Comprehensive pan-cancer molecular characterization of East Asian patients revealed significant levels of genomic diversity as well as clinical actionability and highlights the importance of employing ethnicity-based personalized approaches in cancer therapy.
Abstract
The fundamental principle of precision oncology is centralized on the identification of therapeutically exploitable targets that provides individual patients with cancer an opportunity to make informed decisions on a personalized level. To facilitate and adopt such concepts within clinical practice, we have initiated a nationwide, multi-institutional precision oncology screening program to examine and enroll patients into the most appropriate clinical trial based on their tumor's unique molecular properties. To determine the prevalence of essential major driver mutations and to explore their dynamic associations at both molecular and pathway levels, we present a comprehensive overview on the genomic properties of East Asian patients with cancer. We further delineate the extent of genomic diversity as well as clinical actionability in patients from Western and Eastern cultures at the pan-cancer and single-tumor entity levels. To support fellow oncology communities in future investigations involving large-scale analysis, all data have been made accessible to the public (https://kmportal.or.kr).
Significance:
We present a comprehensive overview of molecular properties of East Asian pan-cancer patients and demonstrate significant diversity in terms of genomic characteristics as well as clinical utility compared with patients with European ancestry. The results of this study will lay the groundwork for designing personalized treatments in the clinical setting.
See related commentary by Moyers and Subbiah, p. 886.
This article is highlighted in the In This Issue feature, p. 873
INTRODUCTION
The demand for personalized treatment has increased substantially among patients with cancer with unmet clinical needs (1–5). In the field of oncology, genomic biomarkers have been widely accepted as a new paradigm for patient treatment as well as drug discovery (6, 7). The significance of molecular profiling via clinical next-generation sequencing (NGS) has been well demonstrated by precedent programs, including MSK-IMPACT and NCI-MATCH (8–14). Previous studies that have enrolled patients across a broad range of different tumor types have made tremendous contributions in understanding the complexity of the cancer genome at the pan-cancer level (15–17). However, as these studies have enrolled patients mainly of European origin, it has been challenging to directly implement such profound insights within clinical practice for East Asian patients with cancer. A substantial number of studies have robustly demonstrated extensive genomic diversity as well as clinical utility among patients from distinct ethnic populations (18–22). Thus, it is of the utmost priority to establish a collection of cancer genomes, focusing on East Asian populations, across a wide spectrum of different cancer types.
Toward this goal, we have initiated the K-MASTER enterprise to collect and characterize the complex genomes of Korean patients with advanced solid tumors. We have leveraged previously established and validated clinical NGS panels to capture and detect major genomic aberrations, including single-nucleotide variants (SNV), small insertions and deletions (indel), copy-number alterations (CNA), and selected structural variations in cancer-related genes. Herein, we report the first phase of the K-MASTER precision oncology initiative, focusing on genomic characteristics of 4,028 pan-cancer patients to identify molecular signatures that constitute unique properties of patients with cancer of East Asian ancestry.
RESULTS
A Schematic Overflow of Prospective Clinical Sequencing in a Korean Pan-Cancer Cohort
Since the June 2017 launch of the K-MASTER enterprise, 4,028 Korean patients with advanced solid tumors have been subjected to prospective clinical sequencing (Supplementary Table S1). All of the patients enrolled in the program were either those whose standard-of-care options had already been exhausted without any other alternatives or will be exhausted in the nearest future. Genomic DNA samples that were isolated from tumor tissue specimens underwent quality control prior to being subjected to deep-coverage sequencing in order to capture potential genomic aberrations, including SNVs, small indels, CNAs, and selected structure variations (Supplementary Fig. S1). All sequencing data were further processed and uploaded into the main database. Final reports were reviewed by oncologists on a weekly basis.
Comprehensive Genomic Landscape of Major Cancer Driver Mutations in a Korean Pan-Cancer Cohort
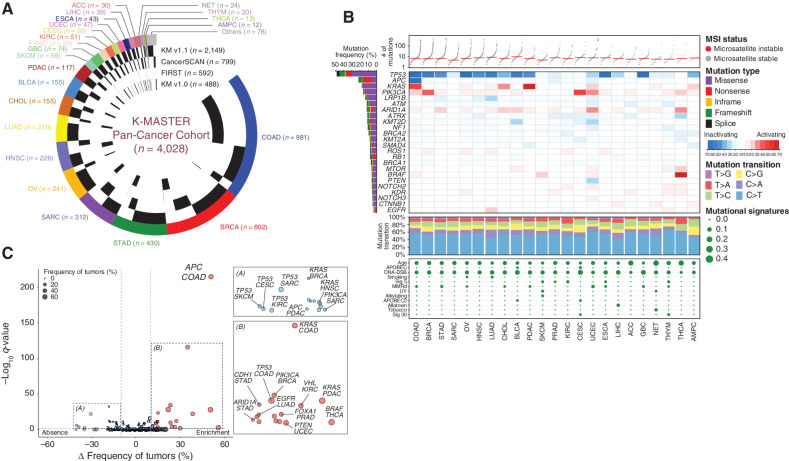
Our pan-cancer cohort at K-MASTER constitutes over 24 major cancer types, primarily including colorectal, breast, gastric, ovarian, lung adenocarcinomas, and sarcoma (Fig. 1A). Three oncology-based sequencing panels that encompass the full coding exons of commonly mutated cancer genes have been used in this study (409, 375, and 183 genes for K-MASTER, CancerSCAN, and FIRST panels, respectively). The average read depth for all clinical panels exceeded more than 650× to ensure that all essential genomic alterations, even at subclonal levels, were covered. A total of 156,233 nonsynonymous mutations, including missense, nonsense, in-frame, frameshift, and splice-site, were detected. Notably, a subset of tumors demonstrated an excessive amount of tumor mutational burden, despite the limited coverage of targeted-exome sequencing. Through various machine-learning algorithms (23, 24), we discovered that several of these tumors harbored tracts of tandemly repeated DNA motifs and were thus classified as microsatellite-instable (MSI) tumors. These MSI-high tumors were mainly detected in colorectal adenocarcinoma (COAD), stomach adenocarcinoma (STAD), and uterine corpus endometrial carcinoma (UCEC). Such results were consistent with previous studies that have reported a high prevalence of microsatellite instability in these cancerous tumors. As these patients could potentially benefit from immune-checkpoint blockades (25–27), our study supported the feasibility of routine clinical sequencing to identify potential benefits of immunotherapy (Fig. 1B).
Figure 1.
Genomic landscape of the K-MASTER pan-cancer cohort. A, Distribution of major tumor types and corresponding clinical sequencing panels from 4,028 pan-cancer patients. B, Genomic landscape of major cancer driver mutations based on distinct tumor types. Top, number of nonsynonymous mutations with microsatellite instability (MSI) status. Top middle, frequency of each mutation in corresponding tumor types. Red indicates activating oncogenes, and blue indicates inactivating tumor suppressors. The left bar indicates the percentage of tumor within the pan-cancer cohort with respect to a different type of mutation. Bottom middle, six classes of base substitution in each mutation type. Bottom, mutational signatures. The size of the node is proportional to the number of patients within each tumor type. C, Volcano plot representation of tumor frequency differences (x-axis) between tumors with the mutation in the corresponding tumor type versus the rest and its significance (y-axis). Mutations that are significantly more enriched in specific tumor types are in colored in red, whereas absences are colored in blue. ACC, adrenal carcinoma; AMPC, ampullary carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast adenocarcinoma; CESC, cervical cancer; CHOL, cholangiocarcinoma; COAD, colorectal adenocarcinoma; DNA-DSB, DNA double-strand break; ESCA, esophageal carcinoma; GBC, gallbladder cancer; HNSC, head and neck cancer; KIRC, kidney renal clear cell carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; MMRd, mismatch repair deficiency; NET, neuroendocrine carcinoma; OV, ovarian carcinoma; PDAC, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma.
The most frequently altered genes in the K-MASTER cohort included mutations in TP53 (48.1%), APC (21.8%), KRAS (17.7%), PIK3CA (16.3%), LRP1B (15.2%), ATM (11.8%), ARID1A (11.1%), and ATRX (10.8%; Fig. 1B). Inactivating mutation in TP53 was the most prevalent genomic event in a number of tumors, including COAD, breast adenocarcinoma (BRCA), STAD, ovarian carcinoma (OV), lung adenocarcinoma (LUAD), and bladder urothelial carcinoma (BLCA). TP53 mutations were also largely observed within the previously identified “hotspots,” including missense mutations in R175, R248, and R273 and nonsense mutations in R196 and R213 (refs. 28, 29; Supplementary Fig. S2A). APC was the second most frequently mutated gene, and it was predominantly found in COAD, prostate adenocarcinoma (PRAD), and UCEC. Activating mutation in KRAS was the third leading genomic aberration, primarily observed in COAD, pancreatic adenocarcinoma (PDAC), cholangiocarcinoma (CHOL), ampullary carcinoma (AMPC), and UCEC. Consistent with previous large-scale genomic profile studies, KRASG12 and KRASG13 mutations were the most common events, accounting for over 66% of all KRAS-mutant tumors (refs. 30–32; Supplementary Fig. S2B). Other major hotspot mutations were PIK3CAE542, PIK3CAH1047, BRAFV600E, ATRXE2246, and NF1S1100. Moreover, we identified a list of mutations that were highly enriched in specific tumor types, such as BRAF mutations in skin cutaneous melanoma (SKCM) and thyroid carcinoma (THCA), KMT2D mutations in BLCA and UCEC, EGFR mutations in LUAD and thymoma (THYM), PTEN mutations in UCEC, and CDH1 mutations in STAD.
To explore and determine the etiology of each tumor lin-eage, we investigated the mutational transition, context, and signature of each disease (33–35). Of the six classes of base substitution in each mutation type, all tumors demonstrated the robust presence of C>T transition at CpG trinucleotides, which has been speculated to be a direct result of the endogenous mutational process via spontaneous deamination of 5-methylcytosine (Fig. 1B). As this phenomenon was strongly associated with the age of the patient at cancer diagnosis, we discovered that mutational signature 1 was one of the predominant signatures that were observed across all tumor types. Other prominent mutational signatures that substantially occupied the overall mutational profiles of each tumor class were signatures that were associated with failure of DNA double-strand break repair through homologous recombination and a defective DNA mismatch repair (MMR) system. As these features are the hallmarks of DNA repair mechanisms that ensure essential cell homeostasis (36), we suspected that a majority of the solid tumors in Korean patients may have largely propagated from MMR deficiency–derived tumorigenesis. We also identified tumor type–specific mutational signatures, including signature 4 (cigarette smoking) in LUAD, alkylating agent and ultraviolet light exposure-associated signatures in SKCM, and aflatoxin-associated signature in liver hepatocellular carcinoma (LIHC). Next, we have assessed the significance of each genomic alteration that may directly affect the tumorigenic mechanism behind individual cancer types. Notably, we have identified previously known associations, including enrichment of APC, TP53, KRAS, and SMAD4 mutations in COAD, PIK3CA and GATA3 mutations in BRCA, and KRAS, ARID1A, EGFR, and PTEN mutations in PDAC, STAD, LUAD, and UCEC, respectively (Fig. 1C; Supplementary Table S2). Conversely, we discovered that mutations in TP53 were relatively scarce in a number of different tumors, including sarcoma (SARC), SKCM, cervical cancer (CESC), and kidney renal clear cell carcinoma (KIRC), compared with other tumor types. KRAS mutations were significantly less frequent in head and neck cancer (HNSC) and BRCA, whereas mutations in APC were rare events in PDAC. Collectively, our results demonstrate the clinical utility of NGS panels to provide a comprehensive understanding of cancer etiology and the unique molecular properties of each individual patient.
Molecular Interactions of Major Canonical Oncogenic Pathways in a Pan-Cancer Model
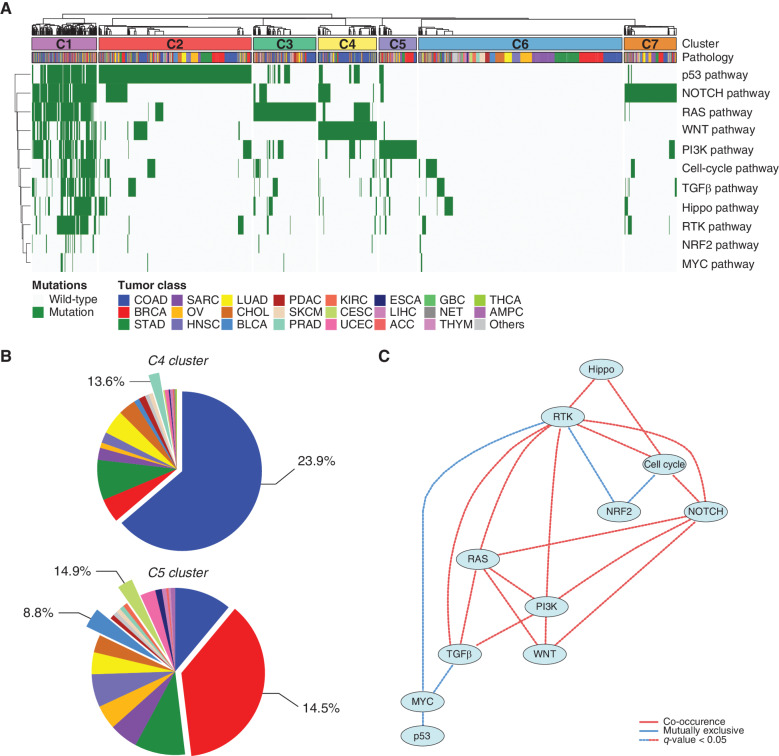
Cancer is a disease of the genome with profound genomic aberrations and complex cellular signaling networks. Although each individual cancer lineage manifests different cellular architecture and hierarchy, dysregulation in the core oncogenic pathways is one of the primary contributing components underlying malignant transformation (15, 37). As such, we sought to determine whether molecular alterations at the cellular signaling pathway level were associated with different tumor origin. When we performed consensus hierarchical clustering of individual tumors based on the presence and absence of genomic alterations that correspond to major canonical pathways, we identified seven distinct clusters with diverse genomic backgrounds (Fig. 2A; Supplementary Fig. S3). Cluster 1 (C1) mainly consisted of tumors harboring multiple genomic aberrations that affect a wide array of different pathways, including p53, NOTCH, RTK–RAS, PI3K, Wnt, and Hippo. Clusters 2, 3, 4, 5, and 7 were defined by enrichments of the p53, RAS, Wnt, PI3K, and NOTCH signaling pathways, respectively. Notably, cluster 6 (C6) was largely composed of tumors lacking any individual mutations that affect previously well-known canonical oncogenic pathways, suggesting potential alternative avenues for these tumors to exploit in terms of tumor propagation. Interestingly, each cluster composition was comprised of different tumor types, albeit both cluster 4 (C4) and cluster 5 (C5) demonstrated large collections of COAD and BRCA, respectively. C4, represented by dysregulation in the Wnt pathway, was largely comprised of COAD and PRAD tumors, whereas C5, marked by genomic aberrations in the PI3K pathway, was more predominantly composed of BRCA, BLCA, and CESC (Fig. 2B).
Figure 2.
Major oncogenic canonical pathways of the K-MASTER cohort. A, Unsupervised hierarchical clustering of pan-cancer patients based on major oncogenic canonical pathways. Patients have been marked with a mutation for each pathway if the patient harbors at least one mutation that belongs to the corresponding pathway. C3, cluster 3; C6, cluster 6; C7, cluster 7. B, Pie chart distribution of patients within each corresponding cluster (C4: top; C5: bottom). The percentage represents the frequency of patients who belong to the respective pathway cluster within the corresponding tumor type (i.e., 23.9% of patients with COAD and 13.6% of patients with PRAD belong to the C4 cluster). C, Bayesian network analysis depicting the co-occurrences and mutual exclusivity of major canonical pathways in the K-MASTER pan-cancer cohort. Only the significant associations are shown. ACC, adrenal carcinoma; ESCA, esophageal carcinoma; GBC, gallbladder cancer; NET, neuroendocrine carcinoma.
Genomic alteration at the pathway level provides a broader perspective on functional synergies within cancer malignancy and may also reflect potential therapeutic resistance or evasion mechanisms. To explore the dynamic associations within molecular pathways, we constructed a Bayesian network–based probabilistic model to identify significant co-occurrences and mutual exclusivity among major cancer driver pathways. Upon construction of multiple networks, we discovered significant co-occurrences of mutations affecting the TGFβ, RAS, PI3K, Hippo, and cell-cycle pathways with RTK mutations, which highlight potential synergistic activations of individual pathways (Fig. 2C). Both RAS and PI3K pathways contained multiple pairs of mutations that were also coenriched. Conversely, we also found several mutually exclusive pairs of mutations, notably in the MYC pathway with RTK, TGFb and p53, and the NRF2 pathway with RTK and cell cycle. Mutual exclusivity of the NRF2 pathway mutations with cell cycle and RTK mutations has also been previously reported in The Cancer Genome Atlas (TCGA) cohorts (15), consistent with our findings. These associations suggest that activation of either pathway was sufficient for tumor propagation and alterations of the two may render tumor cells more susceptible for adverse situations. Our results collectively demonstrate dynamic interactions among major canonical pathways within tumor malignancy and functional interactions, which suggest the use of combinational strategies in clinical settings.
Ethnic Diversification of Pan-Cancer Genomes
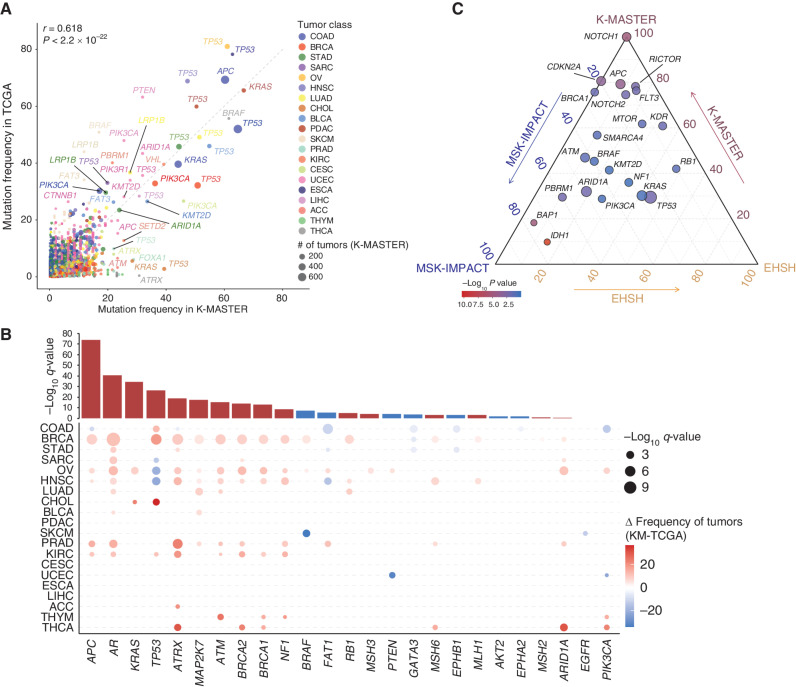
Recent studies have highlighted the existence of genomic diversity based on distinct racial or ethnic populations (18, 20, 38–40). Thus, an assessment of genetic ancestry at the pan-cancer level may provide unprecedented insights into alternative therapeutic vulnerabilities. The identification of molecular pathways that are actively enriched in a specific ethnic subpopulation could potentially facilitate the exploration of new, personalized treatment options. Toward this goal, we leveraged TCGA cohorts, which were comprised mainly of patients from European ancestry, across 20 major cancer types to characterize and compare the prevalence of major cancer driver genomic aberrations at both molecular and pathway levels. Although several major canonical pathways, including TGFb, cell cycle, Myc, and Hippo, constituted highly consistent levels of pathway dysregulation at the pan-cancer level, some oncogenic pathways demonstrated significant levels of genetic diversity (Supplementary Figs. S4 and S5). Surprisingly, although patients from K-MASTER showed significant levels of KRAS mutations, TCGA patients were characterized by recurrent mutations in BRAF. Ethnicity-driven genomic diversity became increasingly more apparent when compared at individual tumor levels. Although the majority of the genomic alterations demonstrated considerable levels of similarity (r = 0.618, P < 2.2 × 10−22), mutations in TP53 differed significantly in several pathologic entities (Fig. 3A). For example, patients who were enrolled in TCGA and were diagnosed with either OV, esophageal carcinoma (ESCA), HNSC, PDAC, or SARC demonstrated higher levels of TP53 ablation, whereas enrichment of TP53 mutation was more evident in K-MASTER patients with COAD, BLCA, BRCA, CHOL, and PRAD. Furthermore, we discovered that TCGA patients with COAD were marked by enrichment of somatic APC and PIK3CA mutations, whereas K-MASTER patients with CESC showed frequent alterations in PIK3CA and ATRX. Additionally, TCGA patients with SKCM showed recurrent dysregulations in major oncogenic drivers, including BRAF, LRP1B, and FAT3.
Figure 3.
Genomic diversity of pan-cancer patients based on ethnicity. A, Mutation frequencies in the TCGA (y-axis) and K-MASTER (x-axis) cohorts. The color corresponds to distinct tumor type, and the size of each node represents the number of tumors within the K-MASTER cohort. B, Pan-cancer meta-analysis on recurrent mutations between the K-MASTER and TCGA cohorts across 20 cancer types. The top bar graph demonstrates the significance of each mutation at the pan-cancer level. The bottom dot plot represents the significance and difference of each mutation at individual cancer lineage levels. The color corresponds to the effect size of K-MASTER patients (KM) compared with that of the TCGA, and the size is proportional to the significance. C, Ternary diagram depicting mutation frequencies in the K-MASTER, EHSH in Shanghai, China, and MSK-IMPACT cohorts. The size of each node represents the number of tumors with respect to the mutation in the K-MASTER cohort, and the color spectrum indicates the significance of relative frequencies. ACC, adrenal carcinoma.
Next, we analyzed all essential mutations that differed significantly between TCGA and K-MASTER at both individual cancer lineage and pan-cancer levels. As a result, we discovered 25 recurrently mutated genes (>200 tumors) that demonstrated significant statistical differences. Among them, APC was the most significantly mutated gene in K-MASTER, followed by AR, KRAS, TP53, ATRX, MAP2K7, and ATM (Fig. 3B). Conversely, TCGA patients demonstrated the predominance of BRAF, FAT1, PTEN, GATA3, EPHB1, AKT2, EPHA2, and EGFR mutations in both pan-cancer and individual tumor types. Furthermore, we discovered that mutations in MMR pathway–encoding genes, including MSH3, MSH6, MLH1, and MSH2, were significantly more frequent in K-MASTER patients, confirming previous observations on the dominance of MMR deficiency–associated mutational signature in our cohort. Furthermore, we have discovered a significant enrichment of IDH1 mutations only in TCGA patients with CHOL, whereas patients from K-MASTER mostly lacked such genomic aberrations; rather they exhibited highly recurrent mutations in TP53 and KRAS (Fig. 3B; Supplementary Fig. S6). Interestingly, one of the earlier studies that took advantage of sequencing technology has identified recurrent mutations in multiple chromatin-remodeling genes, including IDH1 in intrahepatic CHOL (41). However, a majority of the enrolled patients were of European ancestry (93.3%). To further investigate the genomic diversity of CHOL at the ethnicity level, we curated additional mutational profiles of 195 CHOL patients from the Memorial Sloan Kettering (MSK)-IMPACT cohort (42) and 103 patients from the Eastern Hepatobiliary Surgery Hospital (EHSH) in China (43). Remarkably, all key chromatin-remodeling genes, including IDH1, BAP1, and PBRM1, were significantly enriched only in the MSK-IMPACT cohort. Such results further supported our previous findings (Fig. 3C). Taken together, our results demonstrate the significance of genomic diversity at both the pan-cancer and individual cancer lineage levels between populations from European and East Asian ancestries.
Ethnic Diversification of Clinical Actionability
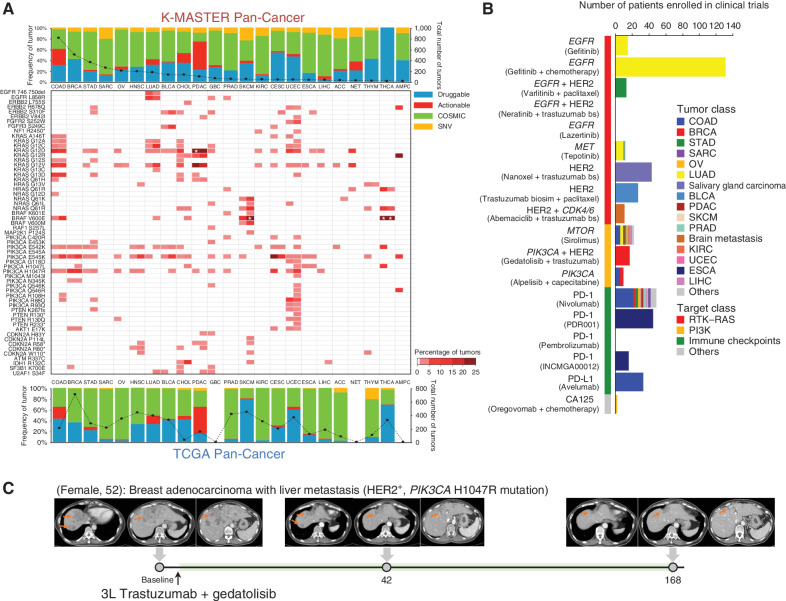
The molecular characterization of tumors enables patient-tailored therapy. Furthermore, a substantial number of studies have demonstrated remarkable clinical success through the employment of essential molecular biomarkers, including trastuzumab against HER2 in BRCA (2), vemurafenib against BRAFV600E in melanoma (44), gefitinib against EGFR in non–small cell lung cancer (NSCLC; ref. 4), and so on. Therefore, numerous institutions have begun to use routine clinical sequencing to make informed decisions in the choice of targeted therapy. In hopes of identifying a subset of patients who may benefit from such therapeutic intervention, we evaluated clinically relevant molecular properties of the K-MASTER cohort compared with TCGA based on different disease backgrounds. Overall, THCA, UCEC, BRCA, and SKCM demonstrated the highest proportion of druggable mutations for both cohorts, whereas tumors with limited levels of targetable alterations included SARC, KIRC, and LIHC. These results urge the exploration of alternative therapeutic avenues (Fig. 4A). Compared with 31.8% of TCGA patients, 28.7% of K-MASTER patients possessed at least one or more molecular targets that were druggable (Supplementary Fig. S7). At the individual molecular level, somatic mutations in KRAS and PIK3CA provided the most therapeutic opportunities for patients with COAD and BRCA, respectively (Supplementary Fig. S8). BRAF mutation was the third leading target, enriched in COAD, CHOL, SKCM, and THCA. Other prospective targets were CDKN2A in HNSC, ERBB2 in STAD, NRAS in SKCM, PTEN in UCEC, AKT1 in BRCA, EGFR in LUAD, BRCA1/2 in OV, FGFR3 and ERCC2 in BLCA, and KIT in SARC.
Figure 4.
Therapeutic landscape of pan-cancer patients based on ethnicity. A, The top bar graph depicts the distribution of major mutations based on the clinical actionability of the mutations in the K-MASTER cohort. The bottom bar graph represents the TCGA pan-cancer cohort. Mutations are categorized as “druggable” if they are FDA-approved biomarkers for FDA-approved drugs. Mutations are labeled as “actionable” if there is substantially compelling evidence to support the use of biomarkers to predict the response, especially the resistance of FDA-approved drugs. Mutations are labeled as “COSMIC” if they have been previously annotated using the COSMIC (Catalogue of Somatic Mutations in Cancer) database. Mutations that do not belong to any of the categories mentioned previously are depicted as “SNV.” Middle, frequencies of clinically actionable mutations across major cancer types in K-MASTER (left portion of the cell) compared with TCGA (right portion of the cell). Genes have been grouped by pathway. The white asterisk represents tumor types with more than 25% frequency (29.1% of KRASG12D mutation in K-MASTER PDAC, 43.6% of BRAFV600E mutation in TCGA SKCM, and 61.5% and 59.3% of BRAFV600E mutations in K-MASTER and TCGA THCA, respectively). B, The number of patients who have been enrolled to matched clinical trials based on unique molecular alterations. Trastuzumab bs, trastuzumab biosimilar. C, Clinical course of third-line (3L) trastuzumab and gedatolisib treatment in a BRCA patient with liver metastasis harboring a PIK3CA H1047R mutation. T1-weighted contrast-enhanced magnetic resonance images are shown for baseline, 6-week, and 24-week posttreatment. Orange arrows indicate measurable tumors. ACC, adrenal carcinoma; GBC, gallbladder cancer; NET, neuroendocrine carcinoma.
Next, we leveraged the OncoKB knowledge database to systematically annotate each molecular alteration based on clinical utility (45). Among 963 pharmacogenomic associations that were previously reported, BRAFV600E mutation was the most prevalent genomic aberration—primarily observed in COAD, SKCM, and THCA—followed by KRASG12D and PIK3CA E545K and H1047R mutations (Fig. 4A; Supplementary Fig. S9A). Strikingly, the level at which BRAFV600E mutation occurred significantly differed between TCGA and K-MASTER, where the patients from TCGA harbored the alteration at a significantly higher rate than patients from K-MASTER (73.3% vs. 25.7%; P = 1.58 × 10−36; Supplementary Fig. S9B). Although the level of treatment opportunity in the K-MASTER and TCGA cohorts was similar among several major solid tumors (COAD, BRCA, BLCA, PDAC, UCEC, and THCA), other cancer types manifested significant differences. Patients with STAD from TCGA were characterized by recurrent mutations in ERBB2 and PIK3CA, whereas K-MASTER patients showed a higher frequency of KRAS and PIK3CA mutations in OV. Moreover, we discovered that patients with both PRAD and KIRC from K-MASTER possessed several clinically targetable aberrations, including EGFR, KRAS, and PIK3CA, whereas patients from TCGA completely lacked such opportunities. In addition, TCGA patients with LUAD exhibited higher activation of various KRAS mutations, whereas K-MASTER patients were marked by enrichments in EGFR exon 19 deletion, which encodes part of the kinase domain and renders higher susceptibility of tumors to various EGFR tyrosine kinase inhibitors, including gefitinib and afatinib.
Patients were enrolled into 20 distinct matched clinical trials depending on the unique molecular and histopathologic properties of each patient. Through such assignment, we were able to target various major molecular aberrations, ranging from receptor tyrosine kinases, including EGFR, c-MET, and HER2, to immune-checkpoint molecules, such as PD-1 and PD-L1 (Fig. 4B). A total of 440 patients have been enrolled on the basis of corresponding molecular targets so far. The most common treatments consisted of EGFR, HER2, PIK3CA, PD-1, and PD-L1 inhibitors, including gefitinib and lazertinib in NSCLC, varlitinib in STAD, a trastuzumab biosimilar in BRCA, STAD, and BLCA, gedatolisib in BRCA, alpelisib in COAD and other solid tumors, nivolumab in solid tumors with homologous recombination deficiency and MMR deficiency, PDR001 in ESCA, pembrolizumab in NSCLC, avelumab in MSI-high/POLE-mutant COAD, and INCMGA00012 in ESCA. Notably, a patient with BRCA with liver metastasis harboring activation of HER2 and a PIK3CA H1047R mutation demonstrated exceptional response to third-line treatment of gedatolisib with trastuzumab (Fig. 4C). The patient was previously treated with docetaxel, trastuzumab, and pertuzumab as first-line therapy and T-DM1 as second-line treatment. However, the patient eventually experienced tumor relapse without any alternative options. Based on the unique molecular property of the patient's tumor, a combinational treatment of gedatolisib, a PI3K inhibitor, with trastuzumab was given, and the patient showed remarkable clinical response within the first 6 weeks, with the response prolonged even after 24 weeks. Collectively, our results demonstrate considerable levels of differences in terms of clinical actionability between East Asian and European American patients with cancer and recommend that molecular-guided treatments should be implemented with caution depending on the ethnic background of the patient.
DISCUSSION
In this study, we report on the clinical utility and significance of systematic prospective sequencing to delineate and identify the major genomic aberrations in East Asian pan-cancer patients. This study, which involved a nationwide, multi-institutional effort, demonstrated clinical feasibility in guiding individual patients to ideally matched clinical trials that may provide maximum therapeutic efficacy over a wide spectrum of different cancer types. Based on this enterprise, we present the first phase of our precision oncology initiative, focusing on a molecular landscape in 4,028 East Asian pan-cancer patients. By utilizing an integrative analytical approach, we have identified key driver alterations and etiologies of 24 major cancer lineages, and dynamic interactions at both molecular and pathway levels. Furthermore, to investigate the impact of ethnic ancestry on both genomic diversity and clinical utility, we leveraged previously established large-scale pan-cancer genomic cohorts, including TCGA and MSK-IMPACT. Our results provided a comprehensive overview of therapeutic opportunities for individual cancer types based on distinct ancestral populations. We have identified considerable levels of differences between Eastern and Western populations in terms of both individual genomic event and pathway levels. Activations of NRF2 and PI3K signals were predominantly observed in TCGA cohort, whereas K-MASTER patients demonstrated recurrent alterations in RTK–RAS, TP53, WNT, and NOTCH pathway genes. This notable distinction was consistently observed in previous studies where patients of European American ancestry showed higher rates of dysregulation in PI3K pathway–encoding genes, including PTEN, compared with African American populations (18). Although African American patients were marked by enrichments of p53 pathway abnormality, specifically through genomic alteration in TP53, our study showed that East Asian patients were constituted by activation of the RTK–RAS and NOTCH pathways. Furthermore, we have discovered higher dysregulation in MMR-encoding genes for K-MASTER patients, which further consolidated previous observations on the enrichment of MMR deficiency–associated mutational signatures. These results collectively suggest that cancer in Korean patients may have largely propagated from impairment of DNA repair mechanisms. Such genomic disparity was further evident when assessed at individual cancer levels. Recurrent mutations of chromatin-remodeling genes such as BAP1, PBRM1, and IDH1 were relatively more scarce in patients with CHOL from Eastern backgrounds. Furthermore, significant differences in molecular-based treatment opportunities at both pan-cancer and individual tumor levels were observed. Notably, we have discovered that BRAFV600E mutation was significantly more enriched in TCGA patients, whereas K-MASTER patients harbored higher levels of KRASG12 mutations. Other key findings included the predominance of mutations in NRAS at both the Q61R and Q61K domains only in TCGA patients and substantial differences of clinical actionability profiles between the K-MASTER and TCGA cohorts in STAD, OV, CHOL, PRAD, KIRC, LIHC, and adrenal carcinoma (ACC). On the contrary, we also found that COAD, BRCA, BLCA, PDAC, UCEC, and THCA tumors shared similar molecular properties across the cohorts.
Although our results have demonstrated profound levels of genomic diversity between patients from different ethnic origins at the pan-cancer level, there were several limitations to this approach. First, TCGA cohort was mainly comprised of tumors that were derived from a single distinct pathologic entity, whereas the K-MASTER cohort originated from more generalized classifications. Furthermore, there were discrepancies in terms of the cohort size at the individual disease level. However, as this was, to the best of our knowledge, the first comprehensive molecular study to evaluate genomic profiles of a pan-cancer cohort in the East Asian population, our results highlight the significance of using an ethnicity-based personalized approach in cancer therapy. Moreover, the current study provides a more appropriate real-world scenario of cancer treatment where the patients have been diagnosed with an advanced stage of tumor and are searching for alternative therapeutic options, whereas other large-scale genomic studies, including TCGA, focused primarily on untreated tumors at a relatively early stage in tumor progression. As such, we speculate that our study provides higher clinical applicability. Furthermore, the primary utility of the K-MASTER program lies in its ability to provide sufficient evidence to make informed decisions in terms of patient treatment. Based on this evidence, 440 patients have been enrolled into 20 different clinical trials targeting major genomic aberrations across a broad range of different tumor types. At this point, a majority of the clinical trials are still ongoing, and thus we were not able to disclose the full results in the present study. However, as a proof of concept, we showed through prospective clinical sequencing that a BRCA-diagnosed patient who harbored a PIK3CA-activating mutation with liver metastasis demonstrated a remarkable response to PIK3CA-targeted therapy.
Both clinical feasibility and adaptation of precision oncology are determined by several key components that need to be recognized and accounted for. The main priorities of patients are the accuracy and timeliness of results, access to treatment, cost–benefit ratio, and degree of improvement in quality of life through new treatments. For clinicians, on the other hand, turnaround time, accuracy of genomic screening tests, reimbursement rate, and possibility of evidence-based clinical decisions are the primary concerns. Through the K-MASTER enterprise, 55 cancer-treating hospitals and centers have participated and experienced a full cycle of personalized care (from NGS testing to making clinically informed decisions) within a short period of time. We strongly believe that the K-MASTER initiative clearly exhibits how quickly and efficiently a public-initiated program can expand the potential of precision oncology, even for latecomers. In the next phase of our study, we aim to explore dynamic interactions of genomic aberrations with therapeutic responses from 20 different clinical trials at the pan-cancer level.
METHODS
K-MASTER Initiative and Tumor Specimen Collection
The K-MASTER initiative is a government-supported precision medicine enterprise that mainly focuses on the diagnosis and treatment of patients with cancer (https://k-master.org/eng.php). The primary objective of the operation is to collect and characterize the complex genome of 10,000 Korean patients with advanced solid tumors who have been enrolled in the master screening protocol, KM-00. Based on patients who were initially screened using the KM-00 protocol and have been identified with at least one actionable therapeutic target of treatment, K-MASTER has initiated 20 distinct clinical trials using single or combination targeting agents. Patients with advanced solid tumors were enrolled to the master screening protocol KM-00 at one of the 55 participating sites after Institutional Review Board approval. After receiving written informed consent from the patients, we collected and archived tumor tissue specimens. Clinical and genomic information is stored in the K-MASTER database and has been released to the public (https://kmportal.or.kr). As of December 2020, more than 7,900 patients have participated and were enrolled in the KM-00 master screening program. Furthermore, we have developed a “Match Master System” that utilizes the OncoKB knowledge database for clinical decision support on the actionability of the genes. We have also leveraged updated results from all relevant clinical trials that have been conducted. The research conformed to the principles of the Declaration of Helsinki.
K-MASTER Sequencing Panels
K-MASTER used two previously established tissue-based NGS panels (FIRST and CancerSCAN) to detect major genomic aberrations, including mutations, CNAs, and small insertions and deletions in cancer-related genes. CancerSCAN has been further upgraded to K-MASTER v1.0 and v1.1. Genomic DNA from formalin-fixed paraffin-embedded (FFPE) samples or plasma was extracted using the QIAamp FFPE tissue kit (Qiagen) or QIAamp circulating nucleic acid kit (Qiagen), respectively. Cell-free DNA purity was measured using an Agilent High Sensitivity DNA Kit and a 2100 Bioanalyzer instrument (Agilent Technologies). When required, additional purification was performed using Agencourt AMPure XP (Beckman Coulter) to further remove contaminating nucleic acid. Centrally isolated genomic DNA samples that underwent quality control were sent to the K-MASTER genomic analysis laboratories.
Mutation Calls
The sequenced reads from the FASTQ files were aligned to the human genome assembly (hg19) using the Burrows–Wheeler aligner. The initial aligned BAM files were further subjected to preprocessing steps, including sorting, removal of duplicated reads, local realignment around small indels, and recalibration of base quality scores using SAMtools, Picard, and Genome Analysis Toolkit (GATK). To make high-confidence predictions on mutation calls, we used MuTect2. The 1000 Genomes, gnomAD, and dbSNP data sets were used as a reference database for known polymorphic sites. We used the variant effect predictor to annotate each variant. Mutations with a minimum depth ≥20 and variant allele frequency of ≥2 were used in this study.
Mutational Signatures
We used deconstructSigs in R to perform mutational signature analysis. It uses a list of mutations based on six substitution classes (C>T, C>A, C>G, T>C, T>A, and T>G) and base contexts immediately before and after the mutated nucleotide within the exome regions. It also generates a composition of a given set of mutational signatures that were previously identified. Thirty different mutational signatures from “signature.cosmic” were used as reference signatures and were represented in the following terms: age (signature 1), APOBEC (signature 2), DNA-DSB (double-strand break; signature 3), smoking (signature 4), sig 5 (signature 5), MMRd (MMR deficiency; signature 6), UV (signature 7), alkylating (signature 11), APOBEC2 (signature 13), aflatoxin (signature 24), tobacco (signature 25), and sig 30 (signature 30). To prevent the overestimation of a mutational signature proportion from patients with a limited number of mutations, we only selected tumors with more than 20 nonsynonymous mutations. We filtered mutation signatures that were present in at least 5% of the samples in each tumor type.
MSI Status
MSIseq in R was used to assess tumor MSI. MSIseq is a decision tree classifier using a list of mutations based on different mutation rates in all sites as well as in simple-sequence repeats. It uses previously established somatic mutation data from the exomes of 361 tumors as a training set and classifies newly generated tumors based on the annotation of locations of simple-sequence repeats and sequence length of each tumor.
Comparison of Genomic Diversity and Pathway Activity between the K-MASTER and TCGA Cohorts
To compare the frequencies of major genomic aberrations based on ethnicity, we acquired TCGA pan-cancer somatic mutation data and the clinical data resource from Genomic Data Commons. Only mutations that were annotated as “PASS” in the “FILTER” column have been retained for all cancer types except for earlier TCGA samples that were sequenced using the whole-genome amplified method. Afterward, we only selected mutation data for patients who were diagnosed with the matching tumor types characterized in the K-MASTER program, including COAD, BRCA, STAD, OV, HNSC, LUAD, CHOL, BLCA, PDAC, SKCM, PRAD, KIRC, CESC, UCEC, ESCA, LIHC, ACC, THYM, and THCA. This resulted in a total of 7,557 patients. Next, we selected for patients who have been annotated as “WHITE” in the “race” column, resulting in the final list of 5,579 patients.
Only genes that were captured from the K-MASTER sequencing panels underwent further selection. Among them, only the protein-coding mutations were considered for comparison analysis. For the pan-cancer comparative analysis, we leveraged mutation profiles of the resulting TCGA patients with the K-MASTER cohort and compared the frequency of each individual mutation corresponding to a major oncogenic canonical pathway. Pathway diagrams and genes were curated from PathwayMapper, a tool that provides visualization and design of major oncogenic pathways from previous TCGA publications. This tool is publicly available online at www.pathwaymapper.org. To perform individual tumor-type comparison, we curated mutation profiles from both TCGA and K-MASTER cohorts based on individual cancer lin-eage and compared the overall mutational frequency. For patients with CHOL, we also acquired mutation profiles for the MSK-IMPACT and EHSH cohorts from cBioPortal (http://www.cbioportal.org). Chi-square tests were performed to compare the frequencies of individual gene-level mutations between K-MASTER and TCGA cohorts, and corrections for multiple-hypothesis testing were performed to account for false discovery rate.
Clinical Trial Enrollment
Genetic alterations, including SNVs, indels, CNAs, or structural rearrangements with clinical actionability were reported in a clinical report format. Treatment options, including clinical trials in the K-MASTER program, were recommended based on the OncoKB knowledge database (OncoKB API; December 2020) and inclusion criteria for each trial.
Statistical Analyses
All statistical analyses were performed using R software 3.4.0 (https://www.r-project.org).
Data Availability
All data have been deposited and are hosted on our portal at https://kmportal.or.kr.
Supplementary Material
Acknowledgments
We would like to thank the following investigators from different institutions for devotional participation: In Keun Park (Gachon University Gil Medical Center), Sang Hoon Chun (The Catholic University of Korea, Bucheon St. Mary's Hospital), Ji Eun Lee (The Catholic University of Korea, Seoul St. Mary's Hospital), Ho Jung An (The Catholic University of Korea, St. Vincent's Hospital), Yoon Ho Ko (The Catholic University of Korea, Eunpyeong St. Mary's Hospital), Der Sheng Sun (The Catholic University of Korea, Uijeongbu St. Mary's Hospital), Jae Ho Byeon (The Catholic University of Korea, Incheon St. Mary's Hospital), Hei Cheul Jeung (Gangnam Severance Hospital), Dong Hoe Koo (Kangbuk Samsung Hospital), Ji Hyun Park (Konkuk University Medical Center), Jong Gwon Choi (Konyang University Hospital), Gyeong Won Lee (Gyeongsang National University Hospital), Sun Kyung Baek (Kyung Hee Univeristy Hospital), Jin Young Kim (Keimyung University Dongsan Hospital), Eun Joo Kang (Korea University Guro Hospital), Jun Sun Kim (Korea University Ansan Hospital), Yeul Hong Kim (Korea University Anam Hospital), Seong Hoon Shin (Kosin University Gospel Hospital), Tak Yun (National Cancer Center), Ha Yeon Lee (National Medical Center), Ja Yoon Heo (National Health Insurance Service Ilsan Hispital), Soon Il Lee (Dankook University Hospital), Yoon Young Cho (Daegu Catholic University Medical Center), Sung Yong Oh (Dong-A University Hospital), Jin Soo Kim (Boramae Medical Center), Hyo Jeong Kim (Pusan National University Hospital), Yu Jung Kim (Seoul National University Bundang Hospital), Yong Wha Moon (CHA Bundang Medical Center), Jee Yun Lee (Samsung Medical Center), Mi So Kim (Seoul National University Hospital), Tae Won Kim (Asan Medical Center), Jung Hye Kwon (Chungnam National University Sejong Hospital), Min Young Lee (Soonchunhyang University Seoul Hospital), Sang Cheol Lee (Soonchunhyang University Cheonan Hospital), Sung Hee Lim (Soonchunhyang University Bucheon Hospital), Mi Sun Ahn (Ajou University Hospital), So Yeon Oh (Pusan National University Yangsan Hospital), Hyo Song Kim (Yonsei Cancer Hospital), Min Kyoung Kim (Yeungnam University Medical Center), Young Ju Min (Ulsan University Hospital), Seung Taek Im (Wonju Severance Christian Hospital), Kyung Eun Lee (Ewha Womans University Medical Center), Moon Young Choi (Inje University Busan Paik Hospital), Young Jin Yuh (Inje University Sanggye Paik Hospital), Seong Yoon Yi (Inje University Ilsan Paik Hospital), Myoung Joo Kang (Inje University Haeundae Paik Hospital), Eun Kee Song (Jeonbuk National University Hospital), In Gyu Hwang (Chung-Ang University Hospital), Tae Kyu Lim (Veterans Health Service Medical Center), Hwan Jung Yun (Chung Nam National University Hospital), Hee Kyung Kim (Chungbuk National University Hospital), Yee Soo Chae (Kyungpook National University Chilgok Hospital), Joo Yeong Jung (Hallym University Dongtan Sacred Heart Hospital), Ho Young Kim (Hallym University Medical Center), Kyung Tae (Hanyang University Medical Center), and Woo Kyun Bae (Chonnam National University Hwasun Hospital).
This research was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, funded by the Ministry of Health and Welfare, Republic of Korea (HI17C2206 to K.H. Park and Y.H. Kim), a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT; NRF-2020R1F1A1076444 to J.K. Sa), the Bio and Medical Technology Development Program of the NRF funded by the Korean government (MSIT; 2020M3A9D8038658 to J.K. Sa), and a Korea Evaluation Institute of Industrial Technology grant funded by the Korean government (MSIT; 20009125 to J.K. Sa).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
Y.H. Kim reports grants from the Ministry of Health and Welfare, Korea, during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
K.H. Park: Conceptualization, resources, data curation, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. J.Y. Choi: Resources, data curation, formal analysis, and investigation. A.-R. Lim: Resources, data curation, formal analysis, and investigation. J.W. Kim: Resources, data curation, formal analysis, and investigation. Y.J. Choi: Resources, data curation, formal analysis, and investigation. S. Lee: Resources, data curation, formal analysis, and investigation. J.S. Sung: Data curation, formal analysis, investigation, and methodology. H.-J. Chung: Resources, data curation, software, formal analysis, investigation, and methodology. B. Jang: Software, formal analysis, validation, and methodology. D. Yoon: Software, for-mal analysis, validation, and methodology. S. Kim: Software, formal analysis, validation, and methodology. J.K. Sa: Conceptualization, data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. Y.H. Kim: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing.
References
- 1. Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med 2010;363:301–4. [DOI] [PubMed] [Google Scholar]
- 2. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde Aet al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783–92. [DOI] [PubMed] [Google Scholar]
- 3. O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes Fet al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 2003;348:994–1004. [DOI] [PubMed] [Google Scholar]
- 4. Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe Het al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380–8. [DOI] [PubMed] [Google Scholar]
- 5. Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko Eet al. Five-year outcomes with dabrafenib plus trame-tinib in metastatic melanoma. N Engl J Med 2019;381:626–36. [DOI] [PubMed] [Google Scholar]
- 6. Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GIet al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 2018;554:189–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Andre T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt Cet al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med 2020;383:2207–18. [DOI] [PubMed] [Google Scholar]
- 8. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HRet al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed Aet al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Penson A, Camacho N, Zheng Y, Varghese AM, Al-Ahmadie H, Razavi Pet al. Development of genome-derived tumor type prediction to inform clinical cancer care. JAMA Oncol 2019;6:84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Flaherty KT, Gray RJ, Chen AP, Li S, McShane LM, Patton Det al. Molecular landscape and actionable alterations in a genomically guided cancer clinical trial: National Cancer Institute Molecular Analysis for Therapy Choice (NCI-MATCH). J Clin Oncol 2020;38:3883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Flaherty KT, Gray R, Chen A, Li S, Patton D, Hamilton SRet al. The Molecular Analysis for Therapy Choice (NCI-MATCH) trial: lessons for genomic trial design. J Natl Cancer Inst 2020;112:1021–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kalinsky K, Hong F, McCourt CK, Sachdev JC, Mitchell EP, Zwiebel JAet al. Effect of capivasertib in patients with an AKT1 E17K-mutated tumor: NCI-MATCH subprotocol EAY131-Y nonrandomized trial. JAMA Oncol 2021;7:271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cancer Genome Atlas Research Network; Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BAet al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KCet al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell 2018;173:321–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe Aet al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;174:1034–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill Eet al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018;173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi Jet al. Integrated analysis of genetic ancestry and genomic alterations across cancers. Cancer Cell 2018;34:549–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang FW, Mosquera JM, Garofalo A, Oh C, Baco M, Amin-Mansour Aet al. Exome sequencing of African-American prostate cancer reveals loss-of-function ERF mutations. Cancer Discov 2017;7:973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Campbell JD, Lathan C, Sholl L, Ducar M, Vega M, Sunkavalli Aet al. Comparison of prevalence and types of mutations in lung cancers among black and white populations. JAMA Oncol 2017;3:801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deng J, Chen H, Zhou D, Zhang J, Chen Y, Liu Qet al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat Commun 2017;8:1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krishnan B, Rose TL, Kardos J, Milowsky MI, Kim WY. Intrinsic genomic differences between African American and white patients with clear cell renal cell carcinoma. JAMA Oncol 2016;2:664–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Niu B, Ye K, Zhang Q, Lu C, Xie M, McLellan MDet al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014;30:1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang MN, McPherson JR, Cutcutache I, Teh BT, Tan P, Rozen SG. MSIseq: software for assessing microsatellite instability from catalogs of somatic mutations. Sci Rep 2015;5:13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sahin IH, Akce M, Alese O, Shaib W, Lesinski GB, El-Rayes Bet al. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br J Cancer 2019;121:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eso Y, Shimizu T, Takeda H, Takai A, Marusawa H. Microsatellite instability and immune checkpoint inhibitors: toward precision medicine against gastrointestinal and hepatobiliary cancers. J Gastroenterol 2020;55:15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JSet al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol 2019;30:1096–103. [DOI] [PubMed] [Google Scholar]
- 28. Baugh EH, Ke H, Levine AJ, Bonneau RA, Chan CS. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ 2018;25:154–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Donehower LA, Soussi T, Korkut A, Liu Y, Schultz A, Cardenas Met al. Integrated analysis of TP53 gene and pathway alterations in The Cancer Genome Atlas. Cell Rep 2019;28:3010. [DOI] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 2017;32:185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega Fet al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell 2018;33:721–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nassar AH, Adib E, Kwiatkowski DJ. Distribution of KRAS (G12C) somatic mutations across race, sex, and cancer type. N Engl J Med 2021;384:185–7. [DOI] [PubMed] [Google Scholar]
- 33. Rosenthal R, McGranahan N, Herrero J, Taylor BS, Swanton C. DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol 2016;17:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AVet al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Yet al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja Ret al. Comprehensive analysis of hypermutation in human cancer. Cell 2017;171:1042–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Way GP, Sanchez-Vega F, La K, Armenia J, Chatila WK, Luna Aet al. Machine learning detects pan-cancer Ras pathway activation in The Cancer Genome Atlas. Cell Rep 2018;23:172–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ademuyiwa FO, Tao Y, Luo J, Weilbaecher K, Ma CX. Differences in the mutational landscape of triple-negative breast cancer in African Americans and Caucasians. Breast Cancer Res Treat 2017;161:491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Powell IJ, Dyson G, Land S, Ruterbusch J, Bock CH, Lenk Set al. Genes associated with prostate cancer are differentially expressed in African American and European American men. Cancer Epidemiol Biomarkers Prev 2013;22:891–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schumacher SE, Shim BY, Corso G, Ryu MH, Kang YK, Roviello Fet al. Somatic copy number alterations in gastric adenocarcinomas among Asian and Western patients. PLoS One 2017;12:e0176045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJet al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet 2013;45:1470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lowery MA, Ptashkin R, Jordan E, Berger MF, Zehir A, Capanu Met al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res 2018;24:4154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JSet al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun 2014;5:5696. [DOI] [PubMed] [Google Scholar]
- 44. Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin Jet al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang Jet al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017;2017:PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data have been deposited and are hosted on our portal at https://kmportal.or.kr.