Abstract
IMPORTANCE
Clinical artificial intelligence (AI) algorithms have the potential to improve clinical care, but fair, generalizable algorithms depend on the clinical data on which they are trained and tested.
OBJECTIVE
To assess whether data sets used for training diagnostic AI algorithms addressing skin disease are adequately described and to identify potential sources of bias in these data sets.
DATA SOURCES
In this scoping review, PubMed was used to search for peer-reviewed research articles published between January 1, 2015, and November 1, 2020, with the following paired search terms: deep learning and dermatology, artificial intelligence and dermatology, deep learning and dermatologist, and artificial intelligence and dermatologist.
STUDY SELECTION
Studies that developed or tested an existing deep learning algorithm for triage, diagnosis, or monitoring using clinical or dermoscopic images of skin disease were selected, and the articles were independently reviewed by 2 investigators to verify that they met selection criteria.
CONSENSUS PROCESS
Data set audit criteria were determined by consensus of all authors after reviewing existing literature to highlight data set transparency and sources of bias.
RESULTS
A total of 70 unique studies were included. Among these studies, 1 065 291 images were used to develop or test AI algorithms, of which only 257 372 (24.2%) were publicly available. Only 14 studies (20.0%) included descriptions of patient ethnicity or race in at least 1 data set used. Only 7 studies (10.0%) included any information about skin tone in at least 1 data set used. Thirty-six of the 56 studies developing new AI algorithms for cutaneous malignant neoplasms (64.3%) met the gold standard criteria for disease labeling. Public data sets were cited more often than private data sets, suggesting that public data sets contribute more to new development and benchmarks.
CONCLUSIONS AND RELEVANCE
This scoping review identified 3 issues in data sets that are used to develop and test clinical AI algorithms for skin disease that should be addressed before clinical translation: (1) sparsity of data set characterization and lack of transparency, (2) nonstandard and unverified disease labels, and (3) inability to fully assess patient diversity used for algorithm development and testing.
Artificial intelligence (AI) algorithms have been widely applied to clinical tasks involving images, ranging from prediction of diagnoses using chest radiographs to assessment of skin lesions for malignant disease. Clinical deployment is the ultimate goal for developing AI algorithms in medicine. To be successful, AI algorithms need to be trained and tested on data that represent clinical scenarios encountered in real-world settings.1 The data that are used to train and test a model can determine its applicability and generalizability.1,2 Therefore, a clear understanding of data set characteristics (eg, how training data were collected, labeled, and processed) is critical.3 Moreover, a description of data demographics is key to understanding the generalizability of any data set to clinical practice.4 The machine learning community has suggested the importance of using standards for describing data sets, building from the idea of standardized data sheets used in the electronics industry, but such standards have not yet been implemented by scientific journals or regulatory authorities.3,4
For nonmedical deep learning applications, large data sets are publicly available and easily evaluated. However, medical data sets are often siloed owing to concerns about patient privacy.5 Because of the inaccessibility to the underlying data used to develop models, data set descriptions are integral to understanding the generalizability, or lack thereof, of a described model. Within the medical community, no broad standards exist at present that are universally applied for describing data sets used in developing AI models. Data set descriptions for specific applications often do not address potential sources of bias.6,7 Most US Food and Drug Administration–approved medical AI devices provide little public information on what data and demographic groups were used for testing.2 Guidelines such as Consolidated Standards of Reporting Trials–AI and Standard Protocol Items: Recommendations for Interventional Trials–AI have addressed transparency in AI clinical trials but do not cover initial AI development and early testing.7,8 Thus, there have been calls for increased transparency in medical AI.9 To better understand the characteristics of data sets used for developing AI algorithms in dermatology, we performed a scoping review that assessed data set availability and data set descriptions such as image labeling and patient diversity. We used this information to assess potential pitfalls in dermatology AI data sets.
Methods
We selected a scoping review approach to allow us to identify the data sets used in existing AI development, to assess data set transparency, and to extract the relevant features that may contribute to bias. We searched PubMed for peer-reviewed articles published between January 1, 2015, and November 1, 2020, that defined a clinically relevant task and developed a deep learning algorithm or that tested an existing deep learning algorithm using clinical or dermoscopic images of skin disease. We constrained the search to English language papers. We used the following pairs of search terms: deep learning and dermatology, artificial intelligence and dermatology, deep learning and dermatologist, and artificial intelligence and dermatologist. We excluded published reviews. The methods and results sections were thoroughly reviewed by one investigator (R.D.) to select studies that met criteria, while another investigator (M.P.S.) independently reviewed the selected abstracts to confirm that they met selection criteria.
Data set audit criteria were determined by consensus of all authors after reviewing existing literature to highlight data set transparency and sources of bias; extracted features are shown in the eTable in the Supplement.4,6 An audit was performed by 2 investigators (R.D. and M.P.S.), with each investigator independently reviewing the full study and supplemental data and reconciling any differences via consensus. Another investigator (M.D.S.) performed a final review to ensure the accuracy of the extracted data. For each data set, we analyzed the sources, the number of images, the number of patients involved, how the data set was used (for training, internal validation, testing, or external validation), whether the data set was clinical or dermoscopic, the diseases studied, how images were labeled and whether the labeling met the gold standard criteria (defined as pathological findings for malignant neoplasms [eMethods in the Supplement]), the descriptions of Fitzpatrick skin type and race/ethnicity, whether the data were publicly available or private, whether image processing was described, and whether the deep learning model was available (eTable in the Supplement).
Data set descriptions were based on how data were presented in the study. For example, if the original study distinguished between different data sets from separate sources in its analysis, then these data sets would also be separated in our audit. Multisource data sets that were presented as combined without indication of how many images came from each source in the original study were considered as a single data set for the audit. Images scraped from internet searches, rather than from defined atlases and repositories, were considered to be private data sets if the images were not shared with their applied labels. Synthetic images were not considered in the image count. To meet criteria for describing data set features in the eTable in the Supplement, the information needed to be included in the main text or supplement, even for data sets that are publicly available. Any discrepancies in reported images are noted in the eTable in the Supplement. The study was not registered with PROSPERO, and a review protocol was not published because such protocols do not apply to scoping reviews.10,11
Results
We identified 70 studies that met our criteria.12–81 Of these studies, 57 developed a new deep learning algorithm and 13 tested an existing algorithm on an additional data set (Table 1 and eTable in the Supplement). Most of the studies were published within the last 2 years (eTable in the Supplement). Among the 57 studies that trained and tested a new algorithm, only 14 had test data that were from a different source than the training set or an additional external validation. For studies training and testing a new deep learning algorithm, the mean (SD) total number of images was 23 222 (47 061) and the median was 10 015 (interquartile range, 2200–14 016), whereas the number of images used in studies testing an existing algorithm had a mean (SD) of 682 (894) and a median of 212 (interquartile range, 100–780).
Table 1.
Summary of Key Findings
| Description | No./total No. of studies (%) |
|---|---|
| Developed a novel algorithm | 57/70 (81.4) |
| With novel algorithms that tested on an external test set | 14/57 (24.6) |
| With tasks involving cutaneous malignant neoplasms | 56/70 (80.0) |
| Involved cutaneous malignant neoplasms and reported biopsy-proven labels for all cutaneous malignant neoplasms | 36/56 (64.3) |
| Reported any race or ethnicity information for ≥1 data set | 14/70 (20.0) |
| Reported any Fitzpatrick skin tone information for ≥1 data set | 7/70 (10.0) |
| Stated that the AI model developed or used was publicly accessible | 21/70 (30.0) |
Abbreviation: AI, artificial intelligence.
In terms of diseases, 56 studies included at least 1 cutaneous malignant neoplasm in their task (eTable in the Supplement). The gold standard for cutaneous malignant neoplasm is histopathological diagnosis.1 Of the 56 studies with a cutaneous malignant neoplasm diagnosis as part of the task, 36 (64.3%) met gold standard criteria, meaning that every cutaneous malignant neoplasm in the data set was confirmed by pathological findings (eTable in the Supplement). This suggests that a substantial number of studies were trained and/or tested on noisy annotations, which could affect diagnostic accuracy in clinical practice.
Among all studies, only 14 of 70 (20.0%) described any information on the ethnicity or race of patients in at least 1 data set and only 7 of 70 (10.0%) included any information on the Fitzpatrick skin types of images used in at least 1 data set (eTable in the Supplement). Studies that used public data sets with previously described demographic information (see Table 2) did not always include this information. Fitzpatrick skin typing represents the skin’s response to UV radiation, and dermatologists often use it as a proxy for skin color.82 Of the 7 studies with Fitzpatrick skin type information, 4 reported having no images of the darkest skin types (Fitzpatrick skin types V and VI) (eTable in the Supplement). We assessed the 3 studies12–14 that included Fitzpatrick skin types V and VI and found the information to be either incomplete or to underrepresent darker skin tones. Dulmage et al12 reported skin tone for just a subset of the test data and grouped Fitzpatrick skin types IV to VI images together, making it impossible to assess how many individual Fitzpatrick skin type V and VI images were included. Liu et al13 had 3.2% type V skin images and 0.3% type VI skin images in the training data and 2.7% type V skin images and 1 type VI skin image in the test data. Phillips et al14 tested an existing algorithm on an external test set that included 2.0% type V skin images and 0.8% type VI skin images. Data on the prevalence of Fitzpatrick skin types in the US population are inadequate. One study83 reported that the general US population includes 3.6% type V skin and 9% type VI skin, although this could be an underrepresentation, because the percentage of racial and ethnic minority group patients included in this study was lower than the reported demographics of the US population. Even where Fitzpatrick skin type was annotated and reported,84,85 the authors did not evaluate the algorithm in person, but rather reviewed post hoc images without information about lighting conditions, which may lead to inaccuracies and additional noise.
Table 2.
Public Data Sets Used in 3 or More Publications
| Data set source | No. of images | No. of patients | Type of image | Diseases | Label | Gold standard for malignant neoplasm diagnosis (pathological finding) | Fitzpatrick skin type description and breakdown | Ethnicity description and breakdown |
|---|---|---|---|---|---|---|---|---|
| ISIC 2016 Challenge | 1279 | Not specified | Dermoscopic | Nonmelanoma and melanoma | Benign (nonmelanoma): expert consensus; malignant (melanoma): pathological findings | Yes | No | No |
| ISIC 2017 Challenge | 2750 | Not specified | Dermoscopic | Benign nevi, seborrheic keratosis, and melanoma | Benign nevi: expert consensus; seborrheic keratosis: expert consensus; melanoma: pathological findings | Yes | No | No |
| ISIC 2018 Challenge (HAM10000) | 10 015 | Not specified | Dermoscopic | Actinic keratosis, intraepithelial carcinoma (Bowen disease), BCC, benign keratosis, dermatofibroma, melanocytic nevi, vascular skin lesions, and melanoma | Actinic keratosis: consensus; intraepithelial carcinoma: pathological findings; BCC: pathological findings; benign keratosis: consensus; dermatofibroma: consensus; melanocytic nevi: consensus; vascular skin lesions: consensus; melanoma: consensus | Yes | No | Yes; nationality breakdown (as a percentage of the 10 015 images in the data set): 2.0% Portuguese (PH2); 22.6% Australian (Rosendahl); Austrian (ViDIR) not specified (Atlas and ISIC 2017) |
| Hellenic Dermatological Atlas |
2663 (as of April 2021) | Not specified | Clinical | Various: 43 broad categories of disease | Not specified | Unable to assess | No | No |
| D@nderm Atlas of Clinical Dermatology | >3000 (as of April 2021) | Not specified | Clinical | Various: Common skin diseases under 9 broad categories | Not specified | Unable to assess | No | No |
| MED-NODE database | 170 | Not specified | Clinical | Melanoma and nevi | Nevi: pathological findings; melanoma: pathological findings | Yes | No | No |
| Edinburgh Dermofit Library | 1300 | Not specified | Clinical | Actinic keratosis, BCC, melanocytic nevus (mole), seborrheic keratosis, SCC, intraepithelial carcinoma, pyogenic granuloma, hemangioma, dermatofibroma, and malignant melanoma | Expert opinion (including dermatologists and dermatopathologists) based on clinical information and pathological findings | Yes | No | No |
| DermNet NZ | >20 000 | Not specified | Clinical | Various: 1000s of categories listed | Not specified | Unable to assess | No | No |
Abbreviations: BCC, basal cell carcinoma; ISIC, International Skin Imaging Collaboration; SCC, squamous cell carcinoma; ViDIR, Vienna Dermatologic Imaging Research.
To the extent that it was possible given the lack of clear labeling in many studies, we estimate that of a total 1 065 291 images, 257 372 (24.2%) were publicly available or available on request, whereas 807 919 (75.8%) were private. The accuracies of these estimates are affected by the fluctuation of available images in public databases and the inability to evaluate the presence of overlap among private databases. For example, some groups published multiple studies but did not indicate how much overlap there was between data sets from the same hospital system or academic center,15,16 whereas some groups did not even specify the source of their data.17 Studies that used publicly available data sets did not always describe what subsets of the data sets they used or reference previously described demographic information or labeling information.22,30,55 In some cases, multisource data sets were created with both private and public data but without indicating the number of images or diagnoses contributed by each source.18
Among the publicly available data sets, the International Skin Imaging Collaboration repository was used in the largest number of studies (n = 28), followed by MED-NODE: University Medical Center Groningen (n = 5), and Hellenic Dermatological Atlas (n = 4).86,87 The features of the publicly available data sets used by 3 or more studies are described in Table 2.19,20,86–92 Most of the AI literature addressing skin conditions is developed from private data sets used once to generate a single study, with no ability for replication (Figure), which greatly limits their effect and generalizability. Public data sets are drivers of model development and publication while allowing greater transparency (Figure). Across all the studies, models were generally not accessible, with only 21 of the 70 studies (30.0%) stating that the model was available on request, publicly, or through a commercial product (eTable in the Supplement).
Figure.
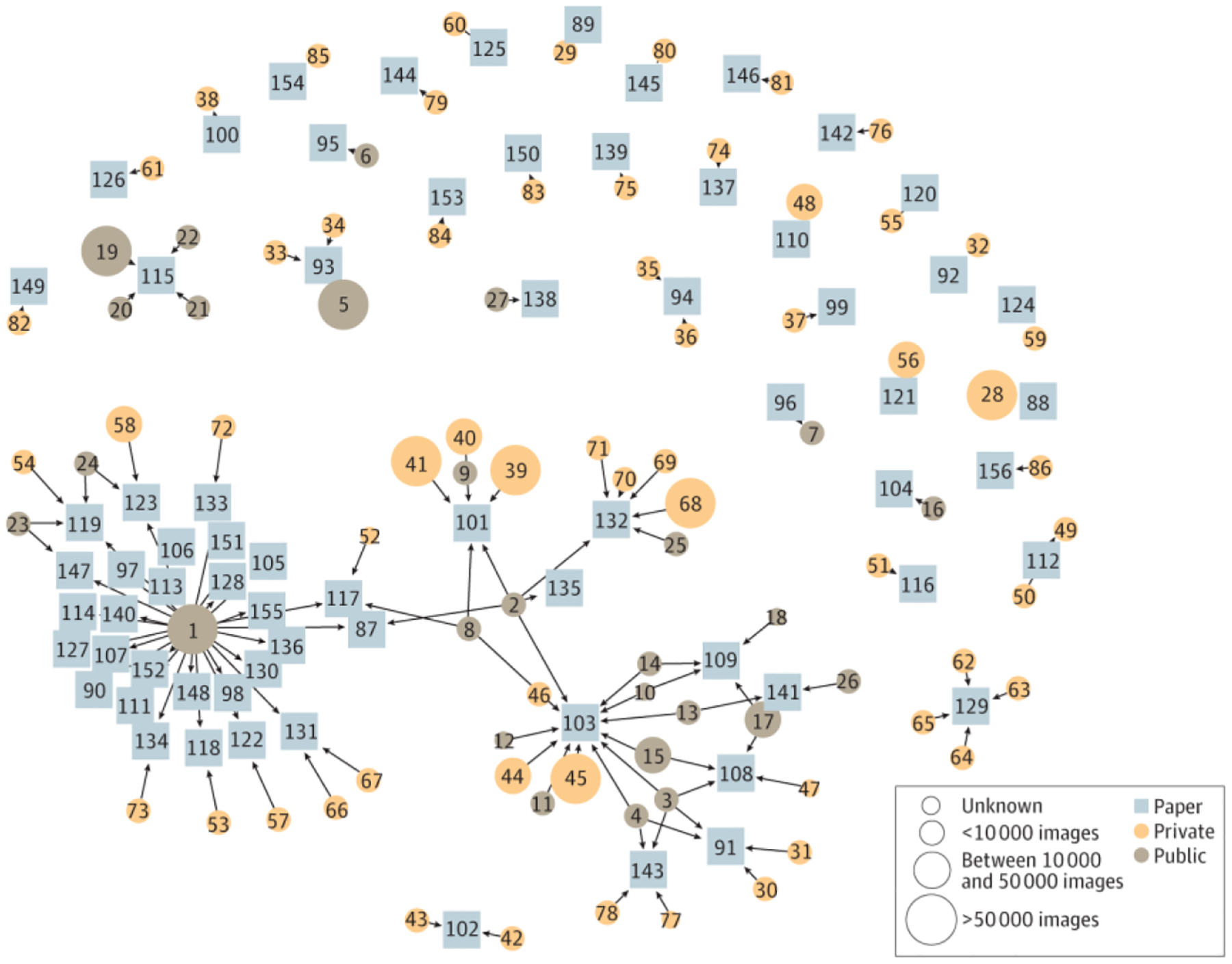
Overview of Data Sets and Studies. Squares represent studies; circles, data sets; and arrows, use of a data set. The number of images in a given data set is represented by the size of the circle. Private data sets are often only connected to 1 study, whereas public data sets help generate multiple studies. A mapping of the corresponding data sets and studies is provided in the eFigure in the Supplement.
Discussion
Developing clinically applicable deep learning models in medicine is predicated on the creation of robust models developed from data sets that are either publicly available for scrutiny or well described. Clinical AI must be held to a high standard owing to the potential for significant medical harm from errors and bias.2 Previous studies93 have shown that direct-to-consumer dermatology AI products have significant performance issues. Algorithmic performance depends significantly on the data on which it is trained and tested. Given the significant effort and cost that it requires to generate a data set for algorithmic development, we provide recommendations that should be implemented prospectively when data sets are being collated and algorithms are being developed. Using skin disease as a case study, we analyzed 70 studies related to AI and dermatology to understand the state of data set transparency and sources for potential bias.
We found that most data used for training and testing AI algorithms in dermatology were not publicly available. Most studies using private data did not describe important demographic information such as ethnicity/race or skin tone. Most developed models were likewise not available for additional evaluation to assess robustness. The lack of details and transparency about data and models limits our ability to systematically assess robustness and potential biases, which is an important direction of future work.
Based on our findings, we provide 3 recommendations for improving transparency and reducing the potential for bias in developed clinical algorithms. First, we recommend that whenever feasible, data sets and/or models be shared at time of publication. Even if the full model cannot be made public owing to intellectual property issues, developers should at least provide application programming interface access to the model to researchers using platforms such as gradio.94 If images are from public data sets or repositories, the images that were used should be clearly delineated. Images scraped from the internet via web search should be shared with their attached labels; these data cannot be considered publicly available if it is impossible to identify which images were used or how they were labeled. Owing to issues around patient privacy, medical data sets are not always readily available; however, repositories such as International Skin Imaging Collaboration have shown how deidentified data cannot only be shared but also catalyze algorithmic development (Figure). Direct access to data sets allows direct assessment of any potential biases. For example, data in International Skin Imaging Collaboration do not include skin tone information, but external groups have attempted to improve labeling for skin tone diversity through their own analyses.85
Second, if data sets cannot be shared, there should be a clear description of important data set characteristics, which include patient populations, skin tones represented, and the image labeling process.4 If available, other metrics that should be reported include data set sources (such as hospital or clinic), cameras used for photograph capture, data set processing steps, and information on photograph quality, because these have also been shown to affect AI performance.21 The aforementioned characteristics are not exhaustive; in fact, a challenge for this study was the lack of dermatology-specific guidelines for defining and describing AI data set characteristics. Through this scoping review, we found that few studies mention patient race or ethnicity race or patient skin tone. Previously, dermatologists have expressed concern about the potential of AI algorithms to perpetuate disparities by the exclusion of diverse skin tones,95 and this effect has been shown in an example where performance significantly differed between lesions on White vs Asian patients.16 However, assessing racial and ethnic diversity among patients in these data sets was limited by sparse reporting of these characteristics. The few studies that include skin tone information have underrepresented darker skin tones when compared with the general US population. Many AI algorithms aim to be used globally, and these algorithms should be trained and tested on data that represent global diversity.
Data label noise is another potential source of algorithm vulnerability. In the machine learning literature,96 mislabeling can lead to algorithms that are good predictors of the mislabeled data but poor predictors of the ground truth. For clinical AI, it is imperative that studies clearly delineate how labels were generated and whether these labels constitute the gold standard for diagnosing that disease. Multiple dermatology AI studies13,18 have relied on consensus panels for the diagnosis of cutaneous malignant neoplasms, such as melanoma. However, studies97 have shown that the number needed to biopsy among dermatologists to identify 1 melanoma is 7.5. This means that a significant portion of images labeled melanoma by a consensus panel without histopathological confirmation may not actually be melanoma.
Third, there should be a clear description of how data sets were used, that is, for training, validation, testing, or additional external validation. Running models on external data sets is an important step for demonstrating algorithmic robustness. Previous studies2,21 have demonstrated that significant performance drops can occur when algorithms trained exclusively at a single site are applied to an external site.
Limitations
Limitations of this study include the sparsity of reported data, which hampered our ability to fully assess some of the factors we were interested in assessing, such as skin tone and racial and ethnic diversity of the data sets. In addition, we focused on PubMed indexed articles; however, the AI literature has many preprints and conference proceedings that may not appear on PubMed. The studies featured in our scoping review are a representative sample of the AI in dermatology literature.
Conclusions
As of early 2021, no US Food and Drug Administration–approved AI devices have addressed cutaneous diseases, yet innovation is occurring at a rapid pace. This scoping review of the data sets used for developing AI algorithms for cutaneous disease reveals concerns about the lack of transparency and inadequate reporting of important data set characteristics, including racial and ethnic diversity among patients. At present, no reporting guidelines are available for AI data sets used to develop algorithms in dermatology. Specific dermatology AI development guidelines could improve data set transparency and the ability to assess bias in AI applications for dermatology.
Supplementary Material
Key Points.
Question
How transparent are the data sets used to develop artificial intelligence (AI) algorithms in dermatology, and what potential pitfalls exist in the data?
Findings
In this scoping review of 70 studies addressing the intersection of dermatology and AI that were published between January 1, 2015, and November 1, 2020, most data set descriptions were inadequate for analysis and replication, disease labels did not meet the gold standard, and information on patient skin tone and race or ethnicity was often not reported. In addition, most data sets and models have not been shared publicly.
Meaning
These findings suggest that the applicability and generalizability of AI algorithms rely on high-quality training and testing data sets; the sparsity of data set descriptions, lack of data set and model transparency, inconsistency in disease labels, and lack of reporting on patient diversity present concerns for the clinical translation of these algorithms.
Funding/Support:
This study was supported by grant T32 5T32AR007422-38 from the National Institutes of Health (NIH) (Dr Daneshjou); the Melanoma Research Alliance (Drs Rotemberg and Smith); Cancer Center Support grant P30 CA008748 from the NIH/National Cancer Institute (Drs Rotemberg and Smith and Ms Sun); the Charina Fund (Dr Rotemberg and Ms Sun); Faculty Early Career Development grant 1942926 from the National Science Foundation (Dr Zou); grant U01MH098953 from the NIH (Dr Zou); and grants from the Silicon Valley Foundation and the Chan-Zuckerberg Initiative (Dr Zou).
Role of the Funder/Sponsor:
The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Conflict of Interest Disclosures:
Dr Daneshjou reports consulting fees from VisualDx and Pfizer Inc and grant support from UCB outside the submitted work. Dr Rotemberg reported non-financial support from Inhabit Brands outside the submitted work. No other disclosures were reported.
REFERENCES
- 1.Daneshjou R, He B, Ouyang D, Zou JY. How to evaluate deep learning for cancer diagnostics—factors and recommendations. Biochim Biophys Acta Rev Cancer. 2021;1875(2): 188515. doi: 10.1016/j.bbcan.2021.188515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu E, Wu K, Daneshjou R, Ouyang D, Ho DE, Zou J. How medical AI devices are evaluated: limitations and recommendations from an analysis of FDA approvals. Nat Med. 2021;27(4):582–584. doi: 10.1038/s41591-021-01312-x [DOI] [PubMed] [Google Scholar]
- 3.Holland S, Hosny A, Newman S, Joseph J, Chmielinski K. The dataset nutrition label: a framework to drive higher data quality standards. arXiv. Preprint posted online May 9, 2018. 1805.03677. [Google Scholar]
- 4.Gebru T, Morgenstern J, Vecchione B, et al. Datasheets for datasets. arXiv. Preprint posted online March 19, 2020. 1803.09010. [Google Scholar]
- 5.Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L. ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. IEEE; 2009:248–255. [Google Scholar]
- 6.Bluemke DA, Moy L, Bredella MA, et al. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers—from the Radiology editorial board. Radiology. 2020;294(3):487–489. doi: 10.1148/radiol.2019192515 [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Cruz Rivera S, Moher D, Calvert MJ, Denniston AK; SPIRIT-AI and CONSORT-AI Working Group. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. Nat Med. 2020;26(9): 1364–1374. doi: 10.1038/s41591-020-1034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor M, Liu X, Denniston A, et al. ; SPIRIT-AI and CONSORT-AI Working Group. Raising the bar for randomized trials involving artificial intelligence: the SPIRIT-Artificial Intelligence and CONSORT-Artificial Intelligence Guidelines. J Invest Dermatol. Published online March 22, 2021. doi: 10.1016/j.jid.2021.02.744 [DOI] [PubMed] [Google Scholar]
- 9.Watson DS, Krutzinna J, Bruce IN, et al. Clinical applications of machine learning algorithms: beyond the black box. BMJ. 2019;364:l886. doi: 10.1136/bmj.l886 [DOI] [PubMed] [Google Scholar]
- 10.Colquhoun HL, Levac D, O’Brien KK, et al. Scoping reviews: time for clarity in definition, methods, and reporting. J Clin Epidemiol. 2014;67 (12):1291–1294. doi: 10.1016/j.jclinepi.2014.03.013 [DOI] [PubMed] [Google Scholar]
- 11.Winters N, Langer L, Geniets A. Scoping review assessing the evidence used to support the adoption of mobile health (mHealth) technologies for the education and training of community health workers (CHWs) in low-income and middle-income countries. BMJ Open. 2018;8(7):e019827. doi: 10.1136/bmjopen-2017-019827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dulmage B, Tegtmeyer K, Zhang MZ, Colavincenzo M, Xu S. A point-of-care, real-time artificial intelligence system to support clinician diagnosis of a wide range of skin diseases. J Invest Dermatol. 2021;141(5):1230–1235. doi: 10.1016/j.jid.2020.08.027 [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Jain A, Eng C, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med. 2020;26(6):900–908. doi: 10.1038/s41591-020-0842-3 [DOI] [PubMed] [Google Scholar]
- 14.Phillips M, Marsden H, Jaffe W, et al. Assessment of accuracy of an artificial intelligence algorithm to detect melanoma in images of skin lesions. JAMA Netw Open. 2019;2(10):e1913436. doi: 10.1001/jamanetworkopen.2019.13436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han SS, Park I, Eun Chang S, et al. Augmented intelligence dermatology: deep neural networks empower medical professionals in diagnosing skin cancer and predicting treatment options for 134 skin disorders. J Invest Dermatol. 2020;140(9): 1753–1761. doi: 10.1016/j.jid.2020.01.019 [DOI] [PubMed] [Google Scholar]
- 16.Han SS, Kim MS, Lim W, Park GH, Park I, Chang SE. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol. 2018; 138(7):1529–1538. doi: 10.1016/j.jid.2018.01.028 [DOI] [PubMed] [Google Scholar]
- 17.Yap J, Yolland W, Tschandl P. Multimodal skin lesion classification using deep learning. Exp Dermatol. 2018;27(11):1261–1267. doi: 10.1111/exd.13777 [DOI] [PubMed] [Google Scholar]
- 18.Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542 (7639):115–118. doi: 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchetti MA, Codella NCF, Dusza SW, et al. Results of the 2016 International Skin Imaging Collaboration International Symposium on Biomedical Imaging challenge: comparison of the accuracy of computer algorithms to dermatologists for the diagnosis of melanoma from dermoscopic images. J Am Acad Dermatol. 2018;78(2): 270–277.e1. doi: 10.1016/j.jaad.2017.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetti MA, Liopyris K, Dusza SW, et al. ; International Skin Imaging Collaboration. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: results of the International Skin Imaging Collaboration 2017. J Am Acad Dermatol. 2020;82 (3):622–627. doi: 10.1016/j.jaad.2019.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tschandl P, Codella N, Akay BN, et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol. 2019; 20(7):938–947. doi: 10.1016/S1470-2045(19)30333-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brinker TJ, Hekler A, Enk AH, et al. ; Collaborators. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer. 2019;111: 148–154. doi: 10.1016/j.ejca.2019.02.005 [DOI] [PubMed] [Google Scholar]
- 23.Wu H, Yin H, Chen H, et al. A deep learning, image based approach for automated diagnosis for inflammatory skin diseases. Ann Transl Med. 2020;8(9):581. doi: 10.21037/atm.2020.04.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin Z, Liu Z, Zhu P, Xue Y. A GAN-based image synthesis method for skin lesion classification. Comput Methods Programs Biomed. 2020;195: 105568. doi: 10.1016/j.cmpb.2020.105568 [DOI] [PubMed] [Google Scholar]
- 25.Pangti R, Mathur J, Chouhan V, et al. A machine learning-based, decision support, mobile phone application for diagnosis of common dermatological diseases. J Eur Acad Dermatol Venereol. 2021;35(2): 536–545. doi: 10.1111/jdv.16967 [DOI] [PubMed] [Google Scholar]
- 26.Chin YPH, Hou ZY, Lee MY, et al. A patient-oriented, general-practitioner-level, deep-learning-based cutaneous pigmented lesion risk classifier on a smartphone. Br J Dermatol. 2020;182(6):1498–1500. doi: 10.1111/bjd.18859 [DOI] [PubMed] [Google Scholar]
- 27.Tschandl P, Kittler H, Argenziano G. A pretrained neural network shows similar diagnostic accuracy to medical students in categorizing dermatoscopic images after comparable training conditions. Br J Dermatol. 2017;177(3):867–869. doi: 10.1111/bjd.15695 [DOI] [PubMed] [Google Scholar]
- 28.Blanco G, Traina AJM, Traina C Jr, et al. A superpixel-driven deep learning approach for the analysis of dermatological wounds. Comput Methods Programs Biomed. 2020;183:105079. doi: 10.1016/j.cmpb.2019.105079 [DOI] [PubMed] [Google Scholar]
- 29.Yu C, Yang S, Kim W, et al. Acral melanoma detection using a convolutional neural network for dermoscopy images. PLoS One. 2018;13(3):e0193321. doi: 10.1371/journal.pone.0193321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maron RC, Utikal JS, Hekler A, et al. Artificial intelligence and its effect on dermatologists’ accuracy in dermoscopic melanoma image classification: web-based survey study. J Med Internet Res. 2020;22(9):e18091. doi: 10.2196/18091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui X, Wei R, Gong L, et al. Assessing the effectiveness of artificial intelligence methods for melanoma: A retrospective review. J Am Acad Dermatol. 2019;81(5):1176–1180. doi: 10.1016/j.jaad.2019.06.042 [DOI] [PubMed] [Google Scholar]
- 32.Winkler JK, Fink C, Toberer F, et al. Association between surgical skin markings in dermoscopic images and diagnostic performance of a deep learning convolutional neural network for melanoma recognition. JAMA Dermatol. 2019;155 (10):1135–1141. doi: 10.1001/jamadermatol.2019.1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burlina PM, Joshi NJ, Ng E, Billings SD, Rebman AW, Aucott JN. Automated detection of erythema migrans and other confounding skin lesions via deep learning. Comput Biol Med. 2019; 105:151–156. doi: 10.1016/j.compbiomed.2018.12.007 [DOI] [PubMed] [Google Scholar]
- 34.Lee S, Lee JW, Choe SJ, et al. Clinically applicable deep learning framework for measurement of the extent of hair loss in patients with alopecia areata. JAMA Dermatol. 2020;156(9): 1018–1020. doi: 10.1001/jamadermatol.2020.2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du-Harpur X, Arthurs C, Ganier C, et al. Clinically relevant vulnerabilities of deep machine learning systems for skin cancer diagnosis. J Invest Dermatol. 2021;141(4):916–920. doi: 10.1016/j.jid.2020.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aggarwal SLP. Data augmentation in dermatology image recognition using machine learning. Skin Res Technol. 2019;25(6):815–820. doi: 10.1111/srt.12726 [DOI] [PubMed] [Google Scholar]
- 37.Goceri E Deep learning based classification of facial dermatological disorders. Comput Biol Med. 2021;128:104118. doi: 10.1016/j.compbiomed.2020.104118 [DOI] [PubMed] [Google Scholar]
- 38.Thomsen K, Christensen AL, Iversen L, Lomholt HB, Winther O. Deep learning for diagnostic binary classification of multiple-lesion skin diseases. Front Med (Lausanne). 2020;7:574329. doi: 10.3389/fmed.2020.574329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brinker TJ, Hekler A, Enk AH, et al. ; Collaborators. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur J Cancer. 2019;113:47–54. doi: 10.1016/j.ejca.2019.04.001 [DOI] [PubMed] [Google Scholar]
- 40.Wang SQ, Zhang XY, Liu J, et al. Deep learning-based, computer-aided classifier developed with dermoscopic images shows comparable performance to 164 dermatologists in cutaneous disease diagnosis in the Chinese population. Chin Med J (Engl). 2020;133(17): 2027–2036. doi: 10.1097/CM9.0000000000001023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucius M, De All J, De All JA, et al. Deep neural frameworks improve the accuracy of general practitioners in the classification of pigmented skin lesions. Diagnostics (Basel). 2020;10(11):969. doi: 10.3390/diagnostics10110969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brinker TJ, Hekler A, Enk AH, et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur J Cancer. 2019; 119:11–17. doi: 10.1016/j.ejca.2019.05.023 [DOI] [PubMed] [Google Scholar]
- 43.Han SS, Park GH, Lim W, et al. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS One. 2018;13(1):e0191493. doi: 10.1371/journal.pone.0191493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujisawa Y, Otomo Y, Ogata Y, et al. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br J Dermatol. 2019;180(2): 373–381. doi: 10.1111/bjd.16924 [DOI] [PubMed] [Google Scholar]
- 45.Minagawa A, Koga H, Sano T, et al. Dermoscopic diagnostic performance of Japanese dermatologists for skin tumors differs by patient origin: A deep learning convolutional neural network closes the gap. J Dermatol. 2021;48(2): 232–236. doi: 10.1111/1346-8138.15640 [DOI] [PubMed] [Google Scholar]
- 46.Phillips M, Greenhalgh J, Marsden H, Palamaras I. Detection of malignant melanoma using artificial intelligence: an observational study of diagnostic accuracy. Dermatol Pract Concept. 2019;10(1):e2020011. doi: 10.5826/dpc.1001a11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seité S, Khammari A, Benzaquen M, Moyal D, Dréno B. Development and accuracy of an artificial intelligence algorithm for acne grading from smartphone photographs. Exp Dermatol. 2019;28 (11):1252–1257. doi: 10.1111/exd.14022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Y, Ge Y, Guo L, et al. Development and validation of two artificial intelligence models for diagnosing benign, pigmented facial skin lesions. Skin Res Technol. 2021;27(1):74–79. doi: 10.1111/srt.12911 [DOI] [PubMed] [Google Scholar]
- 49.Huang HW, Hsu BW, Lee CH, Tseng VS. Development of a light-weight deep learning model for cloud applications and remote diagnosis of skin cancers. J Dermatol. 2021;48(3):310–316. doi: 10.1111/1346-8138.15683 [DOI] [PubMed] [Google Scholar]
- 50.Tschandl P, Argenziano G, Razmara M, Yap J. Diagnostic accuracy of content-based dermatoscopic image retrieval with deep classification features. Br J Dermatol. 2019;181(1): 155–165. doi: 10.1111/bjd.17189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li CX, Fei WM, Shen CB, et al. Diagnostic capacity of skin tumor artificial intelligence-assisted decision-making software in real-world clinical settings. Chin Med J (Engl). 2020;133(17):2020–2026. doi: 10.1097/CM9.0000000000001002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fink C, Blum A, Buhl T, et al. Diagnostic performance of a deep learning convolutional neural network in the differentiation of combined naevi and melanomas. J Eur Acad Dermatol Venereol. 2020;34(6):1355–1361. doi: 10.1111/jdv.16165 [DOI] [PubMed] [Google Scholar]
- 53.Fink C, Jaeger C, Jaeger K, Haenssle HA. Diagnostic performance of the MelaFind device in a real-life clinical setting. J Dtsch Dermatol Ges. 2017; 15(4):414–419. doi: 10.1111/ddg.13220 [DOI] [PubMed] [Google Scholar]
- 54.Hekler A, Kather JN, Krieghoff-Henning E, et al. Effects of label noise on deep learning-based skin cancer classification. Front Med (Lausanne). 2020; 7:177. doi: 10.3389/fmed.2020.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brinker TJ, Hekler A, Enk AH, von Kalle C. Enhanced classifier training to improve precision of a convolutional neural network to identify images of skin lesions. PLoS One. 2019;14(6):e0218713. doi: 10.1371/journal.pone.0218713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tschandl P, Rosendahl C, Akay BN, et al. Expert-level diagnosis of nonpigmented skin cancer by combined convolutional neural networks. JAMA Dermatol. 2019;155(1):58–65. doi: 10.1001/jamadermatol.2018.4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X, Wu J, Chen EZ, Jiang H. From deep learning towards finding skin lesion biomarkers. Annu Int Conf IEEE Eng Med Biol Soc. 2019;2019: 2797–2800. doi: 10.1109/EMBC.2019.8857334 [DOI] [PubMed] [Google Scholar]
- 58.Tschandl P, Rinner C, Apalla Z, et al. Human-computer collaboration for skin cancer recognition. Nat Med. 2020;26(8):1229–1234. doi: 10.1038/s41591-020-0942-0 [DOI] [PubMed] [Google Scholar]
- 59.Han SS, Moon IJ, Lim W, et al. Keratinocytic skin cancer detection on the face using region-based convolutional neural network. JAMA Dermatol. 2020;156(1):29–37. doi: 10.1001/jamadermatol.2019.3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haenssle HA, Fink C, Toberer F, et al. ; Reader Study Level I and Level II Groups. Man against machine reloaded: performance of a market-approved convolutional neural network in classifying a broad spectrum of skin lesions in comparison with 96 dermatologists working under less artificial conditions. Ann Oncol. 2020;31(1): 137–143. doi: 10.1016/j.annonc.2019.10.013 [DOI] [PubMed] [Google Scholar]
- 61.Haenssle HA, Fink C, Schneiderbauer R, et al. ; Reader study level-I and level-II Groups. Man against machine: diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann Oncol. 2018;29(8): 1836–1842. doi: 10.1093/annonc/mdy166 [DOI] [PubMed] [Google Scholar]
- 62.Nasr-Esfahani E, Samavi S, Karimi N, et al. Melanoma detection by analysis of clinical images using convolutional neural network. Annu Int Conf IEEE Eng Med Biol Soc. 2016;2016:1373–1376. doi: 10.1109/EMBC.2016.7590963 [DOI] [PubMed] [Google Scholar]
- 63.Zunair H, Ben Hamza A. Melanoma detection using adversarial training and deep transfer learning. Phys Med Biol. 2020;65(13):135005. doi: 10.1088/1361-6560/ab86d3 [DOI] [PubMed] [Google Scholar]
- 64.Winkler JK, Sies K, Fink C, et al. Melanoma recognition by a deep learning convolutional neural network-Performance in different melanoma subtypes and localisations. Eur J Cancer. 2020;127: 21–29. doi: 10.1016/j.ejca.2019.11.020 [DOI] [PubMed] [Google Scholar]
- 65.Navarrete-Dechent C, Liopyris K, Marchetti MA. Multiclass artificial intelligence in dermatology: progress but still room for improvement. J Invest Dermatol. 2021; 141(5):1325–1328. doi: 10.1016/j.jid.2020.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Masni MA, Kim DH, Kim TS. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput Methods Programs Biomed. 2020;190:105351. doi: 10.1016/j.cmpb.2020.105351 [DOI] [PubMed] [Google Scholar]
- 67.Premaladha J, Ravichandran KS. Novel approaches for diagnosing melanoma skin lesions through supervised and deep learning algorithms. J Med Syst. 2016;40(4):96. doi: 10.1007/s10916-016-0460-2 [DOI] [PubMed] [Google Scholar]
- 68.Sies K, Winkler JK, Fink C, et al. Past and present of computer-assisted dermoscopic diagnosis: performance of a conventional image analyser versus a convolutional neural network in a prospective data set of 1,981 skin lesions. Eur J Cancer. 2020;135:39–46. doi: 10.1016/j.ejca.2020.04.043 [DOI] [PubMed] [Google Scholar]
- 69.Pangti R, Chouhan V, Mathur J, et al. Performance of a deep learning-based application for the diagnosis of basal cell carcinoma in Indian patients as compared to dermatologists and nondermatologists. Int J Dermatol. 2021;60(2): e51–e52. doi: 10.1111/ijd.15242 [DOI] [PubMed] [Google Scholar]
- 70.Muñoz-López C, Ramírez-Cornejo C, Marchetti MA, et al. Performance of a deep neural network in teledermatology: a single-centre prospective diagnostic study. J Eur Acad Dermatol Venereol. 2021;35(2):546–553. doi: 10.1111/jdv.16979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim YJ, Han SS, Yang HJ, Chang SE. Prospective, comparative evaluation of a deep neural network and dermoscopy in the diagnosis of onychomycosis. PLoS One. 2020;15(6):e0234334. doi: 10.1371/journal.pone.0234334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Ke Z, He Z, et al. Real-time burn depth assessment using artificial networks: a large-scale, multicentre study. Burns. 2020;46(8):1829–1838. doi: 10.1016/j.burns.2020.07.010 [DOI] [PubMed] [Google Scholar]
- 73.Saba T, Khan MA, Rehman A, Marie-Sainte SL. Region extraction and classification of skin cancer: a heterogeneous framework of deep CNN features fusion and reduction. J Med Syst. 2019;43(9):289. doi: 10.1007/s10916-019-1413-3 [DOI] [PubMed] [Google Scholar]
- 74.Binol H, Plotner A, Sopkovich J, Kaffenberger B, Niazi MKK, Gurcan MN. Ros-NET: A deep convolutional neural network for automatic identification of rosacea lesions. Skin Res Technol. 2020;26(3):413–421. doi: 10.1111/srt.12817 [DOI] [PubMed] [Google Scholar]
- 75.Zhao S, Xie B, Li Y, et al. Smart identification of psoriasis by images using convolutional neural networks: a case study in China. J Eur Acad Dermatol Venereol. 2020;34(3):518–524. doi: 10.1111/jdv.15965 [DOI] [PubMed] [Google Scholar]
- 76.Hekler A, Utikal JS, Enk AH, et al. ; Collaborators. Superior skin cancer classification by the combination of human and artificial intelligence. Eur J Cancer. 2019;120:114–121. doi: 10.1016/j.ejca.2019.07.019 [DOI] [PubMed] [Google Scholar]
- 77.Maron RC, Weichenthal M, Utikal JS, et al. ; Collabrators. Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks. Eur J Cancer. 2019;119:57–65. doi: 10.1016/j.ejca.2019.06.013 [DOI] [PubMed] [Google Scholar]
- 78.Zhao XY, Wu X, Li FF, et al. The application of deep learning in the risk grading of skin tumors for patients using clinical images. J Med Syst. 2019;43 (8):283. doi: 10.1007/s10916-019-1414-2 [DOI] [PubMed] [Google Scholar]
- 79.Jinnai S, Yamazaki N, Hirano Y, Sugawara Y, Ohe Y, Hamamoto R. The development of a skin cancer classification system for pigmented skin lesions using deep learning. Biomolecules. 2020;10 (8):1123. doi: 10.3390/biom10081123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mahbod A, Tschandl P, Langs G, Ecker R, Ellinger I. The effects of skin lesion segmentation on the performance of dermatoscopic image classification. Comput Methods Programs Biomed. 2020;197:105725. doi: 10.1016/j.cmpb.2020.105725 [DOI] [PubMed] [Google Scholar]
- 81.Zhang X, Wang S, Liu J, Tao C. Towards improving diagnosis of skin diseases by combining deep neural network and human knowledge. BMC Med Inform Decis Mak. 2018;18(suppl 2):59. doi: 10.1186/s12911-018-0631-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gupta V, Sharma VK. Skin typing: Fitzpatrick grading and others. Clin Dermatol. 2019;37(5): 430–436. doi: 10.1016/j.clindermatol.2019.07.010 [DOI] [PubMed] [Google Scholar]
- 83.Keiser E, Linos E, Kanzler M, Lee W, Sainani KL, Tang JY. Reliability and prevalence of digital image skin types in the United States: results from National Health and Nutrition Examination Survey 2003–2004. J Am Acad Dermatol. 2012;66(1): 163–165. doi: 10.1016/j.jaad.2011.02.044 [DOI] [PubMed] [Google Scholar]
- 84.Lester JC, Clark L Jr, Linos E, Daneshjou R. Clinical photography in skin of colour: tips and best practices. Br J Dermatol. 2021;184(6):1177–1179. doi: 10.1111/bjd.19811 [DOI] [PubMed] [Google Scholar]
- 85.Kinyanjui NM, Odonga T, Cintas C, et al. Estimating skin tone and effects on classification performance in dermatology datasets. Paper presented at: NeurIPS 2019 Workshop on Fair ML for Health; Vancouver, Canada; 2019. [Google Scholar]
- 86.Giotis I, Molders N, Land S, Biehl M, Jonkman MF, Petkov N. MED-NODE: a computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Sys Applications. 2015;42(19):6578–6585. doi: 10.1016/j.eswa.2015.04.034 [DOI] [Google Scholar]
- 87.Verros CD. Hellenic Dermatological Atlas. 2011. Accessed April 17, 2021. http://www.hellenicdermatlas.com/en/
- 88.Tschandl P, Rosendahl C, Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. 2018;5:180161. doi: 10.1038/sdata.2018.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Veien NK, Nielsen M. An Atlas of Clinical Dermatology. Accessed April 20, 2021. http://www.danderm-pdv.is.kkh.dk/atlas/index.html
- 90.Ballerini L, Fisher R, Aldridge R, Rees J. A color and texture based hierarchical K-NN approach to the classification of nonmelanoma skin lesions. In: Celebi ME, Schaefer G, eds. Color Medical Image Analysis. Springer; 2013:63–86. doi: 10.1007/978-94-007-5389-1_4 [DOI] [Google Scholar]
- 91.New Zealand Dermatological Society. Dermnet NZ. Updated 2020. Accessed April 17, 2021. https://dermnetnz.org/about-us/
- 92.Codella N, Rotemberg V, Tschandl P, et al. Skin lesion analysis toward melanoma detection 2018: a challenge hosted by the International Skin Imaging Collaboration (ISIC). arXiv. Preprint posted online March 29, 2019. 1902.03368 [Google Scholar]
- 93.Freeman K, Dinnes J, Chuchu N, et al. Algorithm based smartphone apps to assess risk of skin cancer in adults: systematic review of diagnostic accuracy studies. BMJ. 2020;368:m127. doi: 10.1136/bmj.m127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abid A, Abdalla A, Abid A, Khan D, Alfozan A, Zou J. An online platform for interactive feedback in biomedical machine learning. Nat Machine Intelligence. 2020;2(2): 86–88. doi: 10.1038/s42256-020-0147-8 [DOI] [Google Scholar]
- 95.Adamson AS, Smith A. Machine learning and health care disparities in dermatology. JAMA Dermatol. 2018;154(11):1247–1248. doi: 10.1001/jamadermatol.2018.2348 [DOI] [PubMed] [Google Scholar]
- 96.Yun S, Oh SJ, Byeongho H, Han D, Choe J, Chun S. Re-labeling ImageNet: from single to multi-labels, from global to localized labels. arXiv. Preprint posted online July 22, 2021. 2101.05022 [Google Scholar]
- 97.Nelson KC, Swetter SM, Saboda K, Chen SC, Curiel-Lewandrowski C. Evaluation of the number-needed-to-biopsy metric for the diagnosis of cutaneous melanoma: a systematic review and meta-analysis. JAMA Dermatol. 2019;155(10): 1167–1174. doi: 10.1001/jamadermatol.2019.1514 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


