Abstract
The relevance of the mitogen-activated protein (MAP) kinase Hog1p in Candida albicans was addressed through the characterization of C. albicans strains without a functional HOG1 gene. Analysis of the phenotype of hog1 mutants under osmostressing conditions revealed that this mutant displays a set of morphological alterations as the result of a failure to complete the final stages of cytokinesis, with parallel defects in the budding pattern. Even under permissive conditions, hog1 mutants displayed a different susceptibility to some compounds such as nikkomycin Z or Congo red, which interfere with cell wall functionality. In addition, the hog1 mutant displayed a colony morphology different from that of the wild-type strain on some media which promote morphological transitions in C. albicans. We show that C. albicans hog1 mutants are derepressed in the serum-induced hyphal formation and, consistently with this behavior, that HOG1 overexpression in Saccharomyces cerevisiae represses the pseudodimorphic transition. Most interestingly, deletion of HOG1 resulted in a drastic increase in the mean survival time of systemically infected mice, supporting a role for this MAP kinase pathway in virulence of pathogenic fungi. This finding has potential implications in antifungal therapy.
Fungi, like all living organisms, must be able to respond to changes in environmental conditions and hence develop a response which enables their adaptation to the new physiological situation. Signal transduction pathways serve as a molecular mechanism to accomplish this cellular response. In Saccharomyces cerevisiae, a model eukaryotic cell system, some of these pathways involve members of the MAP kinase family (from mitogen-activated protein kinase), a set of enzymes performing essential functions in cell physiology first discovered in mammalian cells but later shown to be also present in lower eukaryotes (3, 12). Among these, the high-osmolarity glycerol (HOG) response pathway (6) allows adaptation to high-osmolarity conditions and seems to be especially important in terms of ecological adaptation. This latter route is triggered in response to high external osmolarity (i.e., low water activity) and results in the accumulation of glycerol as an intracellular compatible solute in S. cerevisiae. Several elements of this cascade have been identified in recent years (see reference 3 for a recent review). The initial triggering events in the cascade are initiated by at least two different pathways (43): the first one involves Sln1p (58), Ypd1 (65), Ssk1, and Ssk2p/Ssk22p kinases (44), while the second involves Sho1p, a putative membrane protein able to interact with, and activate, the Pbs2p MAP kinase kinase via Ste11p (43, 64) and possibly Ste20p/Ste50p (57). Both signals converge at the Pbs2p level, which in turn phosphorylates and activates Hog1p, which mediates the intracellular accumulation of osmolytes such as glycerol (4). This response also involves reorganizations of the cytoskeleton (11) and, presumably, cell wall modifications, as suggested by the involvement of PBS2 in β-(1,6)-glucan assembly (27, 35). Therefore, the HOG pathway participates in a pleiotropic response that enables a correct and rapid adaptation to osmotic stress. Functionally homologue-related cascades have been found in other fungal systems such as the fission yeast Schizosaccharomyces pombe. Interestingly, in this organism, this route not only is restricted to osmoadaptation but also links cell cycle control and sexual development (70, 71).
Candida albicans is a pathogenic yeast of great clinical interest in view of the increasing incidence of the infection that it causes in immunocompromised individuals (55). In addition, its ability to switch between a yeast-like and a hyphal form of growth has long been suspected to play a role in virulence (13, 30, 56). C. albicans has therefore been chosen as a model of pathogenic dimorphic fungi, although its diploid nature and lack of a sexual cycle have hampered molecular genetic studies, which have relied mostly on S. cerevisiae, a nonpathogenic yeast, as a host organism (62, 68). Knowledge of signal transduction pathways in pathogenic fungi is essential not only to understand their mechanisms of adaptation to a complex and changing environment such as the human body (and, therefore, their virulence) but also as a way to identify potential novel targets in antifungal therapy. In C. albicans, some genes homologous to the mating or pseudohyphal pathway genes (32, 36, 46, 72, 73, 77) or PKC1 pathway (53, 60) have been identified in recent years. We have previously described the isolation of the C. albicans gene homologue of the S. cerevisiae HOG1 gene (designated HOG1Ca, previously) and shown its involvement in osmoadaptation by increasing the internal glycerol content upon osmotic stress (67). In the present work, we characterize the phenotype of C. albicans hog1 mutants, showing their defects in the last stages of cytokinesis and cell wall biogenesis and repositioning certain elements of the budding machinery after osmotic stress. In addition, we show that the HOG pathway represses the serum-induced yeast-to-hypha transition in C. albicans and describe its role as a major determinant of virulence. These results suggest an additional role for this MAP kinase pathway in pathogenic fungi.
MATERIALS AND METHODS
Strains and growth conditions.
The yeast and bacterial strains used in this study are listed in Table 1. For clarity and unless otherwise stated, the designation hog1 will always indicate the homozygous hog1/hog1 Ura+ strain (strain CNC13). Although the genotypes of the strains were confirmed by Southern blot analyses, a control strain integrating the HOG1 gene at the LEU2 locus was constructed by homologous recombination using the restriction endonuclease KpnI, thus obtaining strain CNC15-10. Strain CNCH1 was obtained by integrating a p34H derivative (constructed by inserting a 1.59-kbp SspI-SspI fragment from YEP-HISX [63] into the HindII site of p34H [76]) containing the HIS1 marker at the HIS1 locus in the genome of CNC15-10 by using NruI as the restriction enzyme. Yeast strains were grown at 37°C (unless otherwise stated) in YED medium (1% yeast extract, 2% glucose) or SD minimal medium (2% glucose, 0.67% yeast nitrogen base without amino acids) with the appropriate auxotrophic requirements (50 μg/ml). The ability of cells to undergo the yeast-to-hypha transition was tested by using Lee’s medium at pH 6.7 (38), SD medium plus 10% (vol/vol) fetal bovine serum, fetal bovine serum, Spider medium (1% mannitol, 1% nutrient broth, 0.2% K2HPO4, 1.35% agar) (39), SLAD medium (21), or YED medium plus fetal bovine serum at 1, 5, 10, and 20% as well as whole serum. To check the behavior of C. albicans strains with respect to dimorphic transition, cells were inoculated at 105 cells/ml in prewarmed liquid medium. Growth in liquid medium was estimated as the absorbance at 600 nm (A600) or dry weight; in this case, 10 ml of the culture was filtered with a 0.45-μm-pore-size filter (Millipore) and dried at 42°C until a stable weight had been attained. Time lapse photography was performed, with images taken at defined intervals with cells deposited onto a thermostabilized chamber at 37°C containing solid yeast extract-peptone-dextrose (YEPD) medium supplemented with 0.75 M NaCl.
TABLE 1.
Strains used in this study
| Microorganism | Strain | Genotype | Source |
|---|---|---|---|
| E. coli | DH5αF′ | K-12 Δ(lacZYA-argF)U169 supE44 thi1 recA1 endA1 hsdR17 gyrA relA1 (φ80lacZΔM15) F′ | 23 |
| S. cerevisiae | L5366 | MATa/MATα ura3Δ52/ura3Δ52 | 33 |
| YPH499 | MATa ura3 leu2 his3 trp1 lys2 ade2 | M. Gustin | |
| JBY10 | MATa ura3 leu2 his3 trp1 lys2 ade2 hog1-Δ1::TRP1 | M. Gustin | |
| W303 | MATα ura3 leu2 his3 trp1 ade2 can1 | M. Gustin | |
| J134 | MATα ura3 leu2 his3 trp1 ade2 can1 hog1::LEU2 | M. Gustin | |
| C. albicans | SC5314 | Wild type | 20 |
| CAl4 | ura3Δ::imm434/ura3Δ::imm434 | 18 | |
| RM100 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG-URA3-hisG | 54 | |
| RM1000 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG | 54 | |
| CNC11 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG HOG1/hog1::hisG-URA3-hisG | 67 | |
| CNC12 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG HOG1/hog1::hisG | 67 | |
| CNC13 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG hog1::hisG-URA3-hisG/hog1::hisG | 67 | |
| CNC15 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG | This work | |
| CNC15-10 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/his1Δ::hisG hog1::hisG/hog1::hisG LEU2/leu2::HOG1 URA3 | This work | |
| CNCH1 | ura3Δ::imm434/ura3Δ::imm434his1Δ::hisG/HIS1 hog1::hisG/hog1::hisG LEU2/leu2::HOG1 URA3 | This work |
Molecular biology procedures and plasmid constructions.
Standard molecular biology procedures were used (2). C. albicans was transformed as described previously (31). The plasmid YEp352 (a URA3 2μm-derived vector), pHOG1c24.2 (the C. albicans HOG1 gene in YEp352), and pJB30 (the S. cerevisiae HOG1 gene in a 2μm-derived vector) have been described previously (25, 67). The pRM-HOG1 plasmid, an episomic plasmid carrying the C. albicans HOG1 gene and the 5′ regulatory regions, was obtained by inserting a HindIII fragment from pHOG1c24.2 into the SmaI site of pRM1 (63).
Confocal microscopy, flow cytometry, and fluorescent staining methods.
Cells grown in YED medium plus 1 M NaCl were washed twice with 0.2 M NaCl and stained with primuline (Sigma, St. Louis, Mo.) at 50 ng/ml (final concentration) for 30 min at 37°C or calcofluor white (Bayer) at 4 ng/ml (final concentration) for 5 min at room temperature. Cells were briefly sonicated before the fluorescence was quantified with a Bio-Rad Bryte HS flow cytometer (for calcofluor white) or a Becton Dickinson (San José, Calif.) FACScan flow cytometer (for primuline). The same procedure was used for visualization of chitin under a Bio-Rad MRC 1000 confocal microscope. For analyses of DNA content, exponentially growing cells were transferred to YED medium supplemented or not with 1 M NaCl at 37°C. Aliquots were removed at defined intervals, collected by low-speed centrifugation, washed with phosphate-buffered saline (PBS), and resuspended in cold 70% ethanol for 1 min. They were then washed twice with PBS, resuspended in 500 μl of PBS containing 1 mg of RNase/ml, and incubated for 30 min at 37°C. Cells were briefly sonicated, washed twice with PBS, and stained with propidium iodide (Sigma) at a final concentration of 0.005% in PBS, and the DNA content was analyzed with a flow cytometer. Cell viability was assessed by staining the cells with propidium iodide at 0.005% in PBS.
Electron microscopy.
Cells growing in YED medium at 37°C were transferred to YED medium supplemented with 1 M NaCl, and samples were taken at different times. Cells for scanning electron microscopy were prepared as described previously (75, 78) and visualized with a JEOL JSM-6400 microscope. Transmission electron microscopy samples were obtained as described previously (49) and embedded in Epon 812. Eighty-nanometer-thick sections were observed through a Zeiss 902 microscope from the Centro de Microscopía Electrónica Luis Bru (Universidad Complutense de Madrid [UCM]).
Antifungal assays.
MICs were determined by the microdilution method in 96-well plates as described elsewhere (51, 52) by using SD medium without uridine. Drop tests were used to check susceptibility to Congo red and calcofluor white. They were performed by spotting 105, 104, 103, and 102 cells (in a 10-μl volume) onto YEPD solid medium plus Congo red or calcofluor white at 100, 120, and 150 μg/ml and incubated for 24 h at 24, 30, and 37°C for S. cerevisiae or at 30, 37, and 42°C for C. albicans.
Chitinase activity assays.
Chitinase assays were carried out as described before (34). Hydrolysis of the substrate was determined after 1 h of incubation at 30°C. Units of activity are defined as nanomoles of 4-methylumbelliferone (the fluorescent product of hydrolysis of the substrate by chitinase) released per hour.
Virulence assays.
Virulence assays were performed essentially as described previously (17).
RESULTS
Structural alterations in C. albicans hog1 mutants.
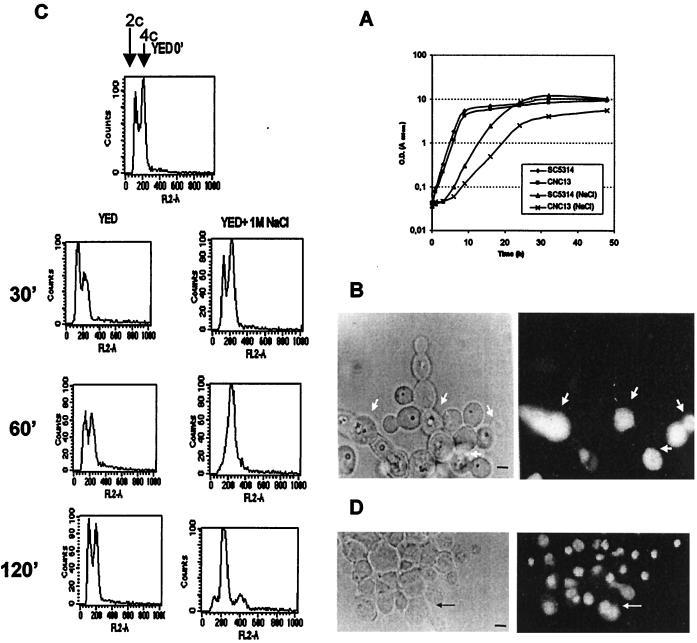
To characterize the influence of high solute concentrations on the structure and morphology of C. albicans and the role of the HOG pathway in C. albicans under these conditions, we performed a detailed characterization of the alterations of wild-type and hog1 mutants. For this purpose, exponentially growing cells in YED medium were diluted and transferred to hyperosmolar conditions (1 M NaCl) and both the optical density (OD) and dry weight were measured at regular intervals. This shift to higher osmolarity caused both wild-type and hog1 cultures to arrest growth (approximately 3 h for the wild type and 5 h for the mutants). Both wild-type and hog1 strains resumed growth, with subsequent doubling times of 2 h for the wild type and 5 h for the hog1 mutants (Fig. 1A). Mutant cells achieved a lower final OD (5.5 versus 10.1) or mass (4.5 versus 8.0 mg/ml) after 48 h of growth and entry into stationary phase. These results indicate that the high osmolarity-induced growth arrest is transient and that despite being osmosensitive and failing to accumulate glycerol intracellularly (67), hog1 mutants are still able to grow in mass.
FIG. 1.
Terminal phenotype of hog1 mutants. (A) Effect of osmostress (1.0 M NaCl) on the growth of wild-type cells (SC5314) or the hog1 mutant (CNC13) in liquid YED medium. The OD (estimated as the A600) is plotted versus time. The y-axis scale is logarithmic. (B) Propidium iodide-stained cells of the hog1 mutant after 24 h of growth under restrictive conditions (right panel). Arrows indicate dead cells in the clusters observed under phase-contrast microscopy (left panel). (C) Flow cytometric analysis of the DNA content of mutant cultures grown in parallel on YED medium or YED medium plus 1 M NaCl. The peaks observed for control cells (labeled YED 0′) represent 2n (left) and 4n (right) DNA content, while the numbers indicate the time in minutes after the transfer to the restrictive conditions. (D) Microscopic analysis of hog1 mutant cells under restrictive conditions (left panel; phase-contrast image), showing nuclei (right panel; fluorescence image). The arrow indicates a detail of a nucleus in the process of segregation to the new bud. A representative cluster of cells is shown. Bars, 1 μm.
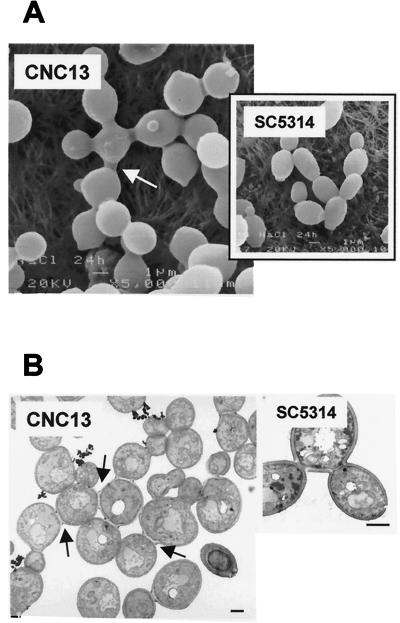
Cell viability was determined by both flow cytometry quantification of dead cells stained with propidium iodide (a fluorochrome able to enter dead cells due to the loss of selective permeability) (16) and direct standard plating (CFU counting) on YED medium (data not shown). Visual microscopic examination was also used to monitor the behavior of the culture. No obvious differences were observed in the morphology of the wild-type or hog1 cells at 2 or 4 h after the change to the 1 M NaCl medium. However, after 24 h, individual mutant cells appeared rounder and smaller than wild-type cells (as determined by flow cytometry). At the same time, several mutant cells remained attached after budding due to an apparent defect in cell separation (Fig. 1B, left panel), a phenotype not observed in the wild-type strain (data not shown). Consistent with the previous data on growth, cell viability in the hog1 culture was high (more than 75% after 24 h under hyperosmotic conditions). Dead cells revealed by propidium iodide uptake had no specific localization within the clusters of cells (Fig. 1B, right panel). The DNA content was also quantified. Under nonrestrictive conditions (YED medium alone), hog1 cells displayed a DNA pattern characteristic of an asynchronous culture (Fig. 1C, left panels), while on YED medium plus 1 M NaCl, the peak containing a 2n DNA content disappeared in 60 min, and after this period, only 4n cells were detected (Fig. 1C, right panels). After this period, the DNA content increased as the result of normal DNA replication but failure of the cells to segregate, therefore appearing as a single cytometric count. Cells under the microscope appeared to have a single nucleus (Fig. 1D). The observed increase in DNA content therefore seems to be the result of impaired cell division and not sensitivity to the high osmolarity of normal DNA replication. This effect was not observed under permissive conditions or in osmostressed wild-type cells (data not shown). Interestingly, some of the hog1 cells also displayed altered budding patterns in which the polarity of bud emergence was lost, some cells budding away from the distal poles (Fig. 2A). Scanning transmission microscopy showed that daughter cells were apparently completely formed but remained attached to the mother cell, unable to complete cytokinesis (Fig. 2A). Transmission electron microscopy (Fig. 2B) revealed that the septum between the mother and daughter cells was physically completed and that the outer cell wall still connected both cells. Sonication for different periods of time or digestion with glusulase [a β-(1,3)-glucanase enriched lytic preparation] did not result in segregation of these clusters, but interestingly, treatment with a commercial preparation of chitinase did (data not shown).
FIG. 2.
Scanning and transmission electron microscopy of hog1 mutants. (A) Scanning electron microscopy of hog1 mutant (CNC13) (left panel) or wild-type (SC5314) (right panel) cells after 24 h of growth under restrictive conditions. The arrow indicates a characteristic cell with a symmetric type of division. (B) Transmission electron microscopy of similar samples. Arrows indicate almost completely separated but still connected cells with well-formed septa. Bars, 1 μm.
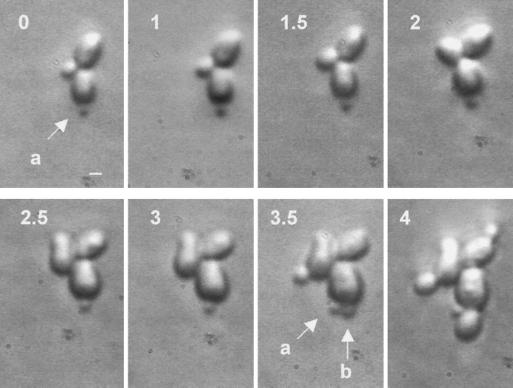
The influence of osmostressing conditions on polarity was investigated in more detail. For this purpose, growing cells were plated onto YEPD solid medium supplemented with 0.75 M NaCl and photographs were taken at different intervals. While the wild-type strain was able to grow normally after a short time of adaptation to the new physiological situation (data not shown), hog1 mutant cells stopped growing for a prolonged time. In this mutant, a small percentage of the population displayed a defect which consisted in small newly formed buds ceasing growth while mother cells emitted a new bud that completed its growth. By contrast, during the same period, the first bud did not resume growth at all (Fig. 3). These observations are consistent with the idea that certain components of the bud positioning or emergence machinery (but not DNA replication) are dependent on a functional HOG pathway (7).
FIG. 3.
Defects in bud site selection after osmotic shock. Time lapse photography of hog1 mutant cells under solid YPD medium supplemented with 0.75 M NaCl. Numbers indicate the time (in hours) after the transfer to restrictive conditions. Arrows labeled “a” indicate small buds, while the arrow labeled “b” indicates a newly formed bud. Bars, 1 μm.
C. albicans hog1 mutants are resistant to certain cell wall inhibitors.
The susceptibility of C. albicans hog1 mutants to antifungals with different structures and mechanisms of action was determined. No differences were found between wild-type and hog1 cell susceptibilities to the following antifungals under nonosmostressing conditions: cilofungine (an inhibitor of β-glucan biosynthesis; MIC, 1 μg/ml), trichodermine (an inhibitor of protein synthesis; MIC, 1 μg/ml), fluconazole and miconazole (inhibitors of ergosterol biosynthesis; MICs, 2.5 and 1 μg/ml, respectively), canavanine (a toxic amino acid analog; MIC, 6.25 μg/ml), 5-fluorocytosine (an inhibitor of nucleic acid synthesis; MIC, 0.0625 μg/ml), or amphotericin B (inhibitor of membrane functionality; MIC, 2.9 μg/ml). However, a drastic difference was observed in the susceptibilities of hog1 and wild-type cells to nikkomycin Z, an inhibitor of chitin biosynthesis. When assayed at 30°C, both wild-type and mutant cells displayed high levels of resistance to nikkomycin Z (MIC, >800 μg/ml). However, when susceptibility was assayed at 37 or 42°C, the wild type became sensitive (nikkomycin Z MICs, 3.12 μg/ml at 37°C and 0.78 μg/ml at 42°C) but the hog1 strain remained resistant to nikkomycin Z (MIC, >800 μg/ml). The MICs of nikkomycin Z for the heterozygous HOG1/hog1 mutant and the wild-type strain, SC5314, were the same. It should be noted that these effects were observed under normal conditions, i.e., nonosmostressed cells.
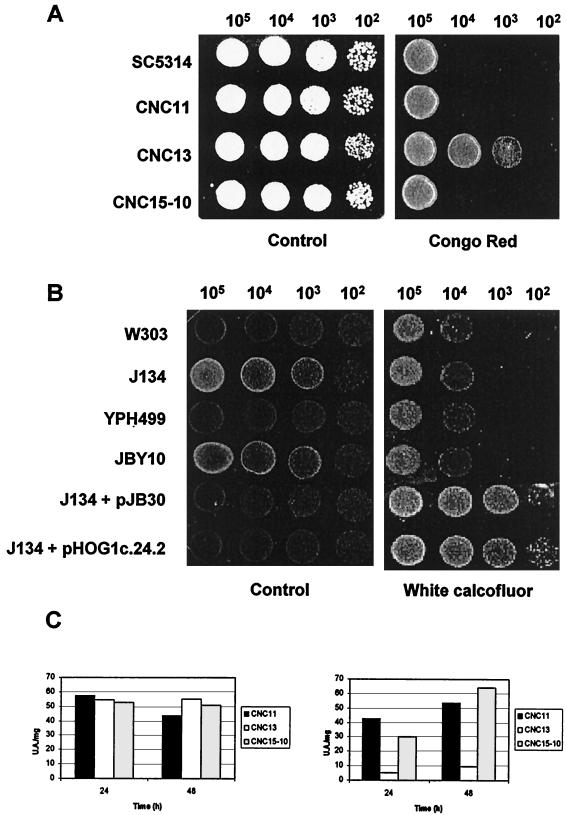
In addition, C. albicans hog1 cells were consistently more resistant than the corresponding heterozygous or wild-type strains to Congo red, a dye which also interacts with the fungal cell wall (Fig. 4A) at all the temperatures tested, and very slightly to calcofluor white (data not shown). These differences were not found in S. cerevisiae hog1 mutants in two different backgrounds. Unexpectedly, overexpression of the C. albicans HOG1 gene (or the homologous S. cerevisiae HOG1 gene) in S. cerevisiae by using an episomal vector also resulted in increased resistance to these compounds, especially to calcofluor white (Fig. 4B).
FIG. 4.
Antifungal susceptibility and cell wall architecture. (A) Different amounts of cells (indicated at the top of the rows) from the indicated C. albicans strains were spotted onto YEPD medium (as a control) or YEPD medium supplemented with Congo red at 150 μg/ml and incubated at 37°C. (B) Experiments similar to those described for panel A were done with the S. cerevisiae strains indicated; cells were spotted onto YEPD medium supplemented with a 150-μg/ml final concentration of calcofluor white and incubated at 30°C for 24 h. (C) Chitinase activity in cell extracts after 24 and 48 h of growth under nonrestrictive (1 M NaCl) (left panel) and restrictive (right panel) conditions. Units of activity (UA) (see Material and Methods) per milligram of dried extract are given in the y axis. Data are the mean value of two independent experiments.
To further explore the relationship between the cell wall and the HOG1 gene, the chitin levels in both wild-type and mutant strains under restrictive (1.0 M NaCl) and nonrestrictive conditions were quantified by flow cytometry with both calcofluor white and primuline, but no significant variation in the chitin content was observed. In addition, confocal microscopy analyses showed the predicted chitin accumulation on scars and mother-bud necks (data not shown). In view of the presence of a well-formed septum between mother and bud but incomplete cytokinesis in mutant cells, we measured chitinase activity in osmostressed cells. As shown in Fig. 4C, a significant reduction in the enzymatic activity of chitinase was observed in total cell extracts. These results suggest that the cell separation defects observed in this mutant could be the result of defective chitinase activity. Chitinase activity has been shown, in fact, to be required for cell separation in S. cerevisiae (34), although no dependence on the HOG pathway has been described. Collectively, our results suggest a link between cell wall metabolism and the HOG pathway in C. albicans.
A role for HOG1 in morphological transitions.
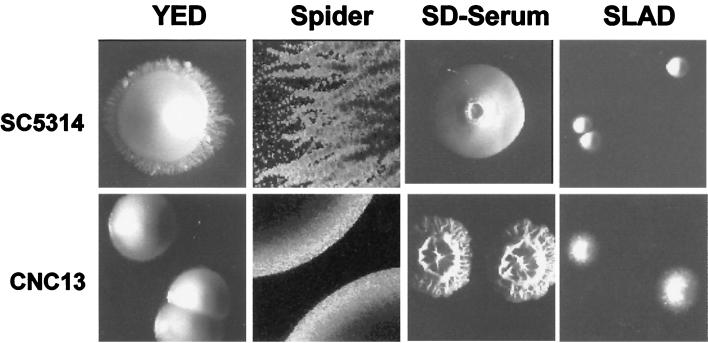
An important biological question to be addressed in C. albicans is its ability to undergo the dimorphic transition, a cell differentiation program that allows yeast cells to generate hyphal forms. Dimorphism, which has long been suspected to play a role in C. albicans pathogenesis (see reference 56 and references therein), can be induced by environmental factors such as the pH or the temperature or can be induced in response to serum, proline, or N-acetylglucosamine. To analyze the role of HOG1 in these transitions, we observed the colony morphology of hog1 cells under different growth conditions. First, on normal YED plates, hog1 cells did not show the limited agar invasion displayed by SC5314 wild-type cells (Fig. 5). A similar difference was also found when both strains were plated in Spider medium, which has been described as inducing hyphal formation (39). Mutant colonies, although able to invade the agar, appeared smooth with small grains, while wild-type colonies showed clear invasive borders. However, the mutant strain observed under an optical microscope appeared as large filaments, similar to the wild type. Most interestingly, mutant cells grown on a nitrogen limiting medium such as SLAD medium (which has been shown to induce pseudohyphal formation in S. cerevisiae [21]) penetrated the agar medium and were hence more invasive than wild-type cells (Fig. 5), frequently appearing under the microscope as short filaments or pseudohyphae. None of these effects were observed in the heterozygous strain (strain CNC11) or the strain in which the wild-type HOG1 gene was reintroduced (data not shown).
FIG. 5.
Colony morphology of hog1 mutants. Colony morphology of wild-type (SC5314) and mutant hog1 (CNC13) cells on different solid media. Approximately 50 CFU were spread onto either YED medium, Spider medium, minimal SD medium plus 10% bovine fetal serum, or SLAD medium on petri dishes and incubated for 7 days at 37°C before photographs were taken. The colony borders are shown for cells on Spider medium.
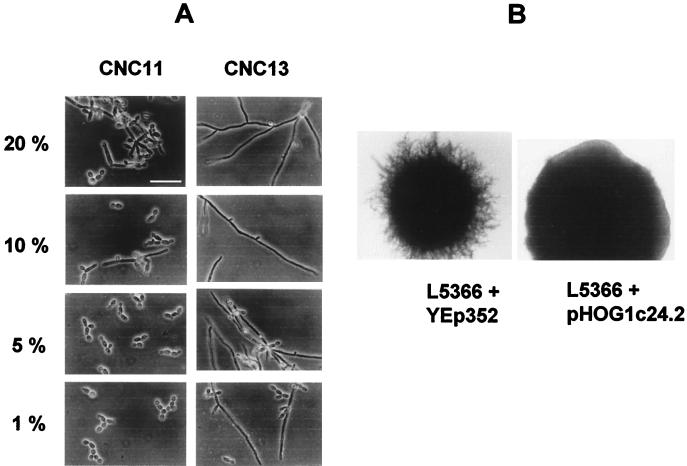
To explore further the filamentation in hog1 cells, we analyzed their behavior in the true dimorphic transition. When assayed on liquid media that induce hyphal formation, such as Lee’s medium at pH 6.7 or serum, no significant differences were found between the mutant (CNC13), the heterozygous (CNC11 and CNC15-10), and the wild-type (SC5314) strains. However, we performed experiments in which the cells were exposed to limiting serum concentrations. On 100% serum, both mutant and wild-type cells generated long filaments and no differences could be observed with respect to the timing of appearance of the germinative tubes. In contrast, on YED medium containing 1, 5, and 10% serum, hog1 mutant cells (strain CNC13) displayed a clear filamentous phenotype, with several cells appearing as long true polynucleated filaments with several septa, the frequency of this occurrence in wild-type cells (data not shown) or the heterozygous strain (CNC11) was much lower (Fig. 6A). Consistent with this, on solid medium containing 10% bovine fetal serum, hog1 cells (strain CNC13) generated wrinkled colonies (Fig. 5) with frequent invaginations towards the inner regions of the colony that were absent in the wild-type cells. This presumably regulatory (repressive) role of HOG1 in morphological transitions was also evidenced by the suppression of pseudohyphal growth in S. cerevisiae. Overexpression of the C. albicans HOG1 gene from a multicopy plasmid partially suppressed the pseudodimorphic transition (invasion) of the diploid S. cerevisiae strain L5366 (Fig. 6B) on nitrogen-deprived medium (SLAD medium). These results clearly support the regulatory role of the HOG pathway in morphogenetic programs in C. albicans.
FIG. 6.
Effect of Hog1p on the serum-induced dimorphic transition. (A) Cells from the indicated strains were inoculated in YED medium plus bovine fetal serum at different concentrations (20, 10, 5, and 1%), and phase-contrast microphotographs were taken after 6 h of incubation at 37°C. (B) Border colony morphologies of S. cerevisiae L5366 transformed with vector YEp352 (left picture) or the multicopy plasmid pHOG1c24.2 (bearing the C. albicans HOG1 gene) (right picture) are shown. Cells were plated onto SLAD medium, and pictures were taken after growth for 6 days at 30°C. Bars, 10 μm.
Virulence of hog1 cells in a mouse model.
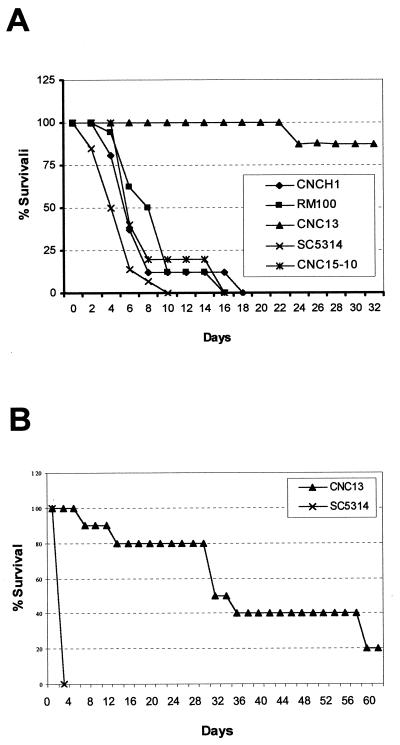
To analyze the role of the HOG pathway in virulence, we checked the behavior of hog1 cells by use of a mouse model of fungal infection. Both BALB/c and DBA/2 mice were challenged with different doses of the parental (SC5314) and a hog1 mutant (CNC13) strain by inoculation into the lateral vein of the tail. These two mouse strains have been shown to differ in their susceptibility to fungal infections, BALB/c mice being more resistant than DBA/2 mice (24). Standard death curves were obtained after the infection, and representative death curves are shown in Fig. 7. In BALB/c mice, a challenge with a dose of 106 cells resulted in a rapid mortality for the wild-type strain (mean survival time [MST] of 3 days). In contrast, mice challenged with hog1 cells showed a drastic decrease in mortality, being able to survive up to 60 days (Fig. 7A). These differences were also observed for mice given a larger inoculum (107 blastospores) (Fig. 7B), with MSTs of 1 day for the wild-type strain and 30 days for the mutant (see also Table 2). A lower dose (105 blastospores) did not lead to any differences in the mortality of mice challenged with either yeast strain.
FIG. 7.
Virulence assays. Standard survival curves of BALB/c mice infected systemically with 106 (A) or 107 (B) cells of the C. albicans strains indicated in the figure. Since strains CNC15-10, RM100, and CNCH1 at a dose of 107 gave curves similar to the one shown for SC5314 in panel B, these results are therefore not shown for clarity.
TABLE 2.
Quantification of virulence in experimental infection assays
| Challenge dose | C. albicans strain | BALB/c mice
|
DBA/2 mice
|
||
|---|---|---|---|---|---|
| MST (days) | No. of dead mice/total no. | MST (days) | No. of dead mice/total no. | ||
| 1 × 107 | SC5314 | 1 | 15/15 | 1 | 8/8 |
| CNC13 | 30 | 8/10 | 1 | 13/13 | |
| 1 × 106 | SC5314 | 3 | 14/14 | 1 | 14/14 |
| CNC13 | >60 | 1/8 | 4 | 11/11 | |
| 1 × 105 | SC5314 | >60 | 3/16 | 4 | 17/17 |
| CNC13 | >60 | 0/12 | >60 | 1/12 | |
| 5 × 104 | SC5314 | >60 | 3/12 | 7 | 12/12 |
| CNC13 | >60 | 0/11 | >60 | 1/17 | |
DBA/2 mice were similarly infected. In this case, as expected, differences at the 107 cell dose were not observed due to the increased susceptibility of DBA/2 mice to C. albicans infections, both strains producing a high mortality. However, differences in MST were observed for the mutant and wild type at a challenge dose of 106 cells (Table 2), and the use of this mouse strain allowed us to detect differences between both C. albicans strains when a small inoculum dose (105) was used (Table 2). In all these experiments, ura3 auxotrophic strains were avoided due to the effect that this nutritional requirement (especially in certain genetic backgrounds) has on C. albicans virulence (29, 69). However, since the hog1 knockout strains were obtained in a his1 background (RM1000), it was confirmed that HIS1 did not play a role in the observed reduction in virulence, as shown for the control strains RM100 and CNCH1 (Fig. 7A). In addition, the hog1 heterozygous strain CNC15-10 (a strain obtained through the reintroduction of the HOG1 gene in the genome of strain CNC15 [see Materials and Methods] to serve as an internal control of the knockout deletion scheme used) displayed virulence similar to that of wild-type (SC5314) cells (Fig. 7A), indicating that a single copy of the HOG1 gene is enough to restore full virulence in this animal model. The fungal burden was quantified in the kidney and brain, representative organs of C. albicans infections (59). Organs were recovered at different times postinfection, and viable cells were quantified. As shown in Table 3, strain CNC13 colonizes tissues less efficiently than SC5314 does and it is cleared from the brains of BALB/c mice in a few days (i.e., 7 days, even with the high dose of 106 cells). Collectively, these data indicate that a functional HOG pathway is essential for the maintenance of full virulence in C. albicans.
TABLE 3.
Virulence in a systemic mouse model of experimental infection in BALB/c and DBA/2 mice
| Challenge dose | Strain | Timea | Fungal burden (log CFU)b in:
|
|||
|---|---|---|---|---|---|---|
| BALB/c mice
|
DBA/2 mice
|
|||||
| Kidney | Brain | Kidney | Brain | |||
| 1 × 107 | SC5314 | 1 | 6.03 ± 0.84 | 4.47 ± 0.13 | NTc | NT |
| CNC13 | 1 | 4.39 ± 0.24 | 3.24 ± 0.2 | 5.27 ± 0.21 | 3.98 ± 0.13 | |
| 1 × 106 | SC5314 | 1 | 5.93 ± 0.18 | 4.04 ± 0.22 | 6.02 ± 0.92 | 3.81 ± 0.16 |
| CNC13 | 1 | 3.26 ± 0.24 | 1.89 ± 0.31* | 4.43 ± 0.09 | 3.16 ± 0.13 | |
| 7 | 3.94 ± 1.13 | * | ||||
| 60 | 5.11 ± 0.98** | * | ||||
| 1 × 105 | SC5314 | 1 | 3.61 ± 0.10 | 3.34 ± 0.36 | 5.17 ± 0.76 | 3.70 ± 0.19 |
| 7 | 4.10 ± 0.84 | * | ||||
| 60 | 4.32 ± 1.51* | * | ||||
| CNC13 | 1 | 2.53 ± 0.21 | 1.78 ± 0.00** | 3.76 ± 0.08 | 2.35 ± 0.18 | |
| 7 | * | * | 3.60 ± 0.98* | 3.34 ± 0.00** | ||
| 60 | 4.98 ± 0.00*** | * | 4.35 ± 0.35** | * | ||
| 5 × 104 | SC5314 | 7 | 6.45 ± 0.56 | 4.53 ± 0.50 | ||
| 60 | 4.87 ± 0.96* | * | ||||
| CNC13 | 1 | * | * | 3.71 ± 0.73 | 3.66 ± 0.00** | |
| 7 | * | * | * | * | ||
| 60 | * | * | 3.92 ± 0.00** | * | ||
Day postchallenge (up to MST) when mice were killed by cervical dislocation for CFU quantification.
Only log CFU values that are statistically significantly different at P = 0.003 (kidney) and P < 0.0001 (brain) are shown (analysis of variance test with C. albicans strain; covariates were time, mice, and dose). *, some or all mice cleared infection; **, more than 50% mice cleared infection.
NT, not tested.
DISCUSSION
In this work, we have addressed the role of the HOG pathway through the characterization of the phenotype of C. albicans mutants defective in the central MAP kinase gene of this pathway, the HOG1 gene, under both restrictive and permissive conditions. DNA replication was insensitive (at least for the period analyzed) to high osmolarity, and the increased DNA content detected by flow cytometry was the result of a block in cytokinesis, similar to that in S. cerevisiae mutants (7). We also found that the defects in cell separation are localized to the last stages of cytokinesis but are, apparently, not due to abnormal septum formation (Fig. 3A and B); instead, we show that chitinase activity is low and may be limiting under these conditions. This is supported not only by the enzymatic analysis of osmostressed cells but also by the effect that externally added chitinase (and not other cell wall lytic enzyme preparations such as zymolyase or glusulase) exerts on cell separation. Also, certain components of the bud polarity machinery appear to be nonfunctional in the mutant cells under restrictive conditions, a phenomenon similar to that of S. cerevisiae, where transfer of the cells to high-osmolarity conditions often results in the selection of a new polarization region for bud emergence to occur (7).
An interesting conclusion from our studies is the suggestion of a link between cell wall metabolism and the HOG pathway. This is inferred from the resistance to compounds which interact with the cell wall of the mutant strain in ways different from those of the wild type. A possible explanation for this result is that hog1 mutants are altered in their permeability by certain compounds. In fact, nikkomycin Z is a nucleoside-peptide antibiotic inhibitor of chitin synthase (8, 19) which is imported into the cell through a peptide transport system (48, 61, 79). Alterations in membrane permeability cannot, in principle, be excluded, although these effects are also obtained with dyes such as calcofluor white or Congo red, which show affinity for external cell wall polymers. The HOG pathway could, therefore, play an as-yet-undefined role in cell wall metabolism. Such a relationship has, in fact, been postulated to occur in S. cerevisiae, since PBS2 (the HOG1 MAP kinase kinase gene) may regulate β-(1,6)-glucan assembly (27, 35). No defects in chitin synthase activity (35) were observed in this study in pbs2Δ mutants, in agreement with our results on quantification of the chitin content in hog1 mutants.
Another aspect of biological relevance that we investigated is dimorphism, a long-suspected mechanism of virulence (see references 13 and 30). The repressive effect that this pathway exerts on pseudohyphal formation is evidenced by the hyperfilamentous phenotype of C. albicans strains on different media such as SLAD medium (21) (a similar phenotype has been observed for S. cerevisiae hog1 mutants [cited in reference 42]) as well as by the suppression of the pseudodimorphic transition in S. cerevisiae when the C. albicans HOG1 gene is overexpressed. Alterations in the colony morphology on different solid media may also support this observation, although the apparently contradictory results observed could be explained by the involvement of different signal transduction pathways in these processes. Alterations in colony morphology are also observed in response to serum, and the conditions used in this assay (1 to 20% serum versus 100% serum) may better reflect the complex environmental conditions that a pathogen finds inside the human body, where different locations may have different concentrations of an inducer(s). Our results indicate the repression that the HOG pathway exerts on the serum-induced dimorphic transition in C. albicans. Given our current knowledge of signal transduction pathways mediated through MAP kinases, it is tempting to speculate about the final targets of this cascade. In C. albicans, hyphal formation seems to be a complex process in which both positive and negative signals do play a role (see reference 45 for an elegant model). For example, the HOG pathway could be involved in the repression of the pathway that leads to a pseudofilamentous or filamentous growth pathway in C. albicans, interacting with those elements of the mating-hyphal pathway presumably involved in dimorphic transition. In fact, in S. cerevisiae, elements of the mating pathway are used for pseudofilamentous growth (32, 36, 40), and it has been shown that the HOG pathway represses the activity of the mating pathway in S. cerevisiae (22). Furthermore, recent studies in S. cerevisiae (57) reveal that HOG1 prevents cross talking between both the mating and HOG pathways. The repressive role of the HOG pathway in C. albicans hyphal formation could be its involvement in the activation of RBF1, a transcription factor whose deletion generates hyphal forms (26), or, alternatively, a putative SSN6-TUP1 complex in C. albicans. In fact, it has been recently shown that the S. cerevisiae Ssn6-Tup1 repressor complex (28) plays a role in the repression of different osmolarity-inducible genes in S. cerevisiae (some of which are HOG1 dependent) and that ssn6 or tup1 mutants partially suppress the characteristic osmotic sensitivity associated with hog1 mutants (47). More interestingly, the C. albicans TUP1 gene has been shown to play a role in hyphal formation in C. albicans since deletion of this gene results in a gene dosage-dependent filamentous growth (5). A possible explanation for our results would be the HOG1-dependent expression or activation of a DNA binding protein able to recruit the Ssn6-Tup1 complex for the repression of specific hyphal genes. In any case, these similarities must be analyzed carefully because of the pleiotropic role of transcription factors like TUP1 in fungal cell physiology and the fact that phenotypes associated with S. cerevisiae and C. albicans mutants may clearly diverge, as occurs with tup1 mutants (5).
It is noteworthy that other elements of this pathway have been found to play a role in hyphal development. For example, deletion of the nik-1+ gene, a Neurospora crassa homologue of the SLN1 gene (58), results, in addition to osmotic sensitivity, in restricted mycelial growth, the loss of conidiophore development, and in aberrant hyphal structures under restrictive conditions (1). Recently, two-component C. albicans kinase gene homologues of SLN1 have been identified (10, 50, 74) and effects on the efficiency of the transition process have been observed (74).
Our results also demonstrate that the HOG pathway plays a major role in C. albicans virulence. The genes involved in virulence currently identified are functionally diverse, probably reflecting the character of a commensal opportunistic pathogen instead of a primary pathogen of C. albicans. Dimorphism provides a morphological switch that has been related to certain features undoubtedly related to pathogenicity, such as adhesion, escape from phagocytic cells, and invasion (9, 30, 56, 66). It is therefore not surprising that strains defective in hyphal formation (under certain conditions) should display a reduced virulence, as has been shown for some signal transduction protein kinases (15, 36, 37). However, although the role that hyphal formation must play in virulence is evident, our results clearly demonstrate that this trait is not enough for virulence, since a functional (and even enhanced) in vitro hyphal development does not necessarily correlate with virulence in this animal model, as suggested recently (14). It should be emphasized that both the dose of cells used in the virulence experiments and the length of the period in which mice infected were followed up indicate the complete avirulence of the mutant strain, in comparison with the standard defined in other recent studies (14, 41), and suggest that the HOG pathway participates in other as-yet-unraveled cellular processes which are essential for virulence. Although it is difficult to define what this role is at this stage, it is tempting to speculate that Hog1p behaves like a general stress kinase, similar to the S. pombe homologue, and that this cellular response is essential for the successful establishment of an infection in the host. In conclusion, we show in this work that the pathway(s) controlling osmotic sensitivity in C. albicans also plays a role in differentiation programs and virulence in this pathogenic fungus, a result which identifies this route of primary importance in the search for novel antifungal targets.
ACKNOWLEDGMENTS
We thank Alistair J. P. Brown for generously providing strain L5366. Calcofluor was a generous gift from Bayer. The excellent assistance of A. Vázquez and A. Álvarez from the Centro de Citometría de Flujo y Microscopía Confocal of the UCM and M. J. Asensio Vela is acknowledged. Electron microscopy was carried out at the Centro de Microscopía Electrónica “Luis Bru” of the UCM. We also thank M. Molina for critical reading of the manuscript.
R. Alonso Monge is recipient of a fellowship from the Comunidad Autónoma de Madrid. This work was supported by FIS grant SAF96-1540 and by grant FISS97/0047-01.
REFERENCES
- 1.Alex L A, Borkovich K A, Simon M I. Hyphal development in Neurospora crassa: involvement of a two-component histidine kinase. Proc Natl Acad Sci USA. 1996;93:3416–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Kingston R E, Brent R, et al., editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1993. [Google Scholar]
- 3.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomberg A, Adler L. Physiology of osmotolerance in fungi. Adv Microb Physiol. 1992;33:145–212. doi: 10.1016/s0065-2911(08)60217-9. [DOI] [PubMed] [Google Scholar]
- 5.Braun B R, Johnson A D. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science. 1997;277:105–109. doi: 10.1126/science.277.5322.105. [DOI] [PubMed] [Google Scholar]
- 6.Brewster J L, de Valoir T, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 7.Brewster J L, Gustin M C. Positioning of cell growth and division after osmotic stress requires a MAP kinase pathway. Yeast. 1994;10:425–439. doi: 10.1002/yea.320100402. [DOI] [PubMed] [Google Scholar]
- 8.Cabib E. Differential inhibition of chitin synthetases 1 and 2 from Saccharomyces cerevisiae by polyoxin D and nikkomycins. Antimicrob Agents Chemother. 1991;35:170–173. doi: 10.1128/aac.35.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calderone R A. Recognition between Candida albicans and host cells. Trends Microbiol. 1993;1:55–58. doi: 10.1016/0966-842x(93)90033-n. [DOI] [PubMed] [Google Scholar]
- 10.Calera J A, Choi G H, Calderone R A. Identification of a putative histidine kinase two-component phosphorelay gene (CaHK1) in Candida albicans. Yeast. 1998;14:665–674. doi: 10.1002/(SICI)1097-0061(199805)14:7<665::AID-YEA246>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 11.Chowdhury S, Smith K W, Gustin M C. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J Cell Biol. 1992;118:561–571. doi: 10.1083/jcb.118.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cid V J, Durán A, del Rey F, Snyder M P, Nombela C, Sánchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corner B E, Magee P T. Candida pathogenesis: unravelling the threads of infection. Curr Biol. 1997;7:R691–R694. doi: 10.1016/s0960-9822(06)00357-5. [DOI] [PubMed] [Google Scholar]
- 14.Csank C, Makris C, Meloche S, Schröppel K, Röllinghoff M, Dignard D, Thomas D Y, Whiteway M. Derepressed hyphal growth and reduced virulence in a VH1 family-related protein phosphatase mutant of the human pathogen Candida albicans. Mol Biol Cell. 1997;8:2539–2551. doi: 10.1091/mbc.8.12.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csank C, Schröppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas D Y, Whiteway M. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 1998;66:2713–2721. doi: 10.1128/iai.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Fuente J M, Alvarez A, Nombela C, Sánchez M. Flow cytometric analysis of Saccharomyces cerevisiae autolytic mutants and protoplasts. Yeast. 1992;8:39–45. doi: 10.1002/yea.320080104. [DOI] [PubMed] [Google Scholar]
- 17.Díez-Orejas R, Molero G, Navarro-García F, Pla J, Nombela C, Sánchez-Pérez M. Reduced virulence of Candida albicans MKC1 mutants: a role for a mitogen-activated protein kinase in pathogenesis. Infect Immun. 1997;65:833–837. doi: 10.1128/iai.65.2.833-837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaughran J P, Lai M H, Kirsch D R, Silverman S J. Nikkomycin Z is a specific inhibitor of Saccharomyces cerevisiae chitin synthase isozyme Chs3 in vitro and in vivo. J Bacteriol. 1994;176:5857–5860. doi: 10.1128/jb.176.18.5857-5860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillum A M, Tsay E Y H, Kirsch D R. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 21.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 22.Hall J P, Cherkasova V, Elion E A, Gustin M C, Winter E. The osmoregulatory pathway represses mating pathway activity in Saccharomyces cerevisiae: isolation of a FUS3 mutant that is insensitive to the repression mechanism. Mol Cell Biol. 1996;16:6715–6723. doi: 10.1128/mcb.16.12.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan D. Techniques for transformation of E. coli. In: Glover D M, editor. DNA cloning. Oxford, United Kingdom: IRL Press; 1988. pp. 109–135. [Google Scholar]
- 24.Hector R F, Domer J E, Carrow E W. Immune responses to Candida albicans in genetically distinct mice. Infect Immun. 1982;38:1020–1028. doi: 10.1128/iai.38.3.1020-1028.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hill J E, Myers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 26.Ishii N, Yamamoto M, Yoshihara F, Arisawa M, Aoki Y. Biochemical and genetic characterization of Rbf1p, a putative transcription factor of Candida albicans. Microbiology. 1997;143:429–435. doi: 10.1099/00221287-143-2-429. [DOI] [PubMed] [Google Scholar]
- 27.Jiang B, Ram A F J, Sheraton J, Klis F M, Bussey H. Regulation of cell wall beta-glucan assembly: PTC1 negatively affects PBS2 action in a pathway that includes modulation of EXG1 transcription. Mol Gen Genet. 1995;248:260–269. doi: 10.1007/BF02191592. [DOI] [PubMed] [Google Scholar]
- 28.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 29.Kirsch D R, Whitney R R. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 1991;59:3297–3300. doi: 10.1128/iai.59.9.3297-3300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi G S, Cutler J E. Candida albicans hyphal formation and virulence: is there a clearly defined role? Trends Microbiol. 1998;6:92–94. doi: 10.1016/s0966-842x(98)01218-9. [DOI] [PubMed] [Google Scholar]
- 31.Köhler G A, White T C, Agabian N. Overexpression of a cloned IMP dehydrogenase gene of Candida albicans confers resistance to the specific inhibitor mycophenolic acid. J Bacteriol. 1997;179:2331–2338. doi: 10.1128/jb.179.7.2331-2338.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Köhler J, Fink G R. Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci USA. 1996;93:13223–13228. doi: 10.1073/pnas.93.23.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kron S J, Styles C A, Fink G R. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuranda M J, Robbins P W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem. 1991;266:19758–19767. [PubMed] [Google Scholar]
- 35.Lai M H, Silverman S J, Gaughran J P, Kirsch D R. Multiple copies of PBS2, MHP1 or LRE1 produce glucanase resistance and other cell wall effects in Saccharomyces cerevisiae. Yeast. 1997;13:199–213. doi: 10.1002/(SICI)1097-0061(19970315)13:3<199::AID-YEA76>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 36.Leberer E, Harcus D, Broadbent I D, Clark K L, Dignard D, Ziegelbauer K, Schmidt A, Gow N A R, Brown A J P, Thomas D Y. Signal transduction through homologs of the Ste20p and Ste7p protein kinases can trigger hyphal formation in the pathogenic fungus Candida albicans. Proc Natl Acad Sci USA. 1996;93:13217–13222. doi: 10.1073/pnas.93.23.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leberer E, Ziegelbauer K, Schmidt A, Harcus D, Dignard D, Ash J, Johnson L, Thomas D Y. Virulence and hyphal formation of Candida albicans require the Ste20p-like protein kinase CaCla4p. Curr Biol. 1997;7:539–546. doi: 10.1016/s0960-9822(06)00252-1. [DOI] [PubMed] [Google Scholar]
- 38.Lee K L, Buckley H R, Campbell C C. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. J Med Vet Mycol. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Köhler J, Fink G R. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science. 1994;266:1723–1726. doi: 10.1126/science.7992058. [DOI] [PubMed] [Google Scholar]
- 40.Liu H, Styles C A, Fink G R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 41.Lo H J, Kohler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 42.Madhani H D, Styles C A, Fink G R. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 43.Maeda T, Takekawa M, Saito H. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 1995;269:554–558X. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 44.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 45.Magee P T. Which came first: the hypha or the yeast? Science. 1998;277:52–53. doi: 10.1126/science.277.5322.52. [DOI] [PubMed] [Google Scholar]
- 46.Malathi K, Ganesan K, Datta A. Identification of a putative transcription factor in Candida albicans that can complement the mating defect of Saccharomyces cerevisiae ste12 mutants. J Biol Chem. 1994;269:22945–22951. [PubMed] [Google Scholar]
- 47.Márquez J A, Pascual-Ahuir A, Proft M, Serrano R. The Ssn6-Tup1 repressor complex of Saccharomyces cerevisiae is involved in the osmotic induction of HOG-dependent and -independent genes. EMBO J. 1998;17:2543–2553. doi: 10.1093/emboj/17.9.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy P J, Troke P F, Gull K. Mechanism of action of nikkomycin and the peptide transport system of Candida albicans. J Gen Microbiol. 1985;131:775–780. doi: 10.1099/00221287-131-4-775. [DOI] [PubMed] [Google Scholar]
- 49.Miret J J, Solari A J, Barderi P A, Goldemberg S H. Polyamines and cell wall organization in Saccharomyces cerevisiae. Yeast. 1992;8:1033–1041. doi: 10.1002/yea.320081206. [DOI] [PubMed] [Google Scholar]
- 50.Nagahashi S, Mio T, Ono N, Yamada-Okabe T, Arisawa M, Bussey H, Yamada-Okabe H. Isolation of CaSLN1 and CaNIK1, the genes for osmosensing histidine kinase homologues, from the pathogenic fungus Candida albicans. Microbiology. 1998;144:425–432. doi: 10.1099/00221287-144-2-425. [DOI] [PubMed] [Google Scholar]
- 51.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Proposed standard M27-P. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1992. [Google Scholar]
- 52.Navarro-García F, Alonso-Monge R, Rico H, Pla J, Sentandreu R, Nombela C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology. 1998;144:411–424. doi: 10.1099/00221287-144-2-411. [DOI] [PubMed] [Google Scholar]
- 53.Navarro-García F, Sánchez M, Pla J, Nombela C. Functional characterization of the MKC1 gene of Candida albicans, which encodes a mitogen-activated protein kinase homolog related to cell integrity. Mol Cell Biol. 1995;15:2197–2206. doi: 10.1128/mcb.15.4.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Negredo A, Monteoliva L, Gil C, Pla J, Nombela C. Cloning, analysis and one-step disruption of the ARG5,6 gene of Candida albicans. Microbiology. 1997;143:297–302. doi: 10.1099/00221287-143-2-297. [DOI] [PubMed] [Google Scholar]
- 55.Odds F C. Candida and candidosis. London, United Kingdom: Bailliére Tindall; 1988. [Google Scholar]
- 56.Odds F C. Candida species and virulence. ASM News. 1994;60:313–318. [Google Scholar]
- 57.O’Rourke S M, Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ota I M, Varshavsky A. A yeast protein similar to bacterial two-component regulators. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 59.Papadimitriou J M, Ashman R B. The pathogenesis of acute systemic candidiasis in a susceptible inbred mouse strain. J Pathol. 1986;150:257–265. doi: 10.1002/path.1711500405. [DOI] [PubMed] [Google Scholar]
- 60.Paravicini G, Mendoza A, Antonsson B, Cooper M, Losberger C, Payton M. The Candida albicans PKC1 gene encodes a protein kinase C homolog necessary for cellular integrity but not dimorphism. Yeast. 1996;12:741–756. doi: 10.1002/(sici)1097-0061(19960630)12:8<741::aid-yea967>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 61.Payne J W, Shallow D A. Studies on drug targetting in the pathogenic fungus C. albicans: peptide transport mutants resistant to polyoxins, nikkomycins and bacilysin. FEMS Microbiol Lett. 1985;28:55–60. [Google Scholar]
- 62.Pla J, Gil C, Monteoliva L, Navarro-García F, Sánchez M, Nombela C. Understanding Candida albicans at the molecular level. Yeast. 1996;12:1677–1702. doi: 10.1002/(SICI)1097-0061(199612)12:16%3C1677::AID-YEA79%3E3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 63.Pla J, Pérez-Díaz R M, Navarro-García F, Sánchez M, Nombela C. Cloning of the Candida albicans HIS1 gene by direct complementation of a C. albicans histidine auxotroph using an improved double-ARS shuttle vector. Gene. 1995;165:115–120. doi: 10.1016/0378-1119(95)00492-o. [DOI] [PubMed] [Google Scholar]
- 64.Posas F, Saito H. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 1997;276:1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 65.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 66.Ryley J F, Ryley N G. Candida albicans—do mycelia matter? J Med Vet Mycol. 1990;28:225–239. [PubMed] [Google Scholar]
- 67.San José C, Alonso R, Pérez-Díaz R M, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J Bacteriol. 1996;178:5850–5852. doi: 10.1128/jb.178.19.5850-5852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scherer S, Magee P T. Genetics of Candida albicans. Microbiol Rev. 1990;54:226–241. doi: 10.1128/mr.54.3.226-241.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shepherd M G. Pathogenicity of morphological and auxotrophic mutants of Candida albicans in experimental infections. Infect Immun. 1985;50:541–544. doi: 10.1128/iai.50.2.541-544.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shiozaki K, Russell P. Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature. 1995;378:739–743. doi: 10.1038/378739a0. [DOI] [PubMed] [Google Scholar]
- 71.Shiozaki K, Russell P. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 1996;10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 72.Singh P, Ganesan K, Malathi K, Ghosh D, Datta A. ACPR, a STE12 homologue from Candida albicans, is a strong inducer of pseudohyphae in Saccharomyces cerevisiae haploids and diploids. Biochem Biophys Res Commun. 1994;205:1079–1085. doi: 10.1006/bbrc.1994.2776. [DOI] [PubMed] [Google Scholar]
- 73.Singh P, Ghosh S, Datta A. A novel MAP-kinase kinase from Candida albicans. Gene. 1997;190:99–104. doi: 10.1016/s0378-1119(96)00758-5. [DOI] [PubMed] [Google Scholar]
- 74.Srikantha T, Tsai L, Daniels K, Enger L, Highley K, Soll D R. The two-component hybrid kinase regulator caNIK1 of Candida albicans. Microbiology. 1998;144:2715–2729. doi: 10.1099/00221287-144-10-2715. [DOI] [PubMed] [Google Scholar]
- 75.Tiedt L R, Jooste W R, Hamilton-Attwell V L. Technique for preserving aerial fungus structure for scanning electron microscopy. Trans Br Mycol. 1987;88:420–422. [Google Scholar]
- 76.Tsang T, Copeland V, Bowden G T. A set of cassette cloning vectors for rapid and versatile adaptation of restriction fragments. Biotechniques. 1991;10:330. [PubMed] [Google Scholar]
- 77.Whiteway M, Dignard D, Thomas D Y. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci USA. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Williams S, Veldkamp C. Preparation of fungi for scanning electron microscopy. Trans Br Mycol. 1974;63:409–412. [Google Scholar]
- 79.Yadan J C, Gonneau M, Sarthou P, Le Goffic F. Sensitivity to nikkomycin Z in Candida albicans: role of peptide permeases. J Bacteriol. 1984;160:884–888. doi: 10.1128/jb.160.3.884-888.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]