Abstract
Mycoplasma pneumoniae cytadherence is mediated by a specialized, polar attachment organelle. Certain spontaneously arising cytadherence mutants (designated class I) lack HMW2, fail to localize the adhesin protein P1 to the attachment organelle, and exhibit accelerated turnover of proteins HMW1, HMW3, and P65. Insertional inactivation of hmw2 by Tn4001 results in a phenotype nearly identical to that of the class I mutants, suggesting that the latter may result from a defect in hmw2. In this study, the recombinant wild-type hmw2 allele successfully complemented a class I mutant when introduced by transposon delivery. Synthesis of recombinant HMW2 at wild-type levels resulted in reacquisition of hemadsorption and normal levels of HMW1, HMW3, and P65. Low-level production of HMW2 in some transformants resulted in only an intermediate capacity to hemadsorb. Furthermore, full restoration of HMW1 and P65, but not that of HMW3, was directly proportional to the amount of recombinant HMW2 produced, reflecting the importance of proper stoichiometry for certain cytadherence-associated proteins. The recombinant class I hmw2 allele did not restore cytadherence, consistent with a defect in hmw2 in this mutant. A frameshift was discovered in different oligoadenine tracts in hmw2 from two independent class I mutants. Finally, protein P28 is thought to be the product of internal translation initiation in hmw2. A transposon excision-deletion mutant produced a truncated HMW2 but no P28, consistent with this conclusion. However, this deletion mutant was hemadsorption positive, indicating that P28 may not be required for cytadherence.
Colonization of the human respiratory epithelium (cytadherence) by the cell wall-less prokaryote Mycoplasma pneumoniae is mediated by a terminal structure termed the attachment organelle. This membrane-bound extension of the mycoplasma cell is characterized by an electron-dense core, which enlarges to form a terminal button (17, 30). The adhesin protein P1 (15) is primarily found densely clustered at the attachment organelle but also occurs scattered elsewhere on the mycoplasma cell surface (1). Cytadherence mutants lacking the high-molecular-weight proteins HMW1, HMW2, and HMW3 (designated class I mutants [Table 1]) fail to localize P1 to the attachment organelle (1, 20). Revertants of class I mutants simultaneously regain HMW1-HMW3 and the ability to cytadhere (19), underscoring the direct relationship between the HMW proteins and P1 localization and function. HMW1-HMW3 are components of a Triton X-100-insoluble, cytoskeleton-like structure in M. pneumoniae (24), suggesting an indirect, scaffolding role in cytadherence (34). HMW3 is an integral part of the terminal button of the attachment organelle (35), while HMW1 is distributed only along the filamentous extensions of the mycoplasma cell (34) and is essential in development of the attachment organelle and localization of P1 to this structure (9).
TABLE 1.
Protein profiles of wild-type, mutant, and class I transformants of M. pneumoniae
| Strain | Relative levela
|
Reference(s) | ||||||
|---|---|---|---|---|---|---|---|---|
| HMW1 | HMW2 | HMW3 | P65 | P41 | P24 | P28 | ||
| Wild type | +++ | +++ | +++ | +++ | +++ | +++ | +++ | 20 |
| Class I | +/− | − | +/− | +/− | +/− | +/− | +/− | 20 |
| crl | +/− | − | +/− | +/− | +/−; − | +/−; − | +/−; − | 11, 21 |
| I-2/P65-HMW2-1 | +++ | +++ | +++ | +++ | +++ | + | ++ | This study |
| I-2/P65-HMW2-2 | +++ | +++ | +++ | +++ | + | + | +++ | This study |
| I-2/P65-HMW2-3 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | This study |
| I-2/P65-HMW2-4 | +++ | +++ | +++ | +++ | + | +++ | +++ | This study |
| I-2/P65-HMW2-5 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | This study |
| I-2/HMW2-1 | + | + | +++ | + | +++ | +++ | ++ | This study |
| I-2/HMW2-2 | + | + | +++ | + | + | +/− | ++ | This study |
| I-2/HMW2-3 | ++ | ++ | +++ | + | +++ | +++ | +++ | This study |
| I-2/HMW2-4 | + | +/− | +++ | + | +/− | + | +/− | This study |
| I-2/HMW2-5 | ++ | +++ | +++ | + | +++ | + | ++ | This study |
+++, protein present at wild-type levels; ++, protein present at near wild-type levels; +, protein present at reduced levels; +/−, protein nearly absent; −, protein absent.
HMW1-HMW3 are encoded by two unlinked genetic loci (Table 2). The genes for HMW1 and HMW3 are part of the hmw locus, along with the genes for the cytadherence-associated protein P30 and six predicted proteins of unknown function (4, 14, 40). The hmw2 gene lies approximately 160 kb from the hmw locus and follows the gene for the mycoplasma surface protein P65 (29) as the second of four genes thought to constitute a single transcriptional unit designated the cytadherence regulatory locus (crl) or P65 operon (21). Protein P28 is thought to result from internal translation initiation in hmw2, while P41 and P24 are the products of downstream genes in the operon. Transposon insertions in hmw2 result in cytadherence mutants phenotypically indistinguishable from the spontaneously arising, class I mutants (11, 21) (Table 1). Loss of HMW1 and HMW3 occurs posttranslationally in both crl and class I mutants by means of accelerated proteolytic turnover (27), indicating that HMW2 is required for the stable maintenance of HMW1 and HMW3 but also suggesting that class I mutants may likewise arise from a mutation in hmw2. In the present study, the wild-type hmw2 allele was introduced into a class I mutant via recombinant transposon delivery, restoring a wild-type phenotype. A recombinant class I hmw2 allele failed to restore cytadherence, localizing the class I mutation to the hmw2 gene. This conclusion was confirmed by nucleotide sequence analysis of two independent class I mutant isolates and a cytadhering revertant thereof. Finally, we present additional evidence indicating that P28 is indeed encoded by the hmw2 gene.
TABLE 2.
Nomenclature used for gene designations in this and previous studies citing the published genome sequence of M. pneumoniae (14)
| Designation used here and previously | Designation from the genome sequence (14) | Protein |
|---|---|---|
| orfp65 | F10_orf405 | P65 |
| hmw2 | F10_orf1818 | HMW2 |
| orfp41 | F10_orf357 | P41 |
| orfp24 | F10_orf218 | P24 |
| hmw1 | H08_orf1018 | HMW1 |
| hmw3 | H08_orf672 | HMW3 |
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli cells were grown in LB broth or on LB agar plates at 37°C (34). Ampicillin was added to the media at a final concentration of 50 μg/ml for plasmid selection. M. pneumoniae cells were grown in Hayflick medium or on PPLO (pleuropneumonia-like organism) agar plates as described previously (8). Gentamicin (18 μg/ml) was included for selection and culture of transformants (12). Individual mycoplasma colonies were visualized on the basis of hemolytic plaques following blood-agar overlay, picked by using sterile Pasteur pipettes (11), and filter cloned as described elsewhere (39).
Molecular cloning and construction of M. pneumoniae strains.
Plasmid DNA was purified from E. coli by using pZ523 (5′-3′, Inc., Boulder, Colo.) or Qiagen-tip 500 (Qiagen, Santa Clarita, Calif.) columns. Alternatively, the alkaline lysis procedure was used for rapid screening of plasmid DNA (34). M. pneumoniae chromosomal DNA was extracted as described previously (26). Standard techniques were used for recombinant DNA construction (33). The region of crl that encompasses the hmw2 gene was cloned from wild-type and mutant M. pneumoniae DNA as an 8.2-kbp PstI-BglII fragment (Fig. 1). Briefly, DNA fragments approximately 7 to 10 kb in length were excised from the agarose gel following restriction endonuclease digestion and electrophoresis, purified by using a Geneclean kit (BIO 101, Vista, Calif.), and ligated with pUC18 previously digested with PstI and BamHI. Ligation mixtures were electroporated (Gene Pulser; Bio-Rad, Richmond, Calif.) into E. coli JM109 (43), and transformants were screened by using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 10 μg/ml). White colonies were picked onto duplicate LB plates containing ampicillin, and colonies containing the desired insert were identified by DNA hybridization to colony lifts on nitrocellulose membranes. Membranes were probed with a 6.3-kbp EcoRI fragment from crl (21) (Fig. 1A) that was labeled by using a Genius kit (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) as specified by the manufacturer. Recombinant plasmids pKV132 and pKV133 contained the 8.2-kbp fragment from wild-type and mutant I-2 M. pneumoniae, respectively. The 8.2-kbp fragment was subcloned by digesting pKV132 or pKV133 with NarI (the site for which lies approximately 165 bp upstream of the HindIII site in the multiple cloning site of pUC18). The 5′ overhang was made double stranded using the Klenow fragment of DNA polymerase, and the DNA was digested with SmaI and ligated into the dephosphorylated SmaI site of pISM2062 (16) (Fig. 1B). Plasmids containing the cloned insert in both orientations were obtained, but for the purposes of this study, only constructs having the insert oriented inward relative to promoter Pin were evaluated further (pKV136 and pKV145, for the wild-type and mutant I-2 alleles, respectively [Fig. 1B]). To subclone only the hmw2 gene into pISM2062, pKV132 (wild type) and pKV133 (mutant I-2) were digested with BstEII (Fig. 1A), and the ends were filled in with Klenow fragment. Following digestion with SmaI, the 5.7-kbp fragment containing hmw2 was purified from an agarose gel and ligated into pISM2062 previously digested with SmaI and treated with calf intestinal phosphatase. Transformants containing these constructs in both orientations were identified, but again only those for which the insert was oriented inward relative to Pin were evaluated further (pKV134 and pKV143, for the wild-type and mutant I-2 alleles, respectively [Fig. 1B]).
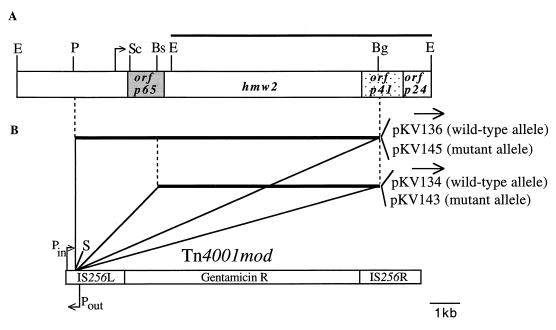
FIG. 1.
(A) Restriction map of the M. pneumoniae P65 operon (crl) region. Open reading frames encoding P65, HMW2, P41, and P24 (orfp65, hmw2, orfp41, and orfp24, respectively) are indicated. The EcoRI fragment used in Southern blot hybridizations is indicated by the bar above the map, and the arrow denotes the predicted transcriptional initiation site based on primer extension studies (21). (B) The recombinant Tn4001mod transposons engineered to contain the PstI-BglII or BstEII-BglII fragment from the P65 operon in the SmaI site of IS256L. Plasmids containing the various recombinant transposons are indicated on the right, and the arrows above each indicate the orientation of the cloned fragment, relative to those of the Pin and Pout promoters. Bs, BstEII; Bg, BglII; E, EcoRI; P, PstI; S, SmaI; Sc, ScaI.
Electroporation of M. pneumoniae was performed as described previously (12). Several independent transformants for each plasmid electroporation were obtained by filter cloning, and the presence of a single copy of the recombinant transposon in the chromosome was confirmed by Southern blot hybridization (33).
Analysis of transformant protein profiles.
Mycoplasma protein profiles were examined by using discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoreses (SDS-PAGE; 3% polyacrylamide stacking and 4.5 or 12% polyacrylamide separating gels). Equivalent amounts of protein as determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.) were loaded onto gels. Following electrophoresis, protein bands were either visualized by silver staining (25) or transferred to nitrocellulose membranes and probed with rabbit antibodies prepared against the following mycoplasma proteins: HMW1 and HMW3 (28, 34, 35), P65 (28, 29), P41 and P24 (21), the HMW2 C-terminal domain (21), and the HMW2 N-terminal domain (31).
Qualitative and quantitative evaluation of HA.
M. pneumoniae hemadsorption (HA) correlates with attachment to respiratory epithelium. Transformants were evaluated for HA by colony screening or by using a quantitative HA assay (18). Briefly, cultures were grown in 15 ml of Hayflick medium containing 200 μCi of [3H]thymidine (6.7 Ci/mmol; Dupont NEN, Boston, Mass.) for 50 to 60 h at 37°C. Cells were harvested and washed three times in cold 10 mM phosphate-buffered saline, pH 7.2 (PBS), suspended in 3 ml of Hayflick medium, dispersed by repeated passage through a 25-gauge needle, and centrifuged for 5 min at 123 × g in a clinical centrifuge (International Equipment Co., Needham Heights, Mass.); 200 μl of the supernatant was mixed with 800 μl of Hayflick medium, divided into six 150-μl aliquots, and incubated for 30 min at 4 or 37°C. Chicken blood cells, previously collected in Alsever’s solution at a 1:1 mixture, were washed two times with PBS and suspended at a dilution of 4% (vol/vol) in PBS; 50 μl of a 4% suspension of chicken blood was added per tube, and the incubations were continued at 4 or 37°C for an additional 30 min. The mixtures were then gently overlaid on 150 μl of 40% sucrose in a 0.65-ml tube and centrifuged at 4,000 × g for 20 s to separate blood cells from unattached mycoplasmas. The blood cell pellets were suspended in 100 μl of PBS, 10 μl of 10% SDS was added per sample, and the samples were incubated overnight at room temperature. After addition of 5 μl of 30% hydrogen peroxide, incubation continued for 2 h at 37°C. Scintillation fluid was then added to each sample, and radioactivity was determined.
PCR and DNA sequencing.
The hmw2 gene was amplified by the method of Cheng et al. (2) for the synthesis of large DNA fragments, using a GeneAmp XL-PCR kit (Perkin-Elmer, Langen, Germany). Mycoplasma DNA was purified as described previously (42). Primers were 22-mers having a melting temperature of approximately 68°C and were designed as described elsewhere (13).
Sequence data were obtained by using a modified enzymatic dideoxy-chain termination method in combination with a fluorescence-based sequence gel reader (model 373A; Applied Biosystems) as described elsewhere (13). Sequence chromatograms were edited with the SeqEd program, version 1.03 (Applied Biosystems). The sequences were assembled and mutations were detected by using the Sequence Project Management program of the Lasergene program package (DNASTAR, Madison, Wis.).
Nucleotide sequence accession numbers.
Accession numbers AF143911, AF143912, and AF143913 have been assigned to the hmw2 sequence of mutants I-2 and I-10.
RESULTS
Genetic complementation with recombinant hmw2.
The class I and crl cytadherence mutants are virtually indistinguishable phenotypically, exhibiting complete loss or, in some cases, truncation of HMW2, reduced levels of P65, P41, P24, HMW1, and HMW3, and loss or reduced levels of P28 (21). The crl phenotype is the result of Tn4001 insertion in hmw2 (21), and it seemed likely that class I mutants also result from a defect in the hmw2 gene. To test this possibility, we evaluated the ability of recombinant hmw2 from wild-type and class I mutant M. pneumoniae to rescue cytadherence in trans in the class I mutant. An 8.2-kb PstI-BglII DNA fragment encompassing p65 and hmw2 (Fig. 1) was isolated from wild-type or mutant I-2 mycoplasma chromosomal DNA and cloned into the SmaI site of the IS256L element of Tn4001mod, generating pKV136 and pKV145 (wild-type and mutant I-2 alleles, respectively [Fig. 1]). Transcription of the cloned DNA in these recombinant transposons is probably directed by a promoter-like region upstream of the p65 gene (21) but might also be directed by promoter Pin in Tn4001mod (Fig. 1), which is functional in M. pneumoniae (6).
Recombinant p65-hmw2 in Tn4001mod was electroporated in M. pneumoniae mutant I-2 cells, and transformants were selected with gentamicin and screened for HA. The plasmid vector is unable to replicate in M. pneumoniae; hence, gentamicin resistance is acquired by insertion of the transposon into the chromosome (12). Transformants of mutant I-2 containing the wild-type recombinant p65-hmw2 allele (I-2/P65-HMW2) were virtually 100% HA+. In contrast, no I-2 transformants receiving the corresponding allele cloned from the class I-2 mutant were HA+. The failure of the mutant allele to restore cytadherence is consistent with a defect within the p65 or hmw2 gene in mutant I-2.
To test whether recombinant hmw2 alone (as opposed to p65 and hmw2 together) restored cytadherence to mutant I-2, the 5.7-kb BstEII-BglII fragment containing only hmw2 (Fig. 1) was subcloned into the SmaI site of Tn4001mod to generate pKV134. This construct was transformed into mutant I-2, and gentamicin-resistant transformants were screened for HA. All transformants screened were HA+, indicating that hmw2 alone was sufficient to restore cytadherence.
Southern blot hybridization of recombinant hmw2 transformants.
Several independent HA+ transformants of mutant I-2 for each construct (I-2/HMW2 and I-2/P65-HMW2) were filter cloned three times. Chromosomal DNA was isolated from each transformant, digested with EcoRI, and analyzed by Southern blot hybridization using a probe consisting of the cloned 6.3-kb EcoRI fragment spanning most of hmw2 (Fig. 1A). Two bands were expected in the transformant profiles: a 6.3-kb restriction fragment corresponding to the resident allele, and a second, larger fragment (>10.8 or >13.3 kb, in transformants with pKV134 or pKV136, respectively), corresponding to the recombinant transposon plus flanking chromosomal DNA. The 6.3-kb band was detected in the transformants and in untransformed M. pneumoniae controls, as anticipated. A band of >10 kb, which varied in size with each transformant, was also observed only in the transformant profiles (data not shown). This second band of variable size in the transformants is consistent with transpositional insertion of the recombinant transposon in diverse sites in the mycoplasma chromosome. None of the transformants exhibited a hybridization pattern indicative of transposon insertion by homologous recombination.
Recombinant hmw2 restores a wild-type protein profile to mutant I-2.
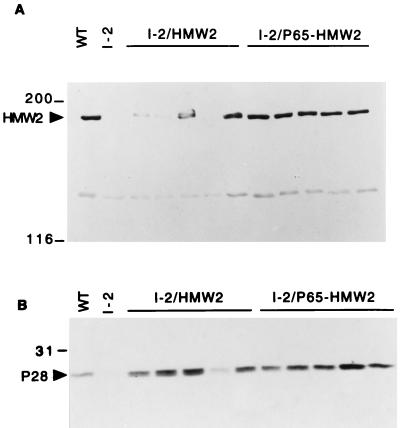
The loss of HMW2 in the class I and crl mutants is associated with accelerated proteolytic turnover of HMW1, HMW3, and P65 and reduced levels or loss of P41 and P24 by an undefined mechanism (7, 21, 27). HA+ transformants were analyzed by SDS-PAGE and silver staining to determine if recombinant wild-type hmw2 restored a normal protein profile to the I-2 transformants. Protein bands corresponding to HMW1, HMW2, and HMW3 were clearly identified in the profiles of wild-type M. pneumoniae and HA+ transformants of I-2/HMW2 and I-2/P65-HMW2 (data not shown). As expected, these proteins were not detected in mutant I-2 cells transformed either with the vector alone or with the recombinant hmw2 allele from the class I-2 mutant (data not shown). Antibodies to the C terminus of HMW2 react with both HMW2 and P28, with the latter thought to arise from internal translation initiation within the hmw2 transcript (21). HMW2 was detected in Western immunoblots of samples from wild-type M. pneumoniae and at various levels in all HA+ transformants examined (Fig. 2A; Table 1). I-2/P65-HMW2 transformants produced HMW2 at near wild-type levels, whereas I-2/HMW2 transformants, lacking the normal promoter for the hmw2 gene, synthesized HMW2 at significantly reduced levels. Similarly, the extent to which P28 was restored in the transformants paralleled that of HMW2 (Fig. 2B), consistent with our previous conclusion that P28 was probably the product of the hmw2 gene (21). To test whether the reduced levels of HMW2 and P28 in the transformants with the subcloned hmw2 gene (I-2/HMW2) were due to loss of the normal promoter for the P65 operon or loss of p65 with subcloning, we generated an internal in-frame deletion (ScaI-BstEII) in p65 in the wild-type PstI-BglII fragment (Fig. 1A). This yielded a recombinant allele that lacked an intact p65 gene but retained the predicted mycoplasma promoter for the operon. When transformed in mutant I-2, once again >90% of the transformants were HA+, and HMW2 production was near wild-type levels (data not shown).
FIG. 2.
Western immunoblot analysis of M. pneumoniae mutant I-2 transformants for production of HMW2 (A) and P28 (B). Mycoplasma protein (20 μg per well) was separated by SDS-PAGE (4.5% [A] or 12% [B] polyacrylamide separating gels), blotted to nitrocellulose, and probed with serum against the C-terminal region of HMW2 (21) at a 1:1,000 dilution. Lanes: WT, wild-type M. pneumoniae; I-2, mutant I-2 transformed with pISM2062 (vector control); I-2/HMW2 and I-2/P65-HMW2, I-2 transformants with pKV134 and pKV136, respectively. HMW2 and P28 are indicated by arrowheads, and protein size standards are shown on the left in kilodaltons.
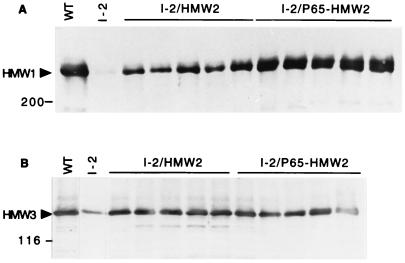
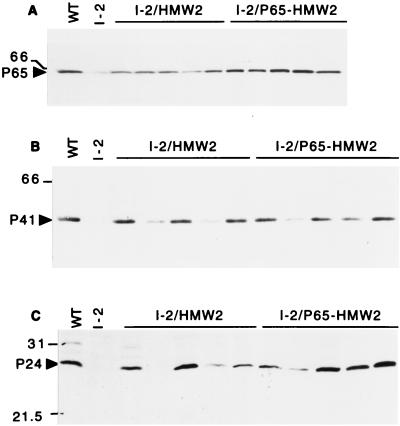
The production of HMW1, HMW3 and P65 was likewise restored in the HA+ transformants of mutant I-2 but to different extents (summarized in Table 1). For example, HMW3 was observed in wild-type quantities in all HA+ transformants examined, regardless of the level of HMW2 (Fig. 3B). In contrast, HMW1 and P65 were present at near wild-type levels in I-2/P65-HMW2 transformants but at significantly reduced levels in I-2/HMW2 transformants (Fig. 3A and 4A, respectively), correlating directly with HMW2 levels. P41 and P24 were detected in wild-type M. pneumoniae and in all transformants (Fig. 4B and C, respectively; Table 1) but, for reasons that are not clear, not in a manner that correlated with HMW2 levels. To summarize, production of recombinant HMW2 in mutant I-2 resulted in increased levels of the cytadherence-associated proteins originally deficient in the mutant. There was a direct correlation between the amount of recombinant HMW2 produced and the extent to which HMW1 and P65 were restored to wild-type levels. HMW3 was less dependent on the amount of HMW2 produced and was observed at near-wild-type levels as long as some HMW2 was present. The variability in P41 and P24 was independent of HMW2 levels, although an increase in the amounts of these two proteins was clearly HMW2 dependent.
FIG. 3.
Western immunoblot analysis of M. pneumoniae mutant I-2 transformants for production of HMW1 (A) and HMW3 (B). Approximately equal amounts of total protein were loaded per well, electrophoresed on a 4.5% polyacrylamide separating gel, blotted to nitrocellulose, and probed with specific antiserum to HMW1 or HMW3 (34, 35). The lanes are the same as indicated in the legend to Fig. 2. HMW1 and HMW3 are indicated by arrowheads, and protein size standards are shown to the left in kilodaltons.
FIG. 4.
Western immunoblot analysis of M. pneumoniae mutant I-2 transformants for production of P65 (A), P41 (B), or P24 (C). Approximately equal amounts of total protein were loaded per well, electrophoresed on a 12% polyacrylamide separating gel, blotted to nitrocellulose, and probed with specific antiserum to P65, P41, or P24 (21, 29). The lanes are the same as indicated in the legend to Fig. 2. P65, P41, and P24 are indicated by arrowheads, and protein size standards are shown to the left in kilodaltons. Previous studies indicated that P24 migrates anonymously (21), but in the current study P24 migrated at the expected size.
Quantitative analysis of HA.
Several I-2/HMW2 and I-2/P65-HMW2 transformants which were HA+ by colony screening were also evaluated quantitatively to determine whether the variable levels of HMW2 in these transformants had a measurable effect on cytadherence. I-2/P65-HMW2 transformants had wild-type levels of HMW2 and exhibited HA activity 56 to 63% of that observed with wild-type M. pneumoniae (Table 3). Perhaps significantly, these two transformants differed in the levels of P41 and P24 present, but this difference did not correlate with the capacity to HA. In contrast, HA activity in the I-2/HMW2 transformants varied from 17 to 44% of the wild-type level, in a manner that correlated directly with the level of HMW2.
TABLE 3.
Relative HA and HMW2 levels of M. pneumoniae transformants
| Strain | % of wild-type HAa | Relative level of HMW2b |
|---|---|---|
| Mutant I-2 | 4.4 | − |
| I-2/HMW2-1 | 44.2 | + |
| I-2/HMW2-2 | 17.4 | + |
| I-2/HMW2-3 | 42.6 | ++ |
| I-2/P65-HMW2-2 | 63.3 | +++ |
| I-2/P65-HMW2-3 | 56.0 | +++ |
Total counts per minute added varied due to differences in the growth rates of some transformants but ranged from 7 × 103 to 3 × 104 in most experiments. Background radioactivity varied proportionally (typically from 1 × 102 to 5 × 102 cpm).
As indicated in Table 1.
Sequence analysis of hmw2 in two class I mutants and an HA+ revertant thereof.
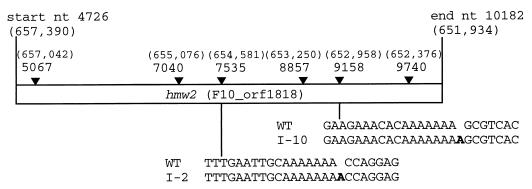
The ability of the recombinant wild-type but not the I-2 hmw2 allele to restore HA in the I-2 mutant suggested that the defect in mutant I-2 lies in the hmw2 gene. The hmw2 alleles from two independent HA mutants (I-2 and I-10 [20]) were analyzed by double-stranded sequencing of the cloned gene and PCR-based sequencing from chromosomal DNA. A single frameshift mutation resulting from the addition of an adenine nucleotide to an oligoadenine region was identified in mutants I-2 and I-10, but at a different site in each mutant (Fig. 5). In both cases, the frameshift resulted in premature termination of HMW2, yielding predicted truncated derivatives 952 and 1483 amino acids long for I-2 and I-10, respectively. Analysis by Western immunoblotting using antibodies against an N-terminal domain of HMW2 revealed a truncated polypeptide of the expected size in low levels in mutant I-2 but not I-10 (data not shown), suggesting that the former is nonfunctional and that both are unstable. Sequence analysis of an HA+ revertant of mutant I-2 (19) established that reacquisition of HMW2 resulted from loss of the additional adenine nucleotide, thus restoring the wild-type reading frame.
FIG. 5.
Identification of frameshift mutations in class I mutants I-2 and I-10. The nucleotide (nt) positions of the start and end of hmw2 and of the first adenine nucleotides of the six oligo(A) tracks (filled arrows) are given according to the numbering of the P65 operon (21). The corresponding nucleotide positions in the M. pneumoniae genome (14) are given in brackets. The extra adenine nucleotide in each isolate is shown in bold lettering. WT, wild type.
P28 is encoded by the 3′ end of hmw2.
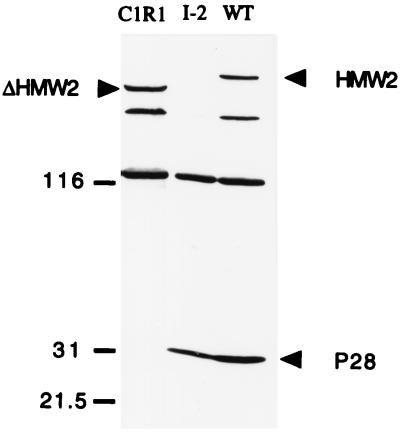
Antibodies prepared against the C terminus of HMW2 detect P28 by Western immunoblotting in wild-type protein profiles (21). P28 is present at greatly reduced levels in class I and some crl mutants and absent from crl mutants with Tn4001 insertion in the 3′ end of hmw2. Taken together, these observations suggest that P28 is a product of internal translation initiation in hmw2 (21). Two different ATG codons near the 3′ end of hmw2 (nucleotides 9460 and 9583 [Fig. 5]) could initiate translation in frame with HMW2 to generate a polypeptide of 28 kDa. C1R1 is a derivative of strain M129 resulting from imprecise transposon excision from a crl mutant and containing an in-frame deletion in hmw2 that spans across the two potential translation initiation codons (21). Western immunoblotting using antibodies against the C terminus of HMW2 demonstrated that C1R1 produces a truncated HMW2 (due to the internal deletion) but no P28 (Fig. 6), suggesting that the sequences deleted in C1R1 are required for P28 synthesis. As C1R1 is HA+, P28 does not appear to be essential for cytadherence.
FIG. 6.
Western immunoblot of M. pneumoniae wild type (WT), mutant I-2, and C1R1. Approximately equal amounts of mycoplasma total protein were electrophoresed on an SDS–5 to 15% polyacrylamide gradient gel, transferred onto nitrocellulose, and probed with antibody against the C-terminal region of HMW2 (1:1,000 dilution). HMW2, truncated HMW2 (ΔHMW2), and P28 are indicated by arrowheads; size standards are indicated in kilodaltons.
DISCUSSION
Despite its minimal genome and no cell wall, M. pneumoniae is a structurally complex bacterium possessing a cytoskeleton-like matrix and a differentiated attachment organelle. Biogenesis of the attachment organelle is poorly understood but appears to be coordinated with cell division and therefore is probably tightly regulated to ensure the proper ordered assembly of its component proteins, including HMW1, HMW2, and HMW3 (17). Detailed analysis of cytadherence mutants is beginning to elucidate form, function, and regulation of attachment organelle proteins in M. pneumoniae. Mutant M6 (22), which lacks the cytadherence-accessory protein HMW1, lacks a morphologically identifiable terminal organelle (9). Genetic complementation in trans appears to restore normal development of the attachment organelle and correct trafficking of the adhesin P1 to this structure, establishing that HMW1 is essential for proper assembly of a fully functional tip structure (9). Likewise, transposon insertion in hmw2 results in loss of cytadherence (11, 21) and accelerated turnover of HMW1 and HMW3 (27). Whether turnover of HMW1 and HMW3 reflects a housekeeping or regulatory function is unclear. Nevertheless, the similarities in phenotypes of spontaneously arising class I mutants and crl mutants predicted that the former also result from a defect in hmw2, as confirmed by using genetic complementation and DNA sequencing in the present study.
Transformation of mutant I-2 with recombinant wild-type hmw2 via transposon delivery restored cytadherence in >90% of the transformants examined. Some transformants remained HA−, probably due to a positional effect of transposon insertion, inactivating other genes required for cytadherence or influencing the expression of the recombinant hmw2 allele, for example. The extents to which transformants exhibited a fully wild-type phenotype varied and were dependent in part on the level of HMW2 production. Thus, transformants producing wild-type levels of HMW2 exhibited the highest levels of HA and near-normal protein profiles, while transformants producing HMW2 at reduced levels displayed intermediate HA activity (Table 3). The extents to which HMW1 and HMW3 were restored to wild-type levels also differed in the transformants (Table 1). HMW3 achieved wild-type steady-state levels even when recombinant HMW2 was produced in very low amounts, while wild-type levels for HMW1 were achieved only when recombinant HMW2 was also produced at wild-type levels. This observation is consistent with the faster turnover of HMW1 than HMW3 in hmw2 mutants (27) and suggests that a threshold level or correct stoichiometry, or both, for HMW2 is required for normal function.
The levels of the other products of the P65 operon (P65, P41, P24, and P28) varied in class I-2 transformants expressing recombinant hmw2, but only the levels of P65 and P28 correlated with the amount of recombinant HMW2 produced. The loss of P65, like loss of HMW1 and HMW3, occurs posttranslationally in hmw2 mutants (7), but it is not known by what mechanism P41 and P24 levels are reduced in the mutants and restored at variable levels in the transformants. However, the restoration of P41 and P24 to wild-type levels in some transformants demonstrates that their loss in mutant I-2 was not due to transcriptional polarity, consistent with the absence of the Rho protein in M. pneumoniae (14, 32).
The loss of HMW2 in mutants I-2 and I-10 resulted from a frameshift in hmw2. This conclusion is based on nucleotide sequence data from the mutants and from a revertant of one of the mutants and is underscored by results from genetic complementation with recombinant wild-type and I-2 hmw2 alleles. For both class I mutants examined, the defect involved the addition of an adenine nucleotide to an oligo(A) tract and probably resulted from slipped-strand mispairing, which has been suggested to occur more frequently in simple repeat sequences (23). Phase variation resulting from reversible frameshift mutations occurs in Neisseria meningitidis (10), Bordetella pertussis (36), and Haemophilus influenzae (41). Furthermore, mutations in hot spots are a mechanism for generating antigenic variation in mycoplasmas in the absence of diverse transcriptional regulators. Examples include mutation in oligo(A) tracts in the genes for the variable adherence-associated antigen lipoprotein of Mycoplasma hominis (44), the P78 lipoprotein of Mycoplasma fermentans (38), and the PMGA gene of Mycoplasma gallisepticum (5). Likewise, in M. pneumoniae the loss of the cytadhesin P1 in a class IV mutant (20) is the result of a similar frameshift in an oligo(A) region of the p1 gene (37).
Examining the frequency and location of heptameric or larger oligo(A) sequences in the M. pneumoniae genome, we determined that 140 genes contain one such oligo(A) sequence, 36 genes contain two, 5 genes contain three, 3 genes contain four, 1 gene contains five, and only hmw2 contains six. The genes containing three or more such oligo(A) sequences are listed in Table 4. The high number found in hmw2 might explain, in part, the frequent appearance of spontaneously arising class I mutants and their reversion to wild type (19, 20). While the number of oligo(A) sequences was not adjusted for gene length, it is noteworthy that the P1 gene is similar in size to hmw2 and yet has only two such oligo(A) sequences. In contrast, a single pentameric oligo(G) sequence and no hexameric or heptameric homo-oligomers of C or G were found in hmw2. Therefore, the occurrence of the oligo(A) sequences in hmw2 seems significant, but it remains unclear whether high-frequency frameshift mutations in hmw2 are important in the host-pathogen interaction; this issue will require additional studies.
TABLE 4.
M. pneumoniae genes having three or more heptameric or larger oligo(A) sequences
| No. of oligo(A) sequencesa | Geneb | Annotationb |
|---|---|---|
| 3 | MP45 (C09_orf718) | Unknown |
| MP185 (K05_orf401) | Hypothetical protein | |
| MP 260 (D02_orf439) | Putative lipoprotein | |
| MP368 (P01_orf293) | degV homolog | |
| MP564 (A65_orf251a) | Putative lipoprotein | |
| 4 | MP102 (D09_orf657) | Putative lipoprotein |
| MP298 (G12_orf664) | Unknown | |
| MP360 (P01_orf838) | Valyl-tRNA synthetase (valS) | |
| 5 | MP496 (H91_orf715) | DNA helicase II (mutB1) |
| 6 | MP527 (F10_orf1818) | hmw2 |
Heptamers or longer.
From reference 14.
HMW1, HMW3, and P65 have distinct subcellular locations, and while HMW2 is clearly required for the stability of each, the mechanism by which this is achieved is not known. The absence of P28 from the HA+ C1R1 revertant (Fig. 6) is consistent with the conclusion that P28 is a product of hmw2 but also demonstrates that unlike HMW2, P28 is not required for cytadherence. We have previously shown that the low levels of HMW1, HMW3, and P65 (7, 27) in hmw2 mutants are due to accelerated turnover, presumably by a proteolytic mechanism. Given that these proteins are components of the mycoplasma cytoskeleton (34), their ordered assembly therein may require HMW2. Failure to incorporate into the cytoskeleton in a timely manner in the absence of HMW2 might render HMW1, HMW3, and P65 more susceptible to turnover. Alternatively, HMW2 may influence the phosphorylation state of HMW1 and HMW3 (3), thereby affecting their stability. Clarification of the roles of HMW2 and P28 will likely require additional analysis of the functional domains of each, using transformation-based techniques described here, as well as identification and characterization of the protease(s) that mediates turnover of HMW1, HMW3, and P65 in the absence of HMW2.
In summary, the class I mutant phenotype results from a frameshift mutation in one of six heptameric oligo(A) regions of hmw2. Furthermore, the class I mutant can be complemented by the recombinant wild-type hmw2 allele. The HA phenotype and the production of several cytadherence-associated proteins appear to be dependent on the level of HMW2, suggesting that proper stoichiometery is important for attachment organelle formation and function. Finally, P28 is probably an internal translation product of the 3′ end of hmw2, but the roles of HMW2 and P28 in the biogenesis of the attachment organelle remain poorly defined.
ACKNOWLEDGMENTS
This work was supported by the Public Health Service research grant AI23362 from the National Institute of Allergy and Infectious Diseases to D.C.K. and by grants He 780/7-2 and He 780/5-2 from the Deutsche Forschungsgemeinschaft to R.H.
We thank R. Frank for the synthesis of oligonucleotides and J. Regula for providing specific antiserum.
REFERENCES
- 1.Baseman J B, Cole R M, Krause D C, Leith D K. Molecular basis for cytadsorption of Mycoplasma pneumoniae. J Bacteriol. 1982;151:1514–1522. doi: 10.1128/jb.151.3.1514-1522.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng S, Fockler C, Barnes W M, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dirksen L B, Krebes K A, Krause D C. Phosphorylation of cytadherence-accessory proteins in Mycoplasma pneumoniae. J Bacteriol. 1994;176:7499–7505. doi: 10.1128/jb.176.24.7499-7505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dirksen L B, Proft T, Hilbert H, Plagens H, Herrmann R, Krause D C. Nucleotide sequence analysis and characterization of the hmw gene cluster of Mycoplasma pneumoniae. Gene. 1996;171:19–25. doi: 10.1016/0378-1119(96)00050-9. [DOI] [PubMed] [Google Scholar]
- 5.Dovc P, Bencina D, Milosevic T, Berlic M D-V, Narat M. Presented at the 12th Congress of the International Organization for Mycoplasmology, Sydney, Australia, July 22 to 28. 1998. Localized changes in Mycoplasma gallisepticum pMGA gene associated with antigenic variants. [Google Scholar]
- 6.Fisseha, M. Unpublished data.
- 7.Fisseha, M., K. M. Birkhead, and D. C. Krause. Unpublished data.
- 8.Hahn T-W, Krebes K A, Krause D C. Expression in Mycoplasma pneumoniae of the recombinant gene encoding the cytadherence-associated protein HMW1 and identification of HMW4 as a product. Mol Microbiol. 1996;19:1085–1093. doi: 10.1046/j.1365-2958.1996.455985.x. [DOI] [PubMed] [Google Scholar]
- 9.Hahn T-W, Willby M J, Krause D C. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J Bacteriol. 1998;180:1270–1276. doi: 10.1128/jb.180.5.1270-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerschmidt S, Muller A, Sillmann H, Muhlenhoff M, Borrow R, Fos A, Putten J, Zollinger W D, Gerardy-Schahn R, Frosch M. Capsule phase variation in Neisseria meningitidis serogroup B by slipped-strand mispairing in the polysialyltransferase gene (sisD): correlation with bacterial invasion and the outbreak of meningococcal disease. Mol Microbiol. 1996;20:1211–1220. doi: 10.1111/j.1365-2958.1996.tb02641.x. [DOI] [PubMed] [Google Scholar]
- 11.Hedreyda C T, Krause D C. Identification of a possible cytadherence regulatory locus in Mycoplasma pneumoniae. Infect Immun. 1995;63:3479–3483. doi: 10.1128/iai.63.9.3479-3483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedreyda C T, Lee K K, Krause D C. Transformation of Mycoplasma pneumoniae with Tn4001 by electroporation. Plasmid. 1993;30:170–175. doi: 10.1006/plas.1993.1047. [DOI] [PubMed] [Google Scholar]
- 13.Hilbert H, Himmelreich R, Plagens H, Herrmann R. Sequence analysis of 56 kb from the genome of the bacterium Mycoplasma pneumoniae comprising the dnaA region, the atp operon and a cluster of ribosomal protein genes. Nucleic Acids Res. 1996;24:628–639. doi: 10.1093/nar/24.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B-C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu P C, Collier A M, Baseman J B. Surface parasitism by Mycoplasma pneumoniae of respiratory epithelium. J Exp Med. 1977;145:1328–1343. doi: 10.1084/jem.145.5.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knudtson K L, Minion F C. Construction of Tn4001lac derivatives to be used as promoter probe vectors in mycoplasmas. Gene. 1993;137:217–222. doi: 10.1016/0378-1119(93)90009-r. [DOI] [PubMed] [Google Scholar]
- 17.Krause D C. Mycoplasma pneumoniae cytadherence: organization and assembly of the attachment organelle. Trends Microbiol. 1998;6:15–18. doi: 10.1016/S0966-842X(97)01168-2. [DOI] [PubMed] [Google Scholar]
- 18.Krause D C, Baseman J B. Inhibition of Mycoplasma pneumoniae hemadsorption and attachment to respiratory epithelium by antibodies to a membrane protein. Infect Immun. 1983;39:1180–1186. doi: 10.1128/iai.39.3.1180-1186.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause D C, Leith D K, Baseman J B. Reacquisition of specific proteins confers virulence in Mycoplasma pneumoniae. Infect Immun. 1983;39:830–836. doi: 10.1128/iai.39.2.830-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause D C, Leith D K, Wilson R M, Baseman J B. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun. 1982;35:809–817. doi: 10.1128/iai.35.3.809-817.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krause D C, Proft T, Hedreyda C T, Hilbert H, Plagens H, Herrmann R. Transposon mutagenesis reinforces the correlation between Mycoplasma pneumoniae cytoskeletal protein HMW2 and cytadherence. J Bacteriol. 1997;179:2668–2677. doi: 10.1128/jb.179.8.2668-2677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Layh-Schmitt G, Hilbert H, Pirkl E. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J Bacteriol. 1995;177:843–846. doi: 10.1128/jb.177.3.843-846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levinson G, Gutman G A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- 24.Meng K E, Pfister R M. Intracellular structures of Mycoplasma pneumoniae revealed after membrane removal. J Bacteriol. 1980;144:390–399. doi: 10.1128/jb.144.1.390-399.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oakley B R, Kirsh D R, Norris N R. A simplified ultrasensitive silver stain for detecting proteins in polyacrylamide gels. Anal Biochem. 1980;105:361–363. doi: 10.1016/0003-2697(80)90470-4. [DOI] [PubMed] [Google Scholar]
- 26.Ogle K F, Lee K K, Krause D C. Cloning and analysis of the gene encoding the cytadherence phase-variable protein HMW3 of Mycoplasma pneumoniae. Gene. 1991;97:69–75. doi: 10.1016/0378-1119(91)90011-y. [DOI] [PubMed] [Google Scholar]
- 27.Popham P L, Hahn T-W, Krebes K A, Krause D C. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc Natl Acad Sci USA. 1997;94:13979–13984. doi: 10.1073/pnas.94.25.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proft T, Herrmann R. Identification and characterization of hitherto unknown Mycoplasma pneumoniae proteins. Mol Microbiol. 1994;13:337–348. doi: 10.1111/j.1365-2958.1994.tb00427.x. [DOI] [PubMed] [Google Scholar]
- 29.Proft T, Hilbert H, Layh-Schmitt G, Herrmann R. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in the strains M129 and FH. J Bacteriol. 1995;177:3370–3378. doi: 10.1128/jb.177.12.3370-3378.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Razin S, Jacobs E. Mycoplasma adhesion. J Gen Microbiol. 1992;138:407–422. doi: 10.1099/00221287-138-3-407. [DOI] [PubMed] [Google Scholar]
- 31.Regula, J. Unpublished data.
- 32.Richardson J P, Grimley C, Lowery C. Transcription termination factor rho activity is altered in Escherichia coli with suA gene mutations. Proc Natl Acad Sci USA. 1975;72:1725–1728. doi: 10.1073/pnas.72.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Stevens M K, Krause D C. Localization of the Mycoplasma pneumoniae cytadherence-associated proteins HMW1 and HMW4 in cytoskeletonlike triton shell. J Bacteriol. 1991;173:1041–1050. doi: 10.1128/jb.173.3.1041-1050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens M K, Krause D C. Cytadherence accessory protein HMW3 of Mycoplasma pneumoniae is a component of the attachment organelle. J Bacteriol. 1992;174:4265–4274. doi: 10.1128/jb.174.13.4265-4274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stibitz S, Aaronson W, Monak D, Falkow S. Phase variation of Bordetella pertussis by frameshift mutation in a gene for a novel two-component system. Nature. 1989;338:266–269. doi: 10.1038/338266a0. [DOI] [PubMed] [Google Scholar]
- 37.Su C-J, Chavoya A, Baseman J B. Spontaneous mutation results in loss of the cytadhesin (P1) of Mycoplasma pneumoniae. Infect Immun. 1989;57:3237–3239. doi: 10.1128/iai.57.10.3237-3239.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Theiss P, Wise K S. Localized frameshift mutation generates selective, high-frequency phase variation in a surface lipoprotein encoded by a mycoplasma ABC transporter operon. J Bacteriol. 1997;179:4013–4022. doi: 10.1128/jb.179.12.4013-4022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tully J G. Cloning and filtration techniques for mycoplasmas. In: Razin S, Tully J, editors. Methods in mycoplasmology. I. San Diego, Calif: Academic Press, Inc.; 1983. pp. 173–177. [Google Scholar]
- 40.Waldo, R. H., P. L. Popham, C. E. Romero-Arroyo, E. A. Mothershed, K. K. Lee, and D. C. Krause. Unpublished data. [DOI] [PMC free article] [PubMed]
- 41.Weiser J N, Love J M, Moxon E R. The molecular mechanism of phase variation of Haemophilus influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- 42.Wenzel R, Herrmann R. Physical mapping of the Mycoplasma pneumoniae genome. Nucleic Acids Res. 1988;16:8323–8336. doi: 10.1093/nar/16.17.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q, Wise K S. Localized reversible frameshift mutation in an adhesin gene confers a phase-variable adherence phenotype in mycoplasma. Mol Microbiol. 1997;25:859–869. doi: 10.1111/j.1365-2958.1997.mmi509.x. [DOI] [PubMed] [Google Scholar]