Abstract
Objectives
An interferon (IFN) gene signature (IGS) is present in approximately 50% of early, treatment naive rheumatoid arthritis (eRA) patients where it has been shown to negatively impact initial response to treatment. We wished to validate this effect and explore potential mechanisms of action.
Methods
In a multicentre inception cohort of eRA patients (n=191), we examined the whole blood IGS (MxA, IFI44L, OAS1, IFI6, ISG15) with reference to circulating IFN proteins, clinical outcomes and epigenetic influences on circulating CD19+ B and CD4+ T lymphocytes.
Results
We reproduced our previous findings demonstrating a raised baseline IGS. We additionally showed, for the first time, that the IGS in eRA reflects circulating IFN-α protein. Paired longitudinal analysis demonstrated a significant reduction between baseline and 6-month IGS and IFN-α levels (p<0.0001 for both). Despite this fall, a raised baseline IGS predicted worse 6-month clinical outcomes such as increased disease activity score (DAS-28, p=0.025) and lower likelihood of a good EULAR clinical response (p=0.034), which was independent of other conventional predictors of disease activity and clinical response. Molecular analysis of CD4+ T cells and CD19+ B cells demonstrated differentially methylated CPG sites and dysregulated expression of disease relevant genes, including PARP9, STAT1, and EPSTI1, associated with baseline IGS/IFNα levels. Differentially methylated CPG sites implicated altered transcription factor binding in B cells (GATA3, ETSI, NFATC2, EZH2) and T cells (p300, HIF1α).
Conclusions
Our data suggest that, in eRA, IFN-α can cause a sustained, epigenetically mediated, pathogenic increase in lymphocyte activation and proliferation, and that the IGS is, therefore, a robust prognostic biomarker. Its persistent harmful effects provide a rationale for the initial therapeutic targeting of IFN-α in selected patients with eRA.
Keywords: inflammation; arthritis, rheumatoid; antirheumatic agents; immune system diseases
What is already known about this topic
Type I interferons (IFNs) and the IFN gene signature (IGS) have received less attention in rheumatoid arthritis (RA) than in other rheumatic diseases such as systemic lupus erythematous.
Nonetheless, emerging evidence hints at a potentially important role for the IGS in early disease although, until now, it was unknown which IFN class was responsible.
What does this study add?
We demonstrate that IFN-alpha levels are transiently elevated in some early RA patients and are responsible for generating the IGS.
We validate the IGS as a robust prognostic biomarker associated with poor 6 month outcomes.
We also implicate IFN-α/IGS in epigenetic modification of circulating B and T lymphocytes, at genes associated with activation and proliferation, providing a potential mechanism for its persistent harmful effects.
How might this impact on clinical practice
Our data provide a strong rationale for the use of therapies that target the IFN-α pathway and the IGS in selected early RA patients.
Our work also has implications for other conditions with high IFN-α levels, such as COVID-19, and the potential for persistent harmful sequelae.
Introduction
An interferon gene signatures (IGS) has been reported in multiple autoimmune conditions, including rheumatoid arthritis (RA).1 It is a composite score of interferon response genes (IRGs) that are classically upregulated in response to type 1 interferons (IFN-I). IFN-I are released on detection of viral/bacterial genetic material by various nucleic acid receptors (NARs),1 2 but the pathway by which IFN production is triggered in RA is unknown. Furthermore, there is an overlap between downstream signalling pathways for all interferon classes with upregulation of common IRGs.3 Historically, the direct measurement of IFN-I has been challenging,3 creating uncertainty around which IFN class drives the IGS in RA thereby limiting understanding of its pathophysiological relevance.
To date, no association has been reported between the IGS and disease activity in established RA. However, longstanding patients with RA are frequently prescribed additional therapies, which modulate the IGS.4 5 By contrast, we previously demonstrated in early RA (eRA) patients (naïve for disease-modifying antirheumatic drugs (DMARDs) and glucocorticoids), the IGS positively associates with baseline disease activity and, independent of conventional markers of disease activity, associates with worse clinical outcomes at 6 months.6 The pathophysiological processes in eRA are distinct from those of established disease and the IGS is more prominent in eRA than in established RA.6 The role of epigenetics in modifying phenotype is increasingly appreciated in autoimmunity7 and RA has an early window of therapeutic opportunity. Thus, understanding and predicting heterogeneity to therapeutic response is important for early precision therapeutics.
In a large multicentre eRA cohort, we sought to: (1) confirm the IGS negatively impacts disease outcomes, (2) clarify which IFN classes are responsible for IGS generation, (3) seek evidence that IFN-α exposure contributes to a harmful epigenetic footprint at disease onset, potentially explaining its negative effect on longer-term outcomes.
Methods
Patient cohorts
DMARD and glucocorticoid naive patients with eRA were recruited from the Newcastle Early Arthritis Clinic (NEAC) as described previously.6 8 Data and samples relating to the ‘Towards A CurE for RA’ (TACERA) cohort,9 an existing additional independent cohort of eRA was obtained from RA-MAP, a multicentre UK industry-academic partnership. All patients with eRA met the 198710 or 201011 RA classification criteria. TACERA patients were included according to the availability of clinical and transcriptome data (quality controlled) and biological samples. In some analyses, missing data sets, particularly longitudinal clinical data, reduced cohort size.
Clinical parameters including autoantibody titres (anticitrullinated protein/peptide antibody (ACPA) and rheumatoid factor (RF)), disease activity score (DAS-28) and its components were recorded. For TACERA at 6 months, DAS-28 was repeated, drug history recorded and additional biological samples were collected. All patients gave informed consent as described in Clark et al and RA-MAP Consortium.8 9
Patient and public involvement
This research was done without formal patient and public involvement.
Serum cytokines
Serum IFN-α was measured using the digital Simoa platform as described.12 Monoclonal antibodies (specifically for all IFN-α subtypes) were isolated from autoimmune polyendocrinopathy candidiasis ecto-dermal dystrophy patients13 and provided to D. Duffy by Immunoqure under a material transfer agreement. IFN-β, IFN-γ and IFN-λ1/IL29 (referred to hereafter as IFN-λ) were measured by MSD technology (Meso Scale Discovery, MD, USA) as per manufacturers’ instructions.
Whole blood and cell-specific transcriptome/methylome
TACERA whole blood analyses used Tempus blood RNA tubes (Applied Biosystems). Peripheral blood mononuclear cells (PBMCs) were isolated using Leucosep separation tubes (Greiner) followed by MagMAX RNA isolation kits (Ambion). For subsequent microarray analysis, amplified RNA was hybridised to beadchips and scanned on an Illumina Beadstation 500 as described further in.9 Full data are available via Gene Expression Omnibus database (GEO), http://www.ncbi.nlm.nih.gov/geo accession number GSE9747638. Existing paired microarray gene expression and DNA methylation data from CD4+ T cells and CD19+ B cells extracted from NEAC eRA patients was preprocessed as described in Clark et al,8 GEO accession number GSE137634. In brief, this involved positive selection of CD4+ T cells/CD19+ B cells, RNeasy Mini kits or AllPrep DNA/RNA Mini kits (Qiagen) and Illumina Whole Genome 6 V.3/12HT BeadChip or a MethylationEPIC BeadChip for RNA and DNA, respectively. Additional method details included in online supplemental file 1.0.
annrheumdis-2022-222370supp001.pdf (128.3KB, pdf)
Whole blood IGS
For both cohorts, the IGS was calculated as an average of whole blood or cell-specific expression of MX1, IFI44L, OAS1, ISG15 and IFI6. IGS scores in the first or fourth quartiles were termed IGS high or low respectively.
Gene expression and DNA methylation analysis of eRA lymphocytes
Analyses included differential gene expression (DEGs) and differential methylation sites (DMSs) between IGS high and low eRA patients, effect of methylation on gene expression, pathway analysis of DEGs and enrichment analysis of DMSs within defined chromatin states. Full methods are provided in online supplemental file 1.0.
Modelling and statistical analysis
GraphPad Prism (V.5.0; GraphPad Software, La Jolla, Calif), JMP Statistical Visualisation (V.14; SAS Institute, Cary, North Carolina) and R Core Team (2020) software was used. Tests included Mann-Whitney U, Wilcoxon matched-pairs signed rank tests, simple linear regression, generalised linear models, multivariable and logistic regression. To adjust for potential confounding variables, tests of a significant association between IGS and 6 month outcomes were performed after adjustment for erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), age, sex, baseline DAS28 and DMARDs, including glucocorticoids. R2 values are reported as a measure of how well the regression model fits the observed data. Statistical significance when p<0.05.
Results
Patient cohorts
The TACERA cohort included 191 seropositive (ACPA and/or RF positive) patients with eRA. A separate NEAC cohort of mixed seropositive and seronegative (ACPA and RF negative) patients with eRA had paired transcriptome and methylome data from circulating B and/or T lymphocytes (n=41 and n=41, respectively, total cohort of n=54) with contemporaneous T and B cell data being available for 28. Patient demographics and clinical characteristics are shown in table 1. The demographic data and relevant methods of an additional smaller validation cohort of patients with NEAC (n=51) who had additional circulating inflammatory cytokines measured in addition to serum-IFN-α and the IGS are shown in online supplemental file 2.0.
Table 1.
Early RA cohort demographics
| RA-MAP TACERA cohort | NEAC | |||
| NEAC lymphocyte methylome and transcriptome cohorts | ||||
| CD4+ T cell | CD19+ B cell | |||
| Number (n) | 191 | 41 | 41 | |
| Age, years | 55 (20–84) | 58 (27–74) | 58 (27–74) | |
| Female, n (%) | 116 (61%) | 26 (63%) | 30 (73%) | |
| RF positive, n (%) | 155* (90%) | 23 (56%) | 26 (63%) | |
| ACPA positive, n (%) | 147* (85%) | 17 (41%) | 22 (54%) | |
| DAS-28-CRP | 5.27 (2.23–8.14) | 4.61 (1.26–6.53) | 4.36 (1.26–6.53) | |
| CRP (mg/L) | 8.65 (1-136) | 9 (5–13) | 9.5 (4–53) | |
| Erythrocyte sedimentation rate (mm/h) | 28 (2–113) | 19 (7–32) | 20.5 (2–86) | |
| DMARDS initiated | Number with available data | 175 | ||
| MTX | 144 (82%) | |||
| SSZ | 10 (6%) | |||
| HCQ | 91 (52%) | |||
| LFU | 0 | |||
| None | 0 | |||
| Glucocorticoid | 124 (71%) | |||
*Missing data for 18 patients.
ACPA, anti-citrullinated protein/peptide antibody; CRP, C reactive protein; DAS-28, disease activity score; DMARDS, disease-modifying anti-rheumatic drugs; HCQ, hydroxychloroquine; LFU, leflunomide; MTX, methotrexate; NEAC, Newcastle Early Arthritis Clinic; RA, rheumatoid arthritis; RF, rheumatoid factor; SSZ, sulfasalazine; TACERA, Towards A CurE for RA.
annrheumdis-2022-222370supp003.pdf (348.2KB, pdf)
Disease-modifying therapy (DMARD) and glucocorticoid naive patients with eRA were recruited at the time of diagnosis from two independent cohorts, RA-MAP TACERA and NEAC. The clinical characteristics and demographics are displayed. Median values with ranges are displayed for continuous variables. For the NEAC-matched methylation and transcription cohort, the total number of patients was 54 with contemporaneous T and B cell data being available for 28.
Baseline IGS but not baseline IFN-α is associated with 6-month clinical outcomes
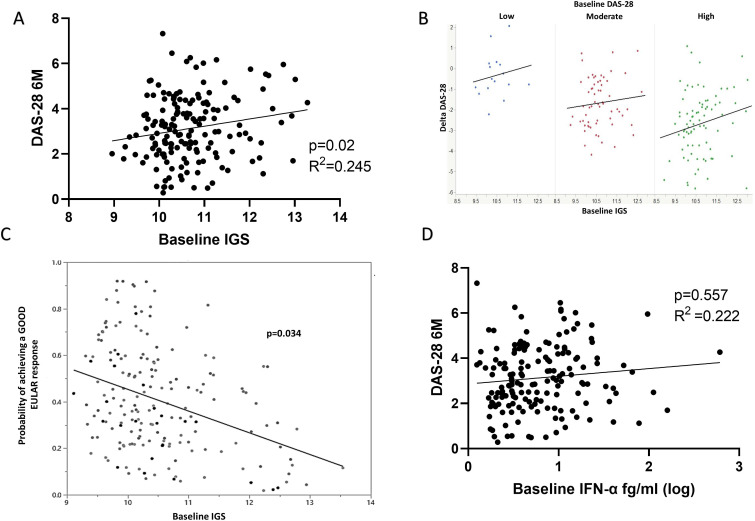
We sought to examine the effect of the IGS and IFN-α on initial clinical outcomes. No significant association between baseline IGS and baseline disease activity (DAS-28, p=0.202) was observed among 171 individuals from the TACERA cohort for whom data were available. Nonetheless, in keeping with our previous findings,6 DAS-28 at 6 months positively associated with the baseline whole blood IGS (n=165, linear regression, p=0.02, R2=0.245), figure 1A. Furthermore, this effect was independent of sex, age and other known confounding variables, including baseline DAS-28, CRP, ESR, glucocorticoids and DMARDs initiated (n=165, multivariable regression analysis, p=0.017). Crucially the interaction term between baseline IGS and baseline DAS-28 is non-significant (p=0.368), indicating that this effect was independent of baseline DAS-28. This is demonstrated graphically by grouping patients according to baseline disease activity (low DAS-28 <3.1; moderate 3.2–5.1; high ≥5.1), where the relationship between baseline IGS and 6-month outcome is consistent across baseline disease activity groups, figure 1B. Smoking and ACPA status similarly demonstrated no significant impact on 6-month outcomes (p=0.399 and p=0.555, respectively) when included in the regression model. In summary, higher baseline IGS scores predicted smaller reductions in DAS-28 (and, therefore, reduced clinical improvements) at 6 months.
Figure 1.
The IGS, circulating IFN-α and clinical outcomes. Early drug naïve RA patients (eRA, n=165) had their IGS calculated from whole blood microarray expression of IFI6, OAS1, MxA, ISG15, IFI44L and the impact of this baseline IGS on 6 month clinical outcomes sought. (A) Linear regression between baseline IGS and DAS-28 at 6 months (6M), p=0.02, R2=0.245. (B) Graphical depiction of baseline IGS consistently impacting on change in DAS-28 at 6 months (Delta DAS-28) regardless of baseline DAS-28 (p=0.017, multivariable regression). Blue dots represent patients with baseline low DAS-28(<3.1), red moderate DAS-28 (3.2–5.1) and green high DAS-28 (>5.1). A negative Delta DAS-28 (Y axis) denotes a fall in DAS-28 and therefore response to therapy. (C) Relationship between the probability of achieving a good EULAR response at 6 months and baseline IGS. Nominal logistic regression, age, sex, DMARD, baseline DAS-28 and glucocorticoid administration corrected, p=0.034. (D) Linear regression between baseline IFN-α and DAS-28 at 6 months (6M), p=0.557, R2=0.222. DAS-28, disease activity score; DMARDS, disease-modifying antirheumatic drugs; IFN, interferon; IGS, gene signature; RA, rheumatoid arthritis.
When classifying/scoring 6-month disease activity into EULAR response outcomes (good, moderate and none) patients with higher baseline IGS scores were less likely to achieve a good EULAR response at 6 months (p=0.034, logistic regression), figure 1C. This was again independent of the above variables.
Baseline IFN-α significantly positively associated with both baseline DAS28 (p=0.018) and ESR (p<0.0001), but not CRP (p=0.053) with similar but less marked associations seen at 6 months (DAS-28 p=0.048, ESR p=0.049, CRP p=0.146), demonstrating that IFN-α levels correlate with disease activity (online supplemental file 3). However, unlike the IGS, baseline IFN-α did not associate with 6-month DAS-28 (p=0.557, linear regression), figure 1D, nor when corrected for the above variables (p=0.57, multivariable regression analysis). IFN-β, -γ or -λ levels did not associate with disease activity at any time point or predict any clinical outcomes (p>0.2 for all, data not shown).
annrheumdis-2022-222370supp004.pdf (165.9KB, pdf)
Circulating IFN-α drives the IGS in eRA
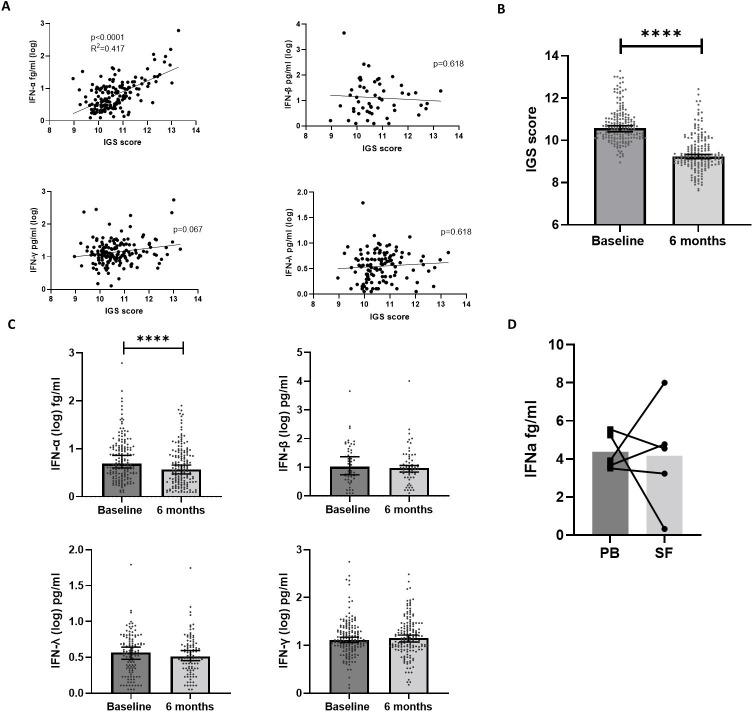
To elucidate which class of IFN is directly responsible for the IGS in eRA, circulating IFN-α, IFN-β, IFN-γ and IFN-λ were examined in relation to the IGS (TACERA cohort).
There was a strong positive association between the IGS and circulating IFN-α (n=164, R2=0.417, p<0.0001, linear regression). Most IFN-β measurements were below the detection threshold but, where detectable, there was no significant association with the IGS (p=0.817, n=53). There was no association between the IGS and IFN-λ (p=0.345, n=117) nor with IFN-γ (p=0.065, n=158), figure 2A. An additional NEAC cohort (n=51) validated the significant association between IFN-α and the IGS (p=0.004), online supplemental file 2.0. In addition, there was no association between the IGS and either TNF-α, IFN-γ, IL6, IL-10, IL12-p70 and IL1β (p>0.1 for all) nor was there any association between IFN-α and any of the above cytokines (p>0.1 for all), online supplemental file 2.0.
Figure 2.
Circulating IFN-I, -II, –III, the IGS and longitudinal expression. (A) Linear regression was performed between the IGS and circulating IFN-α (n=163, p<0.0001, R2=0.29), IFN-β (n=53, p=0.817, R2=0.001), IFN-γ (n=164, p=0.067, R2=0.034), IFN-λ (n=117, p=0.345, R2=0.007). (B) Paired IGS scores between baseline and 6 months in eRA (n=165). Median values are depicted with 95% CIs and statistical analysis using Wilcoxon signed rank test performed on the differences between baseline and 6 months. (C) Comparisons between baseline and 6 month circulating levels of IFN-α, IFN-β, IFN-γ and IFN-λ. Median values are depicted with 95% CIs and statistical analysis using Wilcoxon signed rank test performed on the differences between baseline and 6 months. (D) Comparison of circulating peripheral blood (PB) IFN-α and synovial fluid (SF) IFN-α from five patients with RA, four of whom had established RA and one who had early RA, Wilcoxon signed rank test demonstrated no significant difference. Median values are depicted with paired samples demonstrated. ****p<0.0001. IFN, interferon; IGS, gene signature; RA, rheumatoid arthritis.
As shown previously,6 the IGS significantly fell between baseline and 6 months (n=165, p<0.0001, Wilcoxon signed rank test), figure 2B. Longitudinal serum IFN-α values mirrored the IGS, with a significant fall over 6 months (n=161, p<0.0001). IFN-β, IFN-γ or IFN-λ levels remained static over this period (p=0.275, p=0.819 and p=0.453, respectively), figure 2C. Furthermore, changes in circulating IFN-α correlated with the IGS (p<0.0001, multivariate analysis), but this was not seen for IFN-β, -λ or –γ (p>0.5 for all), online supplemental file 4. Finally, circulating IFN-α itself did not correlate with any other IFN (-β, -γ or –λ) measured at baseline (p>0.7 for all, data not shown). These data, in toto, suggest that the IGS is driven by circulating IFN-α in eRA.
annrheumdis-2022-222370supp005.pdf (75.1KB, pdf)
To compare IFN-α levels in blood and target tissue, IFN-α was measured in matched synovial fluid and serum samples from five patients with RA. Full demographic and descriptive information (not represented in table 1) are shown in online supplemental file 5. There was no significant difference (p=0.8) between serum and synovial fluid IFN-α levels (median 3.93 fg/mL and 4.54 fg/mL, respectively), figure 2D.
annrheumdis-2022-222370supp006.pdf (135.5KB, pdf)
IFN-α/IGS signalling pathways and effect on circulating haematological parameters
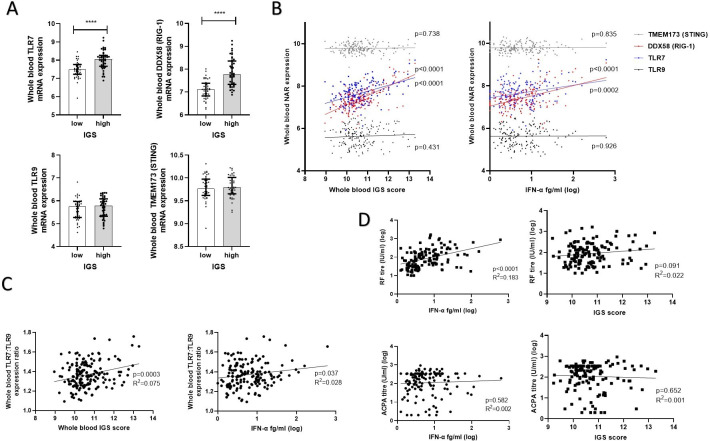
IFN-α production is triggered by NAR ligation. Whole blood mRNA expression of key NARs or their signalling proteins, TLR9, TLR7, TMEM173 (STING) and DDX58 (RIG-1) was, therefore, examined between IGS high and low eRA in the TACERA cohort. Expression of RNA sensing NARs RIG-1 and TLR7 was significantly increased in the IGS high patients (p<0.0001 for both). This was not seen for DNA sensing NARs nor their signalling components, TLR9 and TMEM173 (STING) (p=0.424 and p=0.609, respectively), figure 3A. Furthermore, circulating IFN-α and the IGS positively associated with whole blood expression of TLR7 (p=0.0002, R2=0.191 and p<0.0001 R2=0.216, respectively, linear regression) and RIG-1 (p<0.0001, R2=0.216 and p<0.0001 R2=0.458, respectively). Again this was unique to RNA sensing NARs with no significant association observed between either the IFN-α or the IGS and expression of TLR9 (p=0.926 and p=0.431) or TMEM173 (p=0.835 and p=0.738), figure 3B. TLR7 overexpression, particularly in relation to TLR9 expression, has been linked to autoimmunity and there was a significant positive association between ratio of whole blood TLR7:TLR9 and the IGS (p=0.0003, R2=0.075) as well as with IFN-α (p=0.037, R2=0.027) figure 3C. A similar pattern was observed in circulating PBMCs, online supplemental file 6.
Figure 3.
IFNs, signalling pathways and autoantibody titres. Whole blood expression of nucleic acid receptors (NARs) was examined in early RA TACERA cohort with respect to IFN-α/the IGS. (A) Expression of surface and cytosolic nucleic acid receptors, TLR7, TLR9 DDX58 (RIG-1) and TMEM173 (STING) were examined between IGS high and low patients, n=43 in each cohort. Median values with interquartile ranges are shown. Mann-Whitney U tests were performed. (B) Linear regression between the whole blood IGS or circulating IFN-α and whole blood mRNA expression of TLR7, TLR9 DDX58 (RIG-1) and TMEM173 (STING), n=164. P values are depicted in the figure. (C) Linear regression between whole blood ratio of TLR7: TLR9 mRNA expression and the whole blood IGS score or circulating IFN-α (fg/ml) in 164 eRA patients. (D) Linear regression comparing circulating IFN-α and RF and ACPA titres in seropositive eRA patients (n=132). ****p<0.0001. ACPA, anticitrullinated protein/peptide antibody; eRA, early rheumatoid arthritis; IFN, interferon; IGS, gene signature.
IFN-α is known to affect B cell function so associations with autoantibody titres were sought. RF titre strongly positively correlated with baseline IFN-α (p<0.0001, R2=0.183) but not with baseline IGS (p=0.091). There was no association between RF titre and IFN-β (p=0.379) nor with IFNγ (p=0.230), but there was a weak positive association with IFN-λ (p=0.005, R2=0.069), figure 3D and online supplemental file 7. ACPA titres did not correlate with either the IGS nor any interferon examined (p>0.1 for all), figure 3D and online supplemental file 5.
annrheumdis-2022-222370supp007.pdf (92.4KB, pdf)
IGS correlates with site-specific DNA methylation in B and T cells
Since both IFN-I levels and IGS fall at 6 months in the context of a continued apparent influence on disease activity, we hypothesised that IFN-I-mediated/associated epigenetic alterations may be a plausible mechanism, whereby gene expression programmes in lymphocytes become persistently dysregulated in eRA in response to IFN signalling. We, thus, examined genome-wide transcriptional and methylation data from CD4+ T and CD19+ B lymphocytes isolated from an independent cohort of NEAC patients with eRA (table 1).
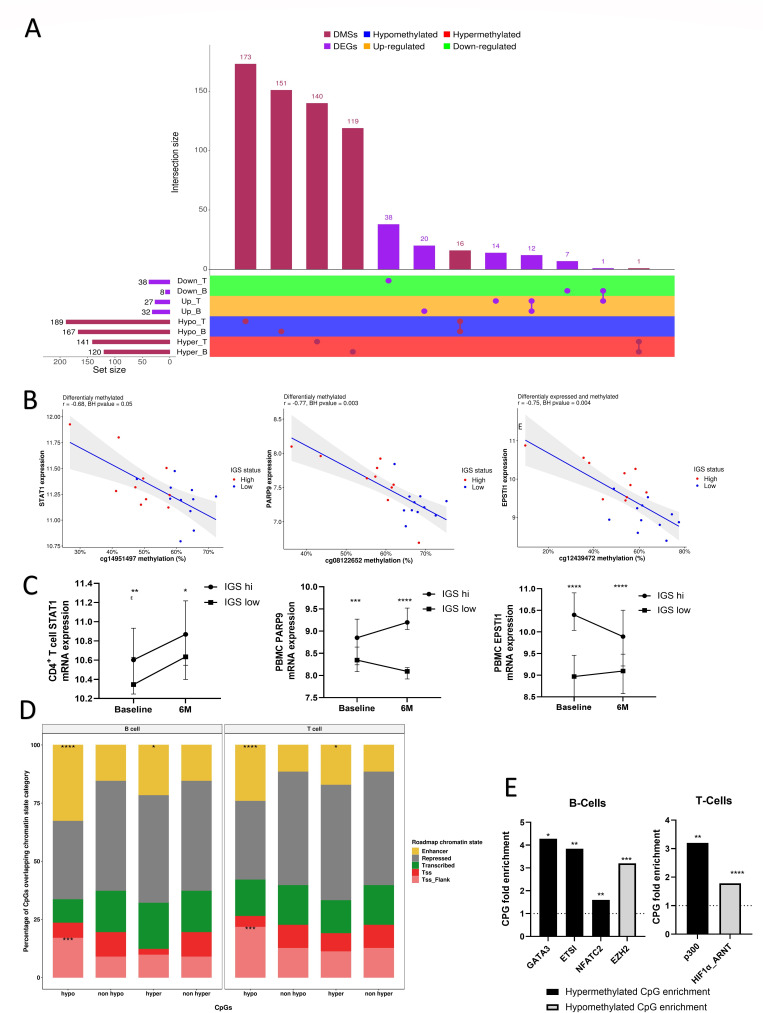
Of 330 CpGs were differentially methylated between IGS high and low CD4+ T cells (57.2% hypomethylated in IGS high, online supplemental file 8) and 287 in CD19+ B cells (58.1% hypomethylated in IGS high, online supplemental file 8). Of the 287 DMSs in CD19+ B cells, 17 (5.9%) showed similar changes in CD4+ T cells, with 16 being hypomethylated in IGS high for both (figure 4A). In addition, 65 DEGs were identified between IGS high and IGS low in CD4+ T cells and 40 in CD19+ B cells. Twelve of these genes were increased for both T and B cells in IGS high patients (figure 4A), online supplemental file 9.
Figure 4.
Differential methylation and expression of pathophysiologically relevant genes in eRA according to IGS. CD4+ T cells and CD19+ B cells were isolated from eRA patients (NEAC cohort) and their cell-specific transcriptome and methylome interrogated according to IGS status. (A) Upset plot45 of differentially methylated sites (DMSs) and differentially expressed genes (DEGs) between IGS high and low early CD19+ B and CD4+ T cells and arranges the co-occurring variables into sets and with a bar chart of their frequency. The horizontal bar graph at the bottom left shows the total number of DEGs/DMSs that are altered in each cell subset between IGS high and IGS low cohorts. Joined red/purples circles to the right of these bar graphs indicate the same DEGs/DMSs were common to the IGS high/IGS low comparisons shown at the left. The vertical bar graph at the top quantitates the number of DEGs/DMSs with similar expression differences in the comparisons. ‘Up’ and ‘Down’ indicate increased expression or reduced expression in the IGS cohort respectively. (B) Scatterplots showing significant correlations (Benjamini-Hochberg (BH) adjusted p value (BHpval) <0.05) between gene expression and DNA methylation of exemplar genes in B and T cells of IGS high and low RA patients. R: Pearson correlation coefficient. (C) Baseline and 6 month (6M) expression of CD4+ T cell STAT1 and peripheral blood mononuclear cell (PBMC) PARP9 and EPSTI1 in IGS high and IGS low patients (n=41 for each) in a separate eRA cohort (RA-MAP TACERA). Median and error bars denoting 95% CI depicted. Mann-Whitney U tests performed between IGS high and IGS low cohorts at each time point. (D) Stacked bar plots indicating the relative distribution of the differentially methylated CPGs (DMS) between IGS high/low eRA patients as previously identified according to their chromatin state annotations. Chromatin states enrichments at DMSs that are hyper- or hypo-methylated in IGS high compared with IGS low RA patients are indicated for both cell types (Fisher’s exact tests) along with standard expression for comparison. TSS: transcription start site; Tss_Flank: flanking a TSS. (E) Exemplar ENCODE and JASPAR transcription factor binding sites (TFBSs) that are significantly enriched (Fisher’s exact test p<0.05) at CD4+ T and CD19+ B cell CPG sites detected as hyper-methylated or hypo-methylated in IGS high RA patients. CPG fold enrichment is displayed. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001. eRA, early rheumatoid arthritis; IGS, gene signature; NEAC, Newcastle Early Arthritis Clinic.
annrheumdis-2022-222370supp008.xlsx (127.6KB, xlsx)
annrheumdis-2022-222370supp009.xlsx (19.9KB, xlsx)
Pathway analysis of DEGs (online supplemental file 10) demonstrated increased expression of genes related to RIG-I and TLR signalling in CD19+ B and CD4+ T cells, respectively, with a significant increase in antiviral pathways and IFN-I signalling. In T cells, pathway analysis also demonstrated significantly increased gene expression linked to RA.
annrheumdis-2022-222370supp010.xlsx (15.8KB, xlsx)
Analysis of genes whose expression correlated with DMSs at relevant loci demonstrated multiple IRGs, such as IFI44L, RSAD2 and Mx1, online supplemental file 11. Of interest to RA pathophysiology, there was increased expression of PARP9 and EPSTI1 in B cells and STAT1 in CD4+ T cells in IGS high patients, which negatively correlated with methylation DMSs (figure 4B). We validated increased expression of CD4+ STAT1 transcript in IGS high patients at baseline in the independent TACERA cohort (p=0.003, Mann-Whitey U test) and showed that this was maintained at 6 months (p=0.02), figure 4C. TACERA PBMC PARP9 and EPSTI1 was examined in lieu of a CD19+ B cell-specific transcriptome, which again confirmed significant gene upregulation in the IGS high cohort at baseline (p=0.0002 and p<0.0001, respectively, Mann-Whitney U tests), again sustained at 6 months (p<0.0001 for both), figure 4C.
annrheumdis-2022-222370supp011.xlsx (17.4KB, xlsx)
To examine the potential effect of these methylation changes on gene regulation and expression, the CD4+ T and CD19+ B cell DMSs were overlapped with chromatin state information for E043 T cell line and E032 B cell line, respectively. DMSs, particularly hypomethylated DMSs, were enriched in putative enhancer regions and regions flanking transcription start sites for both cell types in IGS high patients (figure 4D). In IGS high CD19+ B cells, hypermethylated DMSs were enriched in the binding sites of several transcription factors, including GATA3, ETS1 and NFATC2, whereas hypomethylated DMSs were enriched in binding sites of polycomb protein EZH2. In IGS high CD4+ T cell hypermethylated DMSs were enriched, among others, for p300 TFBSs and hypomethylated DMSs for HIF1αHI (figure 4E). Full list of TFBS enrichment is found in online supplemental file 12.
annrheumdis-2022-222370supp012.xls (61.5KB, xls)
Discussion
In a large, multicentre cohort of DMARD and glucocorticoid nave patients with eRA, we identify, for the first time, IFN-α as primarily responsible for IGS generation. We additionally validate the IGS as a clinically relevant prognostic biomarker in RA for refractory disease.6 14 15 This was independent of conventional markers of disease activity and suggests that IFN-related pathways drive disease persistence. We implicate lymphocyte epigenetic reprogramming as an underpinning mechanism.
Directly linking the IGS and IFN-α has been historically challenging due to difficulties directly measuring circulating IFN-α.3 12 Therefore, our demonstration of the IGS in eRA positively associating with circulating IFN-α protein, and not other classes of IFN or other circulating cytokines, is an important step in dissecting the biological significance of the IGS and is in keeping with what has been reported in other rheumatic diseases.16 Furthermore, both IGS and serum IFN-α levels fall in parallel with clinical response over 6 months, additionally supporting the role of IFN-α driving the IGS in eRA. Like others,17 and in contrast to the IGS, we could not demonstrate an association of baseline IFN-α with longitudinal clinical outcomes. A potential explanation is that the IRG integrates activity over time of IFN-α, which itself has a short half-life.
Our finding of comparable IFN-α levels in serum and synovial fluids implicates a systemic source potentially influencing synovial pathophysiology. Larger studies are required to confirm this, and examination of other inflammatory arthritides will shed further light on the role of IFN-α in synovial pathology. RA is a heterogeneous disease and, despite the association of baseline IGS with clinical outcome reported here, not all patients with a low IGS at inception fared well. This could well reflect the dominance of alternative disease pathways in some patients but does not reduce the prognostic value of a high IGS.
Contrasting with other IFNs, increased baseline levels of IFN-α suggest a pathophysiological role in eRA and, potentially, in disease initiation. The permissive effects of IFN-α on lymphocyte activation and development of autoimmune characteristics are well documented1 and murine transfer of IFN-α secreting dendritic cells propagated a persistent inflammatory arthritis.18 IFN-α associated with raised ESR in our cohort and in other autoimmune conditions12 but is also relevant prior to onset of inflammation as an IGS predicts progression to RA in ‘at risk’ cohorts.19–21 Furthermore, autoantibodies predate clinical presentation, and we demonstrate a clear association between IFN-α and RF titres in eRA consistent with previous observations in autoimmunity.12 17 In contrast, there was no association between IFN-α and ACPA. This dichotomy likely reflects differences in autoantibody sources and generation. RF-producing B cells demonstrate activation of IFN-I pathways, whereas ACPA-producing B cells do not.22 These data, alongside observations in other diseases, highlighting the role of type 1 interferons at disease onset,23 cumulatively suggest IFN-α may promote or accelerate breach of tolerance in susceptible individuals. Indeed, RA twin studies hypothesise that environmental and stochastic factors may be more important than genetic factors in determining development of disease relevant autoantibodies.24
We attempted to identify the receptor(s), and thus potential environmental triggers, responsible for IFN-α release. Although the expression of all examined NARs can be increased following IFN-I exposure,25 only RNA sensing pathways increased in association with a raised IGS/IFN-α, reflecting previous observations in autoimmunity.12 26 Furthermore, the IGS was associated with TLR7:TLR9 imbalance, itself associated with heightened autoimmunity risk and breach of tolerance.2 27 We feel our data are more consistent with RNA sensing pathways triggering IFN-α production and release, but we accept these are associations, and understanding the primary trigger(s) of IFN-α release in eRA remains a pressing priority for future study.
Altered DNA methylation regulates the innate antiviral immune response and hypomethylation of IRGs, such as we report, has also been demonstrated in autoimmunity.7 Pretreatment with IFN-α in vitro enhances subsequent B cell activation28 and pretreatment of macrophages with IFN-α prevented the silencing of NF-κB via effects on chromatin, thereby abolishing TNF-induced tolerance to TLR ligation and potentiating the proinflammatory function of TNF-α.29 Similar chromatin changes were identified in systemic lupus erythematous patients, a condition where IFN-α levels are increased, highlighting the in vivo relevance of IFN-α-related epigenetic modifications.29 In addition, IFN-α treatment of salivary gland tissues reduced DNA methyltransferases, which catalyse DNA methylation, and upregulated TET3 which is involved in demethylation.30 Stratifying eRA T and B cells by IGS, we identified multiple DMSs. These preferentially mapped to enhancers and flank regions, thereby supporting their biological relevance. They also reflected TFBS patterns that favoured proliferative responses. Namely, B cell CpG enrichment inferred increased binding of EZH2, which is increased in cell proliferation and lymphoma31 and reduced binding of (1) GATA3, suggesting increased cell proliferation32, (2) ETS1 which is required to prevent autoimmune responses33 and (3) NFATC2, involved in anergy, which, when reduced, causes a hyperproliferative phenotype.34 In T cells, there was inferred reduction in p300 binding, which would impair Foxp3+ T-regulatory cell function35; and enrichment of HIF1-α, which promotes Th17 differentiation and reduced Foxp3+ expression.36 Additionally, CPG methylation changes in IGS high patients associated with increased gene expression of PARP9, EPSTI1 (B cells) and STAT1 (T-cells) and remained significantly differentially expressed at 6 months between IGS high and low patients in a distinct cohort despite a sustained fall in IGS/circulating IFN-α. PARP9 (BAL-1) can modify B cell proliferation and altered PARP9 methylation, and expression has been implicated in RA pathogenesis37; ETSI promotes pathological B cell activation in Primary Sjogren’s Syndrome (PSS)38; and STAT1 in T cells is a key mediator of inflammatory cytokine signalling and important in focal RA inflammatory infiltrates.39 Our interpretation of these data is that IFN-α-induced perturbation of DNA methylation influences the immune system early in the natural history of an identifiable subpopulation of patients with RA, leading to adverse outcomes.
Further work is needed to support this, namely (1) longitudinal measurements to determine whether the observed methylation changes (and gene expression of correlated transcripts) are sustained over time where the IGS/IFN-α levels are not and (2) ex vivo confirmation of the propensity for IFN-α to induce relevant epigenetic changes in relevant cell populations. Indeed, although we have focused on T and B lymphocytes in view of their known relevance to RA pathophysiology, these effects are likely to extend to other cell subsets.40 41
Cumulatively, these data incriminate IFN-α as a key cytokine underpinning prognosis in RA and support the hypothesis of an IFN-α-driven epigenetic programming at disease onset that perpetuates pathological signalling pathways and refractoriness to therapy. Such phenomena could underpin the well-recognised window of opportunity in eRA by persistent dysregulation of proinflammatory pathways after a period of unopposed activity. JAK inhibitors modify IFN signalling as well as IFN-induced epigenetic programming in PSS.30 Targeted administration of these or similar therapies in the early stages of RA, or in ‘pre-RA’ at-risk groups, may provide a precision medicine approach ultimately reducing clinical progression and patient morbidity. However, before adoption of stratification by IGS, factors to address include IRG selection and interlaboratory standardisation, as recently reviewed in Cooles and Isaacs.42
Sustained immune dysregulation secondary to IFN-I may have additional implications. For example, IFN-I-induced epigenetic modifications29 are also present in patients with COVID-1943 and might have relevance to ‘long-covid’ syndromes as well as autoantibody development and, potentially, autoimmunity.44
In conclusion, we identify, for the first time, IFN-α as primarily responsible for the IGS in eRA and validate a high IGS as a clinically relevant prognostic biomarker in eRA, portending refractory disease and implying a therapeutic window of opportunity for drugs that target IFN-α signalling. We additionally implicate lymphocyte epigenetic reprogramming as an underpinning mechanism with relevance for other IFN-α enriched states.
annrheumdis-2022-222370supp002.pdf (177.3KB, pdf)
annrheumdis-2022-222370supp013.pdf (29.6KB, pdf)
Acknowledgments
JDI is a National Institute for Health and Care Research (NIHR) Senior Investigator. The authors acknowledge the support of TACERA Principal Investigators from all contributing NHS sites and the members of the TACERA Study Steering and Data Monitoring Committee. Additional acknowledgements include patient volunteers.
Footnotes
Handling editor: Josef S Smolen
Twitter: @dennislendrem, @Nicolamaney, @ProfJohnIsaacs
Collaborators: RA-MAP Consortium: Adwoa Hughes-Morley, Alexandra Walker, Alexandru Cuza, Amaya Gallagher-Syed, Amy Anderson, Andrea Haynes, Andrew Filer, Andrew Long, Andrew P Cope, Angela Parke, Anthony Rowe, Arnaud Didierlaurent, Ashley Gilmour, Athula Herath, Ayako Wakatsuki, Pedersen Aysin, Tulunay Virlan, Ben Allen, Benjamin A Fisher, Blerina Kola, Bohdan Harvey, Brian Tom, Carl S Goodyear, Carolyn Cuff, Catharien Hilkens, Catharina Lindholm, Catherine T Mela, Christopher D Buckley, Chris Larminie, Chris Marshall, Christopher John, Christopher M Mela, Claudio Carini, Costantino Pitzalis, Coziana Ciurtin, Dan Baker, Daniel Ziemek, Daniela Dastros-Pitei, Dao Nguyen, David L Scott, David S Watson, Deborah Symmons, Dennis Lendrem, Denny Verbeeck, Desmond Padhji, Donna Finch, Duncan Porter, Emma Vernon, Faye Cooles, Feng Hong, Fiona Clarke, Fiona Stirling, Fowzia Ibrahim, Frances Humby, Francisco Bonachela Capdevila, Frederic Geissmann, Frederique Ponchel, Gemma Molyneux, Gemma Simpson, Georgina Thorborn, Gerry Parker, Gioia Altobelli, Graham R Smith, Hannah Edwards, Hannah Tipney, Hans-Dieter Zucht, Hayley Noble, Heidi Lempp, Humayara AliIain B McInnes, Ian C Scott, Ian N BruceIona Donnelly, Ivana Vranic, James A Butler, James Galloway, Jamie C Sergeant, Jane Worthington, Jehan El-Jawhari, Jessica Tarn, Joanne Ellis, John Casement, John Isaacs, Julie Diboll, Karim Raza, Katriona Goldmann, Kirsty Hicks, Liliane Fossati-Jimack, Lucy Rowell, Marc Levesque, Mark C Coles, Mark Coles, Mark Curran, Martin Hodge, Martin Jenkins, Mateusz Maciejewski, Matt Page, Matthew A Sleeman, Matthew J Loza, Maya Buch, Meilien Ho, Michael Binks, Michael F McDermott, Michael Macoritto, Michael R Barnes, Michael R Ehrenstein, Michele Bombardieri, Myles Lewis, Neil Gozzard, Neil Payne, Neil Ward, Nina Joseph, Paul Emery, Peter C Taylor, Peter Schulz-Knappe, Petra Budde, Philip Jones, Philip Stocks, Rachel Harry, Rafael Henkin, Ravi Rao, Ray Harris, Rekha Parmar, Ruth Toward, Sally Hollis, Samana Schwank, Samantha Lipsky, Samiul Hasan, Sandra Martins, Sandra Ng, Sarah Brockbank, Sarah Keidel, Scott Jelinsky, Sharmila Rana, Simon Read, Stephen Kelly, Stephen Wright, Steve P Young, Sukru Kaymakcalan, Susan Talbot, Suzanne MM Verstappen, Tomi Lazarov, Tony Sabin, Valerie Ludbrook, Vernon Farewell, Wayne Tsuji, Wing Wu, Wivine Burny, Yujie Zhong, Zheng Liu, Zhilong Jia.
Contributors: FAHC devised the experiments, developed the concepts and wrote the first manuscript draft and acts as guarantor. DWL and JT assisted with RA-MAP TACERA data analysis and modelling. NN, AGP and LNR assisted with epigenetic analyses and support. DD, NJM, BM, CMAL and VB assisted with analysis of serum cytokines. AGP, NT, AEA and JD assisted with T and B cell data collection. GRS, MRB, DW, SN, RH assisted with RA-MAP TACERA data processing, QC and curation. APC supervised RA-MAP TACERA data collection. JDI provided direction and oversight to the whole project. All authors approved the final manuscript.
Funding: Newcastle researchers received infrastructural support via the Versus Arthritis Research into Inflammatory Arthritis Centre (Ref 22072), funding from The Medical Research Council; Academy of Medical Sciences; British Society of Rheumatology; The Wellcome Trust; JGW Patterson Foundation; Immune-Mediated Inflammatory Disease Biobank in the UK (IMID-Bio-UK); Connect Immune Research; ANR and RTCure. This work was supported by the National Institute for Health and Care Research (NIHR) Newcastle Biomedical Research Centre for Ageing and Long-Term Conditions; views expressed are the authors’ and not necessarily those of the National Health Service, the National Institute of Health and Care Research or the Department of Health.
Competing interests: JDI discloses research grants from Pfizer, Janssen and GSK; conference support from Eli Lilly and Gilead; speaker/consulting fees from AbbVie, BMS, Gilead, Roche and UCB. FAHC discloses speaker fees from AstraZeneca. The remaining authors have no competing interests.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
Collaborators: RA-MAP Consortium, Adwoa Hughes-Morley, Alexandra Walker, Alexandru Cuza, Amaya Gallagher-Syed, Amy Anderson, Andrea Haynes, Andrew Filer, Andrew Long, Andrew P Cope, Angela Parke, Anthony Rowe, Arnaud Didierlaurent, Ashley Gilmour, Athula Herath, Ayako Wakatsuki, Pedersen Aysin, Tulunay Virlan, Ben Allen, Benjamin A Fisher, Blerina Kola, Bohdan Harvey, Brian Tom, Carl S Goodyear, Carolyn Cuff, Catharien Hilkens, Catharina Lindholm, Catherine T Mela, Christopher D Buckley, Chris Larminie, Chris Marshall, Christopher John, Christopher M Mela, Claudio Carini, Costantino Pitzalis, Coziana Ciurtin, Dan Baker, Daniel Ziemek, Daniela Dastros-Pitei, Dao Nguyen, David L Scott, David S Watson, Deborah Symmons, Dennis Lendrem, Denny Verbeeck, Desmond Padhji, Donna Finch, Duncan Porter, Emma Vernon, Faye Cooles, Feng Hong, Fiona Clarke, Fiona Stirling, Fowzia Ibrahim, Frances Humby, Francisco Bonachela Capdevila, Frederic Geissmann, Frederique Ponchel, Gemma Molyneux, Gemma Simpson, Georgina Thorborn, Gerry Parker, Gioia Altobelli, Graham R Smith, Hannah Edwards, Hannah Tipney, Hans-Dieter Zucht, Hayley Noble, Heidi Lempp, Humayara AliIain B McInnes, Ian C Scott, Ian N BruceIona Donnelly, Ivana Vranic, James A Butler, James Galloway, Jamie C Sergeant, Jane Worthington, Jehan El-Jawhari, Jessica Tarn, Joanne Ellis, John Casement, John Isaacs, Julie Diboll, Karim Raza, Katriona Goldmann, Kirsty Hicks, Liliane Fossati-Jimack, Lucy Rowell, Marc Levesque, Mark C Coles, Mark Coles, Mark Curran, Martin Hodge, Martin Jenkins, Mateusz Maciejewski, Matt Page, Matthew A Sleeman, Matthew J Loza, Maya Buch, Meilien Ho, Michael Binks, Michael F McDermott, Michael Macoritto, Michael R Barnes, Michael R Ehrenstein, Michele Bombardieri, Myles Lewis, Neil Gozzard, Neil Payne, Neil Ward, Nina Joseph, Paul Emery, Peter C Taylor, Peter Schulz-Knappe, Petra Budde, Philip Jones, Philip Stocks, Rachel Harry, Rafael Henkin, Ravi Rao, Ray Harris, Rekha Parmar, Ruth Toward, Sally Hollis, Samana Schwank, Samantha Lipsky, Samiul Hasan, Sandra Martins, Sandra Ng, Sarah Brockbank, Sarah Keidel, Scott Jelinsky, Sharmila Rana, Simon Read, Stephen Kelly, Stephen Wright, Steve P Young, Sukru Kaymakcalan, Susan Talbot, Suzanne MM Verstappen, Tomi Lazarov, Tony Sabin, Valerie Ludbrook, Vernon Farewell, Wayne Tsuji, Wing Wu, Wivine Burny, Yujie Zhong, Zheng Liu, and Zhilong Jia
Data availability statement
Data are available in a public, open access repository. Data are available upon reasonable request. Data included in this article is either already either in the public domain or available on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Ethical approval was authorised by both the National Research Ethics Service London Central Committee (Reference number: 12/LO/0469) for RA-MAP study participants and North East—Newcastle & North Tyneside 2 Research Ethics Committee (REC reference: 12/NE/0251) for NEAC participants. All patients gave full informed written consent.
References
- 1. Muskardin TLW, Niewold TB. Type I interferon in rheumatic diseases. Nat Rev Rheumatol 2018;14:214–28. 10.1038/nrrheum.2018.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lind NA, Rael VE, Pestal K, et al. Regulation of the nucleic acid-sensing Toll-like receptors. Nat Rev Immunol 2022;22:224–35. 10.1038/s41577-021-00577-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cooles FAH, Isaacs JD. The interferon gene signature as a clinically relevant biomarker in autoimmune rheumatic disease. Lancet Rheumatol 2022;4:e61–72. 10.1016/S2665-9913(21)00254-X [DOI] [PubMed] [Google Scholar]
- 4. de Jong TD, Sellam J, Agca R, et al. A multi-parameter response prediction model for rituximab in rheumatoid arthritis. Joint Bone Spine 2018;85:219–26. 10.1016/j.jbspin.2017.02.015 [DOI] [PubMed] [Google Scholar]
- 5. de Jong TD, Snoek T, Mantel E, et al. Dynamics of the type I interferon response during immunosuppressive therapy in rheumatoid arthritis. Front Immunol 2019;10:902. 10.3389/fimmu.2019.00902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cooles FAH, Anderson AE, Lendrem DW, et al. The interferon gene signature is increased in patients with early treatment-naive rheumatoid arthritis and predicts a poorer response to initial therapy. J Allergy Clin Immunol 2018;141:445–8. 10.1016/j.jaci.2017.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barrat FJ, Crow MK, Ivashkiv LB. Interferon target-gene expression and epigenomic signatures in health and disease. Nat Immunol 2019;20:1574–83. 10.1038/s41590-019-0466-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clark AD, Nair N, Anderson AE, et al. Lymphocyte DNA methylation mediates genetic risk at shared immune-mediated disease loci. J Allergy Clin Immunol 2020;145:1438–51. 10.1016/j.jaci.2019.12.910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. RA-MAP Consortium . RA-MAP, molecular immunological landscapes in early rheumatoid arthritis and healthy vaccine recipients. Sci Data 2022;9:196. 10.1038/s41597-022-01264-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arnett FC, Edworthy SM, Bloch DA, et al. The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum 1988;31:315–24. 10.1002/art.1780310302 [DOI] [PubMed] [Google Scholar]
- 11. Aletaha D, Neogi T, Silman AJ, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League against rheumatism collaborative initiative. Ann Rheum Dis 2010;69:1580–8. 10.1136/ard.2010.138461 [DOI] [PubMed] [Google Scholar]
- 12. Reynolds JA, Briggs TA, Rice GI, et al. Type I interferon in patients with systemic autoimmune rheumatic disease is associated with haematological abnormalities and specific autoantibody profiles. Arthritis Res Ther 2019;21:147. 10.1186/s13075-019-1929-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meyer S, Woodward M, Hertel C, et al. AIRE-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell 2016;166:582–95. 10.1016/j.cell.2016.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodríguez-Carrio J, Alperi-López M, López P, et al. Heterogeneity of the type I interferon signature in rheumatoid arthritis: a potential limitation for its use as a clinical biomarker. Front Immunol 2017;8:8. 10.3389/fimmu.2017.02007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Plant D, Maciejewski M, Smith S, et al. Profiling of gene expression biomarkers as a classifier of methotrexate nonresponse in patients with rheumatoid arthritis. Arthritis Rheumatol 2019;71:678–84. 10.1002/art.40810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huijser E, Gopfert J, Brkic Z. Serum IFNalpha2 measured by single-molecule array associates with systemic disease manifestations in Sjogren’s syndrome. Rheumatology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stockfelt M, Lundell A-C, Hetland ML, et al. Plasma interferon-alpha is associated with double-positivity for autoantibodies but is not a predictor of remission in early rheumatoid arthritis-a spin-off study of the NORD-STAR randomized clinical trial. Arthritis Res Ther 2021;23:189. 10.1186/s13075-021-02556-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Narendra SC, Chalise JP, Höök N, et al. Dendritic cells activated by double-stranded RNA induce arthritis via autocrine type I IFN signaling. J Leukoc Biol 2014;95:661–6. 10.1189/jlb.0613320 [DOI] [PubMed] [Google Scholar]
- 19. Lübbers J, Brink M, van de Stadt LA, et al. The type I IFN signature as a biomarker of preclinical rheumatoid arthritis. Ann Rheum Dis 2013;72:776–80. 10.1136/annrheumdis-2012-202753 [DOI] [PubMed] [Google Scholar]
- 20. Macías-Segura N, Castañeda-Delgado JE, Bastian Y, et al. Transcriptional signature associated with early rheumatoid arthritis and healthy individuals at high risk to develop the disease. PLoS One 2018;13:e0194205. 10.1371/journal.pone.0194205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brink M, Lundquist A, Alexeyenko A, et al. Protein profiling and network enrichment analysis in individuals before and after the onset of rheumatoid arthritis. Arthritis Res Ther 2019;21:288. 10.1186/s13075-019-2066-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lu DR, McDavid AN, Kongpachith S, et al. T cell-dependent affinity maturation and innate immune pathways differentially drive autoreactive B cell responses in rheumatoid arthritis. Arthritis Rheumatol 2018;70:1732–44. 10.1002/art.40578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lombardi A, Tsomos E, Hammerstad SS, et al. Interferon alpha: the key trigger of type 1 diabetes. J Autoimmun 2018;94:7–15. 10.1016/j.jaut.2018.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hensvold AH, Magnusson PKE, Joshua V, et al. Environmental and genetic factors in the development of anticitrullinated protein antibodies (ACPAs) and ACPA-positive rheumatoid arthritis: an epidemiological investigation in twins. Ann Rheum Dis 2015;74:375–80. 10.1136/annrheumdis-2013-203947 [DOI] [PubMed] [Google Scholar]
- 25. Rusinova I, Forster S, Yu S. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res 2013;41(Database issue:D1040–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maria NI, Steenwijk EC, IJpma AS, et al. Contrasting expression pattern of RNA-sensing receptors TLR7, RIG-I and MDA5 in interferon-positive and interferon-negative patients with primary Sjögren's syndrome. Ann Rheum Dis 2017;76:721–30. 10.1136/annrheumdis-2016-209589 [DOI] [PubMed] [Google Scholar]
- 27. Fillatreau S, Manfroi B, Dörner T. Toll-Like receptor signalling in B cells during systemic lupus erythematosus. Nat Rev Rheumatol 2021;17:98–108. 10.1038/s41584-020-00544-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Akita K, Yasaka K, Shirai T, et al. Interferon α Enhances B Cell Activation Associated With FOXM1 Induction: Potential Novel Therapeutic Strategy for Targeting the Plasmablasts of Systemic Lupus Erythematosus. Front Immunol 2020;11:498703. 10.3389/fimmu.2020.498703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Park SH, Kang K, Giannopoulou E, et al. Type I interferons and the cytokine TNF cooperatively reprogram the macrophage epigenome to promote inflammatory activation. Nat Immunol 2017;18:1104–16. 10.1038/ni.3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Charras A, Arvaniti P, Le Dantec C, et al. JAK inhibitors suppress innate epigenetic reprogramming: a promise for patients with Sjögren's syndrome. Clin Rev Allergy Immunol 2020;58:182–93. 10.1007/s12016-019-08743-y [DOI] [PubMed] [Google Scholar]
- 31. Li B, Chng W-J. EZH2 abnormalities in lymphoid malignancies: underlying mechanisms and therapeutic implications. J Hematol Oncol 2019;12:118. 10.1186/s13045-019-0814-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu S, Chan HL, Bai F, et al. Gata3 restrains B cell proliferation and cooperates with p18INK4C to repress B cell lymphomagenesis. Oncotarget 2016;7:64007–20. 10.18632/oncotarget.11746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sunshine A, Goich D, Stith A, et al. Ets1 controls the development of B cell autoimmune responses in a cell-intrinsic manner. Immunohorizons 2019;3:331–40. 10.4049/immunohorizons.1900033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teixeira LK, Carrossini N, Sécca C, et al. NFAT1 transcription factor regulates cell cycle progression and cyclin E expression in B lymphocytes. Cell Cycle 2016;15:2346–59. 10.1080/15384101.2016.1203485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu Y, Wang L, Predina J, et al. Inhibition of p300 impairs Foxp3⁺ T regulatory cell function and promotes antitumor immunity. Nat Med 2013;19:1173–7. 10.1038/nm.3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guo X, Chen G. Hypoxia-Inducible factor is critical for pathogenesis and regulation of immune cell functions in rheumatoid arthritis. Front Immunol 2020;11:1668. 10.3389/fimmu.2020.01668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhu H, Wu L-F, Mo X-B, et al. Rheumatoid arthritis-associated DNA methylation sites in peripheral blood mononuclear cells. Ann Rheum Dis 2019;78:36–42. 10.1136/annrheumdis-2018-213970 [DOI] [PubMed] [Google Scholar]
- 38. Sun J-L, Zhang H-Z, Liu S-Y, et al. Elevated EPSTI1 promote B cell hyperactivation through NF-κB signalling in patients with primary Sjögren's syndrome. Ann Rheum Dis 2020;79:518–24. 10.1136/annrheumdis-2019-216428 [DOI] [PubMed] [Google Scholar]
- 39. Kasperkovitz PV, Verbeet NL, Smeets TJ, et al. Activation of the STAT1 pathway in rheumatoid arthritis. Ann Rheum Dis 2004;63:233–9. 10.1136/ard.2003.013276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rodríguez-Ubreva J, de la Calle-Fabregat C, Li T, et al. Inflammatory cytokines shape a changing DNA methylome in monocytes mirroring disease activity in rheumatoid arthritis. Ann Rheum Dis 2019;78:1505–16. 10.1136/annrheumdis-2019-215355 [DOI] [PubMed] [Google Scholar]
- 41. de la Calle-Fabregat C, Rodríguez-Ubreva J, Ciudad L, et al. The synovial and blood monocyte DNA methylomes mirror prognosis, evolution, and treatment in early arthritis. JCI Insight 2022;7:158783. 10.1172/jci.insight.158783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cooles FAH, Isaacs JD. The interferon gene signature as a clinically relevant biomarker in autoimmune rheumatic disease. The Lancet Rheumatology 2022;4:e61–72. 10.1016/S2665-9913(21)00254-X [DOI] [PubMed] [Google Scholar]
- 43. Lee JS, Park S, Jeong HW, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Sci Immunol 2020;5:1554. 10.1126/sciimmunol.abd1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cañas CA. The triggering of post-COVID-19 autoimmunity phenomena could be associated with both transient immunosuppression and an inappropriate form of immune reconstitution in susceptible individuals. Med Hypotheses 2020;145:110345. 10.1016/j.mehy.2020.110345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lex A, Gehlenborg N, Strobelt H, et al. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph 2014;20:1983–92. 10.1109/TVCG.2014.2346248 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2022-222370supp001.pdf (128.3KB, pdf)
annrheumdis-2022-222370supp003.pdf (348.2KB, pdf)
annrheumdis-2022-222370supp004.pdf (165.9KB, pdf)
annrheumdis-2022-222370supp005.pdf (75.1KB, pdf)
annrheumdis-2022-222370supp006.pdf (135.5KB, pdf)
annrheumdis-2022-222370supp007.pdf (92.4KB, pdf)
annrheumdis-2022-222370supp008.xlsx (127.6KB, xlsx)
annrheumdis-2022-222370supp009.xlsx (19.9KB, xlsx)
annrheumdis-2022-222370supp010.xlsx (15.8KB, xlsx)
annrheumdis-2022-222370supp011.xlsx (17.4KB, xlsx)
annrheumdis-2022-222370supp012.xls (61.5KB, xls)
annrheumdis-2022-222370supp002.pdf (177.3KB, pdf)
annrheumdis-2022-222370supp013.pdf (29.6KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available upon reasonable request. Data included in this article is either already either in the public domain or available on reasonable request.