Summary
Triple-negative breast cancer (TNBC) is a subset of breast cancer with an adverse prognosis and significant tumor heterogeneity. Here, we extract quantitative radiomic features from contrast-enhanced magnetic resonance images to construct a breast cancer radiomic dataset (n = 860) and a TNBC radiogenomic dataset (n = 202). We develop and validate radiomic signatures that can fairly differentiate TNBC from other breast cancer subtypes and distinguish molecular subtypes within TNBC. A radiomic feature that captures peritumoral heterogeneity is determined to be a prognostic factor for recurrence-free survival (p = 0.01) and overall survival (p = 0.004) in TNBC. Combined with the established matching TNBC transcriptomic and metabolomic data, we demonstrate that peritumoral heterogeneity is associated with immune suppression and upregulated fatty acid synthesis in tumor samples. Collectively, this multi-omic dataset serves as a useful public resource to promote precise subtyping of TNBC and helps to understand the biological significance of radiomics.
Keywords: triple-negative breast cancer, radiomics, tumor heterogeneity, prognosis, biomarker
Graphical abstract
Highlights
-
•
A radiomic signature identifies TNBC from other subtypes of breast cancer
-
•
Radiomic features are predictive of TNBC molecular subtypes
-
•
A radiomic feature reflecting peritumoral heterogeneity indicates TNBC prognosis
-
•
Peritumoral heterogeneity correlates with metabolic and immune abnormalities in TNBC
Jiang et al. investigate radiomics as a non-invasive approach for evaluating tumor heterogeneity in triple-negative breast cancer (TNBC). Their study utilizes radiomic features to distinguish the intertumoral heterogeneity of TNBC and identify a prognostic radiomic feature reflecting peritumoral heterogeneity to correlate with immune suppression and metabolic reprogramming in TNBC.
Introduction
Breast cancers that lack expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) are classified as triple-negative breast cancers (TNBCs). TNBC, which comprises 15%–20% of newly diagnosed breast cancers,1,2 is characterized by aggressive biological behavior, high incidence of relapse, and unfavorable prognosis.3 Recent years have witnessed increasing recognition of the heterogeneity inside TNBC, while the identification of subtype-specific therapeutic targets is still in urgent need.4, 5, 6, 7, 8, 9 With the largest multi-omic database to date, our previous work unveiled the genomic and transcriptomic landscape of 465 Chinese TNBC patients and classified TNBCs into four molecular subtypes with distinct characteristics: (1) basal-like immune suppressed (BLIS), (2) immunomodulatory (IM), (3) mesenchymal like (MES), and (4) luminal androgen receptor (LAR).7
In the past decade, radiomics has been an emerging field that transforms medical images into mineable data by acquiring multiple quantitative image features.10,11 Compared with conventional invasive biopsies, the radiomic approach has two main advantages. First, radiomics is a non-invasive method to infer tumor characteristics and can be performed several times during the follow-up period.12, 13, 14 In addition, genomic and transcriptomic profiling selects only a small part of the tumors, while radiomics elucidates the landscape of a tumor and is not subject to selection bias, which enables us to explore tumor heterogeneity comprehensively.15, 16, 17 Previous studies focusing on radiomic texture analysis have quantified tumor heterogeneity and suggested its associations with an unfavorable prognosis in breast cancer.18,19 These results warrant further evaluation of tumor heterogeneity using a radiomic approach.
However, a multi-omic TNBC dataset containing radiomic data with a large sample size has yet to be reported, and the correlation between radiomic features and genomic alterations remains largely unknown. In the present study, we performed radiomic profiling based on contrast-enhanced magnetic resonance imaging (CE-MRI) images from 860 Chinese breast cancer patients to distinguish TNBCs from non-TNBCs. We further constructed a TNBC radiogenomic dataset (n = 202) based on our previously developed TNBC multi-omic cohort,7 aiming to build a radiomic model for non-invasive TNBC subtyping and patient outcome stratification. We also integrated the radiomic data with our transcriptomic, metabolomic, and clinical data in this dataset to illustrate the biological basis of prognostic radiomic features in TNBC.
Results
Overview of Fudan University Shanghai Cancer Center (FUSCC) breast cancer radiomic cohort and TNBC radiogenomic cohort
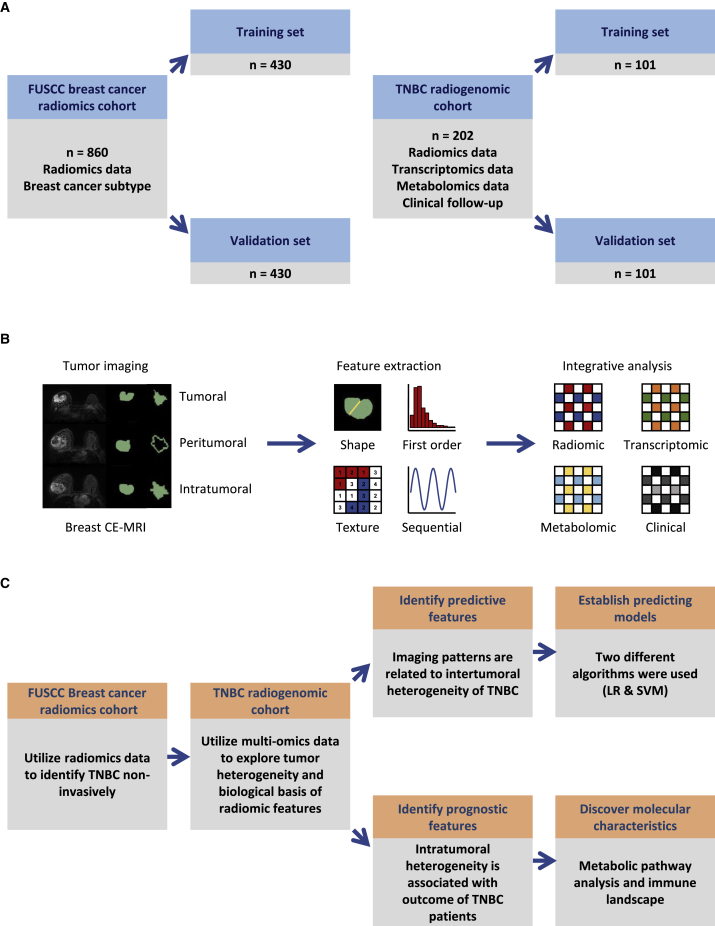
To explore the clinical usefulness of radiomic data in breast cancer, we established FUSCC breast cancer radiomic cohort and TNBC multi-omic cohort, both with high-quality breast CE-MRI images. FUSCC breast cancer radiomic cohort retrospectively enrolled 860 primary breast cancer patients between August 2009 and May 2015 and was utilized to differentiate TNBCs from other breast cancer subtypes (Figure 1A). In this cohort, hormone receptor positivity was observed in 468 patients, HER-2 overexpression was observed in 268 patients, and 246 patients were identified as having TNBC (Figure S1A). In addition, we constructed TNBC radiogenomic cohort consisting of 202 primary TNBC patients based on our previously developed TNBC multi-omic cohort,7 and this cohort was utilized to distinguish TNBC molecular subtypes and explore the biological significance of important radiomic features (Figure 1A). Transcriptomic, metabolomic, and clinicopathological information was matched with radiomic data in this dataset (Figure 1B). The imaging parameters of the CE-MRI machines used in these two cohorts were summarized in Table S1. Tumoral, peritumoral, intratumoral, and tumor-peritumoral regions of interest (ROIs) were delineated (the definitions of these ROIs were included in STAR Methods section). Radiomic features were extracted using the PyRadiomics package based on these ROIs. We proposed our analysis plan as shown in Figure 1C.
Figure 1.
Overview of this integrative radiogenomic study
(A) Description of the radiomic cohorts used in this study.
(B) Generating process of radiomic data and integrative analysis used in TNBC radiogenomic cohort.
(C) Analytical framework of integrative radiogenomic analysis.
CE-MRI, contrast-enhanced magnetic resonance imaging; LR, logistic regression; SVM, support vector machine; TNBC, triple-negative breast cancer. See also Figures S1 and S2 and Tables S1–S3.
Identification of TNBC in FUSCC breast cancer radiomic cohort and external validation cohorts
We first randomly divided FUSCC breast cancer radiomic cohort (n = 860) into 50% training and 50% validation sets (simplified as FUSCC training cohort and FUSCC validation cohort below) to develop a non-invasive radiomic approach to distinguish TNBC from all breast cancers. Using the 10-fold cross validation least absolute shrinkage and selection operator (LASSO) model (α = 0), 11 variables were retained to develop TNBC prediction signature in the training cohort (Figure S1B). These radiomic features were presented in Table S2.
Using logistic regression (LR) to establish prediction models based on the retained features, this radiomic signature could classify TNBC versus non-TNBC with an area under the curve (AUC) of the receiver operator characteristic curve (ROC) of 0.92 (95% confidence interval [CI]: 0.887–0.953) and an AUC of the precision-recall curve (PRC) of 0.819 in the validation set of FUSCC breast cancer radiomic cohort (Figures S1C and S1D). We further validated the efficacy of this prediction model in two external validation datasets generated from Chinese patients. Detailed information of these datasets was listed in STAR Methods section. Two external validation datasets from International Peace Maternal and Children Hospital (IPMCH) (n = 54) and Shanghai Jiaotong University Renji Hospital (RENJI) (n = 110) yielded AUCs of 0.723 (95% CI: 0.552–0.894) and 0.613 (95% CI: 0.461–0.766), respectively (Figures S1E and S1F). In addition, the density distributions of 11 selected features curated from different datasets were approximate, indicating the sound reproducibility of these radiomic features among independent medical centers despite the distinct imaging parameters used (Figure S2; Table S1). These data demonstrated that radiomic features could distinguish TNBC from other types of breast cancers in Chinese patients.
Predictive value of radiomics in distinguishing TNBC molecular subtypes
We further explored whether radiomic signatures could distinguish different TNBC molecular subtypes. As described above, 202 TNBC patients were retrospectively enrolled in our TNBC radiogenomic cohort. The baseline characteristics of this cohort were shown in Table S3. A total of 167 cases with radiomic data had matching transcriptomic data, while 138 cases had matching metabolomic data. Transcriptomic TNBC subtypes were regarded as the ground truth.7
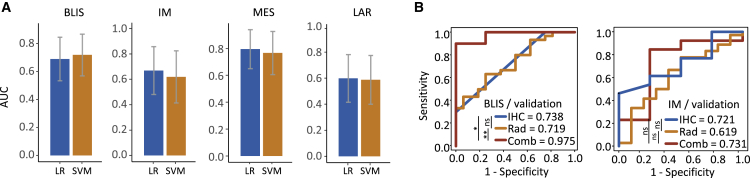
LASSO and Student’s t test retained 4, 11, 2, and 7 radiomic features that were most relevant to BLIS, IM, MES, and LAR subtypes in the training cohort, respectively. These features were presented in Table S4. LR and support vector machine (SVM) were used to construct prediction models in the training and validation cohorts based on the selected features. The AUCs and CIs of the prediction models for each TNBC subtype were shown in Figure 2A. In the validation set, identifying MES, BLIS, IM, and LAR subtypes yielded AUCs of 0.796 (95% CI: 0.650–0.941; LR-based model), 0.719 (95% CI: 0.570–0.867; SVM-based model), 0.669 (95% CI: 0.481–0.858; LR-based model), and 0.598 (95% CI: 0.416–0.781; SVM-based model), respectively.
Figure 2.
Efficacy of predicting TNBC molecular subtypes using radiomics and IHC data with machine learning method
(A) AUC of the radiomic signatures for predicting BLIS, IM, MES, and LAR subtypes. Error bar represented the 95% confidence interval of AUC.
(B) Comparison of combined model, individual radiomic model, and IHC model for predicting BLIS and IM subtypes. ∗∗0.001 < p ≤ 0.01; ∗0.01 < p ≤ 0.05; ns, p > 0.05.
AUC, area under the receiver operating characteristic curve; BLIS, basal-like immune suppressed; IHC, immunohistochemistry; IM, immunomodulatory; LAR, luminal androgen receptor; LR, logistic regression; MES, mesenchymal like; SVM, support vector machine; TNBC, triple-negative breast cancer. See also Tables S3 and S4.
A previous study investigated immunohistochemistry (IHC) as a surrogate approach to distinguish molecular subtypes of TNBC.20 Here, we further explored the discriminatory power of prediction models combining radiomic features and IHC data. Because IHC alone could identify the LAR subtype with outstanding efficacy (AUC = 0.932)20 and a satisfactory radiomic model was established for MES subtype identification, we built combined signatures to identify BLIS and IM subtypes. The AUCs to predict BLIS and IM subtypes were 0.975 (95% CI: 0.906–1; SVM-based model) and 0.731 (95% CI: 0.373–1; LR-based model), respectively, in the validation set (Figure 2B). Combined models showed better performance in the BLIS subtype than individual IHC and radiomics-based models, but no statistical significance was found in the IM subtype. Altogether, these data suggested that radiomics was a promising approach to identify TNBC molecular subtypes, especially when combined with other approaches, including IHC.
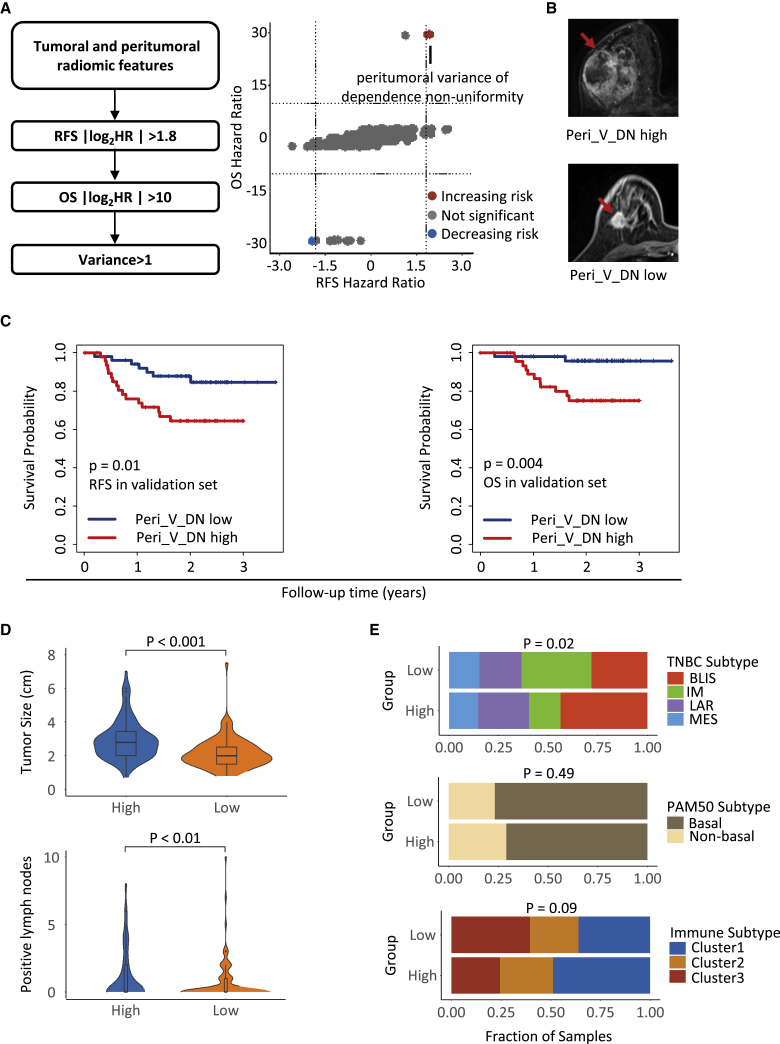
Prognostic value of peritumoral heterogeneity derived from radiomics
With the detailed clinical follow-up data of our TNBC radiogenomic cohort (n = 202), we evaluated robust prognostic radiomic features. According to stringent filtering criteria, variance among the MRI sequences of dependence nonuniformity extracted from peritumoral ROIs (Peri_V_DN), a feature from the gray level dependence matrix group, was identified (Figure 3A). Typical breast CE-MRI images with high and low Peri_V_DN values are shown in Figure 3B. The stratification of patients with survival differences using the median value as the cutoff was verified in the validation set (Figure 3C). The multivariate Cox proportional hazards model also revealed that low Peri_V_DN independently predicted better recurrence-free survival (RFS) and overall survival (OS) in TNBC patients (Table 1). Peri_V_DN represents the variation pattern of peritumoral heterogeneity through different imaging phases, with a lower value indicating less change in peritumoral heterogeneity among the sequences of the image.
Figure 3.
Identification of the prognostic feature Peri_V_DN and its clinicopathological associations
(A) Criteria of prognostic feature selection (left) and hazard ratios for RFS and OS of the radiomic features (right).
(B) Breast CE-MRI images from one patient with high Peri_V_DN (upper) and one patient with low Peri_V_DN (lower).
(C) Kaplan-Meier plots show the prognostic value of Peri_V_DN for RFS and OS in the validation set.
(D) Distribution of tumor size and pathologically confirmed metastatic lymph nodes between Peri_V_DN groups.
(E) Distribution of the TNBC transcriptomic subtypes, PAM50 subtypes, and TNBC microenvironment clusters between Peri_V_DN groups.
HR, hazard ratio; OS, overall survival; Peri_V_DN, peritumoral variance in dependence nonuniformity of peritumoral regions; RFS, recurrence-free survival. See also Figure S3.
Table 1.
Multivariate Cox proportional hazard models for RFS and OS in TNBC radiogenomic cohort
| Variables | RFS |
OS |
|||
|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | ||
| T stage | T1 | ref. | |||
| T2 | 0.58 (0.27–1.25) | 0.17 | 0.70 (0.24–2.02) | 0.51 | |
| T3/T4 | 0.95 (0.24–3.76) | 0.94 | 1.69 (0.29–9.75) | 0.59 | |
| N stage | pN0 | ref. | |||
| pN1 | 2.03 (0.81–5.07) | 0.13 | 2.73 (0.77–9.66) | 0.12 | |
| pN2 | 5.02 (1.88–13.43) | 0.001 | 5.87 (1.66–20.76) | 0.006 | |
| pN3 | 7.62 (2.84–20.43) | 5.38 × 10−5 | 4.85 (1.06–22.24) | 0.04 | |
| TNBC subtype | BLIS | ref. | |||
| IM | 1.12 (0.35–3.52) | 0.85 | 0.67 (0.13–3.49) | 0.63 | |
| MES | 0.93 (0.29–2.96) | 0.90 | 0.82 (0.19–3.46) | 0.78 | |
| LAR | 0.94 (0.34–2.57) | 0.90 | 0.62 (0.16–2.30) | 0.47 | |
| Peri_V_DN | high | ref. | |||
| low | 0.41 (0.18–0.95) | 0.04 | 0.15 (0.03–0.70) | 0.02 | |
CI, confidence interval; HR, hazard ratio; OS, overall survival; Peri_V_DN, variance of dependence nonuniformity extracted of peritumoral regions; RFS, recurrence-free survival; TNBC, triple-negative breast cancer.
Next, we systematically analyzed the correlation between the Peri_V_DN value and tumor characteristics. We observed larger tumor sizes and more pathologically confirmed metastatic lymph nodes (p < 0.001 and p < 0.01, respectively) in the high Peri_V_DN group than in the low Peri_V_DN group (Figure 3D). The high Peri_V_DN group included more patients with the BLIS subtype, and the low Peri_V_DN group comprised more patients with the IM subtype (p = 0.02), while the distribution of the PAM50 subtypes was balanced (Figure 3E). We analyzed the correlation between the Peri_V_DN value and TNBC microenvironment clusters according to TNBC microenvironment subtypes.21 The results revealed a tendency for the high Peri_V_DN group to consist of more “immune-desert” cluster one tumors, while the low Peri_V_DN group included more “immune-inflamed” cluster three tumors (p = 0.09; Figure 3E). Fibrosis and necrosis grades evaluated by hematoxylin and eosin staining sections showed no difference between the two Peri_V_DN groups (Figure S3A). Other molecular biomarkers for precision treatment of TNBC, including stromal tumor-infiltrating lymphocytes (TILs), IHC CD8 readings, tumor mutation burden (TMB), and homologous recombination deficiency (HRD) score, displayed balanced distributions between the Peri_V_DN groups as well (Figures S3B–S3E). Overall, we demonstrated that high Peri_V_DN predicted a poor prognosis for TNBC and more aggressive tumor characteristics.
Integrative analysis elucidated metabolic reprogramming in high Peri_V_DN patients
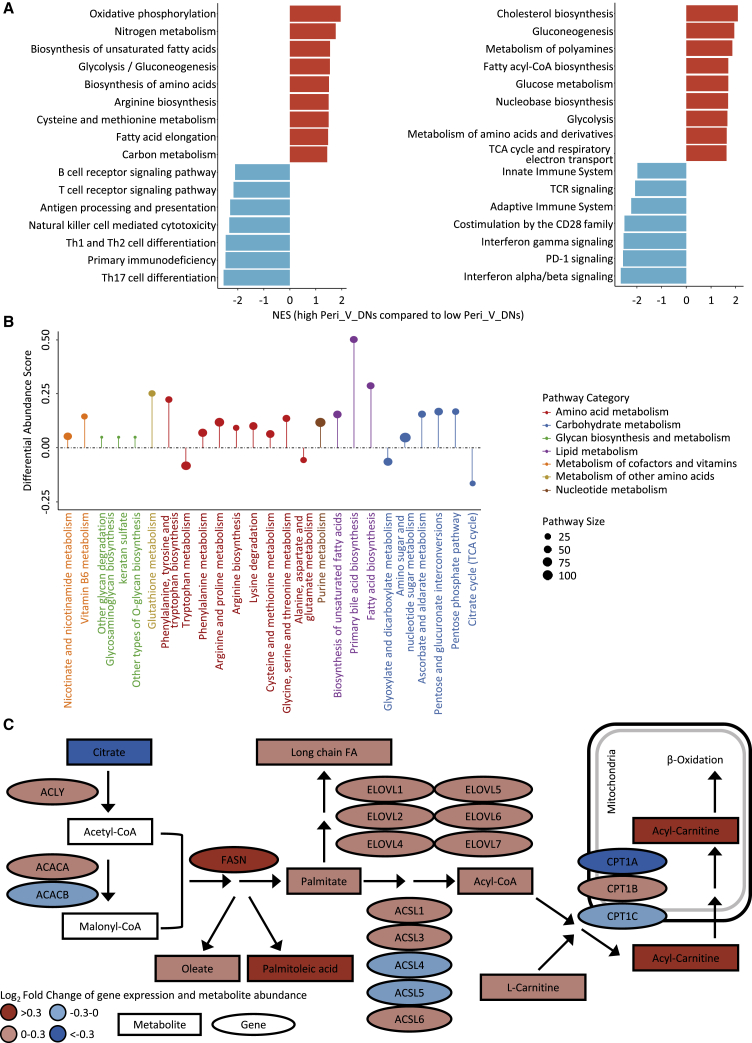
We further investigated the molecular characteristics associated with Peri_V_DN. Using paired transcriptomics and metabolomics data from TNBC radiogenomic cohort, metabolite abundance and gene expression were compared between the Peri_V_DN groups (Figure S4; Tables S5 and S6). Differentially abundant polar metabolites mainly comprised lipids. Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome-based gene set enrichment analysis (GSEA) demonstrated similar results (false discovery rate [FDR] < 0.1) that high Peri_V_DN was significantly associated with aberrant metabolism and suppressed immune-related pathways (Figure 4A; Table S7).
Figure 4.
Identification of differentially expressed pathways and transcriptomic-metabolomic integrative analysis
(A) Enrichment of pathways in high Peri_V_DN group compared with low Peri_V_DN group using GSEA (left panel based on KEGG; right panel based on Reactome).
(B) A pathway-based analysis of metabolomic changes between Peri_V_DN groups. The differential abundance (DA) score captured the overall change in a metabolic pathway. A score of 1 indicated that all metabolites in this pathway increased in high Peri_V_DN group compared with low Peri_V_DN group, and a score of −1 indicated that all metabolites in this pathway decreased.
(C) Transcriptomics and metabolomics distinctions in fatty acid biosynthesis pathway between Peri_V_DN groups. Log2-fold changes of mRNA expression levels and metabolite abundances in high Peri_V_DN tumor samples compared with low Peri_V_DN tumor samples were demonstrated.
CoA, coenzyme A; FA, fatty acid; GSEA, gene set enrichment analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; TCA, tricarboxylic acid; TCR, T cell receptor. See also Figure S4 and Tables S5, S6, S7, and S8.
Previous differentially abundant metabolites and pathway enrichment analyses revealed that metabolic reprogramming was related to high Peri_V_DN. On this basis, we performed differential abundance (DA) score analysis based on metabolomic data between the Peri_V_DN groups. Among 53 pathways in which more than three metabolites were annotated, 21 were upregulated and four were downregulated in high Peri_V_DN patients (Table S8). Among the 21 upregulated pathways, three pathways were upregulated with DA scores of at least 0.25 (Figure 4B). This result was consistent with that of a previous analysis and further highlighted fatty acid metabolism reprogramming in high Peri_V_DN group. We conducted a transcriptomic-metabolomic integrative analysis to depict a more meticulous fatty acid metabolism alteration in this population. Integrative analysis of fatty acid metabolism demonstrated that the initial step of fatty acid synthesis was significantly upregulated (Figure 4C). Taken together, these results demonstrated that vigorous de novo fatty acid synthesis was closely related to a high Peri_V_DN phenotype.
Distinct tumor microenvironments in different Peri_V_DN groups
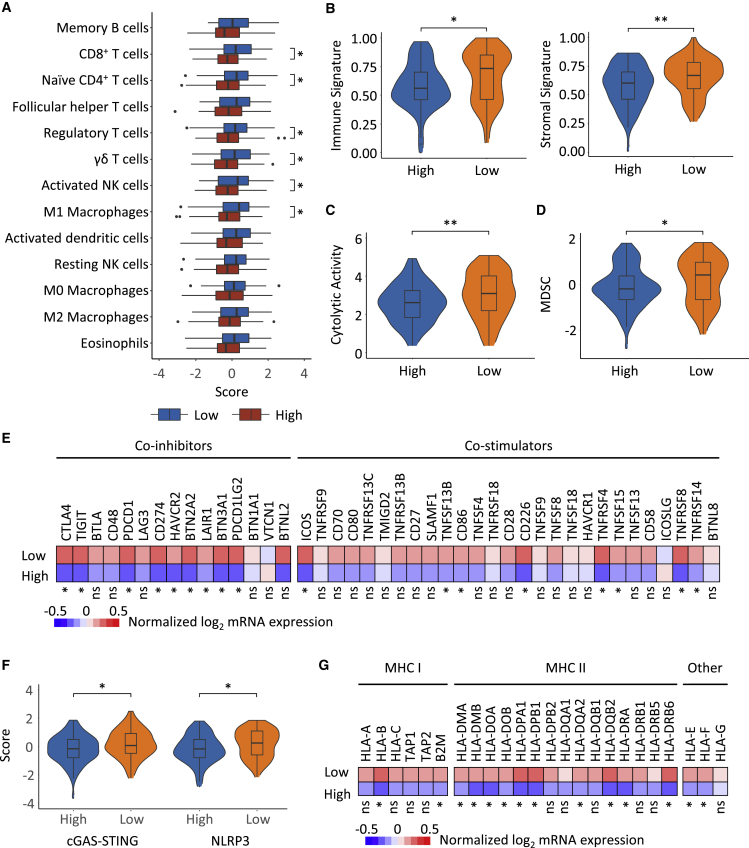
The cell subset composition of the tumor microenvironment was estimated by a published gene signature leveraging transcriptomic data from TNBC radiogenomic cohort.21 The RNA-based immune cell signatures revealed a major difference in the microenvironment between tumors with high and low peritumor heterogeneity (Figures 5A and 5B).22 Low Peri_V_DN was characterized by a higher abundance of CD8+ T cells, naive CD4+ T cells, γδ T cells, activated nature killer (NK) cells, M1 macrophages, and regulatory T cells. Cytolytic activity, which inferred the activity of effector immune cells,23 was lower in high Peri_V_DN cases (p = 0.01; Figure 5C). These results confirmed that high Peri_V_DN was associated with a suppressed immune response.
Figure 5.
Landscape of the tumor microenvironment of the Peri_V_DN groups and distinct escape mechanisms
(A) Differences in the abundance of immune cell types in high Peri_V_DN group compared with low Peri_V_DN group.
(B) Scores of the immune signature (left) and stromal signature (right) inferred by ESTIMATE22 between Peri_V_DN groups.
(C) Comparison of cytolytic activity showed higher effector immune cell activity between Peri_V_DN groups.
(D) Comparison of the abundance of MDSCs between Peri_V_DN groups.
(E) Normalized mRNA expression levels of immune co-inhibitors and co-stimulators between Peri_V_DN groups.
(F) Signature scores of two innate immunity-sensing pathways, cGAS-STING and the NLRP3 inflammasome, between Peri_V_DN groups.
(G) Normalized mRNA expression levels of MHC molecules between Peri_V_DN groups. In total, 167 samples with transcriptomic data were included for analysis.
∗∗0.001 < p ≤ 0.01; ∗0.01 < p ≤ 0.05; ns, p > 0.05. GSVA, gene set variation analysis; MDSC, myeloid-derived suppressor cell; ssGSEA, single-sample gene set enrichment analysis.
Moreover, we investigated the possible immune escape mechanisms of both types of tumors. In addition to the previously described enrichment of regulatory T cells in low Peri_V_DN cases, another inhibitory immune cell type, myeloid-derived suppressor cells (MDSCs), also had a relatively higher abundance in low Peri_V_DN cases (p = 0.05; Figure 5D).24 The expression levels of a wide range of immune co-inhibitors and co-stimulators, including multiple immune checkpoints, were investigated, and a more inhibitory immune context was found in low Peri_V_DN cases (Figure 5E). Overall, the delineation of the tumor microenvironment implied that the low Peri_V_DN group was enriched with hot tumors and might escape immune surveillance by higher inhibitory immune cell infiltration and stronger immune checkpoint molecule expression.
Furthermore, comparison of the two common innate immunity-sensing pathways, cGAS-STING25 and NLRP3 inflammasome,26 demonstrated weaker immunity activation in high Peri_V_DN cases (p = 0.03 and p = 0.02, respectively; Figure 5F). We also analyzed tumor immunogenicity by comparing major histocompatibility complex (MHC) molecules expression. Reduced expression of MHC molecules in the high Peri_V_DN group is demonstrated in Figure 5G. In summary, the high Peri_V_DN group exhibited a cold tumor phenotype, and its potential escape mechanisms included a reduction in innate immune sensing and rejection of immune infiltration.
Discussion
Recent studies have revealed evident tumor heterogeneity among TNBCs, and precision treatment based on molecular profiling has achieved preliminary progress.5,7,27,28 These promising results encouraged molecular subtyping and genomic sequencing for TNBC in clinical practice, which is traditionally conducted by invasive biopsies. Herein, we developed a non-invasive radiomic approach for the identification and molecular classification of TNBC. In addition, we identified a prognostic radiomic feature, which reflected peritumoral heterogeneity, with underlying biological properties. These results demonstrated the potential role of a surrogate radiomic approach in distinguishing TNBC patients and further differentiating TNBC into different subtypes and clinical outcomes.
In this study, we investigated the value of radiomics to distinguish TNBC from other subtypes of breast cancer. We concluded that the non-invasive radiomic approach could identify TNBC with an AUC of 0.922 in the FUSCC cohort, 0.723 in the IPMCH cohort, and 0.613 in the RENJI cohort. This conclusion not only indicated the potential of the MRI-based radiomic approach to identify TNBC but also warned us that the generalization of radiomic signatures remained an important issue to address. These diverse results might be attributed to the redundancy generated by PyRadiomics in the process of feature extraction, and a previous study validated this disadvantage of this widely used open-source tool.29 The significant variation of imaging parameters between different MRI machines, as described in Table S1, also contributed to the suboptimal performance in independent radiomic datasets. Other studies also explored the value of radiomics to classify TNBC. Calstaldo et al.30 found that radiomic signatures identified TNBC with an AUC of 0.91, while Leithner et al.31 identified TNBC with an accuracy of 0.736. In addition, Wu et al.32 conducted a meta-analysis to summarize the efficacy for breast cancer subtype prediction and found that the overall sensitivity and specificity were 0.69 and 0.85, respectively. These results together demonstrated that MRI-based radiomic signatures had the potential to classify TNBC and were consistent with the results of our study.
As radiomic signature could identify TNBC with high accuracy, we further hypothesized that the molecular diversity between TNBC molecular subtypes would lead to different patterns in CE-MRI images, which were quantitatively evaluated using a radiomics approach. Two widely used molecular subtyping systems in TNBC were Lehmann subtype and FUSCC subtype,5,7 which were well correlated with each other. However, considering that FUSCC subtype was developed based on Chinese TNBC patients and was more concise compared with Lehmann subtype, we used FUSCC TNBC subtype as the ground truth in our study. The results revealed a promising approach to identify TNBC molecular subtypes and facilitate precision treatment in a non-invasive way. Combined models including radiomics and IHC staining data demonstrated higher AUCs than individual radiomic and IHC models in the BLIS subtype. However, the combined models did not establish superiority over IHC models regarding the prediction efficacy in the IM subtype. Overall, further studies aiming to optimize the accuracy and simplicity of TNBC classification are warranted.
Besides, we identified prognostic radiomic features and illustrated the underlying molecular pathways. Previous studies have shown that peritumoral heterogeneity can be used to predict the clinical outcomes of several cancer types.33, 34, 35 Herein, we found that Peri_V_DN was strongly associated with adverse clinical outcomes. We further proposed that aberrant metabolism and suppressed immune reactions might be related to peritumoral heterogeneity using transcriptomic and metabolomic data. Several studies have attempted to explore the association between radiomic features and transcriptomic data.17,36 Lee et al.15 found that a four-feature radiomic signature could predict the clinical outcomes of pathological T1 renal cell carcinoma and was associated with the abundance of certain immune cell types. This was consistent with our findings, but the exact mechanisms for the formation of an immunosuppressive microenvironment were not explored in the present study. Wu et al.37 analyzed radiomics and transcriptomics data from TCGA database and found that features extracted from peritumoral regions of CE-MRI were related to clinical outcomes and tumor necrosis factor (TNF) signaling pathway, which were similar to the results of our integrative radiomic analysis.
In conclusion, we presented a radiomic dataset originating from a sizable breast cancer radiomic cohort (n = 860) and a TNBC radiogenomic cohort (n = 202) containing multi-omic data. The radiomic approach showed promising efficacy in identifying TNBC and predicting TNBC molecular subtypes via a non-invasive approach. In addition, peritumoral heterogeneity quantified by radiomics stratified patient outcomes and represented distinct tumor metabolism and immune response patterns. These results demonstrated the potential application of radiomics in the analysis of tumor heterogeneity and clinical management of TNBC.
Limitations of the study
Our study has several limitations. First, more refined models are needed to further improve the prediction efficacy of the radiomic signatures, particularly for predicting TNBC molecular subtypes. These predictive models should also be further verified in a prospective setting. Second, most patients were recruited from a single institution, and the sample size of the independent external validation cohorts was limited. Third, the biological characteristics associated with the Peri_V_DN feature were subjected to the nature of exploratory analysis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| RNA-seq data | Jiang et al.7 | OEP000155; http://www.biosino.org/node |
| Metabolomics data | Xiao et al.38 | OEP000155; http://www.biosino.org/node |
| Software and algorithms | ||
| ANTs toolbox version 2.3.5 | Avants et al.39 | http://stnava.github.io/ANTs/ |
| 3D Slicer version 4.8.1 | Fedorov et al.40 | https://www.slicer.org/ |
| Python version 3.6 | N/A | https://www.python.org/ |
| PyRadiomics version 3.0 | van Griethuysen et al.41 | https://pyradiomics.readthedocs.io/en/latest/ |
| R version 3.6.1 | N/A | https://www.r-project.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Zhi-Ming Shao (zhimingshao@fudan.edu.cn).
Materials availability
This study did not generate novel reagents.
Experimental model and subject details
Patient cohorts
We retrospectively recruited patients diagnosed with malignant breast cancer whose baseline breast CE-MRI images were suitable for radiomics analysis. FUSCC breast cancer radiomic cohort was composed of a total of 860 Chinese patients who were treated at Fudan University Shanghai Cancer Center (FUSCC) from 1 August 2009 to 31 May 2015 and met the following criteria: 1) female patients diagnosed with unilateral invasive ductal carcinoma with known ER, PR and HER2 phenotypes;42 2) no evidence of distant metastasis at diagnosis.
We also generated a TNBC radiogenomic cohort composed of 202 TNBC patients based on our previously developed TNBC multi-omic dataset.7 In this cohort, transcriptomics sequencing (n = 167), metabolomics (n = 138), hematoxylin-eosin stained sections with IHC staining (n = 56) for the expression of AR, CD8A, FOXC1 and DCLK1, and clinical follow-up data (including relapse-free survival and overall survival) were also available. Follow-up within this cohort of patients was completed on 30 June 2017, and the median length of follow-up was 45.8 months. Relapse-free survival was defined as the time from diagnosis to first recurrence or a diagnosis of contralateral breast cancer. Overall survival was defined as the time from diagnosis to death from any cause. Patients without events were censored from the time point of the last follow-up. Additional clinicopathological factors, such as stromal tumor infiltrating lymphocytes (sTILs) and homologous recombination deficiency (HRD) status, were also available. The studies were conducted in accordance with the Declaration of Helsinki. All analyses were approved by the independent ethics committee/institutional review board of Fudan University Shanghai Cancer Center, and written informed consent was obtained from each patient.
To evaluate the ability of radiomic signatures to generalize to additional populations, we collected CE-MRI images and ER, PR and HER2 phenotype information from patients diagnosed with malignant breast cancer from two other independent medical centers. IPMCH dataset comprised 54 patients from International Peace Maternal and Children Hospital from 1 January 2013 to 31 June 2019, and RENJI dataset comprised 110 breast cancer patients from Shanghai Jiaotong University Renji Hospital from 1 August 2018 to 30 November 2020. Data collection was approved by both the IPMCH and Renji Hospital institutional review boards.
Method details
Breast CE-MRI imaging
All the patients had undergone breast MR examination before biopsy, and CE-MRI images were used for radiomics analysis in this study. The imaging parameters are listed in Table S1.
All other phases were co-registered into the first postcontrast phase of CE-MRI through non-linear registration using the symmetric normalization algorithm,39 which was performed using the ANTs toolbox (version 2.3.5), to eliminate the spatial mismatches caused by motion artifacts. Nonparametric nonuniformity normalization algorithm was applied for bias field correction.
ROI delineation and inter- and intra-observer reproducibility
ROIs were delineated semiautomatically on the peak enhanced phase of CE-MRI using 3D Slicer software (version 4.8.1).40 The ROIs were placed on all slices that contained the whole tumor or the largest lesion (in the case of multicentric or multifocal tumors). Two radiologists (C.Y. and D.D.Z. with 9 and 4 years of experience in breast MRI, respectively) were blinded to the pathological and biochemical findings of each patient and were primarily responsible for evaluating the ROIs. The inter- and intra-observer reproducibility of the ROIs and radiomic feature extraction were initially analyzed using the CE-MRI data of 60 randomly selected patients in a blinded fashion by two radiologists. To ensure consistent ROI delineation, one radiologist repeated the ROI delineation twice with an interval of at least 1 month, while another radiologist independently drew the ROIs and generated radiomic features following the same procedure. The agreements of the ROIs between the radiologists and within the same radiologist represent inter- and intra-observer reproducibility, respectively. Intraclass correlation coefficients (ICCs) were used to evaluate the intra- and interobserver agreement in terms of feature extraction. Inter- and intra-observer reproducibility and radiomic feature extraction achieved substantial agreement with ICC > 0.75 both among the ROIs from the two radiologists and between the ROIs from the same radiologist.43
Furthermore, the peritumoral area was obtained by expanding the tumor outward with a 5-mm width and subtracting the tumor area, while the intratumoral area was obtained by shrinking with a 5-mm width. Expanding and shrinking operations were implemented automatically based on dilating and eroding algorithms, with a sphere morphological structuring element (radius = 5 mm). In addition, tumor and peritumoral regions were integrated as another region. In total, four sets of ROIs, including the tumor, peritumor, intratumor and tumor-peritumor regions, were used in the radiomics feature extraction.
Radiomics feature extraction
This study extracted two categories of radiomic features based on CE-MRI images, namely, spatial domain features and sequential features. Spatial domain features included shape features, first-order features, textural features and wavelet domain features, while sequential features comprised enhancement rate features and time-varying curve-based features. These radiomic features were calculated using the PyRadiomics package (voxel size: 0.7 × 0.7 × 1.5 mm3, 'binWidth': 25, version 3.0),41 implemented in Python (version 3.6) and in-house pipelines.
Shape features were common to all phases and included descriptors of the three-dimensional size and shape of the ROIs. First-order features and textural features were calculated from each phase individually. First-order features described the distribution of voxel intensities, and textural features were obtained based on five textural matrices to describe the radiological pattern of the ROIs, including Gray Level Co-occurrence Matrix (GLCM), Gray Level Dependence Matrix (GLDM), Gray Level Run Length Matrix (GLRLM), Gray Level Size Zone Matrix (GLSZM), Neighboring Gray Tone Difference Matrix (NGTDM). Shape, first-order and textural features were extracted using PyRadiomics package. Additionally, wavelet domain features were extracted for each first-order feature and textural feature by applying wavelet filtering to the original images, yielding eight decompositions per level (LLL, LLH, LHL, HLL, LHH, HLH, HHL, HHH).
Sequential features, also known as time domain features, were calculated to consider time dimension information. Sequential features were extracted based on each spatial domain feature, except for shape features (because they were identical between all imaging phases). Enhancement rate features depicted the rate of change of each spatial feature between each two phases during contrast enhancement, which is defined as:
Here, represents the feature value of the former phase, and represents the feature value of the latter phase. Time-varying curve-based features included the mean, variance, skewness, kurtosis and energy of value of each spatial domain feature in its time-varying curve. These features were defined as follows:
-
1.
Mean
-
2.
Variance
-
3.
Skewness
-
4.
Kurtosis
-
5.
Energy
indicated the standard deviation.
Quantification and statistical analysis
Feature selection and radiomics model building
The LASSO method was used to select the most useful predictive features from the training cohort (glmnet R package).44 Tuning parameter (λ) was selected in the LASSO model by 10-fold cross-validation for identifying TNBC and 9-fold cross validation for distinguishing TNBC molecular subtypes. Radiomics scores were calculated for each patient using two different methods: 1) multivariate linear regression (glm R package) and 2) support vector machine (SVM; e1071 R package). The abilities to identify TNBC and distinguish TNBC molecular subtypes were assessed using the area under the curve (AUC) of the receiver operator characteristic curve (ROC) via the pROC R package.45 Confidence intervals of AUCs were calculated using the Delong method. The AUC of the precision recall curve (PRC) was assessed via the PRROC R package. Radiomics data from patients with known transcriptomic TNBC subtype7 and Aurora CE-MRI images were selected to build signatures for distinguishing molecular subtypes inside TNBC.
Radiomics model validation
The radiomics prediction models were validated internally and externally. First, the trained classifiers were assessed by cross-validation via the glmnet R package.44 Next, the trained classifiers were further tested in the validation datasets in terms of the AUC and its confidence intervals of the ROC curve.
Generation and analysis of metabolomics and lipidomics data
The metabolomic and lipidomics data of our study were generated using four steps: sample preparation, metabolite extraction, polar metabolite and lipid detection and mass spectrum (MS) data analysis.
The samples in our TNBC multi-omic cohort with adequate tissues for polar metabolites and lipids were collected. In total, 138 TNBC samples were selected for further metabolomics and lipidomics analysis. Acetonitrile: methanol: water = 2: 2: 1 solution and MTBE: MeOH= 5: 1 solution were applied to extract polar metabolites and lipids, respectively. An equal volume (10 μL) of each sample was mixed for quality control sample preparation. A BEH amide column (2.1 ∗ 100 mm, 1.7 μm, Waters) or Kinetex C18 column (2.1 ∗ 100 mm, 1.7 μm, Phenomen) coupled with a Triple TOF 6600 mass spectrometer or AB triple TOF 5600 mass spectrometer was deployed to conduct LC–MS/MS experiments for polar metabolite and lipid detection. MS raw data files were converted to mzXML format by ProteoWizard software (version 3.0.19282) and processed by R package XCMS (v3.2) and LipidAnalyzer formetabolomics and lipidomics data, respectively.
Detailed information on metabolomics and lipidomics data generation was contained in a metabolomics study published by Xiao et al.38
Analysis of differentially abundant metabolites and differentially expressed genes
The differential abundance of metabolites was calculated by performing Mann–Whitney U tests for all detected metabolites. Metabolites were considered to have significant differences between high and low peritumoral heterogeneity if |log2FC| > 0.3 and p < 0.05.
The differential expression of genes was determined using the edgeR R package. Genes were considered to have significant differences between high and low peritumoral heterogeneity if |log2FC| > 0.5 and FDR < 0.05. Gene set enrichment analysis (GSEA) was performed using the clusterProfiler R package.46 The differential expression analysis outputs of edgeR were used to generate the ranked list file. One thousand total permutations were used.
Differential abundance (DA) score
The DA score was calculated first by determining which metabolites were significantly increased/decreased in abundance, as described above. Then, the DA score was defined as follows:
DA = (Number of metabolites increased - Number of metabolites decreased) / Number of measured metabolites in that pathway47
Thus, the DA score ranges from −1 to 1. A score of −1 indicates that all metabolites in a pathway decreased, while a score of 1 indicates that all metabolites increased in abundance. The components of the metabolic pathways used in the integrative analysis were annotated using the KEGG database.
Calculation of microenvironment cell abundance
A signature containing 364 genes representing 24 microenvironment cell types was obtained from one published immuno-oncology paper.21 This signature modified the CIBERSORT and MCP-Counter signatures and represented a more comprehensive landscape of the TNBC microenvironment. Subsequently, we used single-sample gene set enrichment analysis (ssGSEA, “GSVA” function in GSVA R package)48 to calculate the abundance of each cell subset in each sample with expression data.
Determination of immune checkpoint molecules
To determine which molecules played a critical role in shaping distinct tumor microenvironments, we searched a database of molecules (https://www.rndsystems.com/cn/research-area/co--stimulatory-and-co--inhibitory-molecules) to compare the expression levels of these molecules in tumors with different levels of peritumoral heterogeneity.
Statistical analysis
Student’s t test, Wilcoxon’s test and Kruskal–Wallis test were used to compare continuous variables. Prior to the comparisons, the normality of the distributions was tested with the Shapiro–Wilk test. Pearson’s chi-squared test and Fisher’s exact test were employed for the comparison of unordered categorical variables. To explore the association between radiomics features and survival, Kaplan–Meier analysis and a Cox proportional hazards model were employed in the training and validation sets. Comparison of survival between groups was conducted via the log rank test. All the tests were two-sided, and p < 0.05 was regarded as indicating significance unless otherwise stated. All statistical analyses were performed using R software (version 3.6.1).
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81922048, 81901703, 82071878, 82002792, 91959207, and 92159301), the Shanghai Rising-Star Program (20YF1408600), the Shanghai Key Laboratory of Breast Cancer (12DZ2260100), the Clinical Research Plan of SHDC (SHDC2020CR2008A, SHDC2020CR4002, and SHDC2020CR5005), and the SHDC Municipal Project for Developing Emerging and Frontier Technology in Shanghai Hospitals (SHDC12021103).
Author contributions
Conceptualization, Z.-M.S., Y.-Z.J., and Y.-J.G.; methodology, L.J., C.Y., Y.X., and R.-C.Z.; data curation, C.Y., R.-C.Z., B.-Q.X., H.W., and D.-D.Z.; investigation, L.J., Y.X., and G.-H.S.; writing – original draft, L.J., C.Y., Y.X., and G.-H.S.; writing – review & editing, Y.-Z.J., H.W., Y.-J.G., and Z.-M.S.; supervision, Z.-M.S., Y.-Z.J., and Y.-J.G. All authors approved the final manuscript.
Declaration of interests
L.J. is currently an employee of AstraZeneca.
Published: July 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2022.100694.
Contributor Information
Yi-Zhou Jiang, Email: yizhoujiang@fudan.edu.cn.
Ya-Jia Gu, Email: cjr.guyajia@vip.163.com.
Zhi-Ming Shao, Email: zhimingshao@fudan.edu.cn.
Supplemental information
Data and code availability
-
•
The raw breast CE-MRI images used in this paper are available from the lead contact upon request.
-
•
RNA-seq and metabolomics data can be viewed in The National Omics Data Encyclopedia (NODE) (http://www.biosino.org/node): OEP000155 or through the URL: http://www.biosino.org/node/project/detail/OEP000155.
-
•
Custom codes are available in Supplementary Information.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Waks A.G., Winer E.P. Breast cancer treatment: a review. J. Am. Med. Assoc. 2019;321:288. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 2.Criscitiello C., Azim H.A., Jr., Schouten P.C., Linn S.C., Sotiriou C. Understanding the biology of triple-negative breast cancer. Ann. Oncol. 2012;23 doi: 10.1093/annonc/mds188. vi13-18. [DOI] [PubMed] [Google Scholar]
- 3.Denkert C., Liedtke C., Tutt A., von Minckwitz G. Molecular alterations in triple-negative breast cancer—the road to new treatment strategies. Lancet. 2017;389:2430–2442. doi: 10.1016/s0140-6736(16)32454-0. [DOI] [PubMed] [Google Scholar]
- 4.Metzger-Filho O., Tutt A., de Azambuja E., Saini K.S., Viale G., Loi S., Bradbury I., Bliss J.M., Azim H.A., Jr., Ellis P., et al. Dissecting the heterogeneity of triple-negative breast cancer. J. Clin. Oncol. 2012;30:1879–1887. doi: 10.1200/jco.2011.38.2010. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann B.D., Bauer J.A., Chen X., Sanders M.E., Chakravarthy A.B., Shyr Y., Pietenpol J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 2011;121:2750–2767. doi: 10.1172/jci45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burstein M.D., Tsimelzon A., Poage G.M., Covington K.R., Contreras A., Fuqua S.A., Savage M.I., Osborne C.K., Hilsenbeck S.G., Chang J.C., et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin. Cancer Res. 2015;21:1688–1698. doi: 10.1158/1078-0432.CCR-14-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang Y.Z., Ma D., Suo C., Shi J., Xue M., Hu X., Xiao Y., Yu K.D., Liu Y.R., Yu Y., et al. Genomic and transcriptomic landscape of triple-negative breast cancers: subtypes and treatment strategies. Cancer Cell. 2019;35:428–440.e5. doi: 10.1016/j.ccell.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Dagogo-Jack I., Shaw A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 9.Marusyk A., Janiszewska M., Polyak K. Intratumor heterogeneity: the rosetta stone of therapy resistance. Cancer Cell. 2020;37:471–484. doi: 10.1016/j.ccell.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R.G., Granton P., Zegers C.M., Gillies R., Boellard R., Dekker A., Aerts H.J. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bi W.L., Hosny A., Schabath M.B., Giger M.L., Birkbak N.J., Mehrtash A., Allison T., Arnaout O., Abbosh C., Dunn I.F., et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J. Clin. 2019;69:127–157. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen R.W., van Amstel P., Martens R.M., Kooi I.E., Wesseling P., de Langen A.J., Menke-Van der Houven van Oordt C.W., Jansen B.H.E., Moll A.C., Dorsman J.C., et al. Non-invasive tumor genotyping using radiogenomic biomarkers, a systematic review and oncology-wide pathway analysis. Oncotarget. 2018;9:20134–20155. doi: 10.18632/oncotarget.24893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rios Velazquez E., Parmar C., Liu Y., Coroller T.P., Cruz G., Stringfield O., Ye Z., Makrigiorgos M., Fennessy F., Mak R.H., et al. Somatic mutations drive distinct imaging phenotypes in lung cancer. Cancer Res. 2017;77:3922–3930. doi: 10.1158/0008-5472.CAN-17-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Y.Q., Liang C.H., He L., Tian J., Liang C.S., Chen X., Ma Z.L., Liu Z.Y. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J. Clin. Oncol. 2016;34:2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 15.Lee H.W., Cho H.H., Joung J.G., Jeon H.G., Jeong B.C., Jeon S.S., Lee H.M., Nam D.H., Park W.Y., Kim C.K., et al. Integrative radiogenomics approach for risk assessment of post-operative metastasis in pathological T1 renal cell carcinoma: a pilot retrospective cohort study. Cancer. 2020;12:866. doi: 10.3390/cancers12040866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer S., Tahoun M., Klaan B., Thierfelder K.M., Weber M.A., Krause B.J., Hakenberg O., Fuellen G., Hamed M. A radiogenomic approach for decoding molecular mechanisms underlying tumor progression in prostate cancer. Cancer. 2019;11:1293. doi: 10.3390/cancers11091293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin P., Wen D.Y., Chen L., Li X., Li S.H., Yan H.B., He R.Q., Chen G., He Y., Yang H. A radiogenomics signature for predicting the clinical outcome of bladder urothelial carcinoma. Eur. Radiol. 2020;30:547–557. doi: 10.1007/s00330-019-06371-w. [DOI] [PubMed] [Google Scholar]
- 18.Kim J.H., Ko E.S., Lim Y., Lee K.S., Han B.K., Ko E.Y., Hahn S.Y., Nam S.J. Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology. 2017;282:665–675. doi: 10.1148/radiol.2016160261. [DOI] [PubMed] [Google Scholar]
- 19.Wu J., Gong G., Cui Y., Li R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J. Magn. Reson. Imaging. 2016;44:1107–1115. doi: 10.1002/jmri.25279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao S., Ma D., Xiao Y., Li X.M., Ma J.L., Zhang H., Xu X.L., Lv H., Jiang W.H., Yang W.T., et al. Molecular subtyping of triple-negative breast cancers by immunohistochemistry: molecular basis and clinical relevance. Oncol. 2020:e1481–e1491. doi: 10.1634/theoncologist.2019-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Y., Ma D., Zhao S., Suo C., Shi J., Xue M.Z., Ruan M., Wang H., Zhao J., Li Q., et al. AME Breast Cancer Collaborative Group Multi-omics profiling reveals distinct microenvironment characterization and suggests immune escape mechanisms of triple-negative breast cancer. Clin. Cancer Res. 2019:5002–5014. doi: 10.1158/1078-0432.CCR-18-3524. [DOI] [PubMed] [Google Scholar]
- 22.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angelova M., Charoentong P., Hackl H., Fischer M.L., Snajder R., Krogsdam A.M., Waldner M.J., Bindea G., Mlecnik B., Galon J., Trajanoski Z. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol. 2015;16:64. doi: 10.1186/s13059-015-0620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Woo S.R., Fuertes M.B., Corrales L., Spranger S., Furdyna M.J., Leung M.Y., Duggan R., Wang Y., Barber G.N., Fitzgerald K.A., et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghiringhelli F., Apetoh L., Tesniere A., Aymeric L., Ma Y., Ortiz C., Vermaelen K., Panaretakis T., Mignot G., Ullrich E., et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1β–dependent adaptive immunity against tumors. Nat. Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y.Z., Liu Y., Xiao Y., Hu X., Jiang L., Zuo W.J., Ma D., Ding J., Zhu X., Zou J., et al. Molecular subtyping and genomic profiling expand precision medicine in refractory metastatic triple-negative breast cancer: the FUTURE trial. Cell Res. 2020;31:178–186. doi: 10.1038/s41422-020-0375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garrido-Castro A.C., Lin N.U., Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019;9:176–198. doi: 10.1158/2159-8290.CD-18-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J., Li C., Gensheimer M., Padda S., Kato F., Shirato H., Wei Y., Schönlieb C.B., Price S.J., Jaffray D., et al. Radiological tumour classification across imaging modality and histology. Nat. Mach. Intell. 2021;3:787–798. doi: 10.1038/s42256-021-00377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Castaldo R., Pane K., Nicolai E., Salvatore M., Franzese M. The impact of normalization approaches to automatically detect radiogenomic phenotypes characterizing breast cancer receptors status. Cancer. 2020;12:518. doi: 10.3390/cancers12020518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leithner D., Horvat J.V., Marino M.A., Bernard-Davila B., Jochelson M.S., Ochoa-Albiztegui R.E., Martinez D.F., Morris E.A., Thakur S., Pinker K. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results. Breast. Cancer Res. 2019;21:106. doi: 10.1186/s13058-019-1187-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu J., Mayer A.T., Li R. Integrated imaging and molecular analysis to decipher tumor microenvironment in the era of immunotherapy. Semin. Cancer Biol. 2020:310. doi: 10.1016/j.semcancer.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou T.H., Coroller T.P., van Griethuysen J.J.M., Mak R.H., Aerts H.J.W.L. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. PLoS One. 2018;13:e0206108. doi: 10.1371/journal.pone.0206108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prasanna P., Patel J., Partovi S., Madabhushi A., Tiwari P. Radiomic features from the peritumoral brain parenchyma on treatment-naïve multi-parametric MR imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings. Eur. Radiol. 2017;27:4188–4197. doi: 10.1007/s00330-016-4637-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braman N.M., Etesami M., Prasanna P., Dubchuk C., Gilmore H., Tiwari P., Plecha D., Madabhushi A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast. Cancer Res. 2017;19:57. doi: 10.1186/s13058-017-0846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh A.C., Li H., Zhu Y., Zhang J., Khramtsova G., Drukker K., Edwards A., McGregor S., Yoshimatsu T., Zheng Y., et al. Radiogenomics of breast cancer using dynamic contrast enhanced MRI and gene expression profiling. Cancer Imag. 2019;19:48. doi: 10.1186/s40644-019-0233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J., Li B., Sun X., Cao G., Rubin D.L., Napel S., Ikeda D.M., Kurian A.W., Li R. Heterogeneous enhancement patterns of tumor-adjacent parenchyma at MR imaging are associated with dysregulated signaling pathways and poor survival in breast cancer. Radiology. 2017;285:401–413. doi: 10.1148/radiol.2017162823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao Y., Ma D., Yang Y.S., Yang F., Ding J.H., Gong Y., Jiang L., Ge L.P., Wu S.Y., Yu Q., et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell Res. 2022:477–490. doi: 10.1038/s41422-022-00614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avants B.B., Epstein C.L., Grossman M., Gee J.C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image. Anal. 2008;12:26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fedorov A., Beichel R., Kalpathy-Cramer J., Finet J., Fillion-Robin J.C., Pujol S., Bauer C., Jennings D., Fennessy F., Sonka M., et al. 3D slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging. 2012;30:1323–1341. doi: 10.1016/j.mri.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Griethuysen J.J.M., Fedorov A., Parmar C., Hosny A., Aucoin N., Narayan V., Beets-Tan R.G.H., Fillion-Robin J.C., Pieper S., Aerts H.J., Aerts H. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.Can-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hammond M.E., Hayes D.F., Dowsett M., Allred D.C., Hagerty K.L., Badve S., Fitzgibbons P.L., Francis G., Goldstein N.S., Hayes M., et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010;28:2784–2795. doi: 10.1200/jco.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 44.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Software. 2010;33:1–22. doi: 10.18637/jss.v033.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hakimi A.A., Reznik E., Lee C.H., Creighton C.J., Brannon A.R., Luna A., Aksoy B.A., Liu E.M., Shen R., Lee W., et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw breast CE-MRI images used in this paper are available from the lead contact upon request.
-
•
RNA-seq and metabolomics data can be viewed in The National Omics Data Encyclopedia (NODE) (http://www.biosino.org/node): OEP000155 or through the URL: http://www.biosino.org/node/project/detail/OEP000155.
-
•
Custom codes are available in Supplementary Information.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.