Abstract
Colorectal cancer (CRC) is the leading malignant tumor in terms of morbidity and mortality worldwide, and its pathogenesis involves multiple factors, including environment, lifestyle, and genetics. Continuing evidence suggests that circular RNAs (circRNAs), as a novel non-coding RNA, constitute an important genetic variable in the pathogenesis of CRC. These circRNAs with covalently closed-loop structures exist objectively in organisms. They not only have the biological functions of regulating the expression of target genes, changing the activity of proteins, and translating proteins, but also play a key role in the proliferation, invasion, migration, and apoptosis of tumor cells. CRC is one of the most common cancers in which circRNAs are involved in tumorigenesis, metastasis, and drug resistance, and circRNAs have been demonstrated to function through crosstalk with multiple signaling pathways. Therefore, this review summarizes the biological and carcinogenic functions of circRNAs and their related PI3K/AKT, MAPK, Notch, JAK/STAT, Hippo/YAP, WNT/β-catenin, and VEGF signaling pathways in CRC. We further explore the clinical value of circRNAs and important signaling proteins in the diagnosis, prognosis, and treatment of CRC.
Keywords: colorectal cancer, circular RNA, signaling pathway, biological function, clinical value
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second leading cause of cancer deaths worldwide, with an estimated 1.9 million new cases and 935,000 deaths in 2020 (1). The incidence of CRC has stabilized and declined in highly developed countries as a result of national screening programs and colonoscopy (2). But with economic progress in developing countries, the number of new CRC cases worldwide is expected to reach 2.5 million by 2035 (3). At present, the 5-year survival rate of patients with early CRC is close to 90% (4). However, among newly diagnosed patients with CRC, 20% have already had metastasis, and another 25% will develop metastasis due to locally advanced tumors (5). Moreover, metastatic CRC has a poor prognosis, with a 5-year survival rate of less than 20% (6). Therefore, to seek effective biomarkers for early diagnosis of CRC and new therapeutic targets for advanced and recurrent CRC, it is necessary to further explore and clarify the molecular mechanisms underlying the development and metastasis of CRC.
Circular RNAs (circRNAs) are newly discovered non-coding RNAs (ncRNAs) that exist objectively in living organisms (7). These circRNAs have a covalently closed-loop structure, missing the 5′-3′ terminals and polyadenylate tails (8). The development of high-throughput RNA sequencing and bioinformatic tools has successfully detected thousands of circRNAs distributed in a variety of tissues, cell types, and biological fluids (9, 10). Moreover, researchers have revealed that these RNAs have cell-specific, tissue-specific, and time-specific expression patterns and are conserved across species (11, 12). Recent evidence indicates that circRNAs are not only significantly associated with neurological disorders, cardiovascular diseases, and autoimmune diseases (13–15), but also play a regulatory role in cancer-related processes such as tumorigenesis, progression, and cell apoptosis (16–18). CRC is one of the most commonly reported cancers in which circRNAs are involved in tumorigenesis and metastasis (18–20).
Signal transduction is a common way to regulate basic cellular processes in humans, and abnormal regulation of signal transduction can lead to the occurrence of pathological states such as cancer and autoimmunity (21, 22). It is widely believed that circRNA promotes cancer cell proliferation and metastasis by interacting with key components of major signaling pathways (23, 24). In CRC, the reported crosstalk signaling pathways with abnormally expressed circRNA include phosphatidylinositol 3-kinase (PI3K)/AKT (25), mitogen-activated protein kinases (MAPK) (26), Notch (27), Janus kinase/signal transducers and activators of transcription (JAK/STAT) (28), Hippo/YAP (29), WNT/β-catenin (30), and vascular endothelial growth factor (VEGF) (31). In this review, we summarized the molecular mechanism and role of circRNAs and related signaling pathways in the occurrence and progression of CRC by searching for keywords in Pubmed, Web of Science, ScienceDirect, and Springer SLCC databases. The clinical application value of circRNAs in the diagnosis, prognosis, and treatment of CRC was further discussed.
Genetic and transcriptional characterization of CRC
Recent research suggests that most CRC cells originate from stem cells or stem cell-like cells (32). The accumulation of multiple genetic and epigenetic changes produces these cancer stem cells (CSCs), which ultimately activate oncogenes and inactivate tumor-suppressor genes (33, 34). Two major precursor pathways represent multiple genetic and epigenetic events in a fairly continuous sequence. Most CRCs are chromosomal instability sequences (70-90%), also known as the traditional adenoma-carcinoma pathway (35). In this pathway, tumor development is caused by the sequential accumulation of mutations in the WNT, epidermal growth factor receptor (EGFR), P53, and transforming growth factor-beta (TGF-β) signaling pathways (36). Another is the serrated tumor pathway, which involves activating BRAF mutations or DNA mismatches to repair gene inactivation and accounts for 10-20% of CRCs (37, 38).
In 2015, the International CRC Subtype Consortium proposed a more comprehensive transcriptome classification based on gene expression profiles (39). CRC is divided into four consensus molecular subtypes (CMS): CMS1 (MSI immunity, 14%), CMS2 (canonical, 37%), CMS3 (metabolic, 13%), and the subtype with the worst prognosis, CMS4 (mesenchymal, 23%) (40). These CMS group classifications embody markedly different molecular characteristics associated with biological and clinical stratification and are the basis for targeted interventions (41). There are differences not only in embryology, anatomy, and biology but also in molecular characteristics between right colon cancer (hepatic curvature of transverse colon, ascending colon, and cecum) and left colon cancer (splenic curvature of transverse colon, descending colon, and sigmoid colon) and the rectum (42). Right colon cancer is more common in CMS1 and CMS3 subtypes, while left colon cancer is predominantly in CMS2 subtypes (43).
The biological and oncogenic functions of circRNAs in CRC
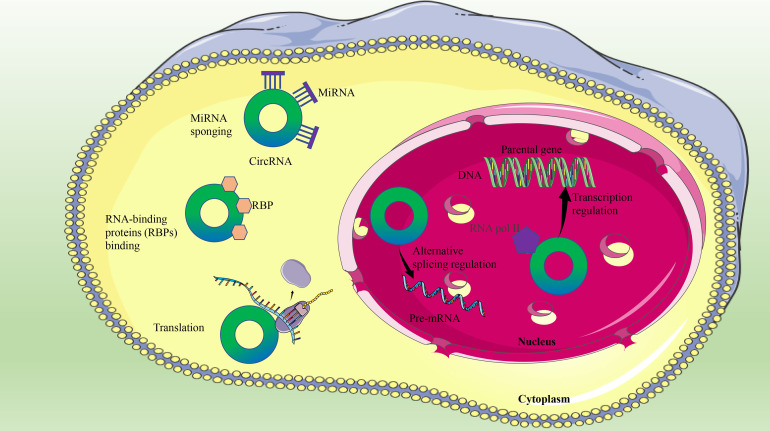
CircRNAs play different biological functions according to their localization in the cytosol or nucleus (44). First of all, circRNAs can regulate gene transcription and alternative splicing, and nuclear circRNAs can also induce parental gene expression (45). Second, some circRNAs, such as circ_0128846 (29) and circ_0106714 (46), influence the expression of target genes by competitively binding microRNA (miRNA) or acting as miRNA sponges, which is the most widely studied mechanism of circRNA in the progression of CRC (47). In addition, another biological function of circRNAs is that their interactions with RNA-binding proteins cause changes in protein activity (48). Recently, two novel circRNAs (circ-BCL2L12-1 and circ-BCL2L12-2) with different protein binding sites have been identified in CRC. Moreover, circ-BCL2L12-1 overexpression was related to shorter OS, while circ-BCL2L12-2 expression was negatively related to TNM staging in CRC (49). Finally, circRNA has the biological function of translating proteins (50), and circ-PPP1R12A is reported to have translation capabilities in CRC (51) (Figure 1).
Figure 1.
The biological functions of circRNAs in CRC.
A large number of studies have shown that circRNA can play an important role in the tumor progression of CRC by regulating tumorigenesis transcription factors or oncogene expression (52). In the study of Li’s team, 448 circRNAs with abnormal expression in CRC were detected by high throughput RNA sequencing. Further in vitro experiments showed that the down-regulation of circDDX17 could enhance the proliferation, invasion, and migration of CRC cells (53). In vivo experiments on nude mice have also confirmed that some circRNAs, such as circ-ERBIN, can promote tumor growth and metastasis of CRC (54). Advanced metastatic CRC is the leading cause of cancer-related death. Epithelial-mesenchymal transition (EMT) is a cellular reprogramming process in which epithelial cells acquire a mesenchymal phenotype, which promotes the development of migrating and invading cells (55). Through GEO data set analysis, circ_101951 was found to be a novel circRNA overexpressed in CRC tissues. In-depth studies on its biological function and mechanism showed that circ_101951 could facilitate the migration and invasion of CRC cells by regulating EMT (56). CSCs are subsets of small cells in tumors that drive tumor progression and metastasis (57). Continuing studies have shown that CSCs in CRC are inherently resistant to treatment and are closely related to cancer regeneration and recurrence after conventional treatment (58). Rengganaten et al. revealed circ_0066631 and circ_0082096 as two abnormally expressed circRNAs in CSC-rich CRC globular cells that play an important role in regulating the stemness properties of CSCs (59).
The role of major signal pathways interacting with circRNAs in CRC
CircRNA/MAPK signaling axis in CRC
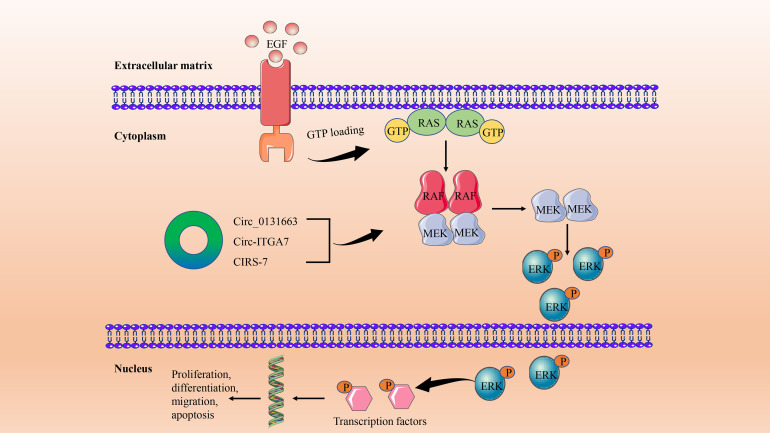
Compared with other intracellular signaling pathways, the MAPK pathway is more important in cell proliferation, differentiation, migration, apoptosis, and angiogenesis. The most critical signal cascade reaction in all MAPK signal transduction pathways is RAS/RAF/MEK/ERK (60). The pathway is initiated by activation of the RAF kinase family (ARAF, BRAF, and CRAF [RAF1]) by members of the RAS family. Activated RAF protein phosphorylates MEK1/2 and then activates and phosphorylates ERK. Finally, ERKs induce phosphorylation of a variety of substrates, such as transcription factors, which are involved in regulating a variety of cellular functions (61) (Figure 2). Abnormal activation of the MAPK signaling pathway in CRC has been reported to occur through activation mutations of RAS and BRAF (62), which are associated with treatment resistance in patients with metastatic CRC (mCRC) (63). Approximately 90% of BRAF mutations are in the V600E series, and although BRAFV600E mutations are rare in CRC (about 10%), their role is important (64). BRAFV600E series mutations in CRC are more common in older women (over 70 years old), right colon tumors, poorly differentiated tumors, and mucous subtypes, and also have a higher frequency of peritoneal metastasis (65). This also predicted poor clinical prognosis (median OS of 11 months) and poor standard treatment response in CRC with BRAFV600E series mutations (66). RAS family is one of the most frequently mutated families in CRC (67). About 40% of mCRCs carry KRAS mutations, mainly in exons 2 (codon 12, 13), 3 (codon 59-61) and 4 (codon 117, 146). Mutations at different points cause different clinical, pathological, and molecular features. Although mutations in NRAS account for only 4% of mCRC, they have similar clinical and pathological features to KRAS mutations. While HRAS mutations are 1%, very few studies have (65, 68).
Figure 2.
The circRNA/MAPK signaling axis in CRC. This signaling pathway is initiated by the binding of activated growth factors, such as epidermal growth factor (EGF), to tyrosine kinase receptors on the cell surface. This causes the downstream RAS to increase the GTP binding state. RAS-GTP dimers recruit RAF or RAF/MEK dimers to the plasma membrane and promote the activation of RAF and the formation of MEK homologous dimers. This is followed by activation and phosphorylation of downstream ERKs. Finally, ERKs induce phosphorylation of transcription factors and other substrates to participate in cell proliferation, differentiation, migration, and apoptosis in the nucleus.
In CRC, circRNA regulation of the MAPK signaling pathway is a topic widely discussed and worthy of study, although there are few relevant research results at present. It is generally believed that the phosphorylation of MAPK14, the core molecule of the MAPK pathway, by upstream signal kinase kinase 3/6 (MKK3/6) promotes nuclear translocation and promotes the progression of CRC (69). Based on the above results, Wang et al. further verified that circ_0131663 (circ-MAPK14) can reduce the nuclear translocation of MAPK14 by competitive binding with upstream MKK6 through a peptide encoding 175 amino acids, and ultimately inhibit the progression and metastasis of CRC (70). In addition, the functions and mechanisms of other novel circRNAs in CRC have also been preliminarily explored. Circ-ITGA7 was significantly underexpressed in CRC tissues and cell lines, and it was found by functional experiments that the expression of circ-ITGA7 prevented the growth and metastasis of CRC cells in vitro and in vivo. Further mechanistic studies have shown that circ-ITGA7 inhibits the growth and metastasis of CRC tumors by inhibiting RAS/RAF/MEK/ERK signaling pathways and promoting ITGA7 transcription (71). Another novel circRNA, CIRS-7, acts as a competitive endogenous RNA (ceRNA) of miR-7 to regulate EGFR/RAF1/MAPK signal transduction and plays an important role in CRC progression (72).
CircRNA/PI3K/AKT signaling axis in CRC
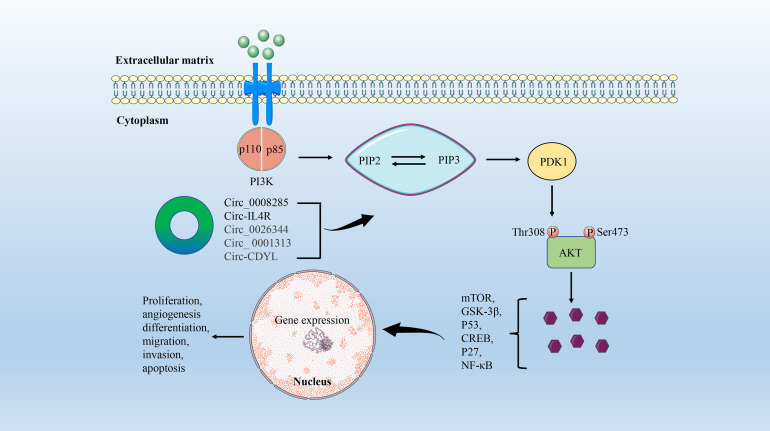
The PI3K/AKT signaling pathway is involved in regulating cell adhesion, growth, survival, migration, and other cellular events (73). PI3K is an intracellular lipid kinase that affects the expression levels of extracellular protein kinase and EGFR, leading to PIP2 phosphorylation to produce PIP3. PIP3 is an important messenger that recruits AKT, which in turn generates mammalian targets of rapamycin (mTOR) or GSK-3β signaling, resulting in a variety of cellular events (74). The PI3K/AKT/mTOR signaling pathway is one of the most critical abnormal regulatory pathways in CRC, and the activation of this pathway is associated with cell proliferation and transformation, tumorigenesis, progression, angiogenesis, and drug resistance (75, 76) (Figure 3).
Figure 3.
The circRNA/PI3K/AKT signaling axis in CRC. PI3K consists of a catalytic (P110) domain and a regulatory (P85) domain. PI3K is activated by a variety of growth factors and signaling complexes. Activated PI3K promotes PIP2 phosphorylation to produce PIP3, which activates PDK1. AKT is then phosphorylated at Thr308 of PDK1. Ultimately, AKT induces cell proliferation, differentiation, migration, and angiogenesis by mediating multiple signaling pathways such as mTOR, GSK-3β, P53, CREB, P27, and NF-κB.
With the continuous development of high-throughput sequencing technology, a large number of circRNAs with abnormal expression in a variety of tumors have been identified (60, 77, 78). The researchers concluded that the expression level of circ-0008285 in CRC tissues and cells was negatively correlated with tumor size and lymphatic metastasis by combining quantitative reverse transcription-PCR (RT-qPCR) and clinicopathological parameter analysis. Further functional and mechanistic studies confirmed that low expression of circ_0008285 promotes the proliferation and migration of CRC cells in vitro by regulating the PI3K/AKT pathway (79). In addition, Jiang et al. found a circRNA that also plays a role in the occurrence and progression of CRC by regulating the PI3K/AKT signaling pathway, named circ-IL4R. However, circIL4R is highly expressed in the serum, tissues, and tumor cell lines of CRC patients, and is positively associated with later clinical stages and poorer prognosis (80).
CircRNA/VEGF signaling axis in CRC
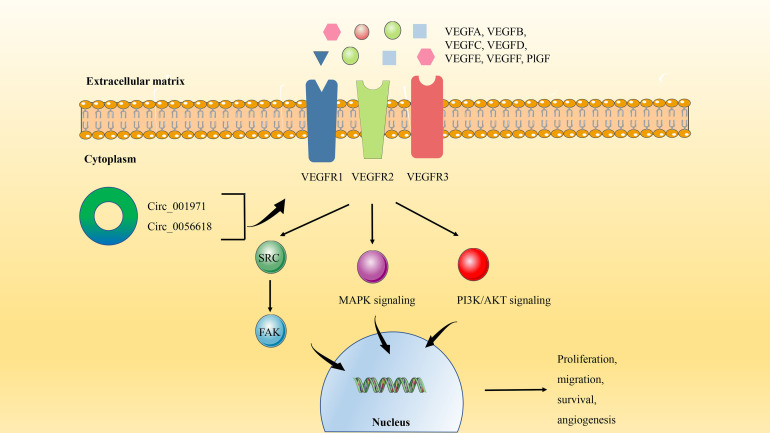
Generally, the most common distant metastasis site of CRC is the liver. According to statistics, about 25% of CRC patients will have liver metastasis, and the prognosis is poor (81). High expression of the VEGF family is often associated with the aggressiveness and metastasis of CRC (82). VEGF protein family includes VEGFA-F and placental growth factor (PlGF). VEGFs bind to tyrosine kinase cell receptors (VEGFR1-3) to activate VEGF signaling in endothelial cells, affecting cell proliferation, migration, survival, and vascular permeability during angiogenesis (83, 84) (Figure 4).
Figure 4.
The circRNA/VEGF signaling axis in CRC. VEGF binds to tyrosine kinase cell receptors (VEGFR1-3) to activate VEGF signaling in endothelial cells. The activation of VEGF signaling can not only induce the protein expression of SRC kinase and FAK Focal adhesion kinase, but also trigger PI3K/AKT and MAPK signal transduction. Finally, it regulates the process of angiogenesis, proliferation, migration, and survival.
VEGFA is often overexpressed in CRC and is considered to be a key factor in inducing tumor angiogenesis, which plays an important role in tumorigenesis, tumor development, and metastasis (85). Besides the previously enumerated signaling pathways, circRNA can also play a tumorigenic role in CRC through the VEGFA signaling axis. Circ_001971 has been observed to act as a ceRNA to mitigate VEGFA inhibition by miR-29C-3p, thereby enhancing the proliferation, invasion, and angiogenesis of CRC (31). In addition, studies have shown that high expression of circ_0056618 not only produces the same effect on CRC by regulating VEGFA as mentioned above, but also is related to the poor overall survival (OS) of CRC patients (86).
CircRNA/JAK/STAT signaling axis in CRC
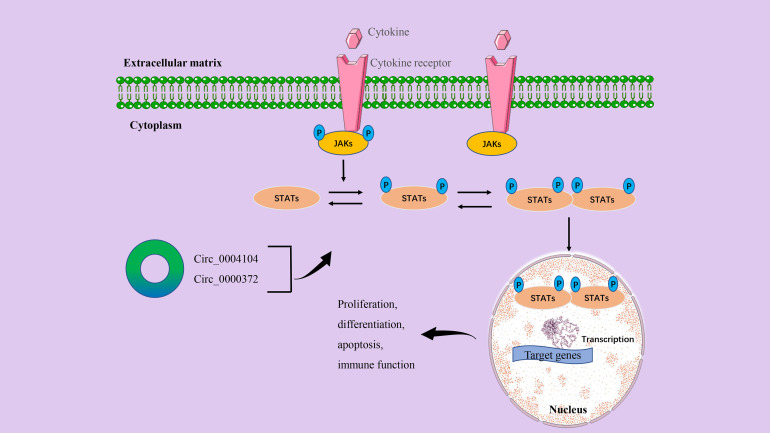
The JAK/STAT signaling pathway is a common intracellular signal transduction pathway that is involved in many biological processes such as cell proliferation, differentiation, apoptosis, and immune regulation (87). In CRC, the JAK/STAT signaling axis has been shown to play a key role in tumor cell genesis, progression, invasion, migration, and chemical tolerance (88). The specific molecular mechanisms by which the JAK/STAT signal transduction pathway regulates CRC progression refer to the expression of multiple proto-oncogenes, tumor suppressor genes, cytokines, and their receptors, including Ras, Src, p27kip1, p16ink4a, interleukin, and EGFR (88, 89). The classic JAK/STAT signaling process is that the connection between the ligand and the receptor activates JAK to form phosphorylation (P)-JAK and forms a docking site for STAT. At this docking site, P-JAK phosphorylates STAT so that it dimers with other members of the STAT family. These dimers will transfer from the cytoplasm to the nucleus and regulate the transcription of target genes (88, 90) (Figure 5).
Figure 5.
The circRNA/JAK/STAT signaling axis in CRC. The binding of the cytokine to the receptor induces receptor dimers and initiates signal transduction. After JAKs are activated and phosphorylated, STATs proteins are recruited to the phosphorylated tyrosine site. STATs are also then activated and phosphorylated. Normally, STATs reside in the cytoplasm and form phosphorylated dimers when activated by upstream signals. STAT-STAT dimers are transferred from the cytoplasm to the nucleus and regulate the transcription of target genes. Finally, it affects the proliferation, differentiation, invasion, inflammation, and immune function of cancer cells.
We have listed a variety of circRNAs that play a key role in the occurrence and development of human CRC, but circRNAs alone may not be enough to promote cancer progression. In a study by Wang et al., the initial RNA sequencing found that circ_0004104 expression levels were significantly upregulated in CRC tissues and were closely related to the prognosis of CRC patients. In-depth mechanistic studies have shown that circ_0004104 modulates the JAK2/STAT3 pathway by acting as a ceRNA binding to miR-485-3p and FUS, ultimately promoting cell proliferation and migration (19). Previous studies have demonstrated that the inflammatory cytokine interleukin-6 (IL6) can mediate the activity of the JAK2/STAT3 signaling pathway to participate in the occurrence and development of CRC (91). Recent evidence also shows that down-regulation of circ_0000372 can inhibit the protein expression of the IL6/AK2/STAT3 signaling axis. Further results confirmed that circ_0000372 may regulate IL6 expression and JAK2/STAT3 signaling pathway activity by acting on miR-495 in CRC (28).
CircRNA/Notch signaling axis in CRC
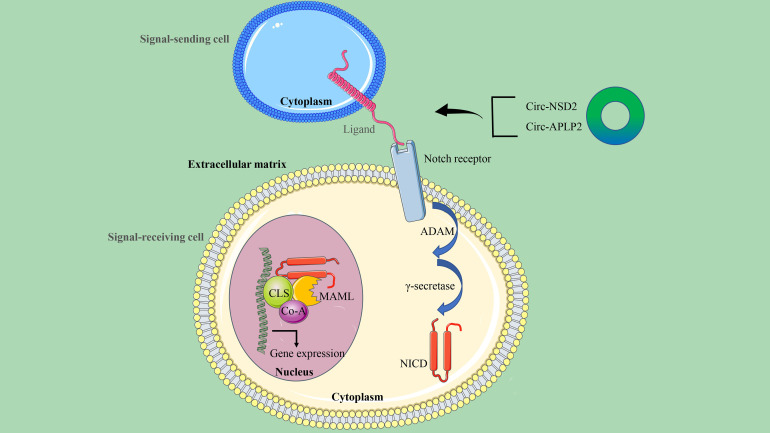
There are generally four Notch receptor subtypes (Notch-1, Notch-2, Notch-3, and Notch-4) and five Notch ligands (Dll-1, Dll-3, Dll-4, Jagged-1, and Jagged-2) in humans (92). The Notch signaling pathway, which is involved in the progression of CRC and the self-renewal and homeostasis of normal intestinal epithelium, is activated when the ligand binds to the receptor (93). Abnormal activation of Notch1 has been reported to initiate CRC and enhance its invasiveness (94). The specific mechanism attributed to Notch1 signaling creates a tumor microenvironment (TME) and promotes CRC metastasis through TGF-β-dependent neutrophil recruitment (95) (Figure 6).
Figure 6.
The circRNA/Notch signaling axis in CRC. This signaling is initiated by ligand-receptor binding between the signal-sending cell and the signal-receiving cell. The receptor-ligand interaction triggers a continuous cleavage mediated by ADAM metalloproteinase and γ-secretase, followed by the release of the intracellular domain NICD by Notch. When NICD is transferred to the nucleus, it recruits MAML and Co-A to CSL to initiate the expression of target genes.
Tumor metastasis is an important factor affecting the survival and prognosis of patients, and circRNA has been confirmed to be involved in the metastasis of CRC cancer. Using RNA transcriptome sequencing, Chen et al. identified a novel highly expressed circRNA, circ-NSD2, in a mouse model of liver metastasis. A series of functional and mechanistic studies revealed that circ-NSD2 may promote the migration, invasion, and metastasis of CRC cells in vitro and in vivo by targeting miR-199b-5p mediated JAG1/Notch1 signaling (27). It can also be seen that more and more experimental results reveal that many signaling pathways, including the Notch signaling pathway, are regulated by miRNA. Therefore, based on the results obtained from bioinformatics analysis, miR-101-3p has binding sites on circ-APLP2 and Notch1. Circ-APLP2 has been proven to act as a miR-101-3p sponge to regulate the Notch1 signaling pathway in CRC and activate proliferation and metastasis-related signals (c-Myc, cyclin D1, MMP-2, and MMP-9), thereby promoting the proliferation and liver metastasis of CRC (96).
CircRNA/Hippo/YAP signaling axis in CRC
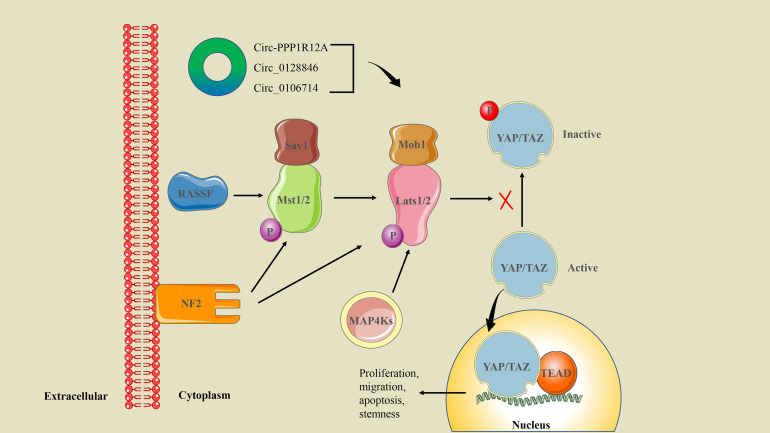
CSCs are the main cause of drug resistance and disease recurrence in CRC treatment. Hippo/YAP is an important signaling pathway involved in the regulation of CSCs, and YAP1 signaling is closely associated with the proliferation and metastasis of CRC cells (58). Hippo pathway core kinases include Mst1/2, Sav1, Lats1/2, and Mob1 (97). When Hippo signaling is activated, the Mst1/2 kinase and Sav1 complex co-phosphorylate and activate Lats1/2 kinase. Subsequently, the downstream transcription coactivators YAP and TAZ are inactivated through cytoplasmic retention and protein degradation, which ultimately regulate the expression of target genes and promote tumor progression (98) (Figure 7).
Figure 7.
The circRNA/Hippo/YAP signaling axis in CRC. Mst1/2 is activated by upstream NF2 and RASSF family proteins. Phosphorylated Mst1/2 and MAP4Ks transmit multiple signals to activate Lats1/2, which inhibit YAP/TAZ phosphorylation. The activated YAP/TAZ enters the nucleus and binds with the transcription factor TEADs to induce gene expression. Ultimately, it regulates cell proliferation, migration, apoptosis, and the stemness properties of CSCs.
Interestingly, there is growing evidence that circRNA can act as an oncogene or tumor suppressor to regulate the CSC-related Hippo/YAP signaling pathway. Recently, circ-PPP1R12A was screened for elevated expression in colon cancer cytoplasm. Circ-PPP1R12A encodes the conserved 73-aa small peptide PPP1R12A-C (but not circ-PPP1R12A itself), which promotes the proliferation, migration, and metastasis of colon cancer in vitro and in vivo by activating the Hippo/YAP signaling pathway (51). In addition, other studies have proved that circ_0128846 and circ_0106714 regulate the proliferation and migration of CRC cells through the Hippo/YAP signaling pathway mediated by miR-1184 and miR-942-5p, respectively (29, 46).
CircRNA/WNT/β−catenin signaling axis in CRC
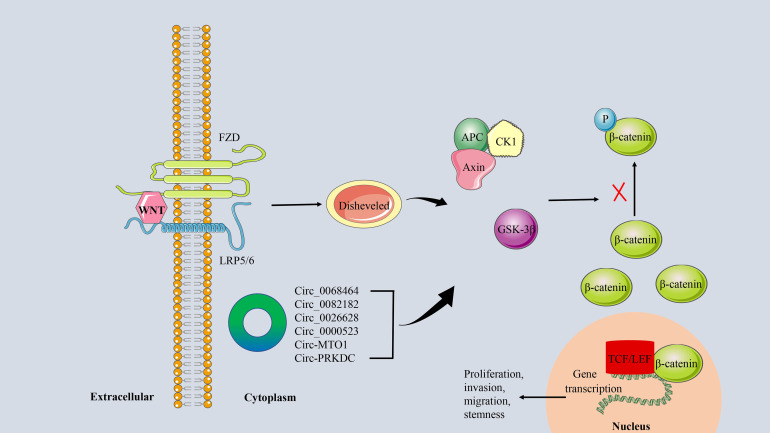
The WNT/β-catenin signaling pathway is a key regulator of normal intestinal stem cell homeostasis (99). Abnormal activation of this pathway is associated with the invasion, migration, proliferation, and differentiation of CRC cells and is a marker of poor prognosis in CRC patients (100). WNT/β-catenin signaling is initiated by binding the WNT protein to the FZD-LRP5/6 receptor complex. This was followed by activation of Disheveled, which further induced dissociation of GSK-3β from Axin (101). This process prevents the WNT-FZD-Axin-LRP5/6 complex from phosphorylating β-catenin. The accumulation of unphosphorylated β-catenin in the cytoplasm translocates to the nucleus, where it binds to transcription factors such as the TCF/LEF family, resulting in the transcription of target genes that enhance CRC stemness and promote CRC progression (102, 103) (Figure 8). Cancer-related deaths in CRC patients are partly due to treatment failure due to chemotherapy resistance. WNT/β-catenin signaling has been shown to mediate chemical resistance to CRC in ncRNA, CSCs, and TME (104).
Figure 8.
The circRNA/WNT/β−catenin signaling axis in CRC. The linking of WNT protein to the FZD-LRP5/6 receptor complex activates the downstream protein Disheveled. It further promoted the dissociation of GSK-3β from Axin, which inhibited the phosphorylation of β-catenin. Unphosphorylated β-catenin is transferred to the nucleus and binds to the transcription factor TCF/LEF to induce transcription of the target genes.
As a special type of ncRNA, circRNA can also influence tumor progression by regulating the WNT/β-catenin signaling pathway in CRC. For example, circ_0068464 (105), circ_0082182 (106), and circ_0026628 (107), which are highly expressed in CRC, can target corresponding miRNAs to regulate the activity of the WNT/β-catenin pathway, thereby promoting tumor progression. In addition, low expression of circRNA, such as circ_0000523 (108) and circ-MTO1 (109), can also participate in the proliferation and invasion of CRC cells in vitro by activating the WNT/β-catenin signaling pathway. 5-FU is a chemotherapeutic drug commonly used in the clinical treatment of CRC, and circRNA has been proven to be a basic regulator of cancer drug resistance (110). Chen et al. found that circ-PRKDC is up-regulated in 5-FU-resistant CRC tissues and cells, and inhibition of circ-PRKDC expression can improve the sensitivity of CRC cells to 5-FU by regulating WNT/β-catenin signaling (111).
Potential clinical application value of circRNAs signal axis in CRC
The diagnostic, prognostic, and therapeutic value of circRNAs in CRC
According to statistics, from 2009 to 2015, the 5-year survival rate of CRC in the United States reached 64%, while the survival rate in many Eastern and Southern European countries was less than 50% (112). In particular, the 5-year survival rate for advanced metastatic CRC is only 14% (113). Therefore, it is necessary to find more effective biomarkers for the early diagnosis and treatment of CRC. Studies have shown that the expression of circ_3823 in serum has high sensitivity and specificity for detecting CRC, suggesting that circ_3823 can be used as a potential biomarker for the diagnosis of CRC (114). In addition, the high expression of circ_0004104 in CRC can not only promote cell proliferation and migration but is also closely related to the prognosis of CRC patients, making it a potential therapeutic target for CRC patients (19). Similarly, high levels of circ-MYH9 predict shorter relapse-free survival and OS in CRC patients, so regulation of circ-MYH9 may lead to an effective treatment for CRC (115). Chemotherapy is one of the main treatment methods for CRC, and oxaliplatin is a more commonly used chemotherapy drug. At present, resistance to CRC treatment is still an important issue for controlling the progression of the disease (116). It has been confirmed that glycolysis and resistance of drug-sensitive cells can be enhanced when exosomes present circ_0005963. The results also showed that the expression level of circ_00059633 in serum exosomes was positively related to the chemoresistance of CRC cells to oxaliplatin, and that silencing of the circRNA could reverse the resistance to oxaliplatin (117). More comprehensive clinical application data of circRNAs in CRC is summarized in Table 1.
Table 1.
Potential clinical application value of circRNAs in CRC.
| CircRNA | Expression | Effect | Potential application | Reference |
|---|---|---|---|---|
| Circ_3823 | Upregulated | Promote growth, metastasis, and angiogenesis | Diagnostic marker or therapeutic target | (114) |
| Circ_0004104 | Upregulated | Promote proliferation, and migration | Diagnostic and prognostic biomarker, and therapeutic target | (19) |
| Circ-MYH9 | Upregulated | Promote proliferation | Therapeutic target | (115) |
| Circ_0005963 | Upregulated | Promote glycolysis and oxaliplatin resistance | Therapeutic target of drug‐resistant patients | (117) |
| CircRNA_0001178 | Upregulated | – | Diagnosing liver metastases from CRC | (18) |
| CircRNA_0000826 | Upregulated | – | Diagnosing liver metastases from CRC | (18) |
| Circ-HERC4 | Upregulated | Promote proliferation and migration/Induce liver and lung metastasis | Prognostic biomarker and therapeutic target | (20) |
| Circ-MAPK14 | Downregulated | Block progression and metastasis | Therapeutic target | (70) |
| Circ-ITGA7 | Downregulated | Suppress growth and metastasis | Therapeutic target | (71) |
| CiRS-7 | Upregulated | Promote proliferation, migration, and invasion | Prognostic biomarker and therapeutic target | (72) |
| Circ_0008285 | Downregulated | Inhibit proliferation and migration | Therapeutic target | (79) |
| Circ-IL4R | Upregulated | Promote proliferation, migration, and invasion | Diagnostic and prognostic biomarker, and therapeutic target | (80) |
| Circ_001971 | Upregulated | Promote proliferation, invasion, and angiogenesis | Therapeutic target | (31) |
| Circ_0056618 | Upregulated | Promote proliferation, migration, and angiogenesis | Therapeutic target | (86) |
| Circ-SPARC | Upregulated | Promote proliferation and migration | Diagnostic and prognostic biomarker, and therapeutic target | (19) |
| Circ_0000372 | Upregulated | Promote proliferation, migration, and invasion | Prognostic biomarker and therapeutic target | (28) |
| Circ-NSD2 | Upregulated | Promote migration and metastasis | Prognostic biomarker and therapeutic target | (27) |
| Circ-APLP2 | Upregulated | Promote proliferation, migration, and invasion/Induce tumor growth and liver metastases | Therapeutic target | (96) |
| Circ-PPP1R12A | Upregulated | Promote proliferation, migration, and invasion | Therapeutic target | (51) |
| Circ_0128846 | Upregulated | Promote proliferation, migration, invasion, and cell cycle progression/Inhibit apoptosis | Therapeutic target | (29) |
| Circ_0106714 | Downregulated | Inhibit proliferation, migration, and invasion/Promote apoptosis | Prognostic biomarker and therapeutic target | (46) |
| Circ_0068464 | Upregulated | Promote proliferation, migration/Induce tumor growth and lung metastasis | Diagnostic biomarker and therapeutic target | (105) |
| Circ_0082182 | Upregulated | Promote proliferation, cell cycle progression, and metastasis/Inhibit apoptosis. | Diagnostic biomarker and therapeutic target | (106) |
| Circ_0026628 | Upregulated | Promote proliferation, migration, EMT, and stemness | Therapeutic target | (107) |
| Circ_0000523 | Downregulated | Inhibit proliferation/Promote apoptosis | Therapeutic target | (108) |
| Circ-MTO1 | Downregulated | Inhibit proliferation and invasion | Therapeutic target | (109) |
| Circ-PRKDC | Upregulated | Enhance 5-FU resistance | Therapeutic target of 5-FU-resistant patients | (111) |
Clinical application of circRNAs related signaling pathways in CRC
Early CRC patients are still mainly treated with surgery, and mCRC patients are based on chemotherapy and targeted therapy. Activation of RAS and BRAF mutations in MAPK signaling is one of the most common mutations in human tumors, and the presence of BRAFV600E mutations is considered to be a marker of poor prognosis in mCRC patients (66). Dabrafenib, encorafenib, and vemurafenib (inhibiting BRAF signaling), and trametinib and binimetinib (inhibiting MEK signaling) have previously been developed for the treatment of mCRC by blocking the MAPK pathway (118). However, some mutations such as KRAS and BRAF make mCRC resistant to these therapies (61). Some preclinical data suggest that abnormal activation of EGFR, PI3K signaling, and WNT signaling pathways may be responsible for resistance to BRAF inhibitor monotherapy in patients with BRAFV600E mCRC (35, 119, 120). Therefore, preclinical studies have been conducted, such as BRAF inhibitor and anti-EGFR antibody dual therapy (vemurafenib + panitumumab) (121), BRAF inhibitor and MEK inhibitor dual therapy (dabrafenib + trametinib) (122), and BRAF inhibitor, anti-EGFR antibody with PI3K or MEK inhibitor triple therapy (encorafenib, cetuximab, and alpelisib or binimetinib) (123, 124). Moreover, these combination therapies involving MAPK pathway blocking have achieved certain efficacy in BRAFV600E mCRC patients. However, the three-drug combination therapy brought more adverse reactions to patients, such as hyperglycemia, nausea, diarrhea, and so on (123). In addition, studies have shown that circ_0131663 (70), circ-ITGA7 (71), and CIRS-7 (72) affect the progression and metastasis of CRC by regulating the MAPK signaling pathway. We believe that the targeted therapies have certain efficacy in specific circRNA patients. Further research is expected. The above data is presented in detail in Table 2.
Table 2.
Clinical trials of circRNAs related signaling pathways in CRC.
| Drug | Signaling pathway | Therapeutic targets | Phase | Reference/NCT number | Identified circRNAs |
|---|---|---|---|---|---|
| Encorafenib | MAPK | BRAF inhibitor | Phase I | (118) | Circ_0131663 (70) Circ-ITGA7 (71) CIRS-7 (72) |
| Vemurafenib | MAPK | BRAF inhibitor | Phase II | NCT00405587 | |
| Trametinib + panitumumab | MAPK | MEK inhibitor and anti-EGFR antibody | Phase II | NCT02399943 | |
| Dabrafenib + trametinib | MAPK | BRAF inhibitor and MEK inhibitor | Phase I/II | (122) | |
| Dabrafenib + panitumumab | MAPK | BRAF inhibitor and anti-EGFR antibody | Phase I | NCT01750918 | |
| Vemurafenib + cetuximab/panitumumab | MAPK | BRAF inhibitor and anti-EGFR antibody | Phase I/Phase II | (121) | |
| Encorafenib + cetuximab | MAPK | BRAF inhibitor and anti-EGFR antibody | Phase III | NCT02928224 | |
| Encorafenib + cetuximab + binimetinib | MAPK | BRAF inhibitor, anti-EGFR antibody with MEK inhibitor | Phase III | NCT02928224 | |
| Dabrafenib + trametinib + panitumumab | MAPK | BRAF inhibitor and MEK inhibitor with anti-EGFR antibody | Phase I | NCT01750918 | |
| PX-866 + cetuximab | PI3K/AKT | PI3K pan-inhibitor and anti-EGFR antibody | Phase II | (125) | Circ_0008285 (79) Circ-IL4R (80) |
| BKM120 (buparlisib) | PI3K/AKT | PI3K pan-inhibitor | Phase I/Phase II | (126)/NCT01833169 | |
| BKM120 + irinotecan/docetaxel | PI3K/AKT | PI3K pan-inhibitor | Phase I/Phase I | NCT01304602/NCT01540253 | |
| BKM120 + panitumumab/paclitaxel/everolimus | PI3K/AKT | PI3K pan-inhibitor and anti-EGFR antibody/mTOR inhibitor/mTOR inhibitor | Phase I/II/Phase III/Phase I | NCT01591421/NCT04338399/NCT01470209 | |
| GDC-0941 | PI3K/AKT | PI3K pan-inhibitor | Phase I | NCT00876109 | |
| GDC-0941 + erlotinib | PI3K/AKT | PI3K pan-inhibitor and anti-EGFR antibody | Phase I | NCT00975182 | |
| MEN1611 | PI3K/AKT | PI3K Selective-inhibitor | Phase I/Ib | NCT04495621 | |
| MEN1611 + cetuximab | PI3K/AKT | PI3K Selective-inhibitor and anti-EGFR antibody | Phase I | NCT04495621 | |
| KRX-0401 | PI3K/AKT | AKT inhibitor | Phase I | (127) | |
| MK-2206 | PI3K/AKT | AKT inhibitor | Phase II | NCT01802320 | |
| GDC-0068 | PI3K/AKT | AKT inhibitor | Phase I | NCT01090960 | |
| PF-05212384 | PI3K/AKT | PI3K/mTOR inhibitor | Phase I | (128) | |
| BEZ235 | PI3K/AKT | PI3K/mTOR inhibitor | Phase I | (129) | |
| GDC-0980 | PI3K/AKT | PI3K/mTOR inhibitor | Phase I | NCT00854152 | |
| DS-7423 | PI3K/AKT | PI3K/mTOR inhibitor | Phase I | NCT01364844 | |
| PKI-587 | PI3K/AKT | PI3K/mTOR inhibitor | Phase I | NCT00940498 | |
| XL-765 | PI3K/AKT | PI3K/mTOR inhibitor | Phase I | NCT00485719 | |
| XL-765 + erlotinib | PI3K/AKT | PI3K/mTOR inhibitor and anti-EGFR antibody | Phase I | NCT00777699 | |
| Temsirolimus + irinotecan/cetuximab | PI3K/AKT | mTOR inhibitor/anti-EGFR antibody | Phase II/Phase I | NCT00827684/NCT00593060 | |
| Everolimus | PI3K/AKT | mTOR inhibitor | Phase II | NCT00419159/NCT01387880/NCT00337545 | |
| Everolimus + BEZ235 | PI3K/AKT | mTOR inhibitor + PI3K/mTOR inhibitor | Phase I/II | NCT01508104 | |
| AZD2014 + paclitaxel | PI3K/AKT | mTORC1/mTORC2 inhibitor | Phase I | NCT02193633 | |
| Encorafenib + cetuximab + alpelisib | MAPK and PI3K/AKT | BRAF inhibitor, anti-EGFR antibody with PI3K inhibitor | Phase I/IIb | (123) | Circ_0131663 (70) Circ-ITGA7 (71) CIRS-7 (72) Circ_0008285 (79) Circ-IL4R (80) |
| BKM120 + binimetinib | PI3K/AKT and MAPK | PI3K pan-inhibitor and MEK inhibitor | Phase Ib | NCT01363232 | |
| BYL719 + LGX818 + cetuximab | PI3K/AKT and MAPK | PI3K Selective-inhibitor, BRAF inhibitor with anti-EGFR antibody | Phase I | NCT01719380 | |
| MK-2206 + AZD6244 | PI3K/AKT and MAPK | AKT inhibitor and MEK inhibitor | Phase II | NCT01333475 | |
| PF-04691502 + PD-0325901 | PI3K/AKT and MAPK | PI3K/mTOR inhibitor and MEK inhibitor | Phase I | (130) | |
| BEZ235 + binimetinib | PI3K/AKT and MAPK | PI3K/mTOR inhibitor and MEK inhibitor | Phase I | NCT01337765 | |
| XL147 + pimasertib/erlotinib | PI3K/AKT and MAPK | PI3K/mTOR inhibitor and MEK inhibitor/anti-EGFR antibody | Phase I | NCT01357330/NCT00692640 | |
| Bevacizumab | VEGF | anti-VEGFA antibody | FDA approved | (131) | Circ_001971 (31) Circ_0056618 (86) |
| Regorafenib | VEGF | anti-VEGFR antibody | FDA approved | (131) | |
| Ramucirumab | VEGF | anti-VEGFR antibody | FDA approved | (131) | |
| Ziv-aflibercept | VEGF | anti-VEGF antibody | FDA approved | (131) | |
| Napabucasin | JAK/STAT | STAT3 inhibitor | Phase III | (132) | Circ_0004104 (19) Circ_0000372 (28) |
| Napabucasin + bevacizumab + FOLFIRI (5‐FU, leucovorin, and irinotecan) | JAK/STAT and VEGF | STAT3 inhibitor and anti-VEGFA antibody | Phase I | NCT02641873 | Circ_001971 (31) Circ_0056618 (86) Circ_0004104 (19) Circ_0000372 (28) |
| Ruxolitinib + regorafenib | JAK/STAT and VEGF | JAK1/2 inhibitor and anti-VEGFR antibody | Phase II | NCT02119676 | |
| OMP-52M51 | Notch | anti-Notch1 antibody | Phase I | (133) | Circ-NSD2 (27) Circ-APLP2 (96) |
| RO4929097 | Notch | Gamma secretase inhibitor | Phase I | NCT01116687 | |
| OMP131R10 | WNT/β-catenin | Wnt-receptor complex inhibitor | Phase I | NCTO2482441 | Circ_0068464 (105) Circ_0082182 (106) Circ_0026628 (107) Circ_0000523 (109) Circ-MTO1 (109) Circ-PRKDC (111) |
| PRI-724 | WNT/β-catenin | Wnt-receptor complex inhibitor | Phase I/II | NCT01764477 | |
| Foxy 5 | WNT/β-catenin | Wnt-receptor complex inhibitor | Phase I/ | NCTO2655952 | |
| LGK974 | WNT/β-catenin | Wnt-receptor complex inhibitor | Phase I/II | NCTO2278133 | |
| ETC-159 | WNT/β-catenin | Wnt-receptor complex inhibitor | Phase I | NCTO2521844 |
Currently, compounds targeting the PI3K/AKT signaling axis are mainly divided into four types: PI3K inhibitors, AKT inhibitors, mTOR inhibitors, and dual PI3K/mTOR inhibitors. Class Ia PI3K pan-inhibitors PX-866 and BKM120 (buparlisib) have shown good anti-tumor effects in preclinical studies of a variety of tumors (126, 134) and some clinical trials in CRC patients are ongoing. These included PX-866 in combination with cetuximab (NCT01252628), BKM120 in combination with panizumab (NCT01591421), and BKM120 in combination with paclitaxel (NCT04338399). However, the results are not satisfactory. In patients with mCRC, the addition of PX-866 to cetuximab failed to improve OS, progression-free survival (PFS), and objective response rates, but resulted in greater toxicity (125). The BURAN Study (NCT04338399) is scheduled for completion in December 2023. Other PI3K inhibitors are specific subtype inhibitors with strong targeted inhibition and low toxicity properties, such as MEN1611 and BYL719. Phase I trials are being conducted in mCRC patients with BRAF mutations (BYL719 and LGX818 [BRAF inhibitor] with cetuximab) (NCT01719380) and CRC patients with PIK3CA mutations (MEN1611 and cetuximab) (NCT04495621) (135). Some AKT inhibitors, such as KRX-0401, have been proven to be effective in patients with mCRC (127), while others, such as MK-2206, are still in clinical trials (NCT01802320) (NCT01333475). In addition, dual PI3K/mTOR inhibitors also showed preliminary tumor regression ability in CRC patients, among which representative ones were PF-05212384 (128), PF-04691502 (130), and NVP-BEZ235 (129). At present, the most common mTOR inhibitors in clinical trials are temsirolimus and everolimus, but the overall efficacy of mTOR inhibition in clinical application is limited except for certain disease-stabilizing effects in patients with refractory mCRC (76). However, these targeted inhibitors may have a better therapeutic effect in circ_0008285 (79) and circ-IL4R (80) patients because these two circRNAs can promote the proliferation and migration of CRC cells by regulating the PI3K/AKT signaling pathway. These clinical applications are also listed in Table 2.
The VEGFA-targeted monoclonal antibody bevacizumab is the first targeted agent to be approved for the treatment of patients with mCRC. After that, three anti-angiogenic drugs, regorafenib, ramucirumab, and ziv-aflibercept, were also approved in the mCRC (131). Antiangiogenic agents do not directly target cancer cells, but rather target the TME like immune checkpoint inhibitors (136). Due to the inevitable problem of drug resistance, it is also a hot topic to explore the combination therapy of antiangiogenic drugs and immune checkpoint inhibitors in addition to developing new antiangiogenic drugs. On the other hand, the combination of targeted regulation of circ_001971 and circ_0056618 may reverse drug resistance. Clinical reports of JAK inhibitors or STAT inhibitors in the treatment of CRC are rare. In a Phase III trial of patients with mCRC, there were no significant differences in OS, PFS, or disease control between the STAT3 inhibitor (napabucasin) group and the placebo group. However, OS was significantly prolonged in py-STAT3-positive patients treated with napabucasin (132). Several natural and small-molecule inhibitors that inhibit Notch signaling have been shown to induce apoptosis in CRC cells in vitro, but they lack target specificity and efficacy in clinical evaluation (94). In addition, monoclonal antibodies targeting Notch1, such as OMP-52M51, did not show significant antitumor efficacy in phase I dose-escalation trials (NCT01778439) (133). Similarly, CRC therapies targeting WNT/β-catenin signaling include natural compounds, small molecules, and biological agents. The drugs currently in clinical trials include vitamin D3, curcumin, genistein, resveratrol, LGK974, and ETC-159 (100). Targeting WNT signaling pathways in CRC seems to be a long and difficult process, and no drugs targeting WNT pathways have been approved at present. These targeted therapies may be beneficial for CRC patients in some identified circRNA patients. The clinical data and potentially identified circRNAs are summarized in Table 2.
Conclusions and perspectives
In conclusion, CRC is still a disease with high morbidity and mortality worldwide. Although with the improvement of people’s health awareness and the continuous improvement of diagnosis and treatment technology, some patients can be detected and treated early with a good prognosis. However, the current treatment methods for patients with advanced metastatic CRC are limited and the prognosis is poor. Therefore, human beings have never stopped exploring the mechanism of CRC occurrence and metastasis. Excitingly, a novel ncRNA, circRNAs, has become the focus of CRC research due to its critical regulatory role in cancer-related processes such as tumorigenesis, development, and apoptosis. To improve patients’ OS, many circRNAs are being developed as potential biomarkers for clinical diagnosis and prognosis of CRC, as well as effective therapeutic targets. More and more studies have shown that circRNA usually promotes the proliferation and metastasis of CRC cells by regulating several important signaling pathways, including PI3K/AKT, MAPK, Notch, JAK/STAT, Hippo/YAP, WNT/β-catenin, and VEGF. Currently, there are very few targeted drugs based on these signaling pathways for the clinical treatment of mCRC patients. This may be related to the lack of a deeper and comprehensive understanding of the biological functions and carcinogenic mechanisms of circRNAs and related signaling pathways in CRC. Therefore, based on the comprehensive elaboration and exploration of the molecular mechanism of CRC occurrence and progression in this review, more circRNA signal axes can be developed as effective targets for clinical diagnosis, prognosis, and treatment of CRC in the future to serve patients.
Author contributions
SW and LC were mainly responsible for literature review and manuscript writing. HW and GL completed the construction pictures and tables. SW designed the ideas of this paper and modified the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by : Wuxi Traditional Chinese Medicine Hospital Inheritance Studio construction project (2020 No.5).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CRC, colorectal cancer; CSCs, cancer stem cells; circRNAs, circular RNAs; CMS, consensus molecular subtypes; ceRNA, competitive endogenous RNA; EGFR, epidermal growth factor receptor; EMT, epithelial-mesenchymal transition; IL6, interleukin-6; JAK, janus kinase; MAPK, mitogen-activated protein kinases; mCRC, metastatic CRC; mTOR, mammalian targets of the rapamycin; miRNA, microRNA; ncRNAs, non-coding RNAs; OS, overall survival; PlGF, placental growth factor; P-JAK, phosphorylation-JAK; PI3K, phosphatidylinositol 3-kinase; PFS, progression-free survival; RT-qPCR, quantitative reverse transcription-PCR; STAT, signal transducers and activators of transcription; TGF-β, transforming growth factor-beta; TME, tumor microenvironment; VEGF, vascular endothelial growth factor.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71(3):209–49. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet (2019) 394(10207):1467–80. doi: 10.1016/S0140-6736(19)32319-0 [DOI] [PubMed] [Google Scholar]
- 3. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut (2017) 66(4):683–91. doi: 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- 4. Long F, Lin Z, Li L, Ma M, Lu Z, Jing L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer (2021) 20(1):26. doi: 10.1186/s12943-021-01318-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA (2021) 325(7):669–85. doi: 10.1001/jama.2021.0106 [DOI] [PubMed] [Google Scholar]
- 6. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin (2016) 66(1):7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 7. Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell (2019) 176(4):869–881 e13. doi: 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Papatsirou M, Artemaki PI, Karousi P, Scorilas A, Kontos CK. Circular RNAs: Emerging regulators of the major signaling pathways involved in cancer progression. Cancers (Basel) (2021) 13(11):2744. doi: 10.3390/cancers13112744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer (2021) 20(1):13. doi: 10.1186/s12943-020-01298-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salami R, Salami M, Mafi A, Vakili O, Asemi Z. Circular RNAs and glioblastoma multiforme: focus on molecular mechanisms. Cell Commun Signal (2022) 20(1):13. doi: 10.1186/s12964-021-00809-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform (2017) 18(6):984–92. doi: 10.1093/bib/bbw081 [DOI] [PubMed] [Google Scholar]
- 12. Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics (2020) 10(8):3503–17. doi: 10.7150/thno.42174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: Biogenesis, function and role in human diseases. Front Mol Biosci (2017) 6(4):38. doi: 10.3389/fmolb.2017.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther (2018) 187:31–44. doi: 10.1016/j.pharmthera.2018.01.010 [DOI] [PubMed] [Google Scholar]
- 15. Zhou Z, Sun B, Huang S, Zhao L. Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis (2019) 10(7):503. doi: 10.1038/s41419-019-1744-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xue C, Li G, Lu J, Li L. Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression. Signal Transduct Target Ther (2021) 6(1):400. doi: 10.1038/s41392-021-00788-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov (2020) 7(6):72. doi: 10.1038/s41420-020-00306-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer (2019) 18(1):8. doi: 10.1186/s12943-018-0932-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J, Zhang Y, Song H, Yin H, Jiang T, Xu Y, et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol Cancer (2021) 20(1):81. doi: 10.1186/s12943-021-01375-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He J, Chu Z, Lai W, Lan Q, Zeng Y, Lu D, et al. Circular RNA circHERC4 as a novel oncogenic driver to promote tumor metastasis via the miR-556-5p/CTBP2/E-cadherin axis in colorectal cancer. J Hematol Oncol (2021) 14(1):194. doi: 10.1186/s13045-021-01210-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park JH, Pyun WY, Park HW. Cancer metabolism: Phenotype, signaling and therapeutic targets. Cells (2020) 9(10):2308. doi: 10.3390/cells9102308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karnell JL, Rieder SA, Ettinger R, Kolbeck R. Targeting the CD40-CD40L pathway in autoimmune diseases: Humoral immunity and beyond. Adv Drug Deliv Rev (2019) 141:92–103. doi: 10.1016/j.addr.2018.12.005 [DOI] [PubMed] [Google Scholar]
- 23. Artemaki PI, Scorilas A, Kontos CK. Circular RNAs: A new piece in the colorectal cancer puzzle. Cancers (Basel) (2020) 12(9):2464. doi: 10.3390/cancers12092464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou R, Wu Y, Wang W, Su W, Liu Y, Wang Y, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett (2018) 425:134–42. doi: 10.1016/j.canlet.2018.03.035 [DOI] [PubMed] [Google Scholar]
- 25. Kong Y, Li Y, Luo Y, Zhu J, Zheng H, Gao B, et al. circNFIB1 inhibits lymphangiogenesis and lymphatic metastasis via the miR-486-5p/PIK3R1/VEGF-C axis in pancreatic cancer. Mol Cancer (2020) 19(1):82. doi: 10.1186/s12943-020-01205-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li C, Zhou H. Circular RNA hsa_circRNA_102209 promotes the growth and metastasis of colorectal cancer through miR-761-mediated ras and rab interactor 1 signaling. Cancer Med (2020) 9(18):6710–25. doi: 10.1002/cam4.3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen LY, Zhi Z, Wang L, Zhao YY, Deng M, Liu YH, et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol (2019) 248(1):103–15. doi: 10.1002/path.5238 [DOI] [PubMed] [Google Scholar]
- 28. Liu X, Qin Y, Tang X, Wang Y, Bian C, Zhong J. Circular RNA circ_0000372 contributes to the proliferation, migration and invasion of colorectal cancer by elevating IL6 expression via sponging miR-495. Anticancer Drugs (2021) 32(3):296–305. doi: 10.1097/CAD.0000000000001002 [DOI] [PubMed] [Google Scholar]
- 29. Wang X, Chen Y, Liu W, Liu T, Sun D. Hsa_circ_0128846 promotes tumorigenesis of colorectal cancer by sponging hsa-miR-1184 and releasing AJUBA and inactivating Hippo/YAP signalling. J Cell Mol Med (2020) 24(17):9908–24. doi: 10.1111/jcmm.15590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (Lond) (2019) 133(10):1197–213. doi: 10.1042/CS20190286 [DOI] [PubMed] [Google Scholar]
- 31. Chen C, Huang Z, Mo X, Song Y, Li X, Li X, et al. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res (2020) 39(1):91. doi: 10.1186/s13046-020-01594-y [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Nassar D, Blanpain C. Cancer stem cells: Basic concepts and therapeutic implications. Annu Rev Pathol (2016) 11:47–76. doi: 10.1146/annurev-pathol-012615-044438 [DOI] [PubMed] [Google Scholar]
- 33. Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauss A, et al. Cancer stem cells-origins and biomarkers: Perspectives for targeted personalized therapies. Front Immunol (2020) 11:1280. doi: 10.3389/fimmu.2020.01280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Munro MJ, Wickremesekera SK, Peng L, Tan ST, Itinteang T. Cancer stem cells in colorectal cancer: a review. J Clin Pathol (2018) 71(2):110–6. doi: 10.1136/jclinpath-2017-204739 [DOI] [PubMed] [Google Scholar]
- 35. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature (2012) 487(7407):330–7. doi: 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. La Vecchia S, Sebastian C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin Cell Dev Biol (2020) 98:63–70. doi: 10.1016/j.semcdb.2019.05.018 [DOI] [PubMed] [Google Scholar]
- 37. Ma H, Brosens LAA, Offerhaus GJA, Giardiello FM, de Leng WWJ, Montgomery EA. Pathology and genetics of hereditary colorectal cancer. Pathology (2018) 50(1):49–59. doi: 10.1016/j.pathol.2017.09.004 [DOI] [PubMed] [Google Scholar]
- 38. Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology (2013) 62(3):367–86. doi: 10.1111/his.12055 [DOI] [PubMed] [Google Scholar]
- 39. Sveen A, Bruun J, Eide PW, Eilertsen IA, Ramirez L, Murumagi A, et al. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res (2018) 24(4):794–806. doi: 10.1158/1078-0432.CCR-17-1234 [DOI] [PubMed] [Google Scholar]
- 40. Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med (2015) 21(11):1350–6. doi: 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Menter DG, Davis JS, Broom BM, Overman MJ, Morris J, Kopetz S. Back to the colorectal cancer consensus molecular subtype future. Curr Gastroenterol Rep (2019) 21(2):5. doi: 10.1007/s11894-019-0674-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Loree JM, Pereira AAL, Lam M, Willauer AN, Raghav K, Dasari A, et al. Classifying colorectal cancer by tumor location rather than sidedness highlights a continuum in mutation profiles and consensus molecular subtypes. Clin Cancer Res (2018) 24(5):1062–72. doi: 10.1158/1078-0432.CCR-17-2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee MS, Menter DG, Kopetz S. Right versus left colon cancer biology: Integrating the consensus molecular subtypes. J Natl Compr Canc Netw (2017) 15(3):411–9. doi: 10.6004/jnccn.2017.0038 [DOI] [PubMed] [Google Scholar]
- 44. Radanova M, Mihaylova G, Nazifova-Tasinova N, Levkova M, Tasinov O, Ivanova D, et al. Oncogenic functions and clinical significance of circular RNAs in colorectal cancer. Cancers (Basel) (2021) 13(14):3395. doi: 10.3390/cancers13143395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer (2017) 16(1):58. doi: 10.1186/s12943-017-0630-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li S, Yan G, Liu W, Li C, Wang X. Circ0106714 inhibits tumorigenesis of colorectal cancer by sponging miR-942-5p and releasing DLG2 via hippo-YAP signaling. Mol Carcinog (2020) 59(12):1323–42. doi: 10.1002/mc.23259 [DOI] [PubMed] [Google Scholar]
- 47. Liu J, Zhang X, Yan M, Li H. Emerging role of circular RNAs in cancer. Front Oncol (2020) 10:663. doi: 10.3389/fonc.2020.00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li F, Yang Q, He AT, Yang BB. Circular RNAs in cancer: Limitations in functional studies and diagnostic potential. Semin Cancer Biol (2021) 75:49–61. doi: 10.1016/j.semcancer.2020.10.002 [DOI] [PubMed] [Google Scholar]
- 49. Karousi P, Artemaki PI, Sotiropoulou CD, Christodoulou S, Scorilas A, Kontos CK. Identification of two novel circular RNAs deriving from BCL2L12 and investigation of their potential value as a molecular signature in colorectal cancer. Int J Mol Sci (2020) 21(22):8867. doi: 10.3390/ijms21228867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mahmoudi E, Kiltschewskij D, Fitzsimmons C, Cairns MJ. Depolarization-associated CircRNA regulate neural gene expression and in some cases may function as templates for translation. Cells (2019) 9(1):25. doi: 10.3390/cells9010025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via hippo-YAP signaling. Mol Cancer (2019) 18(1):47. doi: 10.1186/s12943-019-1010-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer (2019) 10(21):5015–21. doi: 10.7150/jca.30828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA Sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res (2018) 37(1):325. doi: 10.1186/s13046-018-1006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol Cancer (2020) 19(1):164. doi: 10.1186/s12943-020-01272-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang N, Ng AS, Cai S, Li Q, Yang L, Kerr D. Novel therapeutic strategies: targeting epithelial-mesenchymal transition in colorectal cancer. Lancet Oncol (2021) 22(8):e358–68. doi: 10.1016/S1470-2045(21)00343-0 [DOI] [PubMed] [Google Scholar]
- 56. Li YF, Pei FL, Cao MZ. CircRNA_101951 promotes migration and invasion of colorectal cancer cells by regulating the KIF3A-mediated EMT pathway. Exp Ther Med (2020) 19(5):3355–61. doi: 10.3892/etm.2020.8600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Batlle E, Clevers H. Cancer stem cells revisited. Nat Med (2017) 23(10):1124–34. doi: 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- 58. Das PK, Islam F, Lam AK. The roles of cancer stem cells and therapy resistance in colorectal carcinoma. Cells (2020) 9(6):1392. doi: 10.3390/cells9061392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rengganaten V, Huang CJ, Tsai PH, Wang ML, Yang YP, Lan YT, et al. Mapping a circular RNA-microRNA-mRNA-Signaling regulatory axis that modulates stemness properties of cancer stem cell populations in colorectal cancer spheroid cells. Int J Mol Sci (2020) 21(21):7864. doi: 10.3390/ijms21217864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci (2020) 77(9):1661–80. doi: 10.1007/s00018-019-03345-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanz-Garcia E, Argiles G, Elez E, Tabernero J. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann Oncol (2017) 28(11):2648–57. doi: 10.1093/annonc/mdx401 [DOI] [PubMed] [Google Scholar]
- 62. Bellio H, Fumet JD, Ghiringhelli F. Targeting BRAF and RAS in colorectal cancer. Cancers (Basel) (2021) 13(9):2201. doi: 10.3390/cancers13092201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Post JB, Roodhart JML, Snippert HJG. Colorectal cancer modeling with organoids: Discriminating between oncogenic RAS and BRAF variants. Trends Cancer (2020) 6(2):111–29. doi: 10.1016/j.trecan.2019.12.005 [DOI] [PubMed] [Google Scholar]
- 64. Corcoran RB, Andre T, Atreya CE, Schellens JHM, Yoshino T, Bendell JC, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAF(V600E)-mutant colorectal cancer. Cancer Discov (2018) 8(4):428–43. doi: 10.1158/2159-8290.CD-17-1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Afrasanie VA, Marinca MV, Alexa-Stratulat T, Gafton B, Paduraru M, Adavidoaiei AM, et al. KRAS, NRAS, BRAF, HER2 and microsatellite instability in metastatic colorectal cancer - practical implications for the clinician. Radiol Oncol (2019) 53(3):265–74. doi: 10.2478/raon-2019-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Grothey A, Fakih M, Tabernero J. Management of BRAF-mutant metastatic colorectal cancer: a review of treatment options and evidence-based guidelines. Ann Oncol (2021) 32(8):959–67. doi: 10.1016/j.annonc.2021.03.206 [DOI] [PubMed] [Google Scholar]
- 67. Prior IA, Lewis PD, Mattos C. A comprehensive survey of ras mutations in cancer. Cancer Res (2012) 72(10):2457–67. doi: 10.1158/0008-5472.CAN-11-2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Patelli G, Tosi F, Amatu A, Mauri G, Curaba A, Patane DA, et al. Strategies to tackle RAS-mutated metastatic colorectal cancer. ESMO Open (2021) 6(3):100156. doi: 10.1016/j.esmoop.2021.100156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol (1996) 16(3):1247–55. doi: 10.1128/MCB.16.3.1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang L, Zhou J, Zhang C, Chen R, Sun Q, Yang P, et al. A novel tumour suppressor protein encoded by circMAPK14 inhibits progression and metastasis of colorectal cancer by competitively binding to MKK6. Clin Transl Med (2021) 11(10):e613. doi: 10.1002/ctm2.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li X, Wang J, Zhang C, Lin C, Zhang J, Zhang W, et al. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the ras pathway and upregulating transcription of its host gene ITGA7. J Pathol (2018) 246(2):166–79. doi: 10.1002/path.5125 [DOI] [PubMed] [Google Scholar]
- 72. Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, et al. Circular RNA ciRS-7-A promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res (2017) 23(14):3918–28. doi: 10.1158/1078-0432.CCR-16-2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Duan S, Huang W, Liu X, Liu X, Chen N, Xu Q, et al. IMPDH2 promotes colorectal cancer progression through activation of the PI3K/AKT/mTOR and PI3K/AKT/FOXO1 signaling pathways. J Exp Clin Cancer Res (2018) 37(1):304. doi: 10.1186/s13046-018-0980-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res (2019) 38(1):112. doi: 10.1186/s13046-019-1043-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stefani C, Miricescu D, Stanescu S, II RI, Greabu M, Totan AR, et al. Growth factors, PI3K/AKT/mTOR and MAPK signaling pathways in colorectal cancer pathogenesis: Where are we now? Int J Mol Sci (2021) 22(19):10260. doi: 10.3390/ijms221910260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bahrami A, Khazaei M, Hasanzadeh M, ShahidSales S, Joudi Mashhad M, Farazestanian M, et al. Therapeutic potential of targeting PI3K/AKT pathway in treatment of colorectal cancer: Rational and progress. J Cell Biochem (2018) 119(3):2460–9. doi: 10.1002/jcb.25950 [DOI] [PubMed] [Google Scholar]
- 77. Yang X, Ye T, Liu H, Lv P, Duan C, Wu X, et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer (2021) 20(1):4. doi: 10.1186/s12943-020-01300-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang C, Mou Z, Wu S, Ou Y, Zhang Z, Chen X, et al. High-throughput sequencing identified circular RNA circUBE2K mediating RhoA associated bladder cancer phenotype via regulation of miR-516b-5p/ARHGAP5 axis. Cell Death Dis (2021) 12(8):719. doi: 10.1038/s41419-021-03977-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wang J, Luo J, Liu G, Li X. Circular RNA hsa_circ_0008285 inhibits colorectal cancer cell proliferation and migration via the miR-382-5p/PTEN axis. Biochem Biophys Res Commun (2020) 527(2):503–10. doi: 10.1016/j.bbrc.2020.03.165 [DOI] [PubMed] [Google Scholar]
- 80. Jiang T, Wang H, Liu L, Song H, Zhang Y, Wang J, et al. CircIL4R activates the PI3K/AKT signaling pathway via the miR-761/TRIM29/PHLPP1 axis and promotes proliferation and metastasis in colorectal cancer. Mol Cancer (2021) 20(1):167. doi: 10.1186/s12943-021-01474-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Li W, Xu Y, Wang X, Cao G, Bu W, Wang X, et al. circCCT3 modulates vascular endothelial growth factor a and wnt signaling to enhance colorectal cancer metastasis through sponging miR-613. DNA Cell Biol (2020) 39(1):118–25. doi: 10.1089/dna.2019.5139 [DOI] [PubMed] [Google Scholar]
- 82. Deng F, Zhou R, Lin C, Yang S, Wang H, Li W, et al. Tumor-secreted dickkopf2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics (2019) 9(4):1001–14. doi: 10.7150/thno.30056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: Beyond discovery and development. Cell (2019) 176(6):1248–64. doi: 10.1016/j.cell.2019.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med (2003) 9(6):669–76. doi: 10.1038/nm0603-669 [DOI] [PubMed] [Google Scholar]
- 85. Garcia J, Hurwitz HI, Sandler AB, Miles D, Coleman RL, Deurloo R, et al. Bevacizumab (Avastin(R)) in cancer treatment: A review of 15 years of clinical experience and future outlook. Cancer Treat Rev (2020) 86:102017. doi: 10.1016/j.ctrv.2020.102017 [DOI] [PubMed] [Google Scholar]
- 86. Zheng X, Ma YF, Zhang XR, Li Y, Zhao HH, Han SG. Circ_0056618 promoted cell proliferation, migration and angiogenesis through sponging with miR-206 and upregulating CXCR4 and VEGF-a in colorectal cancer. Eur Rev Med Pharmacol Sci (2020) 24(8):4190–202. doi: 10.26355/eurrev_202004_20999 [DOI] [PubMed] [Google Scholar]
- 87. Xin P, Xu X, Deng C, Liu S, Wang Y, Zhou X, et al. The role of JAK/STAT signaling pathway and its inhibitors in diseases. Int Immunopharmacol (2020) 80:106210. doi: 10.1016/j.intimp.2020.106210 [DOI] [PubMed] [Google Scholar]
- 88. Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer (Review). Int J Oncol (2014) 44(4):1032–40. doi: 10.3892/ijo.2014.2259 [DOI] [PubMed] [Google Scholar]
- 89. Yue Y, Zhang Q, Wu S, Wang S, Cui C, Yu M, et al. Identification of key genes involved in JAK/STAT pathway in colorectal cancer. Mol Immunol (2020) 128:287–97. doi: 10.1016/j.molimm.2020.10.007 [DOI] [PubMed] [Google Scholar]
- 90. Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Signal Transduct Target Ther (2021) 6(1):402. doi: 10.1038/s41392-021-00791-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang X, Hu F, Li G, Li G, Yang X, Liu L, et al. Human colorectal cancer-derived mesenchymal stem cells promote colorectal cancer progression through IL-6/JAK2/STAT3 signaling. Cell Death Dis (2018) 9(2):25. doi: 10.1038/s41419-017-0176-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li L, Tang P, Li S, Qin X, Yang H, Wu C, et al. Notch signaling pathway networks in cancer metastasis: a new target for cancer therapy. Med Oncol (2017) 34(10):180. doi: 10.1007/s12032-017-1039-6 [DOI] [PubMed] [Google Scholar]
- 93. Erkasap N, Ozyurt R, Ozkurt M, Erkasap S, Yasar F, Ihtiyar E, et al. Role of notch, IL-1 and leptin expression in colorectal cancer. Exp Ther Med (2021) 21(6):600. doi: 10.3892/etm.2021.10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tyagi A, Sharma AK, Damodaran C. A review on notch signaling and colorectal cancer. Cells (2020) 9(6):1549. doi: 10.3390/cells9061549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jackstadt R, van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell (2019) 36(3):319–336 e7. doi: 10.1016/j.ccell.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wu HB, Huang SS, Lu CG, Tian SD, Chen M. CircAPLP2 regulates the proliferation and metastasis of colorectal cancer by targeting miR-101-3p to activate the notch signalling pathway. Am J Transl Res (2020) 12(6):2554–69. [PMC free article] [PubMed] [Google Scholar]
- 97. Chai Y, Xiang K, Wu Y, Zhang T, Liu Y, Liu X, et al. Cucurbitacin b inhibits the hippo-YAP signaling pathway and exerts anticancer activity in colorectal cancer cells. Med Sci Monit (2018) 24:9251–8. doi: 10.12659/MSM.911594 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98. Si Y, Ji X, Cao X, Dai X, Xu L, Zhao H, et al. Src inhibits the hippo tumor suppressor pathway through tyrosine phosphorylation of Lats1. Cancer Res (2017) 77(18):4868–80. doi: 10.1158/0008-5472.CAN-17-0391 [DOI] [PubMed] [Google Scholar]
- 99. Cho YH, Ro EJ, Yoon JS, Mizutani T, Kang DW, Park JC, et al. 5-FU promotes stemness of colorectal cancer via p53-mediated WNT/beta-catenin pathway activation. Nat Commun (2020) 11(1):5321. doi: 10.1038/s41467-020-19173-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cheng X, Xu X, Chen D, Zhao F, Wang W. Therapeutic potential of targeting the wnt/beta-catenin signaling pathway in colorectal cancer. BioMed Pharmacother (2019) 110:473–81. doi: 10.1016/j.biopha.2018.11.082 [DOI] [PubMed] [Google Scholar]
- 101. Krishnamurthy N, Kurzrock R. Targeting the wnt/beta-catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treat Rev (2018) 62:50–60. doi: 10.1016/j.ctrv.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bian J, Dannappel M, Wan C, Firestein R. Transcriptional regulation of wnt/beta-catenin pathway in colorectal cancer. Cells (2020) 9(9):2125. doi: 10.3390/cells9092125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liu X, Su K, Sun X, Jiang Y, Wang L, Hu C, et al. Sec62 promotes stemness and chemoresistance of human colorectal cancer through activating wnt/beta-catenin pathway. J Exp Clin Cancer Res (2021) 40(1):132. doi: 10.1186/s13046-021-01934-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhu GX, Gao D, Shao ZZ, Chen L, Ding WJ, Yu QF. Wnt/betacatenin signaling: Causes and treatment targets of drug resistance in colorectal cancer (Review). Mol Med Rep (2021) 23(2):105. doi: 10.3892/mmr.2020.11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Zou Y, Liu L, Meng J, Dai M. Circular RNA circ_0068464 combined with microRNA-383 regulates wnt/beta-catenin pathway to promote the progression of colorectal cancer. Bioengineered (2022) 13(3):5113–25. doi: 10.1080/21655979.2022.2036905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Liu R, Deng P, Zhang Y, Wang Y, Peng C. Circ_0082182 promotes oncogenesis and metastasis of colorectal cancer in vitro and in vivo by sponging miR-411 and miR-1205 to activate the wnt/beta-catenin pathway. World J Surg Oncol (2021) 19(1):51. doi: 10.1186/s12957-021-02164-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zhang X, Yao J, Shi H, Gao B, Zhou H, Zhang Y, et al. Hsa_circ_0026628 promotes the development of colorectal cancer by targeting SP1 to activate the wnt/beta-catenin pathway. Cell Death Dis (2021) 12(9):802. doi: 10.1038/s41419-021-03794-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Jin Y, Yu LL, Zhang B, Liu CF, Chen Y. Circular RNA hsa_circ_0000523 regulates the proliferation and apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med Biol Res (2018) 51(12):e7811. doi: 10.1590/1414-431X20187811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ge Z, Li LF, Wang CY, Wang Y, Ma WL. CircMTO1 inhibits cell proliferation and invasion by regulating wnt/beta-catenin signaling pathway in colorectal cancer. Eur Rev Med Pharmacol Sci (2018) 22(23):8203–9. doi: 10.26355/eurrev_201812_16513 [DOI] [PubMed] [Google Scholar]
- 110. Hua X, Sun Y, Chen J, Wu Y, Sha J, Han S, et al. Circular RNAs in drug resistant tumors. BioMed Pharmacother (2019) 118:109233. doi: 10.1016/j.biopha.2019.109233 [DOI] [PubMed] [Google Scholar]
- 111. Chen H, Pei L, Xie P, Guo G. Circ-PRKDC contributes to 5-fluorouracil resistance of colorectal cancer cells by regulating miR-375/FOXM1 axis and wnt/beta-catenin pathway. Onco Targets Ther (2020) 13:5939–53. doi: 10.2147/OTT.S253468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Li N, Lu B, Luo C, Cai J, Lu M, Zhang Y, et al. Incidence, mortality, survival, risk factor and screening of colorectal cancer: A comparison among China, Europe, and northern America. Cancer Lett (2021) 522:255–68. doi: 10.1016/j.canlet.2021.09.034 [DOI] [PubMed] [Google Scholar]
- 113. Mauri G, Sartore-Bianchi A, Russo AG, Marsoni S, Bardelli A, Siena S. Early-onset colorectal cancer in young individuals. Mol Oncol (2019) 13(2):109–31. doi: 10.1002/1878-0261.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer (2021) 20(1):93. doi: 10.1186/s12943-021-01372-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Liu X, Liu Y, Liu Z, Lin C, Meng F, Xu L, et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol Cancer (2021) 20(1):114. doi: 10.1186/s12943-021-01412-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Vaghari-Tabari M, Majidinia M, Moein S, Qujeq D, Asemi Z, Alemi F, et al. MicroRNAs and colorectal cancer chemoresistance: New solution for old problem. Life Sci (2020) 259:118255. doi: 10.1016/j.lfs.2020.118255 [DOI] [PubMed] [Google Scholar]
- 117. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol (2020) 14(3):539–55. doi: 10.1002/1878-0261.12629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ros J, Baraibar I, Sardo E, Mulet N, Salva F, Argiles G, et al. BRAF, MEK and EGFR inhibition as treatment strategies in BRAF V600E metastatic colorectal cancer. Ther Adv Med Oncol (2021) 13:1758835921992974. doi: 10.1177/1758835921992974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov (2012) 2(3):227–35. doi: 10.1158/2159-8290.CD-11-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mao M, Tian F, Mariadason JM, Tsao CC, Lemos R, Jr., Dayyani F, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin Cancer Res (2013) 19(3):657–67. doi: 10.1158/1078-0432.CCR-11-1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yaeger R, Cercek A, O'Reilly EM, Reidy DL, Kemeny N, Wolinsky T, et al. Pilot trial of combined BRAF and EGFR inhibition in BRAF-mutant metastatic colorectal cancer patients. Clin Cancer Res (2015) 21(6):1313–20. doi: 10.1158/1078-0432.CCR-14-2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Corcoran RB, Atreya CE, Falchook GS, Kwak EL, Ryan DP, Bendell JC, et al. Combined BRAF and MEK inhibition with dabrafenib and trametinib in BRAF V600-mutant colorectal cancer. J Clin Oncol (2015) 33(34):4023–31. doi: 10.1200/JCO.2015.63.2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. van Geel R, Tabernero J, Elez E, Bendell JC, Spreafico A, Schuler M, et al. A phase ib dose-escalation study of encorafenib and cetuximab with or without alpelisib in metastatic BRAF-mutant colorectal cancer. Cancer Discov (2017) 7(6):610–9. doi: 10.1158/2159-8290.CD-16-0795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med (2019) 381(17):1632–43. doi: 10.1056/NEJMoa1908075 [DOI] [PubMed] [Google Scholar]
- 125. Bowles DW, Kochenderfer M, Cohn A, Sideris L, Nguyen N, Cline-Burkhardt V, et al. A randomized, phase II trial of cetuximab with or without PX-866, an irreversible oral phosphatidylinositol 3-kinase inhibitor, in patients with metastatic colorectal carcinoma. Clin Colorectal Cancer (2016) 15(4):337–344 e2. doi: 10.1016/j.clcc.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 126. Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, et al. Phase I, dose-escalation study of BKM120, an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol (2012) 30(3):282–90. doi: 10.1200/JCO.2011.36.1360 [DOI] [PubMed] [Google Scholar]
- 127. Bendell JC, Nemunaitis J, Vukelja SJ, Hagenstad C, Campos LT, Hermann RC, et al. Randomized placebo-controlled phase II trial of perifosine plus capecitabine as second- or third-line therapy in patients with metastatic colorectal cancer. J Clin Oncol (2011) 29(33):4394–400. doi: 10.1200/JCO.2011.36.1980 [DOI] [PubMed] [Google Scholar]
- 128. Shapiro GI, Bell-McGuinn KM, Molina JR, Bendell J, Spicer J, Kwak EL, et al. First-in-Human study of PF-05212384 (PKI-587), a small-molecule, intravenous, dual inhibitor of PI3K and mTOR in patients with advanced cancer. Clin Cancer Res (2015) 21(8):1888–95. doi: 10.1158/1078-0432.CCR-14-1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Toyoda M, Watanabe K, Amagasaki T, Natsume K, Takeuchi H, Quadt C, et al. A phase I study of single-agent BEZ235 special delivery system sachet in Japanese patients with advanced solid tumors. Cancer Chemother Pharmacol (2019) 83(2):289–99. doi: 10.1007/s00280-018-3725-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wainberg ZA, Alsina M, Soares HP, Brana I, Britten CD, Del Conte G, et al. A multi-arm phase I study of the PI3K/mTOR inhibitors PF-04691502 and gedatolisib (PF-05212384) plus irinotecan or the MEK inhibitor PD-0325901 in advanced cancer. Target Oncol (2017) 12(6):775–85. doi: 10.1007/s11523-017-0530-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Mody K, Baldeo C, Bekaii-Saab T. Antiangiogenic therapy in colorectal cancer. Cancer J (2018) 24(4):165–70. doi: 10.1097/PPO.0000000000000328 [DOI] [PubMed] [Google Scholar]
- 132. Bharadwaj U, Kasembeli MM, Robinson P, Tweardy DJ. Targeting janus kinases and signal transducer and activator of transcription 3 to treat inflammation, fibrosis, and cancer: Rationale, progress, and caution. Pharmacol Rev (2020) 72(2):486–526. doi: 10.1124/pr.119.018440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ferrarotto R, Eckhardt G, Patnaik A, LoRusso P, Faoro L, Heymach JV, et al. A phase I dose-escalation and dose-expansion study of brontictuzumab in subjects with selected solid tumors. Ann Oncol (2018) 29(7):1561–8. doi: 10.1093/annonc/mdy171 [DOI] [PubMed] [Google Scholar]
- 134. Ihle NT, Paine-Murrieta G, Berggren MI, Baker A, Tate WR, Wipf P, et al. The phosphatidylinositol-3-kinase inhibitor PX-866 overcomes resistance to the epidermal growth factor receptor inhibitor gefitinib in a-549 human non-small cell lung cancer xenografts. Mol Cancer Ther (2005) 4(9):1349–57. doi: 10.1158/1535-7163.MCT-05-0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Zhu M, Jin Q, Xin Y. Recent clinical advances in PI3K inhibitors on colorectal cancer. Pharmazie (2021) 76(12):568–73. doi: 10.1691/ph.2021.1820 [DOI] [PubMed] [Google Scholar]
- 136. Itatani Y, Kawada K, Yamamoto T, Sakai Y. Resistance to anti-angiogenic therapy in cancer-alterations to anti-VEGF pathway. Int J Mol Sci (2018) 19(4):1232. doi: 10.3390/ijms19041232 [DOI] [PMC free article] [PubMed] [Google Scholar]