ABSTRACT
Radical S-adenosylmethionine (SAM) enzymes catalyze an impressive variety of difficult biochemical reactions in various pathways across all domains of life. These metalloenzymes employ a reduced [4Fe-4S] cluster and SAM to generate a highly reactive 5′-deoxyadenosyl radical that is capable of initiating catalysis on otherwise unreactive substrates. Interestingly, the genomes of methanogenic archaea encode many unique radical SAM enzymes with underexplored or completely unknown functions. These organisms are responsible for the yearly production of nearly 1 billion tons of methane, a potent greenhouse gas as well as a valuable energy source. Thus, understanding the details of methanogenic metabolism and elucidating the functions of essential enzymes in these organisms can provide insights into strategies to decrease greenhouse gas emissions as well as inform advances in bioenergy production processes. This minireview provides an overview of the current state of the field regarding the functions of radical SAM enzymes in methanogens and discusses gaps in knowledge that should be addressed.
KEYWORDS: methanogens, radical SAM, archaea, iron-sulfur cluster
INTRODUCTION
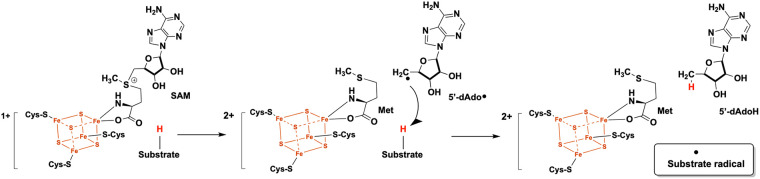
Radical S-adenosylmethionine (SAM) enzymes comprise one of the largest known enzyme superfamilies, with over 700,000 estimated members (1). These enzymes use SAM and a [4Fe-4S] cluster to catalyze complex reactions on inert substrates in various biochemical pathways, including cofactor biosynthesis, secondary metabolite biosynthesis, and nucleic acid modification (2, 3). Radical SAM enzymes are generally defined by the presence of a CX3CX2C motif that coordinates a [4Fe-4S] cluster, with the fourth iron in the cluster being ligated to the amino and carboxylate groups of SAM (Fig. 1) (4, 5). Although the majority of radical SAM enzymes (>90%) contain the canonical CX3CX2C motif, several noncanonical cysteine motifs have also been described in enzymes carrying out radical SAM chemistry (6). In a typical radical SAM reaction, the reduced [4Fe-4S]1+ cluster donates an electron to the sulfonium group of SAM, resulting in homolytic bond cleavage to generate methionine and a highly reactive 5′-deoxyadenosyl radical (5′-dAdo·) (3, 7, 8). The 5′-dAdo· abstracts a hydrogen atom from an otherwise unreactive site to generate 5′-deoxyadenosine (5′-dAdoH) and a substrate radical that can undergo further chemistry (Fig. 1). Established reactions catalyzed by radical SAM enzymes include radical generation on a protein substrate, isomerization, cyclization, carbon backbone rearrangement, sulfur insertion, and methylation (3).
FIG 1.
Common steps in canonical radical SAM enzyme catalysis.
Most radical SAM enzymes are found encoded in bacterial genomes, with the next largest proportion coming from archaea (6). Eukaryotes generally have the fewest radical SAM superfamily members. For example, the human genome encodes eight radical SAM enzymes, all of which have established functions (9–11). Given the extensive suite of natural products that are produced by different bacterial organisms, many radical SAM enzymes in various bacteria are involved in secondary metabolite biosynthesis. Conversely, although secondary metabolites and their biosynthetic pathways have been identified in some archaea (12, 13), most archaeal organisms do not exhibit the potential for robust secondary metabolism, and thus, their radical SAM enzymes are more likely to be involved in essential primary metabolism.
Methanogenic archaea (“methanogens”) are diverse and widespread microbes that are strictly dependent upon methanogenesis, a form of anaerobic respiration that reduces simple carbon compounds to generate methane as an end product. These organisms produce nearly 1 billion tons of methane each year, accounting for about 70% of global methane emissions (14, 15). Given the role of methane as a potent greenhouse gas and as a potential renewable energy source (16, 17), investigating essential enzymes in methanogens could lead to the development of methane mitigation strategies and could have implications for bioenergy applications. Interestingly, methanogens contain an abundance of putative radical SAM enzymes compared to other archaea, eukaryotes, and many bacteria. However, currently, only about half of the putative radical SAM enzymes in methanogens have established or likely functions based on experimental evidence or high sequence similarity to well-characterized radical SAM enzymes (see Tables S1 and S2 in the supplemental material for lists of putative radical SAM enzymes in two model methanogens, Methanococcus maripaludis S2 and Methanosarcina acetivorans C2A). Thus, these organisms represent an underexplored domain for uncovering new functions and mechanisms of radical SAM enzymes.
Some of the radical SAM enzymes with known functions in methanogens are well-characterized and widespread enzymes that are additionally found in bacteria and/or eukaryotes, while others catalyze less common reactions that are unique to only methanogens or a smaller subset of organisms including methanogens (Table 1). This minireview focuses on the latter group of methanogenic radical SAM enzymes to summarize the current state of the field regarding the diverse functions of radical SAM enzymes in methanogens, especially highlighting recent exciting work in this area as well as discussing remaining questions that require further research.
TABLE 1.
Summary of the known functions of radical SAM enzymes in methanogens and their distributions in different organismsa
| Radical SAM enzyme | Function(s) | Distribution(s) | Reference(s) |
|---|---|---|---|
| Tes | Tetraether lipid biosynthesis | Mostly archaea, some bacteria | 22 |
| PylB | Pyrrolysine biosynthesis | Methanosarcinales methanogens | 32 |
| MJ0619 | Methylase in H4MPT biosynthesis | Methanogens and some bacteria | 37 |
| Mmp10 | 5-(S)-Methylarginine synthesis (posttranslational modification of MCR) | Methanogens | 48, 50 |
| QCMT | 2-(S)-Methylglutamine synthesis (posttranslational modification of MCR) | Some methanogens | 47 |
| AhbC | Alternative heme biosynthesis, removal of 2 acetate side chains | Archaea and some bacteria | 58 |
| AhbD | Alternative heme biosynthesis, heme b synthase—conversion of 2 propionate side chains to vinyl groups | Archaea and some bacteria | 58, 62 |
| CofG/CofH | F0 synthase; F420 biosynthesis | Methanogens and some bacteria | 71 |
| NifB | Nitrogenase cofactor biosynthesis | Some archaea and some bacteria | 74, 79 |
| HcgA | [Fe]-hydrogenase cofactor biosynthesis | Hydrogenotrophic methanogens | 89, 91 |
| RaSEA | Archaeosine biosynthesis | Some archaea | 102 |
| Tyw1 (Taw1) | 4-Demethylwyosine synthase; tRNA modification | Archaea and eukaryotes | 108, 110 |
| MtaB | ms2t6A tRNA modification | Widespread distribution in all domains of life | 118 |
| Elp3 | cm5U tRNA modification | Ubiquitous in archaea and eukaryotes, some bacteria | 121, 123 |
| QueE | Biosynthesis of preQ0, precursor of tRNA modifications—queuosine and archaeosine | Ubiquitous in all domains of life | 99 |
| BioB | Biotin biosynthesis | Widespread in all domains of life | 126 |
| ThiC | Thiamine pyrophosphate biosynthesis | Widespread in all domains of life | 127 |
| MoaA | Molybdopterin biosynthesis | Widespread in all domains of life | 128 |
| PFL-AE | Activating enzyme for pyruvate formate lyase | Widespread in archaea and bacteria | 3 |
| RNR-AE | Activating enzyme for anaerobic ribonucleotide reductase | Widespread in archaea and bacteria | 3 |
| KAM | Nε-Acetyl-β-lysine biosynthesis for salt tolerance; l-lysine catabolism | Most methanogens and many bacteria | 7, 129 |
| EAM | β-Glutamate biosynthesis for salt tolerance | Some methanogens and some bacteria | 130 |
| AtsB | Formylglycine generation during sulfatase maturation | Sporadic distribution in all domains of life | 3 |
| Dph2 | Diphthamide biosynthesis (posttranslational modification of Elp2) | Widespread in archaea and eukaryotes | 131 |
QCMT, glutamine C-methyltransferase; PFL-AE, pyruvate formate lyase-activating enzyme; RNR-AE, ribonucleotide reducatse-activating enzyme; KAM, lysine 2,3-aminomutase; EAM, glutamate 2,3-aminomutase.
TETRAETHER LIPID BIOSYNTHESIS
One of the defining features of archaea that distinguishes them from bacteria and eukaryotes is the unusual chemistry of their membrane lipids. As opposed to membranes composed of fatty acids with ester linkages to glycerol backbones, archaea contain isoprenoidal membranes with ether linkages to glycerol backbones. There are two major types of isoprenoid ether lipids: glycerol diphytanyl diethers (archaeol) and glycerol dibiphytanyl glycerol tetraethers (GDGTs) (Fig. 2A) (18). Similar to traditional phospholipid-based membranes, archaeol lipids form a bilayer membrane structure, whereas GDGTs, which consist of two diether lipids fused at their hydrophobic ends (Fig. 2A), result in a membrane-spanning structure to generate monolayer membranes. Different archaea have various proportions of archaeol and GDGTs in their membranes, where the latter are especially prevalent in halophilic and thermophilic organisms (18, 19). The higher rigidity and lower permeability of GDGT lipids likely provide an advantage in extreme environments. Supporting this idea, growth experiments have shown that archaea increase their GDGT/archaeol ratio when cultured at elevated temperatures (20, 21). In some nonmethanogenic archaea, GDGTs are further modified through the introduction of cyclopentane or cyclohexane rings at the C-7 and C-3 positions of each chain (18). Additional GDGT modifications include the addition of hydroxyl or methyl moieties, alteration of the polar head groups, and cross-linking of the two biphytanyl chains. These modifications influence the physicochemical properties of the lipids and thus serve as a mechanism for adaptation to various environmental conditions (18).
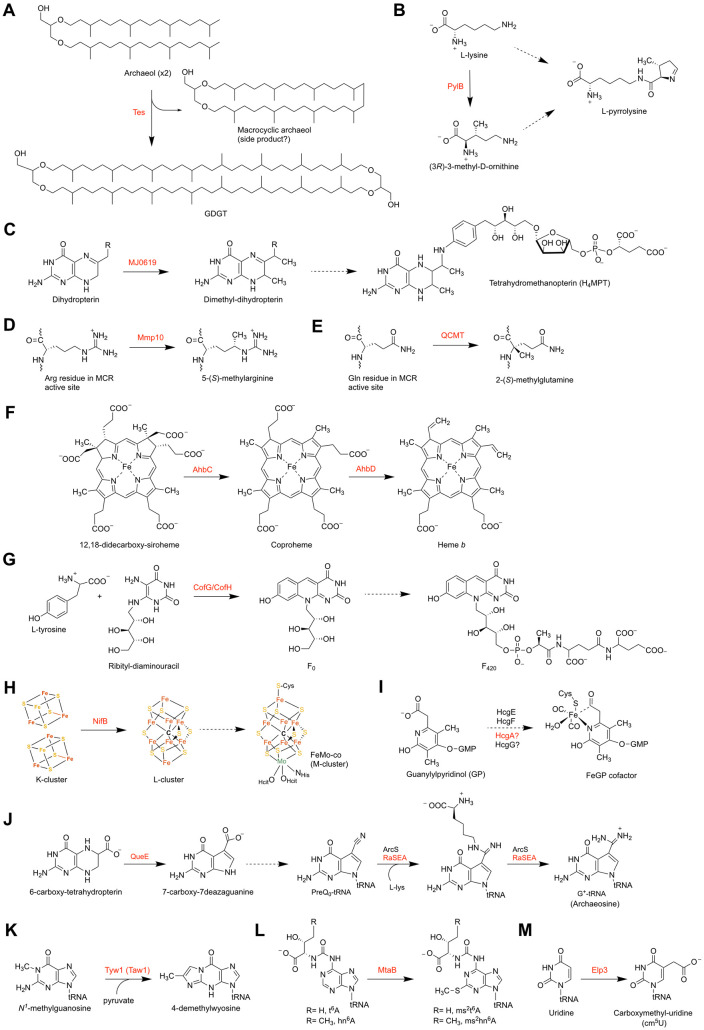
FIG 2.
Summary of reactions catalyzed by methanogenic radical SAM enzymes discussed in this minireview. (A) Proposed reaction catalyzed by tetraether synthase (Tes) in tetraether lipid biosynthesis. (B) Proposed reaction catalyzed by 3-methyl-d-ornithine synthase (PylB) in pyrrolysine biosynthesis. (C) Proposed reaction catalyzed by MJ0619 from M. jannaschii in tetrahydromethanopterin biosynthesis. (D) Reaction catalyzed by Mmp10 in the posttranslational modification of MCR. (E) Reaction catalyzed by glutamine C-methyltransferase (QCMT) in the posttranslational modification of MCR. (F) Reactions catalyzed by AhbC and AhbD in the alternative heme biosynthesis pathway. (G) Reaction catalyzed by F0 synthase (CofG/CofH) in F420 biosynthesis. (H) Reaction catalyzed by NifB in FeMo-co biosynthesis. (I) Proposed involvement of HcgA in FeGP cofactor biosynthesis. (J) Reactions catalyzed by QueE and RaSEA in archaeosine biosynthesis. (K) Reaction catalyzed by Tyw1 (Taw1) in the biosynthesis of wyosine derivatives. (L) Proposed reactions catalyzed by MtaB in the synthesis of methylthiolated tRNAs. (M) Reaction catalyzed by Elp3 in the synthesis of carboxymethyl-uridine. Dotted arrows represent multiple steps.
The enzyme(s) and mechanism involved in the key step of GDGT biosynthesis, the diether-to-tetraether conversion (Fig. 2A), remained a mystery until just this year (22). Through bioinformatic analyses, a putative radical SAM enzyme was identified in the genomes of tetraether lipid-containing archaea, including the model methanogen Methanosarcina acetivorans, which was a likely candidate to catalyze the required C-C bond formation. The heterologous expression of the proposed gene MA_1486 in Methanococcus maripaludis, a non-GDGT-containing methanogen, resulted in the production of GDGT, thus indicating that the MA_1486 gene product is sufficient for GDGT formation, likely using archaeol precursors (22). Thus, MA_1486 was defined as a tetraether synthase (Tes) (Fig. 2A). The M. acetivorans genome encodes another Tes homolog (MA_1114) that, when heterologously expressed in M. maripaludis, does not result in GDGT production. However, the authors noted the production of trace amounts of macrocyclic archaeol (Fig. 2A) in the strain expressing MA_1114. Macrocyclic archaeol was also observed in cells expressing the confirmed Tes (MA_1486 or Maeo_0574) and was proposed to be a side product that occurs during heterologous expression (22). Interestingly, macrocyclic archaeol has been identified in only a select few archaea, all of which are thermophilic methanogens from hydrothermal vents (23–25).
Although not present in methanogenic lipids, the cyclopentane rings in GDGTs found primarily in the phyla Crenarchaeota and Thaumarchaeota are also installed by radical SAM enzymes (26). None of these radical SAM enzymes involved in archaeal lipid biosynthesis have been biochemically characterized, so the mechanism and direct substrates utilized for C-C bond formation in tetraether lipid synthesis and the formation of cyclopentane rings in archaeal lipids remain unclear. Now that the identities of these enzymes have been revealed, in vitro experiments can be pursued to reveal the mechanistic details of these intriguing reactions.
PYRROLYSINE BIOSYNTHESIS
Pyrrolysine is the 22nd genetically encoded amino acid, which was initially described in the active site of the monomethylamine methyltransferase MtmB from the methanogen Methanosarcina barkeri (27). Pyrrolysine is encoded by amber (UAG) codons, which were first identified in the genes encoding the three methyltransferases required for methanogenesis from methylamines (28). Since then, pyrrolysine has been found in other enzymes, including the tRNAHis guanylyltransferases (Thg1) (29) and certain transposases (30) in Methanosarcina, and has also been identified in some bacteria (31). Five pyl genes are necessary for pyrrolysine biosynthesis and its incorporation into proteins (32). The pylT gene encodes tRNAPyl, and pylS encodes pyrrolysyl-tRNA synthetase. The other three genes are required for the biosynthesis of pyrrolysine from 2 molecules of lysine. The first step is carried out by PylB, a radical SAM enzyme that catalyzes a carbon skeleton rearrangement of l-lysine to produce (3R)-3-methyl-d-ornithine (Fig. 2B) (33). The crystal structure of PylB expressed and purified from Escherichia coli revealed both SAM and the product bound to the active site, thus providing strong support for the function of PylB in (3R)-3-methyl-d-ornithine production (34). Despite structural information, an in vitro demonstration of PylB catalytic activity has not yet been reported. The reaction is proposed to occur analogously to that for B12-dependent glutamate mutase (32, 34, 35). Thus, the radical SAM-generated 5′-dAdo· would abstract a hydrogen atom from C-4 of lysine, leading to C-C bond cleavage that results in a glycyl radical intermediate and 4-aminobutene. The glycyl radical can then recombine with the former C-4 position of the substrate to form a new C-C bond followed by reduction and protonation to yield the final (3R)-3-methyl-d-ornithine product (32). Future in vitro studies are necessary to confirm PylB activity and provide experimental evidence for the proposed mechanism.
TETRAHYDROMETHANOPTERIN BIOSYNTHESIS
Tetrahydromethanopterin (H4MPT) (Fig. 2C) is a modified folate cofactor found in methanogens that, like tetrahydrofolate (H4F), functions to carry and transfer C1 groups of various oxidation states (36). H4MPT is used for several steps in the methanogenesis pathway and, in many methanogens, is also used instead of H4F for canonical folate-dependent reactions in amino acid and nucleic acid biosynthesis. One interesting structural feature that distinguishes H4MPT from H4F is the presence of methyl groups at the C-7 and C-9 positions (Fig. 2C). The unreactive nature of the sites of methylation led to the proposal that radical SAM chemistry may be involved in their installation (37). Genomic analyses revealed that in some methanogens, a radical SAM-encoding gene is found near the gene encoding β-ribofuranosylaminobenzene 5′-phosphate synthase, a key enzyme required for H4MPT biosynthesis (38, 39). Thus, the corresponding radical SAM-encoding gene from Methanocaldococcus jannaschii, MJ0619, was used for heterologous-expression experiments in E. coli to gain insight into the potential role of this gene product in H4MPT biosynthesis (37). When MJ0619 was expressed in E. coli, various methylated pterins were detected in the cell extracts by liquid chromatography-mass spectrometry (LC-MS), consistent with this enzyme catalyzing methylation at both the C-7 and C-9 positions of a pterin substrate (Fig. 2C). Since E. coli does not normally produce methylated pterins, this suggested that MJ0619 catalyzes the methylation reactions in H4MPT biosynthesis. Additionally, because 6-hydroxyethyl-7-methylpterin was identified in cell extracts containing MJ0619, but the corresponding dimethyl-folate was not identified, it was concluded that the likely substrate for MJ0619 is 6-hydroxmethyl(dihydro)pterin, a common early intermediate in both H4F and H4MPT biosynthesis (37).
Surprisingly, isotope feeding studies with E. coli expressing MJ0619 revealed that the enzyme likely does not use SAM as a methyl group donor since none of the deuterium atoms from CD3-Met were incorporated into the C-7 pterin methyl group added by MJ0619 (37). This is different from all other characterized radical SAM methylases, which use 2 molecules of SAM, one as the C1 donor for the methylation reaction and one as the source of 5′-dAdo· for radical SAM chemistry (40). Interestingly, further feeding studies with CD3-acetate led to the proposal that MJ0619 uses N5-N10-methylene-tetrahydrofolate (CH2H4F) as a C1 source for the methylation of pterins when it is expressed in E. coli (37). This is analogous to thymidylate synthase, where CH2H4F serves first as a methylene donor and then as a hydride donor to generate the final methyl group for the biosynthesis of dTMP from dUMP (41, 42). Confirmation of the function of MJ0619 and its homologs as well as details of the unique mechanism used for methylation during H4MPT biosynthesis will require in vitro enzymatic studies, which have yet to be reported.
POSTTRANSLATIONAL MODIFICATION OF METHYL-COENZYME M REDUCTASE
Methyl-coenzyme M reductase (MCR) catalyzes the final rate-determining step of methanogenesis. MCR is a dimer of heterotrimers with an α2β2γ2 configuration, harboring two active sites containing the F430 nickel tetrapyrrole prosthetic group. When the first crystal structure of MCR from Methanothermobacter marburgensis was determined, five unusual posttranslational modifications (PTMs) were revealed in the α subunit near the active site: thioglycine, N1-methylhistidine, S-methylcysteine, 5-(S)-methylarginine, and 2-(S)-methylglutamine (43). Except for N1-methylhistidine, the genes/enzymes required to install each of these PTMs have now been identified (44–47), including the identification and biochemical characterization of the radical SAM methylases responsible for the synthesis of 5-(S)-methylarginine (Fig. 2D) (44, 48–50) and 2-(S)-methylglutamine (Fig. 2E) (47). Upon searching the genome neighborhood of MCR operons for potential genes encoding enzymes involved in installing PTMs, a radical SAM-encoding gene annotated as “methanogenesis marker 10” (mmp10) was identified. To elucidate mmp10’s function, it was deleted in M. acetivorans, and the resulting MCR PTMs were analyzed by mass spectrometry (44). The authors found that the α subunit from the mmp10 deletion strain contained an unmodified arginine residue at position 285, thus demonstrating that Mmp10 is required for generating the 5-(S)-methylarginine PTM found in methanogenic MCRs. In terms of the physiological impacts of the unique PTM, it was observed that the lack of methylated arginine impaired the ability of M. acetivorans to grow under oxidative or thermal stress conditions. Furthermore, the purified variant MCR was shown to have a decreased melting temperature (Tm) (74.6°C versus 82.6°C for the wild type), further indicating that the 5-(S)-methylarginine impacts the thermal stability of MCR from M. acetivorans (44). Interestingly, the methylated arginine seems to have a more substantial impact on M. maripaludis MCR, where a deletion strain lacking this modification showed a highly impaired growth rate and the rate of methanogenesis was only about half the rate of the wild type (49). The in vitro enzymatic activities of MCR variants lacking specific PTMs have not yet been reported; thus, any specific role that 5-(S)-methylarginine plays in catalysis remains unclear. Interestingly, the crystal structure of MCR from M. acetivorans lacking the 5-(S)-methylarginine and other unique PTMs was virtually identical to the wild-type MCR structure (45).
Biochemical characterization of Mmp10 revealed that the enzyme is a cobalamin-dependent radical SAM methylase (48) despite the lack of an N-terminal cobalamin binding domain found in other cobalamin-dependent radical SAM methylases (40, 51). Further experiments demonstrated that cobalamin functions as an intermediate methyl group carrier, where SAM is used as the initial methyl group donor to generate methylcobalamin and S-adenosylhomocysteine (SAH) in a nucleophilic substitution reaction with cob(I)alamin (48). Thus, Mmp10 is proposed to employ a mechanism similar to those of other cobalamin-dependent methylases (51), where one SAM is used in nucleophilic chemistry to generate methylcobalamin and the other SAM is used in radical SAM chemistry to generate a substrate radical at the site of methylation that then reacts with methylcobalamin to generate the methylated product and cob(II)alamin. Cob(II)alamin can then be reduced to cob(I)alamin with titanium(III) citrate in vitro to allow remethylation with SAM, followed by subsequent turnovers (48). Recently, the crystal structure of Mmp10 was determined, which supports the proposed mechanism and provides further details of how the enzyme controls the two different reactivities of SAM (50). Interestingly, the radical SAM [4Fe-4S] cluster is coordinated by 3 cysteine residues as well as a strictly conserved tyrosine residue, which excludes SAM binding to the “unique” iron that is normally a defining feature of radical SAM enzymes. It is proposed that this places SAM in a position to react with cob(I)alamin to generate the methylcobalamin intermediate (50). Upon binding another SAM as well as the protein substrate, the active site then appears to reorganize, displacing tyrosine from the cluster, thus allowing coordination with SAM and subsequent radical SAM cleavage to produce 5′-dAdo· that initiates the radical methylation reaction. The structure of Mmp10 also revealed a mononuclear iron site coordinated by 4 cysteine residues. Mutagenesis studies indicated that the intact iron site is required for methylation activity; however, its detailed function in Mmp10 catalysis remains unknown (50).
Like C-5 the arginine methylation, it was proposed that a radical SAM enzyme would be required to catalyze the methylation of the α-carbon of a glutamine residue to generate the 2-(S)-methylglutamine (Fig. 2E) found in the active sites of some MCRs (52), and the identity of this enzyme was recently confirmed (47). Bioinformatic investigations identified a radical SAM enzyme-encoding gene located nearby the mcr gene cluster and mmp10 in some methanogens (44). This gene encodes a predicted N-terminal cobalamin binding domain, as observed in other characterized cobalamin-dependent radical SAM methylases outside Mmp10. The enzyme from Methanoculleus thermophilus was heterologously expressed and purified from E. coli, followed by biochemical characterization (47). The reconstituted protein binds a single [4Fe-4S] cluster as well as cobalamin in a base-off, His-off state. Assays with a 28-mer peptide substrate resulted in the C-2 methylation of the expected glutamine residue and produced 5′-dAdoH and SAH in equal molar ratios. Additionally, the cobalamin cofactor was demonstrated to cycle between cob(I)alamin, methylcob(III)alamin, and cob(II)alamin, consistent with the established mechanism for cobalamin-dependent radical SAM methylases (51). The physiological relevance of the 2-(S)-methylglutamine PTM of MCR is not yet known, but this can now be explored through knockout studies since the identity of the gene required for the modification has been confirmed. Notably, the 2-(S)-methylglutamine modification exhibits a more sporadic distribution compared to the conserved 5-(S)-methylarginine modification in various MCRs for which the PTMs have been elucidated (53).
HEME BIOSYNTHESIS
Heme is an essential prosthetic group for several enzymes involved in a range of fundamental biological processes in most organisms. In methanogens, heme is found in cytochrome-containing methanogens of the order Methanosarcinales, where cytochromes play a key role in energy metabolism (54). In all eukaryotes and most bacteria, heme is synthesized in a well-established canonical pathway (55). In the later steps of heme biosynthesis, two oxidative decarboxylation reactions are required to convert two propionate side chains of coproporphyrinogen III to two vinyl groups in the protoporphyrinogen IX product. In most bacteria, this reaction is catalyzed by oxygen-independent coproporphyrinogen III oxidase, a well-characterized radical SAM enzyme known as HemN that contains two simultaneously bound SAM molecules in the active site (56). Recent mechanistic studies have elucidated the roles of each SAM: the canonical SAM bound to the [4Fe-4S] cluster, SAM1, is the source of 5′-dAdo·, which abstracts a hydrogen atom from the methyl group of SAM2 to generate a SAM-based methylene radical. The latter species is uniquely proposed to be responsible for the key hydrogen atom abstraction from the β-carbon of a propionate side chain to initiate the vinyl conversion reaction (57).
Archaea utilize an alternative heme biosynthesis pathway where, among other differences in later steps of the pathway, the installation of the two vinyl side chains occurs in the last reaction involving the conversion of iron coproporphyrin III to heme b catalyzed by the heme synthase radical SAM enzyme AhbD (Fig. 2F) (58–60). Interestingly, AhbD from M. acetivorans and HemN from E. coli share only 17% identity, and although they catalyze the same reactions, they use different substrates: a porphyrinogen in the case of HemN and an iron-porphyrin for AhbD. In contrast to HemN, there is no evidence for AhbD binding more than one SAM. Furthermore, in addition to the radical SAM cluster, AhbD contains an auxiliary [4Fe-4S], which is coordinated by a cysteine-rich motif in the C terminus (61). In vitro enzymatic and mutagenesis studies on AhbD from M. barkeri indicated that the auxiliary cluster was essential for AhbD heme synthase activity but was not required for SAM cleavage and not directly involved in substrate binding (62). Based on electrochemical studies, the role of the auxiliary cluster was proposed as an electron acceptor necessary for the decarboxylation reaction (62). Thus, the proposed mechanism involves traditional radical SAM chemistry to generate 5′-dAdo· that, as opposed to HemN, abstracts an H atom directly from the propionate side chain of the substrate to initiate the decarboxylation reaction. The single electron resulting from the decarboxylation reaction is proposed to first reduce the heme iron to Fe(II) and then be transferred to the auxiliary cluster, which finally donates the electron to an external acceptor to reset the enzyme for another turnover (62).
The alternative heme biosynthesis pathway contains one additional radical SAM enzyme called AhbC, which catalyzes the penultimate step of the alternative pathway to remove two acetate side chains at C-2 and C-7 of 12,18-didecarboxysiroheme to produce the iron-coproporphyrinogen III substrate for AhbD (Fig. 2F) (58, 60). In M. acetivorans, AhbC has 31% identity and 52% similarity to AhbD and also contains two putative [4Fe-4S] cluster binding motifs, one of which is the canonical radical SAM motif CX3CX2C and the other of which is in the C terminus. This enzyme has not yet been biochemically characterized, and thus, the mechanistic details of the reaction remain unclear.
F420 BIOSYNTHESIS
Cofactor F420 (Fig. 2G) is a deazaflavin hydride carrier found in methanogens and select bacteria. This cofactor was first discovered in mycobacteria (63) and was subsequently structurally characterized from a methanogen (64, 65). The redox-active core ring system of F420, 7,8-didemethyl-8-hydroxy-5-deazariboflavin, with the 5′-ribityl group, is known as F0, while the complete F420 contains an added phosphate as well as a bridging lactyl moiety followed by a γ-linked polyglutamate tail (Fig. 2G). Compared to the isoalloxazine ring of flavin cofactors, the deazaisoalloxazine ring of F0 has a decreased reduction potential and can participate in two-electron chemistry only, as opposed to the characteristic one- or two-electron chemistry catalyzed by flavin adenine dinucleotide (FAD) and flavin mononucleotide (FMN) (66). Bacteria containing F420 use the cofactor in various redox reactions catalyzed by enzymes that often share homology to flavin-utilizing enzymes (67). Methanogens employ F420 for reduction steps in the methanogenesis pathway as well as for other hydride transfer reactions in various pathways.
F0 synthase is a radical SAM enzyme that carries out the key deazaflavin-forming step in F420 biosynthesis to catalyze the oxidative coupling of l-tyrosine to 5-amino-6-ribitylamino-2,4[1H,3H]-pyrimidinedione (ribityl diaminouracil) (Fig. 2G), a common intermediate in the riboflavin biosynthetic pathway. In mycobacteria and other actinobacteria, F0 synthase, encoded by fbiC, is a single bifunctional enzyme with two radical SAM motifs, while in archaea and cyanobacteria, F0 synthase is comprised of two independent radical SAM enzymes known as CofG and CofH, which closely resemble the N- and C-terminal halves of FbiC, respectively (68, 69). In vitro studies demonstrated that CofH catalyzes the first step to produce a stable intermediate, followed by the action of CofG to produce the final F0 molecule (70). Further detailed mechanistic experiments revealed that the first part of the CofH reaction likely proceeds similarly to those of other radical SAM tyrosine lyases such as HydG and ThiH to produce a p-hydroxylbenzyl radical (71), which can then undergo addition to the double bond of diaminouracil, followed by oxidation to produce an intermediate that is the substrate for CofG-catalyzed cyclization and oxidation to produce the final F0 (71). No crystal structures have been reported for any F0 synthase and would be important to obtain in the future to provide structural evidence for the mechanistic details of the two radical SAM reactions necessary for the synthesis of the unique deazaflavin core.
NITROGENASE COFACTOR BIOSYNTHESIS
Found in some bacteria and archaea, nitrogenase plays an essential role in the global nitrogen cycle by reducing molecular nitrogen (N2) to bioavailable ammonia (NH3). The active site where N2 reduction occurs consists of a complex metallocluster [Mo-7Fe-9S-C-(R)-homocitrate], referred to as FeMo-co, which can be described as a [4Fe-3S] cluster combined with a [Mo-3Fe-3S] cluster, connected by three sulfides and one carbide and partially ligated by (R)-homocitrate (Fig. 2H) (72). Molybdenum nitrogenases are the most common and most well characterized, but vanadium- and iron-nitrogenases also exist, where the above-mentioned metal replaces Mo in the unique metallocluster (73).
NifB is a radical SAM enzyme that plays a key role in FeMo-co biogenesis by converting the “K-cluster,” a pair of 4Fe-4S clusters, into the “L-cluster,” containing the bridging carbide carbon and the 9th sulfur atom that closely resembles the Fe-S core of FeMo-co (Fig. 2H) (74–77). Many details of NifB catalysis have been elucidated via in vitro mechanistic studies, and crystal structures have recently been reported (78, 79). Similar to many other radical SAM methylases (40), NifB uses SAM for two distinct reactivities: as a methyl group donor in a nucleophilic substitution reaction to generate SAH as a by-product and as a source of 5′-dAdo· in the radical SAM reaction to produce a substrate radical and 5′-dAdoH as a by-product. Thus, SAM is first used to methylate a sulfide atom in one of the [4Fe-4S] clusters that make up the K-cluster. Next, 5′-dAdo· produced at the radical SAM [4e-4S] cluster abstracts a hydrogen atom from the newly added methyl group to generate a methylene radical (80). Further processing of this species, using either an additional round(s) of radical SAM chemistry or recently proposed deprotonation facilitated by a His residue (81), finally results in the bridging SAM-derived carbide present in FeMo-co. This process is accompanied by the coupling and rearrangement of the two [4Fe-4S] modules of the K-cluster to generate the L-cluster (74) (Fig. 2H). A major remaining question regarding the NifB reaction is the mechanism involved in inserting the 9th sulfur. Sulfite has been shown to be the sulfur source in vitro (82), but the NifB-facilitated chemistry that would be involved in this process remains unclear.
[Fe]-HYDROGENASE COFACTOR BIOSYNTHESIS
Hydrogenases are abundant across various organisms in all domains of life, where they are responsible for the reversible conversion of H2 to protons and electrons. These metalloenzymes are often categorized based on the transition metals present in their active site cofactors: [NiFe], [FeFe], or [Fe]. Methanogens utilize five different types of hydrogenases, four of which are [NiFe] hydrogenases and one of which is an [Fe]-hydrogenase that is found only in methanogens without cytochromes (83). The latter is employed under nickel-limiting conditions to replace the function of the F420-reducing [NiFe] hydrogenase required for the reduction of methenyltetrahydromethanopterin to methylenetetrahydromethanopterin during hydrogenotrophic methanogenesis (84–86). Additionally, when nickel is limiting, the [Fe]-hydrogenase is also required for the production of reduced coenzyme F420, which is involved in several essential processes in methanogens (83).
The [Fe]-hydrogenase utilizes the unique iron-guanylylpyridinol (FeGP) cofactor (Fig. 2I), whose biosynthetic pathway is still not completely resolved. This biosynthesis utilizes genes located in the hcg gene cluster containing seven genes (hcgA–G) that cooccur with the [Fe]-hydrogenase-encoding genes. Recent in vitro studies confirmed the roles of HcgC and HcgB in the production of guanylylpyridinol (GP) in the early stages of FeGP cofactor biosynthesis (87). Furthermore, enzymatic and structural studies demonstrated that HcgE and HcgF catalyze the formation of a thioester-activated acyl group using a two-stage mechanism where HcgE catalyzes the adenylylation of the carboxy group of GP followed by a transesterification reaction to generate a Cys-S-GP thioester on the HcgF protein (88). This is presumably the activated substrate that is then converted to the final FeGP cofactor. HcgA is a radical SAM enzyme present in the hcg gene cluster that contains a noncanonical CX5CX2C motif. Recombinant HcgA has been studied in vitro to demonstrate that the enzyme contains a radical SAM-characteristic [4Fe-4S] cluster and carries out uncoupled SAM cleavage to produce 5′-dAdoH in the presence of dithionite (89). Due to the sequence similarity of HcgA to the well-characterized radical SAM enzyme HydG, which synthesizes the CN and CO ligands of the H-cluster in [FeFe]-hydrogenase (90), it has been proposed that HcgA similarly produces the CO ligands for the FeGP cofactor (89). However, in vivo isotope labeling studies indicated that the CO ligands of the FeGP cofactor are derived from CO2, and no incorporation from pyruvate was observed (91), whereas the CO ligands of the H-cluster are derived from l-tyrosine via dehydroglycine (92, 93). These results indicate that the synthesis of CO ligands for the FeGP cofactor occurs by a different pathway/mechanism compared to the H-cluster, and thus, many open questions remain surrounding the role of HcgA in FeGP cofactor biosynthesis. Future work on HcgA should be facilitated by the recent development of an in vitro system to study the later steps of FeGP cofactor biosynthesis using precursors isolated from Δhcg mutated strains (87).
tRNA MODIFICATIONS
tRNAs are highly modified in all organisms, with up to 10 to 20% of nucleosides modified in a given tRNA and over 170 different types of modifications that have been described (94, 95). The distribution of tRNA modifications can be highly variable, where some are found at the same position in all tRNAs throughout all domains of life and others are found in only one or a few tRNAs in one domain of life (96, 97). These modifications ensure accurate decoding and aminoacylation and stabilize the tertiary structure of tRNAs. Modifications can be simple (e.g., methylation or isomerization) or complex, requiring multiple enzymatic steps. Several radical SAM enzymes in methanogens are involved in tRNA modifications (Table 1).
Archaeosine (7-formamidino-7-deazaguanine [G+]) (Fig. 2J) is a structurally complex modified nucleoside in archaea, where it is found in the D-loop at position 15 of virtually all archaeal tRNAs (98). As a 7-deazaguanosine nucleoside, archaeosine is related to queuosine, the latter of which is ubiquitous throughout eukaryotic and bacterial organisms. The biosynthesis of both of these modified nucleosides involves the common intermediate 7-cyano-7-deazaguanine (preQ0) (Fig. 2J). The biosynthesis of preQ0 from GTP involves the action of four or five enzymes, one of which is QueE, a ubiquitous radical SAM enzyme that catalyzes the key heterocyclic rearrangement reaction to convert 6-carboxy-tetrahydropterin to 7-carboxy-7-deazaguanine, expelling a nitrogen as +NH4 (99). In archaea, preQ0 serves as a substrate for tRNA-guanine transglycosylase, which catalyzes the exchange of guanine at position 15 for preQ0 (100). In most Euryarchaeota, including methanogens, preQ0-tRNA is then converted to G+-tRNA by archaeosine synthase (ArcS), of which the enzyme from M. jannaschii has been investigated in vitro (101). While it was originally thought that ArcS alone synthesizes G+ from PreQ0, it was recently shown that some Euryarchaeota, including M. acetivorans, utilize a complex consisting of ArcS and the radical SAM enzyme for archaeosine formation (RaSEA) (102). Here, ArcS first transfers lysine to preQ0, where the ε-amino group of lysine is added to the cyano group of preQ0 (Fig. 2J). RaSEA is then proposed to abstract a hydrogen atom from the ε-carbon of the lysine moiety, which activates the molecule for C-N bond cleavage to eventually form G+ and 1-piperidine-6-carboxylic acid as a by-product (102). Virtually all sequenced Euryarchaeota contain homologs of both ArcS and RaSEA; thus, this may be the main pathway for G+ synthesis in these organisms (102). In the future, further biochemical and structural studies of ArcS/RaSEA should be carried out to elucidate the mechanistic details of the reaction and to clarify the dependence of ArcS on a radical SAM partner since in vitro experiments with ArcS from M. jannaschii showed that the enzyme alone can produce G+ from preQ0 (101).
Wyosine derivatives are fluorescent modified nucleosides found in eukaryotes and archaea but not bacteria. They are located at position 37 of phenylalanine-specific tRNA and consist of an imidazopurine (tricyclic) core structure. The key ring formation step in the biosynthesis of these wyosine derivatives is catalyzed by wybutosine 4-demethylwyosine synthase (Tyw1), also referred to as Taw1 in archaea (103). This radical SAM enzyme forms the tricyclic ring of 4-demethylwyosine from N1-methylguanosine and pyruvate (Fig. 2K) (104), which is an intermediate in the biosynthesis of several imidazopurine-containing nucleosides, including wyosine and wybutosine and the archaeon-specific isowyosine and 7-methylwyosine (105–107). The Tyw1 reaction is initiated with hydrogen atom abstraction from the methyl group of N1-methylguanosine, which then reacts with pyruvate, followed by decarboxylation and transamination to finally form the tricyclic product (108). Interestingly, eukaryotic Tyw1 contains an N-terminal flavodoxin domain used for reducing the radical SAM [4Fe-4S] cluster to the catalytic +1 state; however, this domain is absent in the archaeal enzyme (109). A crystal structure of Tyw1 from M. jannaschii revealed that pyruvate is bound in the active site via Schiff base formation with a conserved lysine residue (110). Additionally, the structure showed that Tyw1 contains an auxiliary [4Fe-4S] cluster that is partially coordinated by the lysine nitrogen and carboxylate oxygen of the Schiff base intermediate. Thus, the auxiliary cluster is proposed to serve as an electron sink to facilitate the decarboxylation of pyruvate (110).
Residue 37, which is 3′ adjacent to the anticodon, is a frequently modified position in most tRNAs. The modified nucleosides found exclusively in this position include N6-isopentenyladenosine (i6A), N6-threonylcarbamoyladenosine (t6A), and N6-hydroxynorvalylcarbamoyladenosine (hn6A) (95, 97). Depending on the sequence features of the specific tRNA, all three of these can be further modified via the addition of a methylthio group at the 2′ position to generate ms2i6A, ms2t6A, and ms2hn6A, respectively. In methanogens, the threonyl-containing (t6A and ms2t6A) and hydroxynorvalyl-containing (hn6A and ms2hn6A) modifications have been identified in various organisms, but the isopentenyl-containing modifications are absent (105, 107, 111–113). The methylthiolation reaction is catalyzed by a radical SAM methylthiotransferase (MTTase): MiaB for the ms2i6A modification and MtaB for the ms2t6A modification (Fig. 2L). The bacterial MiaB MTTase has been well characterized with detailed mechanistic and structural information available. MTTases contain two iron-sulfur clusters: the radical SAM cluster and an auxiliary cluster that is the source of the sulfur atom that is eventually transferred to the tRNA substrate (114–117). The first stage of MTTase catalysis involves a nucleophilic substitution reaction to transfer the methyl group of SAM to a sulfur species associated with the auxiliary cluster that is proposed to be [3Fe-4S]0 (115). In the second stage, radical SAM-dependent chemistry facilitates the transfer of the resulting methylthio group to C-2 of i6A to produce ms2i6A (114–117). MtaB is expected to utilize analogous chemistry for the methylthiolation of t6A; however, although the enzyme from Bacillus subtilis has been purified and its [4Fe-4S] clusters have been characterized (118), the in vitro catalytic activity has not yet been investigated, and no MtaB structural information is available. Additionally, although MtaB homologs are proposed to be responsible for the methylthiolation of hn6A to produce ms2hn6A (Fig. 2L) (119), this has yet to be experimentally confirmed.
5-Carboxymethyluridine (cm5U) is an intermediate modified nucleoside that leads to a variety of uridine modifications at the wobble position of tRNA in primarily eukaryotes and archaea (120). The synthesis of this modification is carried out by Elp3 (Fig. 2M), a radical SAM enzyme present in the elongator complex, a large protein complex involved in cm5U synthesis and previously implicated in histone acetylation. Enzymatic studies of Elp3 from the methanogen Methanocaldococcus infernus confirmed that the enzyme is solely responsible for the cm5U modification in vitro and utilizes radical SAM-characteristic chemistry (121). Crystal structures of a rare bacterial Elp3 (122) as well as M. infernus Elp3 (123) revealed a radical SAM domain that is highly similar to that of RlmN (124), the radical SAM methylase that modifies specific tRNA and rRNA. Elp3 also contains a C-terminal lysine acetyltransferase (KAT) domain similar to those of the GCN5-like acetyltransferases (125). Based on the available biochemical and structural data, the following mechanistic features have been proposed: (i) tRNA substrate binding initiates a conformational change that leads to acetyl-CoA binding in the KAT domain (123); (ii) acetyl-CoA hydrolysis occurs, and the released acetate is transferred to the radical SAM domain (123); (iii) radical SAM chemistry leads to hydrogen atom abstraction from the methyl moiety of acetate, which then adds to C-5 of the uridine residue to form a C-C bond (121); and (iv) the loss of an electron followed by general base-catalyzed proton abstraction results in the final cm5U product (121). Further detailed mechanistic studies are required to confirm the proposed mechanism, especially with respect to the chemistry occurring in the radical SAM domain. Additionally, the specific roles of the additional proteins present in the elongator complex need to be elucidated.
CONCLUDING REMARKS
Radical SAM enzymes in methanogens catalyze several unique reactions compared to those found in other organisms and often utilize especially intriguing catalytic mechanisms, even among radical SAM superfamily members. Based on the presence of the canonical CX3CX2C motif, the model methanogens M. maripaludis S2 and M. acetivorans C2A have 32 and 58 putative radical SAM proteins (see Tables S1 and S2 in the supplemental material), respectively, with only about half of these having established functions based on experimental evidence or high sequence similarity to well-characterized radical SAM enzymes. Thus, many questions remain regarding the physiological functions and catalytic versatility of radical SAM enzymes in methanogens. Characterizing these enzymes will further expand the catalytic repertoire of the radical SAM superfamily and aid in defining previously unknown aspects of primary metabolism in methanogens. Furthermore, understanding the functions and mechanisms of unique radical SAM enzymes in methanogens could inform methane mitigation strategies as well as bioengineering efforts for biofuel applications.
ACKNOWLEDGMENTS
We thank Anne Brown and Amanda Sharp (Virginia Tech Department of Biochemistry) for bioinformatics assistance to identify radical SAM domain-containing proteins in various methanogenic genomes.
Research on methanogenic radical SAM enzymes in the Allen Lab is funded by the National Science Foundation (CHE-2105598).
Footnotes
Supplemental material is available online only.
Contributor Information
Kylie D. Allen, Email: kdallen@vt.edu.
Julie A. Maupin-Furlow, University of Florida
REFERENCES
- 1.Oberg N, Precord TW, Gerlt JA, Mitchell DA. 2022. RadicalSAM.org: a resource to interpret sequence-function space and discover new radical SAM enzyme chemistry. ACS Bio Med Chem Au 2:22–35. 10.1021/acsbiomedchemau.1c00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 29:1097–1106. 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broderick JB, Duffus BR, Duschene KS, Shepard EM. 2014. Radical S-adenosylmethionine enzymes. Chem Rev 114:4229–4317. 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krebs C, Broderick WE, Henshaw TF, Broderick JB, Huynh BH. 2002. Coordination of adenosylmethionine to a unique iron site of the [4Fe-4S] of pyruvate formate-lyase activating enzyme: a Mossbauer spectroscopic study. J Am Chem Soc 124:912–913. 10.1021/ja017562i. [DOI] [PubMed] [Google Scholar]
- 5.Broderick WE, Broderick JB. 2019. Radical SAM enzymes: surprises along the path to understanding mechanism. J Biol Inorg Chem 24:769–776. 10.1007/s00775-019-01706-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holliday GL, Akiva E, Meng EC, Brown SD, Calhoun S, Pieper U, Sali A, Booker SJ, Babbitt PC. 2018. Atlas of the radical SAM superfamily: divergent evolution of function using a “plug and play” domain. Methods Enzymol 606:1–71. 10.1016/bs.mie.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey PA, Hegeman AD, Ruzicka FJ. 2008. The radical SAM superfamily. Crit Rev Biochem Mol Biol 43:63–88. 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 8.Yang H, McDaniel EC, Impano S, Byer AS, Jodts RJ, Yokoyama K, Broderick WE, Broderick JB, Hoffman BM. 2019. The elusive 5′-deoxyadenosyl radical: captured and characterized by electron paramagnetic resonance and electron nuclear double resonance spectroscopies. J Am Chem Soc 141:12139–12146. 10.1021/jacs.9b05926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landgraf BJ, McCarthy EL, Booker SJ. 2016. Radical S-adenosylmethionine enzymes in human health and disease. Annu Rev Biochem 85:485–514. 10.1146/annurev-biochem-060713-035504. [DOI] [PubMed] [Google Scholar]
- 10.Haskamp V, Karrie S, Mingers T, Barthels S, Alberge F, Magalon A, Muller K, Bill E, Lubitz W, Kleeberg K, Schweyen P, Broring M, Jahn M, Jahn D. 2018. The radical SAM protein HemW is a heme chaperone. J Biol Chem 293:2558–2572. 10.1074/jbc.RA117.000229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gizzi AS, Grove TL, Arnold JJ, Jose J, Jangra RK, Garforth SJ, Du Q, Cahill SM, Dulyaninova NG, Love JD, Chandran K, Bresnick AR, Cameron CE, Almo SC. 2018. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature 558:610–614. 10.1038/s41586-018-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlesworth JC, Burns BP. 2015. Untapped resources: biotechnological potential of peptides and secondary metabolites in archaea. Archaea 2015:282035. 10.1155/2015/282035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang S, Zheng Z, Zou H, Li N, Wu M. 2019. Characterization of the secondary metabolite biosynthetic gene clusters in archaea. Comput Biol Chem 78:165–169. 10.1016/j.compbiolchem.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 14.Lyu Z, Shao N, Akinyemi T, Whitman WB. 2018. Methanogenesis. Curr Biol 28:R727–R732. 10.1016/j.cub.2018.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Jackson RB, Saunois M, Bousquet P, Canadell JG, Poulter B, Stavert AR, Bergamaschi P, Niwa Y, Segers A, Tsuruta A. 2020. Increasing anthropogenic methane emissions arise equally from agricultural and fossil fuel sources. Environ Res Lett 15:071002. 10.1088/1748-9326/ab9ed2. [DOI] [Google Scholar]
- 16.Montzka SA, Dlugokencky EJ, Butler JH. 2011. Non-CO2 greenhouse gases and climate change. Nature 476:43–50. 10.1038/nature10322. [DOI] [PubMed] [Google Scholar]
- 17.Holmes DE, Smith JA. 2016. Biologically produced methane as a renewable energy source. Adv Appl Microbiol 97:1–61. 10.1016/bs.aambs.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Villanueva L, Damste JSS, Schouten S. 2014. A re-evaluation of the archaeal membrane lipid biosynthetic pathway. Nat Rev Microbiol 12:438–448. 10.1038/nrmicro3260. [DOI] [PubMed] [Google Scholar]
- 19.Schouten S, Hopmans EC, Damste JSS. 2013. The organic geochemistry of glycerol dialkyl glycerol tetraether lipids: a review. Org Geochem 54:19–61. 10.1016/j.orggeochem.2012.09.006. [DOI] [Google Scholar]
- 20.Sprott GD, Meloche M, Richards JC. 1991. Proportions of diether, macrocyclic diether, and tetraether lipids in Methanococcus jannaschii grown at different temperatures. J Bacteriol 173:3907–3910. 10.1128/jb.173.12.3907-3910.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai D, Springstead JR, Monbouquette HG. 2008. Effect of growth temperature on ether lipid biochemistry in Archaeoglobus fulgidus. Extremophiles 12:271–278. 10.1007/s00792-007-0126-6. [DOI] [PubMed] [Google Scholar]
- 22.Zeng Z, Chen H, Yang H, Chen Y, Yang W, Feng X, Pei H, Welander PV. 2022. Identification of a protein responsible for the synthesis of archaeal membrane-spanning GDGT lipids. Nat Commun 13:1545. 10.1038/s41467-022-29264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comita PB, Gagosian RB. 1983. Membrane lipid from deep-sea hydrothermal vent methanogen—a new macrocyclic glycerol diether. Science 222:1329–1331. 10.1126/science.222.4630.1329. [DOI] [PubMed] [Google Scholar]
- 24.Comita PB, Gagosian RB, Pang H, Costello CE. 1984. Structural elucidation of a unique macrocyclic membrane lipid from a new, extremely thermophilic, deep-sea hydrothermal vent archaebacterium, Methanococcus jannaschii. J Biol Chem 259:15234–15241. 10.1016/S0021-9258(17)42540-3. [DOI] [PubMed] [Google Scholar]
- 25.Baumann LMF, Taubner R-S, Bauersachs T, Steiner M, Schleper C, Peckmann J, Rittmann SK-MR, Birgel D. 2018. Intact polar lipid and core lipid inventory of the hydrothermal vent methanogens Methanocaldococcus villosus and Methanothermococcus okinawensis. Org Geochem 126:33–42. 10.1016/j.orggeochem.2018.10.006. [DOI] [Google Scholar]
- 26.Zeng Z, Liu XL, Farley KR, Wei JH, Metcalf WW, Summons RE, Welander PV. 2019. GDGT cyclization proteins identify the dominant archaeal sources of tetraether lipids in the ocean. Proc Natl Acad Sci USA 116:22505–22511. 10.1073/pnas.1909306116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. 2002. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296:1462–1466. 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 28.Paul L, Ferguson DJ, Jr, Krzycki JA. 2000. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J Bacteriol 182:2520–2529. 10.1128/JB.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinemann IU, O’Donoghue P, Madinger C, Benner J, Randau L, Noren CJ, Soll D. 2009. The appearance of pyrrolysine in tRNAHis guanylyltransferase by neutral evolution. Proc Natl Acad Sci USA 106:21103–21108. 10.1073/pnas.0912072106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. 2005. Pyrrolysine and selenocysteine use dissimilar decoding strategies. J Biol Chem 280:20740–20751. 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 31.Brugere J-F, Atkins JF, O’Toole PW, Borrel G. 2018. Pyrrolysine in archaea: a 22nd amino acid encoded through a genetic code expansion. Emerg Top Life Sci 2:607–618. 10.1042/ETLS20180094. [DOI] [PubMed] [Google Scholar]
- 32.Krzycki JA. 2013. The path of lysine to pyrrolysine. Curr Opin Chem Biol 17:619–625. 10.1016/j.cbpa.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 33.Gaston MA, Zhang L, Green-Church KB, Krzycki JA. 2011. The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine. Nature 471:647–650. 10.1038/nature09918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quitterer F, List A, Eisenreich W, Bacher A, Groll M. 2012. Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis. Angew Chem Int Ed Engl 51:1339–1342. 10.1002/anie.201106765. [DOI] [PubMed] [Google Scholar]
- 35.Marsh EN. 2009. Insights into the mechanisms of adenosylcobalamin (coenzyme B12)-dependent enzymes from rapid chemical quench experiments. Biochem Soc Trans 37:336–342. 10.1042/BST0370336. [DOI] [PubMed] [Google Scholar]
- 36.Escalante-Semerena JC, Rinehart KL, Jr, Wolfe RS. 1984. Tetrahydromethanopterin, a carbon carrier in methanogenesis. J Biol Chem 259:9447–9455. 10.1016/S0021-9258(17)42721-9. [DOI] [PubMed] [Google Scholar]
- 37.Allen KD, Xu H, White RH. 2014. Identification of a unique radical S-adenosylmethionine methylase likely involved in methanopterin biosynthesis in Methanocaldococcus jannaschii. J Bacteriol 196:3315–3323. 10.1128/JB.01903-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott JW, Rasche ME. 2002. Purification, overproduction, and partial characterization of beta-RFAP synthase, a key enzyme in the methanopterin biosynthesis pathway. J Bacteriol 184:4442–4448. 10.1128/JB.184.16.4442-4448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasche ME, White RH. 1998. Mechanism for the enzymatic formation of 4-(beta-D-ribofuranosyl)aminobenzene 5′-phosphate during the biosynthesis of methanopterin. Biochemistry 37:11343–11351. 10.1021/bi973086q. [DOI] [PubMed] [Google Scholar]
- 40.Bauerle MR, Schwalm EL, Booker SJ. 2015. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J Biol Chem 290:3995–4002. 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carreras CW, Santi DV. 1995. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem 64:721–762. 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 42.Pozzi C, Lopresti L, Tassone G, Mangani S. 2019. Targeting methyltransferases in human pathogenic bacteria: insights into thymidylate synthase (TS) and flavin-dependent TS (FDTS). Molecules 24:1638. 10.3390/molecules24081638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. 1997. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science 278:1457–1462. 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 44.Deobald D, Adrian L, Schone C, Rother M, Layer G. 2018. Identification of a unique radical SAM methyltransferase required for the sp(3)-C-methylation of an arginine residue of methyl-coenzyme M reductase. Sci Rep 8:7404. 10.1038/s41598-018-25716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nayak DD, Liu A, Agrawal N, Rodriguez-Carerro R, Dong SH, Mitchell DA, Nair SK, Metcalf WW. 2020. Functional interactions between posttranslationally modified amino acids of methyl-coenzyme M reductase in Methanosarcina acetivorans. PLoS Biol 18:e3000507. 10.1371/journal.pbio.3000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nayak DD, Mahanta N, Mitchell DA, Metcalf WW. 2017. Post-translational thioamidation of methyl-coenzyme M reductase, a key enzyme in methanogenic and methanotrophic Archaea. Elife 6:e29218. 10.7554/eLife.29218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gagsteiger J, Jahn S, Heidinger L, Gericke L, Andexer JN, Friedrich T, Loenarz C, Layer G. 30 May 2022. A cobalamin-dependent radical SAM enzyme catalyzes the unique Calpha-methylation of glutamine in methyl-coenzyme M reductase. Angew Chem Int Ed Engl 10.1002/anie.202204198. [DOI] [PMC free article] [PubMed]
- 48.Radle MI, Miller DV, Laremore TN, Booker SJ. 2019. Methanogenesis marker protein 10 (Mmp10) from Methanosarcina acetivorans is a radical S-adenosylmethionine methylase that unexpectedly requires cobalamin. J Biol Chem 294:11712–11725. 10.1074/jbc.RA119.007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyu Z, Shao N, Chou CW, Shi H, Patel R, Duin EC, Whitman WB. 2020. Posttranslational methylation of arginine in methyl coenzyme M reductase has a profound impact on both methanogenesis and growth of Methanococcus maripaludis. J Bacteriol 202:e00654-19. 10.1128/JB.00654-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fyfe CD, Bernardo-Garcia N, Fradale L, Grimaldi S, Guillot A, Brewee C, Chavas LMG, Legrand P, Benjdia A, Berteau O. 2022. Crystallographic snapshots of a B12-dependent radical SAM methyltransferase. Nature 602:336–342. 10.1038/s41586-021-04355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang SC. 2018. Cobalamin-dependent radical S-adenosyl-l-methionine enzymes in natural product biosynthesis. Nat Prod Rep 35:707–720. 10.1039/c7np00059f. [DOI] [PubMed] [Google Scholar]
- 52.Buckel W, Thauer RK. 2011. Dual role of S-adenosylmethionine (SAM) in the methylation of sp2-hybridized electrophilic carbons. Angew Chem Int Ed Engl 50:10492–10494. 10.1002/anie.201105076. [DOI] [PubMed] [Google Scholar]
- 53.Gendron A, Allen KD. 2022. Overview of diverse methyl/alkyl-coenzyme M reductases and considerations for their potential heterologous expression. Front Microbiol 13:867342. 10.3389/fmicb.2022.867342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thauer RK, Kaster AK, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 55.Layer G, Reichelt J, Jahn D, Heinz DW. 2010. Structure and function of enzymes in heme biosynthesis. Protein Sci 19:1137–1161. 10.1002/pro.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Layer G, Moser J, Heinz DW, Jahn D, Schubert WD. 2003. Crystal structure of coproporphyrinogen III oxidase reveals cofactor geometry of radical SAM enzymes. EMBO J 22:6214–6224. 10.1093/emboj/cdg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji X, Mo T, Liu WQ, Ding W, Deng Z, Zhang Q. 2019. Revisiting the mechanism of the anaerobic coproporphyrinogen III oxidase HemN. Angew Chem Int Ed Engl 58:6235–6238. 10.1002/anie.201814708. [DOI] [PubMed] [Google Scholar]
- 58.Kuhner M, Haufschildt K, Neumann A, Storbeck S, Streif J, Layer G. 2014. The alternative route to heme in the methanogenic archaeon Methanosarcina barkeri. Archaea 2014:327637. 10.1155/2014/327637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bali S, Palmer DJ, Schroeder S, Ferguson SJ, Warren MJ. 2014. Recent advances in the biosynthesis of modified tetrapyrroles: the discovery of an alternative pathway for the formation of heme and heme d. Cell Mol Life Sci 71:2837–2863. 10.1007/s00018-014-1563-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bali S, Lawrence AD, Lobo SA, Saraiva LM, Golding BT, Palmer DJ, Howard MJ, Ferguson SJ, Warren MJ. 2011. Molecular hijacking of siroheme for the synthesis of heme and d1 heme. Proc Natl Acad Sci USA 108:18260–18265. 10.1073/pnas.1108228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lobo SA, Lawrence AD, Romao CV, Warren MJ, Teixeira M, Saraiva LM. 2014. Characterisation of Desulfovibrio vulgaris haem b synthase, a radical SAM family member. Biochim Biophys Acta 1844:1238–1247. 10.1016/j.bbapap.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 62.Kuhner M, Schweyen P, Hoffmann M, Ramos JV, Reijerse EJ, Lubitz W, Broring M, Layer G. 2016. The auxiliary [4Fe-4S] cluster of the radical SAM heme synthase from Methanosarcina barkeri is involved in electron transfer. Chem Sci 7:4633–4643. 10.1039/c6sc01140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cousins FB. 1960. The prosthetic group of a chromoprotein from mycobacteria. Biochim Biophys Acta 40:532–534. 10.1016/0006-3002(60)91396-2. [DOI] [PubMed] [Google Scholar]
- 64.Cheeseman P, Toms-Wood A, Wolfe RS. 1972. Isolation and properties of a fluorescent compound, factor 420, from Methanobacterium strain M.o.H. J Bacteriol 112:527–531. 10.1128/jb.112.1.527-531.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eirich LD, Vogels GD, Wolfe RS. 1978. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry 17:4583–4593. 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- 66.Walsh C. 1986. Naturally-occurring 5-deazaflavin coenzymes—biological redox roles. Acc Chem Res 19:216–221. 10.1021/ar00127a004. [DOI] [Google Scholar]
- 67.Selengut JD, Haft DH. 2010. Unexpected abundance of coenzyme F420-dependent enzymes in Mycobacterium tuberculosis and other actinobacteria. J Bacteriol 192:5788–5798. 10.1128/JB.00425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi K-P, Kendrick N, Daniels L. 2002. Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F420 and FO biosynthesis. J Bacteriol 184:2420–2428. 10.1128/JB.184.9.2420-2428.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Graham DE, Xu H, White RH. 2003. Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F(420) biosynthesis. Arch Microbiol 180:455–464. 10.1007/s00203-003-0614-8. [DOI] [PubMed] [Google Scholar]
- 70.Decamps L, Philmus B, Benjdia A, White R, Begley TP, Berteau O. 2012. Biosynthesis of Fo, precursor of the F420 cofactor, requires a unique two radical-SAM domain enzyme and tyrosine as substrate. J Am Chem Soc 134:18173–18176. 10.1021/ja307762b. [DOI] [PubMed] [Google Scholar]
- 71.Philmus B, Decamps L, Berteau O, Begley TP. 2015. Biosynthetic versatility and coordinated action of 5′-deoxyadenosyl radicals in deazaflavin biosynthesis. J Am Chem Soc 137:5406–5413. 10.1021/ja513287k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Einsle O, Rees DC. 2020. Structural enzymology of nitrogenase enzymes. Chem Rev 120:4969–5004. 10.1021/acs.chemrev.0c00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jasniewski AJ, Lee CC, Ribbe MW, Hu Y. 2020. Reactivity, mechanism, and assembly of the alternative nitrogenases. Chem Rev 120:5107–5157. 10.1021/acs.chemrev.9b00704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu Y, Ribbe MW. 2016. Maturation of nitrogenase cofactor—the role of a class E radical SAM methyltransferase NifB. Curr Opin Chem Biol 31:188–194. 10.1016/j.cbpa.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiig JA, Hu Y, Ribbe MW. 2011. NifEN-B complex of Azotobacter vinelandii is fully functional in nitrogenase FeMo cofactor assembly. Proc Natl Acad Sci USA 108:8623–8627. 10.1073/pnas.1102773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fay AW, Wiig JA, Lee CC, Hu Y. 2015. Identification and characterization of functional homologs of nitrogenase cofactor biosynthesis protein NifB from methanogens. Proc Natl Acad Sci USA 112:14829–14833. 10.1073/pnas.1510409112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Curatti L, Ludden PW, Rubio LM. 2006. NifB-dependent in vitro synthesis of the iron-molybdenum cofactor of nitrogenase. Proc Natl Acad Sci USA 103:5297–5301. 10.1073/pnas.0601115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fajardo AS, Legrand P, Paya-Tormo LA, Martin L, Pellicer Marti Nez MT, Echavarri-Erasun C, Vernede X, Rubio LM, Nicolet Y. 2020. Structural insights into the mechanism of the radical SAM carbide synthase NifB, a key nitrogenase cofactor maturating enzyme. J Am Chem Soc 142:11006–11012. 10.1021/jacs.0c02243. [DOI] [PubMed] [Google Scholar]
- 79.Kang W, Rettberg LA, Stiebritz MT, Jasniewski AJ, Tanifuji K, Lee CC, Ribbe MW, Hu Y. 2021. X-ray crystallographic analysis of NifB with a full complement of clusters: structural insights into the radical SAM-dependent carbide insertion during nitrogenase cofactor assembly. Angew Chem Int Ed Engl 60:2364–2370. 10.1002/anie.202011367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiig JA, Hu Y, Ribbe MW. 2015. Refining the pathway of carbide insertion into the nitrogenase M-cluster. Nat Commun 6:8034. 10.1038/ncomms9034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rettberg LA, Wilcoxen J, Jasniewski AJ, Lee CC, Tanifuji K, Hu Y, Britt RD, Ribbe MW. 2020. Identity and function of an essential nitrogen ligand of the nitrogenase cofactor biosynthesis protein NifB. Nat Commun 11:1757. 10.1038/s41467-020-15627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanifuji K, Lee CC, Sickerman NS, Tatsumi K, Ohki Y, Hu Y, Ribbe MW. 2018. Tracing the ‘ninth sulfur’ of the nitrogenase cofactor via a semi-synthetic approach. Nat Chem 10:568–572. 10.1038/s41557-018-0029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mand TD, Metcalf WW. 2019. Energy conservation and hydrogenase function in methanogenic archaea, in particular the genus Methanosarcina. Microbiol Mol Biol Rev 83:e00020-19. 10.1128/MMBR.00020-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Buurman G, Shima S, Thauer RK. 2000. The metal-free hydrogenase from methanogenic archaea: evidence for a bound cofactor. FEBS Lett 485:200–204. 10.1016/s0014-5793(00)02225-0. [DOI] [PubMed] [Google Scholar]
- 85.Afting C, Kremmer E, Brucker C, Hochheimer A, Thauer RK. 2000. Regulation of the synthesis of H2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch Microbiol 174:225–232. 10.1007/s002030000197. [DOI] [PubMed] [Google Scholar]
- 86.Afting C, Hochheimer A, Thauer RK. 1998. Function of H2-forming methylenetetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum in coenzyme F420 reduction with H2. Arch Microbiol 169:206–210. 10.1007/s002030050562. [DOI] [PubMed] [Google Scholar]
- 87.Schaupp S, Arriaza-Gallardo FJ, Pan HJ, Kahnt J, Angelidou G, Paczia N, Costa K, Hu X, Shima S. 2022. In vitro biosynthesis of the [Fe]-hydrogenase cofactor verifies the proposed biosynthetic precursors. Angew Chem Int Ed Engl 61:e202200994. 10.1002/anie.202200994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fujishiro T, Kahnt J, Ermler U, Shima S. 2015. Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor. Nat Commun 6:6895. 10.1038/ncomms7895. [DOI] [PubMed] [Google Scholar]
- 89.McGlynn SE, Boyd ES, Shepard EM, Lange RK, Gerlach R, Broderick JB, Peters JW. 2010. Identification and characterization of a novel member of the radical AdoMet enzyme superfamily and implications for the biosynthesis of the Hmd hydrogenase active site cofactor. J Bacteriol 192:595–598. 10.1128/JB.01125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Britt RD, Tao L, Rao G, Chen N, Wang LP. 2022. Proposed mechanism for the biosynthesis of the [FeFe] hydrogenase H-cluster: central roles for the radical SAM enzymes HydG and HydE. ACS Bio Med Chem Au 2:11–21. 10.1021/acsbiomedchemau.1c00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schick M, Xie X, Ataka K, Kahnt J, Linne U, Shima S. 2012. Biosynthesis of the iron-guanylylpyridinol cofactor of [Fe]-hydrogenase in methanogenic archaea as elucidated by stable-isotope labeling. J Am Chem Soc 134:3271–3280. 10.1021/ja211594m. [DOI] [PubMed] [Google Scholar]
- 92.Kuchenreuther JM, George SJ, Grady-Smith CS, Cramer SP, Swartz JR. 2011. Cell-free H-cluster synthesis and [FeFe] hydrogenase activation: all five CO and CN(−) ligands derive from tyrosine. PLoS One 6:e20346. 10.1371/journal.pone.0020346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shepard EM, Duffus BR, George SJ, McGlynn SE, Challand MR, Swanson KD, Roach PL, Cramer SP, Peters JW, Broderick JB. 2010. [FeFe]-hydrogenase maturation: HydG-catalyzed synthesis of carbon monoxide. J Am Chem Soc 132:9247–9249. 10.1021/ja1012273. [DOI] [PubMed] [Google Scholar]
- 94.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. 2011. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 39:D195–D201. 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, Helm M, Bujnicki JM. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46:D303–D307. 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lyons SM, Fay MM, Ivanov P. 2018. The role of RNA modifications in the regulation of tRNA cleavage. FEBS Lett 592:2828–2844. 10.1002/1873-3468.13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jackman JE, Alfonzo JD. 2013. Transfer RNA modifications: nature’s combinatorial chemistry playground. Wiley Interdiscip Rev RNA 4:35–48. 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gregson JM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, McCloskey JA. 1993. Structure of the archaeal transfer RNA nucleoside G*-15 (2-amino-4,7-dihydro-4-oxo-7-beta-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (archaeosine)). J Biol Chem 268:10076–10086. 10.1016/S0021-9258(18)82174-3. [DOI] [PubMed] [Google Scholar]
- 99.McCarty RM, Krebs C, Bandarian V. 2013. Spectroscopic, steady-state kinetic, and mechanistic characterization of the radical SAM enzyme QueE, which catalyzes a complex cyclization reaction in the biosynthesis of 7-deazapurines. Biochemistry 52:188–198. 10.1021/bi301156w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bai Y, Fox DT, Lacy JA, Van Lanen SG, Iwata-Reuyl D. 2000. Hypermodification of tRNA in thermophilic archaea. Cloning, overexpression, and characterization of tRNA-guanine transglycosylase from Methanococcus jannaschii. J Biol Chem 275:28731–28738. 10.1074/jbc.M002174200. [DOI] [PubMed] [Google Scholar]
- 101.Phillips G, Chikwana VM, Maxwell A, El-Yacoubi B, Swairjo MA, Iwata-Reuyl D, de Crecy-Lagard V. 2010. Discovery and characterization of an amidinotransferase involved in the modification of archaeal tRNA. J Biol Chem 285:12706–12713. 10.1074/jbc.M110.102236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yokogawa T, Nomura Y, Yasuda A, Ogino H, Hiura K, Nakada S, Oka N, Ando K, Kawamura T, Hirata A, Hori H, Ohno S. 2019. Identification of a radical SAM enzyme involved in the synthesis of archaeosine. Nat Chem Biol 15:1148–1155. 10.1038/s41589-019-0390-7. [DOI] [PubMed] [Google Scholar]
- 103.de Crecy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, Grosjean H. 2010. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in archaea. Mol Biol Evol 27:2062–2077. 10.1093/molbev/msq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Young AP, Bandarian V. 2011. Pyruvate is the source of the two carbons that are required for formation of the imidazoline ring of 4-demethylwyosine. Biochemistry 50:10573–10575. 10.1021/bi2015053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu N, Jora M, Solivio B, Thakur P, Acevedo-Rocha CG, Randau L, de Crecy-Lagard V, Addepalli B, Limbach PA. 2019. tRNA modification profiles and codon-decoding strategies in Methanocaldococcus jannaschii. J Bacteriol 201:e00690-18. 10.1128/JB.00690-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou S, Sitaramaiah D, Noon KR, Guymon R, Hashizume T, McCloskey JA. 2004. Structures of two new “minimalist” modified nucleosides from archaeal tRNA. Bioorg Chem 32:82–91. 10.1016/j.bioorg.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 107.McCloskey JA, Graham DE, Zhou S, Crain PF, Ibba M, Konisky J, Soll D, Olsen GJ. 2001. Post-transcriptional modification in archaeal tRNAs: identities and phylogenetic relations of nucleotides from mesophilic and hyperthermophilic Methanococcales. Nucleic Acids Res 29:4699–4706. 10.1093/nar/29.22.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Young AP, Bandarian V. 2015. Mechanistic studies of the radical S-adenosyl-L-methionine enzyme 4-demethylwyosine synthase reveal the site of hydrogen atom abstraction. Biochemistry 54:3569–3572. 10.1021/acs.biochem.5b00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Young AP, Bandarian V. 2021. Eukaryotic TYW1 is a radical SAM flavoenzyme. Biochemistry 60:2179–2185. 10.1021/acs.biochem.1c00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Grell TAJ, Young AP, Drennan CL, Bandarian V. 2018. Biochemical and structural characterization of a Schiff base in the radical-mediated biosynthesis of 4-demethylwyosine by TYW1. J Am Chem Soc 140:6842–6852. 10.1021/jacs.8b01493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Noon KR, Guymon R, Crain PF, McCloskey JA, Thomm M, Lim J, Cavicchioli R. 2003. Influence of temperature on tRNA modification in archaea: Methanococcoides burtonii (optimum growth temperature [Topt], 23°C) and Stetteria hydrogenophila (Topt, 95°C). J Bacteriol 185:5483–5490. 10.1128/JB.185.18.5483-5490.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Reddy DM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Stetter KO, Widdel F, McCloskey JA. 1992. Structure determination of two new amino acid-containing derivatives of adenosine from tRNA of thermophilic bacteria and archaea. Nucleic Acids Res 20:5607–5615. 10.1093/nar/20.21.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolff P, Villette C, Zumsteg J, Heintz D, Antoine L, Chane-Woon-Ming B, Droogmans L, Grosjean H, Westhof E. 2020. Comparative patterns of modified nucleotides in individual tRNA species from a mesophilic and two thermophilic archaea. RNA 26:1957–1975. 10.1261/rna.077537.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Landgraf BJ, Arcinas AJ, Lee KH, Booker SJ. 2013. Identification of an intermediate methyl carrier in the radical S-adenosylmethionine methylthiotransferases RimO and MiaB. J Am Chem Soc 135:15404–15416. 10.1021/ja4048448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang B, Arcinas AJ, Radle MI, Silakov A, Booker SJ, Krebs C. 2020. First step in catalysis of the radical S-adenosylmethionine methylthiotransferase MiaB yields an intermediate with a [3Fe-4S](0)-like auxiliary cluster. J Am Chem Soc 142:1911–1924. 10.1021/jacs.9b11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Forouhar F, Arragain S, Atta M, Gambarelli S, Mouesca JM, Hussain M, Xiao R, Kieffer-Jaquinod S, Seetharaman J, Acton TB, Montelione GT, Mulliez E, Hunt JF, Fontecave M. 2013. Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat Chem Biol 9:333–338. 10.1038/nchembio.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Esakova OA, Grove TL, Yennawar NH, Arcinas AJ, Wang B, Krebs C, Almo SC, Booker SJ. 2021. Structural basis for tRNA methylthiolation by the radical SAM enzyme MiaB. Nature 597:566–570. 10.1038/s41586-021-03904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arragain S, Handelman SK, Forouhar F, Wei FY, Tomizawa K, Hunt JF, Douki T, Fontecave M, Mulliez E, Atta M. 2010. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J Biol Chem 285:28425–28433. 10.1074/jbc.M110.106831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. 2008. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc Natl Acad Sci USA 105:1826–1831. 10.1073/pnas.0708608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang B, Johansson MJ, Bystrom AS. 2005. An early step in wobble uridine tRNA modification requires the elongator complex. RNA 11:424–436. 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Selvadurai K, Wang P, Seimetz J, Huang RH. 2014. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat Chem Biol 10:810–812. 10.1038/nchembio.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Glatt S, Zabel R, Kolaj-Robin O, Onuma OF, Baudin F, Graziadei A, Taverniti V, Lin TY, Baymann F, Seraphin B, Breunig KD, Muller CW. 2016. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat Struct Mol Biol 23:794–802. 10.1038/nsmb.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lin T-Y, Abbassi NEH, Zakrzewski K, Chramiec-Głąbik A, Jemioła-Rzemińska M, Różycki J, Glatt S. 2019. The elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat Commun 10:625. 10.1038/s41467-019-08579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schwalm EL, Grove TL, Booker SJ, Boal AK. 2016. Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA. Science 352:309–312. 10.1126/science.aad5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Trievel RC, Rojas JR, Sterner DE, Venkataramani RN, Wang L, Zhou J, Allis CD, Berger SL, Marmorstein R. 1999. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci USA 96:8931–8936. 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jarrett JT. 2015. The biosynthesis of thiol- and thioether-containing cofactors and secondary metabolites catalyzed by radical S-adenosylmethionine enzymes. J Biol Chem 290:3972–3979. 10.1074/jbc.R114.599308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jurgenson CT, Begley TP, Ealick SE. 2009. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem 78:569–603. 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yokoyama K, Li D, Pang H. 2022. Resolving the multidecade-long mystery in MoaA radical SAM enzyme reveals new opportunities to tackle human health problems. ACS Bio Med Chem Au 2:94–108. 10.1021/acsbiomedchemau.1c00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pfluger K, Baumann S, Gottschalk G, Lin W, Santos H, Muller V. 2003. Lysine-2,3-aminomutase and β-lysine acetyltransferase genes of methanogenic archaea are salt induced and are essential for the biosynthesis of Nε-acetyl-β-lysine and growth at high salinity. Appl Environ Microbiol 69:6047–6055. 10.1128/AEM.69.10.6047-6055.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tunckanat T, Gendron A, Sadler Z, Neitz A, Byquist S, Lie TJ, Allen KD. 2022. Lysine 2,3-aminomutase and a newly discovered glutamate 2,3-aminomutase produce beta-amino acids involved in salt tolerance in methanogenic archaea. Biochemistry 61:1077–1090. 10.1021/acs.biochem.2c00014. [DOI] [PubMed] [Google Scholar]
- 131.Su X, Lin Z, Lin H. 2013. The biosynthesis and biological function of diphthamide. Crit Rev Biochem Mol Biol 48:515–521. 10.3109/10409238.2013.831023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2. Download jb.00197-22-s0001.pdf, PDF file, 0.2 MB (182.5KB, pdf)