Abstract
Haploinsufficiency of suppressor of cytokine signaling 1 (SOCS1) is a recently discovered autoinflammatory disorder with significant rheumatologic, immunologic, and hematologic manifestations. Here we report a case of SOCS1 haploinsufficiency in a 5-year-old child with profound arthralgias and immune-mediated thrombocytopenia unmasked by SARS-CoV-2 infection. Her clinical manifestations were accompanied by excessive B cell activity, eosinophilia, and elevated IgE levels. Uniquely, this is the first report of SOCS1 haploinsufficiency in the setting of a chromosomal deletion resulting in complete loss of a single SOCS1 gene with additional clinical findings of bone marrow hypocellularity and radiologic evidence of severe enthesitis. Immunologic profiling showed a prominent interferon signature in the patient’s peripheral blood mononuclear cells, which were also hypersensitive to stimulation by type I and type II interferons. The patient showed excellent clinical and functional laboratory response to tofacitinib, a Janus kinase inhibitor that disrupts interferon signaling. Our case highlights the need to utilize a multidisciplinary diagnostic approach and consider a comprehensive genetic evaluation for inborn errors of immunity in patients with an atypical immune-mediated thrombocytopenia phenotype.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10875-022-01346-x.
Keywords: SOCS1 haploinsufficiency, Interferonopathy, JAK inhibition, Autoinflammatory syndrome, Bone marrow hypocellularity, Immune thrombocytopenia
Introduction
Haploinsufficiency of suppressor of cytokine signaling 1 (SOCS1) is a newly recognized monogenic autoinflammatory and early-onset autoimmune syndrome with wide-ranging rheumatologic, immunologic, and hematologic manifestations [1, 2]. The varied phenotypic findings likely reflect the diverse functions of the SOCS1 protein. Broadly, members of the SOCS family comprise a negative feedback system for pro-inflammatory cytokines. The primary function of SOCS1 is regulation of the JAK-STAT (Janus kinase — signal transducer and activator of transcription protein) pathway via inhibition of JAK1/2 phosphorylation, with additional suppressive activity on TYK2 [3]. Given the role of STAT1 in type I and type II interferon (IFN) signaling, individuals with dysfunction of SOCS1 show enhanced expression of IFN-inducible genes, often called an IFN signature [1]. Additionally, SOCS1 likely impacts the nuclear factor kappa-light-chain enhancer of activated B cells (NFkB), focal adhesion kinase (FAK) 1, and Toll-interleukin-1 receptor domain-containing adaptor protein (TIRAP) pathways, resulting in defective T cell function and aberrant toll-like receptor signaling [4].
Although the disease arises from insufficient protein function, SOCS1 haploinsufficiency is effectively an autosomal dominant condition with a highly variable phenotype with incomplete penetrance. Environmental triggers, such as viral illnesses, may unmask the autoinflammatory clinical phenotype [1]. Based on limited published reports to date, disease onset is typically within the first decade and pathogenic SOCS1 variants are often loss-of-function mutations. Patients may present with rheumatologic findings of polyarthritis, recurrent mucosal ulcerations, fever, glomerulonephritis, alopecia, autoimmune endocrinopathies, psoriasis, and features of systemic lupus erythematosus (SLE). Hepatosplenomegaly, hepatitis, and colitis can affect the gastrointestinal tract. Recurrent infections, as a result of hypogammaglobulinemia, variable T cell lymphopenia, and natural killer cell dysfunction are possible, and secondary granulomatous interstitial lung disease related to immune dysregulation has been described [1, 2]. Abnormal B cell phenotyping, including increased CD21low CD38low B cells, likely leads to inappropriate autoantibody production. Finally, patients may present with hematologic manifestations of generalized lymphadenopathy, elevated IgE, eosinophilia, and severe immune-mediated cytopenias [1, 2, 4, 5]. Treatment of reported cases of SOCS1 haploinsufficiency includes corticosteroids, thrombopoietin receptor agonists, intravenous immunoglobulins (IVIG), and various immunosuppressive therapies, including rituximab, hydroxychloroquine, mycophenolate mofetil, rapamycin, and anti-tumor necrosis factor (TNF) agents [4, 5]. Utilization of JAK inhibition with baricitinib has additionally been reported in a 9-year-old child with the condition [2].
We present a pediatric patient with SOCS1 haploinsufficiency with severe arthralgias and treatment-resistant immune thrombocytopenia (ITP) following apparent SARS-CoV-2 infection with the unique additional features of bone marrow hypocellularity and significant enthesitis on musculoskeletal imaging. A genetic diagnosis was secured via identification of a novel 5 mega-base chromosomal 16p deletion resulting in complete loss of the SOCS1 gene. Immunologic characterizations were consistent with SOCS1 haploinsufficiency and clinical remission was successfully attained via targeted JAK inhibition. Our case expands the genotype and phenotype spectrum of SOCS1 haploinsufficiency and highlights the need for a multi-disciplinary diagnostic approach for patients with immune cytopenias and rheumatologic findings.
Methods
Genetic Analysis
Karyotype chromosome analysis completed via bone marrow aspirate samples. Microarray analysis performed using the Affymetrix Cytoscan HD platform to identify DNA copy number gains/losses and regions of loss of heterozygosity [6]. Peripheral blood chromosomal microarray (CMA) analysis completed using the Illumina CytoSNP-850 K array, with aberrations confirmed by qPCR [7].
Screening for underlying germline inborn errors of immunity variants was completed via next-generation exome sequencing through Blueprint Genetics (642 gene Comprehensive Immune and Cytopenia Panel) [8].
Commercially Available Functional Testing
Telomere length assessment via RepeatDx (Vancouver, British Columbia, Canada). RepeatDx’s flow cytometry-based fluorescence in situ hybridization (FISH) analyzes telomere length on a single cell basis for lymphocytes, granulocytes, B cells, naïve and memory T cells, and natural killer cells [9]. Quantification of plasma chemokines and cytokines using a proximity extension assay was performed commercially by Olink (Olink Target 48 Cytokine; Waltham, MA). The following studies were performed at Cincinnati Children’s Hospital [10]: (1) MHC class I and II expression peripheral blood monocytes and lymphocytes via flow cytometry; (2) Chromosomal breakage via cultured lymphocyte exposed to mitomycin C and diepoxybutane; (3) B cell activating factor level (BAFF) analysis via enzyme-linked immunosorbent assay.
Flow Cytometry
For surface staining of CD169, fresh peripheral blood mononuclear cells (PBMCs) isolated by Ficoll gradient were incubated with anti-human CD14, CD16, and CD169 antibodies for 20 min at room temperature after incubation with Human TruStain FcX (Biolegend). Cells were washed with PBS before analysis. For quantitation of STAT1 phosphorylation, PBMCs were stimulated with recombinant human IFN-α2 or IFN-γ for 20 min before fixation with 4% paraformaldehyde. Cells were permeabilized with Perm/Wash Buffer and incubated with anti-human CD14 and phosphor-STAT1 (S727) antibodies (Biolegend) for 2 h. All flow cytometry experiments were analyzed using a BD Fortessa cytometer.
Transcriptomic Analysis
Total RNA was extracted using Qiagen RNeasy mini kit (Germantown, MD, USA) according to the manufacturer’s instructions. Smart-seq2 transcriptome libraries were prepared and sequenced by the Broad Institute Genomics Platform (Cambridge, MA). Raw sequencing reads were obtained as fastq.gz files. Quality of raw reads was checked by FastQC (v0.11.9). HISAT2 (v2.2.1) were used to align raw reads to the human reference genome (GRCh38—hg38) from Genome Reference Consortium. Resulting SAM files were sorted by Samtools (v1.12). Using HTSeq (v0.13.5), reads were mapped to corresponding annotated gene. To determine differential gene expression and gene set enrichment, the gene count files from HTSeq were further analyzed by preprocessCore package (v1.52.1), limma package (v3.46.0), complexheatmap package (v2.6.2), fGSEA package (v1.16.0), and ggplot2 package (v3.3.3) in R (v4.0.4). Raw data from RNAseq are available from the authors upon request.
Calculation of standardized 28 interferon-regulated gene (28 IRG) score was described previously [11]. Healthy individuals (n = 18; 10 children, 8 adults) were used to determine the mean and standard deviation of the control group. The standardized score for each subject reflects the number of standard deviations away from the mean of the control group. To minimize the impact of outliers, we displayed the median score of the 28 genes, rather than the sum of all 28 genes. Pediatric patients with SLE (n = 5) were incorporated as positive controls.
Real-time Quantitative PCR (qPCR)
Complementary DNA was synthesized from isolated RNA using SuperScript First-Strand Synthesis kit (Thermo Fisher Scientific, Waltham, MA). SYBR Green qPCR was performed using Luna qPCR Mastermix (New England Biolabs, Ipswich, MA) according to the manufacturer’s instructions. Primers for SOCS1 were as follows: Forward-CGCCCTTAGCGTGAAGAT and Reverse-CTCGAAGAGGCAGTCGAAG.
Statistical Analysis
For quantitative variables, differences between 2 groups were analyzed by Student’s t-test. All tests were 2-sided, and P less than 0.05 was considered significant. Statistical analyses were performed using Prism 8.0 software (GraphPad Software).
Results
Clinical Presentation
The patient is a 5-year-old female of Indian and Italian descent who presented to outpatient rheumatology with ambulatory difficulties and limited movement secondary to significant pain. These symptoms began approximately 3-months post-presumed SARS-CoV-2 illness (family member PCR positivity and concurrent symptoms). Musculoskeletal exam showed decreased range of motion and pain in cervical spine and bilateral wrists/hips. Bony overgrowth and synovial thickening with pain in wrists, multiple small joints of hands, and metatarsophalangeal joints of the right foot were also present. Gait was abnormal with pelvic tilt and exaggerated lumbar lordosis. No muscle weakness, Gottron papules, or heliotrope rash were observed.
History was without fevers, morning stiffness, or joint erythema. Laboratory analysis showed mild eosinophilia (absolute count 0.9 K/uL) with otherwise normal blood cell counts and a clinical history without an allergic phenotype. Comprehensive metabolic panel, creatinine phosphokinase, lactate dehydrogenase, CRP, and ESR were normal. Antinuclear antibodies (ANA), cyclic citrullinated peptide, autoimmune myositis, and rheumatoid factor antibody evaluations were also negative. Aldolase was mildly elevated at 12 IU/L. Skeletal survey showed subtle periostitis and patchy demineralization within the hands indicating nonspecific inflammation. Long bones were notable for demineralization and under-tubularization. Family history was without consanguinity or concerns for autoimmunity, hematologic compromise, or inborn errors of immunity. A juvenile idiopathic arthritis diagnosis was given, and naproxen therapy was initiated.
Her condition failed to improve with such therapy though. Repeat CBC showed worsening eosinophilia (now 1.7 K/uL) and mild thrombocytosis (platelet count 466 × 109/L). Ova/parasite testing, somatic variant genetic assessment for hyper-eosinophilic disorders, and skin biopsy for eosinophilic fasciitis were unremarkable. Screening for metabolic disorders was reassuring. Musculoskeletal magnetic resonance imaging demonstrated diffuse tenosynovitis of the hands/wrists symmetrically bilaterally along with tenosynovitis at multiple insertion sites within the pelvis, particularly along the right iliotibial tract and hamstring insertions of ischial tuberosities (Figs. 1, 2, and 3). Given worsening eosinophilia, a bone marrow biopsy was obtained; pathology was notable for hypocellularity (60%) but without concern for malignancy or dysplasia. Karyotype (XX) and flow cytometry panels for evaluation of malignancy were normal. The patient began 1 mg/kg/day oral prednisone with improvement in arthralgias, mobility, and eosinophilia.
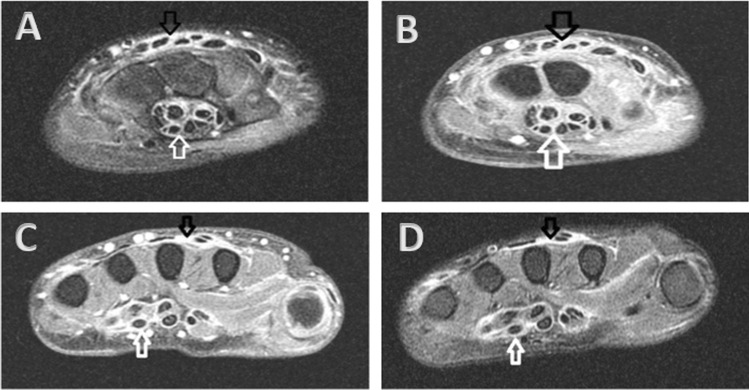
Fig. 1.
Axial MR image of the right wrist demonstrated T2 hyperintense signal (A) with T1 post-contrast enhancement (B) throughout the flexor (white arrow) and extensor (black arrow) tendon sheaths at the level of the carpal tunnel (white arrow). These findings were also seen at the level of the metacarpals on T2 (C) and T1 post-contrast (D) MR images. Findings were bilateral
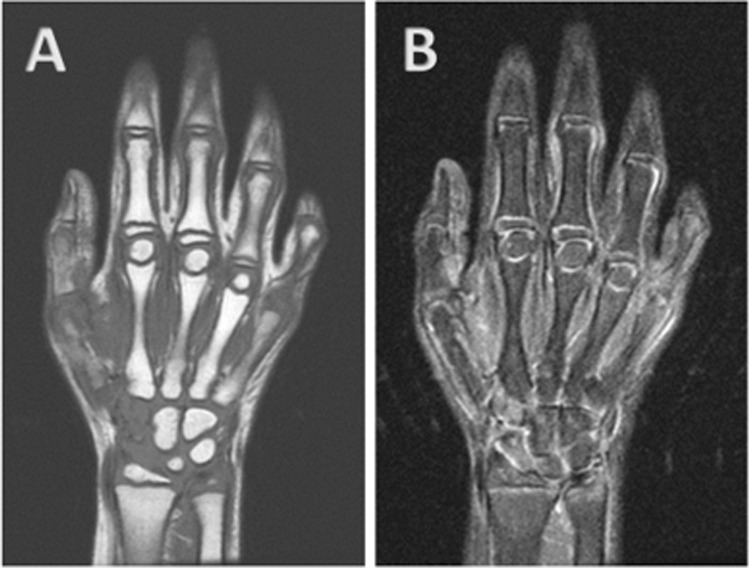
Fig. 2.
Coronal T2 (A) and T1 (B) images of the right hand showed no bone marrow edema, joint effusion, or erosions. Findings were bilateral
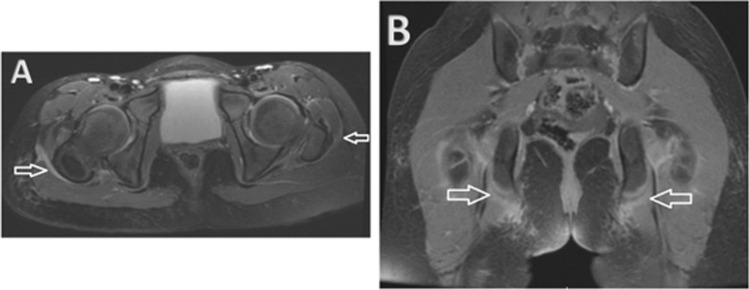
Fig. 3.
(A) Axial T2 images of the pelvis showed thickening and hyperintensity of the iliotibial bands, right greater than left (white arrows), indicating inflammation. (B) Coronal spoiled gradient recalled echo (SPGR) post-contrast sequences showed enhancement at the insertion sites of the hamstring tendons on the ischial tuberosities (white arrows)
Upon weaning of corticosteroids, symptoms unfortunately returned. CBC revealed a white blood cell count 4.7 K/uL, absolute neutrophil count 1.5 K/uL, absolute eosinophil count 0.6 K/uL, hemoglobin 12.6 g/dL, and critically low platelets at 1 K/uL. ESR continued to be normal with mild elevation of aldolase to 17 IU/L. With concern for immune thrombocytopenia (ITP), the patient was admitted and subsequently developed wet purpura, epistaxis, hematuria, and melena with platelets reaching a nadir of 0 K/uL. Oral prednisolone 4 mg/kg/daily and intravenous immunoglobulins 1 g/kg × 2 doses were administered without improvement.
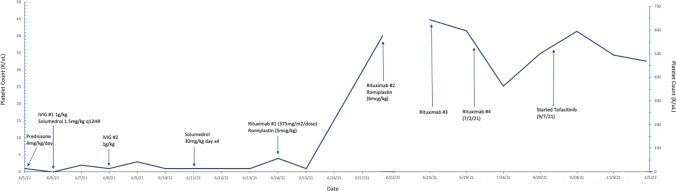
Repeat bone marrow biopsy showed no concern for malignancy/dysplasia, 50% cellularity, and decreased myeloid and erythroid elements but increased megakaryocytes (Fig. 4). Iron stain revealed decreased storage iron with no ring sideroblasts. Flow cytometric analysis revealed no blasts. Cytogenetics microarray was additionally obtained from the bone marrow sample. The patient was subsequently initiated on intravenous methylprednisolone 30 mg/kg/day × 4 days and rituximab 375 mg/m2 to not only treat ITP but also a strongly suspected underlying rheumatologic disorder. Despite aggressive immunosuppressive therapy, her platelet count remained < 2 K/uL. Treatment with the thrombopoietin (TPO) receptor agonist, romiplostim (dosed at 5 mcg/kg), was next given with improvement in platelets to 44 K/uL within days (Fig. 5). Anti-platelet antibody screening revealed strong positivity in IgM-direct platelet antibodies. Direct antiglobulin testing was negative. Anti-phospholipid antibodies were absent with normal C3 but mildly reduced C4 (12 mg/dL). Repeat ANA analysis was positive with additional positivity in anti-SSa/anti-Ro but this evaluation was completed following IVIG therapy. Anti-double-stranded DNA antibody was negative. Nucleocapsid antibodies to SARS-CoV-2 were detected, confirming prior infection. At initial presentation, immunoglobulin profiling revealed normal IgG/IgM/IgA levels but increased IgE (2080 kU/L), while lymphocyte subsets showed normal number of CD4 + and CD8 + T lymphocytes but increased number of NK cells (1168 cells/uL) and B lymphocytes (8548 cells/uL; Table 1). Memory and naïve helper and suppressor T cell absolute values were also found to be normal for age. Following discharge, the patient completed a full course of rituximab therapy (375 mg/m2/week × 4 total doses) with gradual normalization of platelet count. Repeat lymphocyte analysis following rituximab therapy noted near complete depletion of B cells with CD19 and CD20 absolute values of 2 cells/cmm and 0 cells/cmm, respectively.
Fig. 4.
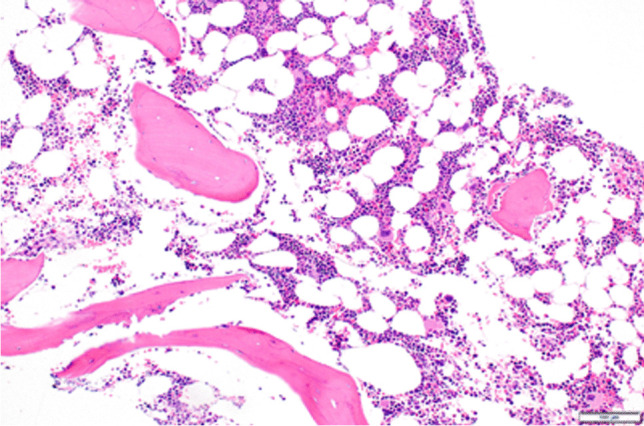
Bone marrow biopsy pathology showing hypocellularity at 50%
Fig. 5.
Patient’s platelet values with medical interventions noted and response in platelet counts
Table 1.
Patient’s rheumatologic, immunologic, hematologic, infectious disease and genetic clinical evaluation findings
| Rheumatologic/immunologic studies | Result | Reference range |
| Angiotensin converting enzyme | 89 (high) | 16–85 U/L |
| Aldolase | 12–17 (high) | 1–7 IU/L |
| Antineutrophil cytoplasmic Ab | Negative | Negative |
| Anti-nuclear Ab | Negative | Negative |
| Rheumatoid factor | < 12 | 0–15 IU/mL |
| C3 | 120 | 83–240 mg/dL |
| C4 | 12 (low) | 13–60 mg/dL |
| B2 glycoprotein Ab | Negative | Negative |
| Cardiolipin Ab | Negative | Negative |
| Lupus anticoagulant | Negative | Negative |
| CCP Ab | < 5 | 0–19 CU |
| Creatine kinase | 30–34 | 26–180 IU/L |
| C-reactive protein | 0.2–0.6 | 0–0.6 mg/dL |
| ESR | 2–12 | 0–20 mm |
| IgA | 101–155 | 35–250 mg/dL |
| IgG | 510–801 | 620–1520 mg/dL |
| IgE | 306–2080 (high) | 0–150 kU/L |
| IgM | 16–94 | 50–240 mg/dL |
| Absolute CD3 count | 3962 | 660–4636 cells/cmm |
| Absolute CD4 count | 1890 | 341–2867 cells/cmm |
| Absolute CD8 count | 1904 | 198–2135 cells/cmm |
| Absolute CD16/56 count | 1168 (high) | 44–1037 cells/cmm |
| Absolute CD19 count | 8548 (high) | 143–1647 cells/cmm |
| Hematologic studies | Results | Reference range |
| Platelet-associated IgM | Strong positive | Negative |
| Cytokine panel | Mildly elevated sIL2R | Normal |
| Bone marrow karyotype | 46, XX | 46, XX |
| Bone marrow immunophenotyping for leukemia/lymphoma | Normal. No aberrant B, T, or mast cells | Normal |
| Bone marrow cancer cytogenetic array | Loss of 5 Mb at 16p13.2p13.11 | Normal |
| B cell activating factor | 6432 (high) | 241–1748 pg/mL |
| Fanconi anemia chromosomal breakage analysis | Normal | Normal |
| CHIC2 FISH | Normal | Normal |
| FGFR1 FISH | Normal | Normal |
| MHC classes I and II | Normal | Normal |
| Telomere length | Very low in lymphocytes and memory T cells. Low in granulocytes, naïve T and NK cells. Insufficient B cells | Normal |
| Genetic studies | Results | Reference range |
| Chromosomal microarray (peripheral blood) | 5 Mb loss at 16p13.2p13.11 | Normal |
| PTEN sequencing | No pathogenic variants | No pathogenic variants |
| PDGFRB/TEL translocation FISH | Negative | Negative |
| Systemic mastocytosis KIT Asp816Val | Negative | Negative |
| Infectious studies | Results | Reference range |
| Cytomegalovirus PCR | Negative | Negative |
| Epstein Barr virus PCR | Negative | Negative |
| HSV1 and 2 PCR | Negative | Negative |
| SARS-CoV-2 Nucleocapsid Ab | Positive | Negative |
Ab, antibody; CCP, cyclic citrullinated peptide; ESR, erythrocyte sedimentation rate; CHIC2, cysteine-rich hydrophobic domain 2; FISH, fluorescent in situ hybridization; FGFR1, fibroblast growth factor receptor 1; MHC, major histocompatability complex; PTEN, phosphatase and tensin homolog; PDGFRB, platelet-derived growth factor receptor beta; HSV, herpes simplex virus; PCR, polymerase chain reaction; QF, QuantiFERON gold
Genetic Testing Results
Bone marrow chromosomal microarray testing subsequently revealed a single 5 mega-base deletion at 16p13.2p13.11, which includes the coding sequence of SOCS1, GRIN2A, EMP2, CIITA, LITAF, ERCC4, and PARN (Table 2). Peripheral blood microarray analysis (obtained given prior concern for development delays and macrocephaly) also was notable for the 16p microdeletion. Additional comprehensive next generation exome sequencing via Blueprint Genetics for inborn errors of immunity was unremarkable.
Table 2.
Genetic testing variants associated with human disease within patient’s chromosomal deletion with accompanying disease associations [12]
| Gene | Inheritance | Associated condition |
|---|---|---|
| SOCS1 | AD | Autoinflammatory syndrome, familial, with or without immunodeficiency |
| GRIN2A | AD | Epilepsy, focal, with speech disorder, and with or without impaired intellectual development |
| EMP2 | AR | Nephrotic syndrome, type 10 |
| CIITA | AR | Bare lymphocyte syndrome, type II, complementation group A |
| LITAF | AD | Charcot-Marie-Tooth disease, type 1C |
| ERCC4 | AR | Fanconi anemia, complementation group Q |
| PARN | AD | Pulmonary fibrosis and/or bone marrow failure, telomere-related, 4 |
AD, autosomal dominant; AR, autosomal recessive
While PARN and ERCC4 variants may be associated with autosomal dominant pulmonary fibrosis and/or bone marrow failure and autosomal recessive Fanconi anemia of complementation group Q, respectively, results from telomere length and chromosomal breakage studies were not consistent with these potential diagnoses. Classically those with telomeropathy conditions/dyskeratosis congenita have very low telomeres (< 1%ile) in most cells analyzed (often all lymphocyte, granulocyte, B, naïve and memory T, and NK cells assessed). Our patient was noted to have only two cell lines < 1%ile. Additionally, with correction of cytopenias via JAK inhibition and lack of additional telomeropathy clinical findings, PARN alteration is less likely to be contributing to our patient’s presentation [13] but continued close monitoring will be required over time to ensure our patient does not develop PARN-associated bone marrow failure or additional phenotypic features. Major histocompatibility complex class I and II epitope expression on peripheral lymphocytes and monocytes assessed via flow cytometry was normal (obtained given CIITA variant within patient’s microdeletion). There are no current clinical concerns for a history of epilepsy or neurologic/developmental delays consistent with pathogenic GRIN2A or LITAF variants with normal brain magnetic resonance imaging and unremarkable electroencephalogram testing, although such a diagnosis cannot be definitively ruled out at this time. Additionally, despite earlier concerns as a toddler, the patient is now developmentally normal with an unremarkable neurologic examination. Urine protein and creatinine assessments were reassuring against nephrotic syndrome secondary to an EMP2 alteration. Parental genetic testing was found to be unremarkable, indicating that the patient’s chromosomal deletion arose de novo.
Functional Analysis of IFN Signaling
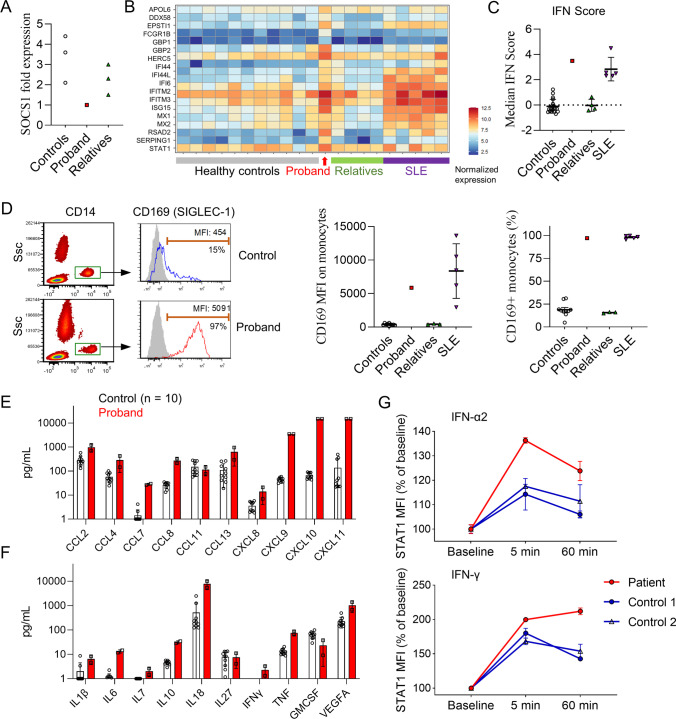
Given the patient’s overlapping findings of musculoskeletal inflammation and autoimmune cytopenia, SOCS1 haploinsufficiency was suspected. Decreased expression of SOCS1 in the proband’s PBMCs was confirmed by quantitative PCR (Fig. 6A). SOCS1 is a negative regulator of type I and type II IFN signaling by inhibiting the activation of Janus kinases (JAK). Accordingly, haploinsufficiency of SOCS1 is associated with enhanced IFN signaling. To interrogate possible disinhibition of IFN signaling, we first examined the IFN gene signature by bulk RNA sequencing of peripheral blood mononuclear cells. Analysis of a panel of IFN-inducible genes (ISG) showed upregulated expression in the proband compared to unaffected family members and healthy controls (Fig. 6B). A validated composite ISG score derived from these data [11] was elevated in the proband at levels comparable to patients with systemic lupus erythematosus (Fig. 6C), an autoimmune disease associated with high type I IFN production.
Fig. 6.
Enhanced interferon response associated with SOCS1 haploinsufficiency. (A) SOCS1 mRNA expression in PBMCs from the proband, unaffected family members (n = 3), and healthy controls (n = 3) measured by quantitative PCR. SOCS1 expression for each individual was normalized to β-actin expression. (B) Heat map display of IFN-inducible genes from bulk RNAseq of PBMC from the proband, healthy controls (n = 11), unaffected family members (n = 4), and patients with active systemic lupus erythematosus (n = 5). Values were normalized to the mean of the healthy control group. (C) Comparison of IFN-inducible gene score derived from bulk RNAseq of PBMC. (D) Flow cytometry plot (left) and comparisons of CD169 expression by mean fluorescence intensity (MFI; middle pane) and by percentage of monocyte with positive staining (right). Gray shade indicates isotype control staining. (E) Quantification of chemokine and (F) cytokine levels in the plasma of healthy controls (n = 10) and the proband (2 replicates) prior to treatment as measured by Olink proximity extension assay. (G) Flow cytometry quantification of phospho-STAT1 induced by IFN-α2 and IFN-γ stimulation in CD14 + in monocytes from healthy controls (n = 2) and the proband (2 replicates each). Data were normalized to baseline MFI. Error bars represent standard deviation in panels (C), (D), (E), (F), and (G)
To confirm the heightened ISG expression at a protein level, we quantified the expression of CD169 (SIGLEC-1) on CD14 + monocytes by flow cytometry. The expression of CD169, analyzed by both mean fluorescence intensity (MFI) or percentage CD14 + monocyte with positive expression, was markedly higher in the proband compared to healthy controls (Fig. 6D). B cell activating factor (BAFF), a cytokine induced by type I and type II IFN [14, 15], was noted to be highly elevated in the proband’s plasma (6432 pg/mL; normal range < 1748 pg/mL). Of note, BAFF levels may be elevated in the setting of SOCS1 haploinsufficiency but may additionally be impacted by rituximab administration (measured following dose three of four of rituximab therapy in our patient). High sensitivity quantification of proinflammatory mediators by proximity-extension assay further showed increased plasma levels of numerous chemokines, including log-fold differences for IFN-induced chemokines (CXCL9, CXCL10, CXCL11), compared to controls (Fig. 6E). In addition to IFN-γ, the proband also displayed higher levels of proinflammatory cytokines including IL-1β, IL-6, IL-18, and TNF in the plasma (Fig. 6F).
Next, we evaluated the patient’s response to IFN stimulation by measuring STAT1 phosphorylation. Compared to healthy controls, monocytes from the proband showed augmented STAT1 phosphorylation by 5 min after stimulation with type I IFN (IFN-α2) or type II IFN (IFN-γ); consistent with an inability to downregulate the IFN signaling associated with SOCS1 haploinsufficiency, the difference was even greater by 60 min post-stimulation (Fig. 6G). Taken together, results from multiple assays confirmed enhanced baseline expression of ISGs and augmented IFN signaling in the proband, supporting SOSC1 haploinsufficiency as the likely cause of disease.
Treatment Response to JAK Inhibition
JAK inhibitors are increasingly used to treat diseases with overt IFN production, termed interferonopathies. Due to our patient’s significant enthesitis-related arthritis and studies supportive of dysregulated IFN signaling, she was started on oral tofacitinib (dosed at 4 mg twice daily), an FDA-approved JAK1/JAK3 inhibitor for the treatment of juvenile idiopathic arthritis. Her enthesitis and arthralgias greatly improved within a month of treatment initiation and her platelet counts remain normal despite repopulation of B cells following rituximab usage. Prior to initiation of tofacitinib, the patient was screened for potential additional thrombotic and infectious risk factors, including assessment for antiphospholipid antibodies and iron deficiency with additional screening via plasma PCR for Epstein-Barr virus (EBV) and Cytomegalovirus (CMV), herpes simplex virus (HSV) serology, QuantiFERON gold, and hepatitis serology. These studies were only remarkable for mild iron deficiency for which the patient is receiving oral iron supplementation.
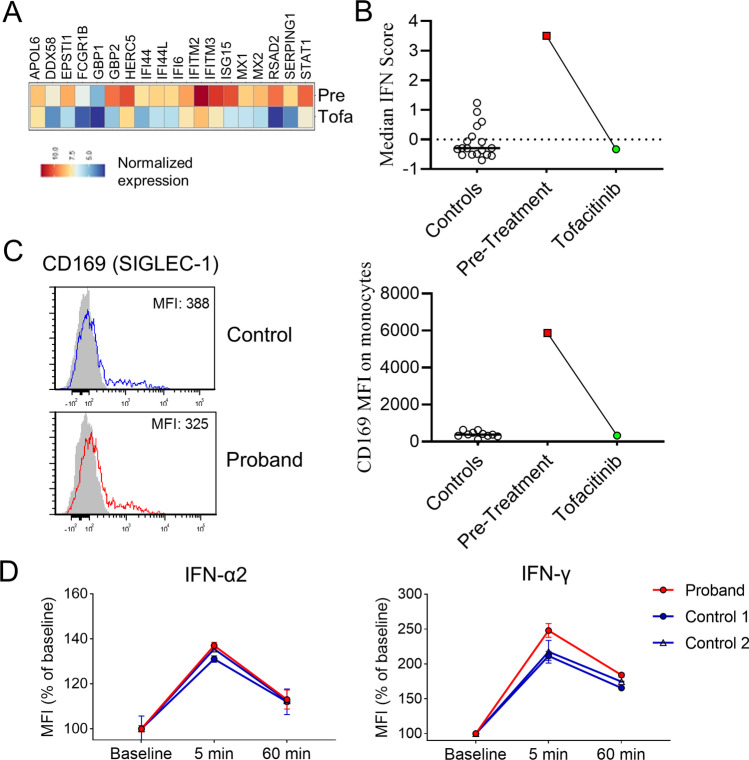
Her clinical response to JAK inhibition was further supported by normalization of ISG expression measured by RNA sequencing and by flow cytometry after 1 month of treatment (Fig. 7A–C). The hyperresponsiveness of monocytes to IFN stimulation also resolved as she displayed normal STAT1 phosphorylation to IFN-α2 and IFN-γ stimulation (Fig. 7D). At the time of manuscript preparation, the patient has taken tofacitinib for 10 months without evidence of disease flare or adverse effects. Her B cells have repopulated with normal absolute counts of CD20 + and CD19 + B cells (no longer elevated as noted during initial presentation with recent values of 264 and 271 cells/cmm, respectively), suggesting that continued disease control is secondary to tofacitinib rather than persistent B lymphocyte depletion after rituximab treatment. Her IgA levels are normal while IgG (612 mg/dL) and IgM (34 mg/dL) levels were only minimally reduced since B cell recovery. Interestingly, the patient also has now normal absolute natural killer cell values (most recently 92 cells/cmm) since the initiation of tofacitinib.
Fig. 7.
Correction of the interferon signature after treatment with the JAK inhibitor tofacitinib. (A) Heat map display of IFN-inducible gene expression in the proband’s PBMC pre- and post-treatment with tofacitinib. (B) Calculation of IFN-inducible gene score and comparison with healthy controls. (C) Flow cytometry plot (left) and comparisons of CD169 expression by MFI on monocytes. (D) Quantification of phospho-STAT1 induced by IFN-α2 and IFN-γ stimulation in CD14 + in monocytes from healthy controls (n = 2) and the proband 4 weeks after initiation of tofacitinib treatment (2 replicates each). Error bars represent standard deviation in panel (D)
Discussion
Here we report a case of SOCS1 haploinsufficiency in a 5-year-old female child with prominent enthesitis and a severe immune-mediated cytopenia. To our knowledge, this case represents the first report of SOCS1 haploinsufficiency in the setting of a de novo chromosomal deletion resulting in the complete loss of a SOCS1 allele [1, 2, 4, 5]. Consistent with the loss of SOCS1 function, PBMC from the patient showed enhanced IFN signaling and augmented response to IFN stimulation. Our work expands the genotype and phenotype spectrum of SOCS1 haploinsufficiency and illustrates the therapeutic efficacy of JAK inhibition.
SOCS1 haploinsufficiency was first described in 2020 and only limited cases have been described in 4 reports to date. Like several prior cases, our patient’s disease onset was possibly unmasked by a viral trigger (SARS-CoV-2). She exhibited evidence of excessive B cell activity, eosinophilia, and elevated IgE while the remainder of her immune evaluation was largely normal, without a history of recurrent infections. While polyarthritis has been mentioned as a manifestation of SOCS1 haploinsufficiency in one case series, our patient’s severe enthesitis on imaging extends the broad spectrum of musculoskeletal findings associated with this disease. Furthermore, bone marrow hypocellularity has not previously been observed in patients with SOCS1 defects. Such a finding raised the concern for a possible congenital bone marrow failure condition or idiopathic aplastic anemia [16], especially with identification of ERCC4 and PARN in the cluster of deleted genes. Reassuringly, our patient’s functional testing for these congenital bone marrow disorders was unremarkable. Of note, excessive interferon production may have various potential deleterious effects on hematopoietic stem cell (HSCs) proliferation, exhaustion, and death. Mice deficient in SOCS1 present with altered hematopoiesis and cytopenias [17]. IFNγ additionally causes STAT1-mediated differentiation resulting in decreased long-term HSC function, promotes apoptosis of HSCs, and changes the HSC niche. Type I interferons have also been associated with bone marrow aplasia, are known to sensitize HSCs to cellular stress, and are clearly stimulated with viral infections [18]. Thus, there may be synergistic activity of type I and II interferons after viral infection in patients with SOCS1 haploinsufficiency, resulting in bone marrow hypocellularity.
JAK-STAT pathways are utilized by multiple cytokines and therefore SOCS1 haploinsufficiency likely contributes to immune-dysregulation beyond IFN signaling. Enhanced IL-4/STAT6 and IL2/STAT5 signaling have been shown in SOCS1-deficient cell lines and excess STAT5 phosphorylation to IL-2 stimulation was confirmed in T cells from patients with SOCS1 haploinsufficiency [2, 19, 20]. Notably, the proband also showed high levels of IL-1β, IL18, IL-6, and TNF. These findings may be explained by additional roles of SOCS1 in restricting NFκB and inflammasome activation [21, p. 1], [22].
JAK inhibition has been used to treat numerous autoinflammatory conditions, including rheumatoid/juvenile idiopathic arthritis, inflammatory bowel disease, psoriasis, alopecia areata, and graft-versus-host disease following allogeneic bone marrow transplantation [23]. Despite the mechanistic link of dysregulated JAK function in the setting of SOCS1 haploinsufficiency, treatment with JAK inhibitor (baricitinib) has only been previously described in one patient with SOCS1 haploinsufficiency (2). We show that beyond achieving clinical and laboratory remission, tofacitinib treatment in our patient led to complete normalization of the IFN signature in PBMCs and reversed the dysregulated STAT1 phosphorylation after IFN stimulation.
Part of the complexity of treatment in those with inborn errors of immunity secondary to JAK-STAT pathway dysregulation includes choosing the appropriate JAK inhibitor. Currently tofacitinib, baricitinib, ruxolitinib, and upadacitinib are all Food and Drug Administration (FDA)-approved agents but differ in selectivity. Type I interferons bind to IFNAR1 and IFNAR2 resulting in a structural change leading to activation of JAK1/TYK2. Type II interferon binds to IFNγR2 resulting in oligomerization and transphosphorylation with IFNγR1 which then activates JAK1/JAK2 [24]. JAK inhibitor selectivity measured with in vitro kinase assays have demonstrated upadacitinib to be more selective for JAK1 and TYK2, baricitinib and ruxolitinib more selective for JAK1 and JAK2, and tofacitinib more selective for JAK1 and JAK3 [25, 26]. Studies comparing the in vitro cellular pharmacology of leukocyte sub-populations at clinically relevant levels have demonstrated some differences between these JAK inhibitors based on distinct cytokine pathways with the most potent inhibitor of IFN-γ being tofacitinib and the most potent inhibitors of IFN-α being upadacitinib and tofacitinib [26]. Based on these results and the fact that tofacitinib recently received approval from the FDA for the treatment of juvenile idiopathic arthritis, we elected to use tofacitinib for treatment.
The long-term impact of JAKi on SOCS1 haploinsufficiency remains to be seen. Long-term safety data for JAK inhibitors in children are lacking but reported risks of thrombosis, cardiac events, and malignancy with JAK-inhibition in adults [27] have not been assessed in children. Prior to initiation of JAK inhibition, it is recommended to screen for factors that may elevate a patient’s thrombosis risk including antiphospholipid antibodies and iron deficiency [28]. Additionally, given the reported increased risk of infections in those utilizing JAK inhibition [23], particularly Herpes zoster and additional viral illnesses, screening for HSV, EBV, CMV, viral hepatitis, and tuberculosis should be considered with further consideration for acyclovir prophylaxis in those with positive HSV serology. Some patients with SOCS1 haploinsufficiency have concurrent immunologic defects that warrant monitoring and initiation of antimicrobial prophylaxis and/or intravenous immunoglobulin administration. The risk of malignancy in patients with SOCS1 haploinsufficiency individuals with or without JAKi treatment is also unclear although a single report of Hodgkin lymphoma was described [2, 4]. Therefore, clinicians caring for patients with SOCS1 haploinsufficiency, especially those that receive JAK inhibitors or other immunosuppression, should be vigilant for clinical signs of malignancy and/or lymphoproliferation.
ITP presents in children typically at 1–4 years with an otherwise unremarkable medical history. The following scenarios should prompt consideration for broadening the diagnostic evaluation of children presenting with thrombocytopenia: lack of response to initial ITP therapies, rheumatologic signs/symptoms, recurrent/abnormal infections, chronic ITP (> 12 months), concurrent immune-mediated hemolysis or neutropenia (Evans Syndrome), or family history of myeloid malignancies, immunodeficiency, or immune-mediated cytopenias. Patients with these findings are at higher risk for harboring an underlying inborn of error of immunity, rheumatologic, malignant, or infectious condition [29, 30]. Patients with Evans Syndrome are particularly at risk for having a germline immunologic defect [31].
Finally, our case highlights the need to utilize a multidisciplinary diagnostic approach and consider comprehensive genetic evaluations in those with atypical ITP. Inborn errors of immunity are increasingly recognized to manifest with varying clinical and laboratory manifestations. Collectively, the prevalence of these monogenic conditions may be as high as 1 in every 1200–2000 individuals [32]. Comprehensive genetic testing is now strongly recommended in those with concerning findings for inborn errors of immunity [5, 33–35]. Timely next generation sequencing and microarray analysis were critical for securing a diagnosis and initiating treatment for our patient. If available, functional studies can further support the diagnosis and delineate the mechanism of disease. Cooperation and expertise of various medical subspecialities including hematology/oncology, genetics, radiology, rheumatology, pharmacy, pathology, and immunology are paramount for the care of patients with inborn errors of immunity.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the patient’s family for allowing us to share their story.
Author Contribution
TFM, KW, LD, NS, MH, MB, SM, KKH, DF, SN, JS, and NB assisted in the clinical care of the patient. PYL, LC, KB, and YD completed the functional SOCS1 studies and provided consultation regarding the case. TFM and PYL drafted the initial manuscript with all additional authors providing review and edits to the work thereafter.
Funding
PYL was supported by the National Institute of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) K08-AR074562, Charles H. Hood Foundation Child Health Research Award, and Arthritis National Research Foundation All Arthritis Grant.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics Approval
All experimental studies were approved by the Institutional Review Board at Boston Children’s Hospital. This study was performed in line with the principles of the Declaration of Helsinki.
Consent to Participate/Consent for Publication
Informed consent was provided by participants or legal guardians. The patient’s family additionally provided authorization to report and publish her case.
Conflict of Interest
No authors have competing interests. Michniacki, Saad, Hannibal, DeMeyer, Brown, Mohan, Lee, Ngo, Simoneau, Chen, Brodeur, Du, Basiaga, Horst: No disclosures. Frame: Research Funding from Jazz Pharmaceuticals. Walkovich: Horizon Pharmaceuticals: Honoraria, Membership on an entity’s Board of Directors or advisory committees; Pharming: Honoraria, Membership on an entity’s Board of Directors or advisory committees; Swedish Orphan Biovitrum AB (Sobi): Consultancy, Honoraria; X4 Pharmaceuticals: Other: Local PI for clinical trial involving mavorixafor and patients with neutropenia.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lee PY, et al. Immune dysregulation and multisystem inflammatory syndrome in children (MIS-C) in individuals with haploinsufficiency of SOCS1. J Allergy Clin Immunol. 2020;146(5):1194–1200.e1. doi: 10.1016/j.jaci.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hadjadj J, et al. Early-onset autoimmunity associated with SOCS1 haploinsufficiency. Nat Commun. 2020;11(1):5341. doi: 10.1038/s41467-020-18925-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.N. P. D. Liau et al, (2018) “The molecular basis of JAK/STAT inhibition by SOCS1,” Nat Commun, vol. 9, no. 1, Art. no. 1, 10.1038/s41467-018-04013-1 [DOI] [PMC free article] [PubMed]
- 4.J Körholz et al 2021 One gene, many facets: multiple immune pathway dysregulation in SOCS1 haploinsufficiency Front Immunol 12 680334 10.3389/fimmu.2021.680334 [DOI] [PMC free article] [PubMed]
- 5.Thaventhiran JED, et al. Whole-genome sequencing of a sporadic primary immunodeficiency cohort. Nature. 2020;583(7814):90–95. doi: 10.1038/s41586-020-2265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.“Pathology handbook,” Cytogenetics, cancer cytogenomic array, blood or bone marrow. https://www.pathology.med.umich.edu/handbook/#/details/888 (accessed Mar. 06, 2022).
- 7.“Pathology handbook,” Chromosomal microarray analysis, blood. https://www.pathology.med.umich.edu/handbook/#/details/4760 (accessed Mar. 06, 2022).
- 8.“Genetic testing for inborn error of immunity,” Blueprint Genetics. https://blueprintgenetics.com/tests/panels/hematology/comprehensive-immune-and-cytopenia-panel/ (accessed Feb. 20, 2022).
- 9.“Flow FISH telomere testing technology | RepeatDx,” Oct. 26, 2015. https://repeatdx.com/flow-fish/ (accessed Mar. 06, 2022).
- 10.“Diagnostic Immunology Lab | Cancer and blood diseases.” https://www.cincinnatichildrens.org/service/c/cancer-blood/hcp/clinical-laboratories/diagnostic-lab (accessed Mar. 06, 2022).
- 11.Kim H, et al. Development of a validated interferon score using NanoString technology. J Interferon Cytokine Res. 2018;38(4):171–185. doi: 10.1089/jir.2017.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.“OMIM - Online Mendelian Inheritance in Man.” https://www.omim.org/. Accessed 27 Apr 2022.
- 13.S. A. Savage and E. Cook, Dyskeratosis congenita and telomere biology disorders: diagnosis and management guidelines, 1st ed. 2015.
- 14.Gorelik L, Gilbride K, Dobles M, Kalled SL, Zandman D, Scott ML. Normal B cell homeostasis requires B cell activation factor production by radiation-resistant cells. J Exp Med. 2003;198(6):937–945. doi: 10.1084/jem.20030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A.-C. Lundell et al. (2017) “IFN type I and II induce BAFF secretion from human decidual stromal cells,” Sci Rep, vol. 7, no. 1, Art. no. 1, 10.1038/srep39904. [DOI] [PMC free article] [PubMed]
- 16.Iwafuchi H. The histopathology of bone marrow failure in children. J Clin Exp Hematop. 2018;58(2):68–86. doi: 10.3960/jslrt.18018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. Metcalf et al. (1999) “Aberrant hematopoiesis in mice with inactivation of the gene encoding SOCS-1,” Leukemia vol 13 no 6 Art no 6. 10.1038/sj.leu.2401440 [DOI] [PubMed]
- 18.J. N. P. Smith, V. S. Kanwar, and K. C. MacNamara, “Hematopoietic stem cell regulation by type I and II interferons in the pathogenesis of acquired aplastic anemia,” Frontiers in Immunology, vol. 7, 2016, Accessed: May 04, 2022. [Online]. Available: https://www.frontiersin.org/article/10.3389/fimmu.2016.00330 [DOI] [PMC free article] [PubMed]
- 19.Dickensheets H, et al. Suppressor of cytokine signaling-1 is an IL-4-inducible gene in macrophages and feedback inhibits IL-4 signaling. Genes Immun. 2007;8(1):21–27. doi: 10.1038/sj.gene.6364352. [DOI] [PubMed] [Google Scholar]
- 20.Sporri B, Kovanen PE, Sasaki A, Yoshimura A, Leonard WJ. JAB/SOCS1/SSI-1 is an interleukin-2-induced inhibitor of IL-2 signaling. Blood. 2001;97(1):221–226. doi: 10.1182/blood.v97.1.221. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Xu C, Chen X, Shi Q, Su W, Zhao H. SOCS-1 suppresses inflammation through inhibition of NALP3 inflammasome formation in smoke inhalation-induced acute lung injury. Inflammation. 2018;41(4):1557–1567. doi: 10.1007/s10753-018-0802-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strebovsky J, Walker P, Lang R, Dalpke AH. Suppressor of cytokine signaling 1 (SOCS1) limits NFkappaB signaling by decreasing p65 stability within the cell nucleus. FASEB J. 2011;25(3):863–874. doi: 10.1096/fj.10-170597. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ. JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov. 2017;17(1):78. doi: 10.1038/nrd.2017.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivashkiv LB. IFNγ: signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat Rev Immunol. 2018;18(9):545–558. doi: 10.1038/s41577-018-0029-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shawky AM, Almalki FA, Abdalla AN, Abdelazeem AH, Gouda AM. A comprehensive overview of globally approved JAK inhibitors. Pharmaceutics. 2022;14(5):1001. doi: 10.3390/pharmaceutics14051001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McInnes IB, et al. Comparison of baricitinib, upadacitinib, and tofacitinib mediated regulation of cytokine signaling in human leukocyte subpopulations. Arthritis Res Ther. 2019;21(1):183. doi: 10.1186/s13075-019-1964-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C. for D. E. and Research, “FDA requires warnings about increased risk of serious heart-related events, cancer, blood clots, and death for JAK inhibitors that treat certain chronic inflammatory conditions,” FDA, Jan. 2022, Accessed: Mar. 06, 2022. [Online]. Available: https://www.fda.gov/drugs/fda-drug-safety-podcasts/fda-requires-warnings-about-increased-risk-serious-heart-related-events-cancer-blood-clots-and-death
- 28.Hung S-H, Lin H-C, Chung S-D. Association between venous thromboembolism and iron-deficiency anemia: a population-based study. Blood Coagul Fibrinolysis. 2015;26(4):368–372. doi: 10.1097/MBC.0000000000000249. [DOI] [PubMed] [Google Scholar]
- 29.A. Schifferli et al., “Understanding immune thrombocytopenia: looking out of the box,” Frontiers in Medicine, vol. 8, 2021, Accessed: Feb. 26, 2022. [Online]. Available: https://www.frontiersin.org/article/10.3389/fmed.2021.613192 [DOI] [PMC free article] [PubMed]
- 30.“The ITP syndrome: pathogenic and clinical diversity | Blood | American Society of Hematology.” https://ashpublications.org/blood/article/113/26/6511/26188/The-ITP-syndrome-pathogenic-and-clinical-diversity (accessed Mar. 06, 2022).
- 31.Hadjadj J, et al. Pediatric Evans syndrome is associated with a high frequency of potentially damaging variants in immune genes. Blood. 2019;134(1):9–21. doi: 10.1182/blood-2018-11-887141. [DOI] [PubMed] [Google Scholar]
- 32.Chinn IK, Orange JS. Immunodeficiency disorders. Pediatr Rev. 2019;40(5):229–242. doi: 10.1542/pir.2017-0308. [DOI] [PubMed] [Google Scholar]
- 33.FA. Bonilla et al. (2015) “Practice parameter for the diagnosis and management of primary immunodeficiency,” J. Allergy Clin. Immunol., vol. 136, no. 5, pp. 1186-1205.e1–78, 10.1016/j.jaci.2015.04.049 [DOI] [PubMed]
- 34.Yska HAF, Elsink K, Kuijpers TW, Frederix GWJ, van Gijn ME, van Montfrans JM. Diagnostic yield of next generation sequencing in genetically undiagnosed patients with primary immunodeficiencies: a systematic review. J Clin Immunol. 2019;39(6):577–591. doi: 10.1007/s10875-019-00656-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heimall JR, et al. Use of genetic testing for primary immunodeficiency patients. J Clin Immunol. 2018;38(3):320–329. doi: 10.1007/s10875-018-0489-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.