Abstract
Stochastic mechanisms diversify cell fates during development. How cells randomly choose between two or more fates remains poorly understood. In the Drosophila eye, the random mosaic of two R7 photoreceptor subtypes is determined by expression of the transcription factor Spineless (Ss). We investigated how cis-regulatory elements and trans factors regulate nascent transcriptional activity and chromatin compaction at the ss gene locus during R7 development. The ss locus is in a compact state in undifferentiated cells. An early enhancer drives transcription in all R7 precursors and the locus opens. In differentiating cells, transcription ceases and the ss locus stochastically remains open or compacts. In SsON R7s, ss is open and competent for activation by a late enhancer, whereas in SsOFF R7s, ss is compact and repression prevents expression. Our results suggest that a temporally dynamic antagonism, in which transcription drives large-scale decompaction and then compaction represses transcription, controls stochastic fate specification.
Keywords: Spineless, stochastic, eye, retina, Drosophila, photoreceptor, enhancer, chromatin, Klumpfuss, Ash2, Lid
eTOC Blurb
Cells sometimes randomly choose between fates during development. In Drosophila, a random mosaic of photoreceptor subtypes is determined by the spineless gene. Voortman et al. find that spineless is regulated by a dynamic interplay between transcription and chromatin compaction during fly eye development.
Graphical Abstract
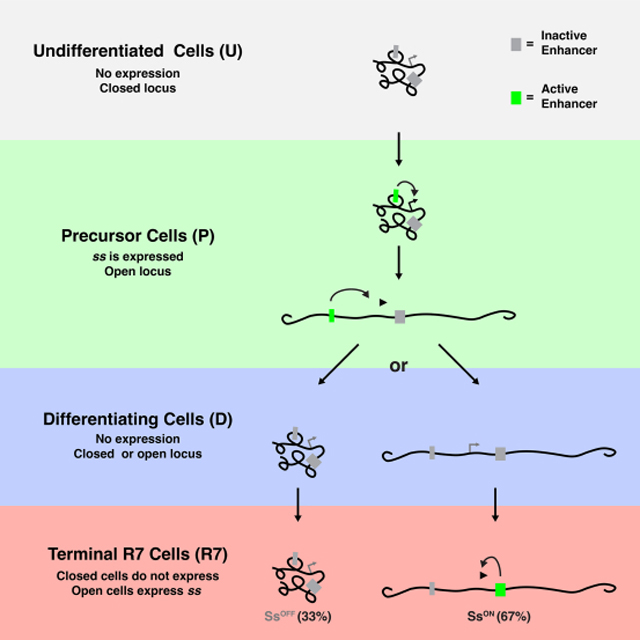
Introduction
Cell fate specification is controlled by lineage, signaling, and stochastic regulatory inputs, leading to highly precise developmental outcomes (Petkova et al., 2019). Stochastic mechanisms promote diversity in populations of photoreceptors, olfactory neurons, motor neurons, and immune cells (Alqadah et al., 2016; Bell et al., 2007; Dasen et al., 2003, 2005; Duffy et al., 2012; Johnston and Desplan, 2010; Miyamichi et al., 2005; Ressler et al., 1993; Vassar et al., 1993). Despite the importance of stochastic cell fate specification, how cells randomly choose between fates is poorly understood.
Stochastic cell fate specification is best understood in prokaryotes. One well-characterized example is the bet-hedging mechanism utilized by Bacillus subtilis. To minimize losses in a changing environment, populations of genetically identical bacteria maintain a subpopulation of cells that are competent for DNA uptake (Dubnau, 1999; Hahn et al., 1998; Nester and Stocker, 1963). The transient and random transition into the competent fate is controlled by expression of the transcriptional regulator ComK (Hahn et al., 1998; Turgay et al., 1997). Though most cells maintain low expression of ComK, a subset will experience a pulse of ComK expression that exceeds a threshold and induces a transition to the competent fate (Maamar et al., 2007; Süel et al., 2006). A similar mechanism occurs in the HIV life-cycle, where transcription of the regulatory factor Trans-Activator of Transcription (Tat) determines the switch from proviral latency to active replication (Hendy et al., 2017; Weinberger et al., 2008). Thus, stochastic cell fate specification often requires a pulse of expression of a critical regulator that determines a fate decision.
In addition to transcriptional dynamics, chromatin-mediated repression is a key mechanism mediating stochastic fate specification. In mice, each olfactory sensory neuron (OSN) expresses only one olfactory receptor (OR) gene from a battery of ~1300 possibilities (Buck and Axel, 1991; Chess et al., 1994; Godfrey et al., 2004). Despite residing in numerous clusters across many chromosomes, all ~1300 OR genes are repressed and coalesce into heterochromatic foci within the nucleus prior to OR selection (Clowney et al., 2012; Magklara et al., 2011; Sullivan et al., 1996; Zhang and Firestein, 2002). In mutants that impact chromatin modifications and nuclear organization, co-expression of multiple ORs is observed (Clowney et al., 2012). While the mechanism of selection remains elusive, a single OR allele escapes the repressive heterochromatic environment and is expressed in each OSN (Armelin-Correa et al., 2014; Lyons et al., 2013). Thus, chromatin-mediated silencing and selective de-silencing are paramount for the stochastic expression of a single OR gene.
The random mosaic of R7 photoreceptors in the fly eye provides a paradigm to study the integration of transcription and chromatin-mediated repression in stochastic cell fate specification. In the fly eye, stochastic expression of the PAS-bHLH transcription factor Spineless (Ss) establishes the random pattern of two R7 subtypes across the retina. SsON R7s express Rhodopsin 4 (Rh4), while SsOFF R7s express Rhodopsin 3 (Rh3)(Fig. 1A–B)(Bell et al., 2007; Duncan et al., 1998; Johnston and Desplan, 2014; Montell et al., 1987; Wernet et al., 2006). In wild type flies, each R7 has a 67% chance of adopting the SsON R7 fate and a 33% chance of assuming the SsOFF R7 fate, yielding a consistent ratio yet unique, random pattern of R7 subtypes across eyes (Fig. 1B). In ss protein null mutants, all R7s express Rh3 (Fig. 1B). The stochastic ON/OFF ss expression is controlled by an enhancer (late enhancer, LE) that drives expression in all R7s and silencers that limit expression to a subset of R7s (Fig. 1C).
Figure 1. ss controls R7 subtype specification.
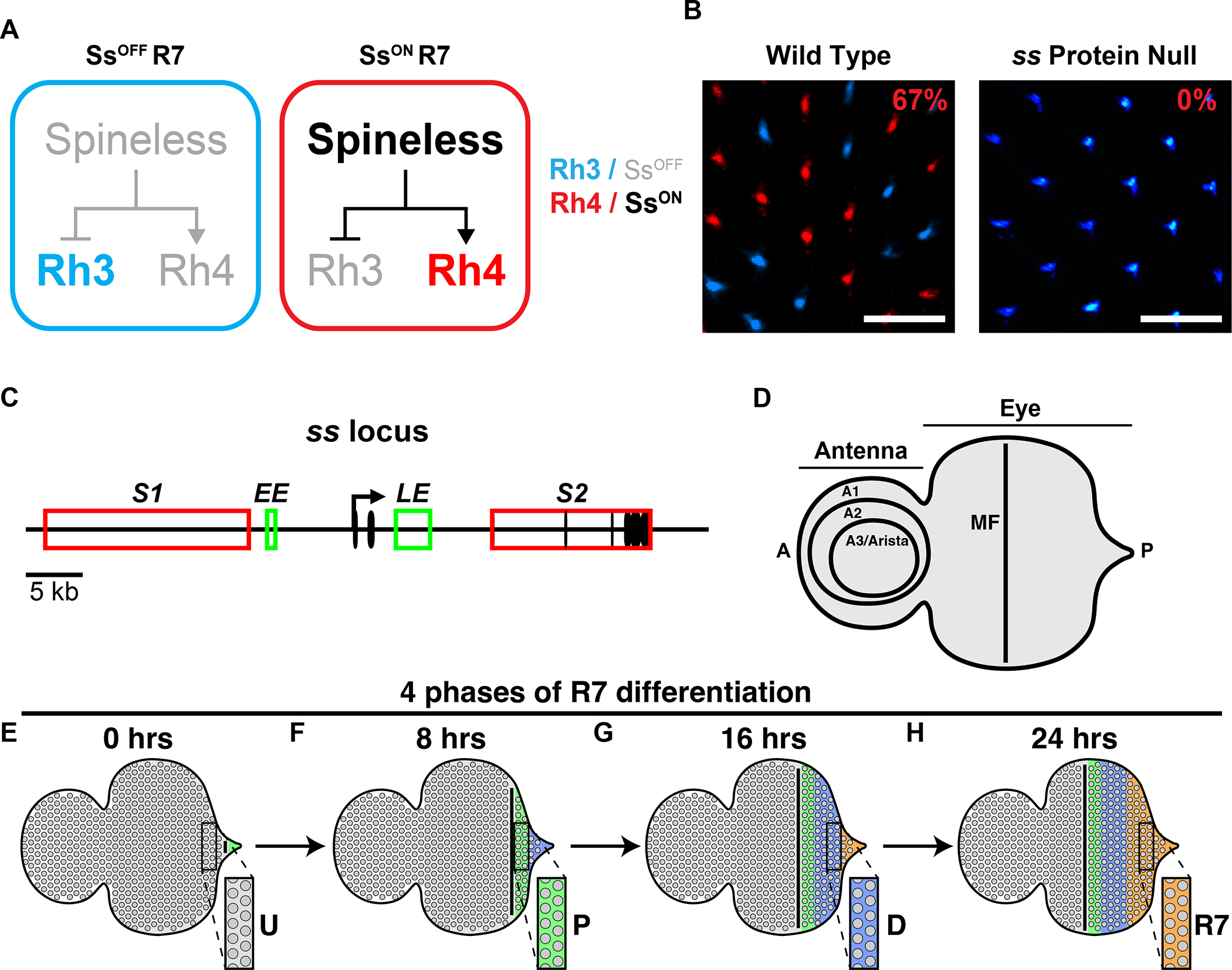
A) R7 subtype specification. Expression of Ss promotes the Rh4-expressing R7 fate. Absence of Ss yields the Rh3-expressing R7 fate.
B) Wild-type retinas contain 33% Rh3/SsOFF R7s and 67% Rh4/SsON R7s in a random pattern (left). ss protein null mutants contain only Rh3/SsOFF R7s and no Rh4/SsON R7s (right). Scalebar=20 μm.
C) ss gene locus. Black oval=exon; black arrow=promoter; red rectangle=silencer; green rectangle=enhancer; S1=silencer 1; S2=silencer 2; EE=early enhancer; LE=late enhancer.
D) Schematized eye-antennal imaginal disc. Antenna is subdivided into the A1, A2, and A3/arista. A=anterior; P=posterior; MF=morphogenetic furrow.
E-H) Schematized depiction of R7 maturation. Insets illustrate how cells proceed through development over time. Gray=undifferentiated cells/U; green=precursors/P; blue=differentiating cells/D; orange=R7.
Here we describe a mechanism that controls stochastic R7 subtype specification. Initially, the ss locus is compact in all undifferentiated cells. An early enhancer (EE) drives ss expression and the ss locus opens in all R7 precursors during larval development. Expression ceases and the ss locus randomly compacts or remains open. In R7s in which ss remains open, the LE drives ss expression and SsON R7 fate. In R7s with compact chromatin, repression prevents expression driven by the LE, yielding the SsOFF R7 fate. Our data suggest that stochastic fate specification is controlled by the dynamic, intertwined relationship of transcription and chromatin: transcription opens chromatin then chromatin compaction represses transcription. We find that transcription is a source of stochasticity as modulating early transcription in precursors alters the proportions of alternative R7 fates.
Results
ss expression is dynamic in developing R7s
Photoreceptor (PR) identity, including R7 subtype, is specified during larval development in the eye-antennal imaginal disc (Fig. 1D). Retinal differentiation begins at the posterior end and progresses in a wave anteriorly. An indentation called the morphogenetic furrow (MF) appears at the posterior end (Fig. S1A). The MF progresses in a developmental wave from posterior to anterior (Fig. S1). Behind the MF, PRs differentiate in a stereotypical progression: R8, R2/R5, R3/R4, R1/R6, and finally R7 (Fig. S1B–F). As the eye develops in this spatiotemporal manner, individual discs provide information on all stages of PR specification, with undifferentiated cells in the anterior and the most differentiated cells in the posterior (Fig. S1F)(Gallagher et al., 2022; Ready et al., 1976; Tomlinson and Ready, 1987a, 1987b; Treisman, 2013; Wolff and Ready, 1991).
We defined four phases that R7s proceed through during development, including undifferentiated (U), precursor (P), differentiating (D), and differentiated R7 (R7) (Fig. 1E–H). In individual discs, we visualized all four phases (Fig. 1H). Undifferentiated cells were anterior to the MF (Fig. 1E–H). Posterior to the MF, precursors were located at 0–10 μm, differentiating cells were located at 10–30 μm, and R7s were located at >30 μm (Fig. 1F–H). In Fig. 1E–H and subsequent figures, we diagram only undifferentiated, precursor, differentiating, and R7 cells in the eye and developing cells in the antenna for simplicity.
To characterize ss expression, we performed nascent RNA fluorescence in situ hybridization (RNA FISH). We generated oligo probes covering the entire ss transcript, including introns and exons (Fig. S2A), and performed single molecule RNA FISH (smFISH)(Beliveau et al., 2012; Little et al., 2013). This strategy yielded single bright fluorescent punctae in ss-expressing nuclei, indicating sites of nascent transcription (Fig. 2A–B). Our observation of one puncta per nucleus is consistent with chromosome pairing in close proximity in somatic cells of Drosophila (Stevens, 1908). This approach enabled quantification of ss transcription in each developmental context. ss is strongly expressed in all cells of the central antennal region, serving as a positive control (Fig. 2A, C–D, A). ss is not expressed in the peripodial membrane that overlies the eye-antennal disc, acting as a negative control (Fig. 2D, S2B, Ctrl).
Figure 2. Two temporally distinct enhancers drive ss expression.
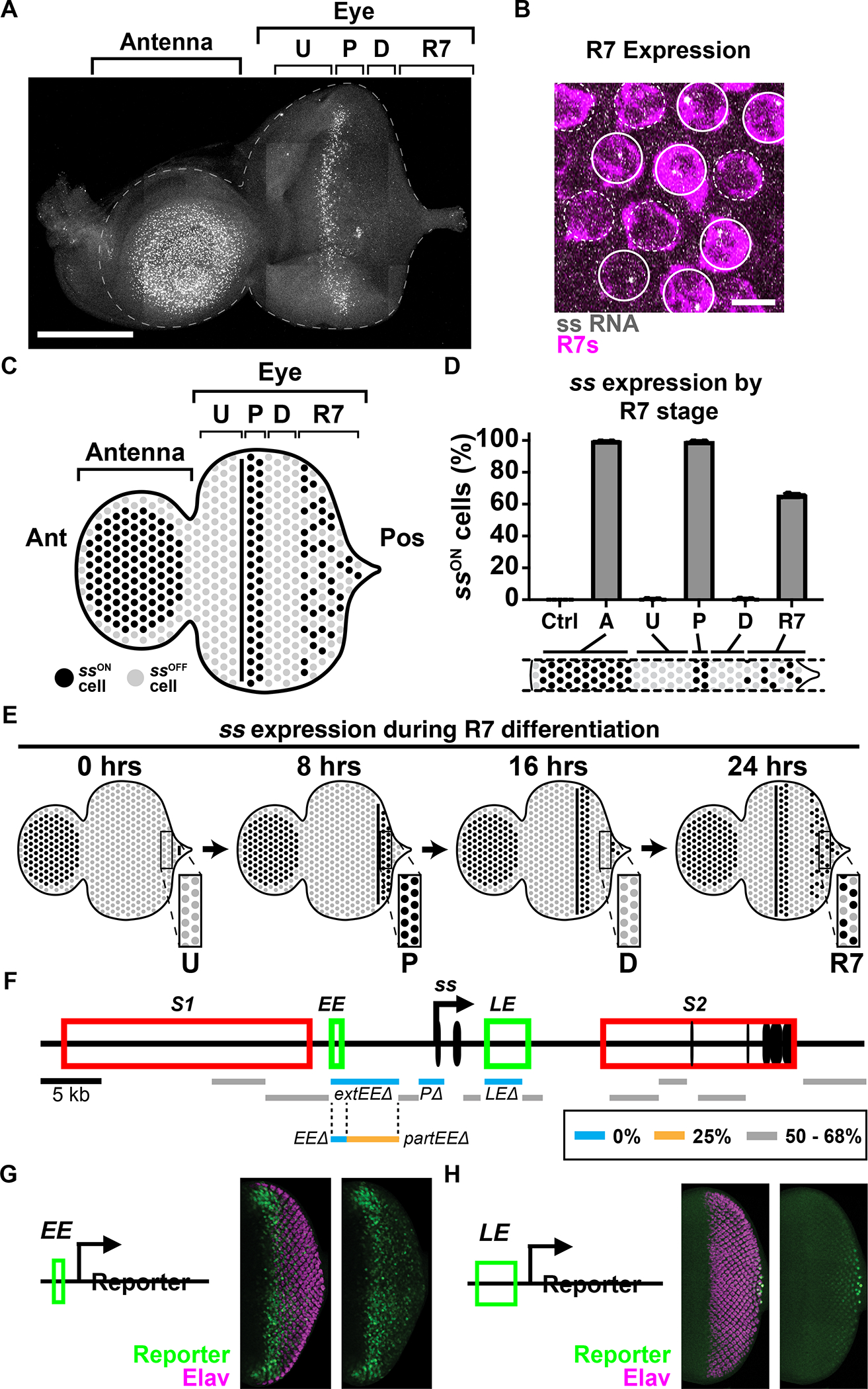
For A-E) Ctrl=peripodial membrane; A=antennal cells; U=undifferentiated cells; P=precursors; D=differentiating cells; R7=R7s. Ant=anterior; Pos=posterior.
A) ss RNA is expressed in antennal cells, precursors, and R7s. Gray=ss RNA. Scalebar=100 μm.
B) Nascent ss RNA transcripts in a subset of R7s distinguished by sev>Gal4, UAS>GFP. Magenta=R7 reporter; gray=ss RNA; sold circles=SsON R7s, dashed circles=SsOFF R7s. Scalebar=5 μm.
C) Schematized eye antennal imaginal disc.
D) % cells expressing ss.
E) Schematized ss expression across time. Insets illustrate ss expression dynamics.
F) ss gene locus and CRISPR deletion screen. Black oval=exon; black arrow=promoter; red rectangle=silencer; green rectangle=enhancer; blue line=0% SsON R7s; orange line=25% SsON R7s; gray line=50–68% SsON R7s; S1=silencer 1; S2=silencer 2; EE=early enhancer; LE=late enhancer.
For G, H) Green=reporter; magenta=Elav (neurons).
G) The EE reporter is expressed in precursors.
H) The LE reporter is expressed in R7s.
In the eye, ss is differentially expressed during R7 specification. ss is not expressed in undifferentiated cells, ss is strongly expressed in all precursors, ss is not expressed in differentiating cells, and ss is expressed in a subset of R7s (Fig. 2A–E, Fig. S2C). We distinguished R7s from other PRs using an R7 reporter line (R7>GFP), allowing quantification of ssON/OFF R7s. The ssON/OFF ratio in larval R7s is similar to the ssON/OFF ratio in adult R7s (Fig. 2D, S2C), consistent with this decision being made in larvae and maintained throughout the lifetime of the organism (Johnston and Desplan, 2014). As expression in an individual disc represents different temporal phases of R7 development, we conclude that ss expression is dynamic as R7s develop: off in the undifferentiated cell phase, on in the precursor phase, off in the differentiating cell phase, and finally, on in a subset of R7s in the differentiated R7 phase (Fig. 2A–E).
Two temporally distinct enhancers drive ss expression in the developing eye
To identify cis-regulatory elements that regulate stochastic ON/OFF expression of ss in R7s, we used CRISPR to make a series of 1–5 kb deletions in the ss locus (Fig. 2F, Table S1). As Rhodopsin expression faithfully reports Ss expression state in adults (Rh4=SsON, Rh3=SsOFF), we examined Rh3 and Rh4 expression and determined the proportions of R7 subtypes. We identified three elements that are required for ss expression in R7s, including the promoter (P), a 5.4 kb upstream element (extEE), and the previously identified late enhancer (LE) (Fig. 2F, Table S1)(Johnston and Desplan, 2014). Deletion of these regions reduced the proportion of SsON R7s to 0% (Fig. 2F, Table S1). Thus, these cis-regulatory regions are required for normal ss expression.
We conducted additional partial deletions of the extEE region to determine a minimal cis-regulatory region required for ss expression. Deletion of the 1.3 kb EE region caused a dramatic decrease of SsON R7s to 0% (Fig. 2F, Table S1), while deletion of the neighboring 4.1 kb partEE caused a partial reduction of SsON R7s to 25% (Fig. 2F, Table S1). As EE was strictly required for ss expression, we interrogated this region further.
To assess the spatiotemporality of EE and LE activities, we generated reporter constructs and examined expression in larval eye-antennal discs. The EE drove expression in precursors similar to ss RNA expression (Fig. 2G). In contrast, the LE drove expression in all R7s (Fig. 2H). We did not observe expression in the antenna for either construct, suggesting that EE and LE are eye-specific enhancers for ss. Thus, EE and LE are sufficient to drive expression in precursors and R7s respectively.
As chromatin accessibility is associated with enhancer activity, ATAC-seq can predict candidate enhancers (Buenrostro et al., 2015). We analyzed published scATAC-seq datasets (Bravo González-Blas et al., 2020). For antennal cells that express ss, accessibility peaks were observed at the promoter, but not at EE or LE (Fig. S2D). For precursors that express ss, peaks occurred at the EE and promoter, but not LE (Fig. S2D). For all PRs, of which only a subset of R7s express ss, peaks were observed at the LE and promoter and were significantly reduced for the EE (Fig. S2D). As a small peak remains at the EE in R7s, some residual chromatin accessibility may remain at this later timepoint. Alternatively, some cells may have been incorrectly clustered into this cell type. These observations support roles for the EE and LE as enhancers that drive expression during distinct temporal phases of R7 development: EE drives early expression in precursors and LE drives late expression in R7s.
Early ss expression in precursors is required for ss expression in R7s
To test how the EE and LE regulate ss expression during PR development, we observed ss expression in mutant conditions. In the fly eye, automated identification and assignment of nascent spots to individual cells is challenging in 3D and not necessary to describe the changes in expression observed here. Therefore, to quantify ss-expressing cells in the eye-antennal disc, we measured the density of nascent RNA spots per unit area (μm2) (Fig. S3A–C). To control for changes in tissue morphology, we measured the density of cells in the antenna and precursors (Fig. S3D–E). We assessed Rh4/SsON and Rh3/SsOFF in R7s in adult retinas (Fig. 3D, W). ss expression in antennal cells in eyeantennal discs served as a positive internal control (Fig. 3B, U). Promoter deletion (PΔ) mutants acted as a negative control, exhibiting a complete loss of ss expression (Fig. 3E–H, U–W).
Figure 3. Decreasing early ss expression decreases % SsON R7s.
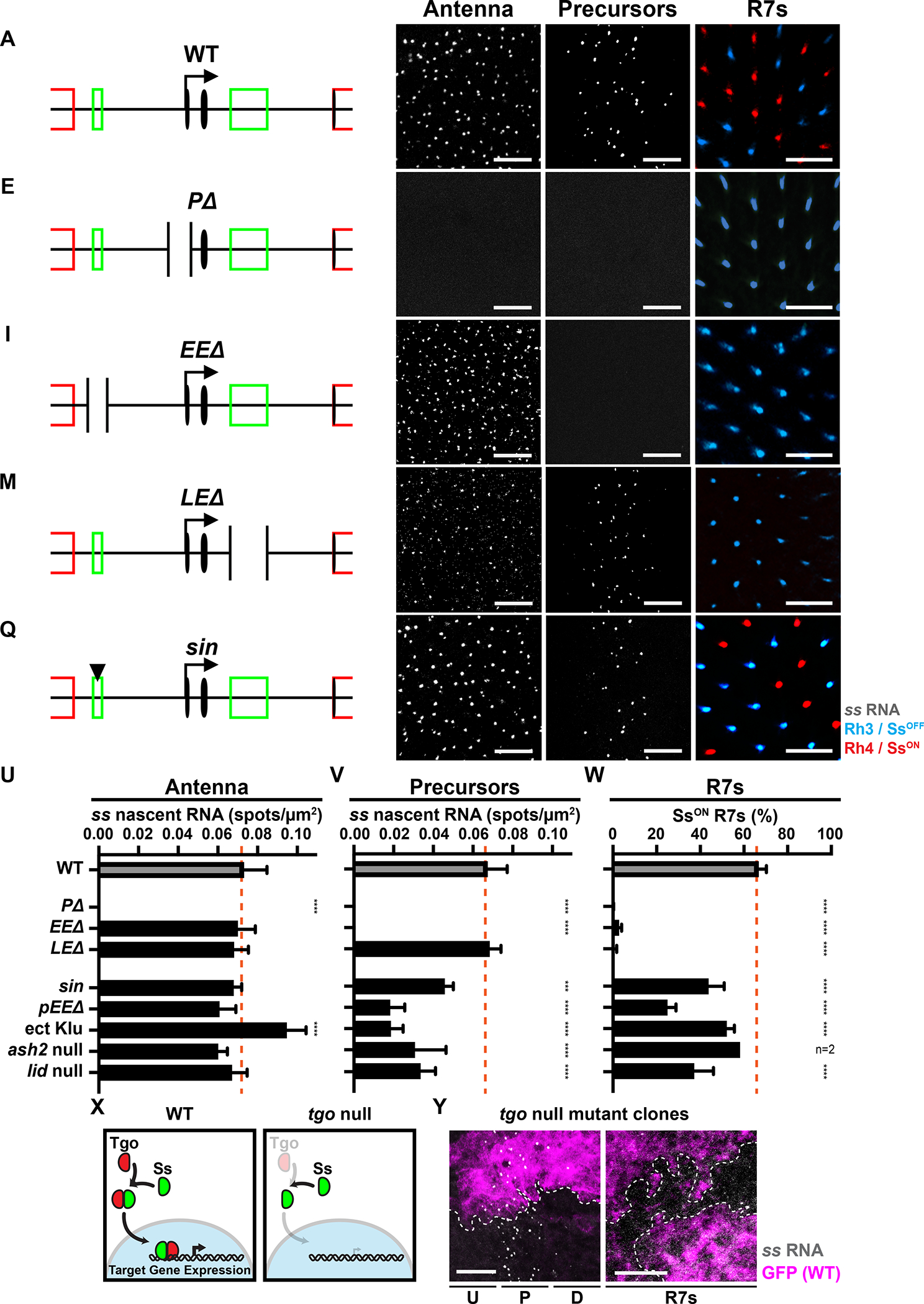
For A, E, I, M, Q) Truncated schematized ss locus.
For B, F, J, N, R) ss RNA in the antenna. Gray=ss RNA. Scalebar=10 μm.
For C, G, K, O, S) ss RNA in precursors. Gray=ss RNA. Scalebar=10 μm.
For D, H, L, P, T) Adult Rh3/SsOFF and Rh4/SsON expression in R7s. Scalebar=20 μm.
A-D) WT.
E-H) PΔ.
I-L) EEΔ.
M-P) LEΔ.
Q-T) Animals with sin variant.
For U-W, Z) Orange line=mean WT expression. n.s. denotes p>0.05; *** denotes p<0.0005; **** denotes p<0.0001.
U) Quantification of ss in antennal cells.
V) Quantification of ss in precursors.
W) Quantification of % SsON R7s.
X) Ss/Tgo mechanism in WT and breakdown in tgo mutants.
Y) ss RNA in precursors and a subset of R7s in tgo null mutant clones. Dashed line=clone boundary. GFP- = tgo null mutant; GFP+ = wild type. Gray=ss RNA; magenta=GFP; dashed line=clone boundary. Scalebar=10 μm.
EEΔ mutants lost ss expression in precursors and Rh4/SsON in R7s, while ss expression was maintained in antennal cells (Fig. 3I–L, U–W). LEΔ mutants displayed a complete loss of Rh4/SsON expression in R7s but showed normal ss expression in precursors and antennal cells (Fig. 3M–P, U–W). PΔ, EEΔ, and LEΔ mutants displayed no differences in antennal or precursor cell densities (Fig S3D–E). Together, the EE is required for ss expression in precursors and R7s, whereas the LE is required for ss expression in R7s.
ss expression does not require Ss protein feedback in precursors or R7s
Early expression often affects later expression from the same gene locus through protein feedback (Maamar et al., 2007; Süel et al., 2006). In the fly eye, no detectable Ss protein is observed in precursors (Johnston and Desplan, 2014). Nevertheless, extremely low levels of Ss protein could trigger regulatory feedback. Ss protein requires heterodimerization with another PAS-bHLH transcription factor, Tango (Tgo) to enter the nucleus and regulate gene expression (Emmons et al., 1999; Thanawala et al., 2013; Ward et al., 1998). To test whether Ss/Tgo feedback activity affects early ss expression, we generated tgo null mutant clones and observed no effect on ss transcription in precursors or R7s (Fig. 3X–Y, S3I). This result suggests that (1) ss regulation in the eye does not require Ss protein feedback, consistent with our previous findings (Johnston and Desplan, 2014) and (2) the early transcription of ss activates late expression by a Ss/Tgo-protein independent mechanism.
Decreasing early ss expression decreases the proportion of SsON R7s
The EE is required for specification of SsON R7s, as knocking out the EE caused a complete loss of ss expression in R7s (Fig. 3I–L, U–W). We hypothesized that reducing activation by the EE would decrease the number of ss-expressing precursors and the proportion of SsON R7s. Because 100% of precursors express ss in wild-type flies, we could identify changes in early and late expression as changes in the density of expression in precursors and the ratio of SsON/OFF R7s in adults (Fig. 2D, S2C).
The EE contains a binding site for the transcriptional repressor, Klumpfuss (Klu), which is expressed during all stages of R7 subtype specification (Fig. S3F)(Anderson et al., 2017). A single base insertion (“sin”) within the EE increases the binding affinity of Klu (Anderson et al., 2017). Flies with sin displayed a reduction in the number of ss-expressing precursors and a decrease in the proportion of SsON R7s (45% SsON) (Fig. 3Q, S–T, V–W). Flies with sin had no change in ss expression in the antenna (Fig. 3R, U). Flies with sin displayed no differences in antennal or precursor cell densities (Fig S3D–E). Ectopic expression of Klu reduced ss expression in precursors and the ratio of SsON R7s (51.8% SsON/Rh4)(Fig. 3V–W, S3Q–R) (Anderson et al., 2017) with no effect on precursor cell density (Fig S3E). Ectopic expression of Klu in precursors caused an increase of ss expression in the antenna (Fig. 3U, S3P) and an increase in antennal cell density (Fig S3D), consistent with differential regulation of ss by Klu across tissues through different enhancers (Klein and Campos-Ortega, 1997; Yang et al., 1997). Additionally, a partial deletion of the EE (pEEΔ) resulted in a reduction in expression in precursors and a reduction of SsON R7s to 25% (Fig. 3U–W, S3M–O). pEEΔ mutants displayed no differences in antennal or precursor cell densities (Fig. S3D–E). This deletion removes the sequence abutting the EE and may disrupt the binding of other trans-acting factors to the EE. These data suggest that decreasing EE activity by genetically altering cis or trans inputs reduces ss expression in precursors and leads to a reduction in SsON R7s.
To identify regulators of R7 subtype specification, we screened flies with mutations or RNAi knockdowns in genes encoding chromatin modifiers for changes in the ratio of SsON and SsOFF R7s (Table S2). Reducing activity of two genes encoding chromatin modifiers, ash2 and lid, caused significant loss of SsON R7s. Knockdown of the trithorax group gene ash2 (Adamson and Shearn, 1996; Papoulas et al., 1998) caused a decrease in SsON R7s in two independent RNAi lines (Table S2). ash21 null mutants displayed a reduction in SsON R7s (38.4%) (Fig. 3W, S3U). ash21 null mutants displayed a cell autonomous decrease in ss expression in precursors (Fig. 3V, S3H, T), and no change in ss expression in antennal cells (Fig. 3U, S3S). Similarly, a null mutation in the histone demethylase gene lid (Eissenberg et al., 2007; Secombe et al., 2007) caused a reduction in ss expression in precursors and the proportion of SsON R7s (36.8% SsON/Rh4), but had no effect on ss expression in antennal cells (Fig. 3U–W, S3V–X). Though ash21 and lid mutants displayed a decrease in precursor cell densities (Fig. S3E), the proportion of cells expressing ss was reduced after normalization to cell density (Fig. S3Y). These data implicate a role for chromatin modifiers in ss regulation and suggest that decreasing ss expression in precursors decreases the proportion of SsON R7s.
To evaluate the relationship of ss expression in precursors to the ratio of SsON R7s, we normalized the densities of ss-expressing precursors and the ratios of SsON R7s for each genotype to wild-type. For expression in precursors and R7s respectively, sin mutants had 67.7% and 65.4%, pEEΔ mutants had 26.7% and 37.1%, ash2 null mutants had 83.2 % and 87.0%, and lid null mutants had 58.3% and 55.4% (Fig. S3Y). These mutants had highly similar proportional changes between precursors and R7s (Fig. S3Y). In contrast, flies with ectopic expression of Klu had 27.3% normalized expression in precursors and 77.9% SsON R7s (Fig. S3Y), suggesting multiple roles for Klu in this process or differences in levels and/or timing caused by the transgenic overexpression. Together, these data suggest that expression early in precursors is required for expression late in R7s.
Derepression of early ss expression increases the proportion of SsON R7s
We next investigated how derepression of ss affected R7 subtype specification. We hypothesized that mutant genotypes with an increase in SsON R7s will have an altered expression pattern earlier in development. For these experiments, we examined ss expression in undifferentiated cells (ssOFF), precursors (ssON), and differentiating cells (ssOFF) in larval eye-antennal discs as well as R7s (mix of ssON and ssOFF) in adult retinas (Fig. 4C–F). As all precursors express ss, we did not observe an increase in the density of expression in these cells in these mutant conditions. Rather, we observed aberrant expression earlier in undifferentiated cells and/or later in differentiating cells during R7 specification.
Figure 4. Derepressing early ss expression increases % SsON R7s.
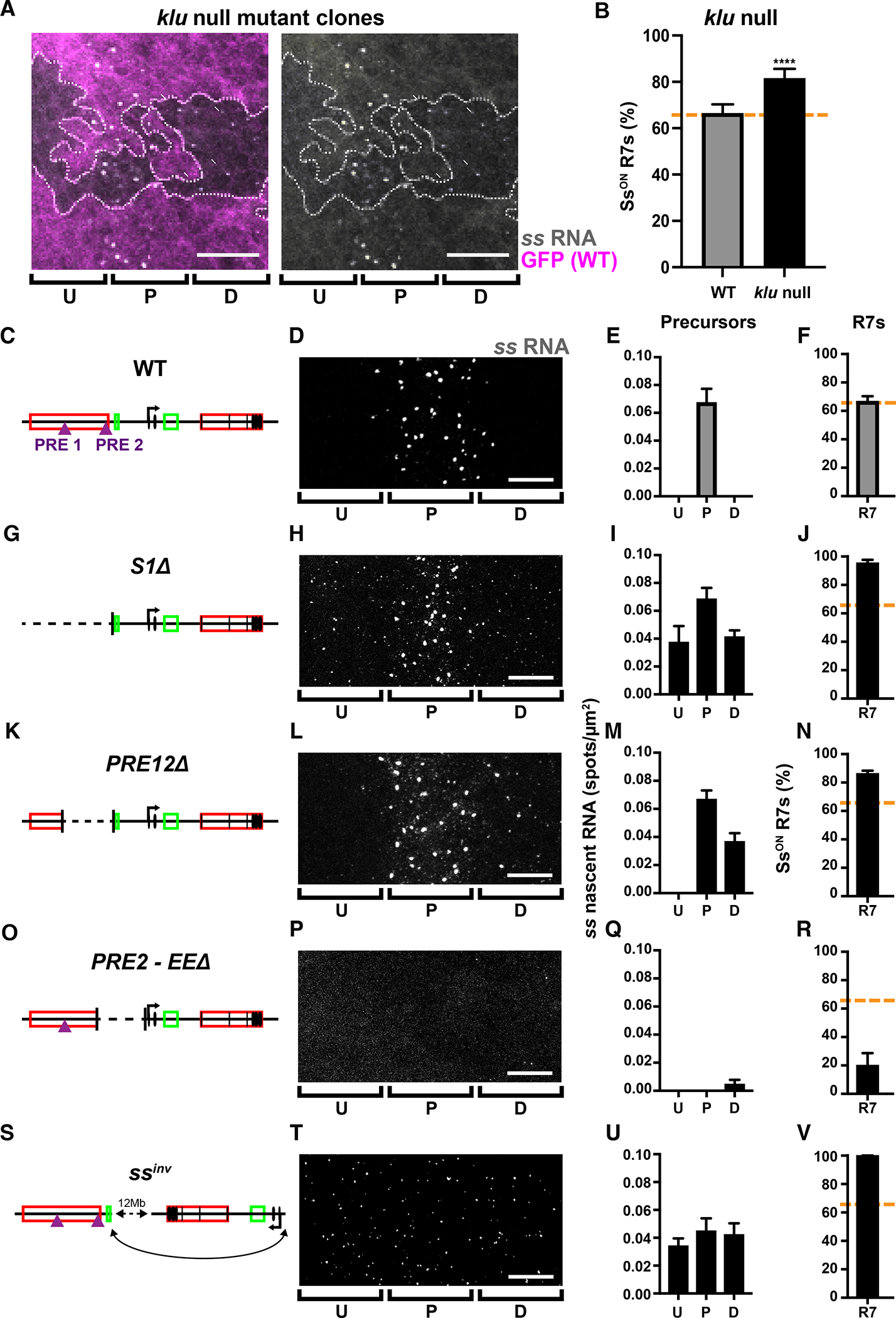
For A-V) U=undifferentiated cells; P=precursors; D=differentiating cells; R7=R7s.
A) ss expression in precursors is extended into differentiating cells in klu null mutant clones. GFP- = klu null mutant; GFP+ = wild type. Gray=ss RNA; magenta=GFP; dashed line=clone boundary; arrows= ss RNA in differentiating cells. Scalebar=10 μm.
For B, F, J, N, R, V) Orange line=mean WT ss expression.
B) % SsON R7s increases in klu null mutants, similar to previous studies (Anderson et al 2017). **** denotes p<0.0001. N=3.
For C, G, K, O, S) Schematized ss locus.
For D, H, L, P, T) ss RNA in undifferentiated cells, precursors, and differentiating cells. Scalebar=10 μm.
For E, I, M, Q, U) Quantification of expression for D, H, L, P, T.
For F, J, N, R, V) % SsON R7s.
C-F) WT.
G-J) S1Δ.
K-N) PRE12Δ.
O-R) PRE2-EEΔ.
S-V) ssinv.
Increasing the binding affinity of a Klu site or increasing Klu levels reduced early ss expression and the proportion of SsON R7s. In contrast, klu null mutant clones displayed a temporal extension of ss expression beyond the precursor state into the differentiating state, when ss is not normally expressed (Fig. 4A). klu null mutants also exhibited an increase in the proportion of SsON R7s (82% SsON) without changing eye morphology (Fig. 4B)(Anderson et al., 2017). These data suggest that Klu is a cell-autonomous off switch for ss expression and that extended expression of ss leads to an increase in the probability of SsON R7 fate.
Repressive silencer elements restrict expression of ss to a subset of R7s (Johnston and Desplan, 2014). We focused on the effects of a 36.4 kb deletion of silencer1 (S1Δ)(Fig. 4G)(Thanawala et al., 2013). Heterozygous S1Δ/+ mutants displayed ss expression in undifferentiated cells prior to the precursor stage and in differentiating cells after the precursor stage (Fig. 4H–I, S4B–D) and an increase in the proportion of SsON R7s (95%)(Fig. 4J, S4E). S1Δ/+ mutants showed low level ss expression in most cells of the eye-antennal disc, including the peripodial membrane, which is normally ssOFF (Fig. S4H). S1Δ/+ mutants did not display changes in cell density (Fig. S4F–G). Together, these data indicate that silencer1 is generally required for repression of ss.
As ss expression was diminished in ash2 and lid mutants, we hypothesized that chromatin is playing a role in ss repression. We examined the region deleted in S1Δ mutants for Polycomb response elements (PREs), DNA elements bound by Polycomb group (PcG) proteins that nucleate repressive heterochromatin (Chan et al., 1994; Paro and Hogness, 1991; Simon et al., 1993; Strutt et al., 1997). ChIP-seq showed distinct peaks for Polycomb group (PcG) proteins, suggesting that two putative PREs (PRE1 and PRE2) fall within the region deleted in the S1Δ mutants (Fig. S4J)(Celniker et al., 2009; Schwartz et al., 2006). These putative PREs correspond to peaks in scATAC-seq datasets (Fig. S2D)(Bravo González-Blas et al., 2020), suggesting that these are binding sites for PcG proteins. We validated the activity of PRE1 and PRE2 using pairing sensitive silencing assays (Fig. S4K–O)(Kassis, 1994; Kassis et al., 1991). Together, these data suggest that PRE1 and PRE2 are functional PREs.
To test the roles for PRE1 and PRE2, we generated a deletion that removed 13 kb containing both PREs (PRE12Δ)(Fig. 4K). Hemizygous PRE12Δ mutants displayed a temporal extension of ss expression into differentiating cells and an increase in the ratio of SsON R7s (86%) but did not exhibit ectopic expression in undifferentiated cells (Fig. 4L–N). These data suggest that the two PREs repress ss expression. Together, chromatin regulation at the ss locus is critical for R7 subtype specification and extending early expression increases the proportion of SsON R7s.
Derepression restores SsON R7 fate in EE mutants
Our data suggest that activation in precursors is necessary and precedes repression in differentiating cells during R7 subtype specification. To test the temporality of these steps, we examined mutants that impaired activation in precursors and repression in differentiating cells. We predicted that derepression in differentiating cells would offset loss of activation in precursors to restore SsON R7 fate. We used imprecise P-element excision to generate an 11.8 kb mutant that deleted one PRE within silencer1 and the EE (PRE2-EEΔ)(Fig. 4O). In PRE2-EEΔ mutants, ss expression in precursors was completely lost (Fig. 4P–Q), consistent with the loss of activation by the EE. PRE2-EEΔ mutants displayed low levels of ss expression in differentiating cells (Fig. 4P–Q), consistent with a loss of repression. PRE2-EEΔ mutants contained 20% SsON R7s (Fig. 4R), suggesting that derepression in differentiating cells restores SsON R7 fate in the absence of activation by the EE.
To further test these interactions, we examined flies with an inversion (ssinv) that moves the ss promoter and LE ~12 Mb away from upstream regions, preventing regulation by the EE and silencer1 (Fig. 4S–V, S4F–G)(Thanawala et al., 2013). ssinv mutants displayed weak ss expression in undifferentiated cells, precursors, and differentiated cells (Fig. 4T–U), consistent with the loss of repression by silencer1. Strong ss expression in precursors was decreased (Fig. 4T–U), consistent with the loss of activation by the EE. ssinv mutants contained 100% SsON R7s (Fig. 4V), suggesting that derepression enables SsON R7 fate when activation by the EE is lost. These data are consistent with activation driven by the EE in precursors before repression by silencer elements in differentiating cells during R7 subtype specification in wild-type conditions.
Repression limits ss expression to a subset of R7s
To test the hypothesis that repression limits ss expression to SsON R7s, we developed a “repression reporter” strategy. A broad PR enhancer reporter ([3×P3]>RFP) drives expression in all PRs of the adult retina when inserted into a control locus on the X chromosome (Fig. 5B–C, F–G, S5A, P3 Ctrl)(Bischof et al., 2007). We used CRISPR to insert this reporter into different locations in the endogenous ss locus (Fig. 5A, inserts 1–4). We hypothesized that, if the local chromatin environment at the ss locus was sufficient to repress expression, expression of the reporter would be limited to SsON R7s. If the ss locus was not sufficient to repress expression, the reporter would be expressed in all PRs, including all R7s.
Figure 5. Repression by the ss locus limits expression to a subset of R7s.
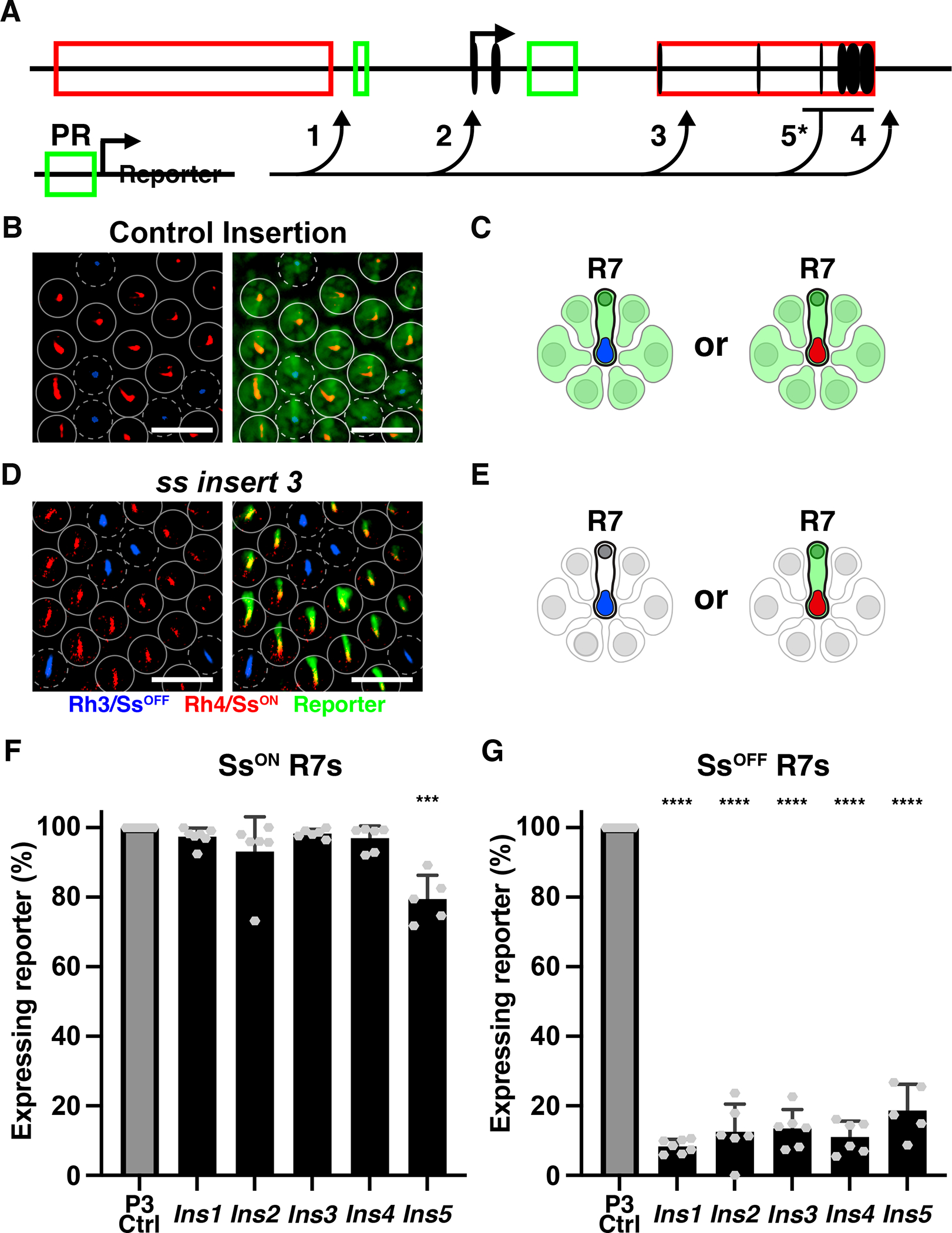
A) Schematic of the PR enhancer reporter construct and insertion sites (arrows). 1–4 inserted using CRISPR. 5* inserted using homologous recombination.
B-C) Control insertion. Scalebar=20 μm.
D-E) Insertion into ss. Scalebar=20 μm.
F-G) *** denotes p<0.0005; **** denotes p<0.0001. F) % SsON R7s with reporter expression.
G) % SsOFF R7s with reporter expression.
When inserted into four different locations in the ss locus, the reporter transgene was nearly perfectly repressed in SsOFF R7s and expressed in SsON R7s in adult retinas (Fig. 5D–E). The reporter was expressed in >93% of SsON R7s and in <14% of SsOFF R7s (Fig. 5F–G, S5A). With the exception of ins2, inserted into the 5’ UTR of ss, the reporter lines did not significantly affect the ratio of SsON/OFF R7s (Fig. S5B). These data suggest that repression at the ss locus limits expression to SsON R7s.
When inserted into control loci, the reporter drives expression in the motion-detecting outer PRs (R1–6) (Fig. 5B–C). When inserted into the ss locus, the reporter was not expressed in outer PRs (Fig. 5D–E), suggesting that the ss locus represses expression in these cells.
In control insertion lines, the reporter drives expression in color-detecting R8 PRs. When inserted into the ss locus, the reporter remained expressed in a subset of R8s (Fig. S5C). In these lines, Ss was expressed in the same subset of R8s (Fig. S5C). These observations suggest that the broad PR enhancer reporter, inserted into the ss locus, was sufficient to overcome repression and ectopically drive ss and reporter expression in R8s.
Regulation of the “repression reporter” may be specific to this enhancer reporter inserted by CRISPR. We previously used homologous recombination to replace the last four exons of ss with a different broad PR enhancer reporter (GMR>GFP, Fig. 5A, insert 5)(Thanawala et al., 2013). For this reporter, we examined ssIns5/+ heterozygous flies. Expression of this reporter was generally expressed in SsON R7s (79.5% co-expressing), repressed in SsOFF R7s (18.7% expressing), repressed in outer PRs, and expressed in R8s (Fig. 5F–G, S5A).
Insertion of two different types of broad PR enhancer reporters by two different methods across five locations in the ss gene locus resulted in repression of the reporter in SsOFF R7s and expression in SsON R7s. These data indicate that the ss gene locus represses expression and suggests a role for the local chromatin environment in repression.
Visualizing chromatin compaction at the ss locus
The repression reporter strategy showed that the ss locus restricts expression to a subset of R7s, likely through chromatin remodeling of the ss locus. Additionally, two silencer elements are required for proper ss expression, consistent with a role for long range repressive interactions possibly through chromatin compaction.
We sought to characterize the compaction state of the ss gene locus during R7 subtype specification. The heterogeneity of cell types in the larval fly eye and limiting quantities of cells impede cell-type-specific analyses through ChIP-seq and ATAC-seq approaches. To examine chromatin compaction with single cell resolution in intact tissue, we developed a 3-color DNA-FISH strategy. We labeled a 50 kb upstream region, a 65 kb region encompassing ss, and a 50 kb downstream region with different fluorescently labeled probes (Fig. 6A). We identified the center of the spheroid for each region and measured the 3-D distance from the upstream region to the ss region (d1) and from the ss region to the downstream region (d2) in individual nuclei. We summed d1 and d2 to generate the total 3-D distance (dt) (Fig. 6B–C, S6A)(Joyce et al., 2012; Rosin et al., 2018; Viets et al., 2019). Larger distances reflect a more open state, while smaller distances indicate a more compact state (Fig. 6B–C). We hypothesized that transcribed ss loci would be more open compared to inactive ss loci which would be more compact.
Figure 6. Dynamic chromatin compaction of the ss locus.
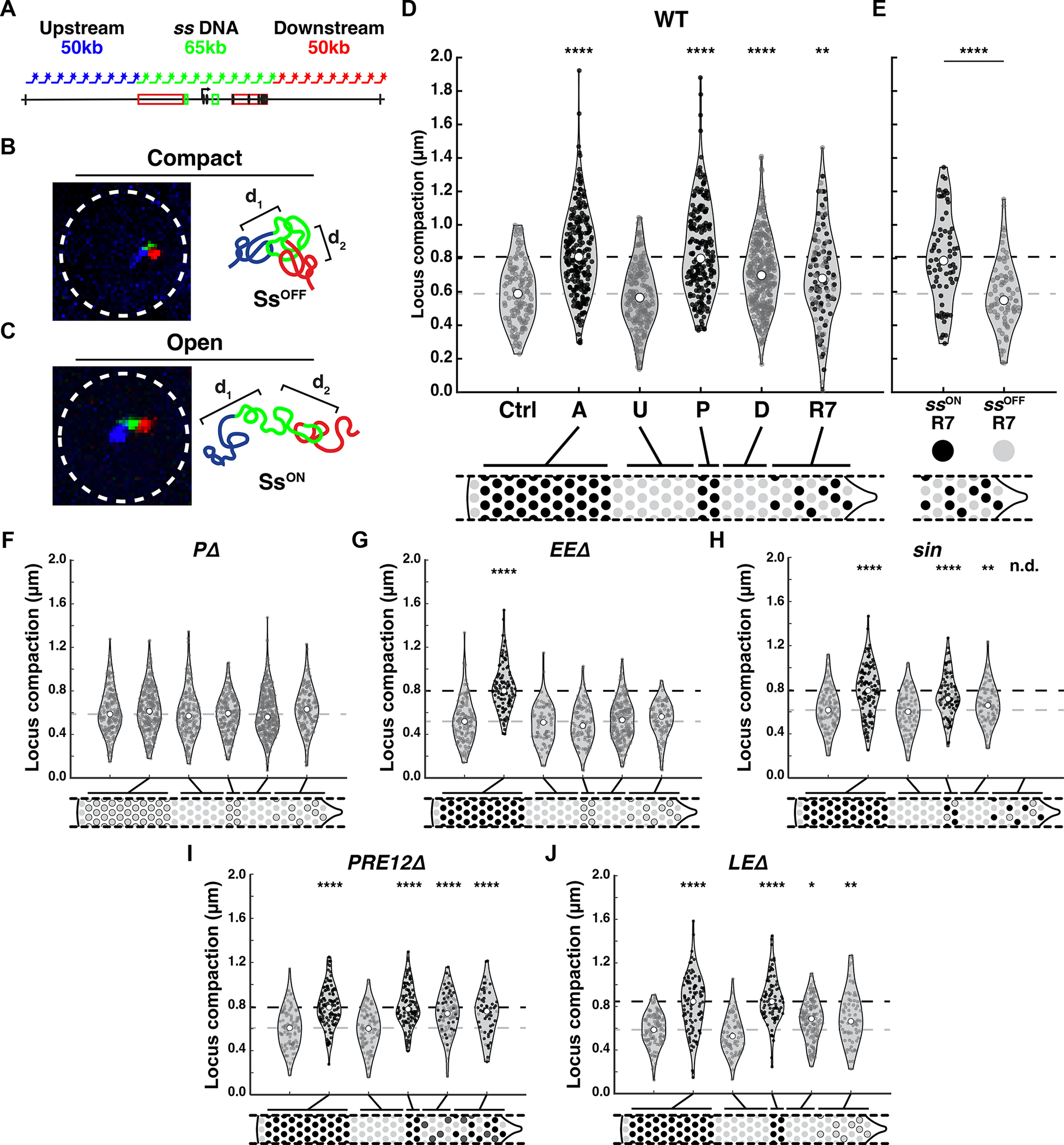
A) Schematic of DNA FISH probes used to label the upstream (blue), ss locus (green), and downstream (red) regions.
For B-C) Left=image; right=schematized model.
B) SsOFF cell with compact chromatin.
C) SsON cell with open chromatin.
For D-J) Quantification. Ctrl=peripodial membrane cells; A=antennal cells; U=undifferentiated cells; P=precursors; D=differentiating cells; R7=R7s. Black circle=ssON cell; gray circle=ssOFF cell; white rectangle=quartile; white circle=median; gray dashed line=ssOFF control median; black dashed line=ssON control median. * denotes p<0.05; ** denotes p<0.005; **** denotes p<0.0001.
For D, F-J) Ctrl cells were compared to A, U, P, D, or R7 cells. For E, SsON R7s were compared to SsOFF R7s.
For D-J) n>70 cells for each region.
D) WT.
E) Compaction in SsON R7s and SsOFF R7s.
F) PΔ.
G) EEΔ.
H) sin.
I) PRE12Δ.
J) LEΔ.
To test our method, we examined compaction in peripodial membrane cells and antennal cells. The ss locus was more compact in peripodial membrane cells (Ctrl) where ss is repressed, with a median compaction of 588 nm (lower quartile=456 nm; upper quartile=720 nm). The ss locus was significantly more open in antennal cells (A) where ss is expressed, with a median compaction of 809 nm (lower quartile=643 nm; upper quartile=985 nm)(Fig. 6D–E). Thus, the DNA FISH method discerned differences in DNA compaction between cells with active or repressed ss.
Chromatin compaction is dynamic during R7 differentiation
We examined compaction at the ss locus in the developing eye. In single eyeantennal discs, we imaged all stages of R7 differentiation. As in previous experiments, we determined the differentiation state of cells based on their positions relative to the MF. Undifferentiated cells were anterior to the MF. Posterior to the MF, precursors were located at 0–10 μm, differentiating cells were located at 10–30 μm, and R7s were located at >30 μm. R7s were also labeled by immunohistochemistry of a GFP reporter expressed in all R7s. The reporter did not alter ss expression in R7s or compaction dynamics (Fig. S6M–O).
The ss locus was more compact in undifferentiated cells (ssOFF), similar to peripodial membrane cells (ssOFF) (Fig. 6D). The ss locus was more open in precursors (ssON), similar to antennal cells (ssON) (Fig. 6D). We predicted that differentiating cells (ssOFF) would be compact but were surprised to observe intermediate compaction (Fig. 6D). This intermediate compaction was also observed in R7s (a mix of ssON and ssOFF) (Fig. 6D). The intermediate compaction measurements suggested two main possibilities: (1) there are two distinct populations of cells, with compact or open chromatin at the ss locus, or (2) the ss locus is at an intermediate compaction state across all cells.
To discern between these hypotheses, we identified ssON and ssOFF R7s using the reporter Ins5. ssON R7s were identified based on Ins5 expression. ssOFF R7s were identified based on the absence of Ins5 expression and their positions. The ss locus was more open in ssON R7s (median=786 nm), similar to other ssON cells, whereas the ss locus was more compact in ssOFF R7s (550 nm), similar to other ssOFF cells (Fig. 6D–E). These data suggested that the intermediate average compaction measurements observed for all R7s represented two distinct populations: ssON R7s with a more open ss locus and ssOFF R7s with a more compacted ss locus.
Differentiating cells do not express ss and the variability in chromatin compaction prevented identification of two distinct cell populations. To characterize changes in compaction over time in differentiating cells, we examined “early” (at 10–20 μm) and “late” (at 20–30 μm) differentiating cells and observed no significant differences in compaction (Fig. S6N–O). Considering (1) the temporal progression of development from differentiating cells to R7s, and (2) the similarity of intermediate compaction between ssOFF differentiating cells and the total population of R7s (including ssON and ssOFF R7s), we surmise that differentiating cells also represented two populations: cells with a more open ss locus and cells with a more compacted ss locus. We cannot rule out that differentiating cells are comprised of cells with intermediate compaction states or a mix of cells with open, compact, and intermediate compaction states.
To determine differential compaction between regions of the locus, we evaluated d1 and d2 distances individually. We did not detect differences in compaction when evaluating d1 or d2 compared to dt (i.e. dt=d1 + d2) (Fig. S6A, C–F), suggesting no asymmetries in local compaction. We also calculated the angles between d1 and d2 and observed similar distributions across the four phases of R7 subtype specification and controls (Fig. S6B, G–L), suggesting no detectable changes in DNA looping at the locus across development.
Comparing the expression and compaction states for developing R7s over time, we find that the ss locus is: (1) inactive and compact in undifferentiated cells, (2) active and open in precursors, (3) inactive and likely a mix of open and compact in differentiating cells, and (4) active and open, or inactive and compact in R7s. We next examined the relationship between transcription and chromatin compaction during R7 subtype specification.
Transcription in precursors is required for large-scale decompaction of the ss locus
The early expression of ss in precursors driven by the EE is required for expression later in R7s driven by the LE. As no discernible Ss protein is generated during the early expression in precursors and Ss does not feedback to regulate its expression, we hypothesized that early expression in precursors was required to open the ss locus. To test this hypothesis, we examined promoter mutants (PΔ) that do not express ss. In PΔ mutants, the ss locus was compact in undifferentiated cells and remained compact in precursors, differentiating cells, and R7s (Fig. 6F), suggesting that transcription plays a role in opening the ss locus in precursors. In PΔ mutants, the ss locus was similarly compact in antennal cells, which normally express ss and have open chromatin (Fig. 6F). These data suggest that ss transcription is required for large-scale decompaction of the ss locus.
To test whether the EE is required to open the ss locus, we examined chromatin compaction in EEΔ mutants. In EEΔ mutants, ss was not expressed in precursors and R7s but remained active in the antenna. In EEΔ mutants, the ss locus was compact in undifferentiated cells, precursors, differentiating cells, and R7s but was open in antennal cells (Fig. 6G). Thus, the EE is required for decompaction of the ss locus in precursors, differentiating cells, and R7s.
Decreasing EE activity reduced the proportion of precursors that expressed ss and the ratio of SsON R7s. Since EE activity is required for chromatin decompaction in precursors, we hypothesized that decreasing ss expression in precursors would decrease the number of open cells in precursors and differentiating cells, resulting in a more compact state.
To test this idea, we assessed ss locus compaction for genetic conditions that reduced the number of ss-expressing precursors and the ratio of SsON R7s. In flies with sin, the ss locus was compact in the control peripodial membrane cells (ssOFF) and undifferentiated cells (ssOFF) and open in antennal cells (ssON), similar to wild-type (Fig. 6H). In sin precursors (mix of ssON and ssOFF), the ss locus displayed intermediate compaction and did not open to the same degree as the antenna (Fig. 6I). The ss locus displayed intermediate compaction in differentiating cells (ssOFF), but the ss locus was more compact than in precursors, consistent with fewer cells opening in precursors. Flies with over-expression of Klu displayed similar effects on ss locus compaction dynamics (Fig. S6P). Though ash2 and lid null mutant flies displayed decreases in cell densities (Fig. S3D–E), limiting our analyses, ss locus compaction trended towards similar effects in these mutants (Fig. S6Q–R). These data suggest that decreasing the proportion of precursors that expressed ss led to more compact chromatin in precursors. The further decrease in compaction in differentiating cells is consistent with the ss locus remaining open in some cells and closing in others.
Repression is required for ss locus compaction in differentiating cells
We next investigated how loss of repression affects ss locus compaction in mutants that delete PRE1 and PRE2 (PRE12Δ). In PRE12Δ mutants, ss was repressed in undifferentiated cells and expressed in precursors. We observed ectopic expression in differentiating cells and the proportion of SsON R7s increased. ss was expressed in antennal cells and repressed in peripodial membrane cells. In homozygous PRE12Δ mutants, the ss locus was compact in undifferentiated cells and peripodial cells (ssOFF) and open in precursors and antennal cells (ssON) (Fig. 6I). The ss locus was more open in differentiating cells and R7s in PRE12Δ mutants compared to wild-type controls (Fig. 6I), suggesting that repression by PREs is required to properly compact the ss locus and repress expression in differentiating cells and R7s.
Compaction is independent of LE activity
Finally, we tested the interaction of chromatin compaction and expression driven by the LE. In LEΔ mutants, ss was expressed in precursors, turned off in differentiating cells, and remained off in all R7s. We hypothesized that either (1) the ss locus would display intermediate compaction in R7s, similar to differentiating cells, or (2) the ss locus would be more compacted similar to other ssOFF cells. In LEΔ mutants, the ss locus was compact in undifferentiated cells, open in precursors, and intermediate in differentiating cells and R7s (Fig. 6J), like in wild-type flies (Fig. 6D), suggesting that chromatin state in maturing R7s is not dependent on transcription driven by the LE.
Together, these data suggest that expression in precursors driven by the EE is required to open the ss locus, repression mediated by PREs is required to compact the ss locus in a subset of differentiating cells, and compaction state is independent of expression driven by the LE.
Discussion
Temporally dynamic antagonism stochastically specifies R7 subtypes
We investigated how regulation of transcription and chromatin compaction at the ss locus controls stochastic R7 patterning in the fly eye. ss is initially in a compact, repressed state in undifferentiated cells. This compacted state is similar in other SsOFF cell types including peripodial cells and SsOFF R7s. As the eye develops, ss is transcribed in precursors and chromatin is opened. ss transcription and large-scale decompaction are lost in mutants deleting the EE or the promoter, suggesting that ss transcription drives the opening of the ss locus in precursors early.
As the cells mature, ss expression ceases and the ss locus compacts during the transition from the precursor to the differentiating cell phase. Our observations that (1) the ss locus is open in SsON R7s and compact in SsOFF R7s and (2) the similarity of median compaction in differentiating cells and all R7s (including SsON R7s and SsOFF R7s), suggest that the ss locus assumes either an open or compact state in differentiating cells that is maintained until terminal R7 subtype specification. Our data are consistent with stable compaction states in differentiating cells, but they cannot rule out changes during this phase of R7 development.
ss expression and compaction during the transition from precursor to differentiating cell phases are critical processes that determine the stochastic R7 fate choice. Decreasing EE activity reduced ss expression in precursors and the proportion of SsON R7s. Extending ss transcription into the differentiating cell phase increased the proportion of SsON R7s. We propose that variable activation and duration of transcription in each precursor determines the probability of recompaction, which ultimately dictates the SsON or SsOFF expression state in R7s.
In the last stage of R7 subtype specification, ss expression driven by the LE is repressed in a subset of R7s. The repression reporter strategy showed that repression at the ss locus limits expression to a subset of R7s. The chromatin compaction assays showed that the ss locus is open in SsON R7s and compact in SsOFF R7s. Deletion of the LE ablated expression but did not alter compaction in R7s. Thus, the chromatin state is set and maintained independent of expression at this stage of R7 maturation. Further, open chromatin is not sufficient to activate ss expression. Together, our data suggest that open chromatin allows activation by the LE whereas compact chromatin represses ss expression.
Based on these findings, we propose a mechanism that controls stochastic R7 subtype specification (Fig. 7, S7). The ss locus is in a compact state in undifferentiated cells (Fig. 7, U). The EE drives transcription and opens the ss locus in precursors (Fig. 7, P). Early expression ceases and the ss locus randomly assumes an open or compact state in differentiating cells (Fig. 7, D). R7s with open chromatin at the ss locus reactivate ss and take on the SsON R7 fate, whereas R7s with compact chromatin at the ss locus repress ss and take on the SsOFF R7 fate (Fig. 7, R7).
Figure 7. Proposed mechanism for stochastic R7 subtype specification.
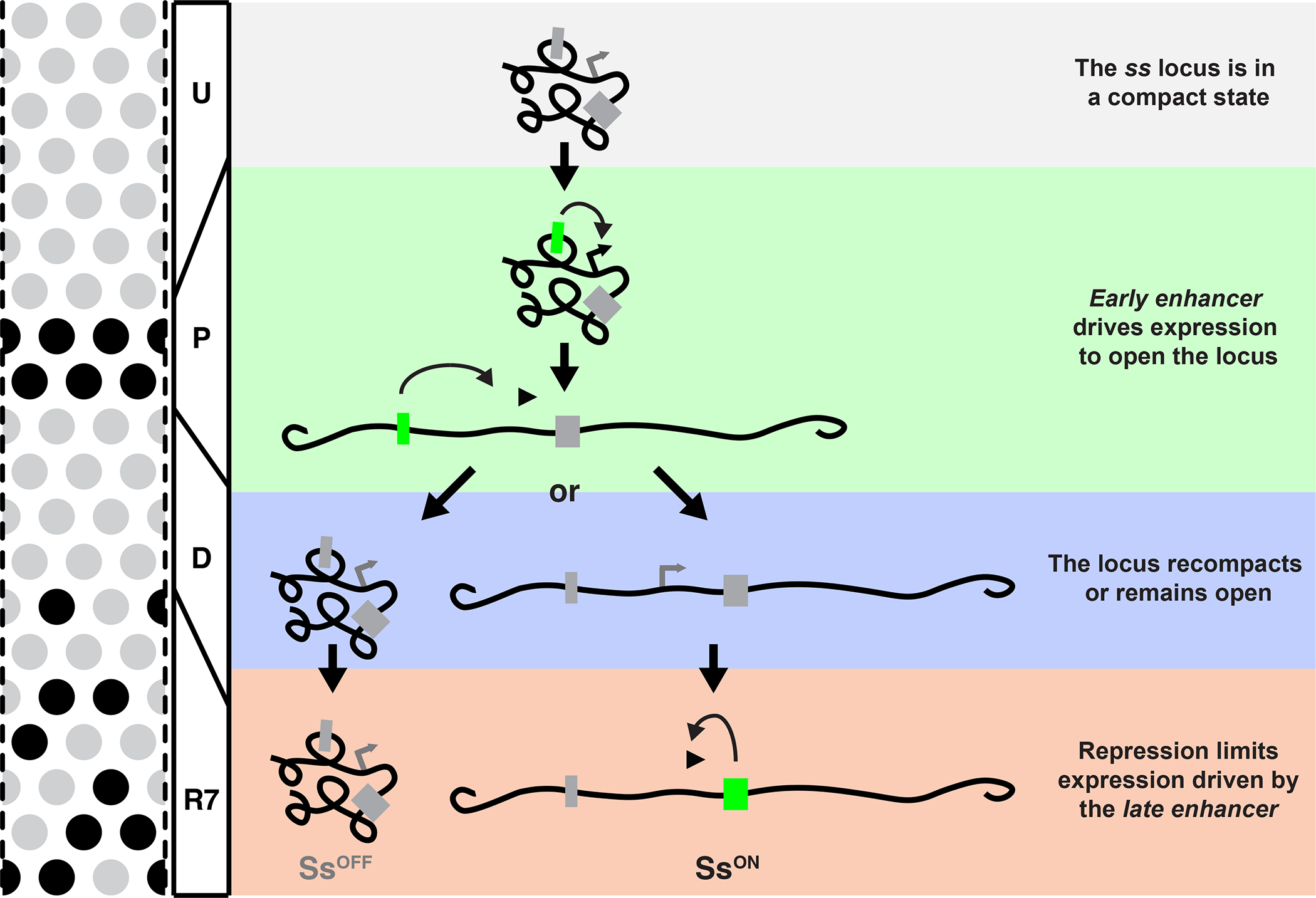
Gray box=inactive enhancer; green box=active enhancer.
Prime and boost mechanisms controlling cell fate specification
A key aspect of this mechanism is the initial “priming” or opening of the ss locus during the early expression in precursors. Transcription-based priming plays important roles in several stereotyped developmental programs (Anderson et al., 2016; Cochella and Hobert, 2012; Greenberg et al., 2017; Kaikkonen et al., 2013; Schmitt et al., 2005). A well-understood example has been described in C. elegans, where the bilateral pair of ASE gustatory neurons display asymmetric gene expression and function (Ortiz et al., 2006, 2009). Stereotyped specification of the left neuron ASEL is dependent upon the asymmetric expression of the microRNA lsy-6 (Johnston and Hobert, 2003), achieved by a “prime and boost” mechanism. Several cell divisions prior to the birth of the terminal ASEL neuron, a pulse of lsy-6 expression in the precursor cell promotes decompaction of the lsy-6 locus. This decompacted state is maintained in the ASEL lineage throughout development, allowing for reactivation of lsy-6 in the terminal ASEL neuron. In the ASER lineage that never experiences the early pulse of lsy-6 expression, the locus remains in a repressed, compacted state, preventing later activation by transcription factors that are expressed in both ASE neurons (Charest et al., 2020; Cochella and Hobert, 2012). Thus, early transcription of a key regulator (lsy-6) promotes one cell fate (ASEL) by antagonizing chromatin-mediated repression important for the specification of the alternative fate (ASER).
The transcription-based prime and boost mechanism controlling ASEL/R sensory neuron specification in C. elegans has many similarities to the mechanism that we have identified for R7 subtype specification. In both systems, early expression of a key regulator in precursor cells opens a locus (prime) so that it can be reactivated later upon terminal specification (boost). A major difference is that the ASEL/R decision requires priming in only the ASEL lineage to reproducibly generate the ASEL fate, whereas the R7 subtype decision utilizes priming in all precursors, which opens the chromatin followed by variable chromatin compaction and repression that ultimately determines the SsON or SsOFF R7 fate.
Both the ASEL/R and R7 subtype decisions also exhibit a window of inactivity between the early and late expression phases. However, this window appears to play two very different roles. In the ASEL/R decision in worms, the early priming of the lsy-6 locus occurs several cell divisions prior to terminal differentiation. The time between the prime and boost is an obstacle that must be overcome to remember the early developmental event. In contrast, the window between the early and late stages of ss expression appears to enable chromatin compaction and repression that determine the SsON or SsOFF expression states in R7s.
Shared features of stochastic fate specification
Though stochastic fate specification is an important feature of many cell fate programs, general features of these mechanisms have not been identified. In the bacterium Bacillus subtilus, transcriptional regulation is critical, as ComK transcription drives a stochastic cell fate switch to the “competent” fate. In both the competence decision in bacteria and R7 subtype specification in flies, all “precursor” cells express the key regulator, yet only a subset undergo the cell fate switch (Maamar et al., 2007; Mugler et al., 2016; Süel et al., 2006).
Stochastic R7 subtype specification in flies also shares mechanistic features with olfactory receptor selection in mice, particularly in the repression of alternative fates. In the olfactory system, OR genes are found in a compact heterochromatic region in the nucleus, with one gene that escapes repression and activates (Clowney et al., 2011; Magklara et al., 2011). Similarly, chromatin compaction and repression play key roles in determining ssON and ssOFF R7 fates. Our studies in flies bridge the roles of transcription in bacteria and chromatin in mice for stochastic cell fate specification.
Stochasticity and the antagonism between transcription and chromatin
Our understanding of the relationship between transcription and chromatin is often a chicken and egg problem: it is unclear whether transcription state dictates large-scale chromatin state or vice versa. Here, we provide evidence that clearly identifies these cause-effect relationships and show how they change during development. The EE drives transcription to open chromatin in precursors. In differentiating cells, the EE ceases to function and transcription stops. Chromatin remains open or closes, marking the stochastic step. Finally, the LE turns on in mature R7s. In cells where the locus is open, transcription reinitiates, while in cells where the locus is closed, transcription is repressed. Thus, initially, transcription state regulates chromatin state and later, chromatin state controls transcription state.
Our studies not only outline this simple mechanism, but also identify how the stochastic step is regulated. The stochastic step occurs as cells cease ss transcription in the precursor phase and assume the open or compact chromatin state in differentiating cells. Decreasing or extending early transcription alters the probability of chromatin closing and ultimately, the proportion of R7 subtypes. Thus, variability in the duration of early transcription is likely a key input that determines the stochastic decision. Our findings provide an important step in understanding how transcription and large-scale chromatin states regulate one another to control how cells randomly assume fates.
Limitations of the Study
Our data suggest that transcription drives large-scale chromatin decompaction and then compaction represses transcription, which controls stochastic cell fate specification. Our study focused on large-scale chromatin remodeling. We hypothesize that local changes of histone modifications at the enhancer and promoter likely precede transcription in precursors. The heterogeneity of the tissue limited testing this hypothesis in a cell-type specific manner in intact tissue. Moreover, we concluded that chromatin compaction represses ss expression in a subset of R7s. The repression reporter experiments showed that the chromatin context at the ss locus is sufficient to repress expression in a subset of R7s. The DNA FISH experiments showed that the ss locus is open in ssON R7s and compact in ssOFF R7s. Our experiments were limited as we could not identify conditions to artificially induce compaction and show sufficiency of compaction to repress ss. Additionally, our studies were conducted in fixed tissue, limiting the observation of the rapid temporal interplay between transcription and chromatin. Future studies could address this challenge with live imaging, enabling assessment of the transcriptional and chromatin dynamics of the ss gene throughout the maturation of individual R7s during subtype specification.
Materials and Methods
RESOURCE AVAILABILITY
Lead Contact
All information queries or requests for resources can be directed to and will be fulfilled by the lead contact, Robert Johnston Jr. (robertjohnston@jhu.edu).
Materials Availability
All reagents and fly lines are available upon request.
Data and Code Availability
The script used to generate 19-bp bar-coding primers for Oligopaints probe design is available at https://github.com/kviets0913/Oligopaints-Primers-Custom-Script. The custom script used to analyze confocal images and determine the density of nascent RNA expression is available at https://zenodo.org/badge/latestdoi/504600646
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila Lines
Flies were raised on standard cornmeal-molasses-agar medium and grown at 25° C. All experiments in this study included both male and female flies. See Table S3 for a full list of fly genotypes used.
METHOD DETAILS
Confocal Image Acquisition
All images were acquired using a Zeiss LSM 700 or LSM 980 confocal microscope. Adult retina images were acquired at a single Z plane at 20x magnification. Immunohistochemistry images at the pupal and larval stages were taken at 40x magnification as minimal Z stacks with a slice thickness of 500 nm. Image acquisition for larval DNA and RNA FISH experiments were taken at 63x magnification as large Z stacks encompassing the tissue with a slice thickness of 300nm.
CRISPR Mediated Deletions
Deletions to the endogenous ss locus were generated using CRISPR (Gratz et al., 2014; Port et al., 2014). Sense and antisense DNA forward and reverse strands of the gRNA were designed to generate BbsI restriction site overhangs. The oligos were annealed and cloned into the pCFD3 cloning vector (Addgene, Cambridge, MA). A single stranded DNA homology bridge was generated with 60 bp homologous regions flanking each side of the predicted cleavage site. The gRNA construct (500 ng/μl) and homology bridge oligo (100 ng/μl) were injected into Drosophila embryos (BestGene, Inc.). Single males were crossed with a balancer stock (yw; +; TM2/TM6B), and F1 female progeny were screened for the deletion via PCR and sequencing. Single F1 males whose siblings were deletion-positive were crossed to the balancer stock (yw; +; TM2/TM6B) and the F2 progeny were screened for the deletion via PCR and sequencing. Information on all CRISPR oligonucleotides used for this study can be found in Table S4.
CRISPR Mediated Insertions
Insertion of the [3XP3]>RFP enhancer reporter construct into the endogenous ss locus was achieved using CRISPR. P3 is a Ey binding site from the rh1 promoter. sgRNAs were designed using Chopchop and cctop (Labuhn et al., 2018; Labun et al., 2016), isolated from injection stocks, and amplified using in vitro transcription. A single stranded DNA homology bridge was generated with homologous regions flanking each side of the predicted cleavage site in the [3XP3]>RFP enhancer reporter construct using Gibson Assembly. The homology bridge was co-injected with Cas9 RNA (2 μg/μl) and sgRNA (1 μg/μl) into 300 Drosophila embryos (Qidong Fungene Biotechnology Co., Ltd.). Single males were crossed with a balancer stock (yw; +; TM2/TM6B), and F1 female progeny were screened for the deletion via PCR and sequencing. Single F1 males whose siblings were deletion-positive were crossed to the balancer stock (yw; +; TM2/TM6B) and the F2 progeny were screened for the deletion via PCR and sequencing. Information on all CRISPR oligonucleotides used for this study can be found in Table S5.
Antibodies
Antibodies and dilutions were as follows: mouse anti-lamin B (DSHB ADL67.10 and ADL84.12), 1:100; rabbit anti-GFP (Invitrogen), 1:500; rabbit anti-RFP (MBL), 1:400 rat anti-Elav (DSHB, 7E8A10), 1:50; rabbit anti-Rh4 (gift from C. Zuker, Columbia University), 1:50; mouse anti-Rh3 (gift from S. Britt, University of Texas at Austin), 1:50; guinea pig anti-Ss (gift from Y.N. Jan, University of California, San Francisco), 1:500; all secondary antibodies (Molecular Probes) were Alexa Fluor-conjugated and used at 1:400.
Immunohistochemistry
Adult, mid-pupal, and larval retinas were dissected as described (Hsiao et al., 2012) and fixed for 15 min with 4% formaldehyde at room temperature. Retinas were rinsed three times in PBS plus 0.3% Triton X-100 (PBX) followed by three 15 min washes in PBX. Retinas were incubated with primary antibodies diluted in PBX >2hrs at room temperature and then rinsed three times in PBX followed by three 15 min washes in PBX. Retinas were incubated with secondary antibodies diluted in PBX >2hrs at room temperature and then rinsed three times in PBX followed by three 15 min washes in PBX. Retinas were mounted in SlowFade Gold Antifade Reagent (Invitrogen). Images were acquired using a Zeiss LSM 700 or LSM 980 confocal microscope at 20x or 40x magnification.
Enhancer Reporter
The early enhancer and late enhancer cassettes were amplified and inserted into the pJR20 plasmid. The early enhancer was amplified from chr3R:16,410,464 – 16,411,045. The late enhancer was amplified from chr3R:16,399,856 −16,396,676. These sequences were inserted upstream of a heat shock gene minimal promoter and the GFP gene coding sequence. All plasmids used were made through standard cloning procedures. Plasmids, plasmid maps, and cloning details are available on request. All constructs were sent to BestGene (Chino Hills, CA) for injection. Constructs were inserted via PhiC31 integration at the attP40 landing site.
Oligopaints Probe Design for RNA and DNA FISH
Probes for RNA and DNA FISH were designed using the Oligopaints technique (Beliveau et al., 2012, 2013, 2015). Target sequences were run through the bioinformatics pipeline available at http://genetics.med.harvard.edu/oligopaints/ to identify sets of 50-bp optimized probe sequences (i.e. “libraries”) tiled across the DNA sequence of interest. Five 19-bp bar-coding primers, gene F and R; universal (univ) F and R, and either sublibrary (sub) F or random (rando) R, were appended to the 5’ and 3’ ends of each probe sequence. To ensure that all probes were the same length, an additional 8-bp random sequence was added to the 3’ end of the probes. The gene F and R primers allowed PCR amplification of a probe library of interest out of the total oligo pool, and the univ F and R primers allowed conjugation of fluorophores, generation of single-stranded DNA probes, and PCR addition of secondary sequences to amplify probe signal. The ss 50-kb left and right extension libraries had a sub F primer between the gene and universal forward primers to allow PCR amplification of probes targeting a specific sub-region of the locus of interest. All other probe libraries had a rando R primer appended at the 3’ end to maintain a constant sequence length between all probes. Bar-coding primer sequences were taken from a set of 240,000 randomly generated, orthogonal 25-bp sequences (Xu et al., 2009) and run through a custom script (available at https://github.com/kviets0913/Oligopaints-Primers-Custom-Script) to select 19-bp sequences with 15-bp homology to the Drosophila genome. Primers were appended to probe sequences using the orderFile.py script available at http://genetics.med.harvard.edu/oligopaints/. Completed probe libraries were synthesized as custom oligo pools by Custom Array (Bothell, WA), and fluorescent FISH probes were generated as described in references (Beliveau et al., 2012, 2013, 2015; Viets et al., 2019).
RNA FISH
RNA FISH was performed using modified versions of the protocols described in references (Beliveau et al., 2012, 2015). 20–50 eye/antennal discs attached to mouth hooks from third instar larvae were collected on ice and fixed in 129 μL ultrapure water, 20 μL 10X PBS, 1 μL Tergitol NP-40, 600 μL heptane, and 50 μL fresh 16% formaldehyde. Tubes containing the fixative and eye discs were shaken vigorously by hand, then fixed for 10 minutes at room temperature with nutation. Eye discs were then given three quick washes in 1X PBX, followed by three five-minute washes in PBX with 0.5% (vol/vol) RNAse inhibitor (Promega) at room temperature with nutation. Eye discs were then removed from the mouth hooks and blocked for 1 hour in 1X PBX:Western Blocking Reagent (Roche) at room temperature with nutation. They were then incubated in primary antibody diluted in 1X PBX with 0.5 U/pL RNAse inhibitor overnight at 4° C with nutation. Next, eye discs were washed three times in 1X PBX for 20 minutes and incubated in secondary antibody diluted in 1X PBX with 0.5 U/pL RNAse inhibitor for two hours at room temperature with nutation. Eye discs were then washed two times for 20 minutes in 1X PBX, followed by a 20-minute wash in 1X PBS. Next, discs were given one 10-minute wash in 20% formamide + 80% 2X SSCT (2X SSC+.001% Tween-20), one 10-minute wash in 40% formamide + 60% 2X SSCT, and two 10-minute washes in 50% formamide + 50% 2X SSCT. Discs were then predenatured by incubating for four hours at 37° C, three minutes at 92° C, and 20 minutes at 60° C. Primary probes were added in 36 μL hybridization buffer consisting of 50% formamide + 50% 2X SSCT+2% dextran sulfate (w/v). All probes were added at a concentration of ≥5 pmol fluorophore/mL. 4 μL of probe was added. After addition of probes, eye discs were incubated at 37° C for 16–20 hours with shaking. Eye discs were then washed for 1 hour at 37° C with shaking in 50% formamide + 50% 2X SSCT. 1 μL of each secondary probe was added at a concentration of 100 pmol/mL in 50 μL of 50% formamide + 50% 2X SSCT. Secondary probes were hybridized for 1 hour at 37° C with shaking. Eye discs were then washed twice for 30 minutes in 50% formamide + 50% 2X SSCT at 37° C with shaking, followed by three 10-minute washes at room temperature in 20% formamide + 80% 2X SSCT and 2X SSCT with nutation. Discs were incubated in 2X SSCT with 300 μM DAPI for 15 minutes at room temperature with nutation, followed by three 10-minute washes at room temperature in 2X SSC with nutation. Discs were mounted in SlowFade Gold immediately after the final 2X SSC wash and imaged using a Zeiss LSM700 or Zeiss LSM980 confocal microscope at 63x magnification.
DNA FISH
DNA FISH was performed using modified versions of the protocols described in references (Beliveau et al., 2012, 2013, 2015; Viets et al., 2019). 20–50 eye/antennal discs attached to mouth hooks from third instar larvae were collected on ice and fixed in 129 μL ultrapure water, 20 μL 10X PBS, 1 μL Tergitol NP-40, 600 μL heptane, and 50 μL fresh 16% formaldehyde. Tubes containing the fixative and eye discs were shaken vigorously by hand, then fixed for 10 minutes at room temperature with nutation. Eye discs were then given three quick washes in 1X PBX, followed by three five-minute washes in PBX at room temperature with nutation. Eye discs were then removed from the mouth hooks and blocked for 1 hour in 1X PBX+1% BSA at room temperature with nutation. They were then incubated in primary antibody diluted in 1X PBX overnight at 4° C with nutation. Next, eye discs were washed three times in 1X PBX for 20 minutes and incubated in secondary antibody diluted in 1X PBX for two hours at room temperature with nutation. Eye discs were then washed two times for 20 minutes in 1X PBX, followed by a 20-minute wash in 1X PBS. Next, discs were given one 10-minute wash in 20% formamide + 80% 2X SSCT (2X SSC+.001% Tween-20), one 10-minute wash in 40% formamide + 60% 2X SSCT, and two 10-minute washes in 50% formamide + 50% 2X SSCT. Discs were then predenatured by incubating for four hours at 37° C, three minutes at 92° C, and 20 minutes at 60° C. Primary probes were added in 36 μL hybridization buffer consisting of 50% formamide + 50% 2X SSCT+2% dextran sulfate (w/v), + 1 μL RNAse A. All probes were added at a concentration of ≥5 pmol fluorophore/mL. For FISH experiments in which a single probe was used, 4 μL of probe was added. For FISH experiments in which three probes were used, 1.3 μL of each probe was added. After addition of probes, eye discs were incubated at 91° C for three minutes and at 37° C for 16–20 hours with shaking. Eye discs were then washed for 1 hour at 37° C with shaking in 50% formamide + 50% 2X SSCT. 1 μL of each secondary probe was added at a concentration of 100 pmol/mL in 50 mL of 50% formamide + 50% 2X SSCT. Secondary probes were hybridized for 1 hour at 37° C with shaking. Eye discs were then washed twice for 30 minutes in 50% formamide + 50% 2X SSCT at 37° C with shaking, followed by three 10-minute washes at room temperature in 20% formamide + 80% 2X SSCT and 2X SSCT with nutation. Discs were incubated in 2X SSCT with 300 μM DAPI for 15 minutes at room temperature with nutation, followed by three 10-minute washes at room temperature in 2X SSC with nutation. Discs were mounted in SlowFade Gold immediately after the final 2X SSC wash and imaged using a Zeiss LSM700 or Zeiss LSM980 confocal microscope at 63x magnification.
scATACseq
Regions of the eye/antennal disc containing open chromatin were obtained from publicly available data, http://genome.ucsc.edu/s/cbravo/Bravo_et_al_EyeAntennalDisc (Bravo González-Blas et al., 2020), and viewed on the UCSC Genome Browser (WJ et al., 2002). We analyzed published scATACseq datasets from developing fly eye-antennal discs that were clustered into cell types by integrating scATAC-seq and scRNA-seq data sets (Fig. S2D). We mapped the cell type clusters expressing ss to the antennal cells, precursors, and mature photoreceptors of the eye/antennal imaginal disc and compared ATAC-seq profiles. We could not map clusters to the undifferentiated, differentiating, and peripodial membrane cells and did not evaluate chromatin accessibility for these cell types based on these datasets. All tracks were scaled to the same parameters for accurate comparisons.
QUANTIFICATION AND STATISTICAL ANALYSIS
Adult Eye Quantifications
The frequencies of Rh4- and Rh3-expressing R7s were scored manually for at least five eyes per genotype. R7s co-expressing Rh3 and Rh4 were scored as Rh4-positive (Mazzoni et al., 2008; Thanawala et al., 2013). 100 or more R7s were scored for each eye. RNAi lines were screened using the Gal4/UAS system, with the an elav driver (elav>Gal4/w ; UAS>Dcr2/+ ; +). The co-expression of reporters and Rh4- or Rh3-expressing R7s were scored manually for at least five adult eyes per genotype. 100 or more R7s were scored for each eye. SsON/Rh4 R7s and SsOFF/Rh3 R7s were scored independently. Due to the binary nature of the cell fate decision, this approach yielded two assessments of R7 subtype fate.
Density of Expression Quantification
The density of ss RNA punctae were calculated computationally using a custom written script in MATLAB. Due to homologous chromosome pairing between both copies of the endogenous chromosome, we observed a single dot for each chromosome. All images were acquired as 3D z-stacks with a slice thickness of 300 nm. The most highly expressed 25 slices within the antenna, and all slices containing punctae in R7 precursors were maximum intensity projected. Undifferentiated, precursor, and differentiating cells were demarcated in 10 μm regions based on distance from the morphogenetic furrow. The punctae were then identified and the area was determined as a bounding box encompassing the identified spots. Nascent RNA spots were distinguished from mature transcripts using an intensity threshold, removing them from density calculations. The density was calculated as the number of punctae per unit area in μm2. To ensure high fidelity identification of spots, four images were quantified manually in parallel, and the number of spots in each image were compared as the percentage of manual IDs. Automated identification had a mean %ID of 100.02% +/− 0.67% when compared to manual quantification, indicating high fidelity identification and density quantification.
Cell Density Quantification
All cell density quantifications were performed in 3D on z-stacks with a slice thickness of 300 nm. Quantifications were performed manually using Fiji (Joyce et al., 2012; Schindelin et al., 2012; Schneider et al., 2012). Three bounding boxes of variable area and a thickness of 33um were drawn, and the number of cells was counted within each boundary. The density was calculated as the number of cells divided by the area demarked by the boundary.
Proportional Expression Changes Quantification
To determine the proportional change from wild-type expression the density of expression in precursors and percentage of SsON/Rh4 in R7s for each genotype was divided by the wild-type average of these values. Normalization first occurred for genotypes showing a significant change in cell density. A normalization factor was determined as the fractional change in cell density from wild-type. This value was multiplied to the raw density values for that genotype.
Compaction Quantification
All compaction quantifications were performed in 3D on z-stacks with a slice thickness of 300 nm. Quantifications were performed manually using Fiji (Joyce et al., 2012; Schindelin et al., 2012; Schneider et al., 2012). Boundaries were drawn for each image to denote cell type. Undifferentiated, precursor, and differentiating cells were demarcated in 10 μm regions based on distance from the morphogenetic furrow. To determine the z position of each FISH dot, an encapsulating box was drawn around the dot and the Plot Profile tool was used to assess the stack in which the dot was brightest. Due to homologous chromosome pairing between both copies of the endogenous chromosome, we observed a single dot for each DNA region. The positional information was scored for several dots in a single channel before scoring dots in a different channel. This method ensured dot position information was gathered blindly relative to the position of other dots within individual cells. To determine the x-y-z distance between FISH dots, we used the multipoint tool to mark the center position for each spot within each nucleus. The distance between the FISH dots was then calculated in 3D as:
Compaction Angle Determination
Compaction angle was determined using the position and length information generated in the compaction quantification. The angle (γ) was calculated with the law of cosines as:
Statistical Analysis
All datasets were tested for a Gaussian distribution using a D’Agostino and Pearson omnibus normality test and a Shapiro-Wilk normality test. If either test indicated a non-Gaussian distribution for any of the datasets in an experiment, datasets were tested for statistical significance using a Wilcoxon rank-sum test (for single comparisons) or a one-way ANOVA on ranks with Dunn’s multiple comparisons test (for multiple comparisons). If both the D’Agostino and Pearson and the Shapiro-Wilk tests indicated a Gaussian distribution for all datasets in an experiment, datasets were tested for statistical significance using an unpaired t-test with Welch’s correction (for single comparisons) or an ordinary one-way ANOVA with Dunnett’s multiple comparisons test (for multiple comparisons). These statistical tests were performed using GraphPad Prism. Statistical tests and p-values are described in the figure legends.
Supplementary Material
KEY RESOURCE TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-Lamin B antibody | DSHB | ADL67.10; RRID: AB_528336 |
| Mouse anti-Lamin B antibody | DSHB | ADL84.12; RRID: AB_528338 |
| Rabbit anti-GFP antibody | Invitrogen | A21311; RRID: AB_221477 |
| Rabbit anti-RFP antibody | MBL International | PM005, RRID: AB_591279 |
| Rabbit anti-Rh4 antibody | gift from C. Zuker, Columbia University | N/A |
| Mouse anti-Rh3 antibody | gift from S. Britt, University of Texas at Austin | N/A |
| Rat anti-Elav antibody | DSHB | 7E8A10; RRID: AB_528218 |
| Guinea pig anti-Ss antibody | gift from Y.N. Jan, University of California, San Francisco | N/A |
| Deposited Data | ||
| Predicted PRE data | modENCODE | FlyBase ID FBlc0000414 |
| scATAC-deq data | UCSC Genome Browser Custom Track | Available at: http://genome.ucsc.edu/s/cbravo/Bravo_et_al_EyeAntennalDisc |
| Experimental Models: Organisms/Strains | ||
| yw ; + ; Df(3R)Exel6269 | Bloomington Stock Center | 7736; RRID: BDSC_7736 |
| yw ; ey>Gal4, UAS>flp/+ ; FRT82B ash21, e, red/GMR>hid | Bloomington Stock Center | 5253; RRID: BDSC_5253 |
| yw ; FRT40A lid140 ; FRT40A GMR>hid ; ey>Gal4, UAS>flp/+ | Bloomington Stock Center | 76954; RRID: BDSC_76954 |
| Ato(384)>Gal4/yw ; UAS>klu/+ ; P{y[+t7.7] w[+mC]=20XUAS-6XGFP}attP2/+ | Bloomington Stock Center | 52262; RRID: BDSC_52262 |
| yw, M{3xP3-RFP.attP}ZH-2A ; + ; + ; M{RFP[3xP3.PB] GFP[E.3xP3]=vas-int.Dm}ZH-102D | Bloomington Stock Center | 24480; RRID: BDSC_24480 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>GFP RNAi/+ | Bloomington Stock Center | 35786; RRID: BDSC_35786 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>Cp190 RNAi/+ | Bloomington Stock Center | 35078; RRID: BDSC_35078 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>su(Hw) RNAi/+ | Bloomington Stock Center | 33906; RRID: BDSC_33906 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>mdg4 RNAi/+ | Bloomington Stock Center | 33907; RRID: BDSC_33907 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>trx RNAi/+ | Bloomington Stock Center | 33703; RRID: BDSC_33703 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>ash2 RNAi v20/+ | Bloomington Stock Center | 64942; RRID: BDSC_64942 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>ash2 RNAi v22/+ | Bloomington Stock Center | 35388; RRID: BDSC_35388 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>Mnn1 RNAi/+ | Bloomington Stock Center | 35150; RRID: BDSC_35150 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>Trl RNAi/+ | Bloomington Stock Center | 67265; RRID: BDSC_67265 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>Pc RNAi/+ | Bloomington Stock Center | 36070; RRID: BDSC_36070 |
| elav>Gal4/w ; UAS>Dcr2/+ ; UAS>lid RNAi/+ | Bloomington Stock Center | 28944; RRID: BDSC_28944 |
| Oligonucleotides | ||
| Oligos for the ss locus deletion CRISPR | See Table S4 for homologous bridge sequence, mutation size, and phenotypic effect | N/A |
| Oligos for the repression reporter insert CRISPR | See Table S5 for homologous bridge sequence, mutation size, and phenotypic effect | N/A |
| Software and Algorithms | ||
| MATLAB_R2019b | MathWorks | https://www.mathworks.com |
| ImageJ | (Schindelin et al., 2012) | https://imagej.nih.gov/ij/ |
| Script used to generate 19-bp barcoding primers for Oligopaints probe design | (Viets et al., 2019) | Available at: https://github.com/kviets0913/Oligopaints-Primers-Custom-Script |
| Custom script used to analyze images and quantify the density of nascent RNA spots | Generated for this study | Available at: https://github.com/lvoortman/Automated_Image_Analysis |
Highlights.
spineless is off and the locus is in a compact state in undifferentiated cells
The early enhancer drives expression and the locus opens
Expression ceases and the locus recompacts or remains open
Repression in a subset of R7s limits expression driven by the late enhancer
Acknowledgements
We thank the JHU integrated Imaging Center. We thank Andre Bedard, Judith Kasis, Kami Ahmed, Jessica Treisman, and Claude Desplan for antibodies and fly lines, Andrew Gordus for input on statistical analysis, Claude Desplan, and Luisa Cochella, for feedback on the manuscript. This work was supported by NIH F31EY032430 (LV), NIH F31EY031963 (EU), NSF PHY-1734030, NIH R01GM097275, U01DA047730 (TG), and NIH R01EY025598 (RJJ).
Footnotes
Declaration of Interests
The authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adamson AL, and Shearn A (1996). Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics 144, 621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqadah A, Hsieh Y-W, Xiong R, and Chuang C-F (2016). Stochastic left-right neuronal asymmetry in Caenorhabditis elegans. [DOI] [PMC free article] [PubMed]
- Anderson C, Reiss I, Zhou C, Cho A, Siddiqi H, Mormann B, Avelis CM, Deford P, Bergland A, Roberts E, et al. (2017). Natural variation in stochastic photoreceptor specification and color preference in Drosophila. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, and Olson EN (2016). Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armelin-Correa LM, Gutiyam LM, Brandt DYC, and Malnic B (2014). Nuclear compartmentalization of odorant receptor genes. Proc. Natl. Acad. Sci. U. S. A. 111, 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang Y, Li JB, Senaratne TN, Williams BR, et al. (2012). Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. U. S. A. 109, 21301–21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Apostolopoulos N, and Wu CT (2013). Visualizing genomes with oligopaint FISH probes. Curr. Protoc. Mol. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliveau BJ, Boettiger AN, Avendaño MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J, et al. (2015). Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat. Commun. 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell ML, Earl JB, and Britt SG (2007). Two types of Drosophila R7 photoreceptor cells are arranged randomly: A model for stochastic cell-fate determination. J. Comp. Neurol. 502, 75–85. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, and Basler K (2007). An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. U. S. A. 104, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo González-Blas C, Quan X, Duran-Romaña R, Taskiran II, Koldere D, Davie K, Christiaens V, Makhzami S, Hulselmans G, Waegeneer M, et al. (2020). Identification of genomic enhancers through spatial integration of single-cell transcriptomics and epigenomics. Mol. Syst. Biol. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, and Axel R (1991). A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65, 175–187. [DOI] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 2015, 21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. (2009). Unlocking the secrets of the genome. Nature 459, 927–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, and Pirrotia V (1994). A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13, 2553–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charest J, Daniele T, Wang J, Bykov A, Mandlbauer A, Asparuhova M, Röhsner J, Gutiérrez-Pérez P, and Cochella L (2020). Combinatorial Action of Temporally Segregated Transcription Factors. Dev. Cell 55, 483–499.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, and Axel R (1994). Allelic inactivation regulates olfactory receptor gene expression. Cell 78, 823–834. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, Magklara A, Colquitt BM, Pathak N, Lane RP, and Lomvardas S (2011). High-throughput mapping of the promoters of the mouse olfactory receptor genes reveals a new type of mammalian promoter and provides insight into olfactory receptor gene regulation. Genome Res. 21, 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowney EJ, Legros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, and Lomvardas S (2012). Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell 151, 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L, and Hobert O (2012). Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell 151, 1229–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Liu JP, and Jessell TM (2003). Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425, 926–933. [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, and Jessell TM (2005). A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123, 477–491. [DOI] [PubMed] [Google Scholar]
- Dubnau D (1999). DNA uptake in bacteria. Annu. Rev. Microbiol. 53, 217–244. [DOI] [PubMed] [Google Scholar]
- Duffy KR, Wellard CJ, Markham JF, Zhou JHS, Holmberg R, Hawkins ED, Hasbold J, Dowling MR, and Hodgkin PD (2012). Activation-induced B cell fates are selected by intracellular stochastic competition. Science (80-. ). 335, 338–341. [DOI] [PubMed] [Google Scholar]
- Duncan DM, Burgess EA, and Duncan I (1998). Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor. Genes Dev. 12, 1290–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, and Shilatifard A (2007). The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat. Struct. Mol. Biol. 14, 344–346. [DOI] [PubMed] [Google Scholar]
- Emmons RB, Duncan D, Estes PA, Kiefel P, Mosher JT, Sonnenfeld M, Ward MP, Duncan I, and Crews ST (1999). The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila. Development 126, 3937–3945. [DOI] [PubMed] [Google Scholar]
- Emmons RB, Duncan D, and Duncan I (2007). Regulation of the Drosophila distal antennal determinant spineless. Dev. Biol. 302, 412–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KD, Mani M, and Carthew RW (2022). Emergence of a geometric pattern of cell fates from tissue-scale mechanics in the Drosophila eye. Elife 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey PA, Malnic B, and Buck LB (2004). The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci. U. S. A. 101, 2156–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, and Oconnor-Giles KM (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC, Glaser J, Borsos M, Marjou F. El, Walter M, Teissandier A, and Bourc’his D (2017). Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat. Genet. 49, 110–118. [DOI] [PubMed] [Google Scholar]
- Hahn J, Burghoorn J, Dubnau D, Turgay K, and Hahn J (1998). Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17, 6730–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendy O, Campbell J, Weissman JD, Larson DR, and Singer DS (2017). Differential context-specific impact of individual core promoter elements on transcriptional dynamics. Mol. Biol. Cell 28, 3360–3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao HY, Johnston RJ, Jukam D, Vasiliauskas D, Desplan C, and Rister J (2012). Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J. Vis. Exp [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, and Desplan C (2010). Stochastic Mechanisms of Cell Fate Specification that Yield Random or Robust Outcomes. Annu. Rev. Cell Dev. Biol. 26, 689–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, and Desplan C (2014). Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science (80-. ). 343, 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, and Hobert O (2003). A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426, 845–849. [DOI] [PubMed] [Google Scholar]
- Joyce EF, Williams BR, Xie T, and Wu C ting (2012). Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genet 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. (2013). Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol. Cell 51, 310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA (1994). Unusual properties of regulatory DNA from the Drosophila engrailed gene: Three “pairing-sensitive” sites within a 1.6-kb region. Genetics 136, 1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA, VanSickle EP, and Sensabaugh SM (1991). A fragment of engrailed regulatory DNA can mediate transvection of the white gene in drosophila. Genetics 128, 751–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T, and Campos-Ortega JA (1997). klumpfuss, a Drosophila gene encoding a member of the EGR family of transcription factors, is involved in bristle and leg development. Development 124, 3123–3134. [DOI] [PubMed] [Google Scholar]
- Labuhn M, Adams FF, Ng M, Knoess S, Schambach A, Charpentier EM, Schwarzer A, Mateo JL, Klusmann JH, and Heckl D (2018). Refined sgRNA efficacy prediction improves largeand small-scale CRISPR-Cas9 applications. Nucleic Acids Res. 46, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Gagnon JA, Thyme SB, and Valen E (2016). CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 44, W272–W276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Herman T, Clandinin TR, Lee R, and Zipursky SL (2001). N-cadherin regulates target specificity in the Drosophila visual system. Neuron 30, 437–450. [DOI] [PubMed] [Google Scholar]
- Little SC, Tikhonov M, and Gregor T (2013). Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell 154, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, and Lomvardas S (2013). An epigenetic trap stabilizes singular olfactory receptor expression. Cell 154, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, and Dubnau D (2007). Noise in gene expression determines cell fate in Bacillus subtilis. Science (80-. ). 317, 526–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. (2011). An epigenetic signature for monoallelic olfactory receptor expression. Cell 145, 555–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzoni EO, Celik A, Wernet MF, Vasiliauskas D, Johnston RJ, Cook TA, Pichaud F, and Desplan C (2008). Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol. 6, 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Serizawa S, Kimura HM, and Sakano H (2005). Continuous and overlapping expression domains of odorant receptor genes in the olfactory epithelium determine the dorsal/ventral positioning of glomeruli in the olfactory bulb. J. Neurosci. 25, 3586–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C, Jones K, Zuker C, and Rubin G (1987). A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J. Neurosci. 7, 1558–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugler A, Kittisopikul M, Hayden L, Liu J, Wiggins CH, Süel GM, and Walczak AM (2016). Noise Expands the Response Range of the Bacillus subtilis Competence Circuit. PLoS Comput. Biol 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nester EW, and Stocker BA (1963). Biosynthetic latency in early stages of deoxyribonucleic acidtransformation in bacillus subtilis. J. Bacteriol. 86, 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Etchberger JF, Posy SL, Frøkjær-Jensen C, Lockery S, Honig B, and Hobert O (2006). Searching for neuronal left/right asymmetry: Genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173, 131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Faumont S, Takayama J, Ahmed HK, Goldsmith AD, Pocock R, McCormick KE, Kunimoto H, Iino Y, Lockery S, et al. (2009). Lateralized Gustatory Behavior of C. elegans Is Controlled by Specific Receptor-Type Guanylyl Cyclases. Curr. Biol. 19, 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, and Tamkun JW (1998). The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 125, 3955–3966. [DOI] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, et al. (2004). Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat. Genet. 36, 288–292. [DOI] [PubMed] [Google Scholar]
- Paro R, and Hogness DS (1991). The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 88, 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova MD, Tkačik G, Bialek W, Wieschaus EF, and Gregor T (2019). Optimal Decoding of Cellular Identities in a Genetic Network. Cell 176, 844–855.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, and Bullock SL (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 111, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, and Benzer S (1976). Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53, 217–240. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, and Buck LB (1993). A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell 73, 597–609. [DOI] [PubMed] [Google Scholar]
- Rosin LF, Nguyen SC, and Joyce EF (2018). Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLoS Genet 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, and Paro R (2005). Intergenic transcription through a Polycomb group response element counteracts silencing. Genes Dev. 19, 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, and Pirrotta V (2006). Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38, 700–705. [DOI] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, and Eisenman RN (2007). The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21, 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, and O’connor M (1993). Elements of the Drosophila bithorax complex that mediate repression by polycomb group products. Dev. Biol. 158, 131–144. [DOI] [PubMed] [Google Scholar]
- Stevens NM (1908). A study of the germ cells of certain diptera, with reference to the heterochromosomes and the phenomena of synapsis. J. Exp. Zool. 5, 359–374. [Google Scholar]
- Stowers RS, and Schwarz TL (1999). A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152, 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strutt H, Cavalli G, and Paro R (1997). Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 16, 3621–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süel GM, Garcia-Ojalvo J, Liberman LM, and Elowitz MB (2006). An excitable gene regulatory circuit induces transient cellular differentiation. Nature 440, 545–550. [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Adamson MC, Ressler KJ, Kozak CA, and Buck LB (1996). The chromosomal distribution of mouse odorant receptor genes. Proc. Natl. Acad. Sci. U. S. A. 93, 884–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanawala SU, Rister J, Goldberg GW, Zuskov A, Olesnicky EC, Flowers JM, Jukam D, Purugganan MD, Gavis ER, Desplan C, et al. (2013). Regional Modulation of a Stochastically Expressed Factor Determines Photoreceptor Subtypes in the Drosophila Retina. Dev. Cell 25, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, and Ready DF (1987a). Cell fate in the Drosophila ommatidium. Dev. Biol. 123, 264–275. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, and Ready DF (1987b). Neuronal differentiation in the Drosophila ommatidium. Dev. Biol. 120, 366–376. [DOI] [PubMed] [Google Scholar]
- Treisman JE (2013). Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay K, Hamoen LW, Venema G, and Dubnau D (1997). Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis. Genes Dev. 11, 119–128. [DOI] [PubMed] [Google Scholar]
- Vassar R, Ngai J, and Axel R (1993). Spatial segregation of odorant receptor expression in the mammalian olfactory epithelium. Cell 74, 309–318. [DOI] [PubMed] [Google Scholar]
- Viets K, Sauria M, Chernoff C, Furlong E, Taylor J, and Johnston R (2019). Characterization of Button Loci that Promote Homologous Chromosome Pairing and Cell-Type-Specific Interchromosomal Gene Regulation. Dev. Cell 51, 341–356.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Mosher JT, and Crews ST (1998). Regulation of bHLH-PAS protein subcellular localization during Drosophila embryogenesis. Development 125, 1599–1608. [DOI] [PubMed] [Google Scholar]
- Weinberger LS, Dar RD, and Simpson ML (2008). Transient-mediated fate determination in a transcriptional circuit of HIV. Nat. Genet. 40, 466–470. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Mazzoni EO, Çelik A, Duncan DM, Duncan I, and Desplan C (2006). Stochastic spineless expression creates the retinal mosaic for colour vision. Nature 440, 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WJ K, CW S, TS F, KM R, TH P, AM Z, Kent D,H, Sugnet WJ, Furey CW, T.S., et al. (2002). The Human Genome Browser at UCSC. Genome Res. 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, and Ready DF (1991). The beginning of pattern formation in the Drosophila compound eye: The morphogenetic furrow and the second mitotic wave. Development 113, 841–850. [DOI] [PubMed] [Google Scholar]
- Xu Q, Schlabach MR, Hannon GJ, and Elledge SJ (2009). Design of 240,000 orthogonal 25mer DNA barcode probes. Proc. Natl. Acad. Sci. U. S. A. 106, 2289–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bahri S, Klein T, and Chia W (1997). Klumpfuss, a putative Drosophila zinc finger transcription factor, acts to differentiate between the identities of two secondary precursor cells within one neuroblast lineage. Genes Dev. 11, 1396–1408. [DOI] [PubMed] [Google Scholar]
- Zhang X, and Firestein S (2002). The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5, 124–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The script used to generate 19-bp bar-coding primers for Oligopaints probe design is available at https://github.com/kviets0913/Oligopaints-Primers-Custom-Script. The custom script used to analyze confocal images and determine the density of nascent RNA expression is available at https://zenodo.org/badge/latestdoi/504600646


