Abstract
Expression of the Tn21 mercury resistance (mer) operon is controlled by a metal-sensing repressor-activator, MerR. When present, MerR always binds to the same position on the DNA (the operator merO), repressing transcription of the structural genes merTPCAD in the absence of Hg(II) and inducing their transcription in the presence of Hg(II). Although it has two potential binding sites, the purified MerR homodimer binds only one Hg(II) ion, employing Cys82 from one monomer and Cys117 and Cys126 from the other. When MerR binds Hg(II), it changes allosterically and also distorts the merO DNA to facilitate transcriptional initiation by ς70 RNA polymerase. Wild-type MerR is highly specific for Hg(II) and is 100- and 1,000-fold less responsive to the chemically related group 12 metals, Cd(II) and Zn(II), respectively. We sought merR mutants that respond to Cd(II) and obtained 11 Cd(II)-responsive and 5 constitutive mutants. The Cd(II)-responsive mutants, most of which had only single-residue replacements, were also repression deficient and still Hg(II) responsive but, like the wild type, were completely unresponsive to Zn(II). None of the Cd(II)-responsive mutations occurred in the DNA binding domain or replaced any of the key Cys residues. Five Cd(II)-responsive single mutations lie in the antiparallel coiled-coil domain between Cys82 and Cys117 which constitutes the dimer interface. These mutations identify 10 new positions whose alteration significantly affect MerR’s metal responsiveness or its repressor function. They give rise to specific predictions for how MerR distinguishes group 12 metals, and they refine our model of the novel domain structure of MerR. Secondary-structure predictions suggest that certain elements of this model also apply to other MerR family regulators.
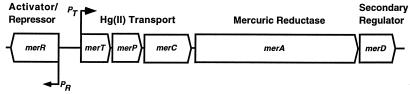
One of the best-characterized mercury resistance (mer) operons is located on transposon Tn21 from the Shigella flexneri IncFII plasmid R100 (26, 35). This operon consists of five structural genes—merT, merP, merC, merA, and merD (49)—and a regulatory gene, merR (72) (Fig. 1). MerT, MerP, and MerC are involved in the transport of Hg(II) into the cell (36). MerA is a cytosolic, NADPH-dependent, flavin adenine dinucleotide-containing oxidoreductase which reduces Hg(II) to Hg(0) (70). The merR gene is transcribed in the direction opposite from that of the structural genes. Its product, MerR, represses expression of the merTPCAD genes in the absence of Hg(II), activates their expression in the presence of Hg(II), and represses its own expression in the presence or absence of Hg(II) (21, 38, 39, 45, 51). MerD plays a minor role in regulation, possibly as an antagonist of MerR, which reestablishes repression of merTPCAD once Hg(II) has been reduced to Hg(0) (50).
FIG. 1.
The Tn21 mer operon. Arrows indicate the direction of transcription.
Presently there is no three-dimensional (3-D) structure for MerR; however, extensive genetic and biochemical data indicate that the protein contains three domains (63, 67, 72): a helix-turn-helix DNA-binding domain from 110 to R29 (14, 44); a “coupling” domain from K30 to H81 which may convey the status of the Hg(II) binding site to the DNA binding site (20, 37); and a long helical region from C82 to C117 which constitutes both the dimer interface and, with the loop containing C126, the Hg(II)-binding domain (84).
MerR binds a palindromic DNA sequence (merO) which lies between the −10 and the −35 recognition sites for ς70 RNA polymerase at the merTPCAD promoter, PT, and which also lies exactly on the start site of its own divergent transcript (39, 54, 55). MerR fosters the binding of ς70 RNA polymerase to PT even in the absence of Hg(II) (39, 43, 44) but still represses transcription, a phenomenon called active repression (24, 29) to distinguish it from repression by simple occlusion of the polymerase binding sites.
The MerR homodimer binds Hg(II) by using the thiols of three conserved cysteines: cysteine 82 (C82) from one monomer and cysteines 117 and 126 (C117 and C126) from the other monomer. These ligands form a novel planar tricoordinate complex with Hg(II) (37, 53, 75, 81). Upon binding Hg(II), MerR undergoes a conformational change that leads to an underwinding of the PT region and thereby enables RNA polymerase to form an open complex (3, 4, 30, 39, 43, 44). Curiously, although the MerR homodimer contains two potential Hg(II) binding sites, the purified protein binds only one Hg(II) per dimer. Moreover, although the two other group 12 metals, Zn(II) and Cd(II), also form stable complexes with protein thiols (12, 33, 64, 76), purified MerR binds Hg(II) preferentially even in the presence of a 1,000-fold excess of Cd(II) or Zn(II) (69) and also requires 100- to 1,000-fold higher concentrations of these metals for transcriptional activation (61).
Although several studies have described mutants altered in either repression or activation (20, 56, 57, 63, 67), no genetic study of merR has explicitly addressed the basis of its metal specificity. This question is especially interesting in light of the growing family of MerR-like regulators, many of which respond not to metals but to hydrophobic cationic drugs (1, 2). Here we report the first variants of MerR with an altered response to a metal; the properties and locations of these mutations shed light on the basis for metal-provoked activation by MerR. They also led us to examine the possible occurrence of similar secondary-structure elements in other members of the MerR family, and our findings in the latter regard indicate conservation of a novel structural domain in a subset of this family.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source | Reference(s) |
|---|---|---|---|
| E. coli strains | |||
| CAG1574 | araD139 Δ(ara leu)7697 ΔlacX74 galU galK hsdR rspL recA56 srl | C. A. Gross | 17, 55, 63 |
| CB806 | ΔlacZ lacY+ galK phoA8 rpsL thi recA56 | C. Beck | 66 |
| DH5α | F′ endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacZYA-argF)U169 (φ80lacZΔM15) | 6 | |
| XL1-Red | endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10 (Tetr) | Stratagene (La Jolla, Calif.) | 25, 34, 59, 65 |
| Plasmids | |||
| pACYC177 AmpΔ | p15A replicon; aph | W. Ross | 63 |
| pACYC184 | p15A replicon; tet cat | 18 | |
| pNH9 | p15A replicon; merR+ merOP+ aph | N. Hamlett | 36 |
| pSJ43 | pMB1 replicon; phoA merRΔ10 merOP+ merT lacZ+bla | S.-J. Park | 55 |
| pWR2 | pMB1 replicon; merR merOP+ merT lacZ+bla | W. Ross | 63 |
| pWR10 | pMB1 replicon; merRΔ10 merOP+ merT lacZ+bla | W. Ross | 63 |
| pJC10 | pMB1 replicon; merRΔ10 merOP+ merT cat bla | This work | This work |
Media and reagents.
Luria-Bertani (LB) broth and M-9 minimal salts medium were prepared as described elsewhere (6), and media were supplemented as needed with 100 μg of ampicillin/ml, 60 μg of kanamycin sulfate/ml, and/or 2 μM metal salts. CdCl2 (Fisher Scientific, Pittsburgh, Pa.) and HgCl2 (J. T. Baker Company, Phillipsburg, N.J.) were dissolved in deionized water. All agar plates containing metal were made with M-9 minimal salts medium and BBL purified agar (Becton Dickinson, Franklin Lakes, N.J.). All other agar plates were made with Bacto Agar (Difco, Detroit, Mich.). The following high-purity metals were used for reporter enzyme induction in liquid medium: CdCl2 (Puratronic grade; 99.998% pure), AuCl3 (65% Au), and ZnCl2 (Puratronic grade; 99.999% pure), all from Alfa Aesar (Ward Hill, Mass.); plasma pure Hg(II) (100 μg/ml in 10% nitric acid) from Leeman Laboratories, Inc. (Lowell, Mass.); potassium antimonyl tartrate (99.95% pure) from Aldrich (Milwaukee, Wis.); AgNO3 (Baker Analyzed Reagent) from the J. T. Baker Company; and CuSO4 · 5H2O (American Chemical Society certified) from Fisher Scientific (Fair Lawn, N.J.).
DNA purification.
Plasmid DNA was isolated by alkaline lysis (10) and further purified by using plasmid preparation kits from Promega Corporation (Madison, Wis.) or Qiagen, Inc. (Santa Clara, Calif.).
PCR.
PCR mixtures contained 20 pmol of each primer, various concentrations of template, 0.2 mM (each) deoxyribonucleotide triphosphate, 2.5 mM MgCl2, 1.25 U of Taq DNA polymerase (Promega), and 1× Promega sample buffer (50 mM KCl, 0.1% [vol/vol] Triton X-100, 10 mM Tris-HCl [pH 9.0]). Target DNA sequences were amplified by a modified hot-start technique employing wax beads (77) and then amplified for 30 cycles of 94°C for 1.0 min, 59°C for 1 min, and 72°C for 2 min. After a final 72°C step for 5 min, the samples were cooled to 15°C and treated with a Wizard PCR Prep kit (Promega). All primers for PCR and sequencing were designed with OLIGO software (National Biosciences, Inc., Plymouth, Minn.) and synthesized by Genosys Biotechnologies, Inc. (The Woodlands, Tex.). For error-prone PCR (16, 31), 0.5 mM MnCl2 · 4H2O (J. T. Baker Company) was added to the PCR mixtures described above.
Plasmid construction.
Plasmid pWR10 (63) was digested with restriction enzymes AvaI and BamHI (New England Biolabs, Beverly, Mass.) and treated with a Wizard Clean-up kit (Promega). The chloramphenicol acetyltransferase (cat) gene was amplified from pACYC184 (18) by PCR using an annealing temperature of 55.7°C and the primers 5′ GCGTTCTCGGGCACCAATAA 3′ (upper primer, containing an AvaI site [underlined]) and 5′ ATCGGGATCCTCAGGAGCTAA 3′ (lower primer, containing a BamHI site). The PCR amplicand was digested with AvaI and BamHI, treated with a Wizard Clean-up kit, and ligated to the AvaI- and BamHI-digested plasmid pWR10 with T4 DNA ligase (Promega). The ligation product was digested with EcoRV (New England Biolabs) to eliminate any remaining parent plasmid (83) and transformed into Escherichia coli DH5α by a CaCl2 method (6). Sequencing of both strands of the resulting mer-cat fusion plasmid, pJC10, indicated recovery of the expected product.
Mutagenesis.
The merR gene was randomly mutagenized by two strategies. The first strategy employed selection or screening for metal-dependent lactose utilization. By error-prone PCR (16, 31) we targeted merR codons 17 to 144, a sequence which includes the presumptive recognition helix of the DNA-binding domain (57, 63), the putative coupling domain, and the Hg(II)-binding–dimerization domain. The merR gene from pWR2 (63) (Table 1) was amplified in the presence of MnCl2, using the upper primer 5′ AACTGCAGAACGGAAAATAAAGCAC 3′ (introducing a PstI restriction enzyme site), and the lower primer 5′ AACTGGAATGGATAGCGTAACCTTA 3′. The amplified DNA was used to replace the corresponding portion of the wild-type merR gene lying between the PstI and EagI sites of pNH9 (36) (New England Biolabs), and the resulting construct was electroporated into E. coli CB806 containing the mer-lac fusion reporter plasmid pSJ43 (Table 1) (55). Note that the EagI site lies within the amplified region, not in the lower primer. For screening of mutants, transformant colonies on LB agar containing kanamycin and ampicillin were screened by replica plating on M-9 agar containing lactose as the sole carbon source and either 2 μM metal chloride or no metal. We defined as Cd(II) responsive those isolates which grew faster on Cd(II)-lactose plates than they did on plates lacking any metal. For example, the Cd(II)-responsive mutant K99T, in which the lysine at position 99 was replaced by threonine, produced good-sized colonies on M-9–lactose–Cd(II) agar in 2 days and on agar lacking metal in 3 days. In comparison, on M-9–lactose–Hg(II) agar, colonies of the wild-type merR strain and of the K99 mutant were fully grown up in 1 day. We also employed metal-dependent lactose utilization as a direct selection technique, plating transformants on ditch gradient plates (71) containing lactose as the sole carbon source and using a 1.0 mM metal salt solution in the ditch (8.0 mm by 8.5 cm), which was located in the center of a square plate (100 by 15 mm). The gradient method was employed so as not to limit our acquisition of mutants to those optimally induced by 2 μM Cd(II). The Cd(II) inducibility of LacZ in colonies that grew on the M-9–lactose–Cd(II) gradient plates was confirmed by replica plating as described above.
The second mutagenesis strategy was based on metal-dependent expression of the cat gene by the mer-cat transcriptional fusion reporter pJC10 described above (Table 1). Mutagenesis targeted all 144 codons of merR and was effected by transforming pNH9 into the mutator strain E. coli XL1-Red (25, 34, 59, 65) (Stratagene, La Jolla, Calif.). Five different XL1-Red(pNH9) transformants were grown in one culture overnight at 37°C, and the organisms were then subcultured overnight three times. Plasmid DNA prepared from the final culture was electroporated into CAG1574(pJC10), and transformants were selected on gradient plates containing 50 μg of chloramphenicol/ml in M-9 medium, with a 1 mM metal chloride solution or water in the ditch. All colonies were screened by replica plating on M-9 agar containing 50 μg of chloramphenicol/ml and either 2 μM metal chloride or no metal. As noted above, the isolates that grew faster on the Cd(II) plates than on the plates lacking metal were defined as Cd(II) responsive.
All mutant derivatives of pNH9 were sequenced on both strands at the University of Georgia Molecular Genetics Instrumentation Facility, using an ABI model 373 Stretch DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Error-prone PCR mutagenesis yielded 50% transitions and 50% transversions. XL1-Red mutagenesis yielded 84.6% transitions and 15.4% transversions. Four transversions (G → C, C → A, C → G, and T → G) were not observed with either method of mutagenesis. As expected, there were no frameshifts or deletions in these gain-of-function selections.
β-Galactosidase (LacZ) assays.
All mutant derivatives of pNH9 were transformed into CAG1574(pWR10) to measure the LacZ activity induced by 2 μM Hg(II), Cd(II), Zn(II), Au(III) (each as the chloride), or Sb(III). Overnight cultures were diluted 1:40 in M-9 medium, grown at 37°C to mid-exponential phase, transferred to nitric acid-washed sterile test tubes, induced for 30 min with 2 μM metal chloride, and assayed for β-galactosidase activity (48). The working concentration of each metal was 0.5 mM. AgNO3, CdCl2, and potassium antimonyl tartrate were dissolved in water. To prevent precipitation, AuCl3 and ZnCl2 stocks were made in 10% (vol/vol) HCl and diluted for use. For assessment of induction with higher Zn(II) concentrations (2, 10, 100, and 500 μM), growth and induction were performed in LB broth buffered with 0.1 M Tris-HCl (pH 7.4) to avoid precipitation of the zinc phosphate. LacZ assays for each mutant were done in duplicate at least twice, and the average and standard deviation of all assays are reported. For assessment of induction with higher Cu(I) concentrations (2, 10, 100, and 500 μM), cells were grown and induced in a modified M-9 minimal medium in which the sodium phosphate was replaced by 100 mM Tris-HCl (pH 7.4) and Casamino Acids were replaced by 0.5% Proteose Peptone no. 3 (Difco) as an amino acid and phosphate source (74). Dithiothreitol (10 mM; Fisher Scientific, Fair Lawn, N.J.) was added immediately before induction to reduce Cu(II) (added as CuSO4 · 5H2O) to Cu(I).
Secondary-structure predictions.
The propensities for formation of a coiled coil by Tn21 MerR (GenBank protein accession no. P07044), Bacillus sp. strain RC607 MerR (P22853), Staphylococcus aureus MerR (P22874), Haemophilus influenzae YBBI-HAEIN (P44617), H. influenzae Y186-HAEIN (P44558), Synechocystis sp. strain PCC6803 Y701-SYNY3 (Q55963), Escherichia coli SoxR (S72675), E. coli ZntR (P36676), Bradyrhizobium japonicum NolA (P22537), Bacillus subtilis BltR (P39842), B. subtilis BmrR (P39075), Thiobacillus ferrooxidans MerR (GenBank nucleotide accession no. X57326 and sequence identification [ID] no. 48151), Streptomyces lividans TipAL (nucleotide accession no. S64314 and sequence ID no. 408223), and B. subtilis YwnD (nucleotide accession no. Z99122 and sequence ID no. 2636185) were determined with the programs COILS (46, 47, 58) and MultiCoil (80). For COILS, the weighted and unweighted MTK (myosin, tropomyosin, and keratin) parameters (47a) were used to detect antiparallel coiled coils. As recommended by the authors of COILS, a window of 28 was used to simply detect the presence of a coiled coil and a window of 21 was used to define the N- and C-terminal ends of a predicted coiled coil. As recommended by the authors of MultiCoil, a window of 28, dimeric scoring distances of 1, 2, and 4, and a trimeric scoring distance of 4 were used to detect antiparallel coiled coils (80a). The very small data set of antiparallel (as opposed to parallel) coiled coils among proteins with defined 3-D structures limits the reliability with which antiparallel coiled coils can be detected by these algorithms. For example, COILS was the only program that correctly predicted the antiparallel coiled coil in the Thermus thermophilus seryl-tRNA synthetase (32, 80).
RESULTS
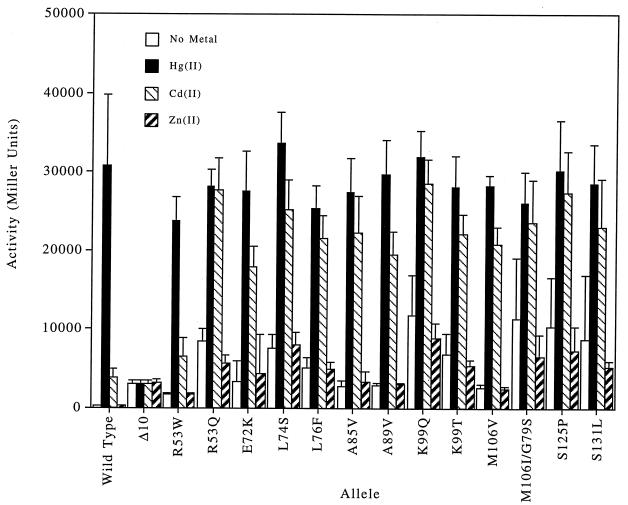
Based on their behavior in LacZ assays, we defined the mutant merR phenotypes obtained in this work as follows (Fig. 2 and 3): (i) a Cd(II)-responsive mutant has Cd(II)-induced LacZ activity which is at least 50% of its Hg(II)-induced activity; (ii) a repression-deficient mutant has uninduced LacZ activity that is no more than 50% of its Hg(II)-induced activity; and (iii) a constitutive mutant has uninduced LacZ activity that is more than 50% of its Hg(II)-induced activity.
FIG. 2.
LacZ activity of Cd(II)-responsive merR mutants in the absence of metal or when treated with Hg(II), Cd(II), or Zn(II).
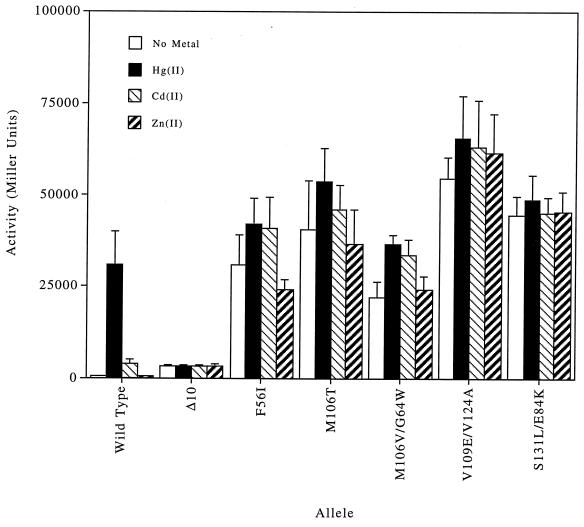
FIG. 3.
LacZ activity of constitutive merR mutants in the absence of metal or when treated with Hg(II), Cd(II), or Zn(II).
Cd(II)-responsive mutants.
Selection or screening for metal-dependent lactose utilization yielded only one Cd(II)-responsive mutant, K99T (Fig. 2), and selection for metal-dependent chloramphenicol resistance yielded 10 Cd(II)-responsive mutants. All were single-residue mutants, and six of them (R53Q, L76F, A85V, K99Q, S125P, and S131L) responded to Hg(II) and Cd(II) almost equally well (Fig. 2). The other Cd(II)-responsive mutants (E72K, L74S, A89V, K99T, and M106V) had Cd(II)-induced LacZ activities ranging from 64 to 79% of their Hg(II)-induced activities.
All 11 Cd(II)-responsive mutants were defective in the active-repression function of the wild-type merR. Four of them (E72K, A85V, A89V, and M106V) had uninduced LacZ activities ranging from 8 to 12% of their Hg(II)-induced activities, comparable to the derepressed (merRΔ) strain, pWR10. The other seven (R53Q, L74S, L76F, K99Q, K99T, S125P, and S131L) were much more leaky, having uninduced LacZ activities ranging from 20 to 37% of their Hg(II)-induced activities. In the presence of 2 μM Sb(III), Au(III) (data not shown), or Zn(II), none had a LacZ activity any greater than that achieved without any metal inducer, so they were considered to be unresponsive to these three metals.
While this work was in progress, a previously noted E. coli chromosomal MerR homolog (formerly YhdM [19]) was defined as ZntR, the regulator (15) of the gene for the Zn(II)/Cd(II) exporter, ZntA (7, 62). ZntR responds only slightly to Hg(II) or Cd(II) at concentrations below 10 or 100 μM, respectively, but responds very strongly to Zn(II) in the 100 to 1,000 μM range (15). Since we had used 2 μM Zn(II) in assessment of our mutants, we revisited the possibility of a Zn(II) response in them by examining the MerR allele A89V, whose change brings it closer to ZntR, which has a valine at the corresponding position (Fig. 4). We found that the MerR mutant A89V responded very slightly at the highest Zn(II) concentration (1,000 μM), with activity only twice its normal repression-defective level (data not shown), indicating that this change alone cannot confer Zn(II) responsiveness on MerR. Moreover, neither wild-type MerR nor the A89V mutant responded at concentrations as high as 500 μM Cu(I) (data not shown), which, like the group 12 dications, also has a filled d shell (28).
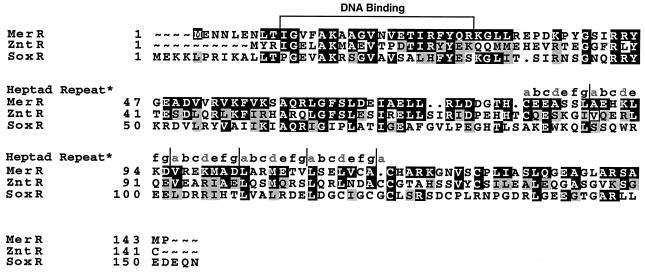
FIG. 4.
Alignment of a metal-binding subset of the MerR family of activator-repressor proteins. Residues highlighted in black are identical, and residues highlighted in gray are functionally similar. ZntR contains a V instead of an A at the position corresponding to residue 89 in MerR (A89V). Gray lowercase letters a and d in the heptad repeats identify residues that make contact between coiled strands. The beginning and end of each protein (∼) and gaps in the alignment (.) are indicated.
Repression-deficient mutants.
On finding that all of these new Cd(II)-responsive mutants were also repression deficient, we examined previously generated repression-deficient mutants A89V, S131L (63), R53W, and M106I/G79S (62a) for Cd(II) responsiveness. Assayed as originally isolated, in cis on the pWR2 plasmid background, A89V and S131L had Cd(II)-induced LacZ activities of 61 and 80% of their Hg(II)-induced activities and uninduced LacZ activities that were 15 and 18% of their Hg(II)-induced LacZ activities (data not shown), quite comparable to the activities of those alleles isolated here (Fig. 2). The double mutant M106I/G79S had Cd(II)-induced activity of 90% of its Hg(II)-induced activity and uninduced activity of 44% of its Hg(II)-induced activity (Fig. 2). The single mutant R53W proved to be only repression deficient, with an uninduced LacZ activity of 7% of its Hg(II)-induced activity and a Cd(II)-induced activity of just 27% of its Hg(II)-induced activity (Fig. 2). None of these earlier mutants responded to Sb(III), Au(III) (data not shown), or Zn(II). Finally, we examined the effect of Cd(II) on the mutants which had first defined the Hg(II) binding site of MerR: C82Y, C117Y, and C126Y (63) (data not shown). Each remained repressed in the presence of 2 μM Hg(II), Cd(II), or Zn(II).
Constitutive mutants.
Three mutants, M106T, M106V/G64W, and V109E/V124A, obtained by mer-lac selection were constitutive; two other constitutive mutants, F56I and S131L/E84K, were obtained by mer-cat selection. The constitutive double mutant M106V/G64W, with uninduced LacZ activity of 60% of its Hg(II)-induced activity, was further activated by Hg(II) or by Cd(II) but not by Zn(II) (Fig. 3). The other four constitutive mutants, F56I, M106T, V109E/V124A, and S131L/E84K, had uninduced LacZ activities ranging from 73 to 92% of their Hg(II)-induced activities, but none of these was further activated by Hg(II), Cd(II), Zn(II), Au(III), or Sb(III). Owing to the stability of the LacZ protein, no direct quantitative comparison of constitutive activity and inducible activity can be made.
Locations of new and old MerR mutations.
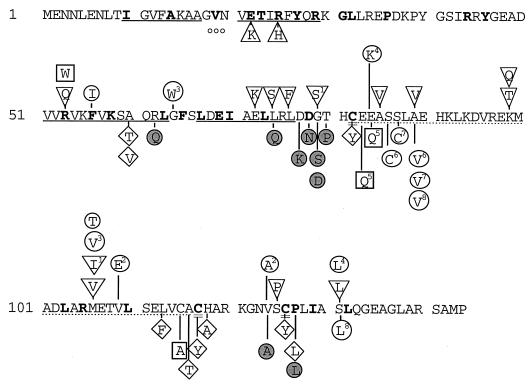
Of the 15 mutagenized sites in this study, only five (R53, E84, A89, M106, and S131) had been identified as significant in previous work (20, 56, 57, 63, 67). Two previously noted alleles, A89V and S131L, appeared again in response to the selection methods used in this work (Fig. 5). Although both mutagenesis strategies would have allowed alterations in all three domains of the merR gene, none of these new mutations occurred in the DNA-binding domain or in the Cys residues which constitute the metal ligands. One Cd(II)-responsive mutation (S125P) was adjacent to a key cysteine residue, but the rest were at least two residues away from these Hg(II)-binding ligands. Only one Cd(II)-responsive mutation (R53Q) and one constitutive mutation (F56I) replaced any of the 32 residues conserved in all known MerRs. However, six Cd(II)-responsive mutations (E72K, L74S, M106V, S125P, S131L, and M106I/G79S) and four constitutive mutations (M106T, M106V/G64W, V109E/V124A, and S131L/E84K) were immediately adjacent to these conserved residues.
FIG. 5.
Amino acid changes in MerR. Mutations described in this work are shown above the amino acid sequence. Previously described mutations (20, 56, 57, 63, 68) are shown below the sequence. The 32 residues conserved in all known MerR proteins in both gram-positive and gram-negative bacteria are shown in boldface. Symbols: double underline, cysteines involved in Hg(II) binding; single underline, predicted helix; ○○○, turn of predicted helix-turn-helix (DNA binding); dotted underline, predicted coiled-coil region; □, repression deficient only; ▿, Cd(II) responsive; ○, fully constitutive; ◊, activation deficient; ▵, activation and repression deficient;  , constitutive mutants constructed in an A89V or S131L background (R62Q and V124A individually combined with S131L, others each with A89V). Superscripted numbers indicate sets of double mutations; e.g., set 2 includes V109E and V124A.
, constitutive mutants constructed in an A89V or S131L background (R62Q and V124A individually combined with S131L, others each with A89V). Superscripted numbers indicate sets of double mutations; e.g., set 2 includes V109E and V124A.
Several of the new mutations enrich clusters of previously identified mutations. The first such cluster includes R53W, R53Q, F56I, and G64W, which flank the previously identified position A60, whose alteration leads to the loss of activation but not of repression (63). The next cluster bridges the coupling domain and the N terminus of the Hg(II)-binding domain (E72K, L74S, L76F, G79S, E84K, and A85V), a region already notable for repression-deficient (typically multiple) mutations (20, 57, 63). The third cluster of old and new mutations lies in or near the loop at the C terminus of the Hg(II)-binding domain (V124A and S125P), a region which has previously yielded repression-deficient mutants. The remaining new mutations occur in a segment of MerR that was not revealed as being functionally significant by prior genetic studies: the center of the long helix between C82 and C117 (K99T, K99Q, M106V, M106I, M106T, and V109E). The possible role of each cluster or region will be discussed below.
Generality of the propensity for coiled-coil formation in members of the MerR family.
We have recently presented biochemical data showing that Tn21 MerR contains a long helical domain from residues C82 to C117 (84) which constitutes a major part of both the dimer interface and the Hg(II)-binding domain. The possibility that this region assumes a coiled-coil structure in the dimer was suggested by the occurrence of five nearly perfect heptad repeats within it (Fig. 4).
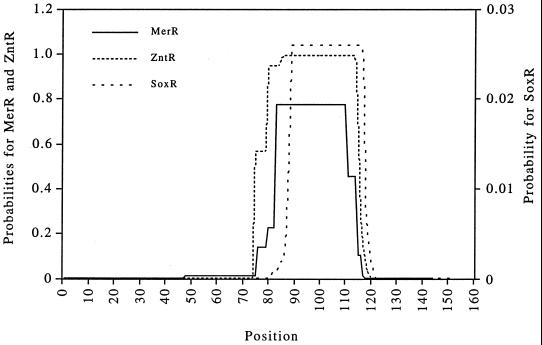
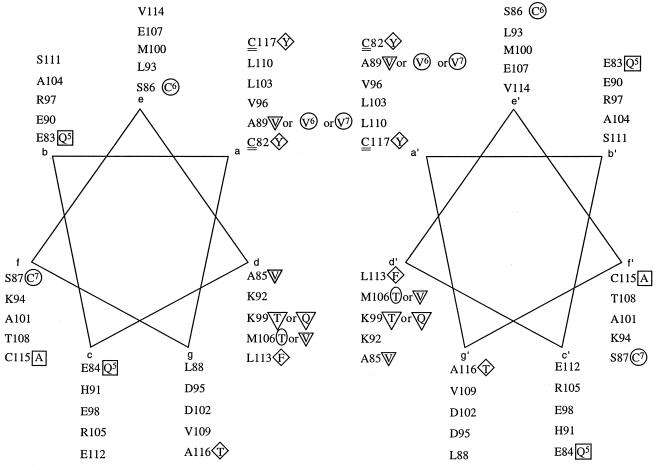
The use of the programs COILS (46, 47, 58) and MultiCoil (80) to test this possibility revealed a clear propensity for coiled-coil formation in MerR and in ZntR (15) and a very low propensity for the formation of coiled coils, albeit in the corresponding region, in the 4Fe-4S redox response regulator SoxR (41) (Fig. 6). Since the two programs employ different algorithms, they differed in their absolute predictions, but the trends were the same for MerR of Tn21, ZntR, and SoxR with both programs. In all three of these proteins, the region with the highest propensity to form coils corresponds to the Hg(II)-binding–dimerization domain of MerR (84). Moreover, the loss- or gain-of-function mutations summarized here cluster strikingly at the interface positions (a, a′ and d, d′ [8]) of two helical-wheel projections of residues C82 through C117 of MerR arranged in an antiparallel orientation (Fig. 7). This representation also highlights the potential juxtaposition of newly identified residues K99 and M106 within this interface. In a conventional alpha-helical projection (not shown), the mutant positions are diffuse and do not fall with such regularity on one face of the helix.
FIG. 6.
Coiled-coil predictions for MerR, ZntR, and SoxR, obtained by using the computer program COILS with unweighted MTK parameters (46, 47, 58). The scale for MerR and ZntR probabilities is located on the left y axis, and the scale for SoxR probability is located on the right y axis.
FIG. 7.
Antiparallel alignment of two helical wheels representing the predicted coiled-coil region between residues C82 and C117 in a MerR dimer. Symbols are described in the legend to Fig. 5.
The possible relevance of this putative coiled-coil structure to the functions of this class of regulatory proteins is testable by examination of its occurrence in three distantly related MerRs and in five non-MerR members of this family (accession numbers are listed in Materials and Methods). The absolute amino acid sequence is not well conserved in the corresponding region of these proteins (similarities range from 39 to 44% for the MerRs and from 27 to 41% for YBBI, Y701, Y186, TipAL, NolA, BltR, BmrR, and YwnD). However, the COILS program predicted coiled-coil propensities ranging from 64.5 to 98.1% in the corresponding regions of Bacillus sp. strain RC607 MerR, known regulators TipAL and BltR, and homologs of unknown function YBBI, Y701, Y186, and YwnD. The MultiCoil algorithm predicted coil propensities of 60% for TipAL and 61.6% for YBBI. While secondary-structure predictions must be treated with caution, the consistency of these predictions for the same region of each protein suggests that these MerR family members may also employ antiparallel alpha-helices, possibly in a coiled-coil structure, as a dimer interface, as has been demonstrated physically for Tn21 MerR.
DISCUSSION
MerR is unusual among metal-binding proteins in that its metal site consists of residues from two distinct protein chains, i.e., Cys82 from one monomer and Cys117 and Cys126 from the other monomer, rather than from a single chain (37, 53, 75, 81). MerR is also unusual among DNA-binding metalloproteins in that unlike the Zn(II) finger proteins (42), the metal is not required to form the DNA binding site. Finally, MerR is unusually specific for its metal ligand (53, 60). By comparison, among metal-binding proteins that have been well examined in vitro, rat liver metallothionein binds Hg(II), Cd(II), Zn(II), Cu(I), Ni(I), Co(I), and other metal ions (52) and rubredoxin binds Fe(III) or Zn(II) in vivo and in vitro (27). The iron-regulatory protein Fur cannot distinguish between Fe(II), Cd(II), Co(II), and Mn(II) (53); the zinc finger domain of SP1 still binds to DNA when Zn(II) is replaced by Co(II), Cd(II), Ni(II), or Mn(II) (73); and the Fe regulator DtxR binds and responds equally well to Fe(II) or to Mn(II) (79). Our efforts to alter MerR’s metal specificity have led us to several conclusions concerning the metal binding site itself, the domain structure of the protein, and the family of proteins of which MerR is a member.
Minimal role for DNA-binding domain in metal recognition.
To be recovered in the screens and selections used here, a MerR mutant protein must undergo a metal-provoked allosteric change to effect open-complex formation and, thus, must retain the ability to bind DNA. Although both mutagenesis strategies used could have altered the DNA-binding domain (residues 10 to 29), we recovered no mutants with variations in this region. Thus, the metal-specific response of MerR is not materially influenced by the DNA-binding domain of the protein.
Metal properties important in discrimination.
The metals examined in this study were chosen on the basis of three properties which might influence their ability to bind to and to provoke an allosteric change in MerR: ionic and covalent size, relative electronegativity, and preferred coordination number. Our working hypotheses were as follows: (i) steric constraints on the protein’s thiol ligands might preclude their stable association with smaller metal ions, which would simply slip through the possible contacts; (ii) electronegativity might influence the length and strength of the bonds formed with the thiol sulfurs and, thus, the consequent conformational changes in the protein; and (iii) coordination preferences might influence the degree to which the metal ion is stably bound by the ligands which the protein can contribute, as opposed to competing buffer or cytosolic ligands, including water or thiols.
The ionic and covalent radii of Hg(II) are 112 and 144 pm, respectively. Wild-type MerR did not respond in vivo to Ag(I) (113 and 134 pm), Sb(III) (89 and 141 pm), Au(III) (91 and 134 pm), or Zn(II) (83 and 125 pm) (28) and responded only weakly to Cd(II) (103 and 141 pm). While conclusions about interactions of metals and MerR in vivo must be tempered by possible (unknown) differences in the cellular uptake of these metals, the fact that others (60) have found similar response patterns for Ag, Au, Zn, Cd, and Hg in vitro suggests that transport bias is negligible for these metals. Thus, these observations suggest that for the metal cations [all but Sb(III), which is likely present as an oxyanion], the covalent radius contributes more than does the ionic radius to provoking a response by MerR. In contrast, the Pauling relative electronegativities (28) of the metals examined were 1.93 for Ag, 2.05 for Sb, 2.54 for Au, 1.65 for Zn, 1.69 for Cd, and 2.0 for Hg. Sb and Ag, whose electronegativities are closest to that of Hg, did not induce a response by MerR, suggesting that electronegativity is of less significance in provoking MerR to undergo an allosteric change. Finally, the preferred coordination geometries of these metals and their ligands differ extensively (22); however, within group 12 (Zn, Cd, and Hg) there is a definite trend from Zn(II), which prefers four to six ligands (9, 23), to Hg(II), which prefers two. (The tricoordinate HgS3 site in MerR is the only biological example of its type.) Although nothing is yet known of the metal binding site in ZntR, many well-defined Zn-binding proteins provide the metal with four nucleophilic protein ligands, as in the Cys4 or Cys2His2 Zn finger proteins (42). The replacement of the electrophile arginine at position 121 in ZntR by the nucleophile histidine [a common Zn(II) ligand] in the loop region between the counterparts of C117 and C126 of MerR might also provide a fourth ligand for Zn(II) in ZntR.
Although many single, widely distributed changes increased the response to Cd(II), none led to greater response even to the closely related metal Zn(II), suggesting that the structure of MerR resists facile changes to accommodate other metals. Additionally, as seen in other work (20, 56, 57, 63), multiple changes, including combinations of those in Cd(II) response variants, led to constitutive activity (i.e., loss of specificity) rather than to a distinct metal specificity. This resilience of MerR’s preference for Hg(II), even under conditions of heavy mutagenesis and selection, was unexpected, given the lack of metal specificity in the several metal-binding proteins noted above, and suggests that the unique intersubunit, tricoordinate metal-binding domain of MerR is poised to prevent the protein from mistaking other metals for Hg(II). Indeed, this makes good biological sense since MerR must ignore the much higher intracellular concentrations of Zn(II) and of Cu(I).
Refinement of the coupling and metal-binding domains.
The clusters of mutations noted above (Fig. 5) indicate five separate regions of MerR (residues 53 to 64, residues 72 to 89, residue 99, residues 106 to 109, and residues 113 to 131) where changes can shift the conformer distribution toward a population that is able to take equal advantage of Cd(II) or Hg(II) chemistry to stabilize the activating conformation of the protein. Moreover, certain single-residue changes in these regions (e.g., F56I) or changes in two of these (often remote) regions (e.g., M106V and G64W or S131L and E84K) can dispose the conformer equilibrium strongly toward the activated configuration, making expression essentially metal independent.
The conservation or divergence in the MerR homologs ZntR and SoxR allows us to discern which regions or residues might be involved in the common (repression and activation) or the unique (metal-binding) functions of these three proteins. MerR and ZntR are identical at 47 positions and functionally similar at 31 positions, while MerR and its more distant cousin, SoxR, are identical in 37 positions and functionally similar in 21 positions (Fig. 4). Most obviously, MerR’s three Hg(II)-binding cysteines (C82, C117, and C126) align with three cysteines (C79, C115, and C124) of ZntR but with only one (C124) of the four Fe(II/III)-binding cysteines of SoxR (13, 40) (Fig. 4), suggesting that the ZntR metal binding site is more similar to the Hg(II) binding site of MerR than either is to the Fe(II/III) binding site of SoxR.
Of 11 positions described here at which single changes led to Cd(II) responsiveness in MerR (Fig. 5), only R53, E72, L74, and M106 are exactly conserved and only L76I and K99R are functionally conserved in ZntR. SoxR also conserves E72 exactly, and K99R and M106L are functionally conserved in this Fe-binding regulator. Of the positions affecting constitutivity in MerR, ZntR retains F56, G64, E84, and V124 exactly and diverges functionally (V→S) at the position 109 equivalent. SoxR also retains G64 and E84 exactly and diverges slightly at F56I and completely at V109E and V124P. Thus, the highly conserved residues G64, E72, and E84, whose alteration alone (E72) or in combination with other changes (G64 and E84) led to Cd(II)-responsive (E72) or constitutive (G64 and E84) behavior in MerR, may stabilize the repressed conformation in these proteins. Only E84, which lies in the core metal-binding domain (84), has been noted previously to have a role in repression (20); mutations at positions 64 and 72 reveal new aspects of the repressor conformation of MerR. Finally, among the highly conserved residues, changes at K99 and M106 occur repeatedly, with distinct alleles in both Cd(II)-responsive and constitutive variants, suggesting that they may have key roles in the repressor-activator equilibrium.
ZntR and SoxR diverge to a greater or lesser degree at positions corresponding to G79, A85, A89, V109, S125, and S131 of MerR; of these positions, G79, A85, A89, and S131 were identified here, and also previously (20, 57, 63), as being functionally important to MerR. It is tempting to theorize that some of these six residues play a role in what is unique in each of these proteins: metal recognition. Indeed, the single nonconservative changes A85V, A89V, S131L, and S125P all enhance MerR’s response to Cd(II). The nonconservative change V109E occurs only with V124A in a constitutive mutant, so its contribution to metal-specific behavior cannot be assessed here. The fact that single changes in four of six divergent positions led to an enhanced Cd(II) response but not to constitutive behavior suggests that although they are not direct metal ligands, A85, A89, S125, and S131 contribute to metal recognition by MerR.
Finally, Eisenberg and colleagues (82) have shown that metal binding sites contain a highly polar metal-binding region surrounded by a shell of hydrophobic residues. If such a hydropathy contrast exists in the tertiary structure of MerR, it likely persists in the mutants described here since changes in the core Hg(II)-binding domain (84) which conferred Cd(II) responsiveness generally maintained the hydropathic character of the replaced residue. The exceptions, S125P and S131L, the two most C-terminal Cd(II)-responsive mutations, lie outside the coiled-coil region, in or just beyond the predicted loop between C117 and C126 (84), where they might affect the orientation of C126 and its interaction with Cd(II).
A novel domain in a subset of MerR-type regulators.
Our biochemical observations (84), the structure predictions for the wild-type MerR and its closest relatives (Fig. 4), and the positions of merR mutations (Fig. 7) suggested that a coiled-coil structure might be characteristic of such repressors-activators. There is growing evidence that the coiled-coil motif occurs in bacterial regulatory proteins (see reference 11 and references cited therein). Indeed, the Bacillus MerR and 7 (ZntR, YBBI-HAEIN, Y701-SYNY3, Y186-HAEIN, TipAL, BltR, and YwnD) of 10 other presently defined MerR family members (all listed in Materials and Methods) were predicted by one or both of the most widely used algorithms to form a coiled coil in their regions which correspond to the coiled region of Tn21 MerR (data not shown). All of these proteins act at operators lying in overly long (19- to 20-bp) spacers between the −10 and −35 hexamers of their target ς70 RNAP promoters. Apart from MerR, nothing is known about the interactions of these proteins with their cognate operators during repression or activation. It remains to be determined what (if any) unique role these putative coiled structures play in controlling expression from these promoters. Interestingly, coil formation propensity does not sort with the type of inducer; i.e., a high propensity for coil formation is predicted for metal-binding MerR and ZntR and also for TipAL, which is not known to bind a metal. Similarly, a low propensity to form coiled coils is seen with metal-binding SoxR and with Bacillus BmrR, which binds organic acids. The latter protein is the only one for which there is a 3-D structure of the ligand-binding domain (85), and observations confirm the prediction that it lacks a coil structure.
In summary, the selections and screens employed here have revealed several positions in MerR, not previously identified, which play a role in repression-activation or in metal recognition. The properties of these mutants and those previously described and comparisons with homologs indicate that as seen with antigen-antibody recognition (5, 78), changes remote from the immediate binding site can subtly influence the specificity of a protein for its ligand. In the case of MerR, the agents of metal recognition lie not only immediately adjacent to the known metal ligand thiols but throughout the core metal-binding domain and the putative coupling region between it and the DNA-binding domain.
ACKNOWLEDGMENTS
We thank Qiandong Zeng for initial modeling of the Hg(II)-binding domain of MerR. We also thank Marly Eidsness, Resham Kulkarni, Don Kurtz, Heather Lumppio, and two anonymous reviewers for valuable comments on the manuscript.
This work was supported by a National Science Foundation fellowship awarded to J.J.C. and by National Institutes of Health grant GM28211 awarded to A.O.S.
REFERENCES
- 1.Ahmed M, Borsch C M, Taylor S S, Vázquez-Laslop N, Neyfakh A A. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J Biol Chem. 1994;269:28506–28513. [PubMed] [Google Scholar]
- 2.Ahmed M, Lyass L, Markham P N, Taylor S S, Vázquez-Laslop N, Neyfakh A A. Two highly similar multidrug transporters of Bacillus subtilis whose expression is differentially regulated. J Bacteriol. 1995;177:3904–3910. doi: 10.1128/jb.177.14.3904-3910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari A Z, Bradner J E, O’Halloran T V. DNA-bend modulation in a repressor-to-activator switching mechanism. Nature. 1995;374:371–375. doi: 10.1038/374370a0. [DOI] [PubMed] [Google Scholar]
- 4.Ansari A Z, Chael M L, O’Halloran T V. Allosteric underwinding of DNA is a critical step in positive control of transcription by Hg-MerR. Nature. 1992;355:87–89. doi: 10.1038/355087a0. [DOI] [PubMed] [Google Scholar]
- 5.Atwell S, Ultsch M, De Vos A M, Wells J A. Structural plasticity in a remodeled protein-protein interface. Science. 1997;278:1125–1128. doi: 10.1126/science.278.5340.1125. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J A, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1994. [Google Scholar]
- 7.Beard S J, Hashim R, Membrillo-Hernandez J, Hughes M N, Poole R K. Zinc(II) tolerance in Escherichia coli K-12: evidence that the zntA gene (o732) encodes a cation transport ATPase. Mol Microbiol. 1997;25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 8.Berger B, Singh M. An iterative method for improved protein structural motif recognition. J Comput Biol. 1997;4:261–273. doi: 10.1089/cmb.1997.4.261. [DOI] [PubMed] [Google Scholar]
- 9.Bertini I, Luchinat C. The reaction pathways of zinc enzymes and related biological catalysts. In: Bertani I, Gray H B, Lippard S J, Valentine J S, editors. Bioinorganic chemistry. Mill Valley, Calif: University Science Books; 1994. pp. 37–106. [Google Scholar]
- 10.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boss A, Nussbaum-Shochat A, Amster-Choder O. Characterization of the dimerization domain in BglG, an RNA-binding transcriptional antiterminator from Escherichia coli. J Bacteriol. 1999;181:1755–1766. doi: 10.1128/jb.181.6.1755-1766.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulanger Y, Goodman C M, Forte C P, Fesik S W, Armitage I M. Model for mammalian metallothionein structure. Proc Natl Acad Sci USA. 1983;80:1501–1505. doi: 10.1073/pnas.80.6.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bradley T M, Hidalgo E, Leautaud V, Ding H, Demple B. Cysteine-to-alanine replacements in the Escherichia coli SoxR protein and the role of the [2Fe-2S] centers in transcriptional activation. Nucleic Acids Res. 1997;25:1469–1475. doi: 10.1093/nar/25.8.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan R G, Matthews B W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 15.Brocklehurst K R, Hobman J L, Lawley B, Blank L, Marshall S J, Brown N L, Morby A P. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol Microbiol. 1998;31:893–902. doi: 10.1046/j.1365-2958.1999.01229.x. [DOI] [PubMed] [Google Scholar]
- 16.Cadwell R C, Gerald F J. Mutagenic PCR. In: Diffenbach C W, Dveksler G S, editors. PCR primer: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 583–589. [Google Scholar]
- 17.Caslake L F, Ashraf S I, Summers A O. Mutations in the alpha and sigma-70 subunits of RNA polymerase affect expression of the mer operon. J Bacteriol. 1996;179:1787–1795. doi: 10.1128/jb.179.5.1787-1795.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christie G E, White T J, Goodwin T S. A merR homologue at 74 minutes on the Escherichia coli genome. Gene. 1994;146:131–132. doi: 10.1016/0378-1119(94)90847-8. [DOI] [PubMed] [Google Scholar]
- 20.Comess K M, Shewchuk L M, Ivanetich K, Walsh C T. Construction of a synthetic gene for the metalloregulatory protein MerR. Biochemistry. 1994;33:4175–4186. doi: 10.1021/bi00180a010. [DOI] [PubMed] [Google Scholar]
- 21.Condee C W, Summers A O. A mer-lux transcriptional fusion for real-time examination of in vivo gene expression kinetics and promoter response to altered superhelicity. J Bacteriol. 1992;174:8094–8101. doi: 10.1128/jb.174.24.8094-8101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotton F A, Wilkinson G. Advanced inorganic chemistry. 5th ed. New York, N.Y: Interscience Publishers; 1988. [Google Scholar]
- 23.Cowan J A. Inorganic biochemistry: an introduction. New York, N.Y: VCH Publishers, Inc.; 1993. [Google Scholar]
- 24.Cowell I G. Repression versus activation in the control of gene transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 25.Cox E C. Bacterial mutator genes and the control of spontaneous mutation. Annu Rev Genet. 1976;10:135–156. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- 26.de la Cruz F, Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982;151:222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eidsness M K, O’Dell S E, Kurtz D M, Jr, Robson R L, Scott R A. Expression of a synthetic gene coding for the amino acid sequence of Clostridium pasteurianum rubredoxin. Protein Eng. 1992;5:367–371. doi: 10.1093/protein/5.4.367. [DOI] [PubMed] [Google Scholar]
- 28.Emsley J, editor. The elements. 2nd ed. Oxford, United Kingdom: Clarendon Press; 1995. [Google Scholar]
- 29.Fondell J D, Roy A L, Roeder R G. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- 30.Frantz B, O’Halloran T V. DNA distortion accompanies transcriptional activation by the metal-responsive gene-regulatory protein MerR. Biochemistry. 1990;29:4747–4751. doi: 10.1021/bi00472a001. [DOI] [PubMed] [Google Scholar]
- 31.Fromant M S, Blanquet S, Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 32.Fujinaga M, Berthet-Colominas C, Yaremchuk A D, Tukalo M A, Cusack S. Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 Å resolution. J Mol Biol. 1993;234:222–233. doi: 10.1006/jmbi.1993.1576. [DOI] [PubMed] [Google Scholar]
- 33.Furey W F, Robbins A H, Clancy L L, Winge D R, Wang B C, Stout C D. Crystal structure of Cd,Zn metallothionein. Science. 1986;231:704–708. doi: 10.1126/science.3945804. [DOI] [PubMed] [Google Scholar]
- 34.Greener A, Callahan M, Jerpseth B. An efficient random mutagenesis technique using an E. coli mutator strain. Mol Biotechnol. 1997;7:189–195. doi: 10.1007/BF02761755. [DOI] [PubMed] [Google Scholar]
- 35.Grinsted J, de la Cruz F, Schmitt R. The Tn21 subgroup of bacterial transposable elements. Plasmid. 1990;24:163–189. doi: 10.1016/0147-619x(90)90001-s. [DOI] [PubMed] [Google Scholar]
- 36.Hamlett N V, Landale E C, Davis B H, Summers A O. Roles of the Tn21 merT, merP, and merC gene products in mercury resistance and mercury binding. J Bacteriol. 1992;174:6377–6385. doi: 10.1128/jb.174.20.6377-6385.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helmann J D, Ballard B T, Walsh C T. The MerR metalloregulatory protein binds mercuric ion as a tricoordinate, metal-bridged dimer. Science. 1990;247:946–948. doi: 10.1126/science.2305262. [DOI] [PubMed] [Google Scholar]
- 38.Heltzel A, Gambill D, Jackson W J, Totis P A, Summers A O. Overexpression and DNA-binding properties of the mer-encoded regulatory protein from plasmid NR1 (Tn21) J Bacteriol. 1987;169:3379–3384. doi: 10.1128/jb.169.7.3379-3384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heltzel A, Lee I W, Totis P A, Summers A O. Activator-dependent preinduction binding of ς-70 RNA polymerase at the metal-regulated mer promoter. Biochemistry. 1990;29:9572–9584. doi: 10.1021/bi00493a011. [DOI] [PubMed] [Google Scholar]
- 40.Hidalgo E, Bollinger J M, Jr, Bradley T M, Walsh C T, Demple B. Binuclear [2Fe-2S] clusters in the Escherichia coli SoxR protein and role of the metal centers in transcription. J Biol Chem. 1995;270:20908–20914. doi: 10.1074/jbc.270.36.20908. [DOI] [PubMed] [Google Scholar]
- 41.Hidalgo E, Leautaud V, Demple B. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 1998;17:2629–2636. doi: 10.1093/emboj/17.9.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klug A, Schwabe J W. Protein motifs. 5. Zinc fingers. FASEB J. 1995;9:597–604. [PubMed] [Google Scholar]
- 43.Lee I W, Livrelli V, Park S-J, Totis P A, Summers A O. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. II. Repressor/activator (MerR)-RNA polymerase interaction with merOP mutants. J Biol Chem. 1993;268:2632–2639. [PubMed] [Google Scholar]
- 44.Livrelli V, Lee I W, Summers A O. In vivo DNA-protein interactions at the divergent mercury resistance (mer) promoters. I. Metalloregulatory protein MerR mutants. J Biol Chem. 1993;268:2623–2631. [PubMed] [Google Scholar]
- 45.Lund P A, Ford S J, Brown N L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986;132:465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- 46.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 47.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 47a.Lupas, A. February 1996, posting date. COILS, version 2.2. [Online.] Max-Planck-Institut für Biochemie, Martinsreid, Germany. http://www.isrec.isb-sib.ch/software/COILS_form.html. [25 February 1999, last date accessed.]
- 48.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 49.Misra T K. Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid. 1992;27:4–16. doi: 10.1016/0147-619x(92)90002-r. [DOI] [PubMed] [Google Scholar]
- 50.Mukhopadhyay D, Yu H, Nucifora G, Misra T K. Purification and functional characterization of MerD: a coregulator of the mercury resistance operon in gram-negative bacteria. J Biol Chem. 1991;266:18538–18542. [PubMed] [Google Scholar]
- 51.Ni’ Bhriain N N, Silver S, Foster T J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983;155:690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielson K B, Atkin C L, Winge D R. Distinct metal-binding configurations in metallothionein. J Biol Chem. 1985;260:5342–5350. [PubMed] [Google Scholar]
- 53.O’Halloran T V. Transition metals in control of gene expression. Science. 1993;261:715–725. doi: 10.1126/science.8342038. [DOI] [PubMed] [Google Scholar]
- 54.O’Halloran T V, Frantz B, Shin M K, Ralston D M, Wright J G. The MerR heavy metal receptor mediates positive activation in a topologically novel transcription complex. Cell. 1989;56:119–129. doi: 10.1016/0092-8674(89)90990-2. [DOI] [PubMed] [Google Scholar]
- 55.Park S-J, Wireman J, Summers A O. Genetic analysis of the Tn21 mer operator-promoter. J Bacteriol. 1992;174:2160–2171. doi: 10.1128/jb.174.7.2160-2171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parkhill J, Ansari A Z, Wright J G, Brown N L, O’Halloran T V. Construction and characterization of a mercury-independent MerR activator (MerRAC): transcriptional activation in the absence of Hg(II) is accomplished by DNA distortion. EMBO J. 1993;12:413–421. doi: 10.1002/j.1460-2075.1993.tb05673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parkhill J, Lawley B, Hobman J L, Brown N L. Selection and characterization of mercury-independent activation mutants of the Tn501 transcriptional regulator, MerR. Microbiology. 1998;144:2855–2864. doi: 10.1099/00221287-144-10-2855. [DOI] [PubMed] [Google Scholar]
- 58.Parry D A D. Coiled coils in alpha-helix-containing proteins: analysis of the residue types within the heptad repeat and the use of these data in the prediction of coiled coils in other proteins. Biosci Rep. 1982;2:1017–1024. doi: 10.1007/BF01122170. [DOI] [PubMed] [Google Scholar]
- 59.Radman M, Wagner R E, Jr, Glickman B W, Meselson M. DNA methylation, mismatch correction and genetic stability. In: Alacevic M, editor. Progress in environmental mutagenesis. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1980. pp. 121–130. [Google Scholar]
- 60.Ralston D M, O’Halloran T V. Metalloregulatory proteins and molecular mechanisms of heavy metal signal transduction. Adv Inorg Biochem. 1990;8:1–31. [PubMed] [Google Scholar]
- 61.Ralston D M, O’Halloran T V. Ultrasensitivity and heavy-metal selectivity of the allosterically modulated MerR transcription complex. Proc Natl Acad Sci USA. 1990;87:3846–3850. doi: 10.1073/pnas.87.10.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rensing C, Mitra B, Rosen B P. The zntA gene of Escherichia coli encodes a Zn(II)-translocating P-type ATPase. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62a.Ross, W. Unpublished data.
- 63.Ross W, Park S-J, Summers A O. Genetic analysis of transcriptional activation and repression in the Tn21 mer operon. J Bacteriol. 1989;171:4009–4018. doi: 10.1128/jb.171.7.4009-4018.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Santos R A, Gruff E S, Koch S A, Harbison G S. Solid-state 199Hg and 113Cd NMR studies of mercury- and cadmium-thiolate complexes. Spectroscopic models for [Hg(SCys)n] centers in the bacterial mercury resistance proteins. J Am Chem Soc. 1991;113:469–474. [Google Scholar]
- 65.Scheuermann R, Tam S, Burgers P M, Lu C, Echols H. Identification of the epsilon subunit of Escherichia coli DNA. Proc Natl Acad Sci USA. 1983;80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider K, Beck C F. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 1986;42:37–48. doi: 10.1016/0378-1119(86)90148-4. [DOI] [PubMed] [Google Scholar]
- 67.Shewchuk L M, Helmann J D, Ross W, Park S-J, Summers A O, Walsh C T. Transcriptional switching by the MerR protein: activation and repression mutants implicate distinct DNA and mercury(II) binding domains. Biochemistry. 1989;28:2340–2344. doi: 10.1021/bi00431a053. [DOI] [PubMed] [Google Scholar]
- 68.Shewchuk L M, Verdine G L, Nash H, Walsh C T. Mutagenesis of the cysteines in the metalloregulatory protein MerR indicates that a metal-bridged dimer via Cys126 activates transcription. Biochemistry. 1989;28:6140–6145. doi: 10.1021/bi00441a002. [DOI] [PubMed] [Google Scholar]
- 69.Shewchuk L M, Verdine G L, Walsh C T. Transcriptional switching by the metalloregulatory MerR protein: initial characterization of DNA and mercury(II) binding activities. Biochemistry. 1989;28:2331–2339. doi: 10.1021/bi00431a052. [DOI] [PubMed] [Google Scholar]
- 70.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 71.Summers A O, Jacoby G A. Plasmid-determined resistance to tellurium compounds. J Bacteriol. 1977;129:276–281. doi: 10.1128/jb.129.1.276-281.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Summers A O. Untwist and shout: a heavy metal-responsive transcriptional regulator. J Bacteriol. 1992;174:3097–3101. doi: 10.1128/jb.174.10.3097-3101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thiesen H, Bach C. Transition metals modulate DNA-protein interactions of SP1 zinc finger domains with its cognate target site. Biochem Biophys Res Commun. 1991;176:551–557. doi: 10.1016/s0006-291x(05)80219-0. [DOI] [PubMed] [Google Scholar]
- 74.Torriani A. Alkaline phosphatase from Escherichia coli. In: Cantoni G L, Davies D R, editors. Procedures in nucleic acid research. Vol. 1. New York, N.Y: Harper & Row; 1966. pp. 224–235. [Google Scholar]
- 75.Utschig L M, Bryson J W, O’Halloran T V. Mercury-199 NMR of the metal receptor site in MerR and its protein-DNA complex. Science. 1995;268:380–385. doi: 10.1126/science.7716541. [DOI] [PubMed] [Google Scholar]
- 76.Utschig L M, Wright J G, O’Halloran T V. Biochemical and spectroscopic probes of mercury(II) coordination environments in proteins. Methods Enzymol. 1993;226:71–97. doi: 10.1016/0076-6879(93)26006-u. [DOI] [PubMed] [Google Scholar]
- 77.Wainwright L A, Seifert H S. Paraffin beads can replace mineral oil as an evaporation barrier in PCR. BioTechniques. 1993;14:34–36. [PubMed] [Google Scholar]
- 78.Wedemayer G J, Patten P A, Wang L H, Schultz P G, Stevens R C. Structural insights into the evolution of an antibody combining site. Science. 1997;276:1665–1669. doi: 10.1126/science.276.5319.1665. [DOI] [PubMed] [Google Scholar]
- 79.White A, Ding X, vanderSpek J C, Murphy J R, Ringe D. Structure of the metal-ion-activated diphtheria toxin repressor/tox operator complex. Nature. 1998;394:502–506. doi: 10.1038/28893. [DOI] [PubMed] [Google Scholar]
- 80.Wolf E, Kim P S, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80a.Wolf, E., P. S. Kim, and B. Berger. June 1997, posting date. MultiCoil. [Online.] Department of Mathematics, Massachusetts Institute of Technology, Cambridge, Mass. http://nightingale.lcs.mit.edu/cgi-bin/multicoil. [25 February 1999, last date accessed.]
- 81.Wright J G, Tsang H-T, Penner-Hahn J E, O’Halloran T V. Coordination chemistry of the Hg-MerR metalloregulatory protein: evidence for a novel tridentate Hg-cysteine receptor site. J Am Chem Soc. 1990;112:2434–2435. [Google Scholar]
- 82.Yamashita M M, Wesson L, Eisenman G, Eisenberg D. Where metal ions bind in proteins. Proc Natl Acad Sci USA. 1990;87:5648–5652. doi: 10.1073/pnas.87.15.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeng Q, Eidsness M K, Summers A O. Near-zero background cloning of PCR products. BioTechniques. 1997;23:412–414. doi: 10.2144/97233bm13. [DOI] [PubMed] [Google Scholar]
- 84.Zeng Q, Stålhandske C, Anderson M C, Scott R A, Summers A O. The core metal-recognition domain of MerR. Biochemistry. 1998;37:15885–15895. doi: 10.1021/bi9817562. [DOI] [PubMed] [Google Scholar]
- 85.Zheleznova E E, Markham P N, Neyfakh A A, Brennan R G. Structural basis of multidrug recognition by BmrR, a transcription activator of a multidrug transporter. Cell. 1999;96:353–362. doi: 10.1016/s0092-8674(00)80548-6. [DOI] [PubMed] [Google Scholar]