Abstract
There exists a tremendous opportunity in identifying and determining the appropriate predictive and prognostic biomarker(s) for risk stratification of patients with colorectal cancers (CRCs). Circulating tumor DNA (ctDNA) has emerged as a promising prognostic and possibly predictive biomarker in the personalized management of patients with CRCs. The disease is particularly suited to a liquid biopsy–based approach since there is a great deal of shedding of circulating tumor fragments (cells, DNA, methylation markers, etc). ctDNA has been shown to have several potential applications, including detecting minimal residual disease (MRD), monitoring for early recurrence, molecular profiling, and therapeutic response prediction. The utility of ctDNA has broadened from its initial use in the advanced/metastatic setting for molecular profiling and detection of acquired resistance mechanisms, toward identifying MRD, as well as early detection. Prospective studies such as CIRCULATE, COBRA, Dynamic II/III, and ACT3 are underway in the MRD setting to further understand how ctDNA may be used to inform clinical decision making using both tumor-informed and tumor-agnostic platforms. These prospective studies use ctDNA to guide management of patients with CRC and will be critical to help guide how and where ctDNA should or should not be used in clinical decision making. It is also important to understand that there are different types of ctDNA liquid biopsy platforms, each with advantages and disadvantages in different clinical indications. This review provides an overview of the current and evolving use of ctDNA in CRC.
BACKGROUND
Liquid biopsies (circulating tumor DNA [ctDNA] testing and circulating tumor cells [CTCs]) have the ability to revolutionize cancer care. Knowledge regarding ctDNA and CTCs has exponentially grown in recent years, although the concept of liquid biopsy is not new. CTCs were described as early as 1869 by Ashworth,1 who then suggested that these cells were shed from the tumor into the bloodstream. It was a century later that cell-free DNA (cfDNA) was discovered in patients with cancer, and it has been an active research entity ever since. The term liquid biopsies is a misnomer since no actual biopsy is performed, but the term encompasses any tumor-derived component detected in a bodily fluid. In the clinic, most of the available platforms are ctDNA-based. It is also important to understand that there are different types of ctDNA liquid biopsy platforms, each with advantages and disadvantages and different clinical indications. Herein, we review the current and evolving practices of using ctDNA in patients with colorectal cancer (CRC) and focus on important ongoing studies of interest.
KEY POINTS
Circulating tumor DNA (ctDNA) has been established as a prognostic biomarker that can define the risk of recurrence after curative-intent surgery.
In practice, ctDNA to assess minimal residual disease status is performed 4 or more weeks after a curative surgery and 2 or more weeks after completion of systemic therapy. For longitudinal monitoring, ctDNA is typically assessed every 8-12 weeks.
For patients who are ctDNA-positive and who have no evidence of disease on routine cross-sectional imaging, a different form of imaging (magnetic resonance imaging with liver-specific gadolinium-based contrast and/or positron emission tomography-computed tomography scan) to find the residual disease that may be amenable to a locoregional approach can be considered to identify the presence of occult disease.
In newly diagnosed metastatic colorectal cancer, liquid biopsy (blood-based next-generation sequencing) testing can provide the relevant molecular profiling (RAS/RAF/HER-status, MSI, and TMB) at diagnosis in place of tissue-based sequencing to inform treatment decision making.
ctDNA can capture intratumoral and temporal heterogeneity in colorectal cancer and provides a platform to detect acquired resistance mechanisms for those exposed to targeted therapies.
It is important to understand that there are different types of ctDNA testing platforms, each with advantages and disadvantages that may make them better suited to certain clinical indications.
CONTEXT
Key Objective
Despite completing appropriate treatment, 15%-30% of the patients with stage II and III colorectal cancer (CRC) experience a recurrence, whereas at least 50% are cured with surgery alone without adjuvant chemotherapy. Circulating tumor DNA (ctDNA) has emerged not only as a prognostic biomarker in CRC but also potentially as a predictive tool to guide treatment decisions in adjuvant and metastatic settings. We reviewed existing literature on the utility of ctDNA to monitor minimal residual disease in resected CRC and to predict therapeutic response in metastatic CRC.
Knowledge Generated
The utility of ctDNA in CRC has broadened from its initial use in the advanced/metastatic setting for molecular profiling and detection of acquired resistance mechanisms, toward identifying minimal residual disease and therapeutic response.
Relevance
ctDNA is arguably the most powerful prognostic biomarker in the adjuvant setting, and ctDNA may be a tool to guide adjuvant therapy decisions. The clearance of ctDNA is also a potential biomarker that could help with early assessment of therapeutic efficacy. ctDNA testing also generally has turnaround time that allows for clinical decision making and could also help with early assessment of clinical efficacy of novel therapeutics. As data emerge from the many ongoing ctDNA CRC clinical trials, we will have more robust data guiding how ctDNA can be incorporated into clinical decision making.
PREANALYTICAL CLINICAL VARIABLES
Before understanding the specifics of any ctDNA assay, it is important to understand the preanalytical and clinical variables that can affect interpretation of results. The types of tubes, reagents, and timing of collection/analysis are beyond the scope of this article. From a clinical standpoint, ctDNA detection can vary by stage, metastatic burden, and the site of metastases.2-5 Kagawa et al6 observed that CRC patients with liver metastases were associated with increased ctDNA detection rates compared with the patients with lung and peritoneal metastases. Across several large studies, CRC has shown a high rate of shedding of circulating tumor fragments (cells, DNA, and methylation markers). The disease, therefore, is particularly suited to a liquid biopsy-approach. Given that the sensitivity of ctDNA detection is high, this has garnered the increasing interest in ctDNA within CRC as a biomarker for various clinical applications.
CLINICAL APPLICATIONS
Molecular Profiling
Although tissue biopsy remains the gold standard for molecular profiling, it has inherent limitations, which include the following.
Insufficient tissue.
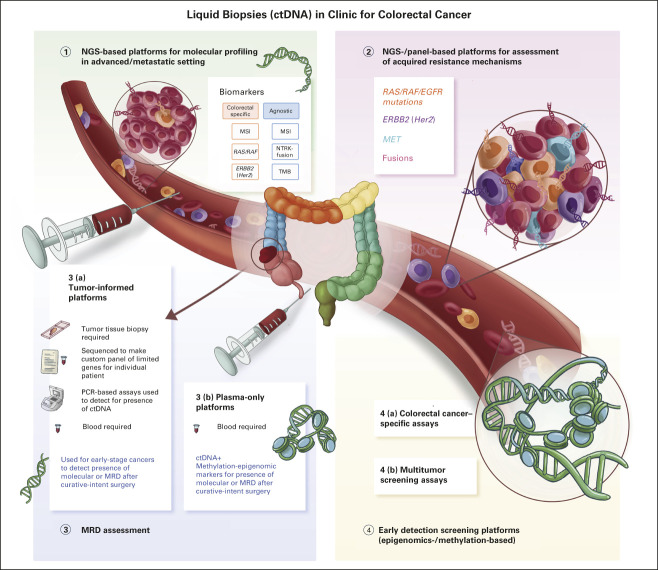
It is underestimated how often tissue-based next-generation sequencing (NGS) fails because of lack of tumor tissue in the specimen. This might be due to the sampling site and/or reflective of tumor histology, eg, mucinous tumors. ctDNA can complement and perhaps replace up front tissue-based genotyping in CRC for identifying key biomarkers such as RAS/BRAF/HER2 status, and detection of microsatellite instability (MSI; Fig 1).
FIG 1.
Types of ctDNA platforms and their various clinical applications for patients with colorectal cancer. The utility of ctDNA has broadened from its initial use in the advanced/metastatic setting for molecular profiling and detection of acquired resistance mechanisms, toward identifying MRD, as well as early detection. ctDNA, circulating tumor DNA; EGFR, epidermal growth factor receptor; MRD, minimal residual disease; MSI, microsatellite instability; NGS, next-generation sequencing; NTRK, neurotrophic tyrosine receptor kinase; PCR, polymerase chain reaction; TMB, tumor mutational burden.
Turnaround time.
Liquid biopsies offer an advantage of a rapid turnaround time (7-10 days as opposed to weeks for tissue-based NGS assays because of inherent delays in accessing the tumor specimen), and promises to be an efficient tool for clinical trial matching and to propel precision medicine.7,8 In a prospective study from Japan, significantly shorter sample accession (median 4 v 14 days, P < .0001) and shorter turnaround time (median 7 v 19 days, P < .0001) resulted in a significant increase in the proportion of patients enrolled in clinical trials (9.5 v 4.1%, P < .0001).9
Sample acquisition.
Traditional tumor sampling is more invasive than phlebotomy and sometimes not feasible (there are procedural complications in up to 20% of cases).10 This also makes liquid biopsies more suited for serial assessments than tissue biopsy.
Intratumoral and temporal heterogeneity.
Tissue NGS testing may not represent intratumoral and/or temporal heterogeneity.11 Tumor heterogeneity has garnered increasing interest.12 This is particularly relevant to patients with CRC who have been exposed to anti-epidermal growth factor receptor (anti-EGFR) or other targeted therapies where detection of resistance may guide future therapeutics (Fig 1).
Monitoring of MRD
With its origins in the world of hematology, the term minimal, molecular, or measurable residual disease (MRD) in the context of CRC often refers to ctDNA detectable after curative-intent surgery. ctDNA has emerged as a powerful biomarker predicting which patients with CRC are likely to relapse on the basis of an occult or persistent state of disease (Fig 2). Its short half-life, ranging from minutes to a few hours, allows for a more accurate and real-time dynamic measure of disease burden.13
FIG 2.
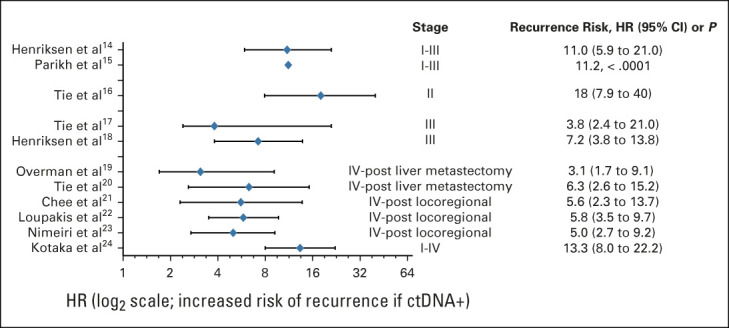
Risk of CRC recurrence in patients who have undergone curative-intent surgery and are ctDNA-positive.14-24. CRC, colorectal cancer; ctDNA, circulating tumor DNA; HR, hazard ratio.
In numerous studies, patients who are MRD-positive (ie, have detectable ctDNA) almost universally relapse if not offered any therapy. It is not just a high-risk marker but an indicator of persistent disease, with 95%-100% of patients with persistently detectable ctDNA postsurgery developing a recurrence, usually within 2 years of follow-up if offered no systemic therapy.16,17,25
Current adjuvant management in resected stage II colon cancers is based on risk stratification using clinical and pathologic prognostic factors. Contrastingly, adjuvant chemotherapy is recommended for every stage III patient, as tolerated, although nearly 50% are cured with surgery alone.26 Furthermore, approximately 15% and 30% of stage II and III patients experience recurrence despite completing appropriate treatment, respectively.27,28 Early detection of MRD could identify a cohort of patients who may benefit from intensification of systemic therapy. It also provides an opportunity to de-escalate care in low-risk patients with no evidence of MRD who may not need toxic systemic therapy.2,29-31 This model is currently being assessed in multiple clinical trials using MRD as an integral biomarker in patients with CRC.
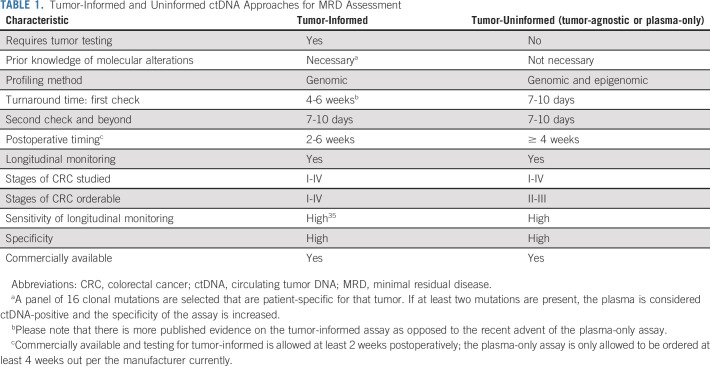
Comparison of tumor-informed and tumor-agnostic (plasma-only) ctDNA testing strategies.
Currently, tumor-informed and tumor-agnostic (also referred to as tumor-uninformed or plasma-only ctDNA) testing approaches are available for MRD assessment and monitoring in CRC (Table 1). Tumor-informed testing, as the name suggests, requires knowledge of particular mutations in a patient's tumor and then designs probes on the basis of the tissue to test the same patient's plasma (Fig 1). Historically, plasma-only approaches using ctDNA were considered less sensitive. However, recent approaches in plasma-only testing have improved sensitivity of these approaches by using additional markers such as methylation or epigenomics markers.15 Plasma-only approaches may also improve turnaround time for the landmark assessment of a patient's MRD status as they do not require tissue to be obtained, sequenced, and a custom ctDNA panel to be created before performing an assessment of MRD status. In the setting of adjuvant therapy, where delays in adjuvant therapy can reduce efficacy, these differences may be important for clinical decision making.32
TABLE 1.
Tumor-Informed and Uninformed ctDNA Approaches for MRD Assessment
Timing of ctDNA for MRD assessment.
The timing of ctDNA testing after resection in the curative-intent setting is cardinal (at least 2-4 weeks from the time of surgery). This is due to elevated levels of circulating normal cfDNA because of surgical procedures leading to background noise that could effectively dilute the ctDNA and lower the sensitivity. Repeat testing is suggested for patients with undetectable ctDNA within the first 4 weeks of resection to avoid false negatives.33,34
Furthermore, ctDNA levels decline very quickly on effective systemic therapy. Ideally, MRD assessment should be before initiation of adjuvant chemotherapy and not during systemic therapy. In the surveillance setting, assessment of ctDNA for MRD at least 2-4 weeks after completion of adjuvant therapy is reasonable.
ctDNA as a predictive biomarker of response to adjuvant therapy.
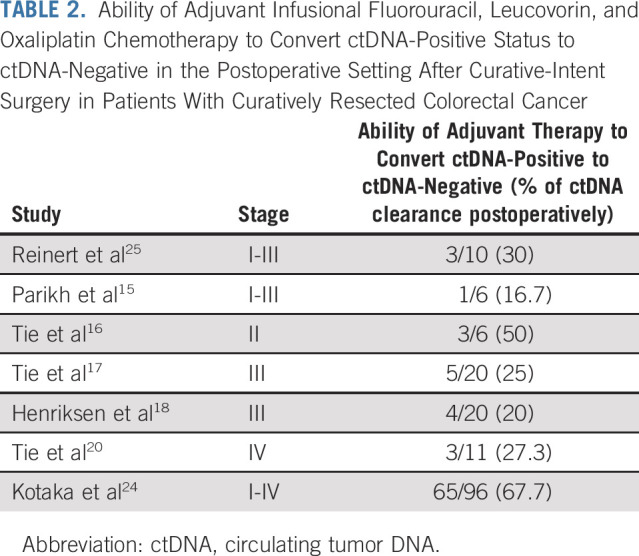
Emerging data demonstrate that aside from a prognostic marker, ctDNA may also be a predictive biomarker of response to adjuvant therapy. Henriksen et al35 studied postadjuvant chemotherapy ctDNA status on risk of recurrence in 160 patients with stage III CRC where 140 patients had postoperative ctDNA available. Postoperative ctDNA was detected in 14% (20/140) of available patients' samples, 90% of these ctDNA-positive patients received adjuvant chemotherapy (18/20), and 13 of them had longitudinal samples available. Only 23% (3/13) of patients showed permanent clearance of ctDNA with adjuvant therapy and had no evidence of relapse at 36 months of follow-up. By contrast, 100% (10/10) of patients who had transient clearance or no clearance relapsed. Significantly shorter relapse-free survival (hazard ratio [HR]: 94.2, 95% CI, 15.74 to 564.30; P < .001) was demonstrated in patients with persistent ctDNA despite adjuvant chemotherapy. The results have been consistent across multiple studies; approximately 25% of patients who have detectable ctDNA postoperatively have clearance with standard infusional fluorouracil, leucovorin, and oxaliplatin adjuvant chemotherapy (Table 2). This high-risk group may benefit from prolonged therapy (longer than the stipulated 3-6 months), intensified therapy (ie, triplet chemotherapy with addition of irinotecan to the infusional fluorouracil, leucovorin, and oxaliplatin adjuvant chemotherapy: FOLFIRINOX), and/or addition of biomarker-based therapy in those with an actionable alteration. ctDNA analysis from the adjuvant IDEA-France study evaluating 3 versus 6 months of chemotherapy was supportive of intensifying therapy in stage III CRC patients with detectable ctDNA postoperatively. A notable improvement in the disease-free survival (DFS) of patients who had detectable ctDNA postoperatively and received a longer duration (6 months) of adjuvant chemotherapy was demonstrated.36
TABLE 2.
Ability of Adjuvant Infusional Fluorouracil, Leucovorin, and Oxaliplatin Chemotherapy to Convert ctDNA‐Positive Status to ctDNA-Negative in the Postoperative Setting After Curative-Intent Surgery in Patients With Curatively Resected Colorectal Cancer
More recently, updated analysis from an observational GALAXY study monitoring MRD in patients with stage I-III and oligometastatic stage IV CRC evaluating the association on ctDNA dynamics with adjuvant chemotherapy efficacy showed the first hints of ctDNA being a predictive biomarker. In this large cohort of nearly 1,000 patients with CRC, 188 were noted to be MRD-positive, and 95 patients received adjuvant chemotherapy. The ctDNA clearance rate at 24 weeks was significantly higher in patients who received adjuvant chemotherapy compared with those who did not (68% v 7%; cumulative HR: 17.1; P < .001). This clearance rate was three times higher than the results obtained in other studies (Table 2). Furthermore, 6-month DFS was also significantly higher in the former group compared with the latter (84% v 34%; HR: 0.15; P < .001).24
ctDNA for early recurrence monitoring.
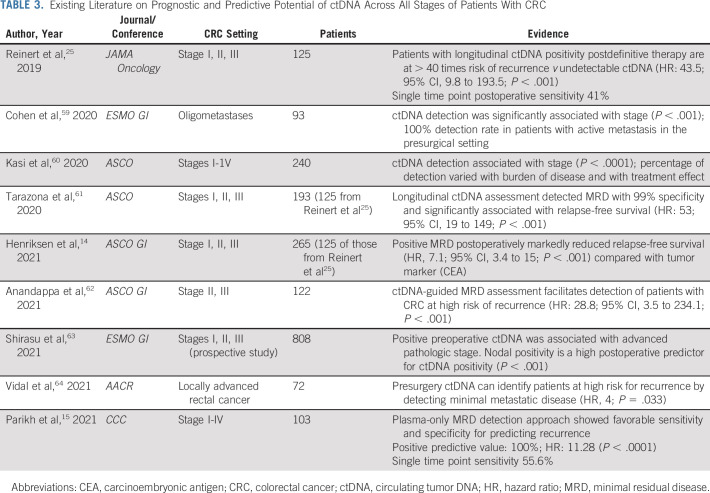
Clinical history and examination, carcinoembryonic antigen testing, imaging, and colonoscopy examinations are periodically performed over a 5-year period to detect recurrence of CRC on the basis of the existing ASCO and National Comprehensive Cancer Network guidelines. Despite the known fact that the sensitivity of carcinoembryonic antigen is limited (< 70%), it continues to be relied on as a tumor marker to predict recurrence in clinical practice.25 Better markers with improved sensitivity as well as positive and negative predictive values are necessary. ctDNA has been shown to predict relapse in CRC with a median lead time of 8.7 months before radiographic assessment.25 Monitoring for recurrence is distinct from MRD assessment at a single time point after definitive therapy. Longitudinal ctDNA monitoring approaches with tumor-informed and tumor-agnostic approaches to predicting recurrence improved sensitivity to 69% and 88%, respectively (Table 3).
TABLE 3.
Existing Literature on Prognostic and Predictive Potential of ctDNA Across All Stages of Patients With CRC
Correlation between rate of ctDNA change and tumor growth was further studied in a subset of 17 patients who relapsed after completion of adjuvant chemotherapy. A bimodal distribution, fast or slow, of the ctDNA growth pattern was revealed with prognostic implications for recurrence prediction.35 For patients who are ctDNA-positive and have no evidence of disease on routine cross-sectional imaging, a different form of imaging (magnetic resonance imaging with liver-specific gadolinium-based contrast and/or positron emission tomography-computed tomography scan) to find the residual disease that may be amenable to a locoregional approach can be considered to identify the presence of occult disease.
Ongoing ctDNA-directed studies in CRC.
With increasing availability in the clinic, prospective randomized trials are needed to help us determine the utility of ctDNA-guided treatment approaches for both MRD assessment and recurrence monitoring. Majority of the ongoing studies are conducted using the two commercial testing platforms that are currently available.65,66
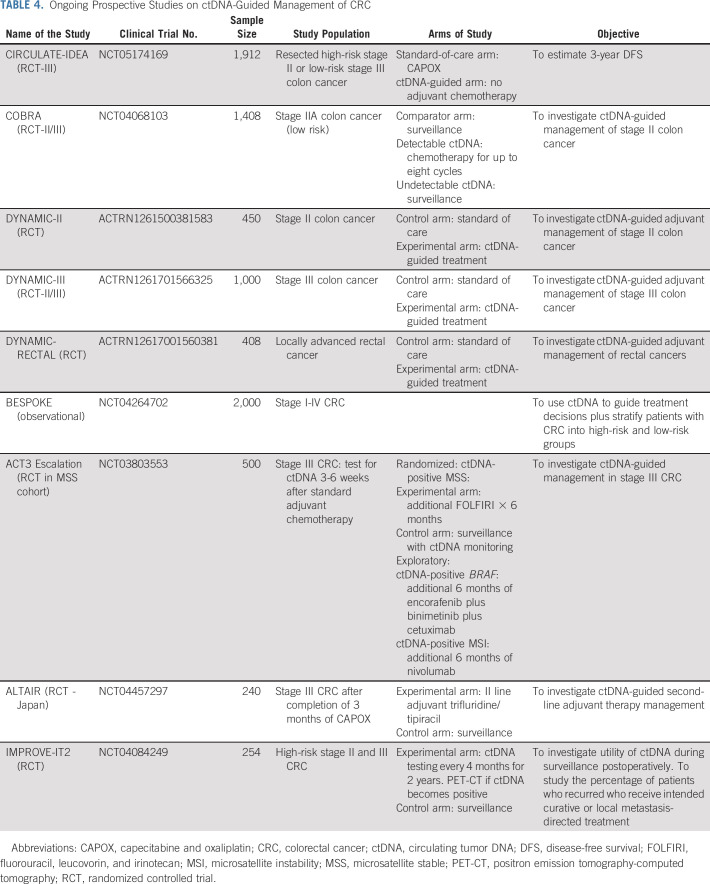
Multiple prospective, observational, and interventional studies are ongoing to appraise the utility of ctDNA-guided management in CRC (Table 4). In the United States, two studies that were among the first to open and are actively recruiting patients are the BESPOKE and COBRA studies. BESPOKE (ClinicalTrials.gov identifier: NCT04264702) is a prospective observational study in patients with stage I-IV CRC who underwent curative surgery and are followed for up to 2 years to examine the impact of tumor-informed ctDNA testing on adjuvant treatment decisions. COBRA (ClinicalTrials.gov identifier: NCT04068103) is a phase II/III NRG Cooperative Group trial evaluating a plasma-only ctDNA detection approach to select high-risk patients to receive infusional fluorouracil, leucovorin, and oxaliplatin chemotherapy in stage-IIA colon cancer following resection. The ongoing Australian/Canadian DYNAMIC-III study (ACTRN1261701566325) is examining the value of ctDNA-guided de-escalation or escalation of chemotherapy in stage III colon cancer. These studies may shed light on the utility of ctDNA as a predictive biomarker in CRC.
TABLE 4.
Ongoing Prospective Studies on ctDNA-Guided Management of CRC
Although serial ctDNA analyses have been shown to detect recurrence with a median lead time ranging from 8.7 to 12 months25,35 compared with imaging assessment, the impact of initiation of therapy as soon as ctDNA is detected on relapse-free survival (on the basis of imaging) remains to be seen. Whether serial ctDNA trends in patients whose treatment is modified on the basis of a change in ctDNA (before scan confirmation of relapse) remains to be explored and will be evaluated in the CIRCULATE studies (CIRCULATE-US: ClinicalTrials.gov identifier: NCT05174169). There are several ongoing prospective studies in CRC directed toward ctDNA-guided second-line adjuvant strategies. ACT-3 (ClinicalTrials.gov identifier: NCT03803553) is a phase III study in microsatellite stable (MSS) stage III CRC that randomly assigns patients who are ctDNA-positive after completion of 3 months of adjuvant capecitabine and oxaliplatin (3-6 weeks after adjuvant chemotherapy) to surveillance or additional fluorouracil, leucovorin, and irinotecan treatment.
Given the unique ability of ctDNA to provide a real-time assessment of therapeutic benefit, ctDNA clearance could be explored as a surrogate end point in the adjuvant setting for drug development as well. In the ACT3 study (ClinicalTrials.gov identifier: NCT03803553), for example, other exploratory cohorts include patients who have MSI‐high, BRAF-V600E mutant, and HER2-amplified tumors who will receive additional nivolumab, encorafenib, binimetinib and cetuximab, and trastuzumab and pertuzumab, respectively. Similarly, the ALTAIR study (ClinicalTrials.gov identifier: NCT04457297) will evaluate the efficacy of adjuvant trifluridine/tipiracil in patients whose ctDNA remain positive despite 3 months of capecitabine and oxaliplatin adjuvant therapy. Many of these studies are using ctDNA clearance as a primary end point in MRD-positive patients after adjuvant therapy.
ctDNA for Response Prediction in Metastatic Disease
Prognostic value of ctDNA in metastatic disease.
The utility of tumor markers as a biomarker for response prediction is sparse because of prolonged half-life37 and limited sensitivity and specificity; however, tumor markers are routinely used. ctDNA is emerging as an adjunct tool in addition to imaging for response prediction. ctDNA correlates with changes in tumor burden and predicts treatment response with some studies showing superiority over standard tumor markers.38-40 A 4-week change in ctDNA was predictive of clinical benefit and prognostic of outcomes with improvement in progression-free survival (PFS) in patients receiving chemotherapy and/or targeted therapy who demonstrated ≥ 30% decrease in ctDNA.40
Quantification of ctDNA.
Mean tumor molecules (MTM) and variant allele frequency (VAF) are metrics commonly used to quantify tumor-specific variants in ctDNA. The majority of existing testing platforms use either of these approaches to estimate quantity of ctDNA. Although VAF measures the ratio of variant alleles to wild-type alleles, MTM accounts for the total amount of cell-free DNA. Tin et al analyzed over 6,000 plasma samples from more than 3,000 patients to evaluate the correlation of VAF and MTM with mean tumor burden in various solid tumors across multiple settings. A positive correlation between VAF and MTM (R2 = 0.91) was demonstrated except when cfDNA background levels were high (≥ 50 ng/mL). VAF values plateaued asymptotically as MTM increased, which could be a limitation of VAF in patients with high cfDNA levels. Furthermore, the level of MTM/ml has been shown to be more prognostic than VAF in patients with advanced solid tumors treated with immunotherapy (rise in MTM v VAF, HR: 4.08 v 2.62).41
Immunotherapy response monitoring.
Akin to response prediction for chemotherapy and/or targeted therapy, another potential ctDNA-guided clinical application could be the identification of nonresponders to immunotherapy recognizing that the kinetics of response is likely different. Although immunotherapy prolonged PFS in MSI-high CRC, it is intriguing that nearly 30% of patients were refractory to pembrolizumab in KEYNOTE-177.42 Early identification of nonresponders using ctDNA monitoring could allow providers the opportunity to switch to chemotherapy or consider addition of an anticytotoxic T-cell lymphocyte-4.43
Response prediction to immunotherapy using ctDNA has shown promise in the INSPIRE study, a prospective phase II trial that followed 94 patients with 25 advanced solid tumor types treated with pembrolizumab with serial ctDNA assessments.44 ctDNA increase at 6 weeks with increasing tumor volume was seen in 42% of the patients and predicted lack of response with 100% accuracy. ctDNA clearance was observed in 16% of patients on immunotherapy, and overall survival (OS) was 100% with a median of > 25 months of follow-up beyond first clearance. Ninety-eight percent of the patients who had an increase in ctDNA at the time of initiation of cycle 3 did not achieve an objective response. This would be a subset of patients for whom ineffective treatment could conceivably be avoided.
Zhang et al45 characterized the prognostic and predictive impact of ctDNA in patients with 16 different solid tumor types across phase I/II trials of either durvalumab alone or in combination with tremelimumab. Higher pretreatment VAF was associated with poor survival but did not correlate with objective response rates (ORRs). By contrast, on-treatment reduction in VAF was associated with prolonged PFS, OS, and ORR, suggestive of predictive benefit with on-treatment ctDNA. ctDNA kinetics in terms of changes in VAF percentage and/or ctDNA clearance is now an integral biomarker for several ongoing clinical trials across tumor types including CRC.
Acquired Resistance and Clonal Evolution
A significant challenge associated with single-lesion biopsy is ineffectiveness in estimating underlying tumoral molecular heterogeneity. Furthermore, discerning acquired resistance to ongoing therapy, typified by the emergence of resistant subclones either within the same lesion or in various metastatic sites in an individual patient, using repeated tumor biopsies is difficult and impractical (Fig 1). ctDNA has the potential to identify emerging genetic alterations and track the clonal evolution of tumors, which could be drivers for resistance, in a noninvasive manner.46,47 ctDNA is cheaper, safer, and more convenient, and provides a more accurate picture of resistance mechanisms compared with repeated tumor biopsies.48 This is particularly relevant to patients with CRC exposed to anti-EGFR therapies or other targets therapies (Fig 1).49
Anti-EGFR antibody rechallenge has been accomplished using a liquid biopsy–driven approach in RAS/BRAF WT mCRC and has led to enhanced response rates (ORR: 30%, 95% CI, 12 to 47) in the CHRONOS trial. In this trial, patients who initially had a response to anti-EGFR therapy were allowed a rechallenge if a ctDNA test did not reveal mutations in the mitogen-activated protein kinase pathway after progression on subsequent line of anti-EGFR therapy.49 Similarly, the CRICKET trial51 demonstrated the benefit from an EGFR rechallenge approach (ORR: 21%, 95% CI: 10 to 40) using liquid biopsies. ctDNA analysis showed that the patients who had a response did not have RAS alterations portending resistance at the time of rechallenge. Extensive research has shown that acquired resistant mutations developed during anti-EGFR antibody therapy diminish with the discontinuation of targeted therapy, a concept referred to as clonal decay.50,51 ctDNA-guided monitoring has allowed for noninvasive molecular selection of patients who may respond again to anti-EGFR therapy in a subgroup of CRC.
CHALLENGES AND LIMITATIONS OF ctDNA
Despite the fact that ctDNA has increasingly become a part of the paradigm for molecular profiling and that high concordance is observed between baseline plasma and tissue testing, tissue biopsy remains the gold standard in general for solid tumors. There are advantages to a tissue-based approach. To date, from a technical standpoint, tissue biopsy is more sensitive in detecting fusions since these are large gene rearrangements.7 Although ctDNA can detect copy-number variations, the efficacy is limited to patients with high tumor content or extreme copy-number amplifications, whereas copy losses are very difficult to detect in plasma. Even for coding variants, sufficient ctDNA may not be detectable in approximately 15% of the patients with metastatic cancers on the basis of tumor type and burden.52
From a clinical standpoint, blood-based ctDNA testing can also have less utility in CRC patients with peritoneal carcinomatosis or brain metastases because of the respective blood-based barriers. Creative strategies to use ctDNA detection in other body fluids (cerebrospinal fluid, ascites, pleural effusion etc) are under investigation.53-57
Steps are needed to standardize ctDNA profiling across platforms and to describe how ctDNA tests relate to a tissue result, as other biomarkers such as plasma tumor mutational burden are being reported. Plasma tumor mutational burden scores trended higher compared with tissue TMB and it is unclear how or whether these should be compared with tissue-based results.58 It is important keep these technical and clinical applications in mind when applying any of the ctDNA platforms for patients with CRC.
In conclusion, ctDNA is emerging as a powerful tool for various applications including molecular profiling, MRD detection, early recurrence monitoring, and treatment response prediction. ctDNA can potentially be used to guide adjuvant therapy, and/or considering additional imaging techniques (magnetic resonance imaging and/or positron emission tomography-computed tomography) to identify sites of occult metastasis potentially amenable to a locoregional therapy approach in the surveillance setting.
ctDNA is highly prognostic and ctDNA clearance is emerging as a biomarker that could help with early assessment of therapeutic efficacy. Whether it can be a surrogate end point for recurrence or DFS is yet to be determined. This approach could expedite clinical trial read out, reduce the cost of trials, and allow us to bring advances to the patients sooner. ctDNA will enhance recruitment to clinical trials and increase the number of patients eligible for a precision medicine approach. It may also allow us to abort approved but ineffective therapies in patients who do not respond to targeted or immunotherapies. With the completion of ongoing clinical trials, we anticipate patients with stage III and high-risk stage II CRC with no evidence of MRD postoperatively can perhaps be monitored under active surveillance without adjuvant chemotherapy and potentially treated at the time of MRD detection.
Keeping in mind the limitations, CRC as a disease is particularly suited to a liquid biopsy–based approach since there is a great deal of shedding of circulating tumor fragments (cells, DNA, methylation markers, etc). The utility of ctDNA has broadened from its initial use in the advanced/metastatic setting for molecular profiling and detection of acquired resistance mechanisms, toward identifying MRD as well as early detection. We are also seeing hints of ctDNA being more than just a prognostic marker in the MRD setting. It is important to reiterate that there are different types of ctDNA liquid biopsy platforms, each with advantages and disadvantages and different clinical indications. Interpretation and application of ctDNA results cannot be done in isolation, and the clinical context needs to be kept in mind besides the assay/technical issues. Enrollment in ongoing clinical trials that use ctDNA as an integral biomarker and harmonizing of reporting across platforms will be key to further advance the field.
ACKNOWLEDGMENT
The authors are deeply indebted to the team at Draw Impacts for their work on Figure 1 that accompanies the article.
Midhun Malla
Honoraria: Omni Health
Consulting or Advisory Role: QED Therapeutics, AstraZeneca
Speakers' Bureau: Natera
Jonathan M. Loree
Consulting or Advisory Role: Taiho Pharmaceutical, Ipsen, Novartis, Bayer, Amgen, Eisai, Pfizer
Research Funding: Ipsen (Inst), Amgen (Inst)
Pashtoon Murtaza Kasi
Consulting or Advisory Role: Taiho Pharmaceutical (Inst), Ipsen (Inst), Natera, Foundation Medicine, MSD Oncology, Tempus, Bayer, Lilly, Delcath Systems, Inflecton Point Biomedical Advisors, QED Therapeutics, Boston Healthcare Associates, Servier, Taiho Oncology, Exact Sciences, Daiichi Sankyo/Astra Zeneca, Eisai
Research Funding: Advanced Accelerator Applications (Inst), Tersera (Inst), Boston Scientific (Inst)
Travel, Accommodations, Expenses: AstraZeneca
Aparna Raj Parikh
Stock and Other Ownership Interests: C2i Genomics
Consulting or Advisory Role: Lilly, Natera, Checkmate Pharmaceuticals, Pfizer, Roche/Genentech, Inivata, Biofidelity, Guardant Health
Research Funding: Plexxikon (Inst), Bristol Myers Squibb (Inst), Genentech (Inst), Guardant Health (Inst), Array BioPharma (Inst), Lilly (Inst), Novartis Pharmaceuticals UK Ltd (Inst), PureTech (Inst), PMV Pharma, Mirati Therapeutics (Inst), Daiichi Sankyo (Inst)
No other potential conflicts of interest were reported.
DISCLAIMER
The content reported in this article is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
SUPPORT
Supported by grant funding from National Institute of General Medical Sciences-NIH (Grant No. 5U54GM104942-05 [M.M.]). J.L. has received a Michael Smith Health Professional Investigator Award, which helps support his research activities. A.R.P. is the recipient of 2019 Conquer Cancer Foundation Career Development Award.
P.M.K. and A.R.P. contributed equally and share co-senior authorship.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Financial support: Pashtoon Murtaza Kasi, Aparna Raj Parikh
Administrative support: Midhun Malla, Pashtoon Murtaza Kasi, Aparna Raj Parikh
Provision of study materials or patients: Pashtoon Murtaza Kasi
Collection and assembly of data: All authors
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Midhun Malla
Honoraria: Omni Health
Consulting or Advisory Role: QED Therapeutics, AstraZeneca
Speakers' Bureau: Natera
Jonathan M. Loree
Consulting or Advisory Role: Taiho Pharmaceutical, Ipsen, Novartis, Bayer, Amgen, Eisai, Pfizer
Research Funding: Ipsen (Inst), Amgen (Inst)
Pashtoon Murtaza Kasi
Consulting or Advisory Role: Taiho Pharmaceutical (Inst), Ipsen (Inst), Natera, Foundation Medicine, MSD Oncology, Tempus, Bayer, Lilly, Delcath Systems, Inflecton Point Biomedical Advisors, QED Therapeutics, Boston Healthcare Associates, Servier, Taiho Oncology, Exact Sciences, Daiichi Sankyo/Astra Zeneca, Eisai
Research Funding: Advanced Accelerator Applications (Inst), Tersera (Inst), Boston Scientific (Inst)
Travel, Accommodations, Expenses: AstraZeneca
Aparna Raj Parikh
Stock and Other Ownership Interests: C2i Genomics
Consulting or Advisory Role: Lilly, Natera, Checkmate Pharmaceuticals, Pfizer, Roche/Genentech, Inivata, Biofidelity, Guardant Health
Research Funding: Plexxikon (Inst), Bristol Myers Squibb (Inst), Genentech (Inst), Guardant Health (Inst), Array BioPharma (Inst), Lilly (Inst), Novartis Pharmaceuticals UK Ltd (Inst), PureTech (Inst), PMV Pharma, Mirati Therapeutics (Inst), Daiichi Sankyo (Inst)
No other potential conflicts of interest were reported.
REFERENCES
- 1.Ashworth TR: A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Australas Med J 14:146-147, 1869 [Google Scholar]
- 2.Bettegowda C, Sausen M, Leary RJ, et al. : Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 6:224ra24, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manca P, Corallo S, Lonardi S, et al. : SO-24 Circulating tumor DNA variant allelic fraction as a surrogate for disease burden estimation in patients with RAS wild-type metastatic colorectal cancer: A secondary endpoint of the VALENTINO study. Ann Oncol 32:S212, 2021 [Google Scholar]
- 4.Vidal J, Muinelo L, Dalmases A, et al. : Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 28:1325-1332, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osumi H, Shinozaki E, Takeda Y, et al. : Clinical relevance of circulating tumor DNA assessed through deep sequencing in patients with metastatic colorectal cancer. Cancer Med 8:408-417, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kagawa Y, Elez E, García-Foncillas J, et al. : Combined analysis of concordance between liquid and tumor tissue biopsies for RAS mutations in colorectal cancer with a single metastasis site: The METABEAM study. Clin Cancer Res 27:2515-2522, 2021 [DOI] [PubMed] [Google Scholar]
- 7.Parikh AR, Leshchiner I, Elagina L, et al. : Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med 25:1415-1421, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshino T: SCRUM-Japan GI-SCREEN: The nationwide cancer genome screening projects for gastrointestinal cancer in Japan. Ann Oncol 26:vii5, 2015 [Google Scholar]
- 9.Nakamura Y, Taniguchi H, Ikeda M, et al. : Clinical utility of circulating tumor DNA sequencing in advanced gastrointestinal cancer: SCRUM-Japan GI-SCREEN and GOZILA studies. Nat Med 26:1859-1864, 2020 [DOI] [PubMed] [Google Scholar]
- 10.Overman MJ, Modak J, Kopetz S, et al. : Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J Clin Oncol 31:17-22, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popper HH: Commentary on tumor heterogeneity. Transl Lung Cancer Res 5:433-435, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris LG, Riaz N, Desrichard A, et al. : Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget 7:10051-10063, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diehl F, Schmidt K, Choti MA, et al. : Circulating mutant DNA to assess tumor dynamics. Nat Med 14:985-990, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriksen TV, Tarazona N, Reinert T, et al. : Circulating tumor DNA analysis for assessment of recurrence risk, benefit of adjuvant therapy, and early relapse detection after treatment in colorectal cancer patients. Clin Cancer Res 28:507-517, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parikh AR, Van Seventer EE, Siravegna G, et al. : Minimal residual disease detection using a plasma-only circulating tumor DNA assay in colorectal cancer patients. Clin Cancer Res 27:5586-5594, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tie J, Wang Y, Tomasetti C, et al. : Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 8:346ra92, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tie J, Cohen JD, Wang Y, et al. : Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 5:1710-1717, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriksen TVV, Tarazona N, Frydendahl A, et al. : Serial circulating tumor DNA analysis to assess recurrence risk, benefit of adjuvant therapy, growth rate and early relapse detection in stage III colorectal cancer patients. J Clin Oncol 39, 2021. (suppl 15; abstr 3540) [Google Scholar]
- 19.Overman MJ, Vauthey J-N, Aloia TA, et al. : Circulating tumor DNA (ctDNA) utilizing a high-sensitivity panel to detect minimal residual disease post liver hepatectomy and predict disease recurrence. J Clin Oncol 35, 2017. (suppl 15; abstr 3522) [Google Scholar]
- 20.Tie J, Wang Y, Cohen J, et al. : Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases : A prospective cohort study. PLoS One 18:e1003620, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chee B, Ibrahim F, Esquivel M, et al. : Circulating tumor derived cell-free DNA (ctDNA) to predict recurrence of metastatic colorectal cancer (mCRC) following curative intent surgery or radiation. J Clin Oncol 39, 2021. (suppl 15; abstr 3565) [Google Scholar]
- 22.Loupakis F, Sharma S, Derouazi M, et al. : Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol 5:1166-1177, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nimeiri H, Young A, Madison R, et al. : Comprehensive genomic profiling (CGP)-informed personalized molecular residual disease (MRD) detection: An exploratory analysis from the PREDATOR study of metastatic colorectal cancer (mCRC) patients undergoing surgical resection. J Clin Oncol 40, 2022. (suppl 4; abstr 187) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kotaka M, Shirasu H, Watanabe J, et al. : Association of circulating tumor DNA dynamics with clinical outcomes in the adjuvant setting for patients with colorectal cancer from an observational GALAXY study in CIRCULATE-Japan. J Clin Oncol 40, 2022. (suppl 4; abstr9) [Google Scholar]
- 25.Reinert T, Henriksen TV, Christensen E, et al. : Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol 5:1124-1131, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson NW, Yothers G, Lopa S, et al. : Long-term survival results of surgery alone versus surgery plus 5-fluorouracil and leucovorin for stage II and stage III colon cancer: Pooled analysis of NSABP C-01 through C-05. A baseline from which to compare modern adjuvant trials. Ann Surg Oncol 17:959-966, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.André T, Boni C, Navarro M, et al. : Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol 27:3109-3116, 2009 [DOI] [PubMed] [Google Scholar]
- 28.Osterman E, Glimelius B: Recurrence risk after up-to-date colon cancer staging, surgery, and pathology: Analysis of the entire Swedish population. Dis Colon Rectum 61:1016-1025, 2018 [DOI] [PubMed] [Google Scholar]
- 29.Chakrabarti S, Peterson CY, Sriram D, et al. : Early stage colon cancer: Current treatment standards, evolving paradigms, and future directions. World J Gastrointest Oncol 12:808-832, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan JCM, Massie C, Garcia-Corbacho J, et al. : Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat Rev Cancer 17:223-238, 2017 [DOI] [PubMed] [Google Scholar]
- 31.Siravegna G, Marsoni S, Siena S, et al. : Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14:531-548, 2017 [DOI] [PubMed] [Google Scholar]
- 32.Biagi JJ, Raphael MJ, Mackillop WJ, et al. : Association between time to initiation of adjuvant chemotherapy and survival in colorectal cancer: A systematic review and meta-analysis. JAMA 305:2335-2342, 2011 [DOI] [PubMed] [Google Scholar]
- 33.Henriksen TV, Reinert T, Christensen E, et al. : The effect of surgical trauma on circulating free DNA levels in cancer patients—implications for studies of circulating tumor DNA. Mol Oncol 14:1670-1679, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dasari A, Morris VK, Allegra CJ, et al. : CtDNA applications and integration in colorectal cancer: An NCI Colon and Rectal–Anal Task Forces whitepaper. Nat Rev Clin Oncol 17:757-770, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henriksen TV, Tarazona N, Frydendahl A, et al. : Circulating tumor DNA in stage III colorectal cancer, beyond minimal residual disease detection, towards assessment of adjuvant therapy efficacy and clinical behavior of recurrences. Clin Cancer Res 14:1748-1753, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taieb J, Taly V, Vernerey D, et al. : Analysis of circulating tumour DNA (ctDNA) from patients enrolled in the IDEA-FRANCE phase III trial: Prognostic and predictive value for adjuvant treatment duration. Ann Oncol 30:v867, 2019 [Google Scholar]
- 37.Dawson SJ, Tsui DW, Murtaza M, et al. : Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368:1199-1209, 2013 [DOI] [PubMed] [Google Scholar]
- 38.Diaz LA, Jr, Williams RT, Wu J, et al. : The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486:537-540, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siravegna G, Mussolin B, Buscarino M, et al. : Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21:795-801, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parikh AR, Mojtahed A, Schneider JL, et al. : Serial CtDNA monitoring to predict response to systemic therapy in metastatic gastrointestinal cancers. Clin Cancer Res 26:1877-1885, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tin A, Aushev V, Kalashnikova E, et al. : Correlation of variant allele frequency and mean tumor molecules with tumor burden in patients with solid tumors. Mol Oncol 15:57-66, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andre T, Shiu KK, Kim TW, et al. : Pembrolizumab versus chemotherapy for microsatellite instability-high/mismatch repair deficient metastatic colorectal cancer: The phase 3 KEYNOTE-177 Study. J Clin Oncol 38, 2020. (suppl 18; abstr LBA4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasi PM, Budde G, Krainock M, et al. : Circulating tumor DNA (ctDNA) serial analysis during progression on PD-1 blockade and later CTLA4 rescue in patients with mismatch repair deficient metastatic colorectal cancer. J Immunother Cancer 10:e003312, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bratman SV, Yang SC, Iafolla MA, et al. : Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer 1:873-881, 2020 [DOI] [PubMed] [Google Scholar]
- 45.Zhang Q, Luo J, Wu S, et al. : Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discov 10:1842-1853, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parseghian CM, Loree JM, Morris VK, et al. : Anti-EGFR-resistant clones decay exponentially after progression: Implications for anti-EGFR re-challenge. Ann Oncol 30:243-249, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murtaza M, Dawson SJ, Tsui DW, et al. : Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497:108-112, 2013 [DOI] [PubMed] [Google Scholar]
- 48.Loree JM, Henry J, Raghav KPS, et al. : Serial circulating tumor DNA (CtDNA) monitoring in metastatic colorectal cancer (mCRC) reveals dynamic profile of actionable alterations. J Clin Oncol 39, 2021. (suppl 15; abstr 3572) [Google Scholar]
- 49.Sartore-Bianchi A, Pietrantonio F, Lonardi S, et al. : Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: The CHRONOS trial. J Clin Oncol 39, 2021. (suppl 15; abstr 3506) [Google Scholar]
- 50.Morelli MP, Overman MJ, Dasari A, et al. : Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 26:731-736, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cremolini C, Rossini D, Dell’Aquila E, et al. : Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: A phase 2 single-arm clinical trial. JAMA Oncol 5:343-350, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma M, Zhu H, Zhang C, et al. : “Liquid biopsy”—CtDNA detection with great potential and challenges. Ann translational Med 3:235, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diehl F, Schmidt K, Durkee KH, et al. : Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology 135:489-498, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Mattos-Arruda L, Mayor R, Ng CKY, et al. : Cerebrospinal fluid- derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 6:8839, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kimura H, Fujiwara Y, Sone T, et al. : EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer 95:1390-1395, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, Springer S, Mulvey CL, et al. : Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med 7:293ra104, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reckamp KL, Melnikova VO, Karlovich C, et al. : A highly sensitive and Quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol 11:1690-1700, 2016 [DOI] [PubMed] [Google Scholar]
- 58.Drusbosky L, Bilen MA, Azzi G, et al. : Blood-based tumor mutational burden from circulating tumor DNA (ctDNA) across advanced solid malignancies using a commercially available liquid biopsy assay. J Clin Oncol 39, 2021. (suppl 15; abstr 3040) [Google Scholar]
- 59.Cohen S, Hook N, Krinshpun S, et al. : SO-34 Clinical experience of a personalized and tumor-informed circulating tumor DNA assay for minimal residual disease detection in oligometastatic colorectal cancer patients. Ann Oncol 31:S229, 2020 [Google Scholar]
- 60.Kasi PM, Dayyani F, Morris V, et al. : Tumor-informed assessment of molecular residual disease and its incorporation into practice for patients with early and advanced stage colorectal cancer (CRC-MRD Consortia). J Clin Oncol 38, 2020. (suppl 15; abstr 4108) [Google Scholar]
- 61.Tarazona N, Henriksen TV, Carbonell-Asins JA, et al. : Circulating tumor DNA to detect minimal residual disease, response to adjuvant therapy, and identify patients at high risk of recurrence in patients with stage I-III CRC. J Clin Oncol 38, 2020. (suppl 15; abstr 4009) [Google Scholar]
- 62.Anandappa G, Starling N, Begum R, et al. : Minimal residual disease (MRD) detection with circulating tumor DNA (CtDNA) from personalized assays in stage II-III colorectal cancer patients in a UK multicenter prospective study (TRACC). J Clin Oncol 39, 2021. (suppl 3; abstr 102) [Google Scholar]
- 63.Shirasu H, Taniguchi H, Watanabe J, et al. : O-11 Monitoring molecular residual disease by circulating tumor DNA in resectable colorectal cancer: Molecular subgroup analyses of a prospective observational study GALAXY in CIRCULATE-Japan. Ann Oncol 32:S222-S223, 2021 [Google Scholar]
- 64.Vidal J, Casadevall D, Bellosillo B, et al. : Clinical impact of presurgery circulating tumor DNA after total neoadjuvant treatment in locally advanced rectal cancer: A biomarker study from the GEMCAD 1402 trial. Clin Cancer Res 27:2890-2898, 2021 [DOI] [PubMed] [Google Scholar]
- 65.MolDX: Minimal Residual Disease Testing for Colorectal Cancer, 2021. https://www.cms.gov/medicare-coverage-database/view/lcd.aspx?LCDId=38290&ContrId=391&ver=5&ContrVer=1&DraftContr=All&bc=AAAAAIAAIAAA&
- 66.Guardant Health Receives New York State CLEP Approval for Guardant Reveal Blood Test to Detect and Monitor Residual Disease in Patients with Early-Stage Cancer, 2021. https://investors.guardanthealth.com/press-releases/press-releases/2021/Guardant-Health-Receives-New-York-State-CLEP-Approval-for-Guardant-Reveal-Blood-Test-to-Detect-and-Monitor-Residual-Disease-in-Patients-with-Early-Stage-Cancer/default.aspx