Abstract
We utilized primer extension analysis to demonstrate that the divergently transcribed regB and senC-regA-hvrA transcripts contain stable 5′ ends 43 nucleotides apart within the regB-senC intergenic region. DNA sequence analysis indicates that this region contains two divergent promoters with overlapping ς70 type −35 and −10 promoter recognition sequences. In vivo analysis of expression patterns of regB::lacZ and senC-regA-hvrA::lacZ reporter gene fusions demonstrates that the regB and senC-regA-hvrA transcripts are both negatively regulated by the phosphorylated form of the global response regulator RegA. DNase I protection assays with a constitutively active variant of RegA indicate that RegA binds between regB and senC overlapping −10 and −35 promoter recognition sequences. Two mutations were also isolated in a regB-deficient background that increased expression of the senC-regA-hvrA operon 10- and 5-fold, respectively. As a consequence of increased RegA expression, these mutants exhibited elevated aerobic and anaerobic photosynthesis (puf) gene expression, even in the absence of the sensor kinase RegB. These results indicate that autoregulation by RegA is a factor contributing to the maintenance of an optimal low level of RegA expression that allows responsiveness to activation by phosphorylation.
Two-component phosphorylation-dependent regulatory systems exist in bacteria to allow adaptation to changes in the environment (28, 29). Among several species of alpha purple bacteria, there exists a highly conserved two-component system comprised of the sensor kinase RegB and the response regulator RegA (2, 3, 15). This regulatory pair was first identified in the nonsulfur photosynthetic bacterium Rhodobacter capsulatus, where it was shown to be required for anaerobic induction of light-harvesting and reaction center structural genes (2, 3, 17, 25, 37). Responding to anaerobiosis, the histidine kinase RegB autophosphorylates itself at a conserved histidine residue. The phosphoryl group is then transferred to a highly conserved aspartate residue of the cognate response regulator RegA (17, 25). Phosphorylated RegA (RegA∼P) then binds to the promoter region of the puf, puc, or puh operon to exert gene activation (7, 11, 12). Recent analyses of the RegB-RegA pair from several species indicate that it is a global signal transduction system involved in the anaerobic induction of many physiological processes. This includes the synthesis of the light-harvesting, reaction center, and cytochrome components of the bacterial photosystem and the assimilation of carbon dioxide and nitrogen (4, 14, 15, 19, 22, 33, 41).
Analysis of regB and regA genes from R. capsulatus, Rhodobacter sphaeroides Rhodovulum sulfidophilum, and Roseobacter denitrificans demonstrated that they are two genes of a highly conserved “photosynthesis regulatory gene cluster” (8, 9, 23, 25). In these species, regB is divergently transcribed from a three-gene operon comprised of senC, regA, and hvrA. Mutational analysis indicated that SenC is involved in the formation of a functional cytochrome c oxidase complex which is required for proper sensing of anaerobiosis by RegB (9), while HvrA appears to be a transcription factor that facilitates RegA binding to DNA under dim-light growth conditions (8, 20).
As the function and mechanism of action of individual components are being unraveled, it is becoming clear that components of the photosynthesis regulatory gene cluster play important roles in controlling anaerobic induction of numerous metabolic processes. Consequently, an investigation of the expression of the regB and senC-regA-hvrA genes can provide important clues to the control hierarchy of anaerobic gene expression in these species. In the present study, we determined the transcription initiation sites of the regB and senC-regA-hvrA genes, which revealed the presence of two divergent overlapping promoters. Reporter gene analyses with lacZ fusions to the regB and senC promoters, as well as DNase I footprint analysis with RegA, indicated that phosphorylated RegA negatively regulates regB and senC-regA-hvrA transcription. Finally, two regB-deleted strains bearing mutations in the overlapping promoter region were also characterized for a thorough understanding of the functional significance of having low-level regulated expression of these regulatory genes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The parent strain R. capsulatus SB1003 (46), the regB-deficient strain SD01 (12), the regA-deficient strain MS01 (37), and the two promoter mutant strains SD01-M52 and SD01-M08 (as described in Results) were grown at 34°C in PY salts medium or in RCV2/3PY (47). Spectinomycin, kanamycin, and gentamicin were all used at a final concentration of 5 μg/ml for the maintenance of plasmids and the construction of stable recombinants in R. capsulatus. Rifampin was used for counterselection of exconjugates at a final concentration of 100 μg/ml. Highly oxygenated and anaerobic growth conditions of R. capsulatus cultures were as described previously (37). Escherichia coli DH5α and S17-1 (λpir) (38) were grown at 37°C in Luria broth medium (35). Kanamycin, spectinomycin, and gentamicin were added to the medium at 50, 50, and 10 μg/ml, respectively.
RNA extraction.
Total cellular RNA was isolated from R. capsulatus cells grown photosynthetically in PY salts medium. Cells from a 300-ml culture were collected after the culture reached approximately 75 Klett units (25). Cell pellets were treated for 20 min at room temperature with 500 μl of lysozyme at 4 mg/ml and then dispersed in 15 ml of a denaturing solution comprised of 4.2 M guanidium thiocyanate, 17 mM Na–N-lauroyl sarcosine, 25 mM Na-acetate, and 100 mM β-mercaptoethanol at pH 5.5 in diethylpyrocarbonate (DEPC)-treated H2O. Total RNA was phenol-chloroform extracted once, precipitated at 4°C overnight with 0.6 volume of isopropanol, and then centrifuged at 17,000 × g for 15 min at 4°C. The crude RNA pellet was then resuspended in 4 ml of denaturing solution supplemented with 2 ml of a 5.7 M CsCl solution. RNA was then purified on a CsCl gradient by using a swinging-bucket rotor at 30,000 × g overnight at 20°C. The pellet was then phenol-chloroform extracted, ethanol precipitated, washed, and finally resuspended in 250 μl of DEPC-treated H2O.
Primer extension.
Two primers, 5′-CCCATCGCAACAGGATCAATGTCCGC-3′ and 5′-GCCGACATGTCGAATTCCGGCCGGTCG-3′, were designed to anneal the regB gene. Similarly, another two primers, 5′-GCGGCAAAGCGATCCGTCTCATGGG-3′ and 5′-GAGATGCCCACCACGACGACGACGGC-3′, were designed to anneal the senC gene. Labeling of the primers was carried at 37°C for 1 h in a 10-μl total reaction volume containing 100 ng of primer, 1× T4 kinase buffer (New England Biolabs), 10 U of T4 polynucleotide kinase (New England Biolabs), and 320 μCi of [γ-32P]ATP (specific activity of 7,000 Ci/mmol; Amersham). Unincorporated label was separated on a Sephadex G-50 fine-nick spin column (Pharmacia) at 500 × g in accordance with the manufacturer’s instructions. Labeled primers were then resuspended in 20 μl of DEPC-treated H2O. Annealing reaction mixtures containing 16.2 μg of total RNA, 2.5 μl of labeled primer, 10 mM Tris-acetate (pH 7.4), and 60 mM NH4Cl in a 10-μl total volume were heated at 80°C for 10 min, cooled to the hybridization temperature of 50°C (hybridization temperature = 29.3°C [0.41% G+C for primer]), and further incubated at the same temperature for an additional 20 min. The premixed extension reaction mixture was composed of 20 mM Tris-acetate (pH 7.4), 20 mM Mg-acetate, 120 mM NH4Cl, actinomycin D at 80 μg/ml, 50 mM dithiothreitol, 750 μM deoxynucleotide triphosphate, and 12.25 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim). Primer extension was performed at 42°C for 1 h and then stopped with 80 μl of 0.3 M Na-acetate (pH 6.0) and 250 μl of 100% ethanol. Reactions were precipitated and resuspended in 10 μl of loading dye (10 mM NaOH, 10 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol in formamide). The sequencing ladders used were obtained as described by the manufacture (thermo-Sequenase sequencing kit; Amersham Pharmacia Biotech) using the same labeled primers. Reactions were heated at 80°C for 3 min and loaded onto a 6% urea denaturing polyacrylamide gel.
DNA sequencing.
DNA fragments containing the 5′ end of regB and the full-length senC and regA genes were amplified by PCR from the wild-type and putative mutant chromosomes by using upstream and downstream primers 5′-CTCTAGAAATAG CCGTAAGCGCAATCAG-3′ and 5′-CGGTACCTCCAGTGAGTGGTTTCATGG-3′, respectively. DNA sequencing was carried out by using several primers that annealed at different sites of the PCR-amplified fragment using an ABI automatic sequencing system (Perkin-Elmer).
Plasmid construction and mobilization.
The wild-type senC-regA-hvrA promoter reporter plasmid pReg-operon and the two promoter mutant reporter plasmids pReg-M52 and pReg-M08 were constructed as follows. DNA fragments containing the regB and senC-regA-hvrA promoter regions were PCR amplified from the chromosomes of the wild-type and mutant strains by using primers 5′-GCGCCGCCAGGTGACCGACGACCG-3′ and 5′-GCATGCACCGCCGATCTGCGCCGA-3′, which contain BstEII and SphI restriction sites (underlined), respectively. The PCR products were first cloned into the BstEII and SphI sites of lacZ shuttle vector pDC400 (21) and subsequently subcloned into the BstEII and SstI sites of lacZ reporter vector pZM400 as described previously (21). A spectinomycin-resistant omega cassette (32) was then cloned into the HindIII site that is located within the kanamycin resistance gene in pZM400.
The regB reporter plasmid pRegB01 was constructed by the procedure described above by using primers 5′-GGTTGCGCCGGTTACCGAGATCAG-3′ and 5′-GGTTCGCATGCACATCGGCAGGTT-3′ to amplify the promoter region, which puts BstEII and SphI sites (underlined) in the opposite orientation to the primers used for construction of the regA promoter reporter vectors.
Promoter mutations were also cloned into suicide vectors and recombined back into regB knockout strain SD01. For this analysis, DNA fragments carrying the full-length senC gene were amplified from the mutant strain genomic DNA preparations by PCR using primers 5′-CTCTAGAAATAGCCGTAAGCGCAATCAG-3′ and 5′-CGAGCTCATCGTCATCGACCAGAAGCAG-3′, which contain XbaI and SacI digestion sites (underlined), respectively. The PCR products were subsequently cloned into the integration plasmid pZJD3 (18), and the resulting constructs were then backcrossed into strain SD01 harboring pufQ::lacZ translational fusion reporter plasmid pCB532Ω (1) by conjugation with plasmid-mobilizing strain S17-1 (λpir) (38). Transconjugates were isolated by restreaking several times onto PY salts plates containing spectinomycin and gentamicin. The resulting recombinants exhibited phenotypes (spectral analysis, photosynthetic growth rates, and puf operon transcription rates) that were indistinguishable from those observed with the original isolates (data not shown).
Spectral analysis and β-galactosidase activity.
In vivo absorption spectra were obtained from crude intracytoplasmic membrane preparations by sonicating cells grown either aerobically or anaerobically that were harvested at 50 to 100 Klett units. Following centrifugation at 10,000 × g for 5 min, the soluble fraction was scanned from 400 to 900 nm with a Beckman DU-50 recording spectrophotometer (47). β-Galactosidase activities of pufQ::lacZ fusion plasmid pCB532Ω in wild-type strain SB1003 and mutant strains SD01, SD01-M52, and SD01-M08, as well as the activities of pReg-operon, pRegB01, pReg-M52, and pReg-M08 in strains SB1003, SD01, and MS01, were measured as described previously (13).
Antibody production and Western blot analysis.
To obtain polyclonal antibodies against RegA, E. coli-expressed recombinant proteins (17) were excised from a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and subsequently injected subcutaneously into a New Zealand White rabbit by Cocalico Biological, Inc. (Reamstown, Pa.). After initial inoculation, booster injections were administrated in the second and the third weeks, followed by a first test bleed in the fourth week. Additional booster and test bleeds were performed on the 49th and 56th days, respectively. The specificity of the antiserum was then tested by Western blot analysis. For Western blot analysis, the wild type and various mutants were grown anaerobically to 50 to 100 Klett units and collected by centrifugation at 7,600 × g for 10 min. The cells were washed once with washing buffer (20 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl, 0.2 mM phenylmethylsulfonium fluoride, pH 7.2), resuspended in washing buffer, and then sonicated for 3 × 20 s. The crude extracts were clarified by centrifugation at 12,000 × g for 10 min, and proteins in the supernatant were then separated by sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis. The proteins were transferred onto a nitrocellulose membrane (Schleicher & Schuell), using a Mini Trans-Blot electrophoretic transfer cell following instructions of the manufacturer (Bio-Rad). The proteins were subsequently detected using the ECL Western blotting analysis system as indicated by the manufacture (Amersham Life Science). A transblotted membrane was blocked with 5% nonfat dry milk in PBS-Tween (80 mM Na2HPO4, 20 mM NaH2PO4, 100 mM NaCl, 0.1% Tween 20, pH 7.5) for 1 h at room temperature and then incubated overnight at 4°C with polyclonal antibody against RegA that was diluted at 1:8,000 in PBS-Tween. The membrane was then washed with PBS-Tween and incubated for 1 h at room temperature with a horseradish peroxidase-labeled anti-rabbit antibody diluted 1:10,000 in PBS-Tween. The membrane was then washed, and the chemiluminescent antigen-antibody complexes were detected by exposure to X-Omat AR film (Kodak).
DNase I footprint analysis.
Using chromosomal DNA from wild-type R. capsulatus as a template, a PCR was performed to generate a 210-bp DNA fragment containing the regB-senC intergenic region by using primers 5′-CCGGACCCATTCCTGATGGCTC-3′ and 5′-CTGCCGTGGCAGCCAGCGCGGC-3′. One of the primers in the PCR mixture was 5′ end labeled with T4 polynucleotide kinase and [γ-32P]ATP prior to amplification as described for primer extension analysis. The 32P-labeled promoter DNA fragments were purified from 5% nondenaturing polyacrylamide gels by electroelution. After precipitation and washing with ethanol, the DNA fragments were resuspended in Tris-EDTA buffer containing 50 mM NaCl.
Footprint assays were initiated with a DNA probe and various amounts of RegA* (RegA* was purified as described in reference 12) in footprint buffer containing 40 mM HEPES (pH7.8), 8 mM Mg-acetate, 75 mM K-acetate, 2 mM CaCl2, 1.5 mM dithiothreitol, bovine serum albumin at 125 μg/ml, and 16% glycerol in a total reaction volume of 20 μl. After 20 min of incubation at room temperature, 2 μl of 0.5-μg/ml DNase I was added to the reaction mixtures and digestion proceeded for 5 min at room temperature before being quenched by the addition of 180 μl of stop solution (0.33 M NH4-acetate, 55 mM EDTA, 14 μg of yeast tRNA per ml). The reaction mixtures were then phenol-chloroform extracted, ethanol precipitated, dried, and resuspended in 3 μl of formamide loading dye. The amount of radioactivity in each sample was determined by Cerenkov scintillation counting to ensure that equal amounts of digested probe were loaded for electrophoresis. Reaction samples were heated at 90°C for 10 min and electrophoresed through a 6% urea denaturing Long Ranger gel (FMC Bioproducts, Rockland, Maine). A modified Maxam-and-Gilbert G+A chemical sequencing reaction was used to determine positions of protected nucleotides relative to the transcription start site (24).
RESULTS
Identification of transcription start sites.
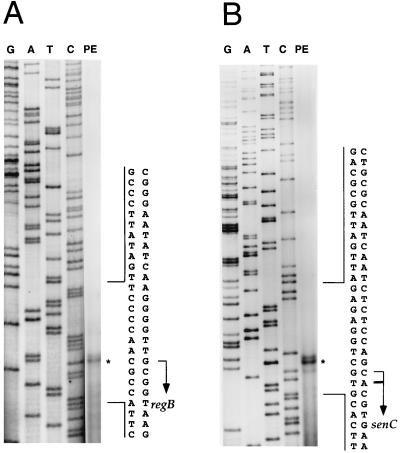
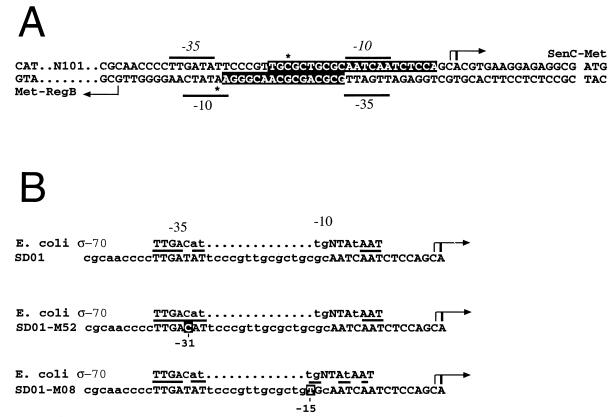
Primer extension analyses were undertaken with total cellular RNA isolated from anaerobically grown wild-type R. capsulatus cells to map the start sites of the regB and senC-regA-hvrA transcripts. When 32P-labeled primers complementary to the regB coding region were used, a stable 5′ end was observed at a G residue 98 nucleotides upstream of the regB start codon (Fig. 1A). When labeled primers for the senC transcript were used, a major 5′-end product was observed at an A residue located 16 nucleotides upstream of the senC start codon, as well as a minor end product at a C residue located at nucleotide 17 (Fig. 1B). As shown in Fig. 2A, the major 5′ ends are 43 nucleotides apart within the regB-senC intergenic region. Inspection of the DNA sequence between the transcription start sites revealed the presence of two divergent promoters that each contain −10 and −35 elements similar to those of E. coli ς70-type promoter recognition sequences. These divergent promoters contain overlapping −10 and −35 sequences (Fig. 2A).
FIG. 1.
Primer extension (PE) analyses to determine the start sites of the regB (A) and senC-regA-hvrA transcripts (B). The G, A, T, and C ladders are as indicated. Primer extension products are indicated by asterisks, and transcription initiation sites are indicated by arrows.
FIG. 2.
(A) Characteristics of the divergent overlapping regB and senC-regA-hvrA operon promoters. The transcription start sites of the regB and senC-regA-hvrA genes are indicated by arrows, with translation start sites of the regB and senC genes also indicated. The −35 and −10 regions are indicated by lines, and the RegA protection regions on both strands are depicted as white letters on a black background. The hypersensitive sites are indicated by asterisks. (B) Comparison of the wild-type and mutant senC-regA-hvrA promoters with the consensus sequence of E. coli ς70-type promoters. The R. capsulatus −35 and −10 regions are in capital letters. The most-conserved nucleotides are represented by large uppercase letters, the less-conserved nucleotides are represented by smaller uppercase letters, and the least-conserved nucleotides for E. coli ς70 are represented by lowercase letters, with conservation between E. coli and B. capsulatus promoters highlighted by lines. The numbers below the white-on-black letters indicate the sites of promoter mutations in strains SD01-M52 and SD01-M08.
The regB and senC-regA-hvrA transcripts are negatively regulated by RegA∼P in vivo.
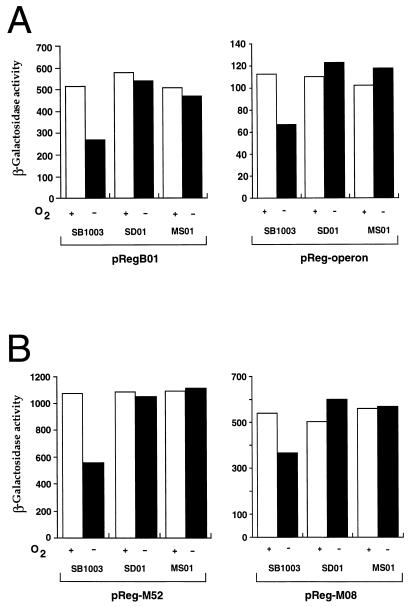
Many transcription factors, including some response regulators, are either positively or negatively autoregulated (6, 10, 16, 26, 27, 34, 36, 39, 40, 42–45). To determine if the RegA protein influences the activity of its own promoter and possibly the regB promoter, we assayed the transcription of the regB gene and the senC-regA-hvrA operon with lacZ transcriptional fusions. By using regB promoter reporter plasmid pRegB01, we reproducibly observed a twofold reduction in β-galactosidase activity when the wild-type strain was grown under anaerobic versus aerobic conditions (Fig. 3A, left). This reduction was caused by phosphorylated RegA, since constitutively high levels of expression were observed in regB-disrupted strain SD01, as well as in regA-disrupted strain MS01.
FIG. 3.
β-Galactosidase activity measurements of wild-type and mutant promoter expression. (A) Activity of the regB reporter plasmid pRegB01 (left) and the wild-type senC reporter plasmid pReg-operon (right) in wild-type (SB1003), regB mutant (SD01), and regA-disrupted (MS01) cells. A plus or minus sign indicates growth in the presence or absence of oxygen. (B) Activities of promoter mutant vectors pReg-M52 (left) and pRegM08 (right) harbored in the same strains as in panel A. Units of activity are micromoles of ONPG (o-nitrophenyl-β-d-galactopyranoside) hydrolyzed per minute per milligram of protein. The results shown are averages of at least three independent assays.
When using the senC-regA-hvrA promoter probe plasmid pReg-operon, we observed a similar pattern of expression, namely, a twofold reduction of activity in anaerobically grown wild-type cells and no effect of oxygen on regB- or regA-disrupted cells (Fig. 3A, right). Thus, both promoters are repressed by phosphorylated RegA.
RegA binds to the regB and senC-regA-hvrA promoters in vitro.
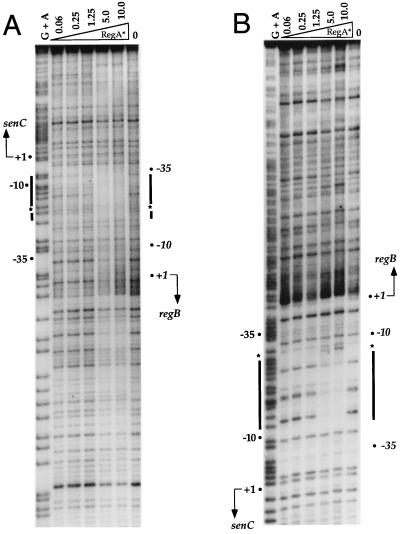
We next addressed the question of whether the negative autoregulation observed with RegA in vivo is a direct consequence of binding of RegA to the overlapping regB gene and senC-regA-hvrA operon promoters. For this analysis, we performed DNase I nuclease protection assays on the overlapping promoter regions by using purified RegA*, which is a previously described variant of RegA that exhibits constitutive (phosphorylation-independent) DNA binding activities in vivo and in vitro (12). As demonstrated by the protection patterns in Fig. 4A and B, RegA* binds to a region that spans the overlapping −10 and −35 promoter regions. Relative to the major start site of the senC-regA-hvrA transcript, RegA* protects a region stretching from −15 to −30 on the bottom strand and a region stretching from −3 to −24 on the top strand. There are also hypersensitive sites located on the top strand at position −22 and on the bottom strand at position −31 (Fig. 2A). Since the regB promoter overlaps the senC-regA-hvrA promoter, protection of this promoter relative to its transcription start site is from −14 to −29 on the top strand and from −20 to −41 on the bottom strand (Fig. 2A). The RegA* protection pattern suggests that RegA∼P directly represses the activity of these divergent overlapping promoters.
FIG. 4.
DNase I footprint analysis of RegA* protection in the regB and senC-regA-hvrA promoter region. (A) Footprint analysis of the top strand. (B) Footprint analysis of the bottom strand. G+A indicates Maxam-and-Gilbert chemical cleavage patterns. Lanes 2 to 6 are DNase I digestions of binding reaction mixtures containing increasing amounts (in micrograms) of purified RegA*. The binding reaction mixture in lane 7 contains no RegA*. The thick lines delineate regions protected by RegA*, and the asterisks indicate hypersensitive sites. The transcription start sites of regB and senC-regA-hvrA are also indicated.
Characterization of promoter mutants.
We next addressed the functional significance of having low-level regulated expression of RegA through the characterization of strains containing point mutations in the overlapping-promoter region. In a previous study (12), we described a genetic screen for the isolation of mutants that exhibit constitutively high-level photosystem synthesis in the absence of the sensor kinase RegB. Two of the 14 isolates were shown to contain point mutations in regA which rendered the response regulator active for initiation of transcription even in the absence of phosphorylation by RegB (7, 12). The remaining 12 “RegB bypass” isolates contained undefined mutations outside of regA. In this study, we further characterized the RegB bypass mutants by performing DNA sequence analysis of the regions flanking regA. The results of this sequence analysis indicated that 10 of the 12 newly sequenced RegB bypass mutants contained base pair substitutions in the intergenic promoter region that drive expression of the senC-regA-hvrA transcript. Among these promoter mutants, eight contained the same T→C transition mutation at position −33 and the other two strains contained the same C→T transition mutation at position −17 relative to the start of the senC-regA-hvrA operon transcript (Fig. 2B). Strain SD01-M52, which contains a T→C mutation at −33, and strain SD01-M08, which contains a C→T mutation at −17, were chosen for further analysis.
Previous in vitro footprint assays indicated that unphosphorylated RegA could bind to the puc promoter with 16-fold lower affinity than phosphorylated RegA (7). It is therefore possible that these putative promoter mutations bypass the requirement for RegB by increasing the intracellular amount of RegA to a level that will allow promoter binding and subsequent activation of photosynthesis gene expression in the absence of phosphorylation. To test whether the intergenic point mutations indeed affect the transcription of the senC-regA-hvrA operon, we constructed promoter probe vectors by using the same PCR primers, cloning techniques, and vectors described above for the wild-type senC-regA-hvrA operon promoter reporter vector pReg-operon. The only difference was the use of SD01-M52 or SD01-M08 chromosomal DNA as the template for PCR amplification, which resulted in the construction of mutant promoter reporter vectors pReg-M52 and pReg-M08, respectively. When the β-galactosidase values obtained with these promoter mutants (Fig. 3B) were compared to β-galactosidase values obtained with the wild-type promoter vector pReg-operon (Fig. 3A, right), we observed that pReg-M52 and pReg-08 had 10- and 5-fold increases in promoter activity, respectively. These promoter mutations were still repressed by phosphorylated RegA, as evidenced by the observation that the activities of both pReg-M52 and pReg-M08 were significantly lower under anaerobic growth conditions than under aerobic conditions in wild-type strain SB1003. This conclusion was confirmed by the fact that a constitutively high level of β-galactosidase activity was observed in regB and regA mutant strains SD01 and MS01, respectively, when they were grown under aerobic or anaerobic conditions (Fig. 3B).
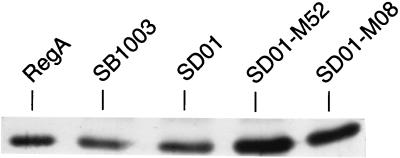
We also directly assayed for RegA protein levels in vivo by performing Western blot analysis on extracts derived from anaerobically grown cells with RegA-specific polyclonal antibodies. As shown in Fig. 5, regB-disrupted strain SD01 exhibited an amount of RegA slightly elevated over that observed with the wild-type strain SB1003. This confirmed the β-galactosidase results described above, which indicated that transcription of senC-regA-hvrA was repressed by phosphorylated RegA. The level of RegA was even higher in SD01-M52 and SD01-M08 cells. This is congruent with the β-galactosidase-based promoter reporter probe results discussed above, which show significantly higher expression for pReg-M52 and pReg-08, which contain the promoter mutations (Fig. 3B), than for the wild-type promoter vector pReg-operon (Fig. 3A).
FIG. 5.
Western blot analysis of crude extracts from strains SB1003, SD01, SD01-M52, and SD01-M08 grown anaerobically. Purified RegA protein (15 ng) was used as a positive control in the first lane, and 20 μg of total cellular protein was added to the other lanes.
Increased expression of the senC-regA-hvrA operon leads to elevated photosynthesis gene expression.
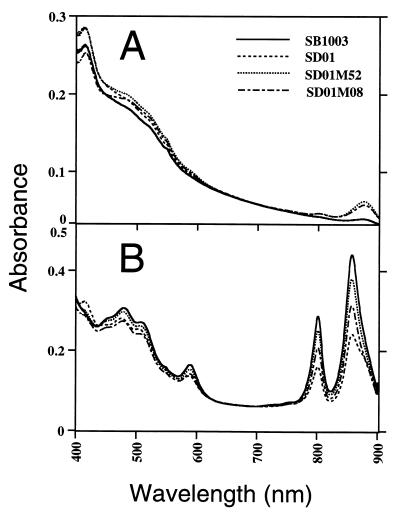
Spectral and puf operon expression analyses were also performed on strains SD01-M52 and SD01-M08 to address the consequence of elevating senC-regA-hvrA operon expression for the synthesis of the photosystem. Spectral analysis demonstrated that both wild-type strain SB1003 and regB-disrupted strain SD01 exhibit characteristic low levels of photopigment synthesis when grown aerobically (Fig. 6A). This is contrasted by a significant increase in pigment levels in aerobically grown SD01-M52 and SD01-M08 cells. As expected, when wild-type strain SB1003 was grown anaerobically, the photopigment levels significantly increased while the regB-disrupted strain SD01 exhibited a lower level of photopigment synthesis as a consequence of the absence of the sensor kinase (Fig. 6B). However, spectral analysis of anaerobically grown SD01-M52 and SD01-M08 cells exhibited significantly higher levels of pigment synthesis than observed with parent strain SD01, with SD01-M52 synthesizing slightly more photopigments than SD01-M08 (Fig. 6B).
FIG. 6.
Spectral analysis of crude membrane fractions from cultures grown aerobically (A) or anaerobically (B). SB1003 is the wild-type strain, SD01 is the regB knockout strain, and SD01-M52 and SD01-M08 are mutant strains harboring mutations in the promoter region of the senC-regA-hvrA operon. Each scan was performed on the same cell mass, as determined by spectral analysis at 660 nm.
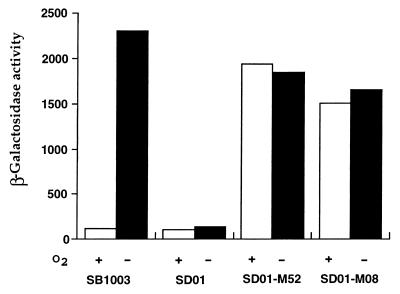
To examine whether the elevated photosystem synthesis observed by spectral analysis is due to the direct effect of RegA on photosynthesis gene expression, measurement of puf operon expression was also undertaken by using the reporter plasmid pCB532Ω in wild-type and mutant strains. As shown in Fig. 7, the β-galactosidase activity levels obtained from the puf reporter plasmid harbored in strains SB1003 and SD01 grown aerobically were essentially the same as reported before, with more than 20-fold anaerobic induction in the wild-type strain and no significant induction in the regB-disrupted strain (25). However, the β-galactosidase activities in strains SD01-M52 and SD01-M08 were constitutively high, with strain SD01-M52 showing a slightly higher level of expression than SD01-M08. These results are in good agreement with the regulation pattern shown by spectral analysis.
FIG. 7.
β-Galactosidase activity measurement of puf::lacZ reporter gene plasmid pCB532Ω in strains SB1003, SD01, SD01-M52, and SD01-M08 (see the legend to Fig. 6 for strain genotypes). Cells were grown either aerobically (open bars) or anaerobically (solid bars), and β-galactosidase activities were then measured as described in the legend to Fig. 3.
DISCUSSION
Divergent promoters for the regB and senC-regA-hvrA operons overlap with promoter activities repressed by RegA∼P.
Prior to this study, there was no investigation of the expression pattern of a photosynthesis regulatory gene from a photosynthetic prokaryote. In order to obtain a better understanding of the RegB-RegA signal transduction pathway, we have characterized the promoters and expression patterns of the regB gene and the senC-regA-hvrA operon. Mapping of transcriptional start sites of the divergently transcribed regB gene and the senC-regA-hvrA operon indicates that the major transcripts are separated by 43 nucleotides delineating two superimposed divergent promoters. These promoters are probably transcribed by an E. coli ς70-RNA polymerase holoenzyme, as indicated by the similarity of the −35 and −10 regions to the ς70 consensus sequence (Fig. 2A and B, upper). Divergent overlapping promoters have been described in many other systems, where they presumably provide some advantage in coregulating transcripts by a common signal. The binding of a regulatory protein to a region that contains overlapping promoters can either activate or repress transcription in both directions or activate transcription in one direction and repress it in the other direction (5, 30, 31, 43). For example, in E. coli K-12, the ilvY and ilvC genes, whose products are involved in isoleucine-valine biosynthesis, are also divergently transcribed from overlapping promoters. The regulator IlvY negatively autoregulates its own expression 2-fold while activating ilvC expression 15-fold (43). In the case of RegA, we observed that phosphorylated RegA negatively influences the transcriptional activity of both regB and senC-regA-hvrA transcripts by approximately twofold. The repressing activity of RegA∼P is presumably mediated by steric interference with RNA polymerase binding to the promoters, since our footprint results indicate that the RegA∼P binding site overlaps the regB and senC-regA-hvrA promoter recognition sequences.
A few additional two-component response regulators have also been demonstrated to be autoregulated either positively or negatively (6, 16, 34, 36, 39, 42, 44, 45). Among them, a situation similar to that described here for RegA was also observed for the response regulator FlbD in Caulobacter crescentus, which is encoded by the last gene in the fliF operon (6, 34, 45). Phosphorylation of FlbD by the sensor kinase FlbE is known to be required for the activation of numerous level III and IV flagellar genes, as well as negative autoregulation of the fliF operon.
Maintaining RegA at an appropriate level of expression is functionally important.
We also isolated and characterized two senC-regA-hvrA promoter mutants in a regB deletion background that significantly affect the expression of photosynthesis genes that are controlled by RegA∼P. Promoter expression analysis indicates that point mutations at positions −33 and −17, relative to the major startsite of the senC-regA-hvrA transcript, resulted in 10- and 5-fold increases in senC-regA-hvrA promoter activity, respectively. Inspection of the DNA sequence indicates that the T-to-C mutation at −33 creates a perfect match to the −35 region of the canonical E. coli ς70 consensus sequence (Fig. 2B, middle). The C-to-T mutation at −17 also results in a better fit to the −10 region of the E. coli ς70 consensus sequence (Fig. 2B, bottom). It should also be noted that the mutation at −17 is well within the region of DNA that is protected by RegA from DNase I digestion, whereas the mutation at position −33 is located just at the boundary of protection (Fig. 2A). However, neither mutation appears to significantly affect RegA∼P binding, as evidenced by the observation that both mutant promoters are anaerobically repressed in vivo by RegA∼P (Fig. 3B). Furthermore, in vitro footprint analysis indicates that the affinity of RegA* binding to mutant DNA templates is similar to that observed with wild-type promoter templates (data not shown). Thus, the increase in promoter activity observed in these mutants may be a result of better promoter binding, or open complex formation, by the RNA polymerase holoenzyme containing a E. coli ς70-type factor rather than due to a reduction in the repressing activity of RegA∼P.
The promoter mutations in strains SD01-M52 and SD01-M08 result in much more photosystem synthesis and puf operon expression than observed with regB-deficient parent strain SD01. This indicates that unphosphorylated RegA activates gene expression in vivo in these strains. Recent footprint analysis has indicated that unphosphorylated RegA exhibits specific binding to the puc operon promoter region with an affinity 16-fold lower than that of phosphorylated RegA (7). Since RegA expression appears to be elevated 5- or 10-fold as a consequence of the promoter mutations, it is reasonable to presume that the RegA concentration has risen in the mutant strains to a level that promotes DNA binding, and subsequent activation of photosynthesis gene expression, in the absence of phosphorylation. Thus, low-level expression of RegA appears to be necessary to promote phosphorylation-dependent activation of target gene expression.
RegA∼P has multiple regulatory roles.
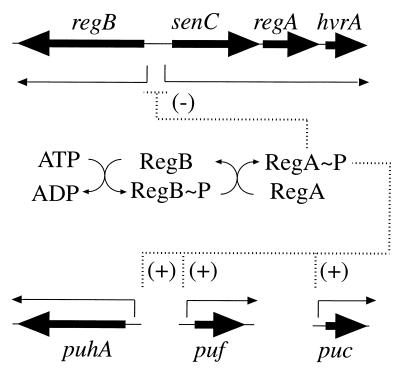
Combining the results of the present study and previous studies (2, 3, 7, 12, 17, 25, 37), the RegB-RegA regulon in R. capsulatus can now be depicted as diagrammed in Fig. 8. At the top of the hierarchy, RegA∼P functions as a transcriptional repressor of regB and senC-regA-hvrA expression. Below that, RegA∼P functions as a transcription activator for the puf, puc, and puh operons, which code for the light-harvesting and reaction center apoproteins, as well as an activator of several nonphotosynthesis genes, such as those involved in CO2 assimilation, nitrogen fixation, and cytochrome c2 expression (4, 7, 12, 14, 15, 19, 22, 33, 41). This study, as well as continued analysis of the RegB-RegA regulon in R. capsulatus and in other species, should provide better understanding of how this regulator controls the expression of these diverse metabolic processes.
FIG. 8.
Activation and repression by the RegA-RegB regulon. Negative and positive regulation of the regulatory gene cluster and photosynthesis genes by RegA∼P is depicted.
ACKNOWLEDGMENTS
We thank Sylvie Elsen for helpful comments regarding the manuscript.
This study was supported by research grant GM40941 (to C.E.B.) from the National Institutes of Health.
REFERENCES
- 1.Bauer C E, Young D A, Marrs B L. Analysis of the Rhodobacter capsulatus puf operon. J Biol Chem. 1988;263:4820–4827. [PubMed] [Google Scholar]
- 2.Bauer C E. Regulation of photosystem gene expression. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1221–1234. [Google Scholar]
- 3.Bauer C E, Bird T H. Regulatory circuits controlling photosynthesis gene expression. Cell. 1996;85:5–8. doi: 10.1016/s0092-8674(00)81074-0. [DOI] [PubMed] [Google Scholar]
- 4.Bauer E, Kaspar T, Fischer H-M, Hennecke H. Expression of the fixR-nifA operon in Bradyrhizobium japonicum depends on a new response regulator, RegR. J Bacteriol. 1998;180:3853–3863. doi: 10.1128/jb.180.15.3853-3863.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck C F, Warren R A J. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988;52:318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson A K, Ramakrishnan G, Ohta N, Feng J, Ninfa A J, Newton A. The Caulobacter crescentus FlbD protein acts at ftr sequence elements both to activate and to repress transcription of cell cycle-regulated flagellar genes. Proc Natl Acad Sci USA. 1994;91:4989–4993. doi: 10.1073/pnas.91.11.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird T H, Du S, Bauer C E. Autophosphorylation, phosphotransfer and DNA-binding properties of the RegB/RegA two-component regulatory system in Rhodobacter capsulatus. J Biol Chem. 1999;274:16343–16348. doi: 10.1074/jbc.274.23.16343. [DOI] [PubMed] [Google Scholar]
- 8.Buggy J J, Sganga M W, Bauer C E. Characterization of a light-responding trans-activator responsible for differentially controlling reaction center and light-harvesting-I gene expression in Rhodobacter capsulatus. J Bacteriol. 1994;176:6936–6943. doi: 10.1128/jb.176.22.6936-6943.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buggy J J, Bauer C E. Cloning and characterization of senC, a gene involved in both aerobic respiration and photosynthesis gene expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:6958–6965. doi: 10.1128/jb.177.23.6958-6965.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casadaban M J. Regulation of the regulatory gene for the arabinose pathway, araC. J Mol Biol. 1976;104:557–566. doi: 10.1016/0022-2836(76)90120-0. [DOI] [PubMed] [Google Scholar]
- 11.Du, S., and C. E. Bauer. Unpublished data.
- 12.Du S, Bird T H, Bauer C E. DNA binding characteristics of RegA*: a constitutively active anaerobic activator of photosynthesis gene expression in Rhodobacter capsulatus. J Biol Chem. 1998;273:18509–18513. doi: 10.1074/jbc.273.29.18509. [DOI] [PubMed] [Google Scholar]
- 13.Elsen S, Richaud P, Colbeau A, Vignais P M. Sequence analysis and interposon mutagenesis of the hupT gene, which encodes a sensor protein involved in repression of hydrogenase synthesis in Rhodobacter capsulatus. J Bacteriol. 1993;175:7404–7412. doi: 10.1128/jb.175.22.7404-7412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elsen, S., and C. E. Bauer. Unpublished data.
- 15.Eraso J M, Kaplan S. PrrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan C-D, Wanner B, Inouye H. Analysis of regulation of phoB expression using a phoB-cat fusion. J Bacteriol. 1983;156:710–717. doi: 10.1128/jb.156.2.710-717.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue K, Kouadio J L K, Mosley C S, Bauer C E. Isolation and in vitro phosphorylation of sensory transduction components controlling anaerobic induction of light harvesting and reaction center gene expression in Rhodobacter capsulatus. Biochemistry. 1995;34:391–396. doi: 10.1021/bi00002a002. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Z-Y, Gest H, Bauer C E. Chemosensory and photosensory perception in purple photosynthetic bacteria utilize common signal transduction components. J Bacteriol. 1997;179:5720–5727. doi: 10.1128/jb.179.18.5720-5727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi H M, Tabita F R. A global two component signal transduction system that integrates the control of photosynthesis, carbon dioxide assimilation, and nitrogen fixation. Proc Natl Acad Sci USA. 1996;93:14515–14520. doi: 10.1073/pnas.93.25.14515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouadio, J.-L. K., and C. E. Bauer. Unpublished data.
- 21.Ma D, Cook D N, O’Brien D A, Hearst J E. Analysis of the promoter and regulatory sequences of an oxygen-regulated bch operon in Rhodobacter capsulatus by site-directed mutagenesis. J Bacteriol. 1993;175:2037–2045. doi: 10.1128/jb.175.7.2037-2045.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda, S., Y. Matsumoto, K. V. P. Nagashima, K. Shimada, K. Inoue, C. E. Bauer, and K. Matsuura. In G. Garan and J. Pusztai (ed.), Proceedings of the XI International Congress on Photosynthesis, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 23.Masuda S, Matsumoto Y, Nagashima K V P, Shimada K, Inoue K, Bauer C E, Matsuura K. Structural and functional analyses of photosynthetic regulatory genes regA and regB from Rhodovulum sulfidophilum, Roseobacter denitrificans and Rhodobacter capsulatus. J Bacteriol. 1999;181:4205–4215. doi: 10.1128/jb.181.14.4205-4215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 25.Mosley C S, Suzuki J Y, Bauer C E. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J Bacteriol. 1994;176:7566–7573. doi: 10.1128/jb.176.24.7566-7573.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunoshiba T, Hidalgo E, Li Z, Demple B. Negative autoregulation by the Escherichia coli SoxS protein: a dampening mechanism for the soxRS redox stress response. J Bacteriol. 1993;175:7492–7494. doi: 10.1128/jb.175.22.7492-7494.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ostrowski J, Kredich N M. Negative autoregulation of CysB in Salmonella typhimurium: in vitro interactions of CysB protein with the cysB promoter. J Bacteriol. 1991;173:2212–2218. doi: 10.1128/jb.173.7.2212-2218.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 29.Parkinson J S. Genetic approaches for signaling pathways and proteins. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 9–23. [Google Scholar]
- 30.Plamann L S, Stauffer G V. Nucleotide sequence of the Salmonella typhimurium metR gene and the metR-metE control region. J Bacteriol. 1987;169:3932–3937. doi: 10.1128/jb.169.9.3932-3937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poteete A R, Ptashne M. Control of transcription by the bacteriophage p22 repressor. J Mol Biol. 1982;157:21–48. doi: 10.1016/0022-2836(82)90511-3. [DOI] [PubMed] [Google Scholar]
- 32.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 33.Qian Y, Tabita F R. A global signal transduction system regulates aerobic and anaerobic CO2 fixation in Rhodobacter sphaeroides. J Bacteriol. 1996;178:12–18. doi: 10.1128/jb.178.1.12-18.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramakrishnan G, Zhao J-L, Newton A. Multiple structural proteins are required for both transcriptional activation and negative autoregulation of Caulobacter crescentus flagellar genes. J Bacteriol. 1994;176:7587–7600. doi: 10.1128/jb.176.24.7587-7600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Scarlato V, Prugnola A, Arico B, Rappuoli R. Positive transcriptional feedback at the bvg locus controls expression of virulence factors in Bordetella pertussis. Proc Natl Acad Sci USA. 1990;87:6753–6757. doi: 10.1073/pnas.87.17.6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sganga M W, Bauer C E. Regulatory factors controlling photosynthetic reaction center and light harvesting gene expression in Rhodobacter capsulatus. Cell. 1992;68:945–954. doi: 10.1016/0092-8674(92)90037-d. [DOI] [PubMed] [Google Scholar]
- 38.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 39.Soncini F C, Vescovi E G, Groisman E A. Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J Bacteriol. 1995;177:4364–4371. doi: 10.1128/jb.177.15.4364-4371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauch M A, Perego M, Burbulys D, Hoch J A. The transition state transcription regulator AbrB of Bacillus subtilis is autoregulated during vegetative growth. Mol Microbiol. 1989;3:1203–1209. doi: 10.1111/j.1365-2958.1989.tb00270.x. [DOI] [PubMed] [Google Scholar]
- 41.Tiwari R P, Reeve W G, Dilworth M J, Glenn A R. Acid tolerance in Rhizobium meliloti strain WSM419 involves a two-component sensor-regulator system. Microbiology. 1996;142:1693–1704. doi: 10.1099/13500872-142-7-1693. [DOI] [PubMed] [Google Scholar]
- 42.van Sinderen D, Venema G. ComK acts as an autoregulatory control switch in the signal transduction route to competence in Bacillus subtilis. J Bacteriol. 1994;176:5762–5770. doi: 10.1128/jb.176.18.5762-5770.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wek R C, Hatfield G W. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. J Biol Chem. 1986;261:2441–2450. [PubMed] [Google Scholar]
- 44.Winans S C, Mantis N J, Chen C-Y, Chang C-H, Han D C. Host recognition of the VirA, VirG two-component regulatory proteins of Agrobacterium tumefaciens. Res Microbiol. 1994;145:461–473. doi: 10.1016/0923-2508(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 45.Wingrove J A, Gober J W. Identification of an asymmetrically localized sensor histidine kinase responsible for temporally and spatially regulated transcription. Science. 1996;274:597–601. doi: 10.1126/science.274.5287.597. [DOI] [PubMed] [Google Scholar]
- 46.Yen H C, Hu N T, Marrs B L. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J Bacteriol. 1976;126:619–629. doi: 10.1128/jb.126.2.619-629.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young D A, Bauer C E, Williams J C, Marrs B L. Genetic evidence for superoperonal organization of the genes for photosynthesis pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol Gen Genet. 1989;218:1–12. doi: 10.1007/BF00330558. [DOI] [PubMed] [Google Scholar]