The chromatin repressor PRC2 induces HLA class II silencing in acute myeloid leukemia leading to immune escape and posttransplantation relapse, while its chemical inhibition rescues HLA expression and triggers T cell–mediated leukemia eradication.
Abstract
Immune escape represents a major driver of acute myeloid leukemia (AML) reemergence after allogeneic hematopoietic cell transplantation (allo-HCT), with up to 40% of relapses prompted by nongenomic loss of HLA class II expression in leukemia cells. By integrative analysis of gene expression, DNA methylation, and chromatin accessibility in paired diagnosis/relapse primary samples and in the respective patient-derived xenografts (PDX), we identify the polycomb repressive complex 2 (PRC2) as a key epigenetic driver of this immune escape modality. We report that loss of expression of HLA class II molecules is accompanied by a PRC2-dependent reduction in chromatin accessibility. Pharmacologic inhibition of PRC2 subunits rescues HLA class II expression in AML relapses in vitro and in vivo, with consequent recovery of leukemia recognition by CD4+ T cells. Our results uncover a novel link between epigenetics and leukemia immune escape, which may rapidly translate into innovative strategies to cure or prevent AML posttransplantation relapse.
Significance:
Loss of HLA class II expression represents a frequent mechanism of leukemia posttransplantation relapse. Here we identify PRC2 as the main epigenetic driver of this immune escape modality and show that its chemical inhibition can reinstate a proficient graft-versus-leukemia effect, providing an innovative rationale for personalized epigenetic immunotherapies.
See related commentary by Köhler and Zeiser, p. 1410.
This article is highlighted in the In This Issue feature, p. 1397
INTRODUCTION
In the last few decades, major breakthroughs have improved the feasibility and safety of allogeneic hematopoietic cell transplantation (allo-HCT) to cure acute myeloid leukemia (AML). Still, posttransplantation relapses remain a major unsolved issue, occurring in up to 50% of patients. It is becoming increasingly recognized that evasion from immune control represents one of the main drivers of posttransplantation relapse (1, 2). Among the mechanisms of immune escape described so far, the selective loss of HLA class II expression on leukemic cells represents one of the most frequent, accounting for up to 40% of posttransplantation relapses (3, 4). The central role of the interaction between CD4+ T cells and HLA class II molecules in driving the graft-versus-tumor effect of allo-HCT has been suggested by both preclinical (5, 6) and clinical (7) studies, and the observation that loss of HLA class II can be sufficient to prompt clinical relapse represents another evidence in this direction.
Of note, genomic profiling of relapses with downregulation of HLA class II molecules did not identify any mutation that could explain the immune escape phenotype, and in particular no specific genetic lesions in either HLA genes or their known regulators were identified (4).
Here, we investigate the cross-talk between epigenetics and cancer immune surveillance, deciphering the molecular mechanisms that control nongenomic loss of HLA class II expression in AML posttransplantation relapses, and propose a new therapeutic rationale to reawaken the immune system by the personalized use of epigenetic therapies.
RESULTS
Patient-Derived Xenografts Recapitulate Relapse-Specific Epigenetic Alterations
To identify and characterize the epigenetic changes responsible for AML immune escape, we conceived an approach based on multiomic profiling of primary AML samples collected longitudinally over treatment and of their respective patient-derived xenografts (PDX; Fig. 1A).
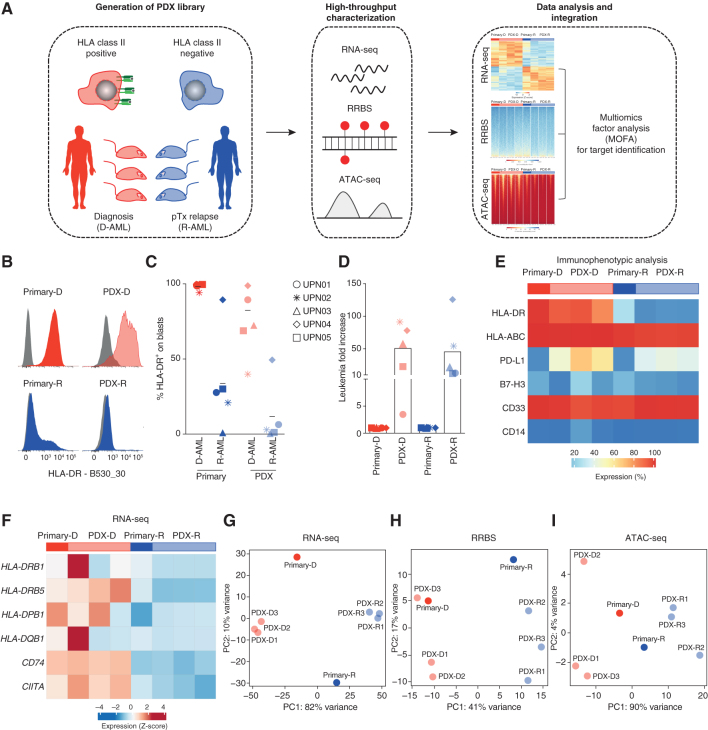
Figure 1.
PDXs recapitulate the immune-related features of primary posttransplantation relapses. A, Overview of the experimental approach to identify epigenetic drivers of AML posttransplantation (pTx) immune escape and relapse. ATAC-seq, Assay for Transposase-Accessible Chromatin using sequencing; RNA-seq, RNA sequencing; RRBS, reduced representation bisulfite sequencing. B, Representative histograms from immunophenotypic analysis of HLA-DR surface expression measured on primary (left, dark colors) and PDX (right, light colors) diagnosis (red, top) and relapse (blue, bottom) samples from UPN01. C, HLA-DR surface expression measured by immunophenotypic analysis on primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples from the different patients. For PDXs, the average of results obtained in the different mice infused with the same primary leukemia is reported. D, Fold change in the number of human leukemic blasts infused in animals (dark colors) and recovered when they were euthanized (light colors) for each of the five cases tested. Shown is the average of results obtained in the different mice infused with the same primary leukemia. E, Heat map representing the surface expression levels of lineage and immune-related markers on UPN01 primary and PDX-derived leukemic cells, measured by immunophenotypic analysis. The bar above the heat map indicates primary (dark colors) and PDX (n = 3 mice for each primary sample, light colors) diagnosis (red) and relapse (blue) samples. F, Heat map representing expression levels of HLA class II transcripts and of their regulators CD74 and CIITA in the UPN01 RNA-seq data set. The bar above the graph indicates primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples. G, PCA of the relative spatial distribution of primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples from the UPN01 RNA-seq data set. H, PCA of the relative spatial distribution of primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples from the UPN01 RRBS data set. I, PCA of the relative spatial distribution of primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples from the UPN01 ATAC-seq data set.
We first identified five patients with downregulation of HLA class II expression at posttransplantation relapse, detected by immunophenotypic analysis and RNA sequencing (RNA-seq; UPN01–05; Supplementary Table S1 and Supplementary Fig. S1A–S1C), and generated PDXs from their paired diagnosis/relapse samples. For all five cases, PDXs largely recapitulated the levels of expression of HLA class II of the original samples (Fig. 1B and C; Supplementary Fig. S2 for gating strategy) and allowed the sizable expansion of the infused leukemia (average 50.7-fold for diagnoses and 45.4-fold for relapses; Fig. 1D).
We selected UPN01 as index case and characterized in more detail the surface expression of immune receptors (HLA class I, PD-L1, and B7-H3) and lineage markers (CD14 and CD33) in primary and PDX samples (Fig. 1E), documenting that all of them were largely unaffected by passage and expansion in PDXs, and consistent among replicates for both diagnoses and relapses (Fig. 1E).
AML blasts from UPN01 PDXs and from the original samples were then profiled by RNA-seq for their transcriptome, reduced representation bisulfite sequencing (RRBS) for DNA methylation, and the Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) for chromatin accessibility. RNA-seq analysis confirmed that PDXs recapitulated the differences in the transcript levels of HLA class II (HLA-DRB1, HLA-DRB5, HLA-DPB1, and HLA-DQB1) and class II–related genes (CD74/invariant chain and CIITA) between diagnosis and relapse (Fig. 1F). Of note, this was true also for their global transcriptional profile, as shown by the principal component analysis (PCA) of our RNA-seq data set (Fig. 1G). Similar observations were also made when analyzing methylation by RRBS (Fig. 1H) and chromatin accessibility by ATAC-seq (Fig. 1I). For all three technologies, the first component (PC1) defined the clear segregation between diagnosis and relapse samples, whereas PC2 (accounting for a minor fraction of the overall variance) captured differences between primary samples and PDXs (Fig. 1G–I). Importantly, genes differentially expressed between primary samples and PDXs belonged mostly to nonimmune-related categories and were mainly involved in the processes of cell proliferation, myeloid differentiation, and protein trafficking, likely reflecting a higher proliferative state of leukemia upon transfer in mice (Supplementary Fig. S3A–S3C).
Our observations indicate that early-passage PDXs recapitulate the immune-related landscape of primary AML samples, allowing their use in combination with the original samples to characterize epigenetic drivers of relapse with loss of expression of HLA class II molecules.
High-Throughput Profiling Identifies Relapse-Specific Chromatin Compaction at the Level of HLA Class II Genes
We next used the entire sample set derived from UPN01 (including primary and PDX samples) to understand if the transcriptional differences between diagnosis and relapse could be explained by differences in DNA methylation and chromatin accessibility. We first analyzed the data from each technology separately. By RRBS we did not identify significant differences in the level of DNA methylation between diagnosis and relapse samples, neither at promoters nor at gene bodies (Fig. 2A and B). Conversely, ATAC-seq showed that relapse samples displayed reduced chromatin accessibility at transcription start sites as compared with their diagnosis counterparts (Fig. 2C and D). In line with these findings, genes differentially methylated (DMG) between diagnosis and relapse were few (n = 130; Supplementary Fig. S4A), were poorly correlated with differentially expressed transcripts (r2 = 0.1121, P = 0.7043; Supplementary Fig. S4B), and were largely nonoverlapping with them (Supplementary Fig. S4C; P = 0.383, representation factor 0.9), whereas differentially accessible genes (DAG) were more numerous (n = 6,085; Supplementary Fig. S4D) and more evidently associated with changes in gene expression (r2 = 0.3352, P = 8.311e−35 for correlation and P = 7.826E−112 for overlap, representation factor 1.8; Supplementary Fig. S4C and S4E).
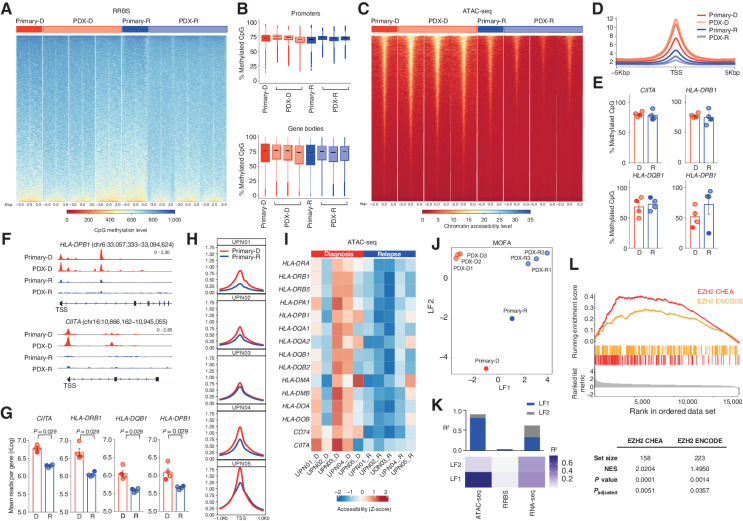
Figure 2.
HLA class II downregulation at posttransplantation relapse is explained by chromatin compaction and heightened EZH2 activity. A, Heat map representing CpG methylation around the transcription start site (TSS) in UPN01 RRBS data set. The bar above the graph indicates primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples. B, Quantitative analysis of CpG methylation at promoters (top) and gene bodies (bottom) in UPN01 primary (dark colors) and PDX (light colors), diagnosis (red) and relapse (blue) samples, measured by RRBS. C, Heat map representing chromatin accessibility around the TSS from the UPN01 ATAC-seq data set. The bar above the graph indicates primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples. D, Average genome-wide accessibility of chromatin around the TSS in UPN01 primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples, measured by ATAC-seq. E, Quantitative analysis of CpG methylation of HLA class II genes and CIITA in UPN01 primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples, measured by RRBS. F, Integrative Genomics Viewer visualization of ATAC-seq tracks from UPN01 primary and PDX representative samples at diagnosis (red, top lanes) and relapse (blue, bottom lanes). Shown are the genomic coordinates for HLA-DPB1 and CIITA. G, Quantitative analysis of chromatin accessibility of HLA class II genes and CIITA in UPN01 primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples, measured by ATAC-seq. P values were calculated by a two-sided paired t test at a 95% confidence interval. H, Average genome-wide accessibility of chromatin around the TSS in diagnosis (red) and relapse (blue) samples from each of the five patients, measured by ATAC-seq. I, Heat map representing chromatin accessibility of HLA class II genes and their regulators measured by ATAC-seq in paired diagnosis/relapse samples from the five patients under study. The bar above the graph indicates diagnosis (red) and relapse (blue) samples. J, PCA of the relative spatial distribution of primary (dark colors) and PDX (light colors) diagnosis (red) and relapse (blue) samples from the UPN01 MOFA data set, integrating information from RNA-seq, RRBS, and ATAC-seq. K, Heat map representing the proportion of the total variance in the UPN01 MOFA data set (R2) explained by individual modalities of analysis for the first two LFs and cumulative proportion of the total variance explained by the first two LFs of these modalities (histograms above the heat map). L, GSEA performed on all genes contributing to the ATAC-seq signal from LF1.
In line with the global epigenetic profiling, we documented that HLA class II genes and their transactivator CIITA also did not differ between diagnosis and relapse in their methylation level (Fig. 2E), whereas they acquired a significantly more closed chromatin conformation status at posttransplantation relapse (Fig. 2F and G). Chromatin compaction at the genome-wide level and in HLA class II genes and their regulators was consistent in all five posttransplantation relapses from our series (Fig. 2H and I).
Multiomics Factor Analysis Identifies the Involvement of Polycomb Repressive Complex 2 in Loss of HLA Class II Expression at Posttransplantation Relapse
To analyze data obtained from the different high-throughput technologies in an integrative and unsupervised manner, we applied multiomics factor analysis (MOFA; ref. 8), a computational method able to extract the major factors of variability [called latent factors (LF)] from input data, concomitantly taking into account all the biological and technical layers of variability of the sample set analyzed in terms of the cell of origin (primary and PDXs) and condition (diagnosis and relapse). MOFA of our data set documented that LF1 captures differences between diagnosis and relapse, whereas LF2 discriminates primary and PDX samples (Fig. 2J), and that major contributors to LF1 were gene expression (as expected, because it represented the starting point for our analysis, r2 = 0.56) and chromatin accessibility (r2 = 0.34), with a very small contribution from DNA methylation (r2 < 0.01; Fig. 2K).
Based on the observation that differences between diagnosis and posttransplantation relapse were mostly explained by changes in chromatin compaction, we further interrogated the list of DAGs from LF1 to characterize its content and identify its epigenetic regulators. Gene set enrichment analysis (GSEA) highlighted a significant overlap between our LF1 and signatures related to the activity of enhancer of zeste homolog 2 (EZH2), the catalytic subunit of the polycomb repressor complex 2 [PRC2; ChEA 2016: normalized enrichment score (NES) = 2.0204, Padjusted = 0.0051; ENCODE: NES = 1.4950, Padjusted = 0.0357; Fig. 2L]. The PRC2 multiprotein complex acts as a chromatin repressor by catalyzing the reaction of trimethylation at the level of lysine 27 of histone 3 (H3K27me3), thus inducing transcriptional repression at its target loci (9).
Consistent with findings obtained by MOFA for UPN01, individual analyses of RNA-seq and ATAC-seq of all five patients also pointed to PRC2 as the main epigenetic regulator of genes altered at relapse (Supplementary Fig. S5A–S5D), and expression of PRC2 subunits was upregulated at relapse (Supplementary Fig. S5E).
As a relevant control, we studied five cases of AML with relapse after sole chemotherapy (UPN06CT–10CT; Supplementary Table S2), detecting levels of HLA class II proteins and transcripts similarly high at diagnosis and relapse (Supplementary Fig. S6A and S6B), paralleled by largely unchanged chromatin accessibility (Supplementary Fig. S6C and S6D). Interestingly, DAGs were enriched for an EZH2-related signature but in the opposite direction compared to posttransplantation relapses (Supplementary Fig. S6E and S6F).
The observation that HLA class II genes and regulators gained a closed conformational state at posttransplantation relapse (Fig. 2F–I), together with the enrichment of PRC2 targets in genes differentially regulated between diagnosis and relapse (Fig. 2L; Supplementary Fig. S5A–S5E), led us to hypothesize that PRC2 might represent the main driver of HLA class II downregulation in posttransplantation leukemia relapses and prompted us to functionally validate this assumption.
Pharmacologic Inhibition of PRC2 Rescues the HLA Class II Expression of Posttransplantation AML Relapses
To experimentally assess the involvement of PRC2 in the silencing of HLA class II at posttransplantation relapse, we tested the effect of its pharmacologic inhibition in relapses from our series. Upon ex vivo treatment with PRC2 inhibitors, we characterized the chromatin changes and the effects on the transcription and surface expression of HLA class II genes and regulators (Fig. 3A). The EZH2 inhibitor EPZ-6438 (tazemetostat) was effective in reducing H3K27me3 protein levels in cell lines (Fig. 3B) and in HLA class II–negative AML relapses (P < 0.0001; Fig. 3C) without affecting blast viability (Fig. 3D). Treatment of relapse samples with EPZ-6438 increased chromatin accessibility of HLA class II genes and their regulators (Fig. 3E) and upregulated the expression of their respective transcripts (Fig. 3F), with the exception of CIITA accessibility in UPN03, which was inconsistent with the other genes and patients. Of interest, most of the genes that were significantly upregulated upon exposure of relapsed leukemia to EPZ-6438 were immune-related (Fig. 3G). We validated these findings by quantitative PCR (Fig. 3H) and immunophenotypic analysis (Fig. 3I): EZH2 inhibition increased the expression of HLA class II genes in all AML relapses analyzed, although surface protein levels did not reach those observed at diagnosis (Fig. 3I; Supplementary Fig. S7).
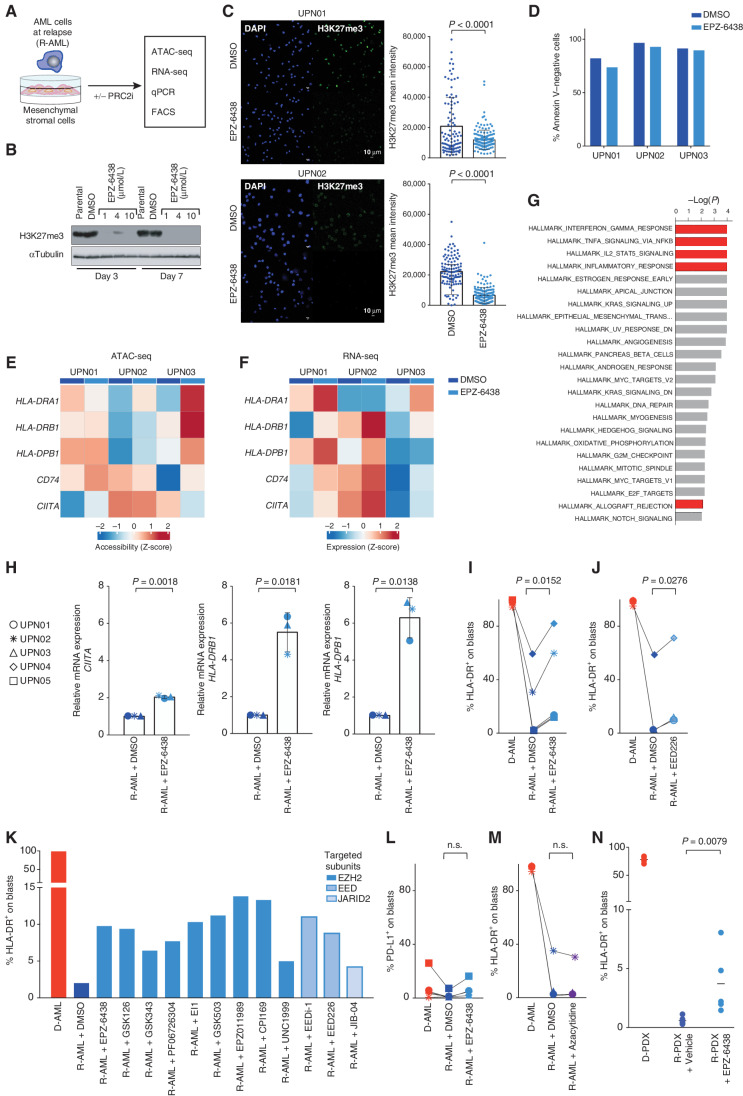
Figure 3.
Inhibition of EZH2 and of the other subunits of PRC2 leads to recovery of HLA class II expression on relapsed leukemia in vitro and in vivo. A, Outline of the experimental layout to test the effects of pharmacologic inhibition of PRC2 in relapsed leukemia. Leukemic cells collected from patients or retrieved from PDXs were cultured on a layer of mesenchymal stromal cells (MS-5) for 7 days and treated or not with PRC2 inhibitors (PRC2i) before the indicated analyses. All PRC2 inhibitors were used at the final concentration of 10 µmol/L and refreshed on day 3 after the first administration. B, Western blot for H3K27me3 performed on Jurkat cells treated or not with EPZ-6438/tazemetostat at the indicated concentrations for 3 or 7 days. Tubulin was used as a loading control. C, Immunofluorescence analysis for H3K27me3 (green) and DAPI (blue) of PDX-derived relapse samples from UPN01 (top) and UPN02 (bottom) cocultured for 7 days with supportive MS-5 stromal cells in the presence or absence of EPZ-6438. Scale bar, 10 µm. Dot plots on the right side of the pictures indicate the mean fluorescence intensity of single cells. Underlying white bars indicate the average fluorescence intensity of all analyzed cells. P values were calculated by a two-sided paired t test at a 95% confidence interval (CI). D, Cell viability, measured as the percentage of Annexin V–negative cells, in PDX-derived relapse samples from UPN01 (top row) and UPN02 (bottom row) cocultured for 7 days with supportive MS-5 stromal cells in the presence or absence of EPZ-6438. E, Heat map representing chromatin accessibility of HLA class II genes and their regulators measured by ATAC-seq in PDX-derived relapse samples from UPN01, UPN02, and UPN03 cultured for 7 days with MS-5 stromal cells alone (dark blue markers in the bar above the heat map) or with the addition of EPZ-6438 (light blue markers in the bar above the heat map). F, Heat map representing levels of expression of transcripts encoding for HLA class II genes and their regulators measured by RNA-seq in PDX-derived relapse samples from UPN01, UPN02, and UPN03 cultured for 7 days with MS-5 stromal cells alone (dark blue markers in the bar above the heat map) or with the addition of EPZ-6438 (light blue markers in the bar above the heat map). G, Histogram outlining the most significantly deregulated biological processes identified from the pairwise comparison of the whole transcriptome of UPN01, UPN02, and UPN03 PDX-derived relapse samples exposed or not to EPZ-6438. The length of each bar is proportional to the significance of enrichment, calculated by a two-sided Fisher exact test. Red bars denote immune-related biological processes. H, mRNA expression level of CIITA, HLA-DRB1, and HLA-DPB1 measured by locus-specific quantitative PCR in PDXs generated from relapses of the indicated patients treated with DMSO alone (dark blue symbols) or with 10 µmol/L of EPZ-6438 (light blue symbols) for 7 days. Underlying white bars indicate the average fold change in expression relative to DMSO control, and whiskers indicate standard deviation. P values were calculated by a two-sided paired t test at a 95% CI. I, Immunophenotypic analysis showing the percentage of HLA-DR+ leukemic blasts from PDXs generated from diagnosis (red) and relapse (blue) samples of different patients cultured for 7 days in the presence of DMSO alone (dark color) or with 10 µmol/L of EPZ-6438 (light color). The corresponding FACS plots are shown in Supplementary Fig. S7. The P value was calculated by a two-sided paired t test at a 95% CI. J, Immunophenotypic analysis showing the percentage of HLA-DR+ leukemic blasts from PDXs generated from diagnosis (red) and relapse (blue) samples of different patients cultured for 7 days in the presence of DMSO alone (dark color) or with 10 µmol/L of EED226. The P value was calculated by a two-sided paired t test at a 95% CI. K, Immunophenotypic analysis showing the percentage of HLA-DR+ leukemic blasts from UPN01 diagnosis (red) or relapse (blue) PDXs cultured for 7 days in the presence of DMSO alone or with different PRC2 inhibitors, all used at 10 µmol/L concentration. The intensity of the blue color of the bars indicates the PRC2 subunits targeted by the compound. L, Immunophenotypic analysis showing the percentage of PD-L1+ leukemic blasts from PDXs generated from diagnosis (red) and relapse (blue) samples of different patients cultured for 7 days in the presence of DMSO alone or with EPZ-6438 10 µmol/L. The P value was calculated by a two-sided paired t test at a 95% CI. M, Immunophenotypic analysis showing the percentage of HLA-DR+ leukemic blasts from PDXs generated from diagnosis (red) or relapse (blue) samples of different patients cultured for 7 days in the presence of DMSO alone or with azacytidine at the 250 nmol/L concentration. The P value was calculated by a two-sided paired t test at a 95% CI. N, Immunophenotypic analysis showing the percentage of HLA-DR+ leukemic blasts circulating in the peripheral blood of mice engrafted with UPN01 diagnosis (red) or relapse (blue) leukemia, treated by oral gavage with vehicle alone or with EPZ-6438 for 21 days. The P value was calculated by a two-sided unpaired t test at a 95% CI. n.s., not significant.
To corroborate our findings and to dissect the contribution of the different PRC2 complex components to the observed phenotype, we tested eight additional inhibitors targeting the catalytic subunit EZH2 (GSK126, GSK343, PF06726304, EI1, GSK503, EPZ011989, CPI169, and UNC1999), as well as other subunits relevant for correct PRC2 assembly and function, such as EED (EEDi-1 and EED226) and JARID2 (JIB-04). We found that all PRC2 chemical inhibitors were able to restore HLA class II expression to a similar extent (with an average fold increase from DMSO control of = 4.6 ± 1.5; Fig. 3J and K), providing further evidence of the involvement of the whole complex in this immune escape modality.
In parallel, we analyzed the transcript (Supplementary Fig. S8A and S8B) and protein level (Supplementary Fig. S8C–S8G) of HLA class I genes, which were already highly expressed at the time of relapse and remained largely unaffected upon PRC2 inhibition.
Of note, the recovery of HLA class II expression driven by PRC2 inhibitors was not accompanied by a concomitant increase in the expression of PD-L1 on leukemia cells (Fig. 3L), which conversely represents one of the major drawbacks of the use of proinflammatory cytokines such as interferons (10).
It has been shown that methylation represents a fundamental layer for regulation of the CIITA promoter activity (11, 12). In a recent study, bisulfite sequencing of two AML relapses with HLA class II downregulation evidenced some extent of methylation of this region (4). In our sample set, we found no difference between the methylation of the CIITA promoter between diagnosis and relapse samples (Fig. 2E), and treatment of the latter with the hypomethylating agent azacytidine had no effect on recovering the expression of HLA class II genes (Fig. 3M).
Finally, we tested the effect of EPZ-6438 on HLA class II expression in vivo by administering the compound to immunocompromised mice engrafted with UPN01 relapsed leukemia. Also in this setting, we observed significant upregulation of HLA class II, consistent with the extent detected in vitro, although still not at the level of the diagnosis PDXs (Fig. 3N).
EZH2 Inhibitor–Mediated Upregulation of HLA Class II Reestablishes T-cell Recognition of Relapsed Leukemia
We next evaluated whether EZH2 inhibition could rescue T cell–mediated recognition of AML blasts. CD4+ T cells from healthy volunteers were cocultured with leukemia blasts collected from UPN01 and UPN05 at the time of diagnosis to enrich them for tumor-specific T cells. After several rounds of stimulation, primed CD4+ T cells were tested against their original stimulators and against their HLA class II–negative relapse counterparts, either left untreated, pretreated with EPZ-6438, or pretreated with IFNγ (shown by previous studies to be able to recover HLA class II expression in this relapse modality; refs. 3, 4, 6). We assessed T cell–mediated IFNγ release, proliferation, degranulation, killing activity, and antigen-specific activation (Fig. 4A) as experimental readouts. In all T-cell donors tested (n = 4), we detected, consistently through all readouts, potent recognition of leukemic blasts at diagnosis, weak recognition of relapse blasts that were left untreated, and a small but consistent and significant increase in T-cell activity and target cell death upon pretreatment of leukemia with the EZH2 inhibitor, similar to the one obtained upon IFNγ pretreatment (Fig. 4B–F).
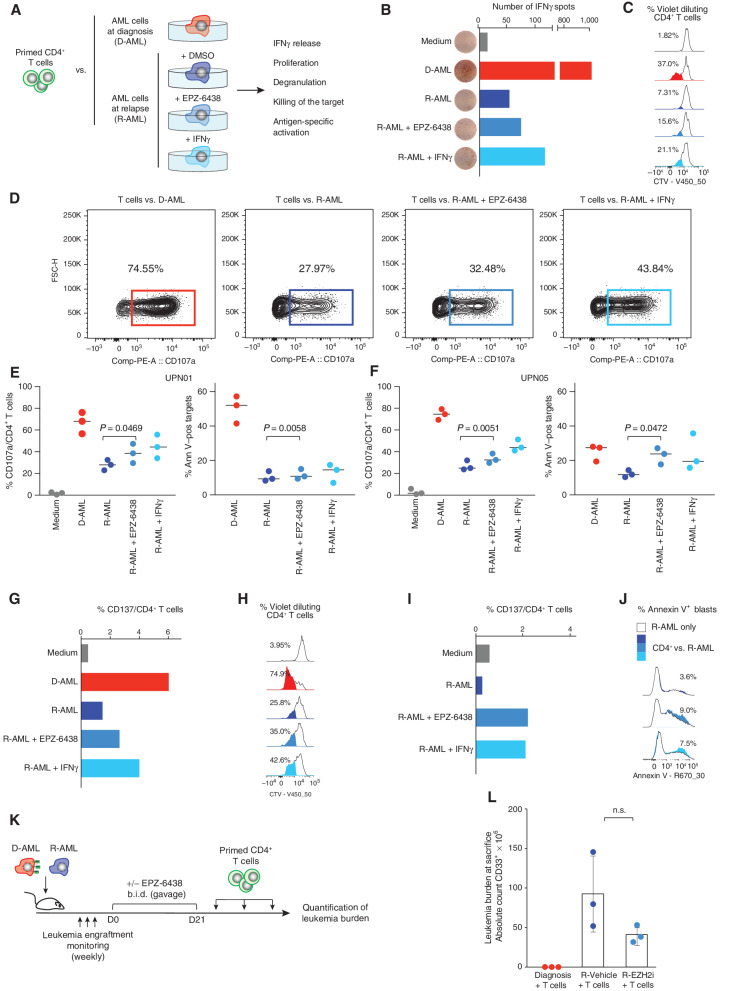
Figure 4.
EZH2 inhibitors reestablish functional recognition of relapsed leukemia by CD4+ T cells. A, Outline of the experimental layout to test the functional effects of pharmacologic inhibition of PRC2 in relapsed leukemia on its CD4+ T cell–mediated recognition. CD4+ T cells, either primed against AML blasts collected from UPN01 at diagnosis or selected for being alloreactive to HLA-DPB1*04:01, were tested against HLA class II–positive AML blasts at diagnosis or against their HLA class II–negative relapsed counterpart, cultured in medium alone or pretreated for 7 days with EPZ-6438 or IFNγ. B and C, CD4+ T cells purified from a healthy individual and primed against UPN01 leukemia cells collected at diagnosis were tested against diagnosis and relapse target cells from the same patient by the IFNγ ELISpot assay (B; showing for each condition the number of IFNγ spots detected from one out of three replicates) or by the CellTrace Violet (CTV)–dilution assay (C; highlighting for each condition the percentage of CD4+ T cells that upon proliferation diluted the vital dye). D, Representative FACS plots showing CD107a degranulation of CD4+ cells from a healthy donor primed against UPN01 AML at diagnosis and tested against the diagnosis (red) and relapse (blue) AML cells from the same patient, pretreated or not with EPZ-6438 or with IFNγ. FSC-H, forward scatter height. E and F, Dot plots summarizing the results of functional experiments performed using CD4+ T cells from three different healthy individuals against AML cells from UPN01 (E) or UPN05 (F). Purified CD4+ T cells were primed against AML blasts collected at diagnosis and tested against their original stimulator (red dots) or leukemia cells collected from the same patient at relapse (blue dots), pretreated or not with EPZ-6438 or with IFNγ. For each panel, the left-side plot shows CD4+ T-cell degranulation and the right-side plot shows the corresponding results in terms of target cell death (subtracting the spontaneous cell death detected in target cells). Ann V–pos, Annexin V–positive. G–J, Third-party CD4+ T cells alloreactive to HLA-DPB1*04:01 were tested against diagnosis and relapse target cells from UPN01 (G and H) or UPN03 (I and J), by a CD137 expression assay (G and I; showing for each condition the percentage of CD4+ T cells that upregulated CD137 in response to the target), by the CTV-dilution assay (H; highlighting for each condition the percentage of CD4+ T cells that upon proliferation diluted the vital dye), and by a cytotoxicity assay (J; highlighting for each condition the percentage of target cells staining positive for Annexin V upon subtraction of spontaneous target cell death, shown as the white histogram profile). K, Outline of the in vivo experiment to test the functional effects of pharmacologic inhibition of PRC2 on reestablishing CD4+ T cell–mediated recognition of relapsed leukemia. Mice were engrafted with UPN01 AML at diagnosis or relapse, treated with EPZ-6438 or vehicle alone, and infused with CD4+ T cells primed against the diagnosis AML blasts. b.i.d., twice a day. L, Absolute counts of human AML blasts recovered from the spleen of the mice at the end of the in vivo experiment. The P value was calculated by a two-sided Mann–Whitney test at a 95% confidence interval. EZH2i, EZH2 inhibitor; n.s., not significant.
To confirm these findings with T cells of known specificity, we took advantage of third-party T cells capable of selectively recognizing allogeneic HLA-DP*04:01 (kindly provided by Prof. Katharina Fleischhauer, University of Essen, and generated as previously described; ref. 13) and tested them against leukemic blasts harvested at posttransplantation relapse from UPN01 (HLA-DP 04:01/04:01; Fig. 4G and H) and UPN03 (HLA-DP 04:01/01:01; Fig. 4I and J). In both cases, we found an increased CD137/4-1BB surface expression when the T cells were challenged with blasts pretreated with EPZ-6438 or IFNγ compared with controls (Fig. 4G and I). T-cell activation was paralleled by antigen-specific proliferation (Fig. 4H) and increased leukemia killing (Fig. 4J).
Because one of the main issues related to the use of immune-modulating drugs in the posttransplantation setting is the potential induction or exacerbation of graft-versus-host disease (GvHD), we next addressed the effect on T cells of exposure to EPZ-6438 (Supplementary Fig. S9A). We documented negligible changes in T-cell phenotype and subset distribution upon culture in the presence of EPZ-6438, except for higher prevalence of CD69+ cells after polyclonal stimulation (CD3/CD28 beads) followed by culture with the compound (Supplementary Fig. S9B). Conversely, T-cell proliferation, in response to both a polyclonal stimulus and allogeneic AML blasts, was consistently reduced in the presence of the compound (Supplementary Fig. S9C–S9F), in line with previous studies that investigated PRC2 inhibition as a strategy to control T-cell alloreactivity (14, 15).
Finally, we explored whether EPZ-6438 could enhance T cell–mediated responses against posttransplantation relapses in vivo. Mice were engrafted with UPN01 AML diagnosis or relapse and treated or not with EPZ-6438 for 3 weeks by oral gavage, followed by multiple infusions of allogeneic CD4+ cells prestimulated with UPN01 AML at diagnosis to enrich in the relevant specificities (Fig. 4K). At sacrifice, mice engrafted with the diagnosis and infused with the T cells were the only ones disease-free, but an interesting, although not significant, reduction in disease burden could also be appreciated in the mice engrafted with the relapsed leukemia who received EPZ-6438 treatment followed by T-cell infusions (Fig. 4L).
Taken together, these results show that alleviating PRC2-mediated repression of HLA class II genes can increase the immunogenicity of relapsed leukemia, improving T cell–mediated recognition and killing.
DISCUSSION
In recent years, relapse has been increasingly recognized as the main obstacle to the full success of allo-HCT for AML (1, 2). This has fueled studies specifically focused on understanding its mechanisms and led to the identification of alterations in antigen processing and presentation as one of its main drivers (3, 4, 16). Defects in this pathway represent an exceptional occurrence in AML at diagnosis but become prevalent when disease reemerges after allo-HCT (1), supporting the idea that relapse occurs when the tumor finds the means to evade T cell–mediated recognition. In particular, AML blasts have been found to alter their HLA asset after transplantation by two modalities: losing at the genetic level one HLA haplotype, which almost invariably encompasses incompatible HLAs (16), or abrogating the surface expression of HLA class II molecules (3, 4).
In particular, this second modality occurs with similar frequency after HLA-compatible and HLA-incompatible transplants (3, 4), and in the latter appears to be largely nonoverlapping with genomic haplotype loss. In fact, of the 14 relapses with class II downregulation from our previous cohort (3), only one had evidence of HLA loss (UPN02 of this series), raising the question of which event occurred first and was dominant in driving immune escape. In addition, we and others found no evidence of new mutations in immune-related genes in cases with HLA class II downregulation (3, 4), prompting our present investigation on the epigenetic basis of this immune escape strategy.
Here, we documented a significant reduction in chromatin accessibility in leukemic cells at relapse, consistent across patients and involving HLA class II genes and their regulators, with negligible changes in their methylation profile. Integrative analysis of the different omics and cases allowed us to pinpoint these changes to a candidate epigenetic regulator, the PRC2 repressor of gene transcription.
Consistent with our findings, recent reports linked PRC2 to the control of expression of HLA molecules and other immune-related molecules in solid tumors and lymphomas (17–19). Specifically, the catalytic subunit of PRC2, EZH2, has emerged as a potent regulator of antigen presentation in genome-wide screenings (18, 19), its mutations have been associated with impaired HLA expression in diffuse large B-cell lymphomas (17), and overexpression of PRC2 in solid tumors has been shown to drive the transcriptional downregulation of HLA class I (18). It has also been reported that EZH2 inhibition can result in derepression of endogenous retroviruses (ERV), which can significantly contribute to boost antitumor responses both by serving as antigens and through the induction of interferon responsive genes (20, 21). In our work, for the first time, we provided evidence in a longitudinal study in patients with AML exposed to immune pressure through allo-HCT of de novo changes driven by PRC2 and responsible for immune escape and clinical relapse. We showed that PRC2 represents a new and promising therapeutic target to revert the mechanism of resistance and reestablish a proficient graft-versus-leukemia effect. A number of compounds that target EZH2 and other subunits of PRC2 are already in clinical development, with EPZ-6438/tazemetostat and GSK126 being the ones in the more advanced stage of testing (22, 23). Of note, the plasma concentrations of tazemetostat measured in patients in the absence of significant toxicities (24, 25) were comparable with the ones used in our experiments, and mice treated with the compound for 3 weeks did not manifest evident side effects. Even though in our in vitro and in vivo experiments the recovery of HLA class II expression upon PRC2 inhibition did not reach the same levels documented at diagnosis, we can speculate that in the clinical setting, longer exposure to the compound and synergistic effect with cytokines released by activated T cells may further enhance HLA class II upregulation. In line with our hypothesis, preclinical and clinical studies with PRC2 inhibitors in solid cancers have already evidenced activation of immune responses as one of the mechanisms of action (26, 27). Moreover, better results in terms of HLA class II recovery may be obtained with more potent EZH2 inhibitors or by simultaneously blocking multiple PRC2 subunits, targeting a larger proportion of AML cells and thus avoiding selection of resistant subclones.
Although EZH2 inhibitors have not yet been tested in transplanted patients, our results on the effect of EPZ-6438 on T cells suggest that this treatment is unlikely to be associated with the risk of inducing GvHD, and are rather in line with preclinical studies that showed that it can tame alloreactive T cells and mitigate the clinical manifestations of GvHD (14, 15). To avoid the risk of hindering antileukemic T cells, already in our preclinical experiments, we have exploited the option of temporally uncoupling the drug effects on cancer and immune cells, administering T cells only after the treatment with PRC2 inhibitors.
Previous studies have shown that exposure of leukemic cells to IFNγ can also recover expression of HLA class II (3, 4). However, the therapeutic index of systemic administration of IFNγ is extremely low, which makes it necessary to devise vehicles for cytokine targeting or strategies to foster cytokine release by immune cells in loco (28). We showed that reestablishing immune recognition of leukemia through PRC2 inhibitors prompts the release of IFNγ by CD4+ T cells, which might amplify the effect of the compound without exogenous administration of the cytokine. Moreover, we did not observe any increase in the expression of PD-L1 on leukemic cells upon exposure to PRC2 inhibitors, a potential advantage of the latter strategy as compared with induction of interferon responses.
Another important consideration relates to the selectivity of the mechanism and therapeutic approach that we have described. It is in fact becoming increasingly recognized that the modalities by which relapse occurs in different patients follow specific and mutually exclusive patterns. In this view, we anticipate that PRC2 inhibitors will yield limited benefit to patients with genomic loss of HLA (16) or increased expression of leukemia inhibitory ligands (3, 29). Therefore, it will become fundamental for clinicians to implement routine diagnostics for relapsing patients, with assays aimed at identifying the distinct relapse modalities, and for pharmaceutical industries to tailor interventional trials to the subset of patients in which the drug has a specific biological rationale. Along the same line, we expect that our bedside-to-bench approach, integrating multiomic analysis and functional studies in primary samples and PDXs, will be applicable to the investigation of other modalities of immune escape and relapse, both after allo-HCT and after cellular immunotherapies, with the ultimate goal to exploit the growing armamentarium of epigenetic therapies in precision medicine approaches.
METHODS
Patient Characteristics and Sample Processing
Primary patient samples were collected and cryopreserved in the San Raffaele Leukemia Biobank upon signing of a specific written informed consent in accordance with the Declaration of Helsinki (Protocol “Banca Neoplasie Ematologiche” approved by the San Raffaele Ethic Committee on October 5, 2010; latest amendment on June 14, 2012). Based on the availability of paired pre- and posttransplantation viable leukemic samples, we selected for our study five patients with AML with reduced HLA class II expression at relapse after allo-HCT, documented by immuophenotypic, genetic, and transcriptional analysis as previously described (ref. 3; UPN01–05; Supplementary Table S1), and five patients with AML who achieved complete remission after intensive chemotherapy and subsequently relapsed (UPN06CT–10CT; Supplementary Table S2). Whenever possible, we used bone marrow samples and matched the same material source for diagnosis and relapse samples, although in selected cases for which we had the opportunity to check both bone marrow and peripheral blood, we observed no differences in terms of HLA class II phenotype in material collected from the two sites.
Analyses pertaining to the present study were approved by the Institutional Ethic Committee according to protocol “ALLO-RELAPSE” (November 3, 2017; latest amendment March 10, 2021).
Primary samples collected from the biobank were thawed and kept for 20 minutes at 37°C in FBS (Euroclone) supplemented with 25 units/mL of Benzonase Nuclease (Sigma-Aldrich, cat. #E1014) at a concentration of 1–2 × 106 cells/mL. Leukemia blasts were labeled in 100 µL of Gartner's medium for 20 minutes at 4°C using fluorescent antibodies according to their leukemia-associated immunophenotype, using CD45-PE-Cy7 (clone HI30, cat. #304016, lot #B229089), CD3-FITC (clone SK7, cat. #344804, lot #B231398), and CD14-APC-Cy7 (clone M5E2, cat. #301820, lot #B251038), all from BioLegend, and CD33-APC (clone WM53, cat. #551378, lot #9008623) from BD Biosciences. Leukemia samples were washed with 10 mL of Gartner's medium, filtered with a cell strainer, and then FACS-purified using a MoFloTM XDP (Beckman Coulter) cell sorter.
PDXs
All in vivo experiments were performed upon approval by the San Raffaele Institutional Animal Care and Use Committee (protocol number 651), by the San Raffaele Ethic Committee (protocol AML-PDX, approved on November 3, 2017), and by the Italian Ministry of Health (authorization number 97/2015-PR on February 18, 2015).
FACS-sorted blasts were engrafted into 4-week-old, nonirradiated male NOD-SCID γ-chain null (NSG) mice by tail-vein infusion of about 5 × 106 cells.
Engraftment was monitored weekly on 50 µL of peripheral blood by flow cytometry. Samples were stained in 100 μL of 1× PBS and 2% FBS plus the relevant mixture of antibodies for 10 minutes at room temperature (RT), using human CD45-PE-Cy7 (clone HI30, cat. #304016, lot #B264588), CD3-FITC (clone SK7, cat. #344804, lot #B231398), and CD14-APC-Cy7 (clone M5E2, cat. #301820, lot #B251038) from BioLegend and CD33-APC (clone WM53, cat. #551378, lot #9008623) from BD Biosciences. After the incubation time, erythrocytes were eliminated upon incubation in ammonium chloride potassium lysis buffer, and samples were washed by centrifugation. For subsequent flow cytometry analysis, a first gate was set to discriminate between mouse and human CD45 cells and the absolute counts of leukemia blasts were quantified upon gating on the leukemia-specific marker CD33 within the gate of human CD45+ cells. We determined the absolute count (cells/µL) by the addition of count beads into each sample (Beckman Coulter).
EPZ-6438 (Selleckchem; cat. #S7128) was resuspended in water with 0.5% PHMC plus 0.5% Tween 80 at a final concentration of 60 mg/mL and administered by oral gavage at a dose of 300 mg/kg every 12 hours for 21 days. Control mice were treated for the same period with vehicle alone. CD4+ T cells purified from a healthy unrelated individual, after three rounds of prestimulation against leukemia cells collected from UPN01 at diagnosis, were infused at a dose of 1 × 106 cells/mouse in animals engrafted with UPN01 diagnosis or relapse AML, pretreated or not with EPZ-6438. T-cell infusions were repeated weekly at the same dose three times.
When animals were euthanized, human leukemia cells were retrieved by processing of spleens, depleting mouse CD45+ cells by immunomagnetic bead selection (Miltenyi Biotec GmbH; cat. #130-045-801).
Multiparametric Flow Cytometry
For immunophenotypic analysis, a maximum of 0.5 × 106 cells per tube were stained in 100 µL of 1× PBS and 2% FBS plus the mixture of antibodies. Staining was performed at RT for 15 minutes, followed by washing with 2 mL of 1× PBS and 2% FBS before the analysis. The complete list of antibodies we used is as follows: CD3 Alexa700 (clone OKT3, cat. #317340, lot #B229089), CD8 APC-H7 (clone SK1, cat. #344714, lot #B341619), CD14 APC-Cy7 (clone M5E2, cat. #301820, lot #B229089), mouse CD45 PerCP-Cy5.5 (clone 30-F11, cat. #103132, lot #B165130), human CD45 PE-Cy7 (clone HI30, cat. #304016, lot #B264588), CD33 BV510 (clone WM53, cat. #303422, lot #B244960), CD45RA Alexa700 (clone HI100, cat. #304120, lot #B257456), CD62 L PerCP/Cy5.5 (clone DREG-56, cat. #304824, lot #B268797), CD69 APC (clone FN50, cat. #310910, lot #B302800), CD95 PE/Cy7 5 (clone DX2, cat. #305622, lot #B251981), HLA-ABC Pacific Blue (clone W6/32, cat. #311418, lot #B191431), HLA-DR FITC (clone L243, cat. #307604, lot #B275368), LAG3 BV605 (clone 11C3C65, cat. #369324, lot #B98854), PD-1 FITC (clone EH12.2H7, cat. #329904, lot #329904), PD-L1 PE (clone 29E2A3, cat. #329706, lot #B282353), and TIM3 BV421 (clone F38-2E2, cat. #345008, lot #B310545) from BioLegend; CD3 BUV737 (clone SK7, cat. #565466, lot #9164571), CD4 BUV395 (clone SK3, cat. #563550, lot #9084515), and CD34 BV605 (clone 8G12, cat. #745247, lot #B256713) from BD Biosciences; B7-H3 APC (clone 85504, cat. #FAB1027A, lot #AAPJ0213061) from R&D Systems; CD25 PE (clone REA570, cat. #563550, lot #5200406623) from Miltenyi Biotec GmbH; and KLRG1 PE-eFluor 610 (clone 13F12F2, cat. #563550, lot #4351541) from e-Biosciences/Thermo Fisher. All antibodies were tested on cell lines reported to be positive for the markers of interest and titrated for optimal on-target/off-target activity. For data analysis of expression on leukemia cells, a first logical gate was based on side scatter and CD45 intensity, followed by a second gate on a leukemia-specific marker (CD33 or CD34) and then analysis of the marker of interest in the CD33+/CD34+ gate. At least 200,000 events in the live cell gate were acquired per sample. Analysis of human samples was performed using an LSRFortessa flow cytometer equipped with 355 nm, 405 nm, 488 nm, 561 nm, and 640 nm lasers and a BD FACS Symphony A5 Sorp flow cytometer equipped with 488 nm, 561 nm, 637 nm, 405 nm, 355 nm lasers (both instruments were from BD Biosciences). Analysis of PDX samples was performed using a Gallios flow cytometer equipped with 488 nm, 638 nm, and 405 nm lasers (Beckman Coulter). Each acquisition was calibrated using Rainbow Calibration Particles (Spherotech; cat. #RCP-30-5A) to correct for day-to-day laser intensity variations. Data were processed using FlowJo version 10.5.2 (Tree Star) or Kaluza (Beckman Coulter).
RNA-seq
Total RNA was extracted using the RNeasy Plus Mini or Micro Kit (Qiagen; cat. #74134 and #74004) following the manufacturer's indications. Concentration and quality of RNA were checked using Invitrogen's Qubit RNA high-sensitivity assay kit (Thermo Fisher Scientific) and the RNA Screen Tape System (Agilent; cat. #5067-5592).
RNA-seq library preparation was performed by starting from 300 ng of total RNA and using the TruSeq stranded mRNA library preparation kit (Illumina; cat. #RS-122-2101) in accordance with a low-throughput protocol. After PCR enrichment (15 cycles) and purification of adapter-ligated fragments, the concentration and length of DNA fragments were measured. Then, RNA-seq libraries were sequenced using the Illumina Next-Seq 500 high platform in order to obtain a minimum of 30 × 106 paired-end reads per sample.
RRBS
Genomic DNA was extracted using the Wizard SV Genomic DNA Purification System (Promega; product ID: A2361) following the manufacturer's instruction and including a step of protein lysis using Proteinase K (New England Biolabs; cat. #P8102S). The integrity of genomic DNA was assessed by running samples on a 1% agarose gel. The concentration and quality of DNA were measured using the Invitrogen's Qubit dsDNA high-sensitivity assay kit (Thermo Fisher Scientific) and Agilent Bioanalyzer technology (Agilent).
A total of 50 ng of purified DNA per sample was processed as previously described (30). Briefly, genomic DNA was digested using methylation-insensitive restriction enzymes to generate short fragments enriched for CpG dinucleotides at the extremities. After end repair, A-tailing, and ligation to methylated adapters, the CpG-rich DNA fragments were subjected to size selection, bisulfite conversion, PCR amplification, and sequencing.
ATAC-seq
A total of 1 × 105 cells from FACS-sorted AML samples were lysed with digitonin (Promega; cat. #G944A) and tagmented with an engineered Tn5 transposase (Illumina; cat. #15027865) at 37°C for 30 minutes, following a protocol optimized for blood cells (31). Tagmented DNA was purified and amplified with 10 cycles of PCR. Before the sequencing, fragments with a 1 to 5 kbp size range were removed by magnetic separation with AMPure XP beads (Beckman Coulter; cat. #A63881). Libraries were then quantified and sequenced on Illumina NextSeq 500 to obtain 40 × 106 paired-end reads per sample.
Bioinformatic Analyses
RNA-seq.
In order to quantify gene-expression levels, input reads were aligned to human transcript annotation (hg38) using hisat (32) with default parameters. Abundancies were then calculated using the Subread featureCounts function (33), and differential gene-expression analysis was performed using the R/Bioconductor package DESeq2 (34), normalizing for library size using DESeq2's median of ratios. In the design matrix, in addition to the tested condition (i.e., diagnosis vs. relapse), a covariate term for the origin of the samples (i.e., primary vs. PDX) was also added to remove the corresponding batch effect. P values were corrected using FDR, and genes having an FDR < 0.05 were selected as differentially expressed.
ATAC-seq.
Input reads were aligned to the human genome assembly (GRCh38) using the BWA software v0.7.17 (35) with standard parameters, and abundancies of regulatory elements were computed using the Subread featureCounts function (33) against the human GeneHancer database (36), which contains annotations of enhancers and promoters, and their inferred target genes. Counts were then translated to genes and analyzed with the R/Bioconductor package DESeq2 (34), normalizing for library size, to obtain differential gene expression results between diagnosis and relapse conditions. P values were corrected using FDR, and genes having an FDR < 0.05 were selected as differentially accessible.
RRBS.
Reads were trimmed using trimmomatic v0.32 (37) and aligned against the hg38 reference genome using BsMAP v2.90 (38). The bisulfite conversion rate of CpG islands was computed using the biseqMethCalling.py script as previously described (39), and the quality check showed data of good quality (bisulfite conversion 99%, mean coverage 7×, more than 3M CpG island covered). Differential methylation analysis was performed using RnBeads (40) using different features (promoters, gene body, tiling regions). Focusing on promoters, P values were corrected using FDR, and genes having an FDR < 0.05 were selected as differentially methylated.
MOFA.
We applied MOFA (8) to integrate data obtained from RNA-seq, ATAC-seq, and RRBS, using an unsupervised approach. Raw counts were normalized using the variant stabilization approach, and low variant features were removed from the analysis. Batch normalization was applied to correct the effect of the tumor expansion in mice in comparison with the primary samples. The model converged in fewer than 5,000 iterations after dropping nonsignificant LFs.
Functional Annotation.
GSEA was performed using the R/Bioconductor package clusterProfiler (41) on preranked gene lists considering the ENCODE and CHEA databases, filtering results with an FDR < 0.05. Enrichment analyses were performed using enrichR (42), exploiting a two-tailed Fisher exact test, considering the Molecular Signatures Database (MSigDB; Gene Ontology and Hallmark signatures) and filtering results with an FDR < 0.05.
Epigenetic Treatment of Leukemic Blasts In Vitro
Leukemic blasts (5 × 105 cells/well) were cocultured with supportive mesenchymal stromal cells (MS-5) for 1 week in Gartner's medium (Alpha-Mem, FBS 12.5%, horse serum 12.5%, TPO, IL3, GCSF, beta-mercaptoethanol, hydrocortisone) in the presence or absence of tazemetostat/EPZ-6438 (Selleckchem; cat. #S7128), GSK126, GSK343, PF06726304, EI1, GSK503, EPZ011989, CPI169, UNC1999, EEDi-1, EED226, and JIB-04, all used at 10 µmol/L and purchased from MedChemExpress, IFNγ (10 ng/µL; PeproTech; cat. #300-02), and azacytidine (250 nmol/L; Celgene) using the appropriate diluent (DMSO or water) at the same concentration as negative control. All compounds were added every 3 days.
Western Blot
Leukemic cells were lysed in a buffer containing 50 mmol/L Tris pH 6.8, 2% SDS, and 20% glycerol. After the addition of reducing 4× Laemmli sample buffer (Bio-Rad; cat. #1610747), 20 µg of proteins were boiled and resolved on 15% Tris-glycine SDS-PAGE gel and transferred to nitrocellulose membranes (Amersham) for 1 hour at 1,000 V at 4°C. Membranes were blocked for 30 minutes with 5% nonfat milk and probed with primary antibodies for 1 hour at RT (H3K27me3 mouse; cat. #ab6002, lot #GR275911-4 from Abcam; α-Tubulin from Sigma). Membranes were incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour at RT before revealing with enhanced chemiluminescence plus western blotting detection kit (Pierce, Thermo Fisher Scientific; cat. #32106).
Immunofluorescence Analysis
Immunofluorescence was performed on relapsed samples treated with tazemetostat or DMSO. A maximum of 1 × 105 cells were seeded on 10-mm round coverslips precoated with 1 mg/mL of poly-L-lysine solution (Sigma-Aldrich; prod. #25988-63-0) for 20 minutes. Cells were fixed with 4% paraformaldehyde (Santa Cruz Biotechnology) for 20 minutes at RT and then permeabilized with 0.3% Triton-X 100 in PBS for 10 minutes. After washing, coverslips were blocked in PBG (0.2% cold water fish gelatin, 0.5% BSA, in PBS) for 30 minutes and stained for 1 hour at RT with primary monoclonal antibody targeting H3K27me3 (mouse; cat. #ab6002, lot #GR275911-4) purchased from Abcam. Alexa488 goat anti-rabbit secondary antibody was then added to probe primary antibody followed by DAPI staining, and slides were mounted with Aqua-Poly-mount (Polysciences, Inc; cat. #18606). Images were obtained with a Leica TCS SP5 confocal laser microscope and visualized with Leica Application Suite software (Leica Microsystems).
qPCR Quantification of Gene Transcripts
qPCR assays and primers for the quantification of HLA-A, HLA-C, HLA-DRB, HLA-DPB1, and CIITA transcripts were developed in- house as previously reported (3). For all reactions, 30 ng of total RNA was retrotranscribed with the iScript cDNA synthesis kit (Bio-Rad; cat. #1708891) according to the manufacturer's instructions. The synthetized cDNA was then preamplified using TaqMan PreAmp Master Mix (Thermo Fisher Scientific; cat. #4488593). Gene expression levels were measured by real-time quantitative PCR (RT-qPCR) on a Viia7 Real-Time PCR System (Applied Biosystems) using Fast SYBR Green chemistry (Thermo Fisher Scientific; cat. #4385618). The relative expression of each target gene was first normalized to RNaseP or GUSB reference genes (ΔCT) and then as fold changes (ΔΔCT) relative to the indicated control conditions.
Mixed Lymphocyte Culture
CD4+ T cells were selected from peripheral blood mononuclear cells of healthy individuals by magnetic bead separation using the CD4+ T-Cell Isolation Kit (Miltenyi Biotec GmbH; cat. #130-096-533). Briefly, purified T cells were stimulated with leukemic blasts at diagnosis at an effector:target ratio of 1:2. Cells were cultured in Iscove's modified Dulbecco's media supplemented with 1% glutamine, 1% penicillin/streptomycin, 10% human serum (Euroclone; cat. #ECB2072L), and IL2 (Novartis; cat. #27131010) at a final concentration of 150 UI/mL. IL2 was replaced every 3 to 4 days, and responders were restimulated every 7 days and tested for functional readouts or infused into PDXs after at least two rounds of stimulation.
Functional Assays for T cell–Mediated Target Recognition and Killing
Cytokine release was measured by means of the IFNγ ELIspot assay. Briefly, T cells were cocultured for 24 hours at 37°C in 5% CO2 at a 1:1 effector:target ratio with target cells pretreated or not with EPZ-6438 or IFNγ in 200 µL of Iscove's modified Dulbecco's media supplemented with 1% glutamine, 1% penicillin/streptomycin, and 10% human serum. Spots were counted by a KS Elispot Reader (Zeiss).
For immunophenotypic assays measuring T-cell activation, proliferation, and target cell killing, conditions for the rechallenge of effectors against targets of interest were the same as described for IFNγ ELISpot, with the appropriate times of coincubation. At the time of readout, immunophenotypic analysis was performed. For all the analyses, a first logical gate was based on side scatter and CD45 intensity, followed by a second gate on CD4+ T cells (CD45hiCD3+CD4+) for measuring T-cell proliferation and target specific recognition, or on leukemic blasts (CD45dimCD33+/CD34+), followed by analysis of the marker of interest.
T-cell activation in response to the relevant target cells was tested after 24 hours of coincubation measuring 4-1BB–positive cells (CD137-PE, BD Biosciences, clone 4B4-1, cat. #555956, lot #B275368).
T-cell degranulation and killing were evaluated after 6 hours of rechallenge with target cells, adding the anti-CD107a antibody at the beginning of the coculture (CD107a-PE, BioLegend, clone H4A3, cat. #328608, lot #B321484) and Monensin A (BD, Protein Transport Inhibitor-Containing Monensin-GolgiStop; cat. #554724) after 3 hours, and measuring the percentage of Annexin V–positive cells at the end of the assay within the population of blasts (BioLegend, APC Annexin V; cat. #640920, lot #B334668). Untreated target cells served as a control to detect the basal proportion of apoptotic cells.
To measure T-cell proliferation, CD4+ T cells were first stained with CellTrace Violet (Thermo Fisher) and then either stimulated with anti-CD3/CD28 beads (Gibco Dynabeads Human T-Activator CD3/CD28, cat. #11141D) or cocultured with patient blasts. After 7 days, cells were collected and stained for immunophenotypic analysis. Proliferating CD4+ T cells were identified by gating on CellTrace Violet diluting cells.
Data Availability
All sequencing data from this study have been deposited in the publicly available Gene Expression Omnibus under accession numbers GSE197416 and GSE197419.
Supplementary Material
Acknowledgments
The authors thank all members of the Vago and Di Micco laboratories at the IRCCS San Raffaele Scientific Institute, Milano, for critical discussion on the experiments and the manuscript. We thank Prof. Katharina Fleischhauer, University of Essen, for kindly providing anti-DPB1*04:01 T cells and for her expert opinion on the study. This work was supported by a San Raffaele Pilot Grant awarded to R. di Micco and L. Vago, by the Italian Ministry of Health (RF-2011-02351998 to F. Ciceri and L. Vago, RF-2011-02348034 to L. Vago, GR-2018-12367860 to L. Vago, and TRANSCAN HLALOSS to L. Vago), by the Associazione Italiana per la Ricerca sul Cancro (Start-Up Grant #14162 and IG #22197 to F. Ciceri; My First AIRC Grant MFAG 2019–PI ID.23321 to R. di Micco), by Telethon (TIGET grant E5 to R. di Micco), by a Career Development Award from Human Frontier Science Program (to R. di Micco), by an Advanced Research Grant from the European Hematology Association (to R. di Micco), by a Hollis Brownstein Research Grant from the Leukemia Research Foundation (to R. di Micco), by the Interstellar Initiative on Healthy Longevity from the New York Academy of Sciences and the Japan Agency for Medical Research and Development (AMED; to R. di Micco), and by the New York Stem Cell Foundation (to R. di Micco). R. di Micco is a New York Stem Cell Foundation Robertson Investigator.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
V. Gambacorta reports personal fees from Merus NV outside the submitted work. D. Gnani reports grants from UniSR and Fondazione Centro San Raffaele outside the submitted work. R. di Micco reports grants from the Leukemia Research Foundation, the European Hematology Association (Advanced Research Grant), the Italian AIRC (under MFAG 2019 ID 23321), American Society of Hematology (Global Research Award), the Telethon Foundation, The New York Stem Cell Foundation (Robertson Stem Cell Investigator Award), the Human Frontier Science Program (Career Development Award), and San Raffaele Hospital (Pilot and Seed Grant) during the conduct of the study, as well as grants from CRACK-IT Challenge Clean-Cut from NC3Rs outside the submitted work. L. Vago reports grants from the Italian Ministry of Health and Associazione Italiana per la Ricerca sul Cancro during the conduct of the study, as well as personal fees from GEN-DX and grants and personal fees from Moderna Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
V. Gambacorta: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. S. Beretta: Data curation, formal analysis, methodology, writing–review and editing. M. Ciccimarra: Data curation, formal analysis, methodology, writing–review and editing. L. Zito: Data curation, formal analysis, investigation, methodology, writing–review and editing. K. Giannetti: Data curation, formal analysis, methodology, writing–review and editing. A. Andrisani: Data curation, formal analysis, methodology, writing–review and editing. D. Gnani: Data curation, formal analysis, methodology, writing–review and editing. L. Zanotti: Data curation, formal analysis, methodology. G. Oliveira: Data curation, formal analysis, methodology, writing–review and editing. M.G. Carrabba: Data curation, supervision, writing–review and editing. D. Cittaro: Conceptualization, formal analysis, supervision, methodology, writing–review and editing. I. Merelli: Data curation, formal analysis, supervision, methodology, writing–review and editing. F. Ciceri: Conceptualization, resources, supervision, funding acquisition, investigation, writing–review and editing. R. di Micco: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing. L. Vago: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Zeiser R, Vago L. Mechanisms of immune escape after allogeneic hematopoietic cell transplantation. Blood 2019;133:1290–7. [DOI] [PubMed] [Google Scholar]
- 2. Rimando JC, Christopher MJ, Rettig MP, DiPersio JF. Biology of disease relapse in myeloid disease: implication for strategies to prevent and treat disease relapse after stem-cell transplantation. J Clin Oncol 2021;39:386–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med 2019;25:603–11. [DOI] [PubMed] [Google Scholar]
- 4. Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stevanović S, van Schie MLJ, Griffioen M, Falkenburg JH. HLA-class II disparity is necessary for effective T cell mediated graft-versus-Leukemia effects in NOD/scid mice engrafted with human acute lymphoblastic leukemia. Leukemia 2013;27:985–7. [DOI] [PubMed] [Google Scholar]
- 6. Matte-Martone C, Liu J, Zhou M, Chikina M, Green DR, Harty JT, et al. Differential requirements for myeloid leukemia IFN-γ conditioning determine graft-versus-leukemia resistance and sensitivity. J Clin Invest 2017;127:2765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fleischhauer K, Shaw BE. HLA-DP in unrelated hematopoietic cell transplantation revisited: challenges and opportunities. Blood 2017;130:1089–96. [DOI] [PubMed] [Google Scholar]
- 8. Argelaguet R, Arnol D, Bredikhin D, Deloro Y, Velten B, Marioni JC, et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol 2020;21:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer 2016;16:803–10. [DOI] [PubMed] [Google Scholar]
- 10. Norde WJ, Maas F, Hobo W, Korman A, Quigley M, Kester MG, et al. PD-1/PD-L1 interactions contribute to functional T-cell impairment in patients who relapse with cancer after allogeneic stem cell transplantation. Cancer Res 2011;71:5111–22. [DOI] [PubMed] [Google Scholar]
- 11. Wright KL, Ting JPY. Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol 2006;27:405–12. [DOI] [PubMed] [Google Scholar]
- 12. Dufva O, Pölönen P, Brück O, Keränen MAI, Klievink J, Mehtonen J, et al. Immunogenomic landscape of hematological malignancies. Cancer Cell 2020;38:380–99. [DOI] [PubMed] [Google Scholar]
- 13. Meurer T, Crivello P, Metzing M, Kester M, Megger DA, Chen W, et al. Permissive HLA-DPB1 mismatches in HCT depend on immunopeptidome divergence and editing by HLA-DM. Blood 2021;137:923–8. [DOI] [PubMed] [Google Scholar]
- 14. He S, Wang J, Kato K, Xie F, Varambally S, Mineishi S, et al. Inhibition of histone methylation arrests ongoing graft-versus-host disease in mice by selectively inducing apoptosis of alloreactive effector T cells. Blood 2012;119:1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. He S, Xie F, Liu Y, Tong Q, Mochizuki K, Lapinski PE, et al. The histone methyltransferase Ezh2 is a crucial epigenetic regulator of allogeneic T-cell responses mediating graft-versus-host disease. Blood 2013;122:4119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vago L, Perna SK, Zanussi M, Mazzi B, Barlassina C, Stanghellini MT, et al. Loss of mismatched HLA in leukemia after stem-cell transplantation. N Engl J Med 2009;361:478–88. [DOI] [PubMed] [Google Scholar]
- 17. Ennishi D, Takata K, Béguelin W, Duns G, Mottok A, Farinha P, et al. Molecular and genetic characterization of MHC deficiency identifies EZH2 as therapeutic target for enhancing immune recognition. Cancer Discov 2019;9:546–63. [DOI] [PubMed] [Google Scholar]
- 18. Burr ML, Sparbier CE, Chan KL, Chan YC, Kersbergen A, Lam EYN, et al. An evolutionarily conserved function of polycomb silences the MHC class I antigen presentation pathway and enables immune evasion in cancer. Cancer Cell 2019;36:385–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dersh D, Phelan JD, Gumina ME, Wang B, Arbuckle JH, Holly J, et al. Genome-wide screens identify lineage- and tumor-specific genes modulating MHC-I- and MHC-II-restricted immunosurveillance of human lymphomas. Immunity 2021;54:116–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morel KL, Sheahan AV, Burkhart DL, Baca SC, Boufaied N, Liu Y, et al. EZH2 inhibition activates a dsRNA-STING-interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat Cancer 2021;2:444–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med 2018;24:1143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gambacorta V, Gnani D, Vago L, Di Micco R. Epigenetic therapies for acute myeloid leukemia and their immune-related effects. Front Cell Dev Biol 2019;7:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morin RD, Arthur SE, Assouline S. Treating lymphoma is now a bit EZ-er. Blood Adv 2021;5:2256–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Italiano A, Soria JC, Toulmonde M, Michot JM, Lucchesi C, Varga A, et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol 2018;19:649–59. [DOI] [PubMed] [Google Scholar]
- 25. Morschhauser F, Tilly H, Chaidos A, McKay P, Phillips T, Assouline S, et al. Tazemetostat for patients with relapsed or refractory follicular lymphoma: an open-label, single-arm, multicentre, phase 2 trial. Lancet Oncol 2020;21:1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou L, Mudianto T, Ma X, Riley R, Uppaluri R. Targeting EZH2 enhances antigen presentation, antitumor immunity, and circumvents anti–PD-1 resistance in head and neck cancer. Clin Cancer Res 2020;26:290–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gounder MM, Zhu G, Roshal L, Lis E, Daigle SR, Blakemore SJ, et al. Immunologic correlates of the abscopal effect in a SMARCB1/INI1-negative poorly differentiated chordoma after EZH2 inhibition and radiotherapy. Clin Cancer Res 2019;25:2064–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gocher AM, Workman CJ, Vignali DAA. Interferon-γ: teammate or opponent in the tumour microenvironment? Nat Rev Immunol 2021;22:158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Noviello M, Manfredi F, Ruggiero E, Perini T, Oliveira G, Cortesi F, et al. Bone marrow central memory and memory stem T-cell exhaustion in AML patients relapsing after HSCT. Nat Commun 2019;10:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 2011;6:468–81. [DOI] [PubMed] [Google Scholar]
- 31. Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019;37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013;41:e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database 2017;2017:bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinf 2009;10:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klughammer J, Datlinger P, Printz D, Sheffield NC, Farlik M, Hadler J, et al. Differential DNA methylation analysis without a reference genome. Cell Rep 2015;13:2621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Müller F, Scherer M, Assenov Y, Lutsik P, Walter J, Lengauer T, et al. RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol 2019;20:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu G, Wang LG, Han YHe QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf 2013;14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data from this study have been deposited in the publicly available Gene Expression Omnibus under accession numbers GSE197416 and GSE197419.