Significance
We identified normative sex differences in the functional topography of personalized association networks, including the ventral attention, default mode, and frontoparietal networks. Furthermore, chromosomal enrichment analyses revealed that sex differences in multivariate patterns of functional topography were spatially coupled to the expression of X-linked genes as well as astrocytic and excitatory neuronal cell–type signatures. These results highlight the role of sex as a biological variable in shaping functional topography.
Keywords: personalized functional networks, functional topography, sex differences, association networks
Abstract
Prior work has shown that there is substantial interindividual variation in the spatial distribution of functional networks across the cerebral cortex, or functional topography. However, it remains unknown whether there are sex differences in the topography of individualized networks in youth. Here, we leveraged an advanced machine learning method (sparsity-regularized non-negative matrix factorization) to define individualized functional networks in 693 youth (ages 8 to 23 y) who underwent functional MRI as part of the Philadelphia Neurodevelopmental Cohort. Multivariate pattern analysis using support vector machines classified participant sex based on functional topography with 82.9% accuracy (P < 0.0001). Brain regions most effective in classifying participant sex belonged to association networks, including the ventral attention, default mode, and frontoparietal networks. Mass univariate analyses using generalized additive models with penalized splines provided convergent results. Furthermore, transcriptomic data from the Allen Human Brain Atlas revealed that sex differences in multivariate patterns of functional topography were spatially correlated with the expression of genes on the X chromosome. These results highlight the role of sex as a biological variable in shaping functional topography.
Significant sex differences have been documented in cognitive domains, including visuospatial processing, social cognition, emotional memory, and executive function (1–4). Prior studies have sought to understand these behavioral differences in the context of sex differences in brain structure and function that emerge in childhood and adolescence (2, 3, 5, 6). Although such studies focus on sex differences—biological differences between males and females due to genetic, hormonal, reproductive, or physical differences—it is important to note that an individual’s experiences based upon societal and cultural concepts of their gender can also shape brain development and lead to gender differences in behavior (7, 8). In this report we focus primarily on sex differences, though we also acknowledge that the effects of sex and gender are difficult to isolate, given the correlational nature of studies in human participants. Understanding normative sex differences in brain structure and function during development not only allows us to learn more about the neurobiology of sex differences in cognition but is also a necessary first step in constructing a framework to study sex differences in psychopathology.
Previous neuroimaging studies have examined sex differences in network connectivity as a contributor to sex differences in cognition and psychiatric disorders (9–11). For example, males on average tend to exhibit greater between-module connectivity and lower within-module connectivity than females (12, 13). These patterns of connectivity have been linked to better performance on spatial and motor tasks, cognitive domains where males outperform females (12). In contrast, females outperform males on semantic decision and verbal recall tasks (14, 15). These findings have been attributed to sex differences in connectivity of functional language processing regions (14, 15). Similarly, females also perform better than males on emotional identification and emotional differentiation tasks (3, 4). These tasks probe social cognition, a domain linked to social and reward networks with significant sex differences (2). Furthermore, depression and anxiety are more prevalent in females (16), and brain connectivity patterns associated with mood disorder symptoms are greater in females (17). In contrast, attention deficit hyperactivity disorder (ADHD) (18) and conduct disorder (19) are more prevalent in male youth and may be related to abnormal functional connectivity in executive systems (20). Although these studies and others suggest that sex differences in network connectivity may underlie diverse behavioral phenotypes, findings have been heterogeneous, raising concerns about reproducibility and potential for clinical translation.
One potential reason for such heterogeneity in findings among prior studies is the use of standardized network atlases. Standardized network atlases assume a stable 1:1 correspondence between structural and functional anatomy across individuals. Such methods assume that by aligning brain structural anatomy across subjects, functional network anatomy across subjects is also brought into alignment. However, evidence from multiple independent groups has shown that there is significant interindividual variation in the spatial distribution of functional networks across the anatomic cortex, or functional topography (21–25). These studies demonstrate that mapping between structure and function varies substantially between individuals in adults (21–24). Building on these studies in adults, our group provided the initial description of the development of the spatial topography of personalized functional networks in youth (25). Interindividual variation in the topography of personalized functional networks is maximal in association networks such as the ventral attention, frontoparietal, and default mode networks (25). Failing to account for functional topography can alias individual differences in topography into measurement of interregional functional connectivity (21). In light of these difficulties, it remains unknown whether sex differences in functional topography exist. Furthermore, it is unknown whether such differences might emerge in youth—a period marked by extensive remodeling of functional networks (25).
Accordingly, here we capitalized upon a large sample of youths imaged as part of the Philadelphia Neurodevelopmental Cohort (26) to evaluate sex differences in functional topography. We used machine learning to define individualized functional networks, hypothesizing that sex differences would be greatest in association networks. Because sex differences in neuroanatomy have previously been linked to sex chromosome gene expression (27, 28), we also evaluated the relationship between sex differences in topography and gene expression. We predicted that cortex with prominent sex differences in functional network topography would also be enriched in the expression of genes on sex chromosomes.
Results
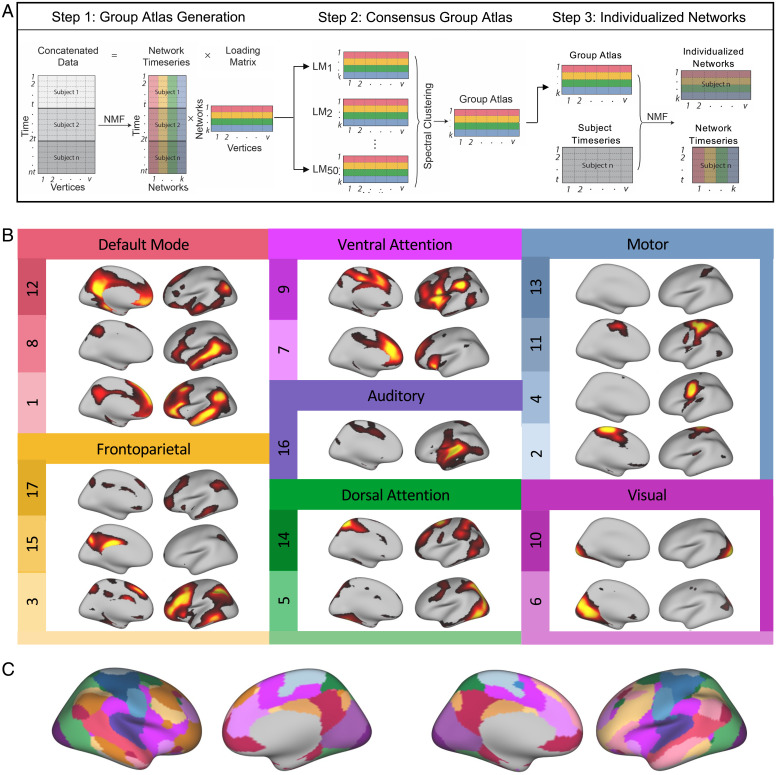
As previously described (23), we used sparsity-regularized non-negative matrix factorization (NMF) (29) to derive individualized functional networks in 693 youth (57% female) ages 8 to 23 y imaged with fMRI as part of the Philadelphia Neurodevelopmental Cohort. In this procedure for defining individualized networks, we first created a consensus atlas for the full sample and then used this consensus atlas to define individualized networks for each participant (Fig. 1A and SI Appendix, Table S1). Use of such a group-consensus atlas ensures spatial correspondence across personalized networks for each individual participant. Seventeen functional networks were identified for each participant (Fig. 1B), which correspond to other commonly used atlases and prior work (22, 25, 30, 31). Networks were named as in Cui et al. (25), and include default mode networks 1, 8, and 12, frontoparietal networks 3, 15, and 17, ventral attention networks 7 and 9, dorsal attention networks 5 and 14, visual networks 6 and 10, somatomotor networks 2, 4, 11, and 13, and auditory network 16. In contrast to hard partitioning methods that assign each vertex to a single network, NMF is a soft partitioning method. NMF yields a probabilistic parcellation such that each location on the cortex (i.e., vertex) receives a loading from each of the networks; these loadings quantify the extent to which a given location belongs to a network. This probabilistic parcellation can be converted into discrete network definitions for display by labeling each vertex according to its highest loading (Fig. 1C).
Fig. 1.
Defining personalized functional networks with non-negative matrix factorization. (A) We used sparsity regularized NMF to derive individualized functional networks. Three fMRI runs were concatenated for each subject, resulting in a 27.4-min time series with 555 time points for each subject. In step 1, time series from 100 randomly selected subjects were concatenated into a matrix with 55,500 time points (rows) and 17,734 vertices (columns). NMF was used to decompose these data into a time series matrix and loading matrix. The loading matrix had 17 rows and 17,734 columns, which encoded the membership of each vertex for each network. This procedure was repeated 50 times, with each run including a different subset of 100 subjects. In step 2, a normalized-cut based spectral clustering method was applied to cluster the 50 loading matrices into one consensus loading matrix, which served as the group atlas and ensured correspondence across individuals. In step 3, NMF was used to calculate individualized networks for each participant, with the group atlas used as a prior. (B) Seventeen functional networks were identified for each participant. Networks identified included default mode networks 1, 8, and 12, frontoparietal networks 3, 15, and 17, ventral attention networks 7 and 9, dorsal attention networks 5 and 14, visual networks 6 and 10, somatomotor networks 2, 4, 11, and 13, and auditory network 16. NMF yields a probabilistic (soft) parcellation such that there are 17 loadings for each vertex that quantify the extent to which it belongs to each network. For each loading map, brighter colors indicate greater loadings. (C) The probabilistic parcellation can be converted into discrete (hard) network definitions for display by labeling each vertex according to its highest loading.
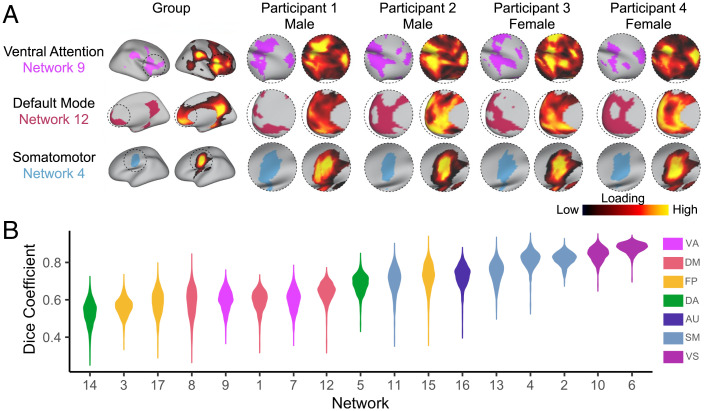
Visual examination of individual participants’ functional networks revealed distinct differences in topographic features (Fig. 2A). This interindividual variation in topography was particularly apparent in association networks such as the ventral attention and default mode networks. In contrast, motor and sensory networks appeared to be much more consistent across individuals. To quantitatively evaluate this variability, we calculated the Dice coefficient between the group atlas and each participant for all 17 networks. When ranking networks by median Dice, we found this measure of similarity was lowest in association networks and greatest in sensorimotor networks, indicating greater interindividual variation in the topography of association networks (Fig. 2B).
Fig. 2.
Functional network topography varies between individuals and by sex. (A) Probabilistic loading map and discrete network parcellations of three networks are displayed for the group and four randomly selected participants. Visual examination of individual participants’ functional networks reveal distinct differences in topographic features. This interindividual variation in topography is particularly apparent in association networks such as the ventral attention and default mode networks. In contrast, motor and sensory networks appear to be more consistent across individuals. (B) To evaluate variability in functional topography across networks, we calculated the Dice coefficient between the group atlas and each subject for all 17 networks. We found this measure of similarity was lowest in association networks and greatest in sensorimotor networks, indicating greater interindividual variation in the topography of association networks. VA, ventral attention; DM, default mode; FP, frontoparietal; DA, dorsal attention; AU, auditory; SM, somatomotor; VS, visual.
Machine Learning Accurately Identifies Sex Using Functional Topography.
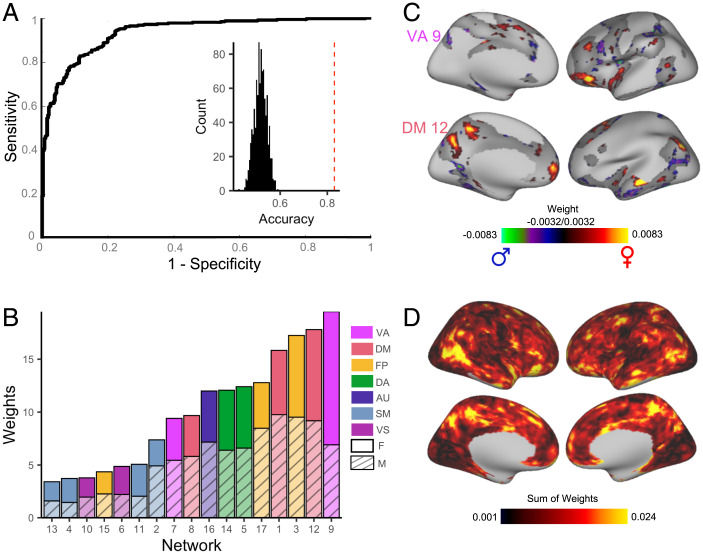
Based on our observation that the spatial distribution of association networks varies across individuals, we hypothesized that sex contributed to this interindividual variation in topography. To test this hypothesis, we first sought to understand the way in which high dimensional patterns of functional topography reflect sex. Multivariate pattern analysis allows for such integration of high dimensional data and can also identify complex patterns of topography that discriminate between males and females. We therefore used a linear support vector machine (SVM) (32) with nested two-fold cross-validation (2F-CV) to construct multivariate models that classified participants as male or female (SI Appendix, Fig. S1). Given our large sample size, using 2F-CV minimizes overfitting while leaving a sufficiently large sample to test model performance. Although there was no significant difference in age between males and females in our sample, we accounted for age and in-scanner head motion in these models by regressing these covariates from each feature in the training datasets (33) and then applied these model parameters directly to the test dataset to avoid leakage. Multivariate models were able to classify unseen participants as male or female with 82.9% accuracy (P < 0.0001; Fig. 3A). Sensitivity and specificity of the model were 0.76 and 0.88, respectively; area under the receiver operating characteristic (ROC) curve (AUC) was 0.94. To understand which networks contributed the most to the prediction, we summed the positive and negative weights separately across all vertices in each network. This revealed that variation in the functional topography of association networks, including the ventral attention, default mode, and frontoparietal network, contributed the most to the model and were therefore relatively more important in predicting participant sex (Fig. 3 B and C). To determine the importance of a given vertex to the predictive model, we summed the absolute weight across all 17 networks to summarize the prediction weight of each vertex. This summary measure highlighted that regions in association cortex, including the temporoparietal junction, superior parietal lobule, and orbitofrontal cortex, were most important in predicting participant sex (Fig. 3D). A conservative spin-based spatial randomization test (34–37) confirmed that feature weights computed from fold 1 were consistent with feature weights computed from fold 2 (pspin < 0.0001 for each of the 100 iterations). Although the summed weights depicted in Fig. 3C identify which networks contribute most to the prediction, this measure does not directly assess the extent to which each network independently contributes to prediction accuracy. Therefore, we ran 17 network-specific models using nested 2F-CV. These network-specific models revealed that topography of association networks most accurately classified participant sex (SI Appendix, Fig. S2).
Fig. 3.
Multivariate pattern analysis using support vector machines predicts subject sex based on functional topography. (A) SVMs with 2F-CV were used to construct multivariate models that classified participants as male or female. The ROC curve of the resulting model is depicted. Area under the ROC curve was 0.94; average sensitivity and specificity of the model were 0.76 and 0.88, respectively. Models classified participants as male or female with 82.9% accuracy. Inset histogram shows distribution of permuted accuracies. Average accuracy from real (nonpermuted) data are represented by the dashed red line. (B) To understand which networks contributed the most to the prediction, positive and negative model feature weights were summed separately across all vertices in each network. The most important topographic features in this model were found in association cortex and were maximal in the ventral attention network and default mode network. (C) The top 25% of vertices in terms of feature importance in the SVM model are displayed for the ventral attention network and default mode network. (D) At each location on the cortex, the absolute contribution weight of each network was summed, revealing that association cortex contributed the most to the multivariate model predicting participant sex.
We next conducted a series of sensitivity analyses. Classification performance remained robust when the sample was split into three age tertiles (SI Appendix and SI Appendix, Fig. S3), when the sample was restricted to postpubertal youths (SI Appendix, Fig. S4), when the analysis was conducted in a sex-balanced subsample (SI Appendix, Fig. S5), when the group atlas was built using a sex-balanced group of subjects that was not involved in model training and testing (SI Appendix, Fig. S6), and when implementing dimensionality reduction prior to classification (SI Appendix, Fig. S7).
Mass Univariate Analyses Yield Convergent Results.
The goal of our multivariate pattern analysis was to classify participant sex using the information contained in all regions jointly. Although multivariate models are optimal for classification problems, their descriptive utility is sometimes limited. The interpretability of features within a multivariate model may be hindered by the inability to determine how the features interact within the model framework due to the high-dimensional nature of the parameter space. In contrast, a traditional mass univariate analysis describes the relationship between a given factor and brain measures of interest on a regional basis, thereby providing descriptive information complementary to multivariate results. Accordingly, we also examined the impact of sex on network topography using traditional mass univariate analyses. We used generalized additive models (GAMs) with penalized splines (38) to account for linear and nonlinear developmental effects. We fit a GAM at each vertex to evaluate the impact of sex on network loadings. Age and in-scanner head motion were included as covariates, and age was modeled using a penalized spline. Multiple comparisons within each network were accounted for by controlling the false discovery rate (FDR; Q < 0.05).
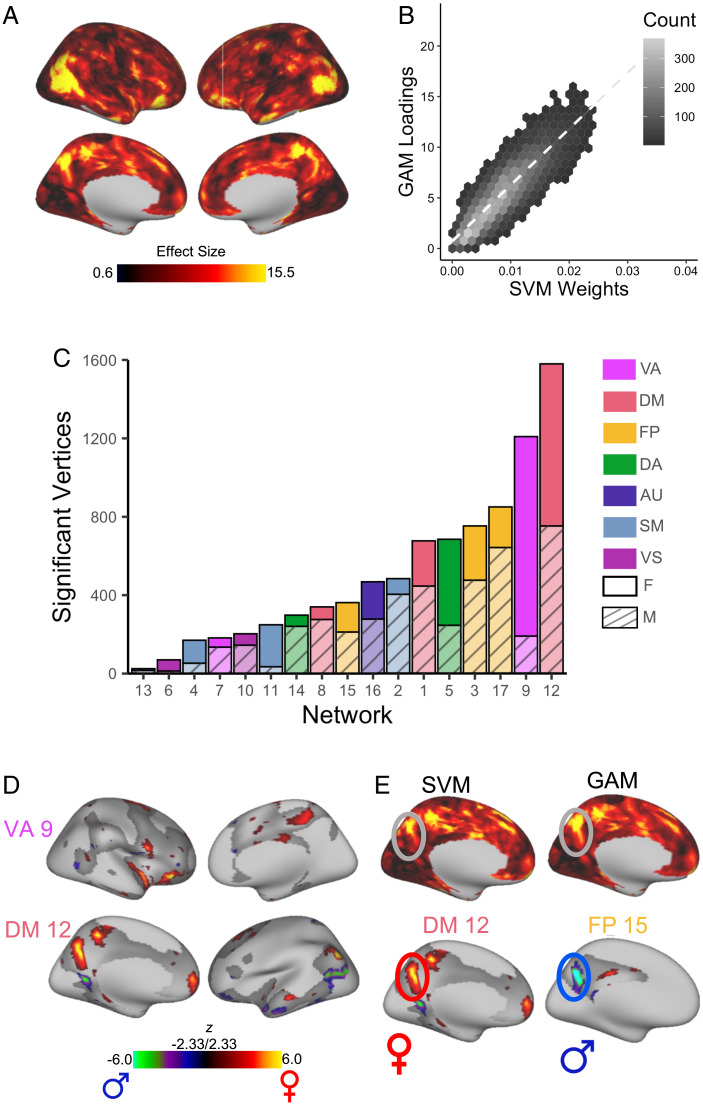
To determine the overall effect of sex at a given vertex, we summed the absolute value of the Z statistic for the effect of sex across all 17 networks. This summary measure highlighted that the impact of sex on topography was greatest in association cortex regions, including the temporoparietal junction, superior parietal lobule, and orbitofrontal cortex (Fig. 4A). Notably, this result from mass univariate models was convergent with our multivariate analysis, which identified the same regions of association cortex as most heavily weighted in classifying participant sex. We evaluated the significance of the correspondence between this mass univariate summary measure and the map of summed absolute prediction weights from our machine learning model (Fig. 3D) using a conservative spin-based spatial randomization test that accounts for spatial autocorrelation (34–37). This analysis revealed a high level of convergence between approaches (r = 0.86, pspin < 0.0001; Fig. 4B). As in our multivariate analysis, the impact of sex estimated using a univariate approach was greatest in association networks, including the ventral attention, default mode, and frontoparietal networks (Fig. 4 C and D and SI Appendix, Figs. S8–S10). Males had more vertices with greater loadings than females in the default mode network and frontoparietal network, though the difference in the default mode network was small. In contrast, females had more vertices with greater loadings than males in the ventral attention network and dorsal attention network. Additionally, both the SVM and GAMs identified the precuneus as a region with large sex differences in topography; loadings in the precuneus were greater in females for the default mode network, but greater in males for the frontoparietal network (Fig. 4E). Analyses evaluating the presence of an age-by-sex interaction revealed no significant effects.
Fig. 4.
Mass univariate analyses provide convergent results, identifying significant sex differences in association networks. A GAM was fit at each vertex to evaluate the impact of sex on network loadings. Age (modeled using a penalized spline) and motion were included as covariates. Multiple comparisons within each network were accounted for by controlling the false discovery rate (Q < 0.05). (A) To determine the overall effect of sex at a given vertex, we summed the absolute sex effect across all 17 networks. This summary measure is depicted and highlights that the impact of sex on topography was greatest in association cortex regions, including the temporoparietal junction, superior parietal lobule, and orbitofrontal cortex. (B) Hexplot shows agreement between univariate summary measure (GAM loadings in A) and multivariate summary measure (SVM weights in Fig. 3D; r = 0.86, pspin < 0.0001). (C) To identify networks with the greatest sex differences, the number of vertices in each network with a significant sex effect was summed separately for males and females. This analysis revealed that sex differences were greatest in association networks. (D) Significant vertices are displayed for the ventral attention network and default mode network, the networks where sex differences were maximal. (E) Both SVM and GAMs identified the precuneus as a region with large sex differences in topography; loadings in this region were greater for females in the default mode network, but greater in the frontoparietal network for males. F, female; M, male.
Gene Enrichment Analyses Link Sex Differences in Topography to X-Chromosome Genes.
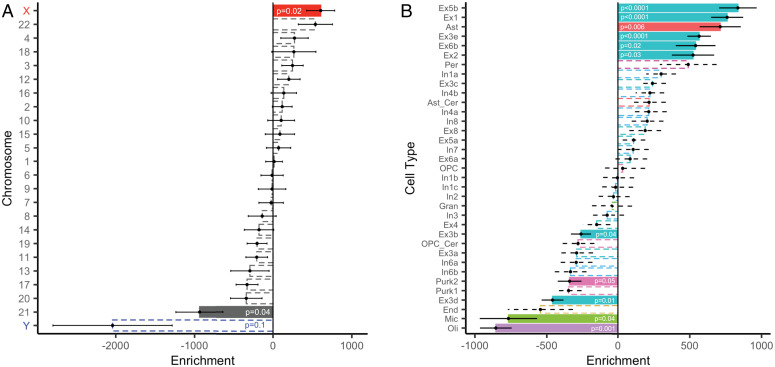
The above findings indicated that there were robust differences in functional topography between males and females. We next sought to understand the biological basis of these sex differences in topography. Although little is currently known about what factors drive interindividual variation in topography, sex differences in neuroanatomy have previously been attributed to differences in sex chromosome gene expression (27, 28). Accordingly, we conducted a chromosomal enrichment analysis to determine whether sex differences in topography were spatially coupled to gene expression. We compared the map of summed absolute prediction weights from our machine learning model to gene expression data from the Allen Human Brain Atlas (across n = 12,986 genes using a 1,000-parcel atlas; Materials and Methods). We first correlated each gene’s spatial pattern of expression with the feature weights from the multivariate (SVM) model. Next, we quantified the degree of spatial correspondence between a chromosomal gene set and the feature weights from the multivariate model as the median rank of the set of correlations between the SVM loading map and expression of genes on that chromosome. As predicted, we observed a significant enrichment of X-chromosome genes (P = 0.02; Fig. 5A)— locations more important in predicting participant sex showed higher correlation with the expression of genes on the X chromosome. This X-chromosome enrichment remained significant in a series of sensitivity analyses that varied the parcellation resolution (P = 0.001); that both varied parcellation resolution and used an independent processing pipeline with alternate methods for annotation, filtering, and sample assignment (39–41) (P = 0.02); that limited the transcriptomic data to male donors only (P = 0.02); and that limited the sample to a sex-balanced subset (P = 0.03); see Materials and Methods and SI Appendix, Materials and Methods for details.
Fig. 5.
Regions exhibiting sex differences in multivariate patterns of functional topography are enriched in expression of X-linked, excitatory neuronal, and astrocytic-related genes. To understand the biological basis of sex differences in topography, we compared the map of summed absolute prediction weights from our machine learning model to gene expression data from the Allen Human Brain Atlas parcellated to the Schaefer1000 atlas. (A) Cortex where sex differences in functional topography were more prominent were significantly correlated with the spatial pattern of expression of genes on the X chromosome. (B) We conducted cell-type–specific enrichment analyses to understand the convergent and divergent patterns of discrete underlying gene sets. We assigned cell types using the neuronal subclass assignments determined by Lake et al. (43). Regions more important in classifying participant sex were enriched in astrocyte-related genes and several excitatory neuron-related gene sets, including Ex5b, Ex1, Ex3e, Ex6b, and Ex2. Point range plots show the median (point) and SE (range) rank of each chromosomal or cell-type gene set. Dashed lines indicate nonsignificant enrichments. All permuted P values were not further corrected for multiple comparisons and determined based on one-sided tests of gene set enrichment. Ast, astrocyte; Ast_cer, cerebellar-specific astrocytes; End, endothelial cells; Ex, excitatory neuron; Gran, cerebellar granule cells; In, inhibitory neuron; Mic, microglia; Oli, oligodendrocytes; OPC, oligodendrocyte progenitor cells; OPC_cer, cerebellar-specific oligodendrocyte progenitor cells; Per, pericytes; and Purk, cerebellar Purkinje cells.
The above results indicate that sex differences in multivariate patterns of functional topography are correlated with the expression of X-linked genes. However, regional differences in cortical gene expression may reflect regional differences in cellular composition of the cortex (42). Therefore, we conducted cell-type–specific enrichment analyses to understand the convergent and divergent patterns of discrete underlying gene sets. Using cell-type–specific gene sets as assigned in prior work (28), we found that regions more important in classifying participant sex were enriched in expression of astrocytic (P < 0.0001) and excitatory neuronal genes (P < 0.0001). To obtain a more nuanced understanding of cytoarchitecture, we then assigned cell types using the finer-grained neuronal subclass assignments determined by Lake et al. (43). Convergent with the coarser cell-type results, regions more important in classifying participant sex were enriched in astrocyte-related genes (P < 0.006; Fig. 5B) as well as several excitatory neuron subclasses, including Ex5b (P < 0.0001), Ex1 (P < 0.0001), Ex3e (P < 0.0001), Ex6b (P = 0.02), and Ex2 (P = 0.03). Notably, these gene sets included numerous X-linked genes (SI Appendix, Table S2). Finally, we conducted a rank-based gene ontology (GO) enrichment analysis using GOrilla (44, 45) to examine functional enrichment. This analysis identified several GO terms relevant to brain anatomy, including “neuron part,” “synapse,” and “glutamatergic synapse” (SI Appendix, Fig. S11).
Discussion
In this study, we leveraged machine learning and a large sample of youths to study sex differences in functional network topography. We first demonstrated that sex differences in topography are greatest in association networks, including the ventral attention and default mode networks. Using complex multivariate patterns of functional topography, we were able to predict an unseen participant’s sex with a high degree of accuracy. Chromosomal enrichment analyses revealed that sex differences in multivariate patterns of functional topography were spatially coupled to the expression of X-linked genes as well as astrocyte and excitatory neuron cell-type expression signatures. These results identify normative sex differences in the topography of personalized association networks and highlight the role of sex as a biological variable in shaping functional topography.
Sex Differences in Functional Topography Are Greatest in Association Networks.
Our principal finding is that there are significant sex differences in functional topography, and that these differences are greatest in association networks. This finding is in line with prior studies of functional topography that show that interindividual variation in topography is greatest in association networks (21–25). Importantly, our findings suggest that some significant portion of interindividual variation in topography is driven by sex. Sex differences in topography were greatest in the default mode network, frontoparietal network, ventral attention network, and dorsal attention network. Variation in the topography of these networks has been linked to emotional, social, and executive functions (22, 25), all of which are behaviors with documented sex differences (2, 46, 47).
Our findings are also generally convergent with studies examining sex differences in functional connectivity, where standardized network atlases may have aliased differences in topography into measurements of connectivity. The sex differences we found in association cortex topography are in line with several prior studies that have found sex differences in association cortex connectivity (11, 48–52). Specifically, prior studies have documented sex differences in default mode network connectivity (48, 49, 53) and have postulated that sex hormones like estrogen and progesterone might impact default mode connectivity (50, 51, 53, 54). Similarly, our findings of sex differences in the topography of frontoparietal, ventral attention, and dorsal attention networks align with prior studies that have reported sex differences in functional connectivity of these networks (48, 52, 55, 56). Our multivariate pattern analysis showed that features from the default mode, frontoparietal, and ventral attention networks were most important in classifying participant sex.
Although no prior studies have classified participant sex based on functional topography, our results generally cohere with findings from a large study using data from the Human Connectome Project that found that functional connectivity features within the default mode network and frontoparietal network were most important in identifying participant sex (48). Further, abnormal patterns of connectivity involving the default mode, frontoparietal, ventral attention, and dorsal attention networks have also been associated with mood, fear, and externalizing symptoms (17), which are all dimensions of psychopathology with well-documented sex differences. Specifically, connectivity abnormalities of default mode, frontoparietal, and ventral attention networks associated with fear symptoms are greater in females (17). Together, these findings suggest that sex differences in topography may contribute to sex differences in cognition and psychopathology, though further work is needed to establish such a relationship.
Sex Differences in Topography Are Associated with X-Linked Gene Expression.
The mechanisms by which sex differences in topography arise are likely multifold. Despite the growing interest in interindividual differences in functional topography, little is currently known about what genetic or environmental factors drive these differences. In prior work, we demonstrated that interindividual variability in topography aligns with fundamental properties of brain organization, including myelin content and cerebral blood flow (25). Here, we built on these findings by linking sex differences in topography to gene expression data. As expected, we found that regions more important in predicting participant sex correlated with the expression of genes on the X chromosome. The correspondence between sex differences in topography and the spatial expression pattern of X-linked genes suggests that the observed sex differences in topography are likely in part driven by gene expression. Although to our knowledge no prior studies have examined the genetic basis of sex differences in functional topography, this finding is globally consistent with prior work that has linked sex differences in brain structure to sex chromosome gene expression (27, 28).
Highly ranked X-linked genes included those related to neuron development [PPEF1 (57) and PCDH19 (58)], neuronal cytoskeletal transport [DYNLT3 (59)], chloride ion channels (CLCN5 and GABRA3), metabolism [GYG2 and PDK3 (60)], and disease states with neuropsychiatric and cognitive symptoms [DMD (61), DYNLT3 (59), and PCDH19 (58)]. Of note, one gene ranking in the top 10 X-linked genes was PCDH19, a gene that encodes a protocadherin protein that supports neuronal organization and migration (58). PCDH19 was also identified as a gene whose spatial expression pattern correlates with sex differences in gray matter volume (27). Mutations in PCDH19 have been associated with intellectual disability, behavioral problems, and autism spectrum disorder (58). Although speculative, it is plausible that these X-linked genes could influence functional topography through their actions on brain development.
Another mechanism by which sex differences in topography may arise is through organizational effects of hormones on cytoarchitecture. Our finding that sex differences in topography were spatially coupled to excitatory neuron cell–type gene expression was robust using two separate cell-type categorizations. This result coheres with the extensive rodent and nonhuman primate literature examining the impact of estradiol on glutamatergic dendritic spine architecture (62) and sex differences in excitatory neurotransmission (63). For example, estrogen is essential to the maintenance of prefrontal cortex dendritic spine density in ovariectomized female rodents and nonhuman primates (62, 64–66). Estrogen also increases the number of spine synapses in the prefrontal cortex of gonadectomized male rats (67). Similarly, compared to male rodents, female rodents show larger α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor synaptic responses (68), greater sensitivity to N-methyl-D-aspartate (NMDA) receptor manipulations (63, 69, 70), and increased expression of NMDA and metabotropic glutamate receptors (63, 71).
We also found that sex differences in topography correlated with astrocytic subtype gene expression. Sex differences in astrocyte structure and astrocytic glutamate release are critical determinants of sex-specific synaptic patterning, which has been implicated in the male-biased risk for autism spectrum disorder (72). Similarly, rodent studies have suggested that estradiol’s differential impact on astrocyte proliferation and apoptosis in males and females may contribute to sex differences in brain organization (73). Additionally, androgen receptors play a role in establishing sex differences in astrocyte number and complexity seen in rodents (74). In the context of this literature, we speculate that hormone effects on cytoarchitecture may contribute to sex differences in functional topography.
Limitations.
Certain limitations of the present study should be noted. First, we concatenated three fMRI runs, two of which were task time series where task effects were regressed from the data. Residuals from task-regressed time series, while similar, are not identical to true resting state data and nonlinear effects associated with performing the task may therefore not have been removed (75). However, several independent studies have shown that functional networks are primarily defined by individual-specific rather than task-specific factors (76) and that networks present during task performance and at rest are similar (75). Including task-regressed data enabled us to generate individualized networks using 27 min of high-quality data. Time series of this length are necessary to reliably detect individual differences in functional networks (77) and sufficient to create parcellations highly similar to those generated using 380 min of data (78). Second, subcortical and cerebellar networks were not evaluated in this study, as individualized parcellation of these networks requires specialized analysis techniques that are distinct from those applied to the cortex (79, 80). Future work should evaluate sex differences in topography in subcortical and cerebellar networks, which are critical for behaviors with known sex differences, including emotional regulation and executive function.
Third, correspondence between sex differences in topography and gene expression were assessed at the group rather than the individual level, though evaluating this relationship on a within-subject basis in a large, developmental sample is precluded by the necessity of postmortem samples for gene expression profiling. Fourth, using the Allen Human Brain Atlas introduces several inherent limitations, including the use of microarray to quantify gene expression, asymmetric sampling, small sample size, donor age, and most notably, donor sex. The Allen Human Brain Atlas includes postmortem samples from five male donors and one female donor. However, our findings regarding the correspondence between sex differences in topography and X-linked gene expression were robust to sensitivity analyses leaving out the female donor, and prior studies have similarly leveraged the Allen to examine sex differences in independent neuroimaging samples (27, 28). Nevertheless, replication of these findings in a sex-balanced sample will be important when such spatially comprehensive maps of cortical gene expression are available. Fifth, sex was assessed using a binary self-report question, and we therefore did not have a sufficient sample to examine functional topography of intersex youths. Furthermore, it should be noted that existing data and theory suggest that binary sex classification may not be useful and that brains may be understood as complex mosaics of male and female characteristics (81).
Conclusions
In summary, we identified normative sex differences in the functional topography of personalized association networks in youth. These results suggest that interindividual variation in functional topography is in part driven by sex. Further, our findings suggest that sex differences in topography are likely in part linked to gene expression. Future work should examine whether the sex differences in the topography of personalized networks explain normative variation in socioemotional or cognitive functions. The relationship between topography and sex differences in psychopathology is also a clear area for future research, as this may identify sex-specific biomarkers of risk for psychiatric disorders.
Materials and Methods
Participants.
In this report, we considered the entire cross-sectional sample of 1,601 subjects imaged as part of the Philadelphia Neurodevelopmental Cohort (26). From this sample, 340 subjects were excluded due to clinical factors, including medical disorders that could affect brain function, current use of psychoactive medications, prior inpatient psychiatric treatment, or an incidentally encountered structural brain abnormality. An additional 568 subjects were excluded because of low-quality or missing structural, resting-state, n-back, or emotion identification imaging data; a functional run was excluded if mean relative root mean square (RMS) framewise displacement was higher than 0.2 mm, or it had more than 20 frames with motion exceeding 0.25 mm (82, 83). The final sample included in the analyses comprised 693 participants of which 301 were male and 392 were female. This sample of participants and their individualized networks are the same as those included in our prior report on individual variation in functional network topography (25). All subjects or their parent/guardian provided informed consent, and minors provided assent. All study procedures were approved by the institutional review boards of both the University of Pennsylvania and the Children’s Hospital of Philadelphia.
Image Acquisition, Preprocessing, and Individualized Network Definition.
Neuroimaging acquisition and preprocessing were as previously described (25); see SI Appendix, Materials and Methods for further details.
Across-Subject Variability of Functional Network Topography.
Prior studies of adults have consistently reported that across-subject variability of functional networks is high in association networks and lower in primary sensorimotor networks (22, 23, 71, 84, 85). In a prior report using the same sample (25), we calculated across-subject variance in network loadings using the median absolute deviation and demonstrated that interindividual variation in topography was greatest in association networks and lowest in sensorimotor networks. To further evaluate variability in functional topography across networks, here we calculated the Dice coefficient between the group atlas and each subject for all 17 networks. Networks were then ranked by the median Dice value.
Multivariate Pattern Analysis.
We used a linear support vector machine (LSVM) as implemented in LIBSVM (86) to construct multivariate models that classified participants as male or female. A free parameter C determines the balance between the training errors and the generalizability of the LSVM classification model. We evaluated the classification using a nested two-fold cross-validation (2F-CV), with the inner 2F-CV determining the optimal parameter C for the SVM classifier and the outer 2F-CV estimating the generalizability of the model (SI Appendix, Fig. S1). Given the large sample size in this study, using two folds minimizes variance and overfitting while leaving a sufficiently large sample to test model performance.
In the outer 2F-CV, the data were randomly divided into two subsets. We initially used subset 1 as the training set, with subset 2 used as the testing set. We accounted for age and in-scanner head motion by regressing these effects from each feature in the training dataset using SurfStat (33) and then applied the acquired coefficients to regress the effects in the testing dataset. Each feature was linearly scaled between zero and one across the training dataset; these scaling parameters were then applied to scale the testing dataset (87, 88).
Within the training dataset of each outer 2F-CV loop, we employed inner 2F-CV loops to determine the optimal C. To do this, the training set of the outer 2F-CV loop was again randomly divided into two subsets; one subset was selected to train the model with a given C in the range [2−5, 2−4, …, 29, 210] (i.e., 16 values in total) (89), and the remaining subset was used to test the model. This procedure was repeated two times such that each subset was used once as the testing subset, resulting in two inner cross-validations in total. The accuracies were calculated for each C value and then averaged across the two inner cross-validations. The C with the highest inner prediction accuracy was chosen as the optimal C (87, 90). Then, we trained a model using the optimal C and all participants in the training set (subset 1) and then used that model to predict the sex of all participants in the testing set (subset 2).
We repeated the above procedure using subset 2 as the training set and subset 1 as the testing set. Because the split between training and testing sets was chosen randomly, the nested twofold cross-validation was performed 100 times to reduce the impact of group assignment. The results of these 100 nested cross-validations were then averaged. This procedure yielded an overall classification accuracy score for classifying unseen males or females on the basis of their functional topography. This procedure also yielded a vector of feature weights for each functional network that described how heavily weighted a given topographic feature was within the multivariate model.
Significance of Prediction Performance.
Permutation testing was used to evaluate whether the prediction performance was significantly better than expected by chance (91). The predictive framework was repeated 1,000 times. In each run, we permuted sex across the training subset without replacement. Significance was determined by ranking the actual prediction accuracy (the average across 100 runs) versus the permuted distribution; the p-value was the proportion of permutations that showed a higher value than the actual accuracy value for the real data.
Interpreting Model Feature Weights.
The nested 2F-CV was repeated 100 times. This yielded a total of 200 weight maps across the 100 iterations. Averaging these 200 weight maps, the features with a nonzero mean weight can be understood as contributing features in the prediction model (87, 91), with the absolute value of the weight quantifying the contribution of the features to the model (91). To understand which networks contributed the most to the prediction, we summed the positive and negative weights separately across all vertices in each network. As vertices had 17 loading values (one for each network), we summed the absolute weight across all 17 networks to summarize the prediction weight of each vertex. This sum represents the importance of a given vertex to the predictive model. To evaluate whether feature weights computed from fold 1 were consistent with feature weights computed from fold 2, we used a conservative spin-based spatial randomization test that accounts for spatial autocorrelation (34–37) [see Spatial Randomization Testing (Spin Test)]. This spin test compared the map of summed absolute weights from fold 1 with the map of summed absolute weights from fold 2 in each of the 100 iterations.
Univariate Associations of Network Topography with Sex.
The goal of a multivariate pattern analysis is to predict an outcome using the information contained in all regions jointly. In contrast, the goal of a traditional mass univariate analysis is to describe the relationship between a given factor and brain measures of interest on a regional basis. Although multivariate models are ideal for classification problems, their descriptive utility is limited. While it is possible to identify the most heavily weighted features within a model, it is not possible to directly visualize their action within the model framework due to the high-dimensional nature of the parameter space. As such, we used both multivariate (predictive) and mass univariate (descriptive) approaches, which are complementary and provide converging evidence.
We evaluated mass univariate associations between sex and network topography using GAMs with penalized splines (38) to account for linear and nonlinear developmental effects. GAMs estimate a smoothing function using restricted maximum likelihood (REML) and penalize nonlinearity in order to avoid overfitting the data. We fit a GAM at each vertex to evaluate the impact of sex on network loadings. This yielded one p-value per vertex where the p-value describes the significance of the sex effect; this p-value was transformed to a signed z-statistic that describes the effect of sex on topography across all subjects in the sample. In-scanner head motion was included as a linear covariate, and age was modeled using a penalized spline. To examine whether sex differences in topography evolve across development, we tested for an age-by-sex interaction on loadings at each vertex using GAMs with penalized splines; these models included motion as a covariate and also accounted for main effects of age and sex. As we considered three functional runs, in-scanner motion was summarized as the grand mean of the mean relative RMS displacement of each functional run. Multiple comparisons within each network were accounted for by controlling the FDR (Q < 0.05).
Spatial Randomization Testing (Spin Test).
To evaluate the significance of the correspondence between our multivariate and univariate results, we compared a map of summed absolute prediction weights from our machine learning model (Fig. 4D) to a map of summed effect sizes from the GAMs (Fig. 5A). We compared these maps using the spin test (34–37) (https://github.com/spin-test/spin-test). The spin test is a spatial randomization method based on angular permutations of spherical projections at the cortical surface. The spin test generates a null distribution of randomly rotated brain maps that preserve spatial features and the spatial covariance structure of the original map. This procedure is therefore far more conservative than randomly shuffling locations, which destroys the spatial covariance structure of the data and produces an unrealistically weak null distribution. The spin-based p-value was calculated as the proportion of times that the observed correlation was higher than the null distribution of correlation values from rotated parcellations.
Gene Enrichment Analysis.
To examine the transcriptomic correlates of sex differences in functional topography, we compared the map of summed absolute prediction weights from our machine learning model to gene expression data from the Allen Human Brain Atlas (92). Publicly available microarray gene expression data processed in line with recent benchmarking recommendations and parcellated to the Schaefer1000 atlas were downloaded from https://figshare.com/articles/dataset/AHBAdata/6852911. Details regarding gene processing, including gene information reannotation, data filtering, probe selection, sample assignment, data normalization, and gene filtering, are described in Arnatkeviciute et al. (93). Of the available parcellations of processed Allen data, the Schaefer1000 parcellation was selected for primary comparison with topography, given the granularity of topographic features. As only two of the six donor brains were sampled from both hemispheres, analyses were restricted to the left hemisphere to minimize variability of samples available across regions (93).
Chromosomal Enrichment Analysis.
We used ranked gene lists to test whether the spatial expression pattern of a given gene set was nonrandomly related to the spatial pattern of sex differences in functional topography. As in prior studies (27, 28, 41), we quantified the degree of spatial correspondence using the median gene set rank. Genes were assigned to chromosomes as annotated in Richiardi et al. (94). We calculated the median ranks for 24 nonoverlapping gene sets: each autosome (chromosomes 1 through 22), chromosome X, and chromosome Y. For each chromosomal gene set, we compared the observed median rank to a null distribution of median ranks calculated from 1,000 same-sized scrambled lists generated by randomly reordering the original ranked list. The p-value from this nonparametric permutation test was the proportion of permutations with a more extreme value than the median rank of the real data. Permuted-based p-values were not further corrected for multiple comparisons.
To test the robustness of our chromosomal enrichments, we conducted a series of sensitivity analyses, including varying the processing strategy and parcellation resolution, as well as leaving the female donor out. Specifically, we replicated our results using publicly available gene expression data processed by Arnatkeviciute et al. (93) and parcellated to the Schaefer300 atlas. For consistency with prior work, we also parcellated Allen data to the Schaefer400 atlas using an independent processing pipeline (39–41) and computed gene expression matrices with and without the female donor.
Cell-Type–Specific Expression Analyses and Gene Ontology.
Because regional differences in cortical gene expression may reflect regional differences in cellular composition of the cortex (37), we conducted cell-type–specific enrichment analyses to understand the convergent and divergent patterns of discrete underlying gene sets. As in chromosomal enrichments, we used ranked gene lists to test whether the spatial expression pattern of a cell-type–specific gene set was nonrandomly related to the spatial pattern of sex differences in functional topography. Nonparametric permutation testing assessed the significance of median ranks of cell-type–specific gene sets. Gene sets for each cell type were first assigned according to categorizations determined by Seidlitz et al. (28). To obtain a more nuanced understanding of cytoarchitecture, we then used the finer-grained neuronal subclass assignments determined by Lake et al. (43). In both cases, only brain-expressed genes (28) were considered, defined by expression levels in the Human Protein Atlas (95, 96).
Finally, we also conducted a rank-based GO enrichment analysis using GOrilla (44, 45) to examine functional enrichment.
Visualization.
Connectome Workbench (version: 1.3.2) provided by the human connectome project (https://www.humanconnectome.org/software/connectome-workbench) (97) was used to visualize the brain surface.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institute of Health: R01MH120482, R37MH125829, R01MH113550, R01EB022573, R01MH107703, RF1MH116920, R01MH112847, P50MH096891, R01MH11186, K01MH102609, R01MH107235, R01MH112070, R01MH123550, K99MH127293, R01NS085211, RC2MH08998, RC2MH089924, R25MH119043, K08MH120564, T32MH014654, T32MH019112, and T32NS091008. This study was also supported by the Penn/CHOP Lifespan Brain Institute.
Footnotes
This work was presented at the 2021 and 2022 Annual Meeting of the Society for Biological Psychiatry.
The authors declare no competing interest.
This article is a PNAS Direct Submission. E.J. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2110416119/-/DCSupplemental.
Data, Materials, and Software Availability
The Philadelphia Neurodevelopmental Cohort (PNC) (26) data are publicly available in the Database of Genotypes and Phenotypes; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000607.v3.p2 (98). All analysis code is available at https://pennlinc.github.io/funcParcelSexDiff1/ (99).
References
- 1.Cahill L., His brain, her brain. Sci. Am. 292, 40–47 (2005). [PubMed] [Google Scholar]
- 2.Alarcón G., et al. , Adolescent gender differences in neural reactivity to a friend’s positive affect and real-world positive experiences in social contexts. Dev. Cogn. Neurosci. 43, 100779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gur R. C., Gur R. E., Complementarity of sex differences in brain and behavior: From laterality to multimodal neuroimaging. J. Neurosci. Res. 95, 189–199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gur R. C., et al. , Age group and sex differences in performance on a computerized neurocognitive battery in children age 8-21. Neuropsychology 26, 251–265 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills K. L., Lalonde F., Clasen L. S., Giedd J. N., Blakemore S. J., Developmental changes in the structure of the social brain in late childhood and adolescence. Soc. Cogn. Affect. Neurosci. 9, 123–131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagliaccio D., et al. , HPA axis genetic variation, pubertal status, and sex interact to predict amygdala and hippocampus responses to negative emotional faces in school-age children. Neuroimage 109, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J. S., Nafziger A. N., Is it sex or is it gender? Clin. Pharmacol. Ther. 68, 1–3 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Bale T. L., Epperson C. N., Sex as a biological variable: Who, what, when, why, and how.Neuropsychopharmacology 42, 386–396 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence K. E., et al. , Sex differences in functional connectivity of the salience, default mode, and central executive networks in youth with ASD. Cereb. Cortex 30, 5107–5120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ernst M., et al. ; IMAGEN Consortium, Pubertal maturation and sex effects on the default-mode network connectivity implicated in mood dysregulation. Transl. Psychiatry 9, 103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs E. G., et al. , Reorganization of functional networks in verbal working memory circuitry in early midlife: The impact of sex and menopausal status. Cereb. Cortex 27, 2857–2870 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satterthwaite T. D., et al. , Linked sex differences in cognition and functional connectivity in youth. Cereb. Cortex 25, 2383–2394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia C. H., et al. , Multi-scale network regression for brain-phenotype associations. Hum. Brain Mapp. 41, 2553–2566 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu M., et al. , Sex differences in functional brain networks for language. Cereb. Cortex 30, 1528–1537 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Wu S., et al. , Interaction of Catechol-O-methyltransferase Val158 Met polymorphism and sex influences association of parietal intrinsic functional connectivity and immediate verbal memory. Brain Behav. 10, e01784 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altemus M., Sarvaiya N., Neill Epperson C., Sex differences in anxiety and depression clinical perspectives. Front. Neuroendocrinol. 35, 320–330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia C. H., et al. , Linked dimensions of psychopathology and connectivity in functional brain networks. Nat. Commun. 9, 3003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramtekkar U. P., Reiersen A. M., Todorov A. A., Todd R. D., Sex and age differences in attention-deficit/hyperactivity disorder symptoms and diagnoses: Implications for DSM-V and ICD-11. J. Am. Acad. Child Adolesc. Psychiatry 49, 217–228.e211-213 (2010). [PMC free article] [PubMed] [Google Scholar]
- 19.Maughan B., Rowe R., Messer J., Goodman R., Meltzer H., Conduct disorder and oppositional defiant disorder in a national sample: Developmental epidemiology. J. Child Psychol. Psychiatry 45, 609–621 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Scofield J. E., Johnson J. D., Wood P. K., Geary D. C., Latent resting-state network dynamics in boys and girls with attention-deficit/hyperactivity disorder. PLoS One 14, e0218891 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bijsterbosch J. D., et al. , The relationship between spatial configuration and functional connectivity of brain regions. Elife 7, e32992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong R., et al. , Spatial topography of individual-specific cortical networks predicts human cognition, personality, and emotion. Cereb. Cortex 29, 2533–2551 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Satterthwaite T. D., Fan Y., Large-scale sparse functional networks from resting state fMRI. Neuroimage 156, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon E. M., et al. , Precision functional mapping of individual human brains. Neuron 95, 791–807.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui Z., et al. , Individual variation in functional topography of association networks in youth. Neuron 106, 340–353.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satterthwaite T. D., et al. , Neuroimaging of the Philadelphia neurodevelopmental cohort. Neuroimage 86, 544–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu S., Seidlitz J., Blumenthal J. D., Clasen L. S., Raznahan A., Integrative structural, functional, and transcriptomic analyses of sex-biased brain organization in humans. Proc. Natl. Acad. Sci. U.S.A. 117, 18788–18798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seidlitz J., et al. , Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nat. Commun. 11, 3358 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee D. D., Seung H. S., Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791 (1999). [DOI] [PubMed] [Google Scholar]
- 30.Wang D., et al. , Parcellating cortical functional networks in individuals. Nat. Neurosci. 18, 1853–1860 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeo B. T., et al. , The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 1125–1165 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burges C. J. C., A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 2, 121–167 (1998). [Google Scholar]
- 33.Worsley K. J., et al. , SurfStat: A Matlab toolbox for the statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. Neuroimage 47, S102 (2009). [Google Scholar]
- 34.Alexander-Bloch A. F., et al. , On testing for spatial correspondence between maps of human brain structure and function. Neuroimage 178, 540–551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon E. M., et al. , Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 26, 288–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sotiras A., et al. , Patterns of coordinated cortical remodeling during adolescence and their associations with functional specialization and evolutionary expansion. Proc. Natl. Acad. Sci. U.S.A. 114, 3527–3532 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vandekar S. N., et al. , Topologically dissociable patterns of development of the human cerebral cortex. J. Neurosci. 35, 599–609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wood S. N., Stable and efficient multiple smoothing parameter estimation for generalized additive models. J. Am. Stat. Assoc. 99, 673–686 (2004). [Google Scholar]
- 39.Romero-Garcia R., Warrier V., Bullmore E. T., Baron-Cohen S., Bethlehem R. A. I., Synaptic and transcriptionally downregulated genes are associated with cortical thickness differences in autism. Mol. Psychiatry 24, 1053–1064 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romero-Garcia R., et al. ; NSPN Consortium, Structural covariance networks are coupled to expression of genes enriched in supragranular layers of the human cortex. Neuroimage 171, 256–267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seidlitz J., et al. ; NSPN Consortium, Morphometric similarity networks detect microscale cortical organization and predict inter-individual cognitive variation. Neuron 97, 231–247.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu Y., et al. , Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lake B. B., et al. , Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z., GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eden E., Lipson D., Yogev S., Yakhini Z., Discovering motifs in ranked lists of DNA sequences. PLOS Comput. Biol. 3, e39 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andreano J. M., Cahill L., Sex influences on the neurobiology of learning and memory. Learn. Mem. 16, 248–266 (2009). [DOI] [PubMed] [Google Scholar]
- 47.Cahill L., Sex influences on brain and emotional memory: The burden of proof has shifted. Prog. Brain Res. 186, 29–40 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Zhang C., Dougherty C. C., Baum S. A., White T., Michael A. M., Functional connectivity predicts gender: Evidence for gender differences in resting brain connectivity. Hum. Brain Mapp. 39, 1765–1776 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conrin S. D., et al. , From default mode network to the basal configuration: Sex differences in the resting-state brain connectivity as a function of age and their clinical correlates. Front. Psychiatry 9, 365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weis S., Hodgetts S., Hausmann M., Sex differences and menstrual cycle effects in cognitive and sensory resting state networks. Brain Cogn. 131, 66–73 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Hidalgo-Lopez E., et al. , Human menstrual cycle variation in subcortical functional brain connectivity: A multimodal analysis approach. Brain Struct. Funct. 225, 591–605 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hjelmervik H., Hausmann M., Osnes B., Westerhausen R., Specht K., Resting states are resting traits--an FMRI study of sex differences and menstrual cycle effects in resting state cognitive control networks. PLoS One 9, e103492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Engman J., Linnman C., Van Dijk K. R., Milad M. R., Amygdala subnuclei resting-state functional connectivity sex and estrogen differences. Psychoneuroendocrinology 63, 34–42 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Petersen N., Kilpatrick L. A., Goharzad A., Cahill L., Oral contraceptive pill use and menstrual cycle phase are associated with altered resting state functional connectivity. Neuroimage 90, 24–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stumme J., Jockwitz C., Hoffstaedter F., Amunts K., Caspers S., Functional network reorganization in older adults: Graph-theoretical analyses of age, cognition and sex. Neuroimage 214, 116756 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Dumais K. M., Chernyak S., Nickerson L. D., Janes A. C., Sex differences in default mode and dorsal attention network engagement. PLoS One 13, e0199049 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Montini E., et al. , A novel human serine-threonine phosphatase related to the Drosophila retinal degeneration C (rdgC) gene is selectively expressed in sensory neurons of neural crest origin. Hum. Mol. Genet. 6, 1137–1145 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Peek S. L., Mah K. M., Weiner J. A., Regulation of neural circuit formation by protocadherins. Cell. Mol. Life Sci. 74, 4133–4157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chu Y., et al. , Alterations in axonal transport motor proteins in sporadic and experimental Parkinson’s disease. Brain 135, 2058–2073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez-Siles G., et al. , Energy metabolism and mitochondrial defects in X-linked Charcot-Marie-Tooth (CMTX6) iPSC-derived motor neurons with the p.R158H PDK3 mutation. Sci. Rep. 10, 9262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thangarajh M., et al. ; Muscle Study Group and TREAT-NMD, Relationships between DMD mutations and neurodevelopment in dystrophinopathy. Neurology 93, e1597–e1604 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shanmugan S., Epperson C. N., Estrogen and the prefrontal cortex: Towards a new understanding of estrogen’s effects on executive functions in the menopause transition. Hum. Brain Mapp. 35, 847–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickens M. M., Bangasser D. A., Briand L. A., Sex differences in psychiatric disease: A focus on the glutamate system. Front. Mol. Neurosci. 11, 197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J. R., et al. , Gonadal hormones modulate the dendritic spine densities of primary cortical pyramidal neurons in adult female rat. Cereb. Cortex 19, 2719–2727 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Hao J., et al. , Interactive effects of age and estrogen on cognition and pyramidal neurons in monkey prefrontal cortex. Proc. Natl. Acad. Sci. U.S.A. 104, 11465–11470 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang Y., et al. , Estrogen replacement increases spinophilin-immunoreactive spine number in the prefrontal cortex of female rhesus monkeys. Cereb. Cortex 14, 215–223 (2004). [DOI] [PubMed] [Google Scholar]
- 67.Hajszan T., MacLusky N. J., Johansen J. A., Jordan C. L., Leranth C., Effects of androgens and estradiol on spine synapse formation in the prefrontal cortex of normal and testicular feminization mutant male rats. Endocrinology 148, 1963–1967 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Monfort P., Gomez-Gimenez B., Llansola M., Felipo V., Gender differences in spatial learning, synaptic activity, and long-term potentiation in the hippocampus in rats: Molecular mechanisms. ACS Chem. Neurosci. 6, 1420–1427 (2015). [DOI] [PubMed] [Google Scholar]
- 69.McDougall S. A., Moran A. E., Baum T. J., Apodaca M. G., Real V., Effects of ketamine on the unconditioned and conditioned locomotor activity of preadolescent and adolescent rats: Impact of age, sex, and drug dose. Psychopharmacology (Berl.) 234, 2683–2696 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wozniak D. F., et al. , Disseminated corticolimbic neuronal degeneration induced in rat brain by MK-801: Potential relevance to Alzheimer’s disease. Neurobiol. Dis. 5, 305–322 (1998). [DOI] [PubMed] [Google Scholar]
- 71.Wang Y., et al. , Prenatal chronic mild stress induces depression-like behavior and sex-specific changes in regional glutamate receptor expression patterns in adult rats. Neuroscience 301, 363–374 (2015). [DOI] [PubMed] [Google Scholar]
- 72.McCarthy M. M., Wright C. L., Convergence of sex differences and the neuroimmune system in autism spectrum disorder. Biol. Psychiatry 81, 402–410 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang L., et al. , Sex-related differences in MAPKs activation in rat astrocytes: Effects of estrogen on cell death. Brain Res. Mol. Brain Res. 103, 1–11 (2002). [PubMed] [Google Scholar]
- 74.Johnson R. T., Breedlove S. M., Jordan C. L., Sex differences and laterality in astrocyte number and complexity in the adult rat medial amygdala. J. Comp. Neurol. 511, 599–609 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fair D. A., et al. , A method for using blocked and event-related fMRI data to study “resting state” functional connectivity. Neuroimage 35, 396–405 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gratton C., et al. , Functional brain networks are dominated by stable group and individual factors, not cognitive or daily variation. Neuron 98, 439–452.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elliott M. L., et al. , General functional connectivity: Shared features of resting-state and task fMRI drive reliable and heritable individual differences in functional brain networks. Neuroimage 189, 516–532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Laumann T. O., et al. , Functional system and areal organization of a highly sampled individual human brain. Neuron 87, 657–670 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buckner R. L., Krienen F. M., Castellanos A., Diaz J. C., Yeo B. T., The organization of the human cerebellum estimated by intrinsic functional connectivity. J. Neurophysiol. 106, 2322–2345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choi E. Y., Yeo B. T., Buckner R. L., The organization of the human striatum estimated by intrinsic functional connectivity. J. Neurophysiol. 108, 2242–2263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fine C., Neuroscience. His brain, her brain? Science 346, 915–916 (2014). [DOI] [PubMed] [Google Scholar]
- 82.Ciric R., et al. , Mitigating head motion artifact in functional connectivity MRI. Nat. Protoc. 13, 2801–2826 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Satterthwaite T. D., et al. , An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64, 240–256 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gordon E. M., Laumann T. O., Adeyemo B., Petersen S. E., Individual variability of the system-level organization of the human brain. Cereb. Cortex 27, 386–399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mueller S., et al. , Individual variability in functional connectivity architecture of the human brain. Neuron 77, 586–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chang C.-C., Lin C.-J., LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2, 27 (2011). [Google Scholar]
- 87.Cui Z., Gong G., The effect of machine learning regression algorithms and sample size on individualized behavioral prediction with functional connectivity features. Neuroimage 178, 622–637 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Erus G., et al. , Imaging patterns of brain development and their relationship to cognition. Cereb. Cortex 25, 1676–1684 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsu C.-W., Chang C.-C., Lin C.-J., A Practical Guide to Support Vector Classification (Department of Computer Science, National Taiwan University) Available at: http://www.csie.ntu.edu.tw/~cjlin/papers.html (2003).
- 90.Cui Z., Su M., Li L., Shu H., Gong G., Individualized prediction of reading comprehension ability using gray matter volume. Cereb. Cortex 28, 1656–1672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mourão-Miranda J., Bokde A. L., Born C., Hampel H., Stetter M., Classifying brain states and determining the discriminating activation patterns: Support Vector Machine on functional MRI data. Neuroimage 28, 980–995 (2005). [DOI] [PubMed] [Google Scholar]
- 92.Hawrylycz M. J., et al. , An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arnatkeviciute A., Fulcher B. D., Fornito A., A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage 189, 353–367 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Richiardi J., et al. ; IMAGEN consortium, BRAIN NETWORKS. Correlated gene expression supports synchronous activity in brain networks. Science 348, 1241–1244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Uhlén M., et al. , Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Sjöstedt E., et al. , An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, eaay5947 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Marcus D. S., et al. ; WU-Minn HCP Consortium, Human Connectome Project informatics: Quality control, database services, and data visualization. Neuroimage 80, 202–219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.S. Shanmugan et al., Neurodevelopmental Genomics: Trajectories of Complex Phenotypes. Database of Genotypes and Phenotypes https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000607.v3.p2. Accession 4 August 2022. [Google Scholar]
- 99.S. Shanmugan et al., Sex Differences in Functional Topography of Association Networks. GitHub https://pennlinc.github.io/funcParcelSexDiff1/. Accessed 4 August 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Philadelphia Neurodevelopmental Cohort (PNC) (26) data are publicly available in the Database of Genotypes and Phenotypes; https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000607.v3.p2 (98). All analysis code is available at https://pennlinc.github.io/funcParcelSexDiff1/ (99).