Abstract
Objectives
To develop and validate a standards-based phenotyping tool to author electronic health record (EHR)-based phenotype definitions and demonstrate execution of the definitions against heterogeneous clinical research data platforms.
Materials and Methods
We developed an open-source, standards-compliant phenotyping tool known as the PhEMA Workbench that enables a phenotype representation using the Fast Healthcare Interoperability Resources (FHIR) and Clinical Quality Language (CQL) standards. We then demonstrated how this tool can be used to conduct EHR-based phenotyping, including phenotype authoring, execution, and validation. We validated the performance of the tool by executing a thrombotic event phenotype definition at 3 sites, Mayo Clinic (MC), Northwestern Medicine (NM), and Weill Cornell Medicine (WCM), and used manual review to determine precision and recall.
Results
An initial version of the PhEMA Workbench has been released, which supports phenotype authoring, execution, and publishing to a shared phenotype definition repository. The resulting thrombotic event phenotype definition consisted of 11 CQL statements, and 24 value sets containing a total of 834 codes. Technical validation showed satisfactory performance (both NM and MC had 100% precision and recall and WCM had a precision of 95% and a recall of 84%).
Conclusions
We demonstrate that the PhEMA Workbench can facilitate EHR-driven phenotype definition, execution, and phenotype sharing in heterogeneous clinical research data environments. A phenotype definition that integrates with existing standards-compliant systems, and the use of a formal representation facilitates automation and can decrease potential for human error.
Keywords: FHIR, CQL, EHR-driven phenotyping, cohort identification
INTRODUCTION
Many studies designed to generate biomedical knowledge begin with cohort identification, also called electronic health record (EHR)-driven phenotyping. This is a resource-intensive process involving many stakeholders that must often be repeated every time the same cohort is studied.1,2 Given the increased focus in EHR-based clinical research, many different approaches and tools have been developed for EHR-driven phenotyping that attempt to reduce duplication of work, decrease the time investment required, and limit the potential for human error.3–12 While these approaches and tools have grown in response to specific needs, the result is a fragmented ecosystem of tools and approaches that are neither interoperable nor portable from one EHR system to another. For example, the electronic Medical Records and Genomics (eMERGE) Network13–15 shares cohort definitions via the Phenotype KnowledgeBase (PheKB),16 but the published knowledge artifacts do not conform to any formal knowledge representation standards, and range from narrative descriptions, to flowcharts, to custom SQL scripts. We present a standards-based representation for cohort definitions (also called phenotype definitions) and an accompanying tool that facilitates interoperability with existing systems. This enables incremental adoption and cross-platform execution, reduces duplication of work, and unifies existing techniques.
BACKGROUND
Computable phenotyping
EHR-driven phenotyping is a 3-step, iterative process: defining the phenotype (authoring), executing the phenotype against some data repository (execution), and evaluating or validating the correctness of the resulting cohort (validation). Authoring is a collaboration between clinical experts and informaticists to elucidate requirements specific enough to proceed to the execution step. Informaticists perform the execution step, sometimes in collaboration with database analysts, in order to extract the cohort from the data source using SQL or other code custom-built for the specific data source. Validation is typically done by experts who review the entire patient medical record. Although recent studies have explored how to identify relevant phenotype logic using automated methods,17,18 the expert-driven authoring step cannot be fully automated as it requires input from clinical domain experts to establish a clinically correct definition. Likewise, validation requires clinical expertise to confirm that the resulting cohort members match the clinical criteria. However, execution is repeated every time the cohort of interest is used in biomedical knowledge generation and can be automated.
Current strategies
The EHR-driven phenotyping research community, and the Phenotype Execution Modeling Architecture (PhEMA) (https://projectphema.org) project, have identified many requirements for the use of computable artifacts to automate EHR-driven phenotyping.19–26 These include: using structured and standardized data representations, using human-readable and computable representations for cohort criteria, and providing interfaces for external software with backwards compatibility. These requirements not only enable automation of the execution step, but they also reduce the risk of human error.
At least 2 strategies have been employed to meet these requirements, namely, the use of common data models (CDMs), and the use of generic formal logic representation and execution environments. The use of CDMs involves transforming and mapping data from existing data repositories into the CDM format and standardized vocabularies. Any computable phenotyping artifacts created at one site can be used at other sites using the same CDM.26 This approach has been successfully used by the Observational Health Data Sciences and Informatics (OHDSI) program,3 the National Patient‐Centered Clinical Research Network (PCORnet),27 and others.26,28,29 However, phenotype definitions created for one CDM cannot be used for another CDM. Generic logic execution environments such as KNIME and Drools have been successful but are not based on any healthcare standard and may require a labor-intensive data transformation step.30–32 Formal representations such as the Health Level Seven International (HL7) Health Quality Measure Format (HQMF), the Clinical Decision Support Knowledge Artifact Specification (CDS KAS), and Arden Syntax have also been used successfully for representing clinical logic.32–34 Yet, only Arden Syntax has a natively human-readable representation, and none have a convenient user interface for authoring and execution.
Existing tools
There are several existing tools for authoring and executing computable phenotype and cohort definitions. One highly mature and widely used tool is the OHDSI Atlas (https://github.com/OHDSI/Atlas) tool. This tool is built on the Observational Medical Outcomes Partnership (OMOP) CDM,3 and provides users with an interface for specifying logical criteria. Additionally, cohort definitions can be exported in JSON format and shared with other OMOP users. However, this format is not a formally defined standard, and cannot contain sub-phenotype definitions. Integrating Biology and the Bedside (i2b2)4 also provides an interface for cohort identification, but similarly does not use a formal standard or have the ability to export or import these definitions natively. A nascent phenotyping method, Phenoflow, supports the development of portable phenotypes using a formal representation.35 While the overall workflow process is based on the Common Workflow Language (CWL) standard,36 the individual steps are implemented using custom programming code.
Building upon these existing systems and lessons learned from their implementations, we built and validated a tool, the PhEMA Workbench, which allows users to author phenotype logic, assemble value sets (collections of standard medical codes) via integration with existing tools, and execute phenotypes without requiring manual translation.
METHODS
Standards-based representation
To represent the phenotype definition, we propose a fully Fast Healthcare Interoperability Resources (FHIR)-based representation. Inclusion and exclusion logic is expressed using the Clinical Quality Language (CQL), which produces an unambiguous and human readable representation. CQL source files are contained in Library resources as defined in the FHIR Clinical Reasoning Module. Each phenotype has one main CQL library containing the “Case” definition, and any number of helper libraries. Value sets are represented using ValueSet resources as defined in the FHIR Terminology Module. A Composition resource is used to collect all Library and ValueSet resources into a single document that fully describes the phenotype definition. By convention, the Composition section entry for the main phenotype Library is titled “Phenotype Entry Point” to indicate to the executing system where to find the “Case” definition. As defined in the FHIR Foundation Module, the Composition and all other resources are contained within a Bundle resource used to persist or transmit the fully specified computable phenotype definition. This representation enables FHIR operations to be used for storage, retrieval, and execution. For example, the $cql operation defined in the Clinical Practice Guidelines (CPG) implementation guide can be used to execute the phenotype, and the $document operation defined in the FHIR Foundation Module can be used to assemble the complete phenotype definition based on the Composition resource.
System description
Architecture
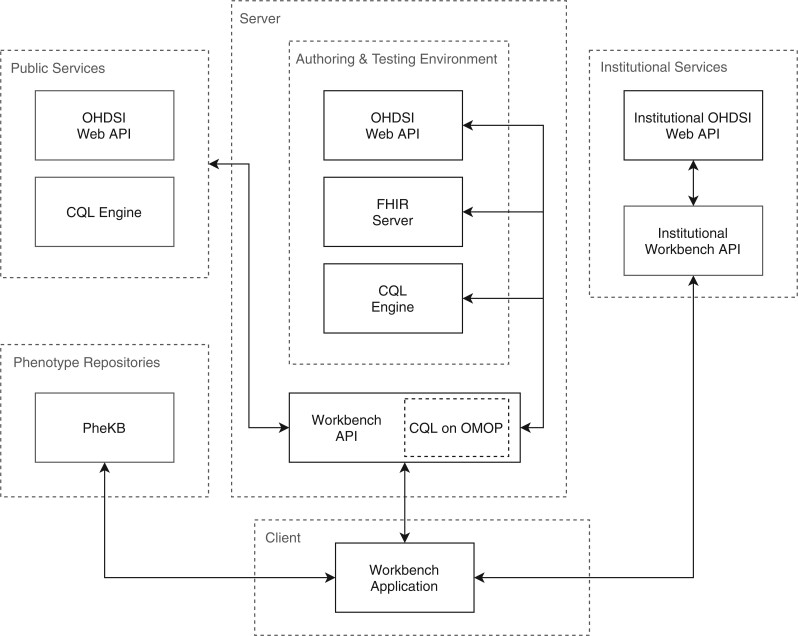
The PhEMA Workbench is designed as a standalone tool that can be used for phenotype authoring, execution, and publishing to a shared phenotype definition repository. It is intended for use by informaticians and research data analysts who routinely work with EHR-based queries. The architecture (Figure 1) is designed to function without requiring any changes to existing phenotyping tools or infrastructure, and integrates with OHDSI out of the box. The components include a web application that runs in the user’s browser, a backend API to support integrating with existing systems, as well as services for phenotype development and testing. The application is written using TypeScript, a strongly typed language that compiles to JavaScript, and the API is written in Java. All code is open-source and available in the PhEMA GitHub organization (https://github.com/PheMA). The tool used for testing during authoring is CQF Ruler (https://github.com/DBCG/cqf-ruler). It consists of a HAPI FHIR server (https://hapifhir.io/), and an implementation of the FHIR Clinical Reasoning Module and CPG implementation guide, which both use the reference implementation of the CQL engine (https://github.com/DBCG/cql_engine).
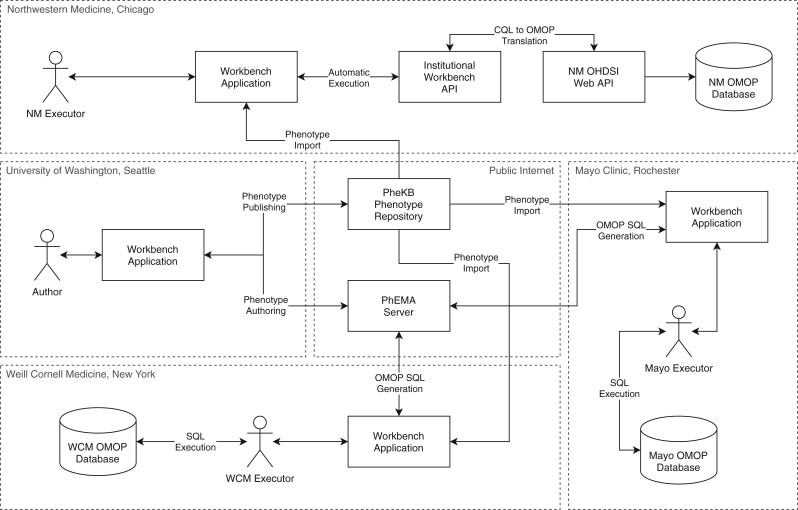
Figure 1.
System architecture. Services in the box labeled Server run on the PhEMA server and are accessible via the public internet. The box labeled Client runs in the browser on the user’s machine and is accessed by navigating to a specific URL on the PhEMA server. Optional publicly accessible third party services such as additional FHIR servers or OHDSI Web API instances are shown in the box labeled Public Services. The Phenotype Repositories box shows repository services (currently only PheKB). The Institutional Services box shows services that run behind institutional firewalls.
Features
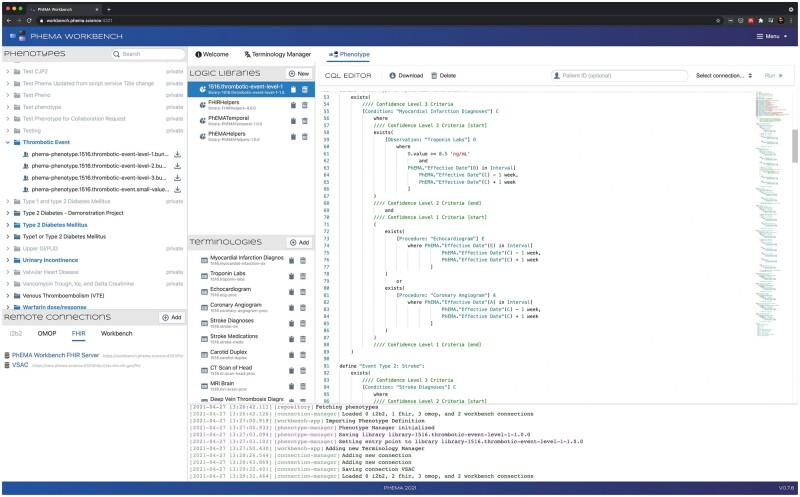
The first component is the CQL editor (Figure 2), for writing CQL expressions. It provides syntax highlighting and allows users to execute CQL against any environment capable of CQL execution, including the provided testing environment.
Figure 2.
CQL editor (right). The Phenotypes box in the top left lists the phenotypes available to import from PheKB. The Remote Connections box on the bottom left lists the configured third-party services. The 3 boxes labeled Logical Libraries, Terminologies, and CQL Editor all show components of the imported phenotype definition. Event logs are shown at the bottom.
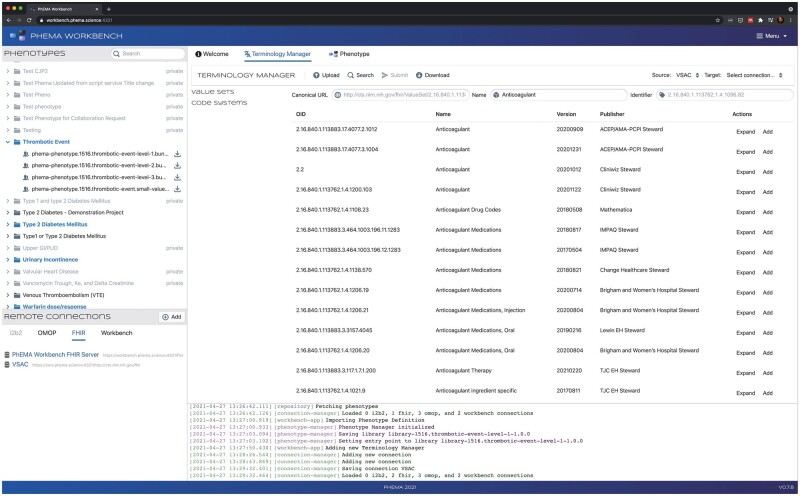
The application also provides a terminology manager (Figure 3), which allows a user to assemble a collection of value sets into an FHIR Bundle of ValueSet resources. Value sets can be uploaded by dragging and dropping them into the application, or by selecting a set of files on the filesystem. Supported formats include FHIR ValueSet or Bundle resources, concept sets exported from OHDSI Atlas, or a custom CSV (comma separated value) format. The user is also able to directly search the NLM’s Value Set Authority Center (VSAC) FHIR server,37 examine the results, and add them into the Bundle if appropriate.
Figure 3.
Terminology manager (right). The Phenotypes box in the top left lists the phenotypes available to import from PheKB. The Remote Connections box on the bottom left lists the configured third-party services. The Terminology Manager box allows the user to import, edit, and assemble value sets. Event logs are shown at the bottom.
To ensure that the expressed logic performs as expected, an author can test their CQL against test data during development. The Workbench provides an FHIR server that can be loaded with synthetic data. This gives the author confidence that the developed phenotype definition is logically and semantically correct. The CQL is executed on the FHIR server using the $cql extended operation, provided by CQF Ruler.
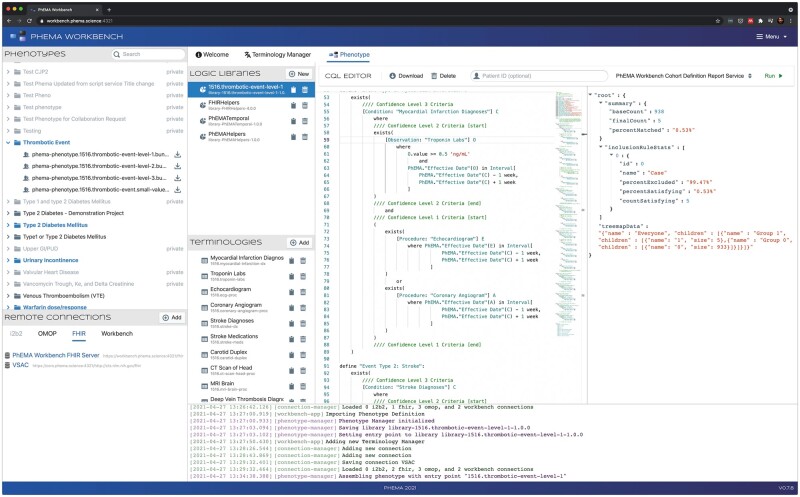
The Workbench integrates with the PheKB phenotype repository (top left panel in Figures 2-4) using the PheKB API, and supports listing all publicly available phenotypes, importing phenotype definitions represented using FHIR, and publishing new phenotype definitions. The Workbench currently supports OHDSI and FHIR as execution targets. At execution time, the Workbench API processes the complete FHIR-based phenotype definition, translates it to the appropriate representation, using CQL on OMOP29 in the case of the OHDSI target, and executes the logic against the target data store (Figure 4); establishing the corresponding cohort. The workbench also supports individual CQL library evaluation outside of the context of a phenotype, and generation of an OMOP compliant SQL script representing the phenotype definition.
Figure 4.
Example of phenotype execution. Results shown in the right-most panel.
Experimental setup
Phenotype definition
To test the PhEMA Workbench, we selected a Thrombotic Event (TE) phenotype developed by clinician collaborators at Weill Cornell Medicine (WCM), which identifies patients who have experienced one or more of 10 different thrombotic events. The definition for each event has up to 3 criteria sets that correspond to different confidence levels. The lowest confidence level (3) only requires that a patient has a single diagnosis code. To meet the Level 2 criteria, patients need to additionally have a specific drug or lab order, depending on the specific thrombotic event. For the highest confidence level (1), patients must meet the Level 2 requirements, and also have an order for a specified procedure, and in some cases an additional lab order as well. All criteria must occur within 1 week. The phenotype definition was shared with the PhEMA collaborators as a textual narrative description (Table 1).
Table 1.
Definition provided for the Thrombotic Event phenotype
| Event type | Confidence level 3 | Confidence level 2 | Confidence level 1 |
|---|---|---|---|
| MYOCARDIAL INFARCTION | ICD-9 code of 410.X or ICD-10 code of I21.X anywhere in the patient's EHR | CL3 + troponin of 0.5 or higher | CL2 + echocardiogram, ECG, or coronary angiogram |
| STROKE | ICD-9 code of 434.11 or ICD-10 code like I63.[0-3]% or ICD-10 code like I63.[5-9]% anywhere in the patient's EHR | CL3 + order for aspirin or clopidogrel + carotid duplex order + echocardiogram order | CL2 + neurology consult order + CT head or MRI brain order |
| DEEP VEIN THROMBOSIS | ICD-9 code of 453.4X or ICD-10 code of I82.4X OR any instance of a DVT sentinel phrase in any note | CL3 + D-Dimer fibrin lab result OR order for anticoagulant | CL2 + LE duplex report with positive DVT sentinel phrase |
| PULMONARY EMBOLISM | ICD-9 code of 415.1X or ICD-10 code of I26.9X anywhere in the patient's EHR | CL3 + new anticoagulant prescription + LE duplex report WITHOUT positive DVT sentinel phrase | CL2 + CT chest or VQ scan order |
| MESENTERIC-SPLANCHNIC THROMBOSIS | ICD-9 code of 557.0X or 444.89 or ICD-10 code of I81.9X/K55.0X/I82.0X/I74.8X anywhere in the patient's EHR | CL3 + new anticoagulant prescription | CL2 + Sonogram order OR CT chest order OR CT abdomen order OR MRI order + D-Dimer fibrin lab result + new order for anticoagulant |
| SUPERFICIAL VEIN THROMBOSIS | ICD-9 code of 453.6X or 451.89 or 453.82 or ICD-10 code of I82.81 or I80.0X or I82.61 | CL2 + new order for anticoagulant | |
| OTHER ARTERIAL THROMBOSIS | ICD-9 code of 444.1X or 444.22 or 453.3X or ICD-10 code of I74.X or I65.1X or I82.3X | CL2 + new order for anticoagulant | |
| PLACENTA THROMBOSIS | ICD-9 code of 663.6X or ICD-10 code of O43.813 | CL2 + new order for anticoagulant | |
| CENTRAL NERVOUS SYSTEM (CNS) THROMBOSIS | ICD-9 code of 437.6X or ICD-10 code of I67.6X | CL2 + new order for anticoagulant | |
| ENDOCARDIAL THROMBOSIS | ICD-9 code of 996.71 or 444.9X or ICD-10 code of I34.8X or I51.3X | CL2 + new order for anticoagulant |
ICD: International Classification of Diseases; CL: confidence level.
Authoring
We used the experimental architecture illustrated in Figure 5. In the first step, authoring and publishing were done by a single author (PSB), referencing the TE phenotype narrative description. We omitted criteria that required natural language processing (NLP), as this data are not directly available to OHDSI cohort definitions. Test data were created using the CQL Testing Framework tool (https://github.com/AHRQ-CDS/CQL-Testing-Framework), which allows users to generate FHIR resources using a light-weight configuration file. Value sets were generated in FHIR format using a custom script that expands a CSV file with extensional and basic intensional (eg, regex) definitions. The value set for anticoagulant drugs was imported directly from the VSAC FHIR server. All value sets were assembled into the final phenotype Bundle using the Workbench terminology manager component. Once all CQL logic was written and tested, and appropriate value sets were assembled, the phenotype was packaged using the proposed FHIR-based representation and published to PheKB.
Figure 5.
Experimental architecture. The Thrombotic Event phenotype was authored by PSB at the University of Washington and published to the PheKB repository in the proposed FHIR-based format using the Workbench application. LVR used the Workbench at Northwestern Medicine (NM) to automatically execute the phenotype using an institutional instance of the Workbench API. At Weill Cornell Medicine (WCM), PA used the Workbench application to generate an SQL script and executed it against the WCM OMOP database manually. The same approach was used at Mayo Clinic by DJS to generate and execute the SQL version of the phenotype definition.
Execution
Phenotypes were executed against EHR data extracted into separate databases to avoid possible performance and patient consent issues. LVR at Northwestern Medicine (NM), PA at WCM, and DJS at the Mayo Clinic (Mayo), imported the phenotype from PheKB into the PhEMA Workbench application. At NM, the phenotype was executed directly against an OMOP instance without any local customization, using the OHDSI Web API, from the Workbench. The OMOP database at NM contains data from a subset of the patient population that has consented to participate in the eMERGE Network with data sourced from the NM EpicCare EHR system. At WCM and Mayo, there was no instance of the OHDSI Web API available; thus, an SQL script was generated by PA and DJS using the SQL execution target available in the Workbench application. This SQL script was then manually executed against the WCM and Mayo OMOP databases, respectively. The WCM OMOP database contains data for patients from both WCM and NewYork-Presbyterian (NYP) hospitals that have at least one visit, condition, and procedure recorded. The EHRs used at WCM and NYP are EpicCare and Allscripts, respectively. The Mayo OMOP database consists of patients that were enrolled in the Mayo Biobank.38
Validation
We defined cases to be only those patients matching the Level 1 criteria. In order to validate edge cases where failures of logic are most likely to occur, we used patients matching the Level 2 criteria (but not matching Level 1) as non-cases. A validation protocol was shared along with an SQL script to randomly select cases and non-cases from the output generated through the Workbench execution step, SQL scripts to extract all relevant data, and a data entry form for reviewers to capture which criteria were met by each cohort member (if any).
At NM, a review of a random selection of 25 cases and 25 non-cases was performed by LVR, who determined whether or not the patient had the appropriate data to meet the confidence Level 1 criteria for at least one of the thrombotic event types. A second author (JAP), blinded to the results of the first reviewer, conducted a confirmatory review of 5 cases and 5 non-cases. This procedure was followed at Mayo, with DMK reviewing 25 cases and 25 non-cases, and DJS performing the blinded confirmatory review of 5 cases and non-cases. At WCM, 2 primary reviewers (ETS and SA) both reviewed and verified the same set of 25 cases and 25 non-cases, with a secondary reviewer (PA) resolving any discordant determinations. We report precision, recall, and inter-rater agreement using Cohen’s kappa39 as performance measures for each site.
RESULTS
Phenotype definition
The resulting thrombotic event phenotype logic consisted of 11 CQL statements, one for each thrombotic event type, and one for the disjunction of the other 10. The phenotype definition had 24 value sets containing a total of 834 codes for the various diagnoses, procedures, labs, and drugs referenced by the phenotype logic. Four different data sources were used: lab values, procedure orders, diagnoses, and drug orders, represented by the Observation, Procedure, Condition, and MedicationRequest FHIR resources, respectively. During the authoring process, 21 test cases were created to test the phenotype definition logic. The resulting phenotype definition, including criteria logic and value sets, as well as the test cases and data, are available in the project repository on GitHub (https://github.com/PheMA/thrombotic-event-phenotype).
Execution and validation
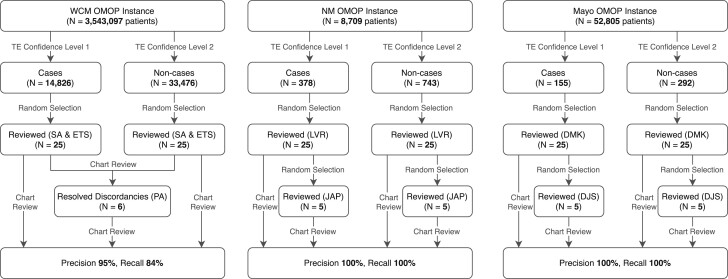
A summary of the phenotype execution and validation process is shown in figure 6.
Figure 6.
Results of the phenotype execution and manual review process.
Northwestern Medicine
There were 8709 total patients in the NM OMOP database, of which 378 (4.34%) met the TE confidence Level 1 criteria, and 743 (8.54%) matched the Level 2 criteria. Of the 25 cases and non-cases randomly selected for review, all were determined by the first reviewer to have been correctly identified, and subsequently confirmed by the secondary reviewer for the 5 random cases and non-cases. Thus, the reviewers were fully concordant (Cohen’s kappa = 1.0) and all patients were correctly identified (precision and recall both 100%), which shows the phenotype was faithfully converted.
Weill Cornell Medicine
The WCM OMOP database contained a total of 3 543 097 patients, of which 14 826 (0.42%) were identified as cases and 33 476 (0.94%) as non-cases. The 2 primary reviewers were discordant in 6 instances (15%), resulting in kappa = 0.74. These discordances were resolved by the secondary reviewer, and of the 25 cases, 22 (88%) were confirmed to match the TE confidence Level 1 criteria, and of the 25 non-cases, 24 (96%) were confirmed as not matching the confidence Level 1 criteria. This results in a precision of 95% and a recall of 84%.
During the CQL to OMOP translation step, cohort members were misclassified at WCM due to how the OHDSI Web API performs concept searches. Concepts are searched based on prefix matches, which in a few cases can return unrelated concepts with matching prefixes. This will be resolved in the next version of the CQL on OMOP tool.
Mayo Clinic
The Mayo Clinic OMOP database instance contained 52 805 total patients, of which 155 (0.29%) patients matched the confidence Level 1 criteria and 292 (0.55%) patients matched the Level 2 criteria. The reviewers were fully concordant (Cohen’s kappa = 1.0). Precision and recall were both 100%, which again shows the phenotype was faithfully converted.
DISCUSSION
Our FHIR-based representation of phenotype definitions and platform-agnostic tool can be used for computable EHR-driven phenotyping, and achieves results comparable to other methods. Furthermore, our approach combines elements of the CDM approach with elements of the generic logic approach, taking advantage of the benefits of both, while mitigating some of the disadvantages. This approach does not require additional data preparation, works across data platforms, uses established healthcare standards, and uses both human-readable and computable phenotype representations Additionally, the Workbench integrates into existing clinical informatics research infrastructure without requiring any changes to currently used tools and can in fact complement them. Table 2 summarizes how the PhEMA Workbench compares to existing phenotyping methods. Since many tools exist to generate FHIR-compliant data (eg, scenario builder (http://clinfhir.com/builder.html) and the CQL Testing Framework), they can be leveraged to populate the testing environment used to validate phenotype logic. This testing data can be used with publicly available standards-based testing tools. Furthermore, since the process is partially (in the case of WCM and Mayo) or completely (in the case of NM) automated, the potential for human error is reduced.
Table 2.
Feature matrix comparing applications and representations used for EHR-driven phenotyping
| Strategy |
Manual |
Common data models |
Formal representations |
Formal logic environments |
Next generation tools |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tool | SQL | OHDSI | i2b2 | HQMF | CDS KAS | Arden Syntax | Drools | KNIME | Phenoflow | PhEMA Workbench | |
| Application | Feature | ||||||||||
| Authoring environment | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Testing environment | ✓ | ||||||||||
| Publishing to central repository | ✓ | ||||||||||
| Importing from repository | (manual) | ✓ | |||||||||
| Multiple execution targets | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| Automated execution | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| No data preparation required | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
| Representation | Formal standards-based definitions | ✓ | ✓ | ✓ | (optional) | (optional) | ✓ | ✓ | |||
| Healthcare standards-based definitions | ✓ | ✓ | ✓ | (optional) | (optional) | ✓ | |||||
| Platform-agnostic definitions | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| Human-readable definitions | ✓ | ✓ | ✓ | ✓ | ✓ | ||||||
OHDSI: Observational Health Data Sciences and Informatics; i2b2: Informatics for Integrating Biology & the Bedside; HQMF: Health Quality Measure Format; CDS KAS: Clinical Decision Support Knowledge Artifact Specification; KNIME: Konstanz Information Miner; PhEMA: Phenotype Execution Modeling Architecture.
Every step of the phenotype authoring process was done using only FHIR and CQL, which are open standards developed by HL7. Furthermore, while CQL is data model agnostic, our choice to use FHIR for representing clinical data elements has several advantages. First, since FHIR is becoming a de facto CDM, much work has been done to generate mappings from the FHIR data model to other widely used data models, such OMOP, i2b2, and others40; thus, our phenotypes can be easily translated to various data models using standardized mappings.
CQL as a logical expression language is highly expressive and is capable of representing clinically validated phenotype definitions. Since CQL is a formal language, it also eliminates ambiguity that may result in variability of implementations. CQL is designed specifically for the clinical domain; thus, it has functionality tailored for representing clinical logic, such as the full set of temporal operators defined by Allen’s interval algebra,41 aggregate operators commonly used for quality measures and decision support, and uncertainty semantics to deal with missing data. Additionally, CQL provides integration points with external systems such as NLP pipelines or ML models.
The use of standards enables decoupling of both technical and conceptual components of EHR phenotyping, which is a widely used strategy in the technology industry to increase scalability, reliability, and extensibility.23,42 For example, the CQL engine focuses on logic execution, and delegates to a data provider module to collect the relevant data; thus, additional modules and execution targets can be developed for different data sources without requiring logic to be rewritten. Different CQL libraries can be assembled in a modular way, allowing for code reuse and easy localization, with site-specific logic contained in a library that can be used as a “drop-in” replacement for more general libraries.
Using a phenotype representation based on published standards decouples phenotype authoring and execution. The phenotype author (informatician or research data analyst) only requires knowledge of documented standards to define the phenotype. No knowledge of the data model where the phenotype definition will be executed is required, nor is access to the data. This means that knowledge artifacts developed by third parties can be executed while maintaining patient privacy, analogous to the model to data approach used for evaluating machine learning models on healthcare data.43
While graphical tools for phenotype authoring do exist, for example, OHDSI Atlas, the i2b2 query interface, and others,21 they have limitations the PhEMA Workbench attempts to address (summarized in Table 2). First, the phenotype definitions produced by these tools do not conform to any healthcare standard. Thus, future changes to how their phenotype definitions are structured could result in unintentional breaking of existing phenotype definitions if not carefully managed. Also, while these definitions can be shared between implementations of the same system, they cannot be shared between systems (ie, they are not cross-platform), except with some recent custom translators which still may lose some information during translation due to the differences of granularity and operations supported by the CDMs.44 Furthermore, the expressivity of the phenotype definitions supported by these tools may be limited. Even directly using SQL to generate cohorts may not be as convenient as using CQL, since SQL lacks clinical operators such as those used to determine patient age (eg, current age or age at date of clinical observation), as well as terminology operators to check whether coded values are part of specific code systems or value sets. Additionally, SQL is very tightly coupled to the data model against which it is executed, which means code is not reusable across databases with different schemas.
Local customization is an important part of EHR-driven phenotyping, as variations in local guidelines and clinical practice can easily result in differences in the downstream observational datasets.26 The PhEMA Workbench supports local customization by allowing users to directly view and edit logic before execution, as well as swap out value sets for ones that are more appropriate for the local context, all while remaining fully standards compliant. Various methods exist to author CQL source code, but only one integrates with a testing environment (the Atom CQL plugin [https://atom.io/packages/language-cql]) and, to our knowledge, no other tool integrates with a phenotype repository or has the ability to assemble value sets from various sources.
Current limitations are as follows. First, we only tested a single phenotype definition, and did not support NLP or machine learning phenotype definitions. Although the TE phenotype we selected includes multiple data elements and temporality, it does not exhaustively demonstrate other complex logic such as medication exposure periods. We have begun to address this limitation by developing a repository of more complex definitions, which we intend to evaluate using the Workbench in the future.45 Additionally, while we demonstrated cross-platform authoring and execution, we only tested a single execution target (OMOP), although we previously demonstrated that cross-platform execution is possible.29 Our support for FHIR as our standard data model is based on the core FHIR specification, and does not currently support specific profiles or extensions. As a result, the FHIR resources and fields we chose to use may not exactly match data in other FHIR data repositories relying on profiles and extensions. Additionally, we do not communicate our data model computationally to other systems using an FHIR implementation guide.
The CQL language also only supports structured data elements out of the box, but the PhEMA collaborators have developed NLP integrations,46,47 to be integrated into the Workbench architecture. There is also an upfront cost involved in implementing a CQL engine for a new data source, which may be prohibitive if engineering resources are not available.
CONCLUSION
We demonstrate that a fully FHIR-based phenotype representation is feasible and enables the decoupling of authoring and execution, leading to several advantages. If such an approach is broadly adopted, it may increase the velocity of biomedical generation by increasing semantic interoperability of phenotype definitions, and facilitating high-throughput cohort identification—reducing the potential for human error.
Our representation of a clinician-developed phenotype definition, executed against 3 OMOP data repositories, achieved results comparable to other methods. Additionally, our modular architecture consisting of existing open-source tools, including the newly developed PhEMA Workbench, can provide an effective and efficient phenotyping environment across institutions. The Workbench complements existing tools, and requires no changes to existing systems, giving end-users additional options without added restrictions.
In the future, we intend to use the PhEMA Workbench in clinical studies, and are planning to extend the capabilities of the system to include NLP (using CQL4NLP47) and more sophisticated machine learning models. We are also planning to continue improving the Workbench user interface, for which we have a user-centered design study currently underway to ensure usefulness for diverse end users of the tool.
FUNDING
This research is funded in part by the NIH grant R01GM105688. PSB is funded by the Fulbright Foreign Student Program and the South African National Research Foundation.
AUTHOR CONTRIBUTIONS
PSB and LVR conceptualized the study and developed the Phenotype Workbench tool. PSB authored the phenotype definition and first draft of the manuscript. LVR, JAP, PA, ETS, SA, DJS, and DMK executed the phenotype definition and/or reviewed the results. All authors helped refine the work and reviewed and edited the manuscript.
ACKNOWLEDGMENTS
The authors wish to thank Rick Kiefer and Dr. Jessica Ancker, whose earlier contributions to the project supported the work presented. The authors also wish to thank Morgan Higby-Flowers and Anthony Ikeogu who developed the PheKB API, as well as Dr. Jodell Jackson and Dr. Josh Peterson for their support of the use of PheKB.
CONFLICT OF INTERESTS STATEMENT
PSB is a consultant for Commure, Inc. The other co-authors have no competing interests to declare.
Contributor Information
Pascal S Brandt, Department of Biomedical Informatics and Medical Education, University of Washington, Seattle, Washington, USA.
Jennifer A Pacheco, Department of Preventive Medicine, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
Prakash Adekkanattu, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Evan T Sholle, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Sajjad Abedian, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Daniel J Stone, Department of Artificial Intelligence and Informatics, Mayo Clinic, Rochester, Minnesota, USA.
David M Knaack, Department of Artificial Intelligence and Informatics, Mayo Clinic, Rochester, Minnesota, USA.
Jie Xu, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Zhenxing Xu, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Yifan Peng, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Natalie C Benda, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Fei Wang, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Yuan Luo, Department of Preventive Medicine, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
Guoqian Jiang, Department of Artificial Intelligence and Informatics, Mayo Clinic, Rochester, Minnesota, USA.
Jyotishman Pathak, Department of Healthcare Policy and Research, Weill Cornell Medicine, New York, New York, USA.
Luke V Rasmussen, Department of Preventive Medicine, Northwestern University Feinberg School of Medicine, Chicago, Illinois, USA.
DATA AVAILABILITY STATEMENT
The phenotype definition used in this work, as well as the application source code is available in the PhEMA GitHub repository (https://github.com/PheMA).
REFERENCES
- 1. Pathak J, Kho AN, Denny JC.. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. J Am Med Inform Assoc 2013; 20 (e2): e206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banda JM, Seneviratne M, Hernandez-Boussard T, Shah NH.. Advances in electronic phenotyping: from rule-based definitions to machine learning models. Annu Rev Biomed Data Sci 2018;1:53–68. doi: 10.1146/annurev-biodatasci-080917-013315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hripcsak G, Duke JD, Shah NH, et al. Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 2015; 216: 574–8. [PMC free article] [PubMed] [Google Scholar]
- 4. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 17 (2): 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Longhurst CA, Harrington RA, Shah NH.. A “green button” for using aggregate patient data at the point of care. Health Aff (Millwood) 2014; 33 (7): 1229–35. [DOI] [PubMed] [Google Scholar]
- 6. Callahan A, Polony V, Posada JD, Banda JM, Gombar S, Shah NH.. ACE: the Advanced Cohort Engine for searching longitudinal patient records. J Am Med Informatics Assoc 2021; 28 (7): 1468–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tao S, Cui L, Wu X, Zhang GQ.. Facilitating cohort discovery by enhancing ontology exploration, query management and query sharing for large clinical data repositories. AMIA Annu Symp Proc 2017; 2017: 1685–94. [PMC free article] [PubMed] [Google Scholar]
- 8. Hurdle JF, Haroldsen SC, Hammer A, et al. Identifying clinical/translational research cohorts: Ascertainment via querying an integrated multi-source database. J Am Med Inform Assoc 2013; 20 (1): 164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dobbins NJ, Spital CH, Black RA, et al. Leaf: an open-source, model-agnostic, data-driven web application for cohort discovery and translational biomedical research. J Am Med Inform Assoc 2020; 27 (1): 109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lowe HJ, Ferris TA, Hernandez PM, Weber SC.. STRIDE—an integrated standards-based translational research informatics platform. AMIA Annu Symp Proc 2009; 2009: 391–5. [PMC free article] [PubMed] [Google Scholar]
- 11. Murphy SN, Barnett GO, Chueh HC.. Visual query tool for finding patient cohorts from a clinical data warehouse of the partners HealthCare system. Proc AMIA Symp 2000; 1174. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2243863/. Accessed May 1, 2021. [PMC free article] [PubMed] [Google Scholar]
- 12. Cui L, Zeng N, Kim M, et al. X-search: an open access interface for cross-cohort exploration of the National Sleep Research Resource 08 Information and Computing Sciences 0806 Information Systems. BMC Med Inform Decis Mak 2018; 18 (1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCarty CA, Chisholm RL, Chute CG, et al. ; eMERGE Team. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4 (1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gottesman O, Kuivaniemi H, Tromp G, et al. ; eMERGE Network. The electronic medical records and genomics (eMERGE) network: past, present, and future. Genet Med 2013; 15 (10): 761–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zouk H, Venner E, Lennon NJ, et al. Harmonizing clinical sequencing and interpretation for the eMERGE III network. Am J Hum Genet 2019; 105 (3): 588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirby JC, Speltz P, Rasmussen LV, et al. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc 2016; 23 (6): 1046–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y, Cai T, Yu S, et al. High-throughput phenotyping with electronic medical record data using a common semi-supervised approach (PheCAP). Nat Protoc 2019; 14 (12): 3426–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zheng NS, Feng Q, Eric Kerchberger V, et al. PheMap: a multi-resource knowledge base for high-throughput phenotyping within electronic health records. J Am Med Inform Assoc 2020; 27 (11): 1675–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mo H, Thompson WK, Rasmussen LV, et al. Desiderata for computable representations of electronic health records-driven phenotype algorithms. J Am Med Inform Assoc 2015; 22 (6): 1220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilcox AB. Leveraging electronic health records for phenotyping. In: Payne PRO, Embi PJ, eds. Translational Informatics: Realizing the Promise of Knowledge-Driven Healthcare. Health Informatics. London: Springer London; 2015: 61–74. doi: 10.1007/978-1-4471-4646-9_4. [DOI] [Google Scholar]
- 21. Xu J, Rasmussen LV, Shaw PL, et al. Review and evaluation of electronic health records-driven phenotype algorithm authoring tools for clinical and translational research. J Am Med Informatics Assoc 2015; 22 (6): 1251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Informatics Assoc 2013; 20 (E1): 147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rasmussen LV, Kiefer RC, Mo H, et al. A modular architecture for electronic health record-driven phenotyping. AMIA Jt Summits Transl Sci Proc 2015; 2015: 147–51. [PMC free article] [PubMed] [Google Scholar]
- 24. Zeng Z, Deng Y, Li X, Naumann T, Luo Y.. Natural language processing for EHR-based computational phenotyping. IEEE/ACM Trans Comput Biol Bioinform 2019; 16 (1): 139–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma H, Mao C, Zhang Y, et al. Developing a portable natural language processing based phenotyping system. BMC Med Inform Decis Mak 2019; 19 (Suppl 3): 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasmussen LV, Brandt PS, Jiang G, et al. Considerations for improving the portability of electronic health record-based phenotype algorithms. AMIA Annu Symp Proc 2019; 2019: 755–64. [PMC free article] [PubMed] [Google Scholar]
- 27. Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS.. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 2014; 21 (4): 578–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The ACT Network. 2018. https://www.actnetwork.us/national. Accessed Mar 29, 2021.
- 29. Brandt PS, Kiefer RC, Pacheco JA, et al. Toward cross-platform electronic health record-driven phenotyping using Clinical Quality Language. Learn Heal Syst 2020; 4 (4): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mo H, Pacheco JA, Rasmussen LV, et al. A prototype for executable and portable electronic clinical quality measures using the KNIME analytics platform. AMIA Jt Summits Transl Sci Proc 2015; 2015 (Icd): 127–31. [PMC free article] [PubMed] [Google Scholar]
- 31. Li D, Endle CM, Murthy S, et al. Modeling and executing electronic health records driven phenotyping algorithms using the NQF Quality Data Model and JBoss® Drools Engine. AMIA Annu Symp Proc 2012; 2012: 532–41. [PMC free article] [PubMed] [Google Scholar]
- 32. Mo H, Jiang G, Pacheco JA, et al. A decompositional approach to executing quality data model algorithms on the i2b2 platform. AMIA Jt Summits Transl Sci Proc 2016; 2016: 167–75. [PMC free article] [PubMed] [Google Scholar]
- 33. Lee PV. Automated Injection of Curated Knowledge Into Real-Time Clinical Systems CDS Architecture for the 21st Century. 2018; (December). https://repository.asu.edu/attachments/209644/content/Lee_asu_0010E_18312.pdf. Accessed Mar 29, 2021.
- 34. Maier C, Kapsner LA, Mate S, Prokosch HU, Kraus S.. Patient cohort identification on time series data using the OMOP common data model. Appl Clin Inform 2021; 12 (1): 57–64. doi: 10.1055/s-0040-1721481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chapman M, Rasmussen LV, Pacheco JA, Curcin V. Phenoflow: A Microservice Architecture for Portable Workflow-based Phenotype Definitions. AMIA Jt Summits Transl Sci Proc. 2021;2021:142–151. [PMC free article] [PubMed]
- 36. Amstutz P, Chapman B, Chilton J, Heuer M. Stojanovic. Common Workflow Language, v1.0 Common Workflow Language (CWL) Command Line Tool Description, v1.0. 2016. doi: 10.6084/m9.figshare.3115156.v2. [DOI]
- 37. Bodenreider O, Nguyen D, Chiang P, et al. The NLM value set authority center. Stud Health Technol Inform 2013; 192: 1224. [PMC free article] [PubMed] [Google Scholar]
- 38. Olson JE, Ryu E, Johnson KJ, et al. The Mayo Clinic Biobank: a building block for individualized medicine [published correction appears in Mayo Clin Proc. 2014 Feb;89(2):276]. Mayo Clin Proc 2013; 88 (9): 952–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas 1960; 20 (1): 37–46. [Google Scholar]
- 40.ONC. Common Data Model Harmonization. https://www.healthit.gov/topic/scientific-initiatives/pcor/common-data-model-harmonization-cdm. Accessed April 21, 2021.
- 41. Allen JF. Maintaining knowledge about temporal intervals. Commun ACM 1983; 26 (11): 832–43. doi: 10.1145/182.358434 [DOI] [Google Scholar]
- 42. Villamizar M, Garcés O, Castro H, Verano M, Salamanca L, Gil S.. Evaluating the monolithic and the microservice architecture pattern to deploy web applications in the cloud (Evaluando el Patrón de Arquitectura Monolítica y de Micro Servicios Para Desplegar Aplicaciones en la Nube). 10th Comput Colomb Conf 2015; 583–90. https://ieeexplore.ieee.org/document/7333476 [Google Scholar]
- 43. Guinney J, Saez-Rodriguez J.. Alternative models for sharing confidential biomedical data. Nat Biotechnol 2018; 36 (5): 391–2. [DOI] [PubMed] [Google Scholar]
- 44. Majeed RW, Fischer P, Günther A. (2021). Accessing OMOP common data model repositories with the i2b2 webclient–algorithm for automatic query translation. In: Röhrig R, Beißbarth T, Brannath W, Prokosch H-U, Schmidtmann I, Stolpe S, Zapf A, eds. German Medical Data Sciences: Bringing Data to Life. Netherlands: IOS Press, 251–259. [DOI] [PubMed] [Google Scholar]
- 45. Brandt PS, Pacheco JA, Rasmussen LV.. Development of a repository of computable phenotype definitions using the clinical quality language. JAMIA Open 2021; 4 (4): ooab094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hong N, Wen A, Stone DJ, et al. Developing a FHIR-based EHR phenotyping framework: a case study for identification of patients with obesity and multiple comorbidities from discharge summaries. J Biomed Inform 2019; 99 (April): 103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wen A, Rasmussen LV, Stone D, et al. CQL4NLP: development and integration of FHIR NLP extensions in clinical quality language for EHR-driven phenotyping. AMIA Jt Summits Transl Sci Proc 2021; 2021: 624–33. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The phenotype definition used in this work, as well as the application source code is available in the PhEMA GitHub repository (https://github.com/PheMA).