Summary
Background
T cells form the major component of anti-tumor immunity. A deeper understanding of T cell exhaustion (TEX) heterogeneity within the tumor microenvironment (TME) is key to overcoming TEX and improving checkpoint blockade immunotherapies in the clinical setting.
Methods
We conducted a comprehensive pan-cancer analysis of TEX subsets from 9564 tumor samples across 30 bulk solid cancer types. Pan-cancer TEX subtypes were identified using literature-derived hierarchical TEX-specific developmental pathway signatures. The potential multi-omics and clinical features involved in TEX heterogeneity were determined.
Findings
Our study yielded a dynamic, progressive roadmap and a hierarchical dysfunction landscape regarding TEX within the TME. In total, we identified five pan-cancer TEX subtypes, revealing tissue/cancer type-specific TEX patterns in low immunogenic tumors. By contrast, highly immunogenic tumors tend to harbor high frequencies of progenitor TEX subsets. In addition, the TEX profile also revealed distinct prognoses, intrinsic molecular subtype distribution, immune microenvironment and multi-omics features among the cancers. Network analysis identified four previously unknown TEX-associated cancer genes (tolloid-like 1, myosin heavy chain 111, P2Y receptor family member 8 and protein kinase D2), the possible association with anti-PD-1 immunotherapy response was validated using a single-cell dataset. Finally, a machine learning-based gene signature was developed to model the hierarchical TEX stages, verified in single-cell and immunotherapy patient cohorts.
Interpretation
Our study provided a TEX-derived system that can be applied for the immune subtyping of cancers and may have implications for the further optimization of personalized cancer immunotherapy.
Funding
This study was supported by the National Natural Science Foundation of China (Grant No. 62072341 and 61973240). The funders had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Keywords: Immunotherapy, Pan-cancer, Tumor microenvironment, T cell exhaustion
Research in context.
Evidence before this study
Accumulating evidence has indicated that T cell exhaustion (TEX) is a dynamic process with a developmental hierarchy, which contains a broad continuum of the phenotypic and functional intermediate states. Therefore, the pool of exhausted CD8+ T cells in the tumor immune microenvironment (TIME) at any given time should not be viewed simply as extremes of this continuum (progenitor and terminally). Instead, it should be considered to contain a more heterogeneous and complex population of CD8+ T cell subsets at different stages of TEX, each with distinct functional and developmental properties. However, the impact of this heterogeneous TEX landscape within the TIME among different tumor types and among tumors arising from the same tissue on clinical outcomes and therapeutic efficacy remains poorly understood.
Added value of this study
Our study characterized the overall pattern of heterogeneous TEX subset distribution within the TIME among different types of cancer whilst revealing the preferences and dependencies of the TEX subtypes in various tumor types and among the same tumor types in other individuals. A TEX-based immunotyping scheme was proposed, which was used to predict prognosis, profile the intrinsic molecular subtype distribution, characterize the immune microenvironment and reveal various multiple omics features, to expand upon previously reported systems for subtyping cancers. Furthermore, the potential underlying genomic, transcriptional and epigenetic mechanisms involved in mediating TEX heterogeneity was preliminarily investigated.
Implications of all the available evidence
The comprehensive characterization of these heterogeneous TEX subsets within the TIME provides insights into cancer immunity and may have important implications for improving personalized cancer immunotherapy by targeting T cells.
Alt-text: Unlabelled box
Introduction
CD8+T lymphocytes form a specialized population of T cells and mediate adaptive cytotoxic T cell responses against chronic infections and cancer.1 CD8+T cells responding to chronic infections or tumor antigens typically differentiate into the phenotype of exhaustion, which is a mechanistically distinct lineage of differentiated T cells.2,3 This process, known as T cell exhaustion (TEX), is characterized by poor effector functions, increased expression of inhibitory receptors (such as programed cell death protein 1 (PD1), programmed death ligand 1 (PD-L1), T-cell immunoglobulin and mucin domain-containing protein 3 (TIM-3), T cell immunoglobulin and ITIM domain (TIGIT) and cytotoxic T-lymphocyte associated protein 4 (CTLA4)), altered usage of transcription factors (such as transcription factors T cell factor (TCF1), thymocyte selection-associated HMG BOX (TOX), and T-bet), metabolic dysfunction and impaired proliferative potential and survival.4,5 Moreover, apart from the transformation of the above molecular factors, TEX is characterized by hierarchical dysfunction of some pathways, such as interferon-γ (IFN-γ), tumor necrosis factor (TNF), cytotoxic potential (CTL) and interleukin 2 (IL-2) production.4, 5, 6 Exhausted T cells were initially characterized into two individual subtypes, namely the progenitor and terminally-differentiated subtypes, based on the expression levels of TCF1 and PD1.7, 8, 9 The progenitor exhausted T cell subtype includes those that are stem cell-like or memory-like, with abilities of effective self-renewal and transforming into the terminally differentiated subtype.3,7, 8, 9, 10 By contrast, the terminally differentiated subtype cannot be functionally rescued and has limited expansion capacity.11,12
The tumor immune microenvironment (TIME) has been extensively recognized to be a crucial regulator of cancer progression, clinical outcome and therapeutic response.13,14 Tumor-infiltrating CD8+T cells form a significant component of the TIME and serve a key role in recognizing and killing tumor cells. However, the majority of infiltrating CD8+T cells become ‘exhausted’ due to the triggering of a complex immunosuppressive signaling network, leading to cancer immune evasion.15, 16, 17 Accumulating evidence has indicated that TEX is a dynamic process with a developmental hierarchy, which contains a broad continuum of phenotypic and functional intermediate states.4,18 Therefore, the pool of exhausted CD8+ T cells presents within the TIME at any given time should not be viewed simply as extremes of this continuum (progenitor and terminally). Instead, it should be considered to contain a more heterogeneous and complex population of CD8+ T cell subsets at different stages of TEX, each with distinct functional and developmental properties. Furthermore, the heterogeneity of TEX has been implicated in determining different clinical outcomes and immunotherapy efficacy.19,20 However, the impact of this heterogeneous TEX landscape within the TIME among different tumor types and tumors arising from the same tissue on clinical outcomes and therapeutic efficacy remains poorly understood.
To address this, we conducted a comprehensive pan-cancer analysis of TEX subsets from 9,564 tumor samples among 30 bulk solid cancer types. This revealed a dynamic roadmap of progressive TEX development and the hierarchical dysfunction landscape of TEX within the TIME. Subsequently, we identified five pan-cancer TEX subtypes on the basis of literature-derived and TEX-specific hierarchical developmental pathway signatures. This was applied to determine the potential molecular and clinical features involved in TEX heterogeneity. Finally, a machine learning-based gene signature was developed to model the hierarchical TEX stages and predict immunotherapy efficacy.
Methods
Molecular and clinical information of the tumor datasets
The pan-cancer multi-omics data, including the clinicopathological information of 9564 tumor samples of 30 bulk solid cancer types excluding leukemia, pheochromocytoma, and paraganglioma from TCGA, were downloaded from the UCSC Xena Database (https://xenabrowser.net/datapages/). They include data regarding copy-number variation produced using the Illumina platform calculated using the GISTIC221 method, somatic mutation produced using the Illumina platform, RNA-sequencing (RNA-seq) data produced using the HiSeq Illumina platform converted through the log(x+1) method and DNA methylation data produced using the Illumina Human Methylation 450K platform. Microsatellite instability data was obtained from Bonneville et al.22 and TCR/BCR data was received from Thorsson et al.23
The 10X single-cell RNA-seq data (GSE159251) of three samples of advanced melanoma, which were comprised of 10518 cells before treatment, was downloaded from Gene Expression Omnibus (GEO) from a previous study.24
Clinicopathological information and RNA-seq data of patients with cancer who underwent anti-PD-1, anti-PD-L1 and anti-CTLA-4 monotherapy produced using the HiSeq Illumina platform were received from previously published prospective ICI clinical trials. They include 41 patients with melanoma from Gide et al. (referred to as the Gide cohort),25 26 patients with metastatic melanoma from Hugo et al.26 49 patients with advanced melanoma from Riaz et al. (referred to as the HugoW/Riaz cohort),27 16 patients with metastatic clear cell renal cell carcinoma from Miao et al. (referred to as the Miao cohort),28 23 patients with glioblastoma from Zhao et al. (referred to as the Zhao cohort),29 80 patients with metastatic urothelial cancer from Mariathasan et al. (referred to as the Mariathasan cohort)30 and 38 patients with melanoma from Van Allen et al. (referred to as the VanAll cohort).31
Activity analysis of TEX pathways
The TEX signaling pathway signatures and hallmark gene sets were received from Molecular Signatures Database (MSigDB, V7.2).4,32 ssGSEA was used to estimate the activity score of the TEX pathways in each patient using the ‘GSVA’ R package.33
Computational index of cancer-related events and immune microenvironment
Immune signatures of 28 immune cells were obtained from a previous genotype/immunophenotype-related study34 before the enrichment score was estimated for each immune cell type and each patient using ssGSEA.33 Infiltrating immune cell fractions were calculated in CIBERSORT using deconvolution methods with the LM22 immune signature matrix.35 To quantify stromal cell activity in solid tumors, the stromal score was calculated using ESTIMATE36 in ssGSEA.37 To characterize the stemness of the exhausted T cells, the stemness activation score (33)was calculated based on the system of stem signature previously obtained by Miranda et al.38 using ssGSEA in each patient. The CYT score33 was calculated to quantify immune effector activity in solid tumors as previously described.39
Development of machine learning-based gene signature for modeling hierarchical TEX stages
The 568 cancer driver genes were obtained from previous studies.40 To identify candidate driver genes involved in TEX, differentially expressed (DE) mRNA between each TEX subgroup were identified using a false discovery rate (FDR)-adjusted P-value <0.01 as the threshold of significance. Pearson's correlation coefficient was then used to measure the correlation between DE mRNA expression and the extent of CD8+ T cell fraction using the threshold of |r| >0.25. Deep neural network modeling was performed using ‘Deep Autoencoders’41,42 to feature selection by cycles of coding and decoding. For each layer, 'tanh' was used as the activation function between the output and input layers, where the hidden layer sizes were 150. Next, machine learning-based gene signature used for predicting hierarchical TEX stages were developed using five artificial intelligence frameworks, including extreme gradient boosting (XGBoost, R package ‘XGBoost’, V1.4.1.1, https://xgboost.readthedocs.io/en/stable/index.html), multi-logistic (R package ‘nnet’, V7.3-16, https://cran.r-project.org/web/packages/nnet/index.html), random forest (RF, R package ‘randomForest’, V4.6-14, https://cran.r-project.org/web/packages/randomForest/index.html), support vector machine (SVM, R package ‘e1071’, V1.7-7, https://cran.r-project.org/web/packages/e1071/index.html) and feedforward neural network (FNN, R package ‘h2o’, V3.32.1.3, https://cran.r-project.org/web/packages/h2o/index.html).
Single-cell RNA-seq analysis
The NormalizeData function was performed to normalize raw counts and FindVariableFeatures function was performed to select the highly variable genes in the R package ‘Seurat’43 (V4.0.3) to address the batch effect and tissue specificity of the single-cell RNA-seq data. The principal component analysis (PCA) was used to dimensionality reduction with the highly variable genes. We used established cell markers44 from a previous study to probe for cell subgroup information before mapping the crucial driver signature genes of exhausted T cells.
Statistical analysis
All statistical analyses were conducted using R version 4.0.3 (https://www.r-project.org/). Hierarchical clustering analysis was performed using the Euclidean distances and ‘Ward. D’ method with the R package ‘stats’ (V4.0.3, https://cran.r-project.org/web/packages/stats/index.html). Wilcoxon rank-sum and Kruskal-Wallis tests were used to estimate the differences between ≥ two groups for continuous variables, whereas Fisher's exact test was performed to test the difference between two categorical variables. The Kaplan-Meier curve and log-rank test were performed to compare survival differences between ≥ two patient groups. Hazard ratios and 95% confidence intervals were estimated by univariate Cox proportional hazard regression analysis using the R package ‘survival’ (V3.2-13, https://cran.r-project.org/web/packages/survival/index.html). Correlations between two variables were measured using Pearson's correlation coefficient and corresponding significance was assessed using a two-sided hypothesis test. Receiver operating characteristic curves and the AUC were applied to assess the predictive performance using the R package ‘pROC’ (V1.18.0, https://cran.r-project.org/web/packages/pROC/index.html).45 Functional annotation analysis of GO and KEGG revealed the potential biological function using the R package ‘clusterprofiler4’ (V4.0.5).46 The protein-protein interaction networks were constructed to show the association among genes using the STRING database (https://cn.string-db.org/). All P-values were two-sided and P<0.05 was considered to indicate a statistically significant difference.
Role of funders
The Funders had no role in study design, data collection, data analyses, interpretation, or writing of report.
Results
Dynamic progressive roadmap of TEX in pan-cancer
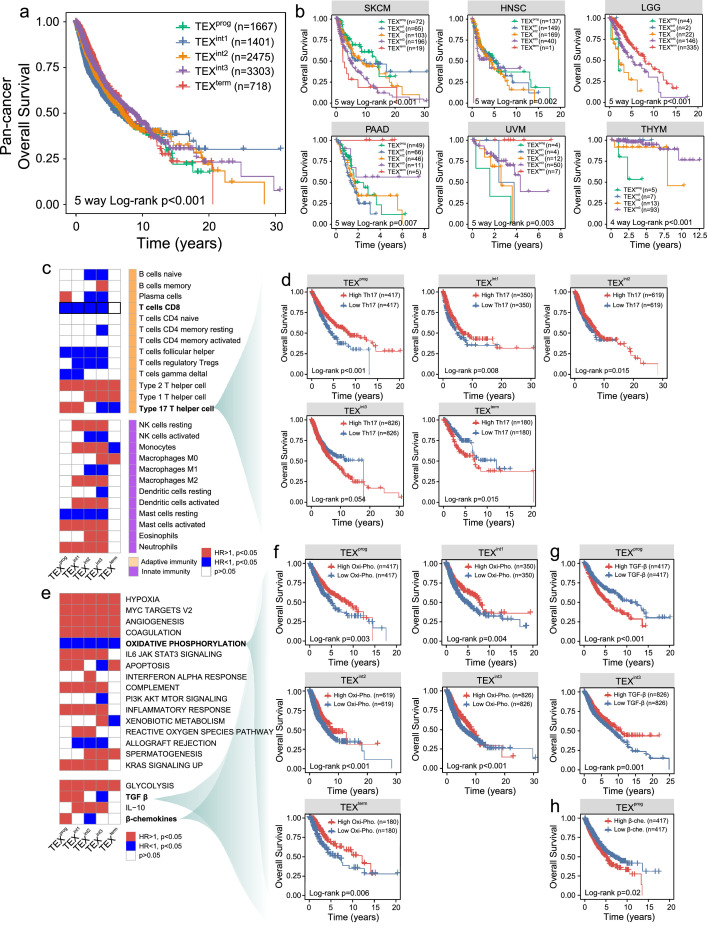
To quantify the degree of TEX in 9,564 bulk tumor samples from 30 solid tumor types in The Cancer Genome Atlas (TCGA), we built a compendium of TEX-specific pathways, including tumor necrosis factor (TNF), interleukin (IL)-2, interferon-γ (IFN-γ) and cytotoxic pathway, through literature mining. We then used single-sample gene set enrichment analysis (ssGSEA) to deconstruct the active TEX-specific pathway profiles from the RNA-sequencing data of individual bulk tumor tissue samples (Supplementary file 1). Hierarchical clustering of the active TEX-specific pathway profiles revealed five main TEX-driven pan-cancer clusters, referred to here as C1-C5 (with 1667, 1401, 2475, 3303 and 718 cases, respectively; Figure 1a). These five resultant patient clusters, named TEX subgroups thereafter, can be characterized using a progressive activity profile of TEX-specific pathways and other molecular pathways implicated in TEX, such as transforming growth factor β (TGF-β), IL-10, glycolysis and chemokine pathways (Figure 1a). We next sought to examine the relationship among the TEX subgroups and their immune cytolytic activity (CYT). On progression from TEX subgroups C1 to C5, the CYT scores were found to be significantly and progressively reduced, with the abundance of infiltrating T lymphocytes, such as CD8+ T cells, Th1, Th2 and Th 17 CD4+ T cells, also decreasing at each advancing stage of TEX from C1 to C5 (Figure 1b). A previous study reported that transcription factors TCF1, T-bet and thymocyte selection-associated high mobility group box can all coordinate the dynamics underlying TEX subset transitions.18 In the present study, we observed significant differences in the expression pattern of TCF1 and T-bet with a decreasing trend among the five TEX subgroups and TOX with a decreasing trend from TEX C1 to C4 but highest in C5 (Figure 1c). Additionally, T cell antigen receptor (TCR) signals and B cell receptor (BCR) signals were progressively increased at each stage of TEX, which has been previously found to be a primary driver of TEX (Figure 1d). Collectively, these results suggest that the five TEX subgroup derived from the present study can accurately represent the biological features of the hierarchical stages of TEX (Figure 1e). Each of these five stages is referred to as ‘TEXprog’, ‘TEXint1’, ‘TEXint2’, ‘TEXint3’ and ‘TEXterm’ hereafter.
Figure 1.
Pan-cancer characterization of TEX heterogeneity within the tumor microenvironment. (a) TEX-specific pathway-based hierarchical clustering of pan-cancer samples taken from TCGA was used to produce five distinct TEX subgroups. The heatmap shows the normalized activity scores (ssGSEA) of four TEX-specific pathways and other molecular pathways implicated in TEX. Curves in the middle represent the distribution of activity scores in the five TEX subgroups, with dashed lines displaying the median. (b) Boxplots showing differences in the immune CYT scores, abundance of infiltrating T lymphocytes of various subsets, (c) expression of TCF1, T-bet, TOX, and (d) TCR and BCR signals among the five TEX subgroups. (e) Schematic description of the features associated with the five pan-cancer TEX subgroups. (f) Bar charts showing the distribution of the five TEX subgroups among the different tumor types. Each bar represents the relative composition of each TEX subgroup (columns) in each cancer type (row). (g) Bar charts showing the distribution of tumor types among the five TEX subgroups. Each bar represents the relative composition of each cancer type (rows) within each TEX subgroup (columns). (h) Overlay of the five TEX subgroups with existing TCGA molecular subgroups. Each row indicated the distribution of TEX subgroups within each molecular subtype. Red revealed a higher proportion, whereas blue revealed a lower proportion. TEX, T cell exhaustion; TCGA, The Cancer Genome Atlas; TCR, T cell receptor; CYT, cytolytic activity; TOX, thymocyte selection-associated high mobility group box; BCR, B cell receptor.
Further examination of the TEX subgroups in relation to tumor types and existing molecular subgroups revealed that although the TEX subgroups spanned various tumor types and anatomical locations, diverse TCGA subgroups and individual solid tumor types varied substantially in their composition of TEX subgroups (Figure 1f-g). The TEXprog and TEXint1 subgroups were particularly enriched in cervical cancer (CESC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), head and neck squamous cell carcinoma (HNSC), lung squamous cell carcinoma (LUSC) and stomach adenocarcinoma (STAD). The TEXint2 subgroup was found to be enriched in breast invasive carcinoma (BRCA), colon adenocarcinoma and HNSC. In addition, the TEXint3 subgroup was enriched in particular subgroups of BRCA, ovarian serous cystadenocarcinoma (OV), prostate adenocarcinoma (PRAD), thymoma (THYM), Kidney Chromophobe (KICH), skin cutaneous melanoma (SKCM) and uterine corpus endometrial carcinoma. The TEXterm subgroup, which represents a state of physical deletion and terminal differentiation, was mainly found in lower-grade gliomas (LGG), Uterine Carcinosarcoma (UCS), OV, PRAD and adrenocortical carcinoma (ACC). Additionally, the CpG island methylator phenotype (CIMP)-low, CIMP-intermediate, CIMP-high subtypes of GBM, LGG and ACC, pilocytic astrocytoma-like subgroups of LGG and GBM were found to be especially enriched of the TEXterm subgroup (Figure 1h). To conclude, these five TEX subgroups represented a dynamic roadmap of hierarchical dysfunction underlying the TEX process, which could be applied to transect traditional cancer clinicopathological classifications to reveal more informative immunological subgroups.
Impact of the hierarchical dysfunction heterogeneity of TEX on cancer prognosis
To examine the effects of hierarchical dysfunction of TEX on clinical outcome, the prognostic value of the TEX subgroups was assessed on a pan-cancer level and within each of the individual cancer types. Pan-cancer survival analysis showed that tumors with different TEX subgroups demonstrated significantly different overall survival (OS) rates (Log-rank test P<0.001; Figure 2a). In addition, the association between the TEX subgroups and OS remained significant even after division into specific subgroups according to different clinical features (Figure S1). Cancer type-specific survival analysis also revealed significant associations between the TEX subgroups and OS in a number of cancer types. As shown in Figure 2b, patients with the TEXprog subgroups have significantly improved survival rates compared with those with other TEX subgroups in SKCM and HNSC (Log-rank test P<0.001 for SKCM and Log-rank test P=0.002 for HNSC). However, in other cancer types, the opposite trend was observed. The TEXterm subgroup was associated with a superior prognosis, whereas TEXprog appeared to be associated with a poor prognosis in LGG, pancreatic adenocarcinoma (PAAD), uveal melanoma (UVM) and THYM (Figure 2b).
Figure 2.
Impact of hierarchical dysfunction heterogeneity during TEX on clinical outcome. The association between each TEX subgroup and OS in (a) pan-cancer and (b) Kaplan-Meier survival plot in several representative tumor types. (c) HRs representing the association between the 25 known immune cell types and OS in each TEX subgroup. Red showed higher risk, blue showed lower risk, whereas white showed no association with risk. (d) Kaplan-Meier curves of OS among the five TEX groups stratified by the degree of type17 T helper cell infiltration. (e) HRs representing the association between the enrichment scores (ssGSEA) of signature hallmark genes and OS in each TEX subgroup. Red showed higher risk, blue showed lower risk, whereas white showed no association with risk. Kaplan-Meier curves of OS among the five TEX subgroups stratified by (f) oxidative phosphorylation, (g) TGF-β and (h) β-chemokines. The P-values were estimated by log-rank test. TEX, T cell exhaustion; OS, overall survival; TCF1, T-cell factor 1; HR hazard ratio; TGF-β, transforming growth factor β.
We then examined the impact of the potential interplay between the TEX subgroups and infiltrating immune cells on patient prognosis. As shown in Figure 2c, most of the infiltrating immune cells were either positively or negatively associated with survival for the different TEX subgroups except for the TEXterm subgroup. A total of five immune cell subpopulations (Th1, Th2, Th17, monocytes and M0 macrophages) were found to be significantly associated with survival within the TEXterm subgroup. In particular, several immune cell subpopulations (plasma cell, Th17 and monocytes) appeared to display dual prognostic roles. For example, high infiltration of Th17 cells was found to be associated with improved survival for the TEXprog, TEXint1 and TEXint2 subgroups, but was associated with poor survival for the TEX int3 and TEXterm subgroups (Figure 2d).
Finally, we explored the impact of the interplay between the TEX subgroups and hallmark cancer-associated signaling pathways on patient prognosis. As shown in Figure 2e, the crosstalk between the TEX subgroups and hallmark cancer signaling pathways can mediate significant effects on the prognosis of patients with cancer. The majority of the cancer-associated pathways tested in the present study demonstrated a significant negative association with survival, whereas only three pathways [oxidative phosphorylation, phosphoinositide-3 kinase/protein kinase B/mammalian target of rapamycin signaling and allograft rejection] were revealed to associate positively with survival significantly. For example, patients with highly active oxidative phosphorylation pathways tended to exhibit a superior prognosis for the different TEX subgroups (Figure 2f).
This apparent dual prognostic function was also found for several other signaling pathways, including apoptosis, xenobiotic metabolism, TGF-β and β-chemokines. For example, patients with highly active TGF-β and β-chemokine pathways tended to have a poorer prognosis in the TEXprog subgroup (Figure 2g and h). To conclude, these results suggest that heterogeneity in the hierarchical dysfunction of TEX may impact cancer prognosis, which can be exploited as a viable predictor of outcomes in patients with cancer.
TEX-based subgroups show distinct immune microenvironments
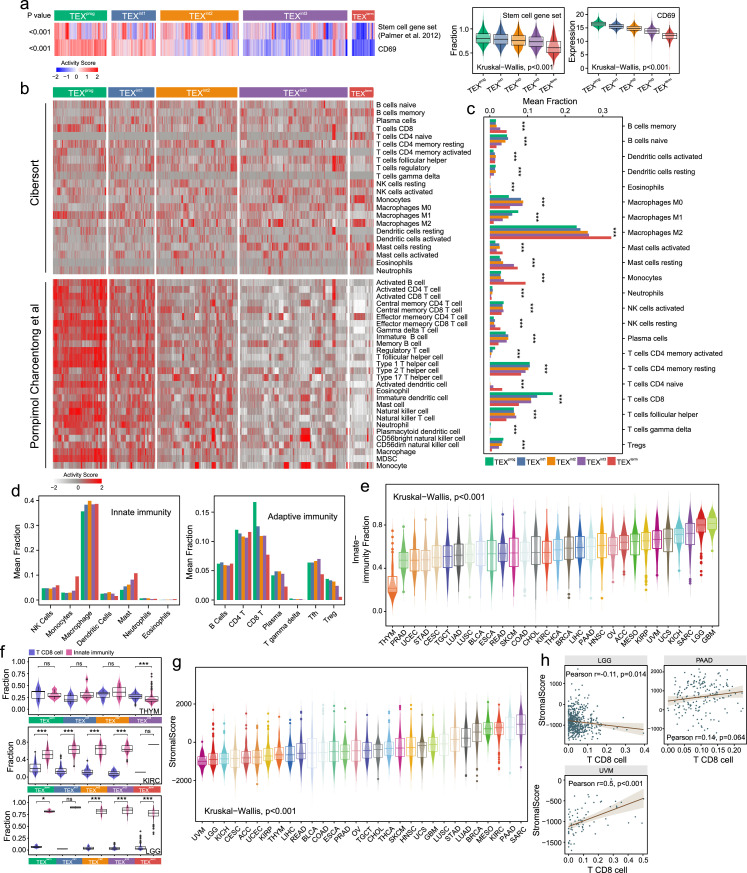
To investigate the stem cell-like properties in each of the five TEX subsets of CD8+ T cells during the exhaustion process, stemness cell signature gene enrichment scores38 and CD69 expression levels were used to estimate their progenitor-like phenotypes and proliferative capacities in individual patients. We found a dynamic decreasing in the gene enrichment scores of stemness cell signature and CD69 expression levels from TEXprog to TEXterm, suggesting that this hierarchical dysfunction occurring during TEX is associated with reduced progenitor-like characteristics and proliferative capacity (Figure 3a).
Figure 3.
Distinct immune microenvironments in the TEX-based pan-cancer subgroups. (a) Heatmap and Box plots showing the stemness cell signature enrichment scores (ssGSEA) and CD69 expression levels among the five TEX subgroups. (b) Heatmap showing the profiles of infiltrating immune cell types from the CIBERSOTR or Charoentong et al. pan-cancer datasets in the five TEX groups. (c) Bar charts showing the proportion of the 22 immune cell types from the CIBERSORT dataset among the five TEX subgroups. (d) The bar chart in (c) was sub-divided into those belonging to innate immunity and adaptive immunity. (e) Box plots displaying the innate immunity cell fraction among the tumor types, as ordered using their median values. (f) Box plots showing the comparison in the fraction between CD8+ T cells and cells of innate immunity within the five TEX groups in three particular cancer types. (g) Box plots displaying the stromal scores (ESTIMATE) among the tumor types were ordered using their median values. (h) Correlation between the stromal scores and the fraction of CD8+ T cells in three selected cancer types. TEX, T cell exhaustion; THYM, thymoma; KIRC, kidney renal clear cell carcinoma; LGG, low-grade glioma; PAAD, pancreatic adenocarcinoma; UVM, uveal melanoma.
Differences among the TEX subgroups distinguished were next examined in accordance with immune cell types using the CIBERSORT cell deconvolution method. Previously reported signatures from Charoentong et al.34 were also applied to verify the detection results for the composition of immune cells among the TEX groups using the ssGSEA method (Figure 3b). ssGSEA analysis revealed that most of the immune cell types in the immune microenvironment exhibited states of decline from immune-enriched to immune-depletion as the hierarchical dysfunction from TEXprog to TEXterm progresses (Figure 3b). As shown in Figure 3c, the top three categories of immune cell types included those from innate and adaptive immunity. They were M2 macrophages, CD8+ T cells and resting CD4+ memory T cells. TEXprog had the highest fractions of CD8+ T cells, activated CD4+ memory T cells, and the lowest fraction of M2 macrophages. The composition of the tumor-promoting M2 phenotype and mast cells (Figure 3c and d) displayed a greater range in TEXterm compared with that in other TEX groups.
Innate-immunity fraction (IF) varied substantially not only among the TEX subgroups, but also among the tumor types. Tumors in the bottom 33% of the IF included tumors most responsive to immune checkpoint inhibitors (ICI), such as LUSC, LUAD, bladder cancer and in particular, LUAD.6 subgroup, LUSC of the secretory subgroup and the BLCA.4 subgroup (Kruskal−Wallis test P<0.001; Figure 3e). LGG and glioblastoma multiforme had higher IF than other tumor types, indicating the absence or presence of microglia in these TIMEs (Figure 3e). In three representative cancer types ranking top, bottom and middle in terms of the IF, named LGG, THYM and KIRC, respectively, the fraction between innate immunity and CD8+ T cells was significantly different in each TEX sub-group (Figure 3f).
The stromal score varied among the tumor types, ranging from stroma-rich tumors, including sarcoma, PAAD and KIRC, to stroma-poor tumors, such as LGG and UVM (Kruskal−Wallis test P<0.001; Figure 3g). In LGG, the stromal score displayed a negative correlation with the quantity of CD8+ T cells (Pearson test r=−0.11; P=0.014), whereas the opposite trend was revealed in PAAD (Pearson test r=0.14; P=0.064) and UVM (Pearson test r=0.5; P<0.001; Figure 3h).
Multi-omic alterations in the pan-cancer subgroups according to TEX classification
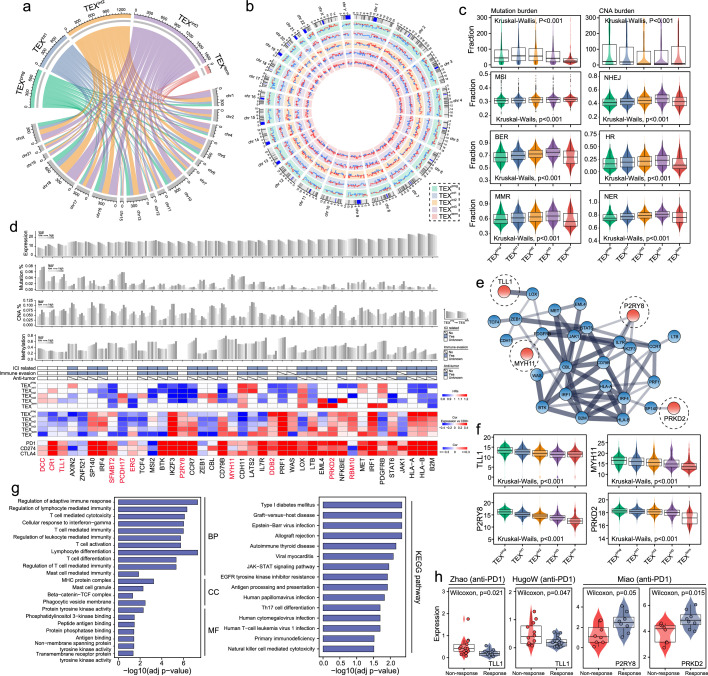
To investigate the molecular features underlying hierarchical dysfunction occurring during TEX, we performed a comparative analysis of the genomic variation, gene expression and DNA methylation profiles among the five TEX subgroups. By comparing genomic alterations, we observed that the TEXterm subset had a lower mutation load, copy number alteration (CNA) burden and genomic instability (Figure 4a-c). These results revealed genomic alterations during hierarchical dysfunction leading up to TEX.
Figure 4.
Featured multi-omic alterations in TEX-based pan-cancer subgroups. (a-b) Circular plots showing the differences in single-nucleotide polymorphism and copy number variations among the five TEX subgroups. (c) Box plots displaying the differences in mutation burden, CNA burden, MSI, BER, MMR, NHEJ, HR and NER among the five TEX subgroups. (d) Gene-level summary of the multi-omics features in the pan-cancer samples. Bar plots show the expression, somatic mutation alterations, DNA copy number alterations and DNA methylation of a list of crucial TEX-associated driver genes. Light gray represents low levels, whereas dark gray represents high levels. Heatmap shows the hazard ratios of key TEX-associated driver genes, the correlation between the expression of these TEX-related driver genes and the fraction of CD8+ T cells, PD-1, PD-L1 and CTLA-4 in the five TEX subgroups. (e) The protein–protein interaction network is formed using the key TEX-associated driver genes. (f) Box plots displaying the expression levels of TLL1, MYH11, P2RY8 and PRKD2 among the five TEX subgroups. (g) Bar charts showing enriched gene ontology terms and KEGG pathways. (h) Box plots showing the expression levels of TLL1, P2RY8 and PRKD2 in responders and non-responders following immunotherapy in the datasets indicated. TEX, T cell exhaustion; CNA, copy number alterations; MSI, microsatellite instable; BER, base excision repair; MMR, mismatch repair; NHEJ, non-homologous end joining; HR, homologous recombination; NER, nucleotide excision repair; PD-1, programmed cell death protein 1; PD-L1, programmed cell death protein ligand 1; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; TLL1, tolloid 1; MYH11, myosin heavy chain 11; P2RY8, P2Y receptor family member 8; PRKD2, protein kinase 2; KEGG, Kyoto Encyclopedia of Genes and Genomes.
We next probed the expression profiles of 568 cancer driver genes in the TEX subgroups, which identified 40 cancer driver genes associated with TEX [false-discovery rate (FDR)-adjusted P<0.01; Figure S2] (Supplementary file 2). Among them, 31 of the 40 TEX-related cancer driver genes showed progressively decreasing expression levels from TEXprog to TEXterm, which is coupled with increasing frequencies of mutation, CNA and DNA methylation. Simultaneously, 22 of the 40 TEX-related cancer driver genes were found to be associated with the ICI, 26 genes were closely related to immune evasion and three genes were associated with anti-tumor immunity (Figure 4d). Furthermore, expression levels of these TEX-related cancer driver genes provided additional prognostic value in the individual TEX subgroups. They were found to be significantly associated with the quantity of CD8+ T cells and the expression of immune checkpoint genes, such as PD1, PD-L1 and CTLA4; Figure 4d).
Protein-protein interaction (PPI) network analysis of the 40 TEX-related cancer driver genes identified a TEX-associated subnetwork consisting of 28 TEX-related cancer driver genes. They include tolloid-like 1 (TLL1), myosin heavy chain 11 (MYH11), P2Y receptor family member 8 (P2RY8) and protein kinase D2 (PRKD2), the roles of which in cancer immunotherapy and immune response regulation remain unknown (Figure 4e). These four genes displayed significantly different expression patterns among five groups whilst also being expressed at progressively decreasing levels during hierarchical dysfunction when TEX occurs (Figure 4f). Functional enrichment analysis of gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) showed that this TEX-associated sub-network was associated with T cell-mediated cytotoxicity, T cell activation and T cell differentiation in addition to viral infection (Figure 4g).
Apart from the broad multi-omic changes found among the five TEX subgroups, downregulation of inhibitory receptor expression was also found during the TEX process, such as PD-1, PDL1 and CTLA4 (Kruskal−Wallis test P<0.001; Figure S3). Therefore, we examined the roles of the four TEX-associated genes found in the present study in mediating the response to anti-PD-1 immunotherapy. Significantly different expression levels for TLL1 with the highest expression in the non-responder group, but for P2RY8 and PRKD2 with the highest expression in the responder group (Figure 4h), suggesting that these genes can regulate the anti-tumor immune response. Taken together, our findings revealed the multi-omic profiles of TEX-related driver genes underlying the hierarchical dysfunction in CD8+ T cells. In addition, the effect of TEX-related cancer driver genes on patient prognosis was found to be specific among the TEX subgroups. However, the TEX-related cancer driver genes mediated inconsistent effects on the tumor immune response.
A gene signature for modeling the hierarchical TEX stages and predicting immunotherapy efficacy
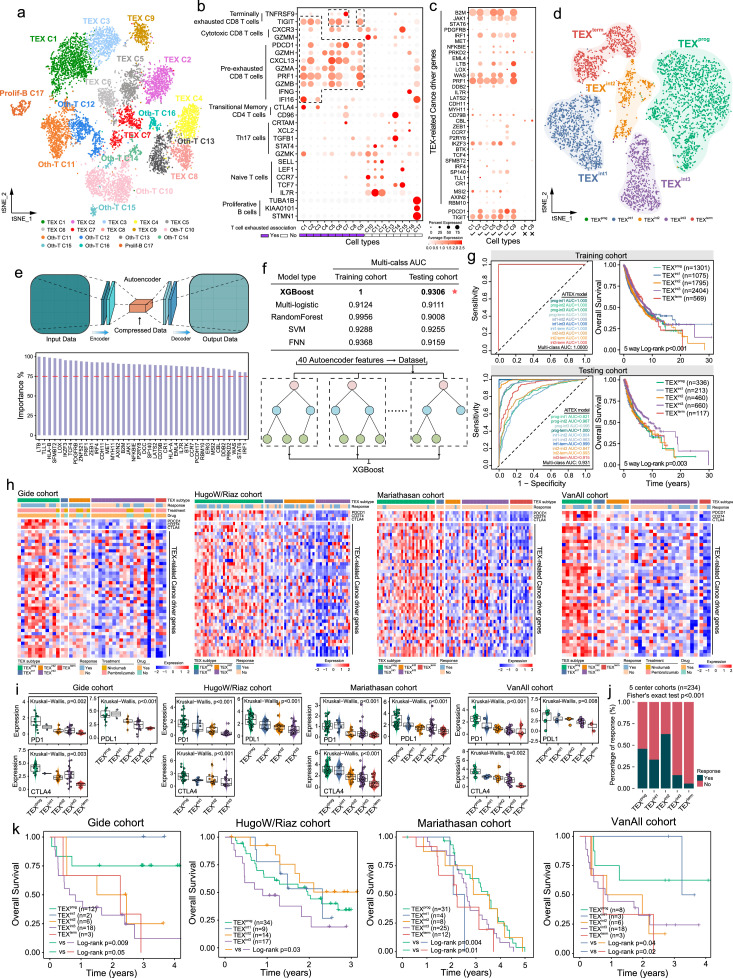
To further explore the association between the 40 TEX-related cancer driver genes identified and the five TEX stages at single-cell resolution, we analyzed the single-cell transcriptomic data of immune cells derived from three patients with melanoma. After quality control, batch effect correction and normalization, 10,518 immune cells were clustered into 17 major immune cell clusters using tSNE. Using established TEX marker genes to annotate the cell types such as PD-1, granzyme A (GZMA), granzyme B (GZMB), C-X-C motif chemokine ligand 13 (CXCL13) and so on, nine of the 17 major immune cell clusters were identified to be associated with TEX, termed TEX-C1-C9; Figure 5a-c). Expression profiles of the TEX-related cancer driver genes displayed hierarchical and differential compositions in these TEX-related cell types (Figure 5c), which were applied to categorize these TEX-related cell types into five TEX subgroups (Figure 5d). A significant and dynamic decline in TEX-related gene expression from TEXprog to TEXterm was observed (Figure S4). These results demonstrated the viability of using TEX-related cancer deriver genes to model hierarchical dysfunction during TEX in the TIME not only on bulk tissue levels but also on a single-cell level. Subsequently, a deep neural network 'autoencoder' was used to evaluate the importance of each TEX-related cancer driver gene. The significance of each TEX-related cancer driver gene surpassed 75% according to autoencoder, implying their indispensability in defining the TEX subgroups (Figure 5e).
Figure 5.
Identification and evaluation of the machine learning-based gene signature for modeling TEX and predicting immunotherapy efficacy. (a) t-SNE plot of 10,518 single melanoma cells sorted into their corresponding colored immune clusters. (b) Bubble heatmap showing expression levels of the selected signature genes and established cell markers. (c) Bubble heatmap showing expression profiles of key TEX-associated driver genes. (d) t-SNE plot of single cells isolated from the exhausted cell types sorted into their corresponding color TEX clusters. (e) The Autoencoder architecture was used to calculate the importance of the key TEX-related driver genes. (f) Comparison of multi-class AUCs calculated using five individual machine learning models in the training and testing cohorts. (g) Receiver operating characteristics of the ML-TEX model in the training and testing cohorts. (h) Heatmaps showing the expression pattern of critical TEX-associated driver genes among the five TEX subgroups as classified using the ML-TEX model. (i) Box plots showing the expression of PD-1, PD-L1 and CTLA-4 in the five TEX subgroups in each of the four immunotherapy dataset cohorts. (j) Proportion of responders in each TEX cluster. (k) Kaplan-Meier survival analysis of TEX subgroups in the Gide, HugoW/Riaz, Mariathasan and VanAll cohorts. TEX, T cell exhaustion; t-SNE, t-distributed stochastic neighbor embedding; AUC, area under the curve; SVM, support vector machine; FNN, feedforward neural network. ML, machine learning; CTLA-4, cytotoxic T-lymphocyte-associated protein 4; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1.
The pan-cancer data was randomly compartmentalized into the training and test cohorts using a ratio of 8:2 for each cancer type. We then developed a machine-learning (ML)-based predictive model, referred to as ML-TEX, to infer the hierarchical TEX stages in bulk tumor samples using the 40 TEX-related driver genes and state-of-the-art models, including XGBoost, multi-logistic, random forest, support vector machine and feedforward neural network, in the training cohort (Figure 5f). Finally, ML-TEX based on XGBoost was found to achieve superior performance for predicting the hierarchical TEX stages in the training cohort, with an area under the curve (AUC) of 1.0 and an AUC of 0.804–1.0 in the test cohort (Figure 5g). Furthermore, there were significant differences in the OS of patients with different predicted severity levels of TEX (Figure 5g), suggesting that ML-TEX can be used to predict pan-cancer survival.
To determine if ML-TEX can be utilized to predict response to immune checkpoint blockade (ICB) therapy, we applied ML-TEX to the different immunotherapy cohorts and classified the patients receiving ICB therapy into different TEX subgroups. As shown in Figure 5h, TEX-related cancer deriver genes exhibited a continuous spectrum of transcriptional states in patients in the different TEX subgroups, which were also associated with the severity levels of TEX. Specifically, there were progressively decreasing expression levels of PD-1, PD-L1 and CTLA4 during the transition from TEXprog to TEXterm (Figure 5i). The percentage of responders to ICB therapy differed significantly among the different TEX subgroups, with the highest percentage of responders in TEXint2 and a decreasing trend of responder percentage in the other four TEX groups from TEXprog to TEXterm (Fisher's exact test P<0.001; Figure 5j). Patients with early stages of TEX were associated with higher rates of favorable responses than those with later stages of TEX. We subsequently evaluated clinical outcomes of ICB therapy in each of the five TEX subgroups and found that in groups showing responses, specifically TEXprog, TEXint1 and TEXint2, longer OS was observed compared with that in the non-responder groups, namely TEXint3 and TEXterm (Log-rank test P=0.05 for Gide cohort; P=0.03 for the HugoW/Riaz cohort; P=0.01 for the Mariathasan cohort and P=0.02 for the VanAll cohort; Figure 5k). Taken together, these results suggested a promising role of ML-TEX as a beneficial tool for predicting responsiveness and survival of patients following ICB treatment.
Discussion
Accumulating evidence suggests that TEX results from a sustained state of hierarchical dysfunction in T cells, which entails a continuum of phenotypic and intermediate functional states, similar to other cellular differentiation processes.4,6,16 Therefore, understanding CD8+T cell dysregulation and exhaustion within the TIME within the same cancer type or in different cancer types is key to overcoming the TEX barrier and improving immune checkpoint blockade therapies in the clinic.47 However, a detailed characterization of TEX dynamics and heterogeneity within the TIME across various cancers is lacking. In the present study, we comprehensively characterized the heterogeneous TEX subset landscape within the TIME of 9564 tumor samples among 30 bulk solid cancer types using a five-stage trajectory as measured using literature-derived, TEX-specific hierarchical developmental signaling pathway signatures. Although expression of several inhibitory receptors, such as TCF1, TOX, T-bet and PD-1, have been reported to define TEX,11,18,48,49 they are not definitive features of exhausted T cells as many highly functional effector T cells also express inhibitory receptors.6 Furthermore, TEX develops with a continuum of phenotypic and functional intermediate states.6,50 Therefore, the single gene-defined dichotomy for TEX remained unsatisfactory, limited power for bulk tissue RNA-seq data, and lacked quantitative indicators of TEX progressive and hierarchical process. Our pathway-based results demonstrated the extent of TEX heterogeneity in many different cancer types, verifying the notion that TEX heterogeneity exists among patients with the same cancer type and patients with other tumor types.
Our study characterized the distribution of heterogeneous TEX subsets within the TIME among the different cancer types and identified five de novo pan-cancer immune subgroups characterized by distinct TEX subsets, which represent another TEX-derived system that can be used for the immune sub-typing of cancers. The TEXprog subgroup was found to be particularly enriched with TCF and T-bet in their constituent tumors and conferred the best prognosis in SKCM, with higher CD8+ T cell quantities and lower degrees of M2 macrophage infiltration. However, a worse prognosis was found in each transcriptional factors (TF) subset. By contrast, the TEXterm subgroup, which was mainly lacking in TCF and T-bet expression, had the most favorable prognosis in terms of LGG, PAAD and UVM. In addition, TEXint1, TEXint2 and TEXint3 demonstrated intermediate distribution among the TF+ and TF- subsets, which further revealed the pattern of dynamic dysfunction occurring during T cell exhaustion in the intermediate active TF subsets, with high M0 macrophage content. The TEXprog subgroup displayed the highest stemness signature score and proliferative capacity, whereas the TEXterm subgroup exhibited the lowest among the five subsets. We further investigated the potential crosstalk among the TEX subgroups, intrinsic molecular subgroups and hallmark cancer signaling pathways, before analyzing their effects on patient prognosis. It was found that this system of TEX subtyping enabled the evaluation of differences in OS among patients with cancer exhibiting varying degrees of TEX within the TIME.
Increasing evidence shows that TEX heterogeneity can be regulated by the TIME.16,51 The main components, such as stromal and inflammatory cells in the TIME play important roles in the TEX development.52 For example, a recent study reported that targeting sialylation in tumor stromal cells could reverse TEX.53 Although recent multi-omics analyses also revealed distinct transcriptional, epigenetic and metabolic characteristics in exhausted T cells, namely functional effector or memory T cells,18,49,54,55 a more comprehensive understanding of the molecular signature underlying TEX heterogeneity within the TIME remained elusive. The possible impact of copy number alteration, somatic alteration and DNA methylation on T cell exhaustion was also assessed in this study. Mutational burden, CNA and methylation in the TEX-related driver genes were abundant in the TEXterm subset but infrequent in the TEXprog subgroup. These mutations in the TEX-related driver genes may alter the immune cell landscape in the tumor through the production of neoantigens. Some TEX-associated genes identified in our study have recently been reported and validated to be involved in T cell function and exhaustion by experiments. Our analysis also constructed a TEX-associated PPI subnetwork consisting of 28 cancer driver genes. Most components in this network have already been previously documented, apart from TLL1, MYH11, P2RY8 and PRKD2. Since the roles in regulating the mechanism of TEX within the TIME remain unknown, additional in vitro and in vivo experiments are needed for validation.
Immunotherapy has demonstrated great potential as a novel clinical strategy for cancer treatment.56 However, responses to immunotherapy typically differ among individuals with the same type of cancer, whereas the circumstances also vary among the different cancer types. Given the association between the TEX subgroups and oncogenic states, the clinicopathological parameters among the TEX subsets in response to tumor immunotherapy were investigated further. An association was revealed between the TEX subgroups identified using machine-learning and response profiles of anti-PD1 immunotherapy for melanoma. In addition, this TEX-derived gene signature displayed an accurate predictive capability compared with traditional immune checkpoint markers (such as PD-1) in both independent and external immunotherapeutic datasets. These findings further demonstrated the robustness and potential of this machine-learning model for the characterization of dynamic dysfunction underlying TEX and prediction of responses to tumor immunotherapy.
However, several limitations exist and should be noted. Firstly, further molecular experiments should be conducted to investigate the functional roles of other candidates identified in the TEX. Second, we only tested and compared several commonly used machine learning methods to infer the hierarchical TEX stages in bulk tumor samples. The ensemble method may be an alternative approach and should be tested in the future. Thirdly, considering the complex interplay between T cell exhaustion and other biological traits across different malignancies and studies, the proposed TEX-derived predictor of immunotherapy efficacy, ML-TEX, should be tested and validated in prospective studies and more cancer types. Meanwhile, the classification of TEX stages in our research was for bulk RNA-seq data, and it was quite necessary to identify whether the expression of TEX-associated genes particularly TLL1, P2RY8, MYH11 and PRKD2 were transformed between non-response and response groups in single-cell RNA-seq data after ICB treatment.
In summary, our study characterized the overall pattern of heterogeneous TEX subset distribution within the TIME among different types of cancer while revealing the preferences and dependencies of the TEX subgroups in various tumor types and among the same tumor types in other individuals. A TEX-based immunotyping scheme was proposed, which was used to predict prognosis, profile the intrinsic molecular subtype distribution, characterize the immune microenvironment and reveal various multiple omics features, to expand upon previously reported systems for subtyping cancers. Furthermore, the potential underlying genomic, transcriptional and epigenetic mechanisms involved in mediating TEX heterogeneity were preliminarily investigated. The comprehensive characterization of these heterogeneous TEX subsets within the TIME provides insights into cancer immunity and may have important implications for improving personalized cancer immunotherapy by targeting T cells.
Contributors
Zicheng Zhang: Data curation, Formal analysis, Methodology, Writing-original draft, Visualization, Implementation. Lu Chen: Data curation, Formal analysis, Methodology. Hongyan Chen: Formal analysis. Jingting Zhao: Formal analysis. Ke Li: Formal analysis. Jie Sun: Conceptualization, Funding acquisition, Supervision. Meng Zhou: Conceptualization, Funding acquisition, Project administration, Writing - review & editing. Zicheng Zhang and Lu Chen have verified the underlying data. All authors read and approved the final version of the manuscript.
Data sharing statement
The datasets used and/or analyzed during the present study are available from the UCSC Xena Database (https://xenabrowser.net/datapages/) and 10X single-cell RNA-seq data of three samples of advanced melanoma are available from the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE159251). (GSE159251).
Declaration of interests
The authors declare that they have no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant No. 62072341 and 61973240). The funders had no roles in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104207.
Contributor Information
Jie Sun, Email: suncarajie@wmu.edu.cn.
Meng Zhou, Email: zhoumeng@wmu.edu.cn.
Appendix. Supplementary materials
Supplementary file 1. The active TEX-specific pathway profiles from the RNA-sequencing data of individual bulk tumor tissue samples.
Supplementary file 2. List of TEX-associated genes identified in this study.
References
- 1.Zhang N, Bevan MJ. CD8(+) T cells: foot soldiers of the immune system. Immunity. 2011;35(2):161–168. doi: 10.1016/j.immuni.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Speiser DE, Utzschneider DT, Oberle SG, Munz C, Romero P, Zehn D. T cell differentiation in chronic infection and cancer: functional adaptation or exhaustion? Nat Rev Immunol. 2014;14(11):768–774. doi: 10.1038/nri3740. [DOI] [PubMed] [Google Scholar]
- 3.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T cell exhaustion during chronic viral infection and cancer. Annu Rev Immunol. 2019;37:457–495. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 4.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12(6):492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 5.Wherry EJ, Kurachi M. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015;15(8):486–499. doi: 10.1038/nri3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blank CU, Haining WN, Held W, et al. Defining 'T cell exhaustion'. Nat Rev Immunol. 2019;19(11):665–674. doi: 10.1038/s41577-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Im SJ, Hashimoto M, Gerner MY, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Utzschneider DT, Charmoy M, Chennupati V, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 2016;45(2):415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Siddiqui I, Schaeuble K, Chennupati V, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T Cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50(1):195–211.e10. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Akbar AN, Henson SM. Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nat Rev Immunol. 2011;11(4):289–295. doi: 10.1038/nri2959. [DOI] [PubMed] [Google Scholar]
- 11.Khan O, Giles JR, McDonald S, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature. 2019;571(7764):211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philip M, Fairchild L, Sun L, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545(7655):452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagaev A, Kotlov N, Nomie K, et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39(6):845–865.e7. doi: 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Crespo J, Sun H, Welling TH, Tian Z, Zou W. T cell anergy, exhaustion, senescence, and stemness in the tumor microenvironment. Curr Opin Immunol. 2013;25(2):214–221. doi: 10.1016/j.coi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Y, Li Y, Zhu B. T-cell exhaustion in the tumor microenvironment. Cell Death Dis. 2015;6:e1792. doi: 10.1038/cddis.2015.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai MC, Zhao X, Cao M, et al. T-cell exhaustion interrelates with immune cytolytic activity to shape the inflamed tumor microenvironment. J Pathol. 2020;251(2):147–159. doi: 10.1002/path.5435. [DOI] [PubMed] [Google Scholar]
- 18.Beltra JC, Manne S, Abdel-Hakeem MS, et al. Developmental relationships of four exhausted CD8(+) T cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity. 2020;52(5):825–841.e8. doi: 10.1016/j.immuni.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barsch M, Salie H, Schlaak AE, et al. T-cell exhaustion and residency dynamics inform clinical outcomes in hepatocellular carcinoma. J Hepatol. 2022;77(2):397–409. doi: 10.1016/j.jhep.2022.02.032. In this issue. [DOI] [PubMed] [Google Scholar]
- 20.Ma J, Zheng B, Goswami S, et al. PD1(Hi) CD8(+) T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J Immunother Cancer. 2019;7(1):331. doi: 10.1186/s40425-019-0814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonneville R, Krook MA, Kautto EA, et al. Landscape of microsatellite instability across 39 cancer types. JCO Precis Oncol. 2017;2017 doi: 10.1200/PO.17.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity. 2018;48(4):812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pauken KE, Shahid O, Lagattuta KA, et al. Single-cell analyses identify circulating anti-tumor CD8 T cells and markers for their enrichment. J Exp Med. 2021;218(4) doi: 10.1084/jem.20200920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gide TN, Quek C, Menzies AM, et al. Distinct immune cell populations define response to anti-PD-1 monotherapy and anti-PD-1/anti-CTLA-4 combined therapy. Cancer Cell. 2019;35(2):238–255.e6. doi: 10.1016/j.ccell.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171(4):934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, Chen AX, Gartrell RD, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mariathasan S, Turley SJ, Nickles D, et al. TGFbeta attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554(7693):544–548. doi: 10.1038/nature25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charoentong P, Finotello F, Angelova M, et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 35.Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259. doi: 10.1007/978-1-4939-7493-1_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Bao S, Yan C, Hou P, Zhou M, Sun J. Computational principles and practice for decoding immune contexture in the tumor microenvironment. Brief Bioinform. 2021;22(3) doi: 10.1093/bib/bbaa07. In this issue. [DOI] [PubMed] [Google Scholar]
- 38.Miranda A, Hamilton PT, Zhang AW, et al. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc Natl Acad Sci USA. 2019;116(18):9020–9029. doi: 10.1073/pnas.1818210116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Jimenez F, Muinos F, Sentis I, et al. A compendium of mutational cancer driver genes. Nat Rev Cancer. 2020;20(10):555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 41.Munir K, Elahi H, Ayub A, Frezza F, Rizzi A. Cancer diagnosis using deep learning: a bibliographic review. Cancers (Basel) 2019;11(9) doi: 10.3390/cancers11091235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24(6):1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Zhang QY, Sun BF, et al. Single-cell transcriptome profiling of the vaginal wall in women with severe anterior vaginal prolapse. Nat Commun. 2021;12(1):87. doi: 10.1038/s41467-020-20358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu T, Hu E, Xu S, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (NY) 2021;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtulus S, Madi A, Escobar G, et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1(-)CD8(+) tumor-infiltrating T cells. Immunity. 2019;50(1):181–194.e6. doi: 10.1016/j.immuni.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Utzschneider DT, Gabriel SS, Chisanga D, et al. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat Immunol. 2020;21(10):1256–1266. doi: 10.1038/s41590-020-0760-z. [DOI] [PubMed] [Google Scholar]
- 49.Jadhav RR, Im SJ, Hu B, et al. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc Natl Acad Sci USA. 2019;116(28):14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi JS, Cox MA, Zajac AJ. T-cell exhaustion: characteristics, causes and conversion. Immunology. 2010;129(4):474–481. doi: 10.1111/j.1365-2567.2010.03255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng L, Qin S, Si W, et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science. 2021;374(6574):abe6474. doi: 10.1126/science.abe6474. [DOI] [PubMed] [Google Scholar]
- 52.Davoodzadeh Gholami M, Kardar GA, Saeedi Y, Heydari S, Garssen J, Falak R. Exhaustion of T lymphocytes in the tumor microenvironment: significance and effective mechanisms. Cell Immunol. 2017;322:1–14. doi: 10.1016/j.cellimm.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Treacy O, Egan H, Lynch K, et al. Stromal cell sialylation suppresses T cells in inflammatory tumour microenvironments: a new tumour stromal cell immune checkpoint? J ImmunoTherapy f Cancer. 2021;9(suppl 2):A1–A1054. [Google Scholar]
- 54.Guo Y, Xie YQ, Gao M, et al. Metabolic reprogramming of terminally exhausted CD8(+) T cells by IL-10 enhances anti-tumor immunity. Nat Immunol. 2021;22(6):746–756. doi: 10.1038/s41590-021-00940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abdel-Hakeem MS, Manne S, Beltra JC, et al. Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nat Immunol. 2021;22(8):1008–1019. doi: 10.1038/s41590-021-00975-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riley RS, June CH, Langer R, Mitchell MJ. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18(3):175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1. The active TEX-specific pathway profiles from the RNA-sequencing data of individual bulk tumor tissue samples.
Supplementary file 2. List of TEX-associated genes identified in this study.