Abstract
Immune-checkpoint inhibitors (ICI), although revolutionary in improving long-term survival outcomes, are mostly effective in patients with immune-responsive tumors. Most patients with cancer either do not respond to ICIs at all or experience disease progression after an initial period of response. Treatment resistance to ICIs remains a major challenge and defines the biggest unmet medical need in oncology worldwide. In a collaborative workshop, thought leaders from academic, biopharma, and nonprofit sectors convened to outline a resistance framework to support and guide future immune-resistance research. Here, we explore the initial part of our effort by collating seminal discoveries through the lens of known biological processes. We highlight eight biological processes and refer to them as immune resistance nodes. We examine the seminal discoveries that define each immune resistance node and pose critical questions, which, if answered, would greatly expand our notion of immune resistance. Ultimately, the expansion and application of this work calls for the integration of multiomic high-dimensional analyses from patient-level data to produce a map of resistance phenotypes that can be utilized to guide effective drug development and improved patient outcomes.
Introduction
The clinical development of immune-checkpoint inhibitors (ICI), specifically those targeting the T cell–inhibitory receptors cytotoxic T lymphocyte antigen-4 (CTLA-4) and programmed death-1 (PD-1), radically shifted the cancer treatment paradigm and transformed oncology practice. Survival outcomes from large, randomized trials with at least 4 to 5 years' follow-up are known for melanoma and non–small cell lung cancer (NSCLC) populations. Reported outcomes from melanoma trials approximate the 4-year survival rate of patients treated with the anti–CTLA-4 therapeutic ipilimumab between 19% and 36%, the anti–PD-1 therapeutic nivolumab at 46%, the anti–PD-1 therapeutic pembrolizumab at 44%, and the combination of nivolumab plus ipilimumab at 53% (1–6). Similar trends are emerging in the NSCLC population treated with anti–PD-1 monotherapy, where the estimated 5-year survival rate of previously treated patients is approximately 16% with nivolumab (CM209-003), 15% with pembrolizumab (KN-001), and of treatment-naïve patients is 23% with pembrolizumab (KN-001; refs. 7, 8). Most recently, it was reported that first-line NSCLC patients with a programmed death ligand 1 (PD-L1) tumor proportion score of at least 50% demonstrated a median overall survival (OS) of 26% (KN-024; ref. 9). Although such outcomes are impressive, most patients with advanced cancer treated with anti–CTLA-4 or anti–PD-1/anti–PD-L1 [anti–PD-(L)1] agents do not achieve durable benefit and show resistance to therapy between 3 months to 3 years after initiating treatment.
Clinical progression on ICIs has been broadly conceptualized into three categories aligned to clinical response scenarios, namely, primary resistance (no response to checkpoint inhibition), adaptive resistance (functional antitumor response that is limited or shaped by immunosuppression), and acquired resistance (initial response followed by eventual disease progression; refs. 10–12). Although these categories closely approximated clinical observations, a deeper understanding of immune resistance necessitates a more expansive framework that can accommodate both dynamic biological complexity and clinical response outcomes.
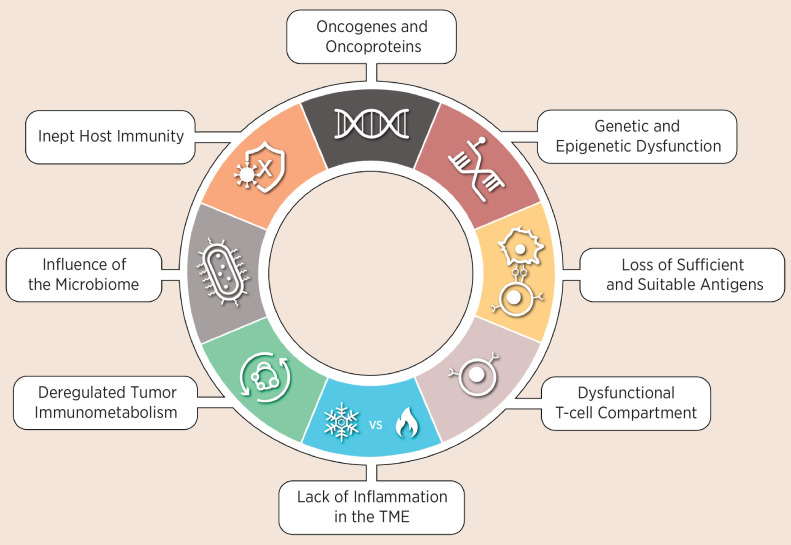
The approach necessary to derive a resistance framework that will guide future research and facilitate reverse translation is likely to include a systematic characterization of resistance to ICIs, where molecular and clinical definitions are integrated into a structure that can be utilized to identify therapeutic interventions. Critical elements of such a resistance framework should include (i) identification of resistance patterns, (ii) characterization of patterns to define discrete resistance phenotypes, (iii) relational mapping between individual resistance mechanisms and resistance phenotypes, and (iv) integrated validation of resistance phenotypes to clinical outcomes. As part of a workshop sponsored by the Parker Institute for Cancer Immunotherapy (PICI), the Cancer Research Institute (CRI), and the Society of Immunotherapy of Cancer (SITC), we constructed a view of immune resistance by aggregating the evidence of known resistance mechanisms that undermine key biological processes and block successful antitumor immunity. We organized the evidence into eight biological converging points and denote them as immune-resistance nodes, collectively representing the emerging hallmarks of resistance to ICIs (Fig. 1). For each immune-resistance node, we examined the seminal discoveries that define each node and posed critical questions (Table 1), which if answered would greatly expand our notion of resistance. We then speculated on how these core immune-resistance nodes might integrate with additional aspects of malignancy, including tumor development, treatment modalities, detection of resistance, and approaches to target resistance (Fig. 2). Although in this Perspective article we interchangeably utilize the terms checkpoint inhibitors, ICIs, immunotherapy, and immune resistance, our focus is centered around resistance to anti–PD-(L)1 and anti–CTLA-4 therapies and predicated primarily on clinical and translational data from blockade of PD-1 and PD-L1. We recognize that anti–PD-(L)1 and anti–CTLA-4 ICIs work through distinct mechanisms but we lack balanced clinical and translational data sets to delineate whether they may be provoking distinct resistance mechanisms and resistance patterns. As monotherapy, anti–PD-(L)1 agents have demonstrated clinical efficacy across a wide range of tumor indications, which makes it possible to examine the range of resistance. Notwithstanding, this question remains vitally important for future work, especially because both anti–PD-(L)-1 and anti–CTLA-4 ICIs are now used in combination regimens with each other and with other drugs that may elicit unique resistance patterns.
Figure 1.
The hallmarks of resistance to ICIs are supported by seminal data sets and organized into key biological processes referred to as resistance nodes.
Table 1.
Critical questions to address in future investigations of resistance to ICIs.
| Resistance node | Critical questions |
|---|---|
| Oncogenes and oncoproteins | How do functional “driver” mutations influence the immune repertoire and tumor immunogenicity? |
| Do driver mutations influence antigen presentation, CD8+ T-cell priming, and CD8+ T-cell tumor infiltration? | |
| How do comutations in tumor suppressors influence response and/or resistance to therapy? | |
| Is there a hierarchy among comutations, and are their effects direct or indirect? | |
| Genetic and epigenetic dysfunction | What is the combined frequency and degree of HLA class I loss across tumor histologies? |
| Are HLA class I loss patterns clustered by mechanisms of action? What are the triggers for loss? | |
| How do tumors subvert HLA class I loss? | |
| What is the timing of HLA class I loss in the context of a primary tumor vs. metastatic disease? | |
| Loss of sufficient and suitable antigens | Given HLA and antigen diversity, are there identifiable patterns that predict ICI resistance? |
| What features of the neoantigen-specific T-cell population are required for tumor clearance? | |
| Do neoantigens need CD4+ (HLA class II) and CD8+ (HLA class I) to circumvent resistance? | |
| Can epigenetic therapies reverse silencing and thereby boost neoantigen expression? | |
| What is the role of mutation-based neoantigens and intra/intertumor heterogeneity? | |
| Dysfunctional T-cell compartment | Which cell subsets are the primary influencers of ICI resistance? |
| How is T-cell “activatability” shaped after exposure to immunotherapy? | |
| Can the “state” of T-cell functionality be monitored continuously in vivo? | |
| Can costimulation be measured in vivo? | |
| What is the role for costimulatory pathways beyond CD28? | |
| What is the role of 4-1BB signaling in T-cell anergy and antitumor immunity? | |
| How can IFNγ sensitivity be utilized to identify patients who are potentially resistant to ICI? | |
| What aspects of IFNγ signaling directly affect ICI resistance and its relationship to disease stage? | |
| How interdependent are fluctuations in IFNγ signaling in the context of ICI resistance? | |
| What characterizes the antigen repertoire against a “successful” CD8+ T-cell memory response? | |
| Is there evidence of continual immunologic memory in long-term survivors? | |
| How is immunologic memory generated and dependent on a persistent T-cell memory clone? | |
| What interactions among immune subsets are needed to generate durable CD8+ T-cell memory? | |
| Lack of inflammation in the TME | Are the immune-excluded and immune-desert phenotypes sufficiently defined? |
| Does the tumor cell of origin affect the different mechanisms underlying the cold phenotypes? | |
| Does the spatial positioning of immune cells dictate or influence resistance to ICI therapies? | |
| What key influencers of trafficking and infiltration can be therapeutically targeted? | |
| Are there dominant chemokine networks within TME that promote ICI resistance? | |
| Are the mechanisms of immune exclusion uniquely influenced by various therapies? | |
| Deregulated tumor immunometabolism | How does therapy induce immune metabolic signature switching toward ICI resistance? |
| Does resistance involve modified mitochondrial biogenesis and immunometabolism plasticity? | |
| Can longitudinal metabolic signatures of circulating cancer and immune cells predict resistance? | |
| Can immunometabolism-based resistance be correlated with TME and immune infiltrate? | |
| Influence of the microbiome | Is the gut microbiome predictive of patient resistance to ICI treatment? |
| Can we modulate the gut microbiome to improve immune capacity and “fitness”? | |
| Can specific clusters of microbes predict, prevent, and/or stop ICI resistance? | |
| Does the gut microbiome affect toxicity to ICI? | |
| Inept host immunity | What is the impact of lymphocyte count and humoral factors (CRP and LDH) on ICI resistance? |
| How do subclinical chronic infections and inflammation shape resistance to immunotherapy? | |
| What are the effects of hormones and prostaglandins? | |
| How is immune capacity defined, and can it serve as a clinically meaningful measure of resistance? |
Abbreviations: CRP, C-reactive protein; LDH, lactate dehydrogenase.
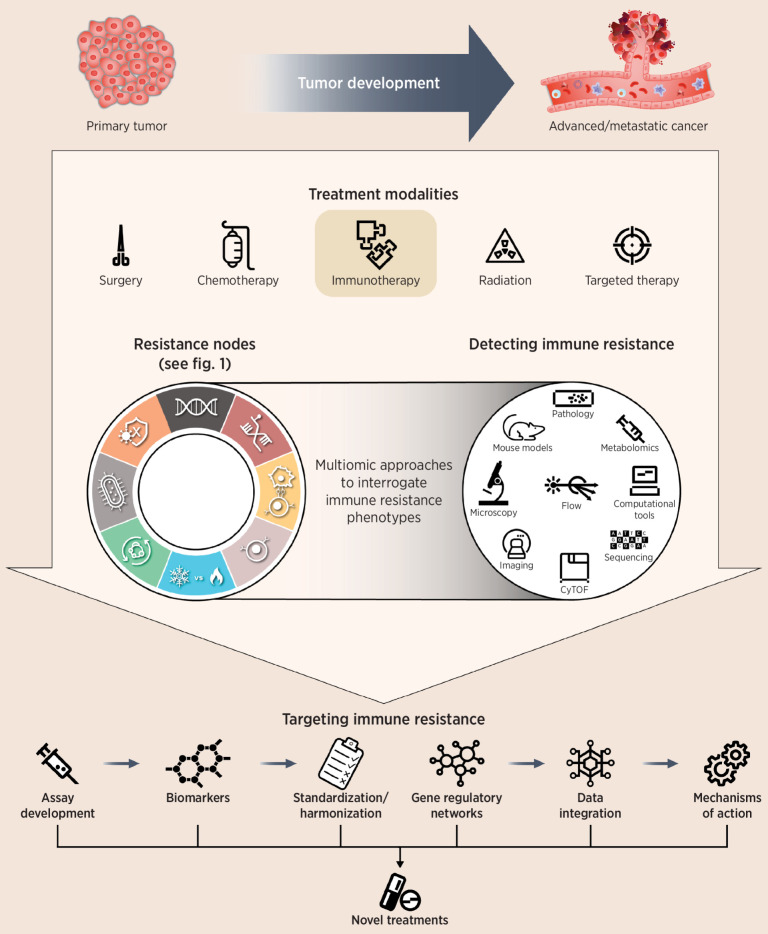
Figure 2.
Efforts to better understand resistance continue to be of great interest to the oncology community. We postulate that resistance mechanisms to immune-checkpoint blockade converge on eight resistance nodes. A deep and systematic characterization of the eight resistance nodes, using integrated multiomic measurements, is likely to inform better clinical trial design, identify clinically actionable insights, discover composite biomarkers, and reveal new drug targets and treatment combinations. Future advancements will likely provide highly accurate prediction capabilities, making it possible to identify patients who might resist immunotherapy so that more meaningful treatments can be applied. CyTOF, cytometry by time of flight.
Impact of Oncogenes and Oncoproteins
Oncogenic signaling is the least understood aspect of functional immunogenicity, but has a dramatic influence on the tumor microenvironment (TME) and normal tissue surrounding the tumor mass. Evidence for key oncogenic processes that can potentially foster resistance to immunotherapy includes: (i) oncogenic cooperation, (ii) exclusion of CD8+ T cells, (iii) induction of regulatory T cells (Treg) and myeloid-derived suppressor cells (MDSC), and (iv) deregulation of IFNγ signaling and PD-L1 expression.
Oncogenic cooperation and the coevolution of tumors and their immune environments were elucidated in recent analysis from The Cancer Genome Atlas (TCGA) database that landscaped 10 canonical cancer pathways (13). This exploration of mutually exclusive and co-occurring pathways indicated the potential influence of oncogenic synergies that can promote treatment resistance and potentially immune evasion. Oncogenic cooperation between Kras and Myc to produce highly proliferative tumors with an inflamed, angiogenic, but immunosuppressed stroma was also demonstrated using a mouse lung cancer model of conditional KrasG12D and inducible Myc (14), indicating the ability of Myc overexpression to rapidly clear the tumor of innate and adaptive immune lymphocytes. Several reports suggesting that Myc gain of function can influence antitumor immunity and immunotherapy response further substantiate these findings (15–18).
Oncogene-driven T-cell exclusion is another process by which tumors can shut down antitumor immunity and is evidenced by gain-of-function alterations of the β-catenin–Wnt pathway. Analyses of human melanoma samples revealed that upregulation of β-catenin–Wnt signaling was associated with a lack of T cells in both the primary tumors and distant metastases (19, 20). Analyses in a conditional BrafV600E and inducible β-catenin mouse model revealed a causal relationship between activation of β-catenin signaling, lower T-cell infiltration, and lack of response to checkpoint inhibition (21–24). Additionally, oncogenic signaling can induce suppressive immune-cell populations as exemplified by the potential of oncogenic KRAS to convert CD4+ T cells to functional Tregs (25) and the inactivation of p53 to induce MDSCs (26). Similarly, the hedgehog pathway can have profound impact on the composition and functionality of tumor-infiltrating immune cells and upregulation of PD-L1 expression (27–29). The activity of oncogenes, particularly members of the receptor tyrosine kinase (RTK)–RAS pathway, has recently been linked more directly to immunosuppression. RAS signaling can upregulate PD-L1 expression on tumor cells by stabilizing PDL1 mRNA (30), an observation that is consistent with the reported association of KRAS expression with increased PD-L1 expression suggesting that the KRAS–IRF3 pathway can support immune suppression and resistance to immunotherapy (31).
Clinical trial observations indicate that patients whose tumors harbor driver mutations do not respond well to ICIs (e.g., KRAS mutations in colorectal carcinoma and EGFR mutations in NSCLC), although melanoma tumors with either BRAFV600E or BRAFWT alleles potentially respond similarly to anti–CTLA-4 or anti–PD-(L)1 agents. Careful analysis of emerging data sets for a better understanding of the local and systemic impact of driver mutations in the context of ICI therapy is key.
Genetic and Epigenetic Dysfunction
Human leukocyte antigen (HLA)–directed antigen presentation is the basis for T cell–mediated antitumor responses. Substantial evidence suggests that tumor cells can evolve to escape immune recognition by reducing HLA proteins on their surface (HLA loss), thus creating an important avenue for ICI resistance (32, 33).
Genetic (“hard”) and epigenetic (“soft”) mechanisms leading to HLA loss can directly affect HLA class I including its transcriptional regulation, or indirectly target genes that are part of the antigen-presentation machinery (APM). In a recent study from the TRACERx consortium analyzing 258 tumor regions from 88 prospectively acquired tumors, 56% of lung adenocarcinomas and 78% of lung squamous cell carcinomas showed evidence of antigen-presentation disruption, through either HLA loss of heterozygosity (LOH) or APM mutations (34). Complete loss of beta-2 microglobulin (B2M) alleles, an essential component of major histocompatibility complex class I (MHC-I) required for antigen presentation, was evident only in ICI nonresponders, whereas permutations of the B2M gene (absence of tumor-specific expression, frameshift mutations, or LOH) were observed in ∼30% of ICI nonresponders (35).
Epigenetically, HLA gene expression in tumor cells can be regulated directly through promoter hypermethylation. HLA class I gene promoter methylation occurs in 49% to 60% of gastric cancer samples, compared with 6% to 19% of adjacent normal tissue, and often results in transcriptional inactivation of MHC-I loci HLA-A, -B, and -C (36). Indirect epigenetic regulation of HLA gene expression, through histone methyltransferases within chromatin remodeling machinery, can subsequently impact a range of potential resistance mechanisms including T-cell infiltration, TNFα responsiveness, PD-L1 expression, and MHC-1 expression (37). Similarly, expression of the HLA-A, -B, and -C genes, and B2M are under at least partial control of the transcription factor DUX4, which is normally suppressed epigenetically after development. In patient subsets across tumor types, genetic variation can trigger dysregulated DUX4 expression, leading to reduced IFNγ responsiveness and decreased MHC-I expression (38).
Although evidence of HLA loss has been documented (32), a systematic evaluation of the combined frequency and contributions of hard and soft HLA loss across tumor types is warranted to expand our understanding of disease etiology. Hard HLA loss, HLA heterogeneity, as well as previously reported HLA types, were not found to be associated with response in a recent meta-analysis (39). It is likely that we need to capture all mechanisms of HLA downregulation to accurately estimate their impact on outcomes across treatment settings and devise strategies to reverse immune resistance, or possibly prevent a tumor from evolving an HLA loss phenotype. Additionally, partial or complete loss of HLA proteins on tumors spotlights natural killer (NK) cells and their role in immunosurveillance. As NK cells detect HLA class I expression loss in infected or transformed cells and initiate target cell killing (40), tumors displaying HLA loss must also have evolved resistance against NK cell–mediated killing (41). Thus, exploration of NK cell–based therapies might extend immunotherapeutic options that do not require neoantigen presentation by HLA.
Lack of Sufficient or Suitable Antigen Production
Tumor antigens can arise from multiple sources (42). The productive engagement of the T-cell receptor (TCR) complex requires not only the presentation of antigens but also immunologically suitable antigens in sufficient quantities to produce effective T-cell targeting and tumor killing. Thus, antigen suitability and antigenic escape can affect resistance to immunotherapies. Recent focus on the evaluation of neoantigens as therapeutic targets has garnered attention from the following observations: (i) lack of thymic deletion of T cells targeting neoantigens relative to deletion of T cells targeting self-antigens (43), (ii) association of predicted neoantigen number with OS and/or clinical response (44–49), (iii) successful therapeutic neoantigen vaccination in mice and evaluation in patients (50–52), and (iv) evidence of clinical responses using neoantigen-specific T cells in adoptive transfer (53, 54).
Lack of ICI response has been associated with heterogeneous neoantigen expression and a lack of truncal neoantigens (34, 46, 55), even though only a minority of predicted neoantigens are truly immunogenic in vivo (56–59). Tumors displaying either few or ample neoantigens can succumb to immune-editing mechanisms selecting for tumor clones with diminished neoantigen expression (60). In fact, it is possible that primary tumors may already display a resistance phenotype determinately characterized by a paucity of immunogenic neoantigens. As discussed above, immunotherapy resistance can be mediated via HLA loss and possibly enriched in tumors with immunogenic neoantigen expression (61, 62). A fascinating observation from preclinical neoantigen vaccination experiments demonstrates that neoantigens predicted based on MHC-I binding characteristics can stimulate CD4+ T-cell responses in excess of CD8+ T-cell responses (50).
A complete view of cancer antigens through tumor antigen expression mapping will enable an assessment of the character and suitability of antigens to trigger productive ICI-directed antitumor immunity. Adoptive transfer studies using tumor-infiltrating lymphocytes (TIL) have shown that T cells enriched for a single dominant neoantigen can elicit responses in patients with metastatic cancer. A neoantigen map together with a computational estimate of the number of neoantigen specificities required to promote immune responses will provide a baseline for understanding antigen suitability. Thus, our ability to better define neoantigens and their role in maximizing antitumor immunity should enable approaches to prevent or reverse ICI resistance and hopefully improve clinical outcomes.
Dysfunctional T-cell Compartment
Impeded T-cell activation and exhaustion
T cell–mediated antitumor immune responses are key to successful immunotherapy, and their dysfunction is central to ICI resistance. In the context of chronic viral infections and malignancies (63, 64), exhausted T cells gradually lose their functional capacities, most notably through chronic antigen exposure, upregulation of immune-checkpoint receptors, and incomplete priming. This T cell–exhaustion program is emerging as a specific differentiation pathway enabled by the HMG-box transcription factor TOX via specific epigenomic remodeling; it normally serves to ensure survival of antigen-experienced T cells during chronic stimulation by preventing TCR overstimulation and activation-induced cell death (65–67). Interestingly, genetic inactivation of TOX in chronically activated T cells does not necessarily promote sustained T-cell functionality, likely due to enhanced cell death and premature loss of TOX-deficient T cells (65, 67). Similarly, in tumors, although it is possible to transiently counteract the exhaustion of chronically stimulated tumor antigen–specific CD8+ T cells, the “rescued” CD8+ T cells often reenter a dysfunctional state (68, 69). PD-1 blockade specifically acts by reactivating partially exhausted progenitor-like TCF-1+PD-1+CD8+ T cells, which can mature and execute their function, but ultimately become terminally exhausted themselves (69–71). Paradoxically, these observations indicate that blocking the PD-1 pathway can accelerate the exhaustion process in progenitor-like tumor-specific T cells, leading to ICI resistance.
A key question is whether dysfunctional T cells can be epigenetically reprogrammed or alternatively eliminated to make space for new, functionally stable, tumor antigen–specific T-cell populations. Additionally, PD-1 blockade of suboptimally primed CD8+ T cells can induce a specific population of dysfunctional T cells (PD-1+CD38hiCD8+), suggesting that ICI resistance can be reversed with functional antigen stimulation and implying a temporal relationship between optimal priming and subsequent PD-1 blockade (72).
Absent or incomplete costimulation
In addition to TCR engagement of a cognate peptide–MHC complex (pMHC), costimulation mediated by canonical CD28–CD80/86 engagement is critical for a productive T-cell response and for long-lasting antitumor immunological memory. Lack of costimulation during TCR–pMHC recognition has been implicated in T-cell anergy and hyporesponsiveness. Evidence for functional T-cell hyporesponsiveness suggests that it occurs under the influence of either chronic antigen stimulation or TCR engagement lacking costimulation (73). It is plausible that inadequate expression of costimulatory molecules in antigen-presenting cells in the presence of a progressing tumor may contribute to ICI resistance. Theoretically, this may be further exacerbated by chronic TCR stimulation of TME-residing T cells, ultimately leading to T-cell anergy (74–76). For example, in a tamoxifen-induced liver cancer model, naïve antigen-reactive T cells displayed hyporesponsiveness 1 to 2 days after transfer, due to upregulation of negative transcriptional regulators, including those associated with T-cell anergy and hyporesponsiveness (Batf, Egr1, Prdm1, Ptpn11, Ptpn12, Dusp1, and Dusp6) and inhibitory cell-surface receptors (Pdcd1, Lag3, Cd160, and Ctla4; ref. 77). The inability of NFAT–AP-1 transcriptional complexes to drive T-cell activation is a common feature of anergy induced by lack of costimulation and checkpoint-induced immune dysfunction (78). NFAT signaling can also drive pathways associated with T-cell exhaustion and anergy in the absence of AP-1 (79, 80), highlighting distinct immune resistance pathways governed by this transcription factor.
Additional costimulatory pathways including 4-1BB have been known to compensate for CD28 signaling (81), and both CD28 and 4-1BB have been used as costimulatory domains in the development of chimeric-antigen receptors (CARs; ref. 82). Ultimately, delineating the role of costimulation from immune agonism will expand our understanding of T-cell activation and possibly identify new strategies to prevent the influence of hyporesponsiveness and anergy on ICI resistance. Ongoing clinical studies, using monoclonal or bispecific antibodies triggering costimulatory targets, will help elucidate the importance of costimulation in the context of treatment resistance.
Dysfunctional IFNγ signaling
IFNγ, the quintessential effector cytokine of Type I immunity, is produced by activated cytotoxic CD8+ T cells, γδ T cells, and NK cells and can exert pleiotropic effects. IFNγ signaling through IFNGR1–IFNGR2 complexed heterodimers recruits JAK1 and JAK2 and leads to STAT1 phosphorylation. Phosphorylated STAT1 modulates IFNγ-regulated gene expression (including genes encoding MHC-I, APM, and costimulatory molecules), induces the expression of CCL9 to recruit CD11b+ dendritic cells, skews macrophages toward an M1 proinflammatory phenotype, and inhibits immune suppressive cells including Tregs, Th2, Th17, and MDSCs (83–86).
Mice lacking IFNγ, IFNγ receptors, or STAT1 develop tumors more rapidly and with increased frequency, demonstrating the central role of IFNγ signaling in tumor surveillance and antitumor immunity (87). IFNγ acts directly on tumor cells to enhance tumor cell immunogenicity through MHC-I upregulation and plays a role in tumor escape by inducing tumor cells to become selectively unresponsive to IFNγ. In melanoma, acquired resistance to PD-1 blockade is associated with IFNγ-pathway defects involving truncating mutations in JAK1 and JAK2 (88). In adaptive resistance, IFNγ can potentially compromise antitumor immunity by directly upregulating both PD-L1 and PD-L2 on tumor and immune cells (48, 89, 90) and can drive resistance to checkpoint blockade by PD-L1–dependent and –independent mechanisms (91). Furthermore, interrupting chronic IFNγ signaling in tumor cells leads to increased IFNγ production by exhausted T cells and enhanced IFN-stimulated gene expression in tumor and immune cells. As expected, tumors with adequate MHC-I and antigen are killed by T cell–mediated mechanisms that produce IFNγ, and tumors with low expression of MHC-I or limited antigen are killed by PD-1+TRAIL+ NK cells receiving maturation signals from IFNγ (92). Despite divergent roles for IFNγ signaling, altering these pathways can enhance response to ICIs (92), and IFNG RNA gene-expression signatures have been shown to predict response and survival with anti–PD-1 therapy (93).
Impaired generation of CD8+ T-cell immune memory
T-cell memory, in particular tissue-resident memory (TRM) CD8+ T cells, may be a crucial mediator of antitumor immunity and ICI resistance. Understanding the mechanisms and conditions needed to improve the generation and maintenance of CD8+ T-cell immune memory against tumors is critical for the improvement of immunotherapy outcomes, especially if immune memory can yield durable complete responses.
The importance of TRM CD8+ T cells is supported by data in patients with melanoma who responded to anti–PD-1 and harbored expanded CD8+ effector memory T-cell populations (94) and in patients with breast cancer whose TRM CD8+ T cells contributed to immunosurveillance (95). Epigenetic and genomic studies characterizing the CD8+ TILs indicated that a subset of these cells have a progenitor-exhausted phenotype, resembling stem cell–like memory cells, characterized by TCF-1 expression and associated with clinical benefit to anti–PD-1 therapy (96). Furthermore, although CD8+ T cells are viewed as key effectors in immune-mediated tumor rejection, as their presence and persistence have been correlated with therapeutic outcome, it is important to recognize that a productive and long-lasting antitumor immune response likely involves the cooperation of multiple immune cells working in concert with CD8+ T cells (97, 98). Thus, fully understanding T-cell memory generation and T-cell persistence will likely help us better select patients suitable for ICI therapy by estimating the quality, depth, and duration of their tumor response and alternatively predict ICI therapy resistance.
Lack of Inflammation in the TME
As we consider the influence of local tumor processes with systemic effects, inflammation is a key culprit influencing pro- and anti-immune signals that can shape a patient's response or resistance to immunotherapy. In the context of the TME, three clinically relevant tumor–immune phenotypes have been described: (i) inflamed tumors, which are regarded as “hot,” and two categories typically referred to as “cold,” (ii) immune-excluded, and (iii) immune-desert tumors. Undoubtedly, a continuum of immunogenic potential exists, broadly bookended by “inflamed” and “noninflamed” phenotypes (99, 100). The noninflamed “cold” phenotypes probably characterize most human tumors; thus, their clinical significance warrants additional probing to specifically elucidate the origin and nature of the inflammation, or lack thereof, needed to direct and sustain antitumor ICI responses. A nuanced systematic approach that expands the classification of cold tumors according to their inflammatory status may identify cancer-promoting inflammation more clearly, an idea supported by evidence from large trials where nonsteroidal anti-inflammatory drugs such as aspirin reduce cancer incidence and/or mortality (101–103). Elucidating complex relationships between tumor heterogeneity, TME plasticity, and the inflammatory process may reveal how systemic or local inflammation can shape the plasticity of tumor, stromal, and immune cells (103), and it may highlight priority areas likely to influence future clinical research (104).
A specific and critical element of the definition of cold tumors is the absence of functional effector T cells in the TME. Dysfunctional T-cell trafficking and infiltration, therefore, impinge on the necessary inflammation that, together with PD-L1 expression, marks the hot or inflamed tumor–immune phenotype and immunotherapy responsiveness (105–110). The process of functional T-cell trafficking and infiltration faces three major hurdles: (i) defective cellular homing, (ii) regulatory immune cells, and (iii) damaging chemokine networks. The successful homing of T cells is controlled spatiotemporally through tethering and rolling, chemokine activation, and transmigration, all of which are disrupted in tumors (110–112). Some examples leading to dysfunctional T-cell migration include upregulation of expression of the adhesion molecule α4β7, induction of various costimulatory molecules, loss of CCR7, and, possibly, downregulation of retinoic acid (113–117). Upon successful intratumoral homing, lymphocytes encounter physical barriers to infiltration via encounters with Tregs, MDSCs, immature dendritic cells, macrophages, and neutrophils, fibroblasts, and abnormal extracellular matrix. Recent data suggest that it may be possible to shift cellular populations in the TME or reprogram cell types that influence ICI resistance. Macrophages, for example, can switch from an antitumorigenic to a protumorigenic phenotype, thus continuing work has examined the reeducation of M2-like macrophages to M1-like antitumor macrophages using LXR agonism, IL8 inhibitors, and CD40 agonism (118, 119). Although ambitious, the reeducation or plasticity of Tregs and other cell types promoting resistance is promising (120).
Deregulated Tumor Immunometabolism
Immunometabolism represents a core process with local and systemic influence that may directly or indirectly modulate response and resistance to ICI treatments. Metabolic signaling dictates the function of immune cells (121–125) and thus presents an avenue for identifying clinically relevant immune resistance patterns (126–129). In the hostile hypoxic TME, oxygen and nutrient constraints lead cancer cells to switch from oxidative phosphorylation to aerobic glycolysis, and they then outcompete T cells for glucose (130–132). Checkpoint blockade mediates reduced glycolysis and increased effector T-cell function (129), indicating that the key metabolic mediators ATP and NAD+ and the ectoenzymes governing their catabolism may play a role to influence immune resistance (133–136).
T-cell metabolic programming determines T-cell lineage commitment and differentiation (137), with important implications for strategies using ex vivo cultures for adoptive transfer. Furthermore, immunometabolic attributes of CAR T cells influence their persistence and lack of exhaustion. For example, CAR T cells with 4-1BB domains promote central memory T-cell growth, optimizing their metabolic fitness in the TME (138), whereas glucose-limiting expansion is counterproductive, leading to more cytotoxic T cells with increased in vivo persistence and longevity (139). 4-1BB costimulation also increases transfer efficiency and PD-1 blockade effectiveness by increasing mitochondrial mass and the respiratory capacity of cytotoxic T cells (140). Conversely, CAR T cells with CD28 domains yield effector memory T cells with enhanced glycolysis (138). Interestingly, antigen-directed asymmetric cell division generating effector and memory T cells (141) may be overcome by infusing patients with differentially weighted immunometabolic T cells, ensuring immediate antitumor activity and long-lived persistence. These and other insights prompt the clinical investigation of immunometabolic targets with the potential to counteract immune resistance. Innovative combinations of immunotherapies with immunometabolic reprogramming agents hold promise for promoting immune activation, response durability, and, perhaps, mitigation of ICI resistance. Distinguishing metabolic signatures of tumor cells from infiltrated T cells may also provide insights to pinpoint metabolic heterogeneity and metabolic signatures that can alter and override ICI resistance.
Influence of the Microbiome
The human microbiome is a complex aggregate of microorganisms that exert influence crucial to the metabolic, immunologic, hormonal, and homeostatic functions of the host. The microbiome's effects on ICI resistance remain underexplored. Recent work in mouse models and human studies highlights the impact of gut and tumor microbiota on the potential for clinical response to anticancer therapeutics (142–148). Early work revealed that antibiotics taken within the month preceding the start of ICIs reduced both progression-free survival (PFS) and OS across many metastatic and stage III malignancies (142, 149). Additional studies revealed that fecal microbiota transplantation of stools from patients who were likely to respond or be unresponsive to ICI-based therapy conferred sensitivity or resistance, respectively, to tumor-bearing mice treated with anti–PD-1 antibodies (150). Efforts to better understand gut microbiome signatures associated with ICI resistance include shotgun metagenomic sequencing of patient stools at diagnosis, with samples currently being evaluated in metastatic melanoma (148), kidney (149), and lung cancers (142).
Conversely, ICIs have been shown to affect the composition of the gut microbiome, with one study reporting that anti–CTLA-4 antibodies resulted in an increase in the relative abundance in Bacteroides spp. (B. fragilis) and Burkholderiaceae family members in the ileum. These bacterial families are involved in the IL12-dependent priming of Th1 cells, hence the immunostimulatory and anticancer effects of CTLA-4 blockade (145). It is possible, therefore, that disruptions in gut homeostasis lead to microbial translocation and augmented function in CD8+ T cells. These data suggest that microbial translocation from the gut results in the release of danger signals (e.g., Toll-like receptor ligands) triggering tumor regression (151–153). This is consistent with research demonstrating that bacterially derived products can activate the innate immune system and trigger tumor regression (154, 155). These studies and others suggest that the intestinal microbiota modulate the effects of ICIs via bacterial translocation of selected species into secondary lymphoid organs (147). Continued research on the microbiome and the application of clinically actionable hypotheses focused on patients showing resistance to ICIs holds great promise.
Inept Host Immunity
Systemic host immunity, not just effector T-cell functionality, inevitably shapes the emergence of a primary tumor and ultimately the potential for response to immunotherapy. In the seminal framework for the role of host immunity in malignant disease, the term “immunoediting” was coined to describe the biological process of immunosurveillance executed through the elimination, equilibrium, and escape phases (156). These principles allow us to explore whether host immunity functions to calibrate responses to checkpoint inhibition and whether there is a relationship between immune capacity and ICI resistance. To better understand all that is at play, a longitudinal systems biology approach in healthy and cancer populations is needed, as well as the means to integrate data sets into meaningful models for testing.
Aged hosts are indicators for declining immune function as measured by redistribution of immune-cell populations, decreases in T-cell functionality, and increases in proinflammatory and immune suppressive cells (157). Additionally, the effects of host germline allelic variation and resistance to therapy suggest that individual immune capacity may be an underexplored variable of immune resistance as patient-specific HLA-I genotypes influence response to anti–PD-1 and anti–CTLA-4 therapy (as described above). Genetic variability conferred by single-nucleotide polymorphisms (SNP) also influences outcomes, with evidence from multiple studies showing that specific SNPs within clinically relevant genes can influence response or resistance to immunotherapy, and even on-treatment hyperprogression. In a study of patients with NSCLC treated with nivolumab, 2 of 7 PDL1 SNP markers were associated with objective response rate and PFS. Both outcomes were improved in patients with G/G and G/T genotypes compared with the T/T genotype at the marker rs2282055/PDL1 (158). By looking for SNPs in multiple genes, including PDL1, PD1, IDO1, and VEGFR2, additional likely scenarios were found where specific SNPs (rs2282055/PDL1 and rs1870377/VEGFR2) might influence accelerated tumor growth in response to ICIs, with perhaps an increased risk of hyperprogression in G/G and G/T genotypes compared with the T/T genotype at rs2282055/PDL1 (159). Although these data carry the limitations of retrospective analyses, they highlight the unfavorable versus favorable alleles in host baseline immunogenetics. Similar approaches investigated SNP analysis in genes associated with autoimmunity but did not find associations between polymorphisms and toxicity. Future work that prospectively and categorically analyzes host immunogenomics will expand our understanding of the systemic immune response and ICI resistance.
Discussion
In this Perspective, we highlight the importance of systematically investigating immune resistance and integrating large data sets into a framework that will allow us to identify resistance patterns that typically interfere with a patient's ability to respond to ICI therapy. Substantial ongoing efforts to address resistance to checkpoint inhibition have highlighted that immune resistance is driven by diverse, complex, interconnected, and evolving mechanisms. The importance of any one individual immune resistance mechanism, however, is isolated and difficult to interpret without biological context linked to clinically meaningful immune resistance. Initial progress has been made following a recent recommendation by the SITC Immunotherapy Resistance Taskforce (160) calling for the implementation of meaningful definitions of PD-(L)1 resistance with harmonized utility across clinical trials. The SITC Immunotherapy Resistance Taskforce is currently engaged in ongoing work to propose working definitions for immunotherapy resistance in the context of combination regimens including anti–PD-(L)1 plus anti–CTLA-4, anti–PD(L1)1 plus chemotherapy, and anti–PD-(L)1 plus VEGF inhibitors. Harmonization of clinical definitions across trials will allow us to better catalog immune resistance and systematically delineate resistance mechanisms and patterns not only for different ICI classes but also for different ICI-based combination therapies. Biological definitions of resistance are equally critical, and a systematic study of how the biology of the immune resistance process is organized will provide meaningful data that can help identify resistance pattens, resistance phenotypes, and, most importantly, a framework that will allow us to map this information to clinical approaches that can be tested in patients.
To break down this complexity, we carried out a thought exercise, assuming that individual resistance mechanisms converge into core biological processes and postulating that immune resistance might follow biological patterns that may be expressed as discrete phenotypes. We identified seminal data that have thus far informed our understanding of immune resistance and organized the data into eight biological processes we named immune resistance nodes. Collectively, we propose that the eight immune-resistance nodes represent the emerging hallmarks of resistance to ICIs (Fig. 1). We then outlined specific questions that, if answered, would greatly expand our view of immune resistance and could possibly generate hypotheses to examine in reverse translational efforts and incorporate prospectively into clinical trials (Table 1). These questions require the application of high-dimensional multiomic measurements matched to patient-derived samples taken at time points representative of both the disease course and signs of treatment resistance (Fig. 2).
Given the resources required to generate, analyze, and integrate such data sets, ensuring broad accessibility to data is critical. Technically, we need data storage, data sharing, and analysis infrastructures that can integrate these data to drive collective insights. A field-wide oncology effort, with robust harmonization and data sharing among all sectors, is needed to establish a data-driven “real-time” resistance framework that can be systematically tested, analyzed, and iterated within the context of clinical scenarios. Although initially difficult, a contained pilot study would greatly inform future steps and chart the path toward achieving improved survival in patients with cancer.
Authors' Disclosures
A.P. Cogdill reports other support from Immunai and personal fees and other support from Vastbiome outside the submitted work. P.B. Robbins reports other support from Pfizer during the conduct of the study; other support from Pfizer outside the submitted work. L.H. Butterfield reports the following unrelated advisory activities: StemImmune/Calidi Scientific and Medical Advisory Board, April 6, 2017–present; Western Oncolytics, Scientific Advisory Board, 2018–2021; Torque Therapeutics, Scientific Advisory Board, 2018–2020; Khloris, Scientific Advisory Board, 2019–present; Pyxis, Scientific Advisory Board, 2019–present; Cytomix, Scientific Advisory Board, 2019–present; DCprime, Scientific Advisory Board meeting, Nov. 2020; RAPT, Scientific Advisory Board, 2020–present; Takeda, Scientific Advisor, 2020–present; EnaraBio scientific advisor, Feb. 2021. A. Cessano reports being a full-time employee and stock owner of ESSA Pharma. Consultant for NanoString, Arch Oncology, and Bayer. None of the above relationships constitute conflict of interest. C. Hammer reports other support from Genentech/Roche outside the submitted work. C.L. Haymaker reports other support from Briacell Therapeutics and personal fees from Nanobiotix outside the submitted work; in addition, C.L. Haymaker has a patent for TLR9 modulators for treating cancer. pending. C.E. Horak reports personal fees from Bristol Myers Squibb outside the submitted work. H.M. McGee reports grants from NIH/NCI K99/R00 CA256526 and Salk Women and Science Special Award during the conduct of the study; personal fees from RefleXion outside the submitted work. R.F. Sweis reports grants and personal fees from BMS, Seattle Genetics, Astellas, Mirati, AstraZeneca, Lilly, Pfizer, Eisai, and Exelixis, grants from Merck, Immunocore, and Moderna, personal fees from Aveo outside the submitted work; in addition, R.F. Sweis has a patent for Neoantigens in Cancer: PCT/US2020/03 pending. B.G. Vincent reports other support from GeneCentric Therapeutics outside the submitted work. R. Zappasodi reports grants from PICI during the conduct of the study; personal fees from Leap Therapeutics outside the submitted work; in addition, R. Zappasodi has a patent for WO2019094352A1 pending, a patent for US20200024350A1 pending and licensed to AGENUS, a patent for US10323091B2 issued and licensed to AGENUS, and a patent for EP3148579B1 issued. V.M. Hubbard Lucey reports personal fees from Bristol Myers Squibb during the conduct of the study; personal fees from Bristol Myers Squibb outside the submitted work. D.K. Wells reports personal fees from Immunai, Rubius Therapeutics, and DeepMind outside the submitted work. T. LaVallee reports personal fees from LisCure Biosciencs, AstraZeneca Stock holder, BiOneCure, Grey Wolf Therapeutics, Immatics, Trex Bio, and Parker Institute of Cancer Immunotherapy outside the submitted work. No disclosures were reported by the other authors.
Acknowledgments
The authors would like to dedicate this work to oncology patients worldwide whose bravery, patience, and dedication is the true inspiration that fuels oncology research. The authors would like to acknowledge Nina C. Flerin and Massimiliano Mazzone for providing insightful materials, Doreen Valentine for scientific review and discussion, and Cathryn Guevarra for graphic design concepts. Editorial and design support was provided by Stuart Rulten, Liam Jewell, and Matthew Weddig, of Spark Medica Inc, funded by Bristol Myers Squibb. This work originated during a workshop organized by the Parker Institute for Cancer Immunotherapy (PICI), the Cancer Research Institute (CRI), and the Society for Immunotherapy of Cancer (SITC). Continuing support for manuscript preparation was provided by Bristol Myers Squibb. Editorial support was provided by Spark Medica Inc, funded by Bristol Myers Squibb. A.P. Cogdill is supported by the CPRIT Research Training Program (RP170067), the Fulbright Commission Franco-Américaine, and the John J. Kopchick Foundation. C.L. Haymaker is supported by NIH/NCI grants PA30CA016672, P50CA221703, R01CA236905, and U24CA224285. H.M. McGee is supported by NIH/NCI grant K99/R00CA256526. L.H. Butterfield and T. LaVallee are supported by PICI. N.-P. Rudqvist is supported by MDACC intramural funding. R.F. Sweis is supported by NIH K08CA234392. R. Zappasodi is supported by the PICI Bridge Fellows Award and NCI SPORE (P50-CA192937).
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
References
- 1. Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 2011;364:2517–26. [DOI] [PubMed] [Google Scholar]
- 3. Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med 2015;372:320–30. [DOI] [PubMed] [Google Scholar]
- 4. Ascierto PA, Long GV, Robert C, Brady B, Dutriaux C, Di Giacomo AM, et al. Survival outcomes in patients with previously untreated BRAF wild-type advanced melanoma treated with nivolumab therapy: three-year follow-up of a randomized phase 3 trial. JAMA Oncol 2019;5:187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hodi FS, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Cowey CL, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol 2018;19:1480–92. [DOI] [PubMed] [Google Scholar]
- 6. Robert C, Ribas A, Schachter J, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE-006): post-hoc 5-year results from an open-label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol 2019;20:1239–51. [DOI] [PubMed] [Google Scholar]
- 7. Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, et al. Five-year survival and correlates among patients with advanced melanoma, renal cell carcinoma, or non-small cell lung cancer treated with nivolumab. JAMA Oncol 2019;5:1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garon EB, Hellmann MD, Rizvi NA, Carcereny E, Leighl NB, Ahn MJ, et al. Five-year overall survival for patients with advanced non-small-cell lung cancer treated with pembrolizumab: results from the phase I KEYNOTE-001 study. J Clin Oncol 2019;37:2518–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaltwasser J. New 5-year KEYNOTE-024 data show pembrolizumab continues to prolong OS in NSCLC. Cancer Network. 2021May 11. Available from:https://www.cancernetwork.com/view/new-5-year-keynote-024-data-show-pembrolizumab-continues-to-prolong-os-in-nsclc.
- 10. Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer 2018;118:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Donnell JS, Long GV, Scolyer RA, Teng MW, Smyth MJ. Resistance to PD1/PDL1 checkpoint inhibition. Cancer Treat Rev 2017;52:71–81. [DOI] [PubMed] [Google Scholar]
- 13. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Brown Swigart L, et al. Myc cooperates with Ras by programming inflammation and immune suppression. Cell 2017;171:1301–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen RC, et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 2017;171:1284–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 2017;7:264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science 2016;352:227–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Topalian SL, Taube JM, Anders RA, Pardoll DM. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer 2016;16:275–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Massi D, Romano E, Rulli E, Merelli B, Nassini R, De Logu F, et al. Baseline beta-catenin, programmed death-ligand 1 expression and tumour-infiltrating lymphocytes predict response and poor prognosis in BRAF inhibitor-treated melanoma patients. Eur J Cancer 2017;78:70–81. [DOI] [PubMed] [Google Scholar]
- 20. Nsengimana J, Muralidhar S, Pozniak J, Laye JP, Bishop DT, Newton-Bishop JA. Immune cell profiles and β-catenin signaling in melanoma. Cancer Res 2017;77:Abstr 4006. [Google Scholar]
- 21. Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 2009;41:544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis 2006;44:262–7. [DOI] [PubMed] [Google Scholar]
- 23. Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, et al. beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell 2011;20:741–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015;523:231–5. [DOI] [PubMed] [Google Scholar]
- 25. Zdanov S, Mandapathil M, Abu Eid R, Adamson-Fadeyi S, Wilson W, Qian J, et al. Mutant KRAS conversion of conventional T cells into regulatory T cells. Cancer Immunol Res 2016;4:354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo G, Marrero L, Rodriguez P, Del Valle L, Ochoa A, Cui Y. Trp53 inactivation in the tumor microenvironment promotes tumor progression by expanding the immunosuppressive lymphoid-like stromal network. Cancer Res 2013;73:1668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanna A, Metge BJ, Bailey SK, Chen D, Chandrashekar DS, Varambally S, et al. Inhibition of Hedgehog signaling reprograms the dysfunctional immune microenvironment in breast cancer. Oncoimmunology 2019;8:1548241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chakrabarti J, Holokai L, Syu L, Steele NG, Chang J, Wang J, et al. Hedgehog signaling induces PD-L1 expression and tumor cell proliferation in gastric cancer. Oncotarget 2018;9:37439–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Spranger S, Gajewski TF. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer 2018;18:139–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Coelho MA, de Carne Trecesson S, Rana S, Zecchin D, Moore C, Molina-Arcas M, et al. Oncogenic RAS signaling promotes tumor immunoresistance by stabilizing PD-L1 mRNA. Immunity 2017;47:1083–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, et al. KRAS-IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 2019;35:559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garrido F. MHC/HLA class I loss in cancer cells. Adv Exp Med Biol 2019;1151:15–78. [DOI] [PubMed] [Google Scholar]
- 33. Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer 2021;21:298–312. [DOI] [PubMed] [Google Scholar]
- 34. Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT, et al. Neoantigen-directed immune escape in lung cancer evolution. Nature 2019;567:479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 2017;8:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ye Q, Shen Y, Wang X, Yang J, Miao F, Shen C, et al. Hypermethylation of HLA class I gene is associated with HLA class I down-regulation in human gastric cancer. Tissue Antigens 2010;75:30–9. [DOI] [PubMed] [Google Scholar]
- 37. Zingg D, Arenas-Ramirez N, Sahin D, Rosalia RA, Antunes AT, Haeusel J, et al. The histone methyltransferase Ezh2 controls mechanisms of adaptive resistance to tumor immunotherapy. Cell Rep 2017;20:854–67. [DOI] [PubMed] [Google Scholar]
- 38. Chew GL, Campbell AE, De Neef E, Sutliff NA, Shadle SC, Tapscott SJ, et al. DUX4 suppresses MHC class I to promote cancer immune evasion and resistance to checkpoint blockade. Dev Cell 2019;50:658–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell 2021;184:596–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shimasaki N, Jain A, Campana D. NK cells for cancer immunotherapy. Nat Rev Drug Discov 2020;19:200–18. [DOI] [PubMed] [Google Scholar]
- 41. Nayyar G, Chu Y, Cairo MS. Overcoming resistance to natural killer cell based immunotherapies for solid tumors. Front Oncol 2019;9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith CC, Selitsky SR, Chai S, Armistead PM, Vincent BG, Serody JS. Alternative tumour-specific antigens. Nat Rev Cancer 2019;19:465–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hacohen N, Fritsch EF, Carter TA, Lander ES, Wu CJ. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer Immunol Res 2013;1:11–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 2014;24:743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, et al. Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 2018;359:582–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, et al. Tumor and microenvironment evolution during immunotherapy with Nivolumab. Cell 2017;171:934–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alspach E, Lussier DM, Miceli AP, Kizhvatov I, DuPage M, Luoma AM, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019;574:696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kreiter S, Vormehr M, van de Roemer N, Diken M, Lower M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015;520:692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547:222–26. [DOI] [PubMed] [Google Scholar]
- 52. Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547:217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016;375:2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 2018;24:724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McGranahan N, Swanton C. Neoantigen quality, not quantity. Sci Transl Med 2019;11:eaax7918. [DOI] [PubMed] [Google Scholar]
- 56. Kalaora S, Wolf Y, Feferman T, Barnea E, Greenstein E, Reshef D, et al. Combined analysis of antigen presentation and T-cell recognition reveals restricted immune responses in melanoma. Cancer Discov 2018;8:1366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith CC, Chai S, Washington AR, Lee SJ, Landoni E, Field K, et al. Machine-learning prediction of tumor antigen immunogenicity in the selection of therapeutic epitopes. Cancer Immunol Res 2019;7:1591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 2014;124:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, et al. The immune landscape of cancer. Immunity 2019;51:411–12. [DOI] [PubMed] [Google Scholar]
- 60. Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol 2004;22:329–60. [DOI] [PubMed] [Google Scholar]
- 61. McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 2017;171:1259–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med 2019;25:603–11. [DOI] [PubMed] [Google Scholar]
- 63. Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med 1998;188:2205–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ahmadzadeh M, Johnson LA, Heemskerk B, Wunderlich JR, Dudley ME, White DE, et al. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009;114:1537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 2019;571:265–69. [DOI] [PubMed] [Google Scholar]
- 66. Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 2019;571:211–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Scott AC, Dundar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 2019;571:270–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature 2017;545:60–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 2016;537:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 2017;545:452–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity 2019;51:840–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Verma V, Shrimali RK, Ahmad S, Dai W, Wang H, Lu S, et al. PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1+CD38hi cells and anti-PD-1 resistance. Nat Immunol 2019;20:1231–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schwartz RH. T cell anergy. Annu Rev Immunol 2003;21:305–34. [DOI] [PubMed] [Google Scholar]
- 74. Macian F, Im SH, Garcia-Cozar FJ, Rao A. T-cell anergy. Curr Opin Immunol 2004;16:209–16. [DOI] [PubMed] [Google Scholar]
- 75. Townsend SE, Allison JP. Tumor rejection after direct costimulation of CD8+ T cells by B7-transfected melanoma cells. Science 1993;259:368–70. [DOI] [PubMed] [Google Scholar]
- 76. Staveley-O'Carroll K, Sotomayor E, Montgomery J, Borrello I, Hwang L, Fein S, et al. Induction of antigen-specific T cell anergy: an early event in the course of tumor progression. Proc Natl Acad Sci U S A 1998;95:1178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schietinger A, Philip M, Krisnawan VE, Chiu EY, Delrow JJ, Basom RS, et al. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity 2016;45:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Macian F, Garcia-Cozar F, Im SH, Horton HF, Byrne MC, Rao A. Transcriptional mechanisms underlying lymphocyte tolerance. Cell 2002;109:719–31. [DOI] [PubMed] [Google Scholar]
- 79. Mohamed AS, Velasco N. Kidneys for sale. Lancet 1990;336:1384. [DOI] [PubMed] [Google Scholar]
- 80. Martinez GJ, Pereira RM, Aijo T, Kim EY, Marangoni F, Pipkin ME, et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity 2015;42:265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bertram EM, Dawicki W, Sedgmen B, Bramson JL, Lynch DH, Watts TH. A switch in costimulation from CD28 to 4-1BB during primary versus secondary CD8 T cell response to influenza in vivo. J Immunol 2004;172:981–8. [DOI] [PubMed] [Google Scholar]
- 82. Lindner SE, Johnson SM, Brown CE, Wang LD. Chimeric antigen receptor signaling: functional consequences and design implications. Sci Adv 2020;6:eaaz3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kaplan G, Luster AD, Hancock G, Cohn ZA. The expression of a gamma interferon-induced protein (IP-10) in delayed immune responses in human skin. J Exp Med 1987;166:1098–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang ZE, Reiner SL, Zheng S, Dalton DK, Locksley RM. CD4+ effector cells default to the Th2 pathway in interferon gamma-deficient mice infected with Leishmania major. J Exp Med 1994;179:1367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Segawa S, Goto D, Iizuka A, Kaneko S, Yokosawa M, Kondo Y, et al. The regulatory role of interferon-gamma producing gamma delta T cells via the suppression of T helper 17 cell activity in bleomycin-induced pulmonary fibrosis. Clin Exp Immunol 2016;185:348–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhan X, Hu S, Wu Y, Li M, Liu T, Ming S, et al. IFN-gamma decreased the suppressive function of CD33(+)HLA-DR(low) myeloid cells through down-regulation of PD-1/PD-L2 signaling pathway. Mol Immunol 2018;94:107–20. [DOI] [PubMed] [Google Scholar]
- 87. Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, et al. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci U S A 1998;95:7556–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 2016;375:819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ribas A. Adaptive immune resistance: how cancer protects from immune attack. Cancer Discov 2015;5:915–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 2016;167:1540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Benci JL, Johnson LR, Choa R, Xu Y, Qiu J, Zhou Z, et al. Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell 2019;178:933–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018;362:eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2016;165:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 2018;24:986–93. [DOI] [PubMed] [Google Scholar]
- 96. Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Menares E, Galvez-Cancino F, Caceres-Morgado P, Ghorani E, Lopez E, Diaz X, et al. Tissue-resident memory CD8(+) T cells amplify anti-tumor immunity by triggering antigen spreading through dendritic cells. Nat Commun 2019;10:4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Jansen CS, Prokhnevska N, Master VA, Sanda MG, Carlisle JW, Bilen MA, et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 2019;576:465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity 2020;52:17–35. [DOI] [PubMed] [Google Scholar]
- 100. Trujillo JA, Sweis RF, Bao R, Luke JJ. T cell-inflamed versus non-T cell-inflamed tumors: a conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol Res 2018;6:990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rothwell PM, Fowkes FG, Belch JF, Ogawa H, Warlow CP, Meade TW. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet 2011;377:31–41. [DOI] [PubMed] [Google Scholar]
- 102. Rothwell PM, Wilson M, Price JF, Belch JF, Meade TW, Mehta Z. Effect of daily aspirin on risk of cancer metastasis: a study of incident cancers during randomised controlled trials. Lancet 2012;379:1591–601. [DOI] [PubMed] [Google Scholar]
- 103. Greten FR, Grivennikov SI. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity 2019;51:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, et al. Chronic inflammation in the etiology of disease across the life span. Nat Med 2019;25:1822–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Wiedle G, Dunon D, Imhof BA. Current concepts in lymphocyte homing and recirculation. Crit Rev Clin Lab Sci 2001;38:1–31. [DOI] [PubMed] [Google Scholar]
- 106. Fu H, Wang A, Mauro C, Marelli-Berg F. T lymphocyte trafficking: molecules and mechanisms. Front Biosci (Landmark Ed) 2013;18:422–40. [DOI] [PubMed] [Google Scholar]
- 107. Butcher EC, Picker LJ. Lymphocyte homing and homeostasis. Science 1996;272:60–6. [DOI] [PubMed] [Google Scholar]
- 108. Picker LJ, Butcher EC. Physiological and molecular mechanisms of lymphocyte homing. Annu Rev Immunol 1992;10:561–91. [DOI] [PubMed] [Google Scholar]
- 109. Moser B, Willimann K. Chemokines: role in inflammation and immune surveillance. Ann Rheum Dis 2004;63:ii84–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sackstein R, Schatton T, Barthel SR. T-lymphocyte homing: an underappreciated yet critical hurdle for successful cancer immunotherapy. Lab Invest 2017;97:669–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Marelli-Berg FM, Cannella L, Dazzi F, Mirenda V. The highway code of T cell trafficking. J Pathol 2008;214:179–89. [DOI] [PubMed] [Google Scholar]
- 112. Garrood T, Lee L, Pitzalis C. Molecular mechanisms of cell recruitment to inflammatory sites: general and tissue-specific pathways. Rheumatology (Oxford) 2006;45:250–60. [DOI] [PubMed] [Google Scholar]
- 113. Okamoto N, Nukada Y, Tezuka K, Ohashi K, Mizuno K, Tsuji T. AILIM/ICOS signaling induces T-cell migration/polarization of memory/effector T-cells. Int Immunol 2004;16:1515–22. [DOI] [PubMed] [Google Scholar]
- 114. Liu VC, Wong LY, Jang T, Shah AH, Park I, Yang X, et al. Tumor evasion of the immune system by converting CD4+CD25− T cells into CD4+CD25+ T regulatory cells: role of tumor-derived TGF-beta. J Immunol 2007;178:2883–92. [DOI] [PubMed] [Google Scholar]
- 115. Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 2003;4:337–42. [DOI] [PubMed] [Google Scholar]
- 116. Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med 2007;204:1775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Woodland DL, Kohlmeier JE. Migration, maintenance and recall of memory T cells in peripheral tissues. Nat Rev Immunol 2009;9:153–61. [DOI] [PubMed] [Google Scholar]
- 118. Ju X, Huang P, Chen M, Wang Q. Liver X receptors as potential targets for cancer therapeutics. Oncol Lett 2017;14:7676–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Guerriero JL. Macrophages: the road less traveled, changing anticancer therapy. Trends Mol Med 2018;24:472–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Eljaszewicz A, Wiese M, Helmin-Basa A, Jankowski M, Gackowska L, Kubiszewska I, et al. Collaborating with the enemy: function of macrophages in the development of neoplastic disease. Mediators Inflamm 2013;2013:831387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. O'Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol 2016;16:553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Hotamisligil GS. Foundations of immunometabolism and implications for metabolic health and disease. Immunity 2017;47:406–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Nieman DC, Lila MA, Gillitt ND. Immunometabolism: a multi-omics approach to interpreting the influence of exercise and diet on the immune system. Annu Rev Food Sci Technol 2019;10:341–63. [DOI] [PubMed] [Google Scholar]
- 124. Van den Bossche J, O'Neill LA, Menon D. Macrophage immunometabolism: where are we (going)? Trends Immunol 2017;38:395–406. [DOI] [PubMed] [Google Scholar]
- 125. Vardhana SA, Hwee MA, Berisa M, Wells DK, Yost KE, King B, et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat Immunol 2020;21:1022–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Legat A, Speiser DE, Pircher H, Zehn D, Fuertes Marraco SA. Inhibitory receptor expression depends more dominantly on differentiation and activation than "exhaustion" of human CD8 T cells. Front Immunol 2013;4:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Odorizzi PM, Pauken KE, Paley MA, Sharpe A, Wherry EJ. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J Exp Med 2015;212:1125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Scharping NE, Menk AV, Moreci RS, Whetstone RD, Dadey RE, Watkins SC, et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 2016;45:701–03. [DOI] [PubMed] [Google Scholar]
- 129. Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 2015;162:1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Barsoum IB, Smallwood CA, Siemens DR, Graham CH. A mechanism of hypoxia-mediated escape from adaptive immunity in cancer cells. Cancer Res 2014;74:665–74. [DOI] [PubMed] [Google Scholar]
- 131. Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol 1927;8:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity 2002;16:769–77. [DOI] [PubMed] [Google Scholar]
- 133. Haag F, Adriouch S, Brass A, Jung C, Moller S, Scheuplein F, et al. Extracellular NAD and ATP: partners in immune cell modulation. Purinergic Signal 2007;3:71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Stark R, Wesselink TH, Behr FM, Kragten NAM, Arens R, Koch-Nolte F, et al. T RM maintenance is regulated by tissue damage via P2RX7. Sci Immunol 2018;3:eaau1022. [DOI] [PubMed] [Google Scholar]
- 135. Chen L, Diao L, Yang Y, Yi X, Rodriguez BL, Li Y, et al. CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov 2018;8:1156–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hammami A, Allard D, Allard B, Stagg J. Targeting the adenosine pathway for cancer immunotherapy. Semin Immunol 2019;42:101304. [DOI] [PubMed] [Google Scholar]
- 137. Sukumar M, Liu J, Ji Y, Subramanian M, Crompton JG, Yu Z, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest 2013;123:4479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Kawalekar OU, RS OC, Fraietta JA, Guo L, McGettigan SE, Posey AD Jr, et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity 2016;44:712. [DOI] [PubMed] [Google Scholar]
- 139. Robbins PF, Dudley ME, Wunderlich J, El-Gamil M, Li YF, Zhou J, et al. Cutting edge: persistence of transferred lymphocyte clonotypes correlates with cancer regression in patients receiving cell transfer therapy. J Immunol 2004;173:7125–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Menk AV, Scharping NE, Rivadeneira DB, Calderon MJ, Watson MJ, Dunstane D, et al. 4-1BB costimulation induces T cell mitochondrial function and biogenesis enabling cancer immunotherapeutic responses. J Exp Med 2018;215:1091–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 2007;315:1687–91. [DOI] [PubMed] [Google Scholar]
- 142. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–97. [DOI] [PubMed] [Google Scholar]
- 143. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The influence of the gut microbiome on cancer, immunity, and cancer immunotherapy. Cancer Cell 2018;33:570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;342:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol 2018;29:1437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Derosa L, Routy B, Fidelle M, Iebba V, Alla L, Pasolli E, et al. Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol 2020;78:195–206. [DOI] [PubMed] [Google Scholar]
- 151. Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol 2004;4:499–511. [DOI] [PubMed] [Google Scholar]
- 152. Reis e Sousa C. Toll-like receptors and dendritic cells: for whom the bug tolls. Semin Immunol 2004;16:27–34. [DOI] [PubMed] [Google Scholar]
- 153. Pasare C, Medzhitov R. Toll-like receptors and acquired immunity. Semin Immunol 2004;16:23–6. [DOI] [PubMed] [Google Scholar]
- 154. Paulos CM, Wrzesinski C, Kaiser A, Hinrichs CS, Chieppa M, Cassard L, et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J Clin Invest 2007;117:2197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Coley WB. The treatment of malignant tumors by repeated inoculations of erysipelas: with a report of ten original cases. 1893. Clin Orthop Relat Res 1991;262:3–11. [PubMed] [Google Scholar]
- 156. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011;331:1565–70. [DOI] [PubMed] [Google Scholar]
- 157. Hurez V, Padron AS, Svatek RS, Curiel TJ. Considerations for successful cancer immunotherapy in aged hosts. Clin Exp Immunol 2017;187:53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Nomizo T, Ozasa H, Tsuji T, Funazo T, Yasuda Y, Yoshida H, et al. Clinical impact of single nucleotide polymorphism in PD-L1 on response to nivolumab for advanced non-small-cell lung cancer patients. Sci Rep 2017;7:45124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Refae S, Gal J, Brest P, Giacchero D, Borchiellini D, Ebran N, et al. Hyperprogression under immune checkpoint inhibitor: a potential role for germinal immunogenetics. Sci Rep 2020;10:3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kluger HM, Tawbi HA, Ascierto ML, Bowden M, Callahan MK, Cha E, et al. Defining tumor resistance to PD-1 pathway blockade: recommendations from the first meeting of the SITC immunotherapy resistance taskforce. J Immunother Cancer 2020;8:e000398. [DOI] [PMC free article] [PubMed] [Google Scholar]