Abstract
The terrorist attacks on the World Trade Center (WTC) created an unprecedented environmental exposure to aerosolized dust, gases and potential carcinogens. Clonal hematopoiesis (CH) is defined as the acquisition of somatic mutations in blood cells and is associated with smoking and exposure to genotoxic stimuli. Here we show that deep targeted sequencing of blood samples identified a significantly higher proportion of WTC-exposed first responders with CH (10%; 48/481) when compared to non-WTC-exposed firefighters (6.7%; 17/255; OR=3.14 (95% CI:1.64–6.03, p-value=0.0006) after controlling for age, sex, and race/ethnicity. The frequency of somatic mutations in WTC-exposed first responders showed an age-related increase and predominantly affected DNMT3A, TET2 and other CH associated genes. Exposure of lymphoblastoid cells to WTC particulate matter (WTC-PM) led to dysregulation of DNA replication at common fragile sites in vitro. Moreover, mice treated with WTC-PM developed an increased burden of mutations in hematopoietic stem and progenitor cell compartments. In summary, the high burden of CH in WTC-exposed first responders provides a rationale for enhanced screening and preventative efforts in this population.
Keywords: Mutations, Firefighters, First Responders, WTC, 9/11, Clonal Hematopoiesis
Editor summary:
First responders to the World Trade Center disaster, who were exposed to particulate matter containing potential carcinogens, have a high burden of somatic mutations in blood cells, raising their risk for cancer and other diseases and highlighting the need for enhanced health screening of these individuals.
World Trade Center (WTC) first responders were exposed to particulate matter containing high levels of potential carcinogens1. Studies have demonstrated an increased risk of prostate cancer, thyroid cancer and other malignancies in WTC-exposed firefighters, rescue workers and civilians 2,3. We demonstrated that WTC exposure is associated with an increased risk of myeloma precursor disease, monoclonal gammopathy of unknown significance (MGUS) in first responders, suggesting that sufficient time has now elapsed post exposure for the manifestation of other premalignant conditions4 Clonal hematopoiesis (CH) is the acquisition of somatic mutations in blood cells and is associated with an increased risk of hematologic malignancies and inflammatory disorders.5.
From December 2013 to October 2015, we collected blood samples from Fire Department of the City of New York (FDNY) WTC-exposed first responders (429 firefighters and 52 emergency medical service (EMS) workers) that were de-identified and used for DNA isolation and sequencing. Control peripheral blood samples were collected from Nashville firefighters (N=203) and from firefighters outside of Nashville belonging to the International Association of Firefighters (IAFF) (N=52), that were not exposed to WTC disaster and had comparable baseline demographics (Extended Data Table 1 showing demographic data). Samples from 481 WTC-exposed and 255 non-WTC exposed responders were analyzed by deep targeted sequencing of 237 genes frequently mutated in hematologic malignancies (Neogenomics, Carlsbad, CA). The majority of the participants were male and identified as Caucasian in both WTC exposed and non-exposed cohorts (Extended Data Table 1). Single nucleotide variations were determined using Mutect and insertion/deletion mutations were determined using Pindel. Variants were excluded from analysis if there were present in ExAC/gnomAD databases at ≥1% minor allele frequency. Variants were analyzed with a minimum sequence depth of 250x and a variant allele frequency of 5% for single nucleotide variants and 10% for insertion/deletion mutations. Sequencing and analysis were conducted in a NY state CLIA certified clinical lab (Neogenomics) and mutations were interpreted for significance using reference databases including Catalog of Somatic Mutations in Cancer (COSMIC), dbSNP database and Exome Aggregation Consortium (ExAC). We considered nonsynonymous mutations reported in these databases as somatic mutations of expected pathogenic potential and also included somatic nonsynonymous mutations affecting driver genes implicated in hematologic malignancies. Putative germline variants (variant allele fraction above 40%) were not included in the somatic mutational analysis.
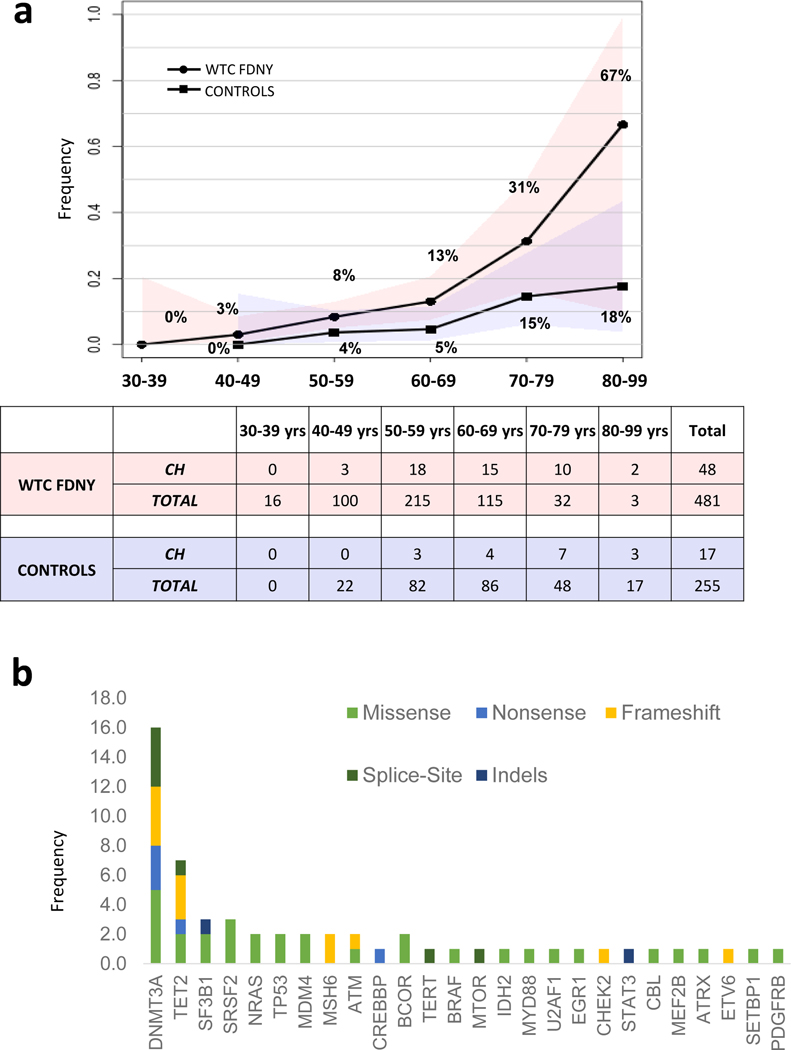
In the WTC-exposed cohort, we identified 48 individuals with 57 unique somatic mutations of expected pathogenic potential for an overall prevalence of 10%. Six of the 480 individuals (13%) carried more than one somatic mutation. The prevalence of CH-associated mutations was 6.7% (17/255) in the non-WTC-exposed firefighter comparison cohort. Among both the WTC-exposed cohort and the non-WTC-exposed cohort, with each subsequent decade, we noted an increase in the prevalence of pathogenic somatic mutations (Figure 1A). In a multivariable logistic model, we observed a significantly increased odds of CH in the WTC-exposed first responders compared with comparison participants (OR=3.14 (95% CI: 1.64–6.03, p-value=0.0006) after controlling for age, sex, and race/ethnicity. Since the WTC-exposed first responders included 52 EMS workers, we next directly compared the 429 WTC-exposed firefighters to the non-WTC-exposed firefighters and again observed an increased odds of CH in WTC-exposed group (OR=2.93, 95% CI: 1.52–5.65, p-value=0.0014) after controlling for age, sex and race. In multivariable analyses restricted to those with smoking information, when controlling for smoking as well as age, sex and race/ethnicity, the association between WTC-exposure and CH remained (OR=3.05, 95% CI: 1.54–6.06, p-value=0.0015 for all WTC-exposed responders vs non-WTC exposed comparison participants and (OR=2.78, 95% CI: 1.39–5.59, p-value=0.004) for WTC-exposed firefighters vs non-WTC-exposed comparison participants); smoking history was not significantly associated with CH in either multivariable model.
Figure 1: Prevalence and characteristics of somatic mutations seen in WTC-exposed first responders:
A: The number of somatic mutations in 481 WTC-exposed first responders is shown as a function of age. 75% and 95% confidence intervals are shown in red shading. B: Percentage of first responders with mutations for specific genes is shown.
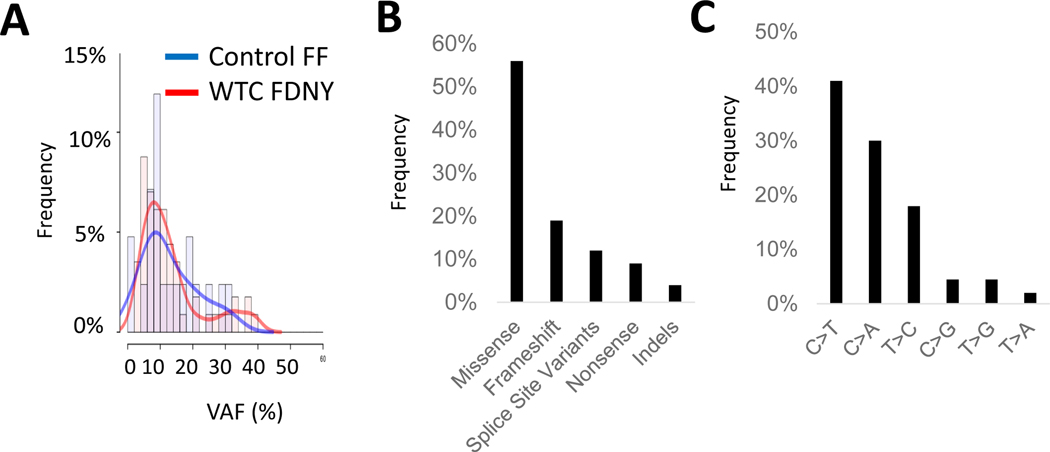
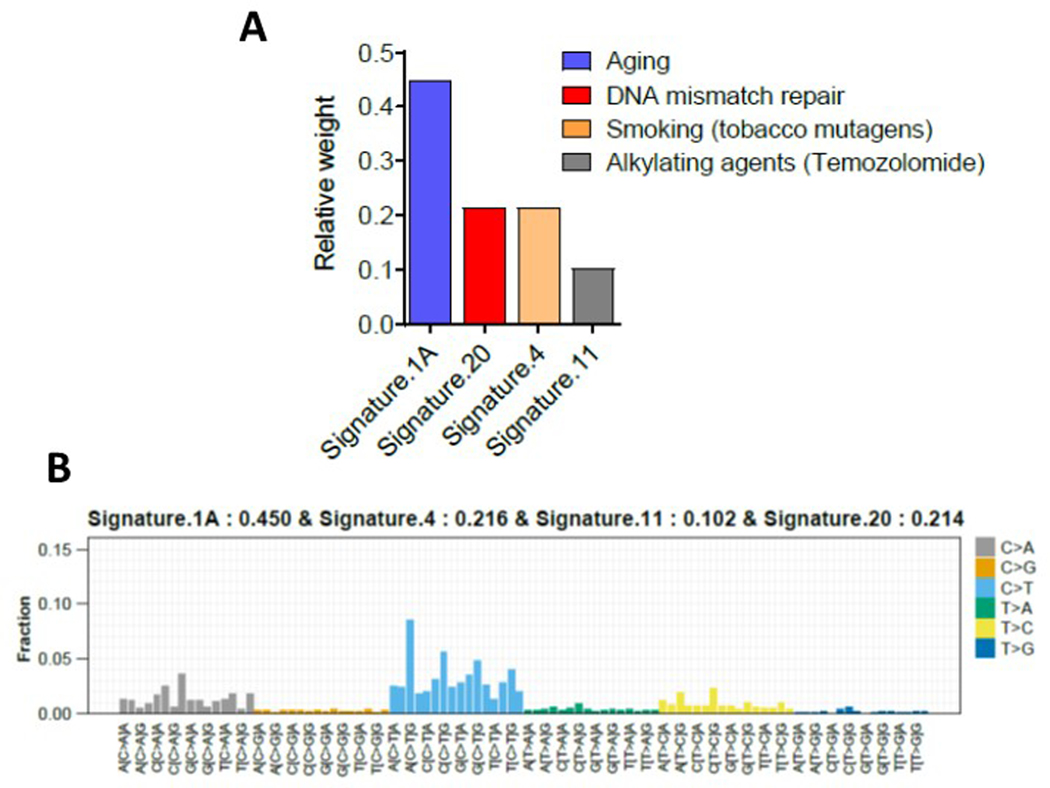
Genes implicated in CH and myeloid malignancies were most frequent mutated in the WTC-exposed cohort. The most commonly identified variants affected DNMT3A (16 of 57) and TET2 (7 of 57). Other genes included those involving the spliceosome machinery, SF3B1, SRSF2 (3 of 57 each), as well as other genes commonly implicated in CH such as NRAS, TP53, MDM4, MSH6 (2 of 57 for each gene, respectively) (Figure 1B, Supplemental Table 1). The pattern of CH mutations was observed to be similar in the non-WTC exposed firefighter cohort (Supplemental Table 2, Extended Data Figure 1). Median VAF of the significant somatic mutations in WTC exposed cohort was 10%, consistent with other published CH studies (Figure 2A). Missense mutations were most frequently identified (Figure 2B). The most common base-pair change in the somatic mutations was a cytosine to thymine transition (C→T), considered a somatic mutational signature of aging (Figure 2C). Analysis of overall mutational signatures in the WTC exposed cohort demonstrated enrichment for aging, DNA mismatch repair, smoking and alkylating agent exposure (Extended Data Figure 2). Examination of annual blood counts for this cohort demonstrated no association of mutations with cytopenia (Supplemental Table 3).
Figure 2: Characteristics of somatic mutations seen in WTC-exposed first responders:
A: The variant allele frequency (VAF; mutant reads/total reads) for each mutation is shown for first responders and controls... B: The frequency at which each of the indicated types of somatic mutations occurred in the WTC-exposed first responders is shown. C: Frequency at which each of the indicated mutation types occurred in WTC exposed first responders is shown
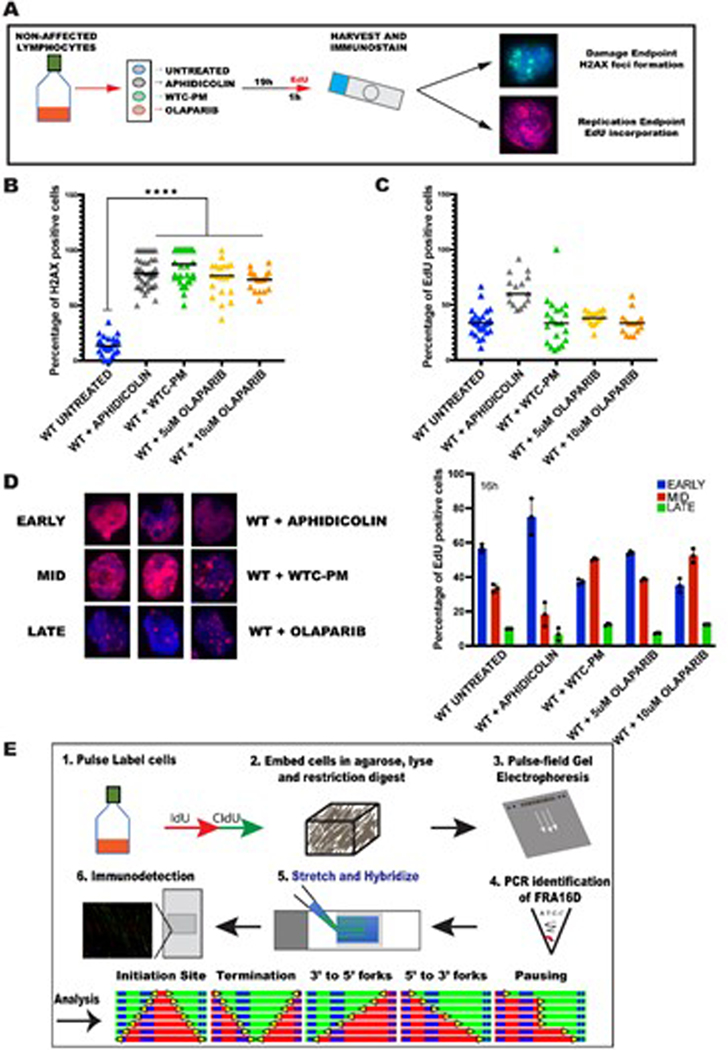
Given the exposure of FDNY first responders to high concentrations of WTC-PM, we wanted to assess the genotoxic effects of WTC-PM collected from Ground Zero in the first 3 days of 9/116. Previously, WTC-PM has been characterized to contain high concentrations of polycyclic aromatic hydrocarbons (PAH) including benzopyrene that have been classified as carcinogens and can form bulky DNA adducts. We exposed lymphoblasts to WTC-PM for 20h and measured DNA damage by fluorescent microscopy for phosphorylated H2AX (p-H2AX) foci accumulation. We included aphidicolin (APH) and olaparib (OLP), that have been previously shown to perturb DNA replication, in the treatment regimen (Extended Data Figure 3A)7. We observed a significant accumulation of p-H2AX in cell treated with WTC-PM or known genotoxic agents as compared to untreated wild-type lymphocytes (Extended Data Figure 3B), indicating elevated levels of DNA damage in treated cells.
Agents that induce DNA damage can block DNA replication, leading to the loss of nucleotide bases, DNA single- and double strand breaks, mismatches, and other physical changes to DNA. To understand the effect of the damage induced by WTC-PM on DNA synthesis, we used Click-it chemistry to measure EdU incorporation and further characterized the progression of cells through S-phase. Exposure of lymphocytes to WTC-PM resulted in a very modest increase in the number of EdU positive cells (Extended Data Figure 3C), indicating that, similar to OLP, the overall number of replicating cells was not altered significantly by WTC-PM exposure. In contrast, APH treatment led to a significant increase in EdU positive cells, indicative of slower progression of cells through S-phase (Extended Data Figure 3C). These results show that WTC-PM exposure elicits a response that is different from isolated replication inhibition.
To further elucidate the effect of WTC-PM exposure on S-phase progression of the cell cycle, we compared the spatiotemporal organization of DNA replication in response to different drug exposures. Cells progressing through S-phase can be further classified as early, mid or late s-phase cells, based on the pattern of EdU incorporation (Extended Data Figure 3D; left). As expected, treatment with APH resulted in a significant increase of cells in early S-phase as compared to non-affected cells. In contrast, WTC-PM treatment caused an accumulation of cells in mid-late S-phase and this increase was more comparable to OLP treatment (Extended Data Figure 3D right). These results show that WTC-PM exposure appears to increase the rate at which cells progress through S-phase.
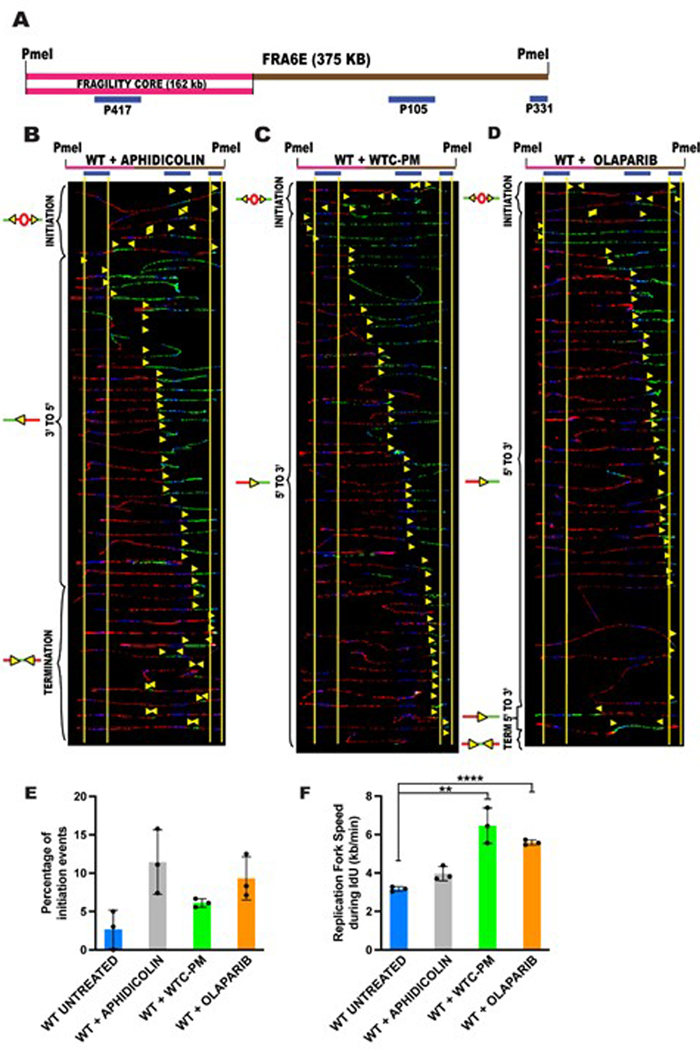
Common fragile sites (CFSs) are genomic hotspots of replication stress, designed to detect basal levels of stress/damage in the cellular environment. To further characterize the mechanism by which WTC-PM exposure affects DNA replication, we profiled DNA replication using CFSs as the model loci. We have previously characterized the replication program of wild-type lymphocytes at CFS-FRA16D and CFS-FRA6E loci using a high resolution, specialized replication assay, the single molecule analysis of replicated DNA (SMARD) (Extended Data Figure 3E) 8. We have established that replication forks at genomic loci can travel bi-directionally, and that both CFSs lack replication initiation sites, a common characteristic of CFSs.
In the current study, cells were treated for 20h, through the first 4 hours of Iododeoxyuridine (IdU) pulse, following which they were released into drug-free chlorodeoxyuridine (CldU) containing media for the remaining 4 hours. This experimental design enabled us to study the direct influence of the treatment during first 4 hours of replication and the cells’ ability to recover in the next 4 hours. The data from two adjacent regions within CFS-FRA16D (Extended Data Figure 4A; ~600kb) showed that treatment with WTC-PM, APH or OLP significantly altered the replication program at both regions 1 and 2 of FRA16D. This is apparent from the replication pausing observed (examples indicated by white rectangles) at multiple sites along the FRA16D locus (Extended Data Figure 4B–D; G–I). Furthermore, there was a striking increase in replication initiation events, characterized as dormant origins that are likely activated to rescue replication pausing (Extended Data Figure 4E, J). Further in depth analysis of the replication dynamics with each treatment measure shows that replication speeds are significantly increased in the presence of WTC-PM and OLP, in contrast to APH treatment which slows down the progression of replication machinery (Extended Data Figure 4F, K). Similar results were obtained at another independent common fragile site (CFS-FRA6E, Extended Data Figure 5). Collectively, these results establish that WTC-PM exposure significantly perturbs DNA replication dynamics in cells.
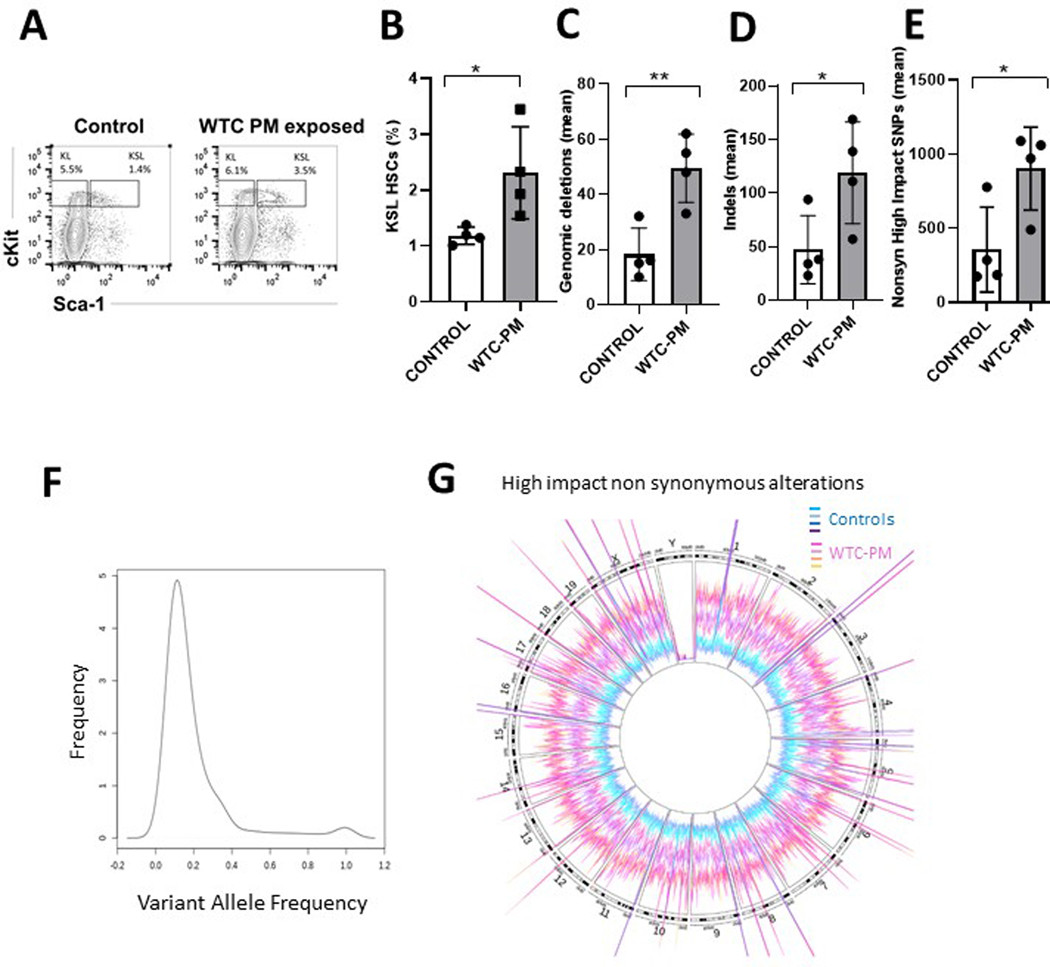
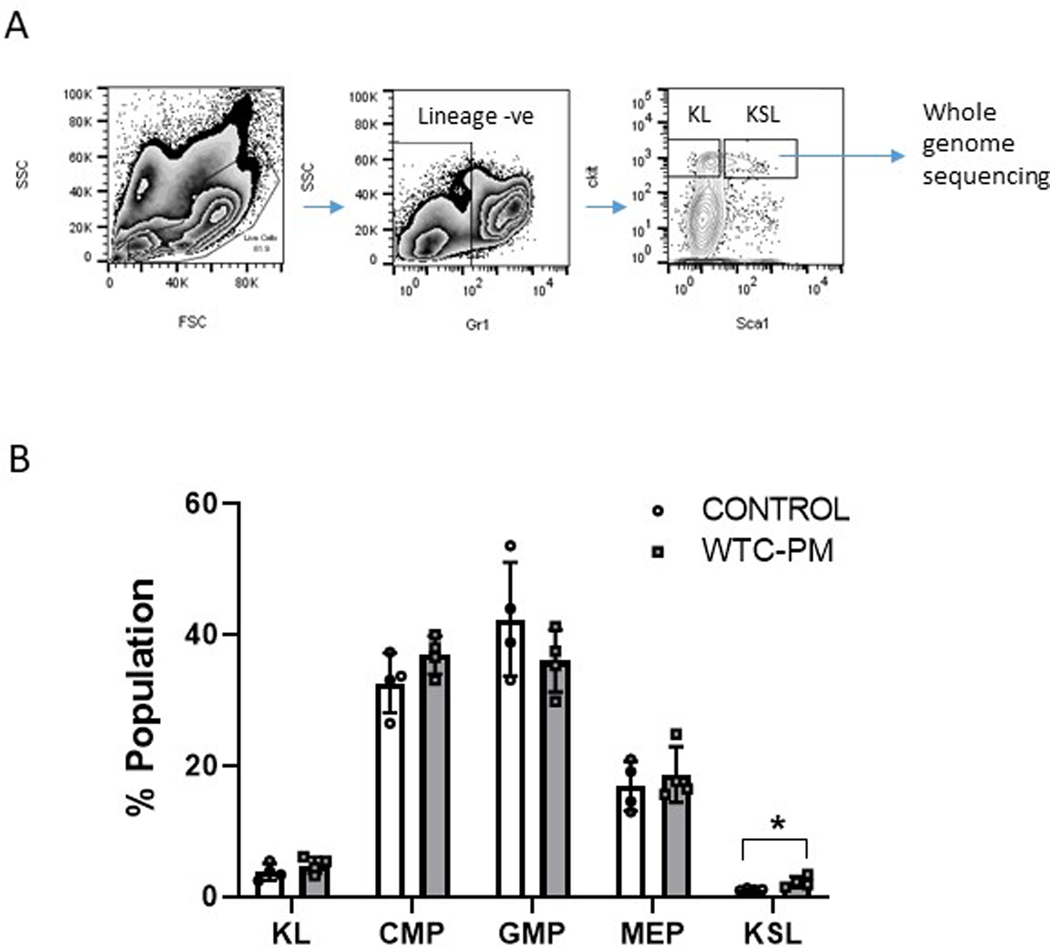
Next, we wanted to determine the effects of WTC-PM exposure on hematopoietic stem cells in vivo. To mimic the respiratory and gastrointestinal exposure to the WTC-exposed first responders, wild type mice were administered WTC-PM in the form of oropharyngeal aspiration with doses of WTC PM thought to be equivalent to the level of exposure experienced by first responders 6,9. Mice were sacrificed 30 days after exposure and bone marrow stem and progenitor compartments were analyzed and sorted. We observed a significant expansion of hematopoietic stem cells (Kit+, Sca1+, Lineage neg, KSL) in WTC-PM treated mice (Extended Data Figure 6A,B) with no significant changes in other populations (Extended Data Figure 7). The KSL+ stem cells from WTC-exposed and littermate controls were isolated by sorting and analyzed by whole genome sequencing. We observed a significant increase in non-synonymous SNPs, deletions as well as indels in stem cells obtained from WTC-PM treated mice when compared to controls (Extended Data Figure 6 C–E, Extended Data Figure 8). We constructed de novo mutational signatures of non-synonymous snps with moderate to high significance from the WTC-PM exposed mice and compared them to known human signatures. We observed that the murine mutational signatures were closely matched to COSMIC signatures that included etiologies associated with tobacco smoking (SBS04) 10 and defective homologous recombination DNA damage repair (SBS03)11 (Extended Data Figure 9). Overall, the genomic alterations observed in WTC-PM exposed mice occurred at low variant allele frequencies (Extended Data Figure 6) throughout the whole genome (Extended Data Figure 6G) suggesting a potential association that should be tested in larger cohorts
In summary, we report that WTC-exposed first responders demonstrate higher rates of CH-associated mutations, with an overall prevalence of 10%. This prevalence is significantly higher than the rates seen in non-WTC-exposed first responders, as well as those reported in the literature 5,12–14. Detection of CH-associated mutations is inherently dependent on the sensitivity of the sequencing technique used and the annotation of somatic mutations in malignancy associated genes. Although deeper sequencing is likely to detect lower frequency clones, the median VAF of clones in our study was 10% compared to the median VAF of 9% reported by Jaiswal et al., indicating that the difference in technique did not greatly bias our study towards increased detection of small, subclinical clones5.
Exposure to WTC-PM in first responders was a potential source of genotoxic stress and inflammation, factors that can potentially be associated with an increased prevalence of CH as noted previously 3,13,15. WTC PM exposure has also been associated with inflammatory respiratory and cardiac disease,16 and recent studies have demonstrated that inflammation can increase the growth of CH clones14. Even though in vitro studies demonstrated DNA replication dysregulation due to WTC-PM, we did not observe an elevated prevalence of TP53, PPM1D, CHEK2 mutations in the WTC cohort, suggesting inflammatory stress as a potential causative factor in first responders. These data demonstrate that environmental exposure to the WTC disaster site is associated with a higher burden of clonal hematopoiesis, exceeding that expected in normal aging, establish a rationale for mutational testing of the larger WTC-exposed population.
ONLINE Methods
World Trade Center exposed participants
The FDNY WTC Health Program provides monitoring exams on its WTC-exposed workforce, approximately every 12–18 months. From December 2013 through October 2015, as part of these exams, whole blood samples were collected from 481 responders who were enrolled the study which was approved by the IRB of Albert Einstein College of Medicine (07–09-320) and who signed informed consent. Data including age at sample collection, race/ethnicity, sex and smoking history were obtained from participants on the day of their exams or from the FDNY employee database.
Non-WTC-exposed comparison population
The non-WTC-exposed comparison population included 52 patients recruited at the annual convention of the International Association of Firefighters (IAFF). An additional 203 Nashville firefighters were extracted from the Synthetic Derivative, a de-identified Electronic Health Record with paired biorepository (BioVU) of Vanderbilt University Medical Center, using natural language processing methodologies. For the selected 203 patient cohort, both firefighter occupation and smoking status were manually validated.
Targeted Sequencing Analysis
The sequencing was done at a NY State CLIA certified lab at Neogenomics, NGS sequences were aligned to genome build hg19 using Burrows-Wheeler aligner and read quality and depth were determined using GATK. Single nucleotide variations were determined using Mutect and insertion/deletion mutations were determined using Pindel. Variants were excluded from analysis if there were present in ExAC/gnomAD databases at ≥1% minor allele frequency. Variants were analyzed with a minimum sequence depth of 250x and a variant allele frequency of 5% for single nucleotide variants and 10% for insertion/deletion mutations. Variants detected between 50–250x depth were analyzed base on a proprietary confidence interval. All variants were annotated by clinically trained molecular geneticists. The median depth of coverage in our study was 839x with a mean coverage of 908x. Variants with VAFs above 40% were excluded as potential germline variants.
Statistical Analyses
We calculated prevalence of somatic mutations and associated 95% confidence intervals of WTC-exposed participants using binomial assumptions for 10-year age groups (i.e., 30–39; 40–49; 50–59; 60–69; 70–79; 80 and older). We then performed a multivariable logistic regression among all responders to evaluate the association of WTC exposure and CH. Models controlled for age at the time of blood collection (as an ordinal variable), race/ethnicity (non-Hispanic white, non-Hispanic black, non-Hispanic Asian, non-Hispanic other races, Hispanic) and sex. Then, we repeated this analysis excluding 52 WTC-exposed EMS workers to allow for appropriate comparison of firefighter cohorts. Finally, two additional analyses that controlled for all the aforementioned confounders as well as self-reported smoking status (ever vs. never) were conducted, first, among the entire WTC-exposed responder cohort and then, among WTC-exposed firefighters. Because IAFF firefighters were not asked smoking status, these two models were restricted to FDNY and Nashville firefighters only.
Cell Culture
Human cell line GM03798 (wild type), Epstein–Barr virus-transformed lymphoblasts were obtained from Coriell Cell Repositories and were grown in RPMI 1640 (Gibco) supplemented with 10% FBS and penicillin/streptomycin (Gibco). Cell lines were regularly tested for mycoplasma contamination. Cells were treated with the different concentrations of the following drugs: olaparib (AZD2281; Astra Zeneca); Aphidicolin (A0781, Sigma). WTC-PM was obtained, as previously described, from 5 locations within 0.5 miles of Ground Zero on 9/13/016,17. Composition was determined by x-ray fluorescence analysis using techniques as previously published17. Cells were exposed to 200μg/ml of WTC-PM for all the experiments.
Immunofluorescence
Suspension cells adhered to Poly-L-lysine slides were fixed and permeabilized simultaneously in PTEMF buffer (20 mM PIPES pH 6.8, 10 mM EGTA, 0.2% Triton X-100, 1 mM MgCl2 and 4% formaldehyde). Slides were then incubated with primary antibody for Phospho-Histone H2A.X (Ser139) (Cell Signaling Technology) overnight at 4°C, and then with an Alexa Fluor 488 secondary antibody (Cell Signaling Technology) for 60 min at RT. After the secondary antibody incubation, slides were washed three times with 1X PBS and Click-It chemistry was utilized to detect EdU incorporation according to the manufacturer’s instructions (Click-IT EdU; Alexa fluor 594 Imaging Kits, Life Technologies). Slides were then mounted with Vecta Shield with DAPI (Vector Laboratories). Images were captured using a Zeiss fluorescence microscope
Single Molecule Analysis of Replicated DNA (SMARD)
SMARD analysis was carried out using a previously described procedure 8. Briefly, exponentially growing cells were cultured in media containing 30 μM 5-iodo-2′-deoxyuridine (IdU) at 37°C for 4h (Sigma-Aldrich, St. Louis, MO). After 4h, the cells were centrifuged at 100 g for 5 min and the media containing IdU was removed. The cells were then cultured in fresh RPMI medium containing 30 μM 5-chloro-2′-deoxyuridine (CIdU) (Sigma-Aldrich, St. Louis, MO) and incubated for an additional 4h. After 4h, the cells were collected by centrifugation, and were resuspended at 3 × 107 cells per ml in PBS. The cells were resuspended in an equal volume of molten 1% InCert agarose (Lonza Rockland, Inc., Rockland, ME) in PBS. DNA gel plugs were made by pipetting the cell-agarose mixture into a chilled plastic mold with 0.5- x 0.2-cm wells with a depth of 0.9 cm. The gel plugs were allowed to solidify on ice for 30 min. The cells in the plugs were lysed in buffer containing 1% n-lauroylsarcosine (Sigma-Aldrich), 0.5 M EDTA, and 20 mg/ml proteinase K. The gel plugs were incubated at 50°C for 3 days and treated with fresh proteinase K at 20 mg/ml concentration (Roche Diagnostics), every 24 h.
The Proteinase K digested plugs were then rinsed in Tris-EDTA (TE) and subjected to phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich) treatment. To prepare the cells for restriction enzyme digestion, the plugs were washed with 10 mM MgCl2 and 10 mM Tris-HCl (pH 8.0) and the genomic DNA in the gel plugs was digested with 70 units of PmeI or SbfI (New England BioLabs Inc.) at 37°C overnight. The digested gel plugs were rinsed with TE and cast into a 0.7% SeaPlaque GTG agarose gel (Lonza Rockland, Inc.) for size separation of DNA by pulse field gel electrophoresis. Gel slices from the appropriate positions in the pulsed-field electrophoresis gel were melted at 72°C for 20 min. The melted agarose was digested with GELase enzyme (Epicentre Biotechnologies 1 unit per 50 μl of agarose suspension) by incubating the GELase-DNA-agarose mixture at 45°C for 4 h. The resulting DNA was pipetted along one side of a coverslip that had been placed on top of a 3-aminopropyltriethoxysilane (Sigma-Aldrich)-coated glass slide and allowed to enter by capillary action. The DNA was denatured with sodium hydroxide in ethanol and fixed with glutaraldehyde.
The slides containing the DNA were hybridized overnight with biotinylated probes (represented as blue bars on the CFS locus maps). The next day, the slides were rinsed in 2 × SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) 1% SDS and washed in 40% formamide solution containing 2 × SSC at 45°C for 5 min and rinsed in 2 × SSC-0.1% IGEPAL CA-630. Following several detergent rinses (4 times in 4× SSC-0.1% IGEPAL CA-630), the slides were blocked with 1% BSA for at least 20 min and treated with Avidin Alexa Fluor 350 (Invitrogen Molecular Probes) for 20 min. The slides were rinsed with PBS containing 0.03% IGEPAL CA-630, treated with biotinylated anti-avidin D (Vector Laboratories) for 20 min, and rinsed again. The slides were then treated with Avidin Alexa Fluor 350 for 20 min and rinsed again, as in the previous step. The slides were incubated with the IdU antibody, a mouse anti-bromodeoxyuridine (Becton Dickinson Immunocytometry Systems), the antibody specific for CldU, a monoclonal rat anti-bromodeoxyuridine (anti-BrdU) (Abcam) and biotinylated anti-avidin D for 1 h. This was followed by incubation with Avidin Alexa Fluor 350 and secondary antibodies, Alexa Fluor 568 goat anti-mouse IgG (H+L) (Invitrogen Molecular Probes), and Alexa Fluor 488 goat anti-rat IgG (H+L) (Invitrogen Molecular Probes) for 1 h. The coverslips were mounted with ProLong gold antifade reagent (Invitrogen) after a final PBS/CA630 rinse. Fluorescence microscopy was carried out using a Zeiss fluorescence microscope to monitor the IdU/CIdU nucleoside.
Murine oropharyngeal aspiration model
Female wild-type C57Bl/6 mice > 12 weeks old (Jackson Laboratory) were used in the study. WTC-PM was obtained, as previously described, from 5 locations within 0.5 miles of Ground Zero on 9/13/016,17. Composition was determined by x-ray fluorescence analysis using techniques as previously published17. Oropharyngeal aspiration, equivalent to intratracheal instillation in deposition efficiency, was used to deliver PM as previously described6,9. Mice aspirated 100μg-WTC-PM suspended in sterile-PBS or isovolemic sterile-PBS (Fisher). Mice that were littermates and cohoused were exposed to both PBS and PM on the same day to avoid batch bias. After 30 days, mice were sacrificed and bone marrow cells were analyzed by FACS analysis. A single, 100μg dose of WTC-PM was chosen due to its estimated ability to cause similar adverse pulmonary effects in mice to those seen in a rescue-worker exposed to 425 μg/m3 of WTC-PM over an 8-hour shift17. This rescue worker exposure level falls within measured concentrations of PM at the 9/11 debris pile
Murine stem cell sorting and whole genome sequencing
Murine bone marrow cells were used for FACS analysis to identify Kit+ Sca1+ Lineage-ve stem cells. The antibody cocktails used are listed in Supplemental Table 4. KSL+ cells were sorted from WTC PM and control treated mice bone marrows and used for DNA isolation. DNA was amplified as previously and then used for whole genome sequencing, as previously described 18
Murine whole genome sequencing:
Genomic DNAs from mouse samples were fragmented and used for libraries preparation using Illumina Compatible Paired-End Sequencing kit by Genewiz. Libraries were sequenced on an Illumina HiSeq platform using High Output mode with a 2×150 PE sequencing configuration. DNA sequences were processed using Illumina HiSeq Analysis Software v2.2 (HAS 2.2). The package consists of a suite of proven algorithms to detect comprehensively and accurately genomic variants including single nucleotide variants (SNV), INDELs, structural variants (SV), and copy number variants (CNV) in tumor and normal samples. After sequencing was completed, base calls and quality scores were stored in bcl format files. Using bcl2fastq v. 2.17, bcl files were converted into fastq files and de-multiplexed per the barcodes used for multiplexing the samples. Data amount and quality were examined and adapter sequences were removed during the process. After fastq files were generated and the adapter removed, the reads were aligned and compared to the Mus musculus GRCm38 reference genome using the Issac Aligner. After the alignment of the reads to the reference genome, a list of SNPs and small INDELs (up to 50bp) were detected in the sample by using the Issac Variant Caller. After the alignment completed, structural variations (SV) including large INDELs (above 50 bp) were detected using the program Manta. All VCF files were annotated against the GRCm38.86 ENSEMBL annotated reference genome using SnpEff to identify genes harboring the mutations and their effects on gene products
Mutational Signatures
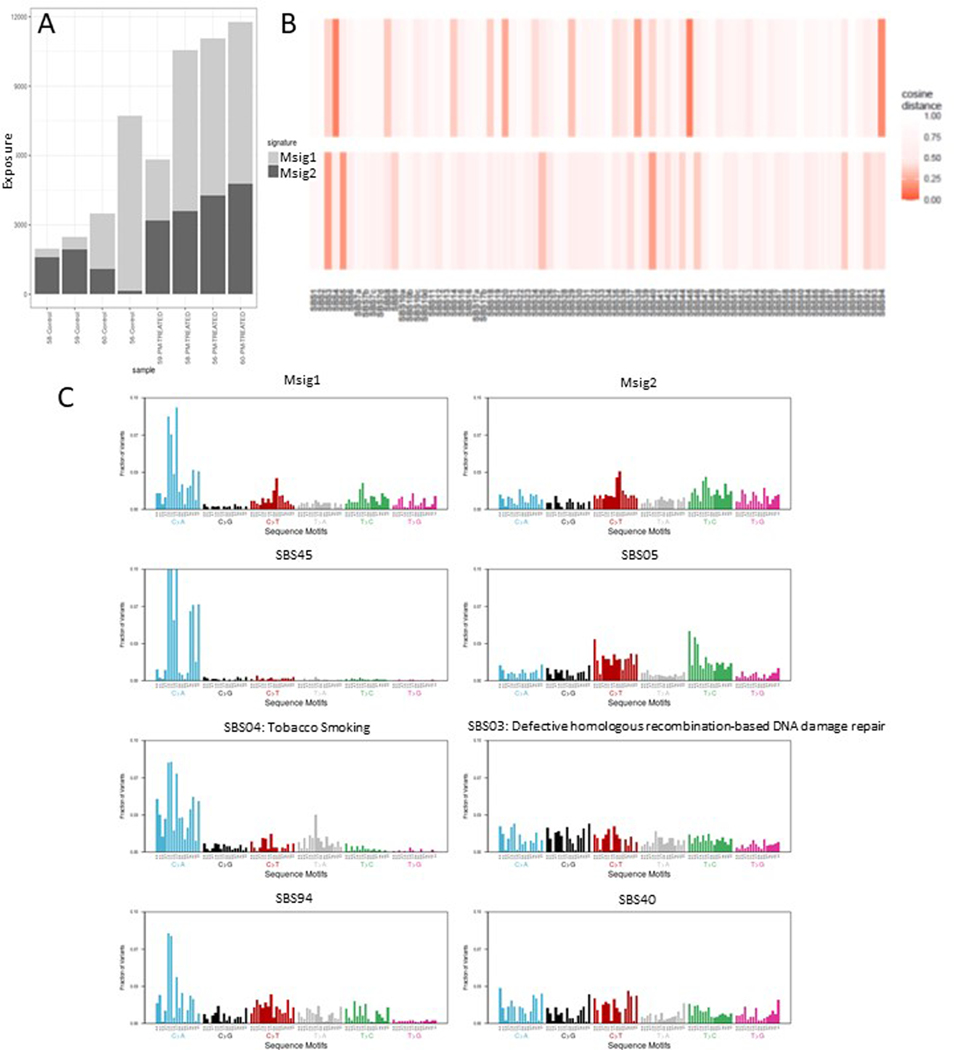
We constructed the trinucleotide matrix of the 8 mouse samples using the chromosomal positions of the high and moderate impact SNPS with adequate read depth. From this 8 × 96 dimension matrix we extracted two denovo mutational signatures (Msig1, Msig2) using an approach based on the WTSI framework by Alexandrov et al.19 relying on non-negative matrix factorization(NMF) and implemented in the R package MutSignatures 20. We plotted the composition of our 8 samples from the two denovo mutational patterns and found higher exposures in treated samples in general over both signatures. We measured the similarity of our denovo signatures to the COSMIC(v3.2 - March 2021) single base substitution (SBS) signatures with cosine distance and reported the closest three to each of our own. Our denovo signature Msig1 was closely matched to COSMIC SBS04 which has an etiology associated with tobacco smoking 10. Msig2 was closely matched to SBS3 which has a known association with defective homologous recombination DNA damage repair 11.
Extended Data
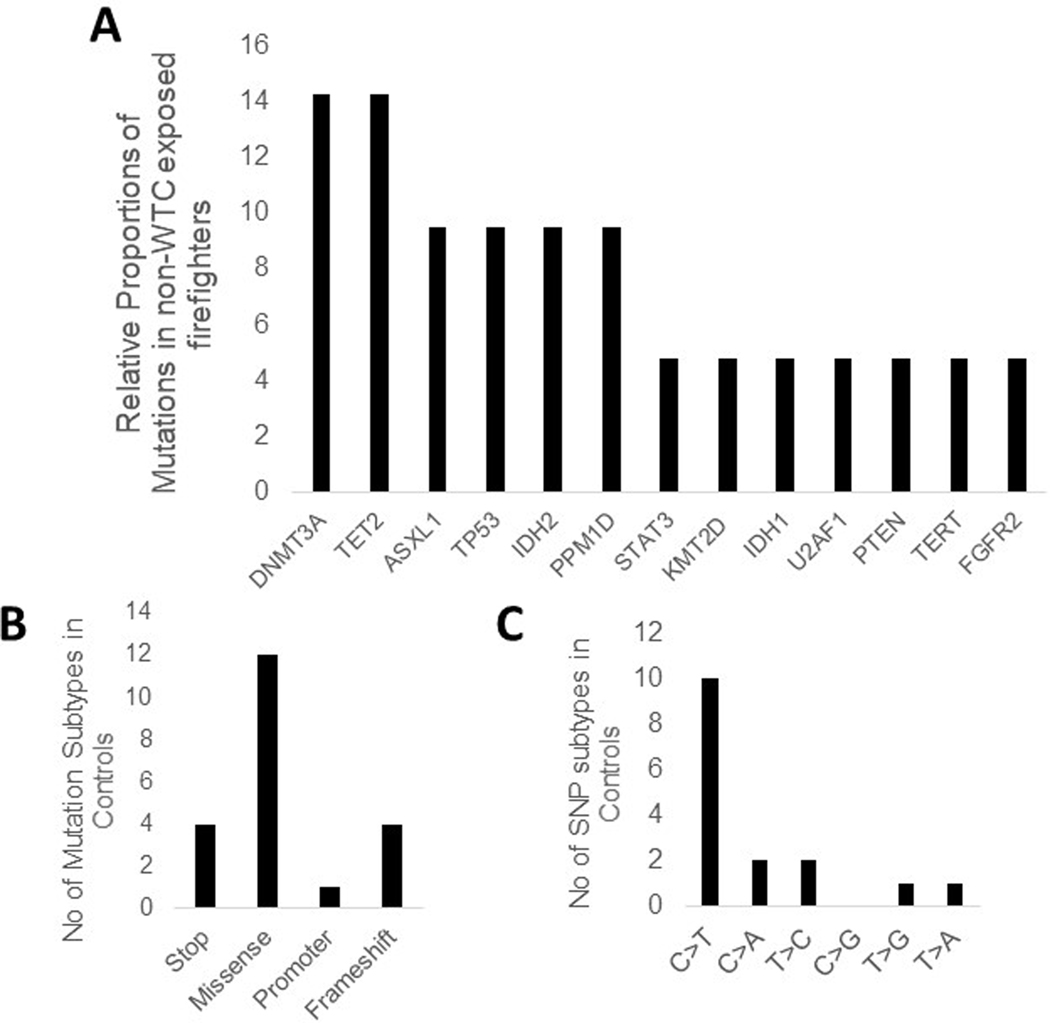
Extended Data Fig. 1. Characteristics of somatic mutations seen in non-WTC exposed first responder controls:
A: Frequency of genes found to be mutated are shown in non-WTC exposed first responder controls
B: Numbers of specific types of non synonymous mutations are shown
C: Frequency of exact nucleotide change for mutations are shown
Extended Data Fig. 2. Mutational signatures for changes seen in WTC-exposed first responders:
A: Relative weight and underlying mechanisms of different mutation signatures inferred from the mutational spectra are shown
B:The 96 trinucleotide mutational spectra of somatic mutations see in WTC exposed first responders. X-axis is showing the 96 combination of nucleotide changes, and their relative weights inferred by deconstructSigs is shown on the Y-axis.
Extended Data Fig. 3. WTC-Particulate matter (WTC-PM) exposure induces DNA damage by promoting faster progression of cells through s-phase.
(A) Schematic of treatment regime
(B) Percentage of cells with H2AX foci in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ 5uM Olaparib (orange bar) and 10uM Olaparib (yellow bar). Approximately 500 cells examined over three independent experiments. Statistical significance was assessed using a two-tailed t-test where ****p<.0001.
(C) Percentage of cells with EdU incorporation in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ 5uM Olaparib (orange bar) and 10uM Olaparib (yellow bar). Approximately 250 cells examined over three independent experiments. Statistical significance was assessed using a two-tailed t-test.
(D) Spatiotemporal pattern of DNA replication. Percentage of cells with EdU incorporation patterns characteristic of early (blue bar), mid (red bar) and late (green bar) s-phase. Error bars represent mean ± s.d. from data collected where n= ~250 cells examined over three independent experiments. Statistical significance was assessed using a two-tailed t-test.
(E) Schematic representation of the various stages of single molecule analysis of replicated DNA (SMARD). Cells are pulsed with nucleoside analogs (IdU-green; CIdU-red) and embedded in agarose plugs. The cells are first lysed; proteins are digested by proteinase K and then subjected to restriction digestion. The restriction digested DNA is resolved by pulse field gel electrophoresis. The slice containing the FRA16D locus is identified by PCR analysis. The agarose from the identified slice is melted and the DNA is stretched onto silanized glass slides. Biotinylated FISH probes are used for identification of the fragment and immunostaining is utilized to visualize the IdU tract in red, the CIdU tract in green and the FISH probes in blue. The resulting molecules are arranged to yield recognizable replication patterns (from the left): initiating molecules, terminating molecules, replication forks progressing in the 3’ to 5’ and 5’ to 3’ direction which are easily interpreted by the IdU incorporation histograms.
Extended Data Fig. 4. WTC-PM exposure perturbs DNA replication at common fragile site FRA16D.
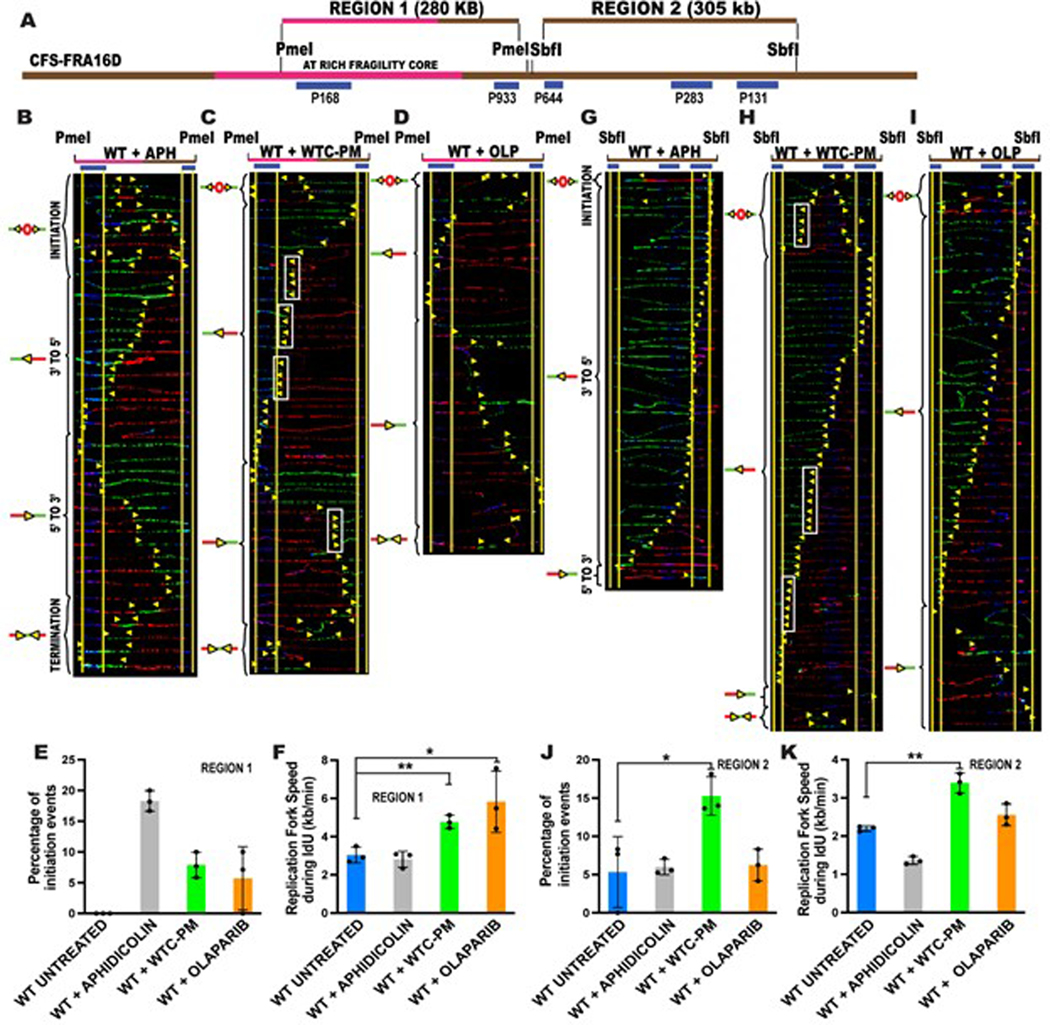
(A) Locus map of the Region 1(R1)-PmeI and Region 2(R2)-SbfI segments, of CFS-FRA16D. The FISH probes that identify the segment are labeled in blue.
(B-D) Top; Locus map of PmeI digested R1 segment. Bottom; Aligned photomicrograph images of labeled DNA molecules from the WILDTYPE (WT) lymphoblastoid cell line treated with 0.2μM Aphidicolin for 20h (B); or treated with 200μg/ml WTC-PM for 20h (C); or treated with 5μM olaparib for 20h (D).
(E) Percentage of molecules with replication initiation sites in Region 1 of CFS-FRA16D in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ olaparib (orange bar). Error bars represent mean ± s.d. from data collected where n=40 molecules analyzed over three independent experiments. Statistical significance was assessed using a two-tailed t-test. Note: Aphidicolin, WTC-PM and olaparib are present during IdU pulse.
(F) Replication fork speed during the IdU pulse of SMARD (first 4 hours of pulsing) in Region 1 of CFS-FRA16D in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ olaparib (orange bar). Error bars represent mean ± s.d. from data collected where n= ~200 DNA molecules analyzed from three independent experiments. Statistical significance was assessed using a two-tailed t-test where *p=0.0439, **p=0.0052. Note: Aphidicolin, WTC-PM and olaparib are present during IdU pulse.
(G-I) Top; Locus map of SbfI digested R2 segment. Bottom; Aligned photomicrograph images of labeled DNA molecules from the WILDTYPE (GM03798) lymphoblastoid cell line treated with 0.2μM Aphidicolin for 20h (G); or treated with 200μg/ml WTC-PM for 20h (H); or treated with 5μM olaparib for 20h (I). The yellow arrows indicate the sites along the molecules where the IdU transitioned to CldU. White rectangles indicate representative sites of replication fork pausing. The molecules are arranged in the following order: molecules with initiation events, molecules with 3’ to 5’ progressing forks, molecules with 5’ to 3’ progressing forks and molecules with termination events.
(J) Percentage of molecules with replication initiation sites in Region 2 of CFS-FRA16D in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ olaparib (orange bar). Error bars represent mean ± s.d. from data collected where n=40 molecules analyzed over three independent experiments. Statistical significance was assessed using a two-tailed t-test where *p=0.0311. Note: Aphidicolin, WTC-PM and Olaparib are present during IdU pulse.
(K) Replication fork speed during the IdU pulse of SMARD (first 4 hours of pulsing) in Region 2 of CFS-FRA16D in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ Olaparib (orange bar). Error bars represent mean ± s.d. from data collected where n= ~200 DNA molecules analyzed from three independent experiments. Statistical significance was assessed using a two-tailed t-test where **p=0.0016 Note: Aphidicolin, WTC-PM and olaparib are present during IdU pulse.
Extended Data Fig. 5. WTC-PM exposure perturbs DNA replication at common fragile site FRA6E.
(A): Locus map of a 375 kb region in the CFS-FRA6E obtained by PmeI digestion. The region includes the fragility core of CFS-FRA6E (pink line – 162 kb). The FISH probes that identify the segment are labeled in blue.
(B-D): Top; Locus map of the PmeI digested FRA6E segment. Bottom; Aligned photomicrograph images of labeled DNA molecules from the WILDTYPE (GM03798) lymphoblastoid cell line treated with 0.2μM Aphidicolin for 20h (D); or treated with 200μg/ml WTC-PM for 20h (E); or treated with 5μM Olaparib for 20h (F).
(E): Percentage of molecules with replication initiation sites in CFS-FRA6E in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ olaparib (orange bar). Error bars represent mean ± s.d. from data collected where n=40 molecules analyzed over three independent experiments. Note: Aphidicolin, WTC-PM and Olaparib are present during IdU pulse.
(F): Replication fork speed during the IdU pulse of SMARD (first 4 hours of pulsing) CFS-FRA6E in wildtype (WT) Untreated (blue bar), WT+ Aphidicolin (grey bar), WT+ WTC-PM (green bar) and WT+ Olaparib (orange bar). Error bars represent mean ± s.d. from data collected where n= ~200 DNA molecules analyzed from three independent experiments. Statistical significance was assessed using a two-tailed t-test where **p=0.0035, ****p<0.0001. Note: Aphidicolin, WTC-PM and olaparib are present during IdU pulse.
Extended Data Fig. 6. Genomic alterations induced by exposure to WTC PM in vivo:
A: Mice were treated with WTC PM and used for bone marrow stem and progenitor FACS analysis. Representative control and WTC PM treated mice samples are shown.
B: Hematopoietic Kit +, Sca1+, Lineage –ve stem cells are shown for WTC PM treated and control mice (N=4 individual mice per group, Means +/− SD, Two tailed TTest, P=0.036)
C: Numbers of high impact deletions are shown for WTC PM and control treated mice within stem and progenitors compartments (N=4 individual mice per group, Means +/− SD, Two tailed TTest, P=0.007)
D: Numbers of high impact indels are shown for WTC PM and control treated mice within stem and progenitors compartments (N=4 individual mice per group, Means +/− SD, Two tailed TTest, P=0.046)
E: Numbers of high impact nonsynonymous SNPs are shown for WTC PM and control treated mice within stem and progenitor compartments. N=4 individual mice per group, Means +/− SD, Two tailed TTest, P=0.03)
F: Variant allele frequency (VAF) of high impact nonsynonymous genomic changes in WTC PM treated stem and progenitors.
G: Circos plot showing magnitude of nonsynonymous genomic changes in 4 WTC PM and 4 control mice.
Extended Data Fig. 7. Mice treated with WTC PM were analyzed for stem and progenitor alterations:
A: Mice were treated with WTC PM were sacrificed at 30 days after oropharyngeal exposure and used for bone marrow stem and progenitor FACS analysis. Representative sorting strategy for KSL stem cells is shown.
B: Relative proportions of stem and progenitor populations are shown. Means +/− SD of 4 mice in each group.
Extended Data Fig. 8. Hematopoietic stem and progenitor cells from mice treated with WTC PM show genomic instability:
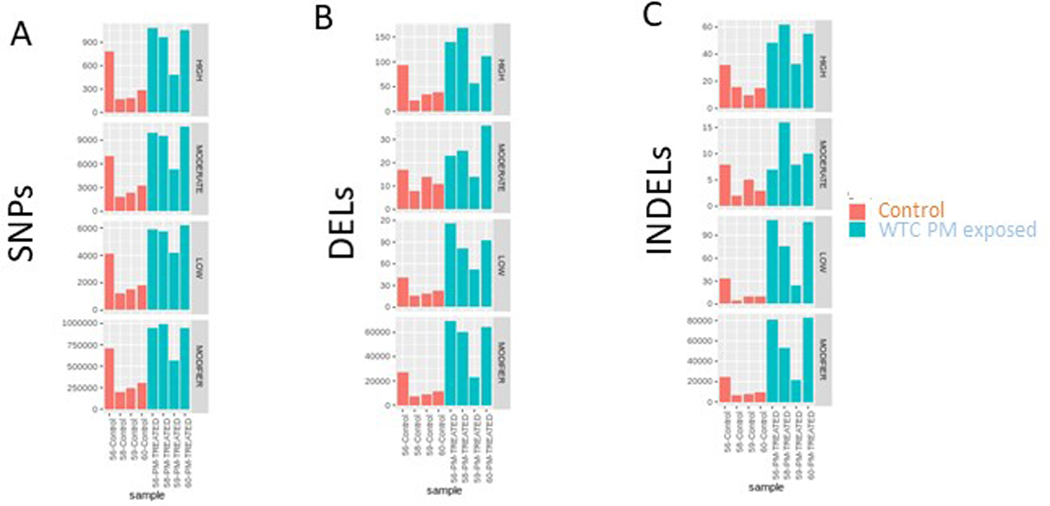
A: Numbers of high, moderate, low and modifier impact SNPs are shown for WTC PM treated and control mice within stem and progenitor compartments. Individual mice are shown.
B: Numbers of high, moderate, low and modifier impact deletions are shown for WTC PM treated and control mice within stem and progenitor compartments. Individual mice are shown.
C: Numbers of high, moderate, low and modifier impact indels are shown for WTC PM treated and control mice within stem and progenitor compartments. Individual mice are shown.
Extended Data Fig. 9. Murine mutational signatures similar to human mutational signatures associated with smoking and defective DNA repair:
A: De novo murine mutational signatures (Msig1 and Msig2) were created from high and moderate impact snps and show greater signature activities in the WTC-PM exposed mice.
B: The murine signatures were compared to known human mutational signatures. Greater similarity was shown with higher intensity of color on the heatmap. Human signatures with most similarity to Msig1 were SBS45, SBS4, SBS94. Human signatures with most similarity to Msig2 were SBS5, SBS3, SBS40.
C: Human signatures with most similarity to murine signatures were SBS 03, 04, 05, 40, 45 and 90.
Extended Data Table 1: Demographics of WTC-exposed and non-WTC-exposed first responders.
(Smoking information was only available for 203 controls)
| WTC Exposed (N=481) N (%) | Non WTC Exposed (N=255) N (%) | |
|---|---|---|
|
| ||
| Age (yrs) | ||
| 30–39 | 16 (3%) | 0 (%) |
| 40–49 | 100(21%) | 22 (9%) |
| 50–59 | 215 (45%) | 82 (32%) |
| 60–69 | 115 (24%) | 86 (34%) |
| 70–79 | 32 (7%) | 48 (19%) |
| 80–89 | 3(1%) | 15(6%) |
| >90 | 0 | 2 (1%) |
| Race | ||
| Caucasians | 402 (84%) | 238 (93%) |
| Other Races | 79 (16%) | 17(7%) |
| Sex | ||
| Male | 468 (97%) | 245 (96%) |
| Female | 13 (3%) | 10(4%) |
| Smoking | ||
| Smoker | 173 (36%) | 102 (50%) |
| Non-Smoker | 308 (64%) | 101 (50%) |
Supplementary Material
ACKNOWLEDGEMENTS
The work was supported by NIH U01OH011933 (AV), U01OH012271 (AV), U01-OH11300 (AN), F30DK127699 (AJS), R01HL119326 (AN) and T32GM007347 (MRS) grants, the Jane and Myles Dempsey Fund (AV), Leukemia Lymphoma Society (AV), the Edward P. Evans Foundation (AV), the Valvano Foundation (AV), the Adventure Alle Fund (MRS), the Biff Ruttenberg Foundation (MRS), the Beverly and George Rawlings Directorship (MRS), and a gift from the Leinbach family (AV). The BioVU dataset used for the analyses described was obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant ULTR000445 from NCATS/NIH. FDNY WTC-Health Program supported by the Clinical Center of Excellence 200-2017-93426 and Data Center 200-2017-93326.
COMPETING INTERESTS STATEMENT
AV has received research funding from Prelude, BMS, GSK, Incyte, Medpacto, Curis and Eli Lilly, is a scientific advisor for Stelexis, Novartis, Acceleron and Celgene, receives honoraria from Stelexis and Janssen and holds equity in Stelexis and Throws Exception. B.L.E. has received research funding from Celgene, Deerfield, Novartis, and Calico and consulting fees from GRAIL. He is a member of the scientific advisory board and shareholder for Neomorph Therapeutics, TenSixteen Bio, Skyhawk Therapeutics, and Exo Therapeutics.
Footnotes
Editor recognition statement:
Michael Basson was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Reviewer recognition statement:
Nature Medicine thanks Duane Hassane Daniel Link, Catherine Metayer, Stanton Gerson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
DATA AVAILABILITY STATEMENT:
All sequencing data for first responders and control has been deposited to the European Variation Archive Project with Accession ID: PRJEB49193. https://www.ebi.ac.uk/eva/
REFERENCES
- 1.Lioy PJ, et al. Characterization of the dust/smoke aerosol that settled east of the World Trade Center (WTC) in lower Manhattan after the collapse of the WTC 11 September 2001. Environ Health Perspect 110, 703–714 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, et al. Cancer Incidence in World Trade Center Rescue and Recovery Workers: 14 Years of Follow-Up. J Natl Cancer Inst (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeig-Owens R, et al. Early assessment of cancer outcomes in New York City firefighters after the 9/11 attacks: an observational cohort study. Lancet 378, 898–905 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Landgren O, et al. Multiple Myeloma and Its Precursor Disease Among Firefighters Exposed to the World Trade Center Disaster. JAMA Oncol 4, 821–827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371, 2488–2498 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caraher EJ, et al. Receptor for advanced glycation end-products and World Trade Center particulate induced lung function loss: A case-cohort study and murine model of acute particulate exposure. PLoS One 12, e0184331 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maya-Mendoza A, et al. High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Madireddy A, et al. FANCD2 Facilitates Replication through Common Fragile Sites. Mol Cell 64, 388–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiden MD, et al. Comparison of WTC dust size on macrophage inflammatory cytokine release in vivo and in vitro. PLoS One 7, e40016 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nik-Zainal S, et al. The genome as a record of environmental exposure. Mutagenesis 30, 763–770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zamborszky J, et al. Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions. Oncogene 36, 5085–5086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bick AG, et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586, 763–768 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371, 2477–2487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trowbridge JJ & Starczynowski DT Innate immune pathways and inflammation in hematopoietic aging, clonal hematopoiesis, and MDS. J Exp Med 218(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombs CC, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell 21, 374–382 e374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen HW, et al. Long-term Cardiovascular Disease Risk Among Firefighters After the World Trade Center Disaster. JAMA Netw Open 2, e199775 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGee JK, et al. Chemical analysis of World Trade Center fine particulate matter for use in toxicologic assessment. Environ Health Perspect 111, 972–980 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat Med 25, 103–110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fantini D, Vidimar V, Yu Y, Condello S.& Meeks JJ MutSignatures: an R package for extraction and analysis of cancer mutational signatures. Sci Rep 10, 18217 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data for first responders and control has been deposited to the European Variation Archive Project with Accession ID: PRJEB49193. https://www.ebi.ac.uk/eva/