SUMMARY
Animals communicate using sounds in a wide range of contexts, and auditory systems must encode behaviorally relevant acoustic features to drive appropriate reactions. How feature detection emerges along auditory pathways has been difficult to solve due to challenges in mapping the underlying circuits and characterizing responses to behaviorally relevant features. Here, we study auditory activity in the Drosophila melanogaster brain and investigate feature selectivity for the two main modes of fly courtship song, sinusoids and pulse trains. We identify 24 new cell types of the intermediate layers of the auditory pathway, and using a new connectomic resource, FlyWire, we map all synaptic connections between these cell types, in addition to connections to known early and higher-order auditory neurons - this represents the first circuit-level map of the auditory pathway. We additionally determine the sign (excitatory or inhibitory) of most synapses in this auditory connectome. We find that auditory neurons display a continuum of preferences for courtship song modes, and that neurons with different song mode preferences and response timescales are highly interconnected in a network that lacks hierarchical structure. Nonetheless, we find that the response properties of individual cell types within the connectome is predictable from its inputs. Our study thus provides new insights into the organization of auditory coding within the Drosophila brain.
eTOC Blurb
Baker et al. discover and functionally characterize over 20 new auditory cell types in the Drosophila brain. They map synaptic connectivity among almost all known auditory neurons, and find that the auditory system is densely interconnected, suggesting that connections between differently tuned neurons are important for shaping auditory responses.
INTRODUCTION
Sounds are integral to the social lives of animals. Accordingly, brains have evolved to recognize behaviorally salient acoustic signals. For instance, courtship songs often contain information about sender status and species, and receivers must decode this information by analyzing patterns within songs 1–4. Several species produce songs comprising multiple acoustic types, often called syllables or modes 5–7. While prior work has examined where selectivity for conspecific sounds emerges across a variety of systems including songbirds 8, primates 9, and mice 10, the circuit mechanisms shaping selectivity are unknown. For example, while a hypothesized circuit for recognizing stereotyped songs has been described in crickets 11,12, connectivity among these neurons has not yet been determined, and cricket songs comprise only a single mode. Understanding how auditory systems establish selectivity for conspecific sounds requires linking responses to different song types with neuronal connectivity, a significant challenge in larger brains. Here we focus on Drosophila melanogaster, a species with a compact brain and both genetic and connectomic tools for circuit dissection.
D. melanogaster males alternate between two major song modes during courtship: pulse and sine song (Figure 1A; 13,14). The composition of song bouts (the duration of each mode and switches between them) are dependent on sensory feedback from the female 15. Receptive females reduce locomotor speed in response to multiple features within conspecific song, from the frequency of individual elements to longer timescale patterns such as the average duration of song bouts 16,17. However, how tuning for these features emerges along the auditory pathway is not known.
Figure 1. Anatomic and functional screen for auditory neurons.
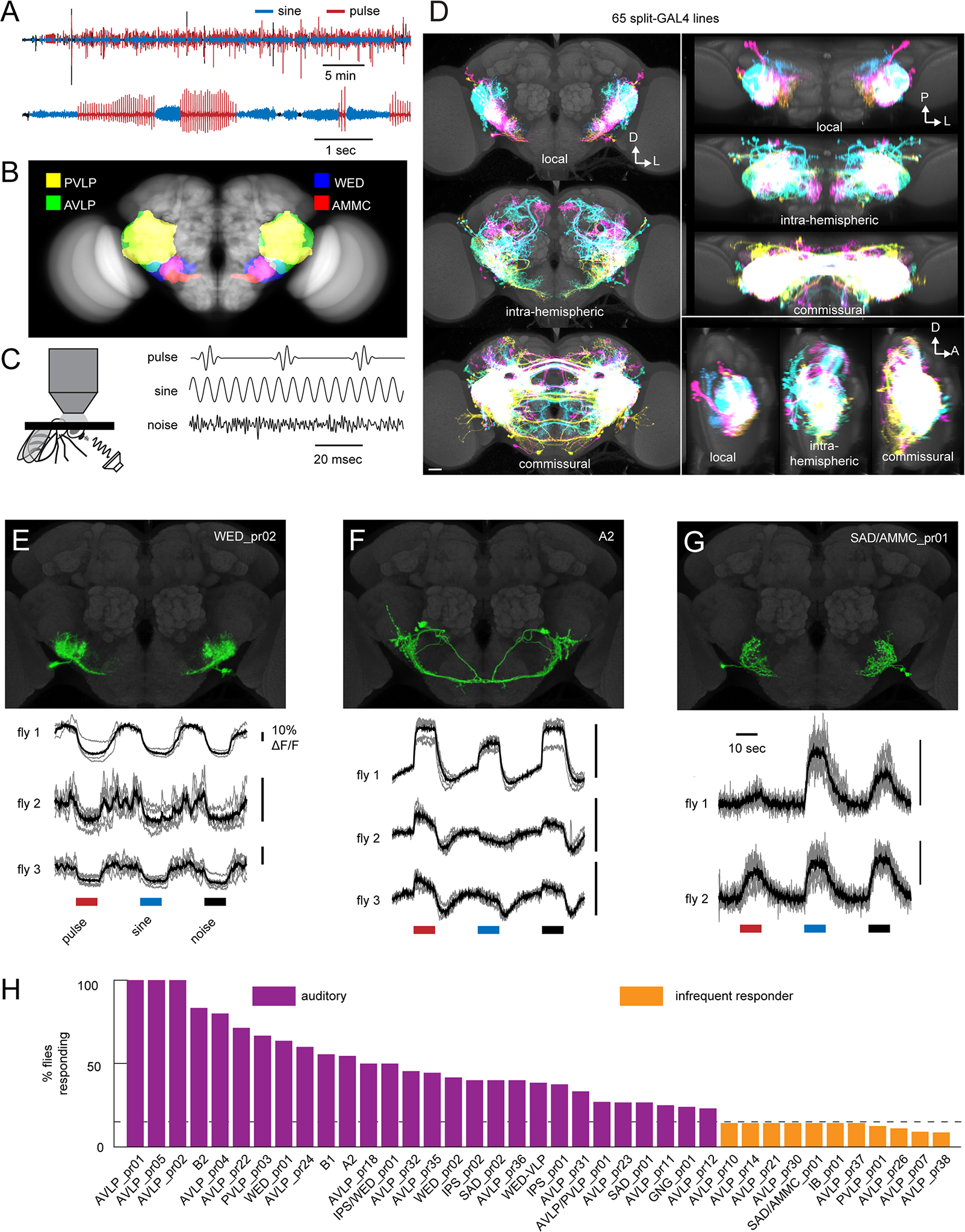
A) Microphone recording from a single wild-type (CS-Tully strain) male fly paired with a virgin female. The top trace shows song over 30 minutes, and the bottom trace shows a close-up of song bouts consisting of switches between the pulse and sine song modes. B) Primary auditory neurons called Johnston Organ neurons in the antenna project to the antennal mechanosensory and motor center (AMMC) in the central brain. Auditory information is then routed to downstream areas including the wedge (WED), anterior ventrolateral protocerebrum (AVLP), and posterior ventrolateral protocerebrum (PVLP). See Table S1 for neuropil abbreviations. C) Schematic showing two-photon calcium imaging set-up with sound delivered to the aristae (left) and calibrated, synthetic acoustic stimuli used to search for auditory responses (right). 100 msec of each stimulus is shown. D) Overlaid images of the split-GAL4 collection’s local interneurons, intra-hemispheric projection neurons, or commissural neurons, segmented from aligned images of the split GAL4 collection (see also Data S1A–B) and shown as maximum projections from the front (left), top (top right), and side (bottom right). Each cell type was colored randomly. D: dorsal; L: lateral; P: posterior; A: anterior. Scale bar: 25 microns. E-G) Calcium responses to pulse, sine, and noise stimuli from three cell classes (see also Figure S1D–E). In the calcium traces, each trial is shown in grey and the mean across trials is shown in black. H) Percentage of imaged flies from each cell class with auditory responses. We defined auditory cell classes as those in which >15% (dotted line) of imaged flies responded to the pulse, sine, or noise stimuli. If at least 1 fly but fewer than 15% of imaged flies responded, we termed the cell class an ‘infrequent responder’. If no flies responded (out of 4–6 total flies), we termed the cell class ‘non-auditory’. Numbers of flies imaged ranged from 4–17 for auditory cell classes, and from 7–23 for infrequently responding cell classes. See Table S2 and Data S1C for cell class images and names. See also Tables S1–2.
Sound detection begins at the Drosophila antenna, within which Johnston’s organ neurons (JONs) detect sound-evoked vibrations of the arista 18. Since fly hearing organs are sensitive to the particle velocity component of sounds, which decreases rapidly with distance 19, flies can only hear nearby sounds - primarily communication signals from conspecifics. JONs are coarsely frequency-tuned and project roughly tonotopically to the antennal mechanosensory motor center (AMMC) in the brain 20. This tonotopy appears to be strengthened in downstream areas, like the wedge (WED) 20 and ventrolateral protocerebrum (VLP) 21 (Figure 1B). The auditory responses of a few AMMC neurons have been characterized 22–25, and one of these, B1, is thought to be part of a major pathway for song processing 26,27. Downstream of the AMMC, only a handful of auditory neurons have been identified 16,17,25,28,29, and for most of these cell types, song coding has not systematically been examined.
Although pan-neuronal imaging found both pulse and sine responses throughout the brain21, we do not yet know how auditory circuits are organized, and whether song feature tuning is sharpened along the pathway hierarchically. In one model of network organization, pulse- and sine-preferring neurons constitute separate pathways, and hierarchical organization within each pathway sharpens tuning to generate strong song mode preference. Support for this model comes from work on higher-order pulse song preferring neurons16,29,30 that connect with descending neurons (neurons with dendrites in the brain and axons in the ventral nerve cord) driving sex-specific behaviors; however no sine-tuned neurons have yet been identified. In a second extreme, pulse and sine preference might arise early in the pathway, and interconnections between neurons with different song mode preferences might act to diversify responses to song features.
We address these outstanding issues here, by uncovering the network organization underlying song processing in the Drosophila brain. We recorded auditory responses from a new collection of split-GAL4 lines, and found 24 novel auditory cell types, as well as new lines for known auditory neurons. We systematically characterized each cell’s preference for sine vs. pulse song, in addition to tuning for sine frequency and pulse rate, discovering a continuum of song mode preferences among auditory neurons. Using FlyWire, a whole brain connectomic resource 31, we mapped synaptic connectivity (in addition to the sign of connections, whether inhibitory or excitatory) among neurons. Rather than being organized into separate streams for sine and pulse processing, we discovered a complex network architecture with dense interconnectivity between cell types with different song responses and without hierarchical structure. Preference for sine and pulse song arises early in the pathway, and using a linear dynamical system model, we showed that neural responses could be predicted by the responses of their inputs. Finally, we found that the time courses of neural responses are matched to natural song statistics. These results reveal how sensory responses are shaped by connectivity within the auditory system of Drosophila.
RESULTS
Identifying cell types of Drosophila auditory circuits
To identify new cell types downstream of known early auditory neurons 31, we focused on three of the five areas to which second-order mechanosensory neurons in the AMMC send projections 32: the wedge (WED), anterior ventrolateral protocerebrum (AVLP), and posterior ventrolateral protocerebrum (PVLP) (Figure 1B). Several higher-order auditory neurons have projections in these areas16,25,28,29. We created sparse and specific split-GAL4 lines targeting such neurons (hereafter referred to as WED/VLP neurons), and then screened the most promising lines for auditory responses (Figure 1C); our goal was not to create a line for every cell type in the WED/VLP (of which over a thousand are estimated to exist 33), but to identify new auditory neuron types. In total, we examined the expression of 1041 split-GAL4 lines, generated stable lines for 117 with the sparsest expression in the brain, and selected 65 for functional recordings. These 65 lines target at least 50 WED/VLP cell types that include local, intra-hemispheric projection, and commissural neurons (Figure 1D, Data S1A).
To uncover cell type diversity within each split-GAL4 line, we collected multicolor flip-out (MCFO) images (see Methods). Most split-GAL4 lines contained morphologically similar neurons, with some heterogeneity within particular lines (Data S1B). As morphologically similar neurons may have different synaptic partners 33, we were unable to disambiguate further any individual subtypes within each line on morphological criteria alone, and consider the WED/VLP neurons in each line a ‘cell class’. We named each cell class by identifying the neuropil with the highest density of neural processes (Table S2). We refer to the neurons imaged in each line by these names, except for auditory neurons previously named (i.e., A2, B1, B2, and WED-VLP) (Table S2) 23,25. To clarify which cells we targeted in each line for calcium imaging, we digitally segmented WED/VLP neurons from the broader expression pattern of each line (Figure 1E–G and Data S1C).
We tested each cell type for auditory responses using GCaMP6s 34 and three stimuli: pulse, sine and broadband noise (Figure 1C; see Methods). Pulse and sine constitute the two major modes of D. melanogaster courtship song, and noise was included to identify responses to acoustic features outside the range of parameters in song. We used intensities within those produced during natural courtship35 (5 mm/sec for pulse and sine, and 2 mm/sec for noise). We found consistent auditory responses in 28 lines (43%) (Figure 1H), including four cell classes representing known auditory neurons: A2, B1, B2, and WED-VLP (Figure 2; Table S2). An additional 11 cell classes (17%) were classified as infrequent responders because responses occurred in fewer than 15% of imaged flies (Figure 1H). The remaining 26 cell classes did not respond to acoustic stimuli. Some auditory neurons were previously classified as lateral horn neurons, but not known to be auditory (e.g., AVLP_pr05 resembles AVLP-PN1 and IPS_pr01 resembles WED-PN136), while others have morphologies similar to neurons that process other mechanosensory stimuli, such as wind (e.g., WED_pr01 is distinct from but morphologically similar to the WPN neurons that encode wind direction 37 and to the WPNBs, whose function has not yet been characterized 38). In sum, our screen identified a total of 24 new auditory cell types. Using FlyWire 39, we identified these cell types in both hemispheres within an electron microscopic (EM) volume of an entire female brain (Figure 2; see Methods), and used the EM data below to examine connectivity between auditory cell types.
Figure 2. Light microscopic (LM) and electron microscopic (EM) images of auditory WED/VLP cell types.
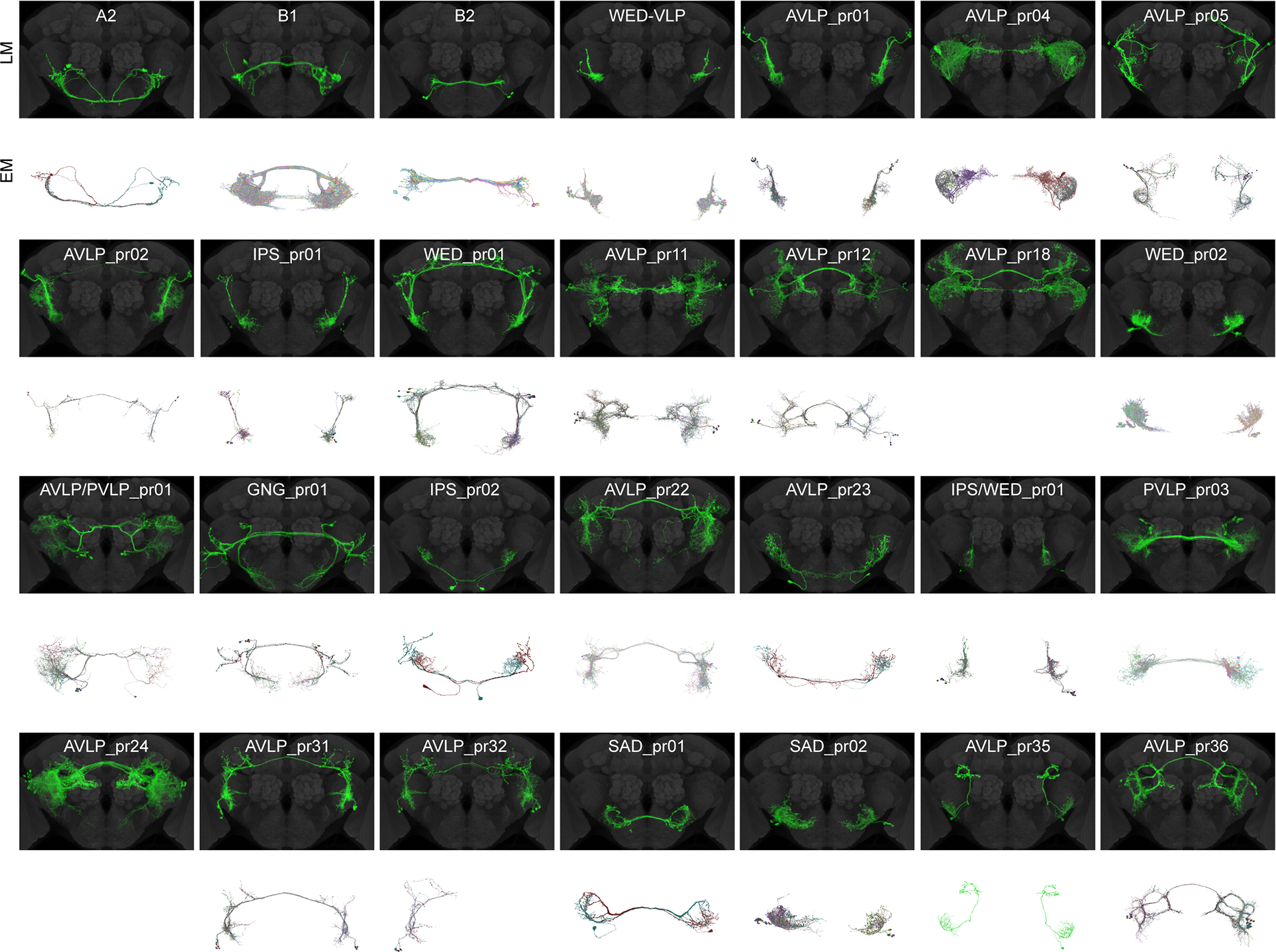
Aligned central brains with expression patterns of WED/VLP neuron classes digitally segmented (see Data S1B–C). Only those cell classes with auditory responses are shown. Gray: nc82. Below each brain expression pattern are the EM reconstructions (identified and proofread in FlyWire.ai, see Table S3) corresponding to each cell class. There was insufficient information in the split-GAL4 and stochastic labeling expression patterns to resolve the EM reconstructions representing two cell classes (AVLP_pr18 and AVLP_pr24). EM reconstructions representing cell type AVLP_pr32 were only found in one hemisphere. AVLP_pr01 and AVLP_pr02 share morphological similarities with vpoINs, which provide sound-evoked inhibition onto descending neurons called vpoDNs that contribute to vaginal plate opening (Wang et al., 2020b). Based on both FlyWire and hemibrain connections, vpoINs (defined as inputs to vpoDN with morphology consistent with vpoINs) consist of two subtypes: one with a commissure, and one with a medial projection that does not cross the midline. This leads us to conclude that AVLP_pr02 are likely the commissural vpoINs, but AVLP_pr01 are a cell type independent from vpoINs. See also Figures S2 and S3 and Tables S2 and S3.
Preferences and tuning for courtship song modes among WED/VLP neurons
We determined the preference of each auditory cell type for three stimuli: sine, pulse, and noise (Figure 3A). We evaluated preference for song (pulse or sine) vs. noise (Figure 3B) and pulse vs. sine (Figure 3C and Figure S1A–B) in each fly imaged (see Methods). Our noise stimulus was filtered to contain frequencies present in song, but lacked the temporal structure of song stimuli (Figure 1C). The overwhelming majority of neurons preferred song stimuli (pulse or sine) over noise (Figure 3B), with no correlation between song vs. noise preference and song mode (pulse vs. sine) preference. Neurons that preferred the noise stimulus (WED_pr01, AVLP_pr05, and WED_VLP) nonetheless produced significant responses to song and were tuned to particular sine frequencies or pulse rates (Figure 4).
Figure 3. Auditory WED/VLP neurons show a continuum of preferences for sine and pulse song modes.
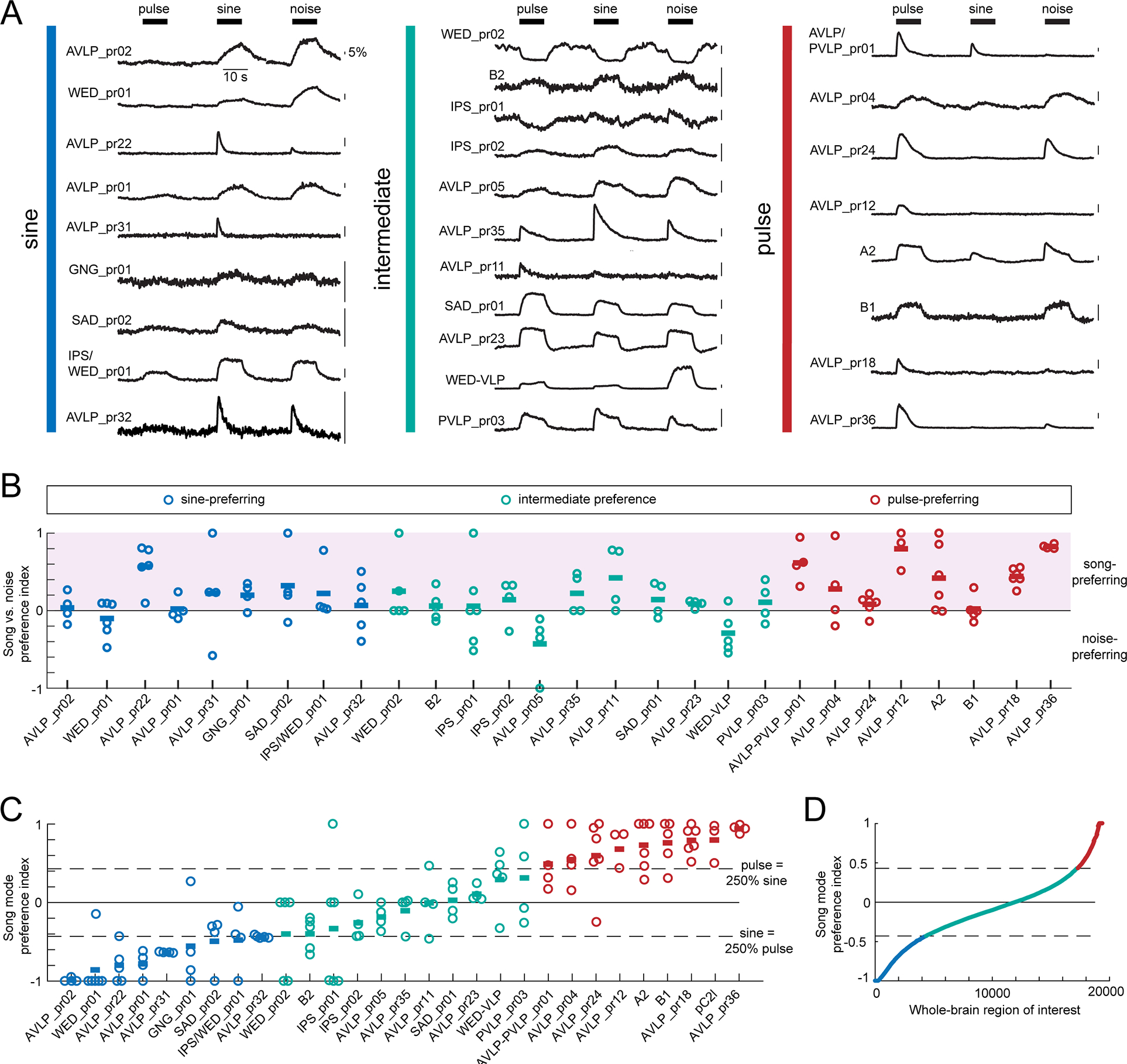
A) Trial-averaged representative calcium traces for a single fly from each cell class in response to pulse, sine, and noise stimuli. Vertical colored bars indicate the pulse- vs. sine-preference of each cell class given in (C). B) The integrals of responses to pulse, sine, and noise were used to calculate a song vs. noise preference index, which ranges from −1 (strongest noise preference) to 1 (strongest song preference) (see Methods and Figure S1A–B). The color of the dots reflect the pulse- vs. sine-preference of each cell class given in (C). Each dot represents the responses of one fly, and horizontal lines represent the mean within each cell type. To identify sine-preferring cell types, we required the mean preference index across flies to be below −0.43, which corresponds to a sine response that is at least 250% that of pulse. To identify pulse-preferring cell types, we required the mean preference index across flies to be above −0.43, which corresponds to a pulse response that is at least 250% that of sine. All other cell types were classified as having intermediate song mode preference. The song mode preference index for pC2l was calculated using data from a previous study 16. Each dot represents the responses of one fly, and horizontal lines represent the mean within each cell type. There was no correlation between song vs. noise preference in (B) and song mode preference in (C) (Spearman’s rho=0.15, p=0.071, n=143 flies). Tan dots indicate cell types that contact JONs (see Figure S4B). See also Figure S1D–F for response variability and Figure S2 for the neuropils innervated by neurons from each song mode preference class. D) The song mode preference index for 19,389 auditory-responsive regions of interest (ROIs) from the entire central brain, obtained via pan-neuronal imaging in a previous study 21. See also Figures S1–3 and Table S2.
Figure 4. Pulse rate (interpulse interval) and frequency tuning.
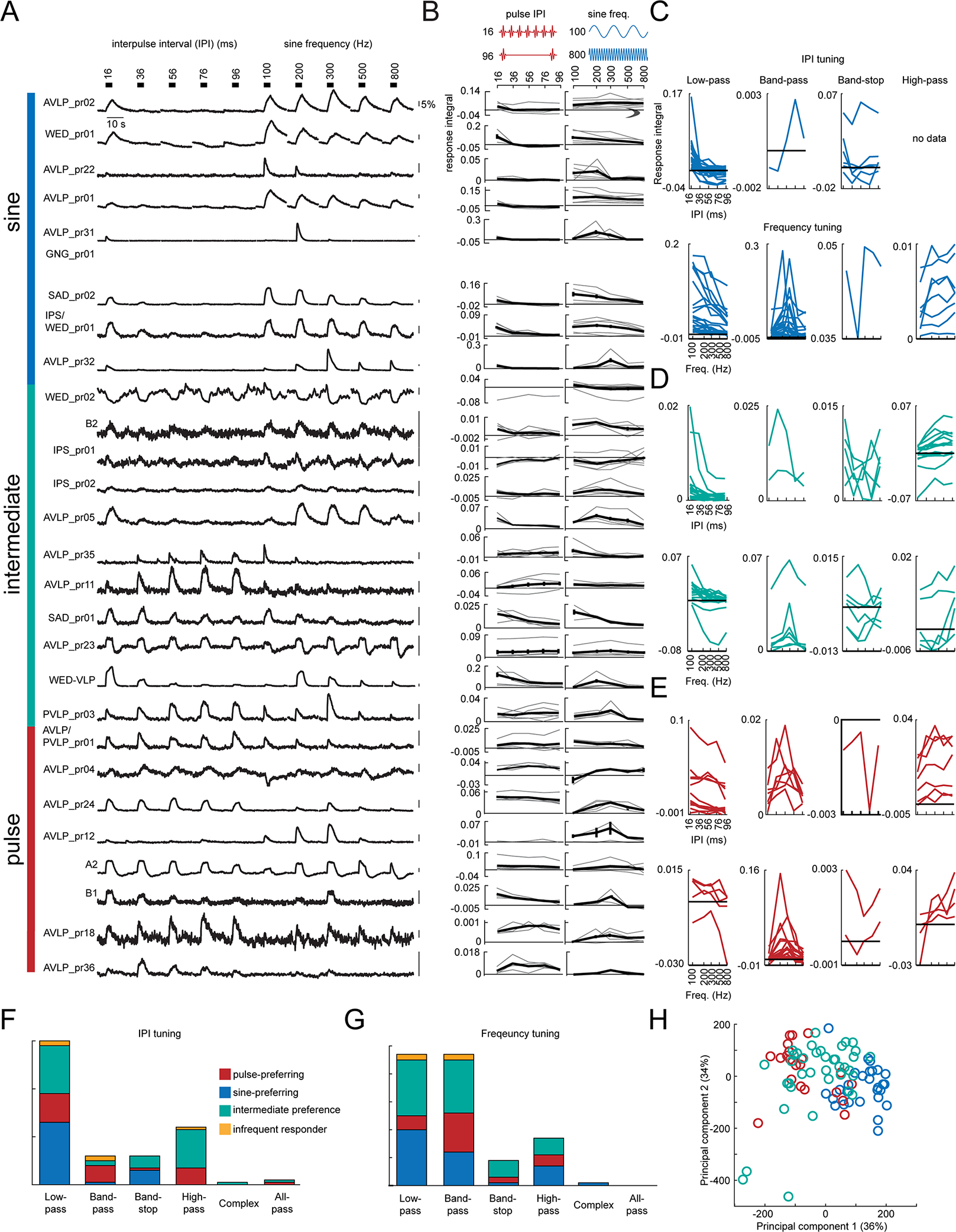
A) Trial-averaged representative calcium traces in response to pulse rate (interpulse interval (IPI)) and sine frequency stimuli. B) Pulse rate (left) and sine frequency (right) tuning curves. The tuning curves from individual flies are shown in grey, and the average across flies is shown in black. Error bars report standard error. C-E) Tuning curves for each fly recorded in the data set. Tuning curves are colored according to the song mode preference of each WED/VLP neuron type. F-G) Histogram of IPI (F) and frequency (G) tuning types across the dataset. Responses that were roughly equal for every stimulus were classified as all-pass, and responses that did not fit any other category were classified as complex (see Methods). H) Principal components analysis (PCA) on the response integrals elicited by IPI and frequency stimuli. Each dot represents one recording, and the color represents the song mode preference for each cell type. PC1 positively correlates with responses to 100 and 200 Hz stimuli, and negatively correlates with responses to 36–96 ms IPI stimuli. PC2 positively correlates with responses to 16 and 36 ms stimuli, and negatively correlates with responses to 100 and 800 Hz stimuli. See also Table S2.
Prior work on Drosophila auditory neurons uncovered largely pulse-preferences 16,28,29, even though males spend the majority of their time singing sine song 15. By sampling a larger population of cell types, we found that song mode preferences across neurons fell along a continuum (Figure 3C), with 31% pulse-preferring, 31% sine-preferring, and the remaining 38% showing intermediate preference (responding to pulse and sine stimuli roughly equally). In the subset of recordings with net inhibition (Figure S1C), pulse-preferring neurons tended to have sine-preferring inhibition, and vice versa, suggesting that inhibition contributes to establishing auditory preferences. We next re-analyzed 19,036 regions of interest spanning the central brain of 33 flies imaged pan-neuronally 21 according to the criteria established here, and uncovered a similar continuum (Figure 3D). This suggests that the song mode preferences of the 28 WED/VLP cell types recorded here may underlie responses brain-wide. In support of this, WED/VLP auditory neurons send projections throughout the central brain, independent of song mode preference (Figs. 3 and 4), including to regions considered primarily visual (ie, posterior lateral protocerebrum) and olfactory (i.e., lateral horn) (Figure S2). Finally, some cell types exhibited more variability in song mode preference than others (Figure 3C), either across-trials or across-animals (Figure S1D–F), but we found a similar range of variability scores within each song mode preference class.
Both males and females change their locomotor speed most in response to conspecific pulse rates (peaked at 35–40 ms interpulse intervals) and sine frequencies (ranging from 150–400 Hz) 16. We therefore examined tuning for pulse rate and sine frequency, using stimuli at an intensity of 4 mm/s, which is within the range produced by males35 (Figure 4A). We observed a diversity in tuning patterns to these stimuli across cell types (Figure 4B). For each preference category (sine-preferring, pulse-preferring, or intermediate), we sorted tuning curves from each recorded fly by tuning type (low-pass, bandpass, high-pass, or band-stop (see Methods)). For pulse rate tuning, sine-preferring neurons were most likely to exhibit low-pass responses (Figure 4C and 4F; consistent with these neurons preferring sinusoids over pulses since the fastest pulse rates begin to approximate sinusoids), while intermediate-preference (Figure 4D) and pulse-preferring (Figure 4E) neurons showed more diversity in pulse rate tuning. Surprisingly, relatively few recordings (2 flies from AVLP_pr36 and 1 fly each from SAD_pr01, SAD/AMMC_pr01, and AVLP_pr35) exhibited band-pass tuning for the conspecific pulse interval (35–40 ms). This is in contrast with cell types tuned for conspecific intervals found in regions downstream from the WED/VLP (e.g., pC2l 16 and vpoEN 29). Our results suggest that WED/VLP responses may serve as a set of building blocks for generating band-pass tuning downstream. For sine frequency stimuli, most responses showed low-pass or band-pass tuning, with roughly equal numbers in these two categories (Figure 4C–E, lower plots and Figure 4G). Recordings from pulse-preferring neurons (Figure 4E) predominantly showed band-pass frequency tuning centered on conspecific pulse carrier frequencies (e.g. around 250 Hz40), whereas recordings from sine-preferring (Figure 4C) or intermediate preference neurons (Figure 4D) showed either low- or band-pass frequency tuning, with individual tuning curves tiling the broad range of frequencies present in conspecific sine song (100–400 Hz) 14. Finally, principal components analysis on responses to pulse rates and sine frequencies showed that, instead of distinct clusters, the population we recorded from (131 recordings from 30 cell types) constituted a continuum from pulse- to sine-preferring, with intermediate preference neurons in the middle (Figure 4H).
The connectome of Drosophila auditory neurons
Several models of network organization would be consistent with the continuum of preferences for sine and pulse song, as well as diversity of tunings for sine frequency and pulse rate (Figure 5A). In one extreme (“Model 1”), pulse- and sine-preferring responses occur in largely separate pathways, with either their own or shared intermediate preference neurons. Model 1 is supported by our analysis of LM images of each cell type (Data S1C), in which we found some spatial segregation of sine- and pulse-preferring neurons in the VLP (Figure S3), suggesting a separation between pathways. In the second extreme, pulse and sine preference already occur early in the pathway, and the continuum of responses arises through extensive interconnectivity between cell types with different preferences. Inhibitory interactions between cell types with different song mode preferences could act to sharpen or diversify tuning. Of course, the real wiring diagram could fall between these two extremes.
Figure 5. The auditory connectome.
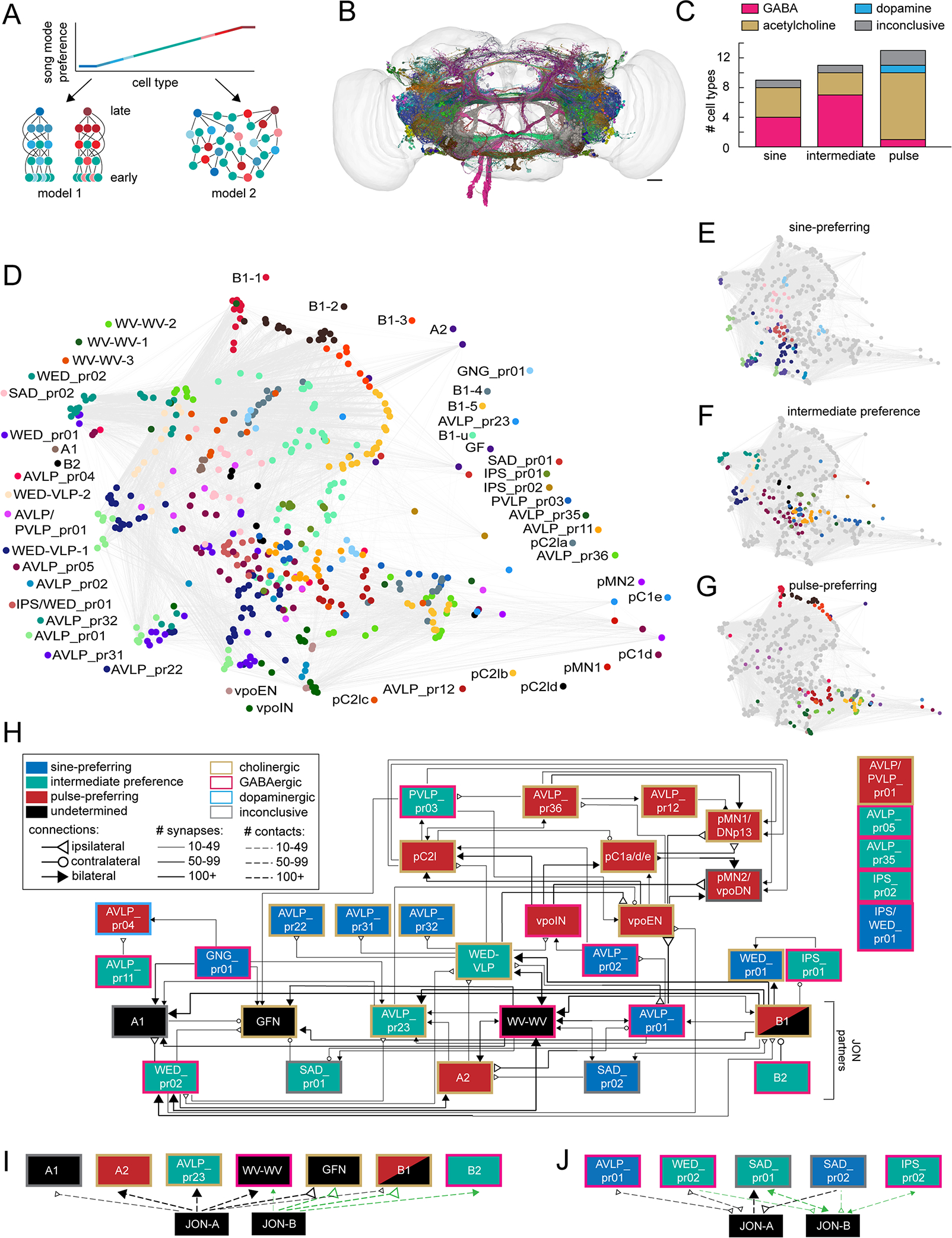
A) Two models for the organization of auditory pathways underlying the observed continuum of song mode preferences. Each circle represents a cell type, and each line represents synaptic connections. Red = pulse-preferring, blue = sine-preferring, and green = intermediate preference (see Figure 3C). The shading of red and blue neurons indicates the strength of pulse or sine preference, respectively. In model 1, neurons selective for pulse and sine are separated into distinct pathways, with neurons of intermediate preferences playing roles in both pathways (left). Within-mode connections sharpen tuning for song features. See Figure S3 for evidence of anatomic separation between pulse and sine pathways, supporting model 1. In model 2, neurons of different song mode preferences are highly interconnected at all levels without hierarchical organization (right). Downstream neurons may pool the responses of diversely tuned neurons to establish selectivity for a variety of song parameters. B) To test these models, we examined synaptic connections among auditory neurons (N=479 neurons from 48 cell types) in an electron microscopic volume of an entire female fly brain (see Figure S4A). D) Representation of the synaptic connectivity of auditory neurons using uniform manifold approximation and projection (UMAP). Each dot represents one neuron, the color of the dot represents the cell type, and lines represent synaptic connections. E-G) Same as D but for only sine-preferring (E), intermediate preference (F), or pulse-preferring (G) neurons. H) Flow-chart diagram of the auditory connectome. Each box represents a cell type, and each line represents a synaptic connection (see Methods for connection criteria). The song mode preference of A2, B2, and WED-VLP come from recordings from the split-GAL4 lines labeling those neurons (Figure 3C; Table S2), and the song preferences of several higher-order and descending neurons (pC2la-c, vpoEN, vpoIN, pC1a,d,e, pMN1/DNp13, and pMN2/vpoDN) come from previous studies 16,28–30. Cell types shown in black boxes have not had their sine/pulse preference determined. B1 shading reflects the observation that the split-GAL4 line we imaged from, while pulse-preferring, labels only a handful of B1 cells, and the tuning of the remaining B1 cells remains to be determined (but see 24. Five cell types formed no connections with other neurons in the dataset. See Figure S5 for neurotransmitter determination and connections of auditory cells with previously reported subtypes (ie, WED-VLP, WV-WV, and B1). See Methods for how the connectivity diagram was formed from identified reconstructions in FlyWire.ai. I) Diagram of connections between JONs and auditory cell types in which JONs are presynaptic (as measured by number of membrane contacts; see Methods and Figure S4B–E). Connections with JON-As are shown in black, and connections with JON-Bs are shown in orange. J) Same as (I) for connections in which auditory cell types are presynaptic to JONs. See also Figures S4–5 and Tables S2–3.
To test these hypotheses, we mapped synaptic connectivity among all auditory neurons (both WED/VLP neurons and previously characterized auditory neurons). We identified and proofread these neurons in both hemispheres of a whole brain electron microscopic volume (FlyWire) 39 (Figure 2, 5B), determined the number of synaptic connections via automated synapse detection 41 (Figure S4A), and used a deep learning-based classifier to predict the neurotransmitter used by each cell type 42 (Figure S5A–C). This classifier used neural networks trained on manually identified synapses to predict with high accuracy the neurotransmitter type of individual synapses. We defined the neurotransmitter of each neuron as the predicted majority neurotransmitter over all its automatically detected synapses43 (see Methods). We found that pulse-preferring neurons were overwhelmingly cholinergic, whereas sine-preferring and intermediate neurons included a mix of GABAergic and cholinergic neurons (Figure 5C). We also found one dopaminergic neuron in the pulse-preferring AVLP_pr04 cell type.
Using the dimensionality reduction method UMAP 44, we clustered auditory neurons based on their connectivity patterns (Figure 5D). Neurons of a cell type tended to cluster, indicating similarity of inputs and outputs, but we found limited clustering between different cell types - instead, we observed dense interconnectivity throughout the network (gray lines), more consistent with model 2. However, there were more sine-preferring and intermediate neurons in the middle of the graph (Figure 5E–F), and pulse-preferring neurons at the ends of the graph (Figure 5G), suggesting some preferential connectivity within a preference category. To examine this, we collapsed neurons by cell type (Figure 5H and Figure S5D–F), which highlighted extensive interconnectivity between cell types with different song mode preferences, again more consistent with model 2. For example, intermediate preference WED-VLP neurons receive inputs from all preference classes, and in turn send outputs to three pulse-preferring (vpoEN, vpoIN, and pC2l) and three sine-preferring (AVLP_pr01, AVLP_pr22, and AVLP_pr31) neurons. While the auditory connectome generated here is still incomplete, these data nonetheless reveal a lack of segregation of neurons into pulse and sine song processing pathways.
We also observed a mix of excitatory and inhibitory connections throughout the network (Figure 5H; Figure S5D–F). Inhibitory connections were most common between intermediate preference and pulse-preferring neurons (Figure S5G), suggesting that this connection type may be especially important for establishing responses to pulse song features. Sine-preferring neurons provided inhibition to both pulse and intermediate preference neurons, suggesting one major role for sine-preferring inhibitory neurons is to oppose responses to sine song (Figure S5G). The most frequent connection type was pulse-preferring neurons exciting other pulse-preferring neurons (Figure S5H) - many of these connections were in the sub-network of pulse-preferring neurons that includes descending neurons pMN1/DNp13 and pMN2/vpoDN 29,45. Although roughly half of the sine-preferring cell types were excitatory (Figure 5C), our screen did not identify any of their downstream targets. As we focused on finding WED/VLP cell types, it is possible that the targets of excitatory sine-preferring neurons have the bulk of their projections in other brain regions. Thus, excitatory pathways for sine processing remain to be further mapped.
To determine whether sine and pulse preference arise early in the pathway (more consistent with model 2), we mapped synaptic connections with 90 JONs previously identified in the EM volume46. Since automated synapse detection only identified a handful of JON synapses (see Methods), we instead detected the number of membrane contacts between JONs and auditory neurons (Figure S4B), with the rationale that membranes must be apposed for a synapse to occur (see Methods). We then manually validated the existence and directionality of synapses between JONs and auditory cells (Figure S4D–E). We discovered that 12 central auditory cell types form synapses with JONs, including both inhibitory and excitatory neurons, and neurons from all song preference classes (Figure 5I–J). This demonstrates that song mode preferences can arise early in the pathway, inconsistent with model 1. Four of these cell types were both pre- and postsynaptic to JONs (Figure 5J, Figure S4C–D), three of which are inhibitory, demonstrating that JONs receive feedback inhibition. One cell type (SAD_pr02) appeared to only be presynaptic to JONs (Figure 5J), though its neurotransmitter remains inconclusive. Overall, these findings are consistent with reports of both pre- and postsynaptic sites on JONs 46.
Taken together, our functional and anatomical results support a novel network organization for auditory processing: sine and pulse preference arise early in the pathway, and synaptic connections between differently tuned cells contribute to generating a continuum of song mode preferences (Figure 3C).
The auditory connectome is non-hierarchical, but nonetheless song responses are predictable from the wiring diagram
We found little evidence of hierarchical organization at a purely structural level. Using the binarized neural connectivity matrix with a threshold of at least 10 synapses between neurons (Figure S4A), we computed three measures of network hierarchy: orderability, feedforwardness, and treeness (Fig 6A). These metrics evaluate the fraction of neurons belonging to network cycles (or recurrent loops in the network), the overall contribution of cycles to paths through the network, and the extent to which the network diverges (fans out) from a set of root nodes, respectively 47. While the network could be condensed into a feedforward network of connected components (Figure 6B), with approximately half of the neurons uniquely orderable (Figure 6C), paths through the network were nonetheless dominated by recurrent connections, and the network tended to neither fan out nor converge from a set of root neurons. The network deviated only marginally in each metric from random networks with matched degree distributions (Figure 6C–E), with values markedly less than those of a perfectly feedforward, tree-like network. Thus, the network structure, even when not considering song mode preferences, exhibits little hierarchical organization beyond that expected from chance.
Figure 6. Evaluating the hierarchical structure of the auditory network and emergence of postsynaptic tuning curves from connectome-weighted presynaptic responses.
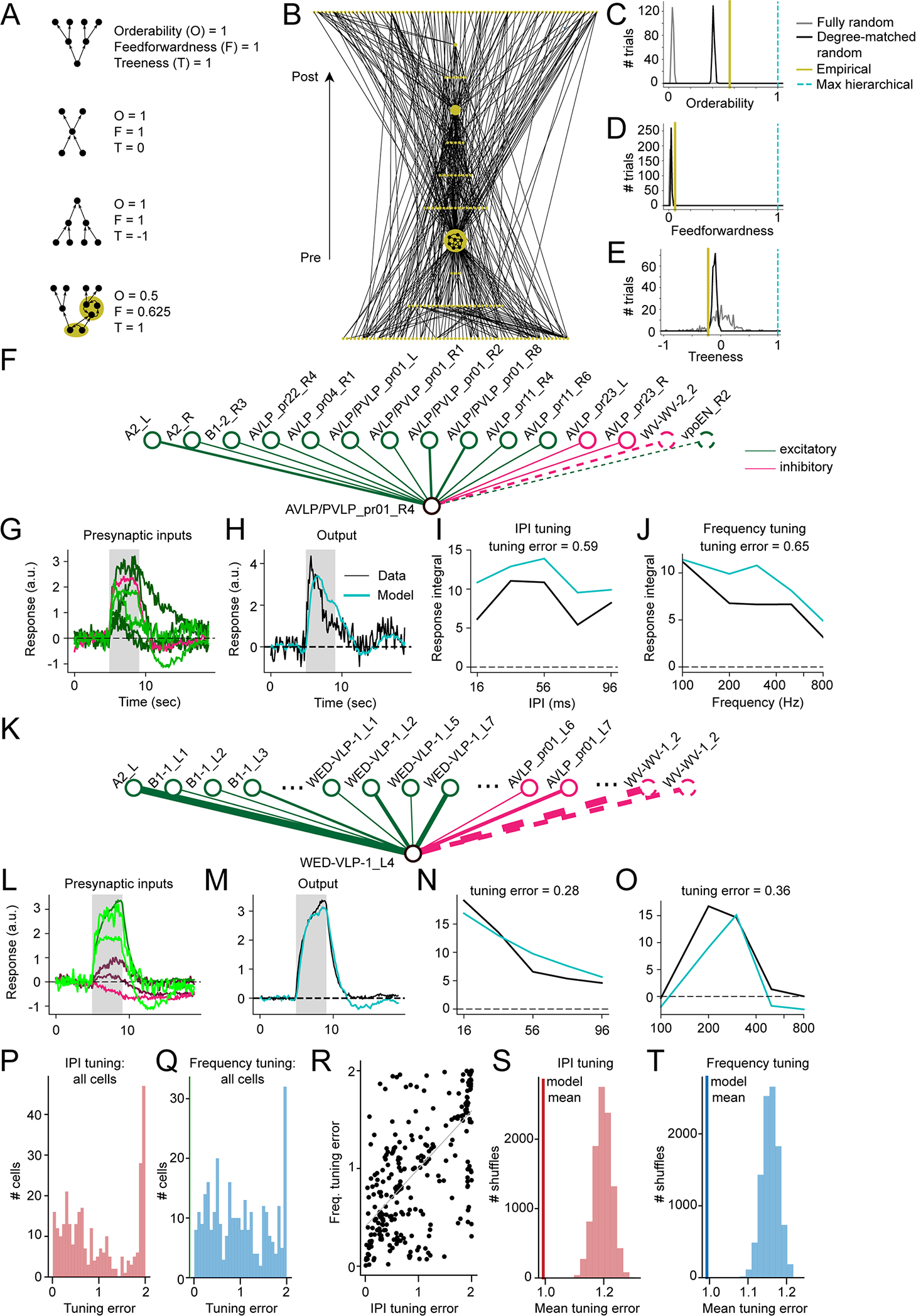
A) Measures of hierarchical structure (orderability, feedforwardness, and treeness) for four simple networks (reproduced from 47); each gold region highlights a strongly connected component (a set of neurons all mutually reachable from one another via at least one directed path). B) Hierarchically displayed node-weighted graph condensation of the empirically measured auditory network. Each node in the condensation is a strongly connected component of the original auditory network, with node size indicating the number of neurons in the component (minimum 1 neuron, maximum 124 neurons). Connections are oriented upward with postsynaptic targets displayed above presynaptic sources. C) Orderability of the auditory network, computed from the original network and the graph condensation (A), as compared to 300 instantiations (trials) of either a fully random network (with only neuron count and connection probability matched to the original auditory network) or degree-matched random network (in which each neuron’s incoming and outgoing connection numbers were inherited from the empirical network but connections were otherwise randomized). D) Feedforwardness of the auditory network, as compared to fully random and degree-matched random networks. The low value arises from most paths passing through the largest connected component. E) Treeness of the auditory network, as compared to fully random and degree-matched random networks. Prior to analysis, connections between two neurons in the original network were only counted if at least 10 synapses were detected (171 out of 476 neurons were unconnected to the main network and left out of the calculations). A maximally hierarchical network has a feedforwardness, orderability, and treeness of 1 (cyan; C-E). F) Schematic of network model used to compute neural responses and tuning for cell AVLP/PVLP_pr01_R4 (lower node). Upper nodes represent input neurons and lines indicate synaptic connections. Solid lines represent inputs from imaged split-GAL4 lines and dotted lines represent inputs from neurons outside our imaging dataset. G) Normalized responses of cells presynaptic to AVLP/PVLP_pr01_R4 (15 total, 2 without tuning data), in response to 36 ms IPI stimulus. Coloring shows weight of presynaptic cell onto postsynaptic cell, with brighter green or red indicating stronger excitatory and inhibitory weights, respectively. The grey box indicates when the stimulus was on. H) Observed postsynaptic response to the same 36 ms IPI stimulus (black) vs response modeled by feeding presynaptic responses through the network model (teal). I) IPI tuning curves from observed and modeled postsynaptic responses, computed by integrating responses from stimulus onset to 4 seconds post stimulus offset. J) Same as (I) for sine frequency tuning. K-O) Same as (F-J) for cell WED-VLP-1_L4 (29 total, 3 without tuning data). Ellipses in (K) indicate additional neurons in the data that we did not have room to show in the diagram. P) Root mean squared error between true and modeled IPI tuning curves across all cells with both post- and presynaptic tuning data (N = 306 cells); tuning curves were z-scored first to compare shape rather than magnitude. Q) Same as (P) for sine frequency tuning curves. R) IPI tuning error vs. sine frequency tuning error across cells (N=306 cells, R = 0.671, p < 10^−39; Wald test with zero-slope null hypothesis). S) Histogram of mean errors computed by shuffling true relative to modeled tuning curves across cells (10000 shuffles, p < 0.0001, computed by counting number of shuffles yielding a mean value less than the observed mean). Vertical line indicates the mean from (P). T) Same as (S) for frequency tuning curves (p < 0.0001). Vertical line indicates the mean from (Q). See also Table S2.
Despite a lack of hierarchical organization, are the responses of individual cell types predictable from their inputs? To address this question, for each neuron belonging to a cell type we imaged, we compared its recorded responses to those predicted by a linear dynamical system model 48,49 (see Methods) whose inputs were the responses of its presynaptic partners, weighted by their synaptic strengths (Figure 6F–H, K–M). In this model, the time derivative of the response at each time point is given by the connectome-weighted sum of the presynaptic neurons’ responses plus a “leak” term that causes the response to decay to zero absent further presynaptic inputs. The model for each neuron is parameterized only by the time constant τ, which determines the decay/leak rate of the postsynaptic response. Since we did not have access to the true membrane time constants, we set τ = 0.5 s for all cells, which roughly recapitulated the observed rise times in the recorded cells (Figure 6H,M).
We computed interpulse interval (IPI or pulse rate) and sine frequency tuning curves of the model responses and compared them to the true tuning curves (Figure 6I–J, N–O). While several neurons’ tuning could be predicted well (Figure 6F–O), this varied across neurons (Figure 6P–Q). This may be due to missing inputs in the model, consistent with our observation that IPI and sine frequency tuning prediction errors were strongly correlated (Figure 6R). Additional causes of mismatches between model predictions and data include nonlinear relationships between synapse number and connection strength, nonlinear transformations of synaptic inputs, and/or response heterogeneity within cell type. Nonetheless, the mean prediction error across the population was significantly less than when we randomly shuffled the predictions relative to true tuning curves (Figure 6S–T). This suggests that even absent recordings from all input neurons and a model of variation in intracellular properties, the connectome meaningfully constrains how the tuning of individual neurons emerges from their inputs.
Evaluation of song response timescales
In addition to diversity in song mode preference, our recordings revealed a diversity of response timescales (Figure 3A, 4A). To quantify this, we computed response and adaptation time constants τr and τa, respectively, by fitting each neuron’s pulse and sine responses with a simple model (Figure 7A; note that these are different time constants from those used to predict neural responses from presynaptic inputs in the previous section, as τr and τa here reflect neural responses to song rather than presynaptic inputs). Response time constants across all recordings were well matched to the duration of male song bouts during natural behavior 15 (Figure 7B), suggesting that the time constants of neural responses are appropriate for analyzing song. Sine song tends to occur in longer contiguous segments than pulse song (Figure 7C), and accordingly, sine-preferring neurons tended to have longer response time constants than pulse-preferring neurons (Figure 7D). Across song mode preference categories, we observed a wide range of response time constants (Figure 7E) and adaptation rates (1/τa) (Figure 7F). Although we found no correlation between these temporal features and pulse vs. sine preferences, we found that stronger preferences for song (compared to noise) were correlated with faster τr (Spearman rank correlation, rho=−0.31, p<0.001) (Figure 7G) and faster adaptation rates (rho=0.35, p<0.0001) (Figure 7H).
Figure 7. Response time courses are matched to song statistics.
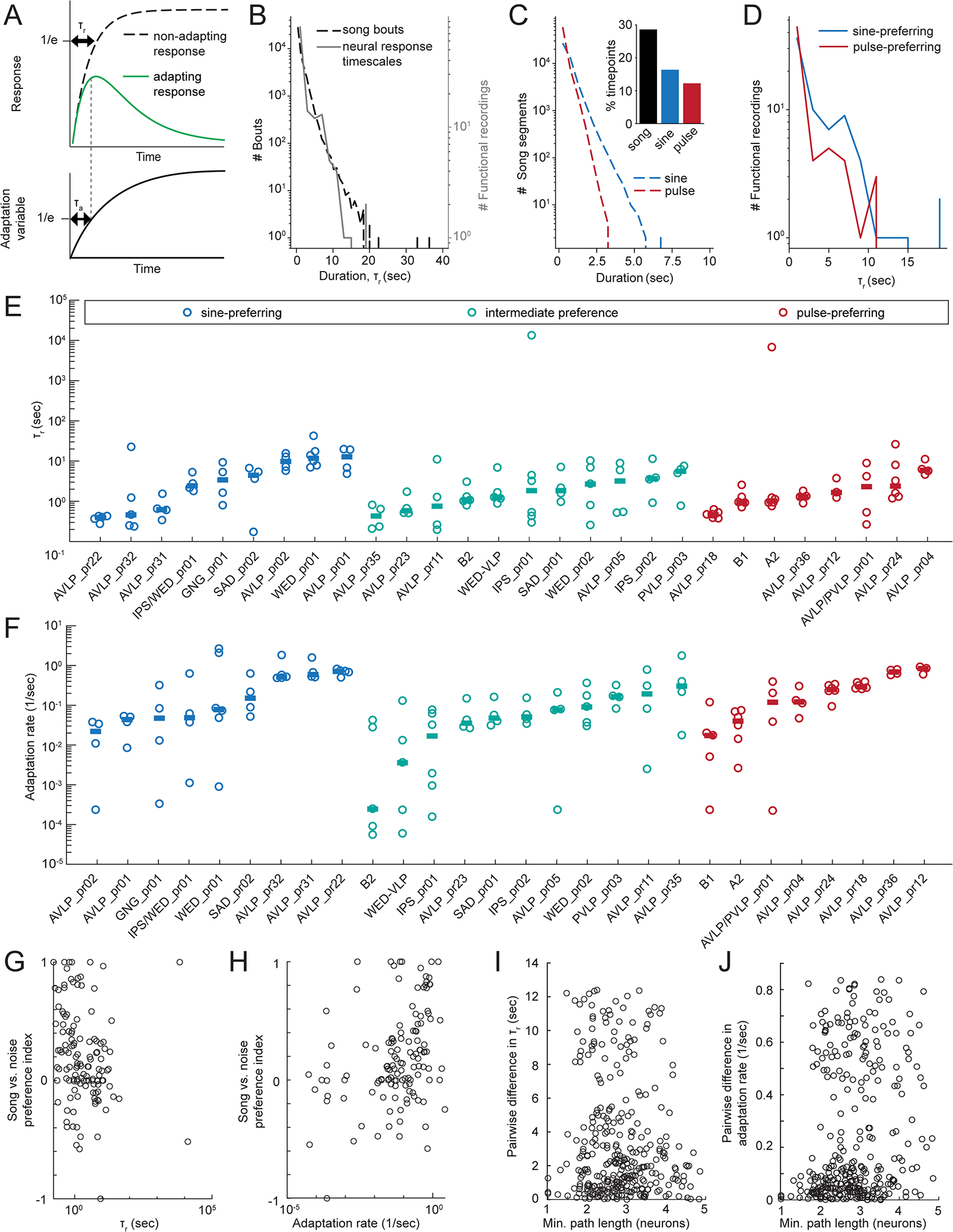
A) Schematic of calculation of response time constant τr and adaptation time constant τa (see Methods). The neuron’s total response is modeled by the green curve. B) Distribution of song bout durations (dotted line) and best fit response timescales τr to imaged neurons (grey). Song data was recorded during a naturalistic courtship assay in a previous experiment 15; data available from 67). Song was recorded with a microphone then downsampled to 30 Hz; each downsampled frame was labeled either sine, pulse, or quiet. Bouts were defined as contiguous singing periods (quiet periods of only 1 frame were ignored in separating bouts). C) Distribution of contiguous pulse and sine song segment durations. Inset shows a histogram of % time with any song, pulse, or sine. D) Distribution of τr for pulse- vs. sine-preferring neurons. E) Response time courses for each cell type. Each dot represents the recording from one fly and horizontal lines indicate across-fly means. F) Same as E for adaptation rate (1/τa). G) Song vs. noise preference vs. τr for all recordings. We found a negative correlation between τr and song vs. noise preference (Spearman rank correlation, rho=−0.31, p<0.001, n=125 flies). H) Song vs. noise preference vs. adaptation rate for all recordings. We found a positive correlation between adaptation rate and song vs. noise preference (Spearman rank correlation, rho=0.35, p<0.0001, n=125 flies). I) Difference in τr between every pair of neurons and the corresponding path length between each pair. There was no significant correlation (Spearman rank correlation, rho=−0.049, p=0.20, n=676 pairs of neurons). J) Same as (I) for adaptation rates. There was a weak but significant correlation between similarity of adaptation rate and path length (Spearman rank correlation, rho=0.24, p<1e-9, n=676 pairs of neurons). See also Table S2.
To determine whether response timescales may be inherited from a neuron’s inputs, we compared the differences in response time constants and adaptation rates between every pair of neurons with the path length between each pair, which is the number of synaptically connected neurons between them. For instance, the path length between a pre- and postsynaptic pair is 1. We found no correlation between the similarity of τr and path length (Figure 7I), and only a weak correlation between the similarity of adaptation rate and path length (Spearman rho=0.24, p<1e-9)(Figure 7J), suggesting that cell-intrinsic, rather than network properties, play the larger role in shaping response time constants. We posit that the diversity of time constants may be important for processing multi-timescale song structure (see Discussion).
DISCUSSION
Here we identified several new auditory neuron types in the Drosophila brain. Prior work had focused on either early or higher-order auditory neurons, leaving open the characterization of intermediate cell types. By systematically characterizing tuning for the two modes of courtship song and examining synaptic connectivity among all new and previously identified auditory neurons using the connectomic resource FlyWire, we discovered extensive interconnectivity between cell classes with different song mode preferences (Figure 5). We found that song mode preference arises in neurons postsynaptic to auditory receptor neurons, and that the connectome lacks hierarchical structure. These results rule out a model in which the two main song modes are processed in separate pathways (and one in which song mode preference is built up within each pathway), but instead support a model in which interconnectivity contributes to generating a continuum of song mode preferences. We tested this by predicting responses at each node in the connectome from the responses of the inputs using a simple linear model (Figure 6), and discovered strong predictability, suggesting that the wiring diagram places a strong constraint on functional responses, consistent with recent studies in the fly olfactory system 50.
Response continuums have been observed in fish 51,52 and rodents 53 54 55, and are thought to support more efficient encoding than would be possible with categorical representations. In the fly auditory system, neurons with intermediate preference (that respond roughly equally to sine and pulse) could encode sound intensity, location, and/or long timescale features, such as the variable sequence of pulses and sines that make up song bouts 15. The continuum of song preferences we discovered among WED/VLP neurons matches that found throughout the entire brain (Figure 3), suggesting that these neurons are representative of the larger auditory population, including those neurons that drive song-responsive behaviors. Many descending neurons (DNs) that control locomotion have dendrites in a number of regions with auditory activity, including WED/VLP 56,57. Sampling from a continuum of responses to sine and pulse would provide these DNs with the ability to respond to a wide array of song patterns. Finally, we also found a continuum of time constants among auditory neurons (Figure 7). This diversity of timescales matches the distribution found in natural song bouts, and female behavior is sensitive to song history across timescales 17. The diversity of neural response time constants could serve as a useful substrate for retaining the multi-timescale structure of song history in a population neural code.
We identified new auditory neurons by targeting cells within the WED/VLP, putative auditory areas 25,32. In the connectome we built using FlyWire 31, which allows proofreading of EM reconstructions in an entire female brain 39, we could examine inter-hemispheric connections, as well as ipsilateral vs. contralateral connection patterns (Figure 5H). 58 By combining functional and connectomic data, we could predict auditory tuning from the wiring diagram, and our findings reveal the importance of the connectome for understanding sensory responses. How many neurons remain unknown in the auditory pathway is difficult to estimate, but continued EM circuit tracing combined with functional imaging of sparse driver lines, as we did here, should ultimately fill out the auditory connectome. Since song representations are present in 33/36 central brain areas 21, we expect auditory circuits to be highly interconnected to circuits of other modalities and functions. Indeed, the auditory neurons we discovered send projections throughout the brain, including to olfactory, visual, and pre-motor areas (Figure S2), suggesting roles in multisensory integration.
Prior pan-neuronal imaging work found tonotopy in JON axons that was maintained in the AMMC and potentially sharpened in the WED 20. Here we uncovered that neurons directly downstream of the JONs exhibit strong preferences for pulse or sine song (Figure 5I–J), suggesting that such preferences may be inherited from the periphery. We also found that multiple cell types provide inhibitory feedback onto JONs (Figure 5I–J). In vertebrates, efferent inhibition protects against damage from loud sounds by modulating the sensitivity and selectivity of the auditory periphery59. Whether any of the connections we describe here play a similar role remains to be determined. Very few WED/VLP neurons showed band-pass tuning for conspecific pulse rate, suggesting that downstream processing generates such selectivity, as is observed in neurons such as pC2l or vpoEN 16,28,29. For instance, the two inputs onto vpoEN with the most synapses are inhibition from AVLP_pr01 and excitation from WED-VLP (Figure 5H), both of which respond most strongly to short IPIs (Figure 4B). This pattern could result in band-pass tuning in vpoEN since the inhibition is sharply tuned to IPIs shorter than the conspecific range, allowing excitation to dominate in the conspecific range. This would be consistent with vertebrate auditory systems, in which frequency tuning arises in the periphery, whereas tuning for temporal features, such as pulse rate, arises through central computations 60. We also found that the responses of some cell types were highly stereotyped across animals, whereas others were more variable. The role of variability in auditory responses is currently unknown, but it may function to generate variation in song preference across animals, potentially useful for diversifying female mate selection preferences.
Mapping synaptic connectivity among auditory neurons revealed a high degree of intermixing between preferences for the two song modes (Figure 5H). One example is inhibitory interactions that refine tuning. For instance, the three sine-preferring inhibitory cell types we described (GNG_pr01, AVLP_pr01, and AVLP_pr02) all send outputs to different pulse-preferring neurons (Figure 5H), suggesting that a major role for these neurons is to enhance or establish pulse preferences.. The use of inhibition to sharpen auditory tuning is widespread. In grasshoppers and katydids, convergence of narrowly-tuned inhibition with broadly-tuned excitation shapes frequency tuning 61,62. Sideband inhibition, in which stimuli just outside of a neuron’s preferred range elicit inhibition, also sharpens frequency tuning in mammalian central neurons 63. Inhibitory connections occurred between almost every possible combination of song-mode preference classes (Figure S5G), suggesting that inhibition may play a number of roles in shaping song tuning in flies.
Prior work on auditory systems has led to ideas about segregation of processing into separate streams for different sound categories 64,65, but these ideas do not address how animals might encode patterns or sequences of sound elements. Further, while separate streams may be useful for detecting different classes of stereotyped acoustic signals, such segregation may not be sufficient for analyzing rapidly varying sequences of elements, as occur in fly and some types of bird songs 15,66. In these cases, encoding longer timescale patterns, such as the recent history of interleaved song modes, may benefit from drawing upon neural responses with a range of auditory preferences. Future work should examine how this network is read out by downstream neurons to drive behavioral responses to song.
STAR METHODS
Resource availability
Lead contact.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mala Murthy (mmurthy@princeton.edu).
Materials availability.
Split-GAL4 lines generated by this study are available from Janelia.
Data and code availability.
Raw data supporting the current study are publicly available: https://doi.org/10.34770/7zem-x425. Code generated by the current study is also publicly available: https://github.com/rkp8000/7UN3_1N_DR0P_0U7. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
Experimental model and subject details.
The split-GAL4 system 68,69 was used to express the activation domain (AD) and the DNA-binding domain (DBD) of GAL4 under the separate control of two genomic enhancers, to obtain the intersection of their expression patterns, with the goal of obtaining a sparser pattern. Split-GAL4 stocks were gifts of Gerald Rubin and Barry Dickson 69–71. 20xUAS-GCaMP6s, td-Tomato/CyO was generated by Diego Pacheco 21. The genotype of imaged flies was 20xUAS-GCaMP6s, td-Tomato/GAL4-AD; GAL4-DBD/+, with the “+” originating from NM91 stocks. For expression pattern staining, split-GAL4s were combined with 20xUAS-CsChrimson-mVenus trafficked in attP18, except that 6 lines (17963, 21914, 23281, 23627, 28822, 29146) were shared from a different project that instead used pJFRC51–3XUAS-IVS-syt::smHA in su(Hw)attP1,pJFRC225–5XUAS-IVS-myr::smFLAG in VK00005, and one (27932) used pJFRC200–10XUAS-IVS-myr::smGFP-HA in attP18, pJFRC216–13XLexAop2-IVS-myr::smGFP-V5 in su(Hw)attP8. For multicolor flip-out (MCFO) staining, all were crossed to MCFO-1 [pBPhsFlp2::PEST (attP3);; pJFRC201– 10XUAS-FRT>STOP>FRT-myr::smGFP-HA (VK0005), pJFRC240–10XUAS-FRT> STOP>FRT-myr::smGFP-V5-THS-10XUASFRT>STOP>FRT-myr::smGFP-FLAG (su(Hw)attP1)]. Flies were kept at 25C with a 12h:12h light:dark cycle. All experiments were performed on female flies.
Method details.
Split-GAL4 creation.
GAL4 images from the Rubin and Dickson collections 69,70,72 were visually screened for lines targeting neurons in the WED and AVLP. For each cell type, a color depth MIP mask search 73 was conducted to find other GAL4 lines with expression in similar cells. Split-GAL4 hemidrivers for these lines were crossed in various combinations to find a combination that targeted the cell type of interest but with sparse expression elsewhere, determined by expression and staining of mVenus in one female fly of genotype 20xUAS-csChrimson::mVenus (attP18)/w; Enhancer-p65ADZp (attP40)/+; Enhancer-ZpGAL4DBD (attP2)/+. 65 combinations were chosen by this method, representing over 50 cell types. In some cases, lines targeting neurons with similar gross morphology revealed one line with a commissure (see SS16374 and SS27885; SS41728 and SS41730 in Data S1A). For these examples, we characterized both lines due to the difficulty in determining whether they were truly similar cell types. Split-GAL4 combinations were then double balanced and combined in the same fly strain to make a stable split-GAL4, for which expression pattern staining was carried out in an additional female to confirm the expression seen in the initial screen. For multiple split-GAL4 lines targeting morphologically similar neurons, one (or sometimes a few) were selected according to the following criteria: least off-target expression in the central brain, largest number of neurons with expression, and/or most reliable calcium responses to acoustic stimuli across flies. Expression was further confirmed in multiple flies by multicolor flip-out staining (see below). All lines generated here and the corresponding image stacks are available via https://splitgal4.janelia.org.
CNS immunohistochemistry & imaging.
Brains and ventral nervous systems were dissected and stained using published methods 74–76. Antibodies used were rabbit anti-GFP (1:500, Invitrogen, #A11122), mouse anti-Bruchpilot (1:50, Developmental Studies Hybridoma Bank, University of Iowa, mAb nc82), Alexa Fluor 488-goat anti-rabbit (1:500, ThermoFisher A11034), and Alexa Fluor 568-goat anti-mouse (1:500, ThermoFisher A11031). Serial optical sections were obtained at 1μm intervals on a Zeiss 700 confocal with a Plan-Apochromat 20x/0.8NA objective. A detailed description of the staining and screening protocol is available at https://www.janelia.org/project-team/flylight/protocols under “IHC - Adult Split Screen.”
Stochastic labeling.
For multicolor flip-out (MCFO) stochastic labeling 74, approximately 8 females per split-GAL4 line received a 15 min heat shock at 37°C at 1–3 days old, and were dissected at 6–8 days. A detailed description of the MCFO protocol can be found at https://www.janelia.org/project-team/flylight/protocols under “IHC - MCFO.”
Image processing.
Images were adjusted for brightness and contrast without obscuring data. Images were processed in ImageJ (https://imagej.nih.gov/ij/) and Photoshop (Adobe Systems Inc.). Where noted, neurons were rendered and segmented from confocal stacks with VVDviewer software (https://github.com/takashi310/VVD_Viewer) 77,78 to visualize them in isolation. For this rendering and for computational alignment of brain images used where noted, brain images were registered using the Computational Morphometry Toolkit 79 to a standard brain template (“JFRC2014”) that was mounted and imaged with the same conditions. Segmented image stacks are available at https://splitgal4.janelia.org.
Neuropil innervation.
We registered the neuropil 80 maps to the unisex JRC2018 template by first bridging from IBNWB to JFRC2 81, and then JFRC2 to unisex JRC2018 82. To determine which neuropils each neuron type targeted, we used the segmented neuron stacks with manually removed somata. We then calculated the percentage of the neuron’s voxels in each neuropil. In Figure S2 we report all neuropils containing at least 1% of each segmented neuron’s total volume.
For naming auditory neurons not previously described as auditory, we used the neuropil in which the segmented neuron had the greatest percentage of expression. If a neuron had nearly equal (<1% difference) expression in two neuropils, we used both neuropil names. A two digit number was added to the neuropil name in sequential order based on the split-GAL4 line number (Table S2). To disambiguate our names from those of the hemibrain project 83 (as our neuron types/cell classes can likely be further segregated into subtypes based on both morphological and connectivity differences within a line), we added a ‘pr’ before each number for Princeton.
EM reconstructions.
We identified individual neurons in the FAFB volume using FlyWire.ai 31. Neurons were proofread using the editing tools in FlyWire to add missing pieces of arbor and remove incorrect pieces, focusing on the main backbone of a neuron, not attempting to add very small missing twigs. Sister cells from the same cell type were used to verify that a cell’s overall morphology appeared generally correct. WED/VLP reconstructions were proofread by 12 proofreaders consisting of both scientists and expert tracers from the Seung and Murthy labs. To find all candidates belonging to a given cell type, we found an EM cross-section of the primary neurite tract, and investigated every cell in the cross-section. We compared candidate reconstructions with LM images of our auditory lines (Data S1A–C). We were conservative with which reconstructions we included for each cell type; if a reconstruction did not match neurons present in MCFO images (Data S1B), we excluded it. Since a limited number of MCFO images were available for each line, it is possible that we may have missed some EM reconstructions belonging to each line. Two auditory cell types (AVLP_pr18 and AVLP_pr24) did not have enough cell-type resolution in MCFO to enable individual EM reconstruction identification and were excluded from the synaptic connectivity mapping. EM reconstructions for pC1a/d/e, pMN1/DNp13, and pMN2/vpoDN came from a previous study 84. We identified vpoEN and vpoIN by inspecting the inputs to pMN2/vpoDN neurons 29. pC2l reconstructions were identified by D. Deutsch (personal communication). A list of all auditory neurons identified in FlyWire, along with their FlyWire coordinates and root ID numbers are listed in Table S3.
There are just under 60 B1 neurons per hemisphere 31, yet our split-GAL4 line targeting B1 neurons labels just a handful. Therefore, to determine which B1 subtypes our split-GAL4 line labeled, we compared MCFO images (Data S1B) to EM reconstructions 31 and estimated that our split-GAL4 line contains a mix of B1–1-3 subtypes.
Synaptic connectivity among auditory neurons.
We used FAFB reconstructions corresponding to previously known auditory neurons 31, higher-order auditory neurons 29,84, and all auditory WED/VLP neurons for which we found EM reconstructions. We then found all automatically detected synapses between every pair of individual neurons 41, using version 11 of the FlyWire synapse table (3rd column of Table S3). We omitted synapses that may have been identified multiple times by eliminating synapses between a given pair for which the presynaptic site was within 150 um of another presynaptic site. We ignored synapses from one neuron onto itself and between individual neurons of the same cell type, due to the high rate of false positives in these cases 31. To generate connectivity diagrams based on cell types, we normalized the total number of synapses between two cell types by the number of postsynaptic neurons, and show all such connections that exceed 10 synapses/postsynaptic neuron. Applying our criteria to early auditory neurons results in the same connectivity results among the neurons in 31, with one exception. Dorkenwald and colleagues found that one automatically indicated connection between cell types was due to a high number of inverted synapses (ie, automatically detected presynaptic partner was determined to be postsynaptic by a human observer) 31. Our connectivity analysis method is unable to confirm synapse directionality, and it is not yet known how often this type of false positive occurs. Five WED/VLP cells (AVLP/PVLP_pr01, AVLP_pr05, AVLP_pr35, IPS_pr02, and IPS/WED_pr01) did not have connections meeting these criteria with any other cell type in our dataset.
A previous study identified 5 subtypes of the B1 neurons (B1–1-4 and B1-u for unidentified) based on connectivity patterns 31. Here we find evidence within B1-u of a 6th subtype we name B1–5. These neurons are distinct from the rest of the B1-u neurons as they have significant outputs onto WED_pr01 and WED_pr02 (Figure S4A).
Contacts between JONs and auditory neurons.
In FlyWire, we proofread the JONs previously identified by Kim and colleagues 46. To map connectivity with auditory neurons, we counted the automatically detected synapses 43 between JONs and auditory neurons using version 274 of the FlyWire synapse table (root ID numbers given in column 4 of Table S3), but found that there were a large number of false negatives compared to previous reports 46. Since the synapse detection algorithm was not trained on JON synapses 43, our results suggest that JON synapses may contain unique morphology at or near the synapse compared to central brain neurons. Therefore, we used the number of contacts between JONs and auditory neurons as a measurement of the number of potential synaptic connections, with the rationale that membranes need to touch for a synapse to occur, but that membrane contact does not ensure synaptic connectivity. We found contacts between neurons by analyzing the PyChunkedGraph representation of the segmentation 31. In the PyChunkedGraph, larger segments are broken down into small segments called supervoxels. All supervoxels are nodes in the supervoxel graph and each touching pair of supervoxels has an edge between them. Edges between supervoxels belonging to the same cell segment are “active” and edges between supervoxels belonging to different cell segments are “inactive”. Therefore, inactive edges in this graph represent contacts between cell segments. To find all contacts between every pair of cell segments, we first identified the inactive supervoxel edges between them. Then, we built a subgraph of all involved supervoxels and all their edges. Finally, we found all contacts by computing the connected components in this subgraph. We compared our membrane contact counts (Figure S4B) to the number of manually quantified synapses for the subset of neurons that appeared in 46, and found a linear relationship with slope near 1 (y = 0.84x +0.40, R2 = 0.90) (Figure S4C). Therefore, we used the number of contacts between JONs and auditory neurons as a measurement of the number of potential synaptic connections. For every pair of cell-type to cell-type connection (n=19) indicated by our membrane contact analysis, we manually verified the existence and directionality of synapses between two randomly selected individual neurons with at least 10 contacts between them. This led us to verify 11 connections in which JONs were presynaptic, 4 connections in which JONs were both pre- and postsynaptic, and 3 connections in which JONs were only postsynaptic (Figure 5I–J); and to exclude 1 connection in which we found only 1 synapse.
Path length measurement.
We used the synaptic connectivity matrix (Figure S4A) to define a graph with directed edges. We then calculated the shortest path between every pair of neurons while ignoring edge weights, and then averaged within cell types.
Analysis of hierarchical network structure.
We binarized the connection matrix between individual neuron pairs (Figure S4A), keeping only connections where at least 10 synapses between the two neurons had been detected in the connectomic analysis, and then computed orderability, feedforwardness, and treeness of the resulting directed binary network G, as described in 47. We first computed the “node-weighted condensed graph,” or graph condensation, GC (Figure 6B) of the original network by identifying all strongly connected components {vi} of G (sets of neurons all mutually reachable from one another via at least one directed path) and retaining whether or not at least one directed connection existed between each pair of components. Thus, GC is a directed, acyclic (no recurrent loops) graph whose nodes are the strongly connected components of G. Any neurons that were not connected to the dominant network were left out of our calculations.
Orderability was calculated as the fraction of neurons that were not included within any recurrent loop in G (i.e. the number of neurons that were their own strongly connected component of size 1, divided by the total number of neurons). Feedforwardness was calculated by examining each path in GC that emanated from a “starting node” (a node with no incoming connections) and finished at an “ending node” (a node with no outgoing connections), dividing the number of nodes in that path by the total number of neurons represented by those nodes, then averaging this ratio over all paths from starting to ending nodes. Treeness was calculated by (1) calculating the forward path entropy hf(vi) of each starting node vi in GC, (2) computing Hf, the mean forward path entropy over all starting nodes, (3) calculating the backward path entropy hb(ui) of each ending node ui in GC, (4) computing Hb, the mean backward path entropy over all ending nodes, and (5) taking the normalized difference between the mean forward and backward path entropies, with the final feedforwardness given by (Hf − Hb)/max(Hf, Hb). The forward path entropy of a starting node is the entropy of all paths emanating from that node and finishing at any ending node, h(vi) = −ΣpathP(path) log2 P(path)) where P(path) is the probability of taking a given path by starting at vi and repeatedly stepping to a downstream node (selecting uniformly at random among all possible downstream nodes) until an ending node is reached. The backward path entropy of an ending node, h(ui), is the equivalent quantity, but starting from the ending node and traversing the network backwards until a starting node is reached. See 47 for further mathematical details.
Fully random networks were generated using the same number of neurons as the empirical network and assigning connections at random with the same connection probability as measured empirically. Degree-constrained random networks were generated by fixing the number of incoming and outgoing connections of each neuron in the random network to match those of a corresponding neuron in the empirical network (with 1-to-1 correspondence), but otherwise assigning connections between neuron pairs at random.
Neurotransmitter predictions.
We predicted the neurotransmitter for 479 neurons within 48 cell types using the method presented in 42. We used FlyWire 31 to retrieve all automatically detected pre-synaptic sites in each neuron segment and filtered out synapses with a cleft score below 50 to remove likely false positives 43. We predicted the neurotransmitter of each pre-synaptic site individually and defined the neurotransmitter of an entire neuron as the predicted majority neurotransmitter over its synapses. We labeled a neuron as inconclusive if its majority transmitter was predicted for less than 65% of its synapses. Using this cutoff leads to a greater than 95% prediction accuracy for the three fast-acting neurotransmitters GABA, acetylcholine and glutamate on the test set presented in 42 (see Figure S5A). We validated whether neurotransmitter predictions from automatically detected pre-synaptic sites were robust by predicting the neurotransmitter of neurons within 6 auditory cell types with known neurotransmitters and found perfect agreement for all neurons with conclusive neurotransmitter predictions (Figure S5B). Neurotransmitter predictions for all cell types are shown in Figure S5B–C.
The classifier predictions for all but 5 cell types (pMN2/vpoDN, SAD_pr02, AVLP_pr04, A1, and SAD_pr01) agreed for at least 60% of individual neurons in each class. Therefore, we did not assign a neurotransmitter identity to these cell classes. The one exception was AVLP_pr04, in which we found evidence for multiple cell types. MCFO results revealed that the split-GAL4 line labeling AVLP_pr04 consisted of two cells per hemisphere, both with diffuse and widespread branching patterns across the AVLP and PVLP, but with one cell type having a projection toward the midline (Data S1B). We identified EM reconstructions matching both these cell types and included them as AVLP_pr04 (Figure 2). The cells with the midline projection in both hemispheres were predicted to be dopaminergic (Figure S5C), whereas the two remaining cells were predicted to be GABAergic or glutamatergic. We also found that synaptic connectivity was different for the dopaminergic vs. non-dopaminergic cells (Figure 5H). Therefore, we conclude that AVLP_pr04 contains one dopaminergic cell per hemisphere and one additional cell with inconclusive neurotransmitter results.
Calcium imaging.
We used GCaMP6s 34 to maximize the signal to noise of our recordings and thus increase the likelihood that we could observe even weak auditory responses. For calcium imaging experiments, we used 3–11 day-old virgin female flies reared at 25C and housed in groups of 1–8 flies/vial after eclosion. Calcium imaging experiments were performed during the fly’s light cycle. Flies were mounted and dissected as described in 85 with the following modifications. Before mounting the fly, we removed the wings and legs with forceps. We removed fat covering the brain with forceps or with mouth suction through a sharp glass pipette. We ensured the aristae were intact and free by gently blowing on the fly before and after each experiment. We also looked for abdominal contractions in response to gentle blowing to indicate the fly was alive and healthy before and after each experiment. We monitored temperature and humidity with a thermometer and hygroscope (Traceable 15–077-963, Webster TX) placed on the air table with the two-photon microscope. Temperature and humidity were stable within an experiment, with fluctuations of <1C and <5% humidity across 8 hours of imaging (~1 fly/hr).
Imaging was performed as previously described 16. Briefly, we used a custom-built two-photon laser scanning microscope controlled in MATLAB by ScanImage 5.1 (Vidrio). We imaged single planes at 8.5 Hz (256 × 256 pixels). Pixel size was 0.75 μm × 0.75 μm. After dissection, a fly was placed under the microscope with continuous saline perfusion delivered to the meniscus.
ROI selection.
To decide which region of interest (ROI) to record from in each neuron, we first sampled from multiple ROIs, depending on the morphology of the neuron and on the level of baseline GCaMP6s expression (ie, only ROIs with some baseline level of GCaMP expression were visible under the two-photon microscope). In some lines, multiple ROIs were visible and responded relatively robustly. In those cases, we narrowed down which ROI to focus on based on response strength and ability to locate roughly the same ROI across flies. In other lines, baseline GCaMP expression was low, so we imaged from the ROI that was visible within a moderate laser power, with the goal of choosing roughly a similar ROI in each fly. We sampled from both the left and the right hemisphere within each line but from different flies. In all of the lines, responses occurred in both hemispheres, suggesting either that we stimulated both aristae or that ROIs at the level of the WED/VLP respond bilaterally.
Stimulus generation and delivery.
Sound stimuli were generated at a sampling frequency of 10kHz using custom MATLAB software and previously published techniques 16. Sound was delivered through an earbud speaker (Koss, 16 Ohm impedance; sensitivity, 112 dB SPL/1 mW), which was attached to a long thin tube (12cm, diameter: 1mm) placed ~2mm from the fly’s head (directed toward the aristae) and controlled by custom software in MATLAB 86. Sound intensity was calibrated by measuring the sound particle velocity component for a range of frequencies (100–800 Hz). The detailed procedures and cross-calibration between the pressure and the pressure gradient microphone were described in 87. To estimate the sound amplitude of each stimulus we placed the calibrated gradient microphone at the same position as the fly (2–3 mm from sound tube outlet) in separate experiments. The recorded voltage was then converted to particle velocity (with units mm/s). The output signal was corrected according to the measured intensities to ensure equivalent intensity across frequencies, as previously described 22.
To characterize a given neuron type as auditory or non-auditory, we used a stimulus set consisting of pulse song, sine song, and broadband noise, each with 10 sec duration and 10 sec pre- and post-stimulus silent periods. The intensity of pulse and sine stimuli was 5 mm/sec and the intensity of the noise stimulus was 2 mm/sec. To further characterize auditory tuning, we used a stimulus set consisting of 5 pure tones at a range of frequencies (100, 200, 300, 500, and 800 Hz) and 5 pulse stimuli with a range of interpulse intervals (16, 36, 56, 79, and 96 ms), all at an intensity of 4 mm/sec. Each of these stimuli were 4 sec in duration, with 5 sec pre-stimulus and 10 sec post-stimulus silent periods. Within a stimulus set, a block was defined as one presentation of each stimulus. Stimulus order was randomized within each block, and 3–7 blocks were presented to each fly.
Analysis.
Data were analyzed using custom software in MATLAB. ROIs were drawn manually based on a z-projection of the td-Tomato signal. We calculated the mean fluorescence within each ROI frame-by-frame. If any stimulus block contained drastic, brief (1–3 sec) increases in fluorescence, indicative of wax or other particles moving into the imaged region, we discarded the entire block. We used only flies in which 3 or more blocks were available for analysis. We corrected for gradual, modest changes in fluorescence during each recording by de-trending the data. To detrend, we concatenated fluorescence traces over an entire recording, used a running percentile filter (20th percentile, 50 sec window, 5 sec shift), and subtracted this long-term trend from the recording. For each stimulus, we calculated dF/F as (F(t)−F0)/F0, where F0 was defined as the mean fluorescence during the 10 sec prestimulus silent period.
Stimulus modulation of calcium signals.
To determine whether the fluorescence within a given ROI was modulated by at least one of the acoustic stimuli, we followed the method previously described in 21. Briefly, we modeled the GCaMP fluorescence trace as a convolution of the stimulus history and a set of filters, one per stimulus (ie, 3 filters for the screen stimuli and 10 filters for the frequency/IPI tuning stimuli), with the filter duration matching the duration of the stimulus. To estimate each filter, we used 80% of the stimulus repetitions as training data and the remaining 20% of the repetitions as test data. Filters were estimated using ridge regression 88. We convolved the estimated filters with the stimulus history to generate the predicted signal for each ROI. All possible combinations of training and test data repetitions were used, which gave a total of 3–15 predicted signals (3–6 total repetitions, respectively) per ROI. We then measured the Pearson correlation coefficient between the raw (ie, test repetitions) and the predicted signals. We used bootstrapping to determine the statistical significance of the resulting correlation coefficients. We randomly shuffled each test fluorescence trace in 10 sec bins, and then calculated the Pearson correlation coefficient between each of 10,000 independently shuffled test signals and the predicted signal. P-values for the correlation coefficients were defined as the fraction of shuffled correlation coefficients > 30th percentile of the estimated correlation coefficients. P-values were corrected for multiple comparisons with the Benjamini-Hochberg procedure, with a false detection rate of 0.05.
Analysis of auditory responses.
We measured the strength of calcium responses to auditory stimuli only in ROIs which we determined to be statistically modulated by the stimuli. In these cases, we calculated the integral of the dF/F signal starting with stimulus onset and ending 4 sec after stimulus offset, and then divided by the total time of the integral window. We set out to image auditory responses from at least 4 flies per cell type. If the percentage of responding flies fell below 15%, we labeled that cell type as an infrequent responder and stopped pursuing recordings from that cell type. We chose the threshold of 15% for defining auditory vs. infrequent responders based on the practicality of pursuing recordings from cells that did not respond reliably. Since we set out to image from multiple flies from the same line, achieving this goal would require many more recordings from cells that respond in only 15% of flies. If no flies from a given cell type responded to acoustic stimuli (out of at least 4 imaged flies), we characterized the cell type as non-auditory.
To quantify within-fly variability, we concatenated z-scored responses to pulse, sine, and noise and then measured the standard deviation. To quantify across-fly variability, we measured the standard deviation between the average z-scored responses for all recordings from a given split-GAL4 line.
We used the integrals elicited by pulse, sine, and noise stimuli to assess each fly’s song vs. noise preference. For each fly, we defined the response to song as the maximum of the integrals in response to pulse and sine. We then calculated a song vs. noise preference index as (song − noise) / (song + noise) after setting all negative integrals to 0. Ignoring instances of net inhibition (ie, negative integrals) allows us to quantify the stimuli that excite a given cell type while limiting the preference index to between −1 and 1. If both integrals were negative, we set the song mode preference to 0 to indicate that the response did not prefer either stimulus. We calculated a song mode preference index as (pulse − sine) / (pulse + sine) after setting all negative integrals to 0. If both integrals were negative, reflective of net inhibition to both stimuli, we set the song mode preference to 0 to indicate that the response did not prefer either stimulus. Next we reasoned that in a pulse-preferring neuron, the pulse integral should be at least 250% of the sine integral, which corresponds to a preference index of 0.43 (Figure 3C). Likewise, we required a sine-preferring neuron to have a sine response that was at least 250% of the pulse integral, which corresponds to a preference index of −0.43. To classify the preference of each cell type, we used the mean preference index across flies. Neurons with a mean preference index between −0.43 and 0.43 were classified as having intermediate preference. We chose the threshold of 250% because it best captured trends in the data. That is, pulse- and sine-selective neurons defined by this threshold had individual fly responses that were almost exclusively pulse- or sine-selective, respectively (Figure 3C). To quantify the stimulus preference of net inhibition (ie, negative integrals), we again calculated a sine vs. pulse preference index after setting all positive integrals to 0. This allowed us to determine which stimulus elicited stronger inhibition.
For principal components analysis (PCA), we measured the integrals of z-scored dF/F traces starting with stimulus onset and ending 4 sec after stimulus offset. Each observation consisted of the integrals evoked by the IPI and frequency tuning stimuli from one recording.
Tuning curve generation and classification.
To generate frequency and interpulse interval tuning curves, we measured the integrals in response to each stimulus. Next we classified frequency and interpulse interval tuning curve types based on previously published methods 89,90. Briefly, if the minimum tuning curve value was >85% of the maximum tuning curve value, we classified the tuning as all-pass (ie, untuned to the tested stimulus parameter). For all other tuning curves, we fit both a sigmoid and a Gaussian function. If the r2 of both the sigmoid and Gaussian fits were <0.5, we classified the tuning as complex. Since high- and low-pass tuning curves can be well fit by both a Gaussian and a sigmoidal curve, we used the r2sigmoid/r2Gaussian ratio. If r2sigmoid/r2Gaussian was <0.85, we classified the tuning curve as band-pass if the Gaussian amplitude was positive and band-stop if the Gaussian amplitude was negative. For frequency tuning curves, if r2sigmoid/r2Gaussian was ≥ 0.85, we classified the tuning as high-pass if the ratio of the sigmoid slope to the sigmoid amplitude was positive, and low-pass if this ratio was negative. For IPI tuning curves, if r2sigmoid/r2Gaussian was ≥ 0.85, we classified the tuning as long-pass if the ratio of the sigmoid slope to the sigmoid amplitude was positive, and short-pass if this ratio was negative.
Prediction of tuning from presynaptic inputs.
For each postsynaptic cell j in the connectome for which we had recorded tuning data (i.e. we had recorded calcium responses to 5 IPIs and 5 sine frequencies from the corresponding split-GAL4 line (Figure 4)), we constructed the following model of its response rj(t) to activity in its presynaptic inputs:
where ri(t) is the activity in the presynaptic neuron i, and wij is the number of detected synapses from neuron i onto neuron j, multiplied by −1 if the synapses were GABAergic. As we did not have enough electrophysiological constraints to set τ differently across individual neurons, we used τ = 0.5 s for all neurons. Each valid cell could be classified into a cell line whose tuning responses we measured and hence assigned a response rj(t) to each of the five IPIs and each of the five sine frequencies that were presented. For each IPI or sine frequency, the response timecourses of the presynaptic cells to that IPI or sine frequency, ri(t), were used as inputs in the model above to produce the model output rj(t). The result was a true and modeled set of IPI and sine frequency response timecourses for each valid cell, where the modeled responses were derived from the presynaptic responses and the connectome. We zeroed the input weights wij of presynaptic cells absent from the connectome or for which we did not have tuning data. Postsynaptic tuning curves of the model to IPI or sine frequency were then computed identically as the tuning curves of the real postsynaptic data. To compare tuning curve shape rather than absolute magnitude, we z-scored both the model and data tuning curves before computing the root mean squared error between the tuning curves.
Analysis of neural response timescales.
We estimated the response and adaptation time constants (τr and τa, respectively; Fig 7A), of all neurons by fitting the following model to each pair of pulse & sine responses:
where Is and It represent sine and pulse inputs (equal to 1 when the corresponding stimulus is on and 0 otherwise), and as and ap are multiplicative adaptation variables for the sine and pulse stimuli, respectively. For each neuron we fit the parameters τr, τa, as, ap that minimized the time-averaged squared difference between the model response r(t) and the recorded response, averaged across sine and pulse stimuli. We define the adaptation rate as 1/τa. Since we used the slow calcium sensor, we may slightly overestimate these time constants due to the slower kinetics of GCaMP6s.
Quantification and statistical analysis
Since the majority of our data were not normally distributed according to Jarque-Bera tests, we used non-parametric tests throughout. Correlation tests were based on Spearman’s rank correlation (Figure 3 and 7) in Matlab and Wald test (Figure 6) in Python. Details on each statistical analysis including exact values of n, what n represents, and definition of center can be found in the figure legends and Results.
Supplementary Material
Data S1. Expression patterns (A), stochastic labeling (B), and segmented cell classes (C) of the WED/VLP split-GAL4 collection, related to Figures 1–2 and STAR Methods.
Table S3. FlyWire coordinates and ID numbers for all auditory neuron reconstructions, related to Figures 2, 5, S4, and S5. Version 11 of the synapse table (column 3) was used for analysis of synaptic connectivity among auditory neurons, and version 274 (column 4) was used for the analysis of contacts between JONs and auditory neurons.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-GFP | Invitrogen | #A11122 |
| mouse anti-Bruchpilot | Developmental Studies Hybridoma Bank | mAb nc82 |
| Alexa Fluor 488-goat anti-rabbit | ThermoFisher | A11034 |
| Alexa Fluor 568-goat anti-mouse | ThermoFisher | A11031 |
| Rat anti-FLAG Tag | Novus Biologicals | NBP1-06712 |
| Rabbit anti-HA tag | Cell Signal Technologies | 3724S |
| Cy2 goat anti-mouse | Jackson Immuno Research | 115-225-166 |
| ATTO647N goat anti-rat IgG | Rockland | 612-156-120 |
| AF594 donkey anti-rabbit | Jackson Immuno Research | 711-585-152 |
| DL550 mouse anti-V5 Tag | AbD Serotec | MCA1360D550GA |
| Bacterial and virus strains | ||
| Biological samples | ||
| Chemicals, peptides, and recombinant proteins | ||
| Critical commercial assays | ||
| Deposited data | ||
| Experimental models: Cell lines | ||
| Experimental models: Organisms/strains | ||
| D. melanogaster: 20xUAS-GCaMP6s, td-Tomato/Cyo | 21 | |
| D. melanogaster: 20xUAS-CsChrimson-mVenus | 91 | |
| D. melanogaster: pJFRC51-3XUAS-IVS-syt::smHA | 74 | |
| D. melanogaster: pJFRC200-10XUAS-IVS-myr::smGFP-HA | 74 | |
| D. melanogaster: pJFRC216-13XLexAop2-IVS-myr::smGFP-V5 | 74 | |
| D. melanogaster: MCFO-1 [pBPhsFlp2::PEST (attP3);; pJFRC201- 10XUAS-FRT>STOP>FRT-myr::smGFP-HA (VK0005), pJFRC240-10XUAS-FRT> STOP>FRT-myr::smGFP-V5-THS-10XUASFRT>STOP>FRT-myr::smGFP-FLAG (su(Hw)attP1) | 74 | |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and algorithms | ||
| Matlab 2019a | Mathworks | Mathworks.com |
| Python | https://www.python.org/ | |
| ScanImage 5.1 | Vidrio | https://vidriotechnologies.com/scanimage/ |
| Photoshop | Adobe | https://www.adobe.com/products/photoshop.html |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| VVDviewer | Takashi Kawase | https://github.com/takashi310/VVD_Viewer) |
| Computational Morphometry Toolkit | SRI International | https://www.nitrc.org/projects/cmtk/ |
| Other | ||
Highlights.
D. melanogaster auditory neurons exhibit a continuum of courtship song preferences
The auditory connectome is densely interconnected without hierarchical organization
Auditory responses can be predicted from the wiring diagram
ACKNOWLEDGEMENTS
We thank Xiao-Juan Guan for help with crosses during split-GAL4 generation; David Deutsch for help with calcium imaging pre-analysis and for identifying pC2l neurons in FlyWire; Sebastian Seung for assistance with FlyWire infrastructure; Greg Jefferis for feedback on the cell type naming scheme and Alex Bates and Greg Jefferis for helpful discussions and ideas with regard to neurotransmitter analysis; Nat Tabris and Arie Matsliah for help with connectivity analysis; Lucas Encarnacion-Rivera, Ben Silverman, Jay Gager, Merlin Moore, Selden Koolman, James Hebditch, Sarah Morejohn, Kyle Willie, Austin Burke, Celia David, Ryan Willie, Sandeep Kumar, Doug Bland, Shirleyjoy Serona, J. Anthony Ocho, Nash Hadjerol, Zhihao Zheng, Szi-chieh Yu, Stefanie Hampel, Lucia Kmecova, Fernando J Figueroa Santiago, Fabianna Szorenyi, Yijie Yin, Imaan Tamimi, Varun Sane, Siqi Fang, Griffin Badalemente, Laia Serratosa Capdevila, Greg Jefferis, Katharina Eichler, Alexander Bates, Irene Salgarella, Marlon Blanquart, A. Javier, Arti Yadav, Joseph Hsu, and Kyle Thieringer for proofreading FlyWire neurons; Marta Costa, Jan Clemens, and Istvan Taisz for comments on the manuscript; and the Janelia Fly Core, FlyLight, and Scientific Computing groups. A.N. thanks Gerry Rubin, in whose lab he performed this work, for his support and encouragement. We acknowledge funding from the Jane Coffin Childs Foundation (to CAB), NIH NINDS DP2 New Innovator Award and HHMI Faculty Scholar Award (to MM), NIH BRAIN Initiative R01 MH117815–01 (to CM, SD, and MM), and the Howard Hughes Medical Institute (to CM, AN, NE, JF, and BJD).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Akre KL, Farris HE, Lea AM, Page RA, and Ryan MJ (2011). Signal perception in frogs and bats and the evolution of mating signals. Science 333, 751–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker CA, Clemens J, and Murthy M (2019). Acoustic Pattern Recognition and Courtship Songs: Insights from Insects. Annu. Rev. Neurosci. 42, 129–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hedwig BG (2016). Sequential Filtering Processes Shape Feature Detection in Crickets: A Framework for Song Pattern Recognition. Front. Physiol. 7, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nieder A, and Mooney R (2020). The neurobiology of innate, volitional and learned vocalizations in mammals and birds. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behr O, and von Helversen O (2004). Bat serenades—complex courtship songs of the sac-winged bat (Saccopteryx bilineata). Behavioral Ecology and Sociobiology 56, 106–115. [Google Scholar]
- 6.Holy TE, and Guo Z (2005). Ultrasonic songs of male mice. PLoS Biol. 3, e386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wohlgemuth MJ, Sober SJ, and Brainard MS (2010). Linked control of syllable sequence and phonology in birdsong. J. Neurosci. 30, 12936–12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore JM, and Woolley SMN (2019). Emergent tuning for learned vocalizations in auditory cortex. Nat. Neurosci. 22, 1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romanski LM, and Averbeck BB (2009). The primate cortical auditory system and neural representation of conspecific vocalizations. Annu. Rev. Neurosci. 32, 315–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts PD, and Portfors CV (2015). Responses to Social Vocalizations in the Dorsal Cochlear Nucleus of Mice. Front. Syst. Neurosci. 9, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schöneich S, Kostarakos K, and Hedwig B (2015). An auditory feature detection circuit for sound pattern recognition. Sci Adv 1, e1500325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemens J, Schöneich S, Kostarakos K, Matthias Hennig R, and Hedwig B (2020). A small, computationally flexible network produces the phenotypic diversity of song recognition in crickets. Cold Spring Harbor Laboratory, 2020.07.27.221655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennet-Clark HC, and Ewing AW (1967). Stimuli provided by Courtship of Male Drosophila melanogaster. Nature 215, 669–671. [Google Scholar]
- 14.Arthur BJ, Sunayama-Morita T, Coen P, Murthy M, and Stern DL (2013). Multi-channel acoustic recording and automated analysis of Drosophila courtship songs. BMC Biol. 11, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coen P, Clemens J, Weinstein AJ, Pacheco DA, Deng Y, and Murthy M (2014). Dynamic sensory cues shape song structure in Drosophila. Nature 507, 233–237. [DOI] [PubMed] [Google Scholar]
- 16.Deutsch D, Clemens J, Thiberge SY, Guan G, and Murthy M (2019). Shared Song Detector Neurons in Drosophila Male and Female Brains Drive Sex-Specific Behaviors. Curr. Biol. 29, 3200–3215.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clemens J, Girardin CC, Coen P, Guan X-J, Dickson BJ, and Murthy M (2015). Connecting Neural Codes with Behavior in the Auditory System of Drosophila. Neuron 87, 1332–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albert JT, Nadrowski B, and Göpfert MC (2007). Mechanical signatures of transducer gating in the Drosophila ear. Curr. Biol. 17, 1000–1006. [DOI] [PubMed] [Google Scholar]
- 19.Göpfert MC, and Robert D (2002). The mechanical basis of Drosophila audition. J. Exp. Biol. 205, 1199–1208. [DOI] [PubMed] [Google Scholar]
- 20.Patella P, and Wilson RI (2018). Functional Maps of Mechanosensory Features in the Drosophila Brain. Curr. Biol. 28, 1189–1203.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pacheco DA, Thiberge SY, Pnevmatikakis E, and Murthy M (2020). Auditory activity is diverse and widespread throughout the central brain of Drosophila. Nat. Neurosci. 24, 93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tootoonian S, Coen P, Kawai R, and Murthy M (2012). Neural Representations of Courtship Song in the Drosophila Brain. Journal of Neuroscience 32, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamikouchi A, Inagaki HK, Effertz T, Hendrich O, Fiala A, Göpfert MC, and Ito K (2009). The neural basis of Drosophila gravity-sensing and hearing. Nature 458, 165–171. [DOI] [PubMed] [Google Scholar]
- 24.Azevedo AW, and Wilson RI (2017). Active Mechanisms of Vibration Encoding and Frequency Filtering in Central Mechanosensory Neurons. Neuron 96, 446–460.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai JS-Y, -Y. Lai JS, -J. Lo S, Dickson BJ, and -S. Chiang A (2012). Auditory circuit in the Drosophila brain. Proceedings of the National Academy of Sciences 109, 2607–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaughan AG, Zhou C, Manoli DS, and Baker BS (2014). Neural pathways for the detection and discrimination of conspecific song in D. melanogaster. Curr. Biol. 24, 1039–1049. [DOI] [PubMed] [Google Scholar]
- 27.Yamada D, Ishimoto H, Li X, Kohashi T, Ishikawa Y, and Kamikouchi A (2018). GABAergic Local Interneurons Shape Female Fruit Fly Response to Mating Songs. J. Neurosci. 38, 4329–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou C, Franconville R, Vaughan AG, Robinett CC, Jayaraman V, and Baker BS (2015). Central neural circuitry mediating courtship song perception in male Drosophila. Elife 4, e08477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Wang F, Forknall N, Yang T, Patrick C, Parekh R, and Dickson BJ (2020). Neural circuit mechanisms of sexual receptivity in Drosophila females. Nature 589, 577–581. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, Wang K, Forknall N, Parekh R, and Dickson BJ (2020). Circuit and Behavioral Mechanisms of Sexual Rejection by Drosophila Females. Current Biology 30, 3749–3760.e3. [DOI] [PubMed] [Google Scholar]
- 31.Dorkenwald S, McKellar CE, Macrina T, Kemnitz N, Lee K, Lu R, Wu J, Popovych S, Mitchell E, Nehoran B, et al. (2022). FlyWire: online community for whole-brain connectomics. Nat. Methods 19, 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuo E, Seki H, Asai T, Morimoto T, Miyakawa H, Ito K, and Kamikouchi A (2016). Organization of projection neurons and local neurons of the primary auditory center in the fruit fly Drosophila melanogaster. J. Comp. Neurol. 524, 1099–1164. [DOI] [PubMed] [Google Scholar]
- 33.Scheffer LK, Xu CS, Januszewski M, Lu Z, Takemura S-Y, Hayworth KJ, Huang GB, Shinomiya K, Maitlin-Shepard J, Berg S, et al. (2020). A connectome and analysis of the adult central brain. eLife 9, e57443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morley EL, Jonsson T, and Robert D (2018). Auditory sensitivity, spatial dynamics, and amplitude of courtship song in Drosophila melanogaster. J. Acoust. Soc. Am. 144, 734. [DOI] [PubMed] [Google Scholar]
- 36.Dolan M-J, Frechter S, Bates AS, Dan C, Huoviala P, Roberts RJV, Schlegel P, Dhawan S, Tabano R, Dionne H, et al. (2019). Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body. eLife 8, e43079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suver MP, Matheson AMM, Sarkar S, Damiata M, Schoppik D, and Nagel KI (2019). Encoding of wind direction by central neurons in Drosophila. Neuron 102, 828–842.E7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coates KE, Calle-Schuler SA, Helmick LM, Knotts VL, Martik BN, Salman F, Warner LT, Valla SV, Bock DD, and Dacks AM (2020). The wiring logic of an identified serotonergic neuron that spans sensory networks. Journal of Neuroscience 40, 6309–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Z, Lauritzen JS, Perlman E, Robinson CG, Nichols M, Milkie D, Torrens O, Price J, Fisher CB, Sharifi N, et al. (2018). A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell 174, 730–743.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clemens J, Coen P, Roemschied FA, Pereira TD, Mazumder D, Aldarondo DE, Pacheco DA, and Murthy M (2018). Discovery of a New Song Mode in Drosophila Reveals Hidden Structure in the Sensory and Neural Drivers of Behavior. Curr. Biol. 28, 2400–2412.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buhmann J, Sheridan A, Gerhard S, Krause R, Nguyen T, Heinrich L, Schlegel P, Lee W-CA, Wilson R, Saalfeld S, et al. (2019). Automatic Detection of Synaptic Partners in a Whole-Brain Drosophila EM Dataset. bioRxiv, 10.1101/2019.12.12.874172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eckstein N, Bates AS, Du M, Hartenstein V, Gregory SX, and Funke J (2020). Neurotransmitter Classification from Electron Microscopy Images at Synaptic Sites in Drosophila. bioRxiv, 10.1101/2020.06.12.148775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buhmann J, Sheridan A, Malin-Mayor C, Schlegel P, Gerhard S, Kazimiers T, Krause R, Nguyen TM, Heinrich L, Lee W-CA, et al. (2021). Automatic detection of synaptic partners in a whole-brain Drosophila electron microscopy data set. Nature Methods 18, 771–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McInnes L, Healy J, and Melville J (2018). UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv [stat.ML]. [Google Scholar]
- 45.Mezzera C, Brotas M, Gaspar M, Pavlou HJ, Goodwin SF, and Vasconcelos ML (2020). Ovipositor Extrusion Promotes the Transition from Courtship to Copulation and Signals Female Acceptance in Drosophila melanogaster. Curr. Biol. 30, 3736–3748.e5. [DOI] [PubMed] [Google Scholar]
- 46.Kim H, Horigome M, Ishikawa Y, Li F, Lauritzen JS, Card G, Bock DD, and Kamikouchi A (2020). Wiring patterns from auditory sensory neurons to the escape and song-relay pathways in fruit flies. J. Comp. Neurol. 528, 2068–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corominas-Murtra B, Goñi J, Solé RV, and Rodríguez-Caso C (2013). On the origins of hierarchy in complex networks. Proc. Natl. Acad. Sci. U. S. A. 110, 13316–13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oja E (1992). Principal components, minor components, and linear neural networks. Neural Netw. 5, 927–935. [Google Scholar]
- 49.Ganguli S, Huh D, and Sompolinsky H (2008). Memory traces in dynamical systems. Proc. Natl. Acad. Sci. U. S. A. 105, 18970–18975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu TX, Davoudian PA, Lizbinski KM, and Jeanne JM (2022). Connectomic features underlying diverse synaptic connection strengths and subcellular computation. Curr. Biol. 32, 559–569.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sproule MKJ, and Chacron MJ (2017). Electrosensory neural responses to natural electro-communication stimuli are distributed along a continuum. PLoS One 12, e0175322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson AW, and Scott EK (2016). Characterisation of sensitivity and orientation tuning for visually responsive ensembles in the zebrafish tectum. Sci. Rep. 6, 34887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minderer M, Brown KD, and Harvey CD (2019). The Spatial Structure of Neural Encoding in Mouse Posterior Cortex during Navigation. Neuron 102, 232–248.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raposo D, Kaufman MT, and Churchland AK (2014). A category-free neural population supports evolving demands during decision-making. Nat. Neurosci. 17, 1784–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saleem AB, Ayaz A, Jeffery KJ, Harris KD, and Carandini M (2013). Integration of visual motion and locomotion in mouse visual cortex. Nat. Neurosci. 16, 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Namiki S, Dickinson MH, Wong AM, Korff W, and Card GM (2018). The functional organization of descending sensory-motor pathways in. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rayshubskiy A, Holtz SL, D’Alessandro I, Li AA, Vanderbeck QX, Haber IS, Gibb PW, and Wilson RI (2020). Neural circuit mechanisms for steering control in walking Drosophila. bioRxiv, 10.1101/2020.04.04.024703. [DOI] [Google Scholar]
- 58.Batchelor AV, and Wilson RI (2019). Sound localization behavior in depends on inter-antenna vibration amplitude comparisons. J. Exp. Biol. 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fuchs PA, and Lauer AM (2019). Efferent Inhibition of the Cochlea. Cold Spring Harb. Perspect. Med. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winer JA, and Schreiner CE (2005). The Central Auditory System: A Functional Analysis. In The Inferior Colliculus, Winer JA and Schreiner CE, eds. (Springer; ), pp. 1–68. [Google Scholar]
- 61.Stumpner A (2002). A species-specific frequency filter through specific inhibition, not specific excitation. Journal of Comparative Physiology A: Sensory, Neural, and Behavioral Physiology 188, 239–248. [DOI] [PubMed] [Google Scholar]
- 62.Romer H, Rheinlaender J, and Dronse R (1981). Intracellular studies on auditory processing in the metathoracic ganglion of the locust. Journal of Comparative Physiology A 144, 305–312. [Google Scholar]
- 63.Woolley SMN, and Portfors CV (2013). Conserved mechanisms of vocalization coding in mammalian and songbird auditory midbrain. Hear. Res. 305, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyttenbach RA, May ML, and Hoy RR (1996). Categorical perception of sound frequency by crickets. Science 273, 1542–1544. [DOI] [PubMed] [Google Scholar]
- 65.Rose GJ, and Brenowitz EA (2002). Pacific treefrogs use temporal integration to differentiate advertisement from encounter calls. Animal Behaviour 63, 1183–1190. [Google Scholar]
- 66.Cohen Y, Shen J, Semu D, Leman DP, Liberti WA 3rd, Perkins LN, Liberti DC, Kotton DN, and Gardner TJ (2020). Hidden neural states underlie canary song syntax. Nature 582, 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calhoun AJ, Pillow JW, and Murthy M (2019). Unsupervised identification of the internal states that shape natural behavior. Nat. Neurosci. 22, 2040–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luan H, Peabody NC, Vinson CR, and White BH (2006). Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron 52, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pfeiffer BD, Ngo T-TB, Hibbard KL, Murphy C, Jenett A, Truman JW, and Rubin GM (2010). Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tirian L, and Dickson BJ (2017). The VT GAL4, LexA, and split-GAL4 driver line collections for targeted expression in the Drosophila nervous system. bioRxiv, 10.1101/198648. [DOI] [Google Scholar]
- 71.Dionne H, Hibbard KL, Cavallaro A, Kao J-C, and Rubin GM (2018). Genetic Reagents for Making Split-GAL4 Lines in. Genetics 209, 31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jenett A, Rubin GM, Ngo T-TB, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. (2012). A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otsuna H, Ito M, and Kawase T (2018). Color depth MIP mask search: a new tool to expedite Split-GAL4 creation. bioRxiv, 10.1101/318006. [DOI] [Google Scholar]
- 74.Nern A, Pfeiffer BD, and Rubin GM (2015). Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proceedings of the National Academy of Sciences 112, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu M, Nern A, Ryan Williamson W, Morimoto MM, Reiser MB, Card GM, and Rubin GM (2016). Visual projection neurons in the Drosophila lobula link feature detection to distinct behavioral programs. eLife 5, e21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo T-TB, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. (2014). The neuronal architecture of the mushroom body provides a logic for associative learning. Elife 3, e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wan Y, Otsuna H, Chien C-B, and Hansen C (2009). An interactive visualization tool for multi-channel confocal microscopy data in neurobiology research. IEEE Trans. Vis. Comput. Graph. 15, 1489–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wan Y, Otsuna H, Chien C-B, and Hansen C (2012). FluoRender: An Application of 2D Image Space Methods for 3D and 4D Confocal Microscopy Data Visualization in Neurobiology Research. IEEE Pac. Vis. Symp, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rohlfing T (2011). User guide to the computational morphometry toolkit. Neuroscience Program, SRI International, Menlo Park, CA 3. [Google Scholar]
- 80.Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, et al. (2014). A systematic nomenclature for the insect brain. Neuron 81, 755–765. [DOI] [PubMed] [Google Scholar]
- 81.Bates AS, Manton JD, Jagannathan SR, Costa M, Schlegel P, Rohlfing T, and Jefferis GS (2020). The natverse, a versatile toolbox for combining and analysing neuroanatomical data. Elife 9, e53350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bogovic JA, Otsuna H, Heinrich L, Ito M, Jeter J, Meissner G, Nern A, Colonell J, Malkesman O, Ito K, et al. (2020). An unbiased template of the Drosophila brain and ventral nerve cord. PLoS One 15, e0236495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu CS, Januszewski M, Lu Z, Takemura S-Y, Hayworth KJ, Huang G, Shinomiya K, Maitin-Shepard J, Ackerman D, Berg S, et al. (2020). A Connectome of the Adult Drosophila Central Brain. bioRxiv, 10.1101/2020.01.21.911859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deutsch D, Pacheco DA, Encarnacion-Rivera LJ, Pereira T, Fathy R, Calhoun A, Ireland EC, Burke AT, Dorkenwald S, McKellar C, et al. (2020). The Neural Basis for a Persistent Internal State in Drosophila Females. bioRxiv, 10.1101/2020.02.13.947952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Murthy M, and Turner G (2013). Dissection of the head cuticle and sheath of living flies for whole-cell patch-clamp recordings in the brain. Cold Spring Harb. Protoc. 2013, 134–139. [DOI] [PubMed] [Google Scholar]
- 86.Clemens J, Ozeri-Engelhard N, and Murthy M (2018). Fast intensity adaptation enhances the encoding of sound in Drosophila. Nat. Commun. 9, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Göpfert MC, Albert JT, Nadrowski B, and Kamikouchi A (2006). Specification of auditory sensitivity by Drosophila TRP channels. Nat. Neurosci. 9, 999–1000. [DOI] [PubMed] [Google Scholar]
- 88.Park M, and Pillow JW (2011). Receptive field inference with localized priors. PLoS Comput. Biol. 7, e1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baker CA, and Carlson BA (2014). Short-term depression, temporal summation, and onset inhibition shape interval tuning in midbrain neurons. J. Neurosci. 34, 14272–14287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Groh JM, Kelly KA, and Underhill AM (2003). A monotonic code for sound azimuth in primate inferior colliculus. J. Cogn. Neurosci. 15, 1217–1231. [DOI] [PubMed] [Google Scholar]
- 91.Klapoetky NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, et al. (2014). Independent optical excitation of distinct neural populations. Nature Meth. 11, 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Expression patterns (A), stochastic labeling (B), and segmented cell classes (C) of the WED/VLP split-GAL4 collection, related to Figures 1–2 and STAR Methods.
Table S3. FlyWire coordinates and ID numbers for all auditory neuron reconstructions, related to Figures 2, 5, S4, and S5. Version 11 of the synapse table (column 3) was used for analysis of synaptic connectivity among auditory neurons, and version 274 (column 4) was used for the analysis of contacts between JONs and auditory neurons.
Data Availability Statement
Raw data supporting the current study are publicly available: https://doi.org/10.34770/7zem-x425. Code generated by the current study is also publicly available: https://github.com/rkp8000/7UN3_1N_DR0P_0U7. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


