Abstract
Objectives
Giant cell arteritis (GCA) is a complex systemic vasculitis mediated by the interplay between both genetic and epigenetic factors. Monocytes are crucial players of the inflammation occurring in GCA. Therefore, characterisation of the monocyte methylome and transcriptome in GCA would be helpful to better understand disease pathogenesis.
Methods
We performed an integrated epigenome-and transcriptome-wide association study in CD14+ monocytes from 82 patients with GCA, cross-sectionally classified into three different clinical statuses (active, in remission with or without glucocorticoid (GC) treatment), and 31 healthy controls.
Results
We identified a global methylation and gene expression dysregulation in GCA monocytes. Specifically, monocytes from active patients showed a more proinflammatory phenotype compared with healthy controls and patients in remission. In addition to inflammatory pathways known to be involved in active GCA, such as response to IL-6 and IL-1, we identified response to IL-11 as a new pathway potentially implicated in GCA. Furthermore, monocytes from patients in remission with treatment showed downregulation of genes involved in inflammatory processes as well as overexpression of GC receptor-target genes. Finally, we identified changes in DNA methylation correlating with alterations in expression levels of genes with a potential role in GCA pathogenesis, such as ITGA7 and CD63, as well as genes mediating the molecular response to GC, including FKBP5, ETS2, ZBTB16 and ADAMTS2.
Conclusion
Our results revealed profound alterations in the methylation and transcriptomic profiles of monocytes from GCA patients, uncovering novel genes and pathways involved in GCA pathogenesis and in the molecular response to GC treatment.
Keywords: Giant Cell Arteritis, Glucocorticoids, Immune Complex Diseases, Inflammation
WHAT IS ALREADY KNOWN ON THIS TOPIC
Giant cell arteritis (GCA) is a complex disease mediated by multiple genetic and epigenetic factors, in which CD14+ monocytes play an important role driving the inflammatory processes occurring in this vasculitis.
The study of the DNA methylation and gene expression profiles of disease-relevant cell types, as well as the integration of omics- datasets, has emerged as a successful approach to better understand the pathogenesis of complex diseases.
WHAT THIS STUDY ADDS
We evaluate for the first time the DNA methylome and transcriptome landscapes of CD14+ monocytes from patients with GCA in three different states of the disease (patients with active disease, patients in remission with or without treatment), identifying profound alterations that provide evidence of novel genes and pathways potentially involved in GCA pathogenesis.
The results of this integrative approach allowed the identification of a significant number of CpG-gene expression interactions, including important genes potentially involved in the molecular mechanisms implicated in the active state of the disease, such as ITGA7 and CD63, as well as genes mediating the molecular response to glucocorticoids, including FKBP5, ETS2, ZBTB16 and ADAMTS2.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE AND/OR POLICY
A better understanding of the pathogenesis of GCA might result in the identification of potential biomarkers that allow advances in early diagnosis, classification and therapy for GCA.
Introduction
Giant cell arteritis (GCA) is a systemic vasculitis with complex aetiology, presenting a wide range of clinical manifestations.1 The most serious complications such as irreversible blindness and stroke can be significantly reduced if patients receive prompt treatment with glucocorticoids (GC).2 However, early recognition of GCA can be challenging due to its clinical heterogeneity, including the presence of non-specific symptoms, along with the absence of specific biomarkers.3
The disease results from dysregulated interactions between the vessel wall and the immune system that lead to inflammation and vascular remodelling of medium and large arteries.4 Most of our current knowledge has been obtained from examination of temporal artery biopsies, where granulomatous infiltrates consist of both innate and adaptive immune cells.5 Besides tissue lesions, GCA is characterised by intense systemic inflammation that is driven by IL-6.6 Although studies focusing on circulating immune cells in GCA are scarce, monocytes are considered major players in the inflammatory process.7 Indeed, circulating monocytes of GCA patients interact with activated endothelial cells in vasa vasorum and neovessels,8 9 and develop tissue invasive capabilities by an aberrant production of matrix metalloproteinase (MMP)-9, thus allowing immune cells to access the vascular wall.10
It is well established that epigenetic modifications may exert a profound influence on cell function by their capacity to modulate gene expression without altering the DNA sequence.11 Despite the high relevance of monocytes in GCA, no study has investigated the DNA methylation landscape of this cell type to date. Here, with the aim of shedding light into GCA pathogenesis and identifying molecular mechanisms that might serve as novel biomarkers or potential drug targets, we analysed for the first time the methylome and transcriptome of GCA monocytes as well as the correlation between DNA methylation and gene expression levels.
Material and methods
Details of the population included in this study, description of the experimental methods, including DNA methylation and RNA sequencing as well as the statistical analyses applied, are provided in online supplemental material.
annrheumdis-2022-222156supp001.pdf (1.5MB, pdf)
Results
Dysregulated DNA methylation and gene expression profiles in CD14+ monocytes of patients with GCA
CD14+ monocytes from patients with GCA showed a global hypermethylation pattern
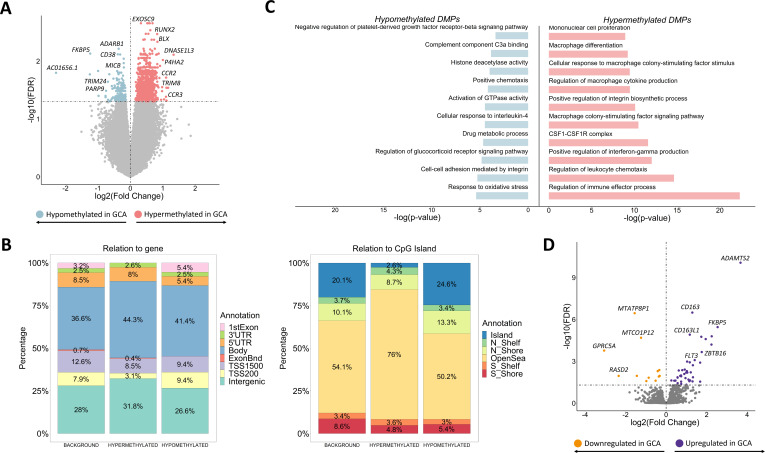
First, the comparison of the DNA methylation landscape of CD14+ monocytes between patients with GCA and controls unveiled the existence of 1371 differentially methylated positions (DMPs), annotated to 1190 unique genes, across the whole genome (figure 1A). Most DMPs were located in intergenic regions (31.8% hypermethylated and 26.6% hypomethylated) and gene bodies (44.3% hypermethylated and 41.4% hypomethylated) and less frequently in promoters, consistent with the findings of studies in other inflammatory conditions,12 13 which suggests that a substantial part of the methylation aberrations might be located in distal regulatory regions. In addition, DMPs, mainly hypermethylated DMPs, were mostly located in open sea regions, outside CpG island and surrounding areas (figure 1B).
Figure 1.
Results from the comparison of both DNA methylation and gene expression patterns of CD14+ monocytes between patients with giant cell arteritis and controls. (A) Volcano plot of the epigenome-wide association study results. False discovery rate (FDR) values are represented on the –log10 scale in the y-axis. Significant threshold (FDR<0.05) is marked by a dashed line. The effect size and direction obtained for each CpG site is depicted in the x-axis. Pink and blue dots represent hypermethylated and hypomethylated differentially methylated positions (DMPs), respectively. (B) Bar plots representing the annotation of the significant hypermethylated and hypomethylated DMPs in relation to CpG island (right panel) and gene location (left panel). (C) Representation of selected gene ontology categories obtained from the DMPs enrichment analysis using the GREAT online tool. (D) Volcano plot of the transcriptome-wide association study results. FDR values are represented on the –log10 scale in the y-axis. Significant threshold (FDR<0.05) is marked by a dashed line. The effect size and direction obtained for each gene is depicted in the x-axis. Purple and orange dots represent upregulated and downregulated differentially expressed genes, respectively. GCA, giant cell arteritis.
Over 85% of the DMPs showed higher DNA methylation levels in patients than in controls. Of note, we identified hypermethylated DMPs located within or close to genes previously associated with immune-mediated diseases, including P4HA2, a susceptibility genetic factor for GCA 14 (figure 1A and online supplemental table 1). Through gene ontology analysis, we observed enrichment in functional pathways of the immune response, such as regulation of interferon-gamma (IFN-γ) production, leucocyte chemotaxis and integrin biosynthesis processes. In addition, we detected a significant enrichment in monocyte cell biology pathways, such as the colony stimulating factor 1 (CSF1)-CSF1 receptor complex, differentiation and proliferation of macrophages and cytokine production like macrophage colony-stimulating factor (figure 1C and online supplemental table 2). The hypomethylated DMPs were also mapped to relevant genes in the context of the immune response, like TRIM24, PRDM16, PARP9, ADARB1, CD38 or MICB (figure 1A and online supplemental table 3), and were enriched in significant biological processes like cellular response to IL-4, oxidative stress response, positive regulation of chemotaxis, complement component C3a binding and negative regulation of the platelet-derived growth factor-beta receptor (PDGF) signalling pathway (figure 1C, online supplemental table 4).
annrheumdis-2022-222156supp002.xlsx (4MB, xlsx)
Global analysis of patients with GCA shows slight alterations of the gene expression profile
Afterwards, we carried out gene expression analysis between CD14+ monocytes from patients with GCA and controls. These results only revealed 54 differentially expressed genes (DEGs), of which 41 were upregulated in GCA patients (figure 1D, online supplemental tables 5-6). In this regard, ADAMTS2, CD163, AMPH, FLT3 and IL1R2 were observed to be among the most significantly upregulated DEGs.
Stratified analysis of patients based on clinical status and treatment display specific DNA methylation patterns in CD14+ monocytes
We further stratified patients with GCA according to the state of the disease at the time of sample collection: patients with active disease, patients in remission with treatment and patients in remission without treatment (detailed description in online supplemental material). To better characterise the methylation alterations driving the molecular mechanisms responsible of the active state of GCA, we compared the methylation landscape of patients with GCA with active disease with those showing no sign or symptoms of this vasculitis (healthy controls and patients in remission with and without GC treatment). In addition, we also aimed to evaluate the influence of GC on the DNA methylation patterns of CD14+ monocytes in disease remission by comparing treated and non-treated patients.
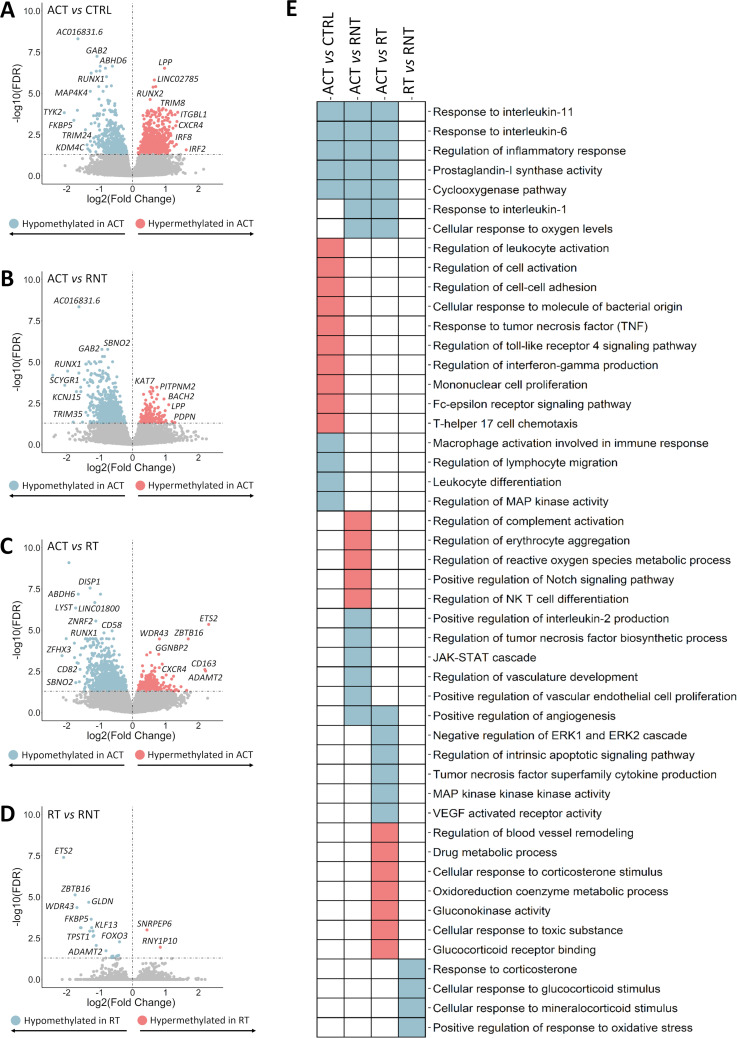
We observed a large number of significant DNA methylation alterations between patients with active disease compared with controls and those patients in remission. In contrast, only several CpGs were differentially methylated between patients in remission with and without treatment. A summary of the results obtained in each analysis is shown in figure 2A.
Figure 2.
Summary of the results obtained from the stratified analysis of patients according to the state of the disease. (A) Number of significant differentially methylated positions (DMPs) and differentially expressed genes (DEGs) obtained in each comparison. (B) Venn diagrams showing the overlap of significant DMPs (left panel) and DEGs (right panel) among the different comparisons performed. ACT, active disease; CRTL, controls; RNT, remission without treatment; RT, remission with treatment.
Global DNA methylation alterations reflect the active state of the disease
We first assessed the differences between the methylation status of CD14+ monocytes from patients with active disease and healthy controls, identifying a total of 1444 DMPs (507 hypomethylated and 937 hypermethylated in active patients) (figure 3A and online supplemental tables 7-8). Interestingly, some of these DMPs mapped to genes encoding chemokine receptors such as CX3CR1, CXCR2P1 and CXCR4, members of the interferon regulatory transcription factor (IRF) family, including IRF2 and IRF8, as well as genes previously described as susceptibility factors for systemic vasculitides, including TYK2 and KDM4C 15 16. In addition, hypermethylated DMPs were enriched in multiple pathways and biological processes, including positive regulation of cell activation and cell–cell adhesion and response to tumour necrosis factor (TNF) (figure 3E and online supplemental table 9). Regarding hypomethylated DMPs, a significant enrichment in interesting pathways in the context of GCA pathogenesis was detected, such as regulation of macrophage activation involved in immune response, positive regulation of lymphocyte migration and regulation of inflammatory response, among others (figure 3E and online supplemental table 10).
Figure 3.
Results of the epigenome-wide association study obtained from the stratified analysis of patients according to the state of the disease. (A–D) Volcano plots showing the results of the epigenome-wide association study for each comparison performed. False discovery rate (FDR) values are represented on the –log10 scale in the y-axis. Significant threshold (FDR<0.05) is marked by a dashed line. The effect size and direction obtained for each CpG site is depicted in the x-axis. Pink and blue dots represent hypermethylated and hypomethylated differentially methylated positions (DMPs), respectively. (E) Scheme summarising the results from the gene ontology enrichment analysis performed using the GREAT online tool. Columns show the different comparisons carried out in the stratified analysis and rows represent selected gene ontology categories. Pink colour denotes statistical significant enrichment of hypermethylated DMPs and blue colour indicates statistical significant enrichment of hypomethylated DMPs. ACT, active disease; CRTL, controls; RT, remission with treatment; RNT, remission without treatment.
Subsequently, we identified substantial alterations in the methylation patterns of monocytes from active disease compared with patients in remission without treatment. Specifically, we identified 820 DMPs (668 hypomethylated and 152 hypermethylated in active patients) (figure 3B and online supplemental tables 11 and 12). Noteworthy, a considerable part of these findings was similar to those obtained in the previous comparison (figure 2B). Aside from the common features, hypomethylated DMPs identified when compared active patients with patients in remission without treatment were enriched in additional biological pathways, such as regulation of reactive oxygen species metabolic processes, positive regulation of IL-2 production, negative regulation of TNF biosynthetic process and regulation of angiogenesis. Moreover, hypermethylated DMPs were enriched in regulatory processes of complement activation, positive regulation of erythrocyte aggregation, regulation of the metabolic process of reactive oxygen species (figure 3E and online supplemental tables 13 and 14).
As expected, CD14+ monocytes from patients with active disease showed large differences in the methylation landscape compared with those from patients in remission with treatment. We identified a total of 688 DMPs, of which 85% presented low levels of methylation in patients with active disease (figure 3C and online supplemental tables 15 and 16). These hypomethylated DMPs were enriched in pathways implicated in the immunopathogenic processes of GCA, including the cellular response to IL-6 as well as response to other members of the IL-6 family, specifically IL-11 (figure 3E and online supplemental table 17).17 18 In contrast, hypermethylated DMPs were enriched in pathways of drug metabolism, cellular response to GC stimulus and regulation of blood vessel remodelling (figure 3E and online supplemental table 18).
Analysis of the transcription factor (TF) binding motifs showed that the hypomethylated DMPs identified in the active patients compared with controls and patients in remission with and without treatment were enriched in the basic region-leucine zipper (bZIP) family, suggesting that factors in this family might play a key role in the regulation of the molecular mechanisms implicated in the active state of the disease (online supplemental figure 1). Particularly, the bZIP TF family has been reported to regulate the expression of genes involved in angiogenesis, fibrosis and Th17 cells plasticity control.19–22 Furthermore, IRF family, known to be involved in monocyte/macrophage polarisation,23 was significantly enriched in the cluster of hypermethylated DMPs in active patients in comparison with controls and patients in remission without treatment.
Remarkably, 65% from the total of DMPs observed in the comparisons between active patients and patients in remission with and without treatment were common (figure 2B). This similarity might be reflecting the success of the GC treatment.
GC treatment greatly affects methylation levels of glucocorticoid receptor target genes
We then compared patients in remission with and without treatment in order to assess the impact of the GC treatment in the DNA methylation landscape of CD14+ monocytes, which revealed 27 CpG sites showing different methylation levels between the two groups (figure 3D and online supplemental table 19). Remarkably, these DMPs, most of which were hypomethylated in patients in remission with treatment, showed high differences in their beta values, for example, ETS2 (ΔBeta=−0.39) and ZBTB16 (ΔBeta=−0.38). It should be mentioned that, despite the low number of DMPs, enrichment of biological processes such as response to corticosterone and cellular response to GC stimulus was significantly detected (figure 3E and online supplemental table 20). Furthermore, the analysis of the TF binding motifs revealed that the cluster of hypomethylated DMPs in patients in remission with GC treatment was enriched in GC response elements (online supplemental figure 1), a family of TF reported to repress the activity of the nuclear factor-kappa B (NF-kB) pathway.24 25
GCA remission leads to reversal of methylation changes
It should be noted that a significant part of the DMPs identified in the comparison between active patients and controls is common to those detected between active patients and patients in remission without treatment (figure 2B), suggesting that the DNA methylation landscapes of these two groups of individuals are similar. Consistently, no significant DMPs were observed between patients in remission without treatment and healthy controls. This evidence indicates that the DNA methylation alterations occurring in monocytes from individuals with active disease are reverted when the disease subsides.
Identification of aberrant gene expression profiles in CD14+ monocytes through stratified analysis of patients based on clinical status and treatment
Following the same reasoning aforementioned, and considering the few differences observed in the comparison between global GCA patients and healthy controls, we also performed a stratified analysis of the gene expression landscape in patients with GCA according to the state of the disease: patients with active disease, patients in remission with treatment and patients in remission without treatment (detailed description in online supplemental material).
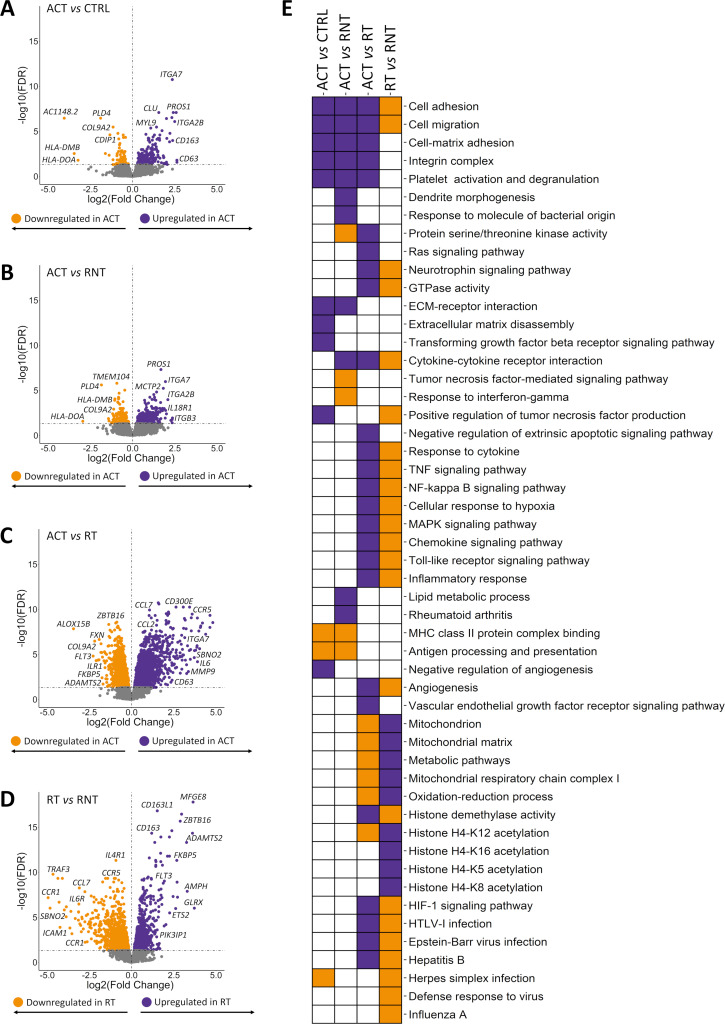
Figure 2A summarises the results obtained through each analysis. Our results revealed that the gene expression profile of CD14+ monocytes largely varies according to the clinical status and treatment. The validity of these results was supported by the high correlation observed for eight deregulated genes (ITGA7, CD63, CCRL2, CD300E, CD163, ETS2, FKBP5 and SBNO2) between ∆Ct values obtained by qPCR and their normalised intensities from the RNA-seq (Spearman rank, |R|=0.79) (online supplemental figure 2). Furthermore, as expected, we have confirmed the significant deregulation of these eight genes observed among subgroups (online supplemental table 21).
Gene expression deregulation in monocytes affects relevant molecular mechanisms during the active state of the disease
The comparison between the expression patterns of CD14+ monocytes from patients with GCA with active disease and controls revealed 292 DEGs (figure 4A and online supplemental tables 22 and 23). The majority (72%) of DEGs presented higher levels of expression in the subgroup of patients with active disease. Interestingly, overexpressed genes were found to be enriched in related biological processes such as cell adhesion, extracellular matrix disassembly, cell–matrix adhesion, integrin complex, among others. The enrichment in negative regulation of angiogenesis and the positive regulation of TNF production should also be mentioned (figure 4E and online supplemental table 24). Remarkably, the downregulated DEGs were enriched in the major histocompatibility complex (MHC) class II protein complex binding including genes like HLA-DMB and HLA-DOA (figure 4E and online supplemental table 25).
Figure 4.
Results of the transcriptome-wide association study obtained from the stratified analysis of patients according to the state of the disease. (A–D) Volcano plots showing the results of the transcriptome-wide association study for each comparison. False discovery rate (FDR) values are represented on the –log10 scale in the y-axis. Significant threshold (FDR<0.05) is marked by a dashed line. The effect size and direction obtained for each gene is depicted in the x-axis. Purple and orange dots represent upregulated and downregulated differentially expressed genes (DEGs), respectively. (E) Scheme summarising the results from the gene ontology enrichment analysis performed using the DAVID online tool. Columns show the different comparisons carried out in the stratified analysis and rows represent selected gene ontology categories. Purple colour denotes statistical significant enrichment of upregulated DEGs and orange colour indicates statistical significant enrichment of downregulated DEGs. ACT, active disease; CRTL, controls; RT, remission with treatment; RNT, remission without treatment.
Subsequently, we compared the gene expression patterns of CD14+ monocytes from active disease and patients in remission without treatment, identifying 551 DEGs (327 upregulated and 224 downregulated in active patients) (figure 4B and online supplemental tables 26 and 27). The cluster of 327 upregulated DEGs was enriched in pathways including platelet degranulation, cell–matrix adhesion, integrin-mediated signalling pathway, lipid metabolic process, rheumatoid arthritis and cytokine–cytokine receptor interaction (figure 4E and online supplemental table 28). Of special mention, HLA-DMB, HLA-DOA and HLA-DRA were underexpressed in active patients (online supplemental table 27). In this context, these results are in accordance with previous studies reporting a lower expression of HLA-DRA in monocytes in inflammatory conditions like sepsis and the immune dysregulation caused by SARS-CoV-2.26 27 Indeed, among the most significant results of the gene ontology analysis, we observed antigen processing and presentation and MHC class II protein complex binding. In addition, it is also worth highlighting the enrichment of important inflammatory pathways such as TNF-mediated and response to IFN-γ (figure 4E and online supplemental table 29).
Afterwards, we observed greater significant differences between the expression profiles of CD14+ monocytes from patients with GCA with active disease and patients in remission with treatment. 5433 DEGs were identified, of which 2701 and 2732 DEGs were upregulated and downregulated in the active disease group, respectively (figure 4C and online supplemental tables 30 and 31). These results were consistent with the previous knowledge of GCA pathogenesis. In this sense, IL-6 and MMP9, as well as other members of the MMP family (MMP2, MMP24, MMP14, MMP19 and MMP25), were upregulated in active patients. We also detected overexpression of several genes of the integrin family, such as ITGA2B, ITGA5, ITGA6, ITGA7, ITGAX, ITGAV, ITGB1, ITGB3, ITGB5, ITGB7 and ITGB8, as well as other remarkable genes that are important in monocyte cell biology like CCR2, CCL2, CCL7, CXCL5, CXCL2 and CXCL3 (online supplemental table 30).28 Of note, important biological processes and pathways involved in GCA pathogenesis such as angiogenesis, TNF signalling pathway, vascular endothelial growth factor (VEGF) receptor pathway, chemokine signalling, mitogen-activated protein kinase (MAPK) cascade, Toll-like receptor signalling (TLR) pathway and cellular response to IL-6, were enriched among the set of upregulated genes in patients with active disease (figure 4E and online supplemental table 32). Among the large number of downregulated DEGs, it is notable the presence of genes related with GC such as FKBP5, a cochaperone that modulates GC receptor (GR) activity, ZBTB16, transcriptional factor contributing to energy balance after GR activation and HPGD which encondes a dehydrogenase expressed on dexamethasone (online supplemental table 31).29–32 Accordingly, mitochondrial metabolic process characteristic of drug metabolism, specifically GC, were significantly enriched (figure 4E and online supplemental table 33). In addition, we also found enrichment in negative regulation of the type I interferon production and the innate response as well as metabolic pathways like oxidative phosphorylation or glucose metabolic process. Of special interest is the enrichment of the apoptotic process that included underexpression of crucial genes in the context of immune-mediated disorders like TNFAIP3, DNASE1, AIM2 and PTK2B, among others. We also detected downregulation of known autoimmunity-related genes, as examples: PLD4, FLT3, ERAP2, BTK, MEFV, DNASE1, PADI14, JAZF1 and GIMAP family members (GIMAP1, GIMAP2, GIMAP4, GIMAP6, GIMAP7 and GIMAP8) (online supplemental table 31).
GC treatment reshapes the gene expression landscape of circulating monocytes
The gene expression landscapes of patients in remission with and without treatment were largely different, with 3550 DEGs when comparing both subgroups (1818 upregulated and 1732 downregulated in treated patients) (figure 4D and online supplemental tables 34 and 35). As reflected in gene ontology analysis, GC treatment might be altering a large number of molecular mechanisms, especially those related with mitochondrial function. Interestingly, as similar to the previous comparison, we also observed enrichment in acetylation of several histones, which could indicate that other epigenetic mechanisms, besides DNA methylation, might be affected (figure 4E and online supplemental table 36). On another side, treated patients showed downregulation in multiple inflammatory-related pathways, apoptotic processes and, notably, defence response to viruses, including pathways such as Herpes simplex infection, Epstein-Barr virus infection, Hepatitis B and Influenza A, among others (figure 4E and online supplemental table 37). Unlike the similarity observed in DNA methylation patterns, the large differences identified in the gene expression patterns between treated and untreated patients are supported by previous evidence describing, in other immune contexts, that the GC treatment has an important effect in reshaping the gene expression landscape but relatively low impact in the DNA methylation profile.13
Aberrant gene expression profile of monocytes in the active state of the disease is lost after remission
Consistent to what we observed in the methylation analyses, the gene expression patterns of patients in remission without treatment and healthy controls were similar. Indeed, 208 DEGs identified in the comparisons of active patients with patients in remission without treatment and controls were common (figure 2B), and only one gene showed significantly different expression levels between patients in remission without treatment and healthy controls. Specifically, higher expression levels of MTCO3P12 were observed in patients in remission without treatment (FDR=0.04, logFC=1.51).
Integrative analysis revealed the existence of relationships between DNA methylation changes and gene expression alterations
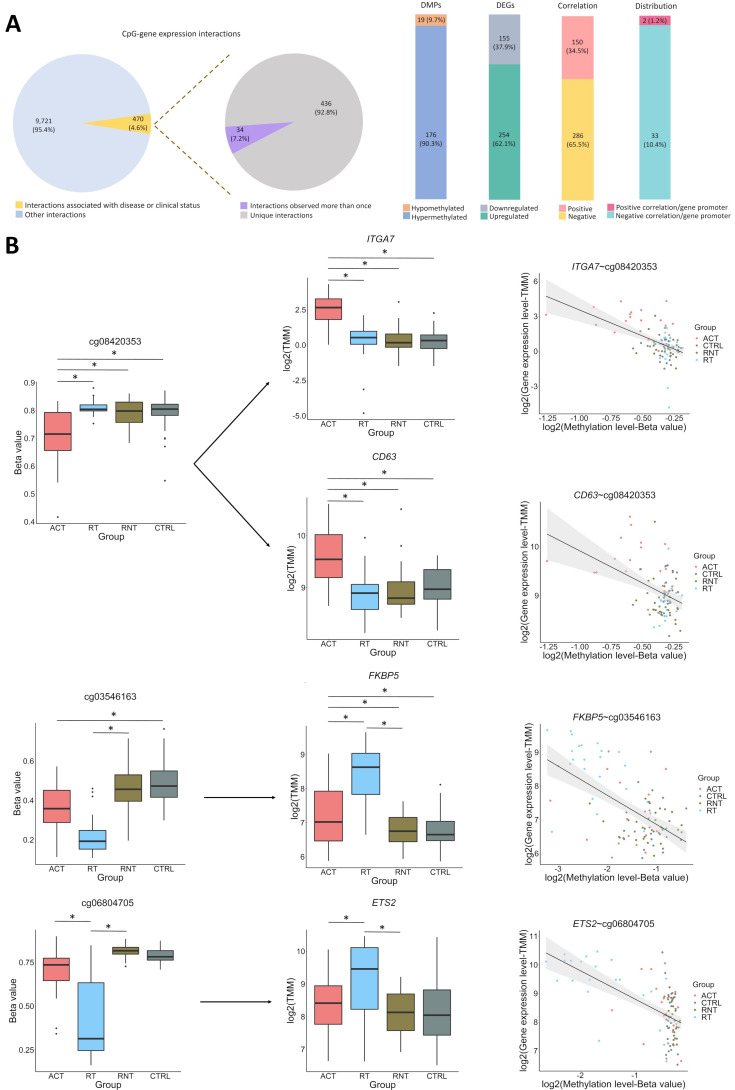
Finally, by performing an integrative analysis, we aimed to investigate the potential relationship between DNA methylation alterations and gene expression in GCA. We found 10 191 significant CpG-gene expression interactions (FDR<0.05). To focus on the interactions that might be potentially relevant for GCA pathogenesis, we selected 470 CpG-gene expression interactions showing both methylation and gene expression levels significantly associated in at least one of the comparisons performed. Of these, 34 CpG–gene expression interactions were significant in more than one comparison. Among the 436 unique interactions, we detected 409 DEGs (254 upregulated and 155 downregulated) and 195 DMPs (176 hypomethylated and 19 hypermethylated). In addition, we found that 65.53% and 34.47% of the total interactions associated with disease or clinical status were negative and positive correlations, respectively. Finally, when studying the distribution of CpGs in relation to the genes that they interact with, we identified that 10.39% of the negative and 1.23% of the positive correlations were located at the gene promoter (figure 5A and online supplemental table 38).
Figure 5.
Integrative analysis of DNA methylation and gene expression. (A) General description of the significant CpG-gene expression interactions identified in our analysis. (B) Selected examples of specific CpG-gene expression interactions. Box plots representing both the differentially methylated positions (DMPs) and the differentially expressed genes (DEGs). Significant differences are marked (*FDR<0.05). Graphical representation of the correlation of DNA methylation and gene expression of DMP-DEG pairs is also shown. DNA methylation and gene expression levels are illustrated in each subset of individuals. ACT, active disease; CRTL, controls; RT, remission with treatment; RNT, remission without treatment.
We detected interactions that involved relevant genes in the context of the pathophysiology of GCA. One example was ITGA7, upregulated in GCA patients with active disease, in which it showed three interactions with different CpGs located in intergenic regions, involving one positive correlation with cg24773560 (r=0.44, FDR=3.03E-02) and two negative correlations with cg08420353 (r=−0.51, FDR=1.85E-03) and cg17016513 (r=−0.41, FDR=4.31E-02). Interestingly, these last two CpGs also showed negative correlation with the CD63 gene (r=−0.41, FDR=4.35E-02 and r=−0.41, FDR=4.92E-02, respectively), which was also upregulated in active disease (figure 5B).
Correlation between DNA methylation and gene expression levels was also evident for several known GR target genes, all of which were upregulated in the subgroup of patients in remission with treatment. These genes include FKBP5, which correlated negatively with cg03546163 (r=−0.51, FDR=1.24E-03); ETS2, which showed a negative interaction with cg06804705 that was located in the promoter region of this gene (r=−0.68, FDR=8.12E-09); ADAMTS2, which correlated negatively with cg14727962 (r=0.57, FDR=7.52E-06) and cg09068128 (r=−0.60, FDR=1.54E-10), positively with cg00854503 (r=0.55, FDR=3.07E-04) and cg02052156 (r=0.51, FDR=1.63E-03); and ZBTB16, which correlated negatively with cg14388315 (r=−0.60, FDR=8.35E-05) and cg25345365 (r=−0.73, FDR=1.23E-11) (figure 5B).
Discussion
The results of the first methylome and transcriptome profiling of GCA monocytes have yielded evidence supporting that the observed widespread alterations are implicated in the molecular mechanisms underlying this disorder. We also found a significant number of genes whose dysregulation in GCA was mediated by an aberrant DNA methylation. In addition, the stratification of patients according to disease activity allowed us to obtain a clearer picture of the changes in both methylation and expression driving the molecular processes involved in disease activity and molecular response to GC treatment.
Monocytes from active patients seemed to have a more proinflammatory phenotype than controls and patients in remission. Supporting the reliability of our results, we observed a dysregulation of pathways involving cytokines and growth factors already known to have a key role in GCA, such as IL-6, TNF, IL-1, IL-4, IL-2, PDGF and VEGF.6 Interestingly, the response to IL-11 pathway was enriched among the DMPs hypomethylated in active patients with respect to controls and patients in remission with and without treatment. Although additional evidence is needed to establish the response to IL-11 as a new molecular mechanism involved in GCA, the potential role that might play in the active state of the disease is intriguing. IL11, a member of the IL-6 family, has been implicated in a range of disease pathologies by exerting diverse immunological roles.33 On the one hand, IL11 inhibits activated macrophages by blocking NF-κB translocation,34 however, several studies have described a proinflammatory function. For example, it has been reported that IL-11 promotes the differentiation of CD4+ T cells into Th17 cells, an important cell type in GCA pathogenesis,35 in multiple sclerosis, and it has been also implicated in angiogenesis in rheumatoid arthritis patients.36 Additionally, it has been reported that IL11 is involved in vascular smooth muscle cell phenotype switching, a mechanism that has been proposed to contribute to vascular remodelling in GCA.37 38 Therefore, further studies will be needed in order to determine the impact of this cytokine in GCA.
Interestingly, functional categories enriched among the set of differential methylated genes and DEGs in active patients suggest a relevant role of monocytes in GCA by recruiting immune cells and through their interaction with lymphocytes, mainly by promoting their differentiation and activation. In this sense, a significant number of genes encoding chemokines, such as CCL2 and CCL7, involved in the recruitment of monocytes, were overexpressed in active patients.39 Furthermore, genes encoding several integrins were also overexpressed. Specifically, the expression level of ITGA7 showed the greatest differences between active patients and healthy controls. Integrins are essential in a wide variety of biological processes, including migration, proliferation, cytokine production and activation, apoptosis and angiogenesis,40 all of which appear to be dysregulated in active patients. Additionally, hypomethylation of both ITGA7 and CCRL2, a chemokine receptor involved in both innate and adaptive immune responses and known to be upregulated in activated cells,41 correlated with higher gene expression levels in active patients. Interestingly, two of the CpGs affecting ITGA7 expression also correlated with overexpression of CD63. Notably, CD63 encodes a tetraspanin family member that interacts with integrins, being crucial for the fusion of monocytes to form multinucleated giant cells, which is the hallmark cell type of GCA.42–45 Finally, CD300E, an immune-activating receptor that promotes the expression of activation markers and the production of proinflammatory cytokines and reactive oxygen species in monocytes as well as the survival of this cell type,46 47 was also among the most significantly upregulated genes in active patients. Interestingly, expression of this gene was proposed to be restricted to CD115+Ly-6Clow/int peripheral blood monocytes in mice, which corresponds to human non-classical (CD14dimCD16+) and intermediate (CD14brightCD16+) monocytes.48 Accordingly, increased levels of CD16+ monocytes have been found in temporal artery biopsies of patients with GCA. This subset of monocytes is characterised to be more proinflammatory than the classical one (CD14brightCD16neg) and shows a higher capacity to adhere to endothelial cells via CX3CR1,7 which, notably, also appeared to be hypermethylated in active patients with respect to controls. Taken together, these results could indicate that, similarly to what has been described in GCA biopsies, increased levels of CD16+ monocytes could be present in peripheral blood of active patients.
It should be noted that similar results were found when active patients were compared with both healthy controls and patients in remission, with and without treatment, which suggests that the proinflammatory methylation and expression profiles observed in the active disease are lost during remission. In fact, no differences were found when DNA methylation and gene expression levels were compared between patients in remission without treatment and healthy controls.
Furthermore, our results suggest that GC therapy remodel the epigenome and, more robustly, the transcriptome, resulting in downregulation of genes involved in pathways with a relevant role in GCA pathogenesis, including cell migration and proliferation, apoptosis, angiogenesis, NF-κB, TNF, IFNγ and TLR signalling pathways and positive regulation of cytokines, such as IL-6 and IL-2. Accordingly, several known target genes for GR that are involved in controlling inflammation, such as ETS2, ZBTB16, FKBP5, and ADAMTS2, 49–51 appeared among the most upregulated genes in patients in remission with treatment. Notably, expression levels of these four genes negatively correlated with methylation levels, suggesting that GCs modify gene expression levels through DNA demethylation of target genes.
Interestingly, two scavenger receptors, CD163 and CD163L1, were the most significant overexpressed genes in the subgroup of patients in remission with treatment compared with non-treated patients. CD163 is considered a phenotypic marker of monocytes with anti-inflammatory potential. Specifically, this receptor binds haemoglobin–haptoglobin complexes triggering endocitosis and activating a signalling cascade that results in the production of anti-inflammatory molecules, such as IL-10,52 another gene upregulated in patients in remission with treatment. Additionally, CD163L1 has been described to have a role in resolution of inflammation.53 It should be also noted that, conversely, both CD163 and CD163L1 presented higher levels of methylation in active patients compared with patients in remission with treatment that correlated with a decreased gene expression, thus supporting the anti-inflammatory role of these molecules. Remarkably, CD163 has been found to be significantly increased in temporal artery biopsies from patients with GCA treated with GC.54
It has been described that GC can act on naïve monocytes inducing monocytes with an anti-inflammatory profile that may suppress T cell activation, present an increased phagocytic capacity and release anti-inflammatory mediators.55 Consistently, our results indicate that, in addition to its role promoting the expression of GR-target genes involved in the suppression of inflammation, GC treatment may also promote the expansion of monocytes with an anti-inflammatory phenotype in GCA. In addition, the large differences observed between patients in remission with and without GC treatment suggest that the alterations derived from GC therapy could be reversed in the absence of treatment.
Besides contributing to the elucidation of the pathogenic mechanisms involved in GCA, our study has revealed the existence of specific methylation and transcription profiles in active and GC-treated patients that could potentially improve the clinical care of this vasculitis. In this regard, evaluation of the molecular pattern of GCA monocytes could be especially relevant for early and differential diagnosis as well as for therapy monitoring, thus avoiding a delay in treatment and the use of ineffective drugs. Moreover, we have provided a significant number of molecules that could be targeted in future functional studies and potentially used as biomarkers. For example, CD163, which is upregulated after GC treatment in circulating monocytes and temporal artery biopsies, represents an interesting candidate to assess the molecular response to this therapy.
In conclusion, we have performed an exhaustive analysis of the methylome and transcriptome of one of the most relevant cell types in GCA, exploring for the first time the contribution of epigenetic to the disease activity and molecular response to GC in a large cohort of patients. Nevertheless, despite our relevant findings, evaluation of methylation and transcription profiles in additional peripheral cell types and, specially, in temporal artery biopsies will be essential to obtain a clearer picture of the molecular network involved in this type of vasculitis.
Acknowledgments
We thank all the patients and control donors who participated in this study and Sofia Vargas and Gema Robledo for her excellent technical assistance. This research is part of the doctoral degree awarded by E.E.M, within the Biomedicine programme from the University de Granada entitled 'Estudio de las causas moleculares de la arteritis de células gigantes mediante una aproximación sistémica'.
Footnotes
Handling editor: Josef S Smolen
EE-M and LO-F contributed equally.
Contributors: EE-M and LO-F: data analysis, interpretation of the results and manuscript drafting. EA-L and LCT-C: data analysis. TL, LC, GE-F and SP-G.: sample and data collection. JH-R and MCC: study design, sample and data collection. AM, EB and JM: study design, interpretation of the results and manuscript drafting. LO-F and JM: guarantors of the study. All authors read and approved the manuscript.
Funding: This work was supported by the HELICAL Innovative Training Network, a European Commission funded project under the Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement number 813 545 and, by the Cooperative Research Thematic Network programme (RD16/0012/0013). AM is recipient of a Miguel Servet fellowship (CP17/00008) from ISCIII (Spanish Ministry of Economy, Industry and Competitiveness). LO-F was supported by Juan de la Cierva Incorporación fellowship (IJC2019-040746-I) funded by MCIN/AEI /10.13039/501100011033.
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. DNA methylation and expression data for this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO SuperSeries accession number GSE201754. The code used for the analyses is available from https://github.com/lterroncamero/GCA_CD14_Analysis.git.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
The study was approved by the ethical committees of all institutions involved in this study. However, these institutions do not provide a reference number or ID for the ethics approval. Participants gave informed consent to participate in the study before taking part.
References
- 1. Jennette JC, Falk RJ, Bacon PA, et al. 2012 revised international chapel Hill consensus conference Nomenclature of vasculitides. Arthritis & Rheumatism 2013;65:1–11. 10.1002/art.37715 [DOI] [PubMed] [Google Scholar]
- 2. Gonzalez-Gay MA, Vazquez-Rodriguez TR, Lopez-Diaz MJ, et al. Epidemiology of giant cell arteritis and polymyalgia rheumatica. Arthritis Rheum 2009;61:1454–61. 10.1002/art.24459 [DOI] [PubMed] [Google Scholar]
- 3. Koster MJ, Matteson EL, Warrington KJ. Large-Vessel giant cell arteritis: diagnosis, monitoring and management. Rheumatology 2018;57:ii32–42. 10.1093/rheumatology/kex424 [DOI] [PubMed] [Google Scholar]
- 4. Weyand CM, Goronzy JJ. Immune mechanisms in medium and large-vessel vasculitis. Nat Rev Rheumatol 2013;9:731–40. 10.1038/nrrheum.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Soriano A, Muratore F, Pipitone N, et al. Visual loss and other cranial ischaemic complications in giant cell arteritis. Nat Rev Rheumatol 2017;13:476–84. 10.1038/nrrheum.2017.98 [DOI] [PubMed] [Google Scholar]
- 6. Terrades-Garcia N, Cid MC. Pathogenesis of giant-cell arteritis: how targeted therapies are influencing our understanding of the mechanisms involved. Rheumatology 2018;57:ii51–62. 10.1093/rheumatology/kex423 [DOI] [PubMed] [Google Scholar]
- 7. van Sleen Y, Wang Q, van der Geest KSM, et al. Involvement of monocyte subsets in the immunopathology of giant cell arteritis. Sci Rep 2017;7:6553. 10.1038/s41598-017-06826-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foell D, Hernández-Rodríguez J, Sánchez M, et al. Early recruitment of phagocytes contributes to the vascular inflammation of giant cell arteritis. J Pathol 2004;204:311–6. 10.1002/path.1660 [DOI] [PubMed] [Google Scholar]
- 9. Cid MC, Cebrián M, Font C, et al. Cell adhesion molecules in the development of inflammatory infiltrates in giant cell arteritis: inflammation-induced angiogenesis as the preferential site of leukocyte-endothelial cell interactions. Arthritis Rheum 2000;43:184–94. [DOI] [PubMed] [Google Scholar]
- 10. Watanabe R, Maeda T, Zhang H, et al. MMP (Matrix Metalloprotease)-9-Producing Monocytes Enable T Cells to Invade the Vessel Wall and Cause Vasculitis. Circ Res 2018;123:700–15. 10.1161/CIRCRESAHA.118.313206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holliday R. Epigenetics: a historical overview. Epigenetics 2006;1:76–80. 10.4161/epi.1.2.2762 [DOI] [PubMed] [Google Scholar]
- 12. McDermott E, Ryan EJ, Tosetto M, et al. Dna methylation profiling in inflammatory bowel disease provides new insights into disease pathogenesis. J Crohns Colitis 2016;10:77–86. 10.1093/ecco-jcc/jjv176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Morante-Palacios O, Ciudad L, Micheroli R, et al. Coordinated glucocorticoid receptor and MafB action induces tolerogenesis and epigenome remodeling in dendritic cells. Nucleic Acids Res 2022;50:108–26. 10.1093/nar/gkab1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carmona FD, Vaglio A, Mackie SL, et al. A genome-wide association study identifies risk alleles in plasminogen and P4HA2 associated with giant cell arteritis. Am J Hum Genet 2017;100:64–74. 10.1016/j.ajhg.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ortiz-Fernández L, López-Mejias R, Carmona FD, et al. The role of a functional variant of Tyk2 in vasculitides and infections. Clin Exp Rheumatol 2020;38:949–55. [PubMed] [Google Scholar]
- 16. Ortiz-Fernández L, Carmona FD, López-Mejías R, et al. Cross-phenotype analysis of Immunochip data identifies KDM4C as a relevant locus for the development of systemic vasculitis. Ann Rheum Dis 2018;77:589–95. 10.1136/annrheumdis-2017-212372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernández-Rodríguez J, Segarra M, Vilardell C, et al. Tissue production of pro-inflammatory cytokines (IL-1beta, TNFalpha and IL-6) correlates with the intensity of the systemic inflammatory response and with corticosteroid requirements in giant-cell arteritis. Rheumatology 2004;43:294–301. 10.1093/rheumatology/keh058 [DOI] [PubMed] [Google Scholar]
- 18. Cook SA, Schafer S. Hiding in plain sight: interleukin-11 emerges as a master regulator of fibrosis, tissue integrity, and stromal inflammation. Annu Rev Med 2020;71:263–76. 10.1146/annurev-med-041818-011649 [DOI] [PubMed] [Google Scholar]
- 19. Hamik A, Wang B, Jain MK. Transcriptional regulators of angiogenesis. Arterioscler Thromb Vasc Biol 2006;26:1936–47. 10.1161/01.ATV.0000232542.42968.e3 [DOI] [PubMed] [Google Scholar]
- 20. Rajasekaran S, Vaz M, Reddy SP. Fra-1/Ap-1 transcription factor negatively regulates pulmonary fibrosis in vivo. PLoS One 2012;7:e41611. 10.1371/journal.pone.0041611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chow FY, Nikolic-Paterson DJ, Ozols E, et al. Monocyte chemoattractant protein-1 promotes the development of diabetic renal injury in streptozotocin-treated mice. Kidney Int 2006;69:73–80. 10.1038/sj.ki.5000014 [DOI] [PubMed] [Google Scholar]
- 22. Renoux F, Stellato M, Haftmann C, et al. The AP1 transcription factor Fosl2 promotes systemic autoimmunity and inflammation by repressing Treg development. Cell Rep 2020;31:107826. 10.1016/j.celrep.2020.107826 [DOI] [PubMed] [Google Scholar]
- 23. Lehtonen A, Veckman V, Nikula T, et al. Differential expression of IFN regulatory factor 4 gene in human monocyte-derived dendritic cells and macrophages. J Immunol 2005;175:6570–9. 10.4049/jimmunol.175.10.6570 [DOI] [PubMed] [Google Scholar]
- 24. Hudson WH, Vera IMSde, Nwachukwu JC, et al. Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements. Nat Commun 2018;9:1337. 10.1038/s41467-018-03780-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xavier AM, Anunciato AKO, Rosenstock TR, et al. Gene expression control by glucocorticoid receptors during innate immune responses. Front Endocrinol 2016;7:31. 10.3389/fendo.2016.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giamarellos-Bourboulis EJ, Netea MG, Rovina N, et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe 2020;27:992–1000. 10.1016/j.chom.2020.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lukaszewicz A-C, Grienay M, Resche-Rigon M, et al. Monocytic HLA-DR expression in intensive care patients: interest for prognosis and secondary infection prediction. Crit Care Med 2009;37:2746–52. 10.1097/CCM.0b013e3181ab858a [DOI] [PubMed] [Google Scholar]
- 28. Gren ST, Rasmussen TB, Janciauskiene S, et al. A single-cell gene-expression profile reveals Inter-Cellular heterogeneity within human monocyte subsets. PLoS One 2015;10:e0144351. 10.1371/journal.pone.0144351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kokkinopoulou I, Moutsatsou P. Mitochondrial glucocorticoid receptors and their actions. Int J Mol Sci 2021;22:6054. 10.3390/ijms22116054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zannas AS, Wiechmann T, Gassen NC, et al. Gene-Stress-Epigenetic regulation of FKBP5: clinical and translational implications. Neuropsychopharmacology 2016;41:261–74. 10.1038/npp.2015.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng H, Pablico SJ, Lee J, et al. Zinc finger transcription factor Zbtb16 coordinates the response to energy deficit in the mouse hypothalamus. Front Neurosci 2020;14:592947. 10.3389/fnins.2020.592947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tong M, Tai H-H. 15-Hydroxyprostaglandin dehydrogenase can be induced by dexamethasone and other glucocorticoids at the therapeutic level in A549 human lung adenocarcinoma cells. Arch Biochem Biophys 2005;435:50–5. 10.1016/j.abb.2004.11.031 [DOI] [PubMed] [Google Scholar]
- 33. Fung KY, Louis C, Metcalfe RD, et al. Emerging roles for IL-11 in inflammatory diseases. Cytokine 2022;149:155750. 10.1016/j.cyto.2021.155750 [DOI] [PubMed] [Google Scholar]
- 34. Schwertschlag US, Trepicchio WL, Dykstra KH, et al. Hematopoietic, immunomodulatory and epithelial effects of interleukin-11. Leukemia 1999;13:1307–15. 10.1038/sj.leu.2401514 [DOI] [PubMed] [Google Scholar]
- 35. Zhang X, Tao Y, Chopra M, et al. Il-11 induces Th17 cell responses in patients with early relapsing-remitting multiple sclerosis. J Immunol 2015;194:5139–49. 10.4049/jimmunol.1401680 [DOI] [PubMed] [Google Scholar]
- 36. Elshabrawy HA, Volin MV, Essani AB, et al. Il-11 facilitates a novel connection between RA joint fibroblasts and endothelial cells. Angiogenesis 2018;21:215–28. 10.1007/s10456-017-9589-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim W-W, Corden B, Ng B, et al. Interleukin-11 is important for vascular smooth muscle phenotypic switching and aortic inflammation, fibrosis and remodeling in mouse models. Sci Rep 2020;10:17853. 10.1038/s41598-020-74944-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Planas-Rigol E, Terrades-Garcia N, Corbera-Bellalta M, et al. Endothelin-1 promotes vascular smooth muscle cell migration across the artery wall: a mechanism contributing to vascular remodelling and intimal hyperplasia in giant-cell arteritis. Ann Rheum Dis 2017;76:1624–34. 10.1136/annrheumdis-2016-210792 [DOI] [PubMed] [Google Scholar]
- 39. Tsou C-L, Peters W, Si Y, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest 2007;117:902–9. 10.1172/JCI29919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mezu-Ndubuisi OJ, Maheshwari A. The role of integrins in inflammation and angiogenesis. Pediatr Res 2021;89:1619–26. 10.1038/s41390-020-01177-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Del Prete A, Bonecchi R, Vecchi A, et al. CCRL2, a fringe member of the atypical chemoattractant receptor family. Eur J Immunol 2013;43:1418–22. 10.1002/eji.201243179 [DOI] [PubMed] [Google Scholar]
- 42. Parthasarathy V, Martin F, Higginbottom A, et al. Distinct roles for tetraspanins CD9, CD63 and CD81 in the formation of multinucleated giant cells. Immunology 2009;127:237–48. 10.1111/j.1365-2567.2008.02945.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tugues S, Honjo S, König C, et al. Tetraspanin CD63 promotes vascular endothelial growth factor receptor 2-β1 integrin complex formation, thereby regulating activation and downstream signaling in endothelial cells in vitro and in vivo. J Biol Chem 2013;288:19060–71. 10.1074/jbc.M113.468199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mantegazza AR, Barrio María Marcela, Moutel S, et al. CD63 tetraspanin slows down cell migration and translocates to the endosomal-lysosomal-MIICs route after extracellular stimuli in human immature dendritic cells. Blood 2004;104:1183–90. 10.1182/blood-2004-01-0104 [DOI] [PubMed] [Google Scholar]
- 45. Pfistershammer K, Majdic O, Stöckl J, et al. CD63 as an activation-linked T cell costimulatory element. J Immunol 2004;173:6000–8. 10.4049/jimmunol.173.10.6000 [DOI] [PubMed] [Google Scholar]
- 46. Borrego F. The CD300 molecules: an emerging family of regulators of the immune system. Blood 2013;121:1951–60. 10.1182/blood-2012-09-435057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brckalo T, Calzetti F, Pérez-Cabezas B, et al. Functional analysis of the CD300e receptor in human monocytes and myeloid dendritic cells. Eur J Immunol 2010;40:722–32. 10.1002/eji.200939468 [DOI] [PubMed] [Google Scholar]
- 48. Isobe M, Izawa K, Sugiuchi M, et al. The CD300e molecule in mice is an immune-activating receptor. J Biol Chem 2018;293:3793–805. 10.1074/jbc.RA117.000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mullick J, Anandatheerthavarada HK, Amuthan G, et al. Physical interaction and functional synergy between glucocorticoid receptor and Ets2 proteins for transcription activation of the rat cytochrome P-450c27 promoter. J Biol Chem 2001;276:18007–17. 10.1074/jbc.M100671200 [DOI] [PubMed] [Google Scholar]
- 50. Zannas AS, Jia M, Hafner K, et al. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB–driven inflammation and cardiovascular risk. Proc Natl Acad Sci U S A 2019;116:11370–9. 10.1073/pnas.1816847116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hofer TPJ, Frankenberger M, Mages J, et al. Tissue-Specific induction of ADAMTS2 in monocytes and macrophages by glucocorticoids. J Mol Med 2008;86:323–32. 10.1007/s00109-007-0284-0 [DOI] [PubMed] [Google Scholar]
- 52. Philippidis P, Mason JC, Evans BJ, et al. Hemoglobin scavenger receptor CD163 mediates interleukin-10 release and heme oxygenase-1 synthesis: antiinflammatory monocyte-macrophage responses in vitro, in resolving skin blisters in vivo, and after cardiopulmonary bypass surgery. Circ Res 2004;94:119–26. 10.1161/01.RES.0000109414.78907.F9 [DOI] [PubMed] [Google Scholar]
- 53. Moeller JB, Nielsen MJ, Reichhardt MP, et al. CD163-L1 is an endocytic macrophage protein strongly regulated by mediators in the inflammatory response. J Immunol 2012;188:2399–409. 10.4049/jimmunol.1103150 [DOI] [PubMed] [Google Scholar]
- 54. Wagner AD, Wittkop U, Thalmann J, et al. Glucocorticoid effects on tissue residing immune cells in giant cell arteritis: importance of GM-CSF. Front Med 2021;8:709404. 10.3389/fmed.2021.709404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ehrchen JM, Roth J, Barczyk-Kahlert K. More than suppression: glucocorticoid action on monocytes and macrophages. Front Immunol 2019;10:2028. 10.3389/fimmu.2019.02028 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
annrheumdis-2022-222156supp001.pdf (1.5MB, pdf)
annrheumdis-2022-222156supp002.xlsx (4MB, xlsx)
Data Availability Statement
Data are available in a public, open access repository. DNA methylation and expression data for this publication have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO SuperSeries accession number GSE201754. The code used for the analyses is available from https://github.com/lterroncamero/GCA_CD14_Analysis.git.