Abstract
Rationale and Objectives:
The objective of this study was to demonstrate improvement in distinguishing between benign lesions and luminal A breast cancers in a large clinical breast magnetic resonance imaging database by using quantitative radiomics over maximum linear size alone.
Materials and Methods:
In this retrospective study, 212 benign lesions and 296 luminal A breast cancers were automatically segmented from dynamic contrast-enhanced breast magnetic resonance images. Thirty-eight radiomic features were extracted. Tenfold cross validation was performed to assess the ability to distinguish between lesions and cancers using maximum linear size alone and lesion signatures obtained with stepwise feature selection and a linear discriminant analysis classifier including and excluding size features. Area under the receiver operating characteristic curve (AUC) was used as the figure of merit.
Results:
For maximum linear size alone, AUC and 95% confidence interval was 0.797 (0.754, 0.835) compared to 0.846 (0.808, 0.875) (P=.005) and 0.848 (0.811, 0.880) (P=.003) for lesion signature feature selection protocols including and excluding size features, respectively. The irregularity feature was chosen in 9 of 10 folds and in all folds when size features were included and excluded, respectively. AUC for the radiomic signature using feature selection from all features was statistically equivalent to using feature selection from all features excluding size features, within an equivalence margin of 2%.
Conclusions:
Inclusion of multiple radiomic features, automatically extracted from magnetic resonance images, in a lesion signature significantly improved the ability to distinguish between benign lesions and luminal A breast cancers, compared to using maximum linear size alone. The radiomic feature of irregularity appears to play an important but not a solitary role within the context of feature selection and computer-aided diagnosis.
Keywords: Breast cancer, MRI, luminal A, radiomics
INTRODUCTION
Breast cancer is a significant health concern for women, with one in eight expected to be diagnosed with the disease during their lifetime (1). Development of image-based computer-assisted diagnosis of breast cancer can support medical decision-making in diagnosis and treatment. Quantitative features of breast lesions can be extracted from medical images acquired using modalities such as mammography, ultrasound, computed tomography, and magnetic resonance imaging (MRI). Machine learning methods can be used to classify lesions by using these features to predict the probability of the lesion’s status in a classification task, such as likelihood of malignancy. This process is known as computer-aided diagnosis (2) or, more recently, radiomics (3,4). In addition to the value of radiomics in diagnosis and the prognosis of breast cancer, the use of radiomics in clinical decision-making may also reduce overdiagnosis (5) and assist in pre- and post-treatment assessment.
Radiomic features, such as those describing size, morphology, and texture, have been shown to be useful in the classification of lesions as benign vs malignant (2,6–8). Other studies have investigated the correlation of radiomic features extracted from MRI with lesion status, such as cancer stage and lymph node involvement (9); luminal B-type cancers (10); human epidermal growth factor 2 (HER2) (11); and luminal A, luminal B, HER2+, and basal-like classifications (12). Additionally, other studies have used these methods to classify lesions according to status as ductal carcinoma in situ and invasive ductal carcinoma (13), triple negative status vs other subtype (14), and all molecular subtypes (15).
In 2012, 74% of diagnosed breast cancers were type luminal A (16), the most of any molecular subtype. Therefore, it is of particular interest to identify radiomic signatures that aid in the diagnosis and prognosis of luminal A breast cancers. Luminal A lesions typically present on images as irregular and spiculated (17,18). Examples of figures from the dataset used in this work are shown, with their radiomic feature values for maximum linear size and irregularity (Fig 1).
Figure 1.

Postcontrast dynamic contrast-enhanced magnetic resonance images of a benign lesion (left) and a luminal A cancer (right). The position of the lesion and the cancer is indicated by a white arrow. Each image was acquired at 1.5T and is 125 × 125 mm2 in size, cropped from the full image. Slice thickness for each image was 2 mm. The maximum linear size for the benign lesion is 20.1 mm and the irregularity is 0.50. The maximum linear size for the luminal A cancer is 13.4 mm and the irregularity is 0.78.
Our investigation was motivated by the frequency of luminal A breast cancers diagnosed, and we aimed to develop a quantitative radiomic method to distinguish between benign lesions and luminal A subtype cancer. We evaluated the classification of a clinical dataset of lesions as benign vs luminal A using three variations of radiomic signatures: using maximum linear size alone, using feature selection from a full set of radiomic features, and using feature selection from radiomic features excluding those describing size. To the best of our knowledge, these methods have not been previously used to evaluate radiomic classification performance for distinguishing between benign lesions and luminal A cancers.
MATERIALS AND METHODS
A large clinical dataset of 508 breast lesions imaged with MRI was used in the present study (Table 1). Dynamic contrast-enhanced magnetic resonance images were collected retrospectively under Health Insurance Portability and Accountability Act (HIPAA) and institutional review board compliance. The benign lesions were either biopsy proven or imaged as part of follow-up care, whereas all luminal A cancers were biopsy proven. Imaging was performed at 1.5 and 3.0 T using Philips scanners. For the group of benign lesions, some subjects were imaged on multiple dates, for example, as a part of a screening program, and some subjects presented with multiple lesions. In these situations, a case was described as the collection of images for a lesion and the features for a case were averaged, resulting in 166 unique cases for the benign lesions and 296 for the luminal A cancers.
TABLE 1.
Overview of the Magnetic Resonance Imaging Database of Benign Lesions and Luminal A Cancers
| Type of Lesion | Number of Lesions | Number of Unique Cases | Mean Age of Subjects (y)* | Age of Subjects (Minimum, Maximum) (y) |
|---|---|---|---|---|
| Benign | 212 | 166 | 49.5±11.8 | (25, 78) |
| Luminal A | 296 | 296 | 58.0±12.5 | (28, 85) |
For some subjects, only the decade of age was available (eg, “40s” or “60s”) as part of the patient information deidentification process. In these situations, the middle of the decade was used for the calculation of the mean subject age. For example, if a subject’s age was given as 40s, the subject’s age was entered as “45” for the calculation. For the benign lesions, the age of 5 subjects was adjusted, whereas for the luminal A lesions, the age of 19 subjects was adjusted in this way. Subject age was not available for 47 benign lesions. In the situation of the same subject being imaged at multiple time points, the patient’s age at the time of imaging was used to calculate the age of the subjects for the group of lesions.
The images were segmented using an automated fuzzy C-means method requiring only the manual indication of a seed point within a lesion (19). Thirty-eight features describing each lesion were extracted from each image, in categories of size, shape, morphology, enhancement texture, kinetics, and enhancement-variance kinetics (7,19–22) (Appendix). The extracted features used in this work were previously used as part of an investigation of deep learning methodologies across multiple modalities in the task of classification of benign lesions and malignant cancers (23). The features of the luminal A cancers used here are part of a larger dataset of features extracted from breast cancers of all molecular subtypes and used in the previous study, and the benign features were used in the previous study as well. We note that the work here differs from the previous work in that this investigation focuses on classification of a single molecular subtype of breast cancers, utilizes features extracted from images from only one modality, and does not implement deep learning methodologies.
The Pearson correlation coefficient was determined for each feature against all other features, with particular attention given to the correlation of size features to morphology features, because of the nature of the proposed protocols. Linear discriminant analysis (LDA) was used as a classifier and we performed 10-fold cross validation in classifier training and testing. Lesions were partitioned to training or testing by case.
We investigated three classification protocols. First, classification was performed using the maximum linear size alone. Second, classification was performed concurrently with stepwise feature selection on all features. Third, classification was performed concurrently with stepwise feature selection on all features except those related to size. In each of the latter two protocols, feature selection was performed for each training fold. We tabulated which sets of features were selected in each fold in the second and third classification variations. Posterior probabilities of malignancy for each lesion in each testing set were scaled to the prevalence of cancer in the entire dataset (approximately 58%) (24). The scaled posterior probabilities of malignancy were then averaged by case.
We used the averaged, scaled posterior probabilities of malignancy by case to compare the classification performance using the area under the receiver operating characteristic curve (AUC) (25) as the figure of merit in assessing the ability to distinguish between benign and luminal A lesions. We used the conventional binormal ROC model (26). The software package ROCkit (27) was used to statistically compare the obtained AUC values using the two-tailed P values for differences in AUCs and the 95% confidence intervals in the difference in AUC for each comparison pair of protocols. The P values were corrected for three comparisons using the Holm-Bonferroni method (28). Classification performance was considered significantly different when the corrected two-tailed P value from the comparisons of AUCs for two protocols was less than .05.
In instances when we failed to reject the null hypothesis (that performances were equal) in superiority testing, equivalence testing was performed based on the same 95% confidence intervals. Equivalence is defined as having been demonstrated when the difference in AUC and the associated confidence interval of this difference falls between ±Δ, where Δ is the equivalence margin (29). The determination of Δ is not well established in medical imaging, but seven studies summarized by Ahn et al. (29) used equivalence margins between 1.5% and 15.0%. In this work, we did not declare an equivalence margin ab initio, but rather observed if the calculated equivalence margin fell within the range of equivalence margins seen in the selected literature reviewed by Ahn et al.
RESULTS
Box plots by cancer status of selected features demonstrate a separation in the median for benign or luminal A cancer, according to lesion status (Fig 2). Of note is the substantial number of outliers for the size feature shown here (maximum linear size) as calculated for this dataset, compared to the more consistent feature values for the irregularity feature by biopsy-proven classification of the lesions as benign or luminal A.
Figure 2.
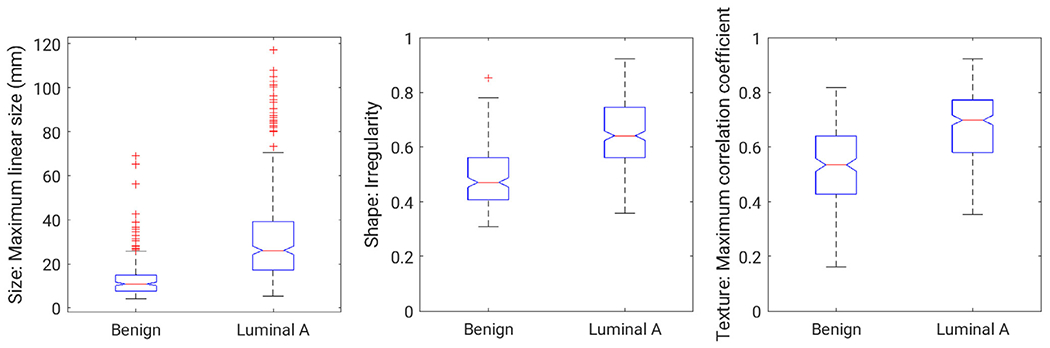
Box plots for three lesion features from the dataset: size (maximum linear size), shape (irregularity), and enhancement texture (maximum correlation coefficient). The width of the notches in a given box is proportional to the interquartile range of the data and provides a visual indication of possible difference in groups. The horizontal lines within the boxes indicate the median of the set, whereas the edges of the boxes indicate the 25th and the 75th percentiles. The crosses indicate outliers.
The Pearson correlation coefficients of the feature of irregularity were 0.42, 0.73 0.60, and 0.80 with the features of lesion volume, effective diameter, surface area, and maximum linear size, respectively.
Feature selections for the scenarios of using all features and all features except those relating to size demonstrate the importance of the lesion shape (irregularity) and the enhancement texture features (Fig 3). The AUCs for each classification protocol demonstrate the performance of the three classification protocols in the clinical task of distinguishing between benign lesions and luminal A breast cancers (Fig 4).
Figure 3.
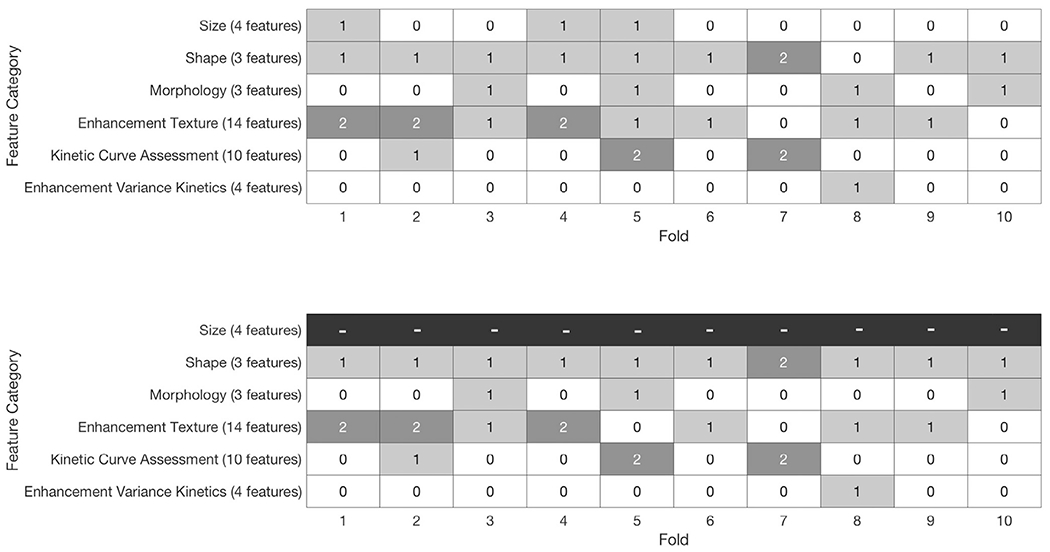
Use of features for each fold, with and without the features of size available to the linear discriminant analysis classifier algorithm for feature selection. The number in each square indicates for a given fold how many features from the associated category were selected for classification. The feature of irregularity was selected in 9 of 10 folds when feature selection drew from all features, including those describing size; the feature of irregularity was chosen in all folds when features describing size were excluded from feature selection.
Figure 4.
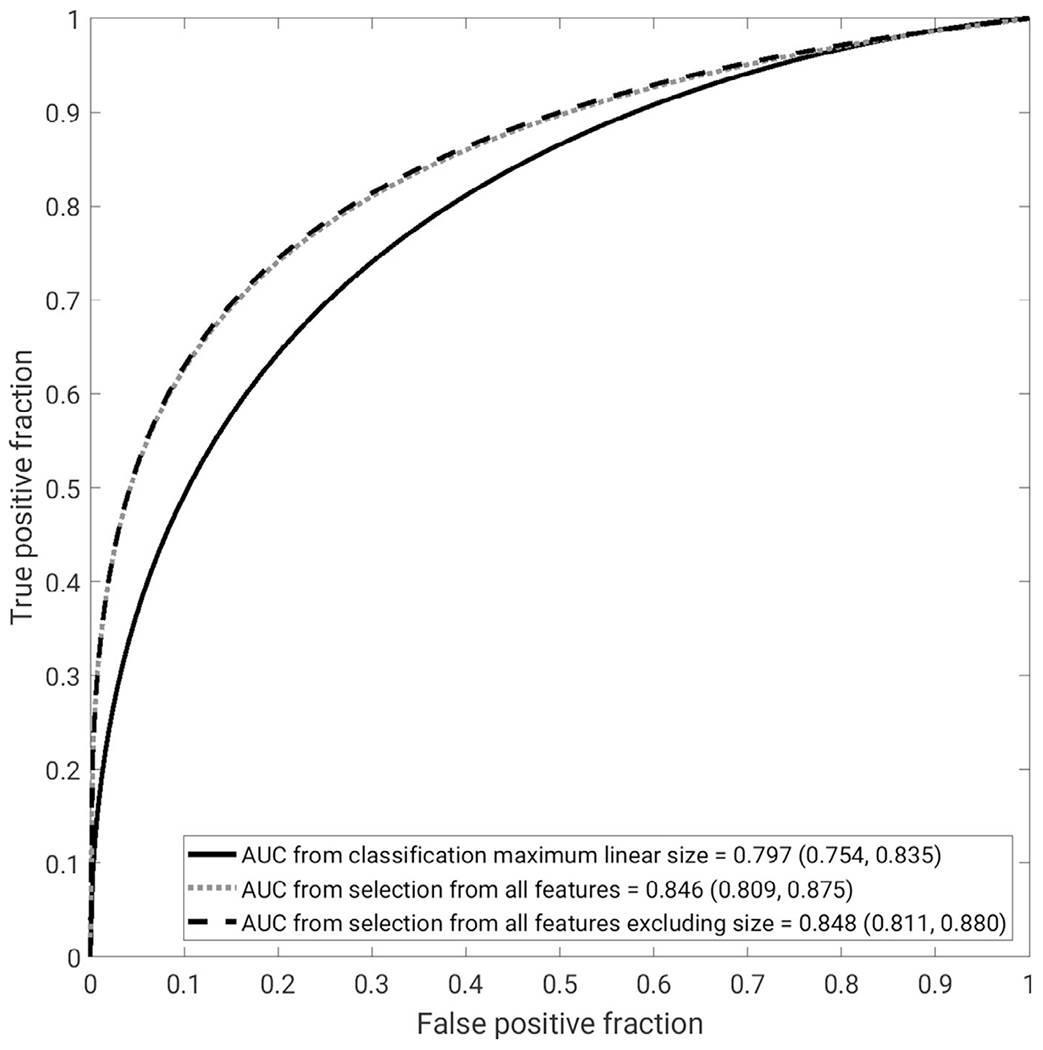
Receiver operating characteristic curves for the three different classification protocols from the 10-fold cross-validation, using maximum linear size alone, radiomics lesion signature obtained through nested feature selection, and radiomics lesion signature obtained through nested feature selection excluding size features, in the task of distinguishing between benign lesions and luminal A cancers. The legend gives the AUC for each with the 95% confidence interval in brackets. AUC, area under the receiver operating characteristic curve.
The AUC for maximum linear size alone was significantly different from the AUCs for both feature selections using all features (P=.005) and without size features included (P=.003). However, the AUC curves for both feature selections with and without size features included failed to show a significant difference from each other (P=.664). The AUC values do not approach nonequivalence until the statistical equivalence margin is less than 2% (Table 2).
TABLE 2.
Difference in Area Under the Receiver Operating Characteristic Curve and 95% Confidence Interval for Comparisons of Classification Protocols
| Classification Protocol Comparison | ΔAUC | 95% Confidence Interval of the Difference | P Value | Statistical Conclusion |
|---|---|---|---|---|
| Signature based on feature selection on all features compared to maximum linear size | −0.049 | (−0.081 to −0.018) | .005 | Superior |
| Signature based on feature selection on all features excluding size compared to maximum linear size | −0.050 | (−0.080 to −0.024) | .003 | Superior |
| Signature based on feature selection on all features excluding size compared to that on all features | −0.002 | (−0.013 to 0.008) | .664 | Equivalent |
AUC, area under the receiver operating characteristic curve.
P values shown are after correction for multiple comparisons.
Statistical conclusion was based upon superiority and equivalence testing.
DISCUSSION
The large number of actual lesions in the present study (over 500) and the emphasis of comparing benign lesions to a specific breast cancer subtype (luminal A) offer a focused investigation into radiomic features useful for the clinical task of malignancy classification, as a majority of breast cancers are of luminal A type. The segmentation of lesions by an algorithm that required only a single seed point per lesion contributed objectivity to the feature extractions, whereas the use of LDA allowed for consideration of multiple features for the classification task, resulting in a lesion signature.
A previous study reported that luminal A cancers were strongly associated with the Breast Imaging Reporting and Data System (BI-RADS) (30) descriptors of “irregular shape,” “spiculation,” “irregular margin,” “rim enhancement,” and “dark internal septation” (31). Using stepwise feature selection and LDA demonstrated that the radiomic feature of irregularity, which describes the so-called roughness of a lesion (20), played a prominent role in the classification of benign lesions and luminal A breast cancers, regardless of whether or not features related to size were included in feature selection. Notably, classification using the radiomic feature of irregularity alone resulted in an AUC of 0.782 (0.760, 0.804) and failed to show a significant difference in performance compared to using maximum linear size alone (P=.38). Despite this and the substantial correlation of the irregularity feature to size features (correlation coefficients between 0.42 and 0.8), feature selection demonstrated the additive benefit of using irregularity and a few other features related to texture over using only size in classification. This work suggests that the degree of roughness is particularly but not uniquely important in the classification of lesions as benign lesions or luminal A cancers. Texture features describe spatial variation in the signal intensity of a collection of pixel values (22). At least one texture feature was selected in 8 of 10 folds when features related to size were considered for feature selection and in 7 of 10 folds when features related to size were excluded from feature selection. The texture features of difference variance and maximum correlation coefficient were selected three and two times, respectively. In all, the increased classification performance compared to using solely the most prominent feature alone identified through a stepwise feature selection, irregularity, emphasizes the utility of using stepwise feature selection and LDA with a collection of radiomic features.
The present study does not separate cases according to their imaging status as pre- or postbiopsy, but previous work by our laboratory (32) found that AUC performance in this classification question compared by biopsy status failed to show a significant difference across all radiomic features.
One limitation of the present study was that, although a large number of clinical cases were used, the cases may not truly represent the full population of luminal A breast cancer cases. However, because all images were acquired at the same medical center, the imaging protocol was likely more consistently administered than if images were collected from multiple institutions, reducing one source of variability in image acquisition. Second, the images used in the study were acquired at two different magnetic field strengths. Our group is currently investigating the impact of field strength difference on this particular classification task (33). At the same time, our inclusion of features extracted from images at both field strengths enabled us to maximize statistical power, and our aim for this work was to describe classification performance for clinical populations, for which imaging can be conducted at the two field strengths used here. Third, in addition to quantitative measurements of lesion size that are produced during case workup, radiologists use the BI-RADS4 lexicon descriptions of lesion appearance, such as margin or shape, in their evaluation of lesions. In this work, we chose to focus on comparing the AUC performance of radiomics of maximum lesion size against feature selection methods, and our work did not compare the performance of radiomics against readings by radiologists for this classification task, nor did it investigate how the availability of radiomic information may affect radiologist performance for this classification task. Such a comparison will be the focus of future studies. Fourth, the 10-fold cross-validation method can, by nature, result in some variability of feature selection, but in our experience, the variability did not show notable differences in the selection of features for classification, particularly in the selection of irregularity. Finally, the present study focused on the issue of classification of benign lesions vs luminal A breast cancers. Although a majority of breast cancers are of subtype luminal A, it would certainly be useful to investigate radiomic signatures for other subtypes and to extend the analysis to make use of deep learning techniques.
CONCLUSIONS
This work demonstrated that in the clinical task of distinguishing between benign lesions and luminal A breast cancers, a radiomic signature using the features described here, quantitatively extracted from MR images, significantly improved the ability to classify the lesions. The radiomic feature of irregularity appears to be particularly useful in classifying lesions as benign or subtype luminal A. Furthermore, excluding features related to size from classification resulted in a radiomic signature that was statistically equivalent in terms of AUC to the radiomic signature using all features. This finding is notable, given the importance of size in the routine visual assessment of lesions on clinical images.
Funding information:
This work was supported by the National Institutes of Health grant U01 CA195564 and the Aldeen Memorial Fund at Wheaton College.
APPENDIX. RADIOMIC FEATURES EXTRACTED FROM BREAST DYNAMIC CONTRAST-ENHANCED MAGNETIC RESONANCE IMAGES
| Image Feature | Description |
|---|---|
| Size | |
| Volume (mm3) | Volume of lesion |
| Effective greatest dimension (mm) | Greatest dimension of a sphere with the same volume as the lesion |
| Surface area(mm2) | Lesion surface area |
| Maximum linear size(mm) | Maximum distance between any two voxels in the lesion |
| Shape | |
| Sphericity | Similarity of the lesion shape to a sphere |
| Irregularity | Deviation of the lesion surface from the surface of a sphere |
| Surface area-to-volume ratio(1/mm) | Ratio of surface area to volume |
| Morphology | |
| Margin sharpness | Mean of the image gradient at the lesion margin |
| Variance of margin sharpness | Variance of the image gradient at the lesion margin |
| Variance of radial gradient histogram | Degree to which the enhancement structure extends in a radial pattern originating from the center of the lesion |
| Enhancement texture | |
| Angular second moment(energy) | Image homogeneity |
| Contrast | Location image variations |
| Correlation | Image linearity |
| Entropy | Randomness of the gray levels |
| Sum of squares(variance) | Spread in the gray-level distribution |
| Difference entropy | Randomness of the difference of neighboring voxels’ gray levels |
| Difference variance | Variations of difference of gray levels between voxel pairs |
| Inverse difference moment | Image homogeneity |
| Sum average | Overall brightness |
| Sum entropy | Randomness of the sum of gray levels of neighboring voxels |
| Sum variance | Spread in the sum of the gray levels of voxel-pair distribution |
| Information measure of correlation 1 | Nonlinear gray-level dependence |
| Information measure of correlation 2 | Nonlinear gray-level dependence |
| Maximum correlation coefficient | Nonlinear gray-level dependence |
| Kinetic curve assessment | |
| Maximum enhancement | Maximum contrast enhancement |
| Time to peak(s) | Time at which the maximum enhancement occurs |
| Uptake rate(1/s) | Uptake speed of the contrast enhancement |
| Washout rate(1/s) | Washout speed of the contrast enhancement |
| Curve-shape index | Difference between late and early enhancements |
| Enhancement at first postcontrast time point | Enhancement at first postcontrast time point |
| Signal-to-enhancement ratio | Ratio of the initial enhancement to the overall enhancement |
| Volume of most enhancing voxels(mm3) | Volume of the most enhancing voxels |
| Total rate variation(1/s2) | How rapidly the contrast will enter and exit from the lesion |
| Normalized total rate variation(1/s2) | How rapidly the contrast will enter and exit from the lesion |
| Enhancement-variance kinetics | |
| Maximum variance of enhancement | Maximum spatial variance of contrast enhancement over time |
| Time to peak at maximum variance(s) | Time at which the maximum variance occurs |
| Enhancement-variance increasing rate(1/s) | Rate of increase of the enhancement variance during uptake |
| Enhancement-variance decreasing rate(1/s) | Rate of decrease of the enhancement variance during washout |
Contributor Information
Heather M. Whitney, Department of Radiology, The University of Chicago, 5841 S. Maryland Avenue, MC 2026 Chicago, IL 60637; Department of Physics, Wheaton College, 501 College Avenue, Wheaton, IL 60187.
Nathan S. Taylor, Department of Physics, Wheaton College, 501 College Avenue, Wheaton, IL 60187
Karen Drukker, Department of Radiology, The University of Chicago, 5841 S. Maryland Avenue, MC 2026 Chicago, IL 60637.
Alexandra V. Edwards, Department of Radiology, The University of Chicago, 5841 S. Maryland Avenue, MC 2026 Chicago, IL 60637.
John Papaioannou, Department of Radiology, The University of Chicago, 5841 S. Maryland Avenue, MC 2026 Chicago, IL 60637.
David Schacht, Department of Radiology, The University of Chicago, 5841 S. Maryland Avenue, MC 2026 Chicago, IL 60637.
Maryellen L. Giger, Department of Radiology, The University of Chicago, 5841 S. Maryland Avenue, MC 2026 Chicago, IL 60637.
REFERENCES
- 1.Siegel R, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017; 67:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Giger ML, Chan H-P, Boone J. Anniversary paper: history and status of CAD and quantitative image analysis: the role of Medical Physics and AAPM. Med Phys 2008; 35:5799–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016; 278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahbar H, McDonald ES, Lee JM, et al. How can advanced imaging be used to mitigate potential breast cancer overdiagnosis? Acad Radiol 2016; 23:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giger ML. Computer-aided detection/computer-aided diagnosis. In: Wolbarst AB, Mossman KL, Hendee WR, eds. Advances in medical physics: 2008, Madison, WI: Medical Physics Publishing; 2008:143–168. [Google Scholar]
- 7.Chen W, Giger ML, Bick U, et al. Automatic identification and classification of characteristic kinetic curves of breast lesions on DCE-MRI. Med Phys 2006; 33:2878–2887. [DOI] [PubMed] [Google Scholar]
- 8.Bickelhaupt S, Paech D, Kickingereder P, et al. Prediction of malignancy by a radiomic signature from contrast agent-free diffusion MRI in suspicious breast lesions found on screening mammography. J Magn Reson Imaging 2017; 46:604–616. [DOI] [PubMed] [Google Scholar]
- 9.Burnside ES, Drukker K, Li H, et al. Using computer-extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer 2016; 122:748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazurowski MA, Zhang J, Grimm LJ, et al. Radiogenomic analysis of breast cancer: luminal b molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 2014; 273:365–372. [DOI] [PubMed] [Google Scholar]
- 11.Blaschke E, Abe H. MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J Magn Reson Imaging 2015; 42:920–924. [DOI] [PubMed] [Google Scholar]
- 12.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging 2015; 42:902–907. [DOI] [PubMed] [Google Scholar]
- 13.Bhooshan N, Giger ML, Jansen SA, et al. Cancerous breast lesions on dynamic contrast-enhanced MR images. Breast Imaging 2010; 254:680–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Kato F, Oyama-Manabe N, et al. Identifying triple-negative breast cancer using background parenchymal enhancement heterogeneity on dynamic contrast-enhanced MRI: a pilot radiomics study. PLoS ONE 2015; 10. e0143308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. Breast Cancer 2016; 2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desantis CE, Fedewa SA, Sauer AG, et al. Breast cancer statistics, 2015: convergence of incidence rates between black and white women. CA Cancer J Clin 2016; 66:31–42. [DOI] [PubMed] [Google Scholar]
- 17.Tamaki K, Ishida T, Miyashita M, et al. Correlation between mammographic findings and corresponding histopathology: potential predictors for biological characteristics of breast diseases. Cancer Sci 2011; 102:2179–2185. [DOI] [PubMed] [Google Scholar]
- 18.Cho N Molecular subtypes and imaging phenotypes of breast cancer. Ultrasonography 2016; 35:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, Giger ML, Bick U. A fuzzy C-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad Radiol 2006; 13:63–72. [DOI] [PubMed] [Google Scholar]
- 20.Gilhuijs KG, Giger ML, Bick U. Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging. Med Phys 1998; 25:1647–1654. [DOI] [PubMed] [Google Scholar]
- 21.Chen W, Giger ML, Lan L, et al. Computerized interpretation of breast MRI: investigation of enhancement-variance dynamics. Med Phys 2004; 31:1076–1082. [DOI] [PubMed] [Google Scholar]
- 22.Chen W, Giger ML, Li H, et al. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med 2007; 58:562–571. [DOI] [PubMed] [Google Scholar]
- 23.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys 2017;44:5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horsch K, Pesce LL, Giger ML, et al. A scaling transformation for classifier output based on likelihood ratio: applications to a CAD workstation for diagnosis of breast cancer. Med Phys 2012; 39:2787–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metz CE. Basic principles of ROC analysis. Semin Nucl Med 1978; 8:283–298. [DOI] [PubMed] [Google Scholar]
- 26.Metz CE, Herman BA, Shen JH. Maximum likelihood estimation of receiver operating characteristic (ROC) curves from continuously-distributed data. Stat Med 1998; 17:1033–1053. [DOI] [PubMed] [Google Scholar]
- 27.Metz CE. ROCkit 0.9b [Internet] Available at: http://metz-roc.uchicago.edu/; 1998. Accessed December 15, 2017.
- 28.Holm S A simple sequentially rejective multiple test procedure. Scand J Stat 1979; 6:65–70. [Google Scholar]
- 29.Ahn S, Park SH, Lee KH. How to demonstrate similarity by using noninferiority and equivalence statistical testing in radiology research. Radiology 2013; 267:328–338. [DOI] [PubMed] [Google Scholar]
- 30.D’Orsi C, Sickles E, Mendelson E, eds. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System, Reston, VA: American College of Radiology; 2013. [Google Scholar]
- 31.Navarro Vilar L, Alandete Germán SP, Medina García R, et al. MR imaging findings in molecular subtypes of breast cancer according to BIRADS system. Breast J 2017; 23:421–428. [DOI] [PubMed] [Google Scholar]
- 32.Whitney H, Drukker K, Edwards A, et al. Effect of biopsy on the MRI radiomics classification of benign lesions and luminal A cancers. In: In: Proceedings of the 14th International Workshop on Breast Imaging; 2018. p. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitney H, Drukker K, Edwards A, et al. Robustness of radiomic breast features of benign lesions and luminal A cancers across MR magnet strengths. In: Mori K, Petrick N, eds. Medical Imaging 2018: Computer-Aided Diagnosis, SPIE; 2018. [Google Scholar]


