Summary
Ovarian aging precedes that of any other mammalian organ and is the primary cause of female age-related infertility. The biological mechanisms responsible for ovarian aging remain unclear. Previous studies have been limited by their use of bulk RNA-sequencing, which masks the dynamic and heterogeneous nature of the ovary. In this study, we spatially resolved the transcriptomic landscape of ovaries from young and aged outbred mice. In total, we defined eight main ovarian cell populations, all of which were characterized by significant transcriptomic changes between young and aged samples. Further sub-cluster analysis revealed separate transcriptomes for distinct granulosa cell populations found in young versus aged mice, in addition to an oocyte sub-cluster population completely absent from aged mouse ovaries. This study provides a new perspective on mammalian ovarian aging using spatial transcriptomics to achieve deeper understanding of the localization and cell-population-specific mechanisms underlying age-related fertility decline.
Subject areas: Physiology, Cellular physiology, Omics, Transcriptomics
Graphical abstract
Highlights
-
•
Spatial transcriptomic analysis of murine ovarian aging characterizes cell populations
-
•
Distinct granulosa cell populations exist in aged mice compared to young
-
•
These are associated with the disruption of inhibins, activins, and gap junction activity
-
•
Oocyte transcriptomes captured in vivo and within follicles of varying maturity
Physiology; Cellular physiology; Omics; Transcriptomics
Introduction
The ovary serves as the primary female reproductive organ and is responsible for the production of oocytes and secretion of sex steroid hormones, which play crucial roles in the regulation of overall female fertility and endocrine activity [Richards, 2018]. The ovary is affected by natural aging more severely and earlier on than any other mammalian tissue, exhibiting significant loss of function by the time a woman enters her mid-thirties [Broekmans et al., 2009]. Ovarian aging is characterized by both quantitative and qualitative losses to the ovarian reserve, and although widely accepted as the chief cause of reproductive decline in women of advanced age (advanced maternal age; AMA ≥35 years), a deeper mechanistic understanding remains to be elucidated [Vollenhoven and Hunt, 2018]. This is increasingly exigent, as more women will suffer from age-related infertility resultant of the ongoing social trend to delay childbearing in the developed world [Shirasuna and Iwata, 2017].
A uniquely structured and highly heterogeneous organ, the ovary comprises many distinctive cell populations and a strictly maintained hierarchy of developing follicles. Each follicle contains an oocyte surrounded by layers of theca, granulosa, and cumulus cells responsible for metabolically supporting the oocyte through growth and maturation [Rimon-Dahari et al., 2016]. The ovarian stroma consists of dynamic extracellular matrices and vasculature, which continuously remodel themselves to allow for follicle growth, ovulation, corpus luteum progression, and regression [Kinnear et al., 2020]. In tissues as diverse and dynamic as the ovary, conventional bulk RNA-sequencing approaches fail to distinguish cell-type-specific changes in gene expression and the signal of rare cell types is often lost. Furthermore, follicular cells and oocytes depleted by ovarian aging create discrepancies in objectively comparing data between age groups.
In this study, we utilized young and naturally aged, outbred female mice to resolve the spatial transcriptomic landscape of mammalian ovarian aging. We identified eight main ovarian cell types based on gene expression and morphological profiling. We further defined these main cell types by characterizing their composition of unique sub-populations. Most sub-populations were significantly different in their relative proportion between young and aged mice, indicating that ovarian aging affects gene expression or cell development within each of the various ovarian compartments, causing divergence in cell population phenotypes. We observed that there are distinct granulosa cell (GC) sub-populations found within young and naturally aged antral follicles, and morphologically identified them as the inner and outer GCs based on their proximity to the oocyte. Interestingly, the identification of differentially expressed genes and pathway analysis revealed dysregulated inhibin and activin expression and aberrant gap junction activity in aged GCs.
To date, spatial transcriptomic technology has not been utilized to study mammalian ovarian biology, or the effect of aging on ovarian function and processes. Our study confirms that ovarian aging has widespread repercussions on gene expression and reveals age-related changes unique to various ovarian compartments in vivo. Additionally, our data provide insight into the relationship between ovarian aging, granulosa cells, and follicle development that may aid in the advancement of therapeutic intervention for AMA women.
Results
Aging phenotype
To confirm an age-related phenotype in our aged mice, ovaries from young (n = 4, 3–4 months) and aged (n = 4, 15–16 months) mice were stained by H&E to visualize developing follicles. Ten tissue sections from each animal were evaluated, and significantly more average follicles were observed in the young sections compared to the aged (young = 269.0 ± 33.35, aged = 77.6 ± 14.93; p < 0.01) (Figure S1). The number of primordial (young = 214.1 ± 26.55, aged = 47.02 ± 9.05; p < 0.001), primary (young = 37.39 ± 4.64, aged = 19.73 ± 3.80; p < 0.01), and secondary follicles (young = 14.55 ± 1.80, aged = 8.614 ± 1.66; p < 0.05) significantly declined in aged ovaries. Follicle attrition in the aged mice indicates that the animals aged in a manner comparable to human ovaries and are a viable model for the analysis of ovarian aging.
Spatial RNA sequencing of the aging ovary
To investigate the molecular repercussions of aging on the mammalian ovary, we performed spatially resolved transcriptomic analysis of young (n = 4, 3–4 months) and aged (n = 4, 15–16 months) mouse ovaries (Figure 1A). The manufacturer-recommended sequencing depth is a minimum of 50,000 reads per tissue-covered spot. We achieved a mean of 125,480 reads per spot, allowing for the detection of lowly expressed transcripts. The subsequent dataset consisted of 4,191 spots (n = 2,566 young, n = 1,625 aged) with >75% tissue coverage and an average of 5,348 genes detected per spot. Each circular spot covered a 55μm2 area of tissue with 100μm center-to-center between spots. Thus, each spot is expected to encompass approximately 5-10 total cells, depending on composition, resulting in approximately 20-40k total cells analyzed (n ≈ 13–26k young cells, n ≈ 8–16k aged cells). In total, 20,434 genes were identified in young ovaries and 20,685 genes were identified in aged ovaries.
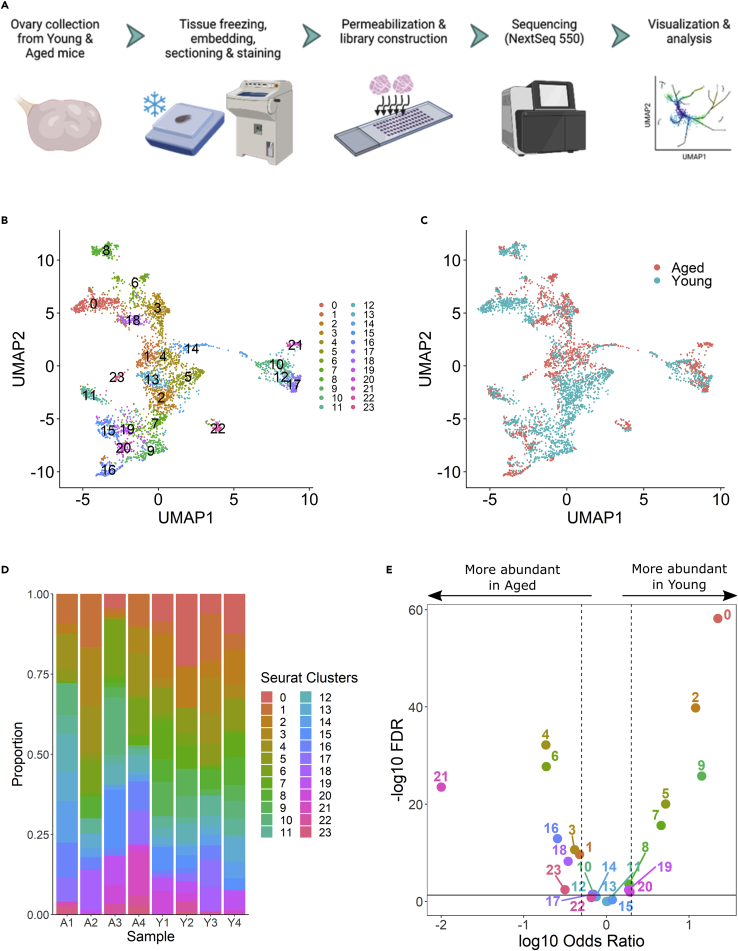
Figure 1.
Spatial transcriptome analysis of mouse aging ovary
(A) Spatial transcriptome profiling workflow depicting collection, tissue handling, library preparation of two sections per capture area (4 capture areas and eight sections total), sequencing, and analysis.
(B and C) Unbiased identification of cell-type heterogeneity and biological variance in young versus aged mouse ovaries. (B) UMAP clustering is colored by identified cell clusters, (C) and sample group (young or aged).
(D) Cluster abundances by individual mouse ovary samples.
(E) Differences of cluster abundance plotted as log10 odds ratio against adjusted p value –log10 FDR. The solid horizontal line represents the significance threshold of 0.05 and the vertical dotted lines represent a 1.5-fold change in either direction.
Clustering and cluster identification in aging ovary
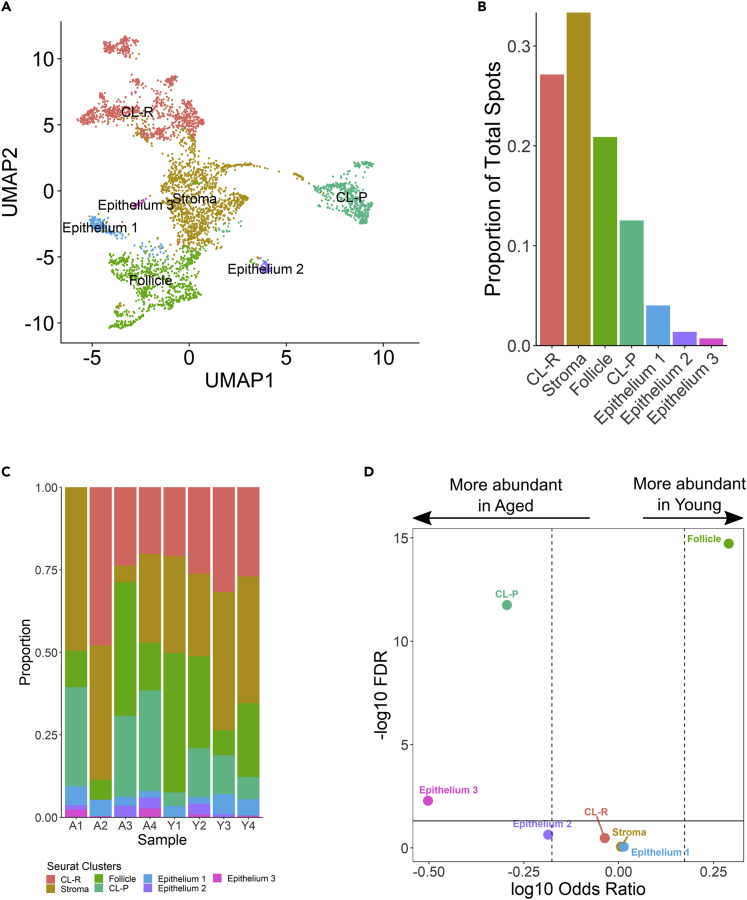
Global gene expression profiles of tissue-covered spots from young and aged ovaries across all four slides were merged and visualized using principal component analysis (PCA) (Figure S2). PCs 1, 2, 3, and 4 demonstrated marked differences in gene expression between young and aged ovaries that were confounded by capture area differences. Therefore, expression profiles for each capture area were integrated for further analysis to minimize technical batch effects. To establish a baseline profile of cell populations, we performed initial UMAP clustering on tissue-covered spots from all eight young and aged tissue sections, ultimately yielding 24 unique clusters (Figures 1B and 1C). Cluster abundances were significantly imbalanced between young and aged ovaries in 13 of 24 clusters (Figures 1D, 1E, and Table S1). Subsequently, we analyzed the top 10 genes expressed per cluster (Figure 2) in order to categorize the clusters into seven major, biologically relevant cell types: corpus luteum-regressing (CL-R), stroma, follicle, corpus luteum-progressing (CL-P), epithelium 1, epithelium 2, and epithelium 3 (Figure 3). The follicle cluster was significantly more abundant in young ovaries; this is indicative of follicle attrition in aged samples and an ovarian aging phenotype consistent with humans (Figures 3D and Table S1). Interestingly, the CL-P and epithelium 3 clusters were significantly more abundant in aged ovaries. Spatial profiling of the newly defined clusters on the original tissue sections aligned strongly with morphology (Figure 4).
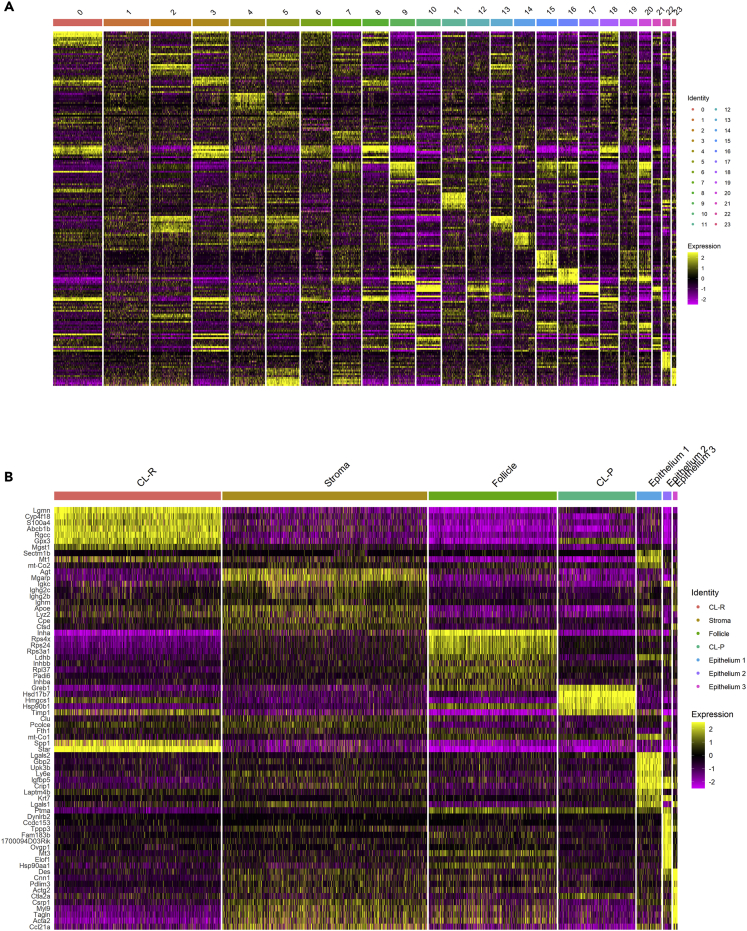
Figure 2.
Cluster-specific gene expression
(A) Heatmap shows the top 10 genes per unbiased cluster.
(B) Heatmap shows the top 10 genes per newly combined cluster. Yellow represents upregulated genes within a cluster, relative to all other clusters and purple represents downregulated genes.
Figure 3.
Major cell types found in the mouse ovary
(A) UMAP plot of new clusters defined by gene expression similarity and subsequently labeled according to their congruence with tissue morphology.
(B) Abundance of major clusters as a proportion of total tissue-covered spots.
(C) Cluster abundances by individual mouse ovary samples.
(D) Differences of cluster abundance plotted as log10 odds ratio against adjusted p value –log10 FDR. The solid horizontal line represents the significance threshold of 0.05 and the vertical dotted lines represent a 1.5-fold change in either direction.
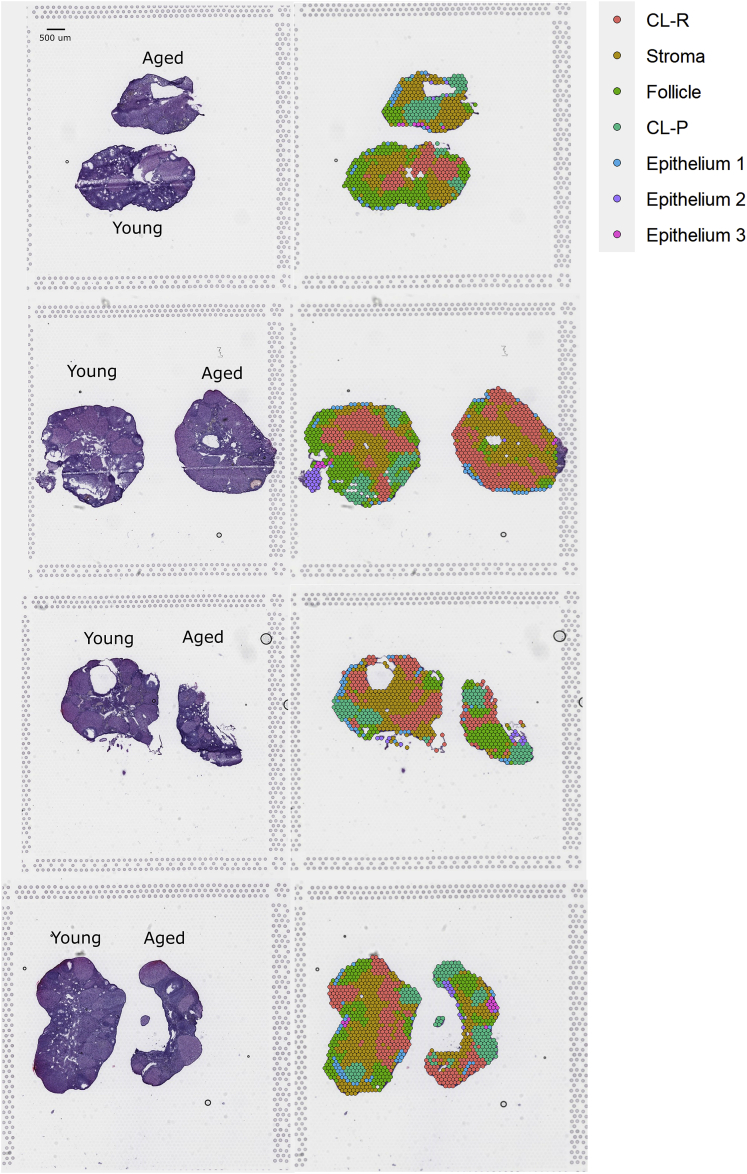
Figure 4.
Spatial mapping of defined clusters
Sections and spatial mapping of defined clusters. Sections stained by hematoxylin and eosin (left side) and spatial distribution of the newly defined, main clusters on the four young and aged ovarian tissue sections (right side). Mapping of clusters on tissue demonstrates appropriate alignment with morphology. Scale bar is 500μm.
Identification of differentially expressed genes in aging ovary
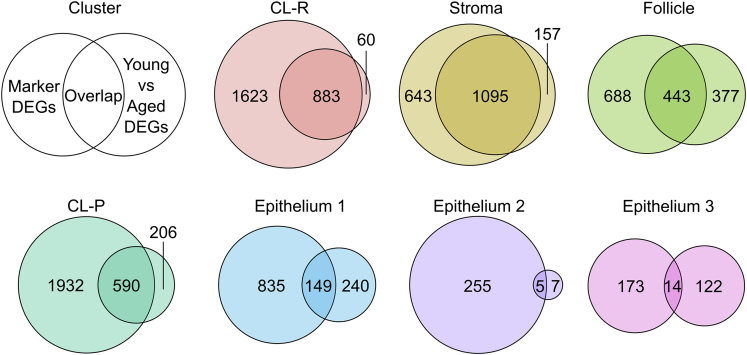
To examine global aging transcriptomic changes within individual cell types, we compared young versus aged gene expression profiles in each defined cluster. Differentially expressed genes (DEGs) were defined as those with an adjusted p value ≤ 0.05, a log2 fold-change > 1 or < −1 and expressed in at least 75% of one of the groups being tested. We identified DEGs between age groups in all clusters: 2,506 in CL-R, 1,252 in stroma, 1,434 in follicle, 796 in CL-P, 984 in epithelium 1, 12 in epithelium 2, and 188 in epithelium 3 (Table 1). The vast majority of DEGs were downregulated in aged spots relative to young spots. To determine what makes each cluster unique, we also compared each cluster to all other clusters to identify cluster-specific marker genes, using the same DEG cutoffs. Interestingly, the majority of marker genes for corpus luteum-R, stroma, follicle, and corpus luteum -P overlap with DEGs. This demonstrates that many of the genes differentially expressed with ovarian aging are the same genes that distinguish that cluster from others; thus, transcriptomic changes related to ovarian aging are largely cell-type specific (Figure 5). The top stroma marker genes coded for genes upregulated in the stroma relative to other clusters and involved in inflammation (Rela, Tnfrsf1a, Ccl21a) and immune response (Igkc, Ighg2c, Ighg2b, Ighm). The top follicle marker genes coded largely for ribosomal proteins (Rps4x, Rps24, Rps3a1, and Rlp37), and inhibin and activin subunits (Inha, Inhbb, and Inhba), all upregulated in the follicle (Table S2). Interestingly, top marker genes upregulated in epithelium 2 and 3, clusters abundant in aged ovaries relative to young, included actins (Acta2 and Actg2), cytoskeleton organizational proteins (Tagln, Pdlim3, and Fam183b), and a number of transcripts implicated in muscle contraction and motor protein function (Cnn1, Des, Dynlrb2, Myl9, and Tppp3) (Table S2).
Table 1.
Differentially expressed genes (DEGs) by cluster
| Cluster | Total genes expressed | Total DEGs (upregulated in aged) |
|---|---|---|
| CL-R | 11,103 | 2,506 (100) |
| Stroma | 10,648 | 1,252 (163) |
| Follicle | 11,434 | 820 (53) |
| CL-P | 11,844 | 796 (104) |
| Epithelium 1 | 12,010 | 984 (3) |
| Epithelium 2 | 13,005 | 12 (2) |
| Epithelium 3 | 12,787 | 188 (25) |
CL-R, Corpus Luteum-Regressing; CL-P, Corpus Luteum-Progressing.
Figure 5.
Cell-type specific differential gene expression in aging ovaries
Differentially expressed genes (DEGs) were identified in all clusters when comparing aged relative to young, defined by an adjusted p value ≤ 0.05, a log2 fold-change > 1 or < −1, and expression in at least 75% of one of the groups being tested. Marker genes for each cluster were identified using the same cutoffs. The majority of marker genes for corpus luteum-regressing (CL-R), stroma, follicle, and corpus luteum-progressing (CL-P) overlap with identified DEGs, demonstrating that many genes differentially expressed with aging are the same genes that distinguish between cell type clusters.
Gene ontology and pathway analysis of differentially expressed genes in aging ovary
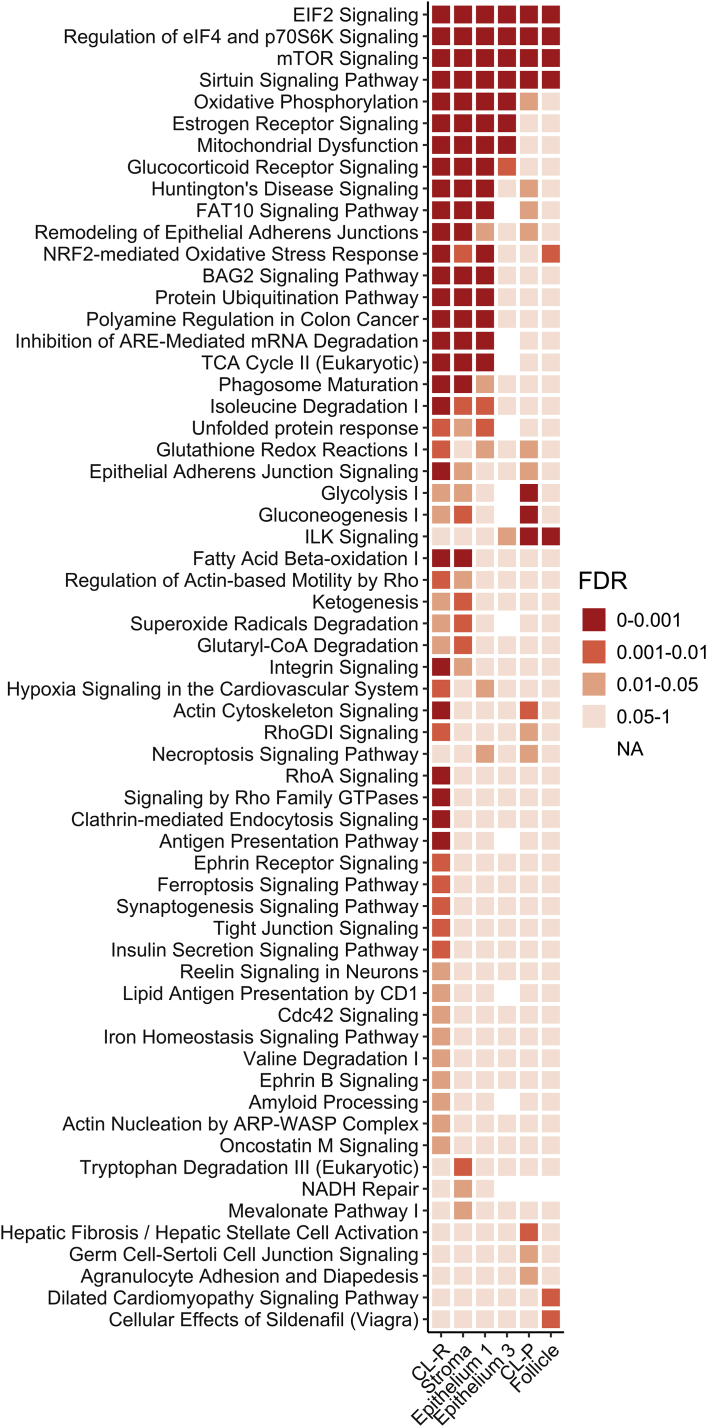
Pathway analysis was performed on DEG lists generated for all clusters using ingenuity pathway analysis (IPA) and demonstrated a shared functional aging signature among the cell types. Pathways for EIF2 signaling, regulation of eIF4 and p70S6K signaling, mTOR signaling, and sirtuin dignaling, major orchestrators of protein translation and cellular metabolism, were significantly enriched (FDR ≤0.05) in the DEGs in all clusters (Figure 6). Additionally, CL-R, stroma, and epithelium 1 shared several other significant pathways including estrogen receptor signaling, oxidative phosphorylation, and mitochondrial dysfunction, remodeling of epithelial adherens junctions, NRF2-mediated oxidative stress response, and BAG2 signaling. Furthermore, gene ontology (GO) analysis for overrepresentation was performed with upregulated and downregulated DEGs in each cell type (Table 2). Regulation of gap junction activity, antagonism of activin by follistatin, and signaling by TGF-beta family members were identified as significantly overrepresented terms in DEGs upregulated in the aged relative to the young follicle (FDR ≤0.05).
Figure 6.
Cell-type specific gene expression changes reveal dysregulated pathways in aging ovaries
Ingenuity Pathway Analysis performed on all cluster DEG lists demonstrated a shared functional aging signature among cell clusters. (corpus luteum-regressing, CL-R; corpus luteum progressing, CL-P). EIF2 signaling, regulation of eIF4 and p70S6K signaling, mTOR signaling, and sirtuin signaling were all significant (FDR ≤0.05) in all clusters. Corpus luteum-R, stroma, and epithelium 1 share several other significant pathways including estrogen receptor signaling, oxidative phosphorylation and mitochondrial dysfunction, remodeling of epithelial adherens junctions, NRF2-mediated oxidative stress response, and BAG2 signaling.
Table 2.
Gene ontology (GO) analysis of differentially expressed genes (DEGs) for overrepresentation
| Cluster | GO terms - upregulated DEGS in aged | FDR | GO terms - downregulated DEGS in aged | FDR |
|---|---|---|---|---|
| CL-R | Immune system | 8.51E-07 | Ubiquitin proteasome pathway | 3.01E-07 |
| Regulation of IGF transport and uptake by IGFBPs | 4.77E-04 | Integrin signaling pathway | 1.96E-05 | |
| Extracellular matrix organization | 1.63E-03 | Cell cycle | 5.47E-03 | |
| Stroma | Respiratory electron transport | 2.24E-14 | Eukaryotic translation initiation | 1.59E-44 |
| Innate Immune System | 6.13E-03 | Mitochondrial biogenesis | 1.79E-09 | |
| ER to Golgi Anterograde Transport | 1.23E-02 | Formation of annular gap junctions | 5.74E-05 | |
| Follicle | Regulation of gap junction activity | 9.40E-03 | Translation | 5.48E-60 |
| Antagonism of activin by follistatin | 1.17E-02 | ER quality control compartment | 1.17E-02 | |
| Signaling by TGF-beta family members | 2.04E-02 | Organelle biogenesis and maintenance | 1.48E-02 | |
| CL-P | Pink/Parkin Mediated Mitophagy | 1.29E-03 | Translation | 6.43E-58 |
| Mitophagy | 3.55E-04 | Eukaryotic translation initiation | 1.05E-50 | |
| Detoxification of Reactive Oxygen Species | 3.83E-03 | Mitochondrion translation | 3.43E-05 | |
| Epithelium1 | N/Aa | N/Aa | Mitotic cell cycle | 3.10E-06 |
| Calnexin/calreticulin cycle | 6.34E-06 | |||
| Collagen formation | 4.07E-02 | |||
| Epithelium2 | Keratan sulfate degradation | 1.01E-02 | N/Sb | N/Sb |
| Epithelium3 | N/Aa | N/Aa | Mitochondrial biogenesis | 2.36E-07 |
| Collagen biosynthesis and modifying enzymes | 8.33E-03 | |||
| Collagen formation | 2.95E-02 |
CL-R, Corpus Luteum-Regressing; CL-P, Corpus Luteum-Progressing.
Not enough upregulated DEGs for analysis.
No significant results.
Heterogeneity in aging ovary sub-populations
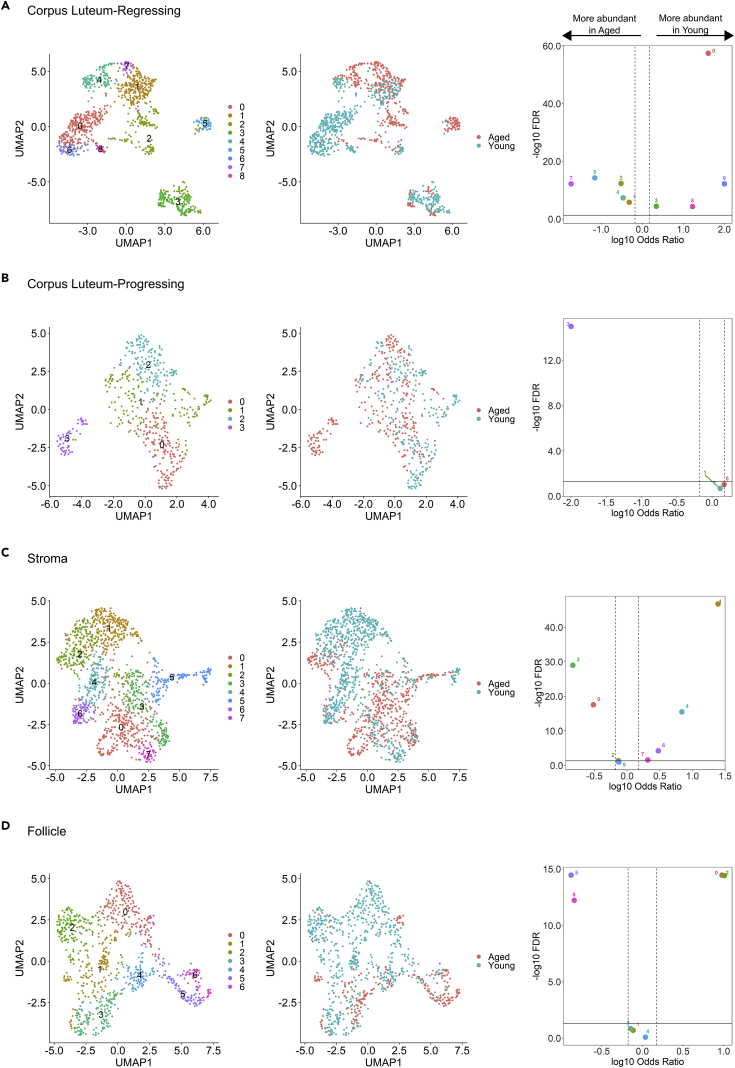
In order to elucidate sub-cell populations and further define the effect of age on major cell types, we performed sub-cluster analysis on each of the four largest cell types originally defined by more than one cluster: CL-R, CL-P, stroma, and follicle. Sub-cluster analysis of CL-R identified nine transcriptionally unique sub-populations (Figure 7A). Relative proportions of all nine populations were significantly different between young and aged (Table S1). Sub-clusters 1, 2, and 3 contained the most DEGs (Table 3); with sub-clusters one and two consisting of primarily aged spots and sub-cluster 3 of primarily young spots. Analysis of CL-P identified four transcriptionally unique sub-populations (Figure 7B), with the relative abundance of sub-cluster 3 being significantly enriched in aged (Table S1). The stroma was found to consist of eight transcriptionally unique sub-populations (Figure 7C); relative proportions of all populations were significantly different between young and aged, except for sub-clusters 2 and 5 (Table S1). Lastly, the analysis of follicles identified seven transcriptionally unique sub-populations (Figure 7D). Relative proportions of the majority of cluster populations, clusters 0, 2, 5, and 6, were significantly different between young and aged (Table S1). These results indicate the widespread impact of aging on gene expression and relative abundance of unique sub-populations within the seven previously established major cell types.
Figure 7.
Analysis of sub-cluster heterogeneity in aged ovaries
UMAP clustering of sub-populations, colored by sub-cluster (left) and age group (middle). Differences in cluster abundance are plotted as log10 odds ratio against adjusted p value –log10 FDR (right).
(A) All corpus luteum-regressing (CL-R) sub-populations are significantly different in terms of relative proportion between young and aged ovary.
(B) There are four sub-populations within the corpus luteum-progressing (CL-P) cluster, of which sub-cluster 3 is significantly more abundant in aged ovaries.
(C) Stroma consists of eight sub-populations, six of which are significantly different in their relative abundance in aged compared to young.
(D) Follicles were found to have seven transcriptionally unique sub-populations; relative proportions of sub-clusters 0, 2, 5, and 6 were significantly different between young and aged ovaries.
Table 3.
Differentially expressed genes (DEGs) within sub-clusters
| Sub-cluster | Total genes expressed | Total DEGs | Upregulated DEGs in aged | Downregulated DEGs in aged |
|---|---|---|---|---|
| Corpus luteum-regressing | ||||
| 0 | 11,617 | 2 | 1 | 1 |
| 1 | 10,819 | 76 | 45 | 31 |
| 2 | 11,098 | 2,531 | 18 | 2,513 |
| 3 | 11,834 | 52 | 4 | 48 |
| 4 | 11,172 | 0 | 0 | 0 |
| 5 | 8,458 | 0 | 0 | 0 |
| 6 | 11,432 | 11 | 0 | 11 |
| 7 | 10,497 | 0 | 0 | 0 |
| 8a | – | – | – | – |
| Corpus luteum-progressing | ||||
| 0 | 11,965 | 0 | 0 | 0 |
| 1 | 11,253 | 134 | 4 | 130 |
| 2 | 12,180 | 17 | 13 | 4 |
| 3 | 11,834 | 52 | 4 | 48 |
| Stroma | ||||
| 0 | 9,750 | 6 | 0 | 6 |
| 1 | 11,561 | 0 | 0 | 0 |
| 2 | 11,013 | 1,052 | 57 | 995 |
| 3 | 10,228 | 1 | 0 | 1 |
| 4 | 10,786 | 41 | 0 | 41 |
| 5 | 11,211 | 71 | 1 | 70 |
| 6 | 11,048 | 26 | 2 | 24 |
| 7 | 10,497 | 0 | 0 | 0 |
| Follicle | ||||
| 0 | 11,946 | 177 | 0 | 177 |
| 1 | 11,063 | 602 | 0 | 602 |
| 2 | 11,826 | 2 | 0 | 2 |
| 3 | 11,962 | 4 | 0 | 4 |
| 4 | 11,665 | 1 | 1 | 0 |
| 5 | 11,610 | 16 | 5 | 11 |
| 6 | 11,432 | 11 | 0 | 11 |
Too few spots to analyze.
Differential impacts of ovarian aging in follicle sub-clusters
Gene expression analysis revealed seven follicle sub-clusters. Of these, sub-clusters 5 and 6 were significantly more abundant in aged ovaries (Figures 7D and Table S1). Analysis of gene expression for known follicular markers, (Esr2, Fshr, Cyp11a1) suggests these sub-clusters represent granulosa cells (GCs) in antral follicles (Figure S3). This was confirmed by morphological analysis of spatial mapping of both sub-clusters (Figure S4). Also, by morphological analysis, sub-cluster 5 appears to represent inner antral GCs with closer proximity to the oocyte, while sub-cluster 6 represents outer antral GCs that make up the perimeter of the follicle. By the same standards, sub-clusters 0 and 2 represent antral follicle GCs in young ovaries with sub-cluster 0 representing the inner GCs and sub-cluster 2 comprising the outer layer of GCs. These sub-clusters are largely only found within their age group (Figure 7D), indicating that GCs within large follicles are significantly impacted by ovarian aging and that young and aged GCs have highly distinct gene expression profiles as a result. Inner GCs in particular were defined by significantly upregulated expression of Inha, Inhba, Inhbb, Fst, and Gja1 compared to all other follicular sub-clusters. Interestingly, Inha was the top marker gene by absolute fold change in both sub-cluster 5 (aged GCs) and sub-cluster 2 (young GCs), but was significantly upregulated in aged relative to young GCs. Sub-clusters 1 and 3 may be representative of early follicles and express many oocyte-specific transcripts known to be essential to early follicle activation and progression, such as Sohlh1, Zar1, and Nobox (Figure S3). Gene expression and age group abundances of these sub-clusters were less severely impacted by aging, suggesting that ovarian aging disturbs follicular growth and oocyte maturation at later stages of development rather than recruitment or quality of resting and early follicles.
Identification of differentially expressed genes in aging oocytes
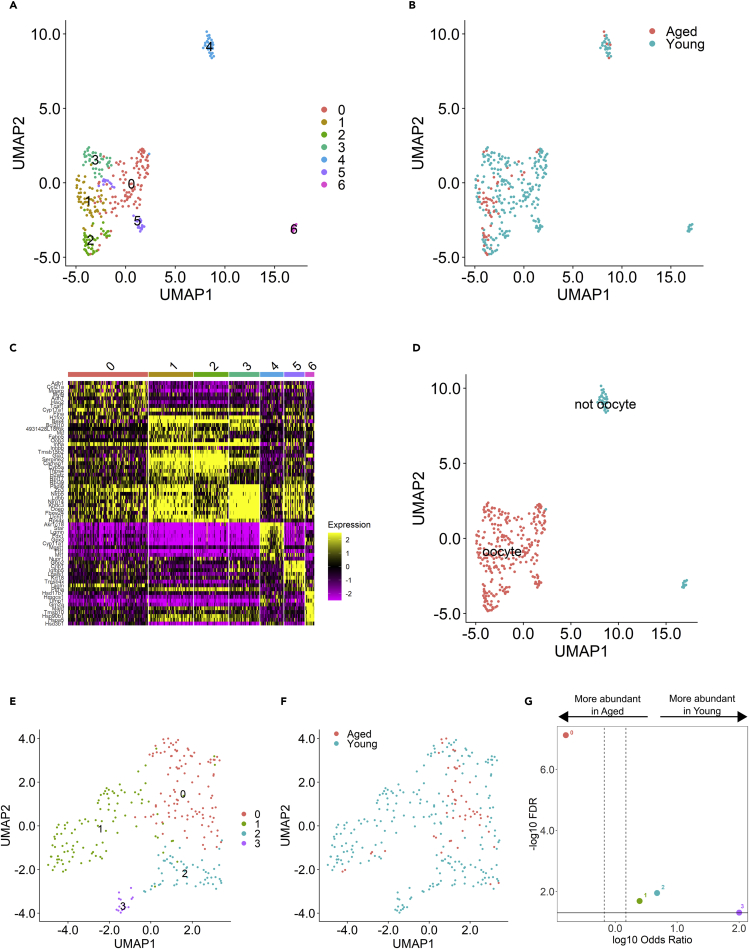
The eighth main cell type, oocyte, was determined by selection for spots expressing oocyte-specific genes, Gdf9 and Zp3. This resulted in the identification of 321 spots (young, n = 265; aged, n = 56) (Figure 8A). The disparity in the number of oocyte-representing spots between age groups confirms a diminished ovarian reserve phenotype in our aged mice (Figure 8B). We further parsed these by selecting clusters with similar gene expression. Specifically, clusters 4 and 6 were removed from the analysis because the majority of their top 10 genes were not expressed in any other cluster, suggesting that they represent a distinct cell type or mixture of cell types (Figure 8C). Spots excluded as non-oocytes may have been contaminated with follicular cell transcripts owing to overlapping morphology within the enclosed spot area (Figure 8D). To examine transcriptomic changes within aged oocytes, we used an adjusted p value ≤ 0.05 and a log2 fold-change > 1 or < −1 to reveal 73 DEGs, all of which were downregulated in aged oocytes relative to young. Pathway analysis of DEGs identified a single enriched pathway, hepatic fibrosis, containing six enriched genes. Five of these genes are targets of transforming growth factor beta-1 (TGF-β1), found to be the top upstream molecular regulator by a regulator effects analytic. TGF-β1 is predicted to be inhibited (Z score = −3.7, FDR = 2.9x10−11) in aged oocytes, and 33/73 DEGs were found to be TGF-β1 targets.
Figure 8.
Transcriptomic profiling reveals heterogeneity in aged oocytes
UMAP clustering of spots selected for expression of Gdf9 and Zp3, colored by identified cell clusters (A) and age group (B).
(C) Heatmap shows the top 10 genes per cluster; yellow represents upregulated genes within a cluster, relative to all other clusters, purple represents downregulated.
(D) Clusters determined to be representative of oocytes versus non-oocyte clusters were eliminated from analysis based on irregular gene expression.
(E-G) Identification of oocyte sub-cluster populations. (E) UMAP clustering of oocyte spots based on gene expression. (F) UMAP clustering of oocyte spots based on age group. (G) Differences of cluster abundance plotted as log10 odds ratio against adjusted p value –log10 FDR. Cluster 0 is significantly more abundant in aged ovaries, while Clusters 1, 2, and 3 are significantly more abundant in young.
Heterogeneity in aging oocyte sub-populations
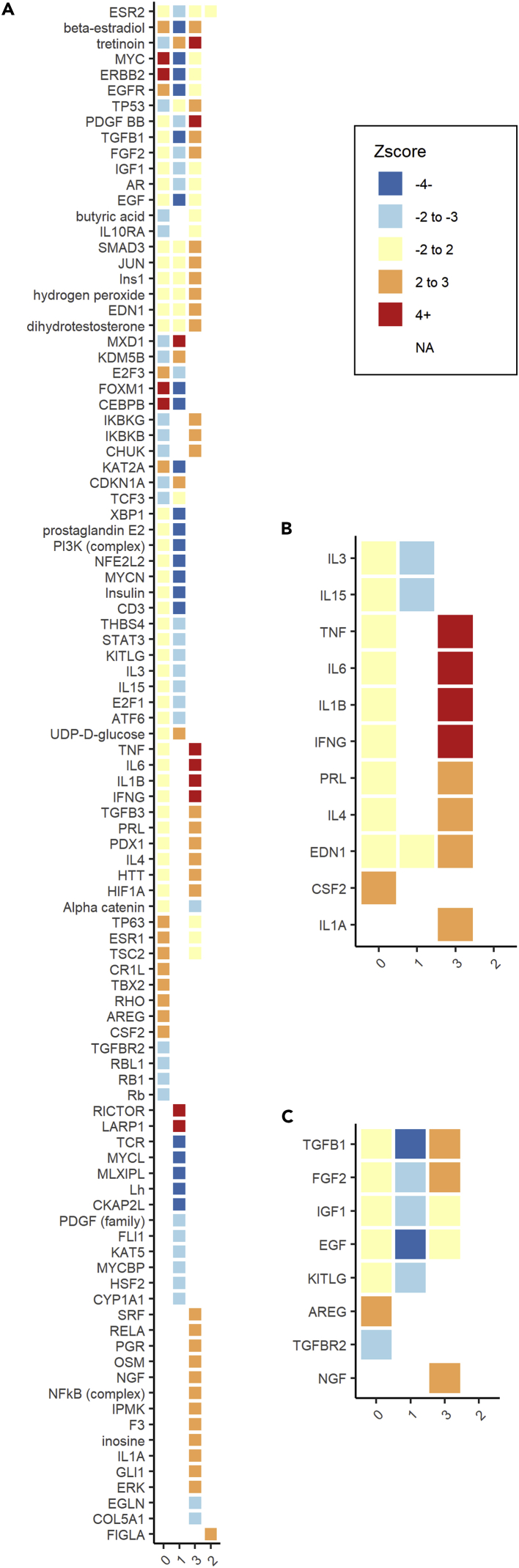
Further analysis of oocyte gene expression revealed four sub-cluster populations (Figure 8E). Three of these sub-populations (Clusters 0–2) were composed of both young and aged spots; however, the fourth sub-cluster (Cluster 3) contained only young spots (Figures 8F and 8G). Because of this, the identification of DEGs and pathway analysis were performed between sub-clusters, versus between age groups within each sub-cluster, to reveal how gene expression defines each oocyte sub-cluster relative to all other sub-clusters. We used an adjusted p value ≤ 0.05 and a log2 fold-change > 1 or < −1 to define DEGs; of which there were 309 in sub-cluster 0, 555 in sub-cluster 1, 90 in sub-cluster 2, and 99 in sub-cluster 3 relative to all the other sub-clusters. Using a regulator effects analytic, we identified upstream regulators, cytokines, and growth factors predicted to be significantly activated or inhibited between the oocyte sub-clusters (Figure 9). Each sub-cluster was highly distinct with many cluster-specific regulators. TGF-β1 was again identified as a significant upstream regulator and predicted to be activated in sub-cluster 3 (Figure 9C). Interestingly, TGF-β1 was previously predicted to be inhibited overall in aged oocytes relative to young, and sub-cluster 3 is composed of only young spots. Sub-cluster 3 also exhibited significant activation of cytokine and growth factors (Figures 9B and 9C). Sub-cluster 0, represented primarily by aged spots, showed activation of CSF2 cytokine, indicating oocyte response to a pro-inflammatory microenvironment.
Figure 9.
Upstream regulator analysis in oocyte sub-clusters
Each significant regulator is assigned a Z score; a positive score (≥2) indicates the regulator is predicted to be activated and a negative score (≤-2) indicates it’s predicted to be inhibited.
(A) Each cluster is highly distinct with cluster-specific regulators and regulators with opposite z-scores from one cluster to another.
(B) Cytokines are activated in Cluster 3.
(C) Growth factors are activated in Cluster 3 but inhibited in Cluster 1.
Discussion
The ovary is prone to rapid and early-onset aging, which is characterized by follicle attrition and diminished oocyte quality and results in loss of reproductive potential. The mechanisms behind the marked reduction in follicle pool and oocyte quality owing to aging are far from being fully understood [Broekmans et al., 2009]. The ovary is also arguably the most dynamic mammalian organ. Tightly regulated molecular mechanisms orchestrate cyclic follicular growth and attrition, oocyte maturation, ovulation, and corpus luteum endocrine function, all simultaneously occurring within the ovarian stroma. The rapidly changing ovarian landscape and vast cell-type heterogeneity have previously limited our ability to effectively study and understand the ovary on a molecular scale. These gaps in knowledge have also restricted the development and success of therapeutics; even artificial reproductive technologies (ART) are often unable to overcome the detriments of maternal age [Ubaldi et al., 2019].
Our spatial transcriptome profiling of ovarian aging in mice advances current knowledge of the relationship between unique ovarian cell types in young fertile ovaries, as well as how gene expression and cellular interactions change between ovarian cell types with age. We originally identified 24 clusters, the majority of which were significantly different in their relative abundance between young and naturally aged ovaries. This may be owing to certain cell populations appearing more predominant overall as other populations die off with age; alternatively, particular cell populations may take on aging phenotypes in the older mice leading to gene expression disparities and separation of young and aged clusters despite representing the same cell type. Ultimately, eight main cell types were defined based on gene expression and spatial mapping; these were further parsed out and defined by their composition of sub-cluster populations to gain a deeper understanding of the distinctive effects of aging within different ovarian compartments.
Specifically, the analysis identified several molecular pathways dysregulated by aging across all ovarian cell populations: EIF2 signaling, Regulation of eIF4 and p70S6K signaling, mTOR signaling, and sirtuins signaling. These pathways are interrelated and have been widely studied in the context of cellular senescence and aging [Lee et al., 2019; Papadopoli et al., 2019; Steffen and Dillin, 2016; Longo and Kennedy, 2006]. Mammalian sirtuins extend organismal lifespan by mediating age-related telomere attrition, genome integrity, DNA damage repair, and metabolism [Lee et al., 2019]. More recently, they have been established as key modulators of antioxidant and redox signaling by providing cellular protection from reactive oxygen species (ROS) [Singh et al., 2018]. Similarly, the mTOR signaling pathway has been widely implicated in processes associated with aging; including cellular senescence, immune response, autophagy, mitochondrial function, and protein homeostasis (proteostasis) [Papadopoli et al., 2019].
The same pathways have been acknowledged in the literature in the context of ovarian aging and serve as validation for the use of spatial transcriptomics in the mammalian ovary. To date, several studies have shown that sirtuins mitigate ROS accumulation otherwise associated with oocyte deterioration, granulosa cell (GC) apoptosis, and accelerated degeneration of the corpus luteum [Yang et al., 2021; Sohel et al., 2019; Wang et al., 2017; Yang et al., 2017; Prasad et al., 2016; Tiwari et al., 2015; Chaube et al., 2014]. Signaling through mTOR is associated with follicle activation, development, and oocyte maturation in response to hormone stimulation [Guo and Yu, 2019]. Overall, spatial transcriptomic profiling indicates that the early onset of the many pathways implicated in general aging and age-related diseases, including neurodegenerative disorders, metabolic diseases, and cancer, are the same ones that characterize ovarian aging and contribute to female reproductive decline [Carafa et al., 2012].
Transcriptome profiling also revealed that ovarian aging is characterized by significant changes in the epithelium and stroma. We observed that the aged stroma takes on an inflammatory state characterized by increased expression of marker genes involved in inflammation (Rela, Tnfrsf1a, Ccl21a) and immune response (Igkc, Ighg2c, Ighg2b, Ighm). These findings are consistent with the occurrence of a recently defined phenotype of mammalian aging known as “inflammaging,” which is characterized by chronic, low-grade inflammation [Huang et al., 2019; Franceschi and Campisi, 2014]. We will not deeply discuss the repercussions of inflammaging here, as recent studies have explored inflammaging in the context of ovarian aging [Zhang et al., 2020]; however, this data provides important validation for the efficacy of spatial transcriptomic technologies in ovarian research. Additionally, organelle localization was enriched in the gene ontology and identified significantly differentially expressed genes involved in mitochondria transport and organization (Mgarp, upregulated in aged; Dnm2, Dynll2, and Vapa, downregulated in aged). Intriguingly, MGARP contributes to steroidogenesis through a feedback loop regulated by hormones derived from the hypothalamic-pituitary-gonadal (HPG) axis [Zhou et al., 2011]. Its aberrant expression leads to severe mitochondrial fragmentation, aggregation, and decrease in cellular content of mtDNA, with demonstrated repercussions for proper steroidogenesis [Matsumoto et al., 2009].
Furthermore, we found ovarian aging to be associated with increased heterogeneity in the epithelium. The ovarian surface epithelium (OSE) is a layer of squamous and cuboidal cells that envelopes the ovary [Edmondson et al., 2002]. Directly beneath the OSE, the extracellular matrix (ECM) is situated to provide the necessary structural and biochemical support. Three unique epithelial cell populations were defined by spatial transcriptome clustering of young and naturally aged ovaries. Two of the three epithelial clusters were more significantly abundant in naturally aged ovaries and morphologically consistent with the thickening of the epithelium [Briley et al., 2016; Mara et al., 2020]. Further analysis of DEGs upregulated in aged revealed Keratan sulfate degradation as the only significantly overrepresented term. Keratan sulfate (KS) is one of the many glycosaminoglycans (GAG) found in ECM [Ahmad and Rosenfeld, 1999]. Degradation of GAGs in the ECM can disrupt the structural integrity of the tissue and result in further degradation. Additionally, nitric oxide (NO), which is generally elevated in response to inflammation, has been shown to mediate GAG degradation [Ahmad and Rosenfeld, 1999]. Thus, in the naturally aged ovary, ubiquitous inflammation leading to elevated NO may contribute to reduced structural integrity and insufficient reconstruction of the ECM and OSE during folliculogenesis and post-ovulation.
Lastly, our investigation of alterations within the spatial transcriptome of follicles and oocytes, the functional units of female reproduction, revealed seven and four sub-populations of follicular cells and oocytes, respectively. Curiously, there were four significant follicle sub-clusters, two more abundant in aged relative to young, and two more abundant in young relative to aged. These sub-populations were highly distinct between age groups and most significantly affected by aging compared to other follicular sub-populations. Gene expression and morphological analysis suggested these sub-clusters represent the inner and outer layers of granulosa cells (GCs), based on proximity to the oocyte, within antral follicles. Previous studies have been limited by their lack of localization data, which prohibited them from distinguishing sub-cell types within a known population.
Interestingly, gene ontology analysis revealed antagonism of activin by follistatin, TGF-beta signaling pathway, Apoptosis signaling pathway, and regulation of gap junction activity to be enriched in aged female GCs. In the ovary, these pathways are responsible for mediating crucial intercellular communications between GCs and the developing oocyte. Specifically, inner GCs of aged antral follicles were enriched for Inha, Inhba, Inhbb, Fst, and Gja1 relative to all other follicular sub-clusters. Oocytes build intimate contacts with their surrounding somatic cells in the follicle; these points of granulosa cell-oocyte contact are crucial for oocyte development and are largely dependent on two specific gap junction proteins, Gja1 and Gja4 [Kidder and Mhawi, 2002]. Connexin-37 (CX37), encoded by Gja1, constitutes gap junctions between the oocyte and immediately surrounding cumulus cells, whereas Gja4 codes for Connexin-43 (CX43) and facilitates communication between layers of GCs. This coupling permits GCs to provide the growing oocyte with nucleotides, amino acids, and energy substrates, as well as allowing the oocyte to dictate its somatic microenvironment via a myriad of oocyte-secreted factors [El-Hayek and Clarke, 2015].
Furthermore, inhibins and activins, encoded by Inha, Inhba, and Inhbb, are evolutionarily conserved members of the TGF-β family also observed to be significantly differentially expressed in aged antral follicles. Inhibin subunits α and βA were significantly overexpressed in aged GCs versus any other follicular sub-populations. Follistatin (Fst), an activin antagonist, was also significantly upregulated in the GCs of aged antral follicles. As major secretory products of GCs, inhibins, and activins exhibit nearly directly opposite biological effects, acting in mammalian females to inhibit and enhance pituitary secretion of follicle-stimulating hormone (FSH), respectively [Knight et al., 2012]. In the ovary, FSH is responsible for stimulating the growth and recruitment of immature follicles, as well as promoting the survival of growing antral follicles through interaction with its GC-expressed receptor [Orlowski and Sarao, 2022]. It has also recently been shown that FSH increases steady-state levels of mRNAs encoding the principal follicular connexins, CX37 and CX43, thus augmenting gap junctional communication between GCs and oocytes during late folliculogenesis [El-Hayek and Clarke, 2015]. Altogether, these data may suggest a new mechanism for the loss of oocyte quality associated with female aging. Age-related dysregulation of the activin-inhibin ratio in the later stages of folliculogenesis may disrupt proper FSH secretion and GC-oocyte gap junctional communications central to oocyte development and competence.
Deeper analysis of oocyte sub-populations revealed cytokine activation in both age groups. The majority of this activation is predicted in young -only Cluster 3 (Figure 9B). Cytokines are crucial regulators of normal ovarian development and function. They are responsible for orchestrating a permissive, supportive environment for follicle growth and selection during folliculogenesis and ovulation [Field et al., 2014]. Given the intricate network of intercellular and paracrine signaling among the oocyte and the surrounding follicle, the spatiotemporal regulation of cytokine processes is likely tightly regulated. Additionally, we found that TGF-β1 signaling was predicted to be significantly inhibited across all identified naturally aged oocytes and upregulated in the oocyte sub-cluster population containing only young oocyte expression spots. It is known that TGF-β1 is not expressed by the oocyte itself, but rather in the surrounding granulosa cells where it mediates signaling cascades via natriuretic peptide type C (NPPC) responsible for maintaining meiotic arrest within the oocyte until activation by luteinizing hormone (LH) surge [Yang et al., 2019]. Aberrant resumption of meiosis prior to cytoplasmic maturation or chromosome alignment may be a contributing factor to the loss of oocyte quality and subsequent reduced reproductive potential as a result of ovarian aging.
In summary, our spatial transcriptomic analysis offers insight into the molecular mechanisms contributing to ovarian aging. Through the comparison of sub-cluster populations within and between age groups, we piece together a more complete picture of the ovary and the consequences of aging for different cell populations in vivo. Our data will aid in the further elucidation of the precise relationship between ovarian aging and female reproductive decline, as well as identifying new targets for the therapeutic restoration of fertility.
Limitations of the study
This study is primarily limited by the sample size (n = 4) of young and aged animals used for spatial transcriptomic analysis. As aged mice have fewer follicles and oocytes within their ovaries, our ability to thoroughly define this population of cells in such a small sample size was limited. We also observed a few limitations with the spatial transcriptomics platform. First, our study is limited by the fact that the ovary is a heterogeneous, three-dimensional organ of which we are only analyzing a single slice from each animal. The use of two sections per capture area revealed the heterogeneity of each capture area, which was highlighted in the initial PCA analysis of the data (Figure S2). Because different mice were used for each capture area, the impact of the capture area relative to the heterogeneity between mice and the ovary cannot be fully evaluated. Additionally, the platform was also considerably limited in its ability to target small and sparsely occurring morphological features, such as follicles and oocytes. Spatial mapping of the oocyte sub-clusters to the tissue sections did not directly align with manually identified germ cells (Figure S5). We used known cell-type specific markers to validate populations identified by the spatial transcriptomics platform; however, further validation of sequencing could be performed by in situ hybridization or similar techniques. Many spots assigned to the oocyte cluster by gene expression appeared morphologically to be follicular cells while some spots visibly containing oocytes were excluded from the oocyte cluster completely. Furthermore, some follicle sub-clusters highly expressed oocyte-specific transcripts, indicating contamination owing to overlapping morphology within the enclosed spot area. These discrepancies are likely owing to the size and spacing of the spots compared to follicles and oocytes; each circular gene expression spot covers 55μm area of tissue, with about 45μm of uncovered space between spots. This presents limitations when assessing follicles, which range from 50μm (primary) to 700μm (pre-ovulatory) in diameter, and even smaller oocytes, falling anywhere between 15 and 80μm diameter depending on development [Xiao et al., 2015]. Without being able to control the size or alignment of gene expression spots on the tissue, it is unavoidable that follicular cells and oocytes were contained within the same spots but assigned to single clusters.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| Visium Spatial Tissue Optimization Slide & Reagents Kit | 10x Genomics | 1,000,193 |
| Visium Spatial Gene Expression Slide & Reagents Kit | 10x Genomics | 1,000,187 |
| Deposited data | ||
| Raw and processed data | This paper | GSE188257 |
| Experimental models: Organisms/strains | ||
| CD-1 Mice | Charles River | CD-1® IGS Mouse |
| Software and algorithms | ||
| Space Ranger 1.2 | 10x Genomics | https://support.10xgenomics.com/spatial-gene-expression/software/downloads/latest |
| Other | ||
| Analysis code | This paper | https://github.com/haywoodm/spatial_transcriptome_ovary.git |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Mandy Katz-Jaffe (mandyk@colocrm.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Animals
All animal experiments were approved by the Fertility Laboratories of Colorado Ethics in Research Committee. Outbred CD-1 female mice were obtained from Charles River Laboratories (Wilmington, MA, USA) at 3–4 weeks of age. Mice were housed at a controlled photoperiod (14h light, 10h dark), temperature (20°C-26°C), and humidity (30%–40%), with food and water available ad libitum. Females were naturally aged to represent young (Young; 3–4 months old) and AMA (Aged; 15–16 months old) populations. Reproductive function in female mice is lost at around 15 months, while in women menopause occurs at an average age of 50. Therefore, we aimed at 13-14 months to reflect human reproductive aging of women in their forties. All animals were sacrificed via cervical dislocation.
Method details
Tissue processing
Ovaries were retrieved from Young and Aged females for spatial transcriptomic analysis for a total of 8 ovary sections from 8 mice (n = 4 per age group). Ovaries were rinsed in cold PBS, cryo-preserved in TissueTek Optimal Cutting Temperature (OCT) compound (VWR International), and stored at −80°C in an airtight container, as recommended by the manufacturer protocol (10x Genomics, Visium Spatial, CG000240 Rev C). Sections were selected for sequencing based on previous optimization of the sectioning depth required to reach the approximate midpoint of each ovary, based on the widest diameter region of the tissue.
Hematoxylin & eosin staining for follicle counting
One ovary per female was fixed in 4% paraformaldehyde (PFA) (Young, n = 4; Aged, n = 5) for 48–72 h and embedded in Formula R paraffin using tissue cassettes (Leica Biosystems). Embedded tissue was cut pole-to-pole in serial 8μm sections and mounted onto positively charged glass slides. Sections were melted for 1 h at 62°C before staining with hematoxylin and counter-staining with eosin. To generate representative follicle counts for Young versus Aged females, ten evenly spaced tissue sections per ovary were selected. A researcher blinded to the treatment groups manually counted and classified all follicles with an oocyte appearing in plane. Follicles were classified as either primordial, primary, secondary, or antral. Total follicle counts and counts per follicular classification were summed and averaged for each ovary. Average counts and classifications for Young versus Aged ovaries were compared by Student’s t test with p < 0.05 considered to be significant.
Spatial transcriptomics
Before proceeding with the full spatial transcriptomics protocol, a tissue optimization experiment was performed to identify the optimal length of permeabilization time for maximum RNA capture (10x Genomics, Visium Spatial, CG000238 Rev A). Briefly, ovaries were sectioned in a pre-cooled cryostat at 10uM thickness onto seven 8 × 8mm capture areas on a Visium Tissue Optimization Slide (10x Genomics) containing oligonucleotides for mRNA capture. Tissue was permeabilized for various amounts of time before captured RNA underwent a reverse transcription (RT) reaction in the presence of fluorescently labeled nucleotides. Fluorescent cDNA footprints were generated and analyzed using the Nikon Ti-E Fluorescent Microscope with an Andor Zyla 4.2 camera to reveal 18 min as the optimum permeabilization time for mouse ovaries (Figure S6).
Young and Aged ovary sections were then processed for complete spatial transcriptomic analysis as per the manufacturer’s instruction (10x Genomics, Visium Spatial, CG000239 Rev D). Tissue sections were cut onto a Visium Gene Expression slide with four 6.5 × 6.5mm capture areas each consisting of 5,000 oligo-barcoded spots. One Young and one Aged ovary section were mounted adjacent to each other within each capture area, effectively increasing the number of technical replicates on each Gene Expression slide (n = 4 per age group). The slide underwent methanol fixation and hematoxylin and eosin (H&E) staining with immediate imaging at 40x magnification on the Aperio AT2 (Leica Biosystems) (Figure S7). Tissue was then subjected to permeabilization with proprietary enzyme (18 min), reverse transcription and second strand synthesis, all performed on the slide. Amplification of cDNA was executed by qPCR using the KAPA SYBR FAST-qPCR kit (KAPA Biosystems) and analyzed on the Veriti Thermal Cycler (Applied Biosystems). Amplified cDNA was quantified using a Bioanalyzer High Sensitivity Chip (Agilent).
Following library construction, as per manufacturer’s instructions, libraries were quantified using the KAPA-Illumina PCR quantification kit (KAPA Biosystems) and pooled at 4nM concentration with a sample ratio corresponding to the slide surface area of tissue coverage obtained from the H&E imaging. Pooled, paired-end libraries were sequenced on a NextSeq 550 (Illumina) at the Massachusetts Institute of Technology using a 75nt NextSeq kit (Illumina) loaded at a concentration of 1.8p.m. A total of four capture areas and eight tissue sections were sequenced on one Visium Gene Expression Slide.
Data processing, clustering, and identification of cell types
Sequencing data was processed using the BMC/BCC 1.8 pipeline (OpenWetware, 2020), which includes Space Ranger 1.2 (10x Genomics). Space Ranger is a set of analysis pipelines for processing Spatial Transcriptomic data. Feature barcode matrices and tissue images were uploaded into R and processed using Seurat version 4.0.2 in general accordance with their integrative guided analysis. Each tissue image slide was divided according to Aged and Young slices. Spots corresponding to less than approximately 25% tissue or to unidentifiable tissue residue were discarded. The mitochondrial content was calculated for each spot and was used as a regression variable in SCTransform normalization. Individual slides were then integrated using 3,000 features. Integration anchors were identified using the “SCT” normalization method. Linear dimension reduction was performed using the following parameters: RunPCA (npcs = 50), FindNeighbors (dims = 1:50, k.param = 10), FindClusters (resolution = 0.9), RunUMAP (dims = 1:50), where the elbow method was used to help determine the number of dimensions to consider. Uniform manifold approximation and projection (UMAP) clustering yielded 24 unique clusters. Significant differences between Young and Aged cluster proportions were calculated using Fisher Exact Test followed by Benjamini-Hochberg p value adjustment. All results are listed in Table S1. Significance was defined as clusters with and FDR ≤0.05 and an odds ratio ≥1.5 or ≤ −1.5. Odds ratios of ±infinity were plotted as ±2. The top 10 positive markers per cluster were identified using FindAllMarkers and logistic regression. The slide was set as a latent variable and genes detected in at least 10% of one population were considered expressed. Seven cell type identities were assigned based on shared gene expression trends.
Differential expression and pathway analysis within and between cell types
Spots within each cell type cluster were stratified by Aged and Young. Differentially expressed genes (DEGs) between the two age groups were calculated using FindMarkers similar to before, with the following parameters: ident.1 – “Aged,” group.by = “group,” subset.ident = “subset,” and logfc.threshold = 0 to output all expressed genes. Statistical significance for differential expression was defined by an adjusted p value ≤ 0.05, a log2 fold change ≥1 or ≤ -1, and ≥75% of spots in one group (pct.1 or pct.2) expressing the gene. Marker genes for each cell cluster were identified similarly, with ident.2 = NULL. Ingenuity Pathway Analysis (IPA, Qiagen) was used to test for biological enrichment, and statistical significance was defined by adjusted p values ≤ 0.05.
Sub-cluster analysis of cell types
Individual cell types were subset from the original dataset and re-clustered using methods similar to before with the following parameters: RunPCA (npcs = 30), FindNeighbors (dims = 1:30, k.param = 20), FindClusters (resolution = 0.7), RunUMAP (dims = 1:30). Cluster proportions, differential gene expression and pathway analysis was completed as described above for initial cell type identification. Oocytes were identified by sub-setting all spots expressing both Gdf9 and Zp3 and FindClusters (resolution = 0.3), RunUMAP (dims = 1:10). Oocyte identity was assigned based on shared gene expression trends. Ingenuity Pathway Analysis (IPA, Qiagen) was used to test for biological enrichment between oocyte sub-cluster populations, and statistical significance was defined by adjusted p values ≤ 0.01 and an upstream regulator Z score ≥2 or ≤ -2.
Gene ontology analysis of DEGs
Gene ontology (GO) overrepresentation analysis was performed using the PANTHER Classification System version 16.0 statistical enrichment test (http://pantherdb.org/). DEG lists generated for all clusters in Young versus Aged ovaries were analyzed by means of their Ensembl gene identifier and fold change, and then annotated using the “Reactome pathways” dataset with Bonferroni correction (FDR≤05).
Acknowledgments
We acknowledge Kathleen Cormier and Dr. Stuart Levine at Massachusetts Institute of Technology for their expertise and assistance in optimizing and executing the 10x Genomics Visium Spatial Transcriptomics protocol. All funding for this research was provided internally by CCRM Fertility.
Author contributions
JER designed and conducted experiments, performed data interpretation, and wrote the article; MEH performed raw sequencing data processing and bioinformatic analysis; SLL assisted in designing and conducting experiments; WBS participated in critical review and discussion; MKJ conceived the study, performed critical discussion, and edited the article.
Declaration of interests
The authors declare no competing interests.
Published: August 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104819.
Supplemental information
and S2
Data and code availability
-
•
Raw sequencing data, images, and processed data have been deposited in the Gene Expression Omnibus (GEO) repository under accession GEO: GSE188257.
-
•
All original code has been deposited at GitHub: https://github.com/haywoodm/spatial_transcriptome_ovary.git.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ahmad S.M., Rosenfeld L. Degradation of keratan sulfate, an extracellular matrix glycosaminoglycan, by derivatives of nitric oxide. Pediatr. Res. 1999;45:180. doi: 10.1203/00006450-199904020-01071. [DOI] [Google Scholar]
- Briley S.M., Jasti S., McCracken J.M., Hornick J.E., Fegley B., Pritchard M.T., Duncan F.E. Reproductive age-associated fibrosis in the stroma of the mammalian ovary. Reproduction. 2016;152:245–260. doi: 10.1530/REP-16-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekmans F.J., Soules M.R., Fauser B.C. Ovarian aging: mechanisms and clinical consequences. Endocr. Rev. 2009;30:465–493. doi: 10.1210/er.2009-0006. [DOI] [PubMed] [Google Scholar]
- Carafa V., Nebbioso A., Altucci L. Sirtuins and disease: the road ahead. Front. Pharmacol. 2012;3:4. doi: 10.3389/fphar.2012.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaube S.K., Shrivastav T.G., Tiwari M., Prasad S., Tripathi A., Pandey A.K. Neem (Azadirachta indica L.) leaf extract deteriorates oocyte quality by inducing ROS-mediated apoptosis in mammals. SpringerPlus. 2014;3:464. doi: 10.1186/2193-1801-3-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson R.J., Monaghan J.M., Davies B.R. The human ovarian surface epithelium is an androgen responsive tissue. Br. J. Cancer. 2002;86:879–885. doi: 10.1038/sj.bjc.6600154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hayek S., Clarke H.J. Follicle-stimulating hormone increases gap junctional communication between somatic and germ-line follicular compartments during murine oogenesis. Biol. Reprod. 2015;93:47. doi: 10.1095/biolreprod.115.129569. [DOI] [PubMed] [Google Scholar]
- Field S.L., Dasgupta T., Cummings M., Orsi N.M. Cytokines in ovarian folliculogenesis, oocyte maturation and luteinisation. Mol. Reprod. Dev. 2014;81:284–314. doi: 10.1002/mrd.22285. [DOI] [PubMed] [Google Scholar]
- Franceschi C., Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A. Biol. Sci. Med. Sci. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- Guo Z., Yu Q. Role of mTOR signaling in female reproduction. Front. Endocrinol. 2019;10:692. doi: 10.3389/fendo.2019.00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Hu C., Ye H., Luo R., Fu X., Li X., Huang J., Chen W., Zheng Y. Inflamm-aging: a new mechanism affecting premature ovarian insufficiency. J. Immunol. Res. 2019:8069898. doi: 10.1155/2019/8069898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder G.M., Mhawi A.A. Gap junctions and ovarian folliculogenesis. Reproduction. 2002;123:613–620. doi: 10.1530/rep.0.1230613. [DOI] [PubMed] [Google Scholar]
- Kinnear H.M., Tomaszewski C.E., Chang F.L., Moravek M.B., Xu M., Padmanabhan V., Shikanov A. The ovarian stroma as a new Frontier. Reproduction. 2020;160:R25–R39. doi: 10.1530/REP-19-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight P.G., Satchell L., Glister C. Intra-ovarian roles of activins and inhibins. Mol. Cell. Endocrinol. 2012;359:53–65. doi: 10.1016/j.mce.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Lee S.H., Lee J.H., Lee H.Y., Min K.J. Sirtuin signaling in cellular senescence and aging. BMB Rep. 2019;52:24–34. doi: 10.5483/BMBRep.2019.52.1.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V.D., Kennedy B.K. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Mara J.N., Zhou L.T., Larmore M., Johnson B., Ayiku R., Amargant F., Pritchard M.T., Duncan F.E. Ovulation and ovarian wound healing are impaired with advanced reproductive age. Aging. 2020;12:9686–9713. doi: 10.18632/aging.103237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T., Minegishi K., Ishimoto H., Tanaka M., Hennebold J.D., Teranishi T., Hattori Y., Furuya M., Higuchi T., Asai S., et al. Expression of ovary-specific acidic protein in steroidogenic tissues: a possible role in steroidogenesis. Endocrinology. 2009;150:3353–3359. doi: 10.1210/en.2008-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OpenWetWare BioMicroCenter:Software BMC-BCC Pipeline. https://openwetware.org/mediawiki/index.php?title=BioMicroCenter:Softwareandoldid=1093116
- Orlowski M., Sarao M.S. StatPearls. StatPearls Publishing; 2022. Physiology, follicle stimulating hormone.https://www.ncbi.nlm.nih.gov/books/NBK535442/ [PubMed] [Google Scholar]
- Papadopoli D., Boulay K., Kazak L., Pollak M., Mallette F., Topisirovic I., Hulea L. mTOR as a central regulator of lifespan and aging. F1000Res. 2019;8 doi: 10.12688/f1000research.17196.1. F1000 Faculty Rev-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S., Tiwari M., Pandey A.N., Shrivastav T.G., Chaube S.K. Impact of stress on oocyte quality and reproductive outcome. J. Biomed. Sci. 2016;23:36. doi: 10.1186/s12929-016-0253-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J.S. The ovarian cycle. Vitam. Horm. 2018;107:1–25. doi: 10.1016/bs.vh.2018.01.009. [DOI] [PubMed] [Google Scholar]
- Rimon-Dahari N., Yerushalmi-Heinemann L., Alyagor L., Dekel N. Ovarian folliculogenesis. Results probl. Cell. Differ. 2016;58:167–190. doi: 10.1007/978-3-319-31973-5_7. [DOI] [PubMed] [Google Scholar]
- Shirasuna K., Iwata H. Effect of aging on the female reproductive function. Contracept. Reprod. Med. 2017;2:23. doi: 10.1186/s40834-017-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh C.K., Chhabra G., Ndiaye M.A., Garcia-Peterson L.M., Mack N.J., Ahmad N. The role of sirtuins in antioxid. Antioxid. Redox. Signal. 2018;28:643–661. doi: 10.1089/ars.2017.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohel M.M.H., Akyuz B., Konca Y., Arslan K., Sariozkan S., Cinar M.U. Oxidative stress modulates the expression of apoptosis-associated microRNAs in bovine granulosa cells in vitro. Cell Tissue Res. 2019;376:295–308. doi: 10.1007/s00441-019-02990-3. [DOI] [PubMed] [Google Scholar]
- Steffen K.K., Dillin A. A ribosomal perspective on proteostasis and aging. Cell Metabol. 2016;23:1004–1012. doi: 10.1016/j.cmet.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Tiwari M., Prasad S., Tripathi A., Pandey A.N., Ali I., Singh A.K., Shrivastav T.G., Chaube S.K. Apoptosis in mammalian oocytes: a review. Apoptosis. 2015;20:1019–1025. doi: 10.1007/s10495-015-1136-y. [DOI] [PubMed] [Google Scholar]
- Ubaldi F.M., Cimadomo D., Vaiarelli A., Fabozzi G., Venturella R., Maggiulli R., Mazzilli R., Ferrero S., Palagiano A., Rienzi L. Advanced maternal age in IVF: still a challenge? The present and the future of its treatment. Front. Endocrinol. 2019;10:94. doi: 10.3389/fendo.2019.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollenhoven B., Hunt S. Ovarian ageing and the impact on female fertility. F1000Res. 2018;7 doi: 10.12688/f1000research.16509.1. F1000 Faculty Rev-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., He G., Chen M., Zuo T., Xu W., Liu X. The role of antioxidant enzymes in the ovaries. Oxid. Med. Cell. Longev. 2017:4371714. doi: 10.1155/2017/4371714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Duncan F.E., Bai L., Nguyen C.T., Shea L.D., Woodruff T.K. Size-specific follicle selection improves mouse oocyte reproductive outcomes. Reproduction. 2015;150:183–192. doi: 10.1530/REP-15-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Xie Y., Yang D., Ren D. Oxidative stress-induced apoptosis in granulosa cells involves JNK, p53 and Puma. Oncotarget. 2017;8:25310–25322. doi: 10.18632/oncotarget.15813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Chen Y., Liu Y., Xing Y., Miao C., Zhao Y., Chang X., Zhang Q. The role of oxidative stress and natural antioxidants in ovarian aging. Front. Pharmacol. 2020;11:617843. doi: 10.3389/fphar.2020.617843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhang Y., Xu X., Li J., Yuan F., Bo S., Qiao J., Xia G., Su Y., Zhang M. Transforming growth factor-β is involved in maintaining oocyte meiotic arrest by promoting natriuretic peptide type C expression in mouse granulosa cells. Cell Death Dis. 2019;10:558. doi: 10.1038/s41419-019-1797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Schlamp F., Huang L., Clark H., Brayboy L. Inflammaging is associated with shifted macrophage ontogeny and polarization in the aging mouse ovary. Reproduction. 2020;159:325–337. doi: 10.1530/REP-19-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Wang Y., Qi S., Wang J., Zhang S. The expression of a mitochondria-localized glutamic acid-rich protein (MGARP/OSAP) is under the regulation of the HPG axis. Endocrinology. 2011;152:2311–2320. doi: 10.1210/en.2011-0050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and S2
Data Availability Statement
-
•
Raw sequencing data, images, and processed data have been deposited in the Gene Expression Omnibus (GEO) repository under accession GEO: GSE188257.
-
•
All original code has been deposited at GitHub: https://github.com/haywoodm/spatial_transcriptome_ovary.git.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.