose at 1:10 ratio, and spread evenly on pre-warmed glass slides (at 37°C). After the agarose solidified at 4°C in the dark, the slides were placed in lysis solution (Trevigen, MD, USA) for 1 h. Successively, slides were then removed from lysis buffer and gently immersed in 50 ml of 1× Neutral Electrophoresis buffer for 30 min. Slides were then subjected to electrophoresis for 45 min at 4°C (set power supply to 21 V), and cells were fixed with 70% (v/v) ethanol and stained with SYBR™ Gold (#S11494, Invitrogen, USA). DNA damage was quantified for 100 cells for each experimental condition by determining tail moment, a function of both tail length and intensity of DNA in the tail relative to total DNA, using the plugin OpenComet v1.3.1. Statistical analysis was done using the Student's t-test.
Chemical regents
VP-16 (#E1383, Sigma-Aldrich, St. Louis, MO); Cisplatin (#PHR1624, Sigma-Aldrich, St. Louis, MO); Camptothecin (#C9911, Sigma-Aldrich, St. Louis, MO); Actinomycin-D (#SBR00013, Sigma-Aldrich, St. Louis, MO); Cycloheximide (# 5087390001, Sigma-Aldrich, St. Louis, MO); Carboplatin (#PHR3417, Sigma-Aldrich, St. Louis, MO); KU-55933 (#SML1109, Sigma-Aldrich, St. Louis, MO); 4EGI-1 (#HY-19831, MedChemExpress, New Jersey, USA); Rapamycin (#HY-10219, MedChemExpress, New Jersey, USA); KU-57788 (#S2638, Selleck, Shanghai, China); Berzosertib (# S7102, Selleck, Shanghai, China).
Laser micro-irradiation and immunofluorescence (IF)
HeLa cells were seeded on 15 mm diameter glass-bottom plates (NEST, Wuxi, China) and presensitized with 10 μM Brdu (Roche, Basel, Switzerland) in phenol red-free medium (Invitrogen, USA) for 24 h at 37°C. Local laser micro-irradiation was performed by a micropoint system (Andor, Belfast, UK) with a 365 nm pulsed nitrogen UV laser (16 Hz pulse, 55% laser output). After irradiation, cells were immunostained as previously described (27) with anti-γH2AX antibody (#9718, Cell Signaling Technology). The images were captured using the Axion Vision Rel.4.6 computerized image analysis system (Carl Zeiss, Jena, Germany).
Immunofluorescence (IF) staining
The indicated cells were plated on cell culture chamber slides (Thermo Fisher Scientific, CA). For IF staining of γH2AX, DDUP, RAD18 foci, the cells were fixed with 4% paraformaldehyde and premeabilized with 0.5% Triton X-100. For IF staining of RAD51C foci, the cells were premeabilized with 0.5% Triton X-100 for 5 min on ice, then fixed with 4% paraformaldehyde. For IF staining of p-ATR and RPA2 foci, the cells were fixed with 4% paraformaldehyde and premeabilized with methanol: acetone (1:1) at -20°C for 20 min. For IF staining of RAD51 foci, the cells were incubated with ice cold buffer (20 mM HEPES, pH7.5; 20 mM NaCl; 5 mM MgCl2; 1 mM DTT; 0.5% NP40) for 20 min on ice, then fixed with 4% paraformaldehyde for 15 min. The cells were incubated with anti-γH2AX antibody (#9718, Cell Signaling Technology), anti-DDUP antibody (Sino Biological, China), Anti-ATR pS428 antibody (#720107, Thermo Fisher Sienctific), anti-DYKDDDDK Tag antibody (#14793, Cell Signaling Technology), anti-RAD18 antibody (#9040, Cell Signaling Technology), anti-RAD51 (#PA5-27195, Invitrogen), anti-RPA2 antibody (ab109084, Abcam) or anti-RAD51C antibody (PA5-77078, Invitrogen). The images were captured using the Axion Vision Rel.4.6 computerized image analysis system (Carl Zeiss, Jena, Germany). Co-localization the fluorescence between molecules was quantified by ImageJ using the Manders' coefficients algorithm.
Patient information and tissue specimens
This study, which complied with all relevant ethical regulations for work with human participants was conducted on a total of 367 paraffin-embedded ovarian cancer samples and four freshly collected ovarian cancer tissues that were histopathologically and clinically diagnosed at the Affiliated Guangzhou Women and Children's Hospital and Sun Yat-sen University Cancer Center. The clinical information regarding the samples is summarized in Supplementary Tables S3 and S4. All patients received standardized platinum-based chemotherapy. Platinum resistance and sensitivity refer to the time-to-relapse within 6 months or after 6 months following completion of platinum-based chemotherapy. The study protocols were approved by the Institutional Research Ethics Committee of Sun Yat-sen University Laboratory Animal Center for the use of these clinical materials for research purposes.
Immunohistochemistry (IHC) analysis
IHC analysis was carried out to determine altered protein expression in indicated paraffin-embedded ovarian cancer tissues and followed by anti-DDUP antibody (Sino Biological, China) overnight at 4°C. According to the histopathological features and patient data of the tissues, the degree of immunostaining of formalin-fixed, paraffin-embedded sections were reviewed and scored separately by two independent pathologists. The scores were determined by the proportion of positively stained tumor cells coupled with the intensity of staining. The scores given by the two independent pathologists were combined into a mean score for further comparative evaluation. Tumor cell proportions were scored as follows: 0, no positive tumor cells; 1, <10% positive cells; 2, 10–35% positive tumor cells; 3, 35–75% positive tumor cells; 4, >75% positive tumor cells. Staining intensity was graded according to the following standard: 1, no staining; 2, weak staining (light yellow); 3, moderate staining (yellow-brown); 4, strong staining (brown). The staining index (SI) was calculated as the product of the staining intensity score and the proportion of positive tumor cells. Using the method of assessment, we evaluated protein expression in ovarian cancer tissues by determining the SI, with scores of 0, 2, 3, 4, 6, 8, 9, 12 and 16. Samples with a SI ≥8 were considered as high expression and samples with a SI <8 were considered as low expression. Cut-off values were determined based on a measure of heterogeneity using the log-rank test for overall survival.
Cell survival assay
The indicated cells (1 × 103) were seeded in 96-well plates and cultured with 5% FBS medium with 10 μM CPT. Cells were stained with 200 μl sterile 3-(4, 5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) dye (0.5 mg/ml, Sigma-Aldrich) at 37°C for 4 h, followed by removal of the culture medium and added to 160 μl of dimethyl sulphoxide (DMSO) (Sigma-Aldrich, USA). The absorbance was measured at 490 nm (Bio-Tek, VT, USA) following the manufacturer's instructions.
Gene conversion assay
HeLa cells (1 × 106) were plated in 10-cm dishes and transfected with DR-GFP plasmid (10 μg; Addgene #26475) coupled with pCBASceI (10 μg; Addgene #26477) and pCherry plasmid (10 μg) using Lipofectamine 3000 (Thermo Fisher Scientific, CA) according to the manufacturer's instructions. The rare-cutting I-Sce I endonuclease located on the DR-GFP plasmids, as a result, will undergo a DSB when I-Sce I is expressed. After 48 h, the indicated cells were subjected to FACS analyses. The HR efficiency were calculated as percentage of GFP positive cells normalized with m-cherry positive cells after 48 h transfection quantified by Gallios™ Flow cytometry (Beckman Coulter, Inc.) using by flow cytometry APC-mCherry and FITC-GFP channel. Results were the mean values of three independent experiments.
NHEJ reporter assay
The integrated DDR consists of a promoter and resistance cassette fused to a T2A peptide and two inverted I-Sce I sites, followed by GFP. Intact or partially cut DSB repair reporter lack GFP expression due to the presence of a STOP codon. Cells were transfected with I-Sce I plasmid (Addgene #98895) and an exogenous donor (Addgene #98896) for NHEJ, and the transfection efficiency was controlled using BFP expression from the repair donor cassette. Repair by NHEJ would lead to GFP expression. The NHEJ efficiency were calculated as percentage of GFP/BFP positive cells after 48 h transfection by flow cytometry.
Fluorescence recovery after photo-bleaching (FRAP)
FRAP was performed using a LSM 880 with Airyscan confocal microscope (Carl Zeiss, Jena, Germany) with a 63× oil (NA 1.4) objective. The indicated stable cells transfected with GFP-RAD18 alone or co-transfected with GFP-RAD18 and DDUP/WT or DDUP/T174A plasmids were cultured on 15 mm glass-bottom dishes (NEST, Wuxi, China). After two prebleach images were obtained, GFP-RAD18 fluorescence was then photo-bleached by scans with a 488 nm argon laser at 100% power. For post-bleached recovery recording, images were captured at 5 min intervals and the fluorescence intensity within a specific region was measured every 30 sec at 20% laser power. The fluorescence intensity was normalized to the pre-bleached signal after subtraction of the background. Data were plotted using GraphPad Prism 8 software.
Molecular docking
The 3D structure of the ATR (PDB ID: 5yz0) and RAD18 (PDB ID:2YBF) was downloaded from RCSB Protein Data Bank. The structure modelling of DDUP and H2A.X were obtained from I-TASSER (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). Protein–protein docking was performed for molecular-docking simulations of DDUP and for predicting the binding affinity to the indicated proteins using the ClusPro server (https://cluspro.org). Molecular graphics were generated using PyMOL Version 1.5.0.5.
Surface plasmon resonance (SPR) analysis
The interaction of recombinant DDUP protein with γ-H2AX (ab15645, abcam) or RAD18 (ab112417, abcam) were detected using a BIAcore T100 instrument (GE Healthcare, UK) following the manufacturer's instructions. SPR equilibrium binding data, consisting of Req values from several concentration series, were analysed by fitting a simple 1:1 binding to yield Rmax and Kd values using BIAcore T100 Evaluation software.
Dual-luciferase reporter system
After the renilla luciferase (Rluc) and the firefly luciferase (Luc) sequences were obtained from a psicheck2 vector (Promega, Beijing, China), Rluc sequence was placed in front of the Luc sequence. The full-length sequences of Rluc and Luc were amplified by homologous recombination kit (ClonExpress, Vazyme, China), and the flanking sequences were connected to pSin-EF2 vector using two restriction enzyme sites (BamHI and EcoRI). The 5′UTR, 5′UTR-ΔuORF, 5′UTR-ΔIRES sequences were inserted in the middle of Rluc and Luc using homologous recombination (ClonExpress, Vazyme, China). Luciferase and renilla signals were measured using the Dual Luciferase Reporter Assay Kit (Promega, Beijing, China) according to a protocol provided by the manufacturer. Three independent experiments were performed, and the data are presented as mean ± SD.
In vitro transcription
The CTBP1-DT-psin-EF2 constructs served as DNA template to synthesize RNA using HiScribe T7 quick high yield RNA synthesis kit (New England Biolabs, MA, USA) according to manufacturer's instructions, in presence of normal m7GpppG cap structure analog or non-functional ApppG cap structure analog (New England Biolabs, MA, USA) or without cap.
In vitro translation
Extracts for in vitro translation were prepared according to the protocol were prepared as described previously (28). Briefly, the HeLa extract treated with 2 μl of 1mg/ml micrococcal nuclease (Promega) was used for in vitro translation assays incubating with translation mix (2M KOAc[pH 7.5], 10 mM Mg(OAc)2, DEPC water) and homogenized with 15 strokes at 4°C. In vitro translation reactions (150 nM in vitro-transcribed RNA was mixed with 4 μl of cell lysates in a final reaction volume of 10 μl containing 100 μM amino acids, 20 μM creatine phosphate, 80 ng/μl creatine kinase, 16 mM HEPES buffer [pH 7.6], 0.8 mM ATP, 0.1 mM GTP, 100 ng/μl calf liver tRNA, 0.1 mM spermidine) were performed at 37°C for 60 min. Aliquots of 10 μl were transferred into a new tube and added an equal volume of stop solution (10 mM EDTA,50 mg/ml RNase A). All aliquots were analysed by SDS-PAGE.
Targeted gene disruption by CRISPR-Cas9
The creation of DDUP heterozygous knockout HeLa cells were conducted using the CRISPR/Cas9 system. GeneChem (Guangzhou, China) designed and cloned the corresponding gRNA1 (GGTTGGTGGAGTGCACAGGCAGG) and gRNA2 (TGCACAGGCAGGGACCTCACTGG) into GV392 plasmid, respectively. Briefly, 3 × 105 HeLa cells were infected with lenti-CRISPR virus. After 24 h, the infected cells were selected for 7 days using 0.5 μg/ml puromycin. Afterward, HeLa/Cas9 cells were re-infected with GV392-GFP-CTBP1-DT gRNA lentivirus at 4 MOI to ensure > 95% cells were positive. Two days later, the infected cells were sorted using flow cytometry and single-cell cloned, the HeLa/DDUP−/−-#1 and DDUP−/−-#2. HeLa/DDUP−/−-#1 and DDUP−/−-#2 referred to single-cell clones 1 and 2, respectively. Genomic DNA was extracted from HeLa/DDUP−/−-#1 and DDUP−/−-#2 cells and using a QIAGEN genomic DNA Kit (QIAGEN, Germany) and the target sequences were amplified. Heterodimerization and digestion were performed using the Knockout and Mutation Detection Kit (GeneChem) according to the manufacturer's instructions. Moreover, the genomic DNA was amplified and then Sanger sequencing was performed to confirm the DDUP knockout status. Primers are listed in Supplementary Table S2.
Statistics and reproducibility
Statistical analysis was carried out using Microsoft Excel 2016 and GraphPad Prism v.8.0.1 for windows. Experimental data are represented as the average ± S.D. of a minimum of three biological replicates. P-values of 0.05 or less were considered statistically significant. For parametric data, unpaired, two-tailed Student's t-tests was used. For non-parametric data, two sided Mann-Whitney test was used. For multiple comparisons, ANOVA with post hoc tests were used. Log-rank (Mantel-Cox) test or Gehan–Breslow–Wilcoxon test was used to determine the statistical differences of the survival data. A chi-squared test was used to analyse the relationship between DDUP expression and the clinicopathological characteristics. The significance of various variables for survival was analysed by univariate and multivariate Cox regression analyses. Statistical analyses were performed using the SPSS 11.0 statistical software package.
Ethics approval and consent to participate
All Patients’ samples were obtained according to the Declaration of Helsinki and each patient signed a written informed consent for all the procedures. All of the animal procedures were approved by the Sun Yet-Sen University Animal Care Committee.
RESULTS
LncRNA CTBP1-DT-encoded microprotein DDUP is upregulated upon DNA damage
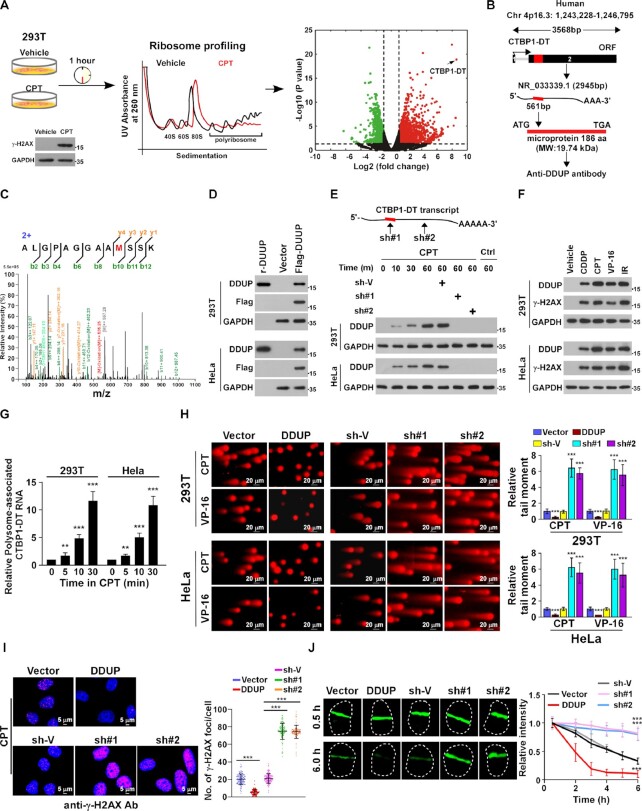
To identify potential new factors involved in DDR, polysome profiling analyses were performed in vehicle- and camptothecin (CPT)-treated 293T cells (1 h; Figure 1A, left panel). CPT-induced DNA damage was determined by increased γ-H2AX expression (Figure 1A, left panel). Analysis of polysome profiling sequencing showed that lncRNA CTBP1 divergent transcript (CTBP1-DT; NR_033339.1) was most enriched RNA associated with ribosome in CPT-treated cells (Figure 1A and B). The human CTBP1-DT gene is located on chromosome Chr 4p16.3 (1 243 228–1 246 795) and the predicted open reading frame (ORF) in the 2nd exon of the human CTBP1-DT transcript is 561 bp, encoding a 186 amino acid (aa) protein (molecular weight:19.74 kDa; Figure 1B; Supplementary Figure S1A). Interestingly, mass spectrometry (LC–MS/MS)-based proteomics revealed that a novel protein, which was only identified in the CPT-treated 293T cells but not in control cells, matched the predicted CTBP1-DT-encoded 186-aa ORF (Figure 1C).
Figure 1.
LncRNA CTBP1-DT-encoded microprotein DDUP promotes DNA damage repair. (A) Left and middle panel: polysome profiling was performed to identify potential new factors involved in DNA damage responses. DNA damage in 293T cells was determined from γ-H2AX expression after CPT (10 μM) treatment for 1 h. Right: volcano plot analysis of dysregulated ribosome-associated RNAs from polysome profiling data; x-axis = log2 fold change in expression of polysome-associated RNAs between CPT- and vehicle-treated cells; y-axis = FDR value (–log10 transformed) of polysome-associated RNAs. (B) Potential ORF of the DDUP protein located in exon 2 of non-coding lncRNA CTBP1-DT in the human genome, and the full-length DDUP protein synthesised to generate a polyclone antibody. Molecular weight (MW):19.74 kDa. (C) Unique CTBP1-DT peptide identified by LC–MS/MS analysis. (D) IB analysis of DDUP expression in Flag-tagged DDUP-transduced 293T and HeLa cells validated by the generated anti-DDUP polyantibody. Synthesised DDUP protein served as a positive control and GAPDH served as a loading control. (E) IB analysis of endogenous DDUP expression in parental and CTBP1-DT-silenced 293T cells treated with CPT (10 μM) at the indicated time-point. GAPDH served as a loading control. F. IB analysis of endogenous DDUP and γ-H2AX expression in cells treated with CPT (10 μM) or VP-16 (10 μM) or CDDP (5 μM) for 1 h or with IR (10 Gy) and were allowed to recover for 6 h. GAPDH served as a loading control. (G) Real-time analysis of polysome-associated CTBP1-DT levels in CPT (10 μM)-treated cells at the indicated time-point. (H) Representative images (left) and quantification (right) of damaged DNA in the indicated cells analysed by comet assay (n = 100). The indicated cells were treated with CPT (10 μM) or VP-16 (10 μM) for 1 h. Scale bar = 20 μm. (I) Representative images (left) and quantification (right) of γ-H2AX foci in the indicated cells with CPT treatment (10 μM, 1 h). At least 100 cells were counted. Scale bar = 5 μm. (J) Representative images (left) and kinetics (right) of γ-H2AX signals in response to laser micro-irradiation in the indicated cells and recovery for the indicated times (n = 100). Each error bar represents the mean ± SD of three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001).
To further confirm that proteomics-identified peptide was translated from CTBP1-DT gene, we then synthesised the corresponding full-length protein to generate a polyclonal antibody. The specificity of antibody was determined by immunoblotting (IB), showing that a specific Flag-tagged protein was expressed in Flag-ORF-transfected cells, and the inductive effect of CPT on endogenous protein was abolished by two CTBP1-DT shRNAs (Figure 1D and E; Supplementary Figure S1B). Interestingly, IB analysis using this antibody revealed that in addition to CPT, treatment with etoposide (VP-16) or cisplatin (CDDP), or ionizing radiation (IR) also resulted in drastic upregulation of the CTBP1-DT-encoded microprotein in both 293T and HeLa cells (Figure 1F). We therefore named this CTBP1-DT-encoded microprotein as DNA damage-upregulated protein (DDUP).
Although no significant alteration in CTBP1-DT transcript levels was observed in CPT-, VP-16, CDDP- or IR-treated cells, real-time PCR analysis showed that the level of polysome-associated CTBP1-DT was dramatically increased upon DNA damage (Supplementary Figure S1C; Figure 1G), suggesting that DNA damage-induced upregulation of DDUP protein involved a post-transcriptional mechanism.
Upregulation of DDUP promotes DNA damage repair
We next investigated whether DDUP contributed to DDR using a series of functional assays. Although dysregulation of DDUP has no effect on DNA-damage in vehicle-treated cells (Supplementary Figure S1D and E), the level of CPT- or VP-16-induced DNA damage was much lower in DDUP-overexpressing cells but significantly higher in DDUP-silenced cells as determined by comet and γ-H2AX IF staining assays (Figure 1H and I; Supplementary Figure S1F). Furthermore, laser micro-irradiation imaging revealed that dysregulation of DDUP had no effect on early DDR, as indicated by the same recruitment level of γ-H2AX to damaged DNA (at 0.5 h), but the laser-induced γ-H2AX signal was rapidly declined in DDUP-overexpressing cells, and remained nearly the same in CTBP1-DT-silenced cells at 6 h (Figure 1J). These results suggest that DDUP contributed to DNA damage repair but not to initiation of DDR. Consistently, overexpressing DDUP significantly enhanced the viability of CPT-treated cells, while CTBP1-DT-silenced cells exhibited increased sensitivity to CPT treatment (Supplementary Figure S1H). However, neither overexpressing DDUP nor silencing CTBP1-DT has any impact on cell survival without DNA damage induction (Supplementary Figure S1G). Taken together, these results indicate that DNA damage-induced upregulation of DDUP promotes DNA damage repair.
LncRNA CTBP1-DT itself does not impact DNA damage repair
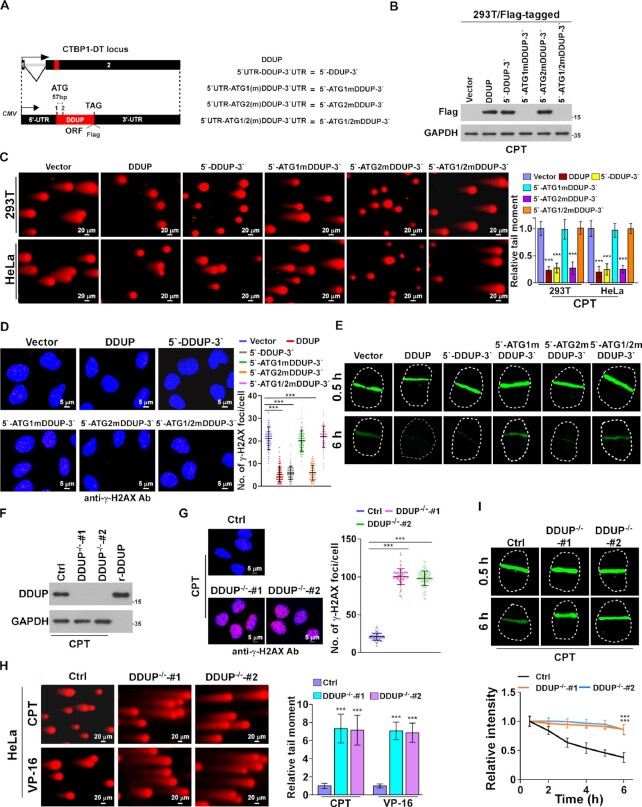
To determine the role of the DDUP protein rather than the lncRNA CTBP1-DT gene in DNA damage repair, a series of lncRNA CTBP1-DT constructs were established, including 5′ untranslated region (UTR)-DDUP-3′UTR (5′-DDUP-3′) wild-type plasmid and three ATG mutated plasmids 5′UTR-ATG1mDDUP-3′UTR (5′-ATG1mDDUP-3′), 5′UTR-ATG2mDDUP-3′UTR (5′-ATG2mDDUP-3′) and 5′UTR-ATG1/2mDDUP-3′UTR (5′-ATG1/2mDDUP-3′), because the DDUP ORF contains two ATG codons (Figure 2A). IB analysis yielded a ∼19.74 kDa band corresponding to Flag-tagged DDUP protein in DDUP-, 5′-DDUP-3′- and 5′-ATG2mDDUP-3′-transduced 293T cells but not in 5′-ATG1mDDUP-3′- and 5′-ATG1/2mDDUP-3′-transduced 293T cells upon CPT treatment (Figure 2B). These results indicate that the first ATG serves as start codon for the initiation of DDUP translation.
Figure 2.
DDUP microprotein, but not CTBP1-DT RNA, promotes DNA damage repair. (A, B) Diagram (A) and expression (B) of the indicated lncRNA CTBP1-DT constructs, including wild-type (WT), ATG1 mutation, ATG2 mutation and double ATG site mutation (ATG mutated to ATT) in CPT (10 μM, 1 h)-treated 293T cells. GAPDH served as a loading control. (C) Representative images (left) and quantification (right) of damaged DNA in the indicated cells analysed by comet assay (n = 100). The indicated cells were treated with CPT (10 μM) for 1 h. Scale bar = 20 μm. (D) Representative images (left) and quantification (right) of γ-H2AX foci in the indicated cells with CPT treatment (10 μM, 1 h). At least 100 cells were counted. Scale bar = 5 μm. (E) Representative images (left) and kinetics (right) of γ-H2AX signals in response to laser micro-irradiation in the indicated cells and recovery for the indicated times (n = 100). (F) IB analysis of endogenous DDUP expression in CPT (10 μM, 1 h)-treated control and DDUP-KO cells. GAPDH served as a loading control. (G) Representative images (left) and quantification (right) of γ-H2AX foci in the indicated cells with CPT treatment (10 μM, 1 h). At least 100 cells were counted. Scale bar = 20 μm. (H) Representative images (left) and quantification (right) of damaged DNA in the indicated cells analysed by comet assay (n = 100). The indicated cells were treated with CPT (10 μM) for 1 h. Scale bar = 5 μm. (I) Representative images (upper) and kinetics (lower) of γ-H2AX signals in response to laser micro-irradiation in the indicated cells and recovery for the indicated times (n = 100). Each error bar represents the mean ± SD of three independent experiments (*P< 0.05, **P< 0.01, ***P< 0.001).
We then probed the effect of the abovementioned four lncRNA CTBP1-DT constructs on DNA-damage repair. Consistent with the stimulatory effect of DDUP on DNA damage repair, 5′-DDUP-3′ and 5′-ATG2mDDUP-3′-transduced cells exhibited less CPT-induced DNA damage, as determined by comet assay and γ-H2AX foci staining (Figure 2C and D; Supplementary Figure S2A). However, transfection of 5′-ATG1mDDUP-3′ and 5′-ATG1/2mDDUP-3′ did not enhance DNA damage repair (Figure 2C and D; Supplementary Figure S2A). Laser micro-irradiation imaging assays further confirmed that overexpression of DDUP-encoding CTBP1-DT plasmids but not DDUP-noncoding CTBP1-DT plasmids promoted the decline in CPT-induced γ-H2AX signal (Figure 2E). These results suggest that lncRNA CTBP1-DT-encoded DDUP protein but not the lncRNA itself contributed to DNA damage repair. In line with this hypothesis, we observed that overexpression of DDUP-coding CTBP1-DT genes but not DDUP-noncoding CTBP1-DT genes promoted cell survival upon CPT treatment (Supplementary Figure S2B).
To further confirm the role of DDUP protein in DNA damage repair, two DDUP-knockout (DDUP-KO) HeLa cell lines were established using CRISPR/Cas9 genome editing technology (Figure 2F; Supplementary Figure S2C and D). As shown in Figure 2G and H, knockout of DDUP further enhanced CPT-induced DNA damage, as indicated by higher tail moments and more γ-H2AX foci, consequently resulting in increased cell death (Supplementary Figure S2E and F). Meanwhile, the irradiation-induced γ-H2AX foci signal remained significantly higher than in control cells at 6 h in irradiated DDUP-KO cells (Figure 2I). Taken together, these results provide further evidence that lncRNA CTBP1-DT-encoded DDUP protein but not lncRNA itself contributes to DNA damage repair.
DNA damage-induced DDUP is translated via the IRES in the 5′UTR of CTBP1-DT
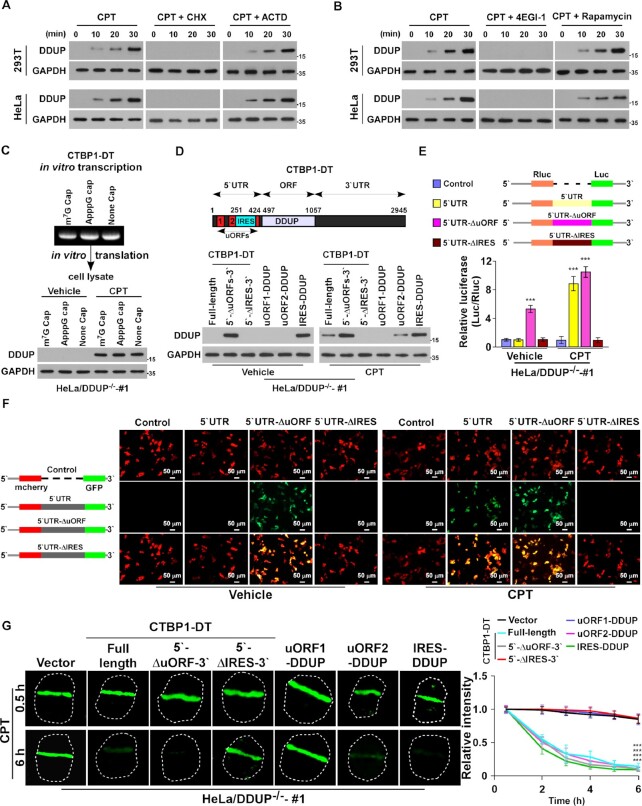
The results in Figure 1D and E, and Supplementary Figure 1C suggested that DNA damage-induced DDUP upregulation might involve a post-transcriptional mechanism. Consistent with this hypothesis, we found that blocking translation using cycloheximide (CHX) completely abrogated CPT-induced DDUP upregulation, whereas inhibiting transcription using actinomycin-D (ACTD) had no such effect (Figure 3A). Furthermore, CPT-induced DDUP upregulation was effectively blocked by 4EGI-1 treatment, which inhibits both cap-dependent and -independent initiation of translation, but not by rapamycin treatment, which inhibits only cap-dependent initiation of translation (Figure 3B), suggesting that cap-independent translation contributes to CPT-induced DDUP upregulation. Consistent with this hypothesis, using m7G-capped, non-functional ApppG-capped, or non-capped CTBP1-DT RNA as template, in vitro translation assays revealed that all three types of CTBP1-DT RNA could be translated to produce DDUP protein via CPT-treated cell lysate but not by vehicle-treated cell lysate (Figure 3C), providing further evidence that CPT-induced DDUP upregulation involved a cap-independent translation mechanism.
Figure 3.
The IRES in the 5′UTR of CTBP1-DT is essential for DNA damage-induced DDUP translation. (A) IB analysis of DDUP expression in 293T and HeLa cells treated with CPT (10 μM), CPT (10 μM) + CHX (5 μg/ml), or CPT (10 μM) + ACTD (5 μg/ml) for the indicated times. GAPDH served as a loading control. (B) IB analysis of DDUP expression in 293T and HeLa cells treated with CPT (10 μM), CPT (10 μM) + 4EGI-1 (25 μM) or CPT (10 μM) + rapamycin (100 nM) for the indicated times. GAPDH served as a loading control. (C) In vitro translation assay analysis of DDUP protein in vehicle- or CPT (10 μM, 1 h)-treated DDUP-KO cell lysates using in vitro-transcribed m7G-capped, non-functional ApppG capped-, or non-capped-CTBP1-DT RNA as template. GAPDH served as a loading control for the levels of cell lysates used for in vitro translation assays. (D) Upper: Graphical representation (upper) of the location of the uORFs, IRES and DDUP ORF, and the 5′UTR and 3′UTR in CTBP1-DT. Lower: expression of DDUP in the vehicle- and CPT (10 μM, 1 h)-treated DDUP-KO cells transfected with the indicated constructs. GAPDH served as a loading control. (E) Luciferase reporter assay analysis of relative Luc/Rluc activity in vehicle- and CPT (10 μM, 1 h)-treated 293T cells transfected with the indicated constructs. (F) Representative fluorescence images of mCherry and GFP signals in vehicle- and CPT (10 μM, 1 h)-treated 293T cells transfected with the indicated constructs. Scale bar = 50 μm. (G) Representative images (left) and kinetics (right) of γ-H2AX signals in response to laser micro-irradiation in the indicated cells and recovery for the indicated times (n = 100). Each error bar represents the mean ± SD of three independent experiments (*P< 0.05, **P< 0.01, ***P< 0.001).
Interestingly, bioinformatics analysis predicted two upstream ORFs (uORFs) spanning 55–132 bp and 185–460 bp that normally inhibit translation of the main ORF (29), and a natural internal ribosome entry segment (IRES, 251–424 bp) that mediates cap-independent translation, within the 5′UTR of CTBP1-DT (Figure 3D; Supplementary Figure S3A). We therefore investigated the contribution of these two elements to DDUP regulation. As shown in Figure 3D, without DNA damage induction, overexpressing the full-length CTBP1-DT gene did not produce DDUP protein, but deletion of the uORFs region in the 5′UTR of the CTBP1-DT gene (5′-ΔuORFs-3′) resulted in DDUP upregulation, which was further increased in response to DNA damage (Figure 3D). Importantly, addition of the uORFs sequence before the DDUP ORF (uORFs-DDUP) completely blocked DDUP expression (Figure 3D) but deletion of the IRES region in the 5′UTR of the CTBP1-DT gene (5′-ΔIRES-3′) abrogated DDUP expression, even in CPT-treated cells (Figure 3D). Therefore, these results indicate that uORFs function as negative regulators by inhibiting DDUP translation, while IRES acts as a positive regulator for DDUP upregulation.
To further confirm the regulatory effects of uORFs and IRES on DDUP expression, full-length 5′UTR and uORFs- and IRES-deleted 5′UTR were inserted into the Rluc-Luc reporter and mCherry/GFP vectors. Consistent with the effects on the regulation of DDUP, deletion of uORFs significantly increased the Luc/Rluc ratio and GFP signals compared with the full-length 5′UTR, while deletion of IRES had the opposite effect (Figure 3E and F). These results further support the notion that the uORFs region functions as a negative regulator while IRES acts as a positive cis-element for DDUP translation.
Furthermore, experiments using comet assays, γ-H2AX staining, and laser micro-irradiation assays showed that deletion of uORFs significantly increased the stimulatory effect of full-length CTBP1-DT on DNA damage repair, whereas ablation of IRES had the opposite effect (Figure 3G; Supplementary Figure S3B and C). Conversely, addition of uORFs reduced the repair capability of DDUP/ORF on damaged DNA, whereas addition of IRES enhanced this capability (Figure 3G; Supplementary Figure S3B and C). Taken together, these results demonstrate that the uORFs and IRES regions within the 5′UTR of the CTBP1-DT gene are involved in DNA damage-induced DDUP translation.
DDUP interacts with multiple putative DDR proteins in DNA-damaged cells
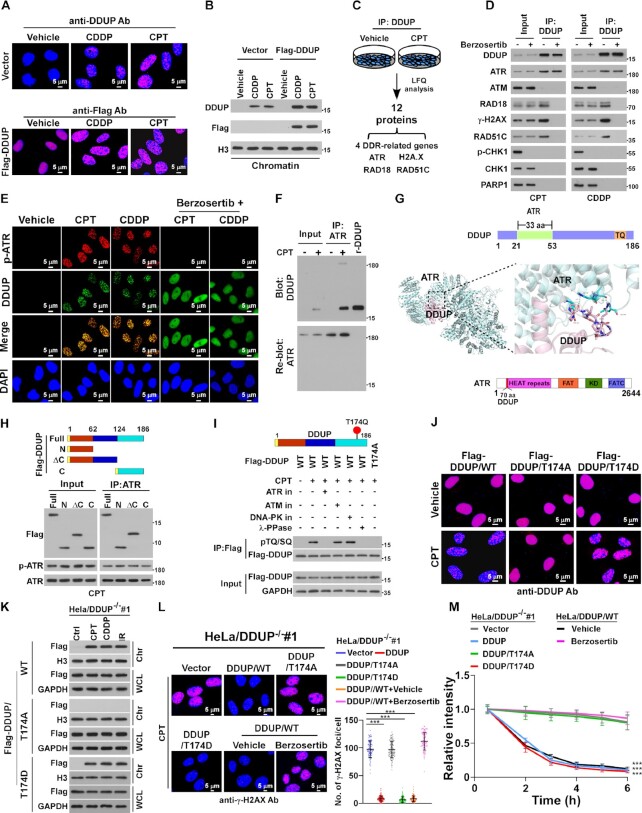
We next investigated the mechanism for DDUP-mediated DNA damage repair. IF staining using anti-DDUP and anti-Flag antibodies revealed that endogenous and Flag-tagged DDUP protein nuclear-localised and formed DNA damage-induced foci (Figure 4A). Since no fluorescence signal was observed in vehicle-treated control and DDUP-KO HeLa cells and in CPT- and VP-16-treated DDUP-KO HeLa cells, suggesting that the anti-DDUP antibody used in the current study was also suitable for IF assay (Supplemental Figure S4A). Furthermore, IB analysis showed that the level of chromatin-associated DDUP was drastically elevated in response to DNA damage (Figure 4B). These results suggest that DNA damage induces DDUP-foci formation.
Figure 4.
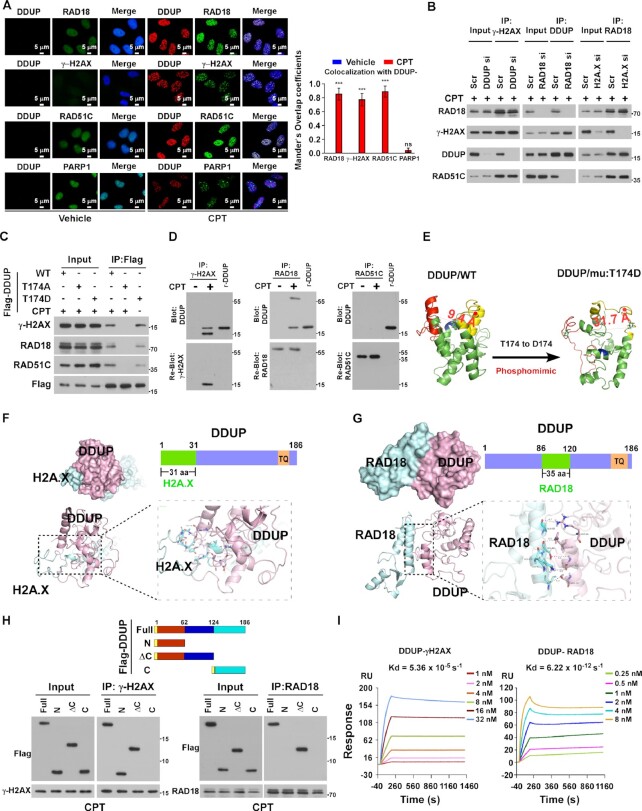
Phosphorylation of DDUP is essential for DDUP-mediated damage repair. (A) IF staining analysis of endogenous DDUP foci using anti-DDUP or Flag-tagged DDUP foci using anti-Flag antibody in vector- or Flag-tagged DDUP-transfected HeLa cells treated with vehicle, CPT (10 μM) or CDDP (5 μM) for 1 h. (B) Chromatin fraction and IB analysis of DNA-bound DDUP in vector- and Flag-tagged DDUP-transfected HeLa cells treated with vehicle, CPT (10 μM) or CDDP (5 μM) for 1 h. H3 served as a loading control. (C) LFQ analysis of potential significantly upregulated DDUP-interacting proteins in vehicle- and CPT (10 μM, 1 h)-treated 293T cells. (D) Co-IP analysis of the interaction of DDUP with ATR, ATM,RAD18, γ-H2AX, RAD51C, p-CHK1, CHK1 and PARP1 in CPT (10 μM, 1 h)—and CDDP (5 μM, 1 h)-treated 293T cells with or without berzosertib (80 nM, 1 h) treatment. (E) IF staining analysis of DNA damage-induced p-ATR foci (red) and endogenous DDUP foci (green) in HeLa cells treated with CPT (10 μM), CDDP (5 μM), or combination with berzosertib (80 nM) for 1 h. (F) Far-western blotting analysis of the direct ATR/DDUP interaction using anti-ATR antibody-immunoprecipitated proteins and detected using anti-DDUP antibody then re-blotting with anti-ATR antibody. Recombinant DDUP protein served as a control. (G) Molecular docking between ATR and DDUP performed using the Cluspro 2.0 web server (https://cluspro.org/help.php). The structure is shown in cartoon representation. The 3D structure of WT DDUP was obtained from the I-TASSER server and the 3D structure of ATR (PDB ID: 5yz0) was downloaded from the RCSB Protein Data Bank. (H) Schematic illustration of full-length and truncated DDUP proteins (upper) and co-IP assay analysis of the ATR-interacting region in DDUP using anti-ATR antibody in for CPT (10 μM, 1 h)-treated HeLa cells transfected with full-length and truncated DDUP fragments (lower). (I) IP assays using anti-Flag antibody performed in DDUP/WT- and DDUP/T174A mutant-transfected cells treated with ATR inhibitor (80 nM), ATM inhibitor (10 μM), or DNA-PKcs inhibitor (2 μM) for 1 h prior to treat with or without CPT (10 μM, 1 h) as indicated, analysed by immunoblotting with anti-pTQ/SQ antibody. (J) IF staining using anti-DDUP antibody performed in DDUP/WT- and DDUP/mutant-transfected cells with or without CPT treatment (10 μM, 1 h), with the image captured by laser confocal microscopy. Scale bar = 5 μm. (K) Chromatin fraction and IB analysis of DNA-bound DDUP/WT, DDUP/T174A and DDUP/T174D in CPT (10 μM, 1 h)-, CDDP (5 μM, 1 h)- and IR (10 Gy)-treated DDUP-KO HeLa cells transfected with DDUP/WT and DDUP/mutant plasmids. (L) Representative images (left) and quantification (right) of γ-H2AX foci in the CPT (10 μM, 1 h)-treated indicated cells with or without berzosertib treatment (80 nM, 1 h). At least 100 cells were counted. Scale bar = 5 μm. (M) Kinetics of γ-H2AX signals in response to laser micro-irradiation in the indicated cells and recovery for the indicated times (n = 100). The indicated cells treated with or without berzosertib (80 nM) for 1 h. Each error bar represents the mean ± SD of three independent experiments (*P< 0.05, **P< 0.01, ***P< 0.001).
Damage-induced foci are generally formed by the recruitment of damage response proteins to chromatin in the vicinity of damaged DNA (30). We next explored potential DDUP-interacting proteins using label free quantification proteomics (LFQ) analysis. As shown in Figure 4C and Supplementary Table S1, among 12 DDUP-interacting proteins, four proteins (ATR, γ-H2AX, RAD18 and RAD51C) were putative DNA damage response proteins. Consistent with the LFQ analysis, co-immunoprecipitation (co-IP) assays showed that DDUP interacted with ATR, γ-H2AX, RAD18 and RAD51C, but not with ATM and PARP1, in CDDP- and CPT-treated cells (Figure 4C and D). These results provide further evidence for the role of DDUP in DNA damage repair.
Phosphorylation of DDUP by ATR is required for DDUP-mediated DNA damage repair
Importantly, treatment with ATR inhibitor Berzosertib, an intravenously administered small molecule exhibiting promising anti-tumour activity in multiple Phase I/II clinical trials, entirely abolished the interaction of DDUP with γ-H2AX, RAD18 and RAD51C, and also inhibited DDUP foci formation (Figure 4D and E). However, deletion of DDUP had no effect on DNA damage-induced ATR foci (Supplementary Figure S4B). These results suggest that ATR activity is required for the role of DDUP in DNA damage repair.
Far-western blotting analysis indicated that DDUP directly interacted with ATR (Figure 4F). Analysis using ClusPro server4-8 (https://cluspro.org) predicted that the three-dimensional (3D) structures of DDUP and ATR could be docked, and residues 21–53 in the N-terminus of DDUP docked with residues 731–801 in the HEAT repeat region of ATR (Figure 4G; Supplementary Figure S4C). This automated prediction of DDUP/ATR association was further confirmed by co-immunoprecipitation (co-IP) assays using serially truncated DDUP and ATR fragments (Figure 4H; Supplementary Figure S4D).
Since T174Q in DDUP is a TQ/SQ motif that is the consensus ATM/ATR substrate site, this promoted us to examine whether DDUP was an ATR substrate. Co-IP assays indicated that CPT treatment induced the phosphorylation of DDUP at T174, but CPT-induced DDUP phosphorylation was abolished by ATR inhibitor Berzosertib, but not ATM inhibitor KU-55933 or DNA-PK inhibitor KU-57788, indicating ATR-mediated DDUP phosphorylation (Figure 4I). Importantly, γ-H2AX IF staining, micro-irradiation, and comet assays showed that mutation of T174 to A, but not D, not only abolished DDUP foci formation and enrichment of DDUP in chromatin upon DNA damage, but also resulted in loss of the DNA damage repair function of DDUP (Figure 4J–M; Supplementary Figure S4E). Taken together, these results demonstrate that ATR-mediated phosphorylation of DDUP is essential for the role of DDUP in DNA damage repair.
Phosphorylated DDUP forms a complex with γ-H2AX and Rad18
In agreement with the LFQ analysis and co-IP results (Figure 4C and D), IF staining assays showed that DNA damage-induced DDUP foci were significantly co-localised with RAD18 foci, γ-H2AX foci and RAD51C foci but not with the PARP1-foci (Figure 5A). Interestingly, silencing RAD18 has no effect on DDUP/γ-H2AX interaction but abolished the DDUP/RAD51C interaction (Figure 5B). Meanwhile, we found that silencing H2AX did not result in reduction of DDUP/RAD18 complex formation (Figure 5B). These results suggested that DDUP might separately interact with RAD18 and γ-H2AX and RAD18 was required for DDUP/RAD51C interaction. Interestingly, co-IP assays showed that the DDUP/T174A mutant in CPT-treated cells could not bind to either γ-H2AX or Rad18, but a complex was formed in DDUP/T174D mutant cells without DNA damage (Figure 5C), which suggested that phosphorylation of DDUP was required for γ-H2AX/DDUP/Rad18 complex formation and association. In line with this suggestion, far-western blotting further confirmed the direct interaction of γ-H2AX and Rad18 but not RAD51C with DDUP/T174D (Figure 5D). However, silencing DDUP had no effect on γ-H2AX/Rad18/RAD51C complex formation, which was consistent with our conclusion that DDUP is not involved in initiation of DDR (Figure 1I and J).
Figure 5.
Phosphorylated DDUP forms a complex with γ-H2AX and RAD18. (A) IF staining analysis of the co-localisation of DDUP foci with RAD18 foci, γ-H2AX foci, RAD51C-foci and PARP1-foci in vehicle- and CPT (10 μM, 1 h)-treated HeLa cells. Scale bar = 5 μm. Co-localization the fluorescence between molecules was quantified using the Manders' overlap coefficients algorithm. (B) Co-IP assay analysis of the formation of the DDUP/γ-H2AX/RAD18/RAD51C complex in the indicated gene-silenced 293T cells treated with CPT (10 μM) for 1 h. (C) Co-IP assay analysis of the interaction of WT and mutated DDUP with γ-H2AX, RAD18 and RAD51C in CPT (10 μM, 1 h) or without CPT-treated 293T cells. (D) Far-western blotting analysis of the direct interaction of DDUP/γ-H2AX using anti-γ-H2AX antibody-immunoprecipitated proteins (left), or DDUP/RAD18 using anti-RAD18 antibody-immunoprecipitated proteins (middle), or DDUP/RAD51C using anti-RAD51C antibody-immunoprecipitated proteins in RAD18-silenced cells (right), then detected using anti-DDUP antibody. Recombinant DDUP/T174D protein served as control. (E) Left: the 3D structure of WT DDUP in the dense state obtained from the I-TASSER server. Right: the 3D structure of the DDUP mutant (T174 to D174), which mimics phosphorylation of DDUP, in the loose state obtained from the I-TASSER server. (F) Molecular docking of H2A.X and DDUP. The 3D structure of H2A.X obtained from the I-TASSER server. The combined surface, cartoon and stick representation shows the predicted interaction interface between H2A.X and DDUP based on the modeled DDUP structure. The DDUP protein is coloured pink and H2A.X is coloured in cyan. (G) Molecular docking of RAD18 and DDUP. The 3D structure of RAD18 (PDB ID: 2YBF) was download from the RCSB Protein Data Bank. The combined surface, cartoon and stick representation shows the predicted interaction interface between RAD18 and DDUP based on the modeled DDUP structure. The DDUP protein is coloured pink and RAD18 is coloured cyan. (H) Upper: Schematic illustration of WT and truncated DDUP proteins. Lower: IP assay analysis of the γ-H2AX-interacting region of DDUP using anti-γ-H2AX antibody (lower left) and the RAD18-interacting region of DDUP using anti-RAD18 antibody in CPT (10 μM, 1 h)-treated 293T cells transfected with full-length and truncated DDUP fragments. (I) SPR analysis of the direct interaction between DDUP and γ-H2AX (left) and the direct interaction between DDUP and RAD18 (right). DDUP protein was immobilised on a Series S Sensor Chip. The Kd value for the DDUP/γ-H2AX and DDUP/RAD18 interaction was calculated as the raw response (RU). Each error bar represents the mean ± SD of three independent experiments (*P< 0.05, **P< 0.01, ***P< 0.001).
The 3D structure analysis using I-TASSER (31) (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) revealed a dense structure for the N- and C-terminal and central region of DDUP protein, but mutation of T174 to D, which mimicked phosphorylation of DDUP, resulted in a drastic ‘dense-to-loose’ change in the 3D structure of DDUP (Figure 5E). These results suggest that ATR-mediated phosphorylation of DDUP promoted interaction of DDUP with other proteins. In line with the molecular docking analysis showing that residues 1–31 of DDUP docked with phosphorylated C-terminal tail, residues 106–143, of H2A.X and residues 86–120 in the central region of DDUP docked with residues 8–88 in the N terminus of RAD18 (Figure 5F and G), while co-IP assays revealed an interaction between the N-terminus of DDUP and γ-H2AX protein, and an interaction between the central region of DDUP and RAD18 protein (Figure 5H). Furthermore, surface plasmon resonance (SPR) analysis showed that the Kd value of DDUP binding to γ-H2AX was 5.36E–5 S−1 and the Kd value of DDUP binding to RAD18 was 6.22E–12 S−1(Figure 5I). Taken together, these results demonstrate a high affinity for binding of DDUP to γ-H2AX and RAD18.
Concordant with the results described above showing that the N-terminus of DDUP bound to γ-H2AX and the central region of DDUP bound to RAD18 (Figure 5F–H), CPT or CDDP treatment induced the foci formation of DDUP/N, DDUP/M and DDUP/ΔC, and drastically increased the level of DDUP/N, or DDUP/M, or DDUP/ΔC mutants, but not the DDUP/C mutant, in chromatin, and overexpressing RAD18- or γ-H2AX-interacting DDUP fragment resulted in a reduction of DNA damage-induced chromatin-bound endogenous DDUP (Supplementary Figure S5A–C). Meanwhile, IB analyses showed that enforced expression of DDUP/N inhibited the DDUP/γ-H2AX interaction, and the DDUP/RAD18 interaction was hampered by DDUP/ΔC mutant overexpression. However, ectopically expressing the C-terminal fragment of DDUP had no effect on DDUP/γ-H2AX/Rad18 complex formation (Supplementary Figure S5D). Taken together, these results indicate that DNA damage induced the formation of DDUP/γ-H2AX/Rad18 complex
DDUP sustains retention of RAD18 at DNA damage sites
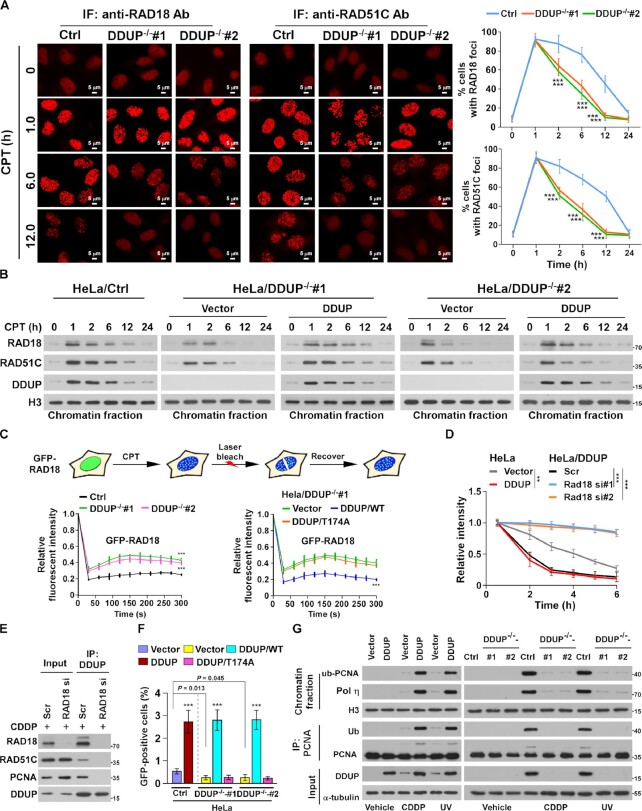
Although our results showed that silencing DDUP had no effect on γ-H2AX/Rad18 interaction (Figure 5B), we found that DNA damage-induced Rad18 foci were rapidly decreased in DDUP-KO cells compared with control cells. As shown in Figure 6A and B, RAD18 foci appeared within 10 min after CPT treatment and reached a plateau around 1 h in both control and DDUP-KO cells, indicating that RAD18 foci formation does not require DDUP protein. Interestingly, in control cells, the number of RAD18 foci slowly decreased after 1 h and returned to background levels after 24 h, whereas DDUP-KO cells displayed a rapid decrease in RAD18 foci to background levels within 12 h (Figure 6A and B). However, restoring DDUP expression in DDUP-KO cells significantly enhanced the retention of RAD18 in chromatin (Figure 6B). Importantly, no significant difference of RPA2- and RAD51-foci was observed in CPT-treated DDUP-KO cells compared with control cells, indicating that depletion of DDUP has no effect on DNA resection (Supplementary Figure S6A). Taken together, these results suggest that DDUP stabilised RAD18 at DNA-damaged regions.
Figure 6.
DDUP enhances the retention of RAD18 at DNA damage sites. (A) Representative images (left) and time course (right) of the formation of CPT (10 μM)-induced RAD18 and RAD51C foci in control and DDUP-KO HeLa cells and allowed to recover for the indicated times. The RAD18- and RAD51C foci was examined every 10 min in the CPT-treated cells within the first 2 h. Cells containing more than 10 RAD18 and RAD51C foci per nucleus were scored. (B) Chromatin fraction and IB analysis of DNA-bound RAD18, RAD51C and DDUP in the indicated CPT (10 μM)-treated cells and allowed to recover for the indicated times. H3 served as a loading control. (C) Quantitative FRAP analysis of GFP-RAD18 in GFP-RAD18-transfected control and DDUP-KO HeLa cells (right), and in DDUP-KO HeLa cells co-transfected with GFP-RAD18 and vector, GFP-RAD18, and DDUP/WT, or GFP-RAD18 and DDUP/T174A, treated with CPT (10 μM) and allowed to recover for the indicated times. (D) Kinetics of γ-H2AX signals in the indicated cells in response to laser micro-irradiation and allowed to recover for the indicated times (n = 100). (E) IP assay analysis of the DDUP/RAD51C and DDUP/PCNA interaction in control and RAD18-silenced 293T cells treated with CDDP (5 μM, 1 h). (F) Homologous recombination repair assays performed in the indicated cells. (G) IP/IB analysis of the regulatory effect of DDUP dysregulation on PCNA monoubiquitination in the indicated cells treated with CDDP (5 μM, 1 h) or UV radiation (60 J/m2). H3 and α-tubulin served as loading control. Each error bar represents the mean ± SD of three independent experiments (*P< 0.05, **P< 0.01, ***P< 0.001).
To further confirm this effect, the dynamics of CPT-induced RAD18 foci were measured using fluorescence recovery after photo bleaching (FRAP), which revealed that ∼20% of the photo-bleached GFP-RAD18 foci signal was regained in control cells, whereas 47% of the GFP-RAD18 foci signal was recovered in DDUP-KO cells (Figure 6C). The signal recovery rate in DDUP-KO cells was significantly inhibited by DDUP/WT overexpression (47–20%) but not by DDUP/T174A mutant overexpression (Figure 6C). Thus, DDUP/Rad18 interaction enhances retention of RAD18 at DNA damage sites.
DDUP participates in two independent DNA damage repair pathways
Next, we examined whether RAD18 was involved in DDUP-mediated DNA damage repair. As shown in Figure 6D and Supplementary Figure S6B and C, silencing RAD18 significantly reduced the stimulatory effect of DDUP on DNA damage repair, as determined by laser micro-irradiation and γ-H2AX assays. These results indicate that RAD18 was a key factor contributing to DDUP-mediated DNA damage repair. It has been previously reported that RAD18-mediated DNA damage repair occurs through homologous recombination repair (HRR) via association with RAD51C and post-replication repair (PRR) via monoubiquitylation of PCNA (13,15). We further examined whether DNA damage could induce the interaction of DDUP with RAD51C and PCNA. As shown in Figure 6E, co-IP assays showed that DDUP interacted with both RAD51C and PCNA in CDDP-treated cells, and this was abolished by RAD18 silencing. These results further support a key role for RAD18 in DDUP-mediated DNA damage repair.
Consistent with the effect of DDUP on retention of RAD18 at damaged DNA, loss of DDUP also resulted in the rapid decrease of RAD51C foci in CPT-treated cells, while recovery of DDUP led to retention (Figure 6A and B). Since RAD51C foci are an indication of HRR efficiency (13,32,33), we then examined the role of DDUP in HRR via a gene conversion assay using the DR-GFP reporter system. As shown in Supplemental Figure S6D and Figure 6F, overexpression of DDUP significantly increased the HRR efficiency but not NHEJ efficiency in HeLa cells, and DDUP-KO/HeLa cells exhibited lower HRR efficiency than control/HeLa cells, which provided further evidence of the role DDUP in HRR. Meanwhile, we also observed that upregulation of DDUP dramatically increased, while loss of DDUP reduced the expression of monoubiquitinated PCNA in CDDP-treated cells (Figure 6G). Importantly, the reduced HRR efficiency and monoubiquitinated PCNA level in DDUP-KO cells was strongly recovered by ectopic expression of DDUP/WT but not DDUP/T174A mutant (Figure 6F and G; Supplementary Figure S6E). More importantly, ablation of DDUP also resulted in the quick dismiss of PCNA foci in CDDP- or UV-treated cells (Supplementary Figure S6F). Taken together, these results indicate that DDUP-mediated DNA damage repair proceeds through two distinct mechanisms.
DDUP expression is inversely correlated with poorer survival of cancer patients
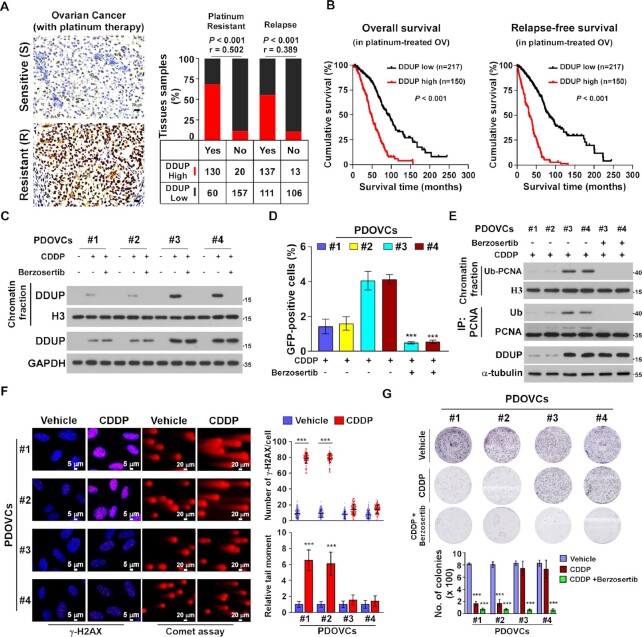
It has been demonstrated that the increased capability of cancer cells to repair therapeutically induced DNA damage contributes to chemoresistance in various cancer types, including ovarian cancer, which results in cancer progression and relapse (34–36). Therefore, immunohistochemistry (IHC) was performed in 367 clinical platinum-treated ovarian cancer samples and statistical analysis revealed that DDUP protein levels were significantly correlated with CDDP resistance (P < 0.001; r = 0.502) and relapse (P < 0.001; r = 0.389) and inversely associated with shorter overall/relapse-free survival in patients with ovarian cancer subjected to platinum-based therapy (P < 0.001; P < 0.001; Figure 7A and B; Supplementary Tables S3 and S4). Taken together, these results suggest that upregulated DDUP protein levels correlate with cancer relapse and poorer patient outcome.
Figure 7.
Upregulation of DDUP confers resistance to cisplatin in ovarian cancer cells in vitro. (A) Representative images of DDUP expression in chemo-sensitive and chemo-resistant ovarian cancer cells (n = 367) (left) and the positive correlation between DDUP levels and platinum resistance (P < 0.001; r = 0.502) and relapse (P < 0.001; r = 0.389) in ovarian cancer tissues (n = 367; right). Scale bar = 20 μm. Chi-square test was used for statistical analysis. (B) Kaplan–Meier analysis of overall survival (left) and relapse-free survival (right) for patients with ovarian cancer stratified by low versus high expression of DDUP (log-rank test; p < 0.001; n = 367). (C) IB analysis of expression of DNA-bound and total DDUP in patient-derived ovarian cancer cells (PDOVCs). H3 and GAPDH served as a loading control. (D) Homologous recombination repair assays performed in PDOVCs transfected with DR-GFP and pCBAScel plasmids following treatment with CDDP (5 μM) alone or CDDP (5 μM) plus Berzosertib (80 nM) for 1 h. (E) IB analysis of expression of DNA-bound and total monoubiquitinated PCNA and DDUP in the indicated PDOVCs treated with CDDP (5 μM) alone or CDDP (5 μM) plus Berzosertib (80 nM) for 1 h. H3 and α-Tubulin served as loading control. (F) Representative images (left) and quantification (right) of γ-H2AX foci determined by IF staining using anti-γ-H2AX antibody and damaged DNA determined by comet assay in the indicated PDOVCs treated with CDDP (5 μM, 1 h). Scale bar = 5 μm (left) and 20 μm (right). (G) Representative images (upper) and quantification (lower) of the number of PDOVCs colonies following the indicated treatment. Each error bar represents the mean ± SD of three independent experiments (*P < 0.05, **P < 0.01, ***P < 0.001).
The correlation between DDUP expression and CDDP chemoresistance was further examined in patient-derived ovarian cancer cells (PDOVCs), which more closely resemble ovarian cancer cells in the clinical tumour mass. Consistent with the results described above, CDDP treatment induced upregulation of DDUP in PDOVCs, which was abolished by ATR inhibitor Berzosertib treatment (Figure 7C). Importantly, #3 and #4 PDOVCs with higher CDDP-induced DDUP displayed greater capability for DNA damage repair, as indicated by higher HRR efficiency, increased monoubiquitinated PCNA level, fewer γ-H2AX foci, shorter tail moment, and stronger resistance to CDDP treatment compared with #1 and #2 PDOVCs with low CDDP-induced DDUP expression (Figure 7D–G). Moreover, we found that the capacity for DNA damage repair was significantly increased in #1 and #2 PDOVCs overexpressing DDUP but reduced in #3 and #4 PDOVCs with silencing of DDUP (Supplementary Figure S7A–C), which provided further evidence that DDUP plays a vital role in DNA damage repair, resulting in CDDP resistance. Taken together, these results demonstrate that combining platinum-based chemotherapy with ATR inhibition improved therapeutic outcome in an ovarian cancer model, which might represent a strategy to overcome clinical ovarian cancer recurrence (Figure 8).
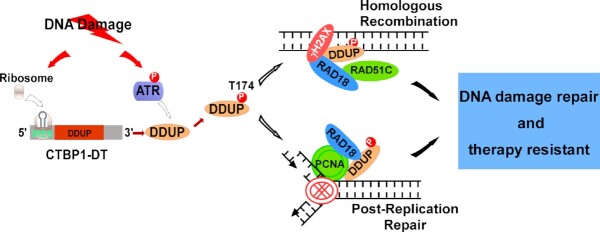
Figure 8.
Hypothetical model. DDUP encoded by an lncRNA orchestrates DNA damage repair by regulating PCNA monoubiquitination and RAD18 retention at the DNA damage sites.
DISCUSSION
Herein, we identified the novel lncRNA-encoded microprotein DDUP, a vital regulator of DDR, and revealed its crucial role in DNA damage repair. We demonstrated that DNA damage-mediated phosphorylation of DDUP resulted in a drastic ‘dense-to-loose’ conformational change in DDUP structure, which enhanced the interaction affinity and sustained the retention of RAD18 at DNA damage sites, thereby promoting DNA damage repair through dependent RAD51C-mediated HRR and monoubiquitinated PCNA-mediated PRR mechanisms. Importantly, treatment with ATR inhibitor Berzosertib, an intravenously administered small molecule, inhibited DDUP foci formation, which elicited rapid elimination of RAD18 foci and PCNA foci, consequently leading to hypersensitivity of ovarian cancer cells to DNA-damaging chemotherapeutics. These findings substantiate the importance of DNA damage-induced DDUP upregulation in chemoresistance, and may represent a new therapeutic strategy for treatment of genotoxic resistant cancers.
Although numerous repair mechanisms specific for many types of lesions have evolved, higher order chromatin structure presents a barrier to the recognition and repair of DNA damage. DNA damage-induced H2A.X phosphorylation overcomes this barrier via recruitment of repair factors to damaged DNA (37). Subsequently, stabilisation or retention of DDR factors recruited to γ-H2AX nucleosomes can be conducive to transmitting DNA damage signals and initiating DNA damage repair (13,14,19,37). For instance, it has been reported that RAD18 promotes HRR-mediated DNA damage repair through augmenting DNA damage signalling by facilitating the loading of RAD51C to damaged sites and the dissolution of Holliday junctions (13), as well as sustaining the retention of 53BP1 at sites of DNA damage (19). Therefore, identification of new factors involved in sustaining the DDR could be helpful for further understanding the DNA damage repair mechanism. Herein, we found that DDUP, encoded by lncRNA CTBP1-DT, was dramatically and rapidly elevated in response to multiple types of DNA damage. We further demonstrated that upregulated DDUP interacted with and sustained the retention of RAD18 foci, thereby promoting DNA damage repair through parallel PRR and HRR mechanisms. Therefore, our results identify a novel DNA damage early response factor that plays a vital role in DNA damage repair through sustaining DDR signalling.
Post-translational modifications, such as ubiquitination, SUMOylation, acetylation, and methylation, play a critical role in DNA damage signalling pathways (37–41). For instance, in response to DNA damage, the histone variant H2AX is immediately and extensively phosphorylated by ATM and ATR kinases (42–44), and phosphorylated H2AX (γ-H2AX) serves as a platform for the recruitment and maintenance of additional DDR factors at sites of DNA damage to transmit DNA damage signals (37,45,46). It has been reported that multiple E3 ligases, such as RNF8, RNF168, HERC2 and BRCA1/BARD1, and the SUMO ligases PIAS1 and PIAS4, are simultaneously recruited to γ-H2AX nucleosomes to elicit modification of themselves, thereby directing repair choices as events unfold (47–51). For example, recruitment of RNF8 to γ-H2AX nucleosomes, through association with RNF8/UBC13-mediated ubiquitin chains on γ-H2AX, initiate either the 53BP1-directed NHEJ pathway (19) or the RAD51C-mediated HRR pathway (13) to repair double-stranded DNA breaks. Herein, we found that DNA damage-activated ATR directly interacted with and phosphorylated DDUP, which induced a drastic ‘dense-to-loose’ conformational change in DDUP structure. Importantly, the conformational change of DDUP dramatically increased its interacting affinity with γ-H2AX and RAD18, resulting in the formation of a stable γ-H2AX/DDUP/RAD18 complex and sustained retention of RAD18 foci at damage sites. Therefore, our results provide further evidence for the role of post-translational modifications in DNA damage repair through a crosslink between ATR and γ-H2AX/DDUP.
Another key finding of the current study is the mechanism by which DNA damage induces DDUP upregulation. Although lncRNA CTBP1-DT was originally annotated as a non-coding RNA, we found that lncRNA CTBP1-DT could act as a template for the biosynthesis of the DDUP protein in response to DNA damage. Analysis of structural features and regulatory sequences predicted that the 5′UTR of CTBP1-DT contains two uORFs and a natural IRES. We demonstrated that the uORFs function as negative regulators via the recruitment and premature dissociation of ribosomes, which subsequently fails to initiate translation of the DDUP-coding ORF under normal conditions. However, under DNA damage conditions, ribosomes can bypass the uORFs region and associate with the IRES sequence, and thereby initiate the translation of DDUP through a cap-independent mechanism. Consistent with our results, accumulating evidence recently confirmed the existence of functional smORF-encoded polypeptides (SEPs) that are produced from non-coding RNAs under stress conditions, such as ultraviolet irradiation, hypoxia, nutrient limitation and temperature changes (52–55). Therefore, our findings strengthen the notion that SEPs driven by unique elements perform regulatory roles in fine-tuned biological systems such as DNA damage responses, and also highlight the potential of DDUP as a biomarker of DNA repair.
DNA damage-based chemotherapy and radiotherapy, which induce severe DNA damage and apoptosis in cancer cells, remain the preferred options for the cure and control of most malignancies, including ovarian cancer, lung cancer and oesophageal carcinoma (2,56,57). Functioning as a positive guardian that maintains genomic integrity to prevent carcinogenesis, DDR also acts as a negative saboteur to resist DNA damage-based cancer therapy, which suggests that DDR plays a ‘double-edged sword’ role in cancer prevention and cancer therapy. Therefore, targeting DDR factors, which are specifically overexpressed in tumours, is a potential strategy for enhancing the anticancer effect of DNA damage-based therapy. Building on our results showing that phosphorylation of DDUP, induced by DNA damage-activated ATR, sustained DDR signalling and promoted DNA damage repair, we further demonstrated that combined treatment with ATR inhibitor Berzosertib, an intravenously administered small molecule, could inhibit DDUP-mediated sustenance of DDR, consequently resulting in hypersensitivity of cancer cells to DNA-damaging chemotherapeutics. Since lncRNA CTBP1-DT is upregulated in multiple cancers, including hepatocellular carcinoma, breast cancer, and gliomas (58–61), CTBP1-DT might function as an onco-lncRNA. However, we found that the DDUP-mediated DNA damage repair function was independent of CTBP1-DT RNA. Taken together, our findings advance the current understanding of the molecular mechanisms underlying DNA damage responses, and represent a promising strategy that improves the efficacy of DNA damage-based anticancer therapies.
DATA AVAILABILITY
All RNA-seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) database with BioProject accession code PRJNA785355. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium and are available via ProteomeXchange with accession number PXD031047.
Cluspro 2.0 web server (https://cluspro.org/help.php).
I-TASSER server (https://zhanggroup.org/I-TASSER/).
Supplementary Material
ACKNOWLEDGEMENTS
Author contributions: R.Y., Y.H. and S.Z. contributed equally to this work. R.Y. designed the experiments and analysed data. Y.H. performed immunoprecipitation, western blot, in vitro translation, and real-time PCR analysis. S.Z. performed immunohistochemical and pathological analysis. X.L. and M.T. performed immunofluorescence staining, molecular docking, SPR analysis and mass spectrometry data analysis. M.T. and M.Y. performed in vitro cell line studies. L.G. performed the NHEJ reporter assay. X.W., Y.X. and Z.L. performed ribosome profiling and analysed RNA-seq data. X.L., S.C. and M.L performed fluorescence recovery after photo-bleaching studies, gene conversion and laser micro-irradiation experiments. L.S. and J.L. supervised the whole study and wrote the paper.
Contributor Information
Ruyuan Yu, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Yameng Hu, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Shuxia Zhang, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Xincheng Li, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Miaoling Tang, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Meisongzhu Yang, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Xingui Wu, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Ziwen Li, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Xinyi Liao, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Yingru Xu, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Man Li, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Suwen Chen, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Wanying Qian, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
Li-Yun Gong, Guangdong Provincial Key Laboratory for Genome Stability and Disease Prevention, Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Health Science Center, Shenzhen University, China.
Libing Song, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, China.
Jun Li, Program of Cancer Research, Affiliated Guangzhou Women and Children's Hospital, Zhongshan School of Medicine, Sun Yat-Sen University, China; Department of Biochemistry, Zhongshan school of medicine, Sun Yat-sen University, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [81830082, 82030078, 82072609, 81621004]. Funding for open access charge: Natural Science Foundation of China [82030078, 81830082, 82072609, 81621004].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lord C.J., Ashworth A.. The DNA damage response and cancer therapy. Nature. 2012; 481:287–294. [DOI] [PubMed] [Google Scholar]
- 2. O’Connor M.J. Targeting the DNA damage response in cancer. Mol. Cell. 2015; 60:547–560. [DOI] [PubMed] [Google Scholar]
- 3. Ciccia A., Elledge S.J.. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010; 40:179–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cleary J.M., Aguirre A.J., Shapiro G.I., D’Andrea A.D. Biomarker-Guided development of DNA repair inhibitors. Mol. Cell. 2020; 78:1070–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao Y., Mutter-Rottmayer E., Zlatanou A., Vaziri C., Yang Y.. Mechanisms of post-replication DNA repair. Genes (Basel). 2017; 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roos W.P., Thomas A.D., Kaina B.. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016; 16:20–33. [DOI] [PubMed] [Google Scholar]
- 8. Surova O., Zhivotovsky B.. Various modes of cell death induced by DNA damage. Oncogene. 2013; 32:3789–3797. [DOI] [PubMed] [Google Scholar]
- 9. Sun X., Wang Y., Ji K., Liu Y., Kong Y., Nie S., Li N., Hao J., Xie Y., Xu C.et al.. NRF2 preserves genomic integrity by facilitating ATR activation and G2 cell cycle arrest. Nucleic Acids Res. 2020; 48:9109–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J.. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007; 131:901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang B., Elledge S.J.. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:20759–20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stucki M., Jackson S.P.. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst.). 2006; 5:534–543. [DOI] [PubMed] [Google Scholar]
- 13. Huang J., Huen M.S., Kim H., Leung C.C., Glover J.N., Yu X., Chen J.. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 2009; 11:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kobayashi S., Kasaishi Y., Nakada S., Takagi T., Era S., Motegi A., Chiu R.K., Takeda S., Hirota K.. Rad18 and Rnf8 facilitate homologous recombination by two distinct mechanisms, promoting Rad51 focus formation and suppressing the toxic effect of nonhomologous end joining. Oncogene. 2015; 34:4403–4411. [DOI] [PubMed] [Google Scholar]
- 15. Durando M., Tateishi S., Vaziri C.. A non-catalytic role of DNA polymerase eta in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 2013; 41:3079–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ting L., Jun H., Junjie C.. RAD18 lives a double life: its implication in DNA double-strand break repair. DNA Repair (Amst.). 2010; 9:1241–1248. [DOI] [PubMed] [Google Scholar]
- 17. Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M.. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004; 23:3886–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S.. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002; 419:135–141. [DOI] [PubMed] [Google Scholar]
- 19. Watanabe K., Iwabuchi K., Sun J., Tsuji Y., Tani T., Tokunaga K., Date T., Hashimoto M., Yamaizumi M., Tateishi S.. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res. 2009; 37:2176–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yanagihara H., Kobayashi J., Tateishi S., Kato A., Matsuura S., Tauchi H., Yamada K., Takezawa J., Sugasawa K., Masutani C.et al.. NBS1 recruits RAD18 via a RAD6-like domain and regulates Pol eta-dependent translesion DNA synthesis. Mol. Cell. 2011; 43:788–797. [DOI] [PubMed] [Google Scholar]
- 21. Day T.A., Palle K., Barkley L.R., Kakusho N., Zou Y., Tateishi S., Verreault A., Masai H., Vaziri C.. Phosphorylated Rad18 directs DNA polymerase eta to sites of stalled replication. J. Cell Biol. 2010; 191:953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu R.L., Dong Y., Deng Y.Z., Wang W.J., Li W.D.. Tumor suppressor miR-145 reverses drug resistance by directly targeting DNA damage-related gene RAD18 in colorectal cancer. Tumour Biol. 2015; 36:5011–5019. [DOI] [PubMed] [Google Scholar]
- 23. Wu B., Wang H., Zhang L., Sun C., Li H., Jiang C., Liu X.. High expression of RAD18 in glioma induces radiotherapy resistance via down-regulating P53 expression. Biomed. Pharmacother. 2019; 112:108555. [DOI] [PubMed] [Google Scholar]
- 24. Schagger H. Tricine-SDS-PAGE. Nat. Protoc. 2006; 1:16–22. [DOI] [PubMed] [Google Scholar]
- 25. Chi H., Liu C., Yang H., Zeng W.F., Wu L., Zhou W.J., Wang R.M., Niu X.N., Ding Y.H., Zhang Y.et al.. Comprehensive identification of peptides in tandem mass spectra using an efficient open search engine. Nat. Biotechnol. 2018; 36:1059–1061. [DOI] [PubMed] [Google Scholar]
- 26. Wang L.H., Li D.Q., Fu Y., Wang H.P., Zhang J.F., Yuan Z.F., Sun R.X., Zeng R., He S.M., Gao W.. pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2007; 21:2985–2991. [DOI] [PubMed] [Google Scholar]
- 27. Kim J.S., Krasieva T.B., Kurumizaka H., Chen D.J., Taylor A.M., Yokomori K.. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 2005; 170:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bergamini G., Preiss T., Hentze M.W.. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA. 2000; 6:1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gebauer F., Hentze M.W.. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004; 5:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lisby M., Rothstein R.. DNA damage checkpoint and repair centers. Curr. Opin. Cell Biol. 2004; 16:328–334. [DOI] [PubMed] [Google Scholar]
- 31. Roy A., Kucukural A., Zhang Y.. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010; 5:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Badie S., Liao C., Thanasoula M., Barber P., Hill M.A., Tarsounas M.. RAD51C facilitates checkpoint signaling by promoting CHK2 phosphorylation. J. Cell Biol. 2009; 185:587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wassing I.E., Esashi F.. RAD51: beyond the break. Semin. Cell Dev. Biol. 2021; 113:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colomer C., Margalef P., Villanueva A., Vert A., Pecharroman I., Sole L., Gonzalez-Farre M., Alonso J., Montagut C., Martinez-Iniesta M.et al.. IKKalpha kinase regulates the DNA damage response and drives Chemo-resistance in cancer. Mol. Cell. 2019; 75:669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang P., Wei Y., Wang L., Debeb B.G., Yuan Y., Zhang J., Yuan J., Wang M., Chen D., Sun Y.et al.. ATM-mediated stabilization of ZEB1 promotes DNA damage response and radioresistance through CHK1. Nat. Cell Biol. 2014; 16:864–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vaughan S., Coward J.I., Bast R.C. Jr, Berchuck A., Berek J.S., Brenton J.D., Coukos G., Crum C.C., Drapkin R., Etemadmoghadam Det al.. Rethinking ovarian cancer: recommendations for improving outcomes. Nat. Rev. Cancer. 2011; 11:719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Celeste A., Petersen S., Romanienko P.J., Fernandez-Capetillo O., Chen H.T., Sedelnikova O.A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M.J.et al.. Genomic instability in mice lacking histone H2AX. Science. 2002; 296:922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou L., Zheng L., Hu K., Wang X., Zhang R., Zou Y., Zhong L., Wang S., Wu Y., Kang T.. SUMOylation stabilizes hSSB1 and enhances the recruitment of NBS1 to DNA damage sites. Signal Transduct. Target Ther. 2020; 5:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elia A.E., Boardman A.P., Wang D.C., Huttlin E.L., Everley R.A., Dephoure N., Zhou C., Koren I., Gygi S.P., Elledge S.J.. Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol. Cell. 2015; 59:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gong F., Miller K.M.. Histone methylation and the DNA damage response. Mutat. Res. Rev. Mutat. Res. 2019; 780:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morozko E.L., Smith-Geater C., Monteys A.M., Pradhan S., Lim R.G., Langfelder P., Kachemov M., Kulkarni J.A., Zaifman J., Hill A.et al.. PIAS1 modulates striatal transcription, DNA damage repair, and SUMOylation with relevance to Huntington's disease. Proc. Natl. Acad. Sci. U.S.A. 2021; 118:e2021836118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lanz M.C., Dibitetto D., Smolka M.B.. DNA damage kinase signaling: checkpoint and repair at 30 years. EMBO J. 2019; 38:e101801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marechal A., Zou L.. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013; 5:a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Blackford A.N., Jackson S.P. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017; 66:801–817. [DOI] [PubMed] [Google Scholar]
- 45. Kuo L.J., Yang L.X.. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo. 2008; 22:305–309. [PubMed] [Google Scholar]
- 46. Huang R.X., Zhou P.K.. DNA damage response signaling pathways and targets for radiotherapy sensitization in cancer. Signal Transduct., Target Ther. 2020; 5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J.. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007; 131:887–900. [DOI] [PubMed] [Google Scholar]
- 48. Mattiroli F., Vissers J.H., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., Sixma T.K.. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012; 150:1182–1195. [DOI] [PubMed] [Google Scholar]
- 49. Hu Q., Botuyan M.V., Zhao D., Cui G., Mer E., Mer G.. Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation. Nature. 2021; 596:438–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K.M., Jackson S.P.. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009; 462:935–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bekker-Jensen S., Rendtlew Danielsen J., Fugger K., Gromova I., Nerstedt A., Lukas C., Bartek J., Lukas J., Mailand N.. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 2010; 12:80–86. [DOI] [PubMed] [Google Scholar]
- 52. Andrews S.J., Rothnagel J.A.. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 2014; 15:193–204. [DOI] [PubMed] [Google Scholar]
- 53. Wang J., Zhu S., Meng N., He Y., Lu R., Yan G.R.. ncRNA-Encoded Peptides or proteins and cancer. Mol. Ther. 2019; 27:1718–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Lecea L., Kilduff T.S., Peyron C., Gao X., Foye P.E., Danielson P.E., Fukuhara C., Battenberg E.L., Gautvik V.T., Bartlett F.S. 2ndet al.. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vale W., Spiess J., Rivier C., Rivier J.. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science. 1981; 213:1394–1397. [DOI] [PubMed] [Google Scholar]
- 56. Klein A.V., Hambley T.W.. Platinum drug distribution in cancer cells and tumors. Chem. Rev. 2009; 109:4911–4920. [DOI] [PubMed] [Google Scholar]
- 57. Goldstein M., Kastan M.B.. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015; 66:129–143. [DOI] [PubMed] [Google Scholar]
- 58. Li Y., Zong J., Zhao C.. lncRNA CTBP1-AS2 promotes proliferation and migration of glioma by modulating miR-370-3p-Wnt7a-mediated epithelial-mesenchymal transition. Biochem. Cell. Biol. 2020; 98:661–668. [DOI] [PubMed] [Google Scholar]
- 59. Wang M., Zhao H.. LncRNA CTBP1-AS2 promotes cell proliferation in hepatocellular carcinoma by regulating the miR-623/Cyclin D1 Axis. Cancer Biother. Radiopharm. 2020; 35:765–770. [DOI] [PubMed] [Google Scholar]
- 60. Mohebi M., Sattari A., Ghafouri-Fard S., Modarressi M.H., Kholghi-Oskooei V., Taheri M.. Expression profiling revealed up-regulation of three lncRNAs in breast cancer samples. Exp. Mol. Pathol. 2020; 117:104544. [DOI] [PubMed] [Google Scholar]
- 61. Liu L.X., Liu B., Yu J., Zhang D.Y., Shi J.H., Liang P.. SP1-induced upregulation of lncRNA CTBP1-AS2 accelerates the hepatocellular carcinoma tumorigenesis through targeting CEP55 via sponging miR-195-5p. Biochem. Biophys. Res. Commun. 2020; 533:779–785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) database with BioProject accession code PRJNA785355. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium and are available via ProteomeXchange with accession number PXD031047.
Cluspro 2.0 web server (https://cluspro.org/help.php).
I-TASSER server (https://zhanggroup.org/I-TASSER/).