Abstract
Given the spasmodic increment in antimicrobial resistance (AMR), world is on the verge of “post-antibiotic era”. It is anticipated that current SARS-CoV2 pandemic would worsen the situation in future, mainly due to the lack of new/next generation of antimicrobials. In this context, nanoscale materials with antimicrobial potential have a great promise to treat deadly pathogens. These functional materials are uniquely positioned to effectively interfere with the bacterial systems and augment biofilm penetration. Most importantly, the core substance, surface chemistry, shape, and size of nanomaterials define their efficacy while avoiding the development of AMR. Here, we review the mechanisms of AMR and emerging applications of nanoscale functional materials as an excellent substitute for conventional antibiotics. We discuss the potential, promises, challenges and prospects of nanobiotics to combat AMR.
Graphical Abstract
Keywords: Antibiotics, Antibiotic resistance, Bacteria, Nanotechnology, Nanomaterials
Introduction
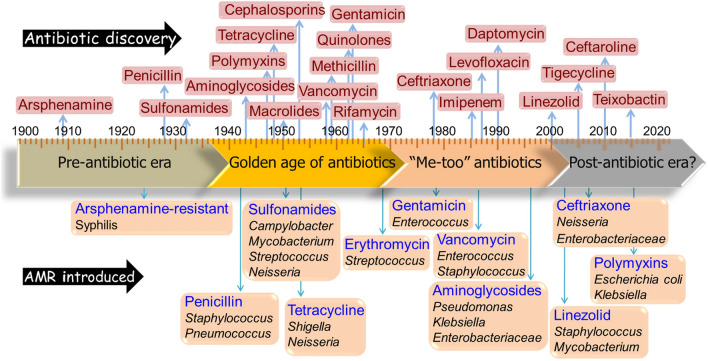
Antibiotics have revolutionized modern medicine for treating bacterial infections. However, indiscriminate use, misuse and often abuse of antibiotics over time have led to rapid emergence of pathogenic strains with antimicrobial resistance (AMR). These bacterial strains can escape conventional treatment modalities. It is disappointing to realize that the development of AMR has outpaced the discovery of new antibiotics (Fig. 1).
Fig. 1.
Timeline depicting the discovery of major antibiotics and subsequent emergence of resistance against them in various bacterial strains. One of the most dramatic events in the field of microbiology was the commercialization of penicillin in mid-1940s during the beginning of the industrial antibiotic era. While millions of human lives have been saved since then, the number of AMR cases continues to increase. AMR bacteria and their genes now circulate among humans, livestock, wildlife, environment, wastewater, and soil
Data from 204 countries and territories revealed that drug-resistant bacterial infections claimed 4·95 million people in 2019 and 1·27 million deaths were directly caused by AMR [1]. With current COVID-19 outbreak, this number is estimated to escalate up as more patients with viral infection are administered with antibiotics to treat secondary infections [2]. Thus, there is an urgent need to tackle AMR with new and innovative approaches. In this context, nanotechnology has opened-up avenues to deal with AMR.
Nanomaterials offer opportunities to access antibacterial modalities novel to bacteria that do not fall in their natural defence arsenal. The therapeutic effect of nanomaterials is largely derived from nanoscale confinement of materials compounded with multivalent interactions and high surface-to-volume ratio. Nano-size metals, metal oxides, organic nanoparticles (NPs), and nanocomposites with potent antibacterial activities are strategically advantageous to safely control superficial infections and infectious diseases. Diverse chemical compositions and intrinsic properties of these antibacterial nanomaterials (or nanobiotics) render multifaceted modes of action against the target bacteria. Notably, physiological states of the bacteria, i.e., planktonic, biofilm, stationary, starved, and logarithmic growth phase, impact their sensitivity to specific nanomaterials. Factors such as aeration, pH, temperature, and many other environmental features greatly influence antimicrobial activities of nanomaterials. These properties of nanobiotics offer opportunities to access unique mechanism of action that selectively and effectively target the bacterial systems.
Here, we review the AMR and provide a comprehensive evaluation of the state-of-art nanotechnology to address the problem of AMR, emphasizing the mechanism of actions of nanobiotics (Fig. 2). We discuss associated challenges of antibiotic discovery as well as prospects of functional nanomaterials as vaccines.
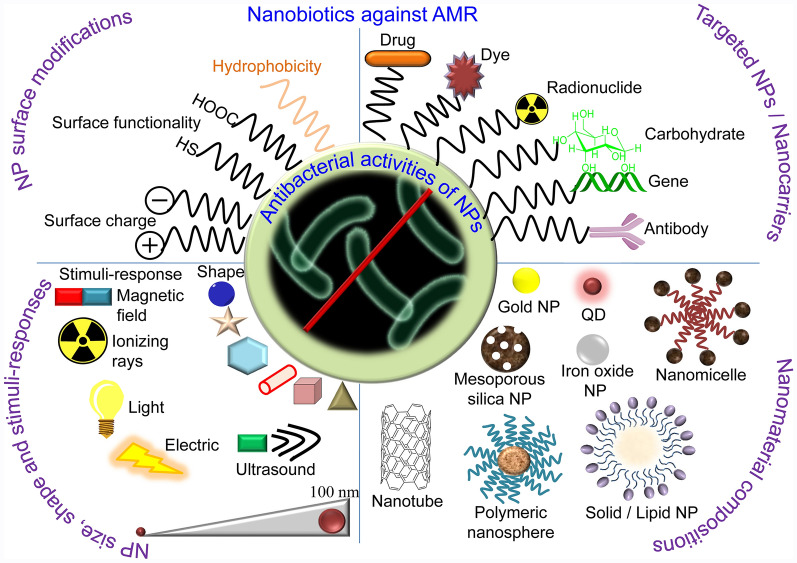
Fig. 2.
Antibacterial nanostrategies. NPs can complement and back antibiotics as a good carrier. The unique small size of nanomaterials grades in novel properties, such as increased interaction with bacteria due to larger surface area-to-mass ratio, and versatile plus controllable applications. Efficacy of the NPs can be increased by tuning sizes, shapes, and chemical compositions of the NPs. Metallic, organic, biomolecular, radio- and antibody modified NPs can effectively distroy bacteria with multiple mechanisms and their potency can be enhanced with addition of ultrasound, magnetic field, light, and ionizing radiation properties
Epidemiology and etiology of AMR
In 2010, BRICS countries (Brazil, Russia, India, China, and South Africa) contributed to 76% antibiotic consumption, with 12.9 billion units consumed by India, and 10 billion units by China [3]. As of 2017, carbapenem-resistant Acetobacter baumannii and Enterobacteriaceae burdened about US$281 million to the US healthcare costs [4]. In general, hospital costs increase significantly for treating patients with AMR infections due to the requirements of higher doses of drugs and longer hospital stays. Moreover, hospital-acquired infections with ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, A. baumannii, Pseudomonas aeruginosa, and Enterobacter species) further complicate the treatment. It is anticipated that the global cost for treating AMR can reach up to US$100 trillion by 2050 [5]. The Centers for Disease Control and Prevention (CDC), USA, list of emerging AMR threats on the basis of level of urgency is summarized in Table 1.
Table 1.
Antimicrobial resistance threat data as reported by CDC in 2019
| Level of concern to human health | Bacteria | Approximate number of deaths/year (year) |
|---|---|---|
| Urgent threat | Carbapenem-resistant Acinetobacter baumannii | 700 (2017) |
| Clostridioides difficile | 12,800 (2017) | |
| Carbapenem-resistant Enterobacterales | 1100 (2017) | |
| Drug-resistant Neisseria gonorrhoeae | 550,000 infections/year (2017) | |
| Serious threat | Drug-resistant Campylobacter | 70 (2017) |
| Extended spectrum β-lactamase (ESBL)-producing Enterobacterales | 9100 (2017) | |
| Vancomycin-resistant Enterococcus | 5400 (2017) | |
| MDR P. aeruginosa | 2700 (2017) | |
| Drug-resistant nontyphoidal Salmonella | 70 (2017) | |
| Drug-resistant Salmonella serotype Typhi | < 5 (2017) | |
| Drug-resistant Shigella | < 5 (2017) | |
| Methicillin-resistant Staphylococcus aureus (MRSA) | 10,600 (2017) | |
| Drug-resistant Streptococcus pneumonia | 3600 (2014) | |
| Drug-resistant Tuberculosis | 62 (2017) | |
| Concerning threats | Erythromycin-resistant Group A Streptococcus (GAS) | 450 (2017) |
| Clindamycin-resistant group B Streptococcus (GBS) | 720 (2016) | |
| Watch list | Drug-resistant Mycoplasma genitalium | – |
| Drug-resistant Bordetella pertussis | – |
Genesis of AMR
Extensive research in the recent past has discovered several illuminating mechanisms of AMR. These include crucial roles of gene mutations, genetic linkage, biochemical compositions in the bacteria, and efflux pumps as described below.
Genetic mechanism of AMR
The prolonged non-judicial usage of antibiotics in clinics and non-clinical use (e.g., in farm animals, aquaculture, poultry, meat, and plants) have eroded their therapeutic efficacy and resulted in increased number of AMR bacterial strains [6]. The selection pressure causes de novo evolution of resistant strains through a stochastic process. A sensitive cell undergoes genomic mutations or acquires gene(s) for resistance through horizontal gene transfer (HGT) using plasmids or phages. The newly acquired resistant cell survives, divides into large number of resistant daughter cells, and establishes as an AMR strain [7]. For example, introduced as the most potent antibiotic, penicillin was widely used for treating patients with Staphylococcal infections. However, soon after its inception, these bacteria developed β-lactamase, the penicillin deactivating enzyme [6]. Streptomyces and Micromonospora acquired resistant genes that produce their own antimicrobial product (aminoglycosides) [6]. Many Enterococci and Mycoplasma species contain the plasmid encoding the gene tetM, responsible for resistance to tetracycline. OtrB and OtrC genes in Streptomyces rimosus express efflux proteins that provide self-resistance to these bacteria [6]. Multi-drug resistant (MDR) phenotypes of Shigella have been transferred to Escherichia coli showing rapid spread of resistance genes in a pool of bacteria [8, 9], further confirms AMR genomic drivers and HGT.
Membrane remodelling mechanism of AMR
By virtue of complexity and diversity of cell surface layers, bacteria can limit the access of antimicrobials at the site of action by multiple mechanisms (Fig. 3). For example, they utilize β-lactams, aminoglycosides, and energy dependent efflux to prevent the accumulation of tetracycline [6]. Outer membrane (OM) remodelling with synchronization of reduced drug uptake and increased active drug efflux facilitate the multiplicative actions of OM in developing MDR [10].
Fig. 3.
The depiction of defense arsenal mechanisms of resistance in bacteria against conventionally used antibiotics. The presence of antibiotic resistance elements in pathogens has made AMR more challenging because of prevalence of HGT. Certain bacterial species are inherently resistant to antibiotics because of an impermeable membrane or lack of the antibiotic targets. Few have MDR efflux pumps that remove antibiotics from the bacteria. Some microbes possess altered genes, target protein, disabling the antibiotic-binding site(s). Inactivation of antibiotic can occur by covalent modification of antibiotics. The AMR mechanism in Gram-positive and Gram-negative bacteria can be different because of morphological and structural differences
The cell envelop architecture of the Gram-positive bacteria is simpler than that of Gram-negative bacteria comprising of a cell membrane surrounded by a thick peptidoglycan cell wall. Despite the absence of OM, modification of peptidoglycan or lysinylation of phosphatidylglycerol in S. aureus decreases the cellular entry of antibiotics with intracellular targets, such as hydrophilic fluoroquinolones. The decreased drug intake can directly contribute to resistance or/and in conjunction with the overexpression of efflux pumps that has been observed in the development of P. aeruginosa imipenem resistance in patients with bloodstream infections in Taiwan [11]. The expulsion of drugs is facilitated by a series of conformational changes in the periplasmic accessory protein AcrA that brings AcrB and To1C in close proximity. This causes the ejection of substrate by To1C from the cytoplasm to exterior of the cell [12]. For example, fluoroquinolone-resistant E. coli overexpresses AcrAB proteins that confer them with high level of resistance against fluoroquinolones [13]. Increased efflux of chlorhexidine and other biocides through the efflux pump AcrAB-TolC has been identified as an important mechanism of AMR in Klebsiella spp. [14]. Tetracycline efflux pumps encoded by tetA, tetB, and tetK genes acquired through HGT provide resistance to tetracycline in both Gram-negative and Gram-positive bacteria [15].
Target modification mechanism of AMR
Antibiotic-target interaction is very specific, and any alteration of the target can reduce the antimicrobial activity. Mutations can alter clinically important targets by multiple mechanisms. These include (i) modification or aberrant enzyme production for folate pathway, transcription, and DNA replication, (ii) reduction in binding affinity of essential cell wall synthesis enzymes, in case of β-lactams or cell wall precursor modification, (iii) changes in ribosomal proteins 16S RNA, and (iv) dimethylation of 23S ribosomal RNA. Modifications of enzymes involved in folate pathway confer “bypass” resistance to sulphonamide and trimethoprim [16].
Mutation in the dhfr gene causing single amino acid substitution in the target enzyme, dihydrofolate reductase, leads to trimethoprim resistance in S. aureus and S. pneumoniae [17, 18]. Alterations in DNA gyrase (gyrA and gyrB genes) and topoisomerases (parC and parE genes) bacterial sensitivity to fluoroquinolones (ciprofloxacin, norfloxacin) [19, 20]. Mutations, insertions, or deletions of amino acids residues in a small fragment of the rpoB gene result in rifampicin-resistance in M. tuberculosis and E. coli [21]. Resistance to methicillin, oxacillin and cephalosporins in S. aureus is credited to the expression and acquisition of the mecA encoding PBP2a, which has reduced affinity for β-lactam antibiotics [22]. Sporadic increase in resistance against glycopeptides in E. faecium and E. faecalis isolates is attributable to the acquisition of van gene clusters, which encode for enzymes ligase, dihydrogenase, and serine racemase [23]. The vanA gene clusters also confer high-level resistance to vancomycin and teicoplanin [24].
Target site for the binding of macrolide, ketolids, lincosamide, streptogramin B (MKLSB) is the large 50S subunit of ribosome consisting of 5S, 23SrRNA and 33 proteins. The MKLSB type resistance is mediated by erm-encoded rRNA methyltransferases, catalysing a post-transcriptional modification of 23S rRNA. Methylation or dimethylation of key adenine bases (e.g., A2058 in E. coli) of domain V within 23S rRNA alters the site of antibiotic binding to ribosomes [25]. High-level linezolid resistance in Clostridium spp. is attributed to Cfr-mediated methylation of 23S rRNA [26].
Drug inactivation mechanism of AMR
The problem of AMR is exacerbated by enzymes that can irreversibly destroy antibiotics. For example, β-lactamase mediated resistance involves the kinetic interaction and subsequent hydrolysis of ester or amide bond of β-lactam substrate, before reaching the site of action. Aminoglycoside-modifying enzymes, such as aminoglycoside phosphotransferases, aminoglycoside acetyltransferases, and aminoglycoside nucleotidyltransferases, catalyse the modifications of amino or hydroxyl group of aminoglycosides (e.g., tobramycin, amikacin, isepamicin, and sisomicin), interfering their antibacterial potential by impeding its binding to the 30S ribosomal subunit [27].
Chloramphenicol resistance is attributed to chemical modification by chloramphenicol acetyl transferases, which acetylate chloramphenicol rendering it incapable of inhibiting bacterial protein synthesis [28]. Flavin-dependent monooxygenase TetX inactivates tetracyclines, which is attributed to mono-hydroxylation of tetracycline resulting in intramolecular cyclization and degradation of the antibiotic. Streptogramin acetyltransferase present in Staphylococci and Enterococci species acetylates the unbound hydroxyl group of streptogramin A and thus inactivates it [29].
Metabolic alteration mechanism of AMR
Antibiotics affect bacterial metabolism in a complex way. Mutations in the 2-oxoglutarate dehydrogenase (sucA) were found in genomes of > 3500 clinical E. coli, implying their role in AMR. Undeniably, metabolic adaptation might embody a discrete class of resistance mechanism beyond conferring tolerance, when cells also amend their metabolic response to palliate downstream toxicity caused by antibiotics [30].
Plasmid-host co-evolution mechanism of AMR
Plasmid-host co-evolution occurs through (i) mutations linked to plasmid DNA replication and host which compensate the interference cost of plasmid, (ii) gaining of a native transposon which carries the toxin–antitoxin system, and (iii) compensatory mutations in the global regulatory pathway of the host. Parasitic plasmid-host association when antibiotics are absent in the environment evolves into mutualism when both bacteria and humans require the presence of antibiotics to survive. This accentuates that plasmid-bacteria pairs are evolving on a rocky adaptive landscape with various evolutionary trajectories, and clonal interference plays a vital role in plasmid-host co-evolutionary dynamics [31].
Antibiotic tolerance mechanism of AMR
There has been increasing incidence where bacterial populations adapted and eventually became tolerant and resistant to the antibiotic [32]. Antibiotic tolerance can be a result of genetic mutation but also arises due to phenotypic factors associated with traumatic environment, such as low temperature, serum levels, pH, host factors, ionicity, size and/or growth phase of inoculum and antibiotic exposure [33, 34]. Antibiotic persistence is observed in a small subpopulation of bacteria (< 1%) that can survive lethal concentrations of the antibiotics [35]. Moreover, current diagnostic systems fail to rapidly detect specific pathogens and delay in diagnosis can lead to AMR development in pathogens. M. tuberculosis is the most antibiotic tolerant bacteria of clinical relevance due to its stochastic and excessively slow growth in the lung. Transition of E. coli between walled (susceptible) and cell wall deficient (tolerant) forms to survive cell wall inhibiting antibiotic has been encountered in patients with high level of recurrence in urinary tract infections (UTIs) [35, 36].
Biofilm formation mechanism of AMR
Biofilms provide considerable survival advantage to pathogenic bacteria and is responsible for recalcitrant infections especially associated with medical devices (catheters, sutures, prosthetic devices, implants, cardiac valves, intrauterine contraceptive devices etc.), skin and soft tissues, endocarditis UTIs and otitis media. Biofilms account for 65–80% of bacterial infections [37]. A biofilm can be described as sessile microbial community composed of one or more species that colonize and irreversibly attach on a surface or to each other encased in an extracellular matrix This extracellular matrix is composed of extracellular polymeric substances (EPS) that include proteins, carbohydrates, glycoproteins, glycolipids, and nucleic acids from biofilm forming bacteria as well as the host [38]. EPS matrix of biofilms provides a physical barrier that limits antibiotic penetration and imparts AMR development by multiple mechanisms. For example, sub-inhibitory antibiotic dosing reaching the colonized bacterial population of biofilm aids in antibiotic tolerance. The glycocalyx layer of EPS serves as an adsorbent for exoenzymes that inhibit the transportation of antibiotics. Biofilms add in the accumulation of enzymes that covalently modify antibiotics, deactivating and impeding their antibacterial potency [39]. In addition, anionic components and extracellular DNA present in the EPS matrix serve as cation chelators, generating a cation-deficient environment and thus promoting AMR [40].
The biofilm formation promotes intra-species associations leading to heterogeneity and subsequently, antibiotic tolerance in polymicrobial biofilms. The nutrient and oxygen concentration gradient across EPS matrix causes bacterial population with varying growth rates. Superficially located bacteria consume surface obtainable oxygen and nutrients before it disperses into the depths of biofilm. Since, antibiotics are more lethal towards metabolically active cells, the deeply located tolerant cells escape the insult.
Swarming motility contributes to AMR
Swarming motility is a form of social behaviour of bacteria that aids to the rapid coordinated movement of bacterial cells on a moist surface. Many bacterial species demonstrate non-genetic adaptive resistance to a broad range of antibiotics during swarming. For example, P. aeruginosa PA14 is more resistant to aminoglycosides, chloramphenicol, ciprofloxacin, β-lactams, macrolides, tetracycline, and trimethoprim under swarming conditions [41]. In contrast to slow metabolic state of AMR species, bacterial swarms are metabolically active and grow vigorously so a new terminology STRIVE (swarming with temporary resistance in various environment), draws a difference between swarming specific adaptive resistance from growth-related resistance. A plausible mechanism involves the release of a necro signal from dead cells that activates the antibiotic survival for the swarmer cells [42].
Host intracellular environment in AMR
Antibiotic tolerance is common in various obligate and facultative pathogens, such as Mycobacterium species, K. pneumonia, Neisseria, Legionella, Brucella, Francisella, Listeria, Salmonella, Rikettsia, Ehrlichia, Coxiella, and Chlamydia [43, 44]. These pathogens survive by either membrane bound or cytosolic environment eventually escaping not only antibiotic exposure but also the host immune defences. Thus, infections caused by intracellular pathogens are difficult to treat since conventional antimicrobials are unable to infiltrate. For example, K. pneumoniae and E. faecalis are able to survive and thrive within intracellular vacuolar compartments upon engulfment by alveolar macrophages [45, 46].
AMR and challenges in traditional antibiotic discovery
Earlier research during 1990s gave birth to bacterial genomics that bared a horde of antibacterial targets, revealing over hundred novel antimicrobial mechanisms. However, companies like Pfizer, AstraZeneca, and GlaxoSmithKline are yet to come up with a novel candidate. Challenges are evident as no Gram-negative bacterial antibiotic with an entirely novel mechanism has been approved in the past 40 years [47]. Most leads could not be conceived into a hit having balance of desired antibacterial, pharmacokinetic, and safety profiles. A large fraction (> 90%) of the identified hit molecules were not suitable for treating AMR infections, whereas the remaining molecules had either low potency against MDR bacteria or were toxic for clinical use [48]. Reasons for the poor success rate in antibiotic development include (i) scientific challenges to discovering new classes of antimicrobial agents, mainly due to insufficient knowledge about AMR origin, diversity and mechanisms, limited understanding of novel targets, technical difficulties in hit-to lead selection, and undesirable pharmacological and safety profiles, (ii) obstacles in conducting clinical trials, such as identifying and recruiting patients, (iii) high competition from approved and currently used antibiotics, (iv) anticipated risk that new antibiotic may become ineffective within a couple of years due to possible resistance development, and (v) requirement for substantial investment during the preclinical research and clinical trials coupled with low approval ratings, numerous challenges on market entry, and much lower profitability than non-antibiotic drugs [48–50]. As of 2017, the average cost of developing an antibiotic was estimated to be about US$1.5 billion [51].
Another major challenge for antibiotic discovery is the ubiquitous presence of AMR genes in the ecosystems. The human intestinal microbiota is a reservoir of MDR genes [52]. AMR genes exist in wastewater that supports the selection of MDR bacteria [53]. These genes have been acquired by many bacterial species that have empowered them to tolerate traditional antibiotics from insects, fungi, plant products, petroleum chemicals, agricultural and industrial wastes [54]. In this context, several questions remain a mystery—are these AMR genes there for discrete genetic/biochemical demands? Do bacteria show resistance because they are exposed to numerous toxins in the environment? How do natural products affect the ecology? Does presence of selective ecosystem pressure lead to resistance in strains? Hence, to stop the propagation of AMR in the environment, scientists must come up with innovative/designer antibiotics that tackle new and increasing AMR, generate less resistance, reduce the risk for cross-resistance, and are self-destroyed post treatment.
Most traditional antibiotics, which are natural or semisynthetic products, are plagued by significant shortfalls, lack of desirable antibacterial efficacy, development of AMR, and challenges associated with monitoring/evaluating antimicrobial activities in a dynamic milieu. They have failed to deliver activity against the most urgent threats from MDR Gram-negative bacteria. Research and development processes for discovering and developing a traditional antibiotic are time-consuming (about 10–12 years), expensive, and fraught with a multitude of barriers [49]. These antibiotics typically target essential processes of survival and/or growth of bacteria, synthesis and maintenance of cell wall/membrane, or DNA replication, transcription and/or translation. Unfortunately, bacteria can acquire the ability to escape the effects of these drugs, irrespective of target functionalities. Another challenge for traditional antibiotic discovery is lack of effective techniques for isolating and purifying naturally occurring antimicrobials against MDR bacteria. Treatment for MDR bacterial infection requires high dosages of traditional antibiotics, often cocktail of multiple drugs or the ‘last resort’ antibiotics. Adding to the therapeutic burden is biofilm-associated resistance, which requires exceptionally high dosages of antibiotics due to difficulty in penetrating the extracellular polysaccharide sheath covering the biofilm and the presence of complex microflora. High dosages of traditionally used antibiotics often result in long and expensive treatment with serious side effects and uncertain outcomes. Furthermore, the utilization of conventional antibiotics carries a high-risk alert for AMR development.
A critical analysis of traditional antibiotics that were approved during 1999–2014 showed lack of novelty and diversity for target bacteria. These antibiotics failed to address AMR issues. It was suggested to apply therapeutic alternatives to antibiotics that would decrease our dependence on traditional antibiotics. The fact that antibiotic effectiveness is reduced over time due the potential development of resistance, new antibiotics will always be needed [55].
Nanotechnology to rescue AMR issues
Emerging nanoscale materials and technologies provide long-lasting answers to the above issues due to the unique mechanism of action of nanomaterials on pathogenic bacteria. The “antibiotic nanocarriers” based on liposomal, solid/lipid, terpenoid, polymeric, dendrimeric, and inorganic materials have shown encouraging results in enhancing the overall antibiotic performance compared to bulk use of antibiotics [56]. This has been achieved by improving the pharmacokinetics of the drug—extending antibiotic half-life in the serum and decreasing the apparent volume distribution attributed to target the site of infection and causing elimination of bacteria even at lower drug dosage. The “combinatorial” or synergistic effect of either multiple drug entrapment in a single nano-construct or two or more NPs constructing hybrid NPs reinforces the pharmacological effects and improves the antibacterial potency while limiting the development of resistance. The modes of actions of nanomaterials to fight AMR strains are novel and unique that are not present in bacterial arsenal of natural defence (Box 1). In the following section, we discuss various types of nanomaterials and nanotechnologies that are being employed or currently investigated to tackle AMR.
Box 1: Essential parameters utilized by NPs to mount antibacterial actions.
|
Physicochemical properties of NPs including surface morphology, size, crystal structure, charge and zeta potential regulate their antibacterial actions. Bacterial strains, exposure time and environmental conditions also impact the potency of antimicrobial drugs. Some of the crucial factors that govern the antibacterial mechanism of NPs are summarized below Size and surface charge of NPs affect the antimicrobial potential. Length and breadth of nanotubes can extend the release of given antimicrobial. Large specific surface area of NPs increases the prospect of closeness, contact, and interaction with microbial membrane [56, 57] Shapes of nanomaterials are the basis of wavering degrees of damage to pathogens through periplasmic enzymes. For example, ZnONPs of various shapes (sphere, pyramid, and plate) exhibit varied photocatalytic activities with β-galactosidase leading to functional and conformational change in the enzyme [58]. Prismatic-shaped Y2O3 NPs show better activity against P. desmolyticum and S. aureus, which may be due to direct interaction of Y2O3 NPs and bacterial cell membrane surfaces, causing leakage of the cellular components [59] |
Roughness of NPs can decrease the adhesion of microbes, as the size and surface area-to-mass ratio stimulate the adsorption of bacterial proteins [60] Zeta potential of NPs resiliently impacts microbial adhesion because of electrostatic attraction generation due to negative charge on bacterial membrane and positively charged NPs [61]. For example, Mg(OH)2_NPs, being positively charged are adsorbed on microbial surfaces and this accumulation at the site of infection ascends the permeability [62] Doping modifications allow proper dispersal of NPs in hydrophilic or aqueous environments. For example, nanocomposites framed with a combination of Au and ZnO show enhanced photocatalytic property and ROS formation as Au allows enriched light absorption (Surface Plasmon Resonance) and ZnO with transformed band gap width increases electron transport efficiency and charge carrier separation [63]. Cr doping on the ZnONPs significantly enhanced their antimicrobial activity against a wide range of pathogenic bacteria [64] Environmental conditions impact the antimicrobial potency of NPs. For example, most bacterial enzymes cannot function beyond optimum temperature and NPs targeting these enzymes would be ineffective. Reduced pH shows high dissolution of ZnONPs and adhesion of NPs on the bacterial membrane of MRSA and E. coli [65] |
Nanoscale antimicrobial substances
NPs can be classified into inorganic/organic, carbon-based, and hybrid structures. The inorganic group comprises of metal/metal oxide NPs and quantum dots (QDs). Organic nanomaterials are apt for drug delivery, antimicrobial use, bio-imaging, and tissue regeneration, as these consist of biocompatible organic components. This class of materials comprises of polymeric NPs, liposomes, and lipid-based NPs. Carbon-based nanomaterials embody carbon black, nanotubes, graphene, nanofibers, nanodots, fullerenes, nano-diamond, carbon onions, carbon rings, etc.
Inorganic NPs are made up of inorganic oxides of Si, Ag, Au, Zn, Mg, Mn, Cu, Se, Al, or Ti and hence they differ in shape, size, solubility and stability. Their characteristics are also defined by pH, temperature, reduction time, concentration of the reducing agent, and aggregation behaviour that impact their antimicrobial potency [66, 67]. For example, iron oxide NPs (FeONPs) are allied with DNA hybridization technique to heighten the capturing of 16S ribosomal RNA gene of bacteria [68]. Silver NPs (AgNPs) attach to cell membrane, interact with membrane proteins, increase membrane porosity, enter and enhance generation of reactive oxygen species (ROS) hampering respiration, including bacterial cell lysis, and evoking inflammatory reactions [69]. A recent study shows that the pH-triggered aggregation of AgNPs favors their penetration into bacterial biofilms [70]. The large aggregated AgNPs remain in the biofilms for a long time, disrupt bacterial biofilm formation, and exhibit better antibacterial activity than traditional AgNPs [70]. Gold NPs (AuNPs) exert bactericidal effects by accumulating on the cell surface credited to heavy electrostatic forces accompanied by cytoplasmic leakage and cell death [71]. Au nanocrystals display facet-dependent antibacterial activities—bacterial membrane damage, inhibition of cellular enzymatic activity, and energy metabolism [72]. QDs are metallic in nature but contain a core of semiconductor materials like Cd or Zn. The antimicrobial activity of QDs can be assigned to their ability to destroy bacterial cell walls or membranes, induce free radicals, bind with genetic material, and inhibit energy production [73]. Their sensitivity can further be increased by functionalization. For example, transferrin-modified silver QDs coupled with zinc and rifampicin exhibit much higher antibacterial activity than the zinc and rifampicin complexes [73]. QDs of custom sizes can be engineered to distinguish various mutants of same microbial species [74]. Photoexcited QDs have been found to inhibit growth of MDR clinical isolates (S. typhimurium, MRSA, K. pneumonia, and carbapenem resistant E. coli) due to redox potential of photogenerated charge carriers that interact with the bacterial environment [75]. Due to their high drug loading capacity and ability to cross barriers, these are employed for intracellular delivery of peptides, drugs and nucleic acids [76].
ZnO and CuO NPs exhibit admirable antibacterial properties but their accumulation has raised safety concerns in the host’s system. Human cells have Cu-transporters for regulation of Cu homeostasis, unlike Au and Ag for which such mechanism has not been reported yet [77]. CaF2 NPs have shown lethal effects against S. mutans due to their adherence on the tooth surfaces and persistent release of fluoride ions which stimulates re-mineralization and suppress virulent S. mutans [78].
Organic NPs such as liposomes, polymeric micelles, polymeric and solid/lipid NPs constitute a class of organic NPs, which can carry both hydrophobic and hydrophilic antibacterial agents [79]. Most of these NPs are biocompatible and can be easily opsonized and degraded quickly due to their hydrophilic/hydrophobic individualities [79].
Liposomes and lipid NPs are spherical vesicles of phospholipid bilayers. They can fuse with microbial membrane and release the given drug into bacteria [80]. They have been developed into a drug delivery system for antimicrobial agents against biofilm-mediated infections. Their unique characteristics, including target-specificity, low toxicity, and ability to fuse biofilm matrix/cell membrane, enhance the efficacy of antibiotics by minimizing recurrent infections [80].
Polymeric NPs, such as nanospheres/nanocapsules, act as drug carriers. They are physically/chemically stable and enhance targeting efficacy. These NPs are prepared by polymerization of monomers (micro-/mini-emulsion) and from polymers via solvent evaporation, salting-out, and dialysis [81]. Lipid-based surface-functionalized PLGA is one such example of drug delivery system that protects antibiotics from degradation and has capability to bind to the components of biofilm [82]. Chitosan NPs can be loaded with antibiotics that exhibit super-antibacterial potential and are framed by ionic gelation of diverse concentrations [83]. Cellulose fibres reformed with AgNPs display sound antimicrobial defence against E. coli and S. aureus [84]. Graphene quantum dots (GQDs), AgNP and silica nano-fabrications enhance ROS production in light-activable GQDs, by conversion of light energy to hyperthermia, resulting in effective bacterial killing [85].
Polymeric micelles are colloidal structures made up of block copolymers. Polystyrene, poly(butyl methacrylate), polylactic acid (PLA), or poly(ethylene oxide), and poly(propylene oxide) have been used to form polymeric micelles [86]. The inner core of micellar structures is generally hydrophobic while outer core remains hydrophilic. These properties define drug release kinetics and charge determines the stability of micelles.
Solid/lipid NPs are lipid-based, solid, colloidal carriers like acetyl palmitate or salts of myristic acid. These are used for their nominal toxicity and stability. These particles show controlled release and high efficiency which differ by the nature of the lipids and milieu’s pH and temperature [87].
Nanozymes are nanomaterials with intrinsic enzyme-like properties [88]. FeO-based artificial peroxidase NPs have been used to treat drug-resistant E. coli and S. aureus [89]. Activity of nanozymes is superior because of the rough surfaces that allow bacterial adhesion, extremely irregular edges that act as active sites, ability to regulate ROS production and knack for photocatalytic activation [90]. In surface-bound state, nanozymes eliminate pathogens and delay the onset of resistance, while in the coated form, they can prevent biofilm formation [91].
Antibacterial surfaces are ways to reduce biofilms by the use of TiO2 and its photocatalytic activity, ensuing ROS generation. Immobilization of surfaces by AgNPs is applied on anti-adhesion surfaces and structured arrays [92, 93]. Collusive with biofilm is quorum sensing, which is used by bacteria to maintain proximity and communicate in order to synchronize virulence causing gene expression [94].
Antimicrobial mechanism of action of NPs
Nanomaterials benefit from their controllable and nanosize structures in comparsion to that of bacterial components [95]. High surface area-to-volume ratio ensures a strong surface chemistry in terms of multivalent interactions with bacterial cells or functionalization for a specific charge or targeted delivery. The forces that majorly dominate the nano-bio interface are van der Walls forces, electrostatic forces, hydrophobic interactions and receptor-ligand interactions. Nanomaterials act along multiple simultaneous or correlated bactericidal mechanisms as described below.
Disruption of bacterial cell wall
Molecular architecture of bacterial cell envelop is the major physical barrier for any antimicrobial. Lipoteichoic/teichoic acid makes diffusion of highly hydrophobic antibiotic moieties across this envelop unfathomable. Materials with positive potential can bind selectively to bacterial surfaces with higher negative potential than mammalian cells. Physical contact of nanomaterials having a proper equilibrium between cationic charge and hydrophobicity (amphiphilicity) accomplish stupendous antimicrobial property with low levels of cytotoxicity and haemolysis [96].
Nanomaterials get anchored electrostatically onto the bacterial envelops and change their membrane potential leading to the depolarization and loss of membrane integrity. As the physical barricade is disrupted, transport discrepancy, respiration impairment and intrusion in metabolic pathways are followed resulting cell death. Typically, graphene-based NPs, regarded as “nano-knives” or MoS2, MnO2 with sharp edges are known to physically disintegrate bacteria cell wall [97]. AgNPs cause jamming of oxidative phosphorylation due to dissipation of proton motive force as a result of anchoring into the bacterial membrane [70, 98]. Fullerenes kill bacteria by physically rupturing their cell wall integrity [99]. Fortification of bacterial resistance against therapeutics that cause damage to the cell wall is limited thus, making these approaches an attractive strategy for long-term usage.
Generation of ROS
ROS are intracellularly originated secondary metabolites of oxidative metabolic activity of a bacterial cell. The cellular ROS levels are maintained by endogenous antioxidant defence system. An excessive production of ROS or redox disequilibrium causes oxidative stress causes the destruction of membrane fatty acid, and macromolecules resulting in peroxidation of lipid molecules, oxidation of proteins which cause inhibition of enzymes and DNA/RNA damage leading to mutation/killing in bacteria.
Nanomaterials can produce four different types of ROS with diverse levels of dynamics and activity—hydrogen peroxide (H2O2), singlet oxygen (1O2), superoxide radical (O2−), and hydroxyl radical (·OH). Nanomaterials, such as carbon nanotubes (CNT), fullerenes, TiO2, ZnO, CeO2 and AgNPs, induce oxidative stress as a major mechanism of their bactericidal property. Major causes for ROS development are (i) pro-oxidant functional groups on the ultra-reactive surface, (ii) multivalent surface of NPs due to involvement of transition metal ions in active redox cycling involving Fenton-type or Haber–Weiss reactions, and (iii) cellular internalization of NPs leading to activation of NADPH oxidase or mitochondrial respiration. Light-induced ROS production is witnessed in AuNPs and TiO2. These are photocatalytic when irradiated with visible light, near UV or UV, proving to exert effective antibacterial potency against spores of Bacillus [100, 101]. AuNPs have been found to impose antibacterial effect against Corynebacterium pseudotuberculosis via ROS generation leading to vacuole formation [102].
Fe3O4@TiO2 core shell magnetic NPs are effective against MDR S. pyogenes and MRSA via cascade of bactericidal effects through the generation of ROS that increases the cell porosity and expulsion of intracellular components [103]. Scanning electron microscopy revealed that nano-ZnO changed the spiral shape of Campylobacter jejuni into spherical shape leading to loss of membrane integrity [104].
ROS-independent oxidative stress
ROS-independent oxidative stress is also a major pathway for bacterial killing. In the absence of light and oxygen, nC60 exerts its antimicrobial actions without ROS production [105]. For example, nC60 directly interacts with cell membrane and disrupts the electron transport system [99, 106]. ROS-independent cytotoxicity exhibited by single walled carbon nanotubes include glutathione disulfide oxidation [107]. Pt-Au bimetallic NPs exert ROS-independent membrane depolarization [108].
Photo-induced antibacterial action
Photo-induced methodologies using nano substrates are being exploited due to minimal invasiveness, localized treatment modality with minimal toxicity. Upon photoexcitation, photothermal agents (PTAs) can transform optical energy into thermal energy upon non-radiative relaxation to the ground state, leading to irreversible damage to bacteria via membrane rupture, protein denaturation and overall collapse of intracellular components [109].
The local hyperthermia is predominantly caused by thermal conduction or generation of vapour bubbles as a consequence of strong heating. The biologically transparent window of near-infrared (NIR) light from 700 to 950 nm is used for photothermal therapy (PTT) due to deep tissue penetration and low absorption by water and haemoglobin. CNTs were the first carbon-based nanomaterial to be exploited for photothermal bacterial killing owing to their high photothermal conversion efficiency and low fluorescence quantum yield [110]. PEG-modified core–shell gold nanorods layered double hydroxide nanomaterials (GNR@LDH-PEG) showed about 99.25% and 88.44% of bacterial killing rates for E. coli and S. aureus, respectively, with 5 min of NIR irradiation attributed to electron deficiency on the gold surface [111]. Wang et al. constructed antibacterial coating on Ti plates by hybridization of MnO2 with CS modified AgNPs [112]. Upon photoexcitation over NIR irradiation, MnO2 NPs were able to produce sufficiently high local heating with photothermal conversion efficiency of 30.79% and exceptional photostability. Combined effects of MnO2 with AgNPs were also observed, aiding to the antibacterial property of the nano-construct [112]. Metal sulphide nanomaterials are also explored for antibacterial PTT. For example, magnetic MoS2, surface functionalized with chitosan, cause bacterial crosslinking along with their photothermic sterilization [113].
Photo-induced generation of ROS causes oxidative stress, which eventually lyse bacterial cells by disrupting the protein function and inhibiting the activity of certain periplasmic enzymes [114]. Under light activation, at energies greater than or equal to the band gap, stimulated electron transduction from valence band to conduction band results in the formation of electron (e−)-hole (H+) pair on the surface and intrinsic region of the semi-conducting materials. H+ adheres to the surface of nanomaterial and on interaction with H2O or OH− gets oxidized to ·OH free radical. ·OH adheres on the surface and electronically interacts with molecular O2 that is subsequently reduced to superoxide free radical O2− [115, 116]. These generated ROS are highly oxidative and disturb the normal physiology and morphology of bacteria under UV or visible light illumination. TiO2 NPs are a classic example of photocatalyst antimicrobial agent when irradiated with near-UV or UVA [117]. The efficiency of TiO2 NPs depends on band gap engineering and thickness of microbial surfaces. Magnetic nanoprobe comprising of Fe3O4@TiO2 core–shell nanostructures are used for photo killing of MDR S. pyogenes and MRSA under UV irradiation [103].
Photo-induced ROS employs appropriate excitation wavelength and O2 as external stimuli factors that selectively activate photosensitizer (PS) at the site of infection producing cytotoxic ROS such as 1O2 and ·OH [118]. Photodynamic therapy (PDT) essentially operates via two pathways—type I and type II. Upon light sensitization, PS in the ground electronic state transits to the short-lived but higher energy singlet excited state (S1). Subsequently, S1 undergoes inter-system crossing to a longer-lived excited triplet state (T1) [119, 120]. In type I PDT, the triplet excited state of PS interacts with biological macromolecules to generate 1O2 and ·O2− mediated through electron transduction, breaking the structural integrity, and altering ionic permeability of the bacterial membrane [121]. In type II PDT, the triplet excited state transfers energy to 3O2 to produce lethal 1O2 species, which in turn interact with immediate biological components causing irreversible oxidative damage to DNA, enzymes, and other cellular components of bacteria [122]. Singlet oxygen has a short lifetime in cellular surroundings and a very small radius of action of ~ 0.2 μm, thus creating a contained response without affecting distant cells. Chlorin e6 (Ce6) loaded into silica NPs showed high antibacterial efficiency against MRSA [123]. Photostability of Ce6 was enhanced with high 1O2 retention capacity upon light irradiation [123]. A lipase sensitive transfersomal nanocarrier with high skin penetration efficiency was tested against Propionibacaterium acnes. Pheophorbide A (PheoA), in absence of lipase gets self-quenched due to intramolecular interactions but PheoA loaded liposomes caused 99% P. acnes elimination due to lipase mediated disruption of transfersomes, the quenching of PheoA was reduced considerably [124].
Controlled release of NP metal ions
Under the influence of external stimuli, the controlled release of metal ions is attributed to the large surface area of NPs. These metal ions get absorbed into the cell membrane leading to direct interface with functional groups of essential proteins and nucleic acids. They infiltrate bacterial cell facilitated by ion channels and biological pump, accumulate above tolerable range causing bacterial cell death [125]. Zn2+ ions are known to bind strongly with –SH functionality of cell membrane, leading to loss in membrane integrity [126]. Silver ions leads to protein coagulation aided by Coulomb gravity via a process called biosorption [127]. The presence of Zn2+ in addition to Ag+ reinforces the bactericidal propensity, providing more active sites resulting in faster adsorption speed and capacity [128]. The solubilization is essential for nanomaterials for production of toxic Zn2+ or Ag+ which increases oxidative stress through Fenton’s reaction. Super-magnetic iron oxide releases Fe3+/Fe2+ which can easily penetrate the cell membrane and interferes in transduction of transmembrane electron to produce O2−, which damages the iron clusters [129]. But the uptake of ZnO NPs is better than Zn2+, which directly impact the antibacterial potency [130]. This can be explained by the uptake of nanomaterials mediated through endocytosis and assist the entry of toxic ions via “trojan horse effect” circumventing the defence mechanisms of cell.
Disruption of protein synthesis
Mindfully engineered nanomaterials arrest bacterial growth and eventually kill bacteria via down regulation of genes, disruption in protein synthesis, and oxidative damage to the DNA [131]. Transcriptomic and proteomic studies to understand the mechanism of action of AuNPs identified two main processes. First barred the merging of ribosomal subunit with the t-RNA and the second, reduction in cellular ATP levels due to disintegration of membrane integrity and loss of ATPase activity [132]. Au-superparamagnetic iron oxide NPs have strong affinity for disulphide bond of bacterial proteins, which affects metabolism and redox homeostasis of the cell [133]. Spores of B. cereus are inhibited by S-nitrosothiol, which is produced by nitric oxide (NO) releasing NPs; where NO reacts with thiols to produce S-nitrosothiol, which nitrosylate thiol residues on bacterial proteins [134]. NO releasing silica NPs are non-toxic to mammary fibroblasts and are also known to stimulate the host immune responses that shrug off the microbial load [135].
DNA damage
Ag+ ions intercalate DNA strands due to complexation with nucleic acid aided by Coulomb interactions and breaking the H-bonds that hold purine and pyrimidines base-pairs together [136]. AgNPs exhibit genotoxic potential by obstructing bacterial DNA unwinding and transcription, infringement of DNA chains and causing chromosomal irregularity [137]. Deamination of cytosine, guanine and adenine is mediated by N-nitrosating agents formed by auto-oxidation of NO [138]. DNA strand breaks are formed by peroxynitrite and NO2 and therefore, NO releasing therapies have been proposed as promising antimicrobial agents [134].
Gene regulation
AgNPs cause upregulation of antioxidant genes and genes encoding metal transport, metal reduction and ATPase pumps [139]. In E. coli, polymer-coated AgNPs were shown to downregulate the aceF and frdB genes that are responsible for tricarboxylic acid cycle and gadB, metL, and argC genes for amino acid metabolism [140]. MgONPs upregulated the expression of weak-thiamine ester-binding and riboflavin metabolic genes and downregulated the expression of metabolite coding genes, disrupting physiological functioning of bacteria [141].
Increased retention of aggregated NPs in biofilms
The large aggregated AgNPs have been shown to exhibit better penetration ability in infected areas and longer retention in the bacterial biofilms, disrupting biofilm formation and effectively eliminating bacterial populations [70]. Aggregated AgNPs would also exhibit longer retention periods in tissues and the exocytosis from the cells would be relatively slower than small non-aggregated particles, adding to their therapeutic implications [70].
Figure 4 summarises possible mechanisms of action of NPs against bacteria.
Fig. 4.
The nano armamentarium to combat AMR. NPs can attach the microbial cell wall and penetrate it, thus causing structural changes in the cell membrane permeability leading to cell death. NPs target at the cell membrane, leading to dissipation of proton motive force thus blocking the oxidative phosphorylation. NPs have ability to produce ROS that can cause DNA damage leading to cell death. They can impel the shape and function of cell membrane, interact with DNA, ribosomes, lysosomes, and enzymes, promoting fluctuations in cell membrane permeability, oxidative stress, heterogeneous alterations, enzyme inhibition, electrolyte balance disorders, protein deactivation, and changes in gene expression leading to bacterial cell death
Antibiotic nanocarriers
In addition to directly targeting bacteria, certain nanomaterials are being exploited as “antibiotic nanocarriers” to encapsulate traditional antibiotics in order to improve pharmacokinetics, stability and bioavailability, and enhance bacteria eradication [142, 143]. This approach improves the efficacy of antibiotics while reducing potential toxicity. Furthermore, nanocarriers decrease the apparent volume distribution, in turn allowing maximum tolerated dose and causing bacterial cell death at much lower antibiotic concentrations and thus expanding the spectrum of action against MDR bacteria. Various biopolymers including proteins, carbohydrates and lipids have been used to synthesize nanocarriers with promising antibacterial activities [144]. For example, linalool-functionalized hollow mesoporous silica spheres efficiently improved the bactericidal activities of the organic component against both Gram-negative (E. coli) and Gram-positive (S. aureus) bacteria by breaking the cell membrane structure [145]. A nano-delivery system constructed by self-assembly of antibacterial berberine and rhein phytochemicals showed good biocompatibility and safety profiles and strong inhibitory effects on S. aureus biofilm [146]. Strategies to encapsulate multiple antibiotics in a single nanocarrier for obtaining a synergistic effect are also being investigated. The delivery of antibiotics using nanocarriers results in highly targeted delivery of huge concentrations of antibiotics to the bacterial cells, increasing the overall efficacy and therapeutic potential. For example, the nanocarrier system, developed using carboxyl-modified mesoporous silica nanoparticles carrying vancomycin and polymyxin B, showed improved biocompatibility and synergistically high antibacterial efficacy [147].
Potential acquisition of bacterial resistance to NPs
NPs attack pathogens with multifunctional mechanisms that are diverse from the contemporary antibiotics. Thus, numerous mutations are required in bacteria to develop resistance toward NPs. However, resistance of microbes to NPs is still a concern [148, 149]. The common resistance patterns encompass ion efflux pumps, electrostatic repulsion, adaptation of biofilms, expression of extracellular matrices, enzyme detoxification, followed by volatilization and mutations [150, 151]. Nano-resistance changes the shape of bacteria and modulates the expression of membrane proteins, which reverses on removal of NPs [152]. The formation of biomolecules corona reduces the binding of metal-based NPs to bacteria, thus imparting resistance [153]. Although rare, but resistance has also been reported against Cu, Au and Ag NPs which can be attributed to high expression of efflux pumps, and alteration in membrane permeability [148, 149, 154].
Defense mechanism against various NP sizes
Increased clinical use of AgNPs is raising concern as resistance has been detected in K. pneumoniae and E. cloacae [154]. E. coli was found to be resistant to AgNPs due to NP aggregation by the altered production of flagellin, diminishing the toxicity of AgNPs [149]. Agglomeration and deactivation of NPs are the mechanisms used by bacteria to prevent the effect of NPs. E. coli acclimatize to AgNPs by releasing ECS. ECS amends the size and zeta potential of NPs through their agglomeration [155]. Bacteria exposed to sub-lethal concentrations of AgNPs showed enhanced resistance against these NPs due to mutations that caused the downregulation of porins, inhibiting the entry of NPs in bacteria [156]. P. putida changes the conformation of unsaturated fatty acids of its membrane, thus modifying the fluidity of the membrane making it less permeable, hence, blocking the cellular entry of NPs and metal ions [157].
Defence mechanisms against surface charge
Charge on the surface of various metal/metal oxide NPs is important for antimicrobial activity [158, 159]. Bactericidal action of MgONPs against endospores, Gram-positive and Gram-negative bacteria is due to electrostatic attraction of positively charged NPs with negatively charged bacteria [160]. Microbes can modulate electrical charge on their surfaces, which can ward off the NPs. Antibacterial potency of positively charged AgNPs is better against Gram-positive (S. pyogenes, S. mutants, S. aureus) and Gram-negative (P. vulgaris and E. coli), in comparison to neutral or negatively charged NPs. However, some bacteria can develop resistance to cationic antimicrobial peptides (CAMP) by changing the surface charge through modification of phospholipids [161].
Envelope stress response (ESR), in both Gram-positive and Gram-negative microbes, safeguards the integrity of microbial cell envelope [162]. It incorporates D-alanine in Gram-positive microbe, decreasing the overall negative net charge and thus protecting against positively charged NPs; whereas in Gram negative bacteria, it adds phospho-ethanolamine (PEA) in lipid-A component of the LPS, which increases positive charge on bacteria [163].
Defence mechanisms against metal ions release
Industrial production of NPs for electronics, computers, food, domestic and automotive products, leads to their accumulation in the environment. These transform the NPs due to change of physicochemical and antimicrobial properties [164]. Transformation can lead to altered dissolution and ion release rate due to ligation, aggregation, redox reaction (oxidation/ interaction with natural organic matter of zero-valent Fe NPs at aerobic condition inhibits antimicrobial properties), adsorption of biomolecules and biotransformation [165, 166]. This allows bacteria to adapt to metal ions through the efflux systems (expressed from metal resistance genes). For example, interaction of P. aeruginosa with CuONPs showed upregulation of cation diffusion facilitator (CDF), RND membrane fusion protein family, and OM factors. These make the bacterial CzcCBA efflux system, transporters, and P-type ATPase efflux complexes [167] that facilitate the efflux of metal ions such as Co2+, Zn2+, Cd2+, Ni2+, Cu+ and Ag+. Cation diffusion facilitators (CDF family) form the second line of defence that mediates the efflux of Zn2+, Co2+, Ni2+, Cd2+, and Fe2+ [168]. P-type ATPases forms the third line of defence and efflux is carried out by ATP hydrolysis. It arbitrates the efflux of H+, Na+, K+, Mg2+, Ca2+, Cu+, Ag+, Zn2+ and Cd2+ [169].
Other mechanisms include pigment production, intra/extra cellular sequestration, morphology alteration bioprecipitation, and biotransformation or enzymatic detoxification [170]. Bacteria also have metal-resistant operons; ion-sequestering proteins like SilG gene in sil operon [171]. Release of pigments by bacteria reduces their exposure to the ion of NPs thus reducing antimicrobial activity, like P. aeruginosa produces pyocyanin which protects bacteria by reducing Ag+ to Ag0 [172]. Through biotransformation, bacterial enzymes transform toxic ions to non-toxic forms [173]. For example, Lactobacillus bulgaricus enzymatically transforms the chitosan-modified selenium NPs into organic seleno-compounds like SeCys2 and SeMet [174]. Bacteria can change their morphology resulting in minor bactericidal effect of NPs. E. coli becomes oval and small to resist ZnONPs. Aforesaid mechanisms are regulated by metallo-regulatory and metal homeostasis systems in microbes [175].
The extensive use of metal/metal oxide NPs can stimulate the co-selection and co-expression of antibiotic resistance genes. Exposure of E. coli at sublethal concentrations of ZnONPs and TiO2NPs facilitated the conjugative transfer of antibiotic resistance plasmids as NPs increases the permeability of cell membranes [176, 177]. ESR also regulates the defence mechanism against ion release, electrical charge, and size of various metal/metal oxide NPs. In Gram-negative bacteria, ESR has an alternative sigma factor (RpoE or σE), three different 2-component regulatory systems [pilus expression (Cpx) response, regulation of capsular synthesis (Rcs) phosphorelay and bacterial adaptive response (Bae)] and phage shock protein response (PSP). RpoE regulates the cell envelope biogenesis, protein folding and degradation, and cell envelope modification. It controls alginate production in P. aeruginosa. RpoE modifies LPS through PhoPQ regulon and MicLs RNAs, which increases the resistance to antibacterial molecules like CAMP and NPs. Cpx regulates the protein export systems associated with virulence. Rcs reaction (second of the three different 2-component regulatory systems) can disrupt cell envelope and control the production of macromolecular envelope structures and its homeostasis. Bae regulates the expression of efflux pumps while PSP sustains proton motive force and determines localized secretin toxicity. ESR in Gram-positive bacteria has 3 systems which regulate the cell wall metabolism, cell envelope charge, and expression of efflux pumps. The first regulates retort to explicit toxic stimuli. Second is activated by dangerous agents existing in the cell wall. Third controls the cell envelope integrity [178].
Defence mechanisms against ROS and oxidative stress
Metal ions affects respiration and scavenging mechanisms resulting in accumulation of singlet oxygen, OH radical, H2O2, superoxide, and other ROS. ROS can cause damage to the internal components of the bacteria such as structural proteins, organelles, enzymes, DNA, respiratory chain and scavenging mechanisms [179]. ROS at sublethal concentrations fuels defence mechanisms in bacteria, a process called as hormesis [180]. First level of adaption through hormesis is enzymatic or short-term response through expression of ROS scavenger enzymes creating balance in bacteria for few seconds or minutes. Second level is long-term adaptation which consists of two sub-levels: Transcriptional and genomic. At transcriptional level, upregulation of antioxidant mechanisms takes place within hours to days [181]. Activation of DNA damage repair mechanisms (excision repair, homologous recombination, and translation DNA synthesis) takes place at genomic level. This creates genome plasticity by repairing the incorrect bases in DNA resulting in resistance to metal/metal oxide NPs [182]. Two regulons: SoxR and OxyR activate the genes responsible for resistance against oxidative stress. OxyR responds to stress induced by H2O2, and SoxR responds to superoxide anion [183].
Defence mechanisms of biofilms
Biofilm is a union of varied microbes within a matrix of extracellular polymers (lipids, DNA, polysaccharides, and proteins), which acts a barricade for NPs to contact bacteria and set of resistance mechanisms are introduced due to various microbial species [184, 185]. ECP modulates size, surface charge, shape, and concentration of NPs. Biofilms need higher concentrations of NPs than planktonic cells because of grouping of microbes. Biofilms of pore size 10–50 nm retain NPs of > 10 nm size, this trapping do not let NPs interact with microbes present deep inside the biofilm, thus rendering NPs ineffective [186]. Whereas in mature biofilm, the pore size is smaller this does not allow the penetration and diffusion of NPs [187]. Sublethal concentrations of NPs to biofilms can generate hormesis.
Clearance and elimination of nanomaterials
The biodegradation and removal of NPs through urinary and biliary pathways is largely low, leading to persistent tissue retention, amassing in liver and spleen and hence augmented toxicity [188, 189]. Charge and size of NPs determine their opsonisation by serum proteins. In this process, the effective size or hydrodynamic diameter (HD) of the NPs can be modulated. Pore size of endothelium is 5 nm, and NPs with HD < 5 nm can get balanced with the extravascular extracellular space (EES), as vascular endothelial layer acts as dynamic and semi-selective barrier. Thus, large sized NPs cannot freely move in the endothelium hence continues prolonged circulation and toxicities [189]. HD has a crucial part in renal clearance as there is a converse relation between HD and rate of glomerular filtration. NPs of HD < 6 nm undergo glomerular filtration and get proficiently filtered but that of > 8 nm do not. Renal filtration rate is determined by charge as well because interactions of charged NPs and serum proteins (improved protein adsorption), leads to increased HD. Cationic, neutral, and anionic is the order in which these NPs get filtered through glomerulus [190]. Prospective communications between charged capillary walls of glomerulus and NPs also monitor the renal filtration rate [191, 192]. NPs (10–20 nm) cannot use renal filtration, but hepatobiliary system. Liver clears NPs through phagocytosis by Kupffer cells followed by intracellular degradation and eradication. NPs which escape these removal processes retains in the body.
Dose optimization and toxicity of antimicrobial nanomaterials
NPs and their toxic by-products can cause lysis of RBCs and impede blood coagulation pathways [193]. In vitro studies depict high toxicity of AgNPs along with dysfunction of vital organs like; lungs, spleen, liver, colon, bone marrow, and lymphatic system in vivo (and via intravenous and inhalation use of NPs in patients) due to their accumulation [194]. Ag can leach out from wound dressings and have been tested in urine and blood. Al2O3NPs interact with various inter/intra cellular biomolecules and cause neurotoxicity [195]. NPs of CuO cause oxidative distress (prompt nephro- and hepatotoxicity) and ZnO/TiO2 roots DNA damage [196].
The judgement for optimum dose is fundamental for therapeutic targets and curtailing toxicity for clinical translation. Realistic and suitable doses should be researched as the statistics from in vitro and in vivo studies may not be directly translated to patients. This brings concern to the table, the interaction of NPs with human tissues/organs and its clinical translation and administration as therapeutic.
Nanomaterial-based vaccines
Immunization by vaccine remains the most effective method to provide protection against infectious diseases [197]. However, clinical applicability of many new potential vaccine candidates is limited due to low immunogenicity and inability to stimulate an effective long-lasting immunity [198, 199]. In particular, subunit vaccines are less immunogenic and often fail to evoke desirable immune reactions. These vaccines require appropriate adjuvants (immune stimulating agents) and innovative delivery systems to increase immunogenicity, intensify innate and adaptive immunity, and provide a long-term memory response; but concerns remain about their safety and tolerability [200]. Several NPs have shown to stimulate immune responses by multiple mechanisms, offering alternatives to currently used adjuvants and vaccine delivery systems [201]. For example, NPs allow the expression of proteins of interest in the correct conformation on their surfaces thus promote a precise immune reaction. NPs are used as delivery vehicles that guard antigens from degradation. In this context, nanomaterials with defined composition, tailorable structures, tunable immunostimulatory properties, and progressive engineering design are excellent candidates for vaccine development to effectively prevent and manage bacterial infections.
Formulation, “cold-chain” storage, stability, route of administration, and long-distance transportation of vaccines are other technical challenges for vaccine development and effective vaccination programs. In this regard, engineered nanomaterials with varying origins, sizes, shapes, surface properties, and biological functionalities, serve as desirable delivery systems, protect vaccine/antigens from degradation, enhance antigen processing and presentation, facilitate vaccine/antigen uptake by professional antigen-presenting cells (APCs), and improve the stability of vaccines [202]. For example, macrophages preferentially ingest anionic particles [203]. Cationic NPs, such as chitosan, are mucoadhesive that allow them to remain in the mucus for longer duration and interact with mucosal immune cells [204, 205]. In addition, nanomaterials offer routes to improve adjuvants’ function, dose reduction by controlled release of antigen/adjuvant near or within APCs, and minimize undesirable systemic or local effects, such as excessive inflammation or toxicity [206–208].
Nanotechnology offers great platform to design novel modern vaccines as well as facilitate their global implementation. Several nanomaterial-based vaccines to control bacterial infections have been approved for human use and there are many being tested in pre-clinical or clinical trials (Table 2). These vaccines use diverse range of nanomaterials, including polymeric materials (e.g., chitosan), polyesters (e.g., PLGA), polyamides (e.g., gamma polyglutamic acid), polyelectrolyte multilayers, and dendrimers. Their main applications in vaccine include transport and deliver varied peptide, nucleic acid, and protein antigens and adjuvants [206]. Antigen can be delivered to the target cells by either encapsulation (this inhibits premature antigen deprivation and attains persistent release) within NPs or by adsorption on the particle surface (this stabilizes the antigen and simplifies receptor-mediated uptake by APCs) [201, 202]. The particulate nature of NPs bestows them with the capacity to manipulate, enhance and optimize antigen density, and stimulate innate immune response while APCs internalize and process the antigen [209]. Nanomaterials can be engineered to simultaneously co-deliver multiple antigens and adjuvants, which is particularly important for controlling the quality and strength of immune responses. They can be tuned to skew the immune polarization towards important subtypes, such as elicitation of Th1 responses. All these fitting properties of NPs can improve vaccine delivery and efficiency when equated to other conventional delivery and adjuvant systems. Thus, NP-based vaccine formulations are an advantageous, favorable and safe strategy to vaccine development to tackle AMR.
Table 2.
Selected nanomaterial-based vaccines against bacterial infections
| Antigen | Nanocarrier used | Disease | References (year) |
|---|---|---|---|
| Antigenic protein | PLGA nanospheres | Anthrax | [210] (2018) |
| DNA encoding T cell epitopes of Esat-6 and FL | Chitosan nanoparticle | Tuberculosis | [210] (2018) |
| Mycobacterium lipids | Chitosan nanoparticle | Tuberculosis | [210] (2018) |
| Polysaccharides | Liposomes | Pneumonia | [210] (2018) |
| Bacterial toxic and parasitic protein | Liposomes | Cholera and malaria | [210] (2018) |
| Fusion protein | Liposomes | H. pylori | [210] (2018) |
| Antigenic protein | Nano-emulsion | Cystic fibrosis, Anthrax | [210] (2018) |
| Mycobacterium fusion protein | Liposome | Tuberculosis | [210] (2018) |
| Flagellin protein | AuNPs | Yersinia pestis, S. pneumoniae | [211] (2012) |
| Antigenic protein | Cationic liposome-based, stabilized with synthetic glycolipid (CAF01) | Tuberculosis | [212] (2014) |
| Plasmid DNA encoding BoNT heavy chain (Hc) | PLGA | Clostridium botulinum | [213] (2016) |
| Capsular polysaccharide serotype 14 and T-helper peptide, ovalbumin 323–339 peptide | AuNPs with branched tetra-saccharide unit b-d-Galp-(1–4)-b-d-Glcp-(1–6)-[b-d-Galp-(1–4)-]b-d-GlcpNAc-(1–) | S. pneumonia | [214] (2012) |
| LomW and EscC | AuNPs | E. coli (EHEC) | [210] (2018) |
| Listeriolysin O (91–99) and glyceraldehyde-3-phosphate-dehydrogenase (1–22) peptide | AuNPs | Listeria monocytogenes | [210] (2018) |
| Hemagglutinin and FIgL | AuNP coated with antigenic capsular LPS | Burkholderia pseudomallei | [215] (2017) |
| N-terminal domains flagellin (1–161) | AuNPs | P. aeruginosa | [216] (2016) |
| Monosialotetrahexosylganglioside (GM1), host receptor for cholera toxin | PLGA | Vibrio cholerae | [217] (2018) |
| Serogroup B | OMV-based vaccine | N. meningitidis | [218] (2004) |
| Membrane proteins | Double-layered membrane vesicles | P. aeruginosa | [218] (2004) |
| Immunodominant antigens (Ag85A & ESAT-6) and IL-21 | Fe2O3 coated plasmid DNA TB vaccine | M. tuberculosis | [219] (2012) |
| Recombinant fusion protein (M72) | Liposomes | M. tuberculosis | [220] (2018) |
| Heat-induced OMV from enterotoxigenic E. coli | Poly(anhydride) NPs | E. coli | [221] (2022) |
Types of nanomaterials used for vaccine production
Various types of NPs of carbon, gold, polymers, dendrimers, liposomes, and immune-stimulating complexes are used to stabilize and deliver vaccines (antigens and adjuvant) that can effectively stimulate immune responses, such as the production of cytokines and antibody responses [222, 223]. However, emerging AMR bacteria create a challenge for new live-attenuated (first generation), synthetic (second generation), DNA/RNA (third generation), and subunit vaccines [224–226]. In this context, NPs provide a viable approach to vaccine delivery, improving vaccine efficacy with slow release, easy antigen uptake, and induction of humoral and cellular responses.
Mechanism of improved immune response by nanomaterial-based vaccines
A major facet in the engineering of nano-vaccines against bacterial diseases implicates their delivery and interaction to key immune cells, like macrophages, dendritic cells, neutrophils, B and T cells. It also ensures that post vaccine-associated antigens have been processed. Multiple factors like, size and shape, in vivo durability, number of antigen copies on/within nanomaterial, precise co-delivery of adjuvants, conformation/orientation of antigens, or complement activation, impact the vaccine delivery to various tissues, with sizeable impression on the quality and strength of upraised immune responses [227].
Encapsulation of antigens within nanomaterials can upsurge the persistence of antigens at injection site in lymphoid cells or in APCs, which in turn can enhance its immunogenicity. This procedure of producing high affinity antibodies to target is dire for causing immunity against infections. For durable cellular immune response, antigens need to be internalized, processed, and presented efficiently by APCs. For efficacious cross-presentation, antigens must escape to the cytosol, for which nanomaterials sensitive to endolysosomal pathway milieu have been travelled [228]. The precise delivery of nano-vaccines to B, T, follicular dendritic cells, and subcapsular sinus macrophages is highly sought for vaccine design. Physical properties, such as charge, shape, and flexibility of NPs influence lymphatic drainage [229]. Hydrophilic polymers have been found to assist transfer of NPs across the mucus layer [230]. Also, cationic NPs are mucoadhesive, permitting them to be reserved in mucus and to interact with mucosal immune cells. Cationic polysaccharide, chitosan has been employed in mucosally delivered vaccine NPs, including TB [231].
Nanomaterial vaccines also mediate improvement of adjuvant functions and minimize negative systemic/local effects, like inflammation or toxicity. Moreover, some NPs have intrinsic adjuvant properties, even without augmentation by TLR ligands or other adjuvants, which might occur by complement activation, inflammasome signalling or B cell activation [223]. These properties of nanovaccines are beneficial because they can limit inflammation and toxicity arising from other adjuvants and simplify the formulation and dosing of a vaccine.
NP-investing companies and clinical trials
Many companies are considering vaccines as favorable preventive approach to handle AMR because they can be used directly as an inhibitory tool against deadly pathogens. Vaccines can reduce the use of antibiotics by decreasing the infection symptoms, which trigger the antibiotic consumption [232]. Most importantly, vaccines can stop the increment of bacterial numbers, and thus reduce the chances of AMR mutations [233]. The advent of recombinant DNA and glycoconjugation techniques drove the possibility of development of better vaccines against resistant pathogens such as H. influenzae type B, Pneumococcus, Meningococcus, group B Streptococcus, E. coli, and Shigella) [234, 235].
The successful cases of nanobiotics in clinical applications are witnessed in recent years. Liposomal amikacin for inhalation (ARIKAYCE®, developed by Insmed Incorporated) is a unique formulation that encapsulates aqueous Amikacin in charge-neutral liposomes composed of dipalmitoyl phosphatidyl choline and cholesterol [236]. This formulation has been approved by USFDA for treating patients with Mycobacterium avium complex lung disease refractory to conventional therapy and with limited or no alternative treatment options [237]. Liposomal encapsulated ciprofloxacin (rapid-release formulations (Lipoquin™ or ARD-3100) and slow-release formulations (Pulmaquin™ or ARD-3150), developed by Aradigm Corporation for non-cystic fibrosis bronchiectasis colonized with P. aeruginosa are commercially available [237]. Moreover, many nano-antibiotics are in clinical use, such as, mupirocin, polymyxin B, fluconazole, gentamicin, and so on. Prevnar, a pneumococcal vaccine developed by Wyeth, consists saccharides of capsular antigens of seven serotypes of S. pneumoniae conjugated to mutant diphtheria toxoid CRM197 [238]. With the unremitting efforts of researchers, the prospect of clinical nano-antibiotics/nanomaterials would be more hopeful.
Challenges of the NPs for clinical applications
The foremost contests associated with clinical translation of nano-vaccine/nanomedicine are safety, biological concerns, biocompatibility, intellectual property, laws/regulations, time and cost-effectiveness, which discourage the usage of nanomaterials in the present markets [239, 240]. Therefore, one should consider nano-pharmaceutical designs for their physical/chemical stability, biocompatibility/biodegradability, and administration route. Determinations for resolving hindrances of large-scale production, batch to batch reproducibility, high cost, quality control assays (polydispersity, scalability complexities, purification from contaminants, consistency and storage stability, morphology, and charge) should be made [241, 242]. Another challenge is preclinical assessment, for example, toxicity detection (administration and interaction of nanomaterials with biological tissues), in vivo estimation, and understanding pharmacokinetics/and pharmacodynamics of the nanomaterial-based therapeutics and vaccines. It is also important to explore clinical examination for commercialization as invention to market is a difficult path to follow for antibiotics.
As nanomaterials embody countless types of nanostructures, challenges in evolving regulatory protocols, approaches for certifying consistent good manufacturing practice (GMP) production, characterization, safety, and economic design of clinical trial are frequently faced. Since nanomaterial-based vaccines patents and intellectual properties are intricate and universalization of them is desirable, it is important to simplify the path from invention to commercialization to reduce the time and expense required for negotiating collaboration and licensing agreements [243]. Thus, there is a long stretch that needs to be followed before plunging nanomaterials into clinical trials and medicine to control AMR.
Conclusion and future directions
It is imperative to comprehend the mechanisms by which nanomaterials can influence microbial viability, as many antimicrobial mechanisms of nanomaterials are still ambiguous. For example, majority of research points the antimicrobial potential to ROS/oxidative stress, whereas for MgONPs, the mechanism may not be associated with regulation of metabolism of microbes. Thus, finding the antibacterial mechanisms of various nanomaterials is noticeably applicable in addressing for future research. The absence of consistent criterions is one limitation of current studies on mechanisms of NPs because there is no solitary scheme which accomplishes all the studies for procurement of evidence of antimicrobial mechanisms of various NPs as each type displays discrete antibacterial effects. Thus, a complete comprehensive analysis is recommended to study the probable mechanisms. More in vivo studies should be incorporated as it is inconsistent to evaluate the antimicrobial action of NPs merely through in vitro studies. It is also insisted that toxicity of NPs on human cells especially neurotoxicity should be conducted as the crossing of NPs by blood brain barrier is less explored. Though few studies find that NPs enter the bacterial cells through porins, it would be important to understand detailed mechanism of NP entry into bacterial cells. Since nanobiology is getting increasing attention from scientists, the production of biogenic NPs by use of safe and eco-friendly natural reducing agents should be the basis of research on NPs synthesis. Multiple desirable properties make nanomaterials useful against widespread MDR bacteria. The translation of nanomaterials into clinical applications is a craft that need to be done by researchers in order to come up with the best and active antibacterial therapeutics.
Acknowledgements
Not applicable.
Abbreviations
- AgNPs
Silver nanoparticles
- AMEs
Aminoglycoside modifying enzymes
- AMR
Antimicrobial resistance
- APCs
Antigen presenting cells
- AuNPs
Gold nanoparticles
- CAMP
Cationic antimicrobial peptides
- CDF
Cation diffusion facilitator
- EES
Extravascular extracellular space
- EPS
Extracellular polymeric substances
- GQDs
Graphene quantum dots
- HD
Hydrodynamic diameter
- HGT
Horizontal gene transfer
- LNPs
Liposomes and lipid nanoparticles
- LPS
Lipopolysaccharide
- MDR
Multi-drug resistant
- MRSA
Methicillin-resistant Staphylococcus aureus
- NO
Nitric oxide
- NPs
Nanoparticles
- OM
Outer membranes
- PBP
Penicillin-binding protein
- PDT
Photodynamic therapy
- PEA
Phospho-ethanolamine
- PLA
Polylactic acid
- PS
Photosensitizer
- PLGA
Polylactic-co-glycolic acid
- PTT
Photothermal therapy
- QDs
Quantum dots
- ROS
Reactive oxygen species
- TA
Toxin-antitoxin
- UTIs
Urinary tract infections
Author contributions
NC and DJ contributed equally. All authors checked different sections of the manuscript. NKV and HKG guided the writing. All authors read and approved the final manuscript.
Funding
This study was supported by research grant from OLP1149. NC is thankful to the Council of Scientific and Industrial Research (CSIR), Government of India for providing research fellowship support. O-TN received salary support from NMRC Clinician Scientist Award (MOH-000276). NKV thanks funding support from the Lee Kong Chian School of Medicine, Nanyang Technological University Singapore (LKC Operating budget). RL thanks funding support from the Singapore Ministry of Health’s National Medical Research Council under its Centre Grant Programme—Optimization of Core Platform Technologies for Ocular Research (INCEPTOR)-NMRC/CG/M010/2017_SERI and R1875/3/2022.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors gave their consent for publication.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nayanika Chakraborty and Diksha Jha contributed equally to this work
Contributor Information
Rajamani Lakshminarayanan, Email: lakshminarayanan.rajamani@seri.com.sg.
Navin Kumar Verma, Email: nkverma@ntu.edu.sg.
Hemant K. Gautam, Email: hemant@igib.res.in
References
- 1.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahoney AR, Safaee MM, Wuest WM, Furst AL. The silent pandemic: emergent antibiotic resistances following the global response to SARS-CoV-2. IScience. 2021;24:102304. doi: 10.1016/j.isci.2021.102304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jani K, Srivastava V, Sharma P, Vir A, Sharma A. Easy access to antibiotics; spread of antimicrobial resistance and implementation of one health approach in India. J Epidemiol Glob Health. 2021;11:444–452. doi: 10.1007/s44197-021-00008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eichenberger EM, Thaden JT. Epidemiology and mechanisms of resistance of extensively drug resistant Gram-negative bacteria. Antibiotics. 2019;8:37. doi: 10.3390/antibiotics8020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Neill J. Review on antimicrobial resistance: tackling drug-resistant infections globally: final report and recommendations. Review on antimicrobial resistance: tackling drug-resistant infections globally. 2016.
- 6.Peterson E, Kaur P. Antibiotic resistance mechanisms in bacteria: relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front Microbiol. 2018;9:1–21. doi: 10.3389/fmicb.2018.02928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nicoloff H, Hjort K, Levin BR, Andersson DI. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol. 2019;4:504–514. doi: 10.1038/s41564-018-0342-0. [DOI] [PubMed] [Google Scholar]
- 8.Baker KS, Dallman TJ, Field N, Childs T, Mitchell H, Day M, et al. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat Commun. 2018;9:1462. doi: 10.1038/s41467-018-03949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worley JN, Javkar K, Hoffmann M, Hysell K, Garcia-Williams A, Tagg K, et al. Genomic drivers of multidrug-resistant shigella affecting vulnerable patient populations in the United States and abroad. MBio. 2021;12:e03188-20. doi: 10.1128/mBio.03188-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosas NC, Lithgow T. Targeting bacterial outer-membrane remodelling to impact antimicrobial drug resistance. Trends Microbiol. 2022;30:544–552. doi: 10.1016/j.tim.2021.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Kao CY, Chen SS, Hung KH, Wu HM, Hsueh PR, Yan JJ, et al. Overproduction of active efflux pump and variations of OprD dominate in imipenem-resistant Pseudomonas aeruginosa isolated from patients with bloodstream infections in Taiwan. BMC Microbiol. 2016;16(1):107. doi: 10.1186/s12866-016-0719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Fan G, Hryc CF, Blaza JN, Serysheva II, Schmid MF, Chiu W, Luisi BF, Du D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. Elife. 2017;6:e24905. doi: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jellen-Ritter AS, Kern WV. Enhanced expression of the multidrug efflux pumps AcrAB and AcrEF associated with insertion element transposition in Escherichia coli mutants selected with a fluoroquinolone. Antimicrob Agents Chemother. 2001;45:1467–1472. doi: 10.1128/AAC.45.5.1467-1472.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wand ME, Darby EM, Blair JMA, Sutton JM. Contribution of the efflux pump AcrAB-TolC to the tolerance of chlorhexidine and other biocides in Klebsiella spp. J Med Microbiol. 2022;71(3):001496. doi: 10.1099/jmm.0.001496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aminov R. Acquisition and spread of antimicrobial resistance: a tet(X) case study. Int J Mol Sci. 2021;22(8):3905. doi: 10.3390/ijms22083905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huovinen P, Sundström L, Swedberg G, Sköld O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother. 1995;39:279–289. doi: 10.1128/AAC.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pikis A, Donkersloot JA, Rodriguez WJ, Keith JM. A conservative amino acid mutation in the chromosome-encoded dihydrofolate reductase confers trimethoprim resistance in Streptococcus pneumoniae. J Infect Dis. 1998;178:700–706. doi: 10.1086/515371. [DOI] [PubMed] [Google Scholar]
- 18.Dale GE, Broger C, D’Arcy A, Hartman PG, DeHoogt R, Jolidon S, et al. A single amino acid substitution in Staphylococcus aureus dihydrofolate reductase determines trimethoprim resistance. J Mol Biol. 1997;266:23–30. doi: 10.1006/jmbi.1996.0770. [DOI] [PubMed] [Google Scholar]
- 19.Redgrave LS, Sutton SB, Webber MA, Piddock LJV. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez-Martínez JM, Cano ME, Velasco C, Martínez-Martínez L, Pascual Á. Plasmid-mediated quinolone resistance: an update. J Infect Chemother. 2010;17:149–182. doi: 10.1007/s10156-010-0120-2. [DOI] [PubMed] [Google Scholar]
- 21.Floss HG, Yu TW. Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev. 2005;105:621–632. doi: 10.1021/cr030112j. [DOI] [PubMed] [Google Scholar]
- 22.Diaz R, Ramalheira E, Afreixo V, Gago B. Methicillin-resistant Staphylococcus aureus carrying the new mecC gene—a meta-analysis. Diagn Microbiol Infect Dis. 2016;84:135–140. doi: 10.1016/j.diagmicrobio.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 23.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 24.Farhadi R, Saffar MJ, Monfared FT, Larijani LV, Kenari SA, Charati JY. Prevalence, risk factors and molecular analysis of vancomycin-resistant Enterococci colonization in a referral neonatal intensive care unit: a prospective study in northern Iran. J Glob Antimicrob Resist. 2022;S2213–7165(22):00122–129. doi: 10.1016/j.jgar.2022.05.019. [DOI] [PubMed] [Google Scholar]
- 25.Roberts MC. Update on macrolide–lincosamide–streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett. 2008;282:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 26.Candela T, Marvaud JC, Nguyen TK, Lambert T. A cfr-like gene cfr(C) conferring linezolid resistance is common in Clostridium difficile. Int J Antimicrob Agents. 2017;50:496–500. doi: 10.1016/j.ijantimicag.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 27.Kishk R, Soliman N, Nemr N, Eldesouki R, Mahrous N, Gobouri A, et al. Prevalence of aminoglycoside resistance and aminoglycoside modifying enzymes in Acinetobacter baumannii among intensive care unit patients, Ismailia, Egypt. Infect Drug Resist. 2021;14:143–150. doi: 10.2147/IDR.S290584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alcala A, Ramirez G, Solis A, Kim Y, Tan K, Luna O, et al. Structural and functional characterization of three Type B and C chloramphenicol acetyltransferases from Vibrio species. Protein Sci. 2020;29:695–710. doi: 10.1002/pro.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morar M, Wright GD. The genomic enzymology of antibiotic resistance. Annu Rev Genet. 2010;44:25–51. doi: 10.1146/annurev-genet-102209-163517. [DOI] [PubMed] [Google Scholar]
- 30.Lopatkin AJ, Bening SC, Manson AL, Stokes JM, Kohanski MA, Badran AH, et al. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science. 2021;371:eaba0862. doi: 10.1126/science.aba0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stalder T, Rogers LM, Renfrow C, Yano H, Smith Z, Top EM. Emerging patterns of plasmid-host coevolution that stabilize antibiotic resistance. Sci Rep. 2017;7:1–10. doi: 10.1038/s41598-017-04662-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westblade LJ, Errington J, Dorr T. Antibiotic tolerance. PLoS Pathog. 2020;16:10. doi: 10.1371/journal.ppat.1008892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Gefen O, Ronin I, Bar-Meir M, Balaban NQ. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science. 2020;367:200–204. doi: 10.1126/science.aay3041. [DOI] [PubMed] [Google Scholar]
- 34.Sulaiman JE, Lam H. Evolution of bacterial tolerance under antibiotic treatment and its implications on the development of resistance. Front Microbiol. 2021;12:617412. doi: 10.3389/fmicb.2021.617412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 36.Yan J, Bassler BL. Surviving as a community: antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe. 2019;26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerra MES, Destro G, Vieira B, Lima AS, Ferraz LFC, Hakansson AP, et al. Klebsiella pneumoniae biofilms and their role in disease pathogenesis. Front Cell Infect Microbiol. 2022;12:877995. doi: 10.3389/fcimb.2022.877995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14:563–575. doi: 10.1038/nrmicro.2016.94. [DOI] [PubMed] [Google Scholar]
- 39.Goel N, Fatima SW, Kumar S, Sinha R, Khare SK. Antimicrobial resistance in biofilms: exploring marine actinobacteria as a potential source of antibiotics and biofilm inhibitors. Biotechnol Rep (Amst) 2021;30:e00613. doi: 10.1016/j.btre.2021.e00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karygianni L, Ren Z, Koo H, Thurnheer T. Biofilm matrixome: extracellular components in structured microbial communities. Trends Microbiol. 2020;28:668–681. doi: 10.1016/j.tim.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Coleman SR, Blimkie T, Falsafi R, Hancock REW. Multidrug adaptive resistance of Pseudomonas aeruginosa swarming cells. Antimicrob Agents Chemother. 2020;64(3):e01999–e2019. doi: 10.1128/AAC.01999-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhattacharyya S, Walker DM, Harshey RM. Dead cells release a ‘necrosignal’ that activates antibiotic survival pathways in bacterial swarms. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Jia Y, Yang K, Wang Z. Heterogeneous strategies to eliminate intracellular bacterial pathogens. Front Microbiol. 2020;11:563. doi: 10.3389/fmicb.2020.00563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanrompay D, Nguyen TLA, Cutler SJ, Butaye P. Antimicrobial resistance in Chlamydiales, Rickettsia, Coxiella, and other intracellular pathogens. Microbiol Spectr. 2018;6(2):485–500. doi: 10.1128/microbiolspec.ARBA-0003-2017. [DOI] [PubMed] [Google Scholar]
- 45.Zou J, Shankar N. The opportunistic pathogen Enterococcus faecalis resists phagosome acidification and autophagy to promote intracellular survival in macrophages. Cell Microbiol. 2016;18:831–843. doi: 10.1111/cmi.12556. [DOI] [PubMed] [Google Scholar]
- 46.Cano V, March C, Insua JL, Aguiló N, Llobet E, Moranta D, et al. Klebsiella pneumoniae survives within macrophages by avoiding delivery to lysosomes. Cell Microbiol. 2015;17:1537–1560. doi: 10.1111/cmi.12466. [DOI] [PubMed] [Google Scholar]
- 47.Krell T, Matilla MA. Antimicrobial resistance: progress and challenges in antibiotic discovery and anti-infective therapy. Microb Biotechnol. 2022;15:70–78. doi: 10.1111/1751-7915.13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shinu P, Mouslem AKA, Nair AB, Venugopala KN, Attimarad M, Singh VA, et al. Progress report: Antimicrobial drug discovery in the resistance era. Pharmaceuticals (Basel) 2022;15(4):413. doi: 10.3390/ph15040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyd NK, Teng C, Frei CR. Brief overview of approaches and challenges in new antibiotic development: a focus on drug repurposing. Front Cell Infect Microbiol. 2021;11:684515. doi: 10.3389/fcimb.2021.684515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altarac D, Gutch M, Mueller J, Ronsheim M, Tommasi R, Perros M. Challenges and opportunities in the discovery, development, and commercialization of pathogen-targeted antibiotics. Drug Discov Today. 2021;26:2084–2089. doi: 10.1016/j.drudis.2021.02.014. [DOI] [PubMed] [Google Scholar]
- 51.Plackett B. Why big pharma has abandoned antibiotics. Nature. 2020;586:S50–S52. doi: 10.1038/d41586-020-02884-3. [DOI] [Google Scholar]
- 52.Lamberte LE, van Schaik W. Antibiotic resistance in the commensal human gut microbiota. Curr Opin Microbiol. 2022;68:102150. doi: 10.1016/j.mib.2022.102150. [DOI] [PubMed] [Google Scholar]
- 53.Kang M, Yang J, Kim S, Park J, Kim M, Park W. Occurrence of antibiotic resistance genes and multidrug-resistant bacteria during wastewater treatment processes. Sci Total Environ. 2022;811:152331. doi: 10.1016/j.scitotenv.2021.152331. [DOI] [PubMed] [Google Scholar]
- 54.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 55.Kållberg C, Salvesen Blix H, Laxminarayan R. Challenges in antibiotic R&D calling for a global strategy considering both short- and long-term solutions. ACS Infect Dis. 2019;5:1265–1268. doi: 10.1021/acsinfecdis.9b00076. [DOI] [PubMed] [Google Scholar]
- 56.Makabenta JMV, Nabawy A, Li CH, Schmidt-Malan S, Patel R, Rotello VM. Nanomaterial-based therapeutics for antibiotic-resistant bacterial infections. Nat Rev Microbiol. 2021;19:23–36. doi: 10.1038/s41579-020-0420-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ndayishimiye J, Kumeria T, Popat A, Falconer JR, Blaskovich MAT. Nanomaterials: the new antimicrobial magic bullet. ACS Infect Dis. 2022;8:693–712. doi: 10.1021/acsinfecdis.1c00660. [DOI] [PubMed] [Google Scholar]
- 58.Sharma S, Kumar K, Thakur N, Chauhan S, Chauhan MS. The effect of shape and size of ZnO nanoparticles on their antimicrobial and photocatalytic activities: a green approach. Bull Mater Sci. 2020;43:20. doi: 10.1007/s12034-019-1986-y. [DOI] [Google Scholar]
- 59.Prasannakumar JB, Vidya YS, Anantharaju KS, Ramgopal G, Nagabhushana H, Sharma SC, et al. Bio-mediated route for the synthesis of shape tunable Y2O3: Tb3+ nanoparticles: photoluminescence and antibacterial properties. Spectrochim Acta A Mol Biomol Spectrosc. 2015;151:131–140. doi: 10.1016/j.saa.2015.06.081. [DOI] [PubMed] [Google Scholar]
- 60.Wu S, Altenried S, Zogg A, Zuber F, Maniura-Weber K, Ren Q. Role of the surface nanoscale roughness of stainless steel on bacterial adhesion and microcolony formation. ACS Omega. 2018;3:6456–6464. doi: 10.1021/acsomega.8b00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ferreyra Maillard APV, Espeche JC, Maturana P, Cutro AC, Hollmann A. Zeta potential beyond materials science: applications to bacterial systems and to the development of novel antimicrobials. Biochim Biophys Acta Biomembr. 2021;1863:183597. doi: 10.1016/j.bbamem.2021.183597. [DOI] [PubMed] [Google Scholar]
- 62.Pan X, Wang Y, Chen Z, Pan D, Cheng Y, Liu Z, et al. Investigation of antibacterial activity and related mechanism of a series of nano-Mg(OH)2. Appl Mater Interfaces. 2013;5:1137–1142. doi: 10.1021/am302910q. [DOI] [PubMed] [Google Scholar]
- 63.He W, Kim HK, Wamer WG, Melka D, Callahan JH, Yin JJ. Photogenerated charge carriers and reactive oxygen species in ZnO/Au hybrid nanostructures with enhanced photocatalytic and antibacterial activity. J Am Chem Soc. 2014;136:750–757. doi: 10.1021/ja410800y. [DOI] [PubMed] [Google Scholar]
- 64.Rajivgandhi GN, Ramachandran G, Alharbi NS, Kadaikunnan S, Khaleed JM, Manokaran N, Li WJ. Substantial effect of Cr doping on the antimicrobial activity of ZnO nanoparticles prepared by ultrasonication process. Mater Sci Eng, B. 2021;263:114817. doi: 10.1016/j.mseb.2020.114817. [DOI] [Google Scholar]
- 65.Saliani M, Jalal R, Goharshadi EK. Effects of pH and temperature on antibacterial activity of Zinc Oxide nanofluid against Escherichia coli O157: H7 and Staphylococcus aureus, Jundishapur. J Microbiol. 2015;8:1–6. doi: 10.5812/jjm.17115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eleraky NE, Allam A, Hassan SB, Omar MM. Nanomedicine fight against antibacterial resistance: an overview of the recent pharmaceutical innovations. Pharmaceutics. 2020;12:142. doi: 10.3390/pharmaceutics12020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shabatina T, Vernaya O, Shumilkin A, Semenov A, Melnikov M. Nanoparticles of bioactive metals/metal oxides and their nanocomposites with antibacterial drugs for biomedical applications. Materials (Basel) 2022;15(10):3602. doi: 10.3390/ma15103602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chung HJ, Castro CM, Im H, Lee H, Weissleder R. A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria. Nat Nanotechnol. 2013;8:369–375. doi: 10.1038/nnano.2013.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tripathi N, Goshisht MK. Recent advances and mechanistic insights into antibacterial activity, antibiofilm activity, and cytotoxicity of silver nanoparticles. ACS Appl Bio Mater. 2022;5:1391–1463. doi: 10.1021/acsabm.2c00014. [DOI] [PubMed] [Google Scholar]
- 70.Cheng X, Pei X, Xie W, Chen J, Li Y, Wang J, Gao H, Wan Q. pH-triggered size-tunable silver nanoparticles: targeted aggregation for effective bacterial infection therapy. Small. 2022;18(22):e2200915. doi: 10.1002/smll.202200915. [DOI] [PubMed] [Google Scholar]
- 71.Okkeh M, Bloise N, Restivo E, De Vita L, Pallavicini P, Visai L. Gold nanoparticles: can they be the next magic bullet for multidrug-resistant bacteria? Nanomaterials (Basel) 2021;11(2):312. doi: 10.3390/nano11020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng Y, Jiang H, Wang X. Facet-dependent antibacterial activity of Au nanocrystals. Chin Chem Lett. 2020;31:3183–3189. doi: 10.1016/j.cclet.2020.05.035. [DOI] [Google Scholar]
- 73.Rajendiran K, Zhao Z, Pei DS, Fu A. Antimicrobial activity and mechanism of functionalized quantum dots. Polymers (Basel) 2019;11(10):1670. doi: 10.3390/polym11101670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Leevy WM, Lambert TN, Johnson JR, Morris J, Smith BD. Quantum dot probes for bacteria distinguish Escherichia coli mutants and permit in vivo imaging. Chem Commun (Camb) 2008;20:2331–2333. doi: 10.1039/b803590c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Courtney CM, Goodman SM, McDaniel JA, Madinger NE, Chatterjee A, Nagpal P. Photoexcited quantum dots for killing multidrug-resistant bacteria. Nat Mater. 2016;15:529–534. doi: 10.1038/nmat4542. [DOI] [PubMed] [Google Scholar]
- 76.Pati R, Sahu R, Panda J, Sonawane A. Encapsulation of zinc-rifampicin complex into transferrin-conjugated silver quantum-dots improves its antimycobacterial activity and stability and facilitates drug delivery into macrophages. Sci Rep. 2016;6:1–14. doi: 10.1038/srep24184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Usman MS, El Zowalaty ME, Shameli K, Zainuddin N, Salama M, Ibrahim NA. Synthesis, characterization, and antimicrobial properties of copper nanoparticles. Int J Nanomed. 2013;8:4467. doi: 10.2147/IJN.S50837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kulshrestha S, Khan S, Hasan S, Khan ME, Misba L, Khan AU. Calcium fluoride nanoparticles induced suppression of Streptococcus mutans biofilm: an in vitro and in vivo approach. Appl Microbiol Biotechnol. 2016;100:1901–1914. doi: 10.1007/s00253-015-7154-4. [DOI] [PubMed] [Google Scholar]
- 79.Fang F, Li M, Zhang J, Lee CS. Different strategies for organic nanoparticle preparation in biomedicine. ACS Mater Lett. 2020;5:531–549. doi: 10.1021/acsmaterialslett.0c00078. [DOI] [Google Scholar]
- 80.Wang Y. Liposome as a delivery system for the treatment of biofilm-mediated infections. J Appl Microbiol. 2021;131:2626–2639. doi: 10.1111/jam.15053. [DOI] [PubMed] [Google Scholar]
- 81.Cano A, Ettcheto M, Espina M, López-Machado A, Cajal Y, Rabanal F, et al. State-of-the-art polymeric nanoparticles as promising therapeutic tools against human bacterial infections. J Nanobiotechnol. 2020;18(1):156. doi: 10.1186/s12951-020-00714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Forier K, Raemdonck K, De Smedt SC, Demeester J, Coenye T, Braeckmans K. Lipid and polymer nanoparticles for drug delivery to bacterial biofilms. J Control Release. 2014;190:607–623. doi: 10.1016/j.jconrel.2014.03.055. [DOI] [PubMed] [Google Scholar]
- 83.Alqahtani F, Aleanizy F, El Tahir E, Alhabib H, Alsaif R, Shazly G, et al. Antibacterial activity of chitosan nanoparticles against pathogenic N. gonorrhoea. Int J Nanomed. 2020;15:7877–7887. doi: 10.2147/IJN.S272736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Smiechowicz E, Niekraszewicz B, Kulpinski P, Dzitko K. Antibacterial composite cellulose fibers modified with silver nanoparticles and nanosilica. Cellulose. 2018;25:3499–3517. doi: 10.1007/s10570-018-1796-1. [DOI] [Google Scholar]
- 85.Yu Y, Mei L, Shi Y, Zhang X, Cheng K, Cao F, et al. Ag-Conjugated graphene quantum dots with blue light-enhanced singlet oxygen generation for ternary-mode highly-efficient antimicrobial therapy. J Mater Chem B. 2020;8:1371–1382. doi: 10.1039/C9TB02300C. [DOI] [PubMed] [Google Scholar]
- 86.Wang Y, Sun H. Polymeric nanomaterials for efficient delivery of antimicrobial agents. Pharmaceutics. 2021;13(12):2108. doi: 10.3390/pharmaceutics13122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Arana L, Gallego L, Alkorta I. Incorporation of antibiotics into solid lipid nanoparticles: a promising approach to reduce antibiotic resistance emergence. Nanomaterials (Basel) 2021;11(5):1251. doi: 10.3390/nano11051251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Korschelt K, Tahir MN, Tremel W. A step into the future: applications of nanoparticle enzyme mimics. Chem Eur J. 2018;24:9703–9713. doi: 10.1002/chem.201800384. [DOI] [PubMed] [Google Scholar]
- 89.Cao F, Zhang L, Wang H, You Y, Wang Y, Gao N, et al. Defect-rich adhesive nanozymes as efficient antibiotics for enhanced bacterial inhibition. Angew Chem Int Ed. 2019;58:16236–16242. doi: 10.1002/anie.201908289. [DOI] [PubMed] [Google Scholar]
- 90.Meng Y, Li W, Pan X, Gadd GM. Applications of nanozymes in the environment. Environ Sci Nano. 2020;7:1305–1318. doi: 10.1039/C9EN01089K. [DOI] [Google Scholar]
- 91.Gao F, Shao T, Yu Y, Xiong Y, Yang L. Surface-bound reactive oxygen species generating nanozymes for selective antibacterial action. Nat Commun. 2021;12:1–18. doi: 10.1038/s41467-020-20314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Santander SA, Vargas AP, Freitas SC, García C. A novel approach to create an antibacterial surface using titanium dioxide and a combination of dip-pen nanolithography and soft lithography. Sci Rep. 2018;8:1–10. doi: 10.1038/s41598-018-34198-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agnihotri S, Mukherji S, Mukherji S. Immobilized silver nanoparticles enhance contact killing and show highest efficacy: elucidation of the mechanism of bactericidal action of silver. Nanoscale. 2013;5:7328–7340. doi: 10.1039/c3nr00024a. [DOI] [PubMed] [Google Scholar]
- 94.Baptista PV, McCusker MP, Carvalho A, Ferreira DA, Mohan NM, Martins M, et al. Nano-strategies to fight multidrugresistant bacteria—“A battle of the titans”. Front Microbiol. 2018;9:1441. doi: 10.3389/fmicb.2018.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gupta A, Mumtaz S, Li CH, Hussain I, Rotello VM. Combatting antibiotic-resistant bacteria using nanomaterials. Chem Soc Rev. 2019;48:415–427. doi: 10.1039/C7CS00748E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huo S, Jiang Y, Gupta A, Jiang Z, Landis RF, Hou S, et al. Fully Zwitterionic nanoparticle antimicrobial agents through tuning of core size and ligand structure. ACS Nano. 2016;10:8732–8737. doi: 10.1021/acsnano.6b04207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lu X, Feng X, Werber JR, Chu C, Zucker I, Kim JH, et al. Enhanced antibacterial activity through the controlled alignment of graphene oxide nanosheets. Proc Natl Acad Sci USA. 2017;114:E9793–E9801. doi: 10.1073/pnas.1710996114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Johnston HJ, Hutchison G, Christensen FM, Peters S, Hankin S, Stone V. A review of the in vivo and in vitro toxicity of silver and gold particulates: particle attributes and biological mechanisms responsible for the observed toxicity. Crit Rev Toxicol. 2010;40:328–346. doi: 10.3109/10408440903453074. [DOI] [PubMed] [Google Scholar]
- 99.Lyon DY, Fortner JD, Sayes CM, Colvin VL, Hughes JB. Bacterial cell association and antimicrobial activity of a C60 water suspension. Environ Toxicol Chem. 2005;24:2757–2762. doi: 10.1897/04-649R.1. [DOI] [PubMed] [Google Scholar]
- 100.Hamal DB, Haggstrom JA, Marchin GL, Ikenberry MA, Hohn K, Klabunde KJ. A multifunctional biocide/sporocide and photocatalyst based on titanium dioxide (TiO2) codoped with silver, carbon, and sulfur. Langmuir. 2010;26:2805–2810. doi: 10.1021/la902844r. [DOI] [PubMed] [Google Scholar]
- 101.Choi JY, Kim KH, Choy KC, Oh KT, Kim KN. Photocatalytic antibacterial effect of TiO(2) film formed on Ti and TiAg exposed to Lactobacillus acidophilus. J Biomed Mater Res B Appl Biomater. 2007;80:353–359. doi: 10.1002/jbm.b.30604. [DOI] [PubMed] [Google Scholar]
- 102.Mohamed MM, Fouad SA, Elshoky HA, Mohammed GM, Salaheldin TA. Antibacterial effect of gold nanoparticles against Corynebacterium pseudotuberculosis. Int J Vet Sci Med. 2017;5:23–29. doi: 10.1016/j.ijvsm.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen WJ, Tsai PJ, Chen YC. Functional Fe3O4/TiO2 core/shell magnetic nanoparticles as photokilling agents for pathogenic bacteria. Small. 2008;4:485–491. doi: 10.1002/smll.200701164. [DOI] [PubMed] [Google Scholar]
- 104.Xie Y, He Y, Irwin PL, Jin T, Shi X. Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Appl Environ Microbiol. 2011;77:2325–2331. doi: 10.1128/AEM.02149-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Q, Mahendra S, Lyon DY, Brunet L, Liga MV, Li D, et al. Antimicrobial nanomaterials for water disinfection and microbial control: potential applications and implications. Water Res. 2008;42:4591–4602. doi: 10.1016/j.watres.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 106.Xin Q, Shah H, Nawaz A, Xie W, Akram MZ, Batool A, et al. Antibacterial carbon-based nanomaterials. Adv Mater. 2019;31:e1804838. doi: 10.1002/adma.201804838. [DOI] [PubMed] [Google Scholar]
- 107.Hsieh HS, Wu R, Jafvert CT. Light-independent reactive oxygen species (ROS) formation through electron transfer from carboxylated single-walled carbon nanotubes in water. Environ Sci Technol. 2014;48:11330–11336. doi: 10.1021/es503163w. [DOI] [PubMed] [Google Scholar]
- 108.Zhao Y, Ye C, Liu W, Chen R, Jiang X. Tuning the composition of AuPt bimetallic nanoparticles for antibacterial application. Angew Chem Int Ed Engl. 2014;53:8127–8131. doi: 10.1002/anie.201401035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Xu JW, Yao K, Xu ZK. Nanomaterials with a photothermal effect for antibacterial activities: an overview. Nanoscale. 2019;11:8680–8691. doi: 10.1039/C9NR01833F. [DOI] [PubMed] [Google Scholar]
- 110.Jaque D, Martínez Maestro L, del Rosal B, Haro-Gonzalez P, Benayas A, Plaza JL, et al. Nanoparticles for photothermal therapies. Nanoscale. 2014;6:9494–9530. doi: 10.1039/C4NR00708E. [DOI] [PubMed] [Google Scholar]
- 111.Ma K, Li Y, Wang Z, Chen Y, Zhang X, Chen C, et al. Core-shell Gold Nanorod@Layered double hydroxide nanomaterial with highly efficient photothermal conversion and its application in antibacterial and tumor therapy. ACS Appl Mater Interfaces. 2019;11:29630–29640. doi: 10.1021/acsami.9b10373. [DOI] [PubMed] [Google Scholar]
- 112.Wang X, Su K, Tan L, Liu X, Cui Z, Jing D, et al. Rapid and highly effective noninvasive disinfection by hybrid Ag/CS@MnO2 nanosheets using near-infrared light. Appl Mater Interfaces. 2019;11:15014–15027. doi: 10.1021/acsami.8b22136. [DOI] [PubMed] [Google Scholar]
- 113.Zhang W, Shi S, Wang Y, Yu S, Zhu W, Zhang X, et al. Versatile molybdenum disulfide based antibacterial composites for in vitro enhanced sterilization and in vivo focal infection therapy. Nanoscale. 2016;8:11642–11648. doi: 10.1039/C6NR01243D. [DOI] [PubMed] [Google Scholar]
- 114.Ansari SA, Nisar A, Fatma B, Khan W, Chaman M, Azam A, et al. Temperature dependence anomalous dielectric relaxation in Co doped ZnO nanoparticles. Mater Res Bull. 2012;47:4161–4168. doi: 10.1016/j.materresbull.2012.08.079. [DOI] [Google Scholar]
- 115.Yu J, Zhang W, Li Y, Wang G, Yang L, Jin J, et al. Synthesis, characterization, antimicrobial activity and mechanism of a novel hydroxyapatite whisker/nano zinc oxide biomaterial. Biomed Mater. 2014;10:015001. doi: 10.1088/1748-6041/10/1/015001. [DOI] [PubMed] [Google Scholar]
- 116.Singh R, Cheng S, Singh S. Oxidative stress-mediated genotoxic effect of zinc oxide nanoparticles on Deinococcus radiodurans. 3 Biotech. 2020;10:66. doi: 10.1007/s13205-020-2054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Depan D, Misra RDK. On the determining role of network structure titania in silicone against bacterial colonization: mechanism and disruption of biofilm. Mater Sci Eng C. 2014;34:221–228. doi: 10.1016/j.msec.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 118.Tavares A, Carvalho CM, Faustino MA, Neves MG, Tomé JP, Tomé AC, et al. Antimicrobial photodynamic therapy: study of bacterial recovery viability and potential development of resistance after treatment. Mar Drugs. 2010;8:91–105. doi: 10.3390/md8010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Abrahamse H, Hamblin MR. New photosensitizers for photodynamic therapy. Biochem J. 2016;473:347–364. doi: 10.1042/BJ20150942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chilakamarthi U, Giribabu L. Photodynamic therapy: past, present and future. Chem Rec. 2017;17:775–802. doi: 10.1002/tcr.201600121. [DOI] [PubMed] [Google Scholar]
- 121.Fan W, Huang P, Chen X. Overcoming the Achilles’ heel of photodynamic therapy. Chem Soc Rev. 2016;45:6488–6519. doi: 10.1039/C6CS00616G. [DOI] [PubMed] [Google Scholar]
- 122.Biel MA. Photodynamic therapy of bacterial and fungal biofilm infections. Methods Mol Biol. 2010;635:175–194. doi: 10.1007/978-1-60761-697-9_13. [DOI] [PubMed] [Google Scholar]
- 123.Lin JF, Li J, Gopal A, Munshi T, Chu YW, Wang JX, et al. Synthesis of photo-excited Chlorin e6 conjugated silica nanoparticles for enhanced anti-bacterial efficiency to overcome methicillin-resistant Staphylococcus aureus. Chem Commun (Camb) 2019;55:2656–2659. doi: 10.1039/C9CC00166B. [DOI] [PubMed] [Google Scholar]
- 124.Park H, Lee J, Jeong S, Im BN, Kim MK, Yang SG, et al. Lipase-sensitive transfersomes based on photosensitizer/polymerizable lipid conjugate for selective antimicrobial photodynamic therapy of acne. Adv Healthc Mater. 2016;5:3139–3147. doi: 10.1002/adhm.201600815. [DOI] [PubMed] [Google Scholar]
- 125.Chang YN, Zhang M, Xia L, Zhang J, Xing G. The toxic effects and mechanisms of CuO and ZnO nanoparticles. Materials. 2012;5:2850–2871. doi: 10.3390/ma5122850. [DOI] [Google Scholar]
- 126.Wang YW, Cao A, Jiang Y, Zhang X, Liu JH, Liu Y, et al. Superior antibacterial activity of zinc oxide/graphene oxide composites originating from high zinc concentration localized around bacteria. ACS Appl Mater Interfaces. 2014;6:2791–2798. doi: 10.1021/am4053317. [DOI] [PubMed] [Google Scholar]
- 127.Jung WK, Koo HC, Kim KW, Shin S, Kim SH, Park YH. Antibacterial activity and mechanism of action of the silver ion in Staphylococcus aureus and Escherichia coli. Appl Environ Microbiol. 2008;74:2171–2178. doi: 10.1128/AEM.02001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jia H, Hou W, Wei L, Xu B, Liu X. The structures and antibacterial properties of nano-SiO2 supported silver/zinc-silver materials. Dent Mater. 2008;24:244–249. doi: 10.1016/j.dental.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 129.Lemire JA, Harrison JJ, Turner RJ. Antimicrobial activity of metals: mechanisms, molecular targets and applications. Nat Rev Microbiol. 2013;11:371–384. doi: 10.1038/nrmicro3028. [DOI] [PubMed] [Google Scholar]
- 130.Su G, Zhang X, Giesy JP, Musarrat J, Saquib Q, Alkhedhairy AA, et al. Comparison on the molecular response profiles between nano zinc oxide (ZnO) particles and free zinc ion using a genome-wide toxicogenomics approach. Environ Sci Pollut Res Int. 2015;22:17434–17442. doi: 10.1007/s11356-015-4507-6. [DOI] [PubMed] [Google Scholar]
- 131.Chatterjee AK, Chakraborty R, Basu T. Mechanism of antibacterial activity of copper nanoparticles. Nanotechnology. 2014;25:135101. doi: 10.1088/0957-4484/25/13/135101. [DOI] [PubMed] [Google Scholar]
- 132.Cui Y, Zhao Y, Tian Y, Zhang W, Lü X, Jiang X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials. 2012;33:2327–2333. doi: 10.1016/j.biomaterials.2011.11.057. [DOI] [PubMed] [Google Scholar]
- 133.Niemirowicz K, Swiecicka I, Wilczewska AZ, Misztalewska I, Kalska-Szostko B, Bienias K, et al. Gold-functionalized magnetic nanoparticles restrict growth of Pseudomonas aeruginosa. Int J Nanomed. 2014;9:2217–2224. doi: 10.2217/nnm.14.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schairer DO, Chouake JS, Nosanchuk JD, Friedman AJ. The potential of nitric oxide releasing therapies as antimicrobial agents. Virulence. 2012;3:271–279. doi: 10.4161/viru.20328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Friedman AJ, Blecher K, Schairer D, Tuckman-Vernon C, Nacharaju P, Sanchez D, et al. Improved antimicrobial efficacy with nitric oxide releasing nanoparticle generated S-nitrosoglutathione. Nitric Oxide. 2011;25:381–386. doi: 10.1016/j.niox.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 136.Monteiro DR, Gorup LF, Takamiya AS, de Camargo ER, Filho AC, Barbosa DB. Silver distribution and release from an antimicrobial denture base resin containing silver colloidal nanoparticles. J Prosthodont. 2012;21:7–15. doi: 10.1111/j.1532-849X.2011.00772.x. [DOI] [PubMed] [Google Scholar]
- 137.Lee SH, Jun BH. Silver nanoparticles: synthesis and application for nanomedicine. Int J Mol Sci. 2019;20:865. doi: 10.3390/ijms20040865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, et al. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 139.Nagy A, Harrison A, Sabbani S, Munson RS, Jr, Dutta PK, Waldman WJ. Silver nanoparticles embedded in zeolite membranes: release of silver ions and mechanism of antibacterial action. Int J Nanomed. 2011;6:1833–1852. doi: 10.2147/IJN.S24019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ashmore D, Chaudhari A, Barlow B, Barlow B, Harper T, Vig K, et al. Evaluation of E. coli inhibition by plain and polymer-coated silver nanoparticles. Rev Inst Med Trop Sao Paulo. 2018;60:e18. doi: 10.1590/s1678-9946201860018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Leung YH, Ng AM, Xu X, Shen Z, Gethings LA, Wong MT, et al. Mechanisms of antibacterial activity of MgO: non-ROS mediated toxicity of MgO nanoparticles towards Escherichia coli. Small. 2014;10:1171–1183. doi: 10.1002/smll.201302434. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y, Lin S, Fu J, Zhang W, Shu G, Lin J, Li H, Xu F, Tang H, Peng G, Zhao L, Chen S, Fu H. Nanocarriers for combating biofilms: advantages and challenges. J Appl Microbiol. 2022 doi: 10.1111/jam.15640. [DOI] [PubMed] [Google Scholar]
- 143.Yeh YC, Huang TH, Yang SC, Chen CC, Fang JY. Nano-based drug delivery or targeting to eradicate bacteria for infection mitigation: a review of recent advances. Front Chem. 2020;8:286. doi: 10.3389/fchem.2020.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liao CC, Yu HP, Yang SC, Alalaiwe A, Dai YS, Liu FC, Fang JY. Multifunctional lipid-based nanocarriers with antibacterial and anti-inflammatory activities for treating MRSA bacteremia in mice. J Nanobiotechnol. 2021;19(1):48. doi: 10.1186/s12951-021-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jina L, Liu X, Bian C, Sheng J, Song Y, Zhu Y. Fabrication linalool-functionalized hollow mesoporous silica spheres nanoparticles for efficiently enhance bactericidal activity. Chin Chem Lett. 2020;31:2137–2141. doi: 10.1016/j.cclet.2019.12.020. [DOI] [Google Scholar]
- 146.Tian X, Wang P, Li T, Huang X, Guo W, Yang Y, Yan M, et al. Self-assembled natural phytochemicals for synergistically antibacterial application from the enlightenment of traditional Chinese medicine combination. Acta Pharm Sin B. 2020;10:1784–1795. doi: 10.1016/j.apsb.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gounani Z, Asadollahi MA, Pedersen JN, Lyngsø J, Skov Pedersen J, et al. Mesoporous silica nanoparticles carrying multiple antibiotics provide enhanced synergistic effect and improved biocompatibility. Colloids Surf B Biointerfaces. 2019;175:498–508. doi: 10.1016/j.colsurfb.2018.12.035. [DOI] [PubMed] [Google Scholar]
- 148.Niño-Martínez N, Salas Orozco MF, Martínez-Castañón GA, Torres Méndez F, Ruiz F. Molecular mechanisms of bacterial resistance to metal and metal oxide nanoparticles. Int J Mol Sci. 2019;20(11):2808. doi: 10.3390/ijms20112808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Panáček A, Kvítek L, Smékalová M, Večeřová R, Kolář M, Röderová M, et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat Nanotechnol. 2018;13:65–71. doi: 10.1038/s41565-017-0013-y. [DOI] [PubMed] [Google Scholar]
- 150.Salas-Orozco M, Niño-Martínez N, Martínez-Castañón GA, Méndez FT, Jasso MEC, Ruiz F. Mechanisms of resistance to silver nanoparticles in endodontic bacteria: a literature review. J Nanomater. 2019;2019:7630316. [Google Scholar]
- 151.Srivastava P, Kowshik M. Mechanisms of metal resistance and homeostasis in haloarchaea. Archaea. 2013;2013:732864. doi: 10.1155/2013/732864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang R, Carlsson F, Edman M, Hummelgård M, Jonsson BG, Bylund D, Olin H. Escherichia coli bacteria develop adaptive resistance to antibacterial ZnO nanoparticles. Adv Biosyst. 2018;2:e1800019. doi: 10.1002/adbi.201800019. [DOI] [PubMed] [Google Scholar]
- 153.Siemer S, Westmeier D, Barz M, Eckrich J, Wünsch D, Seckert C, et al. Biomolecule-corona formation confers resistance of bacteria to nanoparticle-induced killing: implications for the design of improved nanoantibiotics. Biomaterials. 2019;192:551–559. doi: 10.1016/j.biomaterials.2018.11.028. [DOI] [PubMed] [Google Scholar]
- 154.Finley PJ, Norton R, Austin C, Mitchell A, Zank S, Durham P. Unprecedented silver resistance in clinically isolated enterobacteriaceae: major implications for burn and wound management. Antimicrob Agents Chemother. 2015;59:4734–4741. doi: 10.1128/AAC.00026-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Faghihzadeh F, Anaya NM, Astudillo-Castro C, Oyanedel-Craver V. Kinetic, metabolic and macromolecular response of bacteria to chronic nanoparticle exposure in continuous culture. Environ Sci Nano. 2018;5:1386–1396. doi: 10.1039/C8EN00325D. [DOI] [Google Scholar]
- 156.Graves JL, Jr, Tajkarimi M, Cunningham Q, Campbell A, Nonga H, Harrison SH, et al. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front Genet. 2015;6:42. doi: 10.3389/fgene.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hachicho N, Hoffmann P, Ahlert K, Heipieper HJ. Effect of silver nanoparticles and silver ions on growth and adaptive response mechanisms of Pseudomonas putida mt-2. FEMS Microbiol Lett. 2014;355:71–77. doi: 10.1111/1574-6968.12460. [DOI] [PubMed] [Google Scholar]
- 158.Feris K, Otto C, Tinker J, Wingett D, Punnoose A, Thurber A, et al. Electrostatic interactions affect nanoparticle-mediated toxicity to gram-negative bacterium Pseudomonas aeruginosa PAO1. Langmuir. 2010;26:4429–4436. doi: 10.1021/la903491z. [DOI] [PubMed] [Google Scholar]
- 159.Abbaszadegan A, Ghahramani Y, Gholami A, Hemmateenejad B, Dorostkar S, Nabavizadeh M, et al. The effect of charge at the surface of silver nanoparticles on antimicrobial activity against gram-positive and gram-negative bacteria. J Nanomater. 2015;2015:720654. doi: 10.1155/2015/720654. [DOI] [Google Scholar]
- 160.Peter KS, Rosalyn KL, George LM, Kenneth JK. Metal oxide nanoparticles as bactericidal agents. Langmuir. 2002;18:6679–6686. doi: 10.1021/la0202374. [DOI] [Google Scholar]
- 161.Kumariya R, Sood SK, Rajput YS, Saini N, Garsa AK. Increased membrane surface positive charge and altered membrane fluidity leads to cationic antimicrobial peptide resistance in Enterococcus faecalis. Biochim Biophys Acta. 2015;1848:1367–1375. doi: 10.1016/j.bbamem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 162.Jordan S, Hutchings MI, Mascher T. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol Rev. 2008;32:107–146. doi: 10.1111/j.1574-6976.2007.00091.x. [DOI] [PubMed] [Google Scholar]
- 163.Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, et al. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J Bacteriol. 2005;187:5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sharma VK, Sayes CM, Guo B, Pillai S, Parsons JG, Wang C, et al. Interactions between silver nanoparticles and other metal nanoparticles under environmentally relevant conditions: a review. Sci Total Environ. 2019;653:1042–1051. doi: 10.1016/j.scitotenv.2018.10.411. [DOI] [PubMed] [Google Scholar]
- 165.Louie SM, Ma R, Lowry GV. Transformations of nanomaterials in the environment. Front Nanosci. 2014;7:55–87. doi: 10.1016/B978-0-08-099408-6.00002-5. [DOI] [Google Scholar]
- 166.Li Z, Greden K, Alvarez PJJ, Gregory KB, Lowry GV. Adsorbed polymer and NOM limits adhesion and toxicity of nano scale zerovalent iron to E. coli. Environ Sci Technol. 2010;44:3462–3467. doi: 10.1021/es9031198. [DOI] [PubMed] [Google Scholar]
- 167.Guo J, Gao SH, Lu J, Bond PL, Verstraete W, Yuan Z. Copper oxide nanoparticles induce lysogenic bacteriophage and metal-resistance genes in Pseudomonas aeruginosa PAO1. ACS Appl Mater Interfaces. 2017;9:22298–22307. doi: 10.1021/acsami.7b06433. [DOI] [PubMed] [Google Scholar]
- 168.Kolaj-Robin O, Russell D, Hayes KA, Pembroke JT, Soulimane T. Cation diffusion facilitator family: structure and function. FEBS Lett. 2015;589:1283–1295. doi: 10.1016/j.febslet.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 169.Argüello JM, Padilla-Benavides T, Collins JM. Encyclopedia of inorganic and bioinorganic chemistry. New York: Wiley; 2011. Copper(I) ATPases: transport mechanism and cellular functions in bacteria; pp. 1–8. [Google Scholar]
- 170.Imran M, Das KR, Naik MM. Co-selection of multi-antibiotic resistance in bacterial pathogens in metal and microplastic contaminated environments: an emerging health threat. Chemosphere. 2019;215:846–857. doi: 10.1016/j.chemosphere.2018.10.114. [DOI] [PubMed] [Google Scholar]
- 171.Randall CP, Gupta A, Jackson N, Busse D, O’Neill AJ. Silver resistance in Gram-negative bacteria: a dissection of endogenous and exogenous mechanisms. J Antimicrob Chemother. 2015;70:1037–1046. doi: 10.1093/jac/dku523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ellis DH, Maurer-Gardner EI, Sulentic CEW, Hussain SM. Silver nanoparticle antibacterial efficacy and resistance development in key bacterial species. Biomed Phys Eng Express. 2018;5:015013. doi: 10.1088/2057-1976/aad5a7. [DOI] [Google Scholar]
- 173.Ramos-Zúñiga J, Gallardo S, Martínez-Bussenius C, Norambuena R, Navarro CA, Paradela A, Jerez CA. Response of the biomining Acidithiobacillus ferrooxidans to high cadmium concentrations. J Proteom. 2019;198:132–144. doi: 10.1016/j.jprot.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 174.Palomo-Siguero M, Gutiérrez AM, Pérez-Conde C, Madrid Y. Effect of selenite and selenium nanoparticles on lactic bacteria: a multi-analytical study. Microchem J. 2016;126:488–495. doi: 10.1016/j.microc.2016.01.010. [DOI] [Google Scholar]
- 175.Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol. 2017;15:338–350. doi: 10.1038/nrmicro.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang X, Yang F, Zhao J, Xu Y, Mao D, Zhu X, et al. Bacterial exposure to ZnO nanoparticles facilitates horizontal transfer of antibiotic resistance genes. NanoImpact. 2018;10:61–67. doi: 10.1016/j.impact.2017.11.006. [DOI] [Google Scholar]
- 177.Qiu Z, Shen Z, Qian D, Jin M, Yang D, Wang J, et al. Effects of NaNO-TiO2 on antibiotic resistance transfer mediated by RP4 plasmid. Nanotoxicology. 2015;9:895–904. doi: 10.3109/17435390.2014.991429. [DOI] [PubMed] [Google Scholar]
- 178.Flores-Kim J, Darwin AJ. Regulation of bacterial virulence gene expression by cell envelope stress responses. Virulence. 2015;5:835–851. doi: 10.4161/21505594.2014.965580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tang S, Zheng J. Antibacterial activity of silver nanoparticles: structural effects. Adv Healthc Mater. 2018;7:1701503. doi: 10.1002/adhm.201701503. [DOI] [PubMed] [Google Scholar]
- 180.Čáp M, Váchová L, Palková Z. Reactive oxygen species in the signaling and adaptation of multicellular microbial communities. Oxid Med Cell Longev. 2012;2012:976753. doi: 10.1155/2012/976753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rochat T, Nicolas P, Delumeau O, Rabatinová A, Korelusová J, Leduc A, et al. Genome-wide identification of genes directly regulated by the pleiotropic transcription factor Spx in Bacillus subtilis. Nucl Acids Res. 2012;40:9571–9583. doi: 10.1093/nar/gks755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tkachenko AG. Stress responses of bacterial cells as mechanism of development of antibiotic tolerance. Appl Biochem Microbiol. 2018;54:108–127. doi: 10.1134/S0003683818020114. [DOI] [Google Scholar]
- 183.Dale AL, Lowry GV, Casman EA. Modeling nanosilver transformations in freshwater sediments. Environ Sci Technol. 2013;47:12920–12928. doi: 10.1021/es402341t. [DOI] [PubMed] [Google Scholar]
- 184.Sheng Z, Van Nostrand JD, Zhou J, Liu Y. The effects of silver nanoparticles on intact wastewater biofilms. Front Microbiol. 2015;6:680. doi: 10.3389/fmicb.2015.00680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Peulen TO, Wilkinson KJ. Diffusion of nanoparticles in a biofilm. Environ Sci Technol. 2011;45:3367–3373. doi: 10.1021/es103450g. [DOI] [PubMed] [Google Scholar]
- 186.Choi O, Yu CP, Esteban Fernández G, Hu Z. Interactions of nanosilver with Escherichia coli cells in planktonic and biofilm cultures. Water Res. 2010;44:6095–6103. doi: 10.1016/j.watres.2010.06.069. [DOI] [PubMed] [Google Scholar]
- 187.Sahle-Demessie E, Tadesse H. Kinetics and equilibrium adsorption of nano-TiO2 particles on synthetic biofilm. Surf Sci. 2011;605:1177–1184. doi: 10.1016/j.susc.2011.03.022. [DOI] [Google Scholar]
- 188.Lin Z, Monteiro-Riviere NA, Riviere JE. Pharmacokinetics of metallic nanoparticles, WIREs. Nanomed Nanobiotechnol. 2015;7:189–217. doi: 10.1002/wnan.1304. [DOI] [PubMed] [Google Scholar]
- 189.Zaidi S, Misba L, Khan AU. Nano-therapeutics: a revolution in infection control in post antibiotic era. Nanomed Nanotechnol Biol Med. 2017;13:2281–2301. doi: 10.1016/j.nano.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 190.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine. 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Ipe BI, et al. Renal clearance of quantum dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Deen WM, Lazzara MJ, Myers BD. Structural determinants of glomerular permeability. Am J Physiol Renal Physiol. 2001;281:579–596. doi: 10.1152/ajprenal.2001.281.4.F579. [DOI] [PubMed] [Google Scholar]
- 193.Kandi V, Kandi S. Antimicrobial properties of nanomolecules: potential candidates as antibiotics in the era of multi-drug resistance. Epidemiol Health. 2015;37:e2015020. doi: 10.4178/epih/e2015020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ivask A, Juganson K, Bondarenko O, Mortimer M, Aruoja V, Kasemets K, et al. Mechanisms of toxic action of Ag, ZnO and CuO nanoparticles to selected ecotoxicological test organisms and mammalian cells in vitro: a comparative review. Nanotoxicology. 2014;8:57–71. doi: 10.3109/17435390.2013.855831. [DOI] [PubMed] [Google Scholar]
- 195.Ansari MA, Khan HM, Khan AA, Ahmad MK, Mahdi AA, Pal R, et al. Interaction of silver nanoparticles with Escherichia coli and their cell envelope biomolecules. J Basic Microbiol. 2014;54:905–915. doi: 10.1002/jobm.201300457. [DOI] [PubMed] [Google Scholar]
- 196.Lei R, Wu C, Yang B, Ma H, Shi C, Wang Q, et al. Integrated metabolomic analysis of the nano-sized copper particle-induced hepatotoxicity and nephrotoxicity in rats: a rapid in vivo screening method for nanotoxicity. Toxicol Appl Pharmacol. 2008;232:292–301. doi: 10.1016/j.taap.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 197.Poolman JT. Expanding the role of bacterial vaccines into life-course vaccination strategies and prevention of antimicrobial-resistant infections. NPJ Vaccines. 2020;5:84. doi: 10.1038/s41541-020-00232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Bekeredjian-Ding I. Challenges for clinical development of vaccines for prevention of hospital-acquired bacterial infections. Front Immunol. 2020;11:1755. doi: 10.3389/fimmu.2020.01755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Osterloh A. Vaccination against bacterial infections: challenges, progress, and new approaches with a focus on intracellular bacteria. Vaccines (Basel) 2022;10(5):751. doi: 10.3390/vaccines10050751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Marques Neto LM, Kipnis A, Junqueira-Kipnis AP. Role of metallic nanoparticles in vaccinology: implications for infectious disease vaccine development. Front Immunol. 2017;8:239. doi: 10.3389/fimmu.2017.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Curley SM, Putnam D. Biological nanoparticles in vaccine development. Front Bioeng Biotechnol. 2022;10:867119. doi: 10.3389/fbioe.2022.867119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fries CN, Curvino EJ, Chen JL, Permar SR, Fouda GG, Collier JH. Advances in nanomaterial vaccine strategies to address infectious diseases impacting global health. Nat Nanotechnol. 2021;16(4):1–14. doi: 10.1038/s41565-020-0739-9. [DOI] [PubMed] [Google Scholar]
- 203.Fröhlich E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int J Nanomed. 2012;7:5577–5591. doi: 10.2147/IJN.S36111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Singh B, Maharjan S, Cho KH, Cui L, Park IK, Choi YJ, Cho CS. Chitosan-based particulate systems for the delivery of mucosal vaccines against infectious diseases. Int J Biol Macromol. 2018;110:54–64. doi: 10.1016/j.ijbiomac.2017.10.101. [DOI] [PubMed] [Google Scholar]
- 205.Bivas-Benita M, van Meijgaarden KE, Franken KL, Junginger HE, Borchard G, Ottenhoff TH, et al. Pulmonary delivery of chitosan-DNA nanoparticles enhances the immunogenicity of a DNA vaccine encoding HLA-A*0201-restricted T-cell epitopes of Mycobacterium tuberculosis. Vaccine. 2004;22(13–14):1609–1615. doi: 10.1016/j.vaccine.2003.09.044. [DOI] [PubMed] [Google Scholar]
- 206.Feng C, Li Y, Ferdows BE, Patel DN, Ouyang J, Tang Z, Kong N, et al. Emerging vaccine nanotechnology: from defense against infection to sniping cancer. Acta Pharm Sin B. 2022;12:2206–2223. doi: 10.1016/j.apsb.2021.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hanson MC, et al. Nanoparticulate STING agonists are potent lymph node-targeted vaccine adjuvants. J Clin Invest. 2015;125:2532–2546. doi: 10.1172/JCI79915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ilyinskii PO, Roy CJ, O'Neil CP, Browning EA, Pittet LA, Altreuter DH, et al. Adjuvant-carrying synthetic vaccine particles augment the immune response to encapsulated antigen and exhibit strong local immune activation without inducing systemic cytokine release. Vaccine. 2014;32:2882–2895. doi: 10.1016/j.vaccine.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pati R, Shevtsov M, Sonawane A. Nanoparticle vaccines against infectious diseases. Front Immunol. 2018;9:2224. doi: 10.3389/fimmu.2018.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gregory AE, Williamson ED, Prior JL, Butcher WA, Thompson IJ, Shaw AM, et al. Conjugation of Y. pestis F1-antigen to gold nanoparticles improves immunogenicity. Vaccine. 2012;30:6777–6782. doi: 10.1016/j.vaccine.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.van Dissel JT, Joosten SA, Hoff ST, Soonawala D, Prins C, Hokey DA, et al. A novel liposomal adjuvant system, CAF01, promotes long-lived Mycobacterium tuberculosis-specific T-cell responses in human. Vaccine. 2014;32:7098–7107. doi: 10.1016/j.vaccine.2014.10.036. [DOI] [PubMed] [Google Scholar]
- 212.Ruwona TB, Xu H, Li J, Diaz-Arévalo D, Kumar A, Zeng M, et al. Induction of protective neutralizing antibody responses against botulinum neurotoxin serotype C using plasmid carried by PLGA nanoparticles. Hum Vaccines Immunother. 2016;12:1188–1192. doi: 10.1080/21645515.2015.1122147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Safari D, Marradi M, Chiodo F, Th Dekker HA, Shan Y, Adamo R, et al. Gold nanoparticles as carriers for a synthetic Streptococcus pneumoniae type 14 conjugate vaccine. Nanomedicine. 2012;7:651–662. doi: 10.2217/nnm.11.151. [DOI] [PubMed] [Google Scholar]
- 214.Muruato LA, Tapia D, Hatcher CL, Kalita M, Brett PJ, Gregory AE, et al. Use of reverse vaccinology in the design and construction of nanoglycoconjugate vaccines against Burkholderia pseudomallei. Clin Vaccines Immunol. 2017;24:e00206–e217. doi: 10.1128/CVI.00206-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Dakterzada F, Mobarez AM, Roudkenar MH, Mohsenifar A. Induction of humoral immune response against Pseudomonas aeruginosa flagellin(1–161) using gold nanoparticles as an adjuvant. Vaccine. 2016;34:1472–1479. doi: 10.1016/j.vaccine.2016.01.041. [DOI] [PubMed] [Google Scholar]
- 216.Das S, Angsantikul P, Le C, Bao D, Miyamoto Y, Gao W, et al. Neutralization of cholera toxin with nanoparticle decoys for treatment of cholera. PLoS Negl Trop Dis. 2018;12:e0006266. doi: 10.1371/journal.pntd.0006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Danzig L. Meningococcal vaccines. Pediatr Infect Dis J. 2004;23:S285–S292. doi: 10.1097/01.inf.0000147654.03890.b9. [DOI] [PubMed] [Google Scholar]
- 218.Wang N, Qian R, Liu T, Wu T, Wang T. Nanoparticulate carriers used as vaccine adjuvant delivery systems. Crit Rev Ther Drug Carr Syst. 2019;36:449–484. doi: 10.1615/CritRevTherDrugCarrierSyst.2019027047. [DOI] [PubMed] [Google Scholar]
- 219.Yu F, Wang J, Dou J, Yang H, He X, Xu W, et al. Nanoparticle-based adjuvant for enhanced protective efficacy of DNA vaccine Ag85A-ESAT-6-IL-21 against Mycobacterium tuberculosis infection. Nanomedicine. 2012;8:1337–1344. doi: 10.1016/j.nano.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 220.Van Der Meeren O, Hatherill M, Nduba V, Wilkinson RJ, Muyoyeta M, Van Brakel E, et al. Phase 2b controlled trial of M72/AS01E vaccine to prevent tuberculosis. N Engl J Med. 2018;379:1621–1634. doi: 10.1056/NEJMoa1803484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Berzosa M, Pastor Y, Gamazo C, Irache JM. Development of a bacterial nanoparticle vaccine against Escherichia coli. Methods Mol Biol. 2022;2410:357–365. doi: 10.1007/978-1-0716-1884-4_18. [DOI] [PubMed] [Google Scholar]
- 222.Kheirollahpour M, Mehrabi M, Dounighi NM, Mohammadi M, Masoudi A. Nanoparticles and vaccine development. Pharm Nanotechnol. 2020;8:6–21. doi: 10.2174/2211738507666191024162042. [DOI] [PubMed] [Google Scholar]
- 223.Irvine DJ, Hanson MC, Rakhra K, Tokatlian T. Synthetic nanoparticles for vaccines and immunotherapy. Chem Rev. 2015;115:11109–11146. doi: 10.1021/acs.chemrev.5b00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sridhar S, Brokstad KA, Cox RJ. Influenza vaccination strategies: comparing inactivated and live attenuated influenza vaccines. Vaccines. 2015;3:373–389. doi: 10.3390/vaccines3020373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rosini R, Nicchi S, Pizza M, Rappuoli R. Vaccines against antimicrobial resistance. Front Immunol. 2020;11:1048. doi: 10.3389/fimmu.2020.01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Micoli F, Bagnoli F, Rappuoli R, Serruto D. The role of vaccines in combatting antimicrobial resistance. Nat Rev Microbiol. 2021;19:287–302. doi: 10.1038/s41579-020-00506-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Matić Z, Šantak M. Current view on novel vaccine technologies to combat human infectious diseases. Appl Microbiol Biotechnol. 2022;106:25–56. doi: 10.1007/s00253-021-11713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Hirosue S, Kourtis IC, van der Vlies AJ, Hubbell JA, Swartz MA. Antigen delivery to dendritic cells by poly(propylene sulfide) nanoparticles with disulfide conjugated peptides: cross-presentation and T cell activation. Vaccine. 2010;28:7897–7906. doi: 10.1016/j.vaccine.2010.09.077. [DOI] [PubMed] [Google Scholar]
- 229.Kaminskas LM, Porter CJH. Targeting the lymphatics using dendritic polymers (dendrimers) Adv Drug Deliv Rev. 2011;63:890–900. doi: 10.1016/j.addr.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 230.Leleux J, Atalis A, Roy K. Engineering immunity: modulating dendritic cell subsets and lymph node response to direct immune-polarization and vaccine efficacy. J Control Release. 2015;219:610–621. doi: 10.1016/j.jconrel.2015.09.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Singh B, Maharjan S, Cho KH, Cui LH, Park IK, Choi YJ, et al. Chitosan-based particulate systems for the delivery of mucosal vaccines against infectious diseases. Int J Biol Macromol. 2018;110:54–64. doi: 10.1016/j.ijbiomac.2017.10.101. [DOI] [PubMed] [Google Scholar]
- 232.Baker SJ, Payne DJ, Rappuoli R, De Gregorio E. Technologies to address antimicrobial resistance. Proc Natl Acad Sci USA. 2018;115:12887–12895. doi: 10.1073/pnas.1717160115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Rappuoli R, Bloom DE, Black S. Deploy vaccines to fight superbugs. Nature. 2017;552:165–167. doi: 10.1038/d41586-017-08323-0. [DOI] [PubMed] [Google Scholar]
- 234.Bilukha OO, Rosenstein N. Prevention and control of meningococcal disease. Recommendations of the Advisory Committee on Immunization Practices (ACIP) Morb Mort Wkly Rep Recomm Rep. 2005;54:1–21. [PubMed] [Google Scholar]
- 235.Delany I, Rappuoli R, Gregorio ED. Vaccines for the 21st century. EMBO Mol Med. 2014;6:708–720. doi: 10.1002/emmm.201403876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Khan O, Chaudary N. The use of amikacin liposome inhalation suspension (Arikayce) in the treatment of refractory nontuberculous mycobacterial lung disease in adults. Drug Des Devel Ther. 2020;14:2287–2294. doi: 10.2147/DDDT.S146111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Cipolla D, Blanchard J, Gonda I. Development of liposomal ciprofloxacin to treat lung infections. Pharmaceutics. 2016;8(1):6. doi: 10.3390/pharmaceutics8010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bricks LF, Berezin E. Impact of pneumococcal conjugate vaccine on the prevention of invasive pneumococcal diseases. J Pediatr (Rio J) 2006;82:S67–S74. doi: 10.2223/JPED.1475. [DOI] [PubMed] [Google Scholar]
- 239.Narang A, Chang RK, Hussain MA. Pharmaceutical development and regulatory considerations for nanoparticles and nanoparticulate drug delivery systems. J Pharm Sci. 2013;102:3867–3882. doi: 10.1002/jps.23691. [DOI] [PubMed] [Google Scholar]
- 240.Hua S, De Matos MBC, Metselaar JM, Storm G. Current trends and challenges in the clinical translation of nanoparticulate nanomedicines: Pathways for translational development and commercialization. Front Pharmacol. 2018;9:790. doi: 10.3389/fphar.2018.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Tinkle S, McNeil SE, Mühlebach S, Bawa R, Borchard G, Barenholz Y, Tamarkin L, Desai N. Nanomedicines: addressing the scientific and regulatory gap. Ann N Y Acad Sci. 2014;1313:35–56. doi: 10.1111/nyas.12403. [DOI] [PubMed] [Google Scholar]
- 242.Kumar Teli M, Mutalik S, Rajanikant GK. Nanotechnology and nanomedicine: going small means aiming big. Curr Pharm Des. 2010;16:1882–1892. doi: 10.2174/138161210791208992. [DOI] [PubMed] [Google Scholar]
- 243.Murday JS, Siegel RW, Stein J, Wright JF. Translational nanomedicine: status assessment and opportunities. Nanomed Nanotechnol Biol Med. 2009;5:251–273. doi: 10.1016/j.nano.2009.06.001. [DOI] [PubMed] [Google Scholar]