Abstract
DNA polymerase η (Pol η) catalyzes accurate bypass of ultraviolet light-induced cyclobutane pyrimidine dimers, and it also functions in several other related processes, including bypassing DNA with unusual structures. Here, we performed unbiased proteome-wide profiling of Pol η-interacting proteins by using two independent approaches, i.e., proximity labeling and affinity pull-down followed by LC-MS/MS analysis. We identified several helicases, including DHX9, as novel Pol η-interacting proteins. Additionally, ChIP-Seq analysis showed that Pol η is enriched at guanine quadruplex (G4) structure sites in chromatin. Moreover, Pol η promotes the recruitment of DHX9 to G4 structure loci in chromatin and facilitates DHX9-mediated unwinding of G4 structures. Deficiency in Pol η or DHX9 leads to attenuated replication across G4 regions in genomic DNA. Together, we unveiled the interaction between Pol η and DHX9 and demonstrated that the interaction promotes the replicative bypass of G4 structures in chromatin.
Human cells are exposed to various DNA-damaging agents,1 and the resulting DNA lesions can block the progression of replication forks and induce mutations, thereby leading to cell transformation, senescence, and/or apoptosis.2 To cope with unrepaired DNA lesions, cells are equipped with DNA damage tolerance mechanisms, one of which is translesion synthesis (TLS).3
DNA polymerase η (Pol η) displays accurate TLS over UV-induced DNA lesions, especially cyclobutane pyrimidine dimers.4–6 Mutations in the POLH gene, which encodes Pol η, result in the variant form of xeroderma pigmentosum (XPV) syndrome in humans.5,6 Pol η also assumes important functions in several other related processes, including bypassing a broad spectrum of other DNA lesions7 and unusual DNA structures, such as fragile sites8 and guanine quadruplexes (G4s).9
We reason that a comprehensive assessment about Pol η-interacting proteins at the global proteome scale may offer new insights into its functions. Recently, Ting et al.10–13 developed an engineered ascorbate peroxidase (APEX)-based proximity labeling, in combination with LC-MS/MS analysis, to enable temporal and spatial profiling of interaction proteins of a target protein of interest in cultured cells. We fused APEX2 to the C-terminus of Pol η for the biotinylation of proximity proteins of Pol η, where APEX2 fused with a nuclear localization signal (NLS) served as a control (Figure S1). The LC-MS/MS results led to the identification of 279 proximity proteins of Pol η (Figure 1a and Table S1). Gene ontology (GO) and KEGG pathway analyses revealed the enrichment of DNA replication and DNA repair (Figure S2), which is in keeping with the known functions of Pol η and validates our method. Moreover, GO function analysis revealed the enrichment of DNA helicases.
Figure 1.

APEX labeling and affinity pull-down for revealing the Pol η interactome. (a, b) Volcano plots displaying enriched proteins (highlighted in red) for Pol η-APEX versus the control (NLS-APEX) obtained from APEX labeling and LC-MS/MS analysis (a) or anti-Flag pull-down using CRISPR-engineered Pol η-Flag cells versus parental HEK293T cells (b). Proteins with a > 1.5-fold signal over control and a p-value < 0.05 are considered enriched. (c) Venn diagram illustrating the candidate Pol η-interaction proteins obtained from the two proteomic approaches. (d) Western blot results showing Flag pull-down using lysates of Pol η-Flag cells and parental HEK293T cells.
The APEX labeling method entails ectopic overexpression of Pol η, which may introduce artifacts in identifying its interaction proteins. To profile the interactome of endogenous Pol η, we employed CRISPR-Cas9 to introduce three tandem repeats of Flag epitope tag to the C-terminus of endogenous Pol η protein in HEK293T cells (Figure S3).14 After enrichment with anti-Flag beads and LC-MS/MS analysis, we identified 206 proteins with preferential binding toward Pol η (Figure 1b), among which 43 are overlapped with the proximity proteins identified from APEX labeling (Figure 1c), including several helicases, e.g., DHX9, DDX5, and DDX17.
Helicases assume functions in many steps of gene expression by unwinding double-stranded DNA/RNA and more complex nucleic acid structures.15 For instance, DHX9 can unwind noncanonical DNA structures, including intramolecular triplex (H-DNA), DNA- and RNA-containing displacement loops (Dand R-loops), and G4.16–18 Moreover, DDX5 was found to unfold DNA G4 and activate the transcription of the MYC gene in cancer cells.19
G4s are noncanonical secondary structures formed in guanine-rich sequences in DNA and RNA.20 DNA G4s are involved in a number of critical cellular processes, including DNA replication, transcription, and maintenance of genome integrity.20–22 Our quantitative proteomic data unveiled 2- and 4-fold enrichments of DDX5 and DHX9, respectively, in the Pol η pull-down samples relative to the control (Table S1). We also validated the interactions between Pol η and DDX5/DHX9 by immunoprecipitation followed with Western blot analyses (Figure 1d). Both DDX5 and DHX9 are capable of resolving DNA G4 structures,16,19 and Pol η was found to be important in maintaining fork progression past naturally occurring non-B-form DNA structures, such as G4 DNA.23 Thus, Pol η may function together with DNA helicases in promoting replication across G4 structure sites in DNA.
To test the above hypothesis, we asked if Pol η binds to specific DNA structure regions in cells by assessing its genome-wide occupancy with ChIP-Seq analysis. After peak calling against the control, we identified 2049 high-confidence Pol η-binding sites in two biological replicates (Figure S4). The Pol η peaks exhibit enrichment in promoters and 5′-UTR (Figure 2a, Figure S5), which is consistent with the distribution of G4 peaks.24 Importantly, approximately 32% of Pol η peaks overlapped with G4 peaks (Figure 2b). Moreover, the Pol η ChIP-Seq signal was highly enriched and centered at endogenous G4 sites, and vice versa (Figure 2c,d). Integrative Genomics Viewer (IGV) plots also showed a high degree of co-localization between Pol η and BG4 peaks (Figure 2e).
Figure 2.
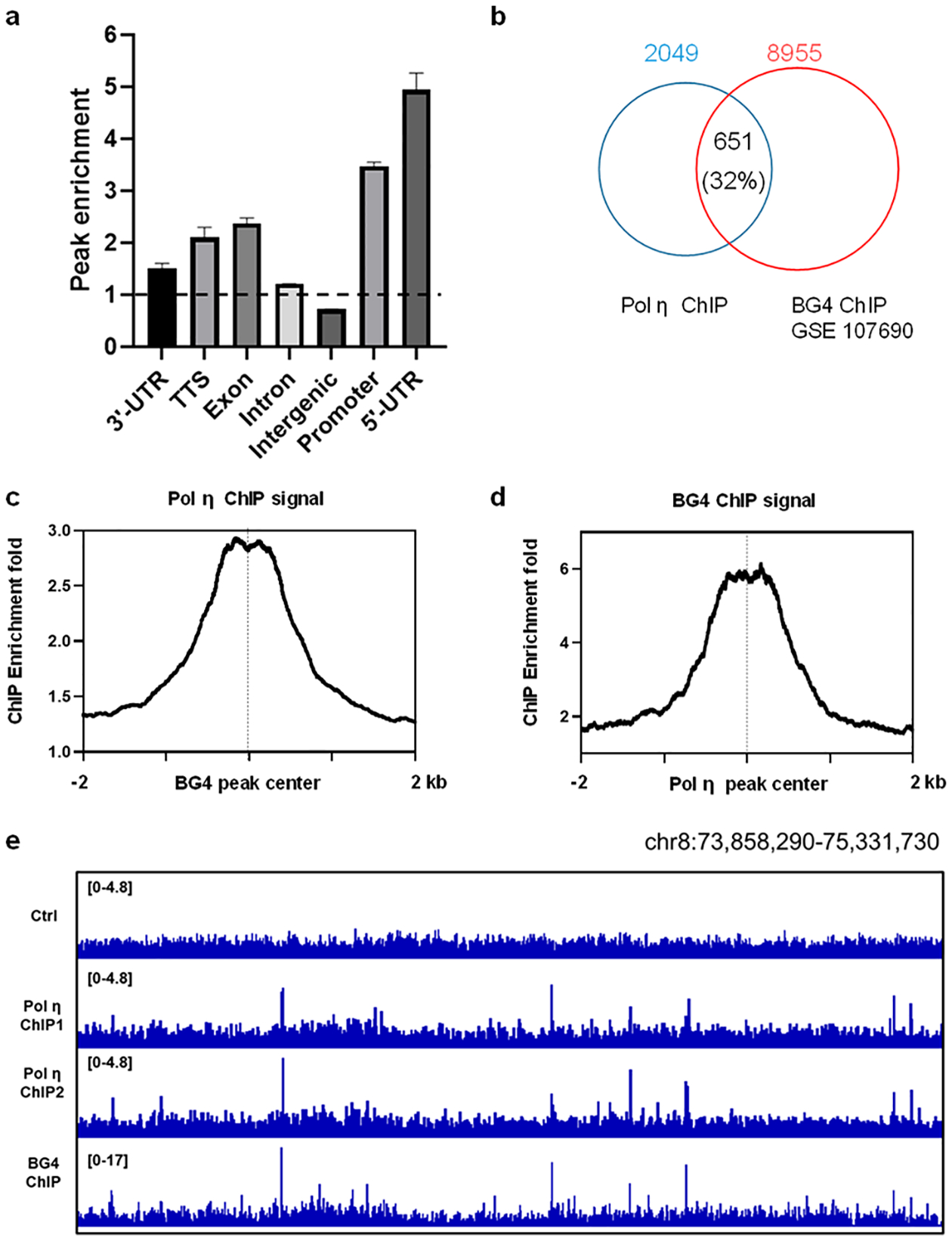
ChIP-Seq revealed enrichment of Pol η at G4 structure loci in chromatin. (a) Relative enrichment of Pol η peaks in different genomic elements. The dashed line represents an enrichment fold of 1.0. Error bars represent standard deviations from two biological replicates. (b) Venn diagram showing the overlap between Pol η ChIP-seq and BG4 ChIP-Seq peaks. (c) Average signal of Pol η ChIP-Seq against the center of the BG4 ChIP-Seq peaks and (d) vice versa. (e) IGV plots showing the comparison of Pol η occupancy and G4 structure loci in the indicated genomic region.
On the basis of the above proteomic and ChIP-Seq data, we hypothesized that Pol η may promote replication across G4 structures by recruiting helicases to unwind these structures. To test this, we conducted a ChIP-qPCR assay to examine how the occupancy of DHX9 at G4 structure loci is modulated by Pol η with the use of patient-derived Pol η-deficient (XPV) human skin fibroblasts and the isogenic cells complemented with human Pol η (XPV+Pol η) (qPCR primers are listed in Table S2).6 Based on the ChIP-Seq data, we selected four genes with both G4 and Pol η peaks in their promoters for DHX9 ChIP-qPCR analysis (Figure 3). Our results showed that the loss of Pol η confers diminished enrichment of DHX9 in G4 regions, but not in non-G4 regions of PMS2, NEAT1, and JUN genes (Figure 3e), suggesting a Pol η-dependent recruitment of DHX9 to these G4 structure sites. In this vein, DHX9 did not display enrichment in the G4 region of the CLSPN gene in Pol η-complemented cells, suggesting the lack of involvement of DHX9 in resolving the G4 structure at this site.
Figure 3.

Pol η recruits DHX9 to promote the replication across G4 structure sites in DNA. Representative IGV plots showing signal tracks for Pol η and BG4 ChIP-Seq results for PMS2 (a), NEAT1 (b), CLSPN (c), and JUN (d) genes in HEK293T cells. (e) ChIP-qPCR analyses at the G4/non-G4 loci using DHX9 antibody. (f) Amount of BrdU incorporation at the G4/non-G4 regions of the indicated genes with or without knocking down DHX9 in XPV and XPV + Pol η cell lines. The data represent mean ± SD from three independent experiments. The p values were calculated by using two-tailed, unpaired Student’s t-test: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Our ChIP-qPCR with the use of BG4 antibody revealed that the knockdown of DHX9 resulted in increased enrichment of G4 structures in the promoter regions of PMS2, NEAT1, and JUN genes, but not that of the CLSPN gene, in XPV + Pol η cells; such increases, however, were abolished in XPV cells (Figure S6). This result demonstrated that the DHX9-mediated unwinding of G4 structures in the promoter regions of these genes requires Pol η.
On the basis of the above findings, we asked whether deficiency in Pol η or DHX9 may impede replication past G4 structure sites. To test this, we labeled newly synthesized DNA with 5-bromo-2′-deoxyuridine (BrdU) in XPV and XPV + Pol η cells, enriched the BrdU-labeled nascent DNA, and subjected the resulting DNA fragments to qPCR analysis. Our results unveiled that the amount of BrdU incorporation across G4 regions is lower in XPV cells than XPV + Pol η cells (Figure 3f), whereas no differences were observed in non-G4 regions. In addition, genetic depletion of DHX9 leads to decreased BrdU incorporation in the promoters of PMS2, NEAT1, and JUN genes, but not that of the CLSPN gene, which is in line with the lack of DHX9 enrichment in the promoter of the CLSPN gene. Genetic depletion of DHX9 did not give rise to further diminutions in the amount of BrdU incorporation in Pol η-deficient cells, suggesting that DHX9 and Pol η function in the same pathway in promoting replication across G4 structure sites. Together, these results suggest that Pol η recruits DHX9 to unwind G4 structures prior to their replicative bypass.
Recently, several studies showed that TLS polymerases have functions beyond their canonical roles in bypassing damaged DNA, where these polymerases are capable of bypassing undamaged structured DNA in vitro and in vivo.25,26 Additionally, Pol η is capable of inserting ribonucleotides opposite template DNA,27 and it enables transcriptional bypass of minor-groove N2-alkyl-dG lesions.28 Moreover, biochemical assay showed that Pol η displays a 6-fold preference toward binding to G4 DNA than non-G4 DNA, and Pol η also exhibits a higher replication efficiency and fidelity than replicative polymerase (polymerase ε) when copying G4 DNA.9 Furthermore, genetic depletion of Pol η renders U2OS cells sensitive to telomestatin, a G4-binding ligand.23 Cellular assays also revealed the function of Pol η in promoting replication across common fragile sites,8 and genetic ablation of Pol η in mice led to senescence in adipose tissues.29
By employing two separate approaches, relying on proximity labeling and affinity pull-down in combination with LC-MS/MS analysis, we explored systematically the interaction proteome of Pol η. Our proteomic data revealed pronounced enrichment of several helicases as interaction partners of Pol η. Additionally, the Pol η ChIP-Seq signal was highly enriched and centered at endogenous G4 sites. Moreover, our DHX9 and BG4 ChIP-qPCR results showed that the co-localization of DHX9 with G4 structure sites in gene promoters and unwinding of G4 structures at these sites require Pol η. Hence, our results support a model where G4 structures in template DNA stall replicative polymerases, which leads to the recruitment of Pol η. The latter further recruits DHX9 to unwind G4 structures, thereby enabling their replicative bypass (Figure 4). This model is in line with the 3′ → 5′ unwinding activity of DHX9.30 While Pol η was previously shown to support the replicative bypass of G4 structures in vitro,9 our work demonstrated this function of Pol η in cells and revealed its relevance at the genome-wide scale. Our study also unveiled that this cellular function of Pol η involves the DHX9 helicase.
Figure 4.

Model of Pol η’s function in promoting replicative bypass of G4 structures through DHX9.
The mechanism through which DHX9 is recruited to Pol η is unclear and warrants further investigation. Along this line, DHX9 was identified as a DNA replication protein interacting with PCNA and WRN helicase, where the latter interaction stimulates WRN’s activity in unwinding Okazaki fragment-like nucleic acid structures.31,32 Pol η is recruited to stalled DNA replication forks through its interaction with monoubiquitylated PCNA.33 Interestingly, Pol η can also be ubiquitylated on its C-terminus by the E3 ubiquitin ligase PIRH2, which inhibits its interaction with PCNA.34,35 In addition, phosphorylation of Pol η at Ser601 and Ser687 assumes crucial roles in facilitating TLS.36,37 It will be important to examine, in the future, whether a similar post-translational mechanism contributes to the recruitment of DHX9 to Pol η and the replicative bypass of G4 structures in DNA. It will also be important to investigate whether this function of Pol η can be extended to other TLS polymerases, whether the Pol η−DHX9 interaction also contributes to replicative bypass of other structured DNA and R-loops, and how other interaction partners of Pol η identified from the present study modulate the functions of this versatile TLS polymerase.
Supplementary Material
■ ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R35 ES031707).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c05312.
Detailed experimental procedures and supplementary data (PDF)
Table S1. Lists of candidate Pol eta-binding proteins identified from APEX labeling and affinity pull-down followed by LC-MS/MS analyses (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.2c05312
The authors declare no competing financial interest.
Contributor Information
Feng Tang, Department of Chemistry, University of California, Riverside, California 92521-0403, United States.
Yinan Wang, Department of Chemistry, University of California, Riverside, California 92521-0403, United States.
Zi Gao, Department of Chemistry, University of California, Riverside, California 92521-0403, United States.
Shiyuan Guo, Genetics, Genomics and Bioinformatics Graduate Program, University of California, Riverside, California 92521-0403, United States;.
Yinsheng Wang, Department of Chemistry, University of California, Riverside, California 92521-0403, United States; Genetics, Genomics and Bioinformatics Graduate Program, University of California, Riverside, California 92521-0403, United States;.
■ REFERENCES
- (1).Lindahl T. J. n. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- (2).Yeeles JT; Poli J; Marians KJ; Pasero P Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol 2013, 5, a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Prakash S; Johnson RE; Prakash L Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem 2005, 74, 317–53. [DOI] [PubMed] [Google Scholar]
- (4).Kannouche PL; Wing J; Lehmann AR Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 2004, 14, 491–500. [DOI] [PubMed] [Google Scholar]
- (5).Johnson RE; Kondratick CM; Prakash S; Prakash L hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 1999, 285, 263–265. [DOI] [PubMed] [Google Scholar]
- (6).Masutani C; Kusumoto R; Yamada A; Dohmae N; Yokoi M; Yuasa M; Araki M; Iwai S; Takio K; Hanaoka F The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 1999, 399, 700–704. [DOI] [PubMed] [Google Scholar]
- (7).Zlatanou A; Despras E; Braz-Petta T; Boubakour-Azzouz I; Pouvelle C; Stewart GS; Nakajima S; Yasui A; Ishchenko AA; Kannouche PL The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol η in response to oxidative DNA damage in human cells. Mol. Cell 2011, 43, 649–662. [DOI] [PubMed] [Google Scholar]
- (8).Twayana S; Bacolla A; Barreto-Galvez A; De-Paula RB; Drosopoulos WC; Kosiyatrakul ST; Bouhassira EE; Tainer JA; Madireddy A; Schildkraut CL Translesion polymerase eta both facilitates DNA replication and promotes increased human genetic variation at common fragile sites. Proc. Natl. Acad. Sci. U.S.A 2021, 118, e2106477118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Eddy S; Maddukuri L; Ketkar A; Zafar MK; Henninger EE; Pursell ZF; Eoff RL Evidence for the kinetic partitioning of polymerase activity on G-quadruplex DNA. Biochemistry 2015, 54, 3218–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hung V; Udeshi ND; Lam SS; Loh KH; Cox KJ; Pedram K; Carr SA; Ting AY Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat. Protoc 2016, 11, 456–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lam SS; Martell JD; Kamer KJ; Deerinck TJ; Ellisman MH; Mootha VK; Ting AY Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 2015, 12, 51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Rhee H-W; Zou P; Udeshi ND; Martell JD; Mootha VK; Carr SA; Ting AY Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yang P; Mathieu C; Kolaitis R-M; Zhang P; Messing J; Yurtsever U; Yang Z; Wu J; Li Y; Pan Q G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell 2020, 181, 325–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Williams P; Li L; Dong X; Wang Y Identification of SLIRP as a G quadruplex-binding protein. J. Am. Chem. Soc 2017, 139, 12426–12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Singleton MR; Dillingham MS; Wigley DB Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem 2007, 76, 23–50. [DOI] [PubMed] [Google Scholar]
- (16).Chakraborty P; Grosse F Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 2011, 10, 654–665. [DOI] [PubMed] [Google Scholar]
- (17).Jain A; Bacolla A; Chakraborty P; Grosse F; Vasquez KM Human DHX9 helicase unwinds triple-helical DNA structures. Biochemistry 2010, 49, 6992–6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jain A; Bacolla A; Del Mundo IM; Zhao J; Wang G; Vasquez KM DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013, 41, 10345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wu G; Xing Z; Tran EJ; Yang D DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. U.S.A 2019, 116, 20453–20461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Bochman ML; Paeschke K; Zakian VA DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet 2012, 13, 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Robinson J; Raguseo F; Nuccio SP; Liano D; Di Antonio M DNA G-quadruplex structures: more than simple roadblocks to transcription? Nucleic Acids Res. 2021, 49, 8419–8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Li L; Williams P; Ren W; Wang MY; Gao Z; Miao W; Huang M; Song J; Wang Y YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol 2021, 17, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Bétous R; Rey L; Wang G; Pillaire MJ; Puget N; Selves J; Biard DS; Shin-ya K; Vasquez KM; Cazaux C Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol. Carcinog 2009, 48, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hänsel-Hertsch R; Spiegel J; Marsico G; Tannahill D; Balasubramanian S Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat. Protoc 2018, 13, 551–564. [DOI] [PubMed] [Google Scholar]
- (25).Sarkies P; Reams C; Simpson LJ; Sale JE Epigenetic instability due to defective replication of structured DNA. Mol. Cell 2010, 40, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wickramasinghe CM; Arzouk H; Frey A; Maiter A; Sale JE Contributions of the specialised DNA polymerases to replication of structured DNA. DNA Repair 2015, 29, 83–90. [DOI] [PubMed] [Google Scholar]
- (27).Su Y; Egli M; Guengerich FP Mechanism of ribonucleotide incorporation by human DNA polymerase η. J. Biol. Chem 2016, 291, 3747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tan Y; Guo S; Wu J; Du H; Li L; You C; Wang Y DNA polymerase η promotes the transcriptional bypass of N2-Alkyl-2′-deoxyguanosine adducts in human cells. J. Am. Chem. Soc 2021, 143, 16197–16205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen YW; Harris RA; Hatahet Z; Chou KM Ablation of XP-V gene causes adipose tissue senescence and metabolic abnormalities. Proc. Natl. Acad. Sci. U.S.A 2015, 112, E4556–E4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Zhang S; Grosse F Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim. Biophys. Sin 2004, 36, 177–183. [DOI] [PubMed] [Google Scholar]
- (31).Loor G; Zhang S-J; Zhang P; Toomey NL; Lee MY Identification of DNA replication and cell cycle proteins that interact with PCNA. Nucleic Acids Res. 1997, 25, 5041–5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Chakraborty P; Grosse F (2010). WRN helicase unwinds Okazaki fragment-like hybrids in a reaction stimulated by the human DHX9 helicase. Nucleic Acids Res. 2010, 38, 4722–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Watanabe K; Tateishi S; Kawasuji M; Tsurimoto T; Inoue H; Yamaizumi M Rad18 guides polη to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004, 23, 3886–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bienko M; Green CM; Sabbioneda S; Crosetto N; Matic I; Hibbert RG; Begovic T; Niimi A; Mann M; Lehmann AR Regulation of translesion synthesis DNA polymerase η by monoubiquitination. Mol. Cell 2010, 37, 396–407. [DOI] [PubMed] [Google Scholar]
- (35).Jung Y-S; Hakem A; Hakem R; Chen X Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Mol. Cell. Biol 2011, 31, 3997–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Göhler T; Sabbioneda S; Green CM; Lehmann AR ATR-mediated phosphorylation of DNA polymerase η is needed for efficient recovery from UV damage. J. Cell Biol 2011, 192, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Dai X; You C; Wang Y The functions of serine 687 phosphorylation of human DNA polymerase η in UV damage tolerance. Mol. Cell. Proteomics 2016, 15, 1913–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


