Abstract
Background
Nasal microbiota is crucial for the pathogenesis of allergic rhinitis (AR), which has been reported to be different from that of healthy individuals. However, no study has investigated the microbiota in nasal extracellular vesicles (EVs). We aimed to compare the microbiome composition and diversity in EVs between AR patients and healthy controls (HCs) and reveal the potential metabolic mechanisms in AR.
Methods
Eosinophil counts and serum immunoglobulin E (IgE) levels were measured in patients with AR (n = 20) and HCs (n = 19). Nasal EVs were identified using transmission electron microscopy and flow cytometry. 16S rRNA sequencing was used to profile the microbial communities. Alpha and beta diversities were analyzed to determine microbial diversity. Taxonomic abundance was analyzed based on the linear discriminant analysis effect size (LEfSe). Microbial metabolic pathways were characterized using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUst2) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses.
Results
Eosinophils, total serum IgE, and IgE specific to Dermatophagoides were increased in patients with AR. Alpha diversity in nasal EVs from patients with AR was lower than that in HCs. Beta diversity showed microbiome differences between the AR and HCs groups. The microbial abundance was distinct between AR and HCs at different taxonomic levels. Significantly higher levels of the genera Acetobacter, Mycoplasma, Escherichia, and Halomonas were observed in AR patients than in HCs. Conversely, Zoogloea, Streptococcus, Burkholderia, and Pseudomonas were more abundant in the HCs group than in the AR group. Moreover, 35 microbial metabolic pathways recognized in AR patients and HCs, and 25 pathways were more abundant in the AR group.
Conclusion
Patients with AR had distinct microbiota characteristics in nasal EVs compared to that in HCs. The metabolic mechanisms of the microbiota that regulate AR development were also different. These findings show that nasal fluid may reflect the specific pattern of microbiome EVs in patients with AR.
Keywords: Allergic rhinitis, Extracellular vesicle, Microbiota, 16S rRNA sequencing
Abbreviations: AR, Allergic rhinitis; EVs, Extracellular vesicles; HCs, Healthy controls; LEfSe, Linear discriminant analysis (LDA) effect size; KEGG, Kyoto Encyclopedia of Genes and Genomes; OMVs, Outer membrane vesicles; MAMPs, Microorganism-associated molecular patterns; LPS, Lipopolysaccharide; PRRs, Pattern recognition receptors; HDM, House dust mite; PBS, Phosphate-buffered saline; TEM, Transmission electron microscopy; FITC, Fluorescein isothiocyanate; ASV, Amplicon sequence variant; PCoA, Principal coordinate analysis; PLS–DA, Partial least squares discriminant analysis; UPGMA, Unweighted pair-group method with arithmetic means; MRPP, Multiple response permutation procedure; GPI, Glycosylphosphatidylinositol
Introduction
Allergic rhinitis (AR) is a prevalent chronic allergic respiratory disease, affecting approximately 10%–25% of the population worldwide.1, 2, 3 Inflammatory responses in the nasal mucosa are typical features of AR and are frequently accompanied by the infiltration of immune cells, including T cells, eosinophils, and basophils.4,5 Previous studies have confirmed that the microbiome directly affects inflammatory responses in allergic diseases such as AR and asthma.6, 7, 8 Therefore, the study of the composition of the microbiota is of great importance for understanding the pathogenesis of AR.
Microbiomes in humans are crucial for health and disease, accounting for 90% of the cells at a ratio of 10:1.8 Recent advances in microbiome research have shown that exposure to indoor environmental microbes is closely associated with the development of AR.9 To our knowledge, gut and nasal microbiota are the 2 most frequently studied microbiota in AR. Several studies have indicated that gut microbiota plays essential roles in the course and symptoms of AR.10,11 For instance, adult AR patients have a unique gut microbiota with reduced microbial diversity and an altered abundance of certain microbes compared to that of healthy subjects.10 Gut microbiota are different between children with AR and healthy controls and are associated with high serum immunoglobulin E (IgE) levels.11 Moreover, there is a stable microbial community in the nasal cavity of AR, and this microbiota can induce crosstalk with the immune system and eliminate pathogens.12 Choi et al13 reported a significant increase in microbial species and bacterial diversity in the nasal tracts of patients with AR. Gan et al14 also indicated that nasal microbiota may exert pivotal effects on the pathogenesis of heterogeneous nasal mucosal inflammation.
Extracellular vesicles (EVs) are nanosized vesicles released from inflammatory and immune cells that are involved in allergic diseases. Extracellular vesicles are considered responsible for communication between cells and are frequently used to investigate the pathogenesis of various diseases, including AR.15 Extracellular vesicles secreted from Gram-negative and Gram-positive bacteria were originally called membrane vesicles (MVs) and outer membrane vesicles (OMVs). Microorganism-associated molecular patterns (MAMPs) such as nucleic acids, peptidoglycan, lipopolysaccharide (LPS), and toxins are present in MVs or OMVs. These MV- or OMV-encapsulated MAMPs trigger intracellular immunomodulatory signaling pathways via pattern recognition receptors (PRRs) in host cells.16,17 Accumulating evidence indicates that microbiota derived from EVs plays a key role in immune function and disease development.18, 19, 20 However, microbiome alteration of nasal EVs in patients with AR remains unknown.
In this study, the microbial composition of nasal EVs in AR patients was characterized using 16S rRNA sequencing. The potential metabolic pathways involved in the detected microbiome were also determined. This study may clarify the microbiome differences in nasal EVs from patients with AR and offer a new clue for investigating the pathogenesis of AR.
Materials and methods
Human subjects sample collection
Patients were recruited from the outpatient clinic of the Department of Otorhinolaryngology of Xiamen Chang Gung Hospital during June 2020 to March 2021. A total of 39 subjects (aged 15–36 years) for the study, including 20 AR patients and 19 healthy controls (HCs). All patients with clinical symptoms suggestive of persistent AR were recruited for the study. Based on the ARIA guidelines,2 the inclusion criteria for AR patients were as follows: patients with a symptom complex that consists any combination of congestion, rhinorrhea, sneezing, nasal itching; and symptoms are present more than 4 days/week and for more than 4 consecutive weeks. Allergen panels of D1 (Dermatophagoides pteronyssinus) + D2 (Dermatophagoides farinae) + I6 (Blattella germanica) were used in all cases. All rhinitis patients had a history of typical rhinitis symptoms relevant to exposure of mites and were diagnosed by in vitro testing for allergen panels of D1, D2, and I6. Only mite-sensitized AR patients were included, excluding AR patients with a seasonal onset tendency which is mostly relevant to outdoor allergen sensitization like pollen. None of the patients had symptoms of chronic rhinosinusitis, nasal polyposis, immunological disease, neurodevelopmental disabilities, or respiratory infections. Age- and sex-matched healthy individuals without allergic disorders or chronic medical conditions were selected as the HCs. In addition, none of the participants had used probiotics, systemic antibiotics, or steroids within 14 d prior to the study. Written informed consent was obtained from all participants.
Eosinophil count and IgE evaluation
The eosinophil count was analyzed using an ABX Pentra 120 Retic (HORIBA, France). IgE test provides an in vitro measure of a patient's specific IgE levels against particular allergens.21 Serum samples collected from the enrolled participants were tested for total IgE levels using the IMX Total IgE assay system (Abbott Laboratories, IL, USA) and specific IgE house dust mite (HDM) aeroallergens such as D1 and D2 using Phadiatop P250 (ThermoFisher Scientific, Uppsala, Sweden). A cutoff of 0.7 KU/L was defined as a positive result (sIgE-D1 > 0.7 KU/L, n = 20; sIgE-D2 > 0.7 KU/L, n = 19, Supplementary Table S1).22
Nasal EVs isolation
In this study, nasal secretions were collected from patients with AR and HCs. To collect the secretion, two small Merocel nasal tampons (Ivalon, ThinPackTM) were inserted into the inferior meatus of each nostril for 10 min. Merocel nasal tampons were weighed before and after application to calculate total secretion weights, and the secretion was eluted by soaking in 3 mL (0.9% w/v) of NaCl at 4 °C for 1 h and collected after centrifugation for 10 min at 3000 × g.23 Viscosity of nasal fluid samples was reduced by phosphate-buffered saline (PBS) dilution, and debris was removed by centrifugation at 2000 × g for 30 min at 4 °C. The supernatant was collected in new Falcon tubes and further centrifuged at 10,000 × g for 45 min at 4 °C. Subsequently, the supernatant was filtered with a 0.45-μm syringe filter, and then ultracentrifuged at 100,000 × g for 70 min at 4 °C (Optima L-100XP, Beckman Coulter, Brea, CA, USA). The exosomal pellet was resuspended in 10 mL of cold PBS, and the ultracentrifugation step was repeated. The final exosomal pellet was resuspended in 50 μL of filtered PBS (0.22 μm) for subsequent analysis.
EV characterization
The morphology of exosomes isolated from the nasal fluid was confirmed by transmission electron microscopy (TEM) with negative staining. The absorbed exosomes were stained with 10 μL of 2% uranyl acetate for 1 min, and excess fluid was removed using a filter paper. The EVs were mounted on the grid and observed using TEM at 80 kV (HT7700, Hitachi High-Technologies Corporation, Minato, Tokyo, Japan). The purity of the exosomes was analyzed using nFCM with a nanoanalyzer, following the manufacturer's instructions. Briefly, 30 μL of each diluted exosome sample (1:4 dilution ratio in cold PBS) was stained with fluorescein isothiocyanate (FITC) mouse anti-human CD9 and CD81 antibody (BD Biosciences, Franklin Lakes, NJ, USA) for 30 min in the dark. Samples positive for CD9 and CD81 were analyzed using a Flow NanoAnalyzer (N30E, NanoFCM Inc., Xiamen, China).
DNA extraction from the nasal EVs
Nasal EV-encapsulated DNA was extracted using a DNA extraction kit (DNeasy Blood & Tissue Kit; Qiagen, Hilden, Germany) according to a previously described method.24 Briefly, the EV samples were boiled at 100 °C for 30 min and centrifuged at 10,000 × g for 30 min at 4 °C. The quality and quantity of the DNA were measured using a NanoDrop assay. The DNA concentration and purity were confirmed on a 1% agarose gel.
PCR amplification and purification
For 16S rRNA gene sequencing, the V3–V4 region was amplified using a specific primer set (319F: 5′-CCTACGGGNGGCWGCAG-3’; 806R: 5′-GACTACHVGGGTAT CTAATCC-3′) according to the 16S metagenomic sequencing library preparation procedure (Illumina, USA). In brief, gDNA was used for the PCR reaction, which was carried out with a KAPA HiFi HotStart ReadyMix (Roche) under the PCR conditions: 95 °C for 3 min; 25 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, 72 °C for 5 min, and hold at 4 °C. The PCR products were monitored on a 1.5% agarose gel. Samples with a bright main strip of approximately 500 bp were chosen and purified using AMPure XP beads for library preparation.
Library preparation and sequencing
The Illumina TruSeq DNA PCR-Free Library Preparation Kit (Illumina, USA) was used to generate sequencing libraries following the manufacturer's recommendations. The PCR product quality was assessed on a Qubit 4.0 Fluorometer (Thermo Scientific) and Qsep100TM system. Finally, the library was sequenced on an Illumina NovaSeq platform and 250 bp paired-end reads were generated.
Microbiome analyses
Amplicon sequencing was performed using 300-bp paired-end raw reads and each sample was demultiplexed based on the barcode identification. After demultiplexing, the QIIME2 cutadapt plugin was used to remove extra primer and adapter sequences from paired-end reads. For the amplicon sequence variant (ASV) construction, a denoising pipeline was performed with the QIIME2 DADA2 plugin (v2020.11), which implements quality filtering, dereplication, dataset-specific error model learning, denoising, paired-end read joining, and chimera removal.25 The feature classifier26 and algorithm in QIIME227 were employed to annotate the taxonomic classification for each representative sequence based on the information retrieved from the NCBI database.
Alpha diversity was indicative of species complexity within individual samples based on the different criteria output from the QIIME pipeline. The unweighted UniFrac belonging to beta diversity was calculated using the QIIME pipeline. Principal coordinate analysis (PCoA) and Partial least squares discriminant analysis (PLS–DA) were performed using the ggplot2 and mixOmics packages in R.
Unweighted pair-group method with arithmetic means (UPGMA) hierarchical clustering was performed to interpret the arithmetic distances based on the average linkage algorithm. Linear discriminant analysis (LDA) effect size (LEfSe) applied LDA to these bacterial taxa and was used to assess the effect size of each differentially significant abundant taxon. In this study, taxa with an LDA score (log 10) of >3 were considered significant. For the functional analysis, functional abundances from 16S rRNA sequencing data were analyzed to predict functional genes using PICRUst2 (v1.1.1).
Statistical analysis
The original observational data in the taxa summary plots were normalized by computing the relative abundance. Differences between the two groups were assessed using a t-test based on clinical characteristics and significance of the taxonomy shown as mean ± standard deviation. Permutational multivariate analysis of variance (ANOVA) (ADONIS) and the multiple response permutation procedure (MRPP) were used to test for significant differences in sampling units between HCs and AR participants. Statistical significance was defined as a P-value less than 0.05. Statistical analysis was performed using GraphPad Prism version 8.0 (GraphPad Software, USA).
Results
Clinical characteristics of participants
A total of 39 subjects (22 men and 27 ± 8 years of age) participated in this study. Among them, 20 subjects were included in the AR group (10 men and 10 women; age, 30 ± 6 years), and 19 subjects were included in the HC group (12 men and 7 women; age, 25 ± 10 years). Eosinophils are the major effector cells of innate immunity and are considered biomarkers of AR.28 As shown in Table 1, the percentage of eosinophils was significantly higher in patients with AR than in HCs (P < 0.01). Moreover, IgE is a molecular component of atopy, and the evaluation of total serum IgE levels has become a diagnostic criterion for AR.29 The results showed that patients with AR had higher total serum IgE levels than that of the HCs (P < 0.01). Dermatophagoides (D) allergens are a risk factor for the development of AR,30 which are reported as the major environmental allergen in house dust in southeast of China.31,32 The levels of serum-specific IgE to D (serum sIgE-D1 and D2) were also dramatically higher in patients with AR than in HCs (P < 0.0001) (Table 1, Supplementary Table S1).
Table 1.
Clinical characteristics.
| Variables | HC | AR | P-value |
|---|---|---|---|
| Subjects (N) | 19 | 20 | – |
| Gender (M/F) | 12/7 | 10/10 | NS |
| Age (years) | 25 ± 10 | 30 ± 6 | NS |
| Eosinophil (%) | 1.90 (0.20–5.10) | 3.30 (1.90–13.00) | <0.01 |
| Total serum IgE (IU/mL) | 25.09 (4.23–92.20) | 188.75 (91.42–1597.00) | <0.01 |
| Serum sIgE-D1 (kUA/L) | 0.03 (0.01–0.55) | 22.65 (0.72–76.60) | <0.0001 |
| Serum sIgE-D2 (kUA/L) | 0.01 (0.00–0.64) | 18.55 (0.33–88.20) | <0.0001 |
HC, healthy control; AR, allergic rhinitis; N, number; M, male; F, female; NS, not significant; IgE, immunoglobulin E; sIgE, specific immunoglobulin E; D, dust mite. Data were shown as mean ± standard deviation (SD), or median (interquartile range). The cutoff of 0.7 KU/L was set for sIgE evaluation. P-value < 0.05 was considered as statistical significance after performing t-test
Phenotypic characterization of nasal EVs
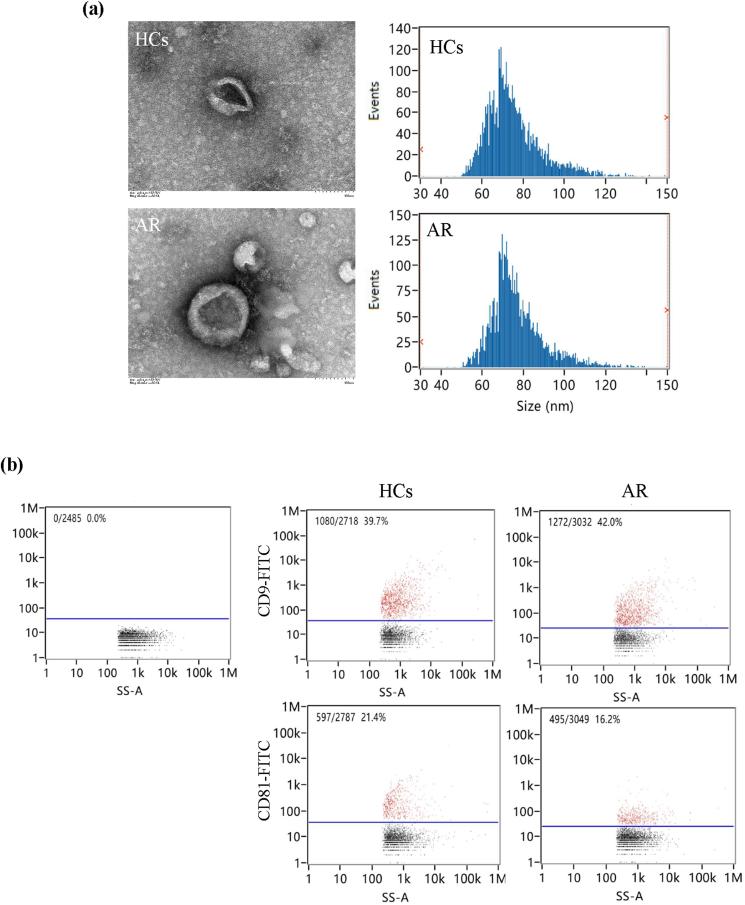
The presence of nasal EVs in patients with AR and HCs was investigated using TEM and flow cytometry. TEM showed that nasal EVs presented a clear spheroid morphology in HCs and AR (Fig. 1a). CD9 and CD81 are commonly used exosomal markers for the identification of EVs.33 Naon-flow cytometry showed positive markers, CD9 and CD81, for isolated EVs compared with IgG as a blank (Fig. 1b). Therefore, a series of centrifugation processes was effective for the isolation of EVs.
Fig. 1.
The characterization of nasal extracellular vesicles (EVs) from allergic rhinitis (AR) patients and health controls (HCs). (a) The morphology of nasal EVs was observed under transmission electron microscopy. Scale bar = 100 nm. (b) Nasal EVs positive for CD9 and CD81 were measured using nano-flow cytometry
Difference of microbiota between AR patients and healthy individuals
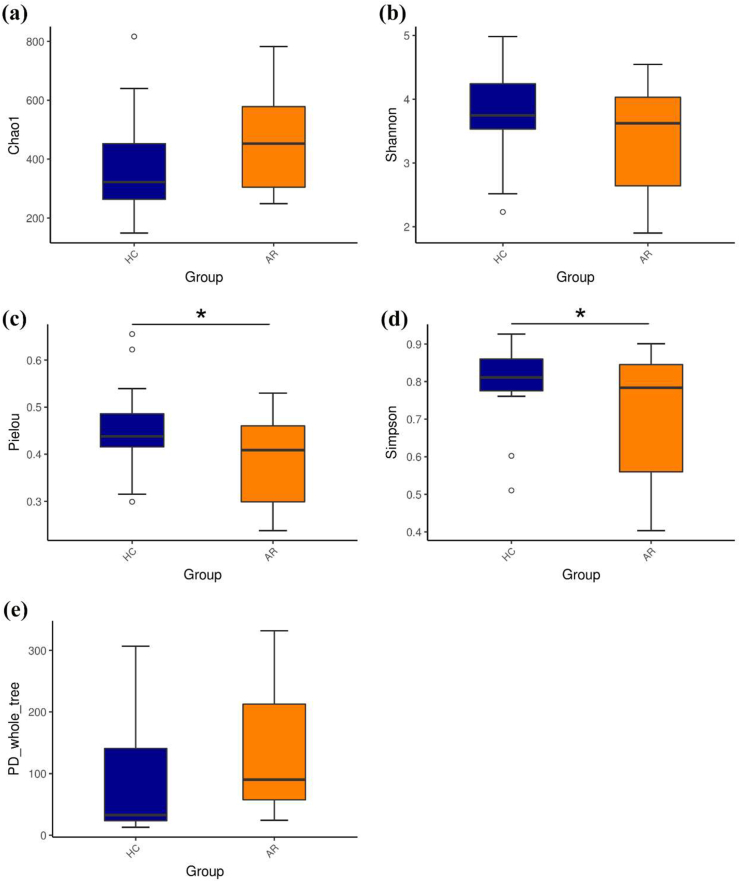
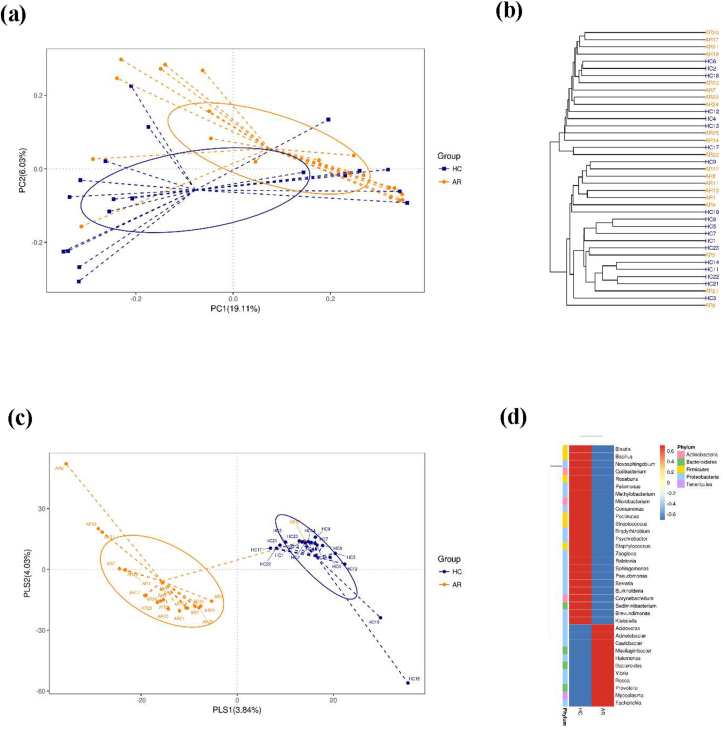
After removing unqualified sequences, each sample yielded a total of 4,001,400 raw reads and an average of 102,600 ± 6785 reads. A total of 2,872,578 effective reads were generated, and each sample produced an average of 73,656 ± 7620 effective reads (range: 761,241–84,396 effective reads). Rarefaction curve analysis showed that more than 60,000 reads captured microbiome diversity across samples as the number of observed species (Supplementary Fig. 1). There were 4084 and 5382 ASVs in healthy controls and AR patients, respectively, with 954 overlapping ASVs in the Venn diagram (Supplementary Fig. 2). Alpha diversity is a pivotal indicator of the abundance and diversity of observed microbes, as assessed by the Chao1, Shannon, PD_whole_tree, Pielou, and Simpson indices. The Chao1 index (P = 0.1107) and Shannon diversity (P = 0.1354) of AR patients and HCs also did not differ significantly, while the Pielou and Simpson indices were dramatically decreased in AR patients compared to those in HCs (P < 0.05) (Fig. 2b–d). The metric PD_whole_tree is the Faith's phylogenetic diversity, which is measured by adding up all branch lengths based on the phylogenetic tree. There was no obvious difference in PD_whole_tree diversity between patients with AR and HCs (P = 0.1417) (Fig. 2e). Unweighted and weighted UniFrac analyses were performed to compare the similarity between the two groups. The two principal component scores were 19.11% and 6.03% of the total variation in the PCoA (Fig. 3a). The distances were clustered using UPGMA on an unweighted UniFrac (Fig. 3b). The significance of beta diversity was estimated using the ADONIS. The results indicated that the microbial community did not differ between the two groups (ADONIS: R2 = 0.05538, P = 0.0555). However, the MRPP analysis showed that the between-group distances were significantly higher than the within-group distances (A = 0.019, P = 0.045). In the observed PLS–DA plots, 3.84% and 4.03% of the total variation in PLS1 and PLS2, respectively, were well separated between the 2 groups (Fig. 3c). Significant differences in microbial community profiles in the HCs and AR groups are presented in a heat map of the top 35 species (Fig. 3d).
Fig. 2.
Alpha diversity metrics for AR and HCs. (a) Chao1 index. (b) Shannon index. (c) Pielou index. (d) Simpson index. (e) PD_whole_tree index. ∗P < 0.05
Fig. 3.
Beta diversity in the microbiota of AR and HCs. (a) β-diversity changes in nasal microbiota across groups by the principal coordinate analysis (PCoA). (b) Unweighted pair group method with arithmetic mean (UPGMA) cluster analysis. (c) Partial least squares discriminant analysis (PLS–DA). (d) Heat map of the top 35 genera among groups
Assessment of relative abundance of microbiota
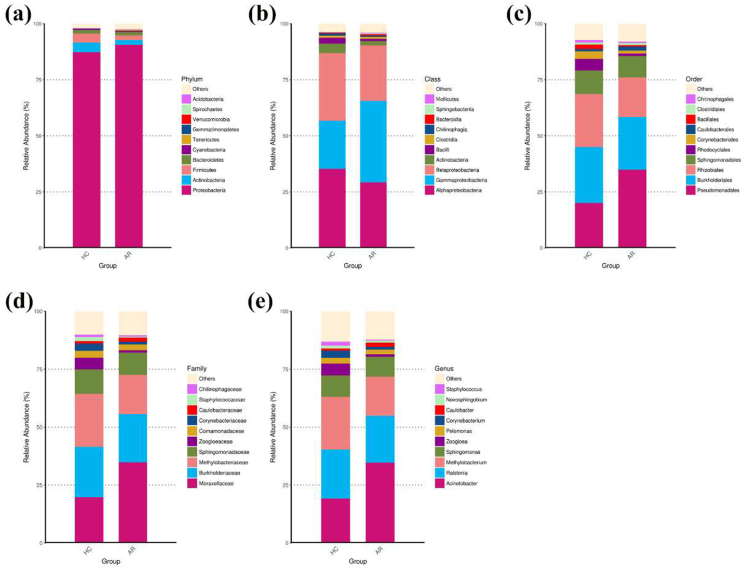
The ASVs were classified into 25 phyla, 55 classes, 108 orders, 205 families, and 351 genera. Assessment of the relative abundance of the top 10 bacteria was considered the predominant bacteria at each taxon level (Fig. 4). At the phylum level, Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes were the dominant bacterial phyla in both the AR patients and HCs (Fig. 4a). Among the top ten bacterial phyla, Tenericutes (0.314% in AR patients vs. 0.041% in HCs, P = 0.002) and Verrucomicrobia (0.067% in AR patients vs. 0.006% in HCs, P = 0.012) were significantly more abundant in AR patients than in HCs (Supplementary Table S2). At the class level, the dominant bacterial classes in the nasal EVs from both AR patients and HCs were Alphaproteobacteria, Gammaproteobacteria, and Betaproteobacteria (Fig. 4b). Among the top 10 bacterial classes, Mollicutes were significantly more abundant in AR patients than in HCs (0.314% in AR patients vs. 0.041% in HCs, P = 0.002) (Supplementary Table S3). Within the order taxonomic rank, the most abundant orders present in both patients with AR and HCs were Pseudomonadales, Burkholderiales, Rhizobiales, and Sphingomonadales (Fig. 4c). Rhodocyclales was dramatically less abundant in AR patients than in HCs (1.077% in AR patients vs. 5.147% in HCs, P = 0.009) (Supplementary Table S4). At the family level, the most abundant families in both patients with AR and HCs were Moraxellaceae, Burkholderiaceae, Methylobacteriaceae, and Sphingomonadaceae (Fig. 4d). Among the top 10 families, Zoogloeaceae were significantly less abundant in AR patients than in HCs (1.060% in AR patients vs. 5.118% in HCs, P = 0.028) (Supplementary Table S5). Within the genus taxonomic rank, Acinetobacter, Ralstonia, Methylobacterium, and Sphingomonas were the dominant bacterial genera in both AR patients and HCs (Fig. 4e). Zoogloea was dramatically less abundant in AR patients than in HCs (1.054% in AR patients vs. 5.098% in HCs, P = 0.00) (Supplementary Table S6). These results indicated distinctive differences in the bacterial communities of the HCs and AR groups.
Fig. 4.
Relative abundance of top 10 bacteria at different taxonomic levels in the nasal EVs from AR patients and HCs. (a) Phylum level. (b) Class level. (c) Order level. (d) Family level. (e) Genus level
Bacterial composition at different taxonomic levels
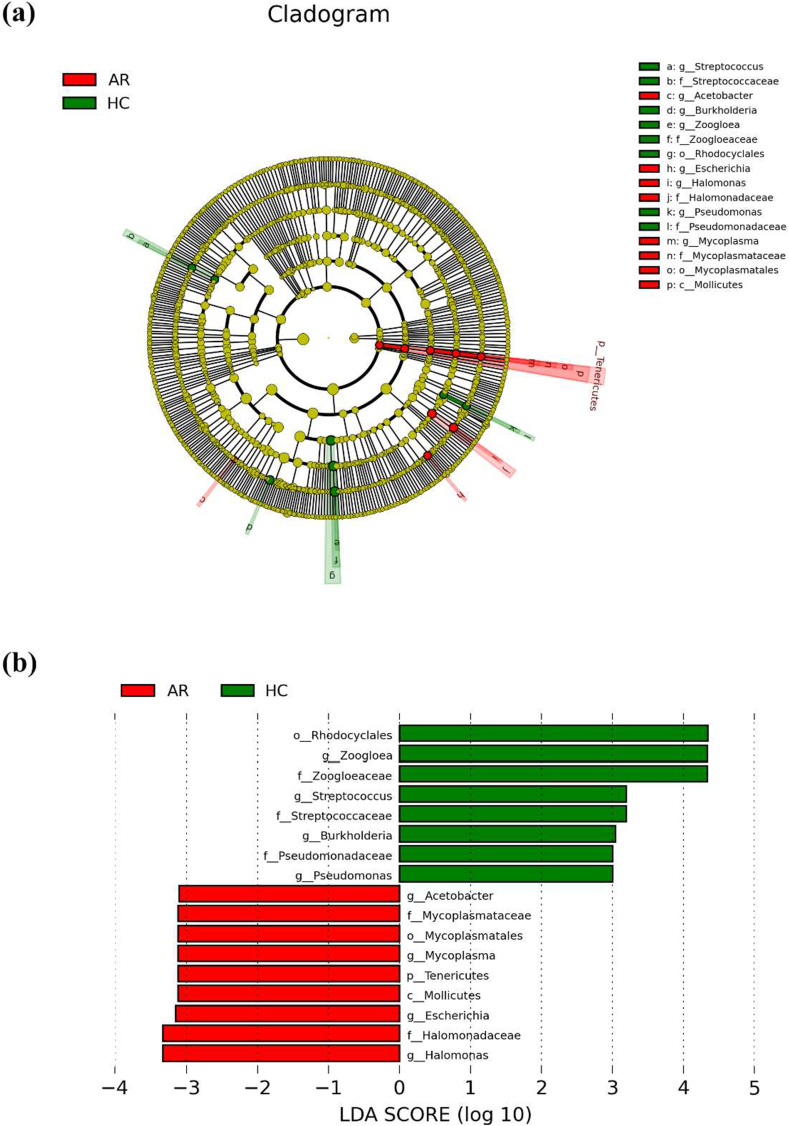
Linear discriminant analysis effect size with a logarithmic LDA score cutoff >3.0 was used to unveil the estimated phylotypes of differential bacterial communities from the HCs and AR groups. The bacterial class that differed markedly between patients with AR and HCs was Mollicutes. At the order level, Mycoplasmatales increased in AR patients, and Rhodocyclales increased in HCs. Analysis at the family level showed ascending levels of Halomonadaceae and Mycoplasmataceae in AR patients, and ascending levels of Streptococcaceae, Zoogloeaceae, and Pseudomonadaceae in HCs. At the genus level, Acetobacter, Escherichia, Halomonas, and Mycoplasma were abundant in patients with AR, and Streptococcus, Burkholderia, Zoogloea, and Pseudomonas were abundant in HCs (Fig. 5a). The 17 taxa mentioned above showed significant differences between patients with AR and HCs (Fig. 5b).
Fig. 5.
Compositional difference of microbiota in nasal EVs from AR patients and HCs. (a) A linear discriminant analysis (LDA) effect size (LEfSe) analysis for AR and HCs. (b) The enriched bacteria in AR (red) and HCs (green) with the logarithmic LDA scores >3.0
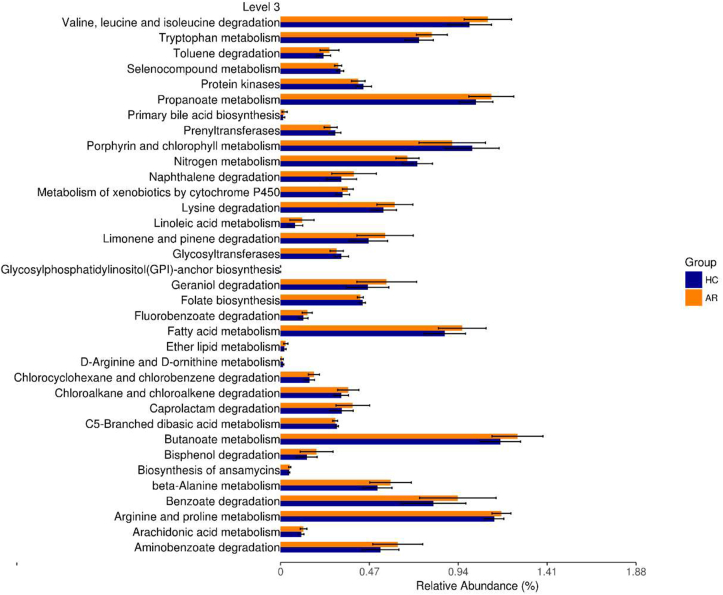
Enrichment of microbial metabolic pathways
To analyze the microbial metabolic function in the nasal EVs of AR, PICRUst2, based on the KEGG database, was used to predict the corresponding microbial metabolic pathways. A total of 35 metabolic pathways with significant differences between patients with AR and HCs were identified (P < 0.05). Among them, 25 microbial metabolic pathways were unregulated in AR, including valine, leucine, and isoleucine degradation; tryptophan metabolism; toluene degradation; propanoate metabolism; and primary bile acid biosynthesis. The rest of the 10 pathways were significantly more abundant in HCs than that in AR patients, including selenocompound metabolism, protein kinases, prenyltransferases, porphyrin and chlorophyll metabolism, nitrogen metabolism, glycosyltransferases, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, folate biosynthesis, d-arginine and d-ornithine metabolism, and C5-branched dibasic acid metabolism (Fig. 6). Accordingly, our data suggest that metabolic functions are related to alterations in the bacterial composition.
Fig. 6.
Microbial metabolic pathways relevant to AR and HCs. These pathways all differed significantly between AR and HCs (P < 0.05)
Discussion
Allergic rhinitis is an inflammatory disorder of the upper airway, and EVs are frequently used as inflammatory drivers to investigate the pathogenesis of allergic diseases.34,35 This study analyzed the microbial communities in nasal EVs from patients with AR and HCs. The AR patients had a different microbiome profile, marked by reduced microbial diversity and altered abundance of specific microbes in nasal EVs compared to HCs. Moreover, different microbial metabolic pathways were identified between the AR cohort and HCs.
Alpha diversity is a key quantity in microbiome research and is defined as the mean diversity of species at a local scale.36 Alpha diversity can be evaluated using Chao1, Shannon, Pielou, Simpson, and PD_whole_tree. Our study found that these Pielou and Simpson presented a significant decrease in the nasal EVs of AR patients, indicating that the diversity of microbes was reduced in AR. Other studies have also confirmed reduced microbial evenness in AR. Watts et al. found that the evenness of the gut microbiome is reduced in AR patients.10 Morin et al. also reported lower evenness of airway microbiota in AR patients than in controls.37 The lack of evenness in the microbial community may impair its capacity to resist exogenous disturbances, thereby inducing AR.38 In addition, the beta diversity metric revealed relative differences in the microbial communities between patients with AR and HCs. These results demonstrated that there were compositional differences in the microbiota of nasal EVs between patients with AR and HCs.
The differential abundance of specific microbial taxa is also a key point in microbiome analysis of AR. In this study, differences in microbial composition and abundance were observed in nasal EVs between patients with AR and HCs. A previous study found that Tenericutes decreased in abundance in mice with allergic asthma.39 However, our study found that Tenericutes at the phylum level were more abundant in nasal EVs from patients with AR than in HCs. This result suggests that Tenericutes is an essential bacterium contributing to AR, but further research is needed to determine its actual role. In addition, LEfSe analysis revealed that Zoogloea, Streptococcus, Burkholderia, and Pseudomonas were enriched in the HCs. A previous study suggested that Streptococcus reduces nasal allergic reactions by producing H2O2 to inhibit IgE-stimulated degranulation.40 Pseudomonas could ameliorate allergic sensitization in asthmatic mice by regulating the T cell response.41 Our results confirmed the potential beneficial role of Streptococcus and Pseudomonas and indicated that Zoogloea and Burkholderia may also be beneficial bacteria. Furthermore, relatively high abundances of Acetobacter, Mycoplasma, Escherichia, and Halomonas were found in nasal EVs from AR patients at the genus level, indicating the potential harmful roles of these bacteria in humans.
Microbiota can influence various metabolic responses in the host, thereby moderating the growth process and chronic disease occurrence.42 According to the PICRUst2 algorithm, we found that 35 microbial metabolic pathways were significantly different between patients with AR and HCs. Among these, 25 pathways were relatively more abundant in patients with AR. Most of these metabolic pathways are associated with allergic sensitization and inflammatory.43, 44, 45, 46, 47 For instance, tryptophan metabolism can attenuate airway inflammation during immunotherapy in a rat asthma model.43 Linoleic acid metabolism may be related to the increased severity of asthma.46 Geraniol has been shown to possess anti-inflammatory properties in AR.47 Our study further suggests that the microbiota in nasal EVs may influence these metabolic pathways, thereby affecting the development of AR.
In conclusion, the microbiome composition and diversity in nasal EVs were significantly different between patients with AR and HCs. Among the bacteria in nasal EVs, Zoogloea, Streptococcus, Burkholderia, and Pseudomonas may be beneficial bacteria, while Acetobacter, Mycoplasma, Escherichia, and Halomonas may be harmful bacteria for AR. Moreover, 25 microbial metabolic pathways may be closely associated with AR. These findings will be useful for investigating the pathogenesis of AR and will provide a basis for clarifying the potential mechanisms of microbial metabolism. However, this study had some limitations. First, the larger sample size and age range are needed to confirm the differences in microbiome composition and diversity between patients with AR and HCs. Second, bacteria taxa may contribute to the development of AR, thus, the association between microbiome composition and severity of AR is needed to be further deciphered. Third, it will be necessary to further study the mechanisms by which core bacterial taxa regulate AR development.
Funding
This study was supported by grants from Xiamen Chang Gung Hospital and Xiamen Municipal Bureau of Science and Technology, China (CMRPG1E0031, CMRPG1E0281, CMRPG1E0885, and CMRPG1G0211).
Availability of data and materials
The data are available upon reasonable request. Data supporting the findings of this study are available from the corresponding author.
Author contributions
Conceptualization, T.Y.C. and W·H·C.; methodology, C·H.F. and T.J.L.; formal analysis, T.Y·C; investigation, T.Y.C. and Y.R.Y.; resources, M.Y.Z, F·Y, and Y·F.Z.; data curation, T.Y·C; writing-original draft preparation, T.Y·C.; writing-review and editing, L.C. and C.J.C.; supervision, C.J.C.; project administration, C.J.C. All authors have read and approved the published version of the paper.
Ethics approval
The study was approved by the Institutional Review Board of Xiamen Chang Gung Hospital (approval number: XMCGIRB2020023) and conducted in accordance with the guidelines of the Declaration of Helsinki.
Authors’ consent for publication
All authors agreed to the publication of this work in the World Allergy Organization Journal.
Informed consent
Informed consent was obtained from all participants.
Declaration of competing interest
None to declare related to this work.
Acknowledgements
The authors thank the nurses for their excellent assistance and careful handling of the serum samples and all patients who participated in this study. We also thank Dr. Chi-Meng Tzeng provide assistance for data analysis and Xiamen LifeInt Technology Co. Ltd. for extracellular vesicle isolation respectively.
Footnotes
Full list of author information is available at the end of the article
Supplementary data to this article can be found online at https://doi.org/10.1016/j.waojou.2022.100674.
Contributor Information
Liang Chen, Email: chenliang700722@hotmail.com.
Chih-Jung Chang, Email: chan.chih.jung@gmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Marshall G.D., Jr. Allergic rhinitis: localized disease with systemic implications. Ann Allergy Asthma Immunol. 2021;127(2):155–156. doi: 10.1016/j.anai.2021.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Brożek J.L., Bousquet J., Agache I., et al. Allergic rhinitis and its impact on asthma (ARIA) guidelines-2016 revision. J Allergy Clin Immunol. 2017;140(4):950–958. doi: 10.1016/j.jaci.2017.03.050. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Hernandez M.C., Dordal M.T., Navarro A.M., et al. Severity and duration of allergic conjunctivitis: are they associated with severity and duration of allergic rhinitis and asthma? Eur Ann Allergy Clin Immunol. 2021 doi: 10.23822/EurAnnACI.1764-1489.231. [DOI] [PubMed] [Google Scholar]
- 4.Steelant B., Seys S.F., Gerven L.V., et al. Histamine and T helper cytokine-driven epithelial barrier dysfunction in allergic rhinitis. J Allergy Clin Immunol. 2018;141(3):951–963. doi: 10.1016/j.jaci.2017.08.039. e8. [DOI] [PubMed] [Google Scholar]
- 5.Koyama T., Miura K., Yamasaki N., et al. Suppressive effect of dexamethasone on murine Th9 cell-mediated nasal eosinophilic inflammation. Asia Pac Allergy. 2021;11(3):e25. doi: 10.5415/apallergy.2021.11.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schaefer M., Enck P. Effects of a probiotic treatment (Enterococcus faecalis) and open-label placebo on symptoms of allergic rhinitis: study protocol for a randomised controlled trial. BMJ Open. 2019;9(10):e031339. doi: 10.1136/bmjopen-2019-031339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirata S.I., Kunisawa J. Gut microbiome, metabolome, and allergic diseases. Allergol Int. 2017;66(4):523–528. doi: 10.1016/j.alit.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Pascal M., Perez-Gordo M., Caballero T., et al. Microbiome and allergic diseases. Front Immunol. 2018;9:1584. doi: 10.3389/fimmu.2018.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu X., Ou Z., Zhang M., et al. Indoor bacterial, fungal and viral species and functional genes in urban and rural schools in Shanxi Province, China-association with asthma, rhinitis and rhinoconjunctivitis in high school students. Microbiome. 2021;9(1):138. doi: 10.1186/s40168-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watts A.M., West N.P., Zhang P., Smith P.K., Cripps A.W., Cox A.J. The gut microbiome of adults with allergic rhinitis is characterised by reduced diversity and an altered abundance of key microbial taxa compared to controls. Int Arch Allergy Immunol. 2021;182(2):94–105. doi: 10.1159/000510536. [DOI] [PubMed] [Google Scholar]
- 11.Lin Y., Rui X., Li Y. [Role of gut microbiota in children with allergic rhinitis with high serum total IgE level] Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2020;34(12):1123–1128. doi: 10.13201/j.issn.2096-7993.2020.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brindisi G., Zicari A.M., Schiavi L., et al. Efficacy of Pidotimod use in treating allergic rhinitis in a pediatric population. Ital J Pediatr. 2020;46(1):93. doi: 10.1186/s13052-020-00859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi C.H., Poroyko V., Watanabe S., et al. Seasonal allergic rhinitis affects sinonasal microbiota. Am J Rhinol Allergy. 2014;28(4):281–286. doi: 10.2500/ajra.2014.28.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan W., Yang F., Meng J., Liu F., Liu S., Xian J. Comparing the nasal bacterial microbiome diversity of allergic rhinitis, chronic rhinosinusitis and control subjects. Eur Arch Oto-Rhino-Laryngol. 2021;278(3):711–718. doi: 10.1007/s00405-020-06311-1. [DOI] [PubMed] [Google Scholar]
- 15.Demkow U., Stelmaszczyk-Emmel A. Extracellular vesicles in allergic rhinitis and asthma and laboratory possibilities for their assessment. Int J Mol Sci. 2021;22(5) doi: 10.3390/ijms22052273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaparakis M., Turnbull L., Carneiro L., et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell Microbiol. 2010;12(3):372–385. doi: 10.1111/j.1462-5822.2009.01404.x. [DOI] [PubMed] [Google Scholar]
- 17.Yang J., Kim E.K., McDowell A., Kim Y.K. Microbe-derived extracellular vesicles as a smart drug delivery system. Transl Clin Pharmacol. 2018;26(3):103–110. doi: 10.12793/tcp.2018.26.3.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macia L., Nanan R., Hosseini-Beheshti E., et al. Host- and microbiota-derived extracellular vesicles, immune function, and disease development. Int J Mol Sci. 2019;21(1) doi: 10.3390/ijms21010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ricci V., Carcione D., Messina S., Colombo G.I., D’Alessandra Y. Circulating 16S RNA in biofluids: extracellular vesicles as mirrors of human microbiome? Int J Mol Sci. 2020;21(23) doi: 10.3390/ijms21238959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C.J., Zhang J., Tsai Y.L., et al. Compositional features of distinct microbiota base on serum extracellular vesicle metagenomics analysis in moderate to severe psoriasis patients. Cells. 2021;10(9) doi: 10.3390/cells10092349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Small P., Keith P.K., Kim H. Allergic rhinitis. Allergy Asthma Clin Immunol. 2018;14(Suppl 2):51. doi: 10.1186/s13223-018-0280-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi S., Chen H., Huang N., et al. Early intervention improves clinical responses to house dust mite immunotherapy in allergic rhinitis patients. Int Arch Allergy Immunol. 2016;171(3–4):234–240. doi: 10.1159/000452333. [DOI] [PubMed] [Google Scholar]
- 23.Segboer C.L., Fokkens W.J., Terreehorst I., Drunen C.M.V. Endotyping of non-allergic, allergic and mixed rhinitis patients using a broad panel of biomarkers in nasal secretions. PLoS One. 2018;13(7):e0200366. doi: 10.1371/journal.pone.0200366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo J.Y., Rho M., You Y.A., et al. 16S rRNA gene-based metagenomic analysis reveals differences in bacteria-derived extracellular vesicles in the urine of pregnant and non-pregnant women. Exp Mol Med. 2016;48:e208. doi: 10.1038/emm.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolyen E., Rideout J.R., Dillon M.R., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalfe D.D., Pawankar R., Ackerman S.J., et al. Biomarkers of the involvement of mast cells, basophils and eosinophils in asthma and allergic diseases. World Allergy Organ J. 2016;9:7. doi: 10.1186/s40413-016-0094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nam J.S., Hwang C.S., Hong M.P., Kim K.S. Prevalence and clinical characteristics of allergic rhinitis in the elderly Korean population. Eur Arch Oto-Rhino-Laryngol. 2020;277(12):3367–3373. doi: 10.1007/s00405-020-06256-5. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y., Xiong L., Yin X.Y., et al. House dust mite allergen levels in households and correlation with allergic rhinitis symptoms. Am J Rhinol Allergy. 2014;28(5):193–196. doi: 10.2500/ajra.2014.28.4095. [DOI] [PubMed] [Google Scholar]
- 31.Huang F.L., Liao E.C., Yu S.J. House dust mite allergy: its innate immune response and immunotherapy. Immunobiology. 2018;223(3):300–302. doi: 10.1016/j.imbio.2017.10.035. [DOI] [PubMed] [Google Scholar]
- 32.Cheng L., Chen J.J., Fu Q.L., et al. Chinese society of allergy guidelines for diagnosis and treatment of allergic rhinitis. Allergy Asthma Immunol Res. 2018;10(4):300–353. doi: 10.4168/aair.2018.10.4.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li D., Luo H., Ruan H., et al. Isolation and identification of exosomes from feline plasma, urine and adipose-derived mesenchymal stem cells. BMC Vet Res. 2021;17(1):272. doi: 10.1186/s12917-021-02960-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soumya S., Adegboyega G., Elhassan H. Surgical approaches for allergic rhinitis: a systematic review protocol. Int J Surg Protoc. 2021;25(1):178–183. doi: 10.29337/ijsp.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu G., Yang G., Zhang R., et al. Altered microRNA expression profiles of extracellular vesicles in nasal mucus from patients with allergic rhinitis. Allergy Asthma Immunol Res. 2015;7(5):449–457. doi: 10.4168/aair.2015.7.5.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amos G.C.A., Logan A., Anwar S., et al. Developing standards for the microbiome field. Microbiome. 2020;8(1):98. doi: 10.1186/s40168-020-00856-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morin A., McKennan C.G., Pedersen C.T., et al. Epigenetic landscape links upper airway microbiota in infancy with allergic rhinitis at 6 years of age. J Allergy Clin Immunol. 2020;146(6):1358–1366. doi: 10.1016/j.jaci.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bäckhed F., Fraser C.M., Ringel Y., et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12(5):611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Banskar S., Detzner A.A., Juarez-Rodriguez M.D., et al. The pglyrp1-regulated microbiome enhances experimental allergic asthma. J Immunol. 2019;203(12):3113–3125. doi: 10.4049/jimmunol.1900711. [DOI] [PubMed] [Google Scholar]
- 40.Okahashi N., Nakata M., Hirose Y., et al. Streptococcal H2O2 inhibits IgE-triggered degranulation of RBL-2H3 mast cell/basophil cell line by inducing cell death. PLoS One. 2020;15(4):e0231101. doi: 10.1371/journal.pone.0231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding F.X., Liu B., Zou W.J., Li Q.B., Tian D.Y., Fu Z. Pseudomonas aeruginosa-derived exosomes ameliorates allergic reactions via inducing the T(reg) response in asthma. Pediatr Res. 2018;84(1):125–133. doi: 10.1038/s41390-018-0020-1. [DOI] [PubMed] [Google Scholar]
- 42.Hoffman D.J., Campos-Ponce M., Taddei C.R., Doa C.M. Microbiome, growth retardation and metabolism: are they related? Ann Hum Biol. 2017;44(3):201–207. doi: 10.1080/03014460.2016.1267261. [DOI] [PubMed] [Google Scholar]
- 43.Hu Q., Jin L., Zeng J., et al. Tryptophan metabolite-regulated Treg responses contribute to attenuation of airway inflammation during specific immunotherapy in a mouse asthma model. Hum Vaccines Immunother. 2020;16(8):1891–1899. doi: 10.1080/21645515.2019.1698900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiu C.Y., Cheng M.L., Chiang M.H., Wang C.J., Tsai M.H., Lin G. Metabolomic analysis reveals distinct profiles in the plasma and urine associated with IgE reactions in childhood asthma. J Clin Med. 2020;9(3) doi: 10.3390/jcm9030887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samra M.S., Lim D.Y., Han M.Y., Jee H.M., Kim Y.K., Kim J.H. Bacterial microbiota-derived extracellular vesicles in children with allergic airway diseases: compositional and functional features. Allergy Asthma Immunol Res. 2021;13(1):56–74. doi: 10.4168/aair.2021.13.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panda L., Gheware A., Rehman R., et al. Linoleic acid metabolite leads to steroid resistant asthma features partially through NF-κB. Sci Rep. 2017;7(1):9565. doi: 10.1038/s41598-017-09869-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y., Yang X.L., Ni Y.H., Xu Z.M. Geraniol suppresses proinflammatory mediators in phorbol 12-myristate 13-acetate with A23187-induced HMC-1 cells. Drug Des Dev Ther. 2018;12:2897–2903. doi: 10.2147/DDDT.S145702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available upon reasonable request. Data supporting the findings of this study are available from the corresponding author.